Abstract
RANKL is a tumor necrosis factor (TNF)-like factor secreted by mesenchymal cells, osteoblast derivatives, and T cells that is essential for osteoclastogenesis. In osteoblasts, RANKL expression is regulated by two major calcemic hormones, 1,25-dihydroxyvitamin D3 [1,25(OH)2D3] and parathyroid hormone (PTH), as well as by several inflammatory/osteoclastogenic cytokines; the molecular mechanisms for this regulation are unclear. To identify such mechanisms, we screened a DNA microarray which tiled across the entire mouse RankL gene locus at a 50-bp resolution using chromatin immunoprecipitation (ChIP)-derived DNA precipitated with antibodies to the vitamin D receptor (VDR) and the retinoid X receptor (RXR). Five sites of dimer interaction were observed on the RankL gene centered at 16, 22, 60, 69, and 76 kb upstream of the TSS. These regions contained binding sites for not only VDR and RXR, but also the glucocorticoid receptor (GR). The most distant of these regions, termed the distal control region (RL-DCR), conferred both VDR-dependent 1,25(OH)2D3 and GR-dependent glucocorticoid (GC) responses. We mapped these activities to an unusual but functionally active vitamin D response element and to several potential GC response elements located over a more extensive region within the RL-DCR. An evolutionarily conserved region within the human RANKL gene contained a similar vitamin D response element and exhibited an equivalent behavior. Importantly, hormonal activation of the RankL gene was also associated with chromatin modification and RNA polymerase II recruitment. Our studies demonstrate that regulation of RankL gene expression by 1,25(OH)2D3 is complex and mediated by at least five distal regions, one of which contains a specific element capable of mediating direct transcriptional activation.
Skeletal remodeling in adults occurs through the coupled actions of bone-forming osteoblasts and bone-resorbing osteoclasts (17). The latter are terminally differentiated, multinucleated cells of the monocyte-macrophage lineage (63). The process of osteoclastogenesis is highly complex and is orchestrated by a number of growth factors, steroidal components, and cytokines, all of which exert their actions in a highly temporal fashion (57). Many of these regulatory factors are produced and secreted by adjacent support cells that include stroma, B and T cells, and cells of the osteoblast lineage. The production of such factors by osteoblasts highlights the extrinsic role that these cells play in the process of bone resorption. Importantly, while many of these secreted components are critical for normal adult bone remodeling, their aberrant secretion can be pathological and lead to either focal or systemic bone disease (45).
Although many factors participate in osteoclastogenesis, the molecule that is now considered to be both necessary and sufficient in vivo and in vitro is the receptor activator of NF-κB ligand (RankL). RankL is a tumor necrosis factor (TNF)-like factor that is produced by stromal cells and osteoblasts as well as a variety of other cell types (38). This factor not only actively promotes the process of osteoclast differentiation, but also is required for the cell's bone-resorbing activity and for its survival (22). The interaction of RankL with receptor activator of NF-κB (Rank), an integral receptor protein located on the surface of osteoclast precursors, triggers a number of signaling cascades that include the IKK/IKβ/NF-κB transduction pathway and the mitogen-activated protein kinase (MAPK), Src, and phosphatidylinositol 3-kinase (PI3K)/AKT pathways as well (64). A novel calcium oscillation pathway that involves ITAM coreceptors is also involved in the downstream effects of RankL (35). Importantly, stimulation of these pathways culminates in the activation of multiple transcription factors, including c-fos, NF-κB, and NFATc1, all of which play strategic roles in the differentiation process at the genetic level (44, 61). Overall, the timely activation of these transcription factors initiates growth arrest and promotes osteoclast differentiation, fusion, activation, and survival (63). The evidence that supports the essentiality of both RankL and its receptor in osteoclast formation is most strongly supported by the skeletal phenotypes of both RankL- and Rank-null mice, neither of which are capable of producing osteoclasts in vivo and thus are phenotypically osteopetrotic (20, 37).
RankL is synthesized and expressed on the surface of regulatory cells in response to a myriad of both local and systemic factors, many of which are essential to physiologic bone turnover. These include the two hormones integral to calcium homeostasis, 1,25-dihydroxyvitamin D3 [1,25(OH)2D3] (60) and parathyroid hormone (PTH) (36, 41). RankL expression can also be influenced by the glucocorticoid (GC) stress hormones (18, 54), inflammatory cytokines such as TNF-α and interleukin-1 (IL-1) (23), the gp130-activating cytokines IL-6 and IL-11 (51, 62), certain prostaglandins (66), and transforming growth factor β (TGF-β) (28). While these factors can function in a physiologic setting, their activities are often manifested during disease. Thus, RankL overproduction can in a number of circumstances lead to the pathological bone resorption associated with age-related and postmenopausal bone disease, rheumatoid arthritis and osteoarthritis, multiple myelomas, diabetic neuropathy, metastatic cancer, general hypercalcemia of malignancy, and a variety of other syndromes that impact the skeleton (19).
The calciotropic hormones 1,25(OH)2D3 and PTH, as well as certain growth factors, cytokines, and prostaglandins, all regulate the expression of RankL from stromal cells and osteoblasts (63). Interestingly, the molecular mechanisms responsible for activation by these regulators remain largely unknown. With respect to vitamin D, 1,25(OH)2D3 is known to induce RankL upregulation primarily through actions initiated by the vitamin D receptor (VDR) (29). Kitazawa and colleagues (34) reported a 1,25(OH)2D3 response in the mouse RankL gene promoter and mapped this activity to a 16-bp vitamin D response element (VDRE) that was located 935 bp upstream of the transcriptional start site (TSS). Inducibility was modest, however, and subsequent studies by a number of laboratories have failed to confirm this finding (11, 48). More recently, Kabe et al. (24) have explored the ability of a potential VDRE located immediately downstream of the RankL TSS to mediate 1,25(OH)2D3 activity. Although this element is similar to that of a consensus VDRE, it is not conserved within RankL genes of other species and is capable of only a modest activity in the context of the RankL proximal promoter. Thus, it remains unclear at this stage whether or how these two elements contribute to the RankL gene's regulation by 1,25(OH)2D3.
PTH also regulates RankL expression (40), although the mechanism through which this peptide acts has remained equally elusive. Recent studies by O'Brien and coworkers (12) have established that PTH is capable of both stabilizing RankL mRNA and inducing its expression, the latter via stimulation of the protein kinase A (PKA) pathway and activation of the CREB transcription factor. It seems likely that activation of this transcription factor may underlie the ability of the prostaglandin PGE2 to stimulate RankL expression as well (25). Despite these insights, however, the mechanism by which CREB induces transcriptional activation at the level of the RankL gene promoter remains undefined. The accompanying article by Fu, Manolagas, and O'Brien delineates the molecular mechanism whereby PTH stimulates RankL gene expression (13).
Finally, the actions of GCs on the skeleton are highly complex. Enhanced exposure to these hormones, however, generally results in osteoporosis (42). At the level of the osteoblast, GCs reduce both the functional capacity of these cells to form bone and the period of time during which this functionality occurs (43, 67). GCs also influence the production of osteoclasts (18, 54). These actions occur through a direct activity on the osteoblast primarily to suppress the expression of osteoprotegerin, a decoy receptor which blocks RankL activity, but also to increase RankL gene expression. Although a role for the glucocorticoid receptor (GR) in RankL induction is likely, the mechanism whereby the stress hormone induces RankL expression remains unexplored.
As outlined above, the mechanism of action of 1,25(OH)2D3 involves a ligand-initiated interaction between the VDR and the regulatory regions of target genes wherein VDR functions together with its retinoid X receptor (RXR) partner to recruit coregulatory complexes that are essential for modulation of transcriptional output (46, 58). Binding sites for the VDR have been found in a number of genes and are generally, although not always, comprised of two hexanucleotide half-sites separated by a short spacer and frequently located within the first kilobase or two upstream of the transcriptional start site (50). The absence of bona fide target sites for VDR action within the first 8 kb of the RankL gene, as reported by us and others (11, 48), prompted a more expansive approach to delineate RankL regulatory regions. We therefore used contemporary chromatin immunoprecipitation (ChIP)/chip analysis (as described below) in this endeavor. We discovered five sites of action located at increasing distances from the mouse RankL gene TSS, the furthest approximately 76 kb upstream. The latter region, which we termed the RankL distal control region (RL-DCR), was transcriptionally active in transfection studies and contained an unusual VDRE sequence. Our studies, and the results described in the accompanying article by O'Brien and colleagues (13), provide essential clues as to how 1,25(OH)2D3 and PTH regulate RankL induction.
MATERIALS AND METHODS
Reagents.
General biochemicals were obtained from Fisher Scientific (Pittsburgh, PA) and Sigma Chemical Co. (St. Louis, MO). 1,25(OH)2D3 was obtained from Solvay (da Weesp, The Netherlands) and Tetrionics (Madison, WI). ZK159222 was kindly provided by Andreas Steinmeyer and Ekkehard May of Schering AG (Berlin, Germany). Alpha minimum essential medium-Earle's medium (α-MEM) was purchased from Mediatech (Herndon, VA), and minimum essential medium alpha (MEM-α) was obtained from Invitrogen Corporation (Carlsbad, CA). Oligonucleotide primers were obtained from IDT (Coralville, IA). Anti-VDR (Sc-1008), -RXR (Sc-774), -C/EBPβ (Sc-150), and -GR (Sc-1004) antibodies were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Anti-acetyl H4 antibody (06-866) was obtained from Upstate (Charlottesville, VA), and anti-RNA polymerase II antibody (8WG16) was obtained from Berkeley Antibody Company (Richmond, CA). Lipofectamine Plus was obtained from Invitrogen Corporation (Carlsbad, CA). [γ-32P]dATP was obtained from NEN Life Science Products, Inc. (Boston, MA). Dexamethasone (D4902), anti-rat immunoglobulin G (IgG; R5128), and RU486 (M8046) were purchased from Sigma Chemical Co. (St. Louis, MO).
Cell culture.
Mouse MC3T3-E1 and ST2 osteoblastic cells were cultured in α-MEM and MEM-α, respectively. Primary calvarial osteoblasts (mOBs) were obtained as previously described (53) and cultured in α-MEM. Human osteosarcoma MG63 cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 1% nonessential amino acids. COS-7 fibroblasts were also cultured in DMEM. Each medium was supplemented with 10% fetal bovine serum (FBS) obtained from HyClone (Logan, UT), 100 U/ml penicillin, and 100 μg/ml streptomycin. All ligands were added in ethanol (0.1% maximum final concentration) or dimethyl sulfoxide.
RNA isolation and analysis.
Total RNA was isolated from cells using Triazol reagent obtained from MRC (Cincinnati, OH). The isolated RNA was reverse transcribed using the SuperScript III RNase H reverse transcriptase kit from Invitrogen (Carlsbad, CA) and then subjected to PCR analysis using standard PCR methods. Primers used include those for the mouse β-actin gene (mβ-actin) (forward, TGTTTGAGACCTTCAACACCC; reverse, CGTTGCCAATAGTGATGACCT), mCyp24a1 (forward, GTGCGGATTTCCTTTGTGATA; reverse, GGTAGCGTGTATTCACCCAGA), mVDR (forward, TCACTGATGTCTCCAGAGCTGGGC; reverse, TGGATAGGCGGTCCTGAATGGC), and mOpn (forward, CTAACTACGACCATGAGATTGGCAG; reverse, CTTTAGTTGACCTCAGAAGATGAA) and mRankL (forward, GAATCCTGAGACTCCATGAAAACGC; reverse, CCATGAGCCTTCCATCATAGCTGG). Primers for the human genes used included those for the human β-actin gene (hβ-actin) (forward, TTAGTTGCGTTACACCCTTTC; reverse, GTCACCTTCACCGTTCCAGTT), hCYP24A1 (forward, CTTTGCTTCCTTTTCCCAGAAT; reverse, CGCCGTAGATGTCACCAGTC), and hRANKL (forward, AACAGGCCTTTCAAGGAGCTGTGC; reverse, AAGAGGACAGACTCACTTTATGGG).
siRNA studies.
All small interfering RNA (siRNA) duplexes were obtained from Dharmacon RNA Technologies (Lafayette, CO). ST2 cells were seeded into six-well plates at a concentration of 1.5 × 105 cells/well and transfected approximately 24 h later using Lipofectamine Plus in serum and antibiotic-free medium. A 20 nM concentration of mVDR siRNA (D-058923-01), nontargeting siRNA pool (D-001206-13), or cyclophilin B siRNA (D-001136-01) was used for transfection. After transfection, the cells were cultured in medium supplemented with 10% FBS for 48 h before they were treated with a routine concentration of 10−7 M 1,25(OH)2D3 for 6 h. RNA isolation and standard PCR analysis were carried out using the primers listed above. Western blot analysis confirmed depletion of VDR protein expression in ST2 cells (data not shown).
