Abstract
Kinase domain (KD) mutations of Bcr-Abl interfering with imatinib binding are the major mechanism of acquired imatinib resistance in patients with Philadelphia chromosome-positive leukemia. Mutations of the ATP binding loop (p-loop) have been associated with a poor prognosis. We compared the transformation potency of five common KD mutants in various biological assays. Relative to unmutated (native) Bcr-Abl, the ATP binding loop mutants Y253F and E255K exhibited increased transformation potency, M351T and H396P were less potent, and the performance of T315I was assay dependent. The transformation potency of Y253F and M351T correlated with intrinsic Bcr-Abl kinase activity, whereas the kinase activity of E255K, H396P, and T315I did not correlate with transforming capabilities, suggesting that additional factors influence transformation potency. Analysis of the phosphotyrosine proteome by mass spectroscopy showed differential phosphorylation among the mutants, a finding consistent with altered substrate specificity and pathway activation. Mutations in the KD of Bcr-Abl influence kinase activity and signaling in a complex fashion, leading to gain- or loss-of-function variants. The drug resistance and transformation potency of mutants may determine the outcome of patients on therapy with Abl kinase inhibitors.
Bcr-Abl, a constitutively active tyrosine kinase, is the defining molecular feature of chronic myeloid leukemia (CML) (8). Biochemical studies and murine models have established that tyrosine kinase activity is essential to the transforming capacity of Bcr-Abl (7, 17). Consistent with this, inhibition of the Bcr-Abl kinase with imatinib, an Abl-specific kinase inhibitor, induces remissions in patients with CML (9, 10). Although responses in the early phases of CML are frequently durable, relapse is common in patients with advanced-phase CML. Mutations in the kinase domain (KD) of Bcr-Abl that impair imatinib binding have been identified as the major mechanism of acquired resistance (1, 3, 13, 15, 34, 37). However, in some patients, drug-resistant mutants were detected prior to therapy (16, 27, 28, 34, 38). The proportion of mutant allele in these samples varied, from being detectable only with allele-specific PCR, which could detect as few as one cell in 100,000 (38), to representing 40% of the BCR-ABL message (34). These data suggest that some mutants may have a proliferative advantage over unmutated Bcr-Abl (herein referred to as native Bcr-Abl) even in the absence of imatinib, while others may be loss-of-function alleles that are selected only in the presence of imatinib. Mutations of the ATP binding loop (P-loop) of Abl are associated with significantly shorter survival than other mutations, regardless of their sensitivity to imatinib (3, 36), suggesting that certain Bcr-Abl mutations may directly contribute to disease progression by conferring a more aggressive phenotype. To test these hypotheses, we performed various biochemical and biological assays to compare five common KD mutants that comprise ca. 60% of Bcr-Abl mutations reported in relapsed patients. These mutants were also selected since they represent several functionally distinct KD regions, including the P-loop (Y253F and E255K), the site of a hydrogen bond with imatinib (T315I), the activation loop hinge (M351T), and the activation loop (H396P) (6, 12).
MATERIALS AND METHODS
Cell culture.
Ba/F3 cells expressing native Bcr-Abl and KD point mutants were maintained in RPMI 1640 (Gibco) supplemented with 2 mM l-glutamine (Gibco), 1 U of penicillin (Gibco)/ml, and 1 μg of streptomycin (Gibco)/ml and either 10 or 1% fetal bovine serum (FBS; HyClone). Bosc23 cells (25) were maintained in Dulbecco modified Eagle medium (DMEM; Gibco) supplemented with 10% FBS, 1 U of penicillin/ml, 1 μg of streptomycin/ml, and 2 mM l-glutamine. All cells were grown at 37°C in 5% CO2.
Immunoblotting.
Cells were collected by centrifugation and lysed in phosphate-buffered saline (PBS) containing 1% NP-40, 1 mM EDTA, 1 mM Na3VO4, 0.5 mM phenylmethylsulfonyl fluoride, and 1 μg of aprotinin/ml. Lysates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (50 to 200 μg of protein/lane). Blots were probed for expression of Bcr-Abl (8E9; Pharmingen), CrkL (C-20; Santa Cruz Biotechnology), Cbl (C-15; Santa Cruz Biotechnology), Akt (Cell Signaling Technology), Erk (Cell Signaling Technology), Jnk (G-7; Santa Cruz Biotechnology), Stat5 (C-17; Santa Cruz Biotechnology), SHIP1 (N-1; Santa Cruz Biotechnology). Cell lysates were also assessed with the phospho-specific antibodies p-CrkL (Tyr207; Cell Signaling Technology), p-Cbl (Tyr774; Cell Signaling Technology), p-Akt (Tyr326, Thr 308, and Ser473; all from Cell Signaling Technology), p-Jnk (Thr183/Tyr185; Santa Cruz Biotechnology), p-Stat5 (Tyr694; Zymed Laboratories), p-Erk (Thr202/Tyr204; Cell Signaling Technology), and p-SHIP1 (Tyr1020; Cell Signaling Technology).
Growth competition experiments.
Ba/F3 cells expressing native Bcr-Abl were mixed at a 1/1 ratio with Ba/F3 expressing Bcr-Abl KD point mutants, Y253F, E255K, T315I, M351T, and H396P at 2 × 105 cells/ml and plated in duplicate. Equal expression of Bcr-Abl was confirmed by immunoblotting. Cells were cocultured for 18 days in RPMI 1640 media containing either 10 or 1% FBS. For growth in 1% FBS, cells were washed twice in RPMI 1640 containing 1% FBS before plating. Cells were fed and split every 2 days to 2 × 105 cells/ml and maintained at a density between 2 × 105 and 5 × 105 cells/ml, ensuring exponential growth. The remaining cells were washed once with PBS and stored as a frozen cell pellet at −20°C. DNA extraction was done by using QIAamp DNA-minikit (QIAGEN). Bcr-Abl was amplified by using the B2A forward primer (15) and the 4065R reverse primer (TTC TCT AGC AGC TCA TAC ACC TG). Automated sequencing was done by Agencourt Bioscience Corp. with the primers F3336 (ACC ACG CTC CAT TAT CCA GCC) and R4000 (ATT TTC CCA AAG TAC TCC). Sequence analysis was performed by using Mutation Surveyor (Soft Genetics).
Production of retrovirus and determination of virus titer.
The MSCV-p210Bcr-Abl-internal ribosome entry site-green fluorescent protein (GFP) construct has been previously described (40). For the production of native and mutant Bcr-Abl and control retrovirus for use in the BM transduction and/or transplantation experiments, Bosc23 cells were transiently transfected by using Fugene6 (Roche). Viral supernatants were harvested at 48 h posttransfection, flash frozen in liquid nitrogen, and stored for the determination of virus titer or use. Relative virus titer was determined by the infection of NIH 3T3 cells. Briefly, 105 cells were plated in 30-mm tissue culture plates. After 24 h the cells were exposed to 100, 200, or 400 μl of viral supernatant in 2-ml total volume of media with 4 μg of Polybrene (Sigma)/ml. At 48 h postinfection NIH 3T3 cells were harvested with trypsin (Gibco) and washed two times in PBS. GFP expression was analyzed by fluorescence-activated cell sorting (FACSAria; BD Biosciences) to determine relative virus titers. Titers were estimated by plotting the proportion of GFP-positive cells versus the proportion of retroviral supernatant contained in the infection mix. Since the resulting plots showed saturation at higher proportions of retrovirus, we chose supernatant proportions from the linear range of the curve and adjusted the volumes accordingly to correct for differences in the concentrations of retrovirus. The required volumes were added to the infection mix, which was then used to infect bone marrow (BM) cells.
B-lymphoid transformation assays.
For the analysis of the transformation of primary BM B-lymphoid progenitors (18), BM from BALB/c mice was used. Erythrocytes were lysed with NH4Cl solution, and the cells were subjected to a single round of transduction and cosedimentation with matched retroviral stock for each mutant, native Bcr-Abl, and vector control in DMEM with 10% FBS and 2 μg of Polybrene/ml. Cells were incubated overnight in the presence of viral supernatant. The cells were then plated for in vitro growth in Whitlock/Witte cultures in RPMI 1640 supplemented with 5% FBS, 200 μM l-glutamine, 50 μM 2-mercaptorethanol, and penicillin-streptomycin (21, 29, 35) in triplicate. A dilution series was created of 1 × 105, 3 × 104, 1 × 104, 3 × 103, and 1 × 103 infected cells/ml in 24-well plates and supplemented with untransduced BM cells for stromal support normalized to 106 cells. Cells were cultured for 3 weeks and fed twice weekly by the removal of 0.5 ml of medium and the addition of 0.5 ml of medium. Cultures were scored as positive for transformation when the number of nonadherent cells exceeded 106 per ml of culture medium.
