Abstract
The Sleeping Beauty (SB) transposon system has generated many transposon-insertional mutant mouse lines, some of which have resulted in embryonic lethality when bred to homozygosity. Here we report one such insertion mapped to the mouse actin-related protein complex subunit 3 gene (Arpc3). Arpc3 is a component of the Arp2/3 complex, which plays a major role in actin nucleation with Y-shaped branching from the mother actin filament in response to migration signaling. Arpc3 transposon-inserted mutants developed only to the blastocyst stage. In vitro blastocyst culture of Arpc3 mutants exhibited severe spreading impairment of trophoblasts. This phenotype was also observed in compound heterozygotes generated using conventional gene-targeted and transposon-inserted alleles. Arpc3-deficient mutants were shown to lack actin-rich structures in the spreading trophoblast. Electron microscopic analysis demonstrated the lack of mesh-like structures at the cell periphery, suggesting a role of Arpc3 in Y-shaped branching formation. These data indicate the importance of Arpc3 in the Arp2/3 complex for trophoblast outgrowth and suggest that Arpc3 may be indispensable for implantation.
Sleeping Beauty (SB) is a Tc1/mariner-like transposon reconstructed from the fish genome and shown to have transposability in the mouse germ line (10, 13, 17) as well as cultured mammalian cells (19, 20, 27). The SB transposon system consists of two components: the transposon, a DNA element flanked by terminal inverted repeats, and transposase, which catalyzes transposition. Transposition occurs when transposases bind to the inverted-repeat sequence, initiating excision of the transposon and reintegrating it into another locus. Highly active transposition has enabled researchers to develop the SB transposon as a novel tool for insertional mutagenesis in mice (4, 18, 23) or as a new vehicle for gene therapy (31, 43). We have extensively characterized chromosomal SB transposition in the mouse germ line and revealed its preferential transposition near the original donor site (DS), called local hopping, in which approximately 50% of total transpositions are clustered in a 3-Mb region around the DS and 80% are located on the same chromosome as the DS (18). This feature of the SB transposon was recently applied as a region-specific saturation mutagenesis screen (23). On the other hand, approximately 20% of the remaining transpositions are randomly distributed throughout the genome, making this system attractive for genome-wide mutagenesis. Moreover, another advantage of utilizing the SB transposon system in mice is that both the transposon and transposase are derived from a different organism, thus preventing any undesired mobilization of endogenous transposable elements. In addition, transposition events can be easily detected using the transposon sequence as a tag. Other cross-species usages for tagged mutagenesis have been successfully applied in (i) the mouse, by using the piggyBac transposon derived from the cabbage looper moth Trichoplusia ni (8), and (ii) Caenorhabditis elegans, by using the Mos transposon derived from Drosophila melanogaster (16). In order to achieve SB transposon-mediated mutagenesis in mice, we employed the gene trap scheme using a novel transposon vector, and we have generated a large number of mutant mice, approximately 30% of which displayed the phenotype (23).
Previously, we reported one mutant line with transposon insertion in the Arpc3 gene (18), a subunit of the Arp2/3 complex initially discovered in Acanthamoeba castellanii (28). The Arp2/3 complex consists of two actin-related proteins (Arp2 and Arp3) and five protein subunits (Arpc1 to Arpc5), conserved from protozoa to mammals. The Arp2/3 complex has an important role for actin nucleation, filament binding, and Y-shaped branching to reorganize filamentous actin (F-actin) in response to extrinsic or intrinsic signaling for cell motility (32, 39). The nucleation activity is activated by the Wiskott-Aldrich syndrome protein (Wasp) family (30, 35). It is thought that this activation occurs through the Wasp family protein's direct interaction with Arpc3, as shown by yeast two-hybrid experiments (29). An in vitro assay for actin-nucleating activity using reconstructed complexes revealed that lack of Arpc3 caused a 12-fold decrease in activity compared to that of the intact complex (15); whether the remaining activity possesses a significant biological function remains to be investigated. Taken together, Arpc3 might function as a modulator of Arp2/3 complex actin-nucleating activity. Our Arpc3-deficient mice generated by the SB transposon system should provide a better insight on this matter.
In this report, we further characterized the phenotype of Arpc3 transposon mutant mice and determined its essential role for mouse development during the peri-implantation stage.
MATERIALS AND METHODS
Generation of Arpc3-deficient mice using the Sleeping Beauty transposon system.
SB transposon-mediated mutagenesis has been described previously (18, 23). One line, TM117, had multiple transposon insertions, of which one insertion was mapped to the Arpc3 gene, located on the same chromosome as the putative DS. This line also contained the SB transposase in another locus. To segregate the transposon integration site in the Arpc3 gene from other transgene loci, the founder mouse was mated with an ICR mouse. Offspring with a single transposon inserted into the Arpc3 gene were identified by competition PCR genotyping using the following primers: 5′-AGCCAGCATTGTTGACAGCAGCGACTAAGG-3′(TM117 IOF2), 5′-CCTCGGTATTGGGATGAGCGATAACTGAGC-3′(TM117 IOR1), and 5′-CTTGTGTCATGCACAAAGTAGATGTCC-3′(T/BAL). PCR conditions were as follows: initial denaturation at 95°C for 15 min; 35 cycles at 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s; and a final extension at 72°C for 7 min. PCR was performed using 0.1 μg of genomic DNA with the HotStarTaq system (QIAGEN). This condition was used for all subsequent PCRs described below, except where otherwise noted. Absence of the SB transposase locus was confirmed by PCR using conditions previously described (17). Single-copy transposon integration into the Arpc3 gene was confirmed by Southern blot analysis using a lacZ probe as described below.
β-Galactosidase staining of embryos.
Embryos taken at 12.5 days postcoitum (dpc) were fixed with 1% paraformaldehyde (PFA)-0.2% glutaraldehyde-0.02% NP-40 in phosphate-buffered saline (PBS) for 30 min, followed by adequate washing in PBS with 0.02% NP-40. The staining protocol has been described previously (18). Genomic DNA was isolated from yolk sac using a standard protocol for competition PCR genotyping with primers TM117 IOF2, TM117 IOR1, and T/BAL.
Targeted disruption of the Arpc3 gene by homologous recombination.