ChIP assay.
Chromatin immunoprecipitation assays were performed as previously described (30). Primer sets used for amplifying mouse and human Cyp24a1, osteopontin (Opn), and RankL gene regions of interest are all listed in Table 1. Densitometric analysis was carried out using Kodak ID Image Analysis (software version 3.5).
TABLE 1.
Primers used for ChIP analysis
| Primer and species | Sequence |
|---|---|
| Mouse | |
| Cyp24a1 | 5′ GGTTATCTCCGGGGTGGAGT |
| 3′ AGTGGCCAATGAGCACGC | |
| Opn | 5′ ACCACCTCTTCTGCTCTATATGGC |
| 3′ TTGACACTTGAACTATGCAGCCGC | |
| IS7 | 5′ CTGAAGCCAAGAGGCAGATT |
| 3′ CGCACATCCTTTCAGGTGCT | |
| IS6 | 5′ GGTACCACATGTGCACATTA |
| 3′ CAGGTGTTGTTTTAAGCTAC | |
| IS5 | 5′ GCTCAGAAAGCAGGACCTCC |
| 3′ CACTTTCTCTTAGAACAGTG | |
| IS4 | 5′ CCTCCTATCTGTTTTACTGACGTT |
| 3′ CACATAGGCAGAAAGTTGAAAAGC | |
| IS2 | 5′ GCTATCATTTATACCTTGGA |
| 3′ CCTGAATTTCTGATCTTCCT | |
| IS1 | 5′ CATGAGTATATGTGTGGAGT |
| 3′ CCTTCATAATGTTTGAGACC | |
| D5 | 5′ GGTCAAGAGGGGCCTGACTT |
| 3′ GCAGTGTGTAAACAAAGAGA | |
| D4 | 5′ GTGCTGTGAAGCAAAAATTC |
| 3′ GCACTACAAATGTGAGGGAA | |
| D3 | 5′ GAGCTGTGTCCTAGAAGAAT |
| 3′ CCCTGCACATAGTAAACAAA | |
| D2 | 5′ TAAGGCTCTGACCATAGGAA |
| 3′ CTGGATTTAAGTGAAACAGT | |
| D1 | 5′ GAGAAAGCCAAGTCCTGGGT |
| 3′ CATTCTGGGGCCGGAACAAA | |
| P1 (TSS) | 5′ CAGAAACCAACCACTGGACCCAA |
| 3′ CAGGAACATGGAGCGGGAGG | |
| Human | |
| CYP24A1 | 5′ CGAAGCACACCCGGTGAACT |
| 3′ CCAATGAGCACGCAGAGGAG | |
| D5-1 | 5′ CAAGTTTCTTTGCTGTCATC |
| 3′ CAGCCACATAAAGCAGTGAG | |
| D5-2 | 5′ GCTCAAGCCTTATGTTTCGG |
| 3′ TAAGCTAGTGTGACCCCTGA |
Tiled oligonucleotide microarray analysis.
ChIP/chip analysis was carried out as described by others (31-33, 47). In brief, DNA was isolated by specific immunoprecipitation using the ChIP methodology described above and then subjected to ligation-mediated PCR as described by Oberley et al. (47). ChIP-purified DNA was blunt-ended using T4 polymerase, ligated to linkers with the sequences 5′ GCGGTGACCCGGGAGATCTGAATTC 3′ and 5′ GAATTCAGATC 3′, and subjected to repeated, low-cycle PCR amplification. The resulting ∼500-bp amplicons were then labeled with Cy3 or Cy5 dyes using an indirect labeling protocol. In this method, biotinylated dUTP is first incorporated into the individual amplicons by standard procedures and the conjugated DNA is labeled subsequently with either Cy5- or Cy3-conjugated streptavidin. Cy3- and Cy5-labeled DNA samples are then mixed in the presence of CoT-1 DNA, denatured, and cohybridized to custom oligonucleotide microarrays (Nimblegen Systems Inc., Madison, WI) as described. The microarrays are washed extensively and scanned using an Axon 4000B scanner at the appropriate wavelengths.
Custom oligonucleotide arrays were synthesized by Nimblegen Systems, Inc (Madison, WI). The microarray probes consisted of maskless-array, in situ-synthesized 50-mer oligonucleotides at 2-bp intervals representing a screen of over 300 kb of the mouse RANKL gene locus from 200 kb upstream of the gene's TSS to 100 kb downstream of the final 3′ noncoding exon. The tiled arrays were synthesized in duplicate in both the forward as well as reverse directions, providing four independent measurements at each site within the gene. In addition, each analysis was carried out using two independently derived ChIP DNA samples. Both the Cyp24a1 and Opn genes (as well as other candidate VDR target genes that are not discussed) were tiled in a similar fashion. A series of comparisons were made between (i) IgG in the presence or absence of hormone, (ii) VDR in the presence or absence of hormone, (iii) VDR in the presence of hormone versus input DNA, (iv) RXR in the presence or absence of hormone, and (v) RXR in the presence of hormone versus input DNA. After sample cohybridization, the logarithmic enrichment ratios of Cy5 to Cy3 hybridization intensities (log2) were plotted as a function of chromosome nucleotide position. While all of the peaks representative of enhanced VDR or RXR binding either in the presence of 1,25(OH)2D3 or as compared to input DNA are presented as the raw data, a peak-finding algorithm was utilized to score the relative levels of binding between the three regions identified (14).
Plasmids.
Full-length hVDR and hRXRα were cloned into the pET-29b vector obtained from Novagen (Darmstadt, Germany) and expressed with C-terminal His6 tags. The pCH110-β-galactosidase (pCH110-βgal) reporter plasmid and the pcDNA-hVDR vector or a mutant (hVDR L417A/E420A) version [pcDNA-hVDR(m)] and pRSV-GRα were previously described (70). The parent thymidine kinase (TK) and luciferase (luc) vectors pTK-luc and pGL3-luc were utilized in subsequent cloning efforts. mRLD1 (−16.4 to −15.2), mRLD2 (−23.1 to −21.5), mRLD3 (−60.4 to −59.3), mRLD4 (−69.0 to −68.1), and mRLD5 (−76045 to −74973) were amplified using primers that contained HindIII, HindIII/SalI, or HindIII/BamHI restriction ends and then cloned into the corresponding sites within the pTK-luc vector. pTK-mRLD5 fragments pTK-mRLD5-5/1 (−75724 to −74973), pTK-mRLD5-5/2 (−75475 to −74973), pTK-mRLD5-5/3 (−75228 to −74973), pTK-mRLD5-3/1 (−75724 to −75228), and pTK-mRLD5-3/2 (−75724 to −75475) were subcloned similarly into the pTK-luc vector using HindIII/SalI restriction sites. The hRLD5 region of the human RANKL gene (−96903 to −95805) was amplified from genomic DNA and cloned into the HindIII/BamHI sites of the TK-luc vector to produce pTK-hRLD5. mRL-VDRE (−75620 to −75590) and the hRL-VDRE (−96467 to −96431) as well as mRL-VDRE1 and mRL-VDRE2, each containing several overhanging 5′ and 3′ nucleotides, were synthesized, annealed, and similarly cloned into the HindIII/BamHI sites of pTK-luc. Triplet mutations in the mRL-VDRE half-sites and in the hRL-VDRE half-sites in the context of pTK-mRLD5 and pTK-hRLD5 were created using the Quikchange mutagenesis kit from Stratagene (San Diego, CA). mRL-VDRE segments containing mutations within each half-site were also synthesized and cloned into the HindIII/BamHI sites of pTK-luc. The pmRL(100) vector was prepared by introducing an amplified segment of the mouse RankL gene promoter (−101 to +54 relative to the RankL TSS) into the pGL3-luc expression vector at the XhoI/HindIII sites. Each of the mRLD regions, mRLD1 (−16.4 to −15.2), mRLD2 (−23.1 to −21.5), mRLD3 (−60.4 to −59.3), mRLD4 (−69.0 to −68.1), and mRLD5 (−76045 to −74973), as well as mRL-VDRE (−75620 to −75590), was then amplified and cloned upstream of pmRL(100) using the MluI/XhoI restriction sites. All plasmid constructs were sequenced to verify successful cloning.
Transfection assays.
MC3T3-E1 and/or ST2 cells were seeded into 24-well plates at appropriate densities and cultured in α-MEM or MEM-α containing 10% FBS. Cells were transfected 24 h later with Lipofectamine Plus in serum and antibiotic-free medium. Individual wells were transfected with 250 ng of a luciferase reporter vector, 50 ng of pCH110-βgal, and 50 ng of pcDNA-hVDR (which was routinely transfected with all luciferase reporters unless otherwise indicated) or 50 ng phRSV-GRα (added only when specifically indicated). Nontargeting, cyclophilin B or VDR siRNA pools (50 nM) were also transfected where indicated. After transfection, the cells were cultured first for 48 h in a medium supplemented with 20% FBS and subsequently for an additional 24 h with or without 1,25(OH)2D3. Cells were then harvested, and the lysates were assayed for luciferase and β-galactosidase activities as previously described (70). Luciferase activity was normalized to β-galactosidase activity in all cases.
Protein purification.
Human VDR and RXRα proteins were produced using the bacterial expression vectors pET-hVDR and pET-hRXRα in BL21(DE3) codon Plus RIL cells obtained from Stratagene (San Diego, CA). Soluble full-length hVDR and hRXRα proteins were purified to homogeneity using sequential Ni-nitrilotriacetic acid (NTA) and SP-Sepharose column chromatography (70). Two forms of RXRα were present due to redundant start sites.
DNA band shift analysis.
The duplex oligonucleotide probes comprised of the mouse osteopontin VDRE, mRL-VDRE, mRL-VDRE1m (M1), mRL-mVDRE2m (M2), and hRL-VDRE, as documented in Table 2, were end labeled using [γ-32P]dATP. Probes were incubated at room temperature with the indicated concentrations of hVDR and hRXRα in 10 mM HEPES, pH 7.4, 1 mM EDTA, 5 mM MgCl2, 10% glycerol, 0.5 mM dithiothreitol, 0.7 mM phenylmethylsulfonyl fluoride, and 50 or 150 mM KCl in the absence or presence of 1,25(OH)2D3 for 30 min. Complexes were resolved on nondenaturing 6% polyacrylamide gels, dried, and then visualized using autoradiography. Densitometric analyses of complex 1 and complex 2 were carried out using Kodak ID Image Analysis (software version 3.5).
TABLE 2.
DNA sequences for EMSA
| DNA type and species | Sequencea |
|---|---|
| Mouse | |
| Opn VDRE | 5′ AACAAGGTTCACGAGGTTCACGTCT |
| RankL VDRE | 5′ GGGTGAACTCAGGCAACCAACAACTCGGTGACTCTGG |
| RankL VDRE1m | 5′ GGGTGAACTCAGGCAACCAACAggaCGGTGAggaTGG |
| RankL VDRE2m | 5′ GGGTGAggaCAGGCAggaAACAACTCGGTGACTCTGG |
| Human | |
| RankL VDRE | 5′ GGGTGAACTCAGACAACCGACAACTTGGTGACTTCGA |
VDRE sequences are underlined. Mutated sequences are indicated in lowercase letters.
Statistical analyses.
All values are expressed as the mean ± standard error of the mean. All statistical calculations were performed with the GraphPad PRISM version 4 statistical software package (GraphPad Software Inc., San Diego, CA). We evaluated differences between groups through one-way analysis of variance or Student's two-tailed t test. Significance was determined at P < 0.05.
RESULTS
1,25(OH)2D3 induces RankL gene expression in osteoblast-like ST2 cells.
