Abstract
The three far-upstream element (FUSE) binding protein (FBP) family members have been ascribed different functions in gene regulation. They were therefore examined with various biochemical, molecular biological, and cell biological tests to evaluate whether their sequence differences reflect functional customization or neutral changes at unselected residues. Each FBP displayed a characteristic profile of intrinsic transcription activation and repression, binding with protein partners, and subcellular trafficking. Although some differences, such as weakened FBP3 nuclear localization, were predictable from primary sequence differences, the unexpected failure of FBP3 to bind the FBP-interacting repressor (FIR) was traced to seemingly conservative substitutions within a small patch of an N-terminal α-helix. The transactivation strength and the FIR-binding strength of the FBPs were in the opposite order. Despite their distinguishing features and differential activities, the FBPs traffic to shared subnuclear sites and regulate many common target genes, including c-myc. Though a variety of functions have been attributed to the FBPs, based upon their panel of shared and unique features, we propose that they constitute a molecular regulatory kit that tunes the expression of shared targets through a common mechanism.
The far-upstream element (FUSE) binding protein (FBP) binds the FUSE of the human c-myc proto-oncogene (1, 10, 16, 32). The DNA-binding domain (DBD) of FBP contains four repeated hnRNPK homology (KH) motifs. Though they are commonly called an RNA-binding motif, some KH domain proteins—including hnRNP K, the prototype of this family—engage single-stranded DNA with an affinity and sequence specificity equal to or greater than those of RNA (3, 4, 10, 16, 32). The carboxyl-terminal domain of FBP stimulates transcription complexes transiting between initiation and promoter escape via activation of the p89/XPB helicase subunit of TFIIH (29). Transcription activation requires at least one copy of a tyrosine-rich motif that is repeated three times in the carboxyl-terminal activation domain (AD) (11). The less well characterized amino terminus of FBP impairs the action of some, but not all, transcription activators (for example, FBP itself and E1a but not VP16) (11).
FBP function is modified by at least two partner proteins. Recruited by FBP, the amino terminus of the FBP-interacting repressor (FIR) blunts AD-mediated stimulation of the p89/XPB helicase activity, permitting only basal transcription (27). Both FBP activation and FIR repression are blocked by mutations in p89/XPB. Associating with the AD, p38/JTV-1/AIMP2 targets FBP for ubiquitinylation and degradation (22). p38 was originally identified as a core protein of a multi-aminoacyl-tRNA synthetase complex (34, 36, 38, 40), and the knockout of p38 in mice indeed dissociates this complex (21); nevertheless, the knockout pups, which develop to full term, support normal levels of protein synthesis (22). Hyperplasia of the lungs and some other organs cause neonatal lethality. FBP and MYC are increased at the RNA and protein levels in these mice, contributing to some or all of the pathology observed (22).
Though the FBP, FBP2, and FBP3 genes are situated upon different chromosomes in both mice and humans, their primary sequences are highly related (7, 41, 42). Each sequence has a four-KH domain DBD that binds FUSE specifically in vitro, and ADs of FBPs 1 to 3, respectively, include three, four, and two highly related tyrosine motifs (7). Because FBP2 and FBP3 are more closely related to FBP than to each other, parsimony designates FBP/FBP1 as the family progenitor. The FBPs are likely to be multifunctional. Besides regulating the transcription of c-myc (and presumably other targets), the FBP family has been shown to bind a variety of RNAs (mostly in vitro). Though considerable evidence indicates a role for FBP2/ KHSRP as a regulator of splicing, it has also been ascribed roles in RNA trafficking, RNA editing, mRNA stabilization, and degradation (5, 6, 14, 24, 25, 30, 33, 39, 43, 44). Functional studies of FBP3 have not been previously reported.
Are the FBPs functionally equivalent or even redundant, tuning the output levels and/or expression patterns among a set of shared targets, or have they diverged to regulate independent targets and to fill distinct physiological niches? To explore this issue, first the molecular parameters governing the FBP-FIR interactions were systematically defined, providing a basis to explain whether or not FBP2 and FBP3 bind FIR. These studies were then extended to compare the three FBPs for their intrinsic activation and repression, subcellular localization, and in vivo targets. The distinctions between the FBP family members suggest that, taken as a set, they usually cooperate to customize the expression profiles among a set of common targets, but they may sometimes act as specialized single agents as well.
MATERIALS AND METHODS
Yeast two-hybrid analysis.
PCR-amplified fragments of FBP or FIR were cloned into pGBT9 for GAL4 DBD fusion or into pGAD424 for GAL4 AD fusion (Clontech). Saccharomyces cerevisiae strain SFY526 or Y190 was transformed using an EZ Yeast transformation kit (Bio 101) and selected using Leu/Trp auxotrophy. Qualitative or quantitative β-galactosidase (β-Gal) assays were done with a yeast β-galactosidase assay kit (Pierce) for at least three different colonies per two-hybrid pair. For quantitative assays, yeast transformants were grown in selective minimal medium until the A595 reached 0.6 to 1.0.
Electrophoretic mobility shift assay (EMSA).
FBP hnRNP K homology motifs KH1+2, KH2+3, and KH3+4 were expressed in Escherichia coli as glutathione S-transferase (GST) fusions and affinity purified using glutathione Sepharose 4B (Pharmacia). The proteins indicated in Fig. 1 were incubated with single-stranded probes in 20 μl of a binding reaction mixture [25 mM HEPES (pH 7.5), 100 mM NaCl, 0.2 mM EDTA, 1 mM dithiothreitol, 10% glycerol, 50 μg/ml poly(dI · dC), and 1 mg/ml bovine serum albumin] for 15 min at room temperature. The complexes were resolved by 8% polyacrylamide gel electrophoresis in 0.5× Tris-borate-EDTA buffer. The 52-mer from the FUSE noncoding strand (5′-GTATA TTCCC TCGGG ATTTT TTATT TTGTG TTATT CCAGG CATGA AAAAC AA-3′) was 5′ end labeled and used as a probe.
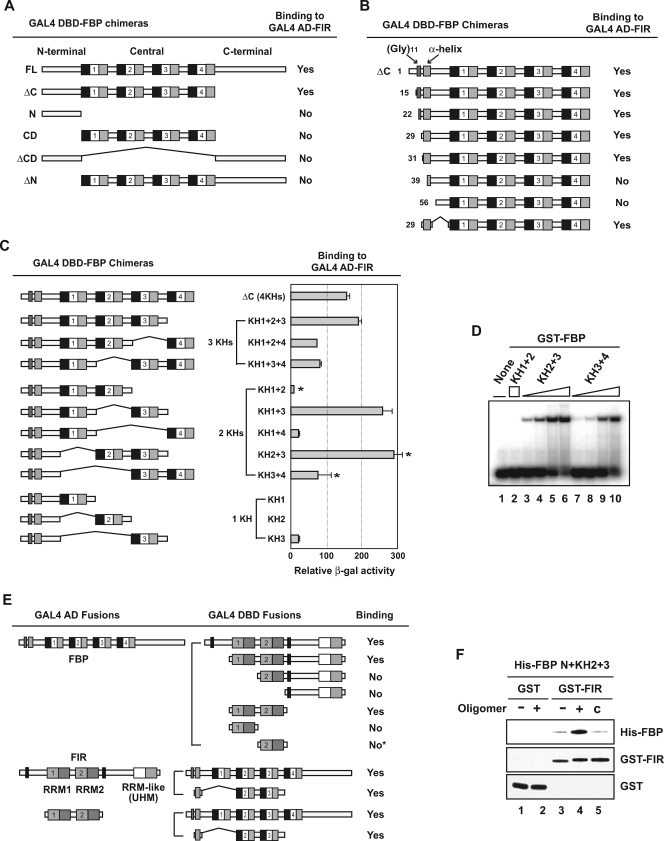
FIG. 1.
Noncontiguous segments of FBP bind FIR. (A) FBP needs both the N-terminal domain and the central domain to bind FIR. Yeast strain SFY526 was cotransformed with pGAD-FIR and the indicated pGBT-FBP constructs, which express GAL4 AD-FIR full-length (FL) and GAL4 DBD-FBP chimeras, respectively. The binding of each pair was monitored by a β-galactosidase assay. Four KH subdomains in the FBP central domain are shown as repeated units of a black box (KH repeat), an open box (spacer), and a gray box (amphipathic α-helix). (B) The α-helix region is the necessary and sufficient feature in the FBP N-terminal domain for FIR binding. A nested deletion series from the N terminus of pGBT-FBP ΔC was cotransformed with pGAD-FIR. The number of each starting amino acid is shown on the left. A direct repeat of 11 glycine residues is indicated in addition to the predicted α-helix-forming region. (C) Different KH domains confer different FIR-binding strengths to the FBP N-terminal domain. pGBT-FBPs containing the full N-terminal domain and the indicated combinations of KH domains were compared for FIR binding using a quantitative β-Gal assay. The three two-KH domain pairs selected for further analysis by EMSA in panel D are marked with asterisks. (D) The DNA binding affinities of different KH domain combinations parallel FIR binding. Ten femtomoles of single-stranded c-myc FUSE 52-mer was incubated with 250 fmol of GST-FBP KH1+2 (lane 2) or with 10, 25, 100, or 250 fmol of GST-FBP KH2+3 (lanes 3 to 6) or GST-FBP KH3+4 (lanes 7 to 10). (E) FBP interacts with the two stereotypical RRM domains of FIR. The indicated parts of FIR were fused to the GAL4 DBD and tested using yeast two-hybrid and β-Gal assays with GAL4 AD-FBP FL (upper half). This result was confirmed by switching the AD-DBD pair combination (lower half). The asterisk indicates that FIR RRM2 alone still displayed, although very weakly, β-Gal activity that was detectable above the background level. Two Ala-rich regions on FIR are shown as black boxes, and three RRM or RRM-like domains are shown as pairs of boxes. The well-conserved RNP2 and RNP1 motifs in RRM1 and RRM2 are shown as light and dark gray, respectively, while the less-conserved ones in the third RRM-like U2AF homology motif (UHM) domain are shown as white and light gray. (F) FBP requires its target nucleic acids for strong FIR interaction. His-tagged FBP N+KH2+3 was incubated with GST (lanes 1 and 2) or GST-FIR RRM1+2 (lanes 3 to 5) in the absence (lanes 1 and 3) or presence (lanes 2 and 4) of the c-myc FUSE 29-mer oligonucleotides. The proportion of His-FBP N+KH2+3 pulled down with GST or GST-FIR RRM1+2 was assessed by Western analysis using anti-FBP or anti-FIR antibody. The non-FBP-binding 40-mer from the complementary FUSE sequence was used as a specificity control (lane 5).
