Abstract
In roughly 5% of cases of acute lymphoblastic leukemia, a chromosomal translocation leads to expression of the oncogenic protein E2A-PBX1. The N-terminal portion of E2A-PBX1, encoded by the E2A gene, is identical in sequence to the corresponding portion of the E proteins E12/E47 and includes transcriptional activation domains. The C terminus consists of most of the HOX interacting transcription factor PBX1, including its DNA-binding homeodomain. Structure-function correlative experiments have suggested that oncogenesis by E2A-PBX1 requires an activation domain, called AD1, at the extreme N terminus. We recently demonstrated that a potentially helical portion of AD1 interacts directly with the transcriptional coactivator protein cyclic AMP response element-binding protein (CBP) and that this interaction is essential in the immortalization of primary bone marrow cells in tissue culture. Here we show that a conserved LXXLL motif within AD1 is required in the interaction between E2A-PBX1 and the KIX domain of CBP. We show by circular dichroism spectroscopy that the LXXLL-containing portion of AD1 undergoes a helical transition upon interacting with the KIX domain and that amino acid substitutions that prevent helix formation prevent both the KIX interaction and cell immortalization by E2A-PBX1. Perhaps most strikingly, substitution of a single, conserved leucine residue (L20) within the LXXLL motif impairs leukemia induction in mice after transplantation with E2A-PBX1-expressing bone marrow. The KIX domain of CBP mediates well-characterized interactions with several transcription factors of relevance to leukemia induction. Circumstantial evidence suggests that the side chain of L20 might interact with a deep hydrophobic pocket in the KIX domain. Therefore, our results serve to identify a potential new drug target.
The neoplastic cells in acute lymphoblastic leukemia (ALL) frequently contain recurrent somatic chromosomal abnormalities that contribute to their abnormal accumulation and function. A reciprocal translocation between chromosome bands 1q23 and 19p13.3 is detectable by conventional cytogenetic techniques in roughly 5% of cases of ALL (10). Of that 5% of cases, in 90 to 95% the presence of t(1;19) leads to the fusion of portions of the E2A gene on chromosome 19 with portions of the PBX1 gene on chromosome 1 and to expression of chimeric transcription factors called E2A-PBX1a and E2A-PBX1b (these isoforms are generated by alternative splicing of the PBX1 portion of the pre-mRNA) (20, 30). Beyond the consistent association with leukemia, the idea of the oncogenic potential of E2A-PBX1 is supported by abundant experimental evidence (reviewed in reference 24). Therefore, it seems likely that elucidating the molecular mechanisms by which E2A-PBX1 contributes to abnormal cellular behavior may uncover novel avenues in the development of better therapies for cases of ALL.
E2A-PBX1 incorporates a PBX1-derived homeodomain near its C terminus and, like wild-type PBX1 proteins, can bind to PBX1 recognition sequences in cooperation with members of the HOX family of transcriptional regulators (11, 26, 31). The wild-type E2A gene products, called E12 and E47, are basic helix-loop-helix transcription factors involved in cell type specification and differentiation (reviewed in reference 35). Mice targeted at the E2A locus manifest a complete lack of B lymphocytes beyond the pro-B stage, indicating a particular role for E12/E47 in B-cell ontogeny (4, 41). Both E2A-PBX1 and wild-type E12/E47 function as transcriptional activators (25, 27, 39). The portion of E2A-PBX1 encoded by E2A, comprising roughly the N-terminal two-thirds of the oncoprotein, is invariant between E12 and E47 and contains two transcriptional activation domains called AD1 and AD2 (3, 18, 34).
Structure-function correlative experiments have implicated AD1, in particular, in oncogenesis by E2A-PBX1 (22, 29). This supports the notion that E2A-PBX1 may contribute to leukemia development through the transcriptional activation of key target genes. Activation domains of transcription factors function essentially as protein-protein interaction modules to recruit transcriptional coregulator complexes to the vicinity of the promoters whose genes they regulate. It follows, therefore, that the identification of proteins or protein complexes that interact with AD1 of E2A could shed light on the mechanism by which E2A-PBX1 induces leukemia.
