Abstract
During liver development, hepatocytes undergo a maturation process that leads to the fully differentiated state. This relies at least in part on the coordinated action of liver-enriched transcription factors (LETFs), but little is known about the dynamics of this coordination. In this context we investigate here the role of the LETF hepatocyte nuclear factor 6 (HNF-6; also called Onecut-1) during hepatocyte differentiation. We show that HNF-6 knockout mouse fetuses have delayed expression of glucose-6-phosphatase (g6pc), which catalyzes the final step of gluconeogenesis and is a late marker of hepatocyte maturation. Using a combination of in vivo and in vitro gain- and loss-of-function approaches, we demonstrate that HNF-6 stimulates endogenous g6pc gene expression directly via a synergistic and interdependent action with HNF-4 and that it involves coordinate recruitment of the coactivator PGC-1α. The expression of HNF-6, HNF-4, and PGC-1α rises steadily during liver development and precedes that of g6pc. We provide evidence that threshold levels of HNF-6 are required to allow synergism between HNF-6, HNF-4, and PGC-1α to induce time-specific expression of g6pc. Our observations on the regulation of g6pc by HNF-6 provide a model whereby synergism, interdependency, and threshold concentrations of LETFs and coactivators determine time-specific expression of genes during liver development.
Liver development is regulated by a dynamic network of liver-enriched transcription factors (LETFs) (5, 8, 34, 35, 51). During this process the two major cell types in the liver, hepatocytes and cholangiocytes, derive from common precursor cells called hepatoblasts (9, 37). Cholangiocytes delineate bile ducts (19, 38), and hepatocytes perform the liver metabolic functions. Differentiating hepatocytes organize into cords and undergo a maturation process during which they progressively acquire their functions. This maturation process relies on the coordinated action of the LETFs, which interact by mutually controlling their expression and by coordinately regulating target genes (1, 10, 15). Transcriptional regulators, including the LETFs, recruit coactivators harboring chromatin-modifying activities to their target promoters. In addition, the activity of LETFs and hepatocyte maturation are dependent on extracellular signals (14). Time- and tissue-specific gene expression is thus achieved through the concerted action of tissue-specific factors and coactivators on their target genes, under the control of extracellular cues. Genome-wide analyses identified target promoters of several LETFs in normal or regenerating adult liver, and mechanisms for coordinated action of LETFs have been proposed with regard to their respective target promoter occupancies (22, 48). In contrast, the molecular dynamics of LETF action in the developing embryo, and how a given transcription factor can exert different functions at different developmental stages, are not well known.
Hepatocyte nuclear factor 6 (HNF-6, also called Onecut-1), a member of the Onecut family of transcriptional activators, belongs to the LETF network (4, 22, 27, 33). It is expressed in the hepatic epithelial cells, i.e., in the hepatoblasts and then in the cholangiocytes and hepatocytes, where it persists in adults. HNF-6 regulates the decision of hepatoblasts to differentiate toward the biliary or hepatocyte lineage, by controlling genes that modulate the response to transforming growth factor β (7). It is also required for bile duct morphogenesis and indirectly regulates B lymphopoiesis (3, 6). In hepatocytes HNF-6 controls the expression of other LETFs (4, 22, 27, 33), and in vivo data indicate that it regulates genes involved in detoxification and glucose metabolism (17, 42, 46). Finally, it has been shown to stimulate transcription by sequence-specific recruitment of the coactivator CBP or p300/CBP-associated factor (pCAF) to its target genes (16).
In the present work we investigated how HNF-6 controls gene expression in differentiating hepatocytes during development. We found that HNF-6 synergistically and interdependently cooperates with HNF-4α1 (referred to below as HNF-4), a LETF critical for hepatocyte differentiation (11, 20, 23, 39, 47), and with the coactivator PGC-1α to induce time-specific expression of the catalytic subunit of glucose-6-phosphatase (g6pc). We propose a model whereby synergism, interdependency, and threshold concentrations of LETFs and coactivators control time-specific expression of genes during hepatocyte differentiation.
MATERIALS AND METHODS
Animals.
All mice, raised in our animal facilities, were treated according to the principles of laboratory animal care of the local Animal Welfare Committee. hnf6 knockout mice and mice homozygous for allele hnf4tm1.1Gonz (hnf4fl/fl) were obtained as described elsewhere (11, 12).
Plasmids.
pCMV-HNF6, pCMV-Flag.HNF6, pCMV-Myc.HNF6, pRL-138, and pEF-GFP have been described elsewhere (16, 24, 25). pCMV-HNF4 and pCMV-Flag.HNF4 were gifts from S. A. Duncan, and pSVSPORT-PGC1α and pSVSPORT-HA.PGC1α were gifts from B. M. Spiegelman. To construct pG6PC(−207/+46)luc, the mouse g6pc promoter was PCR amplified (with primers 5′-GCTCTAGAAATAATTGGCTCTGCCAATG-3′ and 5′-CGGGATCCAGCCCGTGCAGTGAGTCCAG-3′) and subcloned into pBluescript. The fragment was excised by BamHI/XbaI restriction and inserted at the corresponding sites in pGL3-MCS (Promega).
Reverse transcription-PCR (RT-PCR) and real-time quantitative RT-PCR (qRT-PCR).
Total RNA was extracted from embryonic liver, neonate liver, bipotential murine embryonic liver (BMEL) cells, and NIH 3T3 cells with TriPure reagent (Roche). RNA (1 μg) was reverse transcribed as described previously (27). For semiquantitative PCR, the number of cycles corresponded to the mid-logarithmic phase. Primer sequences were 5′-ACCCTTCACCAATGACTCCTATG-3′ and 5′-ATGATGACTGCAGCAAATCGC-3′ for the mRNA coding for TATA-binding protein (tbp), 5′-TGTCTGTGATTGCTGACCTG-3′ and 5′-GTAGAAGTGACCATAACATAG-3′ for g6pc, 5′-CTCAGCTGGCAGCATGGGGTG-3′ and 5′-AACAGCTCCTCCACGTTGACG-3′ for the mRNA coding for phosphoenolpyruvate carboxykinase (pepck), and 5′-GAATTCAGAGCCTCTCGCCCCTCTC-3′ and 5′-CAGGAGCTGTCCGTGGC-3′ for hnf6. Real-time quantitative PCR was performed with the SYBR Green PCR Core kit (Eurogentec) on a MyIQ thermal cycler (Bio-Rad). Threshold cycles were transformed into copy numbers according to the standard calibration curve. The absolute copy number for each mRNA was normalized to the absolute copy number for the mRNA coding for β-actin (β-act). Primer sequences were 5′-TCCTGAGCGCAAGTACTCTGT-3′ and 5′-CTGATCCACATCTGCTGGAAG-3′ for β-act, 5′-TTCCAGCGCATGTCGGCGCTC-3′ and 5′-GGTACTAGTCCGTGGTTCTTC-3′ for hnf6, 5′-TGCAGCCAAGACTCTGTATG-3′ and 5′-CATCAAGTTCAGAAAGGTCAAG-3′ for pgc1α; 5′-GAAAATGTGCAGGTGTTGACCA-3′ and 5′-AGCTCGAGGCTCCGTAGTGTTT-3′ for hnf4, and 5′-TACATCATCGCCCAGTGTGTG-3′ and 5′-AGCTGCAGAGTGCCAATGATC-3′ for the mRNA coding for aquaporin-1 (aqp1). g6pc mRNA was quantified using the Master Mix for Probe Assays (Eurogentec). Primer sequences were generated using Primer3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). Primer sequences were 5′-GCCAGAGGGACTTCCTGGT-3′ and 5′-TCGGAGACTGGTTCAACCTC-3′; the Oligold (Eurogentec) 6-carboxyfluorescein (FAM)/6-carboxytetramethylrhodamine (TAMRA) probe sequence was 5′-GCCCGTATTGGTGGGTCCTGG-3′.
Cell culture and transfection.
