Abstract
NOTCH1 is a large type I transmembrane receptor that regulates normal T-cell development via a signaling pathway that relies on regulated proteolysis. Ligand binding induces proteolytic cleavages in NOTCH1 that release its intracellular domain (ICN1), which translocates to the nucleus and activates target genes by forming a short-lived nuclear complex with two other proteins, the DNA-binding factor CSL and a Mastermind-like (MAML) coactivator. Recent work has shown that human T-ALL is frequently associated with C-terminal NOTCH1 truncations, which uniformly remove sequences lying between residues 2524 and 2556. This region includes the highly conserved sequence WSSSSP (S4), which based on its amino acid content appeared to be a likely site for regulatory serine phosphorylation events. We show here that the mutation of the S4 sequence leads to hypophosphorylation of ICN1; increased NOTCH1 signaling; and the stabilization of complexes containing ICN1, CSL, and MAML1. Consistent with these in vitro studies, mutation of the WSSSSP sequence converts nonleukemogenic weak gain-of-function NOTCH1 alleles into alleles that cause aggressive T-ALLs in a murine bone marrow transplant model. These studies indicate that S4 is an important negative regulatory sequence and that the deletion of S4 likely contributes to the development of human T-ALL.
NOTCH receptors and downstream mediators participate in a signaling pathway that variously regulates the specification of cell fate, proliferation, self-renewal, survival, and apoptosis in a dose- and context-dependent fashion (2). Like other members of the NOTCH receptor family, human NOTCH1 is a large multimodular type I transmembrane glycoprotein (Fig. 1A). Newly synthesized NOTCH1 is cleaved by furin at a site termed S1 just external to the transmembrane domain (21), yielding two noncovalently associated extracellular (NEC) and transmembrane (NTM) subunits (5, 21, 29). Binding of ligands to NEC triggers two sequential proteolytic events within the NTM subunit at sites S2 and S3. S2 cleavage occurs just external to the transmembrane domain and is catalyzed by ADAM-type metalloproteases (6, 24). This creates a short-lived intermediate, NTM*, which is recognized by nicastrin (33), a component of the protease complex called γ-secretase (8, 18, 32). Additional cleavages by γ-secretase free the intracellular domain of NOTCH1 (ICN1), allowing it to translocate to the nucleus, where it activates transcription through the formation of a ternary complex with the DNA-binding factor CSL (16, 19, 36, 44) and coactivator proteins of the Mastermind-like (MAML) family (27, 28, 43).
FIG. 1.
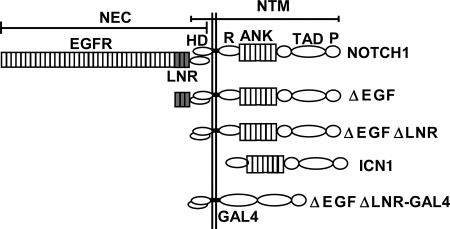
NOTCH1 expression constructs. A schematic representation of the mature full-length human NOTCH1 receptor and of expression constructs bearing N-terminal NOTCH1 deletions is shown. Furin cleavage in the extracellular domain creates NEC (NOTCH1 extracellular) and NTM (NOTCH1 transmembrane) subunits that remain associated through noncovalent interactions between the N-terminal and C-terminal portions of the heterodimerization domain (HD). Other important functional domains include the LNR domain, which comprises the three LIN12/Notch repeats; the transmembrane segment (TM); the intracellular domain (ICN); the RAM domain; the ankyrin repeat domain (ANK); the transactivation domain (TAD); and the PEST domain. Some experiments used chimeric polypeptides, in which the RAM and ANK domains of NOTCH1 were replaced with the DNA-binding domain of the transcription factor GAL4.
Nuclear ICN1 is short-lived. One mechanism that appears to promote the rapid turnover of the CSL/ICN1/MAML transcription complex involves the recruitment of mediator complexes and CycC-CDK8 through the C-terminal tail of MAML1 (13). Phosphorylation of ICN1 on multiple C-terminal serine residues by CycC-CDK8 is hypothesized to create recognition sites for E3 ligases such as FBW7/Sel10 (13), which has been implicated in the ubiquitylation and subsequent degradation of ICN (38, 39). Some of the sites targeted by CycC-CDK8 lie in the far C-terminal portion of NOTCH1 (13), an unstructured region that is enriched for the amino acids proline, glutamate, serine, and threonine (PEST). PEST sequences regulate the degradation of a number of proteins (30), sometimes by serving as substrates for phosphorylation events that mark a protein for degradation (22). In the case of CycC-CDK8, phosphorylation of ICN is hypothesized to couple MAML-dependent transcriptional activation to rapid ICN degradation (12, 13). However, there is evidence that NOTCH stability is also regulated at other levels. For example, phosphorylation by GSKβ appears to promote the degradation of the intracellular domain of NOTCH2 (9), and E3 ligases of the Itch family have been implicated in the ubiquitylation and regulation of membrane-associated NOTCH receptors (23, 34). Thus, inputs from multiple pathways regulate NOTCH at the level of protein stability during different stages of receptor activation and trafficking.
Increased NOTCH1 signaling plays a central part in the pathogenesis of T-cell acute lymphoblastic leukemia (T-ALL), a tumor derived from T-cell progenitors. We observed that human T-ALLs commonly harbor frameshift and stop codon mutations that delete various numbers of C-terminal residues from NOTCH1 (41), a finding that was presaged by the detection of retroviral insertions in murine T-ALLs that cause similar truncations (10, 14). Although these mutations are scattered throughout the 3′ end of exon 34, all of the deletions found to date eliminate at least residues 2524 to 2556, suggesting that this minimal region contains at least one important motif that negatively regulates NOTCH1 signal strength. Here, we analyze the role of a short conserved sequence, WSSSSP (referred to as S4), found within the minimal deleted region that influences not only the function and stability of activated NOTCH1 but also its leukemogenic activity.
MATERIALS AND METHODS
Expression plasmids.
A diagram depicting the various forms of NOTCH1 used in these studies is shown in Fig. 1. Expression constructs that encode full-length human NOTCH1 (residues M1 to K2556); ΔEGF, a form bearing a deletion that removes the coding region for epidermal growth factor (EGF)-like repeats 1 to 36 (residues R23 to I1446); ΔEGFΔLNR, a form bearing a deletion that removes the coding region for EGF-like repeats 1 to 36 and the three Lin12/NOTCH repeats (residues R23 to C1562); and ICN1 (residues 1762 to 2556) have been described (31). Expression constructs for forms of ICN1 bearing a N-terminal FLAG-tag were created by PCR with primers containing a consensus Kozak start codon followed by the coding sequence for the FLAG epitope. A C-terminal deletion removing residues 2473 to 2556 (originally identified in the cell line ALL-SIL) has been described (41). NOTCH1-GAL4 chimeric cDNAs were creating by ligating a PCR product encoding the DNA-binding domain of GAL4 in frame to ICN1 cDNA cut with the restriction enzymes Bsu36I and NcoI, which removes sequences encoding the RAM and ANK domains. In other constructs, premature stop codons and point mutations were introduced by using the QuikChange kit (Stratagene). cDNAs were assembled variously in pcDNA3 (Invitrogen); pcDNA5 (Invitrogen); or the retroviral vector MSCV-GFP, which drives expression of NOTCH1 and green fluorescence protein (GFP) from a single bicistronic RNA containing an internal ribosomal entry sequence (IRES). Expression plasmids for CSL-MYC (4), MAML1-GFP (43), and dominant-negative MAML1-GFP (42) have all been described. A pCMV2 plasmid (Sigma) encoding a “kinase-dead” dominant-negative form of CDK8 was kindly provided by Andrew Rice, Baylor University.
Cell culture.
U2OS and 293T cells (American Type Culture Collection) were maintained in Dulbecco modified Eagle medium (DMEM; Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen), 2 mM l-glutamine (Invitrogen), 100 U of penicillin (Invitrogen)/ml, and 100 μg of streptomycin (Invitrogen)/ml. 293 TRex cells were obtained from Invitrogen. Cells were grown at 37°C under 5% CO2.
Transcriptional activation assays.