In vitro protein binding assay.
FIR with RNA recognition motifs 1 and 2 (RRM1+2) was expressed as a GST fusion protein and affinity purified. The wild type and mutant FBP N+KH2+3 (containing the N terminus fused to KH2+3) were expressed with a six-His tag at their N termini and purified from the insoluble fraction using a His · Bind kit (Novagen) under 6 M-urea-denaturing conditions. Each pair of purified proteins was mixed with glutathione Sepharose in 1× phosphate-buffered saline (PBS) containing 0.2% Triton X-100, incubated at 4°C for 1 h, washed three times using the same buffer, and eluted from beads into glutathione elution buffer (50 mM Tris-HCl [pH 8.0] and 10 mM reduced glutathione). The 29-mer from the FUSE noncoding strand (5′-GTATA TTCCC TCGGG ATTTT TTATT TTGT-3′) or the 40-mer from the FUSE coding strand (5′-AATAA CACAA AATAA AAAAT CCCGA GGGAA TATAC ATTAT-3′) was added to reaction mixtures at 500 nM where indicated in Fig. 1.
Site-directed mutagenesis.
Targeted amino acid residues were changed with a QuikChange site-directed mutagenesis kit (Stratagene) using pGBT-FBP N+KH2+3 as a PCR template. Each substitution was sequence verified and tested for a yeast two-hybrid assay with full-length pGAD-FIR. Selected mutations were transferred to pET-28 for protein expression in E. coli.
Western blotting.
Yeast cell extracts were generated by boiling cell pellets in sodium dodecyl sulfate sample buffer, and human cell lysates were obtained using reporter lysis buffer (Promega) or M-PER reagent (Pierce). Primary antibodies used were custom-generated, affinity-purified rabbit polyclonal anti-FBP/FBP1 and anti-FIR or commercial antibodies (Santa Cruz), anti-GAL4 (DBD; catalog no. sc-577), anti-LexA (2-12; sc-7544), anti-hemagglutinin (HA) (F-7; sc-7392), anti-actin (I-19; sc-1616), anti-FBP3 (I-20; sc-11103), and anti-green fluorescent protein (GFP) (FL protein; sc-8334). FBP2 was detected using a highly cross-reactive batch of anti-FBP1.
Luciferase reporter assay.
The reporter used to compare different FBP subdomains contained one LexA-binding site (5′-GTACTGTATG T CATACAGTAC-3′) and one synthetic GAL4-binding site (5′-CGGAGGAC A GTACTCCG-3′) upstream of a minimal E1B TATA sequence driving firefly luciferase expression in pGL3 (Promega). FBP N-terminal domains were expressed as LexA fusions in pBXL1, and FBP C-terminal domains were expressed as GAL4 DBD fusions in pSG424. HeLa cells were transfected with the amounts of reporter, LexA fusion, and GAL4 fusion constructs indicated in the legend to Fig. 3 using LipofectAmine reagent (Invitrogen) and analyzed after 24 h using a luciferase assay system (Promega). For all cotransfections, the total amounts of plasmid were equalized using the corresponding control vector.
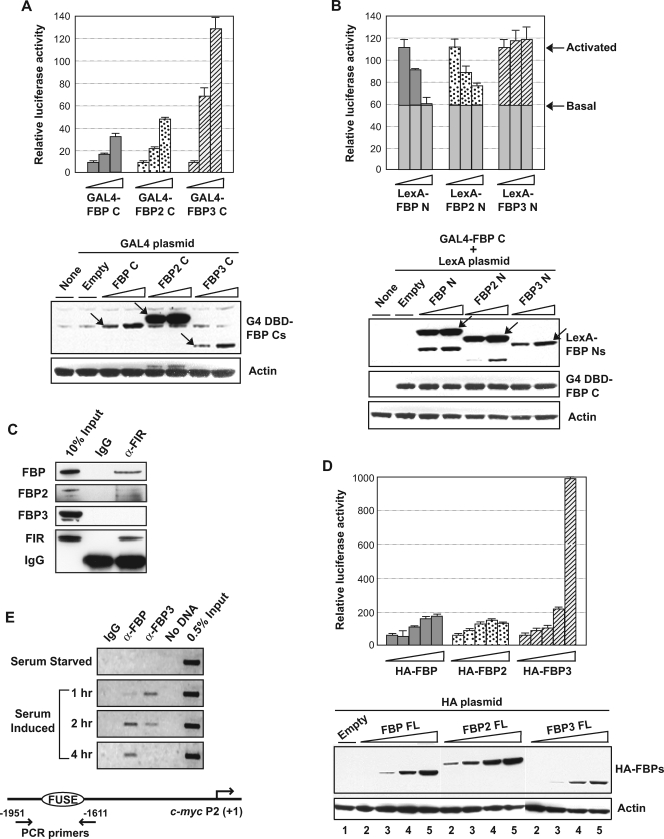
FIG. 3.
The effector output of the FBPs is graded. (A, top) The C terminus of FBP3 is the strongest transactivator among the FBPs. Each FBP C was expressed as a GAL4 DBD fusion in HeLa cells and tested for transactivation of a single GAL4 site-driven firefly luciferase reporter using 0, 100, and 250 ng. The total amount of DNA used per transfection was kept constant by adjustment with an empty GAL4 DBD plasmid, and each point was averaged from duplicate transfections. (Bottom) The expression of each GAL4 DBD fusion protein was monitored using anti-GAL4 (G4) DBD antibody and normalized against the actin signal. Each fusion protein of the expected size is marked with an arrow. (B) Only the FBP3 N terminus is devoid of the repressor activity. Each FBP N was expressed as a LexA fusion, and all were tested for their effect on transactivation by GAL4-FBP C. Arrows mark the luciferase activity levels before and after transactivation by FBP C, demonstrating that the basal transcription (the height of gray bars) is resistant to repression by FBP N. (Bottom) The expression of each LexA fusion protein was monitored using anti-LexA antibody, and GAL4 DBD-FBP C expression from the cotransfected plasmid is shown as an internal control. (C) FBP3 does not interact with FIR in vivo. Extracts of the DSP-cross-linked HeLa cells were immunoprecipitated with anti-FIR and analyzed for the presence of coprecipitated FBPs. FBP2 was monitored through cross-reactivity with anti-FBP/FBP1. IgG, immunoglobulin G; α-FIR, anti-FIR. (D) FBP3 is the strongest c-myc transactivator. Plasmids encoding each HA-tagged full-length FBP were transfected (0, 50, 100, 250, and 500 ng for each) into HeLa cells and compared for their effects on the c-myc promoter luciferase reporter. (Bottom) The expression of each HA-FBP was monitored using anti-HA tag antibody. Lane 1 shows 0 ng of HA-FBP-expressing plasmid transfection, and lanes 2 to 5 show each HA-FBP-expressing plasmid in 50- to 500-ng transfections. (E) Recruitment of FBP3 precedes FBP/FBP1 at the c-myc promoter upon serum stimulation. Serum-starved human fibroblast cells (Hs68) were formaldehyde cross-linked at the indicated time points after serum induction and analyzed for FBP or FBP3 binding to FUSE using chromatin IP. (Bottom) Location of the primer pair is shown relative to the c-myc P2 promoter.
The c-myc-luciferase reporter had the same 3.3-kb c-myc promoter as the previously reported myc-CAT reporter (1) and was cotransfected with each HA-FBP-expressing construct in pCGNM2.
Immunoprecipitation and chromatin IP.
HeLa cells (1 × 107) were washed twice with 1× PBS and resuspended in 0.5 ml PBS. DSP (dithiobis[succinimidyl] propionate; Pierce) was added to cell suspensions to a final concentration of 1.25 mM and incubated at room temperature for 10 min. After the incubation, Tris-HCl was added to 25 mM to stop the cross-linking. Cells were washed twice with PBS and extracted with 600 μl of radioimmunoprecipitation assay buffer (10 mM Tris, 300 mM NaCl, 0.1% sodium dodecyl sulfate, 0.1% sodium deoxycholate, 1% Triton X-100, 1 mM EDTA, 1× protease inhibitor) for 2 h at 4°C. Three-hundred-microliter extracts were used for each immunoprecipitation (IP) with the rabbit polyclonal anti-FIR. Chromatin IP was done with human primary fibroblast cells (Hs68) as described previously (28) using rabbit polyclonal anti-FBP or goat polyclonal anti-FBP3 (I-20; sc-11103; Santa Cruz).
Confocal microscopy.