NIH 3T3 cells were grown in Dulbecco's modified Eagle medium supplemented with 10% bovine serum (Gibco), 5 mM sodium pyruvate, and antibiotics. BMEL cells were cultured in monolayers or as aggregates as described previously (27, 41). For endogenous g6pc transcriptional stimulation assays (see Fig. 1C, 2B and C, 4A and C, and 7B), 105 BMEL cells or 7 × 104 3T3 cells were seeded on 12-well plates (TPP) 18 h prior to transfection. Cells were transfected with 4 μl of Lipofectamine 2000 (Invitrogen) and the following amounts of expression vectors: 750 ng of pCMV-Flag.HNF6, 750 ng of pCMV-Flag.HNF4, and 1.5 μg of pSVSPORT-PGC1α. The total amount of plasmid DNA was adjusted to 3 μg by adding pEF-GFP. For dose-dependent stimulation of g6pc expression (see Fig. 8A), we used the indicated amounts of pCMV-Myc.HNF6 and pCMV-Flag.HNF4, and the total DNA amount was adjusted to 1 μg using pEF-GFP. In all transcriptional stimulation assays, RNA was extracted 24 h after transfection. For luciferase assays, 5.5 × 104 BMEL cells were seeded on 24-well plates (TPP) and transfected with 2 μl of Lipofectamine 2000, 400 ng of pG6PC(−207/+46)luc, 15 ng of pRL-138 as an internal control, and 10 ng of pCMV-Flag.HNF6 and/or pCMV-Flag.HNF4α, and the total amount of plasmid DNA was adjusted to 465 ng by adding pEF-GFP. Luciferase activity was measured 24 h after transfection using the Dual-Luciferase reporter assay system (Promega) and a DLR-ready TD-20/20 luminometer (Turner Design). For small interfering RNA (siRNA) experiments, BMEL cells were transfected with 4 μl of Lipofectamine 2000 in 12-well plates with 750 ng of pCMV-Flag.HNF6, 750 ng of pCMV-Flag.HNF4, and 1.5 μg of pEF-GFP in the presence of 100 nM of a pool of control siRNAs or a pool of siRNAs directed against mouse pgc1α (Dharmacon).
FIG. 1.
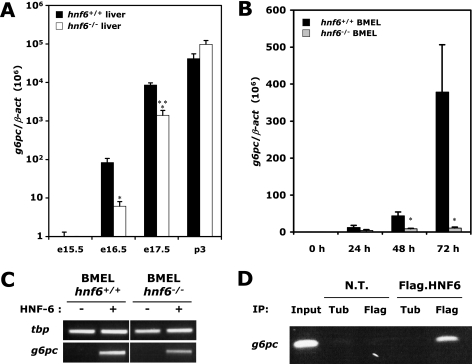
HNF-6 controls expression of g6pc during hepatocyte differentiation. (A) Developmental regulation of g6pc expression is impaired in the hnf6−/− liver. Total-liver RNA was extracted from wild-type (hnf6+/+) and hnf6−/− fetuses at different stages of development, and g6pc mRNA concentrations were measured by qRT-PCR. Data are expressed as the ratio of g6pc mRNA levels to β-act mRNA levels (means ± standard errors of the means; n ≥ 4 for each genotype). Significant differences are indicated (*, P < 0.05; ***, P < 0,001). Note the logarithmic scale. (B) Impaired g6pc expression in in vitro differentiating hnf6−/− BMEL cells. Wild-type and hnf6−/− BMEL cells were cultured in aggregates (38) to induce differentiation toward the hepatocyte lineage. RNA was extracted from those cells before culturing or after 24 h, 72 h, or 120 h of the aggregate culture. g6pc mRNA concentrations were measured as for panel A (means ± standard errors of the means; n ≥ 3 for each genotype). Significant differences between wild-type and hnf6−/− values are indicated (*, P < 0.05). (C) HNF-6 induces g6pc mRNA expression in undifferentiated embryonic liver cells. Wild-type and hnf6−/− BMEL cells were transfected with a vector coding for HNF-6, and g6pc mRNA was amplified by RT-PCR 24 h after transfection. We used the mRNA coding for TATA-binding protein (tbp) as a control. (D) HNF-6 binds endogenous g6pc promoter. BMEL cells were transfected with a vector encoding Flag-tagged HNF-6 and were submitted to a ChIP directed against the Flag epitope 24 h after transfection. A fragment spanning the −207-to-+46 region of g6pc was amplified by PCR. Nontransfected cells (N.T.) and a ChIP against tubulin (Tub) were used as negative controls. The input corresponds to a PCR product obtained from untreated chromatin representing 10% of the amount of chromatin used in each ChIP.
FIG. 2.
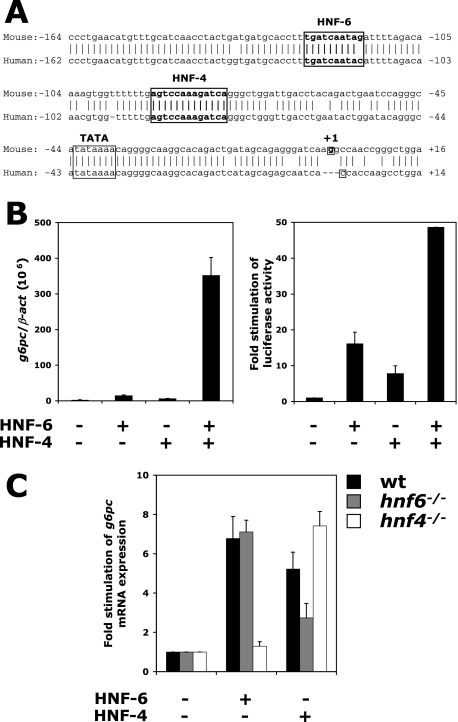
HNF-6 and HNF-4 regulate g6pc expression in a synergistic and interdependent way. (A) Alignment of the human and murine g6pc gene promoters. Binding sites for HNF-6 and HNF-4, as well as the TATA box and the transcription initiation site, are indicated. (B) HNF-6 and HNF-4 synergistically stimulate g6pc expression. (Left) BMEL cells were transfected with combinations of vectors encoding HNF-6 and HNF-4, and g6pc mRNA was quantified by qRT-PCR 24 h after transfection. Data are expressed as the ratio of g6pc mRNA levels to β-act mRNA levels (means ± standard errors of the means for at least four independent transfections performed in duplicate). (Right) HNF-6 and HNF-4 can synergistically stimulate the g6pc promoter. A reporter plasmid containing the g6pc promoter (−207 to +46) upstream of the firefly luciferase gene was transfected into BMEL cells together with combinations of vectors coding for HNF-6 and HNF-4, and luciferase activity was measured 24 h after transfection. Firefly luciferase activity was normalized to Renilla luciferase activity (internal control) and was expressed as n-fold induction of basal promoter activity in control, GFP-transfected cells (means ± standard errors of the means; n ≥ 3). (C) Mutual requirement for HNF-6 and HNF-4 in transcriptional control of g6pc expression. Wild-type, hnf6−/−, and hnf4−/− BMEL cells were transfected with vectors coding either for HNF-6 or for HNF-4, and g6pc mRNA concentrations were measured 24 h after transfection. Values are expressed as relative stimulation of g6pc mRNA expression (means ± standard errors of the means; n ≥ 4).
FIG. 4.
