Abstract
The packaging of DNA into chromatin allows eukaryotic cells to organize and compact their genomes but also creates an environment that is generally repressive to nuclear processes that depend upon DNA accessibility. There are several classes of enzymes that modulate the primary structure of chromatin to regulate various DNA-dependent processes. The biochemical activities of the yeast Isw1 ATP-dependent chromatin-remodeling enzyme have been well characterized in vitro, but little is known about how these activities are utilized in vivo. In this work, we sought to discern genetic backgrounds that require Isw1 activity for normal growth. We identified a three-way genetic interaction among Isw1, the NuA4 histone acetyltransferase complex, and the Swr1 histone replacement complex. Transcription microarray analysis revealed parallel functions for these three chromatin-modifying factors in the regulation of TATA-containing genes, including the repression of a large number of stress-induced genes under normal growth conditions. In contrast to a recruitment-based model, we find that the NuA4 and Swr1 complexes act throughout the genome while only a specific subset of the genome shows alterations in transcription.
Eukaryotic genomes are maintained in chromatin, a compact nucleoprotein complex. Chromatin is generally inhibitory to processes which depend on DNA accessibility. In order to modulate the repressive effects of chromatin, cells have evolved several mechanisms which alter DNA accessibility in chromatin. These mechanisms include histone modification, incorporation of histone variants, and ATP-dependent chromatin remodeling (14, 28, 31, 47, 49).
The posttranslational modification of histones is the most extensively studied of the three mechanisms of chromatin regulation. The majority of the known histone modifications occur on the N-terminal tails of the four core histones and include acetylation, phosphorylation, and methylation (30). These modifications may alter the accessibility of nucleosomal DNA to proteins by altering the charge of the histones or serve as a mechanism to create or destroy epitopes for recognition by particular protein motifs. Lysine residues present in all four core histones can be acetylated. There are several known histone acetyltransferases in yeast which are involved in nuclear processes such as transcription, DNA repair, and chromosome segregation (5, 9, 38). The NuA4 complex, a yeast histone acetyltransferase complex with strong substrate preference for the histone H2A and H4 tails, has been implicated in the positive regulation of transcription at distinct loci (15, 21, 48, 52). However, there is a published example where defects in NuA4-dependent acetylation do not correlate with changes in transcription (53). In addition, high-resolution mapping of histone modifications in budding yeast has shown that NuA4-dependent modifications occur at all transcribed genes in a stereotypical pattern that is independent of the transcription level, in contrast to the prevailing model (41, 50). Therefore, further work is necessary to understand the role of NuA4 and other histone acetyltransferases in transcription.
Histone replacement is a second major mechanism of chromatin modification, in which a variant histone protein, often highly related to its corresponding canonical histone, replaces the conventional histone within the nucleosome (32). The conserved H2A variant H2A.Z (encoded in yeast by the gene HTZ1) is found in a significant proportion of nucleosomes throughout the genome (51, 64). Recently, nucleosome-mapping experiments using high-density oligonucleotide arrays have shown that Htz1 is highly enriched in structurally distinct domains flanking the transcription start site of RNA polymerase II (Pol II) genes, regardless of the transcription state of the gene (51, 64). The mechanisms by which Htz1 is targeted specifically to these promoter regions and the functions of this Htz1-containing chromatin domain remain unclear. However, it appears that acetylation of Htz1 by the NuA4 complex may play an important role in stabilizing these Htz1-containing nucleosomes (1, 34, 43).
The third mechanism, ATP-dependent chromatin remodeling, is carried out by enzymes that utilize the energy of ATP hydrolysis to alter the structure or position of nucleosomes (58, 60). Phylogenetic analysis of the catalytic subunits parses ATP-dependent chromatin-remodeling enzymes into several subfamilies, including the Swi/Snf, Mi-2/CHD1, INO80, and ISWI subfamilies (16). ATP-dependent chromatin-remodeling factors are well conserved across eukaryotes, underscoring their importance in vivo.
The budding yeast Saccharomyces cerevisiae has two members of the ISWI class of ATPases, Isw1 and Isw2 (61). Isw2 slides nucleosomes to repress the transcription of several classes of genes, including early meiotic genes, in parallel with the Sin3/Rpd3 histone deacetylase (18, 19, 23). In contrast, while Isw1 has been well characterized in vitro (61, 62), its in vivo functions remain unclear. isw1 knockout mutants have no significant growth defects under normal culture conditions, and microarray studies using an isw1 deletion mutant showed only subtle effects on gene transcription, including derepression of approximately 140 genes by more than 1.5-fold and deactivation of very few genes (62). Recent data have suggested a role for Isw1 in transcription elongation and termination (46). Chromatin immunoprecipitation (ChIP) experiments at the MET16 gene showed that Isw1 distribution changed upon gene activation. Isw1 localized to the promoter under repressive (+Met) conditions but localized within the open reading frame (ORF) under activating conditions. However, the generality of this model for Isw1 function has not been addressed.
One possible explanation for the lack of an isw1 phenotype is that Isw1 functions in parallel with other factors and the activities of these factors mask the effects of loss of Isw1. To test this hypothesis, we sought to identify genetic backgrounds and conditions that require Isw1 for normal growth. We observed strong genetic interactions between isw1 and mutations affecting the histone H4 tail. This led us to identify three-way genetic interactions among Isw1, the NuA4 histone acetyltransferase, and the Swr1 histone replacement complex. The genetic interactions among these three complexes suggested parallel activities in vivo. Through the analysis of transcription microarray experiments, we identified TATA-containing genes, including a large number of stress-induced genes, as major targets of transcriptional repression by these three chromatin regulators. Unexpectedly, comparison of the transcriptional data with ChIP experiments shows that while the NuA4 and Swr1 complexes act genomewide, only a specific subset of the genome is transcriptionally sensitive to their loss.
MATERIALS AND METHODS
Media and strains.
Yeast strains were grown and manipulated according to standard procedures. All of the strains used in this study were derived from the W303 background, in which a rad5 mutation was repaired (59, 65), and are listed in Table 1. Disruption of genes with the dominant selectable drug resistance markers KanMX, NatMX, and HphMX was mediated by a PCR-based strategy with plasmids pUG6, pAG25, and pAG32, respectively (24, 27). Single-mutant strains were created by single-step deletions in haploid strains. Double- and triple-mutant strains were created either by sequential knockouts in the W1665 diploid strain or by mating, followed by sporulation and tetrad dissection. To test for genetic interactions with the histone N-terminal tails, we successively knocked out either both copies of histones H3 and H4 or both copies of histones H2A and H2B in the wild-type and isw1 backgrounds. The histone deletions were covered by a URA3-marked centromeric plasmid carrying a wild-type copy of the deleted histones. We then used the plasmid shuffle technique to exchange the URA3-marked wild-type copies of these histones for TRP1-marked plasmids carrying a wild-type copy of one of the histones and a mutant copy of the other histone. Plasmids containing point mutations in the histone H4 NH2-terminal tail were derived from pMP3, which contains the HHT2 and HHF2 alleles of histones H3 and H4 (33). Mutations were introduced into pMP3 by site-directed mutagenesis (QuikChange kit; Stratagene). Primers sequences are available upon request.
TABLE 1.