HeLa cells were electroporated with a BTX electroporator (model ECM 830) using 5 μg plasmid DNA and 15 μg sheared salmon sperm carrier DNA in a 2-mm-gap cuvette at 200 V, with four 1-ms pulses at 0.5-s intervals. After electroporation, the cells were plated in Nalgene Lab Tek II chambers, and after ∼6 h, the medium was changed to Dulbecco's modified Eagle's medium with 25 mM HEPES without phenol red (Invitrogen). In cotransfection experiments, the ratio of cyan fluorescent protein (CFP) to yellow fluorescent protein (YFP) constructs was 1:1. Cells were visualized with a Zeiss 510 META confocal microscope with a 63×/1.4-numerical-aperture planapochromat oil objective (12). Cells expressing very high levels of recombinant protein or aggregated material were disregarded.
FRAP.
Fluorescence recovery after photobleaching (FRAP) experiments were performed with a Zeiss LSM 510 confocal microscope with a 100×/1.3-numerical-aperture planapochromat oil objective. GFP was excited with the 488-nm line of an Ar laser, and GFP emission was monitored above 505 nm. Cells were maintained at 37°C with a Nevtek ASI 400 air stream incubator (Nevtek). In transfected cells, a 2.4- by 2.4-μm square spot inside of the nucleoplasm was bleached for ∼0.6 s using the 488-nm laser line at 100% laser power. Cells were monitored at 0.5-s intervals. For quantification, total fluorescent intensities of a region of interest in the bleached area and in the total nuclear area were monitored using Zeiss software. Background fluorescence was measured in a random field outside of the cells. The relative fluorescence intensity double normalized to the prebleach value was calculated as described previously (13). In all FRAP experiments, more than 20 cells from at least two independent experiments were quantified.
RNA interference.
Stealth small interfering RNAs (siRNAs) were synthesized by Invitrogen for the following sequences: siFBP1-990 (5′-CCGAU GUCAA CAUGC UGCAG AAAUU-3′), siFBP2-378 (5′-GGGAG ACUCA AUCAG UUCUC AACUU-3′), and siFBP3-145 (5′-UCAGU AUAUG GAGAC GGAGU ACAAA-3′). Each siFBP was transfected into HeLa or Hs68 cells at a 50 nM concentration using LipofectAmine 2000 (Invitrogen), and cells were harvested after 2 days. Stealth RNA interference-negative control duplexes (Invitrogen) were transfected as the controls.
Microarray analysis.
Total RNAs were isolated using Trizol reagent (Invitrogen) and used to derive fluorescently labeled cDNAs (coupled to Cy3 or Cy5). Expression microarrays were produced by the NCI Microarray Facility at the Advanced Technology Center using the human operon version 3.0 genome oligonucleotide set that contains 34,580 probes representing 24,650 genes and 37,123 gene transcripts as melting temperature-normalized 70-mers. Hybridized arrays were scanned using a model 4000 Axon Scanner and analyzed using the bioinformatic tools supplied by the NCI's CCR Microarray Center (mAdb). At least two independent sets of RNAs were hybridized for each siFBP and compared all together.
Microarray data accession numbers.
The microarray data set was deposited in the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo) under the series submission code GSE4914, which contains six arrays of the accession numbers GSM110526 to GSM110531.
RESULTS
The physiological specialization of transcription factor family members often involves the alternative recruitment of coactivators, corepressors, or other cofactors to effector domains linked with highly homologous and functionally redundant DBDs. The FBPs possess separable repression, DNA binding, and activation domains sequentially arrayed from the amino through the carboxyl termini (7, 11). The interactions of the FBP family members with some of their known partners were compared.
FBPs and FIR play important roles in shaping the pulse of c-myc transcription induced by serum (28). Therefore, the interactions of the FBPs with FIR were studied in detail. FIR was discovered using a yeast two-hybrid screen with full-length GAL4 DBD-FBP as bait and HeLa cDNAs fused to the GAL4 AD as prey (27). A second FBP partner discovered in this same screen was JTV-1/p38/AIMP2, which interacts with FBP's AD to direct ubiquitinylation and degradation (22). p38 bound best with FBP and worst with FBP3 (see Fig. S1A in the supplemental material). A further series of two-hybrid experiments was conducted to define the molecular features enabling or preventing the binding of FIR with the FBPs.
Two segments of FBP are required to bind FIR.
FBP truncated after the central domain (CD) (FBP ΔC) bound FIR as strongly as full-length FBP (FBP FL) (Fig. 1A), predicting that either the N-terminal domain or the central domain would direct this interaction. However, neither the N-terminal domain nor the central domain alone bound with FIR in this assay (Fig. 1A). Inclusion of the C-terminal domain (in constructs FBP ΔCD and FBP ΔN) did not influence these results.
In FBP N, a string of 11 consecutive glycines is followed by a potential amphipathic α-helix (the N-terminal α-helix [αN]) (10). FIR binding remained intact after these glycines were deleted but was abrogated by encroachment upon the predicted α-helix (Fig. 1B). To assess whether other N-terminal sequences were required to bind FIR, just this helical segment was fused to the CD and tested for FIR interaction. FIR binding was restored to the CD by an α-helix as short as 22 amino acids (Fig. 1B).
The central domain of FBP consists of four hnRNP KH domains, each comprised a 30-amino-acid KH repeat followed by an amphipathic α-helix (3, 4, 10). When the KH domains were serially deleted from the C-terminal end of the FBP central domain, removal of the KH3 domain coincided with a profound weakening of the FBP-FIR interaction; further deletion of KH2 completely abolished this interaction (Fig. 1C). To ascertain whether the attenuation and loss of FIR binding related to the number or the quality of the deleted KH domains, FBP derivatives retaining the full amino terminus but having different combinations of KH domains were compared using quantitative two-hybrid assays.
All GAL4 DBD-FBP chimeras tested with three KH domains (KH1+2+3, KH1+2+4, and KH1+3+4) scored strongly positive for FIR interaction, with FBP KH1+2+3 being the strongest (Fig. 1C). Thus, no single KH domain is essential to confer FIR binding capacity. (Although FBP KH2+3+4 was not tested in parallel, subsequent results [see below] assert that it too interacts with FIR). However, when pairs of KH domains were similarly tested, the strength of their FIR interactions varied for different combinations, from very strongly (almost twice as much as FBP ΔC) to quite weakly (only about 5% of FBP ΔC) (Fig. 1C). Interestingly, any two-KH domain combination that included KH3 bound more strongly with FIR (KH1+3, KH2+3, and KH3+4), than those combinations lacking KH3 (KH1+2 and KH1+4). The same relative hierarchy of FIR binding was preserved when the various KH motif pairs were fused directly behind the N-terminal α-helix instead of to the whole N-terminal domain (data not shown). The single strongest determinant of FIR binding is the KH3 motif, which is the only structural element that alone supported low, but still detectable, β-Gal activity (Fig. 1C). Therefore, both the total number and identities of the supporting KH motifs set the strength of the FBP-FIR interaction.
Overlapping determinants for nucleic acid and protein binding by FBP and FIR.
Besides binding with FIR, the four KH domains are directly responsible for sequence-specific nucleic acid binding by FBP (4, 10, 32). What is the relationship between DNA binding and FIR binding for the FBP KH domains? The GST-fused FBP KH3+4 protein has previously been shown using EMSA to bind to the noncoding strand of the c-myc FUSE region with higher affinity than that of FBP KH1+2. (10, 32). As this order of nucleic acid binding affinities paralleled the FIR-binding affinities mentioned above, it seemed that the role of KH domains in FBP-FIR interactions might be related to their nucleic acid binding capacity. A third GST fusion construct for FBP KH2+3, the strongest FIR binder of the various KH domain pairs, in fact, also yielded the strongest complexes by EMSA using c-myc FUSE probes (Fig. 1D, lanes 2, 6, and 10, normalized for protein).
Like FBP, FIR also possesses evolutionarily conserved nucleic acid-binding motifs in its central domain, namely, two copies of the evolutionarily conserved RRM (which can also bind single-stranded DNA) in addition to the distantly related RRM-like U2AF homology motif at its C terminus (20, 35). When the N terminus was serially deleted from FIR and analyzed with yeast two-hybrid assays, the loss of RRM1 abolished FBP-FIR interaction (Fig. 1E). Removal of sequences both amino and carboxyl terminal to RRM1+2 preserved the interaction with FBP, until either RRM was encroached upon. Unlike RRM1, which was totally devoid of FBP-binding capacity when alone, RRM2 fused to GAL4 DBD showed a very weak, but still detectable, interaction with GAL4 AD-FBP. The minimal segment of FIR that bound full-length FBP and the minimal segment of FBP that bound full-length FIR also bound to each other (Fig. 1E). Therefore, FIR RRMs 1 and 2 are necessary and sufficient for full FBP interaction.
Do FBP, FIR, and DNA form a ternary complex, and if so, do FIR and DNA enhance or hinder each other's binding? To answer these questions, the interacting portions of FBP and FIR defined by yeast two-hybrid analysis were expressed in E. coli and affinity purified for in vitro binding assays. When His-tagged FBP N+KH2+3 was incubated with GST-FIR RRM1+2 and recovered on glutathione beads, a small amount of the FBP derivative was recovered (Fig. 1F, lane 3). However, when the same reaction mixture was supplemented with a single-stranded 29-base deoxyoligonucleotide from the c-myc FUSE region, which specifically interacts with the FBP DBD, the amount of FBP pulled down was greatly increased (Fig. 1F, lane 4). This enhancement was seen only with FBP-binding oligonucleotides and not with non-FBP-binding control sequences as long as 40-mers (Fig. 1F, lane 5), arguing against nonspecific DNA bridging. Thus, the interaction of FBP with FIR is either stabilized or augmented by the interaction of FBP and/or FIR with the appropriate nucleic acid sequences.
