Abstract
The repression mechanisms by the nuclear receptor corepressor (N-CoR) of steroid hormone receptor (SHR)-mediated transactivation were examined. Yellow fluorescent protein (YFP)-N-CoR was distributed as intranuclear discrete dots, while coexpression of androgen receptor (AR), glucocorticoid receptor α, and estrogen receptor α ligand-dependently triggered redistribution of YFP-N-CoR. In fluorescence recovery after photobleaching analysis, mobility of the N-CoR was reduced by 5α-dihydrotestosterone (DHT)-bound AR. The middle region of N-CoR mostly contributed to the interaction with agonist-bound SHRs and the suppression of their transactivation function. N-CoR impaired the DHT-induced N-C interaction of AR, and the impaired interaction was dose-dependently recovered by coexpression of SRC-1 and CBP. N-CoR also impaired the intranuclear complete (distinct) focus formation of SHRs. Coexpression of SRC-1 or CBP released YFP-N-CoR or endogenous N-CoR from incomplete foci and simultaneously recovered complete foci of AR-green fluorescent protein. These results indicate that the relative ratio of coactivators and corepressors determines the conformational equilibrium between transcriptionally active and inactive SHRs in the presence of agonists. The intranuclear foci formed by agonist-bound SHRs were completely destroyed by actinomycin D and α-amanitin, indicating that the focus formation does not precede the transcriptional activation. The focus formation may reflect the accumulation of SHR/coactivator complexes released from the transcriptionally active sites and thus be a mirror of transcriptionally active complex formation.
Steroid hormone receptors, including estrogen receptors (ERα and ERβ), progesterone receptors (PR-A and PR-B), androgen receptors (AR), glucocorticoid receptors (GR), and mineralocorticoid receptors (MR), are ligand-inducible transcription factors that specifically regulate expression of target genes involved in metabolism, development, and reproduction. Steroid hormone receptors share a common structural organization containing three functional domains: a variable N-terminal transactivation domain, a highly conserved DNA binding domain, and a moderately well-conserved C-terminal ligand binding domain (5, 57). There are apparent differences in conformation and coregulator binding between the N-terminal region (N) containing a ligand-independent transactivation domain, AF-1, and C-terminal region (C) containing a ligand-dependent transactivation domain, AF-2 (62, 65). It has been reported that an agonist-induced intramolecular interaction, called an N-C interaction, is required for dimerization and full activities of the steroid hormone receptors (18, 31, 54). AR belongs to the steroid hormone receptor superfamily but exhibits some specific characteristics that are different from other members. AR has a particularly long N terminus whose strong autonomous AF-1 transactivation function is dominant compared to its AF-2 function of the C terminus (27, 36). The N terminus of AR is also an important target for coregulator proteins (2, 10).
The activities of nuclear receptors, in general, are modulated by coregulators that are divided into coactivators and corepressors. Two corepressors, nuclear receptor corepressor (N-CoR) and silencing mediator for retinoid and thyroid hormone receptors (SMRT), were initially identified as a component of repression complexes associated with unliganded RAR and TR (8, 21). Both N-CoR and SMRT were also found to interact with antagonist-bound PR and ER to repress their transcriptional activations (1). One model has been proposed that, although corepressors interact with unliganded nuclear receptors and repress their transactivation of target genes, ligand-dependent release of the corepressors leads to recruitment of coactivators and transcriptional activation mediated by ligand-bound receptors (38). The N-CoR and SMRT are thought to recruit histone deacetylase complex (HDAC) in the nuclear receptor complex to reduce the level of histone acetylation and repress transcription (28, 37, 66). Another mechanism has also been advocated in which N-CoR/SMRT may compete with coactivators in interactions with steroid hormone receptors in the presence of antagonists (14, 26, 44).
A highly dynamic and functional architecture that governs gene expression, replication, and repair in the nucleus, including chromatins, proteins, and subnuclear bodies, has emerged in recent years. Some key components are functionally compartmentalized into specialized and punctate subnuclear domains, as documented by biochemical and in situ evidence (47). For example, one of the pre-mRNA splicing factors, SC35, appears as 20 to 40 speckles per nucleus (67). The promyelocytic leukemia gene product controlling aspects of apoptosis, cell proliferation, and transcriptional regulation was found within distinct subnuclear bodies, termed promyelocytic leukemia (PML) nuclear bodies (PML-NBs) (3). Some corepressor complexes were localized in distinct subnuclear bodies called matrix-associated deacetylase nuclear bodies (11, 59). Moreover, in the case of steroid hormone receptors, ligand-dependent subnuclear focus formation is an important characteristic related to the transcriptional activation mediated by the receptors (15, 29, 41, 49, 56). In addition, coactivators, such as SRC-1, TIF2, and CBP, were also recruited into the same subnuclear compartments of the steroid hormone receptors in the presence of ligands (41, 49).
In the present study, the repression mechanism of the N-CoR for agonist-bound steroid hormone receptors was characterized in detail. In the presence of agonists, the N-CoR was redistributed to form intranuclear incomplete foci (speckles) together with steroid hormone receptors, and its mobility was significantly decreased. The middle region of the N-CoR, containing a transcription repression domain, mainly contributed to an interaction with the steroid hormone receptors and repression of the transcriptional activation. In two- and three-dimensional image quantitative analyses, the N-CoR impaired 5α-dihydrotestosterone (DHT)-induced intranuclear distinct/complete focus formation of the AR. The disruption of N-C interaction and/or functional competition with coactivators, such as SRC-1 and CBP, by the N-CoR is considered to be involved in the repression mechanisms of the N-CoR.
MATERIALS AND METHODS
Plasmid constructs.
The full length of the human N-CoR cDNA (for amino acids 1 to 2439) in pEF1-N-CoR-V5His6 (kindly provided by J. Yanagisawa, Tsukuba University, Japan) was inserted into the EcoRI-NotI site of pBS-SK (Stratagene, La Jolla, CA). Next, a SalI enzyme site was added to the C terminus of the N-CoR in the pBS-SK by PCR. An EcoRI-SalI fragment containing the full-length N-CoR was inserted into the cognate sites of pEGFP-C1, pEYFP-C1 (BD Sciences Clontech, Palo Alto, CA), and pFLAG-CMV2 (Sigma-Aldrich Co., St. Louis, MO), respectively, to generate pEGFP-N-CoR, pEYFP-N-CoR, and pFLAG-CMV2-N-CoR. A fragment encoding amino acids 1 to 1798 of the N-CoR, N-CoR(1-1798), was subcloned into the EcoRI-SalI sites of pEGFP-C1, pEYFP-C1, and pFLAG-CMV2 after insertion of a SalI site at the C terminus, and the resulting plasmids were designated pEGFP-N-CoR(1-1798), pEYFP-N-CoR(1-1798), and pFLAG-CMV2-N-CoR(1-1798), respectively. Expression plasmids for the C-terminal amino acids 1803 to 2439 of the N-CoR were generated by PCR amplification of the corresponding region of the N-CoR and subcloned into ApaI-BamHI sites of pEGFP-C1 and pEYFP-C1 or an EcoRI site of pFLAG-CMV2, resulting in pEGFP-N-CoR(1803-2439), pEYFP-N-CoR(1803-2439), and pFLAG-CMV2-N-CoR(1803-2439), respectively. An expression plasmid for yellow fluorescent protein (YFP)-N-CoR(1-1133) was prepared by self-ligation of the expression plasmid for YFP-N-CoR(1-1798) after deletion of the carboxyl-terminal region containing amino acids 1134 to 1798 by KpnI. The fragment encoding amino acids 1134 to 1798 of the N-CoR was subcloned into the KpnI site of pEYFP-C1, and the resulting plasmid was designated pEYFP-N-CoR(1134-1798). Fluorescent proteins were fused to the N termini of these N-CoR fragments. Expression plasmids carrying N-CoR(1-740), N-CoR(742-1798), or N-CoR(1803-2439) were constructed by inserting each N-CoR fragment into the SalI and/or XbaI sites of the pACT vector (Promega Co., Madison, WI) and designated pACT-VP16-N-CoR(1-740), pACT-VP16-N-CoR(742-1798), and pACT-VP16-N-CoR(1803-2439), respectively. Plasmids for analyzing an AR N-C interaction, pACT-VP16-AR-AF-1(1-532) and pBIND-Gal4-AR-AF-2(615-919), were constructed by adding BamHI and XbaI sites into the amino and carboxyl termini of AR fragments and then inserting them into the cognate sites of pACT and pBIND vectors. The validity of each construct was confirmed by DNA sequencing. Plasmids pCMV-AR, pAR-GFP, pCMX-AR-AF-1, pCMV-AR-AF-1-YFP, pCMX-AR-AF-2, pCMX-GRα, pSG5-ERα, pCMV-YFP-SRC-1, pcDNA-SRC-1, pcDNA/mCBP, pGRα-GFP, pERα-GFP, pGL3-PSA, pA3-ERE2, and pGL3-MMTV were prepared as previously described (25, 41, 52, 56, 68). A GRE reporter plasmid was obtained from BD Bioscience Clontech.
Cell culture.
A monkey kidney-derived cell line, COS-7, was obtained from the Riken Cell Bank (Tokyo, Japan). A human prostate cancer cell line, LNCaP, two mouse fibroblast cell lines, 3T3-L1 and NIH 3T3, and a human breast cancer cell line, MCF-7, were obtained from American Type Culture Collection (Manassas, VA). LNCaP, 3T3-L1, and NIH 3T3 cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Invitrogen Corp., Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS) (Cansera International, Inc., Canada) and 100 U/ml penicillin-streptomycin (Invitrogen). MCF-7 cells were cultured in Eagle's minimal essential medium supplemented with 1% nonessential amino acids, 1% sodium pyruvate, 10 μg/ml insulin, 10% FBS, and 100 U/ml of penicillin-streptomycin.
Functional reporter assays.
Luciferase reporter assays were performed as previously described (30). Briefly, cells (1 × 105 cells/well) were split into 12-well plates and cultured at 37°C for 24 h before transfection. The cells were cotransfected with 0.5 μg/well of pGL3-PSA, pGL3-MMTV, or pA3-ERE2 as a reporter, 2 ng/well of pRL-CMV (Promega) as an internal control, and 0.1 to 2.5 μg/well expression plasmids for proteins of interest using 2.7 μl/well of SuperFect transfection reagent (QIAGEN, Hilden, Germany). For coexpression studies, the total amount of vectors added to each well was equalized by the addition of empty vector. At 3 h posttransfection, DMEM containing 10% charcoal-treated FBS was added with or without 10 nM DHT, 100 nM dexamethasone (Dex), 1 μM estradiol (E2), or 10 ng/ml trichostatin A (TSA). After incubation for 20 to 24 h, the cells were lysed in lysis buffer from a luciferase assay kit (Promega) and then subjected to the assay for luciferase activities using the Dual-Luciferase Reporter assay system (Promega) and a Lumat LB 9507 (Berthold Technologies, Bad Wildbad, Germany).
Mammalian two-hybrid assays.
COS-7 and NIH 3T3 cells were seeded into 12-well plates (1 × 105 cells/well). For the mammalian two-hybrid assay, the cells were cotransfected with pG5 (0.1 μg/well), pBIND-Gal4-AR-AF-2 (0.16 μg/well), and pACT-VP16-AR-AF-1 (0.14 μg/well) together with pFLAG-CMV2-N-CoR (0.25, 0.74, or 1.2 μg/well), pFLAG-CMV2-N-CoR(1-1798) (0.2 or 1.0 μg/well), pYFP-N-CoR(1134-1798) (0.14 or 0.7 μg/well), pcDNA-SRC-1 (0.2 or 0.6 μg/well), or pcDNA/mCBP (0.26 or 0.77 μg/well). The total amount of plasmids added to each well was equalized by the addition of empty vectors. After 20 to 24 h of incubation, the cells were lysed and subjected to the assay for luciferase activities.
CoIP.
Coimmunoprecipitation (CoIP) was performed as previously described (25, 30). COS-7 cells were seeded in 100-mm plates (1 × 106 cells/plate). After incubation for 24 h, the cells were cotransfected with 1 μg of pCMV-AR and 5 μg of expression plasmids for green fluorescent protein (GFP)-N-CoR, GFP-N-CoR(1-1798), or GFP-N-CoR(1803-2439). In one experiment, the cells were cotransfected with 1 μg of pCMV-AR and 5 μg of expression plasmids for VP16, VP16-N-CoR(1-740), VP16-N-CoR(742-1798), or VP16-N-CoR(1803-2439). And in another experiment, the cells were cotransfected with 1 μg of pCMV-AR and 5 μg of expression plasmids for YFP, YFP-N-CoR(1-1133), or YFP-N-CoR(1134-1798). Three hours after transfection, the cells were incubated with fresh DMEM in the presence of 10 nM DHT for 20 to 24 h. This was followed by two washes with ice-cold phosphate-buffered saline. The cells were then incubated on ice for 30 min in NEB buffer consisting of 20 mM HEPES-NaOH (pH 7.9), 20% glycerol, 100 mM KCl, 0.2 mM EDTA, and 0.5% NP-40 with a tablet of protease inhibitor cocktail (Sigma). The cells were scraped and then sonicated on ice by Handy Sonic (model UR-20P; Tomyseiko Co. Ltd., Tokyo, Japan) four times for 5 s and then centrifuged at 13,000 rpm for 10 min at 4°C. Supernatants were collected, and the protein concentrations were measured using a BCA protein assay kit (Pierce, Rockford, IL). Each cell lysate was adjusted to 2 mg/ml using NEB buffer. An anti-AR (C19; Santa Cruz Biotechnology, Inc., Santa Cruz, CA) or an anti-VP16 (Santa Cruz) antibody was preincubated with 25 μl of protein A magnetic beads (Amersham Bioscience Corp., Piscataway, NJ) at 4°C for 2 h with agitation. The antibody-bead mixture was added to 200 μl of the cell lysate with 0.05% skim milk. The lysate mixture was incubated at 4°C for 6 h with agitation, and then the beads were washed five times with 500 μl NETN buffer consisting of 20 mM Tris-HCl (pH 7.9), 100 mM NaCl, 1 mM EDTA, 0.5% NP-40, and a tablet of protease inhibitor cocktail. Proteins bound to the antibody were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis using a 6% (for the N-CoR and its mutants) or 10% (for the AR) separating gel and then transferred onto a nitrocellulose membrane (Hybond ECL; Amersham) using a Trans-Blot SD semi-dry transfer cell (Bio-Rad Laboratories, Hercules, CA). The membrane was incubated with the first antibody, anti-GFP (1:200 dilution) or anti-AR (C19, 1:200 dilution) (Santa Cruz), at 4°C overnight. After being washed three times with Tris-buffered saline (TBS)-Tween 20 (10 mM Tris-HCl [pH 8.0], 0.9% NaCl, and 0.05% Tween 20), the membrane was incubated with an appropriate amount of horseradish peroxidase-conjugated anti-rabbit antibody (Amersham) at 25°C for 1 h. After being washed three times with TBS-Tween 20, the membrane was reacted with ECL-plus Western blotting detection reagents (Amersham). The labeled protein bands were visualized and analyzed using a STORM 860 image analyzer (Amersham).