Strains used in this study
| Strain | Description |
|---|---|
| W1588-4C | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 RAD5+ |
| W1665 | MATa/α ade2-1/ade2-1 can1-100/can1-100 his3-11,15/his3-11,15 leu2-3,112/leu2-3,112 trp1-1/trp1-1 ura3-1/ura3-1 RAD5+/ RAD5+ |
| YTT0441a | MATaisw1::KanMX |
| YTT2545 | MATahta1-htb1::HphMX hta2-htb2::NatMX(pRS416-HTA1-HTB1) |
| YTT2568 | MATahta1-htb1::HphMX hta2-htb2::NatMX isw1::KanMX (pRS416-HTA1-HTB1) |
| YTT1969 | MATahht1-hhf1::HphMX hht2-hhf2::NatMX(pRS416-HHT2- HHF2) |
| YTT1971 | MATα hht1-hhf1::HphMX hht2-hhf2::NatMX isw1::KanMX(pRS416-HHT2-HHF2) |
| YTT2256 | MATayng2::NatMX |
| YTT2258 | MATayng2::NatMX isw1::KanMX |
| YTT2329 | MATaeaf1::NatMX |
| YTT2331 | MATaeaf1::NatMX isw1::KanMX |
| YTT2462 | MATaeaf3::KanMX |
| YTT2533 | MATaeaf3::KanMX isw1::NatMX |
| YTT2586 | MATayaf9::KanMX |
| YTT2622 | MATayaf9::KanMX isw1::NatMX |
| YTT3026 | MATα msn2::KanMX msn4::HphMX |
| YTT3067 | MATα msn2::KanMX msn4::HphMX yaf9::KanMX isw1::NatMX |
| YTT3085 | MATaswr1::HphMX |
| YTT3087 | MATaswr1::HphMX isw1::KanMX |
| YTT3119 | MATahtz1::HphMX isw1::KanMX |
| YTT3122 | MATahtz1::HphMX |
| YTT3174 | MATaeaf5::HphMX |
| YTT3178 | MATaeaf5::HphMX isw1::KanMX |
| YTT3182 | MATaeaf6::HphMX |
| YTT3186 | MATaeaf6::HphMX isw1::KanMX |
| YTT3263 | MATaeaf5::NatMX swr1::HphMX |
| YTT3265 | MATaeaf5::NatMX swr1::HphMX isw1::KanMX |
| YTT2323 | MATaesa1::NatMX(pSAPE1-CEN-LEU2-6xHA-ESA1) |
| YTT2434 | MATaesa1::NatMX(pSAPE5-CEN-LEU2-6xHA-esa1-L254P) |
| YTT2435 | MATaesa1::NatMX(pSAPE10-CEN-LEU2-6xHA-esa1-L357H) |
| YTT2436 | MATaesa1::NatMX isw1::KanMX(pSAPE1-CEN-LEU2-6xHA- ESA1) |
| YTT2438 | MATaesa1::NatMX isw1::KanMX(pSAPE5-CEN-LEU2-6xHA- esa1L254P) |
| YTT2439 | MATaesa1::NatMX isw1::KanMX(pSAPE10-CEN-LEU2-6xHA- esa1-L357H) |
All YTT strains were derived from W1558-4C or W1665.
Expression analysis.
RNA was harvested from cultures grown at 30°C in YEPD medium (2% Bacto Peptone, 1% yeast extract, 2% glucose) to an optical density at 600 nm (OD600) of 0.7 by the hot-phenol method. Northern blot assays were run as described previously (23), except that 30 μg of total RNA was loaded into each lane. Northern blot assays were probed with [α-32P]dCTP-labeled probes and quantified with a Phosphorimager and ImageQuant v 1.2 (Amersham Biosciences) software. DNA microarray analysis was performed with 30 μg of total RNA as described previously (18). For each mutant, three independent RNA preparations were independently labeled as dye swap pairs (a total of six arrays per mutant). Image analysis was performed with GenePix Pro v 6.0 (Molecular Devices). The raw data were then filtered for signal quality (3 standard deviations above background) and spot quality (minimum diameter). The data were subjected to Lowess normalization with GeneTraffic v 3.2 (Iobion). The data were then exported for input into Cyber-T (2) to assign Bayes P values to determine for each ORF whether the mutant was significantly different from the wild type. Changes in relative expression were identified as significant by ranking the Bayesian P values and applying a false discovery rate algorithm to account for multiple testing (4). The false discovery rate threshold was set at 5%. In addition, we subsequently required log2(ratio) ≥ 0.585 for all values that met the above criteria. If, for any mutant, an ORF was determined to be significantly different from the wild type, this ORF was included in the cluster analysis. Cluster analysis was performed with Cluster v 2.12 (17) and visualized with Treeview v 1.6 (17). Clusters were analyzed for enrichment of gene classes with FunSpec (54). The complete microarray data set is available at www.fhcrc.org/science/labs/tsukiyama/supplemental_data/Isw1_NuA4-Swr1_transcript_arrays.htm/.
Fluorescence microscopy.
Wild-type and mutant strains were transformed with plasmid pADH1-MSN2-GFP, and transformants were selected on yeast complete (YC)-Leu plates. Transformants were inoculated into YC-Leu liquid medium and grown at 30°C to early log phase. Strains were then switched to YEPD and grown at 30°C for an additional 6 h to an OD600 of 0.6 before being fixed with 4% paraformaldehyde and stained with 4′,6′-diamidino-2-phenylindole (DAPI; 1 μg/ml; Molecular Probes). Green fluorescent protein (GFP) and DAPI fluorescence was visualized with a Nikon E600 microscope. Images were analyzed and merged with MetaMorph (Molecular Devices) software.
ChIP.
ChIP was performed as previously described (23), with the following modifications. Cells were grown at 30°C to mid-log phase before being cross-linked at room temperature with 1% formaldehyde for 5 min. Twenty microliters of protein G Dynabeads (Dynal) was prebound overnight at 4°C with 2 μl of the tetra-acetylated histone H4 antibody (Upstate Biotechnology). A detailed protocol is available at http://www.fhcrc.org/science/labs/tsukiyama/.
RESULTS
The chromatin-remodeling enzyme Isw1 interacts genetically with the acetylatable lysines of the histone H4 tail.
It was previously shown that the Isw2 chromatin-remodeling complex interacts genetically with the Sin3/Rpd3 histone deacetylase complex (18, 23). The parallel function of the Sin3/Rpd3 complex is able to partially mask the effect of an isw2 mutation and therefore obscure aspects of its function (18). A candidate gene approach looking at a number of histone deacetylases did not reveal such genetic interactions with isw1 (data not shown). We therefore sought a forward genetics approach to identify factors that function in parallel with Isw1, with the goal of revealing its in vivo functions.
On the basis of the hypothesis that Isw1 functions in parallel with histone-modifying enzymes, we first asked if isw1Δ showed genetic interactions with deletions of the most common substrates of histone-modifying activity, the N-terminal histone tails. isw1 showed relatively modest genetic interactions with the histone H2A and H3 tails and strong synthetic temperature sensitivity with the histone H4 tail (Fig. 1A).
FIG. 1.
isw1 interacts genetically with mutations of the N-terminal histone tails. The plasmid shuffle technique was used to construct strains carrying either deletions of individual histone N-terminal tails (A) or K-to-R point mutations in the histone H4 tail (B) in both wild-type (WT) and isw1 strains. Cultures were diluted to an OD600 of 0.5, and 10-fold serial dilutions were spotted on rich medium and grown at the indicated temperatures for 3 days.
As histone H4 showed the strongest genetic interaction with isw1, we focused our attention on key lysine residues of the H4 tail to identify histone-modifying enzymes which might collaborate with Isw1 in vivo. The deletion of the histone H4 tail removed residues 4 to 19, covering lysines 5, 8, 12, and 16 of the H4 tail, which are acetylatable in yeast. To determine if these residues were responsible for the genetic interaction with isw1, we tested a panel of lysine-to-arginine point mutants in all possible single, double, and triple combinations. There was no temperature sensitivity when isw1 was combined with single-point mutants. However, isw1 in combination with any of the six possible double-point mutants showed modest synthetic temperature sensitivity at 37°C (Fig. 1B and data not shown). Addition of a third K-to-R substitution resulted in absence of growth at 37°C. The genetic interactions between isw1 and the acetylatable lysines of the histone H4 tail suggested that Isw1 may function in parallel with a histone acetyltransferase complex.