Cyclic AMP response element binding (CREB) protein (CBP) and its close paralog p300 are large, nuclear proteins that function as transcriptional coregulators. They interact with many transcriptional activators, thereby contributing to target gene induction (reviewed in reference 15). We recently mapped the elements required for the interaction in both CBP and E2A-PBX1 and began to address functional correlates of the interaction (5). In CBP, interaction with E2A-PBX1 requires the KIX domain (also called the CREB-binding domain), a well-characterized, globular domain that mediates the prototypic, phosphorylation-dependent interaction with the transcription factor CREB (36). In E2A-PBX1, the interaction with CBP depends largely on a conserved, potentially helical region near the extreme N terminus of the protein, within AD1 of E2A. This portion of the protein includes an LXXLL motif, examples of which are involved in a number of interactions between transcriptional regulators (32). Disrupting the CBP/p300 interaction by deleting 18 amino acids from within the AD1 domain (i.e., residues 11 to 28) markedly impaired the ability of E2A-PBX1b to immortalize a proliferative population of myeloid progenitors in tissue culture from primary, retrovirally transduced bone marrow, suggesting that this region of AD1, and by implication the interaction with CBP/p300, may contribute to E2A-PBX1 oncogenesis.
In the current study we have analyzed the requirements for, and oncogenic consequences of, the interaction between the LXXLL motif of E2A-PBX1 and the KIX domain of CBP. Using circular dichroism (CD) spectroscopy, we demonstrate that this portion of E2A is indeed stabilized in an α-helical conformation on binding to the KIX domain. We demonstrate that helix formation by this domain is required for KIX binding in vitro and, in the context of full-length E2A-PBX1b, for immortalization of hematopoietic cells in tissue culture. We show that a single, critical leucine residue within the LXXLL motif of AD1 is also required in effective KIX binding. Perhaps most strikingly, this same residue is required, in the context of full-length E2A-PBX1b, in the induction of a lethal myeloproliferative disease upon reconstitution of irradiated mice with retrovirally transduced bone marrow cells. The identification of single amino acid side chains that make critical contributions to E2A-PBX1 oncogenesis has important mechanistic implications for understanding E2A-PBX1 function and, potentially, drug development.
MATERIALS AND METHODS
CD spectroscopy.
In order to prepare recombinant KIX protein for CD, GST-CBP 586-673 was bound to glutathione-Sepharose beads and the KIX domain was released by overnight digestion with thrombin (267 units of thrombin in 4 ml of binding buffer) at room temperature. The KIX domain was further purified by size exclusion chromatography using a 16/60 column packed with Superdex 75 matrix (Amersham), concentrated and buffer exchanged into CD buffer (10 mM Na2PO4, 100 mM NaF, pH 7.0) using a Centricon filter unit with a molecular weight cutoff of 5,000 (Millipore, Billerica, MA), and quantified by optical density determination. A 23-residue peptide comprising amino acids 9 to 31 of E2A (Ac-PVGTDKELSDLLDFSMMFPLPVT-NH2) and a S17P, L20P mutant peptide were chemically synthesized and purified at the Protein Function Discovery Facility (Kingston, Canada) and the Alberta Peptide Institute (Edmonton, Canada), respectively. Lyophilized peptide was dissolved in CD buffer, and CD was performed using a rapid-scanning monochrometer fitted with a CD module (RSM 1000; Olis, Inc., Bogart, GA) and a cuvette with a 0.1 cm path length. Each of the datum sets shown for KIX alone and KIX in combination with the E2A peptide represents the average results of three scans, whereas the data for the E2A peptide alone represent the results of an average of nine scans (in consideration of the relatively low signal-to-noise ratio). Molar ellipticity, [θ], was calculated according to the formula [θ] = θ × 100/(nlc), where n represents the number of amino acids in the protein, l represents the path length in centimeters, and c represents the millimolar concentration.