FBP2 also recruits FIR, but FBP3 does not.
FBP's two close relatives, FBP2/KHSRP and FBP3, each share most of FBP's structural features, including αNs, central nucleic acid-binding KH domains, and tyrosine-rich, transactivating C-terminal domains (7, 33). The sequence homology of FBPs 2 and 3 with FBP is especially high within the regions directing FBP-FIR interactions. Therefore, FBPs 2 and 3 were each fused to the GAL4 DBD and checked for interaction with GAL4 AD FIR. Whereas FBP2 bound FIR strongly, interaction with FBP3 was not detectable above background (Fig. 2A). To ascertain the source of this difference, various exchanges were performed between the αNs and the CDs of all three FBPs. When they were combined with FBP1's CD, the αNs of FBP1 and FBP2 supported FIR binding, but the αN of FBP3 did not, following the same pattern displayed by the full-length proteins (Fig. 2A). In contrast, when the three different CDs were fused downstream of the FBPs' αNs, all three hybrids bound strongly with FIR (Fig. 2A). Thus, the CDs of all three FBPs support FIR binding, and so the discriminating determinant of this interaction resides principally within the FBPs' amino termini.
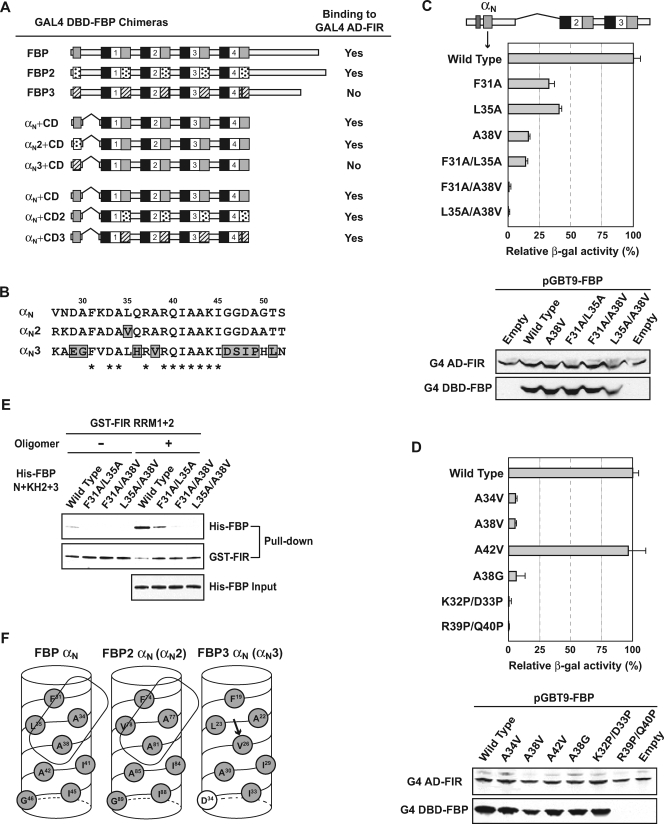
FIG. 2.
An alanine-rich patch on the FBP N-terminal α-helix allows FIR to bind. (A) The N-terminal α-helices of the three FBP homologues license or prohibit their interaction with FIR. Each full-length FBP, from the αN to the last amino acid residue, was fused to the GAL4 DBD and tested for FIR interaction using yeast two-hybrid assays (top). The three different N-terminal α-helices (αN from FBP, αN2 from FBP2, and αN3 from FBP3) fused directly to the same FBP/FBP1 CD were compared for FIR binding (middle). The three different central domains (CD from FBP, CD2 from FBP2, and CD3 from FBP3), each fused to the same FBP/FBP1 αN, were also compared (bottom). α-Helical regions in the N-terminal domain and the central domain are indicated in gray boxes for FBP, dotted boxes for FBP2, and hatched boxes for FBP3. (B) FBP3 has the most divergent N-terminal α-helix. Amino acid sequences are shown for the three αN regions. Numbering is based on the FBP/FBP1 sequence, and the amino acid residues deviating in only one FBP family member are marked in gray boxes. Residues that are the same for all three FBPs are marked with asterisks. (C, top) A hydrophobic patch on the FBP αN composed of Phe-31, Leu-35, and Ala-38 is critical for FIR binding. The GAL4 DBD-FBP derivative shown on the top served as the template for PCR mutagenesis directed at the N-terminal α-helix (αN). Strains with single and double mutations attenuating or eliminating FIR binding were compared with the wild type using yeast two-hybrid and quantitative β-Gal assays. (Bottom) Similar levels of expression of the wild type and the FIR-binding-defective mutant FBPs were verified by immunoblot analysis of the same yeast transformants used for interaction assays. FIR proteins from the cotransformed pGAD-FIR plasmid were used as an internal control. G4, GAL4. (D) Ala-34 also participates in forming the FIR-binding patch on the FBP N-terminal α-helix. Additional Ala residue neighbors of Ala-38 in the predicted three-dimensional α-helix structure (shown in Fig. 2F) were mutated to Val and compared with the wild type and A38V mutant as well as with the helix-breaking Pro substitutions on the hydrophilic surface. In order to test the direct contribution of the alanine methyl group in FIR binding, Ala-38 was also replaced by a smaller residue, Gly. One of the two helix-breaking, double-Pro substitutions failed to express any stably detectable GAL4 DBD fusion protein (R39P Q40P). (E) The same FBP αN mutants faulty for FIR binding in yeast two-hybrid assays failed to bind FIR in vitro. Three purified double mutants as well as wild-type His-FBP N+KH2+3 were incubated with GST-FIR RRM1+2 in the absence (−) or presence (+) of the c-myc FUSE 29-mer, as shown in Fig. 1F. (F) Three-dimensional helical wheel of αNs. The amino acid residues of the three FBP αNs are shown along the hydrophobic face of the helical wheel. Proposed FIR-binding surfaces on αN and αN2 are shown as enclosed boxes. The FBP3 Val that replaces an Ala in FBP1 and FBP2 and disrupts the FBP3-FIR interaction is marked by an arrow. Amino acid numbering is specific for each FBP, and hydrophobic or hydrophilic residues are marked as gray or white, respectively.
Residue-by-residue inspection of the αN sequences of the FBPs revealed that the FBP3 αN (αN3), although 50% identical to FBP1's and FBP2's αNs, diverged more from the FBP1 and FBP2 αNs than FBP1 and FBP2 did from each other (Fig. 2B). In contrast, the central domains were similarly conserved (7). It was not immediately apparent what αN features shared by FBP1 and FBP2, but not by FBP3, give license for the interaction with FIR.
A hydrophobic patch in the αN is required to bind FIR.
The FBP αN sequence, as well as the α3s (the third α-helix within each of the four central domain KH repeats; not αN3), displays a striking amphipathic character, including a broad hydrophobic face (Fig. 2F) (7, 10). In order to map the FIR-binding surface along this helix, all the amino acid residues of FBP1 from Asp-29 to Gly-50, except Ala, Gly, and Ile, were mutated one by one to Ala and examined for FBP-FIR interaction. Though several single amino acid substitutions attenuated β-galactosidase activity by two- to threefold, none of the substitutions eliminated FIR binding (Fig. 2C, F31A and L35A; data not shown for others).
By using a second strategy to generate mutants, the residues targeted in FBP were replaced by residues unique to non-FIR-binding FBP3 (Fig. 2B, from A30G to A49P). The substitution of Ala-38 in FBP with Val as found in FBP3 was the most deleterious for FIR binding among all the single amino acid changes tested (Fig. 2C).
Within the full set of single mutants, A38V, F31A, and L35A most effectively negated FIR binding; these three mutations cluster in a hydrophobic patch on the predicted helix (Fig. 2F). Combining A38V with either F31A or L35A entirely eliminated FBP-FIR interaction (Fig. 2C), supporting a role for this surface in FBP-FIR interaction. In contrast, introducing single or double mutations on the hydrophilic side left FIR binding intact (data not shown).
Inspection of the predicted helices of the three FBP αNs revealed that the single most critical residue for FIR binding, Ala-38, to be in the middle of a strip of four alanines wrapping along the hydrophobic surface (Fig. 2F; results for Ala-30 not shown). Each of the remaining three Ala residues were changed to Val, as was done for A38, and tested for FIR binding. Of the three, only A34V affected FIR binding as much as A38V (Fig. 2D; data for Ala-30 are not shown); thus, the small side chains of A34 and A38, rimmed by the bulky hydrophobic groups of Phe-31 and Leu-35, set the topography enabling FIR binding. Replacement of A38 with G also crippled binding, showing the importance of the hydrophobic character of the FIR-combining surface (Fig. 2D).
The simplest interpretation of this mutational analysis has FIR recognizing a patch on the hydrophobic surface of a predicted α-helix but not contacting residues on the hydrophilic surface. In this model, substitutions along the hydrophilic surface should be inert to FIR (as observed above), unless they disrupted helix formation. Therefore, two double-proline substitutions were placed on the hydrophilic side to break the helical structure. Though one double mutant was unstable and could not be evaluated for FIR binding (Fig. 2D, R39P/Q40P), the other mutant demonstrates that the α-helix formation is a prerequisite for FBP-FIR interaction (Fig. 2D, K32P/D33P). Substituting with alanine rather than proline at these same residues had no effect on FIR binding (K32A, D33A, R39A, and Q40A; [data not shown]).