Isw1 interacts genetically with the NuA4 histone acetyltransferase and the Swr1 histone replacement complexes.
In yeast, there are two known histone H4 histone acetyltransferases, the SAS and NuA4 complexes that contain the catalytic subunits Sas2 and Esa1, respectively. While SAS shows a strong substrate preference for lysine 16, NuA4 is able to acetylate all four lysines of the histone H4 tail (57). Given the broad specificity of the genetic interaction between isw1 and all four lysines of the H4 tail, we predicted that the synthetic phenotypes might be due to diminished histone H4 acetylation by the NuA4 complex.
To test this hypothesis, we created deletions of the genes encoding several nonessential subunits of the NuA4 complex, including EAF1, EAF3, EAF5, EAF6, YNG2, and YAF9, and crossed these with our isw1 deletion strain. isw1 showed synthetic temperature sensitivity in each of the NuA4 double mutants tested (Fig. 2A; data not shown). isw1 also showed modest synthetic temperature sensitivity when combined with point mutations in the essential catalytic subunit of NuA4, esa1-L254P and esa1-L357H (data not shown). These genetic interactions suggest that the synthetic temperature sensitivity seen in strains carrying mutations in both isw1 and the histone H4 tail are due, at least in part, to impaired histone acetylation by the NuA4 complex. Furthermore, the genetic interactions between isw1 and mutations in the NuA4 complex indicate that these two chromatin regulators might have parallel functions in vivo.
FIG. 2.
isw1 interacts genetically with the NuA4 histone acetyltransferase and the Swr1 histone replacement complex. Cultures were diluted to an OD600 of 0.5, and 10-fold serial dilutions were spotted on YEPD medium and grown at the indicated temperatures for 3 days. (A) Single and double mutants affecting Isw1 and the NuA4 complex. (B) Single, double, and triple mutants affecting Isw1 and the NuA4 and Swr1 complexes. WT, wild type.
In addition to its functions in the NuA4 histone acetyltransferase, Yaf9p was also identified as a subunit of the Swr1 histone replacement complex (6, 39, 63). Given the strong synthetic phenotype between isw1 and yaf9, we tested whether this genetic interaction could be attributed solely to Yaf9p function in the NuA4 complex or if there might also be some contribution due to Yaf9p function in the Swr1 complex.
To test for genetic interactions between isw1 and the Swr1 complex, we created double-mutant strains carrying the isw1 deletion in combination with deletions of the nonessential SWR1 and HTZ1 genes. Both the swr1 isw1 and htz1 isw1 strains showed significant synthetic growth defects, indicating that the strong genetic interaction between isw1 and yaf9 is likely due to the role of Yaf9 in both complexes (Fig. 2B). These genetic data are consistent with a recent report which identified genetic interactions between isw1 and members of both the NuA4 and Swr1 complexes with commercially available knockout strains (36). Furthermore, double mutation of the EAF5 and SWR1 genes, specifically affecting the NuA4 and Swr1 complexes, respectively, also showed synthetic temperature sensitivity and that loss of Isw1 in an eaf5 swr1 mutant exacerbated that phenotype. If the severity of a defect in a double mutant exceeds that of either single mutant, it predicts that the two genes likely do not act in a linear pathway, given that at least one of the mutations is a null allele. The isw1, swr1, and htz1 deletions represent null mutations in the Isw1 and Swr1 complexes. Therefore, the genetic interactions predict that the NuA4, Swr1, and Isw1 complexes function in parallel pathways in vivo.
Loss of Isw1, Swr1, and NuA4 activities results in increased sensitivity to the stress-mimetic drug rapamycin.
In phenotypic assays, we observed no isw1-dependent enhancement of many previously reported phenotypes of NuA4 and Swr1 mutants, including sensitivity to the DNA-damaging agents methyl methanesulfonate, camptothecin, and UV, to the microtubule-depolymerizing agent benomyl, or to the replication inhibitor hydroxyurea (data not shown). In contrast, the loss of Isw1 function in an htz1, swr1, or eaf1 mutant background had synthetic effects on the strain's ability to grow in the presence of rapamycin (Fig. 3). While loss of Isw1 function in an eaf5 mutant background had no visible effects on growth at this concentration of rapamycin, the swr1 eaf5 double mutant showed enhanced sensitivity to the drug. Most striking was the sensitivity of the swr1 eaf5 isw1 mutant, which displayed a total absence of colony formation, even 22 days after plating. Rapamycin inhibits the TOR signaling pathway, one of the major pathways regulating cellular response to stress. It has been shown previously that Esa1, the catalytic subunit of NuA4, functions downstream of TOR signaling to positively regulate the transcription of ribosomal protein genes (52) and that NuA4 mutants are sensitive to the drug rapamycin (7). The specificity of the synthetic sensitivities of the NuA4, Swr1, and Isw1 complexes to rapamycin suggests that these three complexes function in parallel for a subset of their respective cellular functions.
FIG. 3.
Mutations affecting the Isw1, NuA4, and Swr1 complexes show synthetic sensitivity to rapamycin. Cultures of single, double, and triple mutants were diluted to an OD600 of 0.5 and then serially diluted 10-fold and plated on rich medium and on rich medium containing 6.25 mM rapamycin. These plates were grown at 23°C. The plate without rapamycin is shown on day 4. The rapamycin plate shown is at day 16. WT, wild type.
Isw1 functions in parallel with the NuA4 and Swr1 complexes to regulate stress-induced gene transcription.
Since the Isw1, NuA4, and Swr1 complexes have each been implicated in the regulation of gene expression (15, 42, 46, 52, 62, 63), we used transcription microarrays to identify gene targets that might be regulated in parallel by these chromatin-modifying complexes. Labeled RNAs from single, double, and triple mutants in the Isw1, NuA4, and Swr1 complexes were competed against labeled RNA from wild-type cells on microarray chips. Statistical methods were used to determine which genes showed significant changes from the wild-type expression pattern (see Materials and Methods for details). Those genes that passed the filtering criteria in any of the 13 mutants, approximately 915 ORFs, were then used in a hierarchical cluster analysis to identify groups of genes which showed common patterns of defective regulation across all of the mutants (Fig. 4A; see Fig. S1 in the supplemental material).
FIG. 4.
Isw1 functions in parallel with the NuA4 and Swr1 complexes to regulate transcription. RNA from three independent RNA preparations from single, double, and triple mutants in the Isw1, NuA4, and Swr1 complexes was labeled and competed against labeled RNA from wild-type (WT) cells on microarray chips. Each labeling was performed in duplicate as a fluor reverse pair. The data were then subjected to Lowess normalization and statistical algorithms to determine which genes showed significant changes from the wild-type expression pattern (see Materials and Methods for details). (A) Genes which passed our filtering criteria for any one of the 13 mutants were used for two-dimensional cluster analysis to identify groups of genes which showed common and dissimilar patterns of defective regulation across all of our mutants. Red indicates derepression, while green indicates deactivation. The black bars to the right indicate the boundaries of cluster I and cluster II, which were selected on the basis of the vertical dendrogram, which represented a >90% correlation coefficient (pink). (B) Venn diagram analysis depicting the overlap of affected genes (derepressed >1.5-fold) in the indicated mutants. Each of the pairwise overlaps is statistically significant by Fisher's exact test. (C) Quantitation of Northern blot analysis on several genes in cluster II, confirming the results of the microarray analysis. Total cellular RNA was prepared from logarithmic cultures of wild-type, isw1, yaf9, and yaf9 isw1 strains. Northern blot assays were probed with labeled DNA fragments of the DIA1, PUT1, YGP1, and SPI1 genes. The signal was quantitated by phosphorimager and normalized to the ACT1 signal. The values are averages of three independent RNA preparations, and error bars indicate standard deviations.