Pull-down experiments and Western blots.
Plasmids conferring bacterial expression of glutathione S-transferase (GST) fusion proteins were constructed by cloning the indicated portions of the human E2A or murine CBP cDNAs into the pGEX-2T expression vector, and the encoded proteins were expressed and purified, as described previously (5). Amino acid substitutions or small deletions were accomplished using a Transformer kit for site-directed mutagenesis (Clontech, Mountain View CA), according to the manufacturer's protocol, and the results were confirmed by nucleotide sequencing. A portion of the mouse CBP cDNA (kindly provided by Gerd Blobel, The Children's Hospital of Philadelphia, Philadelphia, PA) encoding amino acids 1 to 688 was cloned into Bluescript vector for in vitro translation. This restricted portion of the cDNA, which contains the KIX domain, was used in order to reduce the number of truncation products relative to what was observed on in vitro transcription/translation of the full-length CBP cDNA. In vitro transcription/translation reactions were carried out with a TnT quick-coupled transcription/translation system (Promega Corp., Madison, WI), according to the manufacturer's instructions, using 1 μg of Bluescript-based plasmid DNA and a reaction volume of 50 μl in the presence of [35S]methionine and T7 polymerase. For each pull-down reaction, 2 μg of GST fusion protein was bound to 20 μl of preswollen glutathione Sepharose beads, as previously described (5). The beads were resuspended in a binding mixture consisting of 25 μl of the in vitro transcription/translation reaction mixture or 1.0 mg of nuclear extract protein (prepared according to the Dignam method) and made up to 400 μl with HEGN buffer (20 mM HEPES-KOH [pH 7.9], 0.1 mM EDTA, 10% glycerol, and 0.1% NP-40) containing 100 mM KCl. This binding reaction mixture was incubated overnight at 4°C with gentle rocking. The beads were pelleted and washed four times in HEGN buffer containing 0.2 M NaCl and resuspended in sodium dodecyl sulfate-containing electrophoresis sample buffer (New England Biolabs Ltd., Mississauga, Ontario) in preparation for sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Autoradiography and quantification of the radioactive signals in the dried polyacrylamide gels were carried out using a PhosphorImager (Storm 820; Amersham Biosciences, Baie d'Urfe, Canada) and proprietary (ImageQuant) software. In determining relative signal intensities in the various lanes of the gels, the value related to unfused GST was subtracted as “background” from the other results. This was justified in separate experiments, which determined that the signal observed in the GST lane was accounted for entirely by nonspecific binding to the Sepharose beads (data not shown). Values obtained in this manner were normalized according to the quantity of GST fusion protein in the corresponding lane of the gel, as determined by photographing the transilluminated, Coomassie-stained gel followed by digital densitometry using Image-Pro Plus 5.0 software (Media Cybernetics, Silver Spring, MD). Western blot analyses were carried out as previously described using monoclonal antibodies to CBP (clone C-1) and E2A (yae), both purchased from Santa Cruz Biotechnology, Santa Cruz, CA (5).
Retroviral transduction and bone marrow transplantation assays.
Bone marrow was harvested from the femurs and tibias of nine female BALB/c mice, 10 to 12 weeks of age, and transduced with retroviral vectors constructed by cloning the wild-type E2A-PBX1b cDNA and engineered variants into the MSCVneo backbone plasmid. Transduction efficiency was monitored by plating 104 bone marrow cells in semisolid medium containing methylcellulose and the selective drug G418; drug-resistant colonies were counted 6 days later. The details of these procedures have been described previously (5). In the hematopoietic reconstitution experiment, female BALB/c mice of the same age as the donors were exposed to 8.4 Gy of ionizing radiation from a Cesium-137 source, delivered in two equal doses spaced 4 h apart. The next day (i.e., the day of the initial plating in methylcellulose-containing medium), each animal (of six animals each for E2A-PBX1b and the L20A variant) was administered 3.3 × 105 bone marrow cells by tail vein injection. Mice were maintained on acidified water and sterile feed and in isolation cages for 3 weeks following irradiation. Mice were euthanized in a CO2 chamber when they manifested signs of illness or distress, such as reduced mobility, poor grooming, hunched posture, rapid breathing, 20% weigh loss, or hind leg weakness or paralysis.
