Abstract
The Crk adaptor protein, which is encoded by two splice variants termed CrkI and CrkII, contains both SH2 and SH3 domains but no catalytic region. It is thought to function in signal transduction processes involved in growth regulation, cell transformation, cell migration, and cell adhesion. Although the function of Crk has been studied in considerable detail in cell culture, its biological role in vivo is still unclear, and no Crk-knockout mouse model has been available. Therefore, we generated a complete null allele of Crk in mice by using the Cre-loxP recombination approach. The majority of Crk-null mice die at late stages of embryonic development, and the remainder succumb shortly after birth. Embryos lacking both CrkI and CrkII exhibited edema, hemorrhage, and cardiac defects. Immunohistochemical examination suggested that defects in vascular smooth muscle caused dilation and rupturing of blood vessels. Problems in nasal development and cleft palate were also observed. These data indicate that Crk is involved in cardiac and craniofacial development and that it plays an essential role in maintaining vascular integrity during embryonic development.
v-Crk was originally identified as a chicken tumor virus oncoprotein (14). Although it does not contain any known catalytic domains, v-Crk induces a remarkable increase in tyrosine phosphorylation in transformed cells. Mutations in either the SH2 or SH3 domains of Crk abrogate its cell-transforming activity, a finding that suggests that the domains are essential for oncogenesis. CrkII, a cellular counterpart of v-Crk, is located on chromosome 17p13.3 in humans (13, 21). It contains a single SH2 domain and two SH3 domains. Alternative splicing generates CrkI, which lacks the C-terminal SH3 domain, and CrkIII, which has a modified, nonfunctional C-terminal SH3 domain. A similar but distinct Crk-like gene, CrkL, is located on chromosome 22q11.21 in humans. CrkL is similar to Crk in amino acid composition and overall domain structure. Crk and CrkL are expressed ubiquitously (2). The SH2 domain of Crk interacts with many phosphotyrosine-containing proteins, including paxillin (p70), p130Cas, and c-Cbl. The SH3 domain of Crk interacts with proline-rich domains in several proteins, including C3G, DOCK180, and the Abl family. Crk and CrkL contribute to a variety of signaling pathways that influence cell morphology, cell movement, cell proliferation, and differentiation (for review, see reference 5).
CrkL-knockout mice exhibit defects in multiple cranial and cardiac neural crest derivatives, and they do not survive embryogenesis (6). Interestingly, the steady-state level of Crk protein was not altered by the absence of CrkL. This result, together with the embryonic lethality of CrkL-knockout mice, suggests that Crk does not compensate for CrkL functions in vivo. Imaizumi et al. (8) reported a gene-trap mutation in the mouse Crk gene in which truncated CrkI-like proteins lacking the C-terminal SH3 domain were expressed. This CrkII-specific gene disruption did not result in any obvious abnormalities, which suggests that the C-terminal SH3 domain of CrkII is not essential. However, it is still unclear whether CrkI can compensate for the loss of CrkII or whether the loss of both CrkI and CrkII can be compensated for by CrkL. Therefore, it is necessary to generate Crk-knockout mice lacking both CrkI and CrkII to address the in vivo functions of these variants. Here we show that CrkI and CrkII are essential for normal embryonic development and that their in vivo functions are distinct from those of CrkL.
MATERIALS AND METHODS
Construction of the targeting vector and generation of Crk-null mice.
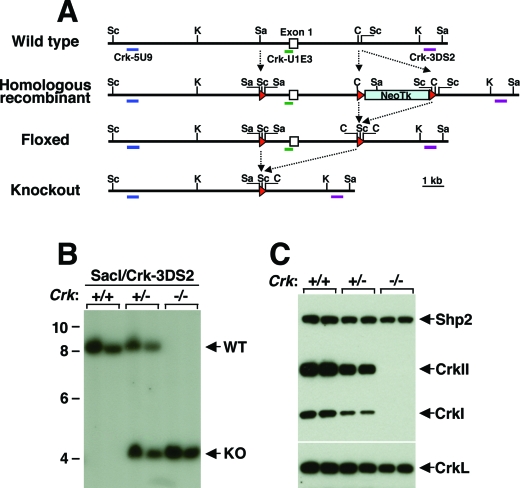
Crk genomic DNA was obtained by screening 129S6/SvEvTac RPCI-22 mouse bacterial artificial chromosome high-density membranes (Invitrogen) using the 273-bp Southern probe Crk-U1E3 (Fig. 1A, green bar). Crk-U1E3 contains 149 bp of the first exon of Crk and the adjacent 124 bp of the 5′ upstream region and was amplified using the following PCR primers: Crk-U1E3F (5′-AGGACTCCGTTTCCCTTCTC-3′) and Crk-U1E3R (5′-GCCCCAGTACCAGCTACTCC-3′), with mouse tail genomic DNA as a template. An 11-kb KpnI genomic fragment containing the first exon of Crk was isolated and subcloned into the pBluescript II KS− vector. Then a 52-bp oligonucleotide containing loxP and a 2.8-kb neomycin resistance-thymidine kinase (NeoTk) cassette flanked by loxP sites were sequentially inserted into the SacI and ClaI restriction enzyme sites, respectively. The gene-targeting vector was linearized with SpeI and electroporated into 129/SvEv mouse embryonic stem (ES) cells (Specialty Media) according to the manufacturer's instructions. Genomic DNA from ES cells was prepared by the conventional phenol-chloroform method, digested with either ScaI or SacI, and probed with Crk-5U9 (Fig. 1A, blue bars) and