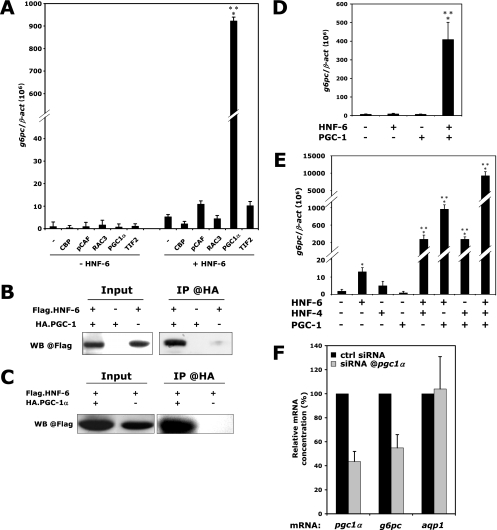
HNF-6 and HNF-4 synergistically stimulate g6pc expression through recruitment of the coactivator PGC-1α. (A) PGC-1α is a coactivator of HNF-6 in stimulation of the g6pc gene. hnf6−/− BMEL cells were transfected with vectors coding for the indicated coactivators, in the absence or the presence of a HNF-6-coding vector, and g6pc mRNA levels, expressed relative to β-act mRNA levels, were quantified by qRT-PCR 24 h after transfection (means ± standard errors of the means; n = 3). Significant differences relative to HNF-6-mediated stimulation of g6pc are indicated (***, P < 0.001). (B) HNF-6 physically interacts with PGC-1α. Protein extracts of BMEL cells expressing Flag.HNF-6 and/or HA.PGC-1α were subjected to an anti-HA coimmunoprecipitation. The presence of Flag.HNF-6 in the eluate was revealed by anti-Flag Western blotting (WB). (C) HNF-6/PGC-1α interaction does not require HNF-4. hnf4−/− BMEL cells were transfected and treated as for panel B. (D) Functional interaction between HNF-6 and PGC-1α is HNF-4 independent. hnf4−/− BMEL cells were transfected with combinations of expression vectors encoding HNF-6 and PGC-1α. g6pc mRNA concentrations were measured by qRT-PCR 24 h after transfection. Results are means ± standard errors of the means from at least four independent transfections. Significant differences are indicated (***, P < 0.001). (E) PGC-1α amplifies HNF6- and HNF-4-mediated stimulation of g6pc mRNA expression. BMEL cells were transfected with all possible combinations of vectors coding for HNF-6, HNF-4, and PGC-1α, and g6pc mRNA levels, expressed relative to β-act mRNA levels, were quantified by qRT-PCR 24 h after transfection (means ± standard errors of the means; n ≥ 4). Significant differences relative to control, GFP-transfected cells are indicated (*, P < 0.05; ***, P < 0.001). (F) PGC-1α is involved in the synergy between HNF-6 and HNF-4. BMEL cells were transfected with vectors coding for HNF-6 and HNF-4, together with control siRNAs or siRNAs directed against pgc1α. Relative mRNA concentrations were measured 24 h after transfection. Aquaporin-1 (aqp1) mRNA was used as a negative control. Values are expressed relative to those for control siRNA-transfected cells, which were set at 100% (means ± standard errors of the means; n = 5).
FIG. 7.
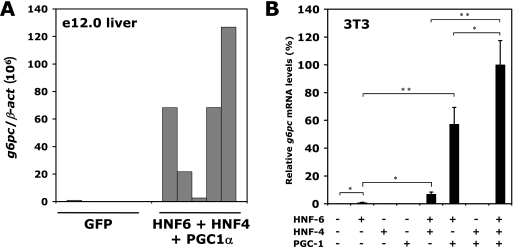
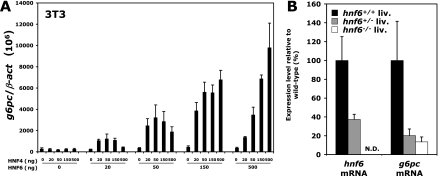
HNF-6, HNF-4, and PGC-1α can induce g6pc expression regardless of the cellular context. (A) Increased expression of HNF-6, HNF-4, and PGC-1α in immature embryonic liver leads to precocious expression of g6pc. Livers from e12.0 mouse embryos were electroporated with the indicated vectors and filter cultured for 48 h. g6pc mRNA concentrations were measured by qRT-PCR. Results of five independent experiments are shown, and data are expressed as the ratio of g6pc mRNA levels to β-act mRNA levels. (B) The activity of the HNF-6/HNF-4/PGC-1α complex is not tissue specific. NIH 3T3 fibroblasts were transfected with combinations of vectors coding for HNF-6, HNF-4, and PGC-1α. g6pc mRNA concentrations were quantified by qRT-PCR 24 h after transfection. Results are expressed as percentages of maximal g6pc mRNA expression ± standard errors of the means (n = 3). Significant differences are indicated (*, P < 0.05; **, P < 0.01).
FIG. 8.
Threshold levels of HNF-6 are required to induce stimulation of g6pc gene expression. (A) Dose-dependent stimulation of g6pc by HNF-4 requires threshold levels of HNF-6. NIH 3T3 fibroblasts were transfected with increasing amounts of HNF-6 and HNF-4 expression vectors as indicated, and g6pc mRNA levels, expressed relative to β-act levels, were measured 24 h after transfection. Data are means ± standard errors of the means from three independent experiments. (B) Time-specific induction of g6pc in the developing liver requires an optimal concentration of HNF-6. hnf6 and g6pc mRNA concentrations were quantified by qRT-PCR in e16.5 livers from hnf6+/+, hnf6+/−, and hnf6−/− embryos (means ± standard errors of the means; n ≥ 7).
Isolation of hnf4−/− BMEL cells from livers of hnf4fl/fl mice.
Mice homozygous for allele hnf4tm1.1Gonz (hnf4fl/fl), in which exons 4 and 5 are flanked by loxP sites (11), were crossed with heterozygous hnf4fl/+ animals. At embryonic day 14.5 (e14.5), embryos were removed and livers dissected. Clonal cell lines from both hnf4fl/fl and hnf4fl/+ livers were isolated as described previously (41). To obtain hnf4 gene disruption, the cells were electroporated with expression vectors coding for an eGFPnlsCre-fusion protein and containing a neomycin resistance cassette. The cells were plated at clonal density. Following selection in G418, isolated colonies were picked, expanded, and subjected to Southern blotting to confirm deletion efficiency. Full details will be given in a future article by G. P. Hayhurst et al.
CoIP.
Wild-type BMEL cells (1.25 × 106) were seeded on 10-cm plates (TPP), grown for 16 h, and transfected with 60 μl of Lipofectamine 2000, 10 μg of pCMV-Flag.HNF6, and/or 10 μg of pSVSPORT-HA.PGC1α, and the total amount of DNA was adjusted to 20 μg by adding pEF-GFP. On the same day, 50 μl of protein G-Sepharose beads per sample was washed four times with ice-cold coimmunoprecipitation assay (CoIP) binding buffer (20 mM Tris [pH 7.4], 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 10% glycerol, 1% Triton X-100, and protease inhibitors) and resuspended in 300 μl CoIP binding buffer. A rabbit polyclonal anti-hemagglutinin (anti-HA) antibody (3 μl per sample; Sigma) was added to the mixture, which was incubated overnight at 4°C with constant rotation. Beads were then washed in CoIP binding buffer. Twenty-four hours after transfection, cells were washed four times in ice-cold phosphate-buffered saline supplemented with protease inhibitors. Cells were then scraped in 300 μl of ice-cold CoIP binding buffer. Following a 10-s sonication, lysates were centrifuged at 4°C and 14,000 × g for 5 min to remove insoluble cell debris. Inputs (50 μl of the supernatant) were stored at −80°C. One-half of the rest of the supernatant was frozen for other applications, and the other half was immediately brought to a volume of 400 μl by adding CoIP binding buffer. The bead-antibody solution (50 μl) was added to the lysates, and the mixture was incubated for 2 h at 4°C with constant rotation. Beads were then washed three times in CoIP wash buffer (20 mM Tris [pH 7.4], 500 mM NaCl, 1 mM EDTA, 1 mM EGTA, 10% glycerol, 1% Triton X-100, and protease inhibitors) and three times in CoIP binding buffer. Beads were boiled for 5 min, and extracts were immediately loaded onto sodium dodecyl sulfate-polyacrylamide gels for Western blotting with a mouse monoclonal anti-Flag M2 antibody (Sigma). The secondary antibody was horseradish peroxidase-conjugated mouse IgG True Blot (eBioscience). The secondary antibody was detected by chemiluminescence (Amersham Pharmacia Biotech).
ChIP.