A targeting vector was constructed to replace exon 2 of the Arpc3 gene with a phosphoglycerate kinase promoter (PGK)-neo selection cassette using Escherichia coli-based bacterial artificial chromosome (BAC) recombineering (24, 26), possibly resulting in a premature truncated protein due to frameshift mutation. A BAC clone (RP23-407I6) carrying the Arpc3 gene was purchased from the BACPAC Resource Center. The Arpc3 exon 2 replaced with PGK-neo and the flanking 2.2-kb short arm and 8.7-kb long arm of Arpc3 were retrieved in a diphtheria toxin-containing vector. For BAC recombineering, flanking sequences of PGK-neo cassette were introduced using the following primers: 5′-CCGTCGACGCAGAACTGGAGGAAGCGCATCCTTCCATA-3′(MR1F1) and 5′-CCGAATTCGGGGTACCGAGTGTGTGTTCATGAATCTGGGTGCTC-3′(MR1R1) for the upstream replacement region and 5′-CCGGATCCAACCTTTCCCCCACCAAAGCTATGACTTCT-3′(MR2F1) and 5′-CCGAGCTCCAGACATAAACTGTTTCAAAGCACTCAGGA-3′(MR2R1) for the downstream replacement region. The retrieving vector was prepared using the following primers: 5′-CCTCTAGACCAGTACCTGGTGACAGGGATTGGGAGAACAGGG-3′(RR1F1) and 5′-GGACTAGTCATCCGGAGATGCGGGGGCTCATTTGG-3′(RR1R1) for the 2.2-kb upstream replacement locus and 5′-GGACTAGTGCCTTTGACAGTGGTGGACTTGCTTTGT-3′(RR2F1) and 5′-GGCTCGAGCAAACCAAACCATAAATAGAAAACCAAGCC-3′(RR2R1) for the 8.7-kb downstream replacement locus. The targeting vector was linearized by PvuI digestion and transfected into v6.5 embryonic stem (ES) cells (11) by electroporation. Genomic DNA isolated from G418-resistant colonies was screened for homologous recombination using PCR and confirmed by Southern blot analyses. PCR primers used for homologous recombination screening were 5′-GAATGGGCTGACCGCTTCCTCGTGCTTTAC-3′(NeoPolAR) and 5′-ACACACATGTACACATGCATGGTCATGTGC-3′(TarCheF).
To detect knockout and wild-type alleles, the following primers were used for competition PCR: 5′-TGACCTCTGACATGTGCACACATGAGCACC-3′(KOGPF1), 5′-TTATAAGAGAGCAGGCTGAGTAAGCCAGG-3′(KOGPR1), and NeoPolAR.
Southern blot analyses of Arpc3 mutant and knockout mice.
To confirm that a single copy of the transposon was inserted into the Arpc3 gene locus, TM117 genomic DNA was digested sequentially with BamHI and PflF1 before being hybridized with either transposon- or Arpc3-specific probes. Probe L, an EcoRV-SacI fragment (827 bp) from a lacZ-containing plasmid vector, was used to detect the transposon-integrated allele. To detect the wild-type allele, probe B (368 bp) was amplified from genomic DNA using the following primers: 5′-TAGGAAGTAGTTTCCTATCTTAACAACTGC-3′(ProbeBF) and 5′-CGTCTTTCATTGAACCCAGAACTTGCTTAT-3′(ProbeBR).
For Arpc3 knockout mice, genomic DNA from ES cells was digested with KpnI before being hybridized with probe E (874 bp), obtained by amplifying Arpc3 exon 1 using the following primers: 5′-GGGAGTCTTCAATTTCAAAATCCAGCCTTT-3′(SPSAF) and 5′-TGTCTACCCGTTAAACTGTGAGCTCCTTGA-3′(SPSAR).
In vitro blastocyst culture.
Blastocysts were flushed from the uterus at 3.5 dpc and cultured independently in 24-well plates with M16 medium for 1 day. Blastocysts were then cultured in ES medium (Dulbecco's modified Eagle's medium supplemented with 2-mercaptoethanol [10−4 M], 20% fetal bovine serum, and 1,000 U/ml leukemia inhibitory factor) on culture dishes coated with 0.1% gelatin.
Tyrode treatment for zona-free blastocysts.
Zonae pellucidae were removed from blastocysts by brief incubation in acidic Tyrode's solution at room temperature, followed by adequate washing with M16 medium. Zona-free blastocysts were cultured in M16 for 1 day before the medium was replaced with ES medium for an additional 3 days. Trophoblast outgrowth on culture dishes was measured using Metacam software (Universal Imaging Corporation).
Analysis of promoter-trapped mRNA by RT-PCR.
Total RNA was isolated from either Arpc3Tp/+ or wild-type adult brains by using TRIzol (Invitrogen). cDNA was synthesized from total RNA (0.5 μg) by using Superscript II (Invitrogen) reverse transcriptase (RT) with random hexamer primers (Promega). To examine the expression of promoter-trapped mRNA (Arpc3 exon 1 to lacZ in the transposon vector), cDNA was amplified using the following primers: 5′-AAACGCTTTCTGAGTTCGGCTTCTCTGGAT-3′(E1F2) and 5′-CCAGGGTTTTCCCAGTCACGACGTTGTAAA-3′(LacZ R1). As a positive control, cDNA was also amplified using the following primers: 5′-TGGGAATGGGTCAGAAGGACTC-3′(β-actin F) and 5′-AGAGGCATACAGGGACAGCACA-3′(β-actin R).
Expression analysis of blastocysts.
Blastocysts in 10 μl of PBS were mixed with 10 μl of 2× RNasin buffer (0.15 M NaCl, 10 mM Tris-HCl [pH 8.0], 5 mM dithiothreitol, 40 U RNasin [Promega]) for a direct RT reaction. The sample was frozen and thawed for the purpose of cell disruption before the RT reaction was performed with an oligo(dT) primer using Superscript II. The cDNA was amplified using the Arpc3-specific primers E1F2 and 5′-TTCATCCACAATGTCCGTGTCTTTGGTCTC-3′(E23R1). The truncated mutant transcript was amplified with primers for green fluorescent protein (GFP) and Arpc3: 5′-GCGATCACATGGTCCTGCTGGAGTTCGTG-3′(GFP-5U) and E23R1. The sample was treated at 95°C for 15 min before 1 μl of lysate was used as a template for competition PCR genotyping with primers TM117 IOF2, TM117 IOR1, and T/BAL. PCR conditions were as described above except that 40 cycles were performed.
Immunostaining and fluorescence microscopy.
Blastocysts were drop-cultured in M16 medium independently for 1 day on untreated dishes before being transferred into ES medium on coverslips coated with 0.1% gelatin for 3 days. These cultured cells were then fixed with 4% PFA-PBS for 10 min and permeabilized for 5 min with 0.2% Triton X-100 (TX-100; Pierce) in PBS. Antibodies against vinculin (hVin-1, immunoglobulin G1 monoclonal antibody; Sigma) and paxillin (BD Transduction Laboratories) and a secondary Alexa Fluor 488-conjugated goat antibody against mouse immunoglobulin G were used for immunostaining analyses. Phalloidin-rhodamine (Invitrogen) for F-actin was used for fluorescence analysis. Images were acquired using an inverted tissue culture microscope (model IX70; Olympus) and captured with a digital charge-coupled device camera (Cool Snap; Photometrics) using Metacam software. Intensity analysis was also performed using the same software. PCR genotyping of blastocysts was performed after immunostaining analyses. Blastocysts were incubated overnight at 56°C in 15 μl of lysis buffer (50 mM KCl, 10 mM Tris-HCl [pH 8.3], 2.5 mM MgCl2, 0.45% NP-40, 0.45% Tween 20) containing proteinase K (final concentration, 0.2 μg/μl). Treated samples were heat inactivated at 95°C for 15 min before 1 μl was used as a template for competitive PCR genotyping with primers TM117 IOF2, TM117 IOR1, and T/BAL.