The isw1 single mutant, as previously described (62), has relatively minor transcriptional defects. However, when combined with a mutation in either the NuA4 or the Swr1 complex, loss of Isw1 function exacerbates the transcriptional defects seen in an NuA4 or Swr1 complex single mutant. The cluster analysis revealed striking similarities between the transcriptional profiles of the mutants in the NuA4 complex and the Swr1 complex. Pairwise comparisons showed statistically significant overlap in the genes affected among the double mutants tested (Fig. 4B and data not shown) and the strongest defects in the eaf5 swr1 isw1 triple mutant, suggesting that these three complexes function in independent, parallel pathways to regulate the transcription of many genes.
Cluster analysis revealed two large clusters with correlation coefficients of greater than 90% containing genes that were either deactivated (cluster I) or derepressed (cluster II) to various degrees in each of the mutants (Fig. 4A). Genes associated with TATA-containing promoters were significantly overrepresented in both cluster I (P value = 0.0052) and cluster II (P value < 0.0001), suggesting that TATA-containing promoters may be strongly affected by the activities of the Isw1, NuA4, and Swr1 chromatin regulators. Cluster I contains 180 genes, enriched for genes involved in metabolism (P < 1e −14) (54), that are deactivated in the NuA4 and Swr1 mutants. Cluster II contains 368 genes that are derepressed in the NuA4 and Swr1 mutants; within this cluster, loss of Isw1 in an NuA4 or Swr1 mutant background resulted in even greater levels of gene derepression (Fig. 3B). Genes classified in the gene ontology as having functions related to the electron transport chain (P < 1e −10) and the stress response (P < 1e −8) were significantly overrepresented in cluster II (54). Specifically, a significant number of genes within this cluster are positively regulated by the general stress-responsive transcription factors Msn2 and Msn4 (P value ≤ 0.0001) (Table 2) (22). In striking contrast to the derepression of many stress-inducible genes, we did not detect significant deactivation of stress-repressed genes in these arrays. This indicates that the transcriptional profile of the mutants is different from that of the general stress response and indicates that the NuA4, Swr1, and Isw1 complexes may play a specific role in the repression of stress-induced genes. In this respect, it is worth noting that TATA-containing promoters are significantly overrepresented among stress-induced genes and significantly underrepresented among stress-repressed genes (3). Importantly, the enrichment of TATA-regulated genes was significant for cluster II even when stress response genes were excluded from the analysis (P value < 0.0001).
TABLE 2.
Derepression of Msn2/Msn4-regulated genes in an swr1 eaf5 isw1 mutanta
| Systematic name | Common name | n-fold change | TATA? |
|---|---|---|---|
| YFL014W | HSP12 | 30.02 | Yes |
| YBR072W | HSP26 | 21.32 | Yes |
| YER150W | SPI1 | 4.61 | Yes |
| YGR088W | CTT1 | 4.153 | Yes |
| YMR090W | 4.094 | Yes | |
| YDL124W | 3.948 | Yes | |
| YCL040W | GLK1 | 3.767 | Yes |
| YJL161W | 3.662 | Yes | |
| YML100W | TSL1 | 3.656 | Yes |
| YGL121C | GPG1 | 3.564 | Yes |
| YLR142W | PUT1 | 3.516 | Yes |
| YLR178C | TFS1 | 3.39 | Yes |
| YDR070C | 3.348 | Yes | |
| YNL160W | YGP1 | 3.127 | |
| YPL223C | GRE1 | 3.12 | Yes |
| YML128C | MSC1 | 3.076 | Yes |
| YKL151C | 3.01 | Yes | |
| YOR289W | 2.988 | ||
| YGR008C | STF2 | 2.72 | Yes |
| YGL037C | PNC1 | 2.674 | Yes |
| YDL024C | DIA3 | 2.658 | |
| YDL204W | RTN2 | 2.657 | Yes |
| YHR139C | SPS100 | 2.651 | Yes |
| YOR173W | DCS2 | 2.597 | Yes |
| YMR169C | ALD3 | 2.583 |
The top 25 Msn2/Msn4-regulated cluster II genes and the n-fold change in the swr1 eaf5 isw1 mutant compared to the wild type are shown. Change values represent the average of three independent biological replicates and two technical (dye swap) replicates. Msn2/Msn4 dependence was determined as reported in reference 22. The presence of a TATA element was determined as reported in reference 3.
NuA4 has previously been linked to transcriptional activation, while ISWI complexes have been implicated in repression of transcription. The synthetic effects of an isw1 mutation in a NuA4 mutant background suggest that Isw1 and NuA4 may have parallel functions in the repression of transcription of genes within cluster II. We compared our transcription data with previously published array data examining the transcription defects in histone H4 lysine to arginine point mutants (13) and found that many of the genes derepressed in the NuA4, Swr1, and Isw1 mutants were also significantly derepressed in the H4 tail mutants (295 of 368 [see Fig. S2 in the supplemental material]). These results support the hypothesis that the genes in cluster II identify a previously undescribed function for NuA4-dependent acetylation in repression of transcription at specific loci. Furthermore, the synthetic effects of mutations affecting Isw1 and the Swr1 complex suggest that all three complexes collaborate to affect this repression.
We confirmed the pattern of expression for genes within cluster II by quantitative Northern blot analysis. With wild-type, isw1, yaf9, and yaf9 isw1 strains, we probed several genes that were identified by cluster analysis as being derepressed across all of the mutants (Fig. 4C). Consistent with the microarray results, each of the genes is derepressed in the yaf9 mutant, and that the derepression was further enhanced by loss of Isw1 activity, confirming that Isw1, NuA4, and Swr1 complexes function independently in the repression of stress-inducible genes.
The upregulation of a large number of stress-induced genes, including a large number of Hsf1-regulated heat shock genes, in the NuA4 and Swr1 complex mutants was unexpected, given the synthetic sensitivity of the mutants to both high temperature and rapamycin. One possible explanation for these results is that derepression of these genes may lead to an adaptation in the mutant strains that leads to a failure of resistance when cells are challenged with heat or rapamycin. However, the yaf9 isw1 mutant shows a normal transcriptional response at both early and late time points (data not shown), indicating that the primary transcriptional response is not disrupted in the mutant. Therefore, it is likely that upregulation of these genes is simply insufficient to protect cells from these stresses.
Derepression of Msn2/Msn4 target genes in the yaf9 isw1 mutant occurs through an Msn2/Msn4-dependent pathway.
In order to understand the pathway by which stress-inducible genes are derepressed in the yaf9 isw1 mutant, we asked whether the increased expression of these genes occurred through their normal transcription factors. On the basis of the observation that a large number of Msn2/Msn4-regulated genes were derepressed, we asked whether these genes were still derepressed in the absence of the Msn2 and Msn4 transcription factors.