Chromatin immunoprecipitation (ChIP) was performed essentially as described previously (10, 25). For HNF-6 and HNF-4 binding assays (see Fig. 3), 1.5 × 106 BMEL cells were transfected with 60 μl of Lipofectamine 2000, 7 μg of pCMV-Flag.HNF6, and/or 7 μg of pCMV-HNF4 or 7 μg of pCMV-Flag.HNF4 and/or 7 μg of pCMV-HNF6. The total amount of plasmid DNA was adjusted to 28 μg by adding pEF-GFP. Binding on the g6pc promoter was measured by qPCR. Primer sequences were 5′-CTGGGTATAGGGGCGAAAGAC-3′ and 5′-GGCCCCAAGACCTCTAATCAT-3′ for 28S genomic DNA and 5′-GTTTTTGTGTGCCTGTTTTG-3′ and 5′-GCTATCAGTCTGCCTTGC-3′ for the g6pc promoter. The Oligold (Eurogentec) FAM/TAMRA probe sequence for the g6pc promoter was 5′-TTGAGTCCAAAGATCAGGGC-3′. Enrichments were calculated as 2exp − [(G6PCChIP − 28SChIP) − (G6PCInput − 28SInput)]. For the PGC-1α recruitment assay (see Fig. 5), BMEL cells were transfected as described above. One or more of the following plasmids were used: pCMV-HNF6 (7 μg), pCMV-HNF4 (7 μg), and pSVSPORT-PGC1α (14 μg). Primer sequences were as indicated above, and binding of PGC-1α was here visualized by an agarose gel. The antibody was monoclonal anti-Flag M2 (10 μg; Sigma), anti-PGC-1 (10 μg; sc-13067; Santa Cruz Biotechnologies), or anti-tubulin (2 μg; Sigma). One-fifth of each ChIP eluate was used in each PCR.
FIG. 3.
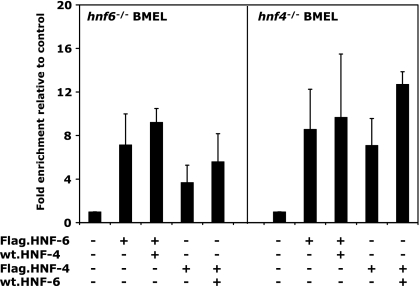
Independent binding of HNF-6 and HNF-4 to the g6pc promoter. hnf6−/− (left) and hnf4−/− (right) BMEL cells were transfected with the indicated combinations of expression vectors and were subjected to ChIP directed against the Flag epitope 24 h after transfection. Binding of Flag-tagged proteins was visualized by qPCR amplification of the g6pc promoter. Enrichments relative to the control were calculated as indicated in Materials and Methods. Data are means ± standard errors of the means from at least three independent experiments.
FIG. 5.
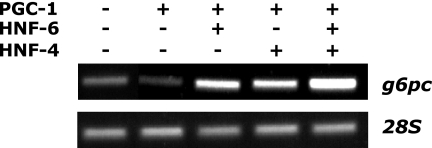
HNF-6 and HNF-4 recruit PGC-1α to the g6pc gene promoter. BMEL cells were transfected with a PGC-1α-expressing vector, either in the presence or in the absence of vectors coding for HNF-6 or HNF-4, and were subjected to ChIP using an antibody against PGC-1α. Binding of PGC-1α to the g6pc promoter was detected by PCR amplification of the coimmunoprecipitated −207-to-+46 g6pc promoter fragment.
Mouse fetal liver electroporation.
The procedure used was an adaptation of the recently described whole-embryo electroporation (26). Livers from e12.0 embryos were dissected in Hanks balanced salt solution (HBSS) buffer (Gibco). Each liver was microinjected (20 pulses, 6 ms per pulse) with a mixture consisting of 20% carboxymethyl cellulose, 0.02% trypan blue stain (Gibco), and 0.5 μg/μl plasmid DNA in HBSS buffer using a Picospritzer III (Parker Instrumentation) microinjector. Plasmid DNA mixtures consisted of 0.5 μg/μl pEF-GFP alone (control explants) or a mix consisting of 0.1 μg/μl pCMV-Flag.HNF6, 0.1 μg/μl pCMV-Flag.HNF4, 0.2 μg/μl pSVSPORT-PGC1α, and 0.1 μg/μl pEF-GFP. Livers in HBSS were then electroporated (3 pulses of 100 V, 50 ms per pulse, 1-s delay between each pulse) with an ECM 830 Electro Square Porator (BTX Genetronics) and filter cultured for 48 h at 37°C in BMEL cell culture medium prior to RNA extraction.
RESULTS
HNF-6 controls the expression of glucose-6-phosphatase during hepatocyte differentiation.
HNF-6 is expressed in hepatocytes throughout liver development and after birth. It is known to control hepatocyte-specific expression of genes in the adult (17, 22, 33, 42, 46) and to control hepatoblast differentiation at early stages of liver development (7), but its function in differentiating hepatocytes, i.e., prior to birth, is not well characterized. Therefore, we compared the expression of genes in hnf6+/+ and hnf6−/− livers at various stages of development and found that glucose-6-phosphatase (g6pc) expression was abnormal in HNF-6-deficient livers (Fig. 1A). Indeed, g6pc expression started around e16.5 in hnf6+/+ livers and increased sharply until postnatal day 3. In contrast, in hnf6−/− livers at e16.5, g6pc expression was reduced about 10-fold, and it reached normal levels only at postnatal day 3. Therefore, HNF-6 controls the time-dependent stimulation of g6pc expression.
To rule out the possibility that the control of g6pc expression depends on nonhepatic cells, we used cell lines derived from e14.5 hnf6+/+ and hnf6−/− livers (27). These BMEL cells behave like undifferentiated hepatoblasts in that they express the complete set of LETFs and can be induced to differentiate toward either the hepatocytic or the biliary lineage (41). Quantitative RT-PCR revealed that undifferentiated BMEL cells did not express detectable amounts of g6pc mRNA (Fig. 1B). When induced to differentiate toward the hepatocytic lineage by culture in aggregates, hnf6+/+ and hnf6−/− BMEL cells started to express g6pc, but this expression was reduced in hnf6−/− BMEL cells, thereby mimicking in vivo liver development.
We next verified if HNF-6 could directly stimulate g6pc expression in embryonic cells. Indeed, previous studies have shown that HNF-6 binds in vivo to the g6pc gene promoter in the adult human liver (22) and activates a g6pc promoter-luciferase reporter construct in transiently transfected HepG2 cells (40). Transient transfection of undifferentiated hnf6+/+ and hnf6−/− BMEL cells with a HNF-6 expression vector stimulated endogenous g6pc expression (Fig. 1C), and ChIP experiments showed that this was associated with binding of HNF-6 to the g6pc promoter (Fig. 1D). These experiments validated the BMEL cells as a model with which to study the control of g6pc expression by HNF-6.
From this set of data we concluded that HNF-6 is required to allow g6pc to become expressed at normal levels at the appropriate stage of hepatocyte differentiation. This control mechanism is independent of nonhepatic cells and is associated with binding of HNF-6 to the g6pc promoter.
HNF-6 and HNF-4 stimulate g6pc expression in a synergistic and interdependent way.
HNF-4, a LETF that is critical for hepatocyte differentiation, regulates g6pc expression in vitro by binding to several sites on the g6pc promoter (30). It also binds to the g6pc promoter in the adult human liver (22). The conserved HNF-4 binding site at −90 relative to the transcription initiation site is of particular importance, since its mutation causes a dramatic decrease in g6pc promoter activity (2). Those observations and the fact that the HNF-4 binding site at −90 is located close to the HNF-6 binding site at −124 (Fig. 2A) led us to hypothesize that HNF-4 and HNF-6 cooperate to regulate g6pc expression. To investigate this possibility, we transfected undifferentiated BMEL cells with HNF-4 and HNF-6 expression vectors and measured endogenous g6pc mRNA by qRT-PCR. The results showed that HNF-6 or HNF-4 alone stimulated endogenous g6pc gene expression but that the two factors together exerted a strong synergistic effect (Fig. 2B, left panel). These data were extended by transfecting BMEL cells with a luciferase reporter construct containing the −207-to-+46 g6pc promoter region, which harbors the HNF-4 and HNF-6 binding sites. HNF-4 and HNF-6 synergistically stimulated this construct in transient transfection experiments (Fig. 2B, right panel), indicating that the −207-to-+46 region of the g6pc promoter can mediate a synergistic effect of the two factors.