Electron microscopy.
Replica electron microscopy (REM) was performed as described previously (40), except for the following modifications for cytoskeleton observation: cells were rinsed briefly with PEM buffer [80 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES) (pH 6.9), 1 mM EGTA, and 1 mM MgCl2] before brief treatment at room temperature with 1% TX-100 and 4% polyethylene glycol in PEM buffer, followed by fixation in 2% PFA-2% glutaraldehyde or 4% PFA.
For actin labeling, cells were rinsed briefly in PEM buffer and treated briefly at room temperature with 1% TX-100 and 4% polyethylene glycol in PEM buffer, followed by fixation in 4% PFA for 5 min. PCR genotyping was performed at this stage by suspending partially picked inner-cell-mass (ICM) cells in 15 μl of lysis buffer supplemented with proteinase K before overnight incubation at 56°C. Templates were heat inactivated at 95°C for 15 min before competitive genotyping PCR with primers TM117 IOF2, TM117 IOR1, and T/BAL.
Fixed samples were treated with biotin-phalloidin (Alexis) for 1 h at room temperature. The samples were rinsed for 5 min before overnight incubation at 4°C in a mixture containing streptavidin-conjugated 15-nm-diameter colloidal gold (British Biocell International). These samples were washed with PBS before postfixation with 2% glutaraldehyde. They were treated in 0.1% tannic acid-distilled water for 20 min. After four washes with distilled water, the samples were treated with 0.1% uranyl acetate in distilled water and dehydrated with gradient ethanol. The samples were then treated overnight in 3-methylbutyl acetate for critical point drying. The samples were then rotary replicated with platinum/carbon at an angle of 25° using a freeze fracture apparatus (BAF 060; BAL-TEC.). The thickness of the replicas (∼2.5 nm) was controlled with a quartz crystal monitor. Replicas were treated with hydrofluoric acid, placed in household bleach, picked up on grids, and examined with an electron microscope (H-7100; Hitachi) at 80 kV.
RESULTS
SB transposon mutagenesis: generation of Arpc3-deficient mutants.
We have previously described one mutant line, TM117, in which multiple transposon integrations were identified and have reported that homozygotes in one integrated locus in the Arpc3 gene were embryonically lethal (18) (Table 1). In order to rule out any effects of other integration loci, DS and/or the SB transposase locus, contributing to the embryonic lethal phenotype, the transposon integration site in the Arpc3 gene was successfully segregated from other transgene loci by breeding the founder male with ICR females. The putative DS for TM117 is located on chromosome 5 by the following observation: another transposon mutant line (TM118) with the same DS has the transposon integrated into the G protein coupled receptor kinase 4 (GPRK4) gene on centromeric chromosome 5 (5qB1). This transposon-inserted locus could not be segregated from the DS, indicating its close proximity to the GPRK4 gene.
TABLE 1.
Homozygous mutation in the mouse Arpc3 gene results in early embryonic lethality
| Cross and time | No. of mice with the following genotype at the indicated time:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| WT | Arpc3Tp/+ | Arpc3Tp/Tp | Arpc3+/KO | Arpc3Tp/KO | ||||||
| Arpc3Tp/+ × Arpc3Tp/+ | ||||||||||
| Postnatal | 36 | 52 | 0 | |||||||
| 10.5 dpc | 5 | 12 | 0 | |||||||
| 8.5 dpc | 11 | 9 | 0 | |||||||
| 7.5 dpc | 11 | 10 | 0 | |||||||
| 5.5 dpc | 16 | 17 | 0 | |||||||
| 3.5 dpc | 31 | 43 | 25 | |||||||
| Arpc3Tp/+ × Arpc3+/KO (genetic complementation test) | ||||||||||
| Postnatal | 10 | 4 | 11 | 0 | ||||||
| 3.5 dpc | 11 | 15 | 6 | 9 | ||||||
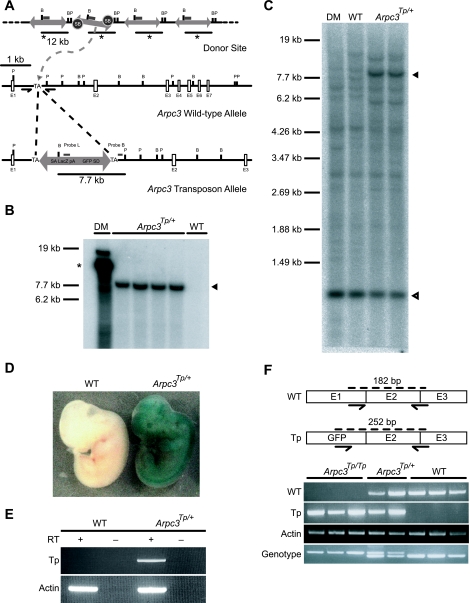
To confirm that a single transposon copy was integrated in the Arpc3 gene, Southern blot analysis was performed using probes L and B, which hybridize to the trap vector and flanking sequence of transposon-integrated loci, respectively (Fig. 1A, B, and C). Heterozygous (Arpc3Tp/+) embryos demonstrated ubiquitous expression at 12.5 dpc (Fig. 1D). Arpc3Tp/+ adult mice appeared normal and fertile and transmitted the mutant allele to approximately 50% of their offspring. However, no live homozygous (Arpc3Tp/Tp) newborns were observed from heterozygous intercrosses (Table 1).
FIG. 1.
Disruption of the Arpc3 gene using the SB transposon system. (A) The transposon insertion site was mapped to intron 1 of the Arpc3 gene located on mouse chromosome 5. SB, Sleeping Beauty transposase; TA, thymidine-adenine dinucleotide; SA, splice acceptor; pA, polyadenylation signal; SD, splice donor; E, exon; B, BamHI; P, PflFI. (B and C) Verification of a single transposon insertion in Arpc3Tp/+ mice by Southern blot analysis using both the transposon-specific probe L and the locus-specific probe B (panels B and C, respectively). A single band in Arpc3Tp/+ mouse lanes indicates a single transposon insertion site segregated from the original donor mouse (DM). Asterisk indicates original DS concatemer in DM. Arrowheads show transposon-inserted allele (7,789 bp), and open arrowhead shows intact Arpc3 locus, in Arpc3Tp/+ mouse lanes (1,072 bp). WT, wild type. (D) Ubiquitous Arpc3 expression as shown by 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) staining of a 12.5-dpc Arpc3Tp/+ embryo. (E) Confirmation of transposon vector promoter trap by RT-PCR analysis of an adult Arpc3Tp/+ brain. Promoter trapped transcripts (Tp; 910 bp) and β-actin as a positive control (Actin; 301 bp) were detected in Arpc3Tp/+ mice. (F) Confirmation of disrupted endogenous Arpc3 transcript. WT (182-bp) and 3′-end mutant (Tp; 252-bp) transcripts of Arpc3 were detected by RT-PCR. β-actin was used as an internal control. The genotype was determined by competition PCR to detect WT (185-bp) and Tp (237-bp) alleles.