Crk-3DS2 (Fig. 1A, purple bars), respectively. Crk-5U9 was amplified using Crk-5U9F (5′-TCCCTACAACCCCTTAACCC-3′) and Crk-5U9R (5′-GCCTTGGTGATGAGAAGCTC-3′). Crk-3DS2 was amplified using Crk-3DS2F (5′-TGGGCATCTTCCTCTATTGC-3′) and Crk-3DS2R (5′-ACAGAAGCCAGCTCCCACTA-3′). ES clones with a normal karyotype in which homologous recombination had occurred were transfected with pMC-Cre plasmid and selected with ganciclovir (Roche) to obtain both floxed and knockout alleles. Three types of Cre recombination products, depending on the extent of recombination, were distinguished using the three Southern probes as described above, in addition to the wild-type and intact homologous recombinant alleles. Finally, ES clones with normal karyotype were selected and microinjected into mouse blastocysts. Chimeric mice, which were identified by coat color, were bred with C57BL/6 mice. Germ line transmission was confirmed initially by PCR (see below) and then by Southern hybridization analysis. Mouse colonies were maintained on a mixed background of C57BL/6 and 129SvEv. All mouse studies were carried out according to protocols approved by the Institutional Animal Care and Use Committee at St. Jude Children's Research Hospital.
FIG. 1.
Targeted disruption of Crk in mice. (A) Schematic diagram of wild-type, homologous recombinant, floxed, and knockout alleles of Crk. Red triangles represent loxP sites. The relevant restriction sites ScaI (Sc), KpnI (K), SacI (Sa), and ClaI (C) and locations of probes (thick colored bars) for Southern blotting are indicated. (B) Southern blot analysis of the genomic DNA from embryos. Two independent DNA samples from each genotype were digested with SacI and then analyzed. WT, wild type. (C) Western blot analysis of total protein lysates from MEFs. An anti-Crk antibody was used to detect the endogenous CrkI and CrkII proteins in two independent samples from each genotype.
PCR genotyping.
Genomic DNA from mouse tails or embryonic yolk sacs was prepared either by the conventional phenol-chloroform extraction method or by using the DNeasy kit (QIAGEN). The following three primers were used to genotype Crk alleles: CGT1 (5′-GGGTGACCTGAGAACTGACC-3′), CGT2 (5′-TCACTTATCCTGGGAATTGGA-3′), and CGT3 (5′-CAGCTCGGACTGCAGAATG-3′). Combination of the CGT1 and CGT3 primers amplifies floxed and wild-type alleles with PCR products of 231 bp and 134 bp, respectively. Combination of the CGT2 and CGT3 primers amplifies only knockout alleles with a 420-bp product. PCR was performed for 30 cycles of 94°C, 55°C, and 72°C (1 min each) using the QIAGEN Taq polymerase and Robocycler (Stratagene).
Preparation of MEFs.
Mouse embryonic fibroblasts (MEFs) were prepared as follows. Embryos derived from intercrosses of Crk+/− mice were harvested at embryonic day 13.5 (E13.5). Heads and viscera were removed and used to extract genomic DNA for Southern hybridization analysis. The remaining embryonic tissue was minced and treated with trypsin-EDTA (Invitrogen), and dissociated cells were cultured in Dulbecco's modified Eagle's medium (Cambrex) supplemented with 10% fetal bovine serum (HyClone), l-glutamine, penicillin, and streptomycin at 37°C under 5% CO2.
Western blot analysis.
Western blotting was carried out as described previously (11). Lysates of MEFs were prepared using 1% NP-40 lysis buffer containing 20 mM Tris-HCl adjusted to pH 7.5, 150 mM NaCl, 1 mM EDTA, 20 mM NaF, 0.5 mM Na3VO4, 10% glycerol, and the protease inhibitor cocktail Complete Mini (Roche). After quantifying the protein content of each cell lysate using the Bio-Rad protein assay kit, total cell lysates containing the same amount of protein were loaded, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and transferred to nitrocellulose membranes (Invitrogen). Membranes were then subjected to Western blot analysis using anti-Crk (BD Pharmingen), anti-CrkL (c-20; Santa Cruz), and anti-Shp2 (sc-7384; Santa Cruz) antibodies. Immunodetection was performed using the WestDura chemiluminescence kit (Pierce).
Morphological and histological analyses.
Embryos at different stages were dissected from mouse uteri and carefully examined under a dissecting microscope to determine whether they were alive by detecting the heartbeat and the blood supply to the yolk sac. Only live embryos were processed. Embryos and placentas were fixed overnight in 4% paraformaldehyde at 4°C, rinsed with phosphate-buffered saline (PBS; Mediatech), and incubated in PBS at 4°C until use. Where indicated, pictures of whole mouse embryos were taken after fixation. Fixed embryos were embedded in paraffin and sectioned at a thickness of 5 μm. Every tenth section was stained with hematoxylin and eosin, and sections with similar anatomical planes were chosen to take images. Slides were photographed using an Axioplan Zeiss microscope (Zeiss) and an RT color SPOT camera (Diagnostic Instruments) to obtain pictures of whole mouse embryos.