To this end, we created strains carrying deletions of MSN2 and MSN4 in the wild-type and yaf9 isw1 backgrounds and performed Northern blot analysis on these strains, probing for Msn2/Msn4-dependent and -independent genes. The Msn2/Msn4-independent gene DIA1 served as a negative control, and as expected, loss of Msn2 and Msn4 had no effect on the level of DIA1 derepression in the yaf9 isw1 background (Fig. 5). In contrast, for the SPI1 and YGP1 genes, which are positively regulated by Msn2 and Msn4 in response to a variety of stresses (22), loss of Msn2 and Msn4 in the yaf9 isw1 background abrogated the derepression of these genes. Additionally, although multiple stress-responsive pathways activate the HSP12 and HSP26 genes, we found that the derepression of these genes in the yaf9 isw1 mutant was fully dependent on the Msn2 and Msn4 transcription factors. These results indicate that derepression of a subset of the stress-inducible genes misregulated in the yaf9 isw1 mutant occurs through a pathway requiring Msn2 and Msn4.
FIG. 5.
Derepression of Msn2/Msn4-regulated genes in the yaf9 isw1 mutant is dependent on Msn2 and Msn4. Total cellular RNA was prepared from logarithmic-phase cultures of wild-type, yaf9 isw1, msn2 msn4, and yaf9 isw1 msn2 msn4 strains. Northern blot assays were probed with labeled DNA fragments of the DIA1, SPI1, and YGP1 genes. The signal was quantitated by phosphorimager and normalized to the ACT1 signal. The values are averages of three independent RNA preparations, and error bars indicate standard deviations. WT, wild type.
The Msn2 transcription factor shows normal localization in the yaf9 isw1 mutant.
One possible model for the derepression of stress-inducible genes is that the loss of Isw1, Swr1, and NuA4 activities cause the mutant strains to experience stress and therefore indirectly activate the expression of Msn2/Msn4 target genes. To examine this model, we sought a system to gauge the relative stress in our mutants.
The localization of the Msn2 and Msn4 transcription factors provides a sensitive readout of the stress load on a given cell. Msn2 and Msn4 are regulated in part through their subcellular localization. Under normal growth conditions, most of the Msn2 and Msn4 protein is localized in the cytoplasm. Upon exposure to stress, however, Msn2 and Msn4 accumulate in the nucleus and activate the transcription of their target genes (25). Therefore, we used Msn2 localization to test whether loss of Yaf9 and Isw1 functions resulted in increased stress on the mutant cells.
We utilized a GFP-tagged MSN2 system to determine the localization of Msn2 protein (25, 26) in both wild-type and yaf9 isw1 cells. We did not observe significant accumulation of Msn2-GFP in either wild-type or yaf9 isw1 strains under normal growth conditions (Fig. 6A). As a positive control, we exposed wild-type and yaf9 isw1 strains to acute osmotic stress, and as expected, Msn2-GFP localized normally in both backgrounds (Fig. 6B).
FIG. 6.
Msn2-GFP is localized normally in yaf9 isw1 mutant cells. A plasmid-borne, GFP-tagged version of the Msn2 protein was overexpressed from the ADH1 promoter in wild-type (WT) and yaf9 isw1 cells. Cells were grown in liquid YEPD for 6 h to an OD600 of 0.6 and then (A) left untreated or (B) exposed to 1 M NaCl for 15 min. Cells were fixed with paraformaldehyde, DAPI stained, and examined by fluorescence microscopy to determine the localization of Msn2-GFP. Representative images are shown. DIC, differential interference contrast.
The cytoplasmic localization of Msn2-GFP under normal growth conditions suggests that the yaf9 isw1 mutant is not subject to increased stress relative to wild-type cells and that derepression of Msn2/Msn4-dependent stress-inducible genes is not simply the result of increased nuclear localization of these factors. These results, taken together with the lack of stress-repressed gene deactivation in the microarray analysis, indicate that the mutants do not display important characteristics of a general stress response.
Defects in histone H4 acetylation occur at promoters throughout the genome in the yaf9 isw1 mutant.
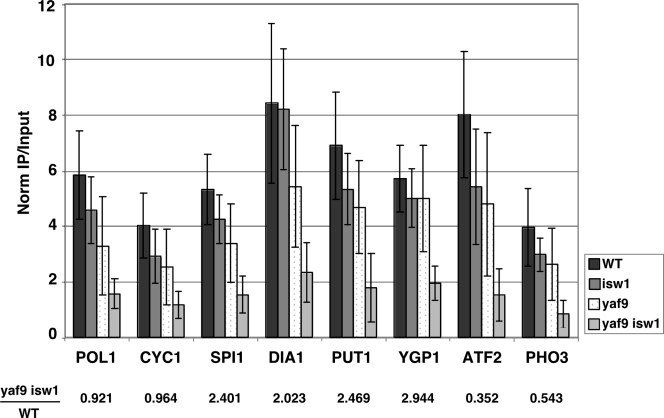
The lack of a detectable stress response in yaf9 isw1 cells suggests that the derepression of Msn2/Msn4-regulated genes may be a direct effect. We therefore used ChIP to assay for defects in the level of histone H4 acetylation in isw1, yaf9, and yaf9 isw1 mutants at several cluster II genes which are derepressed in the mutants (SPI1, DIA1, PUT1, and YGP1), as well as cluster I genes which are deactivated in the mutants (ATF2 and PHO3) and genes which show no defects in transcription in any of the arrays (POL1 and CYC1) (Fig. 7). Surprisingly, we saw that each gene tested showed similar defects in histone H4 acetylation. In each case, the yaf9 mutant strain showed a significant decrease in H4 acetylation, and this effect was exacerbated in the yaf9 isw1 strain. The decrease in H4 acetylation was similar at each locus, regardless of the transcriptional output in the yaf9 isw1 strain. These results suggest that the defects in H4 acetylation are occurring throughout the genome but that decreased H4 acetylation only results in altered transcription at a fraction of the affected genes. Our ChIP results are consistent with microarray data analyzing the effects of H4 lysine-to-arginine point mutations on transcription (13). The point mutations abrogate NuA4-dependent histone H4 acetylation genomewide but show specific transcriptional defects, including the marked derepression of a large number of TATA-containing and stress-induced genes.
FIG. 7.
Defects in histone H4 acetylation occur at promoters throughout the genome in the yaf9 isw1 mutant. The relative enrichment of histone H4 acetylation for wild-type (WT), isw1, yaf9, and yaf9 isw1 strains are shown. The values shown are averages of three independent ChIP experiments. The expression of each gene in the yaf9 isw1 mutant relative to that in the wild type is shown at the bottom.
Similarly, we observed by ChIP that Htz1 was present at the promoter of every gene tested (data not shown), in agreement with recently published global Htz1 localization data (40, 51, 64). This too showed that while the Swr1 complex and Htz1 appear to act globally, the effects of diminished Htz1 incorporation are promoter context dependent.
DISCUSSION
Functional connections among the Isw1, Swr1, and NuA4 complexes.
In this study, we identified a three-way genetic interaction among Isw1, the NuA4 histone acetyltransferase, and the Swr1 histone replacement complex, indicating that these complexes collaborate in vivo. Comparison of our microarray data sets from NuA4 mutants to recently published array data examining transcriptional defects in histone H4 lysine to arginine point mutants (13) showed very similar effects on transcription (see Fig. S2 in the supplemental material). These results alone could be predicted, as the H4 tail is the major substrate for NuA4-dependent acetylation. However, it was unexpected that mutations affecting the Htz1 deposition pathway (swr1 and htz1) also showed similar and additive defects in transcription as the NuA4 and histone H4 mutants, indicating that Htz1 deposition and H4 tail acetylation regulate overlapping sets of genes in parallel pathways. In each Swr1 and NuA4 mutant background tested, loss of Isw1 activity exacerbated the transcriptional defects seen in the single mutant. A triple mutant carrying deletions affecting all three complexes (eaf5 swr1 isw1) showed the most severe transcriptional defects compared to the single and double mutants. The tripartite genetic interactions described in this study closely mirror the three-way genetic interactions previously described for Htz1 with the SAGA histone acetyltransferase and the Swi/Snf ATP-dependent chromatin-remodeling complex (55). This suggests that cells may generally utilize histone modification, histone variants, and ATP-dependent chromatin remodeling together in parallel to regulate transcription.