Primary cell colony formation assays.
For myeloid progenitor colony formation, BM was harvested form 6- to 10-week-old female BALB/c mice. BM was subjected to 24 h of prestimulation in IMDM (Gibco) supplemented with 15% heat-inactivated FBS, 5% WEHI conditioned medium, 6 ng of recombinant mouse interleukin-3 (rm IL-3; StemCell Technologies)/ml, 10 ng of rm IL-6 (StemCell Technologies)/ml, and 50 ng of rm stem cell factor (SCF; StemCell Technologies)/ml (32). After 24 h of prestimulation, equal numbers of cells were transferred to six-well plates and exposed to matched viral supernatants in the presence 2 μg of Polybrene/ml. Cosedimentation of the cells was carried out at 30°C for 90 min at 2,500 rpm. After 4 h the medium was replaced with fresh prestimulation media. At 48 h, a second round of cosedimentation was performed. After an additional 4 h adsorption period the cells were washed twice in IMDM to remove cytokines, counted, and plated in triplicate in MethoCult SFBIT M3236 (StemCell Technologies) supplemented with either 1 or 10% FBS in the presence or absence of cytokines (10 ng of IL-3, 10 ng of IL-6, and 50 ng of SCF/ml). Colony formation was scored at day 10.
Induction of myeloproliferative disease.
BM cell transduction and transplantation was essentially performed as previously described (40). Briefly, BM cells were isolated from the tibias and femurs of 8- to 10-week-old male BALB/c donor mice 4 days after intravenous treatment with 300 mg of 5-fluorouracil (Sigma)/kg. Erythrocytes were lysed in NH4Cl buffer by incubation on ice for 30 min. BM cells pooled from several animals were infected with precisely matched titers of either native, p210Bcr-Abl Y253F, p210Bcr-Abl E255K, p210Bcr-Abl T315I, p210Bcr-Abl M351T, p210Bcr-Abl H396P, or control retroviral supernatant in DMEM containing 1 U of penicillin/ml, 1 μg of streptomycin/μl, 2 mM l-glutamine, 15% FBS, 15% WEHI conditioned media, 12 ng of rm IL-6/ml, 7 ng of rm IL-3/ml, 56 ng of rm SCF/ml, and 3 μg of Polybrene/ml. Cells were plated in six-well plates at 2 × 106 cells/ml, followed by a 90-min cosedimentation (24). Cells were incubated with viral supernatants for 48 h at 37°C in 5% CO2. After infection, the cells were washed twice in cold PBS and resuspended at 4.0 × 105 cells per 100 μl. A total of 100 μl of the cell suspension containing 3 × 105 cells were injected retro-orbitally into recipient mice that had been exposed to two doses of 450-rad whole-body irradiation, administered 4 h apart, in a cesium irradiator. Between 10 and 12 mice were used for native Bcr-Abl, each of the mutants, and the empty vector control. Mice were monitored by daily observation and twice weekly blood counts (animal cell counter; ABC, Heska, CO). The animals were sacrificed when the white blood cell count exceeded 200/nl, if there was greater than 20% loss of body weight, or if they appeared moribund. For secondary transplants, BM and spleens were harvested from leukemic mice and resuspended in DMEM supplemented with 15% FBS, 15% WEHI conditioned medium, 1 U of penicillin/ml, and 1 μg of streptomycin/ml. Cells were washed twice in cold PBS and resuspended at 5 × 105 BM cells and 1 × 106 splenocytes per 100 μl of PBS. A total of 100 μl of the BM-splenocyte suspension was injected retro-orbitally into the sublethally irradiated mice (1 × 450 rads).
Flow cytometry.
Cells were stained with fluorochrome-conjugated monoclonal antibodies to Gr-1 (E-Biosciences), Mac-1 (E-Biosciences), B220 (Pharmingen), Thy1.2 (Pharmingen), and Ter119 (Pharmingen) before data acquisition on a FACSAria flow cytometer.
Purification of Bcr-Abl proteins.
Full-length native, Y253F, E255K, T315I, M351T, and H396P were cloned into the insect donor vector pFastBac (Invitrogen) giving rise to an N-terminal hexahistidine tag to aid in purification. High-titer baculovirus was grown and used to infect Sf9 cells (Invitrogen) grown in Grace's media (Invitrogen) supplemented with 10% FBS. For protein purification, 2 × 109 cells were pelleted by centrifugation at 5,000 × g for 10 min and flash frozen for later purification. Cells were lysed by sonication in 50 mM Tris (pH 8.0), 150 mM NaCl, 0.5% Triton X-100, and 5% glycerol. Lysate was cleared by centrifugation at 15,000 × g for 30 min, and the supernatant loaded directly onto Ni2+-nitrilotriacetic acid (NTA) agrose (QIAGEN) preequilibrated in lysis buffer. Columns were washed in wash buffer I containing 50 mM phosphate buffer (pH 8.0), 300 mM NaCl, 5% glycerol, 0.01% Triton X-100, and 10 mM imidazole and then with wash buffer II containing 50 mM phosphate buffer (pH 8.0), 300 mM NaCl, and 20 mM imidazole. Protein was eluted with a linear gradient from 20 to 300 mM imidazole in wash buffer II. Fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Bcr-Abl-containing fractions were pooled and adjusted to 5 mM EDTA and dialyzed overnight versus wash buffer I. The solution was incubated with Tobacco Etch Virus (TEV) protease (Invitrogen) overnight at 4°C in order to cleave the hexahistidine tag. The solution was passed over a second Ni2+-NTA column to remove the TEV protease and any undigested protein. The flowthrough was collected and dialyzed extensively versus 50 mM Tris-HCl (pH 7.5), 50 mM NaCl, 0.1 mM EDTA, 0.01% Brij35, and 1 mM dithiothreitol. The resultant solution was concentrated and stored for use in kinase assays. The cloning and subsequent cleavage of the hexahistidine tag resulted in the N-terminal addition of the amino acid sequence GAMDPEFRGA to the full-length protein. c-Abl purified from Sf21 cells was purchased from Upstate. Glutathione S-transferase (GST)-Abl fusion proteins were purified as previously described (5).
In vitro kinase assays.
The in vitro kinase activity of GST-Abl KDs, c-Abl, and Bcr-Abl was assessed in triplicate by using a synthetic, N-terminal biotin-linked peptide substrate (biotin-EAIYAAPFAKKK-amide) (30). Assays were carried out at 30°C for 5 or 20 min in 25 μl of reaction mixture consisting of kinase buffer (25 mM Tris-HCl [pH 7.5], 5 mM α-glycerophosphate, 2 mM dithiothreitol, 0.1 mM Na3VO4, 10 mM MgCl2; Cell Signaling Technologies), peptide substrate concentrations of 2.5, 5.0, 10.0, 17.5, 25, 50, 75, and 100 μM with approximately 4 nM enzyme and 100 μM ATP/[γ-32P]ATP (5,000 cpm/pmol). Reactions were terminated by the addition of guanidine hydrochloride to a final concentration of 2.5 M. Then, 10 μl of each terminated reaction mixture was transferred to a streptavidin-coated membrane (SAM2 biotin capture membrane; Promega), washed, and dried according to the manufacturer's instructions, and the phosphate incorporation was determined by scintillation counting. The results were corrected for background binding to the membranes by omitting peptide substrate from the kinase reaction. Plots of velocity as a function of peptide substrate concentration were fit by nonlinear regression to Michaelis-Menten kinetics by using the equation ν = Vmax × [substrate]/(Km + [substrate]) to extract the Km and Vmax. kcat was obtained from the equation kcat = Vmax/[enzyme]. The enzyme concentration was determined by Coomassie gel densitometry and normalized to a concentration of 4 nM for the KD enzyme assays. For the full-length proteins 4 nM native Bcr-Abl was used for the assays. The relative concentration of the remaining Bcr-Abl mutants (as shown) used was determined by Coomassie gel densitometry to calculate the kcat values. Dephosphorylation was carried out by preincubation with LAR (New England Biolabs) for 1 h at 30°C. Reactions were terminated with the addition of Na3VO4 to a final concentration of 1 mM.
Phosphopeptide immunoprecipitation.