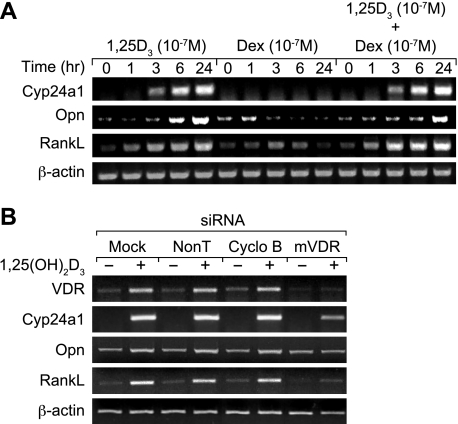
Earlier studies indicated that 1,25(OH)2D3 induces RankL expression in a variety of osteoblast-like cells, including the mouse ST2 cell line (34). To explore this process further and to assess the potential effects of GCs on this induction, we treated ST2 cells with a maximal dose of either 1,25(OH)2D3, dexamethasone (DEX), or the combination and evaluated the level of RankL transcripts produced. As can be seen in Fig. 1A (see Fig. S1 in the supplemental material), 1,25(OH)2D3 was capable of inducing RankL mRNA levels with a time course consistent with that observed for several 1,25(OH)2D3 target genes, including both Opn and Cyp24a1. Interestingly, while DEX was generally inactive on its own, it appeared to potentiate the activity of 1,25(OH)2D3 when used in combination, suggesting a potentially additive or synergistic effect on RankL expression. DEX of course had no effect on either Cyp24a1 or Opn mRNA levels. To establish that the induction of RankL by 1,25(OH)2D3 was mediated via the VDR, we employed an interference assay using VDR siRNA and assessed the effect of this action on RankL expression. ST2 cells were mock transfected or transfected with either nontargeted or cyclophilin B control siRNA or a VDR siRNA pool and treated at 48 h for an additional 6 h with either vehicle or 1,25(OH)2D3. The isolated RNA was then analyzed for target gene expression. As noted in Fig. 1B, exposure to VDR siRNA effectively reduced basal expression of VDR mRNA; it also prevented the well-established upregulation of VDR mRNA seen in response to 1,25(OH)2D3 (as shown in reference 71 and confirmed here) when control siRNAs were administered. Most importantly, knockdown of VDR mRNA with VDR siRNA but not control siRNAs effectively blocked the ability of 1,25(OH)2D3 to stimulate RankL expression. Opn and Cyp24a1 induction was suppressed as well. These experiments demonstrate that 1,25(OH)2D3 is able to induce RankL expression in the ST2 cells in a fashion mediated by the VDR and facilitated in some way by GCs.
FIG. 1.
Induction of mRankL mRNA by 1,25(OH)2D3 in ST2 cells is enhanced by DEX and mediated by the VDR. (A) Induction of RankL expression levels by 1,25(OH)2D3 and DEX in vitro. ST2 cells were treated for periods of up to 24 h with either 1,25(OH)2D3 (10−7 M), DEX (10−7 M), or both (at 10−7 M). Total RNA was isolated and subjected to reverse transcription-PCR (RT-PCR) analysis using primers specific to mouse Cyp24a1 (30 cycles), osteopontin (Opn) (15 cycles), RankL (30 cycles), or β-actin (20 cycles) as documented in Materials and Methods. The results are typical of multiple similar experiments. (B) Effects of mVDR siRNA on 1,25(OH)2D3-induced RankL expression levels in ST2 cells. ST2 cells were transfected with 20 nM nontargeting siRNA, cyclophilin B (Cyclo B) siRNA, or mVDR siRNA. After 48 h, the cells were treated for an additional 6 h with either vehicle or 1,25(OH)2D3 (10−7 M). Total RNA was isolated and subjected to standard RT-PCR analysis using the primers identified in panel A above. The numbers of cycles for amplification of each transcript are as follows: VDR, 20 cycles; Cyp24a1, 25 cycles; Opn, 15 cycles, and β-actin, 15 cycles. These results are typical of several independent experiments.
ChIP/chip analysis of the RankL locus identifies potential VDR binding sites.
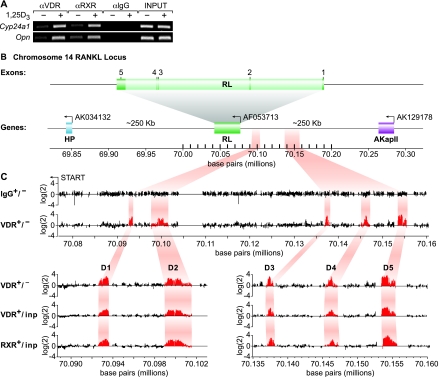
Despite earlier studies (24, 34), regulatory sites within the RankL gene that mediate the actions of 1,25(OH)2D3 remain unclear (11, 48). As a result, we elected to utilize a chromatin immunoprecipitation method combined with DNA microarray analysis (ChIP/chip analysis) to scan the entire mouse RankL gene locus for VDR binding sites. ST2 cells were first treated for 6 h with either vehicle or 1,25(OH)2D3 and then subjected to a standard ChIP using antibodies to VDR, RXR, or a nonspecific IgG. PCR analysis of the resulting immunoprecipitated DNA revealed a significant enrichment of promoter DNA for both Cyp24a1 and Opn (Fig. 2A). This precipitated DNA was then amplified using ligation-mediated PCR, and the individual samples were conjugated directly to either Cy3 or Cy5 (see Materials and Methods). We cohybridized sample pairs to a high-density oligonucleotide DNA microarray which contained the tiled region from 200 kb upstream of the RankL TSS to 100 kb downstream of the final exon at a resolution of 50 bp as seen in Fig. 2B. Figure 2C documents the hybridization signals generated across this tiled array for the following comparisons: (i) IgG in the presence and absence of hormone, (ii) VDR in the absence or presence of hormone, (iii) VDR in the presence of hormone versus input DNA, and (iv) RXR in the presence of hormone versus input DNA. As highlighted in the figure, no preferential enrichment of RankL DNA was evident when a hormone-vehicle comparison was made using DNA derived from immunoprecipitations with control IgG. In contrast, however, five regions of DNA enrichment were observed when similar comparisons were made using ChIP DNA derived from anti-VDR or anti-RXR immunoprecipitations. Interestingly, all of these potential sites of VDR/RXR binding were located at significant distances upstream of the RankL TSS: −16 kb (mRLD1), −22 kb (mRLD2), −60 kb (mRLD3), −69 kb (mRLD4), and −76 kb (mRLD5); no sites were observed within or downstream of the RankL gene itself. Each of these potential binding sites is intergenic in view of the location of AkapII, the annotated gene lying approximately 250 kb upstream. While all of these peaks were obvious to visual inspection, a peak-finding analysis of these data (47) indicated that binding of the VDR and RXR to the mRLD5 region was two- to fivefold more robust than that observed for mRLD1 to mRLD4; mRLD4 generated the weakest signal. These data suggest the locations of at least five potential VDR/RXR binding sites situated long distances upstream of the RankL gene.
FIG. 2.
ChIP/chip analysis reveals five VDR/RXR-interacting regions at significant distances upstream of the mRankL gene TSS. (A) ST2 cells were treated with either vehicle or 1,25(OH)2D3 (10−7 M) for 6 h and then subjected to ChIP analysis using antibodies to VDR, RXR(pan), or control IgG (αVDR, αRXR, and αIgG, respectively). Immunoprecipitated DNA was isolated and then amplified using ChIP primer sets to either Cyp24a1 or Opn as indicated in Materials and Methods and Table 1. Input DNA was obtained prior to precipitation. (B) Schematic diagram of the mouse RankL gene with its five exons and its position relative to adjacent downstream (AK034132) and upstream (AK129178) genes on chromosome 14. The reverse arrow indicates the direction of transcription on the reverse strand. The nucleotide base pairs indicate nucleotide location on chromosome 14 (December 2004 assembly). (C, upper panel) Individual data tracks representing the enrichment ratio of Cy5-to-Cy3 hybridization intensity (log2) for IgG ± hormone or VDR ± hormone. The nucleotide base pairs on the x axis indicate the position on chromosome 14. (C, lower panel) An expanded view of the Cy5/Cy3 signal enrichment ratios for VDR ± hormone, VDR ± versus input, and RXR plus hormone versus input. The highlighted areas indicate the peaks of interest which are designated mRLD1 to mRLD5.
VDR binding to the mRLD1-to-mRLD5 regions of the mouse RankL gene is accompanied by GC-induced GR binding and is associated with RNA pol II recruitment.
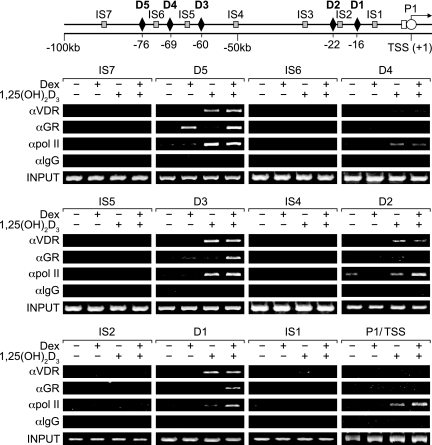
Direct ChIP analysis of immunoprecipitated DNA derived from 1,25(OH)2D3-treated ST2 cells confirmed the presence of VDR and RXR at the mRLD1-to-mRLD5 regions but not at intervening sites within the RankL upstream region (see Fig. S2 in the supplemental material). As anticipated, weak binding of the VDR and RXR was seen at the mRLD4 region. Based upon this direct confirmation, we next explored how DEX might contribute to RankL activation in the presence of 1,25(OH)2D3 (8, 69). ST2 cells were treated with either vehicle, 1,25(OH)2D3, DEX, or the combination and after 3 h subjected to ChIP analysis using antibodies to either VDR or GR. We also attempted to correlate these transcription factor interactions with the recruitment of RNA polymerase II (pol II) at the RankL TSS. The immunoprecipitated DNA was isolated and amplified using the primer sets whose locations are indicated schematically in Fig. 3 and whose nucleotide sequences are documented in Table 1. As can be seen in Fig. 3 (and Fig. S3 in the supplemental material), 1,25(OH)2D3 induced VDR but not GR binding to each of the five regions of the RankL gene, with the weakest binding associated with mRLD4; no binding was observed at the TSS. DEX, on the other hand, induced GR binding to the RankL gene, but had no effect on VDR binding. Interestingly, GR binding in response to DEX was not distributed across the five regions of the RankL gene, however, but rather was restricted to the mRLD5 region alone. The combination of the two hormones led to the accumulation of both VDR and GR to the same sites identified when both ligands were used independently while at the same time promoting some GR binding at mRLD5, mRLD3, and mRLD1. Not unexpectedly, 1,25(OH)2D3 also initiated the recruitment of RNA pol II to the RankL TSS, as observed in Fig. 3; this recruitment was enhanced by DEX. To our surprise, however, 1,25(OH)2D3 and the combination of both 1,25(OH)2D3 and DEX also promoted the recruitment of RNA pol II to the upstream mRLD1-to-mRLD5 regions of the RankL gene. This unanticipated finding suggests that the RankL enhancer regions we have identified may function as recruitment centers for RNA pol II as well (59). Despite this speculation, the results of this experiment provide additional evidence that mRLD1 to mRLD5 may represent important enhancer modules essential to the regulation of RankL gene expression. It is unclear, however, how VDR and GR might collaborate to sensitize the RankL gene to stimulation by both hormones.
FIG. 3.
ChIP analysis of the upstream region of the mRankL gene reveals complex 1,25(OH)2D3- and DEX-stimulated transactivator DNA binding and RNA pol II recruitment. ST2 cells were treated with either vehicle, 1,25(OH)2D3 (10−7 M), DEX (10−7 M), or 1,25(OH)2D3 and DEX (10−7 M) for 6 h and then subjected to ChIP analysis using antibodies to VDR, GR, RNA pol II, or control IgG (αVDR, αGR, αpol II, and αIgG, respectively). The immunoprecipitated DNA was isolated and then amplified using the primer sets whose positions are illustrated in the top panel and whose sequences are documented in Table 1. Amplification utilized 31 cycles for mRLD1 to mRLD5 and mRLIS1 to mRLIS7 and 34 cycles for the TSS. PCR analyses were all performed within the linear range of amplification. These results are typical of several similar studies.
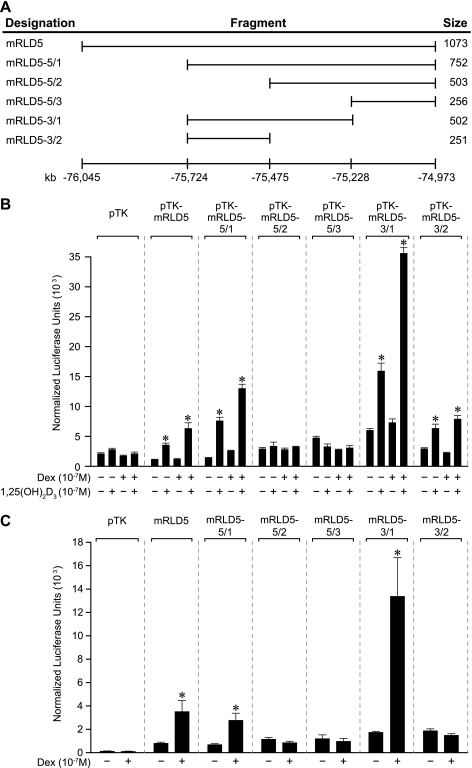
The mRLD5 region of the mouse RankL gene (RankL distal control region) exhibits 1,25(OH)2D3-dependent, GC-modulated enhancer activity in the context of a heterologous promoter.