A number of observations from higher eukaryotes have suggested functional links between the Swr1 and NuA4 complexes. Similar to yeast, the mammalian NuA4-like (TRAAP/Tip60) and presumed Swr1-like (SRCAP) complexes share many of the same subunits (8). The similarity of subunit composition raises the possibility that these complexes make similar protein-protein interactions and may have closely related recruitment mechanisms. More recently, the human Esa1 and Swr1 homologs Tip60 and p400 were shown to copurify as a large multisubunit complex from HeLa cells (20), strengthening the possibility that the histone acetyltransferase activity of NuA4 and the histone replacement activity of the Swr1 complex may be required at common genomic locations. Finally, genetic studies of Caenorhabditis elegans have found that the worm homologs of Esa1 and Swr1, mys-1 and ssl-1, are both part of the class C synMuv pathway, which regulates vulval development (10). The similar genetic interactions and mutant phenotypes again suggest a mechanistic link between histone acetylation and histone variant deposition. The results presented in this paper show that this functional conservation extends to S. cerevisiae and furthermore that Isw1 functions in parallel to these two factors.
Mechanisms of stress-inducible gene repression.
The most striking transcriptional defect observed in this study was the upregulation of a large cluster of approximately 360 genes. Genes involved in energy and the stress response were significantly overrepresented in this cluster. In particular, the cluster contained many genes that are regulated by the stress-responsive transcription factors Msn2 and Msn4. We feel it is unlikely that the derepression of stress-inducible genes is an indirect response to low-grade stress in the mutants. Msn2-GFP, which is excluded from the nucleus under nonstress conditions (25), is localized normally in a yaf9 isw1 mutant. Additionally, we do not detect deactivation of the environmental stress response genes that are normally repressed as part of the general stress response (22). Taken together, these results suggest that under normal growth conditions, NuA4, Swr1, and Isw1 activities combine to antagonize the activities of the Msn2 and Msn4 transcription factors, and this occurs through a pathway independent of Msn2 and Msn4 localization.
Our microarray results agree with a previous report that noted stress response genes, and specifically many Msn2/Msn4-regulated genes, upregulated in an arp4 mutant strain (26). Arp4 is a subunit of the NuA4 and Ino80 complexes (21, 56) and was also recently identified as a subunit of the Swr1 complex (35, 37, 44). In the previous study, it was proposed that derepression of stress response genes in the arp4 mutant is not due to a defect in NuA4 function, as histone acetylation by NuA4 has typically been linked to transcriptional activation. However, our results clearly link the NuA4 complex to transcriptional repression by showing that multiple subunits of NuA4, including the NuA4-specific subunits Eaf5 and Eaf6, are linked to repression of stress-inducible genes.
One possible mechanism by which NuA4, Swr1, and Isw1 could collaborate to repress Msn2/Msn4-regulated gene transcription under normal growth conditions is targeting of their activities to specific loci to create a local chromatin environment that is not conducive to gene expression, for instance, by preventing the binding of the Msn2 and Msn4 transcription factors which are present in the nucleus at low levels under normal growth conditions. However, this recruitment-based model was not supported by the ChIP data examining H4 histone acetylation presented here, which showed global defects in histone H4 acetylation in the yaf9 isw1 mutant, nor by recently published Htz1 localization data (40, 51, 64) which demonstrate that the NuA4 and Swr1 complexes are functioning similarly at promoters genomewide.
The absence of a correlation between defects in histone H4 acetylation and defects in transcription could be explained if NuA4 plays a role in transcription that is independent of its catalytic activities. However, while this is formally possible, the significant overlap of the transcription profiles presented in this study with the transcription defects in histone H4 lysine-to-arginine point mutants (13) indicates that the mechanism by which NuA4 regulates transcription is predominantly through the lysines of the H4 tail. Additionally, we observed stronger genetic interactions between isw1 and mutations affecting the NuA4 complex which show large defects in H4 acetylation, for example, the eaf1 (35) and yng2 (11) deletion mutants, compared to mutations which do not strongly affect global H4 acetylation levels, such as eaf5 (36) and eaf3 (53). Ultimately, we prefer a model in which the relative sensitivity of specific genes to the activity of the NuA4 complex is due to additional gene-specific factors such as promoter structure.
We therefore suggest that the NuA4, Swr1, and Isw1 complexes collaborate to establish normal chromatin structure genomewide, but the consequences of their activities on transcription are promoter context dependent. Detailed mechanistic studies of transcription activation have revealed a number of distinct modes of transcriptional regulation at different model genes (12), suggesting that there are many factors which contribute to the promoter context of individual loci. These factors may include the number and spacing of transcription factor binding sites, nucleosome packing, and location within the nucleus, as well as higher-order chromatin structure. Additionally, the presence or absence of a functional TATA element seems to greatly impact the mechanisms of transcriptional activation. Two recent studies classified yeast promoters into two general categories: a small proportion of the genome which is induced in response to a variety environmental conditions, generally TATA-containing, and strongly regulated by chromatin regulators, and a second, larger class of “housekeeping genes” which is generally TATA-less and less highly regulated (3, 29). TATA-containing promoters generally rely heavily on the activities of the SAGA complex, as well as histone H3 and H4 tails for proper regulation, while TATA-less promoters typically require the activity of TFIID for transcription. The results presented here describe an additional layer of regulation for TATA-containing promoters, specifically, the dependence of TATA-regulated genes on the activities of the NuA4 and Swr1 complexes for normal regulation.
Analysis of the transcription microarray data suggests that Isw1 may play a role in TATA gene regulation distinct from that of the NuA4 and Swr1 complexes. The transcription profile of the isw1 single mutant does not show a bias toward TATA-containing genes, but rather the loss of Isw1 activity enhances the derepression seen in NuA4 and Swr1 mutants. A previously published report indicates that Isw1 functions in steps downstream of transcription initiation through the negative regulation of RNA Pol II elongation (45). Our results are therefore consistent with the possibility that loss of Isw1 contributes to derepression of TATA-containing genes in the NuA4 and Swr1 mutants by allowing enhanced passage of RNA Pol II at the derepressed genes.
Supplementary Material
Acknowledgments
We thank Sue Biggins and members of the Tsukiyama lab for critical reading of the manuscript. We also thank members of the Gottschling, Henikoff, and Biggins labs for helpful discussions during the course of this work, the FHCRC Genomics Facility for processing samples with expedience and care, Chitra Kotwaliwale for assistance with fluorescence microscopy, and Derek Lindstrom for advice and support. We are grateful to Jacques Côté for strains and for providing the plasmids carrying Esa1 point mutants, to Christoph Shüller for generously sending us the pADH1-MSN2-GFP plasmid, and to Mitch Smith for strains. We thank Haiying Zhang and Brad Cairns for sharing array data and primer sequences.