Fluorescence microscopy and image analysis.
Confocal laser scanning microscopy was performed essentially as previously described (12, 30). The COS-7 and LNCaP cells were seeded into 35-mm glass-bottom dishes (2 × 105 cells/dish; Asahi Techno Glass Corp., Chiba, Japan) and transfected with 0.5 to 11 μg/dish of various plasmids using 5 to 20 μl of the SuperFect reagent. After incubation for 24 h, the cells were observed using an LSM 510 META microscope (Carl Zeiss Co. Ltd., Jena, Germany) equipped with a Plan Apochromat 63×, 1.4 numerical aperture oil differential interference contrast immersion objective. For excitation of GFP and YFP, 488-nm and 514-nm argon laser lines were used, respectively. For simultaneous imaging of GFP and YFP, the 488-nm laser line was used for excitation. The detection spectrum range was set from 491 to 576 nm. Images were collected at a 12-bit depth resolution of intensities over 1024 by 1024 pixels. To separate GFP and YFP signals, raw images obtained in a λ mode were subjected to a META unmixing procedure (the emission fingerprinting technique established by Carl Zeiss). During simultaneous multi-imaging, cells showing a similar intensity of each fluorescence protein were selected for further study. Parameters such as laser power, laser line, dichroic beam splitter to separate excitation and emission, and scanning speed were all kept fixed during the same group of experiments. The quantitative analysis of nuclear foci in two-dimensional images was also performed using the Zeiss LSM software (version 3.0) as described previously (12, 43). Cells showing appropriate expression levels of proteins of interest were selected under the microscope, and then fluorescent intensity numerals of each line were scanned and exported to MS Excel for calculation of averages and standard deviations (SD) as well as CV (coefficient of variation) values of the intranuclear fluorescent intensities on each line of interest. The LSM software also constructed fluorescent intensity fluctuation graphs for the line scan of the representative cells. A three-dimensional imaging study was performed essentially in the same manner as previously reported (41, 56) with minor modifications. In brief, a series of 30 to 50 scanning two-dimensional (2D) images were collected for each single nucleus, in which signals for GFP and YFP were simultaneously obtained and subjected to a META unmixing procedure. These tomograms were reconstructed using the three-dimensional analysis software of the TRI graphics program (Ratoc System Engineering, Tokyo, Japan). First, low-intensity background noises were removed from each raw 2D image. Namely, particles became clear as the intensity was decreased from the maximal level and the number of the separated particles became constant at some intensity level, which was set as the cutoff level. This minimum intensity cutoff value was usually 25 to 30% of the dynamic range. Following the treatment with a median filter to remove small background pixels from each 2D image, three-dimensional images were then constructed and the number of foci was calculated. We always put control 2D images of ligand-bound AR foci and confirmed that the constant number was obtained. Both the distribution and calculations of the fluorescent proteins as a distinct volume were thus made possible by removing scattering background fluorescence and lens spherical aberrations and then separating each particle.
FRAP analysis.
Fluorescence recovery after photobleaching (FRAP) analysis was carried out using the LSM 510 META confocal microscope as described previously (12, 50). A region of interest (ROI) representing 20 to 25% of an area of the nucleus was photobleached by a 488-nm laser line for GFP at the maximum power of 50 iterations. After the bleaching, images within the bleached area were taken every 1 s at a resolution of 512 by 512 pixels to monitor the recovery of the fluorescence intensity. The fluorescence recovery was usually incomplete, probably because attenuation of fluorescence occurred during photobleaching and serial scanning. Therefore, we normalized the raw FRAP data (both the intensity and the time) as described by Stenoien et al. (50). The normalized intensities were averaged and plotted against time to make the recovery curve. The logarithmic equation of the recovery curve is y = a · ln(x) + b, where a is the slope, b is the y-axis intercept, ln(x) is the natural logarithm function of the time, and y is the fully normalized intensity at a given time. The half-recovery time (t1/2) was calculated using the following equation: x (when y = 0.5) = e[(0.5 − b)/a]. Each t1/2 was an average of results for at least 20 cells from three independent experiments.
Immunofluorescence staining.
Immunofluorescence staining was performed as previously described (33, 41). Cells were seeded into 8-well Lab-Tek II chamber slide system (2 × 104 cells/well; Nalge Nunc International, Rochester, NY) or 35-mm glass-bottom dishes (2 × 105 cells/dish; Asahi Techno Glass Corp.) and were then transfected with the expression plasmids for proteins of interest. After being washed twice with ice-cold phosphate-buffered saline, the cells were fixed in methanol-acetone (1:1) for 10 min at −20°C. After being incubated in 1× Block-Ace (Dainippon Pharmaceutical Co., Osaka, Japan) for 10 min at 25°C, the cells were incubated with rabbit anti-FLAG antibody (1:200 dilution; Sigma), rabbit anti-AR antibody (1:200 dilution; Santa Cruz), rabbit anti-N-CoR (H303, 1:200 dilution; Santa Cruz), anti-PML (PG-M3, 1:200 dilution; Santa Cruz) or anti-SC35 (1:200 dilution; Sigma) in 0.1× Block-Ace at 25°C for 1 h. After being washed three times with TBS-Tween 20, the cells were treated with Alexa Fluor 546-conjugated goat anti-rabbit immunoglobulin (Ig) antibody or Alexa Fluor 546-conjugated goat anti-mouse Ig antibody (1:200 dilution in 0.1× Block-Ace; Molecular Probes, Inc., Eugene, OR) at 25°C for 1 h. After being washed with TBS-Tween 20, the cells were mounted in Vectashield (Vector Laboratories, Burlingame, CA) and observed with the confocal microscope by excitation with a 543-nm HeNe laser line. For simultaneous imaging of GFP and Alexa Fluor 546, both a 488-nm argon laser line and a 543-nm HeNe laser line were used for excitation. Parameters such as laser power, laser line, and scanning speed were fixed during each group of experiments. The specificity of staining was confirmed by staining with normal rabbit IgG (sc-2027; Santa Cruz) or normal goat IgG (sc-2028; Santa Craz) as the first antibody.
Statistics.
One-way analysis of variance followed by Scheffé's test was used for multigroup comparisons. A P value of <0.05 was considered statistically significant.
RESULTS
Subcellular distributions of N-CoR and truncated mutants.
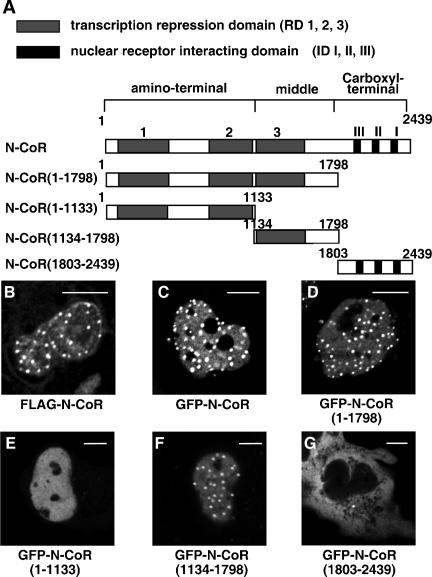
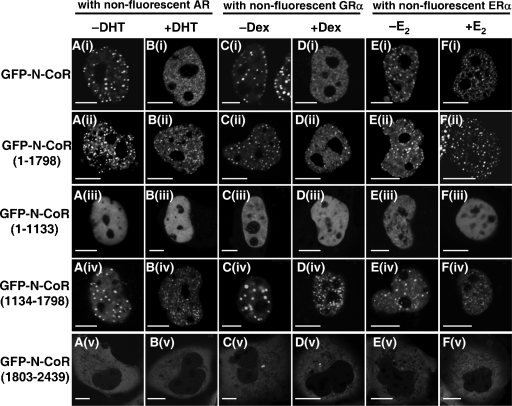
Schematic diagrams of the N-CoR protein and its four truncated mutants used in the present study are illustrated in Fig. 1A. The N-CoR contains three transcription repression domains at the amino-terminal region (RD 1, 2, and 3) and three nuclear receptor-interacting domains at the carboxyl-terminal region (ID I, II, and III). An N-CoR(1-1798) mutant protein consisting of the N-terminal 1,798 amino acid residues lacks the three IDs. The amino-terminal region mutant of the N-CoR, N-CoR(1-1133), contains the N-terminal two RDs (RD1 and RD2), while the middle region mutant, N-CoR(1134-1798), includes RD3. An N-CoR(1803-2439) with the C-terminal 636 amino acid residues carries the three IDs without three RDs.
FIG. 1.
Subcellular distributions of the N-CoR and its truncated mutants. (A) Schematic representation of the full-length human N-CoR and its truncated mutants. RD 1, 2, and 3 represent three transcription repression domains. ID I, II, and III are carboxyl-terminal nuclear receptor-interacting domains. (B) Intranuclear distribution of exogenously expressed FLAG-N-CoR in fixed COS-7 cells. COS-7 cells were transfected with expression vector for FLAG-N-CoR and subjected to immunofluorescence staining using anti-FLAG antibody. (C to G) Intracellular distribution of GFP-N-CoR and its mutants in living COS-7 cells. COS-7 cells were transfected with expression vectors as indicated. Fluorescent signals from expressed proteins were observed under a laser confocal microscope as described in Materials and Methods. Bars, 5 μm.
It has been demonstrated that the GFP-tagged corepressors, such as GFP-N-CoR and GFP-SMRT, showed intranuclear discrete dot patterns (46, 59). As shown in Fig. 1B and C, full-length N-CoRs tagged with FLAG in fixed cells or GFP in living cells were located in the nucleus and showed a discrete dot structure with diffuse background. A similar distribution pattern of GFP-N-CoR was also observed in living 3T3-L1 cells and LNCaP cells derived from prostatic cancer (data not shown).
Then, subcellular distributions of the four GFP-tagged truncated mutants of the N-CoR, as illustrated in Fig. 1A, were observed by the laser confocal microscopy. As shown in Fig. 1D, the GFP-N-CoR(1-1798), which lacks the three IDs, showed an intranuclear discrete dot structure on a diffuse fluorescence background, which was a pattern identical to that of the full-length N-CoR. The distribution pattern of the middle region of N-CoR, GFP-N-CoR(1134-1798) (Fig. 1F) was similar to that of the full-length N-CoR and N-CoR(1-1798), whereas the N-terminal mutant GFP-N-CoR(1-1133) was homogeneously distributed in the nucleus (Fig. 1E). Moreover, the C-terminal mutant GFP-N-CoR(1803-2439), which has been previously reported to be sufficient for the repression effect of the N-CoR (9), was diffusely distributed in the cytoplasm but not in the nucleus (Fig. 1G), as was reported for the C-terminal mutant of SMRT (64).
Subnuclear redistribution of the N-CoR by agonist-bound steroid hormone receptors.
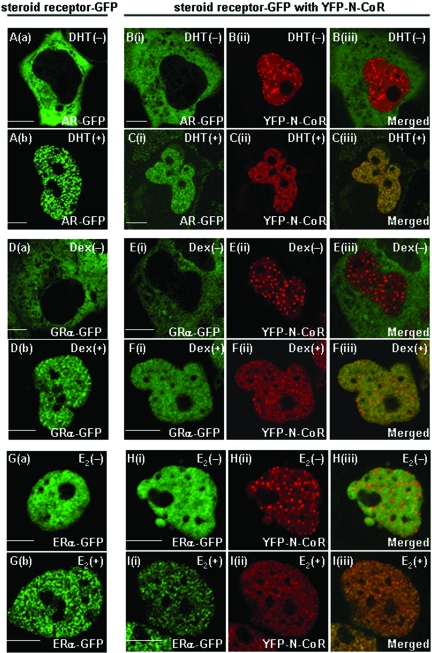
As we previously reported (56), although the AR was initially diffusely distributed in the cytoplasm (Fig. 2Aa), it rapidly moved into the nucleus and made fine foci upon DHT treatment in living COS-7 cells (Fig. 2Ab). Similarly, unliganded GRα-GFP resided in the cytoplasm (Fig. 2Da), while Dex triggered nuclear translocation and focus formation of the receptor (Fig. 2Db). On the other hand, ERα-GFP was homogeneously distributed in the nucleus, even in the absence of the ligand (Fig. 2Ga), and it formed subnuclear foci in the presence of E2 (Fig. 2Gb).
FIG. 2.
Subcellular redistribution of exogenously expressed YFP-N-CoR by agonist-bound steroid hormone receptors. COS-7 cells were transfected with the expression plasmid for AR-GFP (A), GRα-GFP (D), or ERα-GFP (G) alone. Twenty-four hours after the transfection, images were collected using a laser scanning microscope before (a) and after (b) treatment with each ligand (10 nM DHT, 100 nM Dex, or 1 μM E2) at 37°C for 1 h. COS-7 cells were also cotransfected with the expression plasmid for AR-GFP (B and C), GRα-GFP (E and F), or ERα-GFP (H and I) together with the plasmid for YFP-N-CoR. Images were then collected in the absence (B, E, and H) or presence (C, F, and I) of each cognate ligand. Signals from GFP-fused steroid hormone receptors (green, i) and YFP-N-CoR (red, ii) were obtained under the microscope, and the two signals were merged (iii). The molar ratio of transfected amounts of expression plasmids for N-CoRs and steroid hormone receptors was 3:1 in each case. Bars, 5 μm.
The designation of subnuclear compartments of nuclear receptors and cofactors has not been defined concisely in many reported studies. In the present study, we define “foci” as small and clear speckles of agonist-bound steroid hormone receptors and “dots” as relatively large and discrete speckles.
To study spatial relationships of N-CoR with various steroid hormone receptors, we examined subnuclear distributions of YFP-N-CoR in the coexistence of the AR, GRα, and ERα in living COS-7 cells. In the absence of DHT, AR-GFP and YFP-N-CoR were separately located in the cytoplasm and the nucleus, respectively (Fig. 2Bi to 2Biii). These patterns were the same as those observed when the two proteins were individually transfected. In the presence of the ligand, AR-GFP was translocated from the cytoplasm into the nucleus and showed focus formation (Fig. 2Ci). Surprisingly, YFP-N-CoR was also reorganized and formed intranuclear foci (Fig. 2Cii) and colocalized with the DHT-induced AR-GFP foci (Fig. 2Ciii). This phenomenon was also observed in the LNCaP prostate cancer cells (data not shown). As well as the AR and N-CoR, GRα-GFP moved from the cytoplasm into the nucleus and formed foci after treatment with Dex, and YFP-N-CoR was redistributed and colocalized with GRα-GFP (Fig. 2E and F). In the case of ERα and N-CoR, although both proteins resided in the nucleus, even in the absence of E2, they were not completely colocalized because of the discrete dot formation of YFP-N-CoR (Fig. 2H). Again, the addition of E2 induced focus formation of both ERα-GFP and YFP-N-CoR (Fig. 2Ii to 2Iii). Ligand-dependent colocalization of the two proteins was confirmed by a merged image (Fig. 2Iiii).