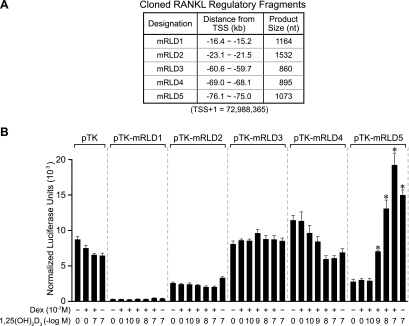
The mRLD1-to-mRLD5 regions of the RankL gene are conserved across several species, including humans, providing further support for the possibility that these regions represent functional control domains for the RankL gene (see Fig. S4 in the supplemental material). We therefore amplified each of the five regions (whose coordinates were defined by both conservation and the tiling array data, and are documented in Fig. 4A), cloned these fragments into a TK promoter-luciferase reporter vector and assessed their transcriptional activity in response to 1,25(OH)2D3, DEX, or a combination of the two ligands. To increase sensitivity, we cotransfected a VDR expression vector together with the RankL regulatory constructs, although this maneuver was not essential, as will be seen later. Strikingly, the results depicted in Fig. 4B reveal that mRLD5 mediated a strong, dose-dependent sensitivity to 1,25(OH)2D3 that was potentiated by DEX. Surprisingly, the regions comprising mRLD1 to mRLD4 were unable to convey 1,25(OH)2D3 response to the TK promoter, although an unusual suppression of activity by the individual hormones was noted with mRLD4. These experimental results suggest that the mRLD5 region is likely an important, if not central, component that mediates the regulation of RankL gene expression by 1,25(OH)2D3 and perhaps the GCs. The interactive and functional properties of this conserved region prompted us to designate this region as part of the RankL distal control region, or RL-DCR (see Discussion for additional details). The significant VDR/RXR binding noted at mRLD1 to mRLD4, however, prevents us from excluding these regions as potential regulators of RankL gene expression.
FIG. 4.
The mRLD5 region of the mRankL gene mediates transcriptional induction by 1,25(OH)2D3 and DEX. (A) Features of the cloned mRLD1-to-mRLD5 regions of the mRankL gene, including the sizes of the fragments cloned for evaluation, the distances from the RankL TSS, and the boundaries of the fragments on chromosome 14 (February 2006 assembly). (B) Basal and hormone-inducible activities of mRLD1 to mRLD5 in ST2 cells. ST2 cells were transfected with pCH110-βgal (50 ng), pcDNA-hVDR (50 ng), and either the pTK control vector (250 ng) or pTK-mRLD1, pTK-mRLD2, pTK-mRLD3, pTK-mRLD4, or pTK-mRLD5. Cells were treated with either vehicle, 1,25(OH)2D3 (10−7 M), DEX (10−7 M), or DEX (10−7 M) plus increasing concentrations of 1,25(OH)2D3 (10−10 to 10−7 M) and evaluated after 24 h for both luciferase and β-galactosidase activities as described in Materials and Methods. Each point represents the normalized relative light unit average ± standard error of the mean for a triplicate set of transfections. These data are representative of three or more similar experiments.
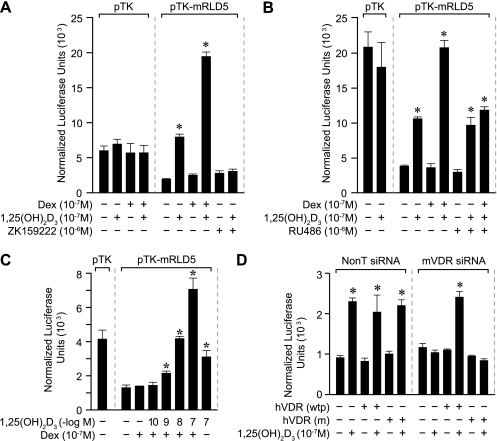
The transcriptional effects of 1,25(OH)2D3 and DEX on mRLD5 are blocked by VDR and GR antagonists.
The ability of 1,25(OH)2D3 and DEX to modulate transcriptional output via the mRLD5 segment of the RankL gene prompted us to explore the roles of VDR and GR in this regulatory process. To test whether both receptors were directly involved, we assessed whether the VDR antagonist ZK159222 and the GR antagonist RU486 could block the activities of the two hormones on mRLD5 transcription. As can be seen in Fig. 5A and B, while 1,25(OH)2D3 alone and both 1,25(OH)2D3 and DEX induced mRLD5-mediated transcription, both ZK159222 and RU486 blocked the transcriptional activities induced by 1,25(OH)2D3 or 1,25(OH)2D3 and DEX, respectively. These results suggest that both VDR and GR activities are integral to the induction process and likely involve the recruitment of coregulators in the process (16, 30). The involvement of the VDR was tested in two additional ways. In the first, we introduced the mRLD5 region into ST2 cells without cotransfection of a VDR expression vector. As seen in Fig. 5C, although this maneuver reduced basal levels of the reporter, the profile of inducibility in the presence of 1,25(OH)2D3 as well as with the combination of hormones was retained. In the second, we utilized VDR siRNA to suppress the level of expression of the VDR and evaluated the effect of this downregulation on induction by 1,25(OH)2D3 alone. As can be seen in Fig. 5D, while cotransfection of the mRLD5 reporter together with control siRNA had no effect on the ability of 1,25(OH)2D3 to induce this DNA construct, the addition of VDR siRNA fully blocked that induction. Importantly, this loss of inducibility could be rescued by cotransfecting a wild-type human VDR expression vector but not a vector expressing a transcriptionally inactive form of the VDR which contained mutations in the transactivating domain (1). Our results support the idea that both the VDR and GR are required for the hormone-inducible activity mediated by the mRLD5 region.
FIG. 5.
Hormone-inducible activity of the mRLD5 region is mediated by the VDR and GR. (A) The VDR antagonist ZK159222 blocks activation of mRLD5 by 1,25(OH)2D3. ST2 cells were transfected with pCH110-βgal (50 ng), pcDNA-hVDR (50 ng), and either the pTK control vector (250 ng) or pTK-mRLD5. Cells were treated with either vehicle, 1,25(OH)2D3 (10−7 M), DEX (10−7 M), DEX (10−7 M) plus 1,25(OH)2D3 (10−7 M), ZK159222 (10−6 M), or 1,25(OH)2D3 (10−7 M) plus ZK159222 (10−6 M) and evaluated after 24 h for both luciferase and β-galactosidase activities as described in Materials and Methods. (B) The GR antagonist blocks activation of mRLD5 by DEX. ST2 cells were treated as in panel A above with either vehicle, 1,25(OH)2D3 (10−7 M), DEX (10−7 M), DEX (10−7 M) plus 1,25(OH)2D3 (10−7 M), RU486 (10−6 M), RU486 (10−6 M) plus 1,25(OH)2D3 (10−7 M), or RU486 (10−6 M) plus 1,25(OH)2D3 (10−7 M) plus DEX (10−7 M) and evaluated after 24 h for both luciferase and β-galactosidase activities as described in Materials and Methods. (C) 1,25(OH)2D3 activates mRLD5 via endogenous VDR. ST2 cells were transfected with pCH110-βgal (50 ng) and either the pTK control vector (250 ng) or pTK-mRLD5 and then treated with either vehicle, DEX (10−7 M), DEX (10−7 M) plus 1,25(OH)2D3 (10−10 to 10−7 M), or 1,25(OH)2D3 (10−7 M) alone. Luciferase and β-galactosidase activities were assessed 24 h later as described in Materials and Methods. (D) The ability of VDR siRNA to block activation of mRLD5-mediated transcription by 1,25(OH)2D3 is rescued through the addition of a functional VDR expression vector. ST2 cells were cotransfected with 10 ng pCH110-βgal, pTK-mRLD5, pcDNA-hVDR(wtp) (wild type), or pcDNA-hVDR(m) (mutant) and 50 nM of either nontargeted siRNA or mVDR siRNA as described in Materials and Methods. Transfected cells were cultured for 48 h and then treated for an additional 24 h with either vehicle or 1,25(OH)2D3 (10−7 M) and processed as in panel B. Each point represents the normalized relative light unit average ± standard error of the mean for a triplicate set of transfections. All studies were repeated with similar results.
Mapping the VDR and GR regulatory regions located within the mRLD5 fragment.
The above ChIP assays together with the transfection studies suggest that the mRLD5 region contains regulatory elements capable of binding both VDR and GR and mediating the activity of 1,25(OH)2D3 and DEX. We therefore created a series of 5′ and 3′ deletion constructs of this region as indicated in Fig. 6A, introduced them together with a VDR expression vector into ST2 cells, and mapped the activity of the two hormones. The results in Fig. 6B reveal that while changes in basal activity are evident within the deletion constructs, the activity of 1,25(OH)2D3 maps directly to a central core fragment of 256 bp (mRLD5-3/2) which is located within mRLD5. The synergistic response to DEX, in contrast, maps to an activity that appears to span two fragments designated mRLD5-3/2 and mRLD5-5/2. Thus, while neither of these fragments is responsive to DEX synergistically, an overlapping fragment termed mRLD5-3/1 that contained the boundary between the above two fragments manifested striking DEX induction. This interpretation is further supported by direct analysis of DEX activity wherein the addition of cotransfected GR provides a direct readout of this receptor's activity on mRLD5 subfragments. These results suggest that inducible activity of mRLD5 is potentially governed by a single regulatory element for 1,25(OH)2D3 and perhaps via several elements for DEX.
FIG. 6.
Deletion analysis of the mRLD5 region reveals the approximate location of transcriptional response to both 1,25(OH)2D3 and DEX. (A) Schematic of the mRankL deletion fragments of the mRLD5 region that were used to map 1,25(OH)2D3 and DEX responses. The numbering at the bottom represents the distance from the RankL TSS (February 2006 assembly). (B) Transcriptional activities of the mRLD5 subfragments in response to 1,25(OH)2D3 and DEX. ST2 cells were transfected with pCH110-βgal (50 ng), pcDNA-hVDR (50 ng), and either the pTK control vector (250 ng) or the pTK-mRLD5 deletion constructs indicated. Cells were treated with either vehicle, 1,25(OH)2D3 (10−7 M), DEX (10−7 M), or DEX (10−7 M) plus 1,25(OH)2D3 (10−7 M) and evaluated after 24 h for both luciferase and β-galactosidase activities as described in Materials and Methods. (C) Transcriptional activities of the mRLD5 subfragments in response to DEX. ST2 cells were transfected with pCH110-βgal (50 ng), pRSV-hGR (50 ng), and either the pTK control vector (250 ng) or the pTK-mRLD5 deletion constructs indicated. Cells were treated with either vehicle or DEX (10−7 M) and evaluated after 24 h as in panel B. These results were confirmed via at least three separate experiments.
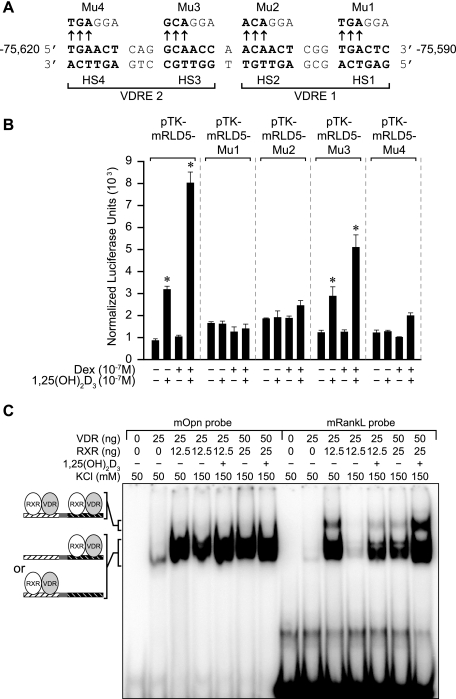
Identification of the mRL-VDRE within the mRLD5 region.