To determine the time of embryonic lethality, embryos were isolated at various gestational ages (3.5 to 10.5 dpc) for PCR genotyping. No live Arpc3Tp/Tp embryos were detected at 5.5 dpc by PCR genotyping. We observed small deciduae containing scarred residues in the uteri of heterozygous intercrossed females (approximately 43% of deciduae were aborted per litter). This frequency is higher than the number of abortions occurring in normal uteri at the same developmental stage. It is presumed that these deciduae contain homozygous embryos. However, this cannot be confirmed by PCR genotyping, due to the progressed state of scarred residue degradation (data not shown). On the other hand, Arpc3Tp/Tp blastocysts were isolated at 3.5 dpc (Table 1). Successful promoter trapping by the transposon vector was confirmed in adult brains (Fig. 1E), and the lack of an intact Arpc3 transcript in Arpc3Tp/Tp blastocysts was confirmed using RT-PCR (Fig. 1F). Arpc3Tp/Tp morulae and early-stage blastocysts were indistinguishable from their wild-type counterparts (data not shown). These results suggest that Arpc3 may be dispensable for embryonic development up to the early blastocyst stage (3.5 dpc) but is essential for development around the peri-implantation stage.
Impaired outgrowth activity in Arpc3Tp/Tp trophoblasts.
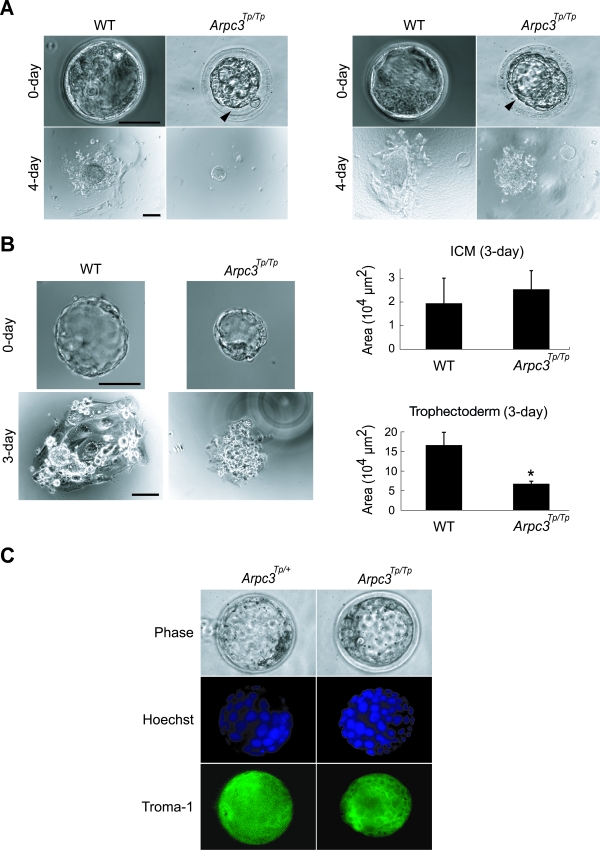
At the early blastocyst stage, Arpc3Tp/Tp blastocysts were macroscopically comparable to their wild-type counterparts (data not shown). However, all Arpc3Tp/Tp blastocysts remained unexpanded at later stages of development, which was seen as a gap between the zona pellucida and trophectoderm (Fig. 2A). To determine whether Arpc3Tp/Tp blastocysts could proliferate and develop normally, 3.5-dpc blastocysts were cultured in vitro with ES medium. Wild-type and Arpc3Tp/+ blastocysts hatched from the zona pellucida and attached to gelatin-coated plates within 24 to 36 h by a process involving outgrowth of their trophoblast cell layers. In contrast, as many as 64% of Arpc3Tp/Tp blastocysts failed to hatch (Fig. 2A), suggesting dysfunction of trophoblasts. To examine the outgrowth activity of trophoblasts, we removed blastocyst zonae pellucidae for in vitro culture. Zona-free treatment resulted in all Arpc3Tp/Tp blastocysts adhering to gelatin-coated dishes. However, trophoblast outgrowth was severely impaired in Arpc3Tp/Tp blastocysts even after 3 days of culture (Fig. 2B). Hoechst staining of Arpc3Tp/Tp trophoblast nuclei displayed comparable cell numbers after 3 days of culture (Arpc3Tp/+, 29.3 ± 4.93 cells; Arpc3Tp/Tp, 27.7 ± 4.93 cells [means ± standard deviations; n = 3; P = 0.70 by Student's t test]), indicating that the defect in mutant trophoblast outgrowth was due to abnormal spreading and not to proliferation. The Arpc3Tp/Tp ICM survived normally on the trophoblasts for as long as 3 days in culture. However, Arpc3Tp/Tp ICM growth was halted, possibly as a result of a trophoblast spreading defect. To verify any autonomous defect of Arpc3Tp/Tp ICM cells, we examined their growth on mouse embryonic fibroblast cells. After 5 days of culture, Arpc3Tp/Tp ICM growth was delayed compared with that of the wild type, indicating that Arpc3Tp/Tp ICM cells may have some autonomous defect (data not shown). Expression of Troma-1, a trophoblast marker (22, 41), was found to be normal in Arpc3Tp/Tp blastocysts, indicating that Arpc3Tp/Tp morulae differentiated normally to trophectoderm (Fig. 2C). These results suggest that Arpc3 is dispensable for blastocyst formation. However, Arpc3 seems to play a critical role in subsequent hatching and trophoblast outgrowth.
FIG. 2.
Trophoblast outgrowth impairment in Arpc3Tp/Tp blastocysts. (A) Unhatched (64%) and hatched (36%) phenotype in Arpc3Tp/Tp blastocysts taken from separate littermates are demonstrated in left and right sets of panels, respectively. Blastocysts were cultured in ES medium for the indicated times. Arrowheads indicate gaps between the zona pellucida and trophectoderm in Arpc3Tp/Tp late-stage blastocysts. WT, wild type. Bars, 50 μm and 100 μm for 0-day and 4-day cultures, respectively. (B) Differences in trophoblast outgrowth efficiency between cultured WT and Arpc3Tp/Tp blastocysts after zona-free treatment. The ordinate represents outgrowth area measurements of the ICM and trophectoderm in culture after 3 days, obtained by using MetaCam software, version 6.0r4. Data are means ± standard deviations from three independent cultures. *, P < 0.05 for comparison with WT (by Student's t test). Bars, 50 μm and 100 μm for 0-day and 3-day cultures, respectively. (C) Immunostaining with Troma-1 and Hoechst staining demonstrated trophectoderm differentiation and nucleus morphology comparable with those of the WT, indicating normal cell growth in Arpc3Tp/Tp trophoblasts up to the blastocyst stage.
Confirmation of the Arpc3Tp/Tp phenotype by genetic complementation analysis.