Functional synergy between HNF-4 and HNF-6 may reflect their mutual requirement to stimulate the g6pc gene. To test this hypothesis, we used wild-type, hnf6−/− (27), and hnf4−/− (this paper; see Materials and Methods) BMEL cells. We transfected the three cell lines with HNF-6- or HNF-4-expressing vectors and measured endogenous g6pc mRNA levels. As in Fig. 2B, each of these two factors stimulated g6pc expression in wild-type cells (Fig. 2C). However, HNF-6 failed to stimulate g6pc expression in the absence of HNF-4 (hnf4−/− cells), and HNF-4 did not significantly stimulate g6pc in the absence of HNF-6 (hnf6−/− cells). Taken together, our data demonstrate that HNF-6 and HNF-4 synergistically and interdependently stimulate g6pc promoter activity.
Independent binding of HNF-6 and HNF-4 to the g6pc promoter.
We next investigated the molecular mechanism underlying the interdependence between HNF-4 and HNF-6 and asked if it is associated with cooperative binding of HNF-6 and HNF-4 to their cognate sites on the g6pc promoter. To test this hypothesis, we performed ChIP experiments on hnf6−/− and hnf4−/− BMEL cells transfected with combinations of vectors encoding either Flag-tagged HNF-6 and untagged HNF-4 or Flag-tagged HNF-4 and untagged HNF-6. We immunoprecipitated the chromatin of those cells with an antibody directed against the Flag epitope. Figure 3 shows that HNF-6 and HNF-4 bound the g6pc promoter in an independent manner. Indeed, Flag.HNF-6 was detected on the g6pc promoter in hnf4−/− BMEL cells, and the addition of excess HNF-4 did not improve HNF-6 binding. We obtained similar results for HNF-4: it bound the g6pc promoter in hnf6−/− BMEL cells, and excess HNF-6 did not significantly enhance HNF-4 binding. These results indicate that the two proteins bind the g6pc promoter independently of each other. Hence, the functional synergy and interdependence between HNF-6 and HNF-4 do not rely on interdependent binding.
HNF-6 and HNF-4 recruit and synergize with PGC-1α.
To investigate if HNF-6 and HNF-4α could synergize by recruiting a common coactivator, we first looked for factors that can coactivate g6pc expression with HNF-6. We expressed various coactivators, namely, CBP, pCAF, RAC-3, PGC-1α, or TIF-2, in hnf6−/− BMEL cells, either in the presence or in the absence of exogenous HNF-6 (Fig. 4A). In the absence of HNF-6, none of these coactivators stimulated g6pc expression. In contrast, in the presence of HNF-6, PGC-1α dramatically stimulated endogenous g6pc gene expression, thereby demonstrating a functional interaction between the two proteins. Such an interaction was supported by coimmunoprecipitation experiments. Indeed, protein extracts from hnf6−/− BMEL cells transfected with vectors coding for Flag.HNF-6 and for HA.PGC-1α were immunoprecipitated using an anti-HA antibody, and the immunoprecipitated proteins were detected by Western blot analysis with an anti-Flag antibody. The results (Fig. 4B) showed that Flag.HNF-6 was coprecipitated with HA.PGC-1α, demonstrating a physical interaction between HNF-6 and PGC-1α. These data identify PGC-1α as a new coactivator of HNF-6.
Since PGC-1α is a well-known coactivator of HNF-4, including in the regulation of g6pc expression (21, 29, 49), we tested whether the physical and functional interactions between HNF-6 and PGC-1α depend on the presence of HNF-4. We performed CoIP experiments on hnf4−/− BMEL cells transfected with Flag.HNF-6 and/or HA.PGC-1α. The results (Fig. 4C) show that HNF-6 and PGC-1α were able to physically interact in a cellular context devoid of HNF-4. In addition, functional interaction between the two proteins did not require HNF-4, since PGC-1α was a strong coactivator of HNF-6 for the stimulation of g6pc expression in hnf4−/− cells (Fig. 4D). Finally, we were unable to coprecipitate HNF-6 and HNF-4 in living cells, suggesting that those two factors do not interact directly (data not shown). These results demonstrate that HNF-6 and PGC-1α are able to interact in the absence of HNF-4.
Given the ability of both HNF-6 and HNF-4 to interact with PGC-1α, we hypothesized that this coactivator is involved in the synergy between HNF-6 and HNF-4 on the g6pc promoter. To test this hypothesis, we transfected BMEL cells with combinations of vectors encoding HNF-6, HNF-4, and PGC-1α and measured g6pc mRNA concentrations by qRT-PCR. Figure 4E shows that the three proteins synergistically stimulated g6pc expression: whereas PGC-1α had no effect on g6pc, it enhanced the effects of HNF-6 and HNF-4, and a very strong effect was observed when the expression of HNF-6, HNF-4, and PGC-1α was combined.
To investigate if endogenous PGC-1α is required for the synergy between HNF-6 and HNF-4, we transfected BMEL cells with HNF-6 and HNF-4 expression vectors in the presence of control or anti-pgc1α siRNAs. As shown in Fig. 4F, anti-pgc1α siRNA caused a 55% decrease in the pgc1α mRNA concentration, and this was associated with a 50% decrease in the HNF-6/HNF-4 synergy for g6pc expression. This effect was specific, since an unrelated mRNA coding for aquaporin-1 was unaffected by anti-pgc1α siRNA. These data indicate that PGC-1α is required for the transcriptional synergy between HNF-6 and HNF-4.
To gain further insight into the molecular mechanism by which PGC-1α participates in the synergism between HNF-6 and HNF-4, we verified if the latter factors corecruit PGC-1α to the g6pc promoter. We transfected BMEL cells with combinations of vectors encoding HNF-6, HNF-4, and PGC-1α and immunoprecipitated their chromatin with an anti-PGC-1α antibody. Figure 5 shows that PGC-1α alone was not associated with the g6pc promoter, consistent with the lack of transcriptional effect induced by overexpression of PGC-1α alone (see Fig. 4A, D, and E). When HNF-6, HNF-4, or both were cotransfected with PGC-1α, the g6pc promoter was readily immunoprecipitated by the anti-PGC-1α antibody. This indicates that HNF-6 and HNF-4 promote the recruitment of PGC-1α to the g6pc promoter.
Developmental regulation of g6pc expression in the liver.
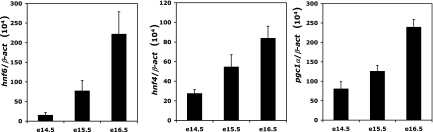
In the fetal liver, g6pc expression starts late in development, i.e., around e16.5 (Fig. 1). In contrast, the expression of HNF-6, HNF-4, and PGC-1α, which we measured by qRT-PCR, starts earlier (Fig. 6). Thus, despite the fact that these three factors are present and play a role at early stages of liver development, they cannot induce g6pc expression at these stages. Since expression of HNF-6, HNF-4, and PGC-1α increases progressively during development (Fig. 6), we hypothesized that sufficient amounts of the three factors have to be present to induce expression of g6pc. To address this hypothesis, we reasoned that increasing the expression of HNF-6, HNF-4, and PGC-1α in early liver would result in precocious expression of g6pc. To test this, we electroporated liver explants from e12.0 embryos, either with a green fluorescent protein (GFP)-coding vector or with a combination of vectors encoding HNF-6, HNF-4, and PGC-1α. The electroporated explants were cultured for 2 days before extraction of RNA. qRT-PCR measurement of the g6pc mRNA concentration revealed that livers electroporated with HNF-6, HNF-4, and PGC-1α expressed g6pc, whereas GFP-electroporated livers did not (Fig. 7A). This indicates that increased expression of HNF-6, HNF-4, and PGC-1α in the embryonic liver can induce precocious expression of g6pc.
FIG. 6.
Expression of hnf6, hnf4, and pgc1α mRNAs increases in the liver between e14.5 and e16.5. Total RNA was extracted from wild-type embryonic livers at e14.5, e15.5, and e16.5, and hnf6 (left), hnf4 (center), and pgc1α (right) mRNA levels were quantified by qRT-PCR and expressed relative to β-act mRNA levels (means ± standard errors of the means; n = 4).
