Abstract
We constructed yeast strains in which rRNA gene repeats are integrated at ectopic sites in the presence or absence of the native nucleolus. At all three ectopic sites analyzed, near centromere CEN5, near the telomere of chromosome VI-R, and in middle of chromosome V-R (mid-V-R), a functional nucleolus was formed, and no difference in the expression of rRNA genes was observed. When two ribosomal DNA (rDNA) arrays are present, one native and the other ectopic, there is codominance in polymerase I (Pol I) transcription. We also examined the expression of a single rDNA repeat integrated into ectopic loci in strains with or without the native RDN1 locus. In a strain with reduced rRNA gene copies at RDN1 (∼40 copies), the expression of a single rRNA gene copy near the telomere was significantly reduced relative to the other ectopic sites, suggesting a less-efficient recruitment of the Pol I machinery from the RDN1 locus. In addition, we found a single rRNA gene at mid-V-R was as active as that within the 40-copy RDN1. Combined with the results of activity analysis of a single versus two tandem copies at CEN5, we conclude that tandem repetition is not required for efficient rRNA gene transcription.
It is now generally recognized that the locations of individual genes on the chromosome, as well as the spatial arrangement of the chromosomes within the nucleus, are often important for the regulation of gene expression. For example, genes brought near or within heterochromatin regions by chromosomal rearrangements or experimental manipulations are often repressed or silenced (for reviews, see references 16 and 44). In addition, recent studies have indicated that individual chromosomes are not randomly arranged within the interphase nucleus, thus suggesting a functional significance of chromosomal locations for gene expression (for reviews, see 49 and 51).
The spatial organization of chromosomes within the interphase nucleus of the yeast Saccharomyces cerevisiae has been studied by analyzing the locations of the centromeres, telomeres, and the nucleolus. Centromeres are clustered around the spindle pole body (SPB), which is localized within the nuclear envelope (4, 14, 18, 21, 22). Telomeres are clustered in a limited number of foci near the nuclear periphery but outside the centromere cluster (4, 13, 15, 33). This clustering of centromeres around the SPB and of telomeres in foci near the nuclear periphery poses some constraints on the mobility of these chromosomal regions, whereas most other chromosomal regions are highly mobile in yeast cells (4, 19, 35). Lastly, the crescent-shaped nucleolus, which contains approximately 150 tandemly repeated rRNA genes on chromosome XII, is positioned along the nuclear envelope opposite to the centromere cluster (4, 22, 56). The positioning of the nucleolus relative to the SPB may be of importance for the functions of centromeres and/or SPB or nucleolar functions, including the efficient transcription of rRNA genes and their regulation. Alternatively, this organization may be simply due to the chromosomal location of the rRNA gene repeats which are ∼300 kb away from the centromere, the length of which is significantly longer than the diameter of the yeast nucleus in its folded chromatin structure in interphase (3, 4).
Earlier studies have analyzed whether an rRNA gene itself without its flanking sequence is able to organize a functional nucleolus at ectopic sites and whether the tandemly repeated structure of rRNA genes is important for efficient expression (see reference 24 and references therein). Experiments investigating these questions most directly in vivo were those carried out by Karpen et al. (24) with Drosophila melanogaster. These experiments established that the ability to organize a nucleolar structure resides in an rRNA gene itself and that a single rRNA gene copy without the tandem structure is functional. The quantitative data indicated some significant (maximum of three- to fourfold) differences among the four sites of integration analyzed, suggesting a position effect on rRNA gene transcription. However, these sites were regions where random integration took place rather than defined and targeted sites, and the nature of the flanking DNA sequences was not characterized. In addition, the analyses of rRNA gene expression were done using laterally amplified polytene chromosomes and not for a truly isolated single-copy rRNA gene. Thus, as the authors commented, the very high copy number of rRNA genes laterally juxtaposed at these ectopic sites as a result of polytenization may have facilitated the recruitment of polymerase I (Pol I) machinery, leading to a high expression of rRNA genes.
In yeast, RNA Pol I expression at ectopic sites has been examined using a single 35S rRNA gene promoter juxtaposed with the E element (referred to as E/P and also as “HOT1” element) but not with an intact rRNA gene repeat. Three unique features for the expression from the E/P element ectopic sites were the requirement of the “enhancer” (or the E element) (23, 50), the requirement of the gene FOB1 (53) and a striking orientation dependency (20). The E element is operationally defined as the ∼320-bp EcoRI-HpaI region following the 35S rRNA coding region. It consists of two regions: the 190-bp EcoRI-HindIII region called “enhancer”(10) and the ∼130-bp HindIII-HpaI region containing the sites where Fob1 binds and DNA replication fork block takes place (2, 28). The enhancer region was originally shown to greatly stimulate rRNA synthesis in Pol I reporter systems carried by plasmids or integrated at ectopic chromosomal sites (10, 11, 23). Our previous studies demonstrated that the enhancer apparently stimulates Pol I transcription of reporter genes at ectopic sites or on plasmids but can be deleted from all of the chromosomal rRNA genes without any effects on Pol I transcription or on cell growth (53). Hence, the enhancer region together with the adjacent replication fork block region was suggested to be involved in Pol I recruitment to the ectopic reporter systems, with the aid of Fob1, perhaps through interactions with the corresponding regions in rRNA gene repeats in the nucleolus (53). Stimulation of Pol I transcription by the E element was also originally discovered in the course of studies of HOT1 which stimulates genetic recombination at nearby regions when inserted at a non-RDN1 site (26, 50). However, in all of these studies demonstrating stimulation of Pol I transcription by E element (or enhancer) at ectopic sites, expression of Pol I was measured by using reporter systems containing both a promoter region and E element or enhancer and not by using an intact rRNA gene copy.
We thought that questions related to position effects on Pol I transcription, nucleolar formation, and functions could be studied using the yeast system we have developed. We have constructed strains of the yeast Saccharomyces cerevisiae (called “rdnΔΔ” strains) in which all of the chromosomal rRNA gene repeats were deleted from the RDN1 locus on chromosome XII (38, 54). These strains are able to grow due to the presence of multicopy plasmids carrying the 35S and 5S rRNA coding regions. The multicopy plasmids were constructed with either the natural Pol I promoter (“Pol I plasmid”) or the GAL7 promoter (“Pol II plasmid”) fused to the 35S coding region. The Pol II plasmid in rdnΔΔ strains allows 35S rRNA to be transcribed by Pol II, and the growth of these strains is dependent on galactose (38, 54).
Our approach was to design DNA fragments to integrate rRNA genes into several desired chromosomal positions and ask the following questions. (i) How does expression of a single copy of 35S rRNA gene at an ectopic site compare to expression of a single copy at RDN1? We thought that this question of expression of a single-copy rRNA gene at an ectopic site should be analyzed in the absence (i.e., in rdnΔΔ strain), as well as in the presence, of rRNA gene repeats at RDN1. (ii) Can rRNA gene repeat expansion take place at ectopic loci and can nucleolar structures with normal functions form at these loci? (iii) If a functional nucleolus can be formed at ectopic loci, how does the expression of 35S rRNA genes at ectopic loci compare to the expression of rRNA genes at RDN1? (iv) When a strain carries rRNA gene repeats at both ectopic and normal loci, is there any phenomenon of nucleolar dominance, as is often observed in cells carrying several rRNA gene arrays such as human cells (43) or in interspecific hybrid cells (reviewed in reference 17).
We used a strategy that allows a single new rRNA gene to be integrated at ectopic chromosomal sites without repeat expansion, followed by expansion of the gene repeats as a separate step. We selected loci close to the centromere on chromosome V, CEN5, and the telomere on the right arm of chromosome VI, TEL VI-R, because telomeres and centromeres occupy unique nuclear locations (Fig. 1A). For the purpose of comparison, a locus between the centromere and telomere was selected as an integration site (“mid-V-R”), as well as the original (RDN1 on chromosome XII) locus (Fig. 1A). We were able to construct yeast strains that carry a single rRNA gene, as well as an expanded number of rRNA genes, at these selected ectopic loci in the absence as well as the in the presence of the native rRNA gene copies at RDN1. The results of experiments using the various yeast strains obtained in this way are discussed in connection with the four questions listed above and related questions.
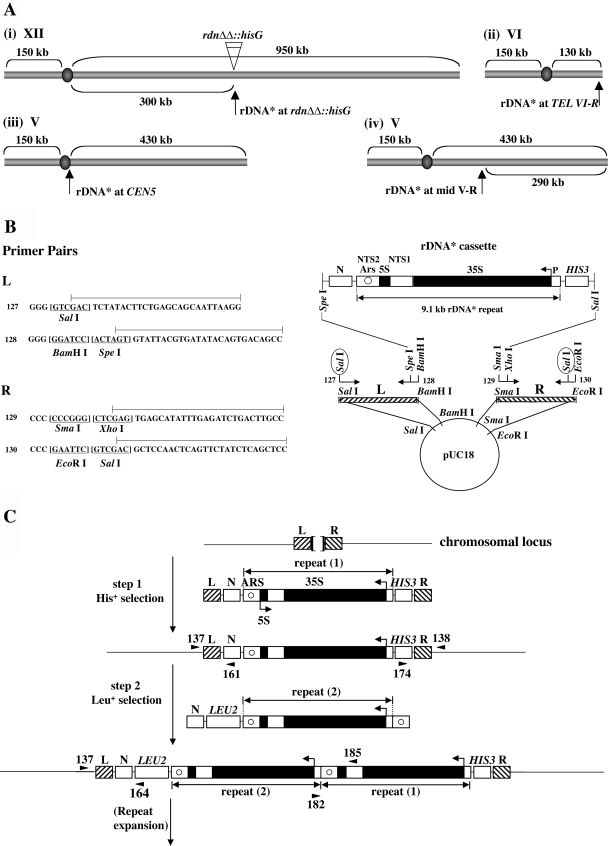
FIG. 1.