NOTCH1 expression plasmids were introduced into U2OS cells by transient transfection with Lipofectamine Plus (Invitrogen) and assessed for their ability to activated a NOTCH-sensitive luciferase reporter gene, as described previously (4). Briefly, cells in 24-well dishes were cotransfected in triplicate with 10 ng of various pcDNA3-NOTCH1 expression constructs, a NOTCH-sensitive firefly luciferase reporter gene (15), and an internal control Renilla luciferase plasmid (Promega). Experiments involving NOTCH1-GAL4 fusion constructs used a GAL4-luciferase reporter gene (Clontech). Total introduced DNA was kept constant by adding empty pcDNA3 plasmid. Normalized firefly luciferase activities were measured in whole-cell extracts prepared 44 to 48 h after transfection using the Dual Luciferase kit (Promega) and a specially configured luminometer (Turner Systems). In some experiments, the cells were treated posttransfection with the γ-secretase inhibitor compound E (kindly provided by Michael Wolfe) at 1 μM or with carrier alone (0.01% dimethyl sulfoxide [DMSO]).
ICN1 immunoprecipitation.
293T cells transfected with pcDNA plasmids encoding various NOTCH signaling components were lysed in 50 mM Tris (pH 8.0), containing 1% NP-40, 100 mM NaCl, 30 mM NaF, 20 mM Na pyrophosphate, 2 mM Na vanadate, 2 mM Na molybdate, and 5 mM Na EDTA (buffer A). ICN1 polypeptides were immunoprecipitated with a rabbit polyclonal antibody raised against the transcriptional activation domain of NOTCH1, as described previously (3). In some experiments, the immunoprecipitation polypeptides were treated with lambda phosphatase (New England Biolabs) according to the manufacturer's recommendations. In other experiments, complexes containing CSL-MYC were immunoprecipitated with the mouse monoclonal antibody 9E10, as described previously (3).
Phosphoamino acid analysis.
293 TRex cells (Invitrogen) were cotransfected with pcDNA5-FLAG-ICN1 plasmids and pOGG44, which expresses Flp recombinase. Isogenic 293 recombinants were selected with hygromycin B and then split into 100-mm dishes. After the induction of ICN1 expression by the addition of tetracycline (1 μg/ml) for 24 h, cells were incubated twice for 1 h in phosphate-free DMEM (Invitrogen) containing 10% dialyzed fetal calf serum (depletion medium) and then grown overnight in a 9:1 mixture of depletion medium and complete medium containing 2.5 mCi of [32P]orthophosphate (New England Nuclear). After three washes with ice-cold phosphate-buffered saline, the cells were lysed in ice-cold buffer A for 15 min and centrifuged at 14,000 × g for 15 min. Proteins in the resulting supernatants were immunoprecipitated by adding FLAG-M2-antibody agarose beads (Sigma), followed by mixing for 2 h at 4°C. The beads were washed four times with buffer A, and bound proteins were released by adding sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer and heating the mixture for 10 min to 100°C. ICN1 proteins were separated by SDS-PAGE in 8% gels, which were dried and analyzed by autoradiography.
After digestion of phosphorylated ICN1 with trypsin-TPCK (tolylsulfonyl phenylalanyl chloromethyl ketone; Worthington), acid hydrolysis was carried out in 6 M HCl at 110°C for 2 h. Phosphoamino acids were separated by thin-layer electrophoresis at pH 1.9 as described previously (17).
Phosphopeptide analysis.
32P-labeled ICN1 polypeptides were prepared from 293 TRex cells by immunoprecipitation, followed by SDS-PAGE in 8% gels as described above. Portions of the dried gels containing phosphorylated ICN1 polypeptides were excised, rehydrated, oxidized with performic acid, and digested with trypsin (Worthington) as described previously (17). Peptides were then resuspended in 5 μl of formic acid (96%) and separated by thin-layer electrophoresis on cellulose plates (Sigma) for 5 h at 400 V in formic acid-acetic acid-water (10:31:359 [pH 1.9]). After drying the cellulose plate, ascending chromatography was performed in butanol-pyridine-acetic acid-water (50:33:10:40). 32P-labeled peptides were visualized by autoradiography.
Pulse-chase analysis.
293 cells in 60-mm dishes were transfected with pcDNA3-FLAG-ICN1 plasmids (1 μg) on day 1, split into six-well dishes on day 2, and then subjected to pulse-chase labeling on day 3 as follows. Cells were incubated twice for 1 h in DMEM without l-methionine containing 10% dialyzed fetal calf serum, followed by incubation in the same medium containing 2.5 mCi of 35S-labeled l-methionine (New England Nuclear) for 30 min. After two washes with Hanks buffered saline, the cells were either harvested immediately or incubated for up to 6 additional hours in replete DMEM containing 10% fetal calf serum and 2 mM cold l-methionine. 35S-labeled ICN1 polypeptides were immunoprecipitated from whole-cell detergent lysates on FLAG-M2-antibody beads (Sigma). After elution from the beads by heating for 10 min at 100°C in SDS-PAGE loading buffer, the proteins were separated by SDS-PAGE in 10% polyacrylamide gels, and detected within dried gels by autoradiography.
Western blot analysis.
Whole-cell extracts and immunoprecipitated polypeptides were resolved by SDS-PAGE in 8% gels and transferred to polyvinylidene difluoride membranes (Millipore) as described previously (3). Membranes were stained with rabbit polyclonal antibodies against the intracellular domain of NOTCH1 (3) or ICN1 (V1744 antibody, Cell Signaling) or with mouse monoclonal antibodies against GFP (Clontech), the MYC epitope (clone 9E10), or the FLAG epitope (Sigma).
Murine bone marrow transplantation assays.
All experiments were performed as described previously (1, 4), in accordance with National Institutes of Health guidelines for the care and use of animals, and with an approved animal protocol from the University of Pennsylvania Animal Care and Use Committee. Briefly, cDNAs cloned into the MigRI vector were packaged into retroviruses by transient transfection of 293T cells. After the virus titers were determined on NIH 3T3 cells, GFP-normalized retroviral supernatants were used to “spinoculate” 5-fluorouracil-treated bone marrow cells from female 4- to 8-week-old C57BL/6 mice (Taconic Farms). Transduction was performed over 48 h in a cocktail consisting of DMEM, 10% heat-inactivated fetal bovine serum (Gibco-BRL, Gaithersburg, MD), 5% WEHI-conditioned medium, 6 U of recombinant mouse interleukin-3 (Genzyme Corp., Cambridge, MA)/ml, 10,000 U of recombinant mouse interleukin-6 (Genzyme)/ml, 5 U of recombinant mouse stem cell factor (Genzyme)/ml, 1 μg of Polybrene (Sigma Chemical Co., St. Louis, MO)/ml, 100 U of streptomycin (Gibco-BRL)/ml, 100 U of penicillin (Gibco-BRL)/ml, and 2 mM l=glutamine (Gibco-BRL). The retrovirally transduced bone marrow cells were then injected into lethally irradiated (900 rads) 4- to 8-week-old female syngeneic recipients.
Flow cytometry.
Peripheral blood samples and tumor cell suspensions were assessed for the presence of GFP+ immature T cells by flow cytometric analysis (FACSCalibur; Becton Dickinson). Cells were incubated with phycoerythrin-labeled anti-CD8α (53-6.7), biotinylated anti-TCRβ (H57-597), and allophycocyanin-labeled anti-CD4 (RM4-5) antibodies (Pharmingen). Biotinylated antibodies were revealed with streptavidin-PerCP. Dead cells, identified by forward scatter and side scatter, were excluded from the analysis. fluorescence-activated cell sorting results were analyzed by using Flowjo software.
Southern blot analysis.
High-molecular-weight DNA was isolated from fresh or snap-frozen spleen tissue. A total of 10 μg of DNA was digested with the appropriate restriction enzymes overnight, fractionated by electrophoresis on a 0.8% agarose gel, and blotted overnight onto Nytran membrane (Schleicher & Schuell, Keene, NH) via alkaline transfer. Blots were hybridized overnight with gel-purified 32P-labeled probes corresponding to the IRES or GFP fragments of MigR1.
Histology and immunohistochemistry.