To test if this inductive effect of the HNF-6/HNF-4/PGC-1α combination is liver specific, we tested NIH 3T3 fibroblasts and compared g6pc mRNA levels in cells transiently transfected with HNF-6-, HNF-4-, and PGC-1α-coding plasmids with levels in control cells transfected with a GFP-coding vector (Fig. 7B). g6pc mRNA levels were very low or undetectable in control NIH 3T3 cells, but high levels of g6pc mRNA were found after overexpression of HNF-6, HNF-4, and PGC-1α. This effect did not result from induction of a hepatic differentiation program, since other markers of hepatic differentiation, namely, albumin, α-fetoprotein (afp), phosphoenolpyruvate carboxykinase (pepck), tyrosine aminotransferase (tat), and transthyretin (ttr), were not detected by RT-PCR (data not shown).
Taken together, our overexpression data in liver and NIH 3T3 cells suggested that threshold levels of HNF-6, HNF-4, and PGC-1α are required to induce g6pc gene expression, regardless of the cellular context. This was tested by expressing increasing amounts of both HNF-6 and HNF-4 in NIH 3T3 fibroblasts (Fig. 8A), which express PGC-1α but not HNF-6 or HNF-4 (data not shown). The results showed that low levels of HNF-6 (20 and 50 ng of expression vector) allow HNF-4 to stimulate g6pc mRNA expression weakly, but this stimulation was not dependent on the dose of HNF-4. In contrast, at high HNF-6 concentrations (150 and 500 ng of expression vector), a strong and dose-dependent stimulation was obtained with HNF-4. These data indicate that threshold levels of HNF-6 are required to induce optimal stimulation of g6pc gene expression.
If this model is correct, one would expect that reduced expression of HNF-6 in the liver at the time when g6pc mRNA becomes detectable would result in impaired expression of g6pc. We compared hnf6 and g6pc mRNA levels in e16.5 livers from wild-type, heterozygous hnf6+/−, and homozygous hnf6−/− embryos (Fig. 8B). We found that a 50% reduction of hnf6 expression in hnf6+/− livers suffices to reduce g6pc mRNA levels to those found in the absence of HNF-6. These data, combined with those above, led us to conclude that threshold levels of HNF-6 allow synergistic and time-specific expression of g6pc in the developing liver.
DISCUSSION
In the present study, we have shown that HNF-6 is required for proper time-specific regulation of g6pc in the liver. This occurs via a synergistic and interdependent action with HNF-4. The synergy and interdependence do not rely on synergistic or interdependent DNA binding but involve interaction with the coactivator PGC-1α. Through a combination of in vitro and in vivo gain- and loss-of-function approaches, which focus on endogenous and not on transfected genes, we also provide arguments that support a model whereby time-specific expression of g6pc occurs only when a threshold level of HNF-6 in the presence of HNF-4 is reached.
The physical and functional interaction between PGC-1α and HNF-6, which occurs on the g6pc gene, identifies PGC-1α as a new coactivator of HNF-6. HNF-6 has been shown previously to interact with CBP and with pCAF (16, 31, 32, 36). Interestingly, CBP and pCAF had no or only a marginal effect on the HNF-6-mediated stimulation of g6pc mRNA expression. Also, PGC-1α was unable to coactivate the Foxa2 and ttr promoters with HNF-6 (data not shown). This highlights the promoter specificity of HNF-6-coactivator interactions. A similar promoter-specific HNF-4-coactivator interaction had been illustrated previously for HNF-4-SMRT interactions (44). On the other hand, PGC-1α is a known coactivator of HNF-4 (49), and we now show that PGC-1α is efficiently recruited to the g6pc promoter by HNF-6 and HNF-4, raising the possibility that a trimeric HNF-6/HNF-4/PGC-1α complex is formed.
The mode of action of HNF-6 and HNF-4 on the g6pc promoter refines our knowledge of the coordinated regulation of common target genes by LETFs. Odom et al. (22) have proposed that such regulation would depend on coordinated binding of LETFs to a common target promoter. However, this model does not take into account the dynamics of such regulations. Our data add a new dimension to the model. Indeed, we show here that the mere presence of transcriptional activators is not sufficient to induce expression of a specific gene but that this induction requires well-adjusted concentrations of the transcriptional activators. This observation is particularly relevant during development. Indeed, under normal conditions HNF-6 and HNF-4 control gene expression in the liver well before they induce g6pc gene expression (4, 6, 7, 20, 23). Our data show that the expression levels of HNF-6 and HNF-4 rise steadily during development and that up- or downregulation of HNF-6 or HNF-4 expression generates premature or delayed expression of the g6pc gene. Therefore, our data show that time- and tissue-specific expression of genes critically depends on the threshold level of transcriptional regulators that coordinately regulate common target genes.
The progressive rise in HNF-6 and HNF-4 concentrations during liver development (the HNF-4α1 isoform was studied throughout this work) has been documented earlier (6, 43), but to our knowledge, that of PGC-1α before the perinatal period was not known (50). The coordinated regulation of g6pc expression during development by HNF-6, HNF-4, and PGC-1α raises the question of their control. The known HNF-6 or HNF-4 regulatory elements are not sufficient to drive their expression during hepatocyte maturation (4, 28), and the developmental mechanisms governing PGC-1α expression have not been analyzed. However, it was shown that signaling molecules originating from the hematopoietic cells in the embryonic liver promote hepatocyte maturation at the end of development. Indeed, oncostatin M-induced signaling in differentiating hepatocytes promotes morphological changes that resemble those associated with hepatocyte maturation; it also induces glycogen accumulation and the expression of several markers of hepatocyte maturation (13). Signaling molecules such as oncostatin M may thus contribute to allowing HNF-6, HNF-4, and PGC-1α to reach threshold levels required to induce g6pc expression.
Our data also reveal that the transcription factor requirements for expression of g6pc differ before and after birth. In hnf6−/− mice the expression of g6pc is perturbed during development but reaches normal levels at birth, suggesting that HNF-6 is dispensable after birth. The difference between pre- and postnatal regulation of g6pc most likely reflects the critical metabolic role of g6pc in postnatal life, when it becomes essential for hepatic glucose production (45). In prenatal life, fetal blood glucose levels depend mainly on maternal supply, whereas after birth glycemia depends on a balance of food intake, glucose consumption, gluconeogenesis, and glycogenolysis. Hormones that control gluconeogenesis and glucose consumption exert a tight regulation of g6pc expression (18). Besides this essential hormonal regulation, postnatal liver-specific regulation must be maintained, and our data suggest that HNF-6 is not required in this process. HNF-4 is, together with PGC-1α, a key mediator of hormonal control of g6pc expression (49). It has been shown to be essential for g6pc expression in the liver before birth (23), but to our knowledge, whether it is essential after birth has not yet been demonstrated through in vivo studies.
During fetal life, the hepatocytes progressively mature, and the expression of gluconeogenic genes at the end of gestation is a hallmark of this maturation process. The regulation of g6pc by HNF-6 positions this transcription factor as a hepatocyte maturation factor. HNF-6 is also required for glucokinase gene expression (17), indicating that HNF-6 is an important regulator of liver glucose metabolism. However, expression of other key enzymes of glucose metabolism, such as phosphoenolpyruvate carboxykinase and glycogen synthase 2, is not affected in the absence of HNF-6 (7) (data not shown). Therefore, HNF-6 participates in hepatocyte maturation and controls glucose metabolism at the transcriptional level but does not seem to drive metabolic pathways toward the anabolic or catabolic orientation.
In conclusion, our data suggest a model whereby the developmental regulation of hepatocyte differentiation relies, in part, on the synergistic action of LETFs and their coactivators on their target genes. Transcriptionally active complex formation requires threshold levels of transcription factors that allow cell- and time-specific regulation of gene expression.
Acknowledgments
We thank B. M. Spiegelman and S. A. Duncan for plasmids, F. Clotman for critical reading of the manuscript, I. Talianidis for advice on chromatin immunoprecipitation, and members of the Lemaigre and Weiss laboratories for discussions.