An evaluation of the DNA sequence within the 1,25(OH)2D3-inducible fragment of the mRLD5 region (mRLD5-3/2) using the CONSITE (http://mordor.cgb.ki.se/cgi-bin/CONSITE/consite) algorithm revealed several nuclear receptor-like regulatory elements. One such region, however, contained two highly conserved VDRE-like sequences separated by a single base pair (see Fig. S5 in the supplemental material). To test whether this unusual element as seen in Fig. 7A might mediate the actions of 1,25(OH)2D3, a set of 3-bp mutations was introduced in the context of the mRLD5 fragment into each of the four half-sites comprising the putative VDRE. The constructs were then transfected into ST2 cells, and their activities were evaluated in response to 1,25(OH)2D3. As can be seen in Fig. 7B, mutations in three of the four half-sites completely abolished hormonal response; activity derived from a fourth construct was compromised. These results suggest that this interesting element does indeed mediate the actions of 1,25(OH)2D3 in the context of mRLD5. We also examined whether the VDR and its RXR partner could bind to this VDRE, particularly as a complex comprised of two heterodimers. To this end, we carried out an electrophoretic mobility shift assay (EMSA) using duplex oligonucleotides of either the Opn-VDRE or the putative mRL-VDRE and purified VDR and RXRα proteins. As can be seen in Fig. 7C, the VDR/RXR heterodimer bound to both the Opn- and the mRL-VDREs in a salt-sensitive, hormone-dependent fashion. Similar binding of endogenous VDR and RXR was also observed in the presence of ST2 nuclear extracts (data not shown). Competition studies suggest that the relative binding affinities of the VDR/RXR heterodimer for these two VDREs were similar (see Fig. S6A in the supplemental material). Interestingly, the results in Fig. 7C also show that incubation of VDR/RXR with the mRL-VDRE but not the Opn-VDRE results in the appearance of a second, more slowly migrating species, suggestive of a specific complex comprised of two VDR/RXR heterodimers. Support for this contention is provided by the observation that the higher-order complex fails to form when purified VDR/RXR is incubated with mRL-VDREs that contain mutations in either VDRE1 or VDRE2 (see Fig. S6B in the supplemental material). Taken together, these results suggest that the element located at −75620 to −75590 upstream of the mouse RankL TSS is capable of both binding two VDR/RXR heterodimers and mediating the transactivation potential of this complex in transfected cells.
FIG. 7.
Mapping the mRL-VDRE. (A) DNA sequence of a putative mRL-VDRE revealed by in silico analysis (http://mordor.cgb.ki.se/cgi-bin/CONSITE/consite). The nucleotide numbering represents the boundaries of the mRL-VDRE relative to the RankL TSS (February 2006 assembly). Nucleotide bases above the arrows represent the triplet alterations introduced by site-directed mutagenesis into each of the half-sites (HS1 to HS4) of the two VDREs (VDRE1 and VDRE2) that constitute the mRL-VDRE. Mutants are designated Mu1 to Mu4. (B) Transcriptional activity of wild-type pTK-mRLD5 or pTK-mRLD5 containing mutation 1 (Mu1), Mu2, Mu3, or Mu4. ST2 cells were transfected with pCH110-βgal (50 ng), pcDNA-hVDR (50 ng), and either the pTK control vector (250 ng), pTK-mRLD5, or pTK-mRLD5 containing Mu1 to Mu4 as illustrated. Cells were treated with either vehicle, 1,25(OH)2D3 (10−7 M), DEX (10−7 M), or the combination and evaluated after 24 h for both luciferase and β-galactosidase activities. These results were repeated with similar findings. (C) Purified VDR and RXR bind to the mRL-VDRE as a pair of heterodimers. Labeled duplex DNA probes from the mouse osteopontin VDRE (mOpn DR3) or the sequence corresponding to the mRL-VDRE depicted in panel A above were incubated with the indicated amounts of purified VDR and RXRα in 50 mM or 150 mM KCl without (−) or with (+) 1,25(OH)2D3 (5 × 10−9 M) as indicated. Complexes were resolved on 6% nondenaturing polyacrylamide gels, dried, and visualized by autoradiography. Complexes 1 and 2 are indicated. These results were confirmed through at least three separate EMSA analyses.
The mRL-VDRE alone confers strong 1,25(OH)2D3 responsiveness to a heterologous promoter.
In a final set of experiments aimed at characterizing the mRL-VDRE, we cloned this DNA sequence as well as each of the half-VDREs that comprise the element as single copies upstream of a TK promoter and examined their ability to mediate 1,25(OH)2D3 response independent of the mRLD5 environment. Constructs were introduced into ST2 cells together with a VDR expression vector, and their activities were assessed in response to increasing concentrations of 1,25(OH)2D3. As can be seen in Fig. 8A, the mRL-VDRE was strongly induced by 1,25(OH)2D3 in a dose-dependent fashion. Interestingly, neither of the two individual VDREs that comprise the mRL-VDRE was capable of mediating any significant response to 1,25(OH)2D3. Further analysis of the mRL-VDRE indicated that the introduction of a 3-bp change in any one of the four mRL-VDRE half-sites (as documented in Fig. 7) fully compromised the response to 1,25(OH)2D3 (Fig. 8B). Only the mutation at the third half-site retained some small activity. The role of the VDR in this induction was further confirmed by a VDR siRNA knockdown and human VDR rescue experiment. Accordingly, while cotreatment of ST2 cells with control siRNA had no effect on the ability of 1,25(OH)2D3 to induce the mRL-VDRE, as documented in Fig. 8C, the introduction of VDR siRNA fully compromised this upregulation. The induction was fully rescued by the addition of wild-type human VDR, although not with a transcriptionally inactive human VDR allele containing mutations in its activation domain as described previously. Importantly, as shown in Fig. S7 in the supplemental material, this mRL-VDRE was also able to mediate 1,25(OH)2D3 inducibility in other mouse osteoblastic cell types, including primary mouse calvarial osteoblasts, as well as in primate cells. Our studies therefore define a functional VDRE within the mRLD5 region that is structurally unique and that likely plays a significant role in mediating the ability of 1,25(OH)2D3 to induce RankL gene expression.
FIG. 8.
The mRL-VDRE confers significant VDR-dependent, 1,25(OH)2D3 response to the heterologous TK promoter. (A) Transcriptional activity of the mRL-VDRE or its two-component VDREs in ST2 cells. ST2 cells were transfected with pCH110-βgal (50 ng), pcDNA-hVDR (50 ng), and either the pTK control vector (250 ng), pTK-mRL-VDRE, pTK-mRL-VDRE1, or pTK-mRL-VDRE2. Cells were treated with either vehicle or increasing concentrations of 1,25(OH)2D3 (10−10 to 10−7 M) and evaluated after 24 h for both luciferase and β-galactosidase activities. (B) Effect of half-site mutations within the mRL-VDRE on response to 1,25(OH)2D3. Duplex mRL-VDRE oligonucleotides containing either the wild-type sequence or individual triplet mutations as depicted in Fig. 7A were cloned into the TK vector and transfected into ST2 cells, and their activity was evaluated in response to 1,25(OH)2D3 (10−7 M) after 24 h. Due to baseline differences, induction (fold) is reported. (C) Ability of mVDR siRNA to block activation of mRL-VDRE-mediated transcription by 1,25(OH)2D3 is rescued through the addition of a functional VDR expression vector. ST2 cells were cotransfected with 10 ng pCH110-βgal, pTK-mRL-VDRE, pcDNA-hVDR(wtp) (wild type), or pcDNA hVDR(m) (mutant) and 50 nM either nontargeted siRNA or mVDR siRNA as described in the legend to Fig. 5. Transfected cells were cultured for 48 h and then treated for an additional 24 h with either vehicle or 1,25(OH)2D3 (10−7 M) and processed as in panel B. Each point represents the normalized relative light unit average ± standard error of the mean for a triplicate set of transfections.
1,25(OH)2D3 stimulates and DEX potentiates the activity of mRLD5 when cloned upstream of the authentic RankL gene promoter.
The studies described above made use of a heterologous viral promoter known to contain elements potentially responsive to glucocorticoids. As a result, we cloned a RankL minimal promoter fragment, mRL(100), into a pGL3 luciferase vector to prepare an authentic RankL promoter reporter, and then introduced the mRL regions mRLD1 to mRLD5 (as well as a single copy of the mRL-VDRE) directly upstream of this TSS. The constructs were introduced into ST2 cells and examined for responses to 1,25(OH)2D3, DEX, and the combination. As can be seen in Fig. S8 in the supplemental material, while neither pGL3 nor the modified pGL3 vector containing the minimal RankL promoter, mRL(100), manifested any response, the profile of regulation of mRLD5 by both 1,25(OH)2D3, DEX, and the combination was virtually identical to that seen when mRLD5 when evaluated in the context of the heterologous TK promoter (Fig. 4). Unfortunately, the basal and hormone-resistant activities of mRLD1 to mRLD4 were also similar to that observed in the context of the TK promoter, suggesting that the individual placement of each of these regions immediately upstream of its native proximal promoter was insufficient to manifest a transcriptional response to 1,25(OH)2D3, DEX, or a combination of the two ligands (data not shown). We therefore conclude that only mRLD5 manifests an autonomous response to the two ligands when examined in the context of a reporter plasmid.
Activation of the five regulatory regions of the mouse RankL gene results in histone acetylation.
While mRLD4 to mRLD1 failed to exhibit inducible transcriptional activity in the context of either the TK promoter or a homologous mRL promoter, the results of the ChIP/chip and direct ChIP assays using VDR, RXR, and RNA pol II strongly support a role for mRLD4 to mRLD1 in mediating the actions of 1,25(OH)2D3 on RankL gene expression. We therefore asked whether 1,25(OH)2D3-induced binding of VDR and RXR to the five RankL gene regions was capable of inducing modifications to histones within those regions of the gene. Previous studies have suggested that this modification is essential to the upregulation of Cyp24a1 (30). ST2 cells were therefore treated with 1,25(OH)2D3, and the cells were subjected to ChIP analysis for increasing times using antibodies to tetra-acetylated histone 4 as well as VDR. 1,25(OH)2D3 induced both a time-dependent accumulation of VDR at the mRLD1-D5 regions and an increase in histone 4 acetylation, as can be seen in Fig. 9 (and see Fig. S9 in the supplemental material). Surprisingly, however, increased H4 acetylation did not occur at mRLD1 to mRLD5 in the RL gene locus (with the possible exception of mRLD3), but rather at sites located between the five enhancers and at the RankL TSS. This finding suggests that while 1,25(OH)2D3 can induce changes in acetylation within the mRankL gene locus, these changes do not appear to correlate directly with regions of VDR/RXR heterodimer binding.
FIG. 9.
1,25(OH)2D3 induces histone acetylation in the upstream regions of the mRankL gene. ST2 cells were treated with 1,25(OH)2D3 (10−7 M) for periods of up to 6 h and then subjected to ChIP analysis using antibodies to VDR, tetra-acetylated histone 4, or IgG (αVDR, αAcH4, and αIgG, respectively). Precipitated DNA was isolated and evaluated by PCR using Cyp24a1 primers (see Materials and Methods) or RankL primers whose locations are depicted in Fig. 3 (upper panel) and whose sequences are delineated in Table 1. Amplifications were carried out as in Fig. 3. Similar results were obtained in at least four separate experiments.
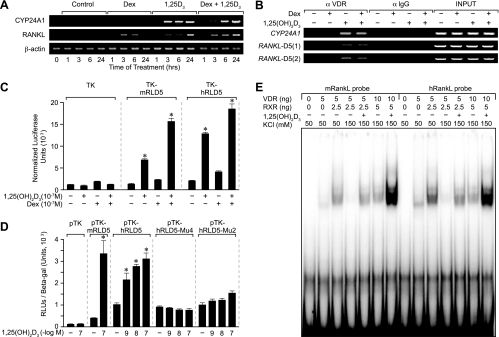
The highly evolutionarily conserved hRLD5 region located upstream of the human RANKL gene is controlled in a similar fashion by 1,25(OH)2D3 and DEX.