To assess histology, paraffin-embedded sections of mouse tissues fixed in 10% phosphate-buffered formalin were stained with hematoxylin and eosin. For immunohistochemistry, sections were deparaffinized in xylene and graded alcohols, subjected to antigen retrieval in citrate buffer using a pressure cooker, and then stained with rabbit polyclonal antibodies specific for the intracellular domain of NOTCH1 (3), CD3 (Dako), and terminal deoxytransferase (Dako). Antibody staining was developed by using the Dako Envision kit, per the manufacturer's instructions, and the horseradish peroxidase substrate diaminobenzamidine.
RESULTS
Identification of WSSSSP as a NOTCH1 negative regulatory sequence.
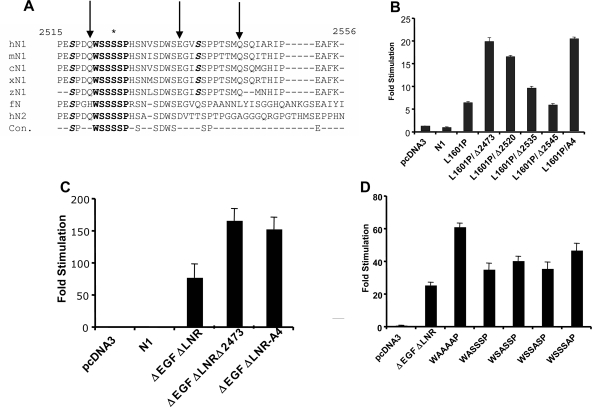
The commonly deleted region in T-ALL contains a number of conserved residues (Fig. 2A). We commenced our functional analysis by creating a series of nested deletions in full-length NOTCH1 cDNAs bearing the leukemia-associated heterodimerization domain mutation L1601P, which causes a modest activation of NOTCH1 signaling (Fig. 2B). We observed that the deletion of residues 2545 to 2556 had little effect on NOTCH1 signaling, whereas larger deletions spanning residues 2535 to 2556, 2520 to 2556, and 2473 to 2556 increased NOTCH1 signal strength in a stepwise fashion. Prior work from Jones' group showed that serine residues at positions 2514, 2517, and 2538 can be phosphorylated by CycC:CDK8, an event that is hypothesized to mark ICN1 for transcription-dependent degradation (13). However, the largest stepwise increase in activity was associated with the deletion of residues 2520 to 2534, a region where none of the serines have been reported to be phosphorylated by CycC-CDK8. This region encompasses the position of the most C-terminal deletion we have yet identified in human T-ALL (a stop codon mutation at residue 2524 in the cell line MOLT-15 [41]; Fig. 2B) and includes a short sequence, WSSSSP (designated S4), that is restricted in the protein sequence database to a subset of NOTCH receptors. Specifically, WSSSSP is 100% conserved within fly NOTCH and vertebrate NOTCH1 and NOTCH2 receptors (Fig. 2A) but has diverged in NOTCH3 (WSDSTP) and is completely absent from NOTCH4.
FIG. 2.
Functional effects of C-terminal NOTCH1 deletions and mutations. (A) Conservation of C-terminal NOTCH sequences. Residues in boldface highlight the highly conserved S4 sequence; amino acids in boldface italics correspond to S residues that are phosphorylated by CycC:CDK8; and the asterisk denotes S2524, which is the site of the most C-terminal mutation yet detected in human T-ALL. The arrows denote the positions of nested deletions engineered to test the function of residues in this region. Key: hN1, human NOTCH1; mN1, mouse NOTCH1; cN1; chicken NOTCH1; xN1, Xenopus NOTCH1; fN, Drosophila NOTCH; hN2, human NOTCH2; Con., consensus. (B to D) Effects of C-terminal deletions and mutations on NOTCH1 signal strength. In each set of experiments, NOTCH1 signaling was assessed by cotransfection of U2OS cells with 10 ng of pcDNA3-NOTCH1 plasmid, 250 ng of CSLx4-luciferase reporter plasmid (15), and 5 ng of pRL-TK-Renilla luciferase internal control plasmid. Normalized luciferase activities were measured in triplicate and expressed relative to an empty plasmid control. Error bars represent standard deviations. (B) Relative effects of deletions removing residues 2473 to 2556 (Δ2473), 2520 to 2556 (Δ2520), 2535 to 2556 (Δ2535), or 2545 to 2556 (Δ2545) and the A4 mutation on signals produced by NOTCH1 polypeptides bearing a point mutation in the heterodimerization domain, L1601P, which causes modest activation of NOTCH1 signaling (41). (C) Relative effects of a deletion removing residues 2473 to 2556 (D2473) and the A4 mutation on signals produced by ΔEGFΔLNR, a form of NOTCH1 bearing a deletion removing the extracellular EGF repeats and LNR domain (31). (D) Relative effects of the indicated point mutations in the S4 sequence on signals produced by ΔEGFΔLNR.
To test the idea that the S4 sequence might regulate NOTCH1 function, we mutated S4 to AAAA (A4). In the context of either full-length NOTCH1 L1601P (Fig. 2B) or a second relatively weak gain-of-function form of NOTCH1, ΔEGFΔLNR (Fig. 2C), the A4 mutation stimulated signaling to an extent comparable to the Δ2473-2556 deletion. We also investigated the effects of mutating individual S4 residues. Each single S-to-A substitution produced a modest stimulation in NOTCH1 activity in the context of the ΔEGFΔLNR polypeptide, but no single mutation was as strong as the A4 substitution (Fig. 2D). Thus, multiple S residues within the S4 sequence appear to contribute to negative regulation of NOTCH1 signaling.
S4 affects ICN1 phosphorylation.
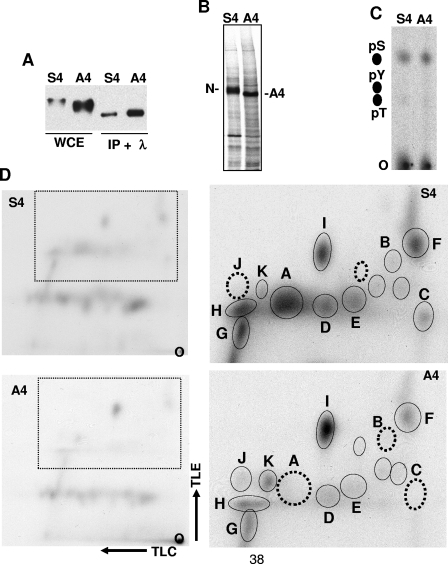
The S4 sequence is distinct from previously identified sites of NOTCH1 phosphorylation. To determine whether the S4 sequence influences ICN1 phosphorylation, we first compared the electrophoretic mobilities of ICN1-S4 and ICN1-A4. The mean electrophoretic mobility of ICN1-A4 was consistently greater than that of ICN1-S4 (Fig. 3A), a finding that could be explained by ICN-A4 being underphosphorylated relative to ICN-S4. In support of this interpretation, the broad bands corresponding to ICN1-S4 and ICN1-A4 on Western blots were resolved into tight bands of faster, but identical, electrophoretic mobility by treatment with lambda phosphatase (Fig. 3A), suggesting that the S4 sequence influences the phosphorylation of ICN1 by one or more protein kinases.
FIG. 3.