Cells were grown in a 5% CO2 incubator at 37°C in RPMI 1640 with 10% FBS and penicillin-streptomycin. Cells (∼2 × 108) were lysed in urea lysis buffer (20 mM HEPES [pH 8.0], 9 M urea, 1 mM sodium vanadate, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate) at 1.25 × 108 cells/ml and sonicated. Sonicated lysates were cleared by centrifugation at 10,000 rpm, and proteins were reduced and alkylated as described previously (31). Soluble trypsin (1 mg/ml in 0.001 M HCl) was added at a 1:100 dilution to the diluted, clarified lysates and digested overnight at room temperature. After digestion, lysates were acidified to a final concentration of 1% trifluoroacetic acid (TFA). Peptide purification was carried out by using Sep-Pak C18 columns as described previously (31). After purification, all elutions were combined and lyophilized. Dried peptides were resuspended in 1.4 ml of morpholinepropanesulfonic acid immunoprecipitation (MOPS IP) buffer (50 mM MOPS-NaOH [pH 7.2], 10 mM Na2HPO4, 50 mM NaCl), and insoluble material was removed by centrifugation at 3,000 rpm for 10 min. The phosphotyrosine monoclonal antibody P-Tyr-100 was coupled at 4 mg/ml to protein G-agarose beads (Roche). Immobilized antibody (50 μl, 200 μg) was added as a 1:1 slurry in MOPS IP buffer to the solubilized peptide fraction, followed by incubation overnight at 4°C. The peptide immunoprecipitations were washed three times with 1 ml of MOPS IP buffer and twice with 1 ml of H2O, all at 4°C. Peptides were eluted from beads by incubation with 60 μl of 0.1% TFA at room temperature for 10 min, followed by a second elution with 40 μl of 0.1% TFA.
Analysis by liquid chromatography tandem mass spectrometry (MS/MS).
Peptides in the immunoprecipitation eluate (53 μl) were concentrated and separated from eluted antibody using ZipTip μC18 columns (Millipore). Peptides were eluted from the microcolumns with 1 μl of 60% MeCN-0.1% TFA in 7.6 μl of 0.4% acetic acid-0.005% heptafluorobutyric acid (HFBA). This sample was analyzed as previously described (31). TurboSequest (ThermoFinnigan) searches were done against the NCBI mouse database released on 25 February 2003 with the addition of the sequence for human Bcr-Abl, allowing for oxidized methionine (M+16) and phosphorylation (Y+80) as dynamic modifications.
Targeted analysis of phosphopeptides was performed by placing the m/z values for the target peptides on the parent mass list. MS/MS spectra of the targeted mass values were collected by using a top-two method, a dynamic exclusion repeat count of 3, and a repeat duration of 0.5 min.
RESULTS
Bcr-Abl KD mutations affect proliferation rate.
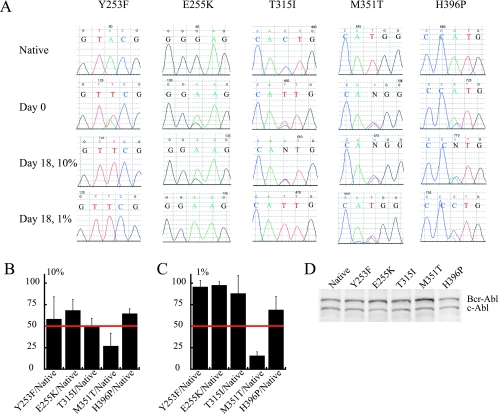
To determine whether the presence of a Bcr-Abl KD mutation affected cell proliferation, equal numbers of Ba/F3 cells expressing native Bcr-Abl and one of the KD mutants Y253F, E255K, T315I, M351T, or H396P were mixed and plated in medium containing either 10 or 1% FBS. Independently derived mass cultures with comparable levels of Bcr-Abl protein expression were used (Fig. 1D), rather than individual clones, to avoid potential bias resulting from the site of retroviral integration. The percentage of mutant clone was estimated from direct sequencing of BCR-ABL amplified from genomic DNA (Fig. 1A). The results of three independent experiments were averaged with the percentage of mutant allele relative to native Bcr-Abl at day 18 as indicated (Fig. 1B and C). In 10% FBS all of the mutants with the exception of M351T grew at rates comparable to native Bcr-Abl (Fig. 1B). In contrast, in 1% FBS, all mutants tested were able to outcompete native Bcr-Abl by day 18, with the exception of M351T (Fig. 1C). Based on proliferation rates of cultures grown in 1% FBS, a hierarchy emerged, with Y253F and E255K being the most aggressive, followed by T315I, H396P, native Bcr-Abl, and lastly M351T.
FIG. 1.
In vitro proliferation competition experiments. (A) Representative chromatograms from the direct sequencing of competition cellular growth experiments. 50/50 mixtures of Ba/F3 cells expressing Bcr-Abl and the indicated KD mutant were grown for 18 days in RPMI medium supplemented with either 10 or 1% FBS as indicated. Row 1 shows native sequence, and row 2 shows the starting conditions with a 50/50 mixture of native and the indicated mutant; rows 3 and 4 show the day 18 chromatograms for the 10 and 1% FBS conditions, respectively. (B) Combined results of the percentage of mutant allele after 18 days in 10% FBS. The percentage of mutant allele was determined by measurement of the area under the curve of mutant allele and native allele taken from chromatograms normalized to a starting condition of 50% mutant. Error bars represent the standard deviation. (C) Combined results of the percentage of mutant allele after 18 days in 1% FBS. The percentage of mutant allele was determined by measurement of the area under the curve of mutant allele, and native allele taken from chromatograms normalized to a starting condition of 50% mutant. Errors bars represent the standard deviation. (D) Anti-Abl immunoblots of Ba/F3 cell lines used for the competition experiments showing comparable expression levels of Bcr-Abl. c-Abl expression is used as an internal control.
Bcr-Abl KD mutations differ in their transformation potency in primary BM B-lymphoid and myeloid progenitors.
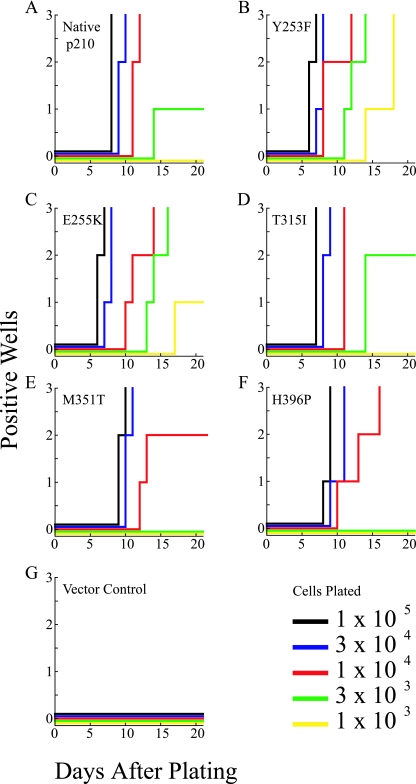
To assess the transformation potency of Bcr-Abl KD mutants in primary B-lymphoid progenitor cells, murine BM was retrovirally infected with the MIGR1 retroviral vector containing native Bcr-Abl and the KD point mutants Y253F, E255K, T315I, M351T, and H396P (40). Cells were plated in Whitlock-Witte cultures at various dilutions in triplicate. The day of initial outgrowth and lowest dilution supporting outgrowth were used as a semiquantitative measure of transformation potency (Fig. 2) (35). Wells were scored as positive when the number of nonadherent cells reached 106 per well. Expression of native Bcr-Abl induced rapid outgrowth of B cells in cultures seeded with as few as 3 × 103 cells, whereas cells infected with empty vector were transformation deficient (Fig. 2A and G). Both Y253F and E255K (Fig. 2B and C) showed more rapid outgrowth than native Bcr-Abl at the highest cell density (105), as well as outgrowth at lower cell densities (103) than native Bcr-Abl. In contrast, the outgrowth of H396P and M351T was observed only at cell densities of 104 and higher (Fig. 2F and E). Lastly, T315I showed no significant difference in transformation potency compared to native Bcr-Abl (Fig. 2D). Comparable results were obtained in two independent experiments with different sets of matched viral stocks to ensure consistency (data not shown).
FIG. 2.
B-lymphoid progenitors outgrowth by Bcr-Abl KD mutants. BM from BALB/c mice was transduced with the indicated BCR-ABL construct and plated at the indicated cell numbers per well in each of three wells. For stromal support, nontransduced cells were added to bring the total number of cells to 106/well. A well was scored as positive when the viable nonadherent cell number reached 106/well. The x axis indicates the number of days after plating, and the y axis indicates the number of wells, from 1 to 3, that are scored as positive. (A) Native Bcr-Abl; (B) Y253F; (C); E255K; (D) T315I; (E) M351T; (F) H396P; (G) vector control.