Immunohistochemistry.
Sections embedded in paraffin were processed for deparaffinization and rehydration followed by antigen retrieval in 10 mM sodium citrate at 80°C, as described previously (22). Slides were treated with 1% H2O2, blocked in PBS supplemented with 3.5% bovine serum albumin, and incubated with anti-smooth muscle actin (SMA) antibody (1A4; DakoCytomation) and anti-CD34 antibody (RAM34; PharMingen) overnight at room temperature. Immunoreactivity was detected using the Vectastain Elite ABC kit (Vector Laboratories) and the diaminobenzidine reagent set (Kirkegaard and Perry Laboratories) according to the manufacturers' instructions. Slides were dehydrated, mounted by standard techniques, and analyzed using an Olympus BX60 microscope. Images were acquired with a Hamamatsu (Bridgewater) C5810 video camera and imported directly into Adobe Photoshop 7.0 (Adobe Systems).
RESULTS
Generation of Crk-deficient mice.
Because it was unclear whether Crk-null mice would be viable, we used the Cre-loxP system to generate both complete null and floxed alleles. The targeting construct contained the first exon of Crk, flanked by two loxP sites, and followed by a NeoTk and another loxP site (Fig. 1A). ES cells harboring the homologous recombinant allele were selected, and Cre excision was induced by transfecting the ES cells with a Cre-expressing plasmid. ES cells carrying either the null or the floxed alleles were successfully obtained. Crk-heterozygous ES cells with a normal karyotype were injected into mouse blastocysts, and a C57BL/6 × 129 chimera that exhibited germ line transmission was obtained. Crk+/− mice born from the mating between the chimera and C57BL/6 wild-type mouse were viable and did not exhibit any gross defects.
Breeding among Crk+/− mice was carried out, and MEFs and genomic DNA were prepared from the embryos. PCR (data not shown) and Southern hybridization analysis (Fig. 1B) of the genomic DNA showed that the wild-type Crk allele was absent in Crk−/− MEFs, whereas Crk+/− MEFs contained both wild-type and Crk-null alleles. Western blot analysis of the MEF lysates clearly indicated that CrkI and CrkII proteins were not detected in Crk−/− MEFs and that their expression was reduced in Crk+/− MEFs (Fig. 1C). On the other hand, there was no obvious change in the expression of a closely related protein, CrkL, or that of an SH2-containing tyrosine phosphatase, Shp-2. These results suggest that deletion of the first exon of the Crk gene leads to the complete loss of CrkI and CrkII proteins without affecting the CrkL expression.
Viability of Crk-null mice and embryos.
Intercrosses among Crk+/− mice were performed, and all pups that died after birth or survived to weaning were genotyped. Of 164 pups examined, 64 (39.0%) were wild type, 97 (59.1%) were Crk+/−, and only 3 (1.8%) were Crk−/− (they died shortly after birth). These results suggest that most Crk−/− mice (about 95%) die before birth and the few that are born die perinatally. Therefore, we examined embryos from intercrosses at different stages and used their yolk sacs to prepare genomic DNA for genotyping (Table 1) to determine the timing of embryonic lethality. Because very few Crk−/− pups were born, we collected embryos from later stages of embryonic development. At E18.5, no Crk−/− embryos examined were alive. At E17.5, 3 viable embryos out of 44 (6.8%) were Crk−/−, and at E15.5, 7 embryos out of 152 (4.6%) were Crk−/−. These results indicate that the survival rate of Crk−/− embryos at E15.5 through E18.5 is far below the expected level of 25%. In contrast, at E13.5 and E11.5, the survival rates for Crk−/− embryos approached 25%. We found 7 live Crk−/− embryos and 1 dead Crk−/− embryo at E13.5 and 7 live and 21 dead Crk−/− embryos at E15.5. This finding suggested that the majority of Crk−/− embryos die during the period from E13.5 to E15.5. However, the time of death was variable as a few embryos died later. (For example, we found four dead Crk−/− embryos at E17.5.) This variation in the timing of lethality may be contributed to, at least in part, by the mixed strain background. When the strain background was shifted to an almost pure FVB background, most Crk−/− embryos died at between E15.5 and E18.5 and they showed similar phenotypes.
TABLE 1.
Genotype analysis of Crk+/− intercrosses
| Stage | Total no. of live embryos or pups examined | No. with genotypea:
|
Survival of Crk−/− embryos (%) | ||
|---|---|---|---|---|---|
| Crk+/+ | Crk+/− | Crk−/− | |||
| Embryonic | |||||
| E11.5 | 29 | 9 | 14 | 6 | 20.7 |
| E12.5 | 27 | 5 | 18 | 4 | 14.8 |
| E13.5 | 29 | 5 | 17 | 7 | 24.1 |
| E15.5 | 152 | 55 | 90 | 7 | 4.6 |
| E17.5 | 44 | 15 | 24 | 3 | 6.8 |
| E18.5 | 9 | 6 | 3 | 0 | 0 |
| Postnatal | 164 | 64 | 97 | 3b | 1.8 |
All embryos were genotyped by analysis of yolk sacs. Crk+/+ and Crk+/− pups were genotyped by tail samples at weaning; Crk−/− pups were genotyped in the same manner after they died.