In vitro binding assays were performed to confirm that these mutations directly weakened the physical association of FBP αN with FIR. When GST-FIR RRM1+2 was used to pull down FBP N+KH2+3 fused with wild-type or mutant αNs, as in Fig. 1F, the partially impaired F31A/L35A double mutant retained residual binding activity, while the severely disabled F31A/ A38V mutant as well as the L35A/A38V mutant displayed no binding, paralleling the yeast two-hybrid results (Fig. 2E).
The outputs of the activation and repression domains of FBP, FBP2, and FBP3 are oppositely graded.
As previously shown using reporter assays with transfected GAL4 DBD fusions, the FBP-FIR complex carries one activation domain (FBP C) and two repression domains (FBP N and the N terminus of FIR) (11, 27); complexes bearing FBP N or FIR neutralize activation by FBP C. All three FBP family members share conserved but nonidentical C-terminal and N-terminal domains. Thus, the net regulatory output of a particular FBP-FIR complex is likely to reflect the relative potencies of its constituent positive and negative effector domains. To study their effector function, singly or in combination, the activation or repression domains of all three FBPs were fused with the GAL4 DBD or the LexA protein, respectively, and the resulting chimeras were tested in vivo using a luciferase reporter carrying a single GAL4 site and a single LexA site upstream of a minimal promoter.
First, when the activation domains of the three FBPs were compared, all FBP C proteins stimulated transcription well, even from a single GAL1 upstream activating sequence (UAS). FBP3 C proved to be by far the most potent activator, with 13-fold-increased activation under the conditions shown (Fig. 3A), whereas under the conditions selected, FBP C and FBP2 C showed 3- to 5-fold-increased activation. The stronger transactivation by FBP2 C than by FBP C most likely reflected a higher expression of GAL4-FBP2 C protein than the other two proteins, despite the transfection of equal amounts of plasmids (Fig. 3A, bottom). Thus, normalizing for protein expression, FBP C transactivates with intermediate strength, while FBP2 C is the weakest transactivator. The activator strength of the three FBPs does not correlate with the number of tyrosine-rich motifs (YMs) borne by each family member (three, four, and two for FBP, FBP2, and FBP3, respectively). FBP and FBP2 harbor proline-rich segments interposed between the CD and the AD, which might modulate YM transactivation, since removal of this segment from FBP C marginally increased its potency (data not shown).
The FBP family N termini were compared for intrinsic repression of FBP C, independent of FIR recruitment. Activation of expression from a single GAL1 UAS by a fixed amount of GAL4-FBP C (Fig. 3B, the level marked as “Activated”) was opposed by cotransfecting increasing doses of LexA-FBP N, -FBP2 N, or -FBP3 N, recruited to the single adjacent LexA-binding site. Whereas FBP N and FBP2 N exhibited the expected reduction of reporter activity, FBP3 N did not (Fig. 3B). Because FBP3 N had no effect on activation by FBP3 C either, the failure to repress FBP C was not due simply to a mismatch between the activation and repression domains (data not shown). Although LexA-FBP3 N was less efficiently expressed than the FBP and FBP2 chimeras (Fig. 3B, bottom), increasing the amount of DNA transfected to much higher levels still failed to unmask FBP3 N's repressor activity; if anything, FBP3 N augmented activation by FBP C or FBP3 C (Fig. 3B and data not shown), which could further potentiate activation by FBP3 in vivo.
Using GAL4 and LexA chimeras to deliver the three FBP Cs and FIR, respectively, to the same reporter carrying a single upstream GAL4 UAS and a LexA operator showed that the three activation domains were repressible by FIR (data not shown); however, because FBP3 is unable to engage FIR, it would escape this repression in vivo. Consistent with this expectation, endogenous FIR from HeLa cells cross-linked in vivo coimmunoprecipitated with native FBP and FBP2 but not with FBP3 (Fig. 3C).
The transactivation strengths of the full-length FBPs were further compared using the c-myc promoter, a well-established in vivo target of FBP. When each HA-tagged FBP-expressing plasmid was cotransfected with the c-myc-luciferase reporter, FBP3 once again proved to be the most potent activator (Fig. 3D). At lower DNA doses, HA-FBP2 was the strongest activator (slightly stronger than FBP). However, FBP2 was also the most highly expressed of the FBPs, whereas FBP3 was the most weakly expressed (Fig. 3D, bottom). After normalization for expression levels, FBP3 is by far the most potent activator of the c-myc reporter, and FBP2 is the weakest of the three.
FBP3 and FBP are recruited sequentially during c-myc activation.
To learn more about the roles of FBP and FBP3 in endogenous c-myc expression in human primary fibroblasts, where both factors abide, chromatin immunoprecipitations were performed throughout a single cycle of serum starvation and restimulation. The kinetics of c-myc induction in this classic system has been well described (9, 19, 31, 45, 47). Recently, FBP and FIR have been shown to act sequentially as a second-stage booster and a late-recruited repressor, respectively, that together help to generate the serum-induced pulse of c-myc transcription (28). Unexpectedly, chromatin IP revealed FBP3 to be the first activator recruited to FUSE during the upsurge of c-myc transcription (Fig. 3E). Approaching peak levels, FBP3 is replaced by FBP (Fig. 3E), and thereafter FBP is displaced by FIR (28). Upon binding to FUSE, with its potent activation domain and its inability to conscript FIR, FBP3 is equipped to ensure an uninterrupted ascent to peak transcription.
Subcellular distributions and trafficking of the FBPs.
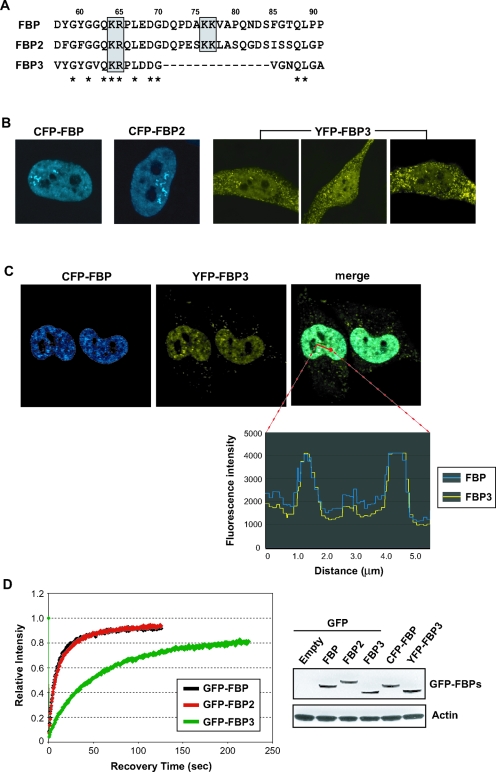
The highest sequence homology in the FBP family members is seen within the central nucleic acid-binding domain, and all three FBP CDs bind the FUSE sequence with great affinity (7). So, if the FBPs cohabit the nucleus and have a common DNA-binding specificity, then they would be expected to coregulate common targets with the relative influence of each family member, determined primarily by intranuclear abundance. Alternatively, belying their sequence homology, they might bind to different targets or they might be partitioned to different regions of the cell; sequestering one of the family members away from chromatin would obviously restrict DNA binding. In the former case of cohabitation, the FBPs would localize to similar intranuclear sites, whereas in the latter instances, independent localization would be expected. Therefore, the intranuclear dispositions of the three FBP family members were compared. It was particularly important to assess the subcellular localization of FBP3, not only because it was functionally disparate but also because it does not share an N-terminal bipartite nuclear localization signal (NLS) conserved in both FBP and FBP2/KHSRP (15, 17). In FBP3, half of this NLS is conspicuously absent (Fig. 4A) Unlike FBP N, the N terminus of FBP3, with its defective NLS, failed to direct its GFP fusion protein to the nucleus (data not shown). Moreover, chimeras of the full-length FBP3 with GFP, CFP, or YFP trafficked to both the cytoplasm and the nucleus (the YFP fusion is shown in Fig. 4B). The relative portion of fluorescent FBP3 inside the nucleus differed from cell to cell, from approximately 30% to 60% of the total. In contrast to FBP3, when expressed the same way, FBP and FBP2 were almost completely nuclear (Fig. 4B). Immunostaining of endogenous FBP and FBP3 in formaldehyde-fixed cells generated exactly the same sorts of patterns as observed in live cell images expressing their GFP-fused counterparts (data not shown). Moreover, similar differential trafficking of GFP-FBP versus GFP-FBP3 occurred in other human cells, including Hs68 primary fibroblasts, MCF-7, and U2-OS (data not shown).
FIG. 4.
Subcellular distributions and intranuclear trafficking of the FBPs. (A) FBP3 lacks half of the bipartite NLS present in FBP and FBP2. Amino acid sequences around the conserved NLS (gray boxes) are compared for the three FBPs. Numbering is based on the FBP/FBP1 sequence, and the residues that are the same for all three FBPs are marked with asterisks. (B) Only FBP3 localizes both inside and outside the nucleus. The three full-length FBPs were expressed as GFP, CFP, or YFP fusions in HeLa cells and observed by confocal microscopy. Representative images are shown for CFP-FBP, CFP-FBP2, and YFP-FBP3. (C) All three FBPs colocalize in the nucleoplasm. Images are of CFP-FBP and YFP-FBP3 expressed in HeLa cells, and the fluorescence intensity of each GFP derivative along the area of the red arrow in the merged image is plotted on the bottom. (D) FBP3 has FRAP kinetics slower than those of the other FBPs. Each GFP-FBP was expressed in HeLa cells, photobleached, and measured for its fluorescence recovery every 0.5 s (left). (Right) The expression of each GFP or its variant fusion protein was monitored by Western analysis using anti-GFP and antiactin antibodies.