Mammalian two-hybrid assay.
Plasmids conferring mammalian expression of GAL4 or VP16 fusion proteins were assembled by excising the various cDNAs from pGEX-2T and, using BamHI and EcoRI restriction sites, ligating them into the pCVM1 mammalian expression plasmid so as to fuse them, in frame, with a portion of cDNA encoding the N-terminal 147 amino acids of GAL4 (i.e., the DNA-binding domain) or two VP16 activation domains arranged in tandem. The VP16 insert was generated by PCR from the plasmid “RSV-2X-VP16-Myb”, kindly provided by Paul Brindle (St. Jude's Children's Hospital, TN). The reporter plasmid p5X GAL4 LUC was a generous gift from Cornelius Murre (University of California San Diego, CA). 293T cells were plated into each well of a 12-well plate in Dulbecco's minimal essential medium containing 10% fetal bovine serum. The next day, the cells were transfected with mammalian expression and reporter plasmids (1.2 μg pCMV-GAL4 fusion plasmid, 0.2 μg pCMV-2xVP16 fusion plasmid, 0.7 μg p5xGAL4 LUC, and 0.1 μg PRL-CVM, a renilla luciferase-expressing plasmid kindly provided by Chris Mueller, Queen's University, Ontario, Canada) by the calcium phosphate precipitation method (25). The cells were lysed 40 h later in passive lysis buffer (Promega), and a portion of the lysate was analyzed using dual-luciferase assay reagents (Promega) and an LB96V MicroLumat Plus luminometer (EG & G Berthold Ltd., Bundoora, Australia). Transfections were carried out and the results analyzed in triplicate. Each luciferase value was normalized using the corresponding renilla result as a standard. The mean value from each triplicate experiment was considered one datum point. The results shown in the figure are from at least three experiments.
RESULTS
LXXLL motif requirements in myeloid cell immortalization by E2A-PBX1b.
We demonstrated recently that deletion of amino acids 11 to 28 (GTDKELSDLLDFSMMFPL), which includes the LXXLL motif (underlined), abrogates the ability of E2A-PBX1b to block myeloid differentiation and stimulate proliferation in cultured, granulocyte-macrophage colony-stimulating factor (GM-CSF)-stimulated primary hematopoietic progenitors (5). In order to elucidate contributions made by individual amino acid side chains in mediating this effect, altered codons relating to the LXXLL motif were engineered into a full-length E2A-PBX1b cDNA inserted in the MSCVneo retroviral backbone. Expression of recombinant proteins in retrovirally transduced NIH 3T3 fibroblasts was confirmed by immunoblotting with an anti-E2A monoclonal antibody (Fig. 1A). Primary bone marrow cells were infected with the retroviruses conferring expression of E2A-PBX1b or with engineered variants. The efficacy of gene transfer was determined by enumerating drug-resistant colonies after 6 days of growth in methylcellulose-containing medium. The average numbers of drug-resistant colonies (per 104 cells plated) for each retroviral construct were as follows: for MSCVneo, 201; for E2A-PBX1b, 35; for Δ 397-405, 216; for Δ 397-405/L20A, 75; for L20A, 28; for S17A, 64; for S17P, 58. Equal numbers of cells from each of these populations were propagated in liquid medium, without drug selection, in the presence of the cytokine GM-CSF.
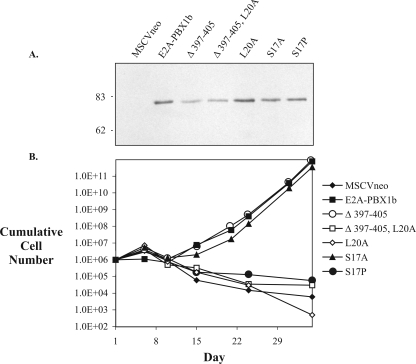
FIG. 1.