Integration of and expansion of a new rDNA repeat at both normal and ectopic chromosomal sites. (A) Sites of integration carried out in the present study. A new rDNA repeat carries a hyg1 mutation and a 26-bp tag (see the text and Fig. 2A) and is designated “rDNA*.” (B) Construction of plasmid pNOY3292 used for integration of rDNA* at the rdnΔΔ::hisG locus [see Fig. 1A(i)]. The rDNA* cassette consists of an entire 9.1-kb rRNA gene copy flanked by the HIS3 gene and a non-yeast sequence N from A. thaliana. The 35S and 5S rRNA coding regions are shown as filled regions of the rDNA* repeat. Nontranscribed sequences 1 and 2 are labeled as NTS1 and NTS2, respectively. ARS is shown as a circle within NTS2. The lines above the primer sequences (primers 127, 128, 129, and 130) show the regions that are identical to the sequences of chromosomal DNA flanking the RDN1 locus (called “L” and “R”). Sequences of restriction enzyme sites are underlined. Digestion of plasmid pNOY3292 with SalI (circled) will produce an ∼13-kb fragment containing L, N, rDNA*, HIS3, and R. This fragment was used for step 1 shown in C. (C) General strategy used for the integration of rDNA* at a given chromosomal site and its subsequent expansion. Two separate steps and DNA fragments used for integration of rDNA* are shown. The sequences flanking the site of integration are shown as L and R. For integration of rDNA* at the original RDN1 site in rdnΔΔ::hisG strains, the Escherichia coli hisG sequence is between L and R flanking sequences (as shown by a bracket at the top of the figure). In the other three cases (see panel A), the sequences between L and R are 0 to 8 bp as described elsewhere (see the supplemental material). The DNA fragment used for the second step is an ∼13-kb fragment obtained after digestion of pNOY3293 with SpeI and SalI. The positions of primers used to confirm correct integration are shown as arrowheads, taking examples for integration at the rdnΔΔ::hisG locus. The primer pairs 137/161 and 174/138 produce 1,036- and 1,263-bp fragments, respectively, for correct integration at the first step. PCRs using primer pairs 137/164 and 182/185 produce 2,007- and 1,800-bp fragments, respectively, for correct integration at the second step. For integration at other ectopic loci, primers at equivalent positions were used for PCRs as described elsewhere (see the supplemental material).
MATERIALS AND METHODS
Yeast strains, plasmids, and media.
Yeast strains and plasmids used are listed in Table 1. The construction of rdnΔΔ strains that carry a single ribosomal DNA* (rDNA*) copy integrated into ectopic loci was as follows. The three ectopic loci selected for integration are shown in Fig. 1Aii, iii, and iv, respectively. Initially, an rDNA cassette was constructed in which the 9.1-kb rRNA gene repeat is flanked with a nonyeast sequence NMD3 (“N”) from Arabidopsis thaliana on the left end and with HIS3 on the right end (Fig. 1B). The rRNA gene repeat used in this cassette (called rDNA*) carries hyg1 mutation in the 18S rRNA coding region (5) and a 26-bp sequence tag (Fig. 2A). For each integration site chosen, primers were designed to separately amplify sequences on the left side and right sides of the site selected for integration (e.g., Fig. 1B for RDN1; for other loci and details, see the supplemental material). The rDNA* cassette was then inserted between the right and left flanking sequences in a plasmid derived from pUC18 (Fig. 1B). After digestion of the resultant plasmid with SalI (Fig. 1B), the DNA fragment containing the rDNA* cassette and flanking L and R sequences for a particular site was isolated. This fragment was transformed directly into our standard strain NOY388 and His+ colonies were selected (Fig. 1C, step 1). The strains with a single rDNA* repeat unit at an ectopic locus were then crossed with the rdnΔΔ strain NOY984 that carried a Pol II helper plasmid. After sporulation and tetrad dissection on YEPG medium, colonies that were galactose dependent and His+ were selected. These rdnΔΔ::hisG strains carried a Pol II helper plasmid and had a single rDNA* repeat (repeat 1) integrated at an ectopic chromosomal locus. Correct integration was confirmed by PCR analysis (see primers shown for the structure after step 1 in Fig. 1C). Three strains—NOY2047, NOY987, and NOY988—carrying a single rDNA repeat at ectopic loci (at or near TEL VI-R, CEN5, and mid-V-R as shown in Fig. 1Aii, iii, and iv, respectively) were obtained in this way. We note that the site of integration in NOY2047 is ∼500 bp from the telomere repeats ([TG1-3]n-end) and, because of the presence of the ∼1.0-kb HIS3 gene, the integrated rDNA* repeat is located ∼1.5 kb from the telomere repeats. (An integration site near TEL VI-R was selected because of the absence of Y′ element at this chromosome end [41].) Similarly, in the case of NOY987, the site of integration is 744 bp to the right of the centromere, and the distance between CEN5 and the 9.1-kb rDNA unit is ∼1.7 kb (744 bp plus ∼960 bp of the N sequence). In carrying out the primer extension experiments to measure 5′-end rRNA transcripts shown in Fig. 2B, we discovered that two (NOY2047 and NOY988, lanes 1 and 3, respectively) of the four strains showed RNA transcripts corresponding to the transcript from the Pol I promoter on the endogenous rDNA repeats even though those repeats are missing. The intensity of these “RDN1” bands was only slightly (20 to 30%) higher than that of rDNA* bands. These endogenous rRNA gene copies are either extrachromosomal DNA similar to 3-μm DNA or extrachromosomal rRNA gene circles (ERC). The 3-μm DNA can be maintained independently of chromosomal rRNA gene repeats (see, for example, references 8, 32, and 41), and the ERC are derived from rRNA gene repeats and accumulate in aging cells because of their strongly biased inheritance by mother cells (48). Since initial attempts to remove the endogenous rRNA gene copies by single-colony purification were not successful, we used NOY988 and NOY2047 as strains carrying a single rDNA* integrated at mid-V-R and near TEL VI-R, respectively, without further attempts to cure the residual endogenous rRNA gene copies. We believe that the presence of these residual rRNA gene copies does not affect any of the conclusions obtained in the present study.
TABLE 1.
Plasmids and yeast strains used in this study
| Plasmid or strain | Description |
|---|---|
| Plasmids | |
| pNOY130 | High-copy-number plasmid carrying GAL7-35S rDNA 5S rDNA URA3 2μ amp (54) |
| pNOY3286 | A derivative of pBluescript SK(+) carrying a single rDNA* (hyg1, 26-bp tag) copy and the nonyeast DNA (N) and HIS3 |
| pNOY3289 | A derivative of pUC18 carrying the DNA fragment used for integration of a single rDNA* copy at the site in the middle of chromosome V-R |
| pNOY3290 | A derivative of pUC18 carrying the DNA fragment used for integration of a single rDNA* copy at the site near TEL VI-R |
| pNOY3291 | A derivative of pUC18 carrying the DNA fragment used for integration of a single rDNA* copy at the site near CEN5 |
| pNOY3292 | A derivative of pUC18 carrying the DNA fragment used for a single integration of rDNA* copy at the site corresponding to the native RDN1 site in rdnΔΔ strains (see Fig. 1B) |
| pNOY3293 | A derivative of pBluescript SK(+) carrying the DNA fragment (∼13-kb SpeI-SalI fragment; Fig. 1C) used for integration of a second rDNA* copy and subsequent rDNA* repeat expansion |
| pNOY3300 | A derivative of pNOY3291 with rDNA* transcribing in the opposite direction (see Fig. 3A) |
| pNOY3313 | A derivative of pNOY3291 with the E element deleted (see Fig. 3A) |
| pNOY3318 | A derivative of pNOY3289 with rDNA* transcribing in the opposite direction |
| pNOY3319 | A derivative of pNOY3290 with rDNA* transcribing in the opposite direction |
| Strains | |
| NOY388 | MATaade2-1 ura3-1 trp1-1 leu2-3,112 his3-11,15 can1-100 |
| NOY396 | MATα ade2-1 ura3-1 trp1-1 leu2-3,112 his3-11,15 can1-100 |
| NOY886 | MATα rpa135Δ-LEU2 ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 fob1Δ::HIS3, pNOY117 [CEN RPA135 TRP1]; rDNA copy no. ∼40 (12) |
| NOY984 | MATα ade2-1 ura3-1 trp1-1 leu2-3,112 his3-11,15 can1-100 rdnΔΔ::hisG, pNOY130 |
| NOY986 | MATaade2-1 ura3-1 trp1-1 leu2-3,112 his3-11,15 can1-100 rdnΔΔ::hisG, pNOY130 |
| NOY987 | MATα ade2-1 ura3-1 trp1-1 leu2-3,112 his3-11,15 can1-100rdnΔΔ::hisG, pNOY130, single rDNA* repeat near CEN5 |
| NOY988 | MATα ade2-1 ura3-1 trp1-1 leu2-3,112 his3-11,15 can1-100rdnΔΔ::hisG, pNOY130, single rDNA* at mid-V-R |
| NOY1071 | MATα ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 fob1Δ::HIS3, rDNA copy no. ∼25 (7) |
| NOY2029 | MATaade2-1 ura3-1 trp1-1 leu2-3,112 his3-11,15 can1-100, rDNA* array substituting for RDN1 |
| NOY2030 | MATα ade2-1 ura3-1 trp1-1 leu2-3,112 his3-11,15 can1-100rdnΔΔ::hisG, rDNA* array near CEN5 |
| NOY2031 | MATα ade2-1 ura3-1 trp1-1 leu2-3,112 his3-11,15 can1-100rdnΔΔ::hisG, rDNA* array at mid-V-R |
| NOY2032 | MATaade2-1 ura3-1 trp1-1 leu2-3,112 his3-11,15 can1-100, rdnΔΔ::hisG, rDNA* array near TEL VI-R |
| NOY2039 | MATa/MATα ade2-1/ade2-1 ura3-1/ura3-1 trp1-1/trp1-1 leu2-3,112/leu2-3,112 his3-11,15/his3-11,15 can1-100/can1-100 RDN1/rDNA* array |
| NOY2040 | Same as NOY2039, but RDN1/rdnΔΔ and rDNA* array near CEN5 on one of two chromosome Vs |
| NOY2041 | Same as NOY2039, but RDN1/rdnΔΔ and rDNA* array at mid-V-R on one of two chromosome Vs |
| NOY2042 | Same as NOY2039, but RDN1/rdnΔΔ and rDNA* array near TEL VI-R on one of two chromosome VIs |
| NOY2047 | MATaade2-1 ura3-1 trp1-1 leu2-3,112 his3-11,15 can1-100rdnΔΔ::hisG, pNOY130, single rDNA* repeat near TEL VI-R |
| NOY2048 | MATaade2-1 ura3-1 trp1-1 leu2-3,112 his3-11,15 can1-100, single rDNA* repeat substituting for RDN1, pNOY130 |
| NOY2050 | Same as NOY388, but carries a single rDNA* near CEN5 |
| NOY2051 | Same as NOY388, but carries a single rDNA* at mid-V-R |
| NOY2052 | Same as NOY388, but carries a single rDNA* near TEL VI-R |
| NOY2053 | MATaade2-1 ura3-1 trp1-1 leu2-3,112 his3-11,15 can1-100, RDN1 and rDNA* array near CEN5 |
| NOY2054 | MAT? ade2-1 ura3-1 trp1-1 leu2-3,112 his3-11,15 can1-100, RDN1 and rDNA* array at mid-V-R |
| NOY2055 | MAT? ade2-1 ura3-1 trp1-1 leu2-3,112 his3-11,15 can1-100RDN1 and rDNA* array near TEL VI-R |
| NOY2056 | MATa/MATα ade2-1/ade2-1 ura3-1/ura3-1 his3-11,15/his3-11,15 trp1-1/trp1-1 leu2-3,112/leu2-3,112 can1-100/can1-100 rdnΔΔ::rDNA* (a single repeat)/RDN1 |
| NOY2063 | Same as NOY886, but fob1Δ::hisG instead of fob1Δ::HIS3 |
| NOY2064 | Same as from NOY2063, but carries a single rDNA* near CEN5 (derived from pNOY3291) |
| NOY2068 | Same as NOY1071, but fob1Δ::hisG instead of fob1Δ::HIS3 |
| NOY2146 | Same as NOY2064, but a single rDNA* near CEN5 is in the opposite orientation (derived from pNOY3300) |
| NOY2147 | Same as NOY2064, but a single rDNA* near CEN5 carries ΔE deletion (derived from pNOY3313) |
| NOY2148 | Same as NOY2063, but carries a single rDNA* copy within rDNA repeats at RDN1 (integration carried out using pNOY3286 digested with MluI, which gives a single cut within the 25S rRNA coding region) |
| NOY2149 | Same as NOY2063, but carries a single rDNA* copy at mid-V-R (derived from pNOY3289) |
| NOY2150 | Same as NOY2149, but a single rDNA* copy at mid-V-R is in the opposite orientation (derived from pNOY3318) |
| NOY2151 | Same as NOY2063, but carries a single rDNA* copy near TEL VI-R (derived from pNOY3290) |
| NOY2152 | Same as NOY2151, but a single rDNA* copy near TEL VI-R is in the opposite orientation (derived from pNOY3319) |
| NOY2158 | Same as NOY2159, but carries a second rDNA* near CEN5 (derived from pNOY3296) |
| NOY2159 | Same as NOY2068, but carries a single rDNA* near CEN5 (derived from pNOY3291) |
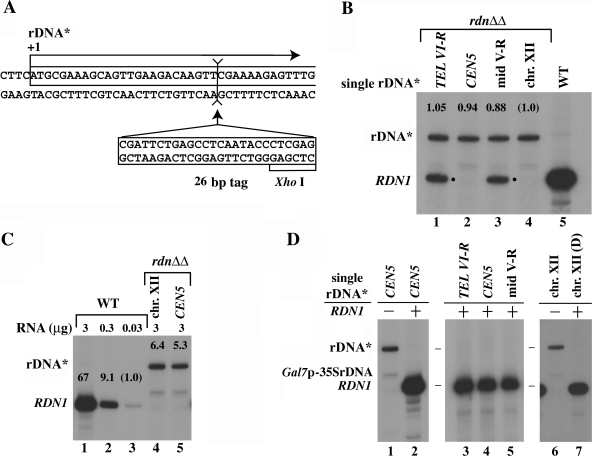
FIG. 2.