The intranuclear distributions of FBP and FBP3 were compared by coexpressing the former as a CFP fusion and the latter as an YFP fusion (Fig. 4C) or vice versa (data not shown); both proteins overlapped perfectly, both in coarse FBP-enriched foci and in a diffuse granular pattern throughout the nucleoplasm. As expected, FBP and FBP2 also colocalized the same way (data not shown). Thus, with the resolution of visible light, all members of the FBP family are recruited to the same nuclear sites. However, once members are recruited to their intranuclear sites, the kinetic behaviors of the FBPs are not all the same; as analyzed by FRAP, FBP3 is decidedly different from FBP and FBP2, which are indistinguishable from each other (Fig. 4D). As reported previously, FBP (and as seen here, FBP2) is a slow-moving protein, recovering fluorescence on the same time scale as HP1 and other chromatin-associated proteins, but slower than most conventional transcription factors (37). The strongest activator of the FBP family is also the least mobile one; FBP3 moves as sluggishly as histone H1. The cytoplasmic fraction of FBP3 recovered faster than the nuclear FBP3, on the same time scale as intranuclear FBP or FBP2 (data not shown). Clearly, the association with FIR is not required to tether the FBPs in the nucleus, since FIR binds only with the two faster-moving family members.
The FBPs coregulate many targets.
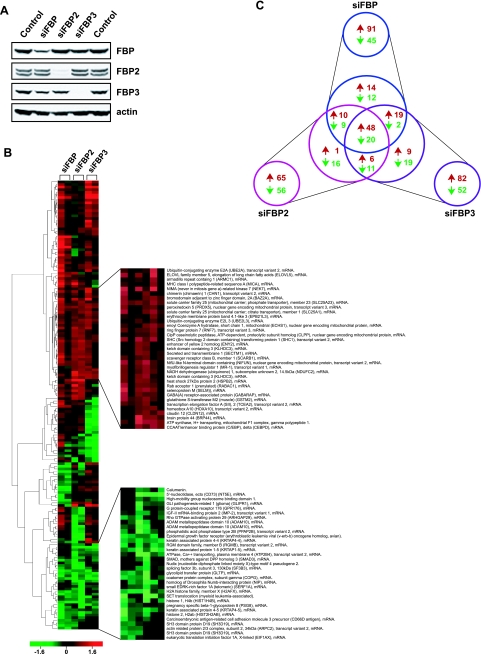
Extensive colocalization of the FBPs might suggest that these molecules coregulate a set of common targets. siRNAs knocking down the levels of each FBP were transfected into fibroblasts to obtain an initial assessment of the degree to which these factors act independently, concertedly, or redundantly. If they act independently, the overlap between sets of target genes would be small; if fully redundant, single knockdowns would perturb few genes because the surviving family members would compensate for the loss of any one FBP. However, if they act in concert, the removal of any family member would disturb the profile of shared target expression. siRNAs were developed and selected for three FBPs that efficiently decreased the steady-state level of each FBP protein in both HeLa and Hs68 cells (Fig. 5A).
FIG. 5.
The FBPs cross-regulate many common targets. (A) Knockdown of each FBP by siRNA transfection. HeLa whole-cell extracts were compared by Western analysis 2 days posttransfection with the indicated siRNAs. Endogenous FBP2 levels were monitored using a cross-reacting anti-FBP/FBP1 primary antibody. (B) Hierarchical cluster analysis of the FBP knockdown microarrays. Each microarray was hybridized with two differentially labeled cDNAs derived from the total human primary fibroblast RNAs, one from the indicated siFBP transfection and the other from the negative-control siRNA transfection. Each column represents an independent experiment, and each row represents an individual gene/feature. Boxes with red or green color indicate the extent of up- or down-regulation as shown in the scale bar at the bottom. One hundred seventy-two selected genes that consistently responded to siFBP transfections are shown on the left, and two representative groups that behaved similarly in all six arrays (either up or down) are shown enlarged with their gene descriptions on the right. The complete list of 172 genes is available in Table S1 in the supplemental material, and the entire microarray data set has been deposited in the GEO database under the accession number GSE4914. MHC, major histocompatibility complex; ADAM, a disintegrin and metalloprotease; RGM, repulsive guidance molecule. (C) A Venn diagram summary of the siFBP microarray. Numbers of up- or down-regulated genes that responded commonly or uniquely in response to the knockdown of each FBP are shown in red and green, respectively.
The genomic response to challenge with the siFBPs revealed several patterns among the 172 unique genes surviving unbiased threshold and quality control filtering (Fig. 5B). This number is likely to underestimate the number of FBP family targets. For some genes, such as c-myc, the influence of the FBP-FIR-TFIIH system is greatest during physiological transitions and of lesser importance under steady-state conditions, so it is likely that they would be missed in a steady-state analysis such as this. Approximately one-third of the total number of targets fell into each of the following categories: (i) targets unique to an individual FBP (or peculiarly regulated by FBPs, i.e., those that were down-regulated by one but up-regulated by the other two), (ii) targets shared between two FBPs toward the same direction of response, and (iii) targets shared among all the FBPs (Fig. 5C). In total, the expression of 89 targets decreased and 107 increased with FBP knockdown (the total 196 exceeds 172 because some targets increased with the knockdown of one FBP while decreasing with the knockdown of another, creating an overlap between the two sets). Although the loss of a pure activator, such as FBP3, would be expected to depress the expression of direct targets, it is more difficult to predict the response of the direct targets of a weaker activator, namely, FBP or FBP2. The rules determining the degree to which activation or repression prevails for these bifunctional proteins remain to be elucidated. The indirect targets for all the FBPs could be either up- or down-regulated depending on the activities of the intermediary direct targets. Regardless of the direct or indirect response, the overall list of the selected genes was reproducible from independent microarray experiments using other siRNAs targeting a different sequence of the FBP coding region (data not included).
The targets of the FBP family include enzymatic, regulatory, and structural components from diverse cellular and extracellular subsystems, as shown in two representative groups in Fig. 5B.
DISCUSSION
The FBPs coregulate targets.
Are the three FBP family members functionally coordinated to impart fine control on common targets, or have they evolutionarily diverged to regulate separate sets of targets? Several lines of evidence from biochemical, molecular biological, cell biological, and physiological studies argue for the former possibility. (i) The knockdown of the individual FBPs modifies the expression of a highly overlapping set of genes. The FBPs must fill distinct roles in the regulation of these common targets, and full redundancy would have been missed in this screen. (ii) During the serum activation cycle at the c-myc promoter, FBP3 is recruited early and then replaced by FBP/FBP1 as expression ascends to peak levels. Superimposing the patterns of FBP and FBP3 binding on the profile of serum-induced c-myc transcription suggests that the weaker activator supersedes the stronger one to modulate the shape of the induced peak at its apex. Indeed, the knockdown of FBP boosts the amplitude of the serum-induced peak of c-myc expression in fibroblasts, as expected if FBP is not available to delimit activation by FBP3 (28). This example dramatizes the functional interplay between the FBPs at common targets (though how the less abundant, slower-moving FBP3 beats FBP to FUSE at an earlier stage of induction remains to be determined). (iii) Within the nucleus, although all three FBPs entirely colocalize, they traffic at different kinetics, again suggesting functional variation at common sites. (iv) Based on their DNA binding specificities, the FBPs are predicted to hit a highly overlapping set of targets.
Among the characteristics discriminating between the FBPs (as summarized in Table 1) are their differing abilities to recruit FIR. Experimental investigation of the αNs of the FBPs has identified the residues that permit or deny FIR binding; these residues would not have been discerned from a strictly in silico or computational analysis of sequences. The comparison of the αNs of the three FBPs and the series of helix-preserving site-directed mutagenesis reveal a hydrophobic patch on the αN as one topographic feature of FBP enabling FIR binding. The KH domains, which also bind nucleic acid, are a second feature that helps to recruit and retain FIR.
TABLE 1.
Comparison of the three FBPs
| FBP | Gene location | Polypeptide length (no. of amino acids) | Codon usage | Presence of 4-KH CD for nucleic acid binding | Transactivator strength | FIR binding | JTV-1/p38/AIMP2 binding | Subcellular localization | FRAP kinetics | Protein abundance |
|---|---|---|---|---|---|---|---|---|---|---|
| FBP1/FBP | 1p31.1 | 644 | Uncommon | Yes | Medium | Yes | Strong | Nuclear | Slow | High |
| FBP2/KHSRP | 19p13.3 | 711 | Common | Yes | Weak | Yes | Weak | Nuclear | Slow | High |
| FBP3 | 9q34.11 | 572 | Relatively common | Yes | Strong | No | Weak | Nuclear and cytoplasmic | Slower | Low |
How do the FBP αN, the KH domains, and nucleic acid cooperate to bind FIR? Although in principle αN could stabilize an FBP-FIR-FUSE ternary complex by interacting with either FIR or DNA, the removal of αN has no significant effect on DNA binding by FBP in the absence of FIR (7, 10), so the αN contributes little to direct nucleic acid recognition. Therefore, the αN should interact with FIR while the KH domains interact with both FIR and DNA. The accommodation of variable spacing between the αN and the KH domains argues against a rigid FIR-binding surface on FBP and for a flexible linkage between these structural elements.