The LXXLL motif of E2A-PBX1b is required for immortalization of proliferative hematopoietic progenitors. (A) Retrovirus-mediated expression of recombinant proteins. NIH 3T3 fibroblasts were infected with retroviruses expressing E2A-PBX1b, or engineered variants, and then drug selected for retroviral integration prior to lysis and immunoblotting with an anti-E2A monoclonal antibody. (B) Proliferation of retrovirally transduced hematopoietic progenitors in GM-CSF. For each construct, 106 bone marrow cells were inoculated immediately after retroviral infection, without drug selection.
Under these conditions, uninfected cells or cells infected with the unmodified retroviral vector proliferate briefly while differentiating into short-lived granulocytes or adherent macrophages. Accordingly, control cells infected with the vector alone had declined in number by day 15; this downward trend continued throughout the remainder of the experiment (Fig. 1B). However, in the cells transduced with E2A-PBX1b, the cell numbers had increased by day 15 and continued to increase exponentially thereafter. These cells have been characterized previously as immature myeloid progenitors (5, 21, 22). They may be propagated for months in culture and appear immortal. Deletion of a conserved, predicted helical domain within AD2 (Δ 397-405) had no evident effect on the ability of E2A-PBX1b to induce the outgrowth of these proliferative cells, whereas replacement of a single, conserved leucine residue at position 20 (i.e., the last leucine in the LXXLL domain) by an alanine (L20A) completely abrogated this proliferative effect (Fig. 1B). Therefore, interactions mediated by the LXXLL motif and, more specifically, a bulky hydrophobic side chain at position 20 appear to be required in mediating the proliferative and cell-immortalizing effect of E2A-PBX1b in this setting.
Interaction with the KIX domain of CBP stabilizes a helical conformation of the LXXLL-containing portion of E2A.
In 1996, Massari and colleagues demonstrated that a synthetic peptide representing amino acids 9 to 31 of E12/E47, encompassing the LXXLL motif in AD1, was unstructured in aqueous solution but assumed a helical conformation in the presence of the organic solvent 2,2,2-trifluoroethanol (TFE) (28). Based on their findings, the authors predicted that this portion of AD1 would be stabilized in a helical conformation upon interacting with an unidentified cognate ligand. Having recently demonstrated that this same portion of E12/E47 interacts directly with the KIX domain of CBP/p300, we used CD spectroscopy to investigate the possibility that interaction with the KIX domain might promote a helical transition in AD1. The KIX domain of murine CBP (i.e., amino acids 586 to 673, including the KIX domain and seven additional amino acids at the C-terminal end) was expressed in Escherichia coli and affinity purified from a bacterial lysate; a peptide comprised of amino acids 9 to 31 of E2A (i.e., the same peptide used by Massari and colleagues) was synthesized and then purified by high-performance liquid chromatography (28).
Evaluation by CD spectroscopy of a solution containing the E2A peptide alone showed negative absorbance with a minimum at 199 nm and an absence of features characteristic of helical structure, consistent with previous results (Fig. 2A) (28). Like others, we observed abundant helical content with the same peptide in the presence of 40% TFE (data not shown). Evaluation of KIX protein alone in aqueous solution (Fig. 2A) produced a spectrum indicative of significant helical content, with pronounced negative absorption and minima at 208 and 222 nm, as shown previously by others (9, 36). To investigate a possible helical transition by the E2A peptide, CD spectroscopy was carried out on a solution containing both KIX and E2A proteins, each present at a concentration of 30 μM. The CD spectrum observed for the protein mixture (Fig. 2A) indicated greater helical content than the spectrum that was calculated based on the spectra produced by the individual proteins and assuming no interaction (Fig. 2A). In contrast, a mixture of a mutant AD1 peptide containing helix-disrupting proline substitutions (S17P, L20P) and the KIX domain gave rise to a CD spectrum equivalent to the spectrum calculated from the sum of the individual protein spectra (Fig. 2B). Furthermore, the mutant peptide failed to adopt a helical conformation in TFE (data not shown). These CD data support the notion that interaction with the KIX domain promotes and requires helical transition of the E2A peptide. Furthermore, the results corroborate our previous pull-down and coimmunoprecipitation findings in indicating a direct interaction between the potentially helical domain of AD1 and the KIX domain of CBP (5).