These data indicate that the corepressor N-CoR is relocated by steroid hormone receptors in the presence of the agonists.
Reduced subnuclear mobility of the N-CoR by DHT-bound ARs.
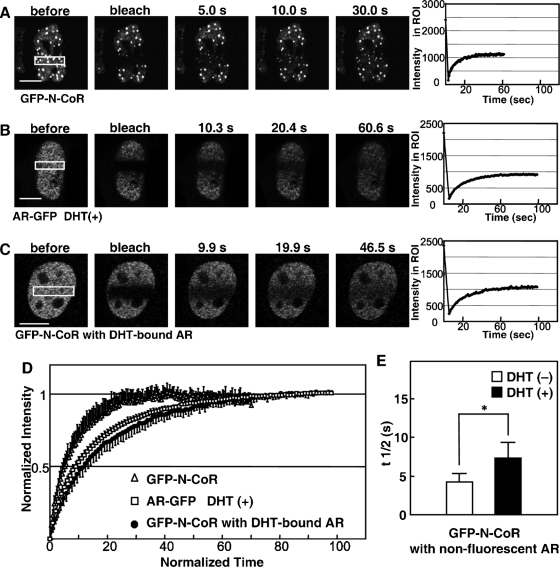
A majority of nuclear proteins, including steroid hormone receptors and cofactors, are now believed to be dynamic in the nucleus (6, 16, 29). Previous FRAP analyses showed that steroid hormone receptors, together with their colocalized coactivators, became less mobile upon treatment with cognate ligands (41, 48, 49). Therefore, we applied the FRAP technique to examine the intranuclear dynamics of N-CoRs and ARs in living cells. The expression plasmids for GFP-N-CoR and AR-GFP were initially transfected into COS-7 cells separately, and FRAP studies were performed for the two proteins individually. As shown in Fig. 3A, a 488-nm laser power bleached GFP-N-CoR fluorescent signals in an ROI of the nucleus, and the fluorescent intensity in the ROI recovered and eventually equilibrated within 30 s. The calculated half-maximal recovery time (t1/2) was 4.0 ± 0.8 s (n = 20). Treatment with 10 nM DHT was unable to alter the recovery speed of GFP-N-CoR (data not shown). In the presence of the ligand, AR-GFP was less dynamic than the N-CoR (Fig. 3B), and the t1/2 for DHT-bound AR-GFP was 7.1 ± 2.0 s (n = 32). Standardized fluorescence recovery curves of both proteins clearly indicate that GFP-N-CoR is more dynamic than AR-GFP (Fig. 3D). Then, GFP-N-CoR was cotransfected with a nonfluorescent AR (pCMV-AR), and the dynamics of GFP-N-CoR were examined by FRAP analysis. After the addition of DHT, cells whose N-CoR patterns were altered to intranuclear foci were subjected to FRAP analysis. These cells were expected to express both GFP-N-CoR and nonfluorescent ARs (Fig. 3C). Interestingly, dynamics of GFP-N-CoR containing DHT-bound ARs apparently decreased compared to that of GFP-N-CoR alone (Fig. 3D). In the absence of DHT, GFP-N-CoR cotransfected with the AR showed a t1/2 of 4.6 ± 0.9 s (n = 20), which was similar to that of single transfected GFP-N-CoR. DHT treatment significantly prolonged the t1/2 of GFP-N-CoR to 7.3 ± 2.0 s (n = 20) (Fig. 3E). These results suggest that the N-CoR interacts with DHT-bound ARs and that the N-CoR is recruited into a complex of ARs.
FIG. 3.
Reduced intranuclear mobility of N-CoRs by DHT-bound ARs. (A and B) FRAP analyses of N-CoRs and DHT-bound ARs when individually expressed. COS-7 cells were transiently transfected with an expression plasmid for GFP-N-CoR (A) or AR-GFP (B) separately and subjected to FRAP analysis 24 h after the transfection. Cells expressing AR-GFP were treated with 10 nM DHT for 1 h prior to FRAP analysis. Each region of interest (ROI) in the nucleus, indicated by a rectangle, was photobleached, and images were then obtained using a laser confocal microscope at the time points indicated in each panel. Graphs on the right represent the recoveries of fluorescent intensities in ROI. (C) FRAP analysis of GFP-N-CoR when coexpressed with nonfluorescent AR. COS-7 cells were cotransfected with expression plasmids for GFP-N-CoR and the wild-type AR. The molar ratio of transfected amounts of expression plasmids for the N-CoR and wild-type AR was 3:1. Twenty-four hours after transfection, the cells were treated with 10 nM DHT for 1 h and then subjected to FRAP analysis. An ROI in the nucleus, indicated by a rectangle, was photobleached, and fluorescent signals were then collected at the time points indicated in each panel. (D) Fluorescent recovery curves for FRAP. Normalized fluorescent signals for GFP-N-CoR (open triangles), AR-GFP [DHT (+)] (open squares), and GFP-N-CoR (with DHT-bound AR) (filled circles) were averaged (n = 10 cells) and plotted to the normalized time points. (E) The half recovery time, t1/2, for GFP-N-CoR with the expression of ARs in the absence (−) or presence (+) of DHT was calculated as described in Materials and Methods, and the values were compared. *, P < 0.05. Bars, 5 μm.
The intranuclear discrete dots of N-CoR are not colocalized with either the splicing factor compartments or PML nuclear bodies.
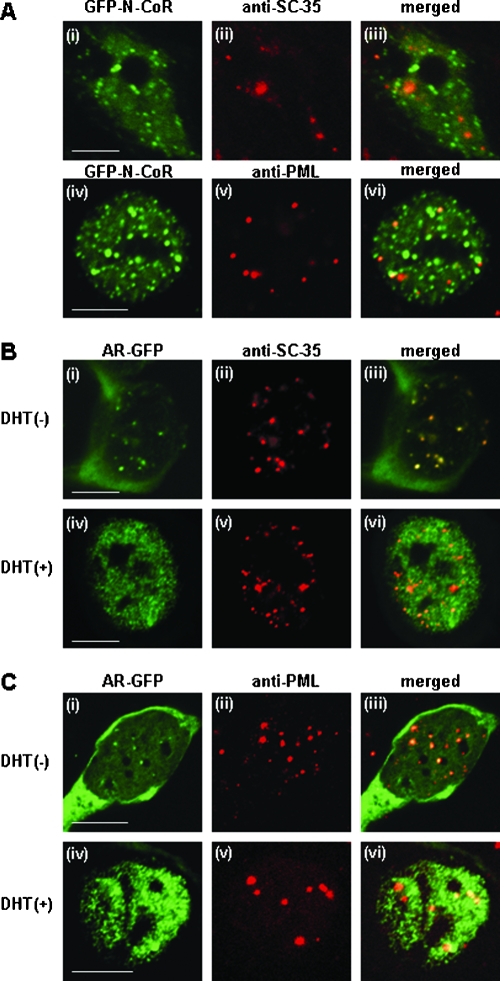
The spatial relationships between N-CoR and two well-characterized subnuclear components, the splicing factor compartments (SFCs) and PML-NBs, were examined by immunofluorescence staining of MCF-7 cells. As previously reported (55, 67), the numbers of intranuclear speckles per nucleus in MCF-7 cells were 20 to 40 for SFCs and 5 to 20 for PML-NBs (data not shown). As shown in Fig. 4A, the discrete intranuclear dots of GFP-N-CoR were not colocalized with either SFCs or PML-NBs. Next, we tested whether the distributions of SFCs and PML-NBs changed in the presence of ligand-bound AR. In the absence of ligand, AR-GFP was diffusely distributed in the cytoplasm, while both SFCs and PML-NBs existed as distinct subnuclear dots. In the presence of DHT, AR-GFP formed distinct nuclear foci, but the intranuclear distributions of SFCs and PML-NBs were not affected by the formation of these foci. These results suggest that redistribution by activated AR is specific to N-CoR.
FIG. 4.
Discrete intranuclear dots of N-CoR differ from those of SFCs and PML-NBs. MCF-7 cells were transfected with expression plasmids for GFP-tagged full-length N-CoR (A) or AR-GFP (B and C) and then subjected to immunofluorescence staining using anti-SC35 or anti-PML antibody. The cells were preincubated in the absence (−, Bi to Biii and Ci to Ciii) or presence (+, Biv to Bvi and Civ to Cvi) of 10 nM DHT for 1 h at 37°C. Signals from GFP-N-CoR (green, Ai and Aiv), AR-GFP (green, Bi, Biv, Ci, and Civ), Alexa Fluor 546-labeled endogenous SC35 (red, Aii, Bii, and Bv), and Alexa Fluor 546-labeled endogenous PML (red, Av, Cii, and Cv) were obtained by laser confocal microscopy, and the two signals were merged (iii and vi). Bars, 5 μm.
The middle region of the N-CoR, N-CoR(1134-1798), is responsible for steroid hormone receptor-induced redistribution of the N-CoR.
To investigate which domain of the N-CoR is responsible for the redistribution by steroid hormone receptors, a GFP-tagged N-CoR or its four truncated mutants were coexpressed with a nonfluorescent AR, GRα, or ERα. As shown in Fig. 5A, the intracellular distribution pattern of each N-CoR variant was unaltered by coexpression of the unliganded AR. As expected, the full-length N-CoR was reorganized and formed subnuclear foci after DHT treatment (panel Bi). Both the N-CoR(1-1798) and the middle region, N-CoR(1134-1798), also formed intranuclear foci after treatment with DHT (panels Bii and Biv). However, the distribution pattern of the amino-terminal region, N-CoR(1-1133), was not changed by DHT-bound AR (panel Biii). Moreover, the carboxyl-terminal region of the N-CoR, N-CoR(1803-2439), remained in the cytoplasm even in the presence of the DHT (panel Bv). Similar results were obtained when the AR was replaced by either the GRα (panels C and D) or ERα (panels E and F). These data suggest that the middle region of the N-CoR is functionally critical for the redistribution of the N-CoR by agonist-bound steroid hormone receptors.
FIG. 5.
The middle region of the N-CoR is responsible for the redistribution by agonist-bound steroid hormone receptors. COS-7 cells were cotransfected with expression plasmids for GFP-tagged full-length N-CoR or its truncated mutants and nonfluorescent wild-type AR (panels A and B), GRα (panels C and D), or ERα (panels E and F). The cells in panels A, C, and E were observed in the absence of ligands, and the cells were incubated with 10 nM DHT (panel B), 100 nM Dex (panel D), or 1 μM E2 (panel F) for 1 h at 37°C before images were taken. The molar ratios of transfected amounts of expression plasmids for GFP-N-CoR and its mutants to steroid hormone receptors were 3:1 in all of the transfection experiments. Bars, 5 μm.
The middle region of the N-CoR plays a major role in suppressing the ligand-dependent transactivation function of steroid hormone receptors.
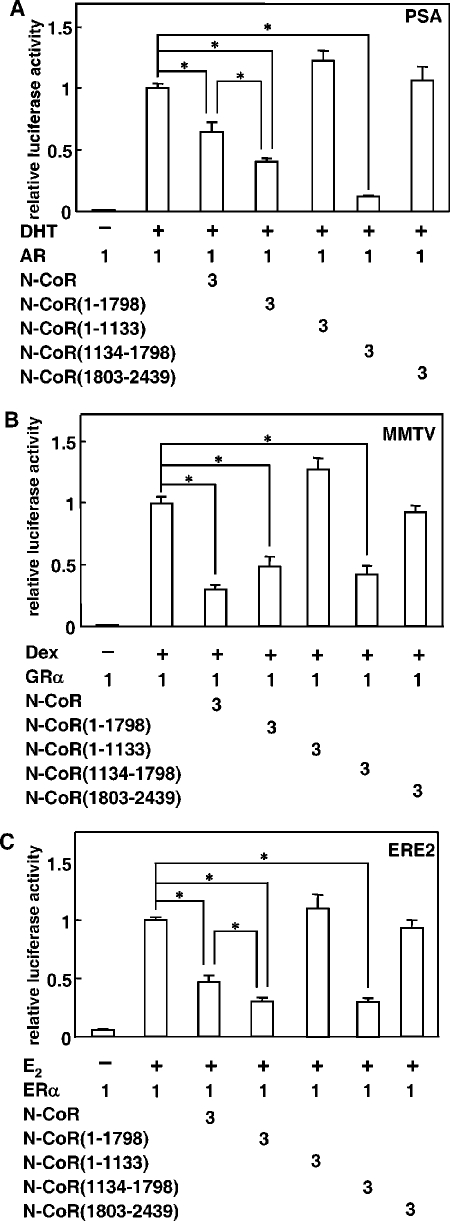
To examine the effect of the middle region of the N-CoR on steroid hormone receptor-mediated transcriptional activation, functional promoter assays were performed using each hormone's responsive element. As shown in Fig. 6, AR-, GRα- and ERα-mediated transcriptional activations were all suppressed by the N-CoR. Transactivation activities of these receptors were suppressed to 30 to 64% of the original activities by a 3 M excess coexpression of the N-CoR. As expected, the N-CoR(1-1798) and the middle region, N-CoR(1134-1798), also exhibited a suppression effect on all three reporters. In the case of AR and ERα, both N-CoR(1-1798) and N-CoR(1134-1798) showed an even more potent suppression than the full-length N-CoR (Fig. 6A and C). In contrast, no significant suppression of promoter activities was observed by coexpression of N-CoR(1-1133) or N-CoR(1803-2439). These results suggest that the N-CoR suppresses steroid hormone receptor-mediated transactivation mainly through its middle region.
FIG. 6.
The middle region of the N-CoR plays a major role in suppressing ligand-dependent transactivation function of steroid hormone receptors. COS-7 cells were cotransfected with expression plasmids for full-length N-CoR or its mutants and AR (A), GRα (B) or ERα (C), with reporter plasmids carrying each hormone responsive element and pRL-CMV. These cells were cultured in the presence (+) or absence (−) of the agonists for 24 h and subjected to the luciferase assay. The luciferase activities are represented as values relative to the activity induced by the full-length steroid hormone receptor alone in the presence of each ligand. Each bar represents the mean ± standard deviation of results from three independent experiments. *, P < 0.05.
Multiple regions of N-CoR are involved in binding with DHT-bound AR.