Each pair of KH domains is connected via a 20- to 30-amino-acid linker that is predicted and, in the case of the segment between KH3 and KH4, demonstrated to be disordered and flexible (4). If only certain configurations of KH3 and KH4 bind FIR, there would be an entropic expense for excluding the others. Similar reasoning argues that FUSE binding by pairs of KH domains would also limit rotation about the linker and so impose an equivalent entropic tax. However, if FBP binds at once to both FIR and DNA, then the entropic expense paid by whichever binds first would finance the binding of the second, thereby yielding cooperativity. Nucleic acid bridging via the DBDs of FBP and FIR without direct protein-protein interaction is unlikely to account for ternary-complex formation because (i) it fails to account for the contribution of the αN, as the entire N terminus is fully dispensable for FBP binding to FUSE; (ii) even a 29-mer oligodeoxynucleotide, barely long enough to bind two KH domains and therefore too short to also bind FIR in tandem efficiently, stimulates ternary-complex formation; and (iii) alone, much larger amounts of FIR than of FBP are required to bind FUSE in vitro (data not shown). Without direct protein-protein interactions, there is no simple way to account for the cooperativity that reduces the levels of FIR necessary to form ternary complexes.
Controlling the effector output of the FBPs.
The cross-regulation of a largely common set of genes by the FBP family suggests that its members exploit the differences in their effector strengths, their abilities to engage partner proteins, and their abundances in order to customize target expression. The influence of each individual FBP on gene expression will be heavily influenced by their absolute and relative intranuclear stoichiometries, as well as by the level of FIR. Under usual conditions, where FBP1 (a weaker activator than FBP3 and, unlike FBP3, easily repressed by FIR) predominates, the dynamic range of FUSE-like elements would be narrow even without FIR and compressed still further with FIR. Thus, FBP1 predominance creates a situation conducive to steady expression. In isolation, FBP2's activation domain is intrinsically even weaker than FBP1's (normalizing for expression levels) and so would support a narrower dynamic range of expression than FBP even without FIR. In the presence of FIR, FBP2-driven expression would likely be damped even further. Where the most potent activator, FBP3, prevails, FIR cannot be recruited and so unopposed FBP3 activation would be expected irrespective of FIR levels.
Once recruited, the FBPs reside at their intranuclear sites longer than most transcription or splicing factors. The persistence of FBP3 at its sites approaches, and even exceeds, the duration characteristic of some intrinsic chromatin proteins, such as HPI and histone H1. Prolonged binding by FBP3 may accelerate the transition from initiation to promoter escape, since the FBPs accelerate several steps in this process (29). Whether the FBPs passively dissociate, are stripped from chromatin, or are actively degraded while DNA bound remains to be determined.
Setting the levels of the FBPs.
The rates of synthesis, degradation, and nuclear import and export are expected to be the key determinants of differential FBP function. Little is known of the pathways regulating the FBPs, but multiple clues imply differential expression at the RNA and protein levels. Though not tissue specific, the relative levels of the FBP family mRNAs vary among a variety of cell lines (7), and the FBPs have been found to be up- or down-regulated by a variety of stimuli. The FBP level generally parallels c-myc expression (2); both are associated with proliferative states and are down-regulated upon differentiation. (Osteoclasts are an exception; during RANK ligand-induced osteoclast differentiation, FBP/FBP1 is up-regulated by over 40-fold [8].) Nuclear run-on demonstrates the transcriptional shutoff of FBP upon the differentiation of HL60 promyelomonocytic leukemia cells (2). FBP2 and FBP3 are also down-regulated during differentiation (7). While FBP1 mRNA is short-lived (2), the half-lives of the FBP2 and -3 mRNAs have not been measured; hence, the influence of RNA stability on differential FBP expression is not known. The translational regulation of FBP family member expression is similarly unexplored despite several clues pointing in this direction, such as strikingly different codon usage distinguishing the family members as shown previously (7) and as summarized here in Table 1.
FBP is a target for ubiquitinylation and degradation by p38/JTV-1/AIMP2, a core component of a multi-aminoacyl-tRNA synthetase complex (7, 21, 22, 38, 40). FBP3 binds JTV-1 more weakly than FBP does. Intriguingly, JTV-1 also binds the E3 ligase Parkin; Parkin is defective in some kindreds of individuals with familial Parkinson's disease (23). Whether Parkin is an E3 targeting FBP is not yet known.
The cytoplasmic fraction of FBP3 compared with those of FBP and FBP2 invites speculation about subcellular localization as another determinant of their differential functions. KH domain proteins with structures similar to those of the FBPs have been proposed to bind RNA zipcodes and shepherd mRNAs to their proper subcellular sites, so regulated trafficking of the FBPs may be expected to alter the profile of target expression (14, 24, 25, 30, 32). The slow recovery of cytoplasmic FBP3 following photobleaching indicates that it is tethered to some substructure or large complex; presumably, FBP3 needs to be released from this cytoplasmic anchor prior to nuclear import.
Diversification or cooperation/coordination?
Gene duplication has been the principal mode used to expand and diversify gene-regulatory proteins in higher metazoans. On the one hand, the duplicated and reduplicated family members, such as the E2Fs, serve to impart a richer regulatory repertoire to a defined, conserved, and common set of shared targets (46, 48). On the other hand, duplicated regulatory factors, such as the steroid hormone receptors or the Rel family members, have diverged to expand the range of independently controlled targets (18, 26). The available data indicate that FBPs are a functionally coordinated set of regulatory molecules like the E2Fs. The intricacies of the FBP family promise to expose new sites with which to control gene expression and provide a new target for biological and pharmacological intervention.
Supplementary Material
Acknowledgments
We thank T. Misteli for helpful discussions and comments, L. Liotta and D. Braddock for critical review, and D. Libutti for preparing GST-FBP KH domain proteins. Imaging experiments were carried out in the Fluorescence Imaging Facility, LRBGE, NCI.
This work was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Avigan, M. I., B. Strober, and D. Levens. 1990. A far upstream element stimulates c-myc expression in undifferentiated leukemia cells. J. Biol. Chem. 265:18538-18545. [PubMed] [Google Scholar]
- 2.Bazar, L., V. Harris, I. Sunitha, D. Hartmann, and M. Avigan. 1995. A transactivator of c-myc is coordinately regulated with the proto-oncogene during cellular growth. Oncogene 10:2229-2238. [PubMed] [Google Scholar]
- 3.Braddock, D. T., J. L. Baber, D. Levens, and G. M. Clore. 2002. Molecular basis of sequence-specific single-stranded DNA recognition by KH domains: solution structure of a complex between hnRNP KKH3 and single-stranded DNA. EMBO J. 21:3476-3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braddock, D. T., J. M. Louis, J. L. Baber, D. Levens, and G. M. Clore. 2002. Structure and dynamics of KH domains from FBP bound to single-stranded DNA. Nature 415:1051-1056. [DOI] [PubMed] [Google Scholar]
- 5.Briata, P., S. V. Forcales, M. Ponassi, G. Corte, C. Y. Chen, M. Karin, P. L. Puri, and R. Gherzi. 2005. p38-dependent phosphorylation of the mRNA decay-promoting factor KSRP controls the stability of select myogenic transcripts. Mol. Cell 20:891-903. [DOI] [PubMed] [Google Scholar]
- 6.Briata, P., C. Ilengo, G. Corte, C. Moroni, M. G. Rosenfeld, C. Y. Chen, and R. Gherzi. 2003. The Wnt/beta-catenin-> Pitx2 pathway controls the turnover of Pitx2 and other unstable mRNAs. Mol. Cell 12:1201-1211. [DOI] [PubMed] [Google Scholar]
- 7.Davis-Smyth, T., R. C. Duncan, T. Zheng, G. Michelotti, and D. Levens. 1996. The far upstream element-binding proteins comprise an ancient family of single-strand DNA-binding transactivators. J. Biol. Chem. 271:31679-31687. [DOI] [PubMed] [Google Scholar]
- 8.Day, C. J., M. S. Kim, S. R. J. Stephens, W. E. Simcock, C. J. Aitken, G. C. Nicholson, and N. A. Morrison. 2004. Gene array identification of osteoclast genes: differential inhibition of osteociastogenesis by cyclosporin a and granulocyte macrophage colony stimulating factor. J. Cell. Biochem. 91:303-315. [DOI] [PubMed] [Google Scholar]
- 9.Dean, M., R. A. Levine, W. Ran, M. S. Kindy, G. E. Sonenshein, and J. Campisi. 1986. Regulation of c-myc transcription and mRNA abundance by serum growth factors and cell contact. J. Biol. Chem. 261:9161-9166. [PubMed] [Google Scholar]