This work was supported by NIH grants GM58465 to T.T. and GM62970 to M.R.P. K.C.L. was supported in part by a predoctoral fellowship from the NSF. T.T. is a Leukemia and Lymphoma Society scholar.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Babiarz, J. E., J. E. Halley, and J. Rine. 2006. Telomeric heterochromatin boundaries require NuA4-dependent acetylation of histone variant H2A. Z in Saccharomyces cerevisiae. Genes Dev. 20:700-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldi, P., and A. D. Long. 2001. A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics 17:509-519. [DOI] [PubMed] [Google Scholar]
- 3.Basehoar, A. D., S. J. Zanton, and B. F. Pugh. 2004. Identification and distinct regulation of yeast TATA box-containing genes. Cell 116:699-709. [DOI] [PubMed] [Google Scholar]
- 4.Benjamini, Y., and Y. Hochberg. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57:289-300. [Google Scholar]
- 5.Bird, A. W., D. Y. Yu, M. G. Pray-Grant, Q. Qiu, K. E. Harmon, P. C. Megee, P. A. Grant, M. M. Smith, and M. F. Christman. 2002. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature 419:411-415. [DOI] [PubMed] [Google Scholar]
- 6.Bittner, C. B., D. T. Zeisig, B. B. Zeisig, and R. K. Slany. 2004. Direct physical and functional interaction of the NuA4 complex components Yaf9p and Swc4p. Eukaryot. Cell 3:976-983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boudreault, A. A., D. Cronier, W. Selleck, N. Lacoste, R. T. Utley, S. Allard, J. Savard, W. S. Lane, S. Tan, and J. Cote. 2003. Yeast enhancer of polycomb defines global Esa1-dependent acetylation of chromatin. Genes Dev. 17:1415-1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai, Y., J. Jin, L. Florens, S. K. Swanson, T. Kusch, B. Li, J. L. Workman, M. P. Washburn, R. C. Conaway, and J. W. Conaway. 2005. The mammalian YL1 protein is a shared subunit of the TRRAP/TIP60 histone acetyltransferase and SRCAP complexes. J. Biol. Chem. 280:13665-13670. [DOI] [PubMed] [Google Scholar]
- 9.Carrozza, M. J., R. T. Utley, J. L. Workman, and J. Cote. 2003. The diverse functions of histone acetyltransferase complexes. Trends Genet. 19:321-329. [DOI] [PubMed] [Google Scholar]
- 10.Ceol, C. J., and H. R. Horvitz. 2004. A new class of C. elegans synMuv genes implicates a Tip60/NuA4-like HAT complex as a negative regulator of Ras signaling. Dev. Cell 6:563-576. [DOI] [PubMed] [Google Scholar]
- 11.Choy, J. S., and S. J. Kron. 2002. NuA4 subunit Yng2 function in intra-S-phase DNA damage response. Mol. Cell. Biol. 22:8215-8225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cosma, M. P. 2002. Ordered recruitment: gene-specific mechanism of transcription activation. Mol. Cell 10:227-236. [DOI] [PubMed] [Google Scholar]
- 13.Dion, M. F., S. J. Altschuler, L. F. Wu, and O. J. Rando. 2005. Genomic characterization reveals a simple histone H4 acetylation code. Proc. Natl. Acad. Sci. USA 102:5501-5506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ehrenhofer-Murray, A. E. 2004. Chromatin dynamics at DNA replication, transcription and repair. Eur. J. Biochem. 271:2335-2349. [DOI] [PubMed] [Google Scholar]
- 15.Eisen, A., R. T. Utley, A. Nourani, S. Allard, P. Schmidt, W. S. Lane, J. C. Lucchesi, and J. Cote. 2001. The yeast NuA4 and Drosophila MSL complexes contain homologous subunits important for transcription regulation. J. Biol. Chem. 276:3484-3491. [DOI] [PubMed] [Google Scholar]
- 16.Eisen, J. A., K. S. Sweder, and P. C. Hanawalt. 1995. Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res. 23:2715-2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genomewide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fazzio, T. G., C. Kooperberg, J. P. Goldmark, C. Neal, R. Basom, J. Delrow, and T. Tsukiyama. 2001. Widespread collaboration of Isw2 and Sin3-Rpd3 chromatin remodeling complexes in transcriptional repression. Mol. Cell. Biol. 21:6450-6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fazzio, T. G., and T. Tsukiyama. 2003. Chromatin remodeling in vivo: evidence for a nucleosome sliding mechanism. Mol. Cell 12:1333-1340. [DOI] [PubMed] [Google Scholar]
- 20.Fuchs, M., J. Gerber, R. Drapkin, S. Sif, T. Ikura, V. Ogryzko, W. S. Lane, Y. Nakatani, and D. M. Livingston. 2001. The p400 complex is an essential E1A transformation target. Cell 106:297-307. [DOI] [PubMed] [Google Scholar]
- 21.Galarneau, L., A. Nourani, A. A. Boudreault, Y. Zhang, L. Heliot, S. Allard, J. Savard, W. S. Lane, D. J. Stillman, and J. Cote. 2000. Multiple links between the NuA4 histone acetyltransferase complex and epigenetic control of transcription. Mol. Cell 5:927-937. [DOI] [PubMed] [Google Scholar]
- 22.Gasch, A. P., P. T. Spellman, C. M. Kao, O. Carmel-Harel, M. B. Eisen, G. Storz, D. Botstein, and P. O. Brown. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11:4241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goldmark, J. P., T. G. Fazzio, P. W. Estep, G. M. Church, and T. Tsukiyama. 2000. The Isw2 chromatin remodeling complex represses early meiotic genes upon recruitment by Ume6p. Cell 103:423-433. [DOI] [PubMed] [Google Scholar]
- 24.Goldstein, A. L., and J. H. McCusker. 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15:1541-1553. [DOI] [PubMed] [Google Scholar]
- 25.Gorner, W., E. Durchschlag, M. T. Martinez-Pastor, F. Estruch, G. Ammerer, B. Hamilton, H. Ruis, and C. Schuller. 1998. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 12:586-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gorzer, I., C. Schuller, E. Heidenreich, L. Krupanska, K. Kuchler, and U. Wintersberger. 2003. The nuclear actin-related protein Act3p/Arp4p of Saccharomyces cerevisiae is involved in transcription regulation of stress genes. Mol. Microbiol. 50:1155-1171. [DOI] [PubMed] [Google Scholar]
- 27.Guldener, U., S. Heck, T. Fielder, J. Beinhauer, and J. H. Hegemann. 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24:2519-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henikoff, S., and K. Ahmad. 2005. Assembly of variant histones into chromatin. Annu. Rev. Cell Dev. Biol. 21:133-153. [DOI] [PubMed] [Google Scholar]
- 29.Huisinga, K. L., and B. F. Pugh. 2004. A genomewide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol. Cell 13:573-585. [DOI] [PubMed] [Google Scholar]
- 30.Iizuka, M., and M. M. Smith. 2003. Functional consequences of histone modifications. Curr. Opin. Genet. Dev. 13:154-160. [DOI] [PubMed] [Google Scholar]
- 31.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 32.Kamakaka, R. T., and S. Biggins. 2005. Histone variants: deviants? Genes Dev. 19:295-310. [DOI] [PubMed] [Google Scholar]