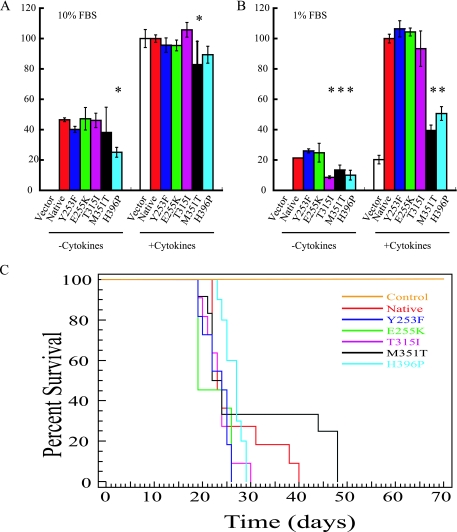
To further assess the transformation potential of Bcr-Abl KD mutants, we compared the ability of native and Y253F, E255K, T315I, M351T, and H396P mutant Bcr-Abl to transform primary murine myeloid cells. Exogenous cytokines are required for normal murine BM to form myeloid colonies in semisolid medium, while Bcr-Abl expression induces cytokine-independent colony formation (11). We plated BM cells in triplicate in the presence or absence of rm SCF, rm IL-3, and rm IL-6 in methylcellulose supplemented with either 1 or 10% FBS. Subtle differences were observed in 10% serum. In the presence of cytokines M351T induced fewer colonies than native Bcr-Abl (P = 0.04; Fig. 3A), and in the absence of cytokines H396P formed fewer colonies than native Bcr-Abl (P < 0.0001; Fig. 3A). Growth in 1% FBS accentuated the defects in colony formation by M351T and H396P (Fig. 3B). Similarly, colony formation by cells transduced with the T315I mutant was significantly reduced compared to native Bcr-Abl, but only in the absence of cytokines (P = 0.003; Fig. 3B). These results show that the reduced transforming potential of M351T and H396P is not limited to B-cell progenitors but also occurs in the myeloid lineage. In contrast to the B-cell assays, however, the transformation potency of the P-loop mutants Y253F and E255K was not significantly increased over native Bcr-Abl.
FIG. 3.
Myeloid colony formation by Bcr-Abl kinase domain mutants, and Kaplan-Meyer survival curves. (A) BM from BALB/c mice infected with the indicated BCR-ABL construct was plated in triplicate in methylcellulose supplemented with 10% FBS in the presence or absence of IL-3, IL-6, and SCF in two independent trials with the results combined. Colony formation was scored on day 10 with colonies containing 50 or more cells scored as positive. The results are plotted as a percentage of WT BCR-ABL colonies that formed in the presence of cytokines. An asterisk indicates a statistically significant (P < 0.05) deviation from native Bcr-Abl. (B) BM from BALB/c mice infected with the indicated BCR-ABL construct was plated in triplicate in methylcellulose supplemented with 1% FBS in the presence or absence of IL-3, IL-6, and SCF in two independent trials with the results combined. Colony formation was scored on day 10 with colonies containing 50 or more cells scored as positive. The results are plotted as a percentage of native BCR-ABL colonies that formed in the presence of cytokines. An asterisk indicates a statistically significant (P < 0.05) deviation from native Bcr-Abl. (C) Kaplan-Meyer survival curve for recipients of BM from 5-fluoruracil-treated donors transduced with MSCV-p210Bcr-Abl-internal ribosome entry site-GFP harboring native Bcr-Abl, Y253F, E255K, T315I, M351T, H396P, or control retrovirus. None of the mutants tested exhibited statistical difference in survival compared to the wild type (P > 0.05).
Bcr-Abl KD mutants show only trends towards survival differences in a murine model of CML.
To compare the potency of the mutants for induction of myeloproliferative disease in vivo, lethally irradiated BALB/c mice were transplanted with BM transduced with native or mutant Bcr-Abl. Mice transplanted with BM expressing native Bcr-Abl had a median survival of 23 days. There was a trend toward longer survival in mice transplanted with BM transduced with H396P mutant (median survival, 27 days) and shorter survival in recipients of E255K (median survival, 19 days). However, these differences did not reach statistical significance in Kaplan-Meyer analysis (Fig. 3C). The median survival periods of animals transplanted with Y253F, T315I, or M351T mutants were 24, 24, and 23 days, respectively, which is similar to recipients of native Bcr-Abl. Similar to the survival data, no significant differences in latency (the time from transplant to the day of the first abnormal white blood cell count) were seen (data not shown). Expression of the lineage specific markers Gr-1, Mac-1, B220, Thy1.2, and Ter119 was not significantly different between the mutants and native Bcr-Abl in the peripheral blood, BM, spleen, and liver, and no significant differences were seen in the spleen or liver sizes (see Table S1 in the supplemental material). Autopsy demonstrated lung hemorrhage in all animals, a finding consistent with previously reported MIGR1-p210Bcr-Abl induced CML (40). Spleen cells and BM harvested from leukemic primary recipient mice and injected retro-orbitally into sublethally irradiated secondary recipients induced a CML-like myeloproliferative syndrome between 40 and 60 days after injection, with no appreciable differences in phenotype (data not shown).
Bcr-Abl KD mutants exhibit significant differences in enzymatic activity compared to native Bcr-Abl.
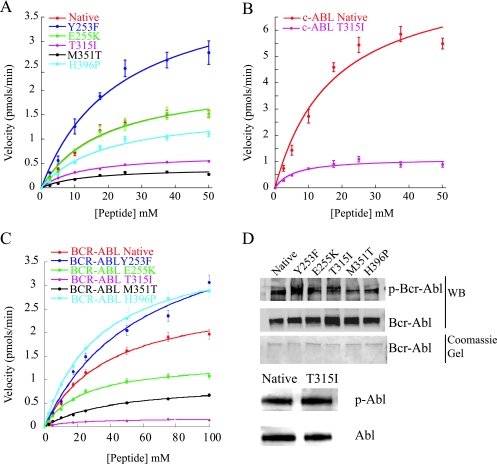
The reproducible differences in transformation potency of the various mutants compared to Bcr-Abl prompted us to determine whether these differences could be explained by increased or reduced intrinsic kinase activity. In a first set of experiments, we analyzed the kinetic properties of the purified KDs of the various Abl mutants using in vitro kinase assays with a peptide substrate (4). Using nonlinear regression fits the Michaelis-Menten parameters describing substrate binding (Km), the maximum rate (Vmax), the catalytic rate constant (kcat), and the catalytic efficiency (kcat/Km) were determined (Fig. 4A and Table 1). In these assays the Y253F mutant exhibited significantly increased activity compared to native Bcr-Abl, as demonstrated by an ∼1.5-fold increase in kcat/Km, both in the case of the isolated KD and in the case of the full-length Bcr-Abl protein (Table 1). Although the kcat/Km of the E255K mutant was not significantly different from that of the native Abl KD, the kcat/Km values of the M351T, T315I, and H396P mutants were reduced to 27, 47, and 70%, respectively, of the native Abl (Table 1). The apparent Km values of T315I and M351T were ∼1.5-fold lower than the Km of native Abl KD (Table 1), indicating stronger substrate binding to the active site. These data suggest that the differences in the transformation potency may in part be due to differences in kinase activity, particularly for the Y253F and M351T mutants. To investigate whether the kinase activity may be further influenced by regulatory mechanisms operational only in the full-length Bcr-Abl proteins, we purified native Bcr-Abl and the various mutants from insect cells (Fig. 4B and C). The relative phosphorylation state of the proteins was comparable as evidenced by immunoblotting with phospho-tyrosine specific antibodies (Fig. 4D). kcat/Km was increased in the case of Y253F, comparable in the case of H396P, slightly reduced in the case of E255K and very considerably reduced in the case of T315I and M351T (Table 1). Analogous assays were carried out with commercially available recombinant Abl and T315I mutant Abl, which confirmed the reduced kcat/Km. In addition, assays were performed to examine the effect of phosphorylation using dephosphorylated native and T315I full-length c-Abl. In both cases, the Km and kcat/Km values were not affected by dephosphorylation relative to the phosphorylated enzymes. However, both proteins exhibited an ∼1.5-fold increase in Vmax relative to the phosphorylated enzyme (Table 1).
FIG. 4.
In vitro kinase assays. (A) Plots of velocity versus peptide concentration using isolated KDs of the indicated construct fused to GST. Assays were performed in triplicate, with the standard deviation of the velocities shown as error bars. (B) Plots of velocity versus peptide concentration using native and T315I expressed as full c-Abl constructs. Assays were performed in triplicate with the standard deviation of the velocities shown as error bars. (C) Plots of velocity versus peptide concentration using native and mutants expressed in full-length Bcr-Abl. Assays were performed in triplicate with the standard deviation of the velocities shown as error bars. (D) Anti-Abl and anti-phospho-tyrosine blots of purified c-Abl and Bcr-Abl and a Coomassie gel of purified full-length Bcr-Abl.
TABLE 1.