S4 affects ICN1 phosphorylation. (A) Mutation of S4 affects the electrophoretic mobility of ICN1. 293T cells transfected with the plasmids pcDNA3-ICN1-S4 or pcDNA3-ICN1-A4 were lysed 2 days posttransfection. Immunoprecipitates prepared from these lysates with an antibody specific for ICN1 were treated with lambda phosphatase (Sigma). Untreated whole-cell extracts (WCE) and phosphatase-treated immunoprecipitates (IP + λ) were analyzed on a Western blot stained with anti-NOTCH1 antibody. (B) Metabolic labeling of ICN1-S4 and ICN1-A4 with [32P]orthophosphate. Isogenic 293 Flp-in cells engineered to express FLAG-ICN1-S4 and FLAG-ICN1-A4 were metabolically labeled with [32P]orthophosphate. 32P-labeled ICN1-S4 and ICN1-A4 were immunoprecipitated on FLAG-M2-antibody beads (Sigma) and electrophoresed in an 8% SDS-PAGE gel, which was dried and exposed to X-ray film. The resultant autoradiogram is shown. (C) Phosphoamino acid analysis. Immunoprecipitated 32P-labeled ICN1-S4 and ICN1-A4 were excised from SDS-PAGE gels and converted to amino acids by proteolysis, followed by acid hydrolysis. 32P-labeled amino acids were spotted onto thin-layer cellulose plates (O, origin) along with phosphoserine (pS), phosphothreonine (pT), and phosphotyrosine (pY) amino acid standards and separated by electrophoresis. After staining with ninhydrin to detect the positions of the amino acid standards, the 32P-labeled amino acids in the test samples were detected by autoradiography. (D) Two-dimensional phosphopeptide analysis. Immunoprecipitated 32P-labeled ICN1-S4 and ICN1-A4 were excised from SDS-PAGE gels, digested exhaustively with trypsin-TPCK, and spotted onto thin-layer cellulose plates (O, origin). The 32P-labeled peptides were subjected to thin-layer electrophoresis (TLE), followed by thin-layer chromatography (TLC) in the second dimension. After drying, the positions of 32P-labeled peptides were determined by autoradiography. The left pair of autoradiograms shows the complex, but similar, patterns of phosphopeptides that are produced from ICN1-S4 and ICN1-A4. The right pair of autoradiograms focuses in on the most pronounced differences in the observed patterns of spots, which lie in the area outlined by the boxes in the left pair of autoradiograms. Individual spots are identified arbitrarily as A to I. The data are representative of two independent experiments. Dotted circles denote the position of spots that are absent from the map shown and present in the corresponding map prepared from the other form of ICN1.
To further characterize the effect of the S4 sequence on phosphorylation, we compared the phosphoamino acid content and the phosphopeptide maps of ICN1-S4 and ICN1-A4 prepared from metabolically labeled isogenic 293 TRex cells. 32P-labeled ICN1-S4 and ICN-A4 again demonstrated a difference in electrophoretic mobility consistent with underphosphorylation of the A4 mutant (Fig. 3B). Phosphoserine was the major phosphoamino acid in both ICN1-S4 and ICN1-A4 (Fig. 3C), a finding consistent with previous analysis (7). Two-dimensional tryptic maps prepared from ICN1-S4 and ICN1-A4 revealed a complex, but reproducible, set of 32P-labeled peptides (Fig. 3D). Of these phosphopeptides, one major peptide (designated peptide A) and two minor peptides (peptides B and C) were absent from digests prepared from ICN1-A4. We also noted that the labeling of a number of other peptides (such as peptides D to H in Fig. 3D) was reduced in ICN1-A4 compared to ICN1-S4. This tendency toward a global decrease in labeling of ICN1-A4 was consistent with the results of counting of the excised ICN1-S4 and ICN1-A4 bands prior to tryptic digestion, which typically revealed that ICN1-A4 incorporated 40 to 50% less 32P (data not shown). On the other hand, while the overall effect of the A4 mutation was to decrease ICN1 phosphorylation, one major phosphopeptide (designated I) and several minor phosphopeptides (designated J and K) reproducibly showed relatively increased labeling in ICN1-A4 compared to ICN1-S4 (Fig. 3D). Thus, these data indicate that the A4 mutation has complex effects on ICN1 phosphorylation that are generally, but not uniformly, inhibitory.
S4 influences ICN1 stability.
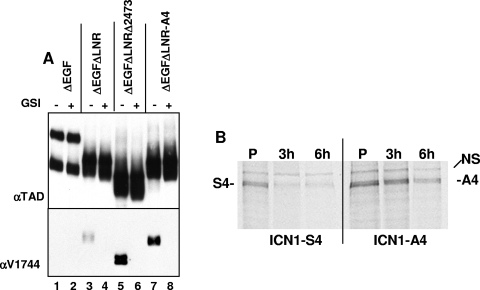
The simplest way for the A4 mutation to stimulate ICN1 activity is by increasing ICN1 protein levels. We first addressed this by comparing the levels of ICN1 that are present in cells expressing various forms of ΔEGFΔLNR, a NOTCH1 polypeptide that is susceptible to ligand-independent S2 and S3 cleavages due to the absence of the protective LNR domain (31). Relative to intact ΔEGFΔLNR, ΔEGFΔLNR polypeptides bearing the deletion Δ2473-2556 or the A4 mutation generated substantially higher steady-state levels of ICN1 in a fashion that was sensitive to a γ-secretase inhibitor, compound E (Fig. 4A).
FIG. 4.
S4 influences ICN1 stability. (A) U2OS cells in 6-well dishes were transfected in duplicate with 1 μg of pcDNA3 plasmids encoding inactive ΔEGF (lanes 1 and 2), constitutively active ΔEGFΔLNR (lanes 3 and 4), or ΔEGFΔLNR bearing the deletion of residues 2473 to 2556 (lanes 5 and 6) or the A4 mutation (lanes 7 and 8). Immediately posttransfection, cells were refed medium containing 0.1% DMSO or 0.1% DMSO plus 1 μM compound E, a γ-secretase inhibitor. At two days posttransfection, whole-cell extracts were prepared and analyzed on Western blots stained with a polyclonal antibody specific for an epitope in the NOTCH1 transcriptional activation domain (TAD) or anti-V1744 (Cell Signaling), a polyclonal antibody that is specific for a neo-epitope created at the N-terminal end of ICN1 by γ-secretase cleavage. (B) ICN1-A4 is more stable than ICN1-S4. 293 cells in 60-mm dishes were transfected with 1 μg of the pcDNA3 plasmids FLAG-ICN1-S4 or FLAG-ICN1-A4. The cells were split 24 h posttransfection into six-well dishes. Two days posttransfection, the cells were depleted twice for 1 h in medium lacking methionine, “pulsed” for 30 min with medium containing 2.5 mCi of [35S]methionine, and then “chased” for the indicated time periods with complete medium supplemented with 2 mM unlabeled l-methionine. 35S-labeled ICN1 polypeptides were immunoprecipitated from whole-cell lysates on FLAG-M2-antibody beads, separated by SDS-PAGE, and detected by autoradiography. NS, nonspecific band.
To determine directly whether the A4 mutation stabilized ICN1, pulse-chase experiments were performed with FLAG-tagged ICN1-S4 and ICN1-A4 (Fig. 4B). This revealed that the A4 mutation has a modest, but appreciable, stabilizing effect on ICN1. Together with experiments, such as those in Fig. 4A, showing that the A4 mutation allows ICN1 to accumulate to higher levels in cells, these results provide a likely explanation for the stimulatory effect of the A4 mutation on ICN1 function.
S4 is unlikely to be a target sequence for CDK8.
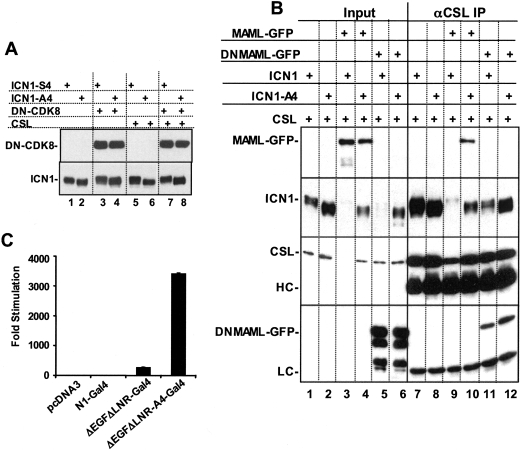
Three sites in the C terminus of ICN1 near the S4 site (S2514, S2517, and S2538) can be phosphorylated by CDK8, which appears to be recruited to ICN1 through the C terminus of MAML cofactors (13). We thus performed a series of experiments to explore the relationship of the S4 site to CDK8. Most directly, we first investigated whether a “kinase-dead” dominant-negative form of CDK8 altered the difference in phosphorylation between ICN1-S4 and ICN1-A4, as judged by electrophoretic mobility. We observed that ICN1-A4 remained underphosphorylated relative to ICN1-S4 in whole-cell extracts and in CSL complexes even in the presence of a 10-fold excess of dominant-negative CDK8 plasmid (Fig. 5A). Additional experiments were performed in cells overexpressing CSL and full-length MAML1 (which should enhance CDK8 recruitment), or CSL and a dominant-negative form of MAML1 (DN-MAML1). DN-MAML consists of a 62-amino-acid kinked α-helix that forms a stable ternary complex through contacts on both CSL and the ankyrin repeats of NOTCH1 (25) but which lacks the C-terminal portions of MAML1 that are responsible for recruitment of p300 and CycC:CDK8 (12, 13, 40). Thus, if S4-targeted phosphorylation is dependent on CycC:CDK8, DN-MAML should render ICN1-S4 equivalent to ICN1-A4, both in terms of stability and phosphorylation, by preventing the recruitment of CycC:CDK8. In these experiments, we adjusted the input of the ICN1-A4 and ICN1-S4 plasmids, relative to the CSLmyc and MAML1 plasmids, to make ICN1 the limiting factor for complex assembly (Fig. 5B). Whether judged by Western blots of whole-cell extracts (lanes 1 to 6) or CSL immunoprecipitates (lanes 7 to 12), we observed differences in electrophoretic mobility consistent with underphosphorylation of ICN1-A4 in the presence of endogenous MAMLs (compare lanes 1 and 2 and lanes 7 and 8), overexpressed full-length MAML1 (compare lanes 3 and 4 and lanes 9 and 10), and DN-MAML1 (compare lanes 5 and 6 and lanes 11 and 12). We also observed a stabilizing effect of the A4 mutation on full-length MAML1 and CSL when these proteins were coexpressed (compare the recovery of MAML1 in CSL complexes immunoprecipitated in lanes 9 and 10). The ability of overexpressed MAML1 to promote degradation of CSL and ICN1 is consistent with prior data implicating assembly of the CSL/ICN/MAML ternary complex in ICN1 turnover (12, 13, 40).