Synthesis of rRNA in strains carrying single rRNA* integrated at ectopic and the native sites. (A) Nucleotide sequence of rDNA* around the transcription start site (+1) showing the site of insertion of the 26-bp tag. The XhoI site in the 26-bp tag is also indicated. (B) RNA was prepared from exponentially growing cells of the four single rDNA* integrants (NOY2047, NOY987, NOY988, and NOY2048; lanes 1 to 4) and NOY388 (WT; lane 5). A total of 1 μg of RNA was analyzed by primer extension, and an autoradiogram is shown. The amounts of radioactive rRNA in relevant bands were measured, and normalized values are shown above the bands. The bands labeled by a dot are those corresponding to transcripts from native rDNA copies presumably on 3-μm circles or ERCs (see Materials and Methods). (C) RNA was prepared from rdnΔΔ strains carrying a single rDNA* copy at the native site (lane 4) or the CEN5 site (lane 5) and from NOY388 (lanes 1 to 3). The indicated amounts of RNA were used to measure the amounts of the 5′ end of 35S rRNA by primer extension, and the autoradiogram is shown The measured values were normalized to that obtained for 0.03 μg of RNA from the WT, and normalized values are shown above the respective bands. (D) An autoradiogram showing transcription of rDNA* analyzed by primer extension. RNA samples (3 μg) from NOY987 and NOY2050 (lanes 1 and 2, respectively) and those from NOY2052, NOY2050, NOY2051, NOY2048, and NOY2056 (lanes 3 to 7, respectively) were analyzed. We note that, for samples corresponding to lanes 2 to 5 and lane 7, bands for rDNA* transcripts were not recognized even after longer exposures and/or using higher amounts of RNA for primer extension.
For integration into the original RDN1 locus, the DNA fragment containing the rDNA* cassette together with flanking L and R sequences was transformed into a diploid strain (formed by crossing NOY396 with NOY986 [see Table 1]) carrying one chromosome XII with RDN1 and another with rdnΔΔ::hisG. His+ transformants that carried a single rDNA* repeat replacing rdnΔΔ::hisG were sporulated, and galactose-dependent His+ haploid segregants were selected. NOY2048 obtained in this way carried helper Pol II plasmid, and its growth was galactose dependent.
The rDNA* cassette for the second integration and expansion step carried a 9.1-kb rDNA* repeat with an additional “NTS2” region (between the 5S RNA gene and the SmaI site) on the right side and an N-LEU2 fragment on the left side (Fig. 1C, step 2). Recombination at the two regions, N and NTS2, resulted in transformants carrying two (or more) tandem rDNA* repeats that were selected as Leu+. The correct integration was confirmed by PCR analysis of the left end of the insertion site and the junction of the two rDNA* repeats (see primer pairs shown for the structure after step 2 in Fig. 1C). Additional information regarding strain construction and PCR primers is provided in the supplemental material.
Yeast extract-peptone-galactose (YEPG), YEP-glucose (YEPD), synthetic galactose (SG), and glucose (SD) media were as described previously (45, 47).
Other methods.
Immunofluorescence microscopy (IFM) was done as described previously (38). The rabbit anti-A190 antibody was described previously (55). The mouse monoclonal anti-Nop1 antibody (28F2) was obtained from EnCor Biotechnology, Inc. (Alachua, FL). To localize SPB in interphase, YOLI/34 monoclonal rat anti-yeast tubulin antibody (obtained from Accurate Chemical and Scientific Corp., Westbury, NY) was used (22).
Contour-clamped homogeneous electric field electrophoresis (CHEF) was also carried out as described previously (39). Approximate rDNA copy numbers were calculated from the size of chromosomes carrying integrated rDNA* array, as was done previously (39). Isolation of DNA and Southern hybridization analysis were carried out as described by Maniatis et al. (34). For Southern analysis, DNA was digested with XhoI and EcoRV, which gave 783 bp (8760 to +406) for rDNA and 427 bp (8760 to the XhoI site within the 26-bp insert; see Fig. 2A) for rDNA*. These two fragments were separated by agarose gel electrophoresis and detected by 32P-labeled 784-bp SphI-SmaI (8145 to −209) DNA fragment as a probe. Relative rDNA copy numbers were then determined by using a PhosphorImager (Bio-Rad, Hercules, CA).
Analysis of 5′ ends of precursor rRNA by primer extension was carried out as described previously (25), using the primer 5′-ACACGCTGTATAGAGACTAGGC-3′, which hybridizes to the 35S precursor rRNA 130 nucleotides downstream of the Pol I start site. Quantification was done with a PhosphorImager.
RESULTS
In our previous study (54), reintegration of a new rDNA copy into the original RDN1 site in a rdnΔΔ strain and its subsequent expansion were performed in a single step. In the present study, we carried out integration and expansion separately, at three ectopic sites shown in Fig. 1Aii, iii, and iv, respectively. Various strains carrying a single or expanded tagged-rDNA (rDNA* [see Fig. 2A]) at these ectopic loci either in the absence or presence of the endogenous rDNA repeats were constructed as described in Materials and Methods.
Expression of a single copy of 35S rRNA gene at an ectopic site compared to expression of a single copy at RDN1.
The expression of the integrated 35S rRNA gene was analyzed by measuring the amounts of the 5′ end of unstable precursor 35S rRNA by primer extension. Transcripts from the integrated rDNA* repeats detected by primer extension migrate more slowly than the control RDN1 transcripts because of the presence of the 26-bp insert near the 5′ start site of the 35S rRNA (Fig. 2A).
Strains with a single rDNA* repeat in a rdnΔΔ background (with Pol II helper plasmid pNOY130) were grown in galactose (YEPG) medium and then shifted to glucose (YEPD) for 1 h prior to RNA sample preparation. As shown in Fig. 2B, there was no significant difference in Pol I transcription activity among four strains carrying a single rDNA* copy at one of the three ectopic loci or at the native RDN1 locus. Thus, a single rDNA* copy at ectopic loci or at the site corresponding to the native RDN1 locus is equally accessible to Pol I and presumably to other Pol I transcription factors in rdnΔΔ strains. We conclude that Pol I transcription of a single 35S rRNA gene is not dependent on any particular characteristics specific for chromosome XII, nor is it affected by the locations in ectopic chromosomal regions analyzed.
Additional experiments were carried out to compare quantitatively the transcription activity of a single copy of the 35S rRNA gene in rdnΔΔ strains relative to the total rRNA transcription activity of rDNA repeats on chromosome XII in the standard strain, NOY388. An example is shown in Fig. 2C. Quantification from the results of this and other experiments showed that the Pol I transcription of a single copy (at any of the four sites) is ca. 6% (6.0 ± 2.2; from four independent experiments and comparison based on a unit amount of cellular RNA) of that seen in the control strain. After correction for the difference in cellular levels of RNA per cell mass (A600), this value corresponds to ∼3.4% of the rRNA synthesis rate (per cell mass) of the control strain NOY388 and may represent the maximum activity possible for transcription of a single 35S rRNA gene.
The experiments described above were performed in a rdnΔΔ background where Pol I is diffused throughout the nucleus and there is no normal crescent-shaped nucleolus (38). Using strains carrying both a single rDNA* copy at an ectopic site and the native RDN1, which were constructed initially (NOY2050, NOY2051, and NOY2052), we examined transcription of a single ectopic rDNA* relative to total transcription of the native genes at RDN1. As shown in Fig. 2D (lanes 2 to 5 compared to lane 1), none of the three ectopic rDNA* copies gave any detectable Pol I transcripts. Similarly, we also failed to detect the expression of a single rDNA* copy at the native site in the presence of the native RDN1 on the other chromosome XII in a diploid strain (Fig. 2D, lane 7 compared to lane 6). If the efficiency of expression of a single rDNA* copy at an isolated site is the same as the average efficiency of a single copy at the native RDN1 which contains ∼200 copies, we would expect the expression of rDNA* relative to the total rRNA transcription rate to be ∼0.5%. Since the limit of detection in these experiments was ∼0.5 to 1% of the RDN1 band (Fig. 2C), we conclude that the level of expression of a single rDNA* copy at ectopic sites is much less than 1% of rRNA synthesis in wild-type (WT) strains. We therefore examined the expression of a single ectopic rDNA* (integrated near CEN5) in strains, in which rRNA gene repeat number at RDN1 is reduced significantly less than ∼200 present in the original strain.