Km, Vmax, kcat, and kcat/Km of Bcr-Abl, c-Abl, and KD mutants
| Mutant | Mean ± SDa
|
|||
|---|---|---|---|---|
| Km (μM) | Vmax (pmol/min) | kcat (min−1) | kcat/Km (106 M−1 min−1) | |
| Native KD-GST | 18.8 ± 3.9 | 2.22 ± 0.19 | 0.56 ± 0.05 | 0.03 ± 0.007 |
| Y253F KD-GST | 23.1 ± 5.0 | 4.25 ± 0.42 | 1.06 ± 0.1 | 0.046 ± 0.01 |
| E255K KD-GST | 19.4 ± 4.2 | 2.24 ± 0.21 | 0.56 ± 0.05 | 0.029 ± 0.007 |
| T315I KD-GST | 12.1 ± 0.8 | 0.69 ± 0.02 | 0.17 ± 0.005 | 0.014 ± 0.001 |
| M351T KD-GST | 12.6 ± 4.7 | 0.41 ± 0.05 | 0.1 ± 0.01 | 0.008 ± 0.003 |
| H396P KD-GST | 18.9 ± 4.6 | 1.59 ± 0.16 | 0.4 ± 0.04 | 0.021 ± 0.006 |
| Native c-Abl | 17.3 ± 5.3 | 8.22 ± 1.02 | 2.06 ± 0.26 | 0.12 ± 0.04 |
| T315I c-Abl | 5.64 ± 1.96 | 1.11 ± 1.00 | 0.28 ± 0.25 | 0.05 ± 0.05 |
| Native Bcr-Abl | 39.6 ± 4.9 | 2.84 ± 0.15 | 0.71 ± 0.04 | 0.018 ± 0.002 |
| Y253F Bcr-Abl | 61 ± 14 | 4.67 ± 0.54 | 1.87 ± 0.22 | 0.03 ± 0.008 |
| E255K Bcr-Abl | 23.9 ± 2.3 | 1.39 ± 0.05 | 0.24 ± 0.04 | 0.01 ± 0.002 |
| T315I Bcr-Abl | 19.4 ± 3.4 | 0.18 ± 0.01 | 0.064 ± 0.004 | 0.003 ± 0.0006 |
| M351T Bcr-Abl | 43.3 ± 3.4 | 0.94 ± 0.03 | 0.18 ± 0.006 | 0.004 ± 0.0003 |
| H396P Bcr-Abl | 33.5 ± 1.4 | 3.88 ± 1.45 | 1.04 ± 0.39 | 0.03 ± 0.01 |
| Dephospho Native c-Abl | 19.6 ± 3.6 | 12.4 ± 0.7 | 3.1 ± 0.175 | 0.16 ± 0.03 |
| Dephospho T315I c-Abl | 7.99 ± 1.87 | 1.73 ± 0.12 | 0.43 ± 0.03 | 0.05 ± 0.01 |
Km and Vmax are given as values obtained from the fit of the equation Vmax × [substrate]/(Km + [substrate]) ± the error in the fit. The kcat was obtained from the equation kcat = Vmax/[enzyme].
Ba/F3 cells expressing Bcr-Abl KD mutants exhibit differences in tyrosine phosphorylation compared to native Bcr-Abl.
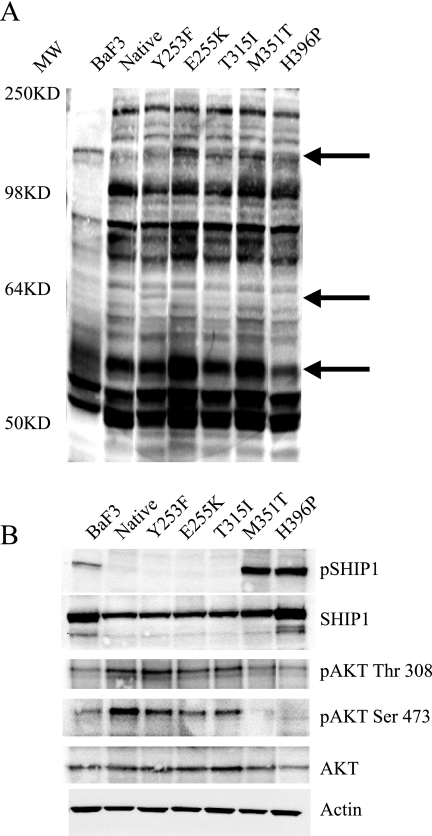
The data presented above suggested that differences in kinase activity only partially explain the differences in transformation potency between native and KD mutant Bcr-Abl. We thus reasoned that expression of various mutants may result in the differential utilization of signaling pathways downstream of Bcr-Abl. We compared substrate phosphorylation and the activation of various signaling pathways in Ba/F3 cells expressing mutant and native Bcr-Abl (see Fig. S1 in the supplemental material). Bcr-Abl activates numerous signaling pathways, including mitogen-activated protein, STAT5, and phosphatidylinositol 3-kinase (8). Although no appreciable differences between native Bcr-Abl and the various mutants were seen in these Bcr-Abl-driven pathways, whole-cell lysates probed with an anti-phosphotyrosine antibody consistently yielded reproducible differences between native Bcr-Abl and KD mutants (Fig. 5A). To identify potential differences in tyrosine phosphorylation, we applied a novel and robust method to evaluate the phosphotyrosine proteome of biological samples (31). The global phosphotyrosine profile from control Ba/F3 cells was compared to those expressing either native Bcr-Abl or the KD mutants Y253F, E255K, T315I, M351T, and H396P. Utilizing a phosphotyrosine antibody, tyrosine phosphorylated peptides were immunoprecipitated directly from protease-digested cellular protein extracts. The immunoprecipitated peptides were then identified by MS/MS (31). In the six cell lines we identified a total of 120 unique phosphorylation sites within 87 proteins (see Table S2 in the supplemental material), many of which are important signaling molecules implicated in Bcr-Abl signaling. Interestingly, we observed tyrosine phosphorylation sites unique to the various Bcr-Abl mutants, not observed in the native Bcr-Abl expressing cells (Table 2). For example, phosphorylation of the adaptor protein Fyn binding protein (Y559) was found in all of the mutants except the M351T- and H396P-expressing cell lines. Cbl (Y698) was present in all mutant cell lines except the E255K- and H396P-expressing cells. Phosphorylation of the Src substrate, Scap2 at tyrosine 260, was only observed in T315I and Y253F mutants. The adaptor protein Cbl-b (Y845) is only phosphorylated in the E255K, M351T, and T315I mutant cell lines. Vinculin (Y822) and Nck (Y105) are both only seen in the E255K and M351T mutant cell lines. The delta polypeptide of the lipid kinase phosphatidylinositol 3-kinase (Y936) is only phosphorylated in the M351T mutant cell line. Finally, the adaptor protein Dok2 (Y142) and the inositol phosphatase SHIP1 (Y1021) were only observed in the M351T and H396P mutant cell lines. To validate the initial observations, we carried out a “targeted” analysis of these peptides in a completely independent experiment, focusing on proteins with unique phosphorylation sites (Table 2). As during data-dependent analysis, the mass spectrometer chooses the four most intense peptides eluting from the LC column for MS/MS fragmentation, peptides may not be identified during this initial analysis due to sample complexity. Thus, phosphotyrosine peptides from the Bcr-Abl expressing cell lines were subjected to a targeted MS/MS analysis of specific phosphopeptides. This targeted strategy expands the effective dynamic range of the mass spectrometer by focusing analysis time exclusively on the phosphopeptides of interest, resulting in a greater chance of identification. Using this approach we were able to confirm the initial analysis (Table 2). In addition, we confirmed by Western blotting that SHIP1 (Y1021) is strongly phosphorylated only in the M351T- and H396P-expressing Ba/F3 cells (Fig. 5B). In addition, a decrease in the phosphorylation of Akt at serine 473 and to a lesser extent tyrosine 308 was also observed in these two mutants (Fig. 5B).
FIG. 5.
Signaling of Bcr-Abl in cell lines expressing wild type or KD mutants. Ba/F3 cells were serum starved for 24 h, and lysates were immunoblotted with the indicated phospho-specific antibody. Specific antibodies were used to assure equal loading. (A) Whole-cell lysates probed with anti-phosphotyrosine (4G10); arrows indicate potential differences in phosphorylation. All lanes contained 50 μg of total cell lysate as measured by a bicinchoninic acid assay. The molecular weight (MW) is indicated. (B) A total of 200 μg of the same lysates were probed with SHIP1, phospho-SHIP1, Akt, phospho-Akt (Thr 308) and phospho-Akt (Ser 473), with actin used as a loading control.
TABLE 2.