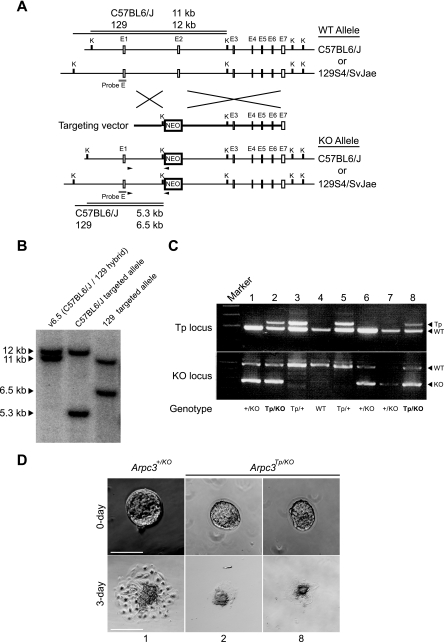
Although Arpc3Tp/Tp contains only a single transposon insertion site (confirmed by Southern blot analysis in Fig. 1B and C), other, unexpected genomic mutations may have occurred elsewhere besides the Arpc3 locus. Therefore, to confirm that the phenotype was a result of Arpc3 disruption by the SB transposon system, we generated an Arpc3-null allele (Arpc3KO) by homologous recombination in ES cells (Fig. 3A). The targeted clone was confirmed by Southern blot analysis (Fig. 3B). The ES cell line (v6.5) used in this study is a hybrid of C57BL6/J and 129S4/SvJae (11). We found that restriction fragment length polymorphism exists between C57BL6/J and 129S4/SvJae in the Arpc3 locus (Fig. 3B, left lane). Although homologous recombination occurred in both alleles (Fig. 3B, center and right lanes), the ES clone with the targeted C57BL6/J allele (Fig. 3B, center lane) was used for generating Arpc3+/KO mice because its sequence is known in public databases. Arpc3+/KO mice were mated with Arpc3Tp/+ mice to produce compound heterozygotes (Arpc3Tp/KO). PCR genotyping for compound transposon and knockout heterozygotes was performed under conditions described in Material and Methods (Fig. 3C). Blastocysts of compound heterozygotes determined from Fig. 3C were used for in vitro culture analyses (Fig. 3D). They were used to determine whether the knockout allele could complement the phenotype observed in Arpc3Tp/Tp mice. As expected, compound heterozygosity (Arpc3Tp/KO) was lethal to mice (Table 1), and their blastocysts(Arpc3Tp/KO) exhibited a phenotype comparable to the Arpc3Tp/Tp phenotype (compare Fig. 2B with Fig. 3D). Hatching failure was evident in approximately 22% of Arpc3Tp/KO embryos, while the remaining Arpc3Tp/KO embryos displayed impaired outgrowth activity. Genetic complementation analysis confirms that the phenotype is a direct result of SB transposon insertion into and disruption of the Arpc3 locus.
FIG. 3.
Genetic complementation test between the mutated transposon and knockout alleles of the Arpc3 gene. (A) Schematic representation of the targeting strategy to replace Arpc3 exon 2 with a neo cassette in ES cells. Probe E for Southern blot analysis is shown as a filled box. Arrowheads show primers used for homologous recombination screening. K, KpnI; E, exon; WT, wild type; KO, knockout. (B) Southern blot analysis of targeted ES cell lines. Left lane, control v6.5 ES cell line with C57BL6/J and 129S4/SvJae alleles; middle and right lanes, C57BL6/J and 129S4/SvJae targeted alleles, respectively. The C57BL6/J targeted ES cell line was used for generating Arpc3+/KO mice and for subsequent analyses. Polymorphism between C57BL6/J and 129S4/SvJae exists upstream of Arpc3 exon 1. (C) PCR genotyping for compound heterozygotes (Arpc3Tp/KO) obtained from intercrosses between Arpc3Tp/+ and Arpc3+/KO mice. Two independent genotypings were determined by competition PCR to detect WT (185-bp) and transposon (Tp) (237-bp) alleles for the Tp locus and WT (493-bp) and KO (278-bp) alleles for the KO locus. Sample numbers shown in PCR genotyping (top) correspond with samples used for culture in panel D. (D) Comparable phenotypes were detected for compound heterozygous blastocysts (Arpc3Tp/KO) and Arpc3Tp/Tp blastocysts by using in vitro culture analysis (see Fig. 2A). Bar, 100 μm.
In a separate experiment to confirm the Arpc3Tp/Tp phenotype, we tried to remobilize the SB transposon vector from the Arpc3 locus by an SB transposase rescue experiment. However, after screening approximately 400 offspring from Arpc3Tp/+,SB+mice mated with wild-type mice, no remobilization was detected by PCR and sequencing analyses. We have previously shown that heterochromatin conformation enhances SB transposition (44). Since Arpc3 is ubiquitously expressed, as shown by lacZ staining (Fig. 1D), it is assumed that this locus attains a highly euchromatic conformation, making remobilization of a single transposon copy extremely difficult.
Dynamics of actin accumulation are defective in Arpc3Tp/Tp blastocysts.
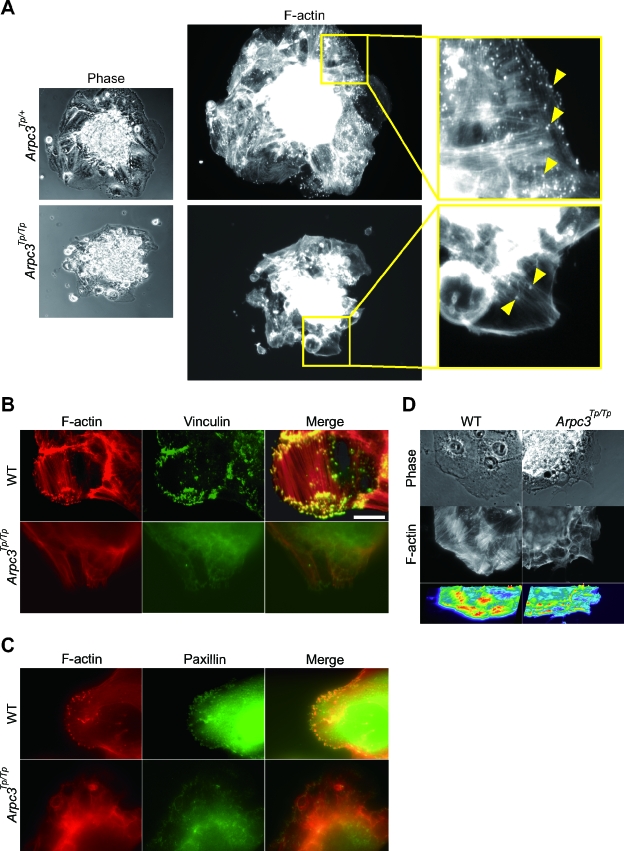
To further investigate the impaired outgrowth phenotype in Arpc3Tp/Tp trophoblasts, we examined actin morphology using rhodamine-phalloidin staining. Since Arp2/3 complex-dependent actin remodeling has been shown to be important for membrane protrusion in the leading edge during cell migration in other cell types (33), we proceeded to analyze the state of actin accumulation in Arpc3Tp/Tp trophoblasts. Abundant actin-rich structures were observed in normal trophoblasts (Fig. 4A, top panels). Immunostaining revealed that these actin-rich structures also stained for vinculin and paxillin, both known adherent structure markers (Fig. 4B and C, top panels). It has been shown that vinculin transiently interacts with the Arp2/3 complex during cell migration (7). These actin-rich adherent structures observed during cell migration were drastically reduced at the cell periphery of Arpc3Tp/Tp trophoblasts (Fig. 4A, B, and C, lower panels). It has been shown that β1 integrin was involved in the trophoblast migration signalling and accumulation of focal adhesion kinase and other focal adhesion components (14). A similar reduction in actin-rich adherent structures was also observed in mutant trophoblasts by using β1 integrin immunostaining (data not shown), suggesting reduced adhesion affinity with extracellular matrix components. These data suggest that total actin-nucleating activity was impaired in Arpc3Tp/Tp trophoblasts, as shown by intensity analysis of F-actin fibers (Fig. 4D).