Crk-null pups died perinatally.
General phenotype of Crk-null embryos.
The E11.5 Crk−/− embryos did not show any obvious differences compared to wild-type littermates (data not shown). In contrast, at E12.5, Crk−/− embryos exhibited small focal edema in the midline of the craniofacial region (Fig. 2). The focal edema was generally located on the tip of the snout and nasion, which is the surface area between the eyes. At E13.5, focal edema on the snout and nasion became enlarged and sometimes was filled with blood cells. At E15.5, hemorrhagic edema was observed on the snout and the nasion of all the live Crk−/− embryos. Some mutants exhibited hemorrhagic edema only on the snout, whereas others had lesions both on the snout and the nasion (Fig. 2, red arrows). In addition to focal and hemorrhagic edema, extensive edema was observed on the head and back of some Crk−/− embryos at E13.5 and E15.5 (Fig. 2, yellow arrowheads). Hematoxylin and eosin staining of Crk−/− embryos at E12.5 clearly showed focal edema (Fig. 3B and C) that was not seen in wild-type embryos (Fig. 3A). In addition, in some E15.5 Crk−/− embryos, there was marked dilation of the left common carotid artery of the heart (Fig. 3E and F, green arrows) and hemorrhage into the lateral ventricle (Fig. 3F, orange arrow), compared to what was seen in wild-type embryos (Fig. 3D).
FIG. 2.
General morphology of Crk−/− embryos. Embryos at E12.5, E13.5, and E15.5 were dissected from mouse uteri and fixed in 4% paraformaldehyde at 4°C overnight. Images of whole mouse embryos were taken before paraffin processing. Red arrows indicate hemorrhagic edema on the snout and the nasion. Yellow arrowheads indicate edema.
FIG. 3.
Histological analysis of Crk−/− embryos. Wild-type and Crk−/− embryos at E12.5 (A and B) and E15.5 (D, E, and F) were embedded in paraffin, and sagittal sections were obtained at a thickness of 5 μm. (C) High magnification of the focal edema (blue arrowheads) in panel B. (E and F) Arrows indicate aberrant accumulation of blood cells.
The heart of Crk−/− embryos looked different from that of wild-type embryos (Fig. 3D to F); therefore, we examined the E15.5 heart in greater detail. As shown in Fig. 4A and C, the ventricle of the wild-type heart was surrounded by a thick muscular wall consisting of tightly packed cells. In contrast, the muscular wall of the Crk−/− heart was thinner and contained fewer loosely packed cells (Fig. 4B and D). Transverse sections of wild-type embryos indicated well-developed ventricular walls and an interventricular septum with a thick muscle layer (Fig. 3E and G). In contrast, the ventricles in the Crk−/− embryonic heart were severely dilated, probably due to the extremely thin and poorly developed muscular wall (Fig. 4F and H).
FIG. 4.
Histological analysis of the Crk−/− embryonic heart. Wild-type (A, C, E, and G) and Crk−/− (B, D, F, and H) embryos at E15.5 were embedded in paraffin, and sagittal sections were obtained at a thickness of 5 μm. Sections were then stained with hematoxylin and eosin, and anatomically matching images of heart tissue were taken. The ventricles (V) in the Crk−/− embryos were markedly dilated, and the ventricular walls (double-headed arrows) and interventricular septum (VS) were strikingly thin compared to those structures in the wild-type littermates. Panels C, D, G, and H are high-magnification images of boxes in panels A, B, E, and F, respectively. Bar, 100 μm.
Defects in blood vessels and vascular smooth muscle cells.
Crk−/− embryos have obvious blood loss; therefore, we examined the integrity of their vascular endothelial and smooth muscle cells. Hemorrhagic edema on the snout (Fig. 5A, arrow) was analyzed by staining blood vessel endothelial cells with an anti-CD34 antibody. As shown in Fig. 5B, the edema was framed by CD34-positive endothelial cells, suggesting that it occurred as a consequence of dilation of blood vessels. On close examination, we found locally scattered, irregular distribution of CD34-positive cells (Fig. 5B and C, arrows), which indicated a rupture of blood vessels. However, there was no obvious difference in the pattern of endothelial cell staining in neighboring areas such as the mesenchyme below the epidermal layer in Crk−/− embryos, compared with that of wild-type littermates (Fig. 5D and E). In contrast, staining of vascular smooth muscle cells in the head of E15.5 embryos revealed a clear difference.
FIG. 5.
CD34 immunostaining of Crk−/− embryos. Sagittal paraffin sections from E15.5 Crk−/− and wild-type littermates were stained with anti-CD34 antibody and counterstained with hematoxylin. (A) Two hemorrhagic edemas are present on the nasion (arrowhead) and tip of the snout (arrow). (B) High-magnification view of the hemorrhagic edema on the tip of snout. CD34-labeled endothelial cells are stained brown. The hemorrhagic edema is not lined with endothelial cells (arrow), which suggests the presence of a ruptured blood vessel. The ruptured blood vessel can be seen clearly at higher magnification (C). CD34 immunostaining of the region between the two areas of hemorrhagic edema in a Crk−/− embryo shown in the box in panel A (E) was compared with that of a corresponding region in a wild-type littermate (D). Hemorrhagic edema (he), mesenchyme (mc), and epidermal layer (ep) are marked. Bar, 50 μm.