- 33.Kelly, T. J., S. Qin, D. E. Gottschling, and M. R. Parthun. 2000. Type B histone acetyltransferase Hat1p participates in telomeric silencing. Mol. Cell. Biol. 20:7051-7058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keogh, M. C., T. A. Mennella, C. Sawa, S. Berthelet, N. J. Krogan, A. Wolek, V. Podolny, L. R. Carpenter, J. F. Greenblatt, K. Baetz, and S. Buratowski. 2006. The Saccharomyces cerevisiae histone H2A variant Htz1 is acetylated by NuA4. Genes Dev. 20:660-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kobor, M. S., S. Venkatasubrahmanyam, M. D. Meneghini, J. W. Gin, J. L. Jennings, A. J. Link, H. D. Madhani, and J. Rine. 2004. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A. Z into euchromatin. PLoS Biol. 2:E131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krogan, N. J., K. Baetz, M. C. Keogh, N. Datta, C. Sawa, T. C. Kwok, N. J. Thompson, M. G. Davey, J. Pootoolal, T. R. Hughes, A. Emili, S. Buratowski, P. Hieter, and J. F. Greenblatt. 2004. Regulation of chromosome stability by the histone H2A variant Htz1, the Swr1 chromatin remodeling complex, and the histone acetyltransferase NuA4. Proc. Natl. Acad. Sci. USA 101:13513-13518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krogan, N. J., M. C. Keogh, N. Datta, C. Sawa, O. W. Ryan, H. Ding, R. A. Haw, J. Pootoolal, A. Tong, V. Canadien, D. P. Richards, X. Wu, A. Emili, T. R. Hughes, S. Buratowski, and J. F. Greenblatt. 2003. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell 12:1565-1576. [DOI] [PubMed] [Google Scholar]
- 38.Kurdistani, S. K., and M. Grunstein. 2003. Histone acetylation and deacetylation in yeast. Nat. Rev. Mol. Cell. Biol. 4:276-284. [DOI] [PubMed] [Google Scholar]
- 39.Le Masson, I., D. Y. Yu, K. Jensen, A. Chevalier, R. Courbeyrette, Y. Boulard, M. M. Smith, and C. Mann. 2003. Yaf9, a novel NuA4 histone acetyltransferase subunit, is required for the cellular response to spindle stress in yeast. Mol. Cell. Biol. 23:6086-6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li, B., S. G. Pattenden, D. Lee, J. Gutierrez, J. Chen, C. Seidel, J. Gerton, and J. L. Workman. 2005. Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc. Natl. Acad. Sci. USA 102:18385-18390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu, C. L., T. Kaplan, M. Kim, S. Buratowski, S. L. Schreiber, N. Friedman, and O. J. Rando. 2005. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol. 3:e328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meneghini, M. D., M. Wu, and H. D. Madhani. 2003. Conserved histone variant H2A. Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell 112:725-736. [DOI] [PubMed] [Google Scholar]
- 43.Millar, C. B., F. Xu, K. Zhang, and M. Grunstein. 2006. Acetylation of H2AZ Lys 14 is associated with genomewide gene activity in yeast. Genes Dev. 20:711-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mizuguchi, G., X. Shen, J. Landry, W. H. Wu, S. Sen, and C. Wu. 2004. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303:343-348. [DOI] [PubMed] [Google Scholar]
- 45.Morillon, A., N. Karabetsou, A. Nair, and J. Mellor. 2005. Dynamic lysine methylation on histone H3 defines the regulatory phase of gene transcription. Mol. Cell 18:723-734. [DOI] [PubMed] [Google Scholar]
- 46.Morillon, A., N. Karabetsou, J. O'Sullivan, N. Kent, N. Proudfoot, and J. Mellor. 2003. Isw1 chromatin remodeling ATPase coordinates transcription elongation and termination by RNA polymerase II. Cell 115:425-435. [DOI] [PubMed] [Google Scholar]
- 47.Narlikar, G. J., H. Y. Fan, and R. E. Kingston. 2002. Cooperation between complexes that regulate chromatin structure and transcription. Cell 108:475-487. [DOI] [PubMed] [Google Scholar]
- 48.Nourani, A., R. T. Utley, S. Allard, and J. Cote. 2004. Recruitment of the NuA4 complex poises the PHO5 promoter for chromatin remodeling and activation. EMBO J. 23:2597-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peterson, C. L., and M. A. Laniel. 2004. Histones and histone modifications. Curr. Biol. 14:R546-R551. [DOI] [PubMed] [Google Scholar]
- 50.Pokholok, D. K., C. T. Harbison, S. Levine, M. Cole, N. M. Hannett, T. I. Lee, G. W. Bell, K. Walker, P. A. Rolfe, E. Herbolsheimer, J. Zeitlinger, F. Lewitter, D. K. Gifford, and R. A. Young. 2005. Genomewide map of nucleosome acetylation and methylation in yeast. Cell 122:517-527. [DOI] [PubMed] [Google Scholar]
- 51.Raisner, R. M., P. D. Hartley, M. D. Meneghini, M. Z. Bao, C. L. Liu, S. L. Schreiber, O. J. Rando, and H. D. Madhani. 2005. Histone variant H2A. Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell 123:233-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reid, J. L., V. R. Iyer, P. O. Brown, and K. Struhl. 2000. Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Esa1 histone acetylase. Mol. Cell 6:1297-1307. [DOI] [PubMed] [Google Scholar]
- 53.Reid, J. L., Z. Moqtaderi, and K. Struhl. 2004. Eaf3 regulates the global pattern of histone acetylation in Saccharomyces cerevisiae. Mol. Cell. Biol. 24:757-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Robinson, M. D., J. Grigull, N. Mohammad, and T. R. Hughes. 2002. FunSpec: a web-based cluster interpreter for yeast. BMC Bioinformatics 3:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Santisteban, M. S., T. Kalashnikova, and M. M. Smith. 2000. Histone H2A. Z regulates transcription and is partially redundant with nucleosome remodeling complexes. Cell 103:411-422. [DOI] [PubMed] [Google Scholar]
- 56.Shen, X., G. Mizuguchi, A. Hamiche, and C. Wu. 2000. A chromatin remodelling complex involved in transcription and DNA processing. Nature 406:541-544. [DOI] [PubMed] [Google Scholar]
- 57.Shia, W. J., S. Osada, L. Florens, S. K. Swanson, M. P. Washburn, and J. L. Workman. 2005. Characterization of the yeast trimeric-SAS acetyltransferase complex. J. Biol. Chem. 280:11987-11994. [DOI] [PubMed] [Google Scholar]
- 58.Smith, C. L., and C. L. Peterson. 2005. ATP-dependent chromatin remodeling. Curr. Top. Dev. Biol. 65:115-148. [DOI] [PubMed] [Google Scholar]
- 59.Thomas, B. J., and R. Rothstein. 1989. The genetic control of direct-repeat recombination in Saccharomyces: the effect of rad52 and rad1 on mitotic recombination at GAL10, a transcriptionally regulated gene. Genetics 123:725-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tsukiyama, T. 2002. The in vivo functions of ATP-dependent chromatin-remodelling factors. Nat. Rev. Mol. Cell. Biol. 3:422-429. [DOI] [PubMed] [Google Scholar]
- 61.Tsukiyama, T., J. Palmer, C. C. Landel, J. Shiloach, and C. Wu. 1999. Characterization of the imitation switch subfamily of ATP-dependent chromatin-remodeling factors in Saccharomyces cerevisiae. Genes Dev. 13:686-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vary, J. C., Jr., V. K. Gangaraju, J. Qin, C. C. Landel, C. Kooperberg, B. Bartholomew, and T. Tsukiyama. 2003. Yeast Isw1p forms two separable complexes in vivo. Mol. Cell. Biol. 23:80-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang, H., D. O. Richardson, D. N. Roberts, R. Utley, H. Erdjument-Bromage, P. Tempst, J. Cote, and B. R. Cairns. 2004. The Yaf9 component of the SWR1 and NuA4 complexes is required for proper gene expression, histone H4 acetylation, and Htz1 replacement near telomeres. Mol. Cell. Biol. 24:9424-9436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang, H., D. N. Roberts, and B. R. Cairns. 2005. Genomewide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell 123:219-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhao, X., E. G. Muller, and R. Rothstein. 1998. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol. Cell 2:329-340. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.