Observed tyrosine phosphorylation sites unique to the various Bcr-Abl mutants
| Protein | Site | Sequencea | NCBIb | PSDBc | T315I | M351T | Y253F | E255K | H396P |
|---|---|---|---|---|---|---|---|---|---|
| Cbl | Y698 | LPPGEQGESEEDTEY*MTPTSRPVGVQKPEPK | NP_031645 | 1708-6 | +d | + | + | ||
| Btk | Y223 | KVVALY*DYMPMNANDLQLR | NP_038510 | 875-6 | + | + | + | + | |
| Cbl-b | Y845 | ASQDY*DQLPSSSDGSQAPARPPKPRPR | XP_156257 | 3296-6 | + | + | + | ||
| Fyb | Y559 | TTAVEIDY*DSLK | NP_035945 | 2479-5 | + | + | + | ||
| Scap2 | Y260 | SQPIDDEIY*EELPEEEEDTASVK | NP_061243 | 2628-6 | + | + | |||
| Nck | Y105 | RKPSVPDTASPADDSFVDPGERLY*DLNMPAFVK | NP_035008 | 2163-6 | + | + | |||
| Vinculin | Y822 | SFLDSGY*R | NP_033528 | 3317-5 | + | + | |||
| Dok2 | Y142 | SGSPCMEENELY*SSSTTGLCK | NP_034201 | 1502-6 | + | + | |||
| PI3K p110 delta | Y936 | ERVPFILTY*DFVHVIQQGK | NP_032866 | 3382-6 | + | ||||
| SHIP1 | Y1021 | VEALLQEDLLLTKPEMFENPLY*GSVSSFPK | NP_034696 | 1481-6 | + | + |
An asterisk indicates phosphorylation of the preceding tyrosine within the peptide.
NCBI is the number given to each protein in the protein database.
PSDB is the number given to each protein in the PhosphoSite database.
+, Phosphorylation sites were found in Bcr-Abl KDs but not in native Bcr-Abl.
DISCUSSION
Point mutations in the KD of Bcr-Abl that impair drug binding have been established as the major mechanism of acquired resistance to imatinib (1, 3, 13, 15, 37). Here we provide evidence that, in addition to conferring drug resistance, KD mutations significantly affect the transformation potency of Bcr-Abl. The transformation potency of five common KD mutants compared to native Bcr-Abl was found to depend not only on the type of mutant but also on the specific growth conditions and cell type. For example, the P-loop mutants E255K and Y253F showed a pronounced increase of potency in primary B cells and Ba/F3 cells grown in low serum, while there was no significant difference compared to native Bcr-Abl in myeloid colony formation assays. T315I was equipotent to native Bcr-Abl in Ba/F3 cells, primary B cells, and clonogenic assays performed in the presence of either 10% FBS or growth factors but had a severe transformation defect in the absence of both. This suggests that T315I, unlike native Bcr-Abl, may be unable to activate pathways that compensate for the lack of cytokines and serum. These observations indicate that the competitiveness of a given Bcr-Abl KD mutant is influenced in a complex fashion by intrinsic and extrinsic factors. Based on all in vitro assays, an approximate consensus ranking of transformation potency is Y253F > E255K > native Bcr-Abl ≥ T315I > H396P > M351T (Table 3).
TABLE 3.
Qualitative summary of resultsa
| Mutant | Growth competition (low serum) | B-lymphoid transformation | Colony formation, primary cells (low serum, no cytokines) |
kcat/Km
|
|
|---|---|---|---|---|---|
| KD | Bcr-Abl-FL | ||||
| Y253F | > | > | ∼ | > | > |
| E255K | > | > | ∼ | ∼ | < |
| T315I | > | ∼ | < | < | < |
| M351T | < | < | < | < | < |
| H396P | ∼ | < | < | ∼ | > |
FL, full length. Changes are listed as greater than (>), less than (<), or approximately the same (∼) relative to native Bcr-Abl.
In contrast to the in vitro assays, no statistically significant differences were observed in a transduction/transplantation model of CML, although there were trends for longer survival in mice transplanted with BM expressing the H396P mutant and for shorter survival in the recipients of BM transduced with E255K. The lack of significant survival differences may reflect the aggressive nature of the murine CML model that may obscure more subtle effects of the mutations on disease biology. This finding is consistent with the Ba/F3 cell proliferation and the clonogenic assays, where differences were decreased or even abrogated under optimal growth conditions.
To investigate whether differences in intrinsic kinase activity may account for the differences in transformation potency, we determined kinetic parameters in substrate phosphorylation assays. In an initial series of experiments, we used the isolated KDs of Abl. Here, the kcat/Km value of the Y253F mutant was increased over native Bcr-Abl, the kcat/Km value of E255K was similar, and the kcat/Km value of the other mutants were slightly (H396P) or significantly (M351T and T315I) reduced. Analysis of the full-length proteins confirmed the findings for Y253F, T351I, and M351T but in contrast to the isolated KD showed a slight reduction of kcat/Km value for E255K and a slight increase of kcat/Km for H396P. The subtle differences between the isolated KD and the full-length Bcr-Abl constructs point to regulatory mechanisms that are present only in the full-length proteins. The results of our in vitro kinase assays are in contrast to data reported by Yamamoto et al., who found that both E255K and T315I show increased kinase activity toward recombinant GST-CrkL relative to native Bcr-Abl (39). However, there are several factors in their experiments that would not allow precise determinations of kinase activity. The constructs used contained only a small fragment of Bcr fused N terminally to the Abl SH3 domain, lacking both the N-terminal “cap” region of Abl, critical to the autoinhibition of the kinase, and the coiled-coil motif of Bcr that mediates dimerization of Bcr-Abl (23, 26). Thus, the mechanism of kinase activation in these constructs may differ from full-length Bcr-Abl, similar to our experiments using the isolated kinase domains. In addition, anti-Flag immunoprecipitates were used in the kinase assays, which may contain additional proteins that could influence kinase activity. Similar limitations apply to data recently published by another group who failed to detect differences in kinase activity in anti-Abl immunoprecipitates of cell lines expressing T315I and native Bcr-Abl, respectively (19).
Autophosphorylation of Abl on tyrosines 226 (phosphorylated in all mutants and native Bcr-Abl) and 393 (phosphorylated in native Bcr-Abl and all mutants except H396P) influence kinase activity as well as imatinib sensitivity (20, 30). This regulation appears to require full-length protein, as Nagar et al. demonstrated that in the isolated KD of Abl, phosphorylation affected imatinib binding but not kinase activity (22). Using dephosphorylated proteins, Roumiantsev et al. found no difference in kinase activity between native and Y253F mutant c-Abl, although they observed increased levels of phosphotyrosine in Ba/F3 cells expressing Y253F mutant Bcr-Abl compared to native Bcr-Abl, a finding consistent with increased intracellular kinase activity (30). In contrast to their results, we were unable to document consistent differences in total cellular phosphotyrosine levels between the various mutants and native Bcr-Abl. The reason for this discrepancy is unclear. It is possible that the relatively subtle differences in intrinsic kinase activity may not be detectable on the relatively insensitive total phosphotyrosine blots. In addition, total phosphotyrosine levels reflect additional factors, such as the activity of phosphatases that operate only in intact cells.
Overall, it is evident that the differences in intrinsic kinase activity only partially explain the differences in transformation potency. For example, the E255K mutant exhibits increased transformation potency, although its kinase activity is slightly reduced compared to native Bcr-Abl. Similarly, the T315I mutant is equipotent to native Bcr-Abl in the majority of assays, although it has consistently much lower kinase activity. These data raise the question of additional factors impacting transformation potency. Guided by reproducible differences observed on total phosphotyrosine blots among the mutants tested, we carried out a detailed analysis of protein tyrosine phosphorylation using mass spectroscopy and identified differentially phosphorylated tyrosine residues on several proteins. Importantly, for a subset of these phosphopeptides, differences were confirmed in an independent “targeted” experiment. However, we cannot exclude that some differences would not be reproducible if the entire analysis was repeated, since some phosphorylation sites may be specific to the individual cell lines rather than the Bcr-Abl mutant. Experiments are under way to validate the results in a series of independently derived BaF/3 and 32D cell lines expressing native Bcr-Abl or the various mutants.