The mRLD5 region of the mouse RankL gene is highly conserved within vertebrate genomes, including that of humans. In humans, the corresponding RANKL D5 region (hRLD5) is also similarly located upstream of the TSS, although at a slightly greater distance of approximately −96 kb. To explore whether this region in the human RANKL gene might similarly mediate the actions of 1,25(OH)2D3 and DEX on RANKL gene expression, we first established a model for RANKL expression using the human MG63 osteoblastic cell line. We treated MG63 cells with either 1,25(OH)2D3, DEX, or both ligands and then evaluated the effects of these ligands as a function of time on RANKL induction. CYP24A1 was used as a positive control for 1,25(OH)2D3 induction. As can be seen in Fig. 10A, while both 1,25(OH)2D3 and DEX were able to modestly induce RANKL expression in this cell line, the combination was slightly more effective. Next, we treated MG63 cells with either vehicle or 1,25(OH)2D3 for 6 h and subjected the harvested cells to a ChIP analysis using antibodies to either VDR or an IgG control. The precipitated DNA was evaluated by PCR using primers to either CYP24A1 or two separate primer sets to the hRLD5 region as indicated in Materials and Methods and documented in Table 1. As depicted in Fig. 10B, 1,25(OH)2D3 induced VDR binding to both control CYP24A1 and to the human hRLD5 region as well. DEX did not appear to potentiate or influence the extent of VDR binding. Based upon these results, we cloned approximately 0.8 kb of the human hRLD5 region into the TK promoter, transfected this plasmid or the mouse mRLD5 TK plasmid into MG63 cells, and evaluated the transcriptional response to 1,25(OH)2D3, DEX, or both ligands. The results in Fig. 10C reveal that the hRLD5 region is comparable to the mouse version in its capacity to mediate response to both 1,25(OH)2D3 and DEX. DEX, however, appears able to exert a small but measurable effect directly on the hRLD5 region even in the absence of 1,25(OH)2D3. Importantly, mutagenesis of two of the putative human VDRE half-sites within the context of hRLD5 abrogated the response to 1,25(OH)2D3, as seen in Fig. 10D, thus confirming the role of these elements in 1,25(OH)2D3-dependent induction. In a final set of experiments, we assessed the capacities of VDR and RXR to bind directly to the hRL-VDRE using mRL-VDRE as a control. As can be seen in Fig. 10E, both VDR and RXR bind to each of the labeled RL-VDREs in a salt-sensitive, hormone-dependent fashion. The low input levels of VDR and RXR limit the binding of two VDR/RXR heterodimers. These results clearly establish the ability of the hRLD5 region to mediate the actions of 1,25(OH)2D3 and DEX on the human RANKL gene and provide additional supportive data for the hypothesis that the conserved enhancer activity of RLD5 is essential to the regulation of both mouse and human RANKL genes.
FIG. 10.
The highly evolutionarily conserved hRLD5 region mediates the transcriptional activity of 1,25(OH)2D3 in the human RANKL gene. (A) RANKL mRNA is induced by 1,25(OH)2D3 and enhanced by DEX in MG63 cells. MG63 cells were treated for periods up to 24 h with either vehicle, DEX (10−7 M), 1,25(OH)2D3 (10−7 M), or both (at 10−7 M). Total RNA was isolated and subjected to reverse transcription-PCR (RT-PCR) analysis using primers specific to human CYP24A1 (27 cycles), RANKL (30 cycles), or β-actin (20 cycles) as documented in Materials and Methods. The results are typical of multiple similar experiments. (B) 1,25(OH)2D3 induces VDR binding to the hRLD5 region of the human RANKL gene. MG63 cells were treated with either vehicle, DEX (10−7 M), 1,25(OH)2D3 (10−7 M), or 1,25(OH)2D3 and DEX (10−7 M) and then subjected to ChIP analysis using antibodies to VDR or control IgG (αVDR and αIgG, respectively). The immunoprecipitated DNA was isolated and then amplified using the primer sets documented in Table 1. PCR was performed for 31 cycles. Two separate primer sets were used for validation. (C) The hRLD5 region of the hRANKL gene mediates transcriptional induction by 1,25(OH)2D3 and DEX. MG63 cells were transfected with pCH110-βgal (50 ng), pcDNA-hVDR (50 ng), and either the pTK control vector (250 ng), pTK-mRLD5, or pTK-hRLD5. Cells were treated with either vehicle, 1,25(OH)2D3 (10−7 M), DEX (10−7 M), or DEX (10−7 M) plus 1,25(OH)2D3 (10−7 M) and evaluated after 24 h for both luciferase and β-galactosidase activities. Each point represents the normalized relative light unit (RLU) average ± standard error of the mean for a triplicate set of transfections. (D) Transcriptional activities of wild-type pTK-hRLD5 or mutant pTK-hRLD5 containing triplet changes in the hRL-VDRE. MG63 cells were transfected with pCH110-βgal (50 ng), pcDNA-hVDR (50 ng), and either the pTK control vector (250 ng), pTK-mRLD5, pTK-hRLD5, or pTK-hRLD5 containing triplet mutations in half-site 2 (Mu2) or 4 (Mu4). Cells were treated with either vehicle or increasing concentrations of 1,25(OH)2D3 (10−9 to 10−7 M) and evaluated after 24 h for both luciferase and β-galactosidase activities. Each point represents the normalized RLU average ± standard error of the mean for a triplicate set of transfections. (E) VDR and RXR bind directly to the hRL-VDRE in a salt- and hormone-dependent fashion. Labeled duplex DNA probes comprising the mRL-VDRE or the hRL-VDRE were incubated with the indicated amounts of purified VDR and RXRα in 50 mM (−) or 150 mM (+) KCl without or with 1,25(OH)2D3 (10−9 M) as indicated. Complexes were resolved on 6% nondenaturing polyacrylamide gels, dried, and visualized using autoradiography. The results of these separate studies were reproduced at least three times.
DISCUSSION
The importance of RankL as a key osteoclastogenic factor in bone remodeling is now well established (63). Perhaps most persuasive is the finding through deletion studies in mice that the loss of either RankL or its receptor (Rank) results in a significant deficiency in osteoclast production (20, 37). Indeed, several osteoporotic states in humans have been ascribed to genetic defects in the RANKL/RANK/OPG signaling pathway (68). Thus, although factors separate from RankL are known to play roles in the osteoclastogenic process (35, 63), it is clear that this gene product and its signal transduction pathway are key components. An understanding of the mechanisms whereby this gene is regulated is therefore of paramount importance.
Our studies using ChIP/chip analysis, direct ChIP, and more traditional molecular approaches identified five distinct regions located at significant distances upstream of the TSS that were responsible for regulating the expression of the RankL gene. VDR and RXR localize to each of these regions in response to 1,25(OH)2D3 and appear to be accompanied by other factors as well, including GR. This supports the idea that each of these regions may represent enhancer modules integral to RankL gene expression. A typical consequence of transactivation at enhancers is the rapid recruitment of coregulatory enzymes and the subsequent modification of local chromatin structure, in part through acetylation (5, 52, 55, 56). It is believed that these epigenetic modifications function to promote the chromatin decondensation that is often necessary for increased transcription. In our studies, while it is clear that 1,25(OH)2D3 induced histone 4 acetylation at the RankL gene locus, these modifications did not occur at sites of VDR/RXR binding, but rather at sites located in between. Thus, although direct recruitment of histone-modifying enzymes by the VDR/RXR heterodimer may be involved, we believe it more likely that the VDR precipitates events at these sites that lead in turn to the increased acetylation seen between the enhancer regions. The nature of these events remains to be determined. Interestingly, 1,25(OH)2D3 also induced the recruitment of RNA pol II to the mRLD1-to-mRLD5 regions of the RankL gene. This finding suggests that the mRLD1-to-mRLD5 enhancers may also act as recruitment centers for components of the transcriptional apparatus as well (59). Although speculative, it is possible that the recruitment of transcriptional components to the five enhancer regions and the consequences of this recruitment may represent the initiating event for the histone modifications seen above. Regardless, our data suggest that the ChIP/chip and ChIP approaches taken here were strategic in identifying key regions within the RankL locus that are responsible for transcriptional regulation of this gene by 1,25(OH)2D3 and DEX.
While all of the sites appear to contribute to the response initiated by 1,25(OH)2D3, only the mRLD5 region was capable of conferring sensitivity to 1,25(OH)2D3 in a transcriptional reporter plasmid assay. The respective responses to 1,25(OH)2D3, DEX, and the combination are consistent with that seen at the level of RankL mRNA, thereby validating further the relevance of mRLD5 to RankL gene expression. The roles of VDR and RXR in this regulation were further established using both selective antagonists as well as siRNA, which reduced VDR mRNA levels and compromised the capacity of 1,25(OH)2D3 to induce transcription via the mRLD5 segment. Because ZK159222 and RU486 do not prevent receptor DNA binding, but rather alter their ability to recruit coregulators (16, 30), these results provide additional support for cofactor involvement in enhanced RankL gene expression. We defined the boundaries of mRLD5 based upon the VDR and RXR binding activities seen in the ChIP/chip analysis. The fragment was comprised of approximately 1,100 bp of highly conserved sequence present across multiple RankL genes. An additional highly conserved region of 800 bp immediately upstream of the mRLD5 region was also noted. This segment corresponds directly to the region that mediates PTH response, as described in the accompanying article by Fu, Manolagas, and O'Brien (13). Our additional studies using ChIP analysis, transient transcription assays, and mutagenesis fully confirm these observations (S. Kim and J. W. Pike, unpublished observations). Thus, we propose that this entire conserved region be designated the RankL distal control region, or RL-DCR.
The transcriptional activity of the mRLD5 region allowed us both to map the mRL-VDRE and to characterize a specific GC response. The mRL-VDRE retains a unique structure comprised of two separate VDRE sequences linked via a single base pair. This element is clearly capable of the simultaneous binding of two VDR/RXR heterodimers and is likewise responsible for the 1,25(OH)2D3 sensitivity observed within the mRLD5 region. Interestingly, the half-elements alone bind single VDR/RXR heterodimers, but do not elicit a significant transcriptional response when cloned and analyzed independently. We did not map the existing GREs within this region of the RankL gene, however. The absence of activity in two contiguous but nonoverlapping fragments and the DEX sensitivity observed in an mRLD5 fragment which spanned the boundary between these two fragments suggest the presence of several GREs that may function synergistically to mediate DEX activation. Further studies will be necessary to define the putative GREs that we observed using in silico analysis (data not shown). Although DEX activity on mRLD5 appears to be synergistic, the addition of exogenous GR enabled us to detect a direct response to the GC. This effect was also noted when the human RLD5 region (hRLD5) was evaluated in the context of MG63 cells. Regardless of the nature of these overall effects, it seems clear that 1,25(OH)2D3 action is mediated via a single VDRE, whereas DEX activity is likely mediated by several independent GREs.
The appearance of VDR and RXR, histone acetylation, and RNA pol II recruitment all support the idea that mRLD1 to mRLD5 are active in the regulation of the RankL gene by 1,25(OH)2D3. It is therefore curious as to why the mRLD1-to-mRLD4 upstream regions of the gene are incapable of mediating a transcriptional response to 1,25(OH)2D3 in transfection studies. We believe that this resistance to hormonal induction highlights the importance of context, wherein the presence of both positive as well as negative cis elements and their respective transregulators exert significant influence on transcriptional readouts obtained during transient transfection assays. Interestingly, even the activity of a natural promoter in a plasmid context may be deceptive. The large distances that exist between mRLD1 to mRLD5 and the location of the most distance element (−76 kb) made it difficult for us to assess the transient activity of a “full-length” RankL promoter by these means. Fu, Manolagas, and O'Brien overcame this problem, however, by using recombineering methods to produce large, bacterial artificial chromosome (BAC) clone-derived RankL constructs which could be stably transfected into host cells (13). Interestingly, despite the differences in the two approaches, both of our groups were able to identify the same highly conserved distal region within the RankL gene that displayed sensitivity to the two hormones 1,25(OH)2D3 and PTH. While the above results clearly demonstrate the overall dominance of the RL-DCR, the ability of 1,25(OH)2D3 to induce a large RankL DNA fragment that no longer contained the mRLD5 (13) provides further evidence that the mRLD1-to-mRLD4 regions are also important contributors to 1,25(OH)2D3 response. Therefore, we believe that ChIP analysis may represent a more reliable method of assessing the presence of functional enhancers than the alternative analysis using plasmid transfection. Studies are ongoing in our laboratory, however, to identify the precise locations of the VDREs lodged within the mRLD1-to-mRLD4 regions that regulate RankL gene expression.
Regardless of the properties of mRLD1 to mRLD4, the studies of both Fu, Manolagas, and O'Brien (13) as well as our own highlight the important role of the mRL-DCR in mediating both PTH and 1,25(OH)2D3 responses. It is therefore noteworthy that in the accompanying article, Fu et al. (13) provide in vivo evidence that genetic deletion of the mRL-DCR in mice leads to a loss of PTH-mediated RankL induction. We would predict that these mice will also display a similar resistance to 1,25(OH)2D3, although whether this resistance will be only partial or complete remains to be determined. Collaborative studies are currently under way to more fully investigate the properties of this altered RankL gene locus in vivo and, more importantly, perhaps, to determine the nature of the skeletal phenotype that results from this genetic change.