FIG. 5.
CDK8 is unlikely to be the S4 kinase. (A) Dominant-negative CDK8 fails to affect the difference in ICN1-S4 and ICN1-A4 phosphorylation. 293 cells in 60-mm dishes were transfected with the indicated combinations of pcDNA3-ICN1-S4 or pcDNA3-ICN1-A4 (100 ng), pCMV2-FLAG-dominant-negative CDK8 (DN-CDK8) (1 μg), and pcDNA3-CSL-MYC (1 μg) plasmids. At 2 days posttransfection, whole-cell detergent extracts were prepared from each dish of transfected cells. Extracts prepared from cells cotransfected with CSL-MYC (lanes 5 to 8) were further subjected to immunoprecipitation with anti-MYC 9E10 antibody. The upper panel shows a Western blot containing whole-cell extracts that was stained with the FLAG M2 antibody, which recognizes FLAG-DN-CDK8. The lower panel shows a blot stained with anti-NOTCH1. In this blot, lanes 1 to 4 contain whole-cell extracts, while lanes 5 to 8 contain NOTCH1 polypeptides that were coprecipitated with CSL-MYC. (B) Altered MAML1 function fails to abrogate the difference in ICN1-S4 and ICN1-A4 phosphorylation. 293T cells in six-well format were cotransfected with pcDNA3-CSL-MYC (1 μg), pEGFP-MAML1-GFP or dominant-negative MAML1-GFP (0.5 μg), and pcDNA3-ICN1 or ICN1-A4 (0.1 μg). At 2 days posttransfection, whole-cell detergent extracts and CSL-MYC immunoprecipitates were prepared as described previously (3). Lanes 1 to 6 contain 1% of total protein inputted into the immunoprecipitates; lanes 7 to 12 show 20% of the proteins in the corresponding immunoprecipitates. The blot was sequentially stained with anti-MYC (9E10), anti-GFP (Clontech), and anti-NOTCH1, using an antibody against the intracellular transcriptional activation domain (3). HC and LC, immunoglobulin heavy chain and light chains, respectively. (C) The A4 mutation increases the function of NOTCH1-GAL4 fusion proteins lacking the ankyrin repeats. U2OS cells in 24-well format were cotransfected with 10 ng of empty pcDNA3 plasmid or pcDNA3 plasmids encoding GAL4 fusion proteins in which the RAM and ankyrin repeat domains of NOTCH1 were replaced with the DNA-binding domain of GAL4. N1 corresponds to a full-length NOTCH1-GAL4 construct, whereas ΔEGFΔLNR corresponds to a construct bearing a deletion removing the EGF and LNR repeat coding regions of NOTCH1. ΔEGFΔLNR-A4 is a GAL4 fusion construct in which the S4 site has been mutated to A4. These plasmids were cotransfected with 250 ng of a GAL4-luciferase reporter plasmid bearing four GAL4 binding sites and 5 ng of the pRL-TK-Renilla luciferase internal control plasmid. Normalized luciferase activities were measured in triplicate and are expressed relative to an empty plasmid control. Error bars represent standard deviations.
We also studied whether the stabilizing effect of the A4 mutation could be transferred to other types of transcriptional activation complexes. Experiments were performed with chimeric ΔEGFΔLNR polypeptides in which the RAM and ankyrin repeat domains of ICN1 were replaced with the DNA-binding domain of GAL4 (an approach used first by Struhl and Adachi [35]). This form of NOTCH1 activates GAL4 reporter genes in a γ-secretase-dependent fashion but fails to assemble a ternary complex and cannot stimulate transcription from CSL-dependent promoters (data not shown). As seen in Fig. 5C, the A4 mutation strongly stimulated the activity of ΔEGFΔLNR-GAL4 on a GAL4-reporter gene. Taken together, these data suggest that the S4 site is phosphorylated by a kinase or kinases other than CycC-CDK8 and that this phosphorylation event does not depend on the entry of ICN1 into the ternary complex.
S4 influences leukemogenesis.
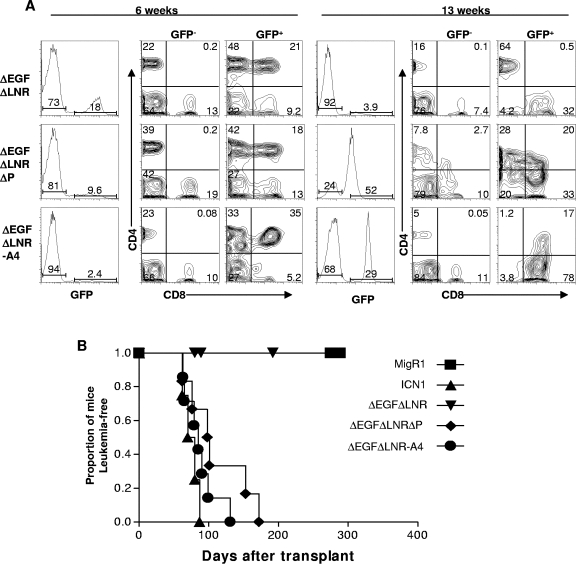
A striking feature of the NOTCH1 tumor-associated mutations is that extracellular HD mutations frequently occur in cis with deletions of the intracellular PEST region (41). The data described above suggested that loss of S4-associated regulation of ICN1 might enhance the leukemogenic activity of weak gain-of-function forms of activated NOTCH1, which are not themselves leukemogenic, and might thus mimic the effect of tumor-associated mutations found in cis. To assess this possibility, we compared the leukemogenic activity of the weak gain-of-function ΔEGFΔLNR form of NOTCH1 with ΔEGFΔLNR-A4 in a murine bone marrow transplantation assay. The selection of ΔEGFΔLNR for these experiments was based on the observations showing that although this form of NOTCH1 generated signals of sufficient strength to drive T-cell development (31), it did not induce T-ALL (Fig. 6A). Typically, mice reconstituted with bone marrow cells expressing ΔEGFΔLNR developed a GFP+ CD4+ CD8+ immature “double-positive” T-cell population by 6 weeks after transplantation that disappeared by 13 weeks posttransplantation. None of these animals have developed leukemia at times greater than 1 year posttransplantation (Fig. 6B and data not shown).
FIG. 6.
S4 influences the development of T-ALL. (A) ΔEGFΔLNRΔP(Δ2473-2556) and ΔEGFΔLNR-A4 cause the development of leukemia, whereas ΔEGFΔLNR causes only a transient lymphocytosis of CD4+ CD8+ T cells. Representative flow cytometric analyses of peripheral blood samples drawn from mice at 6 and 13 weeks posttransplant with bone marrow cells transduced with the indicated MigRI retroviruses are shown. (B) Kaplan-Meier curve showing that leukemia develops in mice reconstituted with bone marrow cells expressing ΔEGFΔLNRΔP(Δ2473-2556), ΔEGFΔLNR-A4, and ICN1 NOTCH1 polypeptides but not in ΔEGFΔLNR or MigRI animals. Each group in this experiment contained at least five mice.