FIG. 4.
Reduced actin-rich adherent structures in Arpc3Tp/Tp trophoblasts. (A) Impaired actin-rich structures in Arpc3Tp/Tp trophoblasts. By using rhodamine-phalloidin staining for F-actin, reduction of actin-rich structures in cultured Arpc3Tp/Tp trophoblasts was observed (bottom center panel). In contrast, Arpc3Tp/+ trophoblasts displayed abundant actin-rich structures (top center panel). (Left panels) Phase-contrast images of the same trophoblasts at lower magnification. (Right panels) Magnified images of boxed regions in center panels. (B and C) Double staining comprising rhodamine-phalloidin, followed by anti-vinculin (B) or anti-paxillin (C). WT, wild type. Bar, 20 μm. (D) Reduction of total F-actin in Arpc3Tp/Tp trophoblast as shown by intensity image analysis. Calculations were performed using MetaCam (version 6.0r4) fluorescence microscope management software.
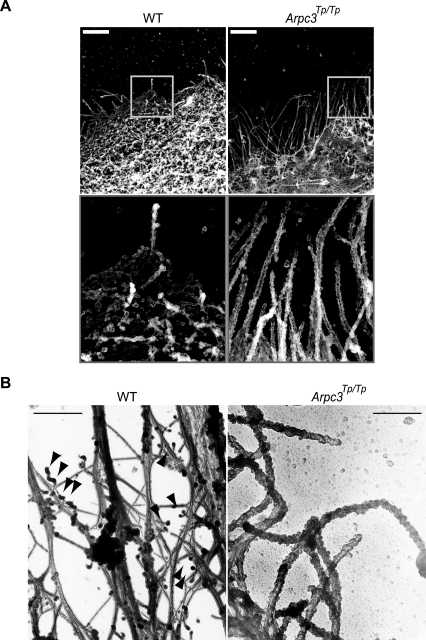
To further investigate the cytoskeletal organization in Arpc3Tp/Tp trophoblasts, REM analysis was performed. Arpc3Tp/Tp trophoblasts demonstrated impaired actin net assembly and aberrant cytoskeleton at the cell periphery (Fig. 5A, right panels), compared with the wild type (Fig. 5A, left panels). Wild-type fibers were approximately 8 nm in diameter, consistent with the expected size of actin filaments. In contrast, aberrant filaments in Arpc3Tp/Tp trophoblasts were more than 50 nm in diameter, indicating that these aberrant structures consisted of bundled cytoskeletal components such as microtubules or other intermediate filaments. To clarify whether the aberrant cytoskeleton consists of actin or other fibers, we performed phalloidin-labeled electron microscopy (Fig. 5B). The aberrant Arpc3Tp/Tp cytoskeleton did not interact with phalloidin (Fig. 5B, right panel), in contrast with the wild-type cytoskeleton (Fig. 5B, left panel). These data suggest that Arpc3 is essential for actin assembly at the cell periphery of migrating cells.
FIG. 5.
Apparent anomaly of cell-peripheral cytoskeletal structure in Arpc3Tp/Tp mutant trophoblast. (A) Impairment of cytoskeleton in the cell periphery of an Arpc3Tp/Tp trophoblast as shown by replica electron microscopy. Boxed areas are enlarged in lower panels. Bars, 2 μm. WT, wild type. (B) Aberrant cytoskeleton structure in Arpc3Tp/Tp trophoblast not labeled with phalloidin compared with WT. Arrowheads show representative streptavidin-gold labeling. Bar, 500 nm.
DISCUSSION
In this study, we have provided further evidence that the SB transposon system is a useful tool for phenotypic analysis and rapid identification of the phenotype causative gene in mice. Analysis of the mouse genome is important for elucidating molecular mechanisms of pathological and physiological phenomena because it highly resembles the human genome, and such information will be beneficial in searching for new drug targets. The mouse is the only mammalian model whose genome and transcriptome sequence have been almost completely determined. Therefore, mouse insertional mutagenesis can further advance research elucidating the human genome.
During the process of phenotypic analysis of the transposon mutant mouse, we have to consider the possibility of compound phenotypes caused by the effects of the transposase gene integration site, DS, and “footprints” (addition of a 5-bp sequence,TAC(A/T)G, which occurs as a result of remobilization). We have shown that a single transposon copy was inserted into the Arpc3 gene and that the Arpc3Tp/Tp phenotype was not complemented by the Arpc3 knockout allele, indicating that disruption of the Arpc3 gene is genuinely responsible for the aberrant trophoblast phenotype. In germ line analysis, the transposase has to be segregated before phenotypic analyses in order to avoid additional transpositions in somatic cells (6, 9).
The present paper revealed that Arpc3, a component of the Arp2/3 complex, does not have any obvious phenotype, at least up to compaction and blastocoel formation. However, a severe outgrowth defect was observed in the trophoblast cell. In Caenorhabditis elegans, Arp2/3 complex function is dispensable for cytokinesis, probably due to compensatory roles of formin and profilin for actin nucleation, but indispensable for ventral enclosure associated with cell migration (12, 36, 38). These data are consistent with phenotypic analyses of Arpc3 mutant mouse embryos. Although we did not detect any Arpc3 maternal mRNA by using RT-PCR at the early-blastocyst stage (Fig. 1F), we cannot exclude the possibility of residual maternal protein, which may have supported the growth of mutants up to the early blastocyst stage.
We have observed that 64% of Arpc3Tp/Tp blastocysts failed to hatch, while the remaining 36% hatched and adhered to the plate. Hatching is regulated by embryonic tension due to actin filaments (5) and by embryonic proteases (37). It is certain that Arpc3Tp/Tp embryos displayed impaired expansion at later blastocyst stages, which was seen as a gap between the blastocyst and zona pellucida (Fig. 2A). This is the first morphological difference between wild-type and Arpc3Tp/Tp blastocysts. The hatching event for 36% of Arpc3Tp/Tp embryos could be the result of residual extracellular enzymatic lysis factors or embryonic proteases.