Differentially phosphorylated proteins included known substrates of Bcr-Abl such as Cbl, as well as proteins that had not been previously associated with Bcr-Abl-positive leukemia, such as Scap2 (Table 2). Phosphorylation of tyrosine 1021 of SHIP1 was detected in Ba/F3 cells expressing the M351T and H396P mutants of Bcr-Abl but not in cells expressing the Y253F, E255K, and T315I mutants or native Bcr-Abl. Immunoblot analysis confirmed this mass spectroscopy finding. In accord with published data (33), SHIP1 levels were significantly reduced in Ba/F3 cells expressing native Bcr-Abl compared to the parental cells. SHIP1 has diverse functions in hematopoietic cells. Mice with targeted disruption of SHIP1 develop a myeloproliferative syndrome (14), a finding consistent with a negative regulatory role in hematopoiesis. In Ba/F3 cells transformed by Bcr-Abl, SHIP1 negatively regulates motility. Thus, re-expression of SHIP1 reduces the enhanced migration of these cells, and this is dependent on Y1021, since a Y1021F mutant of SHIP1 failed to inhibit motility (33). Thus, the phosphorylation of Y1021 in Ba/F3 cells expressing M351T and H396P may attenuate Bcr-Abl-mediated transformation by reducing cell motility. In addition, SHIP1 has been shown to negatively regulate Akt activity in B cells after cross-linking of FcγRIIB1 (2). It is not known whether these effects are dependent on Y1021, but the reduction in the phosphorylation of Akt on threonine 308 and serine 473 in cells expressing the M351T and H396P mutants suggests that Y1021 may be involved in mediating the negative regulation of Akt. Further analysis of these differences in signaling should allow insight into pathways that enhance or antagonize transformation by Bcr-Abl.
The detection of Bcr-Abl KD mutations prior to commencing therapy with imatinib would be predicted to be primarily dependent on the transformation potency of the mutant. Consistent with this, we detected Y253F (gain of function) but not M351T (loss of function) in an unbiased analysis of imatinib-naive patients (38). In contrast, the prevalence of a given Bcr-Abl mutant in a patient undergoing therapy with imatinib will be determined by several factors. The primary factor is its degree of drug resistance. However, other factors, such as the levels of Bcr-Abl expression, may also contribute to the prevalence of a given Bcr-Abl mutant. In addition, we showed that several of the mutant clones demonstrate improved survival compared to native Bcr-Abl under suboptimal growth conditions. As such, it is conceivable that mutant clones may gain a survival advantage in certain niches with lower concentrations of growth factors and cytokines.
Clinical observations suggest that Bcr-Abl kinase mutations may have biological significance beyond conferring drug resistance. The expansion of a clone carrying a somatic mutation may have two explanations. First, the mutation may not be causal to the clone's expansion but solely comigrate with it. If this were the case, one would expect to see random somatic mutations in genes other than BCR-ABL, in addition to KD mutations. Although this has not been exhaustively investigated, we and others (1, 34) failed to detect mutations in c-Kit and the platelet-derived growth factor receptors in CML patients with primary or acquired resistance to imatinib, arguing against comigration. Second, the mutation may endow the clone with a proliferative advantage over unmutated cells, which would represent a mechanism of disease progression. Our finding that the P-loop mutants E255K and Y253F have increased transformation potency is in accord with the clinical observation that mutations of the P-loop confer a poor prognosis compared to other mutation types, regardless of their imatinib sensitivity (3, 36).
In summary, we provide evidence that KD mutations modulate the biology of Bcr-Abl-induced leukemia irrespective of sensitivity to imatinib. Gain-of-function mutants may independently contribute to disease progression, while loss-of function mutants are selected only in the presence of drug. Intrinsic kinase activity, substrate specificity, and extrinsic factors, such as cytokines, ultimately determine the outgrowth of a given mutant. This information, together with the sensitivity of the various mutants to imatinib and other Abl kinase inhibitors, may be useful to rationally design the optimal therapeutic approach for individual patients.
Supplementary Material
Acknowledgments
We thank Julie Nardone for assistance with bioinformatics analysis and Chris Koontz for editorial assistance with the manuscript.
This study was sponsored by funding from the Howard Hughes Medical Institute and grants from The Leukemia and Lymphoma Society, the National Cancer Institute (R01 CA65823), and the Burroughs Wellcome Fund. M.W.D. is a Junior Faculty Scholar of the American Society of Hematology.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Al-Ali, H. K., M. C. Heinrich, T. Lange, R. Krahl, M. Mueller, C. Muller, D. Niederwieser, B. J. Druker, and M. W. Deininger. 2004. High incidence of BCR-ABL kinase domain mutations and absence of mutations of the PDGFR and KIT activation loops in CML patients with secondary resistance to imatinib. Hematol. J. 5:55-60. [DOI] [PubMed] [Google Scholar]
- 2.Aman, M. J., T. D. Lamkin, H. Okada, T. Kurosaki, and K. S. Ravichandran. 1998. The inositol phosphatase SHIP inhibits Akt/PKB activation in B cells. J. Biol. Chem. 273:33922-33928. [DOI] [PubMed] [Google Scholar]
- 3.Branford, S., Z. Rudzki, S. Walsh, A. Grigg, C. Arthur, K. Taylor, R. Herrmann, K. P. Lynch, and T. P. Hughes. 2002. High frequency of point mutations clustered within the adenosine triphosphate-binding region of BCR/ABL in patients with chronic myeloid leukemia or Ph-positive acute lymphoblastic leukemia who develop imatinib (STI571) resistance. Blood 99:3472-3475. [DOI] [PubMed] [Google Scholar]
- 4.Corbin, A. S., E. Buchdunger, F. Pascal, and B. J. Druker. 2002. Analysis of the structural basis of specificity of inhibition of the Abl kinase by STI571. J. Biol. Chem. 277:32214-32219. [DOI] [PubMed] [Google Scholar]
- 5.Corbin, A. S., P. La Rosee, E. P. Stoffregen, B. J. Druker, and M. W. Deininger. 2003. Several Bcr-Abl kinase domain mutants associated with imatinib mesylate resistance remain sensitive to imatinib. Blood 101:4611-4614. [DOI] [PubMed] [Google Scholar]
- 6.Cowan-Jacob, S. W., V. Guez, G. Fendrich, J. D. Griffin, D. Fabbro, P. Furet, J. Liebetanz, J. Mestan, and P. W. Manley. 2004. Imatinib (STI571) resistance in chronic myelogenous leukemia: molecular basis of the underlying mechanisms and potential strategies for treatment. Mini Rev. Med. Chem. 4:285-299. [DOI] [PubMed] [Google Scholar]
- 7.Daley, G. Q., R. A. Van Etten, and D. Baltimore. 1990. Induction of chronic myelogenous leukemia in mice by the P210bcr/abl gene of the Philadelphia chromosome. Science 247:824-830. [DOI] [PubMed] [Google Scholar]
- 8.Deininger, M. W., J. M. Goldman, and J. V. Melo. 2000. The molecular biology of chronic myeloid leukemia. Blood 96:3343-3356. [PubMed] [Google Scholar]
- 9.Druker, B. J., C. L. Sawyers, H. Kantarjian, D. J. Resta, S. F. Reese, J. M. Ford, R. Capdeville, and M. Talpaz. 2001. Activity of a specific inhibitor of the BCR-ABL tyrosine kinase in the blast crisis of chronic myeloid leukemia and acute lymphoblastic leukemia with the Philadelphia chromosome. N. Engl. J. Med. 344:1038-1042. [DOI] [PubMed] [Google Scholar]
- 10.Druker, B. J., M. Talpaz, D. J. Resta, B. Peng, E. Buchdunger, J. M. Ford, N. B. Lydon, H. Kantarjian, R. Capdeville, S. Ohno-Jones, and C. L. Sawyers. 2001. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N. Engl. J. Med. 344:1031-1037. [DOI] [PubMed] [Google Scholar]
- 11.Gishizky, M. L., and O. N. Witte. 1992. Initiation of deregulated growth of multipotent progenitor cells by bcr-abl in vitro. Science 256:836-839. [DOI] [PubMed] [Google Scholar]
- 12.Goldman, J. M., and J. V. Melo. 2003. Chronic myeloid leukemia: advances in biology and new approaches to treatment. N. Engl. J. Med. 349:1451-1464. [DOI] [PubMed] [Google Scholar]