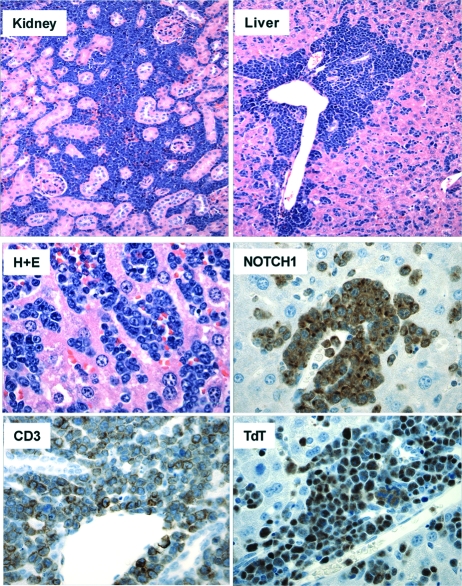
In contrast, the double-positive T-cell count in the peripheral blood of mice reconstituted with bone marrow cells expressing the ΔEGFΔLNR-A4 continued to rise (Fig. 6A), and all of these animals eventually became moribund and succumbed to disseminated leukemia (Fig. 6B). The disease latency in ΔEGFΔLNR-A4 mice was similar to that seen in animals reconstituted with bone marrow cells expressing ΔEGFΔLNRΔPEST (which bears a deletion removing the 73 C-terminal amino acids of NOTCH1) and ICN1, a strong gain-of-function form of NOTCH1 (Fig. 6B). At necropsy, leukemic blasts replaced the bone marrow and heavily infiltrated the spleen, liver, lymph nodes, and viscera such as the kidneys (Fig. 7). Immunohistochemical stains confirmed that these blasts were immature T cells expressing CD3, terminal deoxytransferase (TdT), and NOTCH1 (Fig. 7). Western blots prepared from heavily infiltrated spleens confirmed the presence of NOTCH1 polypeptides of the expected size of the precursor and furin-processed forms of ΔEGFΔLNR-A4 (Fig. 8). Further workup of ΔEGFΔLNR-A4 tumors included flow cytometry, which revealed that the tumors expressed surface CD3 and variable levels of CD4 and CD8 (data not shown). As anticipated given the results of Western blotting, the tumors contained intact proviruses and were monoclonal or oligoclonal based on the presence of one or several dominant proviral insertions (Fig. 9 and data not shown).
FIG. 7.
Pathology of ΔEGFΔLNR-A4-induced T-ALL. Sections stained with hematoxylin and eosin (H+E) show lymphoblasts infiltrating the liver and the kidney. In the bottom four panels, tumor cells are shown in sections of liver stained with H+E or antibodies specific for CD3, terminal deoxytransferase (TdT), and NOTCH1. Immunostaining was developed by method that produces a brown color (hematoxylin counterstain).
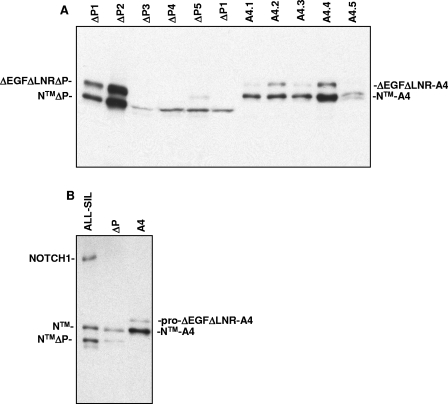
FIG. 8.
Detection of NOTCH1 polypeptides in ΔEGFΔLNRΔP(Δ2473-2556) and ΔEGFΔLNR-A4-induced T-ALLs. Detergent extracts from spleens heavily involved by T-ALLs arising in ΔEGFΔLNRΔP(Δ2473-2556) and ΔEGFΔLNR-A4 mice were analyzed on Western blots stained with a polyclonal antibody specific for the intracellular domain of NOTCH1. (A) Cross-reactive NOTCH1 polypeptides are observed in both types of tumors of the expected size for pro-ΔEGFΔLNRΔ, pro-ΔEGFΔLNRA4, and the furin-processed mature forms of each of these polypeptides (NTMΔPEST and NTM-A4, respectively). (B) The size of the Notch1 polypeptides in panel A is compared to NOTCH1 polypeptides expressed in the cell line ALL-SIL, which has a normal NOTCH1 allele that gives rise to NTM, and a mutated NOTCH1 allele containing the same PEST deletion (residues 2473 to 2556) that has been engineered into the ΔEGFΔLNRΔ expression plasmid (41). The deleted allele gives rise to NTMΔP after furin processing.
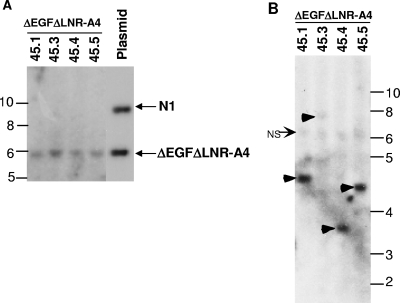
FIG. 9.
Analysis of proviral integrity and integration in A4-induced T-ALLs. (A) Southern blot of EcoRV-digested genomic DNAs isolated from four spleens involved by A4-induced T-ALLs. EcoRV, which cleaves twice within the provirus at sites flanking the ΔEGFΔLNR-A4 cDNA, is expected to release a proviral fragment of 5.9 kb from the intact ΔEGFΔLNR-A4 provirus when hybridized to a probe specific for the IRES sequence. The correctly sized fragment is shown in the right-hand lane, which contains MigRI-ΔEGFΔLNR-A4 and MigRI-NOTCH1 plasmid DNAs digested with Eco RV. Each of the left-hand four lanes contain genomic DNA cut with EcoRV obtained from spleens heavily infiltrated by different MigRI-ΔEGFΔLNR-A4 T-ALLs. (B) Genomic DNAs from the same four spleens shown in panel A were analyzed by digestion with EcoRI, which cleaves once within the provirus; thus, each EcoR1-digested provirus will produce a different sized DNA fragment. The Southern blot hybridized to a 592-bp ECMV IRES probe. Single proviruses are readily seen in tumors 1, 4, and 5, whereas a single faint proviral band is seen in tumor 3 (arrowheads). NS, nonspecific band. In both panels A and B, the numbers correspond to sizes in kilobases.
DISCUSSION
Our studies indicate that the sequence WSSSSP (S4), which spans residues 2521 to 2526 of human NOTCH1, exerts a functionally important restraint on NOTCH1 signal strength, and influences the development of T-ALL in a murine model. It follows that the C-terminal NOTCH1 deletions that are often seen in human T-ALL contribute to leukemogenesis, at least in part, by removing this sequence. Consistent with this possibility, the most C-terminal deletion that we have yet observed in T-ALL starts within this sequence at residue 2524 (41). The gains in ICN1 function that occur when the S4 sequence is mutated are correlated with changes in ICN1 phosphorylation, making it probable that this sequence acts by altering the recognition of ICN1 by one or more kinases. Because it is necessary to mutate multiple S residues within S4 to maximally activate NOTCH1, it is possible that one or more kinases phosphorylate more than one site within this sequence. More broadly, mutation of the S4 site appears to affect the phosphorylation of multiple ICN1 tryptic peptides, suggesting that the phosphorylation status of S4 influences the recognition of ICN1 by other kinases. NOTCH signaling is tightly coordinated with other signaling pathways, and it is possible that S4 phosphorylation may serve to prime ICN1 for recognition by other kinases. The existence of multiple kinases that target ICN1 for degradation through S4 and adjacent residues such as those recognized by CDK8 provides a reasonable explanation for why deletions involving the C terminus of NOTCH1 are common in T-ALL, whereas point mutations are rare (41).