This work was supported by grants from the Belgian State Program on Interuniversity Poles of Attraction, the Actions de Recherche Concertées of the French Community of Belgium, and the Belgian Fund for Scientific Medical Research and by a Marie Curie Fellowship of the European Community program “Improving Human Research Potential and the Socio-Economic Knowledge Base” (contract HPMF-CT-2001-01405). J.-B.B. holds a fellowship from the Fonds pour la Formation à la Recherche dans l'Industrie et l'Agriculture, and N.P.-R. held a fellowship from Télévie.
REFERENCES
- 1.Bailly, A., G. Spath, V. Bender, and M. C. Weiss. 1998. Phenotypic effects of the forced expression of HNF4 and HNF1α are conditioned by properties of the recipient cell. J. Cell Sci. 111:2411-2421. [DOI] [PubMed] [Google Scholar]
- 2.Boustead, J. N., B. T. Stadelmaier, A. M. Eeds, P. O. Wiebe, C. A. Svitek, J. K. Oeser, and R. M. O'Brien. 2003. Hepatocyte nuclear factor-4α mediates the stimulatory effect of peroxisome proliferator-activated receptor gamma co-activator-1 alpha (PGC-1α) on glucose-6-phosphatase catalytic subunit gene transcription in H4IIE cells. Biochem. J. 369:17-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bouzin, C., F. Clotman, J. C. Renauld, F. P. Lemaigre, and G. G. Rousseau. 2002. The Onecut transcription factor hepatocyte nuclear factor-6 controls B lymphopoiesis in fetal liver. J. Immunol. 171:1297-1303. [DOI] [PubMed] [Google Scholar]
- 4.Briançon, N., A. Bailly, F. Clotman, P. Jacquemin, F. P. Lemaigre, and M. C. Weiss. 2004. Expression of the α7 isoform of HNF4 is activated by HNF6/OC2 and HNF1, and repressed by HNF4α1 in the liver. J. Biol. Chem. 279:33398-33408. [DOI] [PubMed] [Google Scholar]
- 5.Cereghini, S. 1996. Liver-enriched transcription factors and hepatocyte differentiation. FASEB J. 10:267-282. [PubMed] [Google Scholar]
- 6.Clotman, F., V. J. Lannoy, M. Reber, S. Cereghini, D. Cassiman, P. Jacquemin, T. Roskams, G. G. Rousseau, and F. P. Lemaigre. 2002. The Onecut transcription factor HNF6 is required for normal development of the biliary tract. Development 129:1819-1828. [DOI] [PubMed] [Google Scholar]
- 7.Clotman, F., P. Jacquemin, N. Plumb-Rudewiez, C. E. Pierreux, P. Van der Smissen, H. C. Dietz, P. J. Courtoy, G. G. Rousseau, and F. P. Lemaigre. 2005. Control of liver cell fate decision by a gradient of TGFβ signaling modulated by Onecut transcription factors. Genes Dev. 19:1849-1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Costa, R. H., V. V. Kalinichenko, A. X. Holterman, and X. Wang. 2003. Transcription factors in liver development, differentiation, and regeneration. Hepatology 38:1331-1347. [DOI] [PubMed] [Google Scholar]
- 9.Germain, L., M. J. Blouin, and N. Marceau. 1988. Biliary epithelial and hepatocytic cell lineage relationships in embryonic rat liver as determined by the differential expression of cytokeratins, alpha-fetoprotein, albumin, and cell surface-exposed components. Cancer Res. 48:4909-4918. [PubMed] [Google Scholar]
- 10.Hatzis, P., and I. Talianidis. 2001. Regulatory mechanisms controlling human hepatocyte nuclear factor 4α gene expression. Mol. Cell. Biol. 21:7320-7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayhurst, G. P., Y. H. Lee, G. Lambert, J. M. Ward, and F. J. Gonzalez. 2001. Hepatocyte nuclear factor 4α is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol. Cell. Biol. 21:1393-1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacquemin, P., S. M. Durviaux, J. Jensen, C. Godfraind, G. Gradwohl, F. Guillemot, O. D. Madsen, P. Carmeliet, M. Dewerchin, D. Collen, G. G. Rousseau, and F. P. Lemaigre. 2000. Transcription factor hepatocyte nuclear factor-6 regulates pancreatic endocrine cell differentiation and controls expression of the proendocrine gene ngn3. Mol. Cell. Biol. 20:4445-4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kamiya, A., T. Kinoshita, Y. Ito, T. Matsui, Y. Morikawa, E. Senba, K. Nakashima, T. Taga, K. Yoshida, T. Kishimoto, and A. Miyajima. 1999. Fetal liver development requires a paracrine action of oncostatin M through the gp130 signal transducer. EMBO J. 18:2127-2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kinoshita, T., and A. Miyajima. 2002. Cytokine regulation of liver development. Biochim. Biophys. Acta 1592:303-312. [DOI] [PubMed] [Google Scholar]
- 15.Kuo, C. J., P. B. Conley, L. Chen, F. M. Sladek, J. E. Darnell, Jr., and G. R. Crabtree. 1992. A transcriptional hierarchy involved in mammalian cell-type specification. Nature 355:457-461. [DOI] [PubMed] [Google Scholar]
- 16.Lannoy, V. J., A. Rodolosse, C. E. Pierreux, G. G. Rousseau, and F. P. Lemaigre. 2000. Transcriptional stimulation by hepatocyte nuclear factor-6. Target-specific recruitment of either CREB-binding protein (CBP) or p300/CBP-associated factor (p/CAF). J. Biol. Chem. 275:22098-22103. [DOI] [PubMed] [Google Scholar]
- 17.Lannoy, V. J., J. F. Decaux, C. E. Pierreux, F. P. Lemaigre, and G. G. Rousseau. 2002. Liver glucokinase gene expression is controlled by the Onecut transcription factor hepatocyte nuclear factor-6. Diabetologia 45:1136-1141. [DOI] [PubMed] [Google Scholar]
- 18.Lemaigre, F. P., and G. G. Rousseau. 1994. Transcriptional control of genes that regulate glycolysis and gluconeogenesis in adult liver. Biochem. J. 303:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lemaigre, F. P. 2003. Development of the biliary tract. Mech. Dev. 120:81-87. [DOI] [PubMed] [Google Scholar]
- 20.Li, J., G. Ning, and S. A. Duncan. 2000. Mammalian hepatocyte differentiation requires the transcription factor HNF-4α. Genes Dev. 14:464-474. [PMC free article] [PubMed] [Google Scholar]
- 21.Lin, J., P. H. Wu, P. T. Tarr, K. S. Lindenberg, J. St-Pierre, C. Y. Zhang, V. K. Mootha, S. Jager, C. R. Vianna, R. M. Reznick, L. Cui, M. Manieri, M. X. Donovan, Z. Wu, M. P. Cooper, M. C. Fan, L. M. Rohas, A. M. Zavacki, S. Cinti, G. I. Shulman, B. B. Lowell, D. Krainc, and B. M. Spiegelman. 2004. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1α null mice. Cell 119:121-135. [DOI] [PubMed] [Google Scholar]
- 22.Odom, D. T., N. Zizlsperger, D. B. Gordon, G. W. Bell, N. J. Rinaldi, H. L. Murray, T. L. Volkert, J. Schreiber, P. A. Rolfe, D. K. Gifford, E. Fraenkel, G. I. Bell, and R. A. Young. 2004. Control of pancreas and liver gene expression by HNF transcription factors. Science 303:1378-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parviz, F., C. Matullo, W. D. Garrison, L. Savatski, J. W. Adamson, G. Ning, K. H. Kaestner, J. M. Rossi, K. S. Zaret, and S. A. Duncan. 2003. Hepatocyte nuclear factor 4α controls the development of a hepatic epithelium and liver morphogenesis. Nat. Genet. 34:292-296. [DOI] [PubMed] [Google Scholar]