Actin-rich adherent structures and stress fibers were also drastically reduced in Arpc3Tp/Tp trophoblasts (Fig. 4A, B, and C). We hypothesize that these Arpc3-dependent actin-rich adherent structures are podosomes. Podosomes are highly dynamic actin-rich adherent structures that are thought to contribute to tissue invasion and matrix remodeling (25). Arp2/3-dependent actin polymerization is required for podosome formation at the stress fiber-focal-adhesion interface (21). Additional evidence to reinforce our hypothesis is that Wasp (Arp2/3 activator)-defective dendritic cells (2) and osteoclasts (3) exhibit diminished podosomes. Taken together, these observations indicate that the Arpc3 gene is essential for podosome-like structures in trophoblasts.
The assembly of stress fibers in Arpc3Tp/Tp was reduced with loss of cytoskeletal alignment (Fig. 4B, C, and 4D). REM demonstrated that actin net assembly in Arpc3Tp/Tp trophoblasts was impaired (Fig. 5A) within 1 μm from the leading edge, where the Arp2/3 complex is distributed (1, 39) and actin assembly rates are elevated (34). These data suggest that Arp2/3-dependent actin-rich adherent and peripheral mesh structures could stabilize stress fiber and regulate its alignment.
Finally, Arpc3-deficient mouse embryos show an earlier stage of developmental arrest than other known mutants of Arp2/3 complex activators, such as the Wasp/Scar family proteins (42). Since several activators are known to functionally associate with the Arp2/3 complex, their compensatory function among themselves may account for the milder phenotype compared with Arpc3-defective mutants. Therefore, blockage of Arpc3 or another subunit of the Arp2/3 complex may lead to a severe loss of mobility of invasive cells in vivo, including growing tumors. Arpc3 may be a potential candidate for drug target design to inhibit metastatic signals.
Acknowledgments
We thank M. Nozaki for providing the Troma-1 antibody. We are grateful to E. Oiki, R. Ikeda, E. S. Saito, T. Hayakawa, R. Hashimoto, C. Kokubu, H. Fukui, K. Kuratani, K. Yokota, K. Yoshino, M. Yamamoto, M. Okabe, M. Ikawa, and H. Hamada for helpful advice and excellent technical support.
This work was supported by grants from the New Energy and Industrial Technology Development Organization of Japan; the Uehara Memorial Foundation; the Preventure Program, Japan Science and Technology Agency; and RIKEN, The Institute of Physical and Chemical Research. It was also supported by a grant-in-aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
REFERENCES
- 1.Bailly, M., F. Macaluso, M. Cammer, A. Chan, J. E. Segall, and J. S. Condeelis. 1999. Relationship between Arp2/3 complex and the barbed ends of actin filaments at the leading edge of carcinoma cells after epidermal growth factor stimulation. J. Cell Biol. 145:331-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Calle, Y., H. C. Chou, A. J. Thrasher, and G. E. Jones. 2004. Wiskott-Aldrich syndrome protein and the cytoskeletal dynamics of dendritic cells. J. Pathol. 204:460-469. [DOI] [PubMed] [Google Scholar]
- 3.Calle, Y., G. E. Jones, C. Jagger, K. Fuller, M. P. Blundell, J. Chow, T. Chambers, and A. J. Thrasher. 2004. WASp deficiency in mice results in failure to form osteoclast sealing zones and defects in bone resorption. Blood 103:3552-3561. [DOI] [PubMed] [Google Scholar]
- 4.Carlson, C. M., A. J. Dupuy, S. Fritz, K. J. Roberg-Perez, C. F. Fletcher, and D. A. Largaespada. 2003. Transposon mutagenesis of the mouse germline. Genetics 165:243-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheon, Y. P., M. C. Gye, C. H. Kim, B. M. Kang, Y. S. Chang, S. R. Kim, and M. K. Kim. 1999. Role of actin filaments in the hatching process of mouse blastocyst. Zygote 7:123-129. [DOI] [PubMed] [Google Scholar]
- 6.Collier, L. S., C. M. Carlson, S. Ravimohan, A. J. Dupuy, and D. A. Largaespada. 2005. Cancer gene discovery in solid tumours using transposon-based somatic mutagenesis in the mouse. Nature 436:272-276. [DOI] [PubMed] [Google Scholar]
- 7.DeMali, K. A., C. A. Barlow, and K. Burridge. 2002. Recruitment of the Arp2/3 complex to vinculin: coupling membrane protrusion to matrix adhesion. J. Cell Biol. 159:881-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ding, S., X. Wu, G. Li, M. Han, Y. Zhuang, and T. Xu. 2005. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell 122:473-483. [DOI] [PubMed] [Google Scholar]
- 9.Dupuy, A. J., K. Akagi, D. A. Largaespada, N. G. Copeland, and N. A. Jenkins. 2005. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature 436:221-226. [DOI] [PubMed] [Google Scholar]
- 10.Dupuy, A. J., S. Fritz, and D. A. Largaespada. 2001. Transposition and gene disruption in the male germline of the mouse. Genesis 30:82-88. [DOI] [PubMed] [Google Scholar]
- 11.Eggan, K., H. Akutsu, J. Loring, L. Jackson-Grusby, M. Klemm, W. M. Rideout III, R. Yanagimachi, and R. Jaenisch. 2001. Hybrid vigor, fetal overgrowth, and viability of mice derived by nuclear cloning and tetraploid embryo complementation. Proc. Natl. Acad. Sci. USA 98:6209-6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evangelista, M., S. Zigmond, and C. Boone. 2003. Formins: signaling effectors for assembly and polarization of actin filaments. J. Cell Sci. 116:2603-2611. [DOI] [PubMed] [Google Scholar]
- 13.Fischer, S. E., E. Wienholds, and R. H. Plasterk. 2001. Regulated transposition of a fish transposon in the mouse germ line. Proc. Natl. Acad. Sci. USA 98:6759-6764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gleeson, L. M., C. Chakraborty, T. McKinnon, and P. K. Lala. 2001. Insulin-like growth factor-binding protein 1 stimulates human trophoblast migration by signaling through α5 β1 integrin via mitogen-activated protein kinase pathway. J. Clin. Endocrinol. Metab. 86:2484-2493. [DOI] [PubMed] [Google Scholar]
- 15.Gournier, H., E. D. Goley, H. Niederstrasser, T. Trinh, and M. D. Welch. 2001. Reconstitution of human Arp2/3 complex reveals critical roles of individual subunits in complex structure and activity. Mol. Cell 8:1041-1052. [DOI] [PubMed] [Google Scholar]