In addition to previously recognized retroviral insertions (10, 14), several recent reports describe acquired frameshift or stop codon mutations in diverse murine T-ALL models that produce C-terminal truncations of NOTCH1 (20, 26). The most C-terminal deletion noted in murine tumors to date falls at amino acid residue 2490 in murine NOTCH1 (20), which is the equivalent of residue 2516 in human NOTCH1; thus, the mutations that occur in murine T-ALL also consistently delete the S4 sequence. While these correlations (and the studies directed at the S4 sequence described here) do not preclude additional important contributions of other sequences in the C-terminal tail to the negative regulation of normal and pathophysiologic NOTCH1 signaling, they are consistent with a major role for the S4 sequence in T-ALL.
Several questions arise from this work, the most immediate of which concerns the identity of the kinase(s) that targets ICN1 through S4. After MAML-dependent recruitment to the NOTCH1 transcription complex on DNA, CycC-CDK8 can phosphorylate at least three serine residues in the far C-terminal region of NOTCH1, including one within the minimal deleted region (41). However, ICN1-A4 is still underphosphorylated relative to ICN1-S4 even under a variety of conditions in which ternary complex formation and recruitment of CDK8 are defective. These findings suggest that the S4 kinase is likely to be different than CycC-CDK8. Other candidates include the GSK3β (9, 11) and MEK/ERK kinases, based on the well-recognized, but complex, functional interactions between the RAS and NOTCH signaling pathways (37). However, experiments conducted with pharmacologic inhibitors of MEK and GSK3β, RNAi directed against GSK3β, and constitutively active forms of MEK1, have failed to detect evidence of epistasis between these kinases and the S4 sequence (J. C. Aster, data not shown). It is likely that unbiased functional screens will be necessary to identify the kinase(s) that is responsible for S4-dependent ICN1 phosphorylation.
We have also failed to see evidence of epistasis between dominant-negative forms of FBW7 (a homolog of Sel-10) and S4 (J. C. Aster, data not shown). In this regard, it is relevant that the stability of mammalian NOTCH4 is regulated by FBW7/Sel-10 (38, 39) despite the absence of the S4 sequence from this Notch receptor. We thus favor the idea that S4-dependent modulation of ICN1 levels depends on factors other than FBW7/Sel-10. It will be of interest to determine whether mutations in the kinases or the destruction machinery responsible for S4-dependent ICN1 degradation will be identified in human T-ALLs lacking C-terminal NOTCH1 deletions.
The C-terminal NOTCH1 deletions that are found in human T-ALL often occur in concert with mutations involving the heterodimerization domain of NOTCH1 that lead to increased ICN1 production (41). We have also further characterized here a form of NOTCH1 with a mutation affecting the ectodomain, ΔEGFΔLNR, which drives abnormal double positive T-cell development (31) without causing T-ALL. This demonstrates for the first time that the threshold dose of NOTCH1 signals that is required for efficient induction of T-ALL development is higher than that which is required to induce T-cell development. This raises the question of whether certain heterodimerization domain mutations will suffice to create signals that are strong enough to induce T-ALL, or whether they will require additional events in cis or in trans. The “hypoleukemic” phenotype induced in vivo by weak gain-of-function NOTCH1 alleles such as ΔEGFΔLNR should be useful in identifying not only cis-acting elements such as the S4 sequence but also elements that act in trans. Such trans-acting factors could either elevate NOTCH1 signaling tone directly or, by complementing NOTCH1 functions that require strong signals, lower the dose of NOTCH1 that is required for efficient induction of T-ALL.
Acknowledgments
We thank Hong Sai and Zhigang Li for excellent technical assistance. We are also grateful to the John Morgan and Stemmler ASU (University of Pennsylvania), the Abramson Cancer Center Flow Cytometry Core (University of Pennsylvania), the AFCRI Core (University of Pennsylvania), and the Hematopathology Core of the Dana Farber/Harvard Cancer Center.
This study was supported by grants from the National Institutes of Health to W.S.P. (CA93615 and AI47833), J.C.A. (CA82308), S.C.B. (CA92433), and D.B.S. (CA75205) and from the Leukemia and Lymphoma Society SCOR Program. M.L.X. was the recipient of an American Society of Hematology Medical Student Trainee award. M.C. was supported by a training grant from the NIH/NIDDK (T32-DK007780-07).
REFERENCES
- 1.Allman, D., F. G. Karnell, J. A. Punt, S. Bakkour, L. Xu, P. Myung, G. A. Koretzky, J. C. Pui, J. C. Aster, and W. S. Pear. 2001. Separation of Notch1 promoted lineage commitment and expansion/transformation in developing T cells. J. Exp. Med. 194:99-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Artavanis-Tsakonas, S., M. D. Rand, and R. J. Lake. 1999. Notch signaling: cell fate control and signal integration in development. Science 284:770-776. [DOI] [PubMed] [Google Scholar]
- 3.Aster, J. C., E. S. Robertson, R. P. Hasserjian, J. R. Turner, E. Kieff, and J. Sklar. 1997. Oncogenic forms of NOTCH1 lacking either the primary binding site for RBP-Jκ or nuclear localization sequences retain the ability to associate with RBP-Jκ and activate transcription. J. Biol. Chem. 272:11336-11343. [DOI] [PubMed] [Google Scholar]
- 4.Aster, J. C., L. Xu, F. G. Karnell, V. Patriub, J. C. Pui, and W. S. Pear. 2000. Essential roles for ankyrin repeat and transactivation domains in induction of T-cell leukemia by notch1. Mol. Cell. Biol. 20:7505-7515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blaumueller, C. M., H. Qi, P. Zagouras, and S. Artavanis-Tsakonas. 1997. Intracellular cleavage of Notch leads to a heterodimeric receptor on the plasma membrane. Cell 90:281-291. [DOI] [PubMed] [Google Scholar]
- 6.Brou, C., F. Logeat, N. Gupta, C. Bessia, O. LeBail, J. R. Doedens, A. Cumano, P. Roux, R. A. Black, and A. Israel. 2000. A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol. Cell 5:207-216. [DOI] [PubMed] [Google Scholar]
- 7.Cagan, R. L., and D. F. Ready. 1989. Notch is required for successive cell decisions in the developing Drosophila retina. Genes Dev. 3:1099-1112. [DOI] [PubMed] [Google Scholar]
- 8.De Strooper, B., W. Annaert, P. Cupers, P. Saftig, K. Craessaerts, J. S. Mumm, E. H. Schroeter, V. Schrijvers, M. S. Wolfe, W. J. Ray, A. Goate, and R. Kopan. 1999. A presenilin-1-dependent γ-secretase-like protease mediates release of Notch intracellular domain. Nature 398:518-522. [DOI] [PubMed] [Google Scholar]
- 9.Espinosa, L., J. Ingles-Esteve, C. Aguilera, and A. Bigas. 2003. Phosphorylation by glycogen synthase kinase-3β down-regulates Notch activity, a link for Notch and Wnt pathways. J. Biol. Chem. 278:32227-32235. [DOI] [PubMed] [Google Scholar]
- 10.Feldman, B. J., T. Hampton, and M. L. Cleary. 2000. A carboxy-terminal deletion mutant of Notch1 accelerates lymphoid oncogenesis in E2A-PBX1 transgenic mice. Blood 96:1906-1913. [PubMed] [Google Scholar]
- 11.Foltz, D. R., M. C. Santiago, B. E. Berechid, and J. S. Nye. 2002. Glycogen synthase kinase-3β modulates notch signaling and stability. Curr. Biol. 12:1006-1011. [DOI] [PubMed] [Google Scholar]