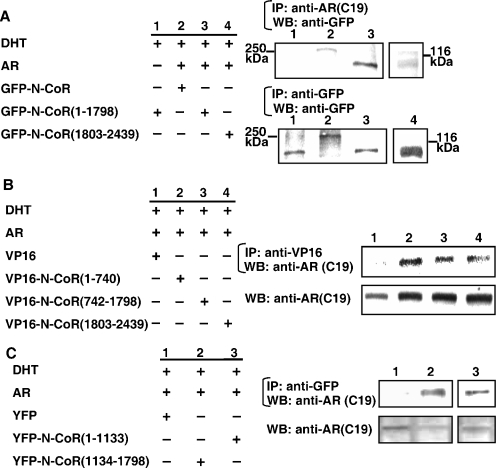
To further determine which region of the corepressor N-CoR is important for the interaction with steroid hormone receptors, we carried out a CoIP study. GFP-fused N-CoR variants were subjected to the CoIP experiments using the anti-AR antibody (Fig. 7A). Although GFP-N-CoR, GFP-N-CoR(1-1798), and GFP-N-CoR(1803-2439) were all coimmunoprecipitated with ligand-bound ARs by an anti-AR antibody (right upper panel), GFP-N-CoR(1-1798) (lane 3) manifested a much denser band than GFP-N-CoR(1803-2439) (lane 4). Without AR coexpression, GFP-N-CoR(1-1798) was not precipitated (lane 1). Proper expression levels of the GFP-fused N-CoR variants were confirmed by Western blot analysis using an anti-GFP antibody (Fig. 7A, right lower panel).
FIG. 7.
Multiple regions of the N-CoR are involved in the interaction with DHT-bound AR. (A to C) Coimmunoprecipitation of the full-length and truncated N-CoR with DHT-bound AR. COS-7 cells were cotransfected with expression plasmids for the wild-type AR and GFP-fused full-length N-CoR or its truncated mutants, VP16 or VP16-fused N-CoR mutants, or YFP or YFP-fused N-CoR mutants, as indicated. After 24 h incubation with 10 nM DHT, the cells were lysed and immunoprecipitation (IP) was performed using antibodies against AR, GFP, or VP16, as indicated. The precipitates were then subjected to Western blotting (WB) analysis using antibodies as indicated. +, present; −, absent.
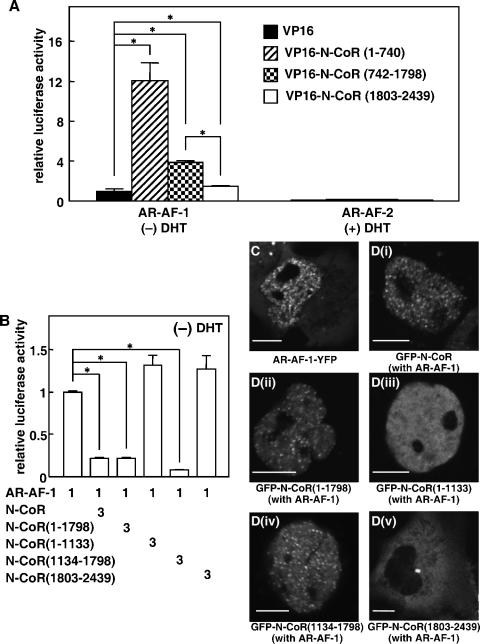
We also generated three VP16-fused N-CoR mutants. VP16-N-CoR(1-740) contained the amino-terminal 1 to 740 amino acid residues of the N-CoR, VP16-N-CoR(742-1798) contained amino acid residues 742 to 1798 of the N-CoR, and VP16-N-CoR(1803-2439) carried the carboxyl-terminal amino acid residues containing the three IDs. These VP16-fused N-CoR variants were coexpressed with the AR in COS-7 cells (Fig. 7B). After treatment with DHT, protein complexes were precipitated with an anti-VP16 antibody, and ARs in the precipitant were detected by an anti-AR antibody (right upper panel). AR bands were detected when VP16-N-CoR(1-740) or VP16-N-CoR(742-1798) was coexpressed (lanes 2 and 3). A relatively smaller amount of AR protein was also precipitated in the cells cotransfected with the C-terminal VP16-N-CoR(1803-2439) (lane 4). No AR protein was detectable when the VP16 empty vector was cotransfected (lane 1). Proper expressions of AR proteins were validated by Western blotting of whole-cell extracts using an anti-AR antibody (Fig. 7B, right lower panel). Further deletion analysis of N-CoR was performed and N-CoR(1134-1798), which contains the transcription repression domain 3 (RD3) (Fig. 1A), was able to interact with AR in CoIP (Fig. 7C). This N-CoR(1134-1798) mutant also repressed the AR-mediated transactivation as well as the full-length N-CoR and N-CoR(1-1798) (Fig. 6A), whereas N-CoR(1-1133), which contains RD1 and 2, interacted with AR (Fig. 7C) but failed to repress the transactivation (Fig. 6A). These results suggest that N-CoR(1134-1798) can bind with AR and would be enough to repress the AR-mediated transactivation function.
The intranuclear complete/distinct focus formation of agonist-bound steroid hormone receptor is an indicator of the transcription activation status.
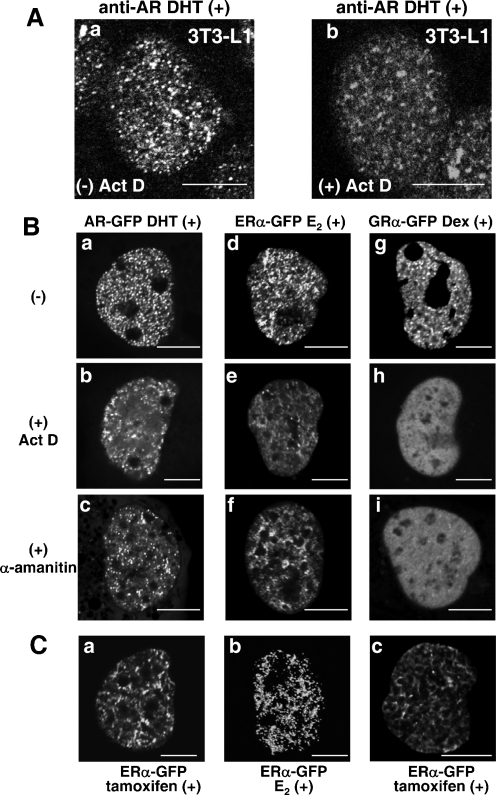
Intranuclear compartmentalization of steroid hormone receptors are so far mostly demonstrated using the exogenously expressed GFP-tagged receptors by laser confocal microscopy (13, 17, 22, 30, 33, 34, 42, 48-50, 52, 53, 56). In the present study, we examined the intracellular distribution of endogenous ARs in fixed 3T3-L1 cells, which were found to contain a relatively high quantity of ARs by immunofluorescence staining. The endogenous ARs were found to form intranuclear foci in the presence of DHT (Fig. 8Aa). Ligand-dependent subnuclear focus formation of steroid hormone receptor is considered to be closely associated with the transcriptional activation mediated by the receptors (41, 56). To clarify the mechanism of intranuclear compartmentalization, an effect of a transcription inhibitor, actinomycin D (Act D), on the intranuclear focus formation of endogenous AR was investigated in fixed 3T3-L1 cells. Unexpectedly, distinct endogenous AR foci were not detected when treated with Act D (Fig. 8Ab). Then the effects of transcription inhibitors on the exogenously expressed steroid hormone receptors were also investigated in living COS-7 cells. Comparing to the fine foci of DHT-bound AR-GFP (Fig. 8Ba), the treatment of Act D or α-amanitin markedly impaired the intranuclear focus formation of DHT-bound AR-GFPs, which were accumulated as vague speckles close to the nuclear membrane (Fig. 8Bb and 8Bc). The distribution pattern of E2-bound ERα-GFPs was changed from focal to reticular (Fig. 8Bd to f). Moreover, the intranuclear foci of Dex-bound GRα-GFPs totally disappeared (Fig. 8Bg to i). These results strongly suggested that focus formation does not precede transcriptional activation and also that an appearance of intranuclear foci of steroid hormone receptors reflects the transactivation function status.
FIG. 8.
Intranuclear foci of agonist-bound steroid hormone receptors were destroyed in the presence of transcription inhibitors or tamoxifen. (A) Endogenous ARs formed intranuclear foci in the presence of 10 nM DHT (a) which were impaired by treatment with 10 μg/ml Act D in 3T3-L1 cells. Signals from Alexa Fluor 546-labeled endogenous AR were obtained by laser confocal microscopy. (B) The intranuclear foci of agonist-bound steroid hormone receptors were destroyed by treatments with transcription inhibitors, Act D and α-amanitin, in living COS-7 cells. COS-7 cells were transfected with the expression plasmids for AR-GFP (a to c), ERα-GFP (d to f), or GRα-GFP (g to i). Twenty-four hours later, the cells were treated with 10 nM DHT (a to c), 1 μM E2 (d to f), or 100 nM Dex (g to i) at 37°C for 1 h, and then fluorescent signals from the expressed proteins were observed. The cells were pretreated with 10 μg/ml Act D (b, e, and h) or with 100 μg/ml α-amanitin (c, f, and i) at 37°C for 2 h before the addition of each cognate agonist. (C) Intranuclear distributions of tamoxifen-bound and E2-bound ERα-GFP in living COS-7 cells. COS-7 cells were transfected with the expression plasmid for ERα-GFP. Twenty-four hours later, the cells were treated with 1 μM tamoxifen (a and c) or 1 μM E2 (b) at 37°C for 1 h. (a) Two-dimensional image. (b and c) Three-dimensional images as a surface view on the z axis. Bars, 5 μm.
Additionally, tamoxifen did not activate the transcription of the ERE-containing luciferase reporter by ERα (data not shown), but tamoxifen-bound ERα was reported to form concentrated subnuclear regions similar to foci (49). Then we compared the intranuclear distribution patterns of E2- or tamoxifen-bound ERα-GFP in both two- and three-dimensional images. In two-dimensional images, these two distribution patterns seemed similar (Fig. 8Bd and Ca). However, in three-dimensional images, E2-bound ERα-GFP formed distinct foci, whereas tamoxifen-ERα-GFP showed a reticular pattern (Fig. 8Cb and Cc).
The N-CoR impairs intranuclear complete/distinct focus formation of ligand-bound steroid hormone receptors via its middle region.
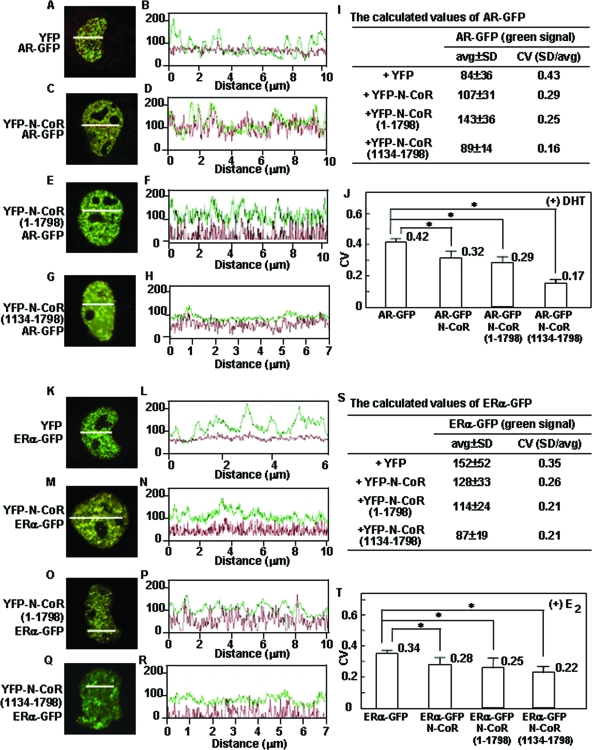
The results shown in Fig. 2 to 5 demonstrated that the agonist-induced intranuclear compartmentalization of steroid hormone receptors was accompanied by the recruitment of N-CoRs, while the N-CoR firmly suppressed the transactivation function of steroid hormone receptors (Fig. 6). These observations were apparently inconsistent with our current hypothesis. Accordingly, we precisely analyzed the AR focus pattern when coexpressed with the N-CoR using two- and three-dimensional quantitative image analyses (Fig. 9 and 10). A quantitative analysis in a two-dimensional image for fluctuation of fluorescent intensity in the nucleus was previously established by Htun et al. (22). A straight line was drawn across a target nucleus, and fluorescent intensities along the line were recorded by Zeiss LSM software. An average and a SD value of fluorescent intensities were calculated. A CV, which was equal to the SD divided by the average, was used as a degree of heterogeneity of nuclear protein distribution, namely, if the CV value was high, the distribution of the protein was heterogeneous. This method has been applied to analyses of the nuclear distribution of several nuclear receptors, such as the hGRα (43) and Ad4BP/SF-1 (12). The CV values of these nuclear receptors significantly increased after treatment with cognate ligands, which meant a change in subnuclear distribution from a homogeneous to a heterogeneous pattern. In the case of AR-GFP coexpressed with YFP in the presence of DHT, the fluorescence intensity curve for AR-GFP manifested about 9 to 11 clear wave peaks (Fig. 9B). The YFP signal was diffusely distributed in the nucleus and cytoplasm and showed no wave peak in the fluorescence intensity curve (Fig. 9B), which represented a diffuse homogeneous distribution pattern. The calculated CV value of DHT-bound AR-GFP, 0.43, represented the intranuclear heterogeneous distribution (Fig. 9I). Interestingly, when YFP-N-CoR was coexpressed, the wave peaks of AR-GFP became unclear (Fig. 9D). Furthermore, the number of wave peaks in the fluorescence intensity curve was apparently decreased. The decreased CV value (0.29) for AR-GFP (Fig. 9I) suggested that the fluctuation of fluorescence intensity was reduced and the distribution pattern of AR-GFP became less heterogeneous with the coexpression of YFP-N-CoR. Similarly, when coexpressed with YFP-N-CoR(1-1798) or YFP-N-CoR(1134-1798), the wave peaks in the fluorescence intensity curve for AR-GFP became unclear (Fig. 9F and H) and the CV value decreased to 0.25 or 0.16 (Fig. 9I). As shown in Fig. 9J, the CV value of AR-GFP significantly decreased with the coexpression of YFP-N-CoR, YFP-N-CoR(1-1798), or YFP-N-CoR(1134-1798). As well as the AR, the fluorescence intensity curve for E2-bound ERα-GFP manifested clear wave peaks (Fig. 9L) and the CV value was 0.35 (Fig. 9S). As expected, when coexpressed with YFP-N-CoR, YFP-N-CoR(1-1798), or YFP-N-CoR(1134-1798), the wave peaks in the fluorescence intensity curve for ERα-GFP became unclear (Fig. 9N, P, and R) and the CV values significantly decreased (Fig. 9S and T), representing a relatively homogeneous distribution of ERα-GFP.