- 13.Gorre, M. E., M. Mohammed, K. Ellwood, N. Hsu, R. Paquette, P. N. Rao, and C. L. Sawyers. 2001. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science 293:876-880. [DOI] [PubMed] [Google Scholar]
- 14.Helgason, C. D., J. E. Damen, P. Rosten, R. Grewal, P. Sorensen, S. M. Chappel, A. Borowski, F. Jirik, G. Krystal, and R. K. Humphries. 1998. Targeted disruption of SHIP leads to hemopoietic perturbations, lung pathology, and a shortened life span. Genes Dev. 12:1610-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hochhaus, A., S. Kreil, A. S. Corbin, P. La Rosee, M. C. Muller, T. Lahaye, B. Hanfstein, C. Schoch, N. C. Cross, U. Berger, H. Gschaidmeier, B. J. Druker, and R. Hehlmann. 2002. Molecular and chromosomal mechanisms of resistance to imatinib (STI571) therapy. Leukemia 16:2190-2196. [DOI] [PubMed] [Google Scholar]
- 16.Kreuzer, K. A., P. Le Coutre, O. Landt, I. K. Na, M. Schwarz, K. Schultheis, A. Hochhaus, and B. Dorken. 2003. Preexistence and evolution of imatinib mesylate-resistant clones in chronic myelogenous leukemia detected by a PNA-based PCR clamping technique. Ann. Hematol. 82:284-289. [DOI] [PubMed] [Google Scholar]
- 17.Lugo, T. G., A. M. Pendergast, A. J. Muller, and O. N. Witte. 1990. Tyrosine kinase activity and transformation potency of bcr-abl oncogene products. Science 247:1079-1082. [DOI] [PubMed] [Google Scholar]
- 18.McLaughlin, J., E. Chianese, and O. N. Witte. 1987. In vitro transformation of immature hematopoietic cells by the P210 BCR/ABL oncogene product of the Philadelphia chromosome. Proc. Natl. Acad. Sci. USA 84:6558-6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miething, C., S. Feihl, C. Mugler, R. Grundler, N. von Bubnoff, F. Lordick, C. Peschel, and J. Duyster. 2006. The Bcr-Abl mutations T315I and Y253H do not confer a growth advantage in the absence of imatinib. Leukemia 20:650-657. [DOI] [PubMed] [Google Scholar]
- 20.Miething, C., C. Mugler, R. Grundler, J. Hoepfl, R. Y. Bai, C. Peschel, and J. Duyster. 2003. Phosphorylation of tyrosine 393 in the kinase domain of Bcr-Abl influences the sensitivity toward imatinib in vivo. Leukemia 17:1695-1699. [DOI] [PubMed] [Google Scholar]
- 21.Million, R. P., N. Harakawa, S. Roumiantsev, L. Varticovski, and R. A. Van Etten. 2004. A direct binding site for Grb2 contributes to transformation and leukemogenesis by the Tel-Abl (ETV6-Abl) tyrosine kinase. Mol. Cell. Biol. 24:4685-4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagar, B., W. G. Bornmann, P. Pellicena, T. Schindler, D. R. Veach, W. T. Miller, B. Clarkson, and J. Kuriyan. 2002. Crystal structures of the kinase domain of c-Abl in complex with the small molecule inhibitors PD173955 and imatinib (STI-571). Cancer Res. 62:4236-4243. [PubMed] [Google Scholar]
- 23.Nagar, B., O. Hantschel, M. A. Young, K. Scheffzek, D. Veach, W. Bornmann, B. Clarkson, G. Superti-Furga, and J. Kuriyan. 2003. Structural basis for the autoinhibition of c-Abl tyrosine kinase. Cell 112:859-871. [DOI] [PubMed] [Google Scholar]
- 24.Pear, W. S., J. P. Miller, L. Xu, J. C. Pui, B. Soffer, R. C. Quackenbush, A. M. Pendergast, R. Bronson, J. C. Aster, M. L. Scott, and D. Baltimore. 1998. Efficient and rapid induction of a chronic myelogenous leukemia-like myeloproliferative disease in mice receiving P210 bcr/abl-transduced bone marrow. Blood 92:3780-3792. [PubMed] [Google Scholar]
- 25.Pear, W. S., G. P. Nolan, M. L. Scott, and D. Baltimore. 1993. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. USA 90:8392-8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pluk, H., K. Dorey, and G. Superti-Furga. 2002. Autoinhibition of c-Abl. Cell 108:247-259. [DOI] [PubMed] [Google Scholar]
- 27.Roche-Lestienne, C., J. L. Lai, S. Darre, T. Facon, and C. Preudhomme. 2003. A mutation conferring resistance to imatinib at the time of diagnosis of chronic myelogenous leukemia. N. Engl. J. Med. 348:2265-2266. [DOI] [PubMed] [Google Scholar]
- 28.Roche-Lestienne, C., V. Soenen-Cornu, N. Grardel-Duflos, J. L. Lai, N. Philippe, T. Facon, P. Fenaux, and C. Preudhomme. 2002. Several types of mutations of the Abl gene can be found in chronic myeloid leukemia patients resistant to STI571, and they can preexist to the onset of treatment. Blood 100:1014-1018. [DOI] [PubMed] [Google Scholar]
- 29.Roumiantsev, S., I. E. de Aos, L. Varticovski, R. L. Ilaria, and R. A. Van Etten. 2001. The src homology 2 domain of Bcr/Abl is required for efficient induction of chronic myeloid leukemia-like disease in mice but not for lymphoid leukemogenesis or activation of phosphatidylinositol 3-kinase. Blood 97:4-13. [DOI] [PubMed] [Google Scholar]
- 30.Roumiantsev, S., N. P. Shah, M. E. Gorre, J. Nicoll, B. B. Brasher, C. L. Sawyers, and R. A. Van Etten. 2002. Clinical resistance to the kinase inhibitor STI-571 in chronic myeloid leukemia by mutation of Tyr-253 in the Abl kinase domain P-loop. Proc. Natl. Acad. Sci. USA 99:10700-10705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rush, J., A. Moritz, K. A. Lee, A. Guo, V. L. Goss, E. J. Spek, H. Zhang, X. M. Zha, R. D. Polakiewicz, and M. J. Comb. 2005. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat. Biotechnol. 23:94-101. [DOI] [PubMed] [Google Scholar]
- 32.Sattler, M., M. G. Mohi, Y. B. Pride, L. R. Quinnan, N. A. Malouf, K. Podar, F. Gesbert, H. Iwasaki, S. Li, R. A. Van Etten, H. Gu, J. D. Griffin, and B. G. Neel. 2002. Critical role for Gab2 in transformation by BCR/ABL. Cancer Cell 1:479-492. [DOI] [PubMed] [Google Scholar]
- 33.Sattler, M., S. Verma, Y. B. Pride, R. Salgia, L. R. Rohrschneider, and J. D. Griffin. 2001. SHIP1, an SH2 domain containing polyinositol-5-phosphatase, regulates migration through two critical tyrosine residues and forms a novel signaling complex with DOK1 and CRKL. J. Biol. Chem. 276:2451-2458. [DOI] [PubMed] [Google Scholar]
- 34.Shah, N. P., J. M. Nicoll, B. Nagar, M. E. Gorre, R. L. Paquette, J. Kuriyan, and C. L. Sawyers. 2002. Multiple BCR-ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer Cell 2:117-125. [DOI] [PubMed] [Google Scholar]
- 35.Smith, K. M., R. Yacobi, and R. A. Van Etten. 2003. Autoinhibition of Bcr-Abl through its SH3 domain. Mol. Cell 12:27-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soverini, S., G. Martinelli, G. Rosti, S. Bassi, M. Amabile, A. Poerio, B. Giannini, E. Trabacchi, F. Castagnetti, N. Testoni, S. Luatti, A. de Vivo, D. Cilloni, B. Izzo, M. Fava, E. Abruzzese, D. Alberti, F. Pane, G. Saglio, and M. Baccarani. 2005. ABL mutations in late chronic phase chronic myeloid leukemia patients with up-front cytogenetic resistance to imatinib are associated with a greater likelihood of progression to blast crisis and shorter survival: a study by the GIMEMA Working Party on Chronic Myeloid Leukemia. J. Clin. Oncol. 23:4100-4109. [DOI] [PubMed] [Google Scholar]
- 37.von Bubnoff, N., F. Schneller, C. Peschel, and J. Duyster. 2002. BCR-ABL gene mutations in relation to clinical resistance of Philadelphia-chromosome-positive leukaemia to STI571: a prospective study. Lancet 359:487-491. [DOI] [PubMed] [Google Scholar]
- 38.Willis, S. G., T. Lange, S. Demehri, S. Otto, L. Crossman, D. Niederwieser, E. P. Stoffregen, S. McWeeney, I. Kovacs, B. Park, B. J. Druker, and M. W. Deininger. 2005. High sensitivity detection of Bcr-Abl kinase domain mutations in imatinib-naive patients: correlation with clonal cytogenetic evolution but not response to therapy. Blood 106:2128-2137. [DOI] [PubMed] [Google Scholar]
- 39.Yamamoto, M., T. Kurosu, K. Kakihana, D. Mizuchi, and O. Miura. 2004. The two major imatinib resistance mutations E255K and T315I enhance the activity of BCR/ABL fusion kinase. Biochem. Biophys. Res. Commun. 319:1272-1275. [DOI] [PubMed] [Google Scholar]
- 40.Zhang, X., and R. Ren. 1998. Bcr-Abl efficiently induces a myeloproliferative disease and production of excess interleukin-3 and granulocyte-macrophage colony-stimulating factor in mice: a novel model for chronic myelogenous leukemia. Blood 92:3829-3840. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.