- 12.Fryer, C. J., E. Lamar, I. Turbachova, C. Kintner, and K. A. Jones. 2002. Mastermind mediates chromatin-specific transcription and turnover of the Notch enhancer complex. Genes Dev. 16:1397-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fryer, C. J., J. B. White, and K. A. Jones. 2004. Mastermind recruits CycC:CDK8 to phosphorylate the Notch ICD and coordinate activation with turnover. Mol. Cell 16:509-520. [DOI] [PubMed] [Google Scholar]
- 14.Hoemann, C. D., N. Beaulieu, L. Girard, N. Rebai, and P. Jolicoeur. 2000. Two distinct Notch1 mutant alleles are involved in the induction of T-cell leukemia in c-myc transgenic mice. Mol. Cell. Biol. 20:3831-3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsieh, J. J., T. Henkel, P. Salmon, E. Robey, M. G. Peterson, and S. D. Hayward. 1996. Truncated mammalian Notch1 activates CBF1/RBPJk-repressed genes by a mechanism resembling that of Epstein-Barr virus EBNA2. Mol. Cell. Biol. 16:952-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jarriault, S., C. Brou, F. Logeat, E. H. Schroeter, R. Kopan, and A. Israel. 1995. Signalling downstream of activated mammalian Notch. Nature 377:355-358. [DOI] [PubMed] [Google Scholar]
- 17.Joyal, J. L., and D. B. Sacks. 1994. Insulin-dependent phosphorylation of calmodulin in rat hepatocytes. J. Biol. Chem. 269:30039-30048. [PubMed] [Google Scholar]
- 18.Kimberly, W. T., W. P. Esler, W. Ye, B. L. Ostaszewski, J. Gao, T. Diehl, D. J. Selkoe, and M. S. Wolfe. 2003. Notch and the amyloid precursor protein are cleaved by similar γ-secretase(s). Biochemistry 42:137-144. [DOI] [PubMed] [Google Scholar]
- 19.Kopan, R., E. H. Schroeter, H. Weintraub, and J. S. Nye. 1996. Signal transduction by activated mNotch: importance of proteolytic processing and its regulation by the extracellular domain. Proc. Natl. Acad. Sci. USA 93:1683-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin, Y. W., R. A. Nichols, J. J. Letterio, and P. D. Aplan. 2006. Notch1 mutations are important for leukemic transformation in murine models of precursor-T leukemia/lymphoma. Blood 107:2540-2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Logeat, F., C. Bessia, C. Brou, O. LeBail, S. Jarriault, N. G. Seidah, and A. Israel. 1998. The Notch1 receptor is cleaved constitutively by a furin-like convertase. Proc. Natl. Acad. Sci. USA 95:8108-8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.MacKichan, M. L., F. Logeat, and A. Israel. 1996. Phosphorylation of p105 PEST sequence via a redox-insensitive pathway up-regulates processing of p50 NF-κB. J. Biol. Chem. 271:6084-6091. [DOI] [PubMed] [Google Scholar]
- 23.McGill, M. A., and C. J. McGlade. 2003. Mammalian numb proteins promote Notch1 receptor ubiquitination and degradation of the Notch1 intracellular domain. J. Biol. Chem. 278:23196-23203. [DOI] [PubMed] [Google Scholar]
- 24.Mumm, J. S., E. H. Schroeter, M. T. Saxena, A. Griesemer, X. Tian, D. J. Pan, W. J. Ray, and R. Kopan. 2000. A ligand-induced extracellular cleavage regulates γ-secretase-like proteolytic activation of Notch1. Mol. Cell 5:197-206. [DOI] [PubMed] [Google Scholar]
- 25.Nam, Y., P. Sliz, L. Song, J. C. Aster, and S. Blacklow. 2006. Structural basis for cooperativity in recruitment of the mastermind coactivator to Notch transcription complexes. Cell 124:973-983. [DOI] [PubMed] [Google Scholar]
- 26.O'Neil, J., J. Calvo, K. McKenna, V. Krishnamoorthy, J. C. Aster, C. H. Bassing, F. W. Alt, M. Kelliher, and A. T. Look. 2006. Activating Notch1 mutations in mouse models of T-ALL. Blood 107:781-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petcherski, A. G., and J. Kimble. 2000. LAG-3 is a putative transcriptional activator in the Caenorhabditis elegans Notch pathway. Nature 405:364-368. [DOI] [PubMed] [Google Scholar]
- 28.Petcherski, A. G., and J. Kimble. 2000. Mastermind is a putative activator for Notch. Curr. Biol. 10:R471-R473. [DOI] [PubMed] [Google Scholar]
- 29.Rand, M. D., L. M. Grimm, S. Artavanis-Tsakonas, V. Patriub, S. C. Blacklow, J. Sklar, and J. C. Aster. 2000. Calcium depletion dissociates and activates heterodimeric notch receptors. Mol. Cell. Biol. 20:1825-1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rechsteiner, M. 1989. PEST regions, proteolysis, and cell cycle progression. Revis. Biol. Celular. 20:235-253. [PubMed] [Google Scholar]
- 31.Sanchez-Irizarry, C., A. C. Carpenter, A. P. Weng, W. S. Pear, J. C. Aster, and S. C. Blacklow. 2004. Notch subunit heterodimerization and prevention of ligand-independent proteolytic activation depend, respectively, on a novel domain and the LNR repeats. Mol. Cell. Biol. 24:9265-9273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schroeter, E. H., J. A. Kisslinger, and R. Kopan. 1998. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature 393:382-386. [DOI] [PubMed] [Google Scholar]
- 33.Shah, S., S. F. Lee, K. Tabuchi, Y. H. Hao, C. Yu, Q. LaPlant, H. Ball, C. E. Dann III, T. Sudhof, and G. Yu. 2005. Nicastrin functions as a γ-secretase-substrate receptor. Cell 122:435-447. [DOI] [PubMed] [Google Scholar]
- 34.Shaye, D. D., and I. Greenwald. 2005. LIN-12/Notch trafficking and regulation of DSL ligand activity during vulval induction in Caenorhabditis elegans. Development 132:5081-5092. [DOI] [PubMed] [Google Scholar]
- 35.Struhl, G., and A. Adachi. 1998. Nuclear access and action of notch in vivo. Cell 93:649-660. [DOI] [PubMed] [Google Scholar]
- 36.Struhl, G., and I. Grenwald. 1999. Presenilin is required for activity and nuclear access of Notch in Drosophila. Nature 398:522-525. [DOI] [PubMed] [Google Scholar]
- 37.Sundaram, M. V. 2005. The love-hate relationship between Ras and Notch. Genes Dev. 19:1825-1839. [DOI] [PubMed] [Google Scholar]
- 38.Tetzlaff, M. T., W. Yu, M. Li, P. Zhang, M. Finegold, K. Mahon, J. W. Harper, R. J. Schwartz, and S. J. Elledge. 2004. Defective cardiovascular development and elevated cyclin E and Notch proteins in mice lacking the Fbw7 F-box protein. Proc. Natl. Acad. Sci. USA 101:3338-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsunematsu, R., K. Nakayama, Y. Oike, M. Nishiyama, N. Ishida, S. Hatakeyama, Y. Bessho, R. Kageyama, T. Suda, and K. I. Nakayama. 2004. Mouse Fbw7/Sel-10/Cdc4 is required for notch degradation during vascular development. J. Biol. Chem. 279:9417-9423. [DOI] [PubMed] [Google Scholar]
- 40.Wallberg, A. E., K. Pedersen, U. Lendahl, and R. G. Roeder. 2002. p300 and PCAF act cooperatively to mediate transcriptional activation from chromatin templates by notch intracellular domains in vitro. Mol. Cell. Biol. 22:7812-7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weng, A. P., A. A. Ferrando, W. Lee, J. P. T. Morris, L. B. Silverman, C. Sanchez-Irizarry, S. C. Blacklow, A. T. Look, and J. C. Aster. 2004. Activating mutations of NOTCH1 in human T-cell acute lymphoblastic leukemia. Science 306:269-271. [DOI] [PubMed] [Google Scholar]
- 42.Weng, A. P., Y. Nam, M. S. Wolfe, W. S. Pear, J. D. Griffin, S. C. Blacklow, and J. C. Aster. 2003. Growth suppression of pre-T acute lymphoblastic leukemia cells by inhibition of notch signaling. Mol. Cell. Biol. 23:655-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu, L., J. C. Aster, S. C. Blacklow, R. Lake, S. Artavanis-Tsakonas, and J. D. Griffin. 2000. MAML1, a human homologue of Drosophila mastermind, is a transcriptional coactivator for NOTCH receptors. Nat. Genet. 26:484-489. [DOI] [PubMed] [Google Scholar]
- 44.Ye, Y., N. Lukinova, and M. E. Fortini. 1999. Neurogenic phenotypes and altered Notch processing in Drosophila Presenilin mutants. Nature 398:525-529. [DOI] [PubMed] [Google Scholar]