The regulatory regions for RankL are widely dispersed across a rather large segment of upstream DNA, the furthest some 76 kb from the TSS. It is therefore clear that an understanding of the function of each of these regions and their individual roles in modulating the expression of the RankL gene will require significant additional work. In that vein, there is increasing evidence that many genes contain distant regulatory elements such that this mechanism of regulation may be more frequent than previously believed (2, 21, 39). The extended pattern and distance of these elements within the RankL gene as well as in other genes almost certainly highlight the crucial impact of chromatin structure and organization on the expression of these genes such that the elements can directly impact activity at the TSS. One might speculate that these regions converge directly on the proximal RankL promoter by virtue of extensive chromatin looping (6, 9, 49, 65). New technologies are currently available or in development to test distance relationships between regulatory regions and their functional promoters, thus making it possible to explore this intriguing hypothesis (7).
A final question that emerges from these studies is why RNA pol II might be recruited to each of the five enhancer domains within the RankL gene. One might imagine that the presence of RNA pol II at these sites might also require the simultaneous recruitment of basal transcription factors such as TF-IIA, TF-IIB and TAF-II250 as well (59). Indeed, we have seen in preliminary studies that 1,25(OH)2D3 can induce the recruitment of TAF-II250 to the mRLD5 region (S. Kim and J. Pike, unpublished data). One frequent consequence of the assembly of such factors at upstream regulatory regions, however, is the production of nascent noncoding mRNAs (3, 4, 10, 26, 27). The role of this transcription and these transcripts is currently unknown, although it has been suggested that their production may be essential to the maintenance of an open chromatin state necessary for gene regulation (15). An alternative proposal is that upstream enhancers may function as centers for the recruitment of basal transcriptional machinery, thereby providing a source of such factors for authentic promoter-driven transcription (59). Now that some of the fundamentals of hormonal regulation have been established within the RankL gene locus, future studies will focus on these issues involving RNA pol II recruitment and on understanding the spatial arrangement that likely exists between the five regulatory regions and the TSS.
In summary, we have shown that 1,25(OH)2D3 and its receptor induce the expression of RankL via five regulatory domains located at significant distances upstream of the TSS. This regulation is facilitated by DEX and the GR, the latter appearing to localize to several but not all of these regions. Transcription factor binding within these regions is associated with the recruitment of cofactors such as RNA pol II and adjacent histone acetylation. Mapping studies of mRLD5, perhaps the dominant control region for the RankL gene, led to the delineation of an unusual VDRE. Support for mRLD5 as a primary enhancer of mouse RankL gene expression was increased by the discovery that an analogous component is located within the human RANKL gene.
Supplementary Material
Acknowledgments
We thank members of the Pike laboratory for helpful discussion. We thank Adam Steinberg and Laura Vanderploeg for preparing the figures for this article.
This work was supported by National Institutes of Health grant DK-74993 (to J.W.P.).
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Bettoun, D. J., T. P. Burris, K. A. Houck, D. W. Buck II, K. R. Stayrook, B. Khalifa, J. Lu, W. W. Chin, and S. Nagpal. 2003. Retinoid X receptor is a nonsilent major contributor to vitamin D receptor-mediated transcriptional activation. Mol. Endocrinol. 17:2320-2328. [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 2.Carroll, J. S., X. S. Liu, A. S. Brodsky, W. Li, C. A. Meyer, A. J. Szary, J. Eeckhoute, W. Shao, E. V. Hestermann, T. R. Geistlinger, E. A. Fox, P. A. Silver, and M. Brown. 2005. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122:33-43. [DOI] [PubMed] [Google Scholar]
- 3.Cawley, S., S. Bekiranov, H. H. Ng, P. Kapranov, E. A. Sekinger, D. Kampa, A. Piccolboni, V. Sementchenko, J. Cheng, A. J. Williams, R. Wheeler, B. Wong, J. Drenkow, M. Yamanaka, S. Patel, S. Brubaker, H. Tammana, G. Helt, K. Struhl, and T. R. Gingeras. 2004. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell 116:499-509. [DOI] [PubMed] [Google Scholar]
- 4.Cheng, J., P. Kapranov, J. Drenkow, S. Dike, S. Brubaker, S. Patel, J. Long, D. Stern, H. Tammana, G. Helt, V. Sementchenko, A. Piccolboni, S. Bekiranov, D. K. Bailey, M. Ganesh, S. Ghosh, I. Bell, D. S. Gerhard, and T. R. Gingeras. 2005. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science 308:1149-1154. [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 5.Cosgrove, M. S., J. D. Boeke, and C. Wolberger. 2004. Regulated nucleosome mobility and the histone code. Nat. Struct. Mol. Biol. 11:1037-1043. [DOI] [PubMed] [Google Scholar]
- 6.de Bruin, D., Z. Zaman, R. A. Liberatore, and M. Ptashne. 2001. Telomere looping permits gene activation by a downstream UAS in yeast. Nature 409:109-113. [DOI] [PubMed] [Google Scholar]
- 7.Dekker, J., K. Rippe, M. Dekker, and N. Kleckner. 2002. Capturing chromosome conformation. Science 295:1306-1311. [DOI] [PubMed] [Google Scholar]
- 8.Dhawan, P., X. Peng, A. L. M. Sutton, P. N. MacDonald, C. M. Croniger, C. Trautwein, M. Centrella, T. L. McCarthy, and S. Christakos. 2005. Functional cooperation between CCAAT/enhancer-binding proteins and the vitamin D receptor in regulation of 25-hydroxyvitamin D3 24-hydroxylase. Mol. Cell. Biol. 25:472-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drissen, R., R. J. Palstra, N. Gillemans, E. Splinter, F. Grosveld, S. Philipsen, and W. de Laat. 2004. The active spatial organization of the beta-globin locus requires the transcription factor EKLF. Genes Dev. 18:2485-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.ENCODE Project Consortium. 2004. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science 306:636-640. [DOI] [PubMed] [Google Scholar]
- 11.Fan, X., E. M. Roy, T. C. Murphy, M. S. Nanes, S. Kim, J. W. Pike, and J. Rubin. 2004. Regulation of RANKL promoter activity is associated with histone remodeling in murine bone stromal cells. J. Cell Biochem. 93:807-818. [DOI] [PubMed] [Google Scholar]
- 12.Fu, Q., R. L. Jilka, S. C. Manolagas, and C. A. O'Brien. 2002. Parathyroid hormone stimulates receptor activator of NFkappa B ligand and inhibits osteoprotegerin expression via protein kinase A activation of cAMP-response element-binding protein. J. Biol. Chem. 277:48868-48875. [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 13.Fu, Q., S. C. Manolagas, and C. A. O'Brien. Parathyroid hormone controls receptor activator of NF-κB ligand gene expression via a distant transcriptional enhancer. Mol. Cell. Biol. 26:6453-6468. [DOI] [PMC free article] [PubMed]
- 14.Glynn, E. F., P. C. Megee, H. G. Yu, C. Mistrot, E. Unal, D. E. Koshland, J. L. Derisi, and J. L. Gerton. 2004. Genome-wide mapping of the cohesion complex in the yeast Saccharomyces cerevisiae. PLoS Biol. 2:E259. [Online.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gribnau, J., K. Diderich, S. Pruzina, R. Calzolari, and P. Fraser. 2000. Intergenic transcription and developmental remodeling of chromatin subdomains in the human beta-globin locus. Mol. Cell 5:377-386. [DOI] [PubMed] [Google Scholar]
- 16.Groyer, A., G. Schweizer-Groyer, F. Cadepond, M. Mariller, and E. E. Baulieu. 1987. Antiglucocorticosteroid effects suggest why steroid hormone is required for receptors to bind DNA in vivo but not in vitro. Nature 328:624-626. [DOI] [PubMed] [Google Scholar]
- 17.Harris, W. H., and R. P. Heaney. 1969. Skeletal renewal and metabolic bone disease. N. Engl. J. Med. 280:193-202. [DOI] [PubMed] [Google Scholar]
- 18.Hofbauer, L. C., F. Gori, L. Riggs, D. L. Lacey, C. R. Dunstan, T. C. Spelsberg, and S. Khosla. 1999. Stimulation of osteoprotegerin ligand and inhibition of osteoprotegerin production by glucocorticoids in human osteoblastic lineage cells: potential paracrine mechanisms of glucocorticoid-induced osteoporosis. Endocrinology 140:4382-4389. [DOI] [PubMed] [Google Scholar]
- 19.Hofbauer, L. C., S. Khosla, C. R. Dunstan, D. L. Lacey, W. J. Boyle, and B. L. Riggs. 2000. The roles of osteoprotegerin and osteoprotegerin ligand in the paracrine regulation of bone resorption. J. Bone Miner. Res. 15:2-12. [DOI] [PubMed] [Google Scholar]
- 20.Hsu, H., D. L. Lacey, C. R. Dunstan, I. Solovyev, A. Colombero, E. Timms, H. L. Tan, G. Elliott, M. J. Kelley, I. Sarosi, L. Wang, X. Z. Xia, R. Elliott, L. Chiu, T. Black, S. Scully, C. Capparelli, S. Morony, G. Shimamoto, M. B. Bass, and W. J. Boyle. 1999. Tumor necrosis factor receptor family member RANK mediates osteoclast differentiation and activation induced by osteoprotegerin ligand. Proc. Natl. Acad. Sci. USA 96:3540-3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Impey, S., S. R. McCorkle, H. Cha-Molstad, J. M. Dwyer, G. S. Yochum, J. M. Boss, S. McWeeney, J. J. Dunn, G. Mandel, and R. H. Goodman. 2004. Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell 119:1041-1054. [DOI] [PubMed] [Google Scholar]
- 22.Jimi, E., S. Akiyama, T. Tsurukai, N. Okahashi, K. Kobayashi, N. Udagawa, T. Nishihara, N. Takahashi, and T. Suda. 1999. Osteoclast differentiation factor acts as a multifunctional regulator in murine osteoclast differentiation and function. J. Immunol. 163:434-442. [PubMed] [Google Scholar]
- 23.Jimi, E., I. Nakamura, L. T. Duong, T. Ikebe, N. Takahashi, G. A. Rodan, and T. Suda. 1999. Interleukin 1 induces multinucleation and bone-resorbing activity of osteoclasts in the absence of osteoblasts/stromal cells. Exp. Cell Res. 247:84-93. [DOI] [PubMed] [Google Scholar]
- 24.Kabe, Y., J. Yamada, H. Uga, Y. Yamaguchi, T. Wada, and H. Handa. 2005. NF-Y is essential for the recruitment of RNA polymerase II and inducible transcription of several CCAAT box-containing genes. Mol. Cell. Biol. 25:512-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaji, H., T. Sugimoto, M. Kanatani, M. Fukase, M. Kumegawa, and K. Chihara. 1996. Prostaglandin E2 stimulates osteoclast-like cell formation and bone resorbing activity via osteoblasts: role of cAMP-dependent protein kinase. J. Bone Miner. Res. 11:62-71. [DOI] [PubMed] [Google Scholar]
- 26.Kamp, D., J. Cheng, P. Kapranov, M. Yamanaka, S. Brubaker, S. Cawley, J. Drenkow, A. Piccolboni, S. Bekiranov, G. Helt, H. Tammana, and T. R. Gingeras. 2004. Novel RNAs identified from an in-depth analysis of the transcriptome of human chromosomes 21 and 22. Genome Res. 14:331-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kapranov, P., S. E. Cawley, J. Drenkow, S. Bekiranov, R. L. Strausberg, S. P. Fodor, and T. R. Gingeras. 2002. Large-scale transcriptional activity in chromosomes 21 and 22. Science 296:916-919. [DOI] [PubMed] [Google Scholar]