Our reduced rRNA gene copy strains are in a fob1Δ background in order to maintain the rRNA gene repeats at a reduced number. NOY886 has ∼40 rRNA gene copies and grows as well as control strains carrying ∼140 rRNA gene copies (12, 29). NOY1071 has ∼25 rRNA gene copies and grows slightly slower than control strains carrying ∼190 rRNA gene copies (7). We integrated a single rDNA* fragment (obtained from pNOY3291) at the CEN5 locus of a strain (NOY2063) derived from NOY886 and were indeed able to detect expression of the single rDNA* copy at this ectopic site (Fig. 3B, lanes 2 and 4, and C, lane 1). We then analyzed several factors that may influence expression of an ectopic rRNA gene.
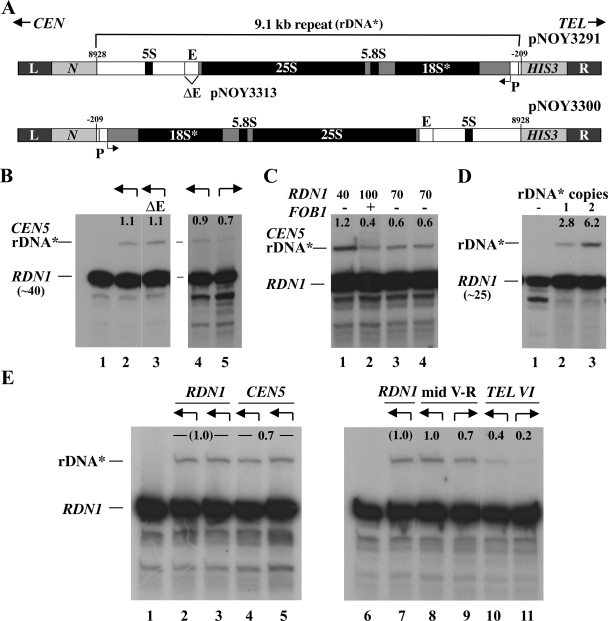
FIG. 3.
Transcription of a single rRNA gene at an ectopic site in the presence of reduced copy number of rRNA genes at RDN1. (A) Structures of the DNA fragment carried by pNOY3291 used for integration of a single copy of rDNA* near CEN5 and that carried by pNOY3300 used for integration of a single-copy rDNA* in the opposite orientation near CEN5. The L and R sequences are those flanking the sites of integration, as explained in Fig. 1B and C. The structure of the first DNA fragment is the same as that shown in Fig. 1B, with more details identified. The Pol I transcription start site is defined as +1, and the position of the E element (the 319-bp EcoRI-HpaI region) is indicated. pNOY3313 is the same as the pNOY3291 used for integration near CEN5 of rDNA* but has a deletion of the E element (ΔE; see lane 3 in panel B). (B) RNA samples (5 μg) from NOY2063, NOY2064, and NOY2147 (lanes 1 to 3, respectively) were analyzed by primer extension. In a separate experiment, RNA samples (10 μg) from NOY2064 (lane 4) and NOY2146 (lane 5) were similarly analyzed. The amounts of transcripts from ectopic rDNA* and those from the 40 copy repeats at RDN1 were measured by using a PhosphorImager, and the ratios of the ectopic ones to those from the RDN1 are given as percentage values above the rDNA* bands. The arrows above each lane indicate the direction of transcription of the rRNA* gene integrated at the CEN5 ectopic site as shown in panel A. (C) RNA samples (10 μg) from NOY2064 (lane 1), NOY2064 after introduction of FOB1 (lane 2), and after introduction followed by the removal of FOB1 (lanes 3 and 4 for two independent isolates) were analyzed by primer extension. The rDNA copy numbers at RDN1 of the same cultures used for primer extension were measured by Southern blot analysis and are indicated above the lanes. The expression of rDNA* relative to that of rDNA repeats at RDN1 was calculated and the values are indicated above the rDNA* bands as in panel B. (D) An autoradiogram showing transcripts of rDNA* compared to RDN1 (∼25-copy strain). RNA samples (5 μg) from NOY2068 (lane 1), NOY2159 (lane 2), and NOY2158 (lane 3) were analyzed by primer extension. The expression of rDNA* relative to that of rRNA gene repeats at RDN1 was calculated, and the values are indicated above the rDNA* bands as in panel B. The results shown are from one of several independent transformants. The values from this and other experiments averaged 2.8 ± 0.2 for NOY2159 and 6.2 ± 0.4 for NOY2158. (E) Transcription of a single rRNA* gene at an ectopic site compared to the expression of a single rDNA* copy integrated into native copies at RDN1. Arrows above each lane indicate the directions of transcription of the rDNA* gene shown in the way similar to that shown in panel B. Autoradiograms show the transcription of rDNA* analyzed by primer extension. RNA samples (5 μg) from NOY2063 (lane 1); NOY2148 (two independent rDNA* integrants [lanes 2 and 3]); NOY2064 (two independent rDNA* integrants [lanes 4 and 5]); and NOY2063, NOY2148, NOY2149, NOY2150, NOY2151, and NOY2152 (lanes 6 to 11, respectively) were analyzed. The ratios of expression of rDNA* to that of the native rRNA gene repeats (∼40 copies) at RDN1 were measured. The average of values for lanes 4 and 5 were normalized to the average of those for lanes 2 and 3. The average of the normalized value from this and other experiments was 0.68 ± 0.09 and is shown as 0.7. For lanes 8 to 11, the ratios of rDNA* to rRNA gene repeats at RDN1 transcripts were measured and normalized to the value for NOY2148 (lane 7). Two independent integrants were used for each ectopic site and both orientations. The normalized values from this and other experiments were averaged, yielding values for samples corresponding to lanes 8, 9, 10, and 11 to be 1.03 ± 0.17, 0.68 ± 0.21, 0.40 ± 0.17, and 0.21 ± 0.17, respectively. The rounded numbers are given above the rDNA* bands.
Transcription from the Pol I promoter was previously studied using the E/P and related elements (see the introduction). To our surprise, we discovered that the expression of a single rDNA* (Fig. 3A, pNOY3291 versus pNOY3313) at the CEN5 site in our ∼40 rRNA copy strain is different from that observed for the E/P element and does not require the E element (Fig. 3B, lanes 2 and 3). In addition, the expression is, if any, only weakly orientation dependent (Fig. 3B, lanes 4 and 5). The ratio of the expression of rDNA* at the opposite orientation to that of rDNA* at the “normal” orientation was found to be 0.70 ± 0.28. Lastly, the expression is FOB1 independent. In order to examine whether the presence of the FOB1 gene stimulates the expression of a single rDNA* copy at the CEN5 ectopic site, we introduced FOB1 on a URA3-based CEN plasmid (pRS316) into the 40-copy strain carrying a single rDNA* copy near CEN5 (NOY2064) by transformation. One of the transformants was grown in YEPD, and a portion of the culture was used for primer extension analysis of transcription of rDNA* relative to that of rRNA gene repeats at RDN1. Simultaneously, another portion was used for determination of rRNA gene copy number at RDN1. It was found that, because of the introduction of the FOB1 gene, the rRNA gene copy number at RDN1 increased to ∼100 and that transcription of the ectopic rDNA* was ∼0.4% of that of the genes at RDN1 (Fig. 3C, lane 2). Separately, the cells that received the FOB1 gene were streaked on plates containing 5-fluoorotic acid to eliminate the FOB1 gene on the CEN plasmid. Two independently obtained fob1Δ clones were then analyzed for rRNA gene copy number at RDN1 (∼70 copies in both), as well as for relative expression of a single ectopic rDNA* copy at the CEN5 site (∼0.6% of the total activity of the genes at RDN1) (Fig. 3C, lanes 3 and 4). Thus, we can formally calculate that the activity for single rDNA* expression relative to a single copy at RDN1 (for more accurate estimates of transcription activities at ectopic loci, see below) in fob1Δ strains with 40 copies and 70 copies of rRNA genes at RDN1 was 48% (1.2/[100/40] = 0.48) and 42% (0.6/[100/70] = 0.42), respectively. These values are not very different from (and certainly not less than) the 40% value (0.4/[100/100] = 0.4) calculated for the FOB1 strain with 100 copies at RDN1. Thus, we conclude that, in contrast to the transcription from the E/P fragment at an ectopic site (53), transcription from the single rDNA* copy at this ectopic site is independent of FOB1. The results also support the notion of competition between the rRNA gene repeats at RDN1 and a single rDNA* copy at ectopic sites and explains why expression of the latter was not observed in the original strains carrying ∼200 rRNA gene copies at RDN1.
We should note that the single rRNA gene copy we used in the present study is a 9.1-kb repeat unit obtained after digestion of the rRNA gene repeats with SmaI, which cleaves rRNA genes at −209 (see Fig. 3A for repeat details). This choice was rationalized because of the known upstream promoter element extending to approximately −155 (6, 27, 30, 37), but not beyond, combined with a convenience of the use of SmaI to generate a single repeat unit. However, the promoter region was defined using reporter plasmids or in vitro extracts but not in a chromosome context. For this reason, we constructed a fob1Δ strain carrying two tandemly repeated rDNA* copies stably integrated at the CEN5 ectopic site in addition to the ∼25 rRNA copies at RDN1 (NOY2158). The expression from two copies of rDNA* was compared to the control strain carrying a single rDNA* copy at the same CEN5 site (NOY2159). The former strain showed a twofold higher value (within experimental errors) than the latter (Fig. 3D, compare lanes 2 and 3). This result confirms the absence of any significant stimulation of transcription of a rRNA gene by hypothetical cis elements further “upstream” of SmaI site (−209) or by transcription of the upstream rRNA gene as proposed by earlier investigators (“polymerase hand-over model” [1, 36, 42; see also the Discussion]).
In order to overcome difficulty inherent in the quantitative comparison of a very weak activity of a single copy of rDNA* at the CEN5 ectopic site with the total transcription activity at RDN1, we compared the expression of a single copy of rDNA* at the CEN5 ectopic site with a single copy of rDNA* integrated at RDN1. As shown in Fig. 3E, the expression of rDNA* at the CEN5 ectopic site was slightly weaker than that of an rDNA* at RDN1 (Fig. 3E, compare lanes 4 and 5 with lanes 2 and 3). The ratio of the expression at CEN5 to that at RDN1 was 0.68 ± 0.09.