SMA staining in wild-type embryos was evident as solid ring structures (Fig. 6A, C, and E). SMA staining was much weaker in Crk−/− embryos (Fig. 6B, D, and F) than in wild-type littermates, when anatomically similar regions were compared. Smooth muscle in Crk−/− embryos was sometimes discontinuous (arrows in Fig. 6D), suggestive of the initial stage of blood vessel disruption. Interestingly, most of the defective smooth muscles were detected near the nasal cavity (Fig. 6G and H). These observations indicate that smooth muscle cells in Crk−/− embryos, especially those in the head area, have a defect in their ability to support blood vessels, making the blood vessels prone to dilation and rupture.
FIG. 6.
SMA immunostaining of the head region in Crk−/− embryos. Sagittal sections from E15.5 Crk−/− and wild-type littermates were stained with anti-SMA antibody and counterstained with hematoxylin. (A to F) High-magnification views of SMA staining. A, C, and E (wild-type) anatomically correspond to B, D, and F (Crk−/−), respectively. (G and H) Locations of B, D, and F are indicated as boxes. Defective regions (arrows), hemorrhagic edema (*), nasal cavity (n), upper lip (u), and tongue (t) are marked. Bar, 25 μm.
Defective nasal development and cleft palate.
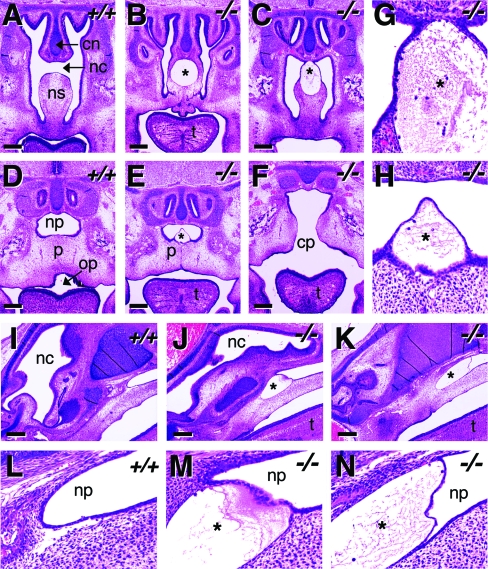
Focal edema and hemorrhagic edema developed at the tip of the snout, and most of the defective smooth muscle was observed near the nasal cavity; therefore, we further examined nasal development in E15.5 coronal and sagittal sections. In wild-type E15.5 embryos, the nasal septum formed as a columnar structure, through the midline, in the anterior of the nasal cavity. The posterior nasal septum was divided into upper and lower parts, leaving a gap in the midline (Fig. 7A). The bottom of the nasal septum fused with the palatal shelves and the primary palate above the oropharynx. However, in all Crk−/− embryos examined, the separation was hampered due to edema in the middle of the nasal septum, although the fusion of the nasal septum with the palatal shelves and the primary palate was retained (Fig. 7B, C, and G). In wild-type embryos, the gap posterior of the nasal cavity in the midline becomes the nasopharynx and it is separated from the oropharynx by the palate (Fig. 7D). In contrast, in Crk−/− embryos, either edema developed in the midline above the fused palate (Fig. 7E and H) or a cleft palate was observed (Fig. 7F).
FIG. 7.
Defective nasal development and cleft palate in Crk−/− embryos. E15.5 Crk−/− and wild-type littermates were fixed and embedded in paraffin, and coronal and sagittal sections were obtained. Paraffin sections were stained with hematoxylin and eosin, and matching images of the nasal parts were taken. (A to F) Panels A, B, and C are anatomically matching coronal sections that are anterior to panels D, E, and F, respectively. (G and H) High-magnification views of the edema formed in panels C and E, respectively. (I to N) Panels I, J, and K are anatomically matching sagittal sections, and their high-magnification views of the nasopharynx are shown in panels L, M, and N, respectively. Nasal cavity (nc), nasal septum (ns), cartilage primordium of nasal septum (cn), edema (*), palate (p), cleft palate (cp), nasopharynx (np), and oropharynx (op) are marked. Bar, 250 μm.
Sagittal sections of E15.5 embryos clearly indicated edema in the nasopharynx. The inside of the nasopharynx in wild-type embryos was clear (Fig. 7I and L), but that of Crk−/− embryos showed edema at the anterior end, where the primary palate fuses with the nasal septum (Fig. 7J and K). Close comparison of the nasopharynges of Crk−/− embryos (Fig. 7M and N) and wild-type embryos (Fig. 7L) revealed that in the mutant embryos, the mucosal epithelium separates from the submucosa, resulting in a space filled by extracellular fluid.