- 16.Granger, L., E. Martin, and L. Segalat. 2004. Mos as a tool for genome-wide insertional mutagenesis in Caenorhabditis elegans: results of a pilot study. Nucleic Acids Res. 32:e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horie, K., A. Kuroiwa, M. Ikawa, M. Okabe, G. Kondoh, Y. Matsuda, and J. Takeda. 2001. Efficient chromosomal transposition of a Tc1/mariner-like transposon Sleeping Beauty in mice. Proc. Natl. Acad. Sci. USA 98:9191-9196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horie, K., K. Yusa, K. Yae, J. Odajima, S. E. Fischer, V. W. Keng, T. Hayakawa, S. Mizuno, G. Kondoh, T. Ijiri, Y. Matsuda, R. H. Plasterk, and J. Takeda. 2003. Characterization of Sleeping Beauty transposition and its application to genetic screening in mice. Mol. Cell. Biol. 23:9189-9207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ivics, Z., P. B. Hackett, R. H. Plasterk, and Z. Izsvak. 1997. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell 91:501-510. [DOI] [PubMed] [Google Scholar]
- 20.Izsvak, Z., Z. Ivics, and R. H. Plasterk. 2000. Sleeping Beauty, a wide host-range transposon vector for genetic transformation in vertebrates. J. Mol. Biol. 302:93-102. [DOI] [PubMed] [Google Scholar]
- 21.Kaverina, I., T. E. Stradal, and M. Gimona. 2003. Podosome formation in cultured A7r5 vascular smooth muscle cells requires Arp2/3-dependent de-novo actin polymerization at discrete microdomains. J. Cell Sci. 116:4915-4924. [DOI] [PubMed] [Google Scholar]
- 22.Kemler, R., P. Brulet, M. T. Schnebelen, J. Gaillard, and F. Jacob. 1981. Reactivity of monoclonal antibodies against intermediate filament proteins during embryonic development. J. Embryol. Exp. Morphol. 64:45-60. [PubMed] [Google Scholar]
- 23.Keng, V. W., K. Yae, T. Hayakawa, S. Mizuno, Y. Uno, K. Yusa, C. Kokubu, T. Kinoshita, K. Akagi, N. A. Jenkins, N. G. Copeland, K. Horie, and J. Takeda. 2005. Region-specific saturation germline mutagenesis in mice using the Sleeping Beauty transposon system. Nat. Methods 2:763-769. [DOI] [PubMed] [Google Scholar]
- 24.Lee, E. C., D. Yu, J. Martinez de Velasco, L. Tessarollo, D. A. Swing, D. L. Court, N. A. Jenkins, and N. G. Copeland. 2001. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics 73:56-65. [DOI] [PubMed] [Google Scholar]
- 25.Linder, S., and M. Aepfelbacher. 2003. Podosomes: adhesion hot-spots of invasive cells. Trends Cell Biol. 13:376-385. [DOI] [PubMed] [Google Scholar]
- 26.Liu, P., N. A. Jenkins, and N. G. Copeland. 2003. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 13:476-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo, G., Z. Ivics, Z. Izsvak, and A. Bradley. 1998. Chromosomal transposition of a Tc1/mariner-like element in mouse embryonic stem cells. Proc. Natl. Acad. Sci. USA 95:10769-10773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Machesky, L. M., S. J. Atkinson, C. Ampe, J. Vandekerckhove, and T. D. Pollard. 1994. Purification of a cortical complex containing two unconventional actins from Acanthamoeba by affinity chromatography on profilin-agarose. J. Cell Biol. 127:107-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Machesky, L. M., and R. H. Insall. 1998. Scar1 and the related Wiskott-Aldrich syndrome protein, WASP, regulate the actin cytoskeleton through the Arp2/3 complex. Curr. Biol. 8:1347-1356. [DOI] [PubMed] [Google Scholar]
- 30.Machesky, L. M., R. D. Mullins, H. N. Higgs, D. A. Kaiser, L. Blanchoin, R. C. May, M. E. Hall, and T. D. Pollard. 1999. Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc. Natl. Acad. Sci. USA 96:3739-3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mikkelsen, J. G., S. R. Yant, L. Meuse, Z. Huang, H. Xu, and M. A. Kay. 2003. Helper-independent Sleeping Beauty transposon-transposase vectors for efficient nonviral gene delivery and persistent gene expression in vivo. Mol. Ther. 8:654-665. [DOI] [PubMed] [Google Scholar]
- 32.Mullins, R. D., J. A. Heuser, and T. D. Pollard. 1998. The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc. Natl. Acad. Sci. USA 95:6181-6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pollard, T. D., and G. G. Borisy. 2003. Cellular motility driven by assembly and disassembly of actin filaments. Cell 112:453-465. [DOI] [PubMed] [Google Scholar]
- 34.Ponti, A., M. Machacek, S. L. Gupton, C. M. Waterman-Storer, and G. Danuser. 2004. Two distinct actin networks drive the protrusion of migrating cells. Science 305:1782-1786. [DOI] [PubMed] [Google Scholar]
- 35.Rohatgi, R., L. Ma, H. Miki, M. Lopez, T. Kirchhausen, T. Takenawa, and M. W. Kirschner. 1999. The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell 97:221-231. [DOI] [PubMed] [Google Scholar]
- 36.Sawa, M., S. Suetsugu, A. Sugimoto, H. Miki, M. Yamamoto, and T. Takenawa. 2003. Essential role of the C. elegans Arp2/3 complex in cell migration during ventral enclosure. J. Cell Sci. 116:1505-1518. [DOI] [PubMed] [Google Scholar]
- 37.Sawada, H., K. Yamazaki, and M. Hoshi. 1990. Trypsin-like hatching protease from mouse embryos: evidence for the presence in culture medium and its enzymatic properties. J. Exp. Zool. 254:83-87. [DOI] [PubMed] [Google Scholar]
- 38.Severson, A. F., D. L. Baillie, and B. Bowerman. 2002. A formin homology protein and a profilin are required for cytokinesis and Arp2/3-independent assembly of cortical microfilaments in C. elegans. Curr. Biol. 12:2066-2075. [DOI] [PubMed] [Google Scholar]
- 39.Svitkina, T. M., and G. G. Borisy. 1999. Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J. Cell Biol. 145:1009-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Svitkina, T. M., and G. G. Borisy. 1998. Correlative light and electron microscopy of the cytoskeleton of cultured cells. Methods Enzymol. 298:570-592. [DOI] [PubMed] [Google Scholar]
- 41.Tamai, Y., T. Ishikawa, M. R. Bosl, M. Mori, M. Nozaki, H. Baribault, R. G. Oshima, and M. M. Taketo. 2000. Cytokeratins 8 and 19 in the mouse placental development. J. Cell Biol. 151:563-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vartiainen, M. K., and L. M. Machesky. 2004. The WASP-Arp2/3 pathway: genetic insights. Curr. Opin. Cell Biol. 16:174-181. [DOI] [PubMed] [Google Scholar]
- 43.Yant, S. R., L. Meuse, W. Chiu, Z. Ivics, Z. Izsvak, and M. A. Kay. 2000. Somatic integration and long-term transgene expression in normal and haemophilic mice using a DNA transposon system. Nat. Genet. 25:35-41. [DOI] [PubMed] [Google Scholar]
- 44.Yusa, K., J. Takeda, and K. Horie. 2004. Enhancement of Sleeping Beauty transposition by CpG methylation: possible role of heterochromatin formation. Mol. Cell. Biol. 24:4004-4018. [DOI] [PMC free article] [PubMed] [Google Scholar]