Expression of a single-copy rDNA* at mid-V and that at TEL VI-R was also analyzed in a similar way. At the mid-V ectopic site, expression of a single rDNA* copy in the same orientation as that in RDN1 was comparable to that of a single rDNA* copy within RDN1 but expression in the opposite orientation appeared to be weakly reduced (Fig. 3E, lanes 8 and 9, compared to lane 7; see the legend). Finally, expression of a single-copy rDNA* at the TEL VI-R site was found to be considerably less, especially in its opposite orientation, than that of a single rDNA* copy within RDN1. Accurate estimate of the weak expression at the TEL VI-R site was difficult because of background problems. Nevertheless, it is clear that expression at this ectopic site is significantly weaker than expression at RDN1, especially in the orientation opposite to that at RDN1 (see the legend to Fig. 3E).
Repeat expansion can occur at ectopic loci and nucleolar structures with normal functions can be formed at these loci.
Since we successfully integrated a single rDNA* at various ectopic loci, we proceeded to examine whether expansion of rRNA gene copy number into an array was possible. Expansion of rRNA gene repeats was observed after the integration of a second rDNA* repeat into strains carrying a single rDNA* repeat at the ectopic loci in rdnΔΔ strains carrying the Pol II plasmid. Insertion of the second rDNA* repeat initiated expansion of rDNA* repeats which led to appearance of cells that are able to grow on glucose and thus were able to grow in the absence of the helper Pol II plasmid. Thus, at each of the three ectopic loci, the rDNA* array was functional in the absence of the native nucleolus at RDN1.
Extensive expansion of rDNA* repeats was evident by ∼100 generations of growth (first on YEPG, followed by growth on YEPD). There were no differences in growth rate (∼2.5-h doubling time in YEPD) observed between the strains carrying ectopic expanded rDNA* repeats and the strain carrying expanded rDNA* at the original RDN1 locus. This observed doubling time, ∼2.5 h, is significantly longer than the ∼1.7 h observed for the control strain, NOY388, and can be explained by the presence of the hygromycin-resistant mutation within the 18S coding region of the rRNA gene (54).
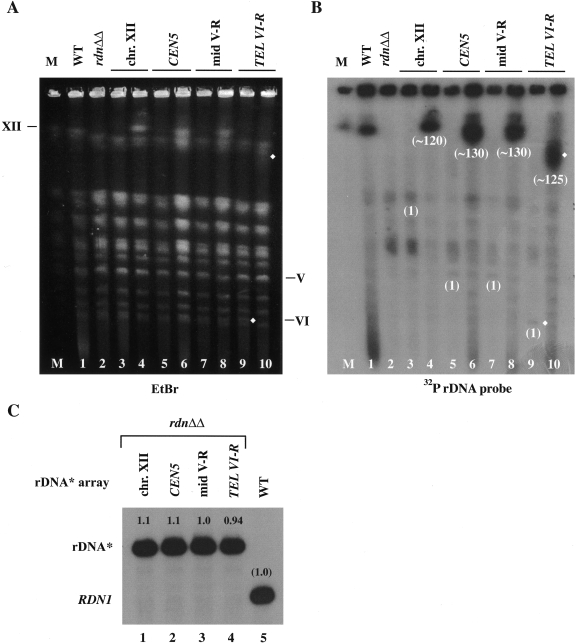
We performed CHEF analyses of chromosomes prepared from the single rDNA* integrants and the expanded rDNA* integrants. Upon the second integration and subsequent expansion of rDNA* repeats, chromosomes carrying the ectopic rDNA* array grew in length (Fig. 4A and B; compare lanes 3, 5, 7, and 9 for strains with a single integrated rDNA* copy to lanes 4, 6, 8, and 10 for strains after repeat expansion, respectively). We analyzed at least three independent transformants from the second integration of rDNA* at each ectopic site. The number of repeats after expansion varied between ∼120 and ∼130, and there were no significant differences in the ability to expand between copies integrated at the native locus and those at the ectopic loci (examples given in Fig. 4, lanes 4, 6, 8, and 10).
FIG. 4.
CHEF analysis of chromosomes from strains carrying single and expanded rDNA* integrated at the native (RDN1) and ectopic sites and expression of rRNA from these ectopic loci. Chromosomal DNA was isolated from NOY388 (WT, lane 1); NOY986 (rdnΔΔ, lane 2); single rDNA* integrants NOY2048, NOY987, NOY988, and NOY2047 (lanes 3, 5, 7, and 9, respectively); and expanded rDNA* integrants NOY2029, NOY2030, NOY2031, and NOY2032 (lanes 4, 6, 8, and 10, respectively). Lane M, marker yeast chromosomes (obtained from Bio-Rad, Richmond, CA). (A) Chromosomal pattern revealed by staining with EtBr. The positions of native chromosomes V, VI, and XII are indicated. (B) Autoradiogram obtained after hybridization with a radioactive rRNA gene probe. Copy numbers of integrated rDNA* after expansion estimated from the chromosomal sizes are shown below chromosome bands carrying rDNA*. Expansion of the rDNA* integrated near TEL VI-R can be clearly recognized by comparing the bands indicated with diamonds in lanes 9 and 10 (in both panels A and B). The chromosome XII bands in rdnΔΔ with or without a single rDNA* copy at the native site (in lanes 2 and 3) overlap with the chromosome XV and VII bands, and the chromosome V bands with a single rDNA* (in lanes 5 and 7) overlap with the chromosome VIII band. Thus, the positions of chromosome XII or V before expansion of rDNA* at the native (rdn1) site, CEN5, or mid V-R cannot be clearly recognized in lanes 3, 5, or 7. C. RNA was prepared from the four expanded rDNA* integrants and NOY388 (WT), and equal amounts were analyzed by primer extension as in Fig. 2B. An autoradiogram is shown. The values obtained were normalized to the value obtained for the control WT strain (NOY388) and are indicated above the bands.
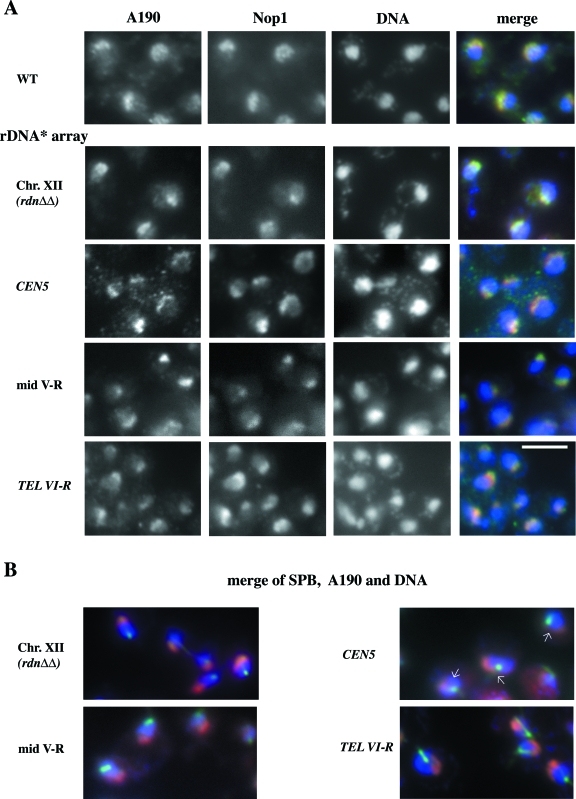
Strains carrying ectopic expanded rDNA* (NOY2030, NOY2031, and NOY2032) were analyzed for nucleolar morphology using IFM and compared to the strain carrying expanded rDNA* at the native RDN1 locus (NOY2029) and the control strain (NOY388). Antibodies against the A190 subunit of Pol I, and the nucleolar protein Nop1 were used to visualize the nucleolus (Fig. 5A). No significant difference was observed in nucleolar morphology among these five strains; a crescent-shaped nucleolar structure was observed at nuclear periphery with partial overlap with the DAPI-stained nuclear region rich in DNA.
FIG. 5.
IFM analysis of nucleolus in yeast strains carrying expanded rDNA* repeats at ectopic sites. (A) Yeast strains NOY388 (wild type [WT]), NOY2029 (rDNA* array at the native site, “Chr. XII”), NOY2030 (rDNA* array near CEN), NOY2031 (rDNA* array at mid-V-R), and NOY2032 (rDNA* array near TEL VI-R) were analyzed for the A190 subunit of Pol I and for Nop1 using IFM. DNA was stained with DAPI. Images of A190 (green), Nop1 (red), and DAPI (blue) were overlaid. Individual images are shown in black and white, and the overlaid one is shown in color (“merge”). (B) IFM analysis of location of the nucleoli relative to the location of SPB. Strains carrying expanded rDNA* analyzed in panel A were examined. The YOL1/34 monoclonal antibody to yeast tubulin showed the area to be slightly larger than the SPB in interphase cells (green), and antibodies to A190 revealed the nucleolus containing Pol I (red). Overlays of these two images and DAPI-stained DNA (blue) are shown in color. Arrows indicate the positions of contact of the nucleolus with CEN5 that is close to SPB.
Because the nucleolus is usually localized opposite to SPB in interphase and centromeres are known to be clustered near the SPB in the interphase (see the introduction), we were particularly interested in the strain which had the expanded rDNA* at the locus very close to CEN5 (NOY2030). By visualizing the location of SPB in the interphase by the use of anti-yeast-tubulin antibodies, we found that the location of the nucleolus as visualized by staining with anti-A190 antibodies appears to be different from the normal location. The ectopic nucleolus was adjacent to SPB but then spread along the nuclear periphery away from SPB (Fig. 5B; see arrows showing “contacts” of the nucleolus with SPB). We also carried out RNA-fluorescence in situ hybridization (FISH)/immunofluorescence experiments using probes specific to the 5′ end of 35S precursor rRNA to localize the site of rRNA transcription and anti-yeast-tubulin antibodies. We obtained the results suggesting that the nucleolar location observed by the A190 staining reflects the site of rRNA transcription (data not shown). Because the strain with the expanded rDNA* repeats near CEN5 showed no obvious defects in growth or rRNA transcription, as described above, we conclude that a functional nucleolus can be formed at the site close to CEN5, which is localized near SPB in the interphase, without an obvious growth defect. (We did not directly localize the expanded rDNA* at ectopic sites by FISH and, hence, cannot exclude the possibility that the expanded rDNA* is not fully associated with the nucleolus visualized by IFM using anti-A190. However, since the expanded rDNA* is as active as the native rDNA repeats, we assume that the expanded rDNA* is mostly associated with the nucleolus visualized by IFM as has been shown for the native rDNA repeats.)