DISCUSSION
Our study clearly demonstrates that Crk is essential for embryonic development and provides some insight into the in vivo functions of CrkI and CrkII. Although Crk and CrkL have similar primary structures and a high level of amino acid sequence identity (60%), the observation of embryonic lethality of both Crk−/− and CrkL−/− mice suggests that each gene plays a unique biological role during development. Although ablation of CrkII in a gene-trap mutant mouse that still expressed CrkI (8) showed no obvious phenotype, the majority (about 95%) of Crk−/− embryos died at late stages of embryonic development. The most likely explanation for this difference is that CrkI and CrkII play distinct biological roles and that only CrkI is necessary for development. Another possibility is that CrkI and CrkII have redundant functions and therefore one is enough to secure normal development. In either case, it appears that the common region of CrkI and CrkII, including the SH2 domain and the N-terminal SH3 domain, is crucial for development, whereas the C-terminal SH3 domain of CrkII is not.
Most of the proteins that interact with the SH3 domains of Crk, including C3G, DOCK180, the Abl family, HPK1, KHS, and phosphatidylinositol 3-kinase, bind to the N-terminal SH3 domain (reviewed in reference 5). The function of the C-terminal SH3 domain of CrkII is still poorly understood. In overexpression studies, CrkII is significantly less active than CrkI at inducing tyrosine phosphorylation of p130Cas, morphological alterations, and cell transformation (13, 16). This finding suggests that the C-terminal SH3 domain plays a negative regulatory role.
Recently, the C-terminal SH3 domain and an adjacent linker region were shown to be necessary for transactivation of the Abl tyrosine kinase (20). In addition, the C-terminal SH3 domain of CrkII was found to be required for engulfment of apoptotic cells and cell spreading on extracellular matrix in C. elegans (1). Therefore, our study, together with these previous reports, suggests that CrkI plays essential roles in development, whereas the C-terminal SH3 domain of CrkII contributes regulatory functions under certain circumstances. Mice lacking C3G, a guanine nucleotide exchange factor that binds to Crk and CrkL, die before E7.5 (17), which is earlier than the major period of lethality of Crk- or CrkL-mutant mice. Therefore, we cannot rule out the possibility that Crk and CrkL have both redundant and nonredundant functions during development, and it is still unclear whether CrkII is sufficient to replace CrkI.
The edema and hemorrhagic edema observed on the snout of Crk−/− embryos were similar to those observed in C3Ggt/gt mutant mice, which express a fusion protein encoding the first 19 amino acids of C3G followed by β-galactosidase and neomycin phosphotransferase (27), although the reported hemorrhage near the hindbrain of C3Ggt/gt embryos was not observed in Crk−/− embryos. Furthermore, both mutants exhibit similar defects in smooth muscle cells. These results are consistent with observations indicating that Crk and C3G are closely connected in many signaling pathways. Interestingly, in vascular smooth muscle cells, Crk and its substrate p130Cas function in angiotensin II signaling to mediate smooth muscle contraction (23, 24). Recently, Crk and p130Cas were shown to play similar roles in regulating actin dynamics and smooth muscle contraction (25, 26). In addition, mice lacking p130Cas exhibited dilated blood vessels (7). Therefore, the p130Cas-Crk-C3G pathway most likely contributes to the maintenance of vascular integrity by regulating smooth muscle contraction.
The placenta of Crk−/− embryos developed normally (data not shown); thus, it is quite possible that the cardiovascular failure in Crk−/− embryos such as the poorly developed heart and defects in blood vessels results in a significant compromise of the circulatory system and leads to embryonic lethality. However, given the broad role of Crk in a variety of biological processes, we cannot rule out the possibility that other, undetected defects contribute to the death of Crk-null embryos.
Cleft palate was observed occasionally in Crk−/− embryos. Cleft palate is a multifactorial disease that often results from a combination of genetic and environmental factors (reviewed in reference 9). Mice deficient in growth factors and growth factor receptors such as transforming growth factor β3 (10, 18), platelet-derived growth factor C (PDGF-C) (3), and epidermal growth factor receptor (EGFR) (15) also exhibit cleft palate. Because Crk functions in growth factor receptor signaling pathways, it may participate in palate formation by mediating the signals downstream of multiple growth factor receptors or non-receptor tyrosine kinases. The partial penetration of the defect in Crk−/− mice indicates that other factors can compensate for the loss of Crk.
In contrast to the low frequency of the cleft palate phenotype, edema at the anterior end of the nasopharynx, where nasal septum and the primary palate fuse, was detected in all Crk−/− embryos examined. We do not yet understand the biological significance of edema in the nasopharynx; the precipitating cause of edema on the face and in the nasopharynx is also unclear. One possible explanation is that a defect in the mucosa/submucosal layers leads to separation of the two layers and provides a space that can fill with extracellular fluids. Interestingly, cleft palate and hemorrhagic edema on the face have also been seen in PDGF-C-deficient mice (3). Most of the defects found in Crk−/− embryos, including hemorrhagic edema, defective vascular smooth muscle cells, and edema in the nasopharynx, occurred primarily at the midline of the head. Since EGFR signaling is required for formation of head midline structures in Drosophila (4), Crk may specifically contribute to the formation of some midline structures by signaling downstream of EGFR.