FIG. 9.
The N-CoR impairs the intranuclear complete focus formation of agonist-bound AR-GFP and ERα-GFP through the middle region of the N-CoR. COS-7 cells were transfected with the expression plasmids for AR-GFP and YFP (A), AR-GFP and YFP-N-CoR (C), AR-GFP and YFP-N-CoR(1-1798) (E), AR-GFP and YFP-N-CoR(1134-1798) (G), ERα-GFP and YFP (K), ERα-GFP and YFP-N-CoR (M), ERα-GFP and YFP-N-CoR(1-1798) (O), or ERα-GFP and YFP-N-CoR(1134-1798) (Q). The molar ratio of the transfected amounts of each pair was 1:3. The cells were treated with 10 nM DHT or 1 μM E2 at 37°C for 1 h and then subjected to laser confocal microscopy. A representative cell from each group is shown, and a white line for analysis of fluorescent intensity drawn through the nucleus is also indicated. The fluorescent intensity fluctuation graphs for the white lines are shown in panels B, D, F, H, L, N, P, and R. The x axis depicts the distance from the start point of the line, and the y axis shows the fluorescent intensity of each point. Green curves represent GFP signals from AR-GFP (B, D, F, and H) or ERα-GFP (L, N, P, and R), and red curves represent YFP signals from YFP (B and L), YFP-N-CoR (D and N), YFP-N-CoR(1-1798) (F and P), or YFP-N-CoR(1134-1798) (H and R). The calculated values (average [avg], standard deviation [SD], and coefficient of variation [CV]) of these representative cells are shown in panels I and S. The CV values of 10 lines per cell and at least 20 cells from three independent experiments were averaged and are shown in panels J and T. *, P < 0.05.
FIG. 10.
The three-dimensional image analyses of intranuclear foci of agonist-bound AR-GFP and ERα-GFP coexpressed with YFP-N-CoR. COS-7 cells were transfected with the expression plasmids for AR-GFP and YFP, AR-GFP and YFP-N-CoR, AR-GFP and YFP-N-CoR(1-1798), AR-GFP and YFP-N-CoR(1134-1798), ERα-GFP and YFP, ERα-GFP and YFP-N-CoR, ERα-GFP and YFP-N-CoR(1-1798), or ERα-GFP and YFP-N-CoR(1134-1798) (A and B). The molar ratio of the transfected amounts of each pair was 1:3. The cells were treated with 10 nM DHT or 1 μM E2 at 37°C for 1 h, and then the confocal images of the nuclei were collected to reconstruct three-dimensional images. The three-dimensional images for AR-GFP and ERα-GFP were displayed as a surface view on the z axis. The number of AR-GFP and ERαGFP foci is indicated in each panel. Bar, 5 μm.
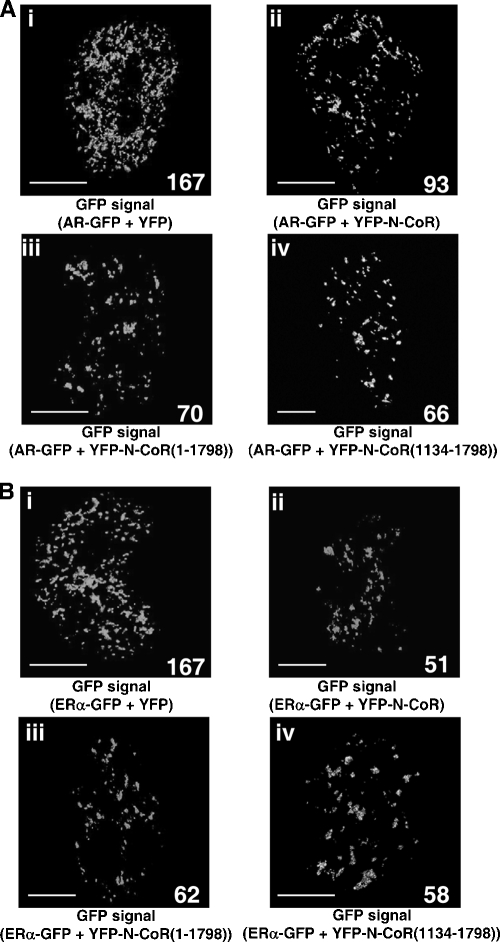
The impairment of complete/distinct focus formation by the N-CoR was further confirmed by three-dimensional image analyses (Fig. 10). The three-dimensional construction method for confocal images allowed us to observe intranuclear fluorescent proteins at a high resolution and to quantify the number of fluorescent spots in the nucleus (41, 56). This method can detect only a distinct volume (complete foci) by removing background scattering fluorescence and relatively diffusely distributed fluorescence and then clearly showing a difference in the intranuclear spatial distribution of the foci. With the coexpression of YFP, 167 intranuclear foci of AR-GFP were detected as a distinct volume in one nucleus (147 ± 24, n = 12) (Fig. 10Ai). However, coexpression of YFP-N-CoR clearly decreased the number of intranuclear foci of AR-GFP to 93, as shown in Fig. 10Aii (80 ± 31, n = 12, P = 0.0001 versus AR-GFP with YFP). Moreover, as well as the full-length N-CoR, both N-CoR(1-1798) and N-CoR(1134-1798) destroyed the DHT-induced AR focus formation (Fig. 10Aiii and Aiv). In addition, the impairment of the complete/distinct ERα focus formation by the N-CoR via its middle region was also confirmed by three-dimensional image analyses. As shown in Fig. 10B, the intranuclear foci of ERα-GFP were clearly destroyed by the coexpression of YFP-N-CoR, YFP-N-CoR(1-1798), or YFP-N-CoR(1134-1798). Therefore, these results suggest that the N-CoR is recruited to the intranuclear compartment of agonist-bound steroid hormone receptors, but simultaneously, the N-CoR inhibits the completion of focus formation.
The N-CoR interacts with the N terminus of the AR and inhibits the AR-AF-1-mediated transcriptional activation.
To clarify which region of the AR is associated with the N-CoR, we performed the mammalian one-hybrid assay using VP16-fused N-CoR mutants with AR-AF-1 (N terminus) or AR-AF-2 (C terminus). As shown in Fig. 11A, interactions were observed between N-CoR mutants and AR-AF-1 in the absence of DHT but not AR-AF-2, even in the presence of DHT. The N-terminal N-CoR variants, N-CoR(1-740) and N-CoR(742-1798), showed a strong interaction with AR-AF-1, while an interaction between the C-terminal N-CoR(1803-2439) and AR-AF-1 was quite weak. These results suggest that the N-terminal region of the AR and N-CoR are major domains for the interaction between these two proteins. In the functional promoter assay, the middle region of the N-CoR was able to repress the AR-AF-1-mediated transactivation to the same level as that of the full-length N-CoR, whereas the amino-terminal and carboxyl-terminal regions of the N-CoR could not repress the transactivation (Fig. 11B). As we previously reported (41), AR-AF-1 was located in the nucleus and formed foci even in the absence of DHT (Fig. 11C). Interestingly, similar to GFP-N-CoR, both GFP-N-CoR(1-1798) and GFP-N-CoR(1134-1798) (Fig. 11Di), when coexpressed with nonfluorescent AR-AF-1, formed intranuclear foci (Fig. 11Dii and Div). However, N-CoR(1-1133) was still homogeneously distributed in the nucleus, and N-CoR(1803-2439) remained in the cytoplasm even when AR-AF-1 was coexpressed (Fig. 11Diii and Dv). Colocalization of AR-AF-1 and the N-CoR was also confirmed by observing cells coexpressing GFP-tagged N-CoR mutants with AR-AF-1-YFP (data not shown). These results suggest that the interaction between the N terminus of the AR and the middle region of the N-CoR is responsible for the colocalization and repression.
FIG. 11.
The N-CoR interacts with the N-terminal domain of the AR and inhibits the AR-AF-1-mediated transcriptional activation. (A) The N-CoR interacted with the N-terminal domain of the AR. COS-7 cells were cotransfected with expression plasmids for AR-AF-1 or AR-AF-2 and VP16 or VP16-fused N-CoR mutants with pGL3-MMTV and pRL-CMV for the mammalian one-hybrid assay. The molar ratio of the transfected amount of expression plasmids for AR-AF-1 or AR-AF-2 and N-CoR was 1:5. The cells were incubated for 24 h in the absence (for AR-AF-1) or presence (for AR-AF-2) of 10 nM DHT. The luciferase activities are represented as values relative to the activity induced by AR-AF-1 alone in the absence of ligand. (B) Inhibition of AR-AF-1-mediated transactivation by the N-CoR. COS-7 cells were cotransfected with expression plasmids for AR-AF-1 and the full-length N-CoR or its mutants with pGL3-MMTV and pRL-CMV. The cells were incubated for 24 h in the absence of DHT. The luciferase activities are represented as values relative to the activity induced by AR-AF-1 alone in the absence of ligand. Each bar in panels A and B represents the mean ± standard deviation of results from three independent experiments. *, P < 0.05. (C) Subcellular distribution of AR-AF-1-YFP in COS-7 cells. (D) Redistribution of GFP-N-CoR by nonfluorescent AR-AF-1 in the absence of DHT. An expression plasmid for GFP-N-CoR (i), GFP-N-CoR(1-1798) (ii), GFP-N-CoR(1-1133) (iii), GFP-N-CoR(1134-1798) (iv), or GFP-N-CoR(1803-2439) (v) was cotransfected into COS-7 cells with the plasmid for nonfluorescent AR-AF-1. Bars, 5 μm.
The N-CoR disturbs the N-C interaction of the AR and functionally competes with SRC-1 and CBP on AR-mediated transactivation.
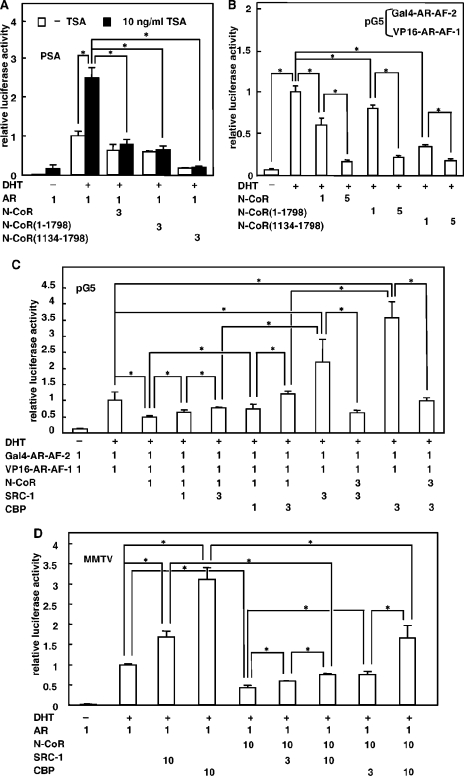
A modulation of chromatin structure by HDACs, which directly or indirectly interact with the amino-terminal region of the N-CoR, plays a central role in N-CoR-mediated transcriptional repression of an unliganded nuclear hormone receptor (28). AR-mediated activation of the prostate-specific antigen (PSA) promoter was apparently enhanced by the addition of a specific HDAC inhibitor, TSA (Fig. 12A) (19). However, suppression of AR-mediated transactivation by the full-length N-CoR, N-CoR(1-1798), and N-CoR(1134-1798) was not significantly recovered by TSA. Similar results were obtained in case of the ERα (data not shown). In addition, TSA had no effect on the subcellular distribution of the AR, GRα, or ERα in the presence of cognate ligands when coexpressed with the N-CoR (data not shown). These results suggest that there would be some mechanisms for repression of AR-mediated transactivation by the N-CoR other than the recruitment of HDAC.
FIG. 12.
Impairment of DHT-induced AR N-C interaction by the N-CoR and functional competition between the N-CoR and coactivators, SRC-1 or CBP, on AR-mediated transactivation. (A) TSA could not relieve the repression of AR-mediated transactivation by N-CoR. COS-7 cells were cotransfected with expression plasmids for the wild-type AR and the full-length N-CoR, N-CoR(1-1798), or N-CoR(1134-1798) with pGL3-PSA and pRL-CMV. The cells were treated with 10 nM DHT in the absence or presence of TSA (10 ng/ml) for 24 h. The luciferase activities are represented as values relative to the activity induced by DHT-bound ARs, which was set to 1.0. (B) N-CoRs dose-dependently inhibited the DHT-induced AR N-C interaction. In the mammalian two-hybrid assay, COS-7 cells were transfected with pG5, expression plasmids for Gal4-AR-AF-2, VP16-AR-AF-1, and N-CoR or its mutants. (C) The inhibition of DHT-induced AR N-C interaction by the N-CoR was dose-dependently reversed by coexpressing with SRC-1 or CBP. In the mammalian two-hybrid assay, the expression plasmids for Gal4-AR-AF-2, VP16-AR-AF-1, the N-CoR, and SRC-1 or CBP were cotransfected into COS-7 cells as indicated. The luciferase activities are represented as values relative to the activity induced by Gal4-AR-AF-2 and VP16-AR-AF-1 in the presence of 10 nM DHT in panels B and C. (D) Functional competition between N-CoRs and the coactivators, SRC-1 or CBP, on AR-mediated transactivation. COS-7 cells were transiently transfected with the expression plasmids for the wild-type AR, the full-length N-CoR, and SRC-1 or CBP, together with the pGL3-MMTV reporter in the presence of 10 nM DHT where indicated by a plus sign (+). The luciferase activities are represented as values relative to the activity induced by DHT-bound ARs alone, which was set to 1.0. The molar ratio of the transfected amounts of plasmids are shown at the bottom of each panel. Each bar represents the mean ± standard deviation of results from three independent experiments in panels A to D. *, P < 0.05.
It has been suggested that the agonist-induced N-C interaction is required for the full activity of the AR (2, 24, 41). To examine an effect of the N-CoR on the AR N-C interaction, we performed a mammalian two-hybrid assay using VP16-fused AR-AF-1 and Gal4-fused AR-AF-2 (Fig. 12B). In the absence of the ligand, VP16-AR-AF-1 and Gal4-AR-AF-2 exhibited almost no interaction. As a result of the interaction between Gal4-AR-AF-2 and VP16-AR-AF-1, the addition of DHT strongly stimulated the activation of the pG5 reporter. Coexpression of equimolar amounts of the N-CoR suppressed this ligand-dependent activation by 40%, indicating that N-CoRs suppress the AR N-C interaction. N-CoR(1-1798) and N-CoR(1134-1798) also suppressed the N-C interaction by 20% and 60%. The disturbance of the AR N-C interaction by these N-CoR mutants was more evident at increasing amounts (Fig. 12B). In contrast, N-CoR(1803-2439) did not inhibit the AR N-C interaction (data not shown). Similar results were obtained with NIH 3T3 cells (data not shown). These results strongly suggest that the N-CoR inhibits the transactivation activity of the AR by disturbing N-C interactions mainly through its middle region.