Expression of 35S rRNA genes at ectopic loci compared to the expression of rRNA genes at RDN1.
The rdnΔΔ strains with expanded rDNA* repeats at ectopic sites had the same growth rate as the control strain that had reintegrated and expanded rDNA* at the original RDN1 site. Therefore, we expected total rRNA transcription rates in these three strains to be similar to that in the control strain. The results of primer extension analysis shown in Fig. 4C confirm the expectation; there was no significant difference in total rRNA transcription rate among the four strains, each carrying expanded rDNA* at different chromosomal loci.
Analysis of expression of rRNA genes at both ectopic and normal loci for nucleolar dominance.
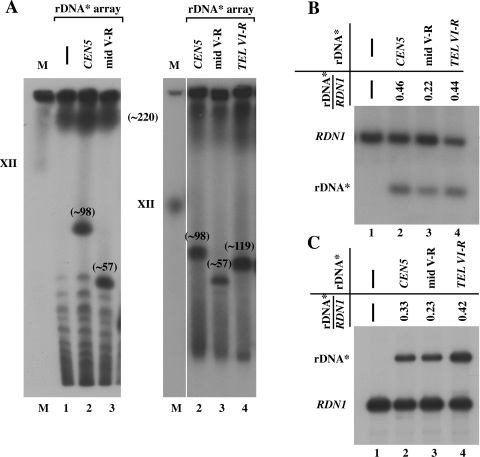
Strains carrying ectopic expanded rDNA* repeats were crossed with a WT RDN1, NOY388 (MATa), or NOY396 (MATα) strain and sporulated, and we obtained haploid segregants carrying two rRNA gene arrays (i.e., two nucleolar organizing regions [NORs]) on different chromosomes. Analyses by CHEF indicated that the strains were able to accommodate two rRNA gene arrays (Fig. 6A). The approximate copy number of rRNA gene repeats on chromosome XII was 220 and that of ectopic rDNA* repeats was 98, 57, and 119 for CEN5, mid-V-R, and TEL VI-R sites, respectively, in these strains (Fig. 6A).
FIG. 6.
Transcription activities of rDNA* arrays at ectopic sites relative to that at the native site in haploid strains carrying an rDNA* array in addition to the native rRNA gene array. (A) CHEF analysis of control NOY388, NOY2053 (RDN1, rDNA* array near CEN5), NOY2054 (RDN1, rDNA* array at mid-V-R), and NOY2055 (RDN1, rDNA* array near TEL VI-R) is shown in lanes 1, 2, 3, and 4, respectively. Lane M, marker yeast chromosomes. Autoradiograms of gels from two independent CHFF analyses are shown, and approximate rDNA* copy numbers are indicated as in Fig. 2B. (B) Southern analysis of rRNA gene repeat numbers of the nucleolus at ectopic sites relative to that of the native nucleolus in the four strains in panel A. An autoradiogram and ratios of repeat numbers calculated are shown. (C) Primer extension analysis to compare the levels of transcription of two rRNA gene arrays in the strains given above. An autoradiogram and the ratios calculated are shown.
We then sought to determine whether there is any preference in the use of the two rRNA gene repeats for transcription, e.g., a possible preferential use of the native rRNA gene repeats over ectopic rDNA* repeats. The ratio of copy number of ectopic rDNA* to that of the native rRNA gene in these strains was determined more quantitatively by Southern analysis (Fig. 6B). The ratios found (0.46, 0.22, and 0.44 for the strains shown in lanes 2, 3, and 4, respectively) were similar to those calculated from the approximate copy numbers obtained by CHEF (0.45, 0.26, and 0.54 calculated from the values given in Fig. 6A). The expression of the ectopic rDNA* compared to the native rRNA gene was then examined by primer extension analysis. We found that the ratios of rDNA* to rRNA gene transcripts (0.33, 0.23, and 0.42 as shown in Fig. 6C, lanes 2, 3, and 4, respectively, for strains indicated) are roughly similar to ratios of rRNA gene repeat numbers obtained from the southern analysis (values given in Fig. 6B). Thus, we conclude that there is no significant bias in rRNA gene expression based on specific chromosomes or locations on a chromosome, and cells maintain the rRNA synthesis rate appropriate for given growth conditions by using both native and ectopic rRNA genes without any preference of one over the other.
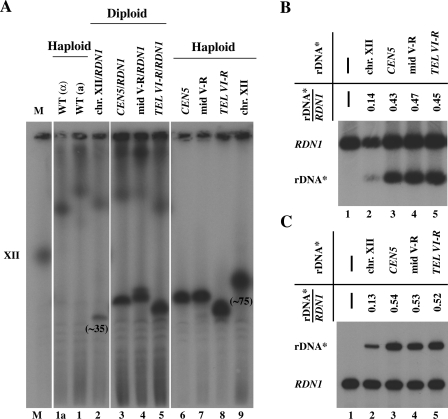
Similar experiments were also done using diploid strains carrying two rRNA gene repeat arrays: one at an ectopic locus and the other at the native RDN1 locus on chromosome XII. Four diploid strains were constructed. Three of them contained, in addition to a copy of the native rDNA array on chromosome XII (RDN1/rdnΔΔ), an expanded ectopic rDNA* array on one copy of another chromosome. One of the diploids contained the native rRNA gene repeat array on one chromosome XII and the expanded rDNA* repeat array on another copy of chromosome XII. The results are shown in Fig. 7. The conclusion obtained was the same as that obtained for the haploid strains, namely, that expression of rRNA genes from an ectopic rDNA* array relative to the native rRNA gene array was approximately the same as the ratio of the ectopic rDNA* repeat number to the repeat number of the native rRNA genes. No preferential expression of rRNA genes in the native RDN1 locus was observed.
FIG. 7.
Transcription activities of rRNA gene arrays at ectopic sites relative to that at the native site in diploid strains carrying a pair of chromosome XII with or without the native RDN1 and a pair of another chromosome with or without rDNA* array at an ectopic locus. (A) CHEF analysis of diploid strains, NOY2039 (RDN1 together with rDNA* array at the native site), NOY2040 (one rDNA* array near CEN5), NOY2041 (one rDNA* array at mid V-R), and NOY2042 (one rDNA* array near TEL VI-R) (lanes 2 to 5, respectively) and haploid strains used to construct the diploid strains, NOY396, NOY388, NOY2030, NOY2031, NOY2032, and NOY2029 (lanes 1a, 1, 6, 7, 8, and 9, respectively). An autoradiogram after probing with radioactive rRNA gene probe is shown. Chromosome XII carrying an rDNA* array in NOY2029 (lane 9) and that in diploid NOY2056 (lane 2) are marked by giving approximate copy numbers below the chromosome bands to indicate a significant alteration in copy numbers upon construction of the diploid strain. (The copy number of rDNA* in such diploid varied depending on the diploid clones analyzed and ranged from ∼35 to ∼98. In diploid cells formed from two haploids, one with high and one with low rDNA repeat numbers at RDN1, by CHEF analysis, the size of chromosome XII derived from the latter is observed to be more homogeneous, i.e., rDNA repeat expansion and contraction appears to be inhibited. A similar phenomenon was previously observed, and a possible explanation was presented [39].) (B) Southern analysis of relative rRNA gene repeat numbers. (C) Primer extension analysis of relative transcription levels. In panels B and C, the diploid strains given in panel A (lanes 2 to 5) and haploid control strain NOY388 (lane 1) were analyzed, and the ratios of the value for rDNA* to that for RDN1 in respective diploid strains are shown.
In higher eukaryotic cells that carry several NORs on different chromosomes, it is known that several NORs are often found within a single morphologically defined nucleolus, a phenomenon known as nucleolar fusion. In the above-mentioned strains which carry rRNA genes on two different chromosomes, a nucleolar fusion appears to be taking place because we observed a single nucleolus in most of the interphase cells as examined by IFM using anti-A190 antibodies. Nevertheless, we occasionally observed two apparently separated nucleoli within a single interphase cell (unpublished observations). We have not studied the question of whether there is any difference in the frequencies of occurrence of two nucleoli relative to single fused nucleoli among the strains carrying two rRNA gene repeats on different chromosomes relative to control diploid strains with two RDN1 loci.
DISCUSSION
Functional nucleolus can be formed at ectopic sites.
Using a rdnΔΔ strain carrying a Pol II helper plasmid, we were able to construct strains carrying a single rDNA* repeat at one of the three ectopic sites selected, as well as the native site on chromosome XII. Upon introducing a second rDNA* copy adjacent to the first one, expansion of the number of rDNA* copies occurred. Copy numbers comparable to those (100 to 200) observed in the native nucleolus were attained at these ectopic sites as in the case of expansion at the native locus on chromosome XII (54).
The transcription activity of a single 35S rRNA gene in the rdnΔΔ strain background was essentially the same at three ectopic sites as rRNA transcription of a single copy at the control native locus. Likewise, the transcription activity of the 35S rRNA genes in expanded rDNA* repeats was also essentially the same at the three ectopic loci as that of expanded rDNA* repeats at the native site on chromosome XII. In addition, strains with a nucleolus at ectopic sites showed the same growth rate as the control strain with a nucleolus at the native site. Clearly, no cis elements outside the rRNA gene repeats are required for efficient Pol I transcription of rRNA genes nor for the ability of rRNA gene units to expand and form a functional nucleolus. We conclude that a functional nucleolus can be formed at ectopic sites in the absence of the native nucleolus. In addition, there are no significant position effects on nucleolar functions related to rRNA transcription and ribosome production or other functions required for optimal cell growth under the standard laboratory conditions used.
The nucleolus formed near TEL VI-R or at the mid V-R was crescent-shaped and localized at the nuclear periphery opposite to the SPB in interphase nuclei, as was the nucleolus in control strains. However, the nucleolus formed at the site near CEN5 appeared to show a crescent-shaped structure at the nuclear periphery starting from a position close to the SPB and spreading away from it at the nuclear periphery. The position relative to SPB is thus different from the typical one, but the main portion of the nucleolus appears to be formed at a region away from the SPB. As noted above, Pol I transcription is not affected by the centromere present very close to the end of this ectopic nucleolus. However, it is possible that the nucleolus present at such a position might give some negative effects on functions of the nearby CEN5; for example, faithful segregation of chromosome V might be affected. More specific experiments have to be carried out to examine such possible effects and thereby assess the significance of the location of the nucleolus opposite to the SPB in normal cells.
Codominance of two NORs, one native and the other ectopic, in Pol I transcription of rRNA genes.