Both Crk and CrkL have been reported to be ubiquitously expressed during development (2). Furthermore, Prosser et al. (19) reported mRNA and protein expression for all Crk proteins in all mouse tissues tested (including the heart). We confirmed the broad expression pattern of Crk and CrkL in early embryos, including the developing heart and the craniofacial region affected by the absence of Crk, in our recently published Gene Expression Atlas (GENSAT) project (http://www.stjudebgem.org/web/view/probe/viewProbeDetails.php?id=401 and http://www.stjudebgem.org/web/view/probe/viewProbeDetails.php?id=402) (12). We have been able to grow fibroblasts from E13.5 Crk−/− embryos, and so far they exhibit normal growth properties. At present, it is hard to pinpoint exactly which cells cause the developmental abnormalities we report here and it is possible they are consequences of early developmental errors that could either be cell extrinsic or cell intrinsic. The best way to address these interesting possibilities and to investigate the complex biological functions of both Crk and CrkL will be to use conditional alleles.
Acknowledgments
The authors would like to thank the following: St. Jude Children's Research Hospital shared core facilities for their help, the Cancer Center Core Cytogenetics Laboratory for karyotyping, the Transgenic Gene Knockout Facility for ES cell injection and germ line transmission, the Animal Resources Center for mouse husbandry, and Biomedical Communications for imaging of embryos. We also thank Michele Connelly and Jennifer DeBeauchamp for their technical assistance and Curran lab members and Angela J. McArthur for critical reading of the manuscript. Plasmids for Cre and NeoTk cassettes were kindly provided by Peter McKinnon.
This work was supported in part by grant 5R37NS036558 from NINDS, Cancer Center Support CA21765 from NCI, and the American Lebanese Syrian Associated Charities.
REFERENCES
- 1.Akakura, S., B. Kar, S. Singh, L. Cho, N. Tibrewal, R. Sanokawa-Akakura, C. Reichman, K. S. Ravichandran, and R. B. Birge. 2005. C-terminal SH3 domain of CrkII regulates the assembly and function of the DOCK180/ELMO Rac-GEF. J. Cell Physiol. 204:344-351. [DOI] [PubMed] [Google Scholar]
- 2.de Jong, R., L. Haataja, J. W. Voncken, N. Heisterkamp, and J. Groffen. 1995. Tyrosine phosphorylation of murine Crkl. Oncogene 11:1469-1474. [PubMed] [Google Scholar]
- 3.Ding, H., X. Wu, H. Bostrom, I. Kim, N. Wong, B. Tsoi, M. O'Rourke, G. Y. Koh, P. Soriano, C. Betsholtz, T. C. Hart, M. L. Marazita, L. L. Field, P. P. Tam, and A. Nagy. 2004. A specific requirement for PDGF-C in palate formation and PDGFR-alpha signaling. Nat. Genet. 36:1111-1116. [DOI] [PubMed] [Google Scholar]
- 4.Dumstrei, K., C. Nassif, G. Abboud, A. Aryai, A. Aryai, and V. Hartenstein. 1998. EGFR signaling is required for the differentiation and maintenance of neural progenitors along the dorsal midline of the Drosophila embryonic head. Development 125:3417-3426. [DOI] [PubMed] [Google Scholar]
- 5.Feller, S. M. 2001. Crk family adaptors—signalling complex formation and biological roles. Oncogene 20:6348-6371. [DOI] [PubMed] [Google Scholar]
- 6.Guris, D. L., J. Fantes, D. Tara, B. J. Druker, and A. Imamoto. 2001. Mice lacking the homologue of the human 22q11.2 gene CRKL phenocopy neurocristopathies of DiGeorge syndrome. Nat. Genet. 27:293-298. [DOI] [PubMed] [Google Scholar]
- 7.Honda, H., H. Oda, T. Nakamoto, Z. Honda, R. Sakai, T. Suzuki, T. Saito, K. Nakamura, K. Nakao, T. Ishikawa, M. Katsuki, Y. Yazaki, and H. Hirai. 1998. Cardiovascular anomaly, impaired actin bundling and resistance to Src-induced transformation in mice lacking p130Cas. Nat. Genet. 19:361-365. [DOI] [PubMed] [Google Scholar]
- 8.Imaizumi, T., K. Araki, K. Miura, M. Araki, M. Suzuki, H. Terasaki, and K. Yamamura. 1999. Mutant mice lacking Crk-II caused by the gene trap insertional mutagenesis: Crk-II is not essential for embryonic development. Biochem. Biophys. Res. Commun. 266:569-574. [DOI] [PubMed] [Google Scholar]
- 9.Jugessur, A., and J. C. Murray. 2005. Orofacial clefting: recent insights into a complex trait. Curr. Opin. Genet. Dev. 15:270-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaartinen, V., J. W. Voncken, C. Shuler, D. Warburton, D. Bu, N. Heisterkamp, and J. Groffen. 1995. Abnormal lung development and cleft palate in mice lacking TGF-beta 3 indicates defects of epithelial-mesenchymal interaction. Nat. Genet. 11:415-421. [DOI] [PubMed] [Google Scholar]