Some coactivators can enhance AR-mediated transcriptional activation through stabilization of the AR N-C interaction by direct binding to the N-terminal AF-1 domain (7, 24). In the mammalian two-hybrid assay, SRC-1 or CBP dose-dependently recovered the suppression of N-C interaction by the N-CoR (Fig. 12C). Conversely, coexpression of an increasing amount of N-CoRs suppressed the enhancement effects of SRC-1 or CBP. A similar tendency was observed in the functional reporter assay using the mouse mammary tumor virus (MMTV) promoter (Fig. 12D). The repression of AR-mediated transactivation by the N-CoR was dose-dependently recovered by coexpressing an increasing amount of coactivator, SRC-1 or CBP. Moreover, the N-CoR strongly repressed the coactivation effects of SRC-1 or CBP. These results suggest that the functional competition with the coactivaors by the N-CoR may be one of the mechanisms of N-CoR-mediated repression.
Release of the N-CoR from intranuclear foci of AR-GFP by coexpression of SRC-1 and CBP.
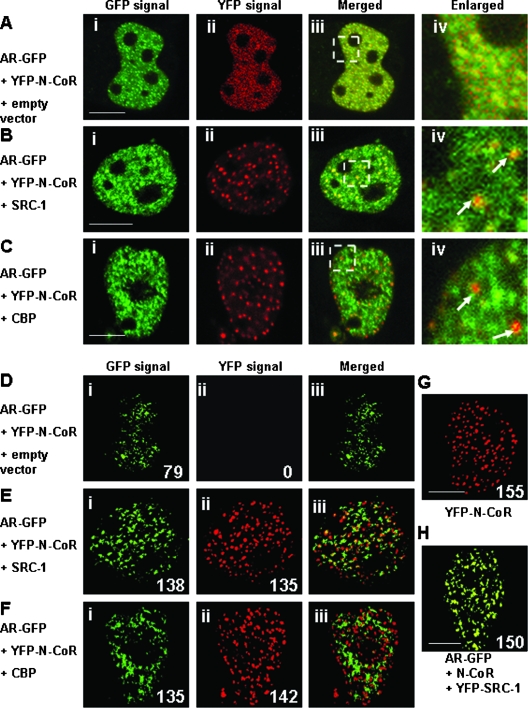
We then investigated the intranuclear distribution of the YFP-N-CoR and DHT-bound AR-GFP in living cells when the cells were cotransfected with relatively higher levels of the expression plasmids for coactivators, SRC-1 and CBP, compared with that for N-CoRs. The N-CoR was originally located in the nucleus with a discrete dot structure (Fig. 1 and 2). In the presence of DHT, localization of AR-GFP and YFP-N-CoR almost overlapped due to the redistribution of YFP-N-CoR and discrete dots of YFP-N-CoR disappeared (Fig. 2C and 13A). Interestingly, if the cells were cotransfected with nonfluorescent SRC-1 (Fig. 13B) or CBP (Fig. 13C), the red dots of YFP-N-CoR reappeared (Fig. 13Bii and Cii). In the merged and enlarged images, these red dots were observed separated from the green signal of AR-GFP (Fig. 13Biv and Civ). This strongly suggests that YFP-N-CoR was released from the AR compartment when coactivators were coexpressed.
FIG. 13.
The two- and three-dimensional image analyses of the effects of YFP-N-CoRs and coactivators on the intranuclear focus formation of agonist-bound ARs. COS-7 cells were cotransfected with the expression plasmids for AR-GFP and YFP-N-CoR together with the empty vector (A and D), nonfluorescent SRC-1 (B and E), or CBP (C and F). The molar ratio of transfected amounts of expression plasmids for these three proteins was 1:3:10. The cells were also transfected with the expression vectors for YFP-N-CoR alone (G), or AR-GFP and YFP-SRC-1 together with nonfluorescent N-CoR (H), and the transfected amounts were the same as those in panels B. Images were then collected in the presence of 10 nM DHT. Signals from AR-GFP (green, i) and YFP-N-CoR (red, ii) were obtained under the microscope, and the two signals were merged (iii) in panels A to F. (A to C) Two-dimensional image analyses. The area indicated in the white rectangle in panel iii is magnified as shown in panel iv. The white arrows indicate the intranuclear discrete dots in red, which were derived from YFP-N-CoR. (D to F) Three-dimensional image analyses. The confocal images of the nuclei in living cells were collected to reconstruct three-dimensional images. The merged images were displayed as surface views on the z axis. (G) Three-dimensional image for YFP-N-CoR. (H) Three-dimensional image for AR-GFP and YFP-SRC-1 coexpressed with nonfluorescent N-CoR. The merged images were displayed as a surface view on the z axis. The number of AR-GFP foci or YFP-N-CoR dots was indicated in each panel. The number of discrete dots of YFP-N-CoR was 155 ± 46 (n = 10). Bars, 5 μm.
Using the three-dimensional imaging technique, the number of discrete YFP-N-CoR dots was 155 (Fig. 13G) when the cells were transfected with YFP-N-CoR only. The number of distinct foci of AR-GFP was decreased to 79 by coexpression with YFP-N-CoR (Fig. 13Di), as shown in Fig. 10. No YFP-N-CoR signal was detected after processing of the three-dimensional reconstruction (Fig. 13Dii), suggesting that the original discrete dots of YFP-N-CoR disappeared by redistribution (Fig. 13Aii) and the relatively homogeneously redistributed YFP-N-CoR was not included in distinct foci of AR-GFP. When SRC-1 or CBP was coexpressed, the number of distinct foci of AR-GFP was recovered to 138 and 135 (Fig. 13Ei and Fi). Moreover, discrete dots of YFP-N-CoR were detected (Fig. 13Eii and Fii) and localized separately from the distinct foci of AR-GFP in the merged images (Fig. 13Eiii and Fiii). The number of dots of YFP-N-CoR was approximately 140 per nucleus, which was similar to that observed with transfection of YFP-N-CoR alone. These results further confirmed that YFP-N-CoR was released from intranuclear compartments of agonist-bound AR-GFP and then formed discrete dots by coexpression of coactivators SRC-1 and CBP. When AR-GFP and YFP-SRC-1 were transfected with the nonfluorescent N-CoR, SRC-1 was found to form distinct foci with AR-GFP, showing a yellow signal in the merged three-dimensional image (Fig. 13H).
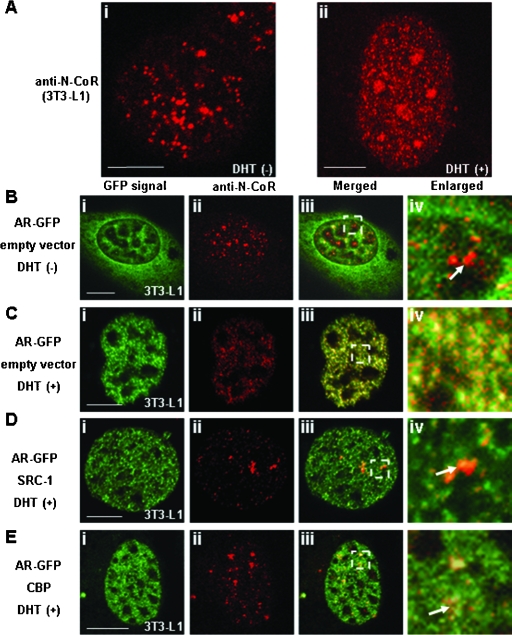
To determine whether endogenous N-CoR shows the same intranuclear distribution pattern as GFP-N-CoR, we performed an immunofluorescence staining study using anti-N-CoR antibody. In the absence of DHT, N-CoR was distributed in the form of discrete intranuclear dots in a diffuse background signal (Fig. 14Ai). Similar to the results obtained with GFP-tagged N-CoR, the endogenous N-CoR dots disappeared in the presence of DHT to form intranuclear foci (Fig. 14ii). These results suggest that endogenous N-CoR is also redistributed by ligand-bound endogenous AR.
FIG. 14.
Intranuclear redistribution of endogenous N-CoR by DHT-bound AR and release of the endogenous N-CoR from the AR compartment by exogenously expressed coactivators. 3T3-L1 cells were treated with ethanol (Ai) or 10 nM DHT (Aii) for 1 h and then subjected to immunofluorescence staining using anti-N-CoR antibody. 3T3-L1 cells were transfected with the expression plasmids for AR-GFP with empty vector (B and C), with nonfluorescent SRC-1 (D), or with nonfluorescent CBP (E). The molar ratio of transfected amounts of expression plasmids for two proteins was 1:3. Twenty-four hours after the transfection, the cells were subjected to immunofluorescence staining in the absence (B) or presence (C, D, and E) of 10 nM DHT, and then images were collected. Signals from AR-GFP (green, i)- and Alexa Fluor 546 (red, ii)-labeled endogenous N-CoR were obtained by laser confocal microscopy, and these two signals were merged (iii). The area indicated in the white rectangle in panel iii is magnified in panel iv. The white arrows indicate the intranuclear discrete dots in red, which were derived from the labeled endogenous N-CoR proteins. Bars, 5 μm.
We further examined the redistribution of endogenous N-CoRs by coexpressing AR-GFP in 3T3-L1 cells. In the absence of DHT, endogenous N-CoR, which was detected by immunostaining, and AR-GFP were separately located in the cytoplasm and the nucleus (Fig. 14B). These distribution patterns were similar to those observed when the two proteins were cotransfected (Fig. 2Bi to iii). In the presence of DHT, AR-GFP was translocated into the nucleus and formed intranuclear foci (Fig. 14Ci). Simultaneously, the intranuclear discrete dots of endogenous N-CoRs disappeared (Fig. 14Cii) and endogenous N-CoR proteins were recruited into the DHT-bound AR foci (Fig. 14Ciii and Civ). When the cells were cotransfected with nonfluorescent SRC-1 (Fig. 14D) or CBP (Fig. 14E), the red dots of endogenous N-CoRs reappeared (Fig. 14Dii and Eii) and were separated from the green signals of AR-GFP (Fig. 14Diii to iv and Eiii to iv). Expression of endogenous N-CoR, which was estimated by the intensity of fluorescence on laser confocal microscopy, was not affected by the overexpression of SRC-1 or CBP detected in this assay (data not shown). These results strongly suggest that the endogenous N-CoR is also recruited into AR foci in a ligand-dependent manner and released from the AR compartment by overexpression of coactivators.
DISCUSSION
The present study provides novel findings about mechanisms of repression of steroid hormone receptor function by the N-CoR. The N-CoR was recruited to subnuclear foci formed by agonist-bound steroid hormone receptors, but the recruited N-CoR impaired complete focus formation, reflecting its inhibitory effect on transactivation. The middle region of the N-CoR, N-CoR(1134-1798), was mostly responsible for both the interaction with steroid hormone receptors and the repression of their activities. The repression mechanism included disruption of the N-C interaction of the receptors and/or competition with the coactivators, such as SRC-1 and CBP, rather than recruitment of HDACs.
We demonstrated that both the endogenous and exogenous N-CoR proteins formed discrete dot structures with a diffuse background and were redistributed and colocalized with agonist-bound steroid hormone receptors. Similar redistributions of corepressors have been observed by other researchers. For example, Dex-bound endogenous GRα redistributed a corepressor, receptor-interacting protein 140, from its intranuclear dots to a more diffuse pattern, and the redistribution of receptor-interacting protein 140 was correlated with its repression of GR-mediated transactivation (53). GFP-SMRT was reorganized to more reticular, microspeckled pattern by ligand-bound ERα, which was colocalized with ligand-bound ERα (58). Furthermore, the redistribution of corepressors was also induced by other classes of transcription factors, such as pituitary-gland-specific Pit-1. Both SMRT and N-CoR repressed Pit-1-mediated transcription, and the intranuclear dot structures of SMRT and N-CoR were changed to more diffuse nucleoplasmic compartments with the coexpression of Pit-1 (59). In contrast, in the present study, the distributions of two well-characterized nuclear compartments, SFCs and PML-NBs, were not affected by ligand-bound AR, indicating that the redistribution of N-CoR by agonist-bound AR is not a secondary event induced by the possible alteration of nuclear architecture. Thus, this widely observed redistribution of corepressors may be a common and specific process during the regulation of transcription activity by the corepressors.
The redistribution of N-CoR by steroid hormone receptors and the interaction of N-CoR with those receptors occurred in an agonist-dependent manner. Interactions between the N-CoR and AR or GRα have so far been demonstrated by binding assays such as glutathione S-transferase pull-down and two-hybrid assay (9, 33), and previous studies were mostly focused on ligand-independent interactions between the domains of N-CoRs and steroid hormone receptors. We also demonstrated the interaction between the N-CoR and agonist-bound AR in living cells by FRAP analysis. The N-CoR became less mobile by coexpression of ARs in the presence of DHT. It has been revealed that, although most steroid hormone receptors are highly dynamic in the nucleoplasmic space, the mobility of the receptors decreases in the presence of ligand (6, 17, 40, 42, 48). Steroid receptors were detected in a nuclear matrix preparation after treatment with ligands (4), and such an interaction would be necessary for transactivation. The decrease in mobility is presumed to come from the interaction of ligand-bound receptors with the nuclear matrix. Therefore, the decreased mobility of the N-CoR was thus considered to reflect a direct and functional interaction of N-CoRs with liganded ARs.
Previous studies showed that the carboxyl-terminal region of N-CoR/SMRT, containing a CoRNR motif (I/LXXII), was enough for the repression of the transactivation of nuclear receptors (9, 23, 44, 63). However, our results found that the middle region of the N-CoR, N-CoR(1134-1798), exerted a significant inhibitory effect on the transactivation function of steroid hormone receptors but not the amino- or carboxyl-terminal region of the N-CoR (Fig. 6). In previous studies, strong interactions between the C terminus of N-CoR and steroid hormone receptors were detected in the absence of the ligand (9) or in the presence of antagonists (44, 60). Although Cheng et al. showed a repression of DHT-induced AR transactivation by the C terminus of N-CoR, the interaction between the N-CoR C terminus and AR was ligand independent. Moreover, in the mammalian two-hybrid assay for AR-N-CoR interaction analysis, the authors used AR-ligand binding domain, not the full-length of the receptor. In fact, in a recent study by Hodgson et al., DHT-dependent interaction between the C terminus of N-CoR and the full-length AR was not detected (20), which corresponds with our present results (Fig. 1 and 6). Therefore, it is supposed that the middle region may be important for the agonist-dependent interaction between N-CoR and AR, whereas the C terminus of N-CoR may be important for the antagonist-induced interaction. In the present study, the middle region of the N-CoR, N-CoR(1134-1798), but not the amino- or carboxyl-terminal region of the N-CoR, was recruited by agonist-bound AR, GRα, and ERα in living cells (Fig. 5). Furthermore, coimmunoprecipitation experiments revealed that N-CoR(1134-1798) containing RD3 was able to interact with the AR. Therefore, N-CoR(1134-1798) containing RD3 is strongly suggested to be the major region for the repression of transactivation function of agonist-bound steroid hormone receptors. The N-CoR(1-1133) also contributes to the interaction with agonist-bound receptors but does not seem to be responsible for the repression of their transactivation function (Fig. 6 and 7).