- 10.Duncan, R., L. Bazar, G. Michelotti, T. Tomonaga, H. Krutzsch, M. Avigan, and D. Levens. 1994. A sequence-specific, single-strand binding protein activates the far upstream element of c-myc and defines a new DNA-binding motif. Genes Dev. 8:465-480. [DOI] [PubMed] [Google Scholar]
- 11.Duncan, R., I. Collins, T. Tomonaga, T. Zhang, and D. Levens. 1996. A unique transactivation sequence motif is found in the carboxyl-terminal domain of the single-strand-binding protein FBP. Mol. Cell. Biol. 16:2274-2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dundr, M., M. D. Hebert, T. S. Karpova, D. Stanek, H. Xu, K. B. Shpargel, U. T. Meier, K. M. Neugebauer, A. G. Matera, and T. Misteli. 2004. In vivo kinetics of Cajal body components. J. Cell Biol. 164:831-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dundr, M., U. Hoffmann-Rohrer, Q. Hu, I. Grummt, L. I. Rothblum, R. D. Phair, and T. Misteli. 2002. A kinetic framework for a mammalian RNA polymerase in vivo. Science 298:1623-1626. [DOI] [PubMed] [Google Scholar]
- 14.Gherzi, R., K. Y. Lee, P. Briata, D. Wegmuller, C. Moroni, M. Karin, and C. Y. Chen. 2004. A KH domain RNA binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery. Mol. Cell 14:571-583. [DOI] [PubMed] [Google Scholar]
- 15.Hall, M. P., S. Huang, and D. L. Black. 2004. Differentiation-induced colocalization of the KH-type splicing regulatory protein with polypyrimidine tract binding protein and the c-src pre-mRNA. Mol. Biol. Cell 15:774-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He, L., J. Liu, I. Collins, S. Sanford, B. O'Connell, C. J. Benham, and D. Levens. 2000. Loss of FBP function arrests cellular proliferation and extinguishes c-myc expression. EMBO J. 19:1034-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He, L. S., A. Weber, and D. Levens. 2000. Nuclear targeting determinants of the far upstream element binding protein, a c-myc transcription factor. Nucleic Acids Res. 28:4558-4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoffmann, A., T. H. Leung, and D. Baltimore. 2003. Genetic analysis of NF-kappa B/Rel transcription factors defines functional specificities. EMBO J. 22:5530-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kelly, K., and U. Siebenlist. 1986. The regulation and expression of c-myc in normal and malignant cells. Annu. Rev. Immunol. 4:317-338. [DOI] [PubMed] [Google Scholar]
- 20.Kielkopf, C. L., S. Lucke, and M. R. Green. 2004. U2AF homology motifs: protein recognition in the RRM world. Genes Dev. 18:1513-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim, J. Y., Y. S. Kang, J. W. Lee, H. J. Kim, Y. H. Ahn, H. Park, Y. G. Ko, and S. Kim. 2002. P38 is essential for the assembly and stability of macromolecular tRNA synthetase complex: implications for its physiological significance. Proc. Natl. Acad. Sci. USA 99:7912-7916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim, M. J., B. J. Park, Y. S. Kang, H. J. Kim, J. H. Park, J. W. Kang, S. W. Lee, J. M. Han, H. W. Lee, and S. Kim. 2003. Downregulation of FUSE-binding protein and c-myc by tRNA synthetase cofactor p38 is required for lung cell differentiation. Nat. Genet. 34:330-336. [DOI] [PubMed] [Google Scholar]
- 23.Ko, H. S., R. von Coelln, S. R. Sriram, S. W. Kim, K. K. Chung, O. Pletnikova, J. Troncoso, B. Johnson, R. Saffary, E. L. Goh, H. Song, B. J. Park, M. J. Kim, S. Kim, V. L. Dawson, and T. M. Dawson. 2005. Accumulation of the authentic parkin substrate aminoacyl-tRNA synthetase cofactor, p38/JTV-1, leads to catecholaminergic cell death. J. Neurosci. 25:7968-7978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kroll, T. T., W. M. Zhao, and P. W. Huber. 2001. KSRP/FBP2 binds to the localization element of Xenopus Vg1 mRNA and to Prrp. Mol. Biol. Cell 12:115A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kroll, T. T., W. M. Zhao, C. Jiang, and P. W. Huber. 2002. A homolog of FBP2/KSRP binds to localized mRNAs in Xenopus oocytes. Development 129:5609-5619. [DOI] [PubMed] [Google Scholar]
- 26.Leung, T. H., A. Hoffmann, and D. Baltimore. 2004. One nucleotide in a kappaB site can determine cofactor specificity for NF-kappaB dimers. Cell 118:453-464. [DOI] [PubMed] [Google Scholar]
- 27.Liu, J., L. He, I. Collins, H. Ge, D. Libutti, J. Li, J. M. Egly, and D. Levens. 2000. The FBP interacting repressor targets TFIIH to inhibit activated transcription. Mol. Cell 5:331-341. [DOI] [PubMed] [Google Scholar]
- 28.Liu, J., F. Kouzine, Z. Nie, H. J. Chung, Z. Elisha-Feil, A. Weber, K. Zhao, and D. Levens. 2006. The FUSE/FBP/FIR/TFIIH system is a molecular machine programming a pulse of c-myc expression. EMBO J. 25:2119-2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, J. H., S. Akoulitchev, A. Weber, H. Ge, S. Chuikov, D. Libutti, X. W. Wang, J. W. Conaway, C. C. Harris, R. C. Conaway, D. Reinberg, and D. Levens. 2001. Defective interplay of activators and repressors with TFIIH in xeroderma pigmentosum. Cell 104:353-363. [DOI] [PubMed] [Google Scholar]
- 30.Lorenz, M., S. Huttelmaier, J. Condeelis, and R. Singer. 2002. Localization and interaction of PTB/hnRNP I, Raver1 and ZBP2/KSRP in the perinucleolar compartment determined by FRET microscopy. Mol. Biol. Cell 13:375A. [Google Scholar]
- 31.Marcu, K. B., S. A. Bossone, and A. J. Patel. 1992. myc function and regulation. Annu. Rev. Biochem. 61:809-860. [DOI] [PubMed] [Google Scholar]
- 32.Michelotti, G. A., E. F. Michelotti, A. Pullner, R. C. Duncan, D. Eick, and D. Levens. 1996. Multiple single-stranded cis elements are associated with activated chromatin of the human c-myc gene in vivo. Mol. Cell. Biol. 16:2656-2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Min, H. S., C. W. Turck, J. M. Nikolic, and D. L. Black. 1997. A new regulatory protein, KSRP, mediates exon inclusion through an intronic splicing enhancer. Genes Dev. 11:1023-1036. [DOI] [PubMed] [Google Scholar]
- 34.Nicolaides, N. C., K. W. Kinzler, and B. Vogelstein. 1995. Analysis of the 5′-region of pms2 reveals heterogeneous transcripts and a novel overlapping gene. Genomics 29:329-334. [DOI] [PubMed] [Google Scholar]
- 35.Page-McCaw, P. S., K. Amonlirdviman, and P. A. Sharp. 1999. PUF60: a novel U2AF65-related splicing activity. RNA 5:1548-1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park, S. G., K. L. Ewalt, and S. Kim. 2005. Functional expansion of aminoacyl-tRNA synthetases and their interacting factors: new perspectives on housekeepers. Trends Biochem. Sci. 30:569-574. [DOI] [PubMed] [Google Scholar]
- 37.Phair, R. D., P. Scaffidi, C. Elbi, J. Vecerová, A. Dey, K. Ozato, D. T. Brown, G. Hager, M. Bustin, and T. Misteli. 2004. Global nature of dynamic protein-chromatin interactions in vivo: three-dimensional genome scanning and dynamic interaction networks of chromatin proteins. Mol. Cell. Biol. 24:6393-6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quevillon, S., J. C. Robinson, E. Berthonneau, M. Siatecka, and M. Mirande. 1999. Macromolecular assemblage of aminoacyl-tRNA synthetases: identification of protein-protein interactions and characterization of a core protein. J. Mol. Biol. 285:183-195. [DOI] [PubMed] [Google Scholar]
- 39.Rehbein, M., K. Wege, F. Buck, M. Schweizer, D. Richter, and S. Kindler. 2002. Molecular characterization of MARTA1, a protein interacting with the dendritic targeting element of MAP2 mRNAs. J. Neurochem. 82:1039-1046. [DOI] [PubMed] [Google Scholar]
- 40.Rho, S. B., M. J. Kim, J. S. Lee, W. G. Seol, H. Motegi, S. Kim, and K. Shiba. 1999. Genetic dissection of protein-protein interactions in multi-tRNA synthetase complex. Proc. Natl. Acad. Sci. USA 96:4488-4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ring, H. Z., V. Vameghi-Meyers, H. S. Min, J. M. Nikolic, D. L. Black, and U. Francke. 1999. The mouse Fubp gene maps near the distal end of chromosome 3. Genomics 56:357-358. [DOI] [PubMed] [Google Scholar]
- 42.Ring, H. Z., V. Vameghi-Meyers, J. M. Nikolic, H. S. Min, D. L. Black, and U. Francke. 1999. Mapping of the KHSRP gene to a region of conserved synteny on human chromosome 19p13.3 and mouse chromosome 17. Genomics 56:350-352. [DOI] [PubMed] [Google Scholar]
- 43.Rydziel, S., A. M. Delany, and E. Canalis. 2004. AU-rich elements in the collagenase 3 mRNA mediate stabilization of the transcript by cortisol in osteoblasts. J. Biol. Chem. 279:5397-5404. [DOI] [PubMed] [Google Scholar]
- 44.Snee, M., G. J. Kidd, T. P. Munro, and R. Smith. 2002. RNA trafficking and stabilization elements associate with multiple brain proteins. J. Cell Sci. 115:4661-4669. [DOI] [PubMed] [Google Scholar]
- 45.Spencer, C. A., and M. Groudine. 1991. Control of c-myc regulation in normal and neoplastic cells. Adv. Cancer Res. 56:1-48. [DOI] [PubMed] [Google Scholar]
- 46.Takahashi, Y., J. B. Rayman, and B. D. Dynlacht. 2000. Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 14:804-816. [PMC free article] [PubMed] [Google Scholar]
- 47.Waters, C. M., T. D. Littlewood, D. C. Hancock, J. P. Moore, and G. I. Evan. 1991. c-myc protein expression in untransformed fibroblasts. Oncogene 6:797-805. [PubMed] [Google Scholar]
- 48.Wells, J., K. E. Boyd, C. J. Fry, S. M. Bartley, and P. J. Farnham. 2000. Target gene specificity of E2F and pocket protein family members in living cells. Mol. Cell. Biol. 20:5797-5807. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.