As described in the introduction, one of the goals of the present studies was to construct, if possible, yeast strains carrying both active and inactive NORs in order to study mechanisms for the “differential silencing” of NORs (or “nucleolar dominance”) observed in many organisms such as human cells (43). Our results have demonstrated that the 35S rRNA genes in an array near or at CEN5, TEL VI-R, or mid V-R have an equal efficiency to be transcribed relative to those in the native array both in the haploid and in the diploid strains analyzed. No significant position effect was observed in Pol I transcription of rRNA genes in ectopic relative to native loci, that is, the native and the ectopic NORs were codominant in these strains.
Transcription activity of a single rRNA gene.
Using rdnΔΔ strains carrying a single rRNA gene at ectopic sites or at the native site on chromosome XII, we found no position effects on rRNA synthesis rate as measured by primer extension analysis of unstable 35S pre-rRNA. The value obtained was ∼3.4% ± 1.2% of the rRNA synthesis rate (per cell density) of the control strain carrying ∼200 rDNA copies. Because of the presence of excess Pol I machinery spread through the nucleoplasm in rdnΔΔ strains (38), this value may represent the maximum activity that could be obtained for transcription of a single 35S rRNA gene. Consequently, one can calculate that ∼30 copies of rRNA genes would be sufficient for attaining the rRNA synthesis rate in the control strain growing at a maximum rate in a rich medium such as YEPD. This estimate is roughly consistent with our previous observations that strains with ∼40 rDNA copies (stabilized in the fob1 background) showed the same growth rate and rRNA synthesis rate as a control strain with ∼140 rDNA copies (12). However, strains with ∼25 copies showed a slightly reduced growth rate relative to a control strain with ∼190 copies (7; unpublished observations). The reason for the presence of ca. 150 to 200 copies in most wild-type yeast strains has not been clearly established. In addition to the function of ribosome synthesis, the nucleolus is known to play roles in other important cellular activities (for a review, see reference 40), such as the regulation of mitotic exit (46, 52). Thus, it is possible that excess rRNA gene copies might be required under some special conditions not investigated thus far. The question may deserve future studies.
In eukaryotes, rRNA genes exist in tandemly repeated structures. Because of this unique structure combined with the observations of transcription of intergenic regions by read-through Pol I molecules in some organisms (9, 31) and mutational analyses using reporter systems, it was previously suggested that tandemly repeated structures might play an important role in efficient transcription of rRNA genes. It was proposed that the initiation of transcription is coupled with termination events coming from the transcription of upstream genes (or transcription from spacer promoters) (called “readthrough enhancement” or “polymerase hand-over” model [see reviews in references 1, 36, and 42]). Even though such coupling might exist in vivo, there has not been any convincing demonstration of the importance of such a mechanism for the efficiency of transcription of intact rRNA genes. Using the 40-copy strain, we have shown that a single rDNA copy at the mid V-R is as active as that within the 40 tandem rDNA repeats at RDN1. This result, together with that of activity analysis of a single copy versus two copies at CEN5, demonstrates that tandem repetition is not required for achieving high transcription rates of rRNA genes.
Recruitment of Pol I machinery to a single rDNA* copy at ectopic sites in the presence of a native nucleolus at RDN1.
The three single ectopic rDNA* copies showed the same Pol I transcription in rdnΔΔ strains where Pol I machinery is spread through the nucleoplasm. Therefore, the inefficient transcription of a single rDNA* copy at TEL VI-R relative to that integrated at RDN1 (or at mid-V-R or CEN5) in the presence of the ∼40 copies of rDNA repeats at RDN1 was somewhat unexpected. The observed differences in expression must be due to differences in the efficiency of the recruitment of Pol I machinery to the ectopic rRNA* when Pol I machinery is mostly or entirely sequestered in the native nucleolus formed at RDN1 on chromosome XII.
We originally thought that the movement of a single rDNA* copy near CEN5 is constrained in a region near the SPB. Since the SPB is localized at a nuclear position opposite to the native nucleolus (4, 22, 56), transcription of a single rDNA* at CEN5 by recruitment of the Pol I machinery from the nucleolus might be difficult. In contrast, the chromosome VI-R telomere is much closer to the nucleolus and is considerably mobile, even though there is some constraint in its movement (4). Telomeres, as well as the nucleolus, are present preferentially at the nuclear periphery, so recruitment of the Pol I machinery from the nucleolus could be easier for rDNA* at TEL VI-R. Thus, the observed difference in Pol I transcription between the two ectopic rDNA* copies, one near CEN5 and the other near TEL VI-R, cannot be explained by their nuclear positions relative to the nucleolus in interphase cells.
Although transcriptionally active rRNA genes and telomere chromatins share common features in that Sir2 protein is an associated protein and both regions silence transcription of reporter Pol II genes (reviewed in reference 44), they clearly have some very different features. For example, it is well known that Sir3 and Sir4 are associated with telomeres and are required for silencing at telomeres, whereas they are not associated with rRNA genes and are not required for silencing of reporter genes in RDN1 (44). Such telomere-specific chromatin structures might inhibit the recruitment of Pol I machinery from the native nucleolus, for example, perhaps by the spreading of these structures into the nearby rDNA* region. Nevertheless, once the rDNA* is expanded in copy number at this locus near TEL VI-R, the resultant nucleolus is as active as the native one, apparently recruiting and retaining the Pol I machinery at this ectopic site. After repeat expansion most of the repeats may escape the negative effect of the telomere on recruitment of Pol I machinery seen for a single copy rDNA*, possibly because distances from the telomere are increased for a majority of the rDNA* repeats or the telomere-proximal repeat acts as an insulator.
In contrast to the single rDNA* at TEL VI-R, the single rDNA* at CEN5 is transcribed fairly efficiently by Pol I, even though it is physically well separated from the nucleolus in interphase. Two possibilities can be considered. One is that not all of the Pol I machinery is sequestered in the nucleolus and that Pol I recruitment to the CEN5 locus is possible without direct contact with the nucleolus. Several observations make this model unattractive. The concentrations of the Pol I machinery in the nucleoplasm outside the nucleolus are significantly lower than that in the nucleolus as judged by IFM (see, for example, Fig. 5 and reference 38). In addition, the equal efficiency of Pol I recruitment to a single rDNA* at the CEN5 and TEL VI-R sites in rdnΔΔ strains suggests that when cells have an intact nucleolus the concentration of the Pol I machinery outside the nucleolus may be significantly lower (see, for example, reference 38). Finally, a weak but perhaps significant orientation dependence in rDNA* expression seen for this CEN5 site, as well as for mid-V and TEL VI-R sites, might be difficult to explain by this model. Another possibility is that centromeres have a close contact with the nucleolus during mitosis when the centromeres of a set of newly duplicated chromosomes, together with one of the duplicated SPBs, migrates back toward the nucleolus in the mother cells and reaches a position opposite the other SPB (4). According to this hypothesis, the Pol I machinery recruited to the ectopic rDNA* at CEN5 during mitosis is postulated to remain associated with this single rDNA* copy. A mininucleolus is formed, and rRNA transcription occurs throughout the subsequent interphase and in the absence of a significant physical contact with the native nucleolus.
Role of E element in Pol I transcription of rRNA genes at ectopic sites.
In the present study, we have found that when we integrated a single rRNA gene copy at the CEN5 site, Pol I transcription was not affected by deletion of the E element and was also FOB1 independent. The typical E/P (HOT1) integrated at the same ectopic site showed Pol I transcription to be dependent on FOB1 and the presence of the E element (our unpublished experiments). It may be interesting to identify cis elements within rRNA gene responsible for the difference between the E/P system and the intact rRNA gene unit in this respect.
In summary, we have successfully constructed several yeast strains in which a single rRNA gene copy is integrated at a defined ectopic site and its function in rRNA transcription, as well as its repeat expansion at such a site, can be studied. This system now allows us to study the effects of various mutations on the formation of the nucleolus, as well as on transcription or other nucleolar functions at ectopic and endogenous chromosomal sites. In addition to some of the questions discussed above, there are other unsolved questions related to nucleolar functions such as silencing of Pol II reporter genes and boundary elements or factors that determine conversion between active (or open) and inactive (or closed) states of rRNA genes. Various yeast strains and the strategy for rRNA gene integration at ectopic sites described in the present study may be useful for studies of these questions.
. .
Supplementary Material
Acknowledgments
We thank R. Steele for critical reading of the manuscript and S. VanAmburg for help in preparation of the manuscript.