Many coactivators have been shown to regulate AR transactivation function through an interaction with the N-terminal domain of the AR. The p160 coactivators such as SRC-1 and TIF2 directly interacted with the N-terminal domain of the AR (2). It has previously been reported that SRC-1 and TIF2 were recruited into the AR-AF-1 foci (41). The N-CoR protein was also shown to interact with the N-terminal domain of steroid hormone receptors (20, 44, 60). In the present study, the N-CoR was also recruited to the AR-AF-1 foci, and the interaction between N-CoR and AR-AF-1 was confirmed by mammalian one-hybrid assay. The N-CoR is thus suggested to repress AR-mediated transactivation through an interaction with the N-terminal domain of the AR.
We demonstrated that the N-CoR inhibited the AR N-C interaction in the mammalian two-hybrid assay and also in the AR-mediated transactivation. A specific HDAC inhibitor, TSA, could not recover the N-CoR-mediated suppression of AR-dependent transactivation (Fig. 12A). It has been recently reported that SMRT-mediated inhibition of the steroid hormone receptor function is not recovered by TSA (1, 23) and is largely due to the blockage of the AR N-C interaction (1, 34). In the present study, we confirmed that N-CoR also had such inhibitory effects on the AR N-C interaction. In a recent paper (32), it has been reported that the intramolecular N-C interaction controls AR chromatin binding and is required for the recruitment of SWI/SNF, which remodels chromatin. Inhibition of intramolecular interactions by N-CoR might affect chromatin remodeling, resulting in repression of the transactivation function.
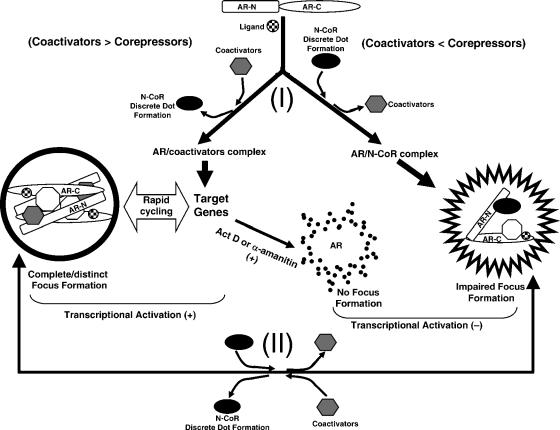
Recently, it was proposed that coactivators and corepressors exert equilibrium interactions with nuclear receptors (61) and that the properties of nuclear receptors are determined by the intracellular ratio of coactivators and corepressors (45, 51, 61). It was also proposed that the transcriptional regulation by nuclear receptors requires the corepressor-coactivator exchange via the TBL1/TBLR1-containing exchange complex (39). Competition between N-CoR and coactivators to regulate N-C interaction and the transactivation activity of AR was revealed in the present study, which seems consistent with the above hypothesis.
The appearance of intranuclear focus formation has been shown to be closely related to the transcriptionally active conformation of steroid hormone receptors with coactivators (13, 30, 41, 56). Upon treatment with DHT, AR-GFP alone formed complete foci with recruitment of coactivators such as SRC-1, TIF2, and CBP (41). Such focus formation was clearly demonstrated for endogenous AR in the present study (Fig. 8A). Treatment with some antagonists, which inhibit the transcriptional activation, translocates the receptors from the cytoplasm into the nucleus; however, the translocated receptors are not able to form intranuclear foci and are diffusely distributed (13, 56). The N-C interaction is also required for complete focus formation of ARs (41). However, the present study clearly demonstrated that the quality of AR foci or ERα foci containing the N-CoR was different from that of AR foci or ERα foci without N-CoRs. The focus pattern of AR-GFP and ERα-GFP without N-CoRs (Fig. 2Ab and Gb and 9A and K) seemed much more distinct than the speckle pattern of AR-GFP and ERα-GFP coexpressed with the N-CoR (Fig. 2Ci and Ii and 9C and M), but such differences in the appearance of the intranuclear compartments were not confirmed only by simple two-dimensional images. However, a quantitative analysis of fluorescence intensity fluctuation in two-dimensional images and a three-dimensional imaging method we previously developed (41, 56) clearly demonstrated impaired formation of AR or ERα foci (incomplete focus formation) in the presence of the N-CoR (Fig. 9 and 10). The transactivation activities of AR and ERα were parallel at the level of their distinct (complete) focus formation (Fig. 6 and 10). It was thus suggested that only complete focus formation is related to the full transactivation function of ARs. An antagonist, tamoxifen, was shown to induce ERα focus formation, which was similar to foci induced by estradiol (Fig. 8Bd and Ca). However, the present study demonstrated that the tamoxifen-induced foci of ERα are not complete foci (Fig. 8Cb and Cc). The biochemical analyses using the two-hybrid and reporter gene assays revealed the functional competition between the N-CoR and CBP/SRC-1 on transactivation activity of the AR. This finding would further conceive that the N-CoR and coactivators mutually and exclusively bind to ARs, namely, binding of CBP and SRC-1 leads to a change in molecular conformation of the AR that hinders further binding of N-CoRs and vice versa. The present three-dimensional imaging analysis provided evidence for the above concept. Additional expression of SRC-1 or CBP in DHT-bound ARs coexpressed with N-CoRs led to release of N-CoRs from incomplete foci containing DHT-bound ARs and N-CoRs and to formation of distinct/complete foci with the coactivators (Fig. 13). Essentially the same phenomena were observed for endogenous N-CoR (Fig. 14). We speculate that there is a conformational equilibrium between transcriptionally active and inactive steroid hormone receptors in the presence of agonists, determined by the relative ratio of coactivators and corepressors.
The present observations that transcription inhibitors, actinomycin D and α-amanitin, disrupt intranuclear foci of steroid hormone receptors (Fig. 8B) revealed that the focus formation is considered not to precede the transcriptional activation. The steroid hormone receptors and coactivators show multiphasic on and off switching for binding to promoter elements of the target genes. The receptor-coactivator complexes also undergo a rapid exchange between the target genes and their subnuclear compartments (34, 35, 48). Based on these reported observations and the present findings that complete focus formation is related to the transcriptional activation, it is suggested that complete focus formation of steroid hormone receptors reflects the accumulation of steroid hormone receptor-coactivator complexes released from their target genes. Therefore, it is further suggested that focus formation is a mirror of transcriptionally active complex formation of steroid hormone receptors and coactivators at transcription sites. Thus, investigation of focus formation would be useful for the elucidation of the mechanism of steroid hormone receptor-dependent transactivation in living cells. A physiological role for focus formation still remains to be clarified. One speculated possibility is as follows. Numerous important molecules are functioning in the nucleus. Some regulatory mechanisms for these molecules should be present. Focus formation might be one of these mechanisms, and foci function as temporary storing and buffering sites that are related to steroid hormone receptor cycling. Figure 15 summarizes the speculation of the intranuclear compartmentalization and transactivation function of the AR based on the present findings.
FIG. 15.
Speculation of N-CoR action on intranuclear focus formation and transactivation function of the AR. DHT induces a conformational change of the AR and binding domains for coactivators, and the corepressors, N-CoRs, in the amino termini of the AR are unmasked. Under the intranuclear environmental conditions in which the content of the N-CoR is considerably low compared with that of coactivators, DHT-bound AR takes proper N-C interaction with bound coactivators and forms a transcriptionally active conformation, and the AR-coactivator complexes transactivate on the target genes. The transcriptionally active AR-coactivator complexes undergo a rapid cyclic exchange between the subnuclear compartment (complete/distinct foci) and transcriptionally active sites of the target genes. When transcription is prevented by a transcription inhibitor, actinomycin D or α-amanitin, a rapid exchange may be inhibited and then the focus formation is destroyed. The appearance of complete/distinct foci would be an indicator for the transcription active status. Under the environmental conditions in which the N-CoR content is relatively high, the N-CoR binds to liganded AR (apparent overlapping) and interferes with the N-C interaction, resulting in a transcriptionally inactive conformation, which shows the intranuclear impaired focus formation. There are several possible phases (I and II) for the conformational equilibrium between transcriptionally active and inactive DHT-bound AR dependent on the relative ratio of coactivators and N-CoR.
Acknowledgments
We thank Mitoshi Toki for excellent technical assistance in performing the three-dimensional imaging analyses.
This study was supported in part by grants-in-aid for Science Research (B) and Exploratory Research (C) and by a grant for the 21st Century Center of Excellence (COE) Program (Kyushu University) from the Japanese Ministry of Education, Culture, Sports, Science, and Technology.
REFERENCES
- 1.Agoulnik I. U., W. C. Krause, W. E. Bingman III, H. T. Rahman, M. Amrikachi, G. E. Ayala, and N. L. Weigel. 2003. Repressors of androgen and progesterone receptor action. J. Biol. Chem. 278:31136-31148. [DOI] [PubMed] [Google Scholar]
- 2.Alen, P., F. Claessens, G. Verhoeven, W. Rombauts, and B. Peeters. 1999. The androgen receptor amino-terminal domain plays a key role in p160 coactivator-stimulated gene transcription. Mol. Cell. Biol. 19:6085-6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ariumi, Y., T. Ego, A. Kaida, M. Matsumoto, P. P. Pandolfi, and K. Shimotohno. 2003. Distinct nuclear body components, PML and SMRT, regulate the trans-acting function of HTLV-1 Tax oncoprotein. Oncogene 22:1611-1619. [DOI] [PubMed] [Google Scholar]
- 4.Barrack, E. R. 1987. Specific association of androgen receptors and estrogen receptors with the nuclear matrix: summary and perspectives, p. 85-107. In V. K. Moudgil (ed.), Recent advances in steroid hormone action. Walter de Gruijter, Berlin, Germany.