- 28.Karst, M., G. Gorny, R. J. Galvin, and M. J. Oursler. 2004. Roles of stromal cell RANKL, OPG, and M-CSF expression in biphasic TGF-beta regulation of osteoclast differentiation. J. Cell Physiol. 200:99-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kato, S., K. Sekine, T. Matsumoto, and T. Yoshizawa. 1998. Molecular genetics of vitamin D receptor acting in bone. J. Bone Miner. Metab. 16:65-71. [Google Scholar]
- 30.Kim, S., N. K. Shevde, and J. W. Pike. 2005. 1,25-Dihydroxyvitamin D3 stimulates cyclic vitamin D receptor/retinoid X receptor DNA-binding, co-activator recruitment, and histone acetylation in intact osteoblasts. J. Bone Miner. Res. 20:305-317. [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 31.Kim, T. H., L. O. Barrera, M. Zheng, C. Qu, M. A. Singer, T. A. Richmond, Y. Wu, R. D. Green, and B. Ren. 2005. A high resolution map of active promoters in the human genome. Nature 436:876-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim, T. H., L. O. Barrera, C. Qu, S. Van Calcar, N. D. Trinklein, S. J. Cooper, R. M. Luna, C. K. Glass, M. G. Rosenfeld, R. M. Myers, and B. Ren. 2005. Direct isolation and identification of promoters in the human genome. Genome Res. 15:830-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kirmizis, A., S. M. Bartley, A. Kuzmichev, R. Margueron, D. Reinberg, R. Green, and P. J. Farnham. 2004. Silencing of human polycomb target genes is associated with methylation of histone H3 lys 27. Genes Dev. 18:1592-1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kitazawa, S., R. Kitazawa, and S. Maeda. 1999. Promoter structure of mouse RANKL/TRANCE/ODF gene. Biochim. Biophys. Acta 1445:134-141. [DOI] [PubMed] [Google Scholar]
- 35.Koga, T., M. Inui, K. Inoue, S. Kim, A. Suematsu, E. Kobayashi, T. Iwata, H. Ohnishi, T. Matozaki, T. Kodama, T. Taniguchi, H. Takayanagi, and T. Takai. 2004. Costimulatory signals mediated by the ITAM motif cooperate with RANKL for bone homeostasis. Nature 428:758-763. [DOI] [PubMed] [Google Scholar]
- 36.Kondo, H., J. Guo, and F. R. Bringhurst. 2002. Cyclic adenosine monophosphate/protein kinase A mediates parathyroid hormone/parathyroid hormone-related protein receptor regulation of osteoclastogenesis and expression of RANKL and osteoprotegerin mRNAs by marrow stromal cells. J. Bone Miner. Res. 17:1667-1679. [DOI] [PubMed] [Google Scholar]
- 37.Kong, Y. Y., H. Yoshida, I. Sarosi, H. L. Tan, E. Timms, C. Capparelli, S. Morony, A. J. Oliveira-dos-Santos, G. Van, A. Itie, W. Khoo, A. Wakeham, C. R. Dunstan, D. L. Lacey, T. W. Mak, W. J. Boyle, and J. M. Penninger. 1999. OPGL is a key regulator of osteoclastogenesis, lymphocyte development and lymph-node organogenesis. Nature 397:315-323. [DOI] [PubMed] [Google Scholar]
- 38.Lacey, D. L., E. Timm, H. L. Tan, M. J. Kelley, C. R. Dunstan, T. Burgess, R. Elliott, A. Colombero, G. Elliott, S. Scully, H. Hsu, J. Sullivan, N. Hawkins, E. Davy, C. Capparelli, A. Eli, Y. X. Qian, S. Kaufman, I. Sarosi, V. Shalhoub, G. Senaldi, J. Guo, J. Delaney, and W. J. Boyle. 1998. Osteoprotegerin ligand is a cytokine that regulates osteoclast differentiation and activation. Cell 93:165-176. [DOI] [PubMed] [Google Scholar]
- 39.Laganiere, J., G. Deblois, C. Lefebvre, A. R. Bataille, F. Robert, and V. Giguere. 2005. Location analysis of estrogen receptor alpha target promoters reveals that FOXA1 defines a domain of the estrogen response. Proc. Natl. Acad. Sci. USA 102:11651-11656. [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee, S. K., and J. A. Lorenzo. 1999. Parathyroid hormone stimulates TRANCE and inhibits osteoprotegerin messenger ribonucleic acid expression in murine bone marrow cultures: correlation with osteoclast-like cell formation. Endocrinology 140:3552-3561. [DOI] [PubMed] [Google Scholar]
- 41.Liu, B. Y., J. Guo, B. Lanske, P. Divieti, H. M. Kronenberg, and F. R. Bringhurst. 1998. Conditionally immortalized murine bone marrow stromal cells mediate parathyroid hormone-dependent osteoclastogenesis in vitro. Endocrinology 139:1952-1964. [DOI] [PubMed] [Google Scholar]
- 42.Manolagas, S. C. 2000. Birth and death of bone cells: basic regulatory mechanisms and implications for the pathogenesis and treatment of osteoporosis. Endocr. Rev. 21:115-137. [DOI] [PubMed] [Google Scholar]
- 43.Manolagas, S. C., R. S. Weinstein, R. L. Jilka, and A. M. Parfitt. 1998. Parathyroid hormone and corticosteroid-induced osteoporosis. Lancet 352:1940. [DOI] [PubMed] [Google Scholar]
- 44.Matsuo, K., D. L. Galson, C. Zhao, L. Peng, C. Laplace, K. Z. Wang, M. A. Bachler, H. Amano, H. Aburatani, H. Ishikawa, and E. F. Wagner. 2004. Nuclear factor of activated T-cells (NFAT) rescues osteoclastogenesis in precursors lacking c-Fos. J. Biol. Chem. 279:26475-26480. [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 45.Mundy, G. R. 2000. Pathogenesis of osteoporosis and challenges for drug delivery. Adv. Drug Delivery Rev. 42:165-173. [DOI] [PubMed] [Google Scholar]
- 46.Nagpal, S., S. Na, and R. Rathnachalam. 2005. Noncalcemic actions of vitamin D receptor ligands. Endocr. Rev. 26:662-687. (First published 29 March 2005; doi: 10.1210/er.2004-0002.) [DOI] [PubMed] [Google Scholar]
- 47.Oberley, M. J., J. Tsao, P. Yau, and P. J. Farnham. 2004. High throughput screening of chromatin immunoprecipitates using CpG-island microarrays. Methods Enzymol. 376:315-334. [DOI] [PubMed] [Google Scholar]
- 48.O'Brien, C. A., B. Kern, I. Gubrij, G. Karsenty, and S. C. Manolagas. 2002. Cbfa1 does not regulate RANKL gene activity in stromal/osteoblastic cells. Bone 30:453-462. [DOI] [PubMed] [Google Scholar]
- 49.O'Sullivan, J. M., S. M. Tan-Wong, A. Morillon, B. Lee, J. Coles, J. Mellor, and N. J. Proudfoot. 2004. Gene loops juxtapose promoters and terminators in yeast. Nat. Genet. 36:1014-1018. [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 50.Pike, J. W., and N. K. Shevde. 2005. The vitamin D receptor, p. 167-191. In D. Feldman, J. W. Pike, and F. H. Glorieux (ed.), Vitamin D, 2nd ed. Elsevier, San Diego, Calif.
- 51.Romas, E., N. Udagawa, H. Zhou, T. Tamura, M. Saito, T. Taga, D. J. Hilton, T. Suda, K. W. Ng, and T. J. Martin. 1996. The role of gp130-mediated signals in osteoclast development: regulation of interleukin 11 production by osteoblasts and distribution of its receptor in bone marrow cultures. J. Exp. Med. 183:2581-2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schubeler, D., D. M. MacAlpine, D. Scalzo, C. Wirbelauer, C. Kooperberg, F. van Leeuwen, D. E. Gottschling, L. P. O'Neill, B. M. Turner, J. Delrow, S. P. Bell, and M. Groudine. 2004. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 18:1263-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shevde, N. K., L. A. Plum, M. Clagett-Dame, H. Yamamoto, J. W. Pike, and H. F. DeLuca. 2002. A potent analog of 1alpha,25-dihydroxyvitamin D3 selectively induces bone formation. Proc. Natl. Acad. Sci. USA 99:13487-13491. [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sivagurunathan, S., M. M. Muir, T. C. Brennan, J. P. Seale, and R. S. Mason. 2005. Influence of glucocorticoids on human osteoclast generation and activity. J. Bone Miner. Res. 20:390-398. [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 55.Smith, C. L., and B. W. O'Malley. 2004. Coregulator function: a key to understanding tissue specificity of selective receptor modulators. Endocr. Rev. 25:45-71. [DOI] [PubMed] [Google Scholar]
- 56.Spotswood, H. T., and B. M. Turner. 2002. An increasingly complex code. J. Clin. Investig. 110:577-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Suda, T., N. Takahashi, N. Udagawa, E. Jimi, M. T. Gillespie, and T. J. Martin. 1999. Modulation of osteoclast differentiation and function by the new members of the tumor necrosis factor receptor and ligand families. Endocr. Rev. 20:345-357. [DOI] [PubMed] [Google Scholar]
- 58.Sutton, A. L., and P. N. MacDonald. 2003. Vitamin D: more than a “bone-a-fide” hormone. Mol. Endocrinol. 17:777-791. [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 59.Szutorisz, H., N. Dillon, and L. Tora. 2005. The role of enhancers as centres for general transcription factor recruitment. Trends Biochem. Sci. 30:593-599. [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 60.Takahashi, N., T. Akatsu, T. Sasaki, G. C. Nicholson, J. M. Moseley, T. J. Martin, and T. Suda. 1993. Induction of calcitonin receptors by 1,25-dihydroxyvitamin D in osteoclast-like multinucleated cells formed from mouse bone marrow cells. Endocrinology 123:1504-1510. [DOI] [PubMed] [Google Scholar]
- 61.Takayanagi, H., S. Kim, T. Koga, H. Nishina, M. Isshiki, H. Yoshida, A. Saiura, M. Isobe, T. Yokochi, J. Inoue, E. F. Wagner, T. W. Mak, T. Kodama, and T. Taniguchi. 2002. Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts. Dev. Cell 3:889-901. [DOI] [PubMed] [Google Scholar]
- 62.Tamura, T., N. Udagawa, N. Takahashi, C. Miyaura, S. Tanaka, Y. Yamada, Y. Koishihara, Y. Ohsugi, K. Kumaki, and T. Taga. 1993. Soluble interleukin-6 receptor triggers osteoclast formation by interleukin 6. Proc. Natl. Acad. Sci. USA 90:11924-11928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Teitelbaum, S. L. 2000. Bone resorption by osteoclasts. Science 289:1504-1508. [DOI] [PubMed] [Google Scholar]
- 64.Teitelbaum, S. L., and F. P. Ross. 2003. Genetic regulation of osteoclast development and function. Nat. Rev. Genet. 4:638-649. [DOI] [PubMed] [Google Scholar]
- 65.Tolhuis, B., R. J. Palstra, E. Splinter, F. Grosveld, and W. de Laat. 2002. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell 10:1453-1465. [DOI] [PubMed] [Google Scholar]
- 66.Wani, M. R., K. Fuller, N. S. Kim, Y. Choi, and T. Chambers. 1999. Prostaglandin E2 cooperates with TRANCE in osteoclast induction from hemopoietic precursors: synergistic activation of differentiation, cell spreading, and fusion. Endocrinology 140:1927-1935. [DOI] [PubMed] [Google Scholar]
- 67.Weinstein, R. S., R. L. Jilka, A. M. Parfitt, and S. C. Manolagas. 1998. Inhibition of osteoblastogenesis and promotion of apoptosis of osteoblasts and osteocytes by glucocorticoids. Potential mechanisms of their deleterious effects on bone. J. Clin. Investig. 102:274-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Whyte, M. P., and S. Mumm. 2004. Heritable disorders of the RANKL/OPG/RANKL signaling pathway. Musculoskelet. Neuronal Interact. 4:254-267. [PubMed] [Google Scholar]
- 69.Yamada, K., D. T. Duong, D. K. Scott, J. C. Wang, and D. K. Granner. 1999. CCAAT/enhancer-binding protein beta is an accessory factor for the glucocorticoid response from the cAMP response element in the rat phosphoenolpyruvate carboxykinase gene promoter. J. Biol. Chem. 274:5880-5888. [DOI] [PubMed] [Google Scholar]
- 70.Yamamoto, H., N. K. Shevde, A. Warrier, L. A. Plum, H. F. DeLuca, and J. W. Pike. 2003. 2-Methylene-19-nor-(20S)-1,25-dihydroxyvitamin D3 potently stimulates gene-specific DNA binding of the vitamin D receptor in osteoblasts. J. Biol. Chem. 278:31756-31765. [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 71.Zella, L. A., S. Kim, N. K. Shevde, and J. W. Pike. 2006. Enhancers located within two introns of the vitamin D receptor gene mediate transcriptional autoregulation by 1,25-dihydroxyvitamin D3. Mol. Endocrinol. 20:1231-1247. (First published 23 February 2006; 10.1210/me. 2000-0015.) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.