- 11.Keshvara, L., S. Magdaleno, D. Benhayon, and T. Curran. 2002. Cyclin-dependent kinase 5 phosphorylates disabled 1 independently of Reelin signaling. J. Neurosci. 22:4869-4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Magdaleno, S., P. Jensen, C. L. Brumwell, A. Seal, K. Lehman, A. Asbury, T. Cheung, T. Cornelius, D. M. Batten, C. Eden, S. M. Norland, D. S. Rice, N. Dosooye, S. Shakya, P. Mehta, and T. Curran. 2006. BGEM: an in situ hybridization database of gene expression in the embryonic and adult mouse nervous system. PLoS Biol. 4:e86. [Online.] doi: 10.1371/journal.pbio.0040086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsuda, M., S. Tanaka, S. Nagata, A. Kojima, T. Kurata, and M. Shibuya. 1992. Two species of human CRK cDNA encode proteins with distinct biological activities. Mol. Cell. Biol. 12:3482-3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mayer, B. J., M. Hamaguchi, and H. Hanafusa. 1988. A novel viral oncogene with structural similarity to phospholipase C. Nature 332:272-275. [DOI] [PubMed] [Google Scholar]
- 15.Miettinen, P. J., J. R. Chin, L. Shum, H. C. Slavkin, C. F. Shuler, R. Derynck, and Z. Werb. 1999. Epidermal growth factor receptor function is necessary for normal craniofacial development and palate closure. Nat. Genet. 22:69-73. [DOI] [PubMed] [Google Scholar]
- 16.Ogawa, S., H. Toyoshima, H. Kozutsumi, K. Hagiwara, R. Sakai, T. Tanaka, N. Hirano, H. Mano, Y. Yazaki, and H. Hirai. 1994. The C-terminal SH3 domain of the mouse c-Crk protein negatively regulates tyrosine-phosphorylation of Crk associated p130 in rat 3Y1 cells. Oncogene 9:1669-1678. [PubMed] [Google Scholar]
- 17.Ohba, Y., K. Ikuta, A. Ogura, J. Matsuda, N. Mochizuki, K. Nagashima, K. Kurokawa, B. J. Mayer, K. Maki, J. Miyazaki, and M. Matsuda. 2001. Requirement for C3G-dependent Rap1 activation for cell adhesion and embryogenesis. EMBO J. 20:3333-3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Proetzel, G., S. A. Pawlowski, M. V. Wiles, M. Yin, G. P. Boivin, P. N. Howles, J. Ding, M. W. Ferguson, and T. Doetschman. 1995. Transforming growth factor-beta 3 is required for secondary palate fusion. Nat. Genet. 11:409-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prosser, S., E. Sorokina, P. Pratt, and A. Sorokin. 2003. CrkIII: a novel and biologically distinct member of the Crk family of adaptor proteins. Oncogene 22:4799-4806. [DOI] [PubMed] [Google Scholar]
- 20.Reichman, C., K. Singh, Y. Liu, S. Singh, H. Li, J. E. Fajardo, A. Fiser, and R. B. Birge. 2005. Transactivation of Abl by the Crk II adapter protein requires a PNAY sequence in the Crk C-terminal SH3 domain. Oncogene 24:8187-8199. [DOI] [PubMed] [Google Scholar]
- 21.Reichman, C. T., B. J. Mayer, S. Keshav, and H. Hanafusa. 1992. The product of the cellular crk gene consists primarily of SH2 and SH3 regions. Cell Growth Differ. 3:451-460. [PubMed] [Google Scholar]
- 22.Romer, J. T., H. Kimura, S. Magdaleno, K. Sasai, C. Fuller, H. Baines, M. Connelly, C. F. Stewart, S. Gould, L. L. Rubin, and T. Curran. 2004. Suppression of the Shh pathway using a small molecule inhibitor eliminates medulloblastoma in Ptc1(+/−)p53(−/−) mice. Cancer Cell 6:229-240. [DOI] [PubMed] [Google Scholar]
- 23.Sayeski, P. P., M. S. Ali, J. B. Harp, M. B. Marrero, and K. E. Bernstein. 1998. Phosphorylation of p130Cas by angiotensin II is dependent on c-Src, intracellular Ca2+, and protein kinase C. Circ. Res. 82:1279-1288. [DOI] [PubMed] [Google Scholar]
- 24.Takahashi, T., Y. Kawahara, T. Taniguchi, and M. Yokoyama. 1998. Tyrosine phosphorylation and association of p130Cas and c-Crk II by ANG II in vascular smooth muscle cells. Am. J. Physiol. 274:H1059-H1065. [DOI] [PubMed] [Google Scholar]
- 25.Tang, D. D., and J. Tan. 2003. Role of Crk-associated substrate in the regulation of vascular smooth muscle contraction. Hypertension 42:858-863. [DOI] [PubMed] [Google Scholar]
- 26.Tang, D. D., W. Zhang, and S. J. Gunst. 2005. The adapter protein CrkII regulates neuronal Wiskott-Aldrich syndrome protein, actin polymerization, and tension development during contractile stimulation of smooth muscle. J. Biol. Chem. 280:23380-23389. [DOI] [PubMed] [Google Scholar]
- 27.Voss, A. K., P. Gruss, and T. Thomas. 2003. The guanine nucleotide exchange factor C3G is necessary for the formation of focal adhesions and vascular maturation. Development 130:355-367. [DOI] [PubMed] [Google Scholar]