- 5.Beato, M., and J. Klug. 2000. Steroid hormone receptors: an update. Hum. Reprod. Update 6:25-36. [DOI] [PubMed] [Google Scholar]
- 6.Belmont, A. 2003. Dynamics of chromatin, proteins, and bodies within the cell nucleus. Curr. Opin. Cell Biol. 15:304-310. [DOI] [PubMed] [Google Scholar]
- 7.Bubulya, A., S. Y. Chen, C. J. Fisher, Z. Zheng, X. Q. Shen, and L. Shemshedini. 2001. c-Jun potentiates the functional interaction between the amino and carboxyl termini of the androgen receptor. J. Biol. Chem. 276:44704-44711. [DOI] [PubMed] [Google Scholar]
- 8.Chen, J. D., K. Umesono, and R. M. Evans. 1996. SMRT isoforms mediate repression and anti-repression of nuclear receptor heterodimers. Proc. Natl. Acad. Sci. USA 93:7567-7571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng, S., S. Brzostek, S. R. Lee, A. N. Hollenberg, and S. P. Balk. 2002. Inhibition of the dihydrotestosterone-activated androgen receptor by nuclear receptor corepressor. Mol. Endocrinol. 16:1492-1501. [DOI] [PubMed] [Google Scholar]
- 10.Dotzlaw, H., U. Moehren, S. Mink, A. C. Cato, J. A. Iniguez Lluhi, and A. Baniahmad. 2002. The amino terminus of the human AR is target for corepressor action and antihormone agonism. Mol. Endocrinol. 16:661-673. [DOI] [PubMed] [Google Scholar]
- 11.Downes, M., P. Ordentlich, H. Y. Kao, J. G. Alvarez, and R. M. Evans. 2000. Identification of a nuclear domain with deacetylase activity. Proc. Natl. Acad. Sci. USA 97:10330-10335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fan, W., T. Yanase, Y. Wu, H. Kawate, M. Saitoh, K. Oba, M. Nomura, T. Okabe, K. Goto, J. Yanagisawa, S. Kato, R. Takayanagi, and H. Nawata. 2004. Protein kinase A potentiates adrenal 4 binding protein/steroidogenic factor 1 transactivation by reintegrating the subcellular dynamic interactions of the nuclear receptor with its cofactors, general control nonderepressed-5/transformation/ transcription domain-associated protein, and suppressor, dosage-sensitive sex reversal-1: a laser confocal imaging study in living KGN cells. Mol. Endocrinol. 18:127-141. [DOI] [PubMed] [Google Scholar]
- 13.Fejes-Tóth, G., D. Pearce, and A. Náray-Fejes-Tóth. 1998. Subcellular localization of mineralocorticoid receptors in living cells: effects of receptor agonists and antagonists. Proc. Natl. Acad. Sci. USA 95:2973-2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graham, J. D., D. L. Bain, J. K. Richer, T. A. Jackson, L. Tung, and K. B. Horwitz. 2000. Nuclear receptor conformation, coregulators, and tamoxifen-resistant breast cancer. Steroids 65:579-584. [DOI] [PubMed] [Google Scholar]
- 15.Hager, G. L., A. K. Nagaich, T. A. Johnson, D. A. Walker, and S. John. 2004. Dynamics of nuclear receptor movement and transcription. Biochim. Biophys. Acta 1677:46-51. [DOI] [PubMed] [Google Scholar]
- 16.Hager, G. L., C. Elbi, and M. Becker. 2002. Protein dynamics in the nuclear compartment. Curr. Opin. Genet. Dev. 12:137-141. [DOI] [PubMed] [Google Scholar]
- 17.Hager, G. L. 1999. Studying nuclear receptors with green fluorescent protein fusions. Methods Enzymol. 302:73-84. [DOI] [PubMed] [Google Scholar]
- 18.He, B., and E. M. Wilson. 2002. The NH(2)-terminal and carboxyl-terminal interaction in the human androgen receptor. Mol. Genet. Metab. 75:293-298. [DOI] [PubMed] [Google Scholar]
- 19.Heltweg, B., and M. Jung. 2002. A microplate reader-based nonisotopic histone deacetylase activity assay. Anal. Biochem. 302:175-183. [DOI] [PubMed] [Google Scholar]
- 20.Hodgson, M. C., I. Astapova, S. Cheng, L. J. Lee, M. C. Verhoeven, E. Choi, S. P. Balk, and A. N. Hollenberg. 2005. The androgen receptor recruits nuclear receptor corepressor (N-CoR) in the presence of mifepristone via its N and C termini revealing a novel molecular mechanism for androgen receptor antagonists. J. Biol. Chem. 280:6511-6519. [DOI] [PubMed] [Google Scholar]
- 21.Horlein, A. J., A. M. Naar, T. Heinzel, J. Torchia, B. Gloss, R. Kurokawa, A. Ryan, Y. Kamei, M. Soderstrom, C. K. Glass, and M. G. Rosenfeld. 1995. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature 377:397-404. [DOI] [PubMed] [Google Scholar]
- 22.Htun, H., L. T. Holth, D. Walker, J. R. Davie, and G. L. Hager. 1999. Direct visualization of the human estrogen receptor alpha reveals a role for ligand in the nuclear distribution of the receptor. Mol. Biol. Cell 10:471-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu, X., Y. Li, and M. A. Lazar. 2001. Determinants of CoRNR-dependent repression complex assembly on nuclear hormone receptors. Mol. Cell. Biol. 21:1747-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ikonen, T., J. J. Palvimo, and O. A. Janne. 1997. Interaction between the amino- and carboxyl-terminal regions of the rat androgen receptor modulates transcriptional activity and is influenced by nuclear receptor coactivators. J. Biol. Chem. 272:29821-29828. [DOI] [PubMed] [Google Scholar]
- 25.Ishizuka, M., H. Kawate, R. Takayanagi, H. Ohshima, R. H. Tao, and H. Hagiwara. 2005. A zinc finger protein TZF is a novel corepressor of androgen receptor. Biochem. Biophys. Res. Commun. 331:1025-1031. [DOI] [PubMed] [Google Scholar]
- 26.Jackson, T. A., J. K. Richer, D. L. Bain, G. S. Takimoto, L. Tung, and K. B. Horwitz. 1997. The partial agonist activity of antagonist-occupied steroid receptors is controlled by a novel hinge domain-binding coactivator L7/SPA and the corepressors N-CoR or SMRT. Mol. Endocrinol. 11:693-705. [DOI] [PubMed] [Google Scholar]
- 27.Jenster, G., H. A. van der Korput, J. Trapman, and A. O. Brinkmann. 1995. Identification of two transcription activation units in the N-terminal domain of the human androgen receptor. J. Biol. Chem. 270:7341-7346. [DOI] [PubMed] [Google Scholar]
- 28.Jones, P. L., and Y. B. Shi. 2003. N-CoR-HDAC corepressor complexes: roles in transcriptional regulation by nuclear hormone receptors. Curr. Top. Microbiol. Immunol. 274:237-268. [DOI] [PubMed] [Google Scholar]
- 29.Kawata, M. 2001. Subcellular steroid/nuclear receptor dynamics. Arch. Histol. Cytol. 64:353-368. [DOI] [PubMed] [Google Scholar]
- 30.Kawate, H., Y. Wu, K. Ohnaka, H. Nawata, and R. Takayanagi. 2005. Tob proteins suppress steroid hormone receptor-mediated transcriptional activation. Mol. Cell. Endocrinol. 230:77-86. [DOI] [PubMed] [Google Scholar]
- 31.Langley, E., Z. X. Zhou, and E. M. Wilson. 1995. Evidence for an anti-parallel orientation of the ligand-activated human androgen receptor dimer. J. Biol. Chem. 270:29983-29990. [DOI] [PubMed] [Google Scholar]
- 32.Li, J., J. Fu, C. Toumazou, H. G. Yoon, and J. Wong. 2006. A role of the amino-terminal (N) and carboxyl-terminal (C) interaction in binding of androgen receptor to chromatin. Mol. Endocrinol. 20:776-785. [DOI] [PubMed] [Google Scholar]
- 33.Liao, G., L. Y. Chen, A. Zhang, A. Godavarthy, F. Xia, J. C. Ghosh, H. Li, and J. D. Chen. 2003. Regulation of androgen receptor activity by the nuclear receptor corepressor SMRT. J. Biol. Chem. 278:5052-5061. [DOI] [PubMed] [Google Scholar]
- 34.McNally, J. G., W. G. Müller, D. Walker, R. Wolford, and G. L. Hager. 2000. The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science 287:1262-1265. [DOI] [PubMed] [Google Scholar]
- 35.Metivier, R., G. Penot, M. R. Hubner, G. Reid, H. Brand, M. Kos, and F. Gannon. 2003. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell 115:751-763. [DOI] [PubMed] [Google Scholar]
- 36.Moilanen, A., N. Rouleau, T. Ikonen, J. J. Palvimo, and O. A. Janne. 1997. The presence of a transcription activation function in the hormone-binding domain of androgen receptor is revealed by studies in yeast cells. FEBS Lett. 412:355-358. [DOI] [PubMed] [Google Scholar]
- 37.Nagy, L., H. Y. Kao, D. Chakravarti, R. J. Lin, C. A. Hassig, D. E. Ayer, S. L. Schreiber, and R. M. Evans. 1997. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell 89:373-380. [DOI] [PubMed] [Google Scholar]
- 38.Nagy, L., H. Y. Kao, J. D. Love, C. Li, E. Banayo, J. T. Gooch, V. Krishna, K. Chatterjee, R. M. Evans, and J. W. Schwabe. 1999. Mechanism of corepressor binding and release from nuclear hormone receptors. Genes Dev. 13:3209-3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perissi, V., A. Aggarwal, C. K. Glass, D. W. Rose, and M. G. Rosenfeld. 2004. A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell 116:511-526. [DOI] [PubMed] [Google Scholar]
- 40.Phair, R. D., and T. Misteli. 2000. High mobility of proteins in the mammalian cell nucleus. Nature 404:604-609. [DOI] [PubMed] [Google Scholar]
- 41.Saitoh, M., R. Takayanagi, K. Goto, A. Fukamizu, A. Tomura, T. Yanase, and H. Nawata. 2002. The presence of both the amino- and carboxyl-terminal domains in the AR is essential for the completion of a transcriptionally active form with coactivators and intranuclear compartmentalization common to the steroid hormone receptors: a three-dimensional imaging study. Mol. Endocrinol. 16:694-706. [DOI] [PubMed] [Google Scholar]
- 42.Schaaf, M. J., and J. A. Cidlowski. 2003. Molecular determinants of glucocorticoid receptor mobility in living cells: the importance of ligand affinity. Mol. Cell. Biol. 23:1922-1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schaaf, M. J., L. J. Lewis-Tuffin, and J. A. Cidlowski. 2005. Ligand-selective targeting of the glucocorticoid receptor to nuclear subdomains is associated with decreased receptor mobility. Mol. Endocrinol. 19:1501-1515. [DOI] [PubMed] [Google Scholar]
- 44.Schulz, M., M. Eggert, A. Baniahmad, A. Dostert, T. Heinzel, and R. Renkawitz. 2002. RU486-induced glucocorticoid receptor agonism is controlled by the receptor N terminus and by corepressor binding. J. Biol. Chem. 277:26238-26243. [DOI] [PubMed] [Google Scholar]
- 45.Smith C. L., Z. Nawaz, and B. W. O'Malley. 1997. Coactivator and corepressor regulation of the agonist/antagonist activity of the mixed antiestrogen, 4-hydroxytamoxifen. Mol. Endocrinol. 11:657-666. [DOI] [PubMed] [Google Scholar]
- 46.Söderström, M., A. Vo, T. Heinzel, R. M. Lavinsky, W. M. Yang, E. Seto, D. A. Peterson, M. G. Rosenfeld, and C. K. Glass. 1997. Differential effects of nuclear receptor corepressor (N-CoR) expression levels on retinoic acid receptor-mediated repression support the existence of dynamically regulated corepressor complexes. Mol. Endocrinol. 11:682-692. [DOI] [PubMed] [Google Scholar]
- 47.Stein, G. S., S. K. Zaidi, C. D. Braastad, M. Montecino, A. J. van Wijnen, J. Y. Choi, J. L. Stein, J. B. Lian, and A. Javed. 2003. Functional architecture of the nucleus: organizing the regulatory machinery for gene expression, replication and repair. Trends Cell Biol. 13:584-592. [DOI] [PubMed] [Google Scholar]
- 48.Stenoien, D. L., K. Patel, M. G. Mancini, M. Dutertre, C. L. Smith, B. W. O'Malley, and M. A. Mancini. 2001. FRAP reveals that mobility of oestrogen receptor-alpha is ligand- and proteasome-dependent. Nat. Cell Biol. 3:15-23. [DOI] [PubMed] [Google Scholar]
- 49.Stenoien, D. L., M. G. Mancini, K. Patel, E. A. Allegretto, C. L. Smith, and M. A. Mancini. 2000. Subnuclear trafficking of estrogen receptor-alpha and steroid receptor coactivator-1. Mol. Endocrinol. 14:518-534. [DOI] [PubMed] [Google Scholar]
- 50.Stenoien, D. L., M. Mielke, and M. A. Mancini. 2002. Intranuclear ataxin1 inclusions contain both fast- and slow-exchanging components. Nat. Cell Biol. 4:806-810. [DOI] [PubMed] [Google Scholar]
- 51.Szapary, D., Y. Huang, and S. S. Simons, Jr. 1999. Opposing effects of corepressor and coactivators in determining the dose-response curve of agonists, and residual agonist activity of antagonists, for glucocorticoid receptor-regulated gene expression. Mol. Endocrinol. 13:2108-2121. [DOI] [PubMed] [Google Scholar]
- 52.Tao, R. H., H. Kawate, Y. Wu, K. Ohnaka, M. Ishizuka, A. Inoue, H. Hagiwara, and R. Takayanagi. 2006. Testicular zinc finger protein recruits histone deacetylase 2 and suppresses the transactivation function and intranuclear foci formation of agonist-bound androgen receptor competitively with TIF2. Mol. Cell. Endocrinol. 247:150-165. [DOI] [PubMed] [Google Scholar]
- 53.Tazawa, H., W. Osman, Y. Shoji, E. Treuter, J. A. Gustafsson, and J. Zilliacus. 2003. Regulation of subnuclear localization is associated with a mechanism for nuclear receptor corepression by RIP140. Mol. Cell. Biol. 23:4187-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tetel, M. J., S. Jung, P. Carbajo, T. Ladtkow, D. F. Skafar, and D. P. Edwards. 1997. Hinge and amino-terminal sequences contribute to solution dimerization of human progesterone receptor. Mol. Endocrinol. 11:1114-1128. [DOI] [PubMed] [Google Scholar]
- 55.Tian, B., Z. Mei, Y. Song, J. Dian, Y. Dong, H. Liu, B. Liu, G. Zhao, and Z. Sun. 2006. Knocking down PML impairs p53 signaling transduction pathway and suppresses irradiation induced apoptosis in breast carcinoma cell MCF-7. J. Cell. Biochem. 97:561-571. [DOI] [PubMed] [Google Scholar]
- 56.Tomura, A., K. Goto, H. Morinaga, M. Nomura, T. Okabe, T. Yanase, R. Takayanagi, and H. Nawata. 2001. The subnuclear three-dimensional image analysis of androgen receptor fused to green fluorescence protein. J. Biol. Chem. 276:28395-283401. [DOI] [PubMed] [Google Scholar]
- 57.Tsai M. J., and B. W. O'Malley. 1994. Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Annu. Rev. Biochem. 63:451-486. [DOI] [PubMed] [Google Scholar]
- 58.Voss, T. C., I. A. Demarco, C. F. Booker, and R. N. Day. 2005. Corepressor subnuclear organization is regulated by estrogen receptor via a mechanism that requires the DNA-binding domain. Mol. Cell. Endocrinol. 231:33-47. [DOI] [PubMed] [Google Scholar]
- 59.Voss, T. C., I. A. Demarco, C. F. Booker, and R. N. Day. 2005. Functional interactions with Pit-1 reorganize corepressor complexes in the living cell nucleus. J. Cell Sci. 118:3277-3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang, D., and S. S. Simons, Jr. 2005. Corepressor binding to progesterone and glucocorticoid receptors involves the activation function-1 domain and is inhibited by molybdate. Mol. Endocrinol. 19:1483-1500. [DOI] [PubMed] [Google Scholar]
- 61.Wang, Q., J. A. Blackford, Jr., L. N. Song, Y. Huang, S. Cho, and S. S. Simons, Jr. 2004. Equilibrium interactions of corepressors and coactivators with agonist and antagonist complexes of glucocorticoid receptors. Mol. Endocrinol. 18:1376-1395. [DOI] [PubMed] [Google Scholar]
- 62.Warnmark, A., E. Treuter, A. P. Wright, and J. A. Gustafsson. 2003. Activation functions 1 and 2 of nuclear receptors: molecular strategies for transcriptional activation. Mol. Endocrinol. 17:1901-1909. [DOI] [PubMed] [Google Scholar]
- 63.Webb, P., C. M. Anderson, C. Valentine, P. Nguyen, A. Marimuthu, B. L. West, J. D. Baxter, and P. J. Kushner. 2000. The nuclear receptor corepressor (N-CoR) contains three isoleucine motifs (I/LXXII) that serve as receptor interaction domains (IDs). Mol. Endocrinol. 14:1976-1985. [DOI] [PubMed] [Google Scholar]
- 64.Wu, X., H. Li, E. J. Park, and J. D. Chen. 2001. SMRTe inhibits MEF2C transcriptional activation by targeting HDAC4 and 5 to nuclear domains. J. Biol. Chem. 276:24177-24185. [DOI] [PubMed] [Google Scholar]
- 65.Yeh, S., E. R. Sampson, D. K. Lee, E. Kim, C. L. Hsu, Y. L. Chen, H. C. Chang, S. Altuwaijri, K. E. Huang, and C. Chang. 2000. Functional analysis of androgen receptor N-terminal and ligand binding domain interacting coregulators in prostate cancer. J. Formos. Med. Assoc. 99:885-894. [PubMed] [Google Scholar]
- 66.Yoon, H. G., D. W. Chan, Z. Q. Huang, J. Li, J. D. Fondell, J. Qin, and J. Wong. 2003. Purification and functional characterization of the human N-CoR complex: the roles of HDAC3, TBL1 and TBLR1. EMBO J. 22:1336-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zeng, C., E. Kim, S. L. Warren, and S. M. Berget. 1997. Dynamic relocation of transcription and splicing factors dependent upon transcriptional activity. EMBO J. 16:1401-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhao, Y., K. Goto, M. Saitoh, T. Yanase, M. Nomura, T. Okabe, R. Takayanagi, and H. Nawata. 2002. Activation function-1 domain of androgen receptor contributes to the interaction between subnuclear splicing factor compartment and nuclear receptor compartment. Identification of the p102 U5 small nuclear ribonucleoprotein particle-binding protein as a coactivator for the receptor. J. Biol. Chem. 277:30031-30039. [DOI] [PubMed] [Google Scholar]