This study was supported by Public Health Service grant GM-35949.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Baker, S. M., and T. Platt. 1986. Pol I transcription: which comes first, the end or the beginning? Cell 47:839-840. [DOI] [PubMed] [Google Scholar]
- 2.Brewer, B. J., and W. L. Fangman. 1988. A replication fork barrier at the 3′ end of yeast rRNA genes. Cell 55:637-643. [DOI] [PubMed] [Google Scholar]
- 3.Bystricky, K., P. Heun, L. Gehlen, J. Langowski, and S. M. Gasser. 2004. Long-range compaction and flexibility of interphase chromatin in budding yeast analyzed by high-resolution imaging techniques. Proc. Natl. Acad. Sci. USA 101:16495-16500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bystricky, K., T. Laroche, G. van Houwe, M. Blaszczyk, and S. M. Gasser. 2005. Chromosome looping in yeast: telomere pairing and coordinated movement reflect anchoring efficiency and territorial organization. J. Cell Biol. 168:375-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chernoff, Y. O., A. Vincent, and S. W. Liebman. 1994. Mutations in eukaryotic 18S rRNA affect translational fidelity and resistance to aminoglycoside antibiotics. EMBO J. 13:906-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choe, S. Y., M. C. Schultz, and R. H. Reeder. 1992. In vitro definition of the yeast RNA polymerase I promoter. Nucleic Acids Res. 20:279-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cioci, F., L. Vu, K. Eliason, M. Oakes, I. Siddiqi, and M. Nomura. 2003. Silencing in yeast rDNA chromatin: reciprocal relationship in gene expression between RNA polymerase I and II. Mol. Cell 12:135-145. [DOI] [PubMed] [Google Scholar]
- 8.Clark-Walker, G. D., and A. A. Azad. 1980. Hybridizable sequences between cytoplasmic rRNAs and 3 micron circular DNAs of Saccharomyces cerevisiae and Torulopsis glabrata. Nucleic Acids Res. 8:1009-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Winter, R. F., and T. Moss. 1986. The ribosomal spacer in Xenopus laevis is transcribed as part of the primary rRNA. Nucleic Acids Res. 14:6041-6051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elion, E. A., and J. R. Warner. 1984. The major promoter element of rRNA transcription in yeast lies 2 kb upstream. Cell 39:663-673. [DOI] [PubMed] [Google Scholar]
- 11.Elion, E. A., and J. R. Warner. 1986. An RNA polymerase I enhancer in Saccharomyces cerevisiae. Mol. Cell. Biol. 6:2089-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.French, S. L., Y. N. Osheim, F. Cioci, M. Nomura, and A. L. Beyer. 2003. In exponentially growing Saccharomyces cerevisiae cells, rRNA synthesis is determined by the summed RNA polymerase I loading rate rather than by the number of active genes. Mol. Cell. Biol. 23:1558-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galy, V., J. C. Olivo-Marin, H. Scherthan, V. Doye, N. Rascalou, and U. Nehrbass. 2000. Nuclear pore complexes in the organization of silent telomeric chromatin. Nature 403:108-112. [DOI] [PubMed] [Google Scholar]
- 14.Goh, P. Y., and J. V. Kilmartin. 1993. NDC10: a gene involved in chromosome segregation in Saccharomyces cerevisiae. J. Cell Biol. 121:503-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gotta, M., T. Laroche, A. Formenton, L. Maillet, H. Scherthan, and S. M. Gasser. 1996. The clustering of telomeres and colocalization with Rap1, Sir3, and Sir4 proteins in wild-type Saccharomyces cerevisiae. J. Cell Biol. 134:1349-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grewal, S. I., and S. C. Elgin. 2002. Heterochromatin: new possibilities for the inheritance of structure. Curr. Opin. Genet. Dev. 12:178-187. [DOI] [PubMed] [Google Scholar]
- 17.Grummt, I., and C. S. Pikaard. 2003. Epigenetic silencing of RNA polymerase I transcription. Nat. Rev. Mol. Cell. Biol. 4:641-649. [DOI] [PubMed] [Google Scholar]
- 18.Guacci, V., D. Koshland, and A. Strunnikov. 1997. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in Saccharomyces cerevisiae. Cell 91:47-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heun, P., T. Laroche, K. Shimada, P. Furrer, and S. M. Gasser. 2001. Chromosome dynamics in the yeast interphase nucleus. Science 294:2181-2186. [DOI] [PubMed] [Google Scholar]
- 20.Huang, G. S., and R. L. Keil. 1995. Requirements for activity of the yeast mitotic recombination hotspot HOT1: RNA polymerase I and multiple cis-acting sequences. Genetics 141:845-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin, Q., E. Trelles-Sticken, H. Scherthan, and J. Loidl. 1998. Yeast nuclei display prominent centromere clustering that is reduced in nondividing cells and in meiotic prophase. J. Cell Biol. 141:21-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jin, Q. W., J. Fuchs, and J. Loidl. 2000. Centromere clustering is a major determinant of yeast interphase nuclear organization. J. Cell Sci. 113(Pt. 11):1903-1912. [DOI] [PubMed] [Google Scholar]
- 23.Johnson, S. P., and J. R. Warner. 1989. Unusual enhancer function in yeast rRNA transcription. Mol. Cell. Biol. 9:4986-4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karpen, G. H., J. E. Schaefer, and C. D. Laird. 1988. A Drosophila rRNA gene located in euchromatin is active in transcription and nucleolus formation. Genes Dev. 2:1745-1763. [DOI] [PubMed] [Google Scholar]
- 25.Keener, J., C. A. Josaitis, J. A. Dodd, and M. Nomura. 1998. Reconstitution of yeast RNA polymerase I transcription in vitro from purified components. TATA-binding protein is not required for basal transcription. J. Biol. Chem. 273:33795-33802. [DOI] [PubMed] [Google Scholar]
- 26.Keil, R. L., and G. S. Roeder. 1984. Cis-acting, recombination-stimulating activity in a fragment of the ribosomal DNA of S. cerevisiae. Cell 39:377-386. [DOI] [PubMed] [Google Scholar]
- 27.Keys, D. A., B. S. Lee, J. A. Dodd, T. T. Nguyen, L. Vu, E. Fantino, L. M. Burson, Y. Nogi, and M. Nomura. 1996. Multiprotein transcription factor UAF interacts with the upstream element of the yeast RNA polymerase I promoter and forms a stable preinitiation complex. Genes Dev. 10:887-903. [DOI] [PubMed] [Google Scholar]
- 28.Kobayashi, T. 2003. The replication fork barrier site forms a unique structure with Fob1p and inhibits the replication fork. Mol. Cell. Biol. 23:9178-9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kobayashi, T., D. J. Heck, M. Nomura, and T. Horiuchi. 1998. Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev. 12:3821-3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kulkens, T., D. L. Riggs, J. D. Heck, R. J. Planta, and M. Nomura. 1991. The yeast RNA polymerase I promoter: ribosomal DNA sequences involved in transcription initiation and complex formation in vitro. Nucleic Acids Res. 19:5363-5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Labhart, P., and R. H. Reeder. 1986. Characterization of three sites of RNA 3′ end formation in the Xenopus ribosomal gene spacer. Cell 45:431-443. [DOI] [PubMed] [Google Scholar]
- 32.Larionov, V. L., A. V. Grishin, and M. N. Smirnov. 1980. 3 micron DNA: an extrachromosomal ribosomal DNA in the yeast Saccharomyces cerevisiae. Gene 12:41-49. [DOI] [PubMed] [Google Scholar]
- 33.Laroche, T., S. G. Martin, M. Gotta, H. C. Gorham, F. E. Pryde, E. J. Louis, and S. M. Gasser. 1998. Mutation of yeast Ku genes disrupts the subnuclear organization of telomeres. Curr. Biol. 8:653-656. [DOI] [PubMed] [Google Scholar]
- 34.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 35.Marshall, W. F., A. Straight, J. F. Marko, J. Swedlow, A. Dernburg, A. Belmont, A. W. Murray, D. A. Agard, and J. W. Sedat. 1997. Interphase chromosomes undergo constrained diffusional motion in living cells. Curr. Biol. 7:930-939. [DOI] [PubMed] [Google Scholar]
- 36.Moss, T., and V. Y. Stefanovsky. 1995. Promotion and regulation of ribosomal transcription in eukaryotes by RNA polymerase I. Prog. Nucleic Acids Res. Mol. Biol. 50:25-66. [DOI] [PubMed] [Google Scholar]
- 37.Musters, W., J. Knol, P. Maas, A. F. Dekker, H. van Heerikhuizen, and R. J. Planta. 1989. Linker scanning of the yeast RNA polymerase I promoter. Nucleic Acids Res. 17:9661-9678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oakes, M., J. P. Aris, J. S. Brockenbrough, H. Wai, L. Vu, and M. Nomura. 1998. Mutational analysis of the structure and localization of the nucleolus in the yeast Saccharomyces cerevisiae. J. Cell Biol. 143:23-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oakes, M., I. Siddiqi, L. Vu, J. Aris, and M. Nomura. 1999. Transcription factor UAF, expansion and contraction of ribosomal DNA (rDNA) repeats, and RNA polymerase switch in transcription of yeast rDNA. Mol. Cell. Biol. 19:8559-8569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Olson, M. O. J. 2004. Nontraditional roles of the nucleolus, p. 329-342. In M. O. J. Olson (ed.), The nucleolus. R.G. Landes & Co., Austin, Tex.
- 41.Olson, M. V. 1991. Genome structures and organization in Saccharomyces cerevisiae, p. 1-39. In J. R. Broach, J. R. Pringle, and P. Jorgensen (ed.), The molecular and cellular biology of the yeast Saccharomyces. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 42.Reeder, R. H. 1989. Regulatory elements of the generic ribosomal gene. Curr. Opin. Cell Biol. 1:466-474. [DOI] [PubMed] [Google Scholar]
- 43.Roussel, P., C. Andre, L. Comai, and D. Hernandez-Verdun. 1996. The rDNA transcription machinery is assembled during mitosis in active NORs and absent in inactive NORs. J. Cell Biol. 133:235-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rusche, L. N., A. L. Kirchmaier, and J. Rine. 2003. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 72:481-516. [DOI] [PubMed] [Google Scholar]
- 45.Sherman, F., G. R. Fink, and J. B. Hicks. 1986. Laboratory course manual for methods in yeast genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 46.Shou, W., J. H. Seol, A. Shevchenko, C. Baskerville, D. Moazed, Z. W. Chen, J. Jang, A. Shevchenko, H. Charbonneau, and R. J. Deshaies. 1999. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell 97:233-244. [DOI] [PubMed] [Google Scholar]
- 47.Siddiqi, I. N., J. A. Dodd, L. Vu, K. Eliason, M. L. Oakes, J. Keener, R. Moore, M. K. Young, and M. Nomura. 2001. Transcription of chromosomal rRNA genes by both RNA polymerase I and II in yeast uaf30 mutants lacking the 30-kDa subunit of transcription factor UAF. EMBO J. 20:4512-4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sinclair, D. A., and L. Guarente. 1997. Extrachromosomal rDNA circles: a cause of aging in yeast. Cell 91:1033-1042. [DOI] [PubMed] [Google Scholar]
- 49.Spector, D. L. 2003. The dynamics of chromosome organization and gene regulation. Annu. Rev. Biochem. 72:573-608. [DOI] [PubMed] [Google Scholar]
- 50.Stewart, S. E., and G. S. Roeder. 1989. Transcription by RNA polymerase I stimulates mitotic recombination in Saccharomyces cerevisiae. Mol. Cell. Biol. 9:3464-3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taddei, A., F. Hediger, F. R. Neumann, and S. M. Gasser. 2004. The function of nuclear architecture: a genetic approach. Annu. Rev. Genet. 38:305-345. [DOI] [PubMed] [Google Scholar]
- 52.Visintin, R., E. S. Hwang, and A. Amon. 1999. Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature 398:818-823. [DOI] [PubMed] [Google Scholar]
- 53.Wai, H., K. Johzuka, L. Vu, K. Eliason, T. Kobayashi, T. Horiuchi, and M. Nomura. 2001. Yeast RNA polymerase I enhancer is dispensable for transcription of the chromosomal rRNA gene and cell growth, and its apparent transcription enhancement from ectopic promoters requires Fob1 protein. Mol. Cell. Biol. 21:5541-5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wai, H. H., L. Vu, M. Oakes, and M. Nomura. 2000. Complete deletion of yeast chromosomal rDNA repeats and integration of a new rDNA repeat: use of rDNA deletion strains for functional analysis of rDNA promoter elements in vivo. Nucleic Acids Res. 28:3524-3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wittekind, M., J. M. Kolb, J. Dodd, M. Yamagishi, S. Memet, J. M. Buhler, and M. Nomura. 1990. Conditional expression of RPA190, the gene encoding the largest subunit of yeast RNA polymerase I: effects of decreased rRNA synthesis on ribosomal protein synthesis. Mol. Cell. Biol. 10:2049-2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang, C. H., E. J. Lambie, J. Hardin, J. Craft, and M. Snyder. 1989. Higher order structure is present in the yeast nucleus: autoantibody probes demonstrate that the nucleolus lies opposite the spindle pole body. Chromosoma 98:123-128. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.