- 24.Pierreux, C. E., B. Urso, P. De Meyts, G. G. Rousseau, and F. P. Lemaigre. 1998. Inhibition by insulin of glucocorticoid-induced gene transcription: involvement of the ligand-binding domain of the glucocorticoid receptor and independence from the phosphatidylinositol 3-kinase and mitogen-activated protein kinase pathways. Mol. Endocrinol. 12:1343-1354. [DOI] [PubMed] [Google Scholar]
- 25.Pierreux, C. E., V. Vanhorenbeeck, P. Jacquemin, F. P. Lemaigre, and G. G. Rousseau. 2004. The transcription factor hepatocyte nuclear factor-6/Onecut-1 controls the expression of its paralog Onecut-3 in developing mouse endoderm. J. Biol. Chem. 279:51298-51304. [DOI] [PubMed] [Google Scholar]
- 26.Pierreux, C. E., A. V. Poll, P. Jacquemin, F. P. Lemaigre, and G. G. Rousseau. 2005. Gene transfer into mouse prepancreatic endoderm by whole embryo electroporation. J. Pancreas 6:128-135. [PubMed] [Google Scholar]
- 27.Plumb-Rudewiez, N., F. Clotman, H. Strick-Marchand, C. E. Pierreux, M. C. Weiss, G. G. Rousseau, and F. P. Lemaigre. 2004. Transcription factor HNF-6/OC-1 inhibits the stimulation of the HNF-3α/Foxa1 gene by TGF-β in mouse liver. Hepatology 40:1266-1274. [DOI] [PubMed] [Google Scholar]
- 28.Poll, A. V., C. E. Pierreux, L. Lokmane, C. Haumaitre, Y. Achouri, P. Jacquemin, G. G. Rousseau, S. Cereghini, and F. P. Lemaigre. 2006. A vHNF1/TCF2-HNF6 cascade regulates the transcription factor network that controls generation of pancreatic precursor cells. Diabetes 55:61-69. [PubMed] [Google Scholar]
- 29.Puigserver, P., and B. M. Spiegelman. 2003. Peroxisome proliferator-activated receptor-gamma coactivator 1 alpha (PGC-1α): transcriptional coactivator and metabolic regulator. Endocr. Rev. 24:78-90. [DOI] [PubMed] [Google Scholar]
- 30.Rajas, F., A. Gautier, I. Bady, S. Montano, and G. Mithieux. 2002. Polyunsaturated fatty acyl coenzyme A suppress the glucose-6-phosphatase promoter activity by modulating the DNA binding of hepatocyte nuclear factor 4α. J. Biol. Chem. 277:15736-15744. [DOI] [PubMed] [Google Scholar]
- 31.Rausa, F. M., Y. Tan, and R. H. Costa. 2003. Association between hepatocyte nuclear factor 6 (HNF-6) and FoxA2 DNA binding domains stimulates FoxA2 transcriptional activity but inhibits HNF-6 DNA binding. Mol. Cell. Biol. 23:437-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rausa, F. M., D. E. Hughes, and R. H. Costa. 2004. Stability of the hepatocyte nuclear factor 6 transcription factor requires acetylation by the CREB-binding protein coactivator. J. Biol. Chem. 279:43070-43076. [DOI] [PubMed] [Google Scholar]
- 33.Rubins, N. E., J. R. Friedman, P. P. Le, L. Zhang, J. Brestelli, and K. H. Kaestner. 2005. Transcriptional networks in the liver: hepatocyte nuclear factor-6 function is largely independent of Foxa2. Mol. Cell. Biol. 25:7069-7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schrem, H., J. Klempnauer, and J. Borlak. 2002. Liver-enriched transcription factors in liver function and development. Part I: the hepatocyte nuclear factor network and liver-specific gene expression. Pharmacol. Rev. 54:129-158. [DOI] [PubMed] [Google Scholar]
- 35.Schrem, H., J. Klempnauer, and J. Borlak. 2004. Liver-enriched transcription factors in liver function and development. Part II: the C/EBPs and D site-binding protein in cell cycle control, carcinogenesis, circadian gene regulation, liver regeneration, apoptosis, and liver-specific gene regulation. Pharmacol. Rev. 56:291-330. [DOI] [PubMed] [Google Scholar]
- 36.Sheng, W., H. Yan, F. M. Rausa, R. H. Costa, and X. Liao. 2004. Structure of the hepatocyte nuclear factor 6α and its interaction with DNA. J. Biol. Chem. 279:33928-33936. [DOI] [PubMed] [Google Scholar]
- 37.Shiojiri, N., J. M. Lemire, and N. Fausto. 1991. Cell lineages and oval cell progenitors in rat liver development. Cancer Res. 51:2611-2620. [PubMed] [Google Scholar]
- 38.Shiojiri, N. 1997. Development and differentiation of bile ducts in the mammalian liver. Microsc. Res. Tech. 39:328-335. [DOI] [PubMed] [Google Scholar]
- 39.Späth, G. F., and M. C. Weiss. 1998. Hepatocyte nuclear factor 4 provokes expression of epithelial marker genes, acting as a morphogen in dedifferentiated hepatoma cells. J. Cell Biol. 140:935-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Streeper, R. S., L. A. Hornbuckle, C. A. Svitek, J. K. Goldman, J. K. Oeser, and R. M. O'Brien. 2001. Protein kinase A phosphorylates hepatocyte nuclear factor-6 and stimulates glucose-6-phosphatase catalytic subunit gene transcription. J. Biol. Chem. 276:19111-19118. [DOI] [PubMed] [Google Scholar]
- 41.Strick-Marchand, H., and M. C. Weiss. 2002. Inducible differentiation and morphogenesis of bipotential liver cell lines from wild-type mouse embryos. Hepatology 36:794-804. [DOI] [PubMed] [Google Scholar]
- 42.Tan, Y., G. Adami, and R. H. Costa. 2002. Maintaining HNF6 expression prevents AdHNF3β-mediated decrease in hepatic levels of Glut-2 and glycogen. Hepatology 35:790-798. [DOI] [PubMed] [Google Scholar]
- 43.Torres-Padilla, M. E., C. Fougere-Deschatrette, and M. C. Weiss. 2001. Expression of HNF-4α isoforms in mouse liver development is regulated by sequential promoter usage and constitutive 3′ end splicing. Mech. Dev. 109:183-193. [DOI] [PubMed] [Google Scholar]
- 44.Torres-Padilla, M. E., and M. C. Weiss. 2003. Effects of interactions of hepatocyte nuclear factor 4α isoforms with coactivators and corepressors are promoter-specific. FEBS Lett. 539:19-23. [DOI] [PubMed] [Google Scholar]
- 45.van Schaftingen, E., and I. Gerin. 2002. The glucose-6-phosphatase system. Biochem. J. 362:513-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang, M., Y. Tan, R. H. Costa, and A. X. Holterman. 2004. In vivo regulation of murine CYP7A1 by HNF-6: a novel mechanism for diminished CYP7A1 expression in biliary obstruction. Hepatology 40:600-608. [DOI] [PubMed] [Google Scholar]
- 47.Watt, A. J., W. D. Garrison, and S. A. Duncan. 2003. HNF4: a central regulator of hepatocyte differentiation and function. Hepatology 37:1249-1253. [DOI] [PubMed] [Google Scholar]
- 48.White, P., J. E. Brestelli, K. H. Kaestner, and L. E. Greenbaum. 2005. Identification of transcriptional networks during liver regeneration. J. Biol. Chem. 280:3715-3722. [DOI] [PubMed] [Google Scholar]
- 49.Yoon, J. C., P. Puigserver, G. Chen, J. Donovan, Z. Wu, J. Rhee, G. Adelmant, J. Stafford, C. R. Kahn, D. K. Granner, C. B. Newgard, and B. M. Spiegelman. 2001. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature 413:131-138. [DOI] [PubMed] [Google Scholar]
- 50.Yubero, P., E. Hondares, M. C. Carmona, M. Rossell, F. J. Gonzalez, R. Iglesias, M. Giralt, and F. Villarroya. 2004. The developmental regulation of peroxisome proliferator-activated receptor-gamma coactivator-1α expression in the liver is partially dissociated from the control of gluconeogenesis and lipid catabolism. Endocrinology 145:4268-4277. [DOI] [PubMed] [Google Scholar]
- 51.Zhao, R., and S. A. Duncan. 2005. Embryonic development of the liver. Hepatology 41:956-967. [DOI] [PubMed] [Google Scholar]