FIG. 2.
An LXXLL-containing E2A peptide assumes a helical conformation upon interacting with the KIX domain of CBP. (A) CD spectroscopy was carried out using a synthetic peptide consisting of amino acids 9 to 31 of E2A and bacterially expressed recombinant protein representing the KIX domain of CBP. Each protein was present at 30 μM. Dotted black line, E2A peptide; dashed gray line, KIX domain; solid black line, measured results for E2A plus KIX; solid gray line, calculated results for E2A plus KIX; [θ], molar ellipticity. (B) Repetition of the CD experiment using a mutant E2A 9-31 peptide, S17P:L20P, bearing two helix-disrupting proline substitutions.
In order to investigate whether adoption of a helical conformation and, by implication, interaction with a helix-stabilizing ligand is required in cell immortalization by E2A-PBX1b, we replaced the serine residue at position 17 of full-length E2A-PBX1b with a helix-disrupting proline residue. The proline mutant (S17P) inactivated E2A-PBX1b in the immortalization assay (Fig. 1B). Substituting an alanine at the same location (S17A) had no evident effect, indicating that it was introduction of the proline, rather than removal of the serine, that resulted in the functional defect. Therefore, these results support the idea of a requirement for a helical LXXLL motif in mediating protein-protein interactions involved in the immortalization of primary myeloid cells by E2A-PBX1b.
LXXLL motif requirements in the KIX interaction.
In order to elucidate in greater detail the requirements for KIX binding, we established a pull-down assay in which bacterially expressed GST-E2A fusion proteins were evaluated for their ability to bind to a 35S-labeled, in vitro-translated portion of CBP (amino acids 1 to 688) that included the KIX domain. Amino acid substitutions were inserted in the context of E2A-encoded amino acids 1 to 483, the portion present in E2A-PBX1, in order to evaluate their effects on KIX binding.
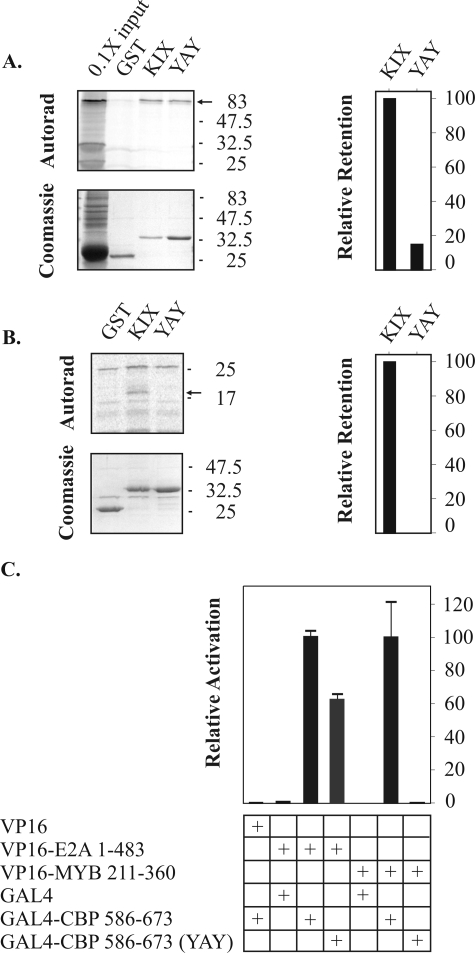
Deletion of a conserved, predicted helical domain within AD2 (Δ 397-405) resulted in a modest reduction of KIX binding (71% binding relative to wild-type E2A 1-483, Fig. 3A). Subsequent mutations were carried out in the context of the 397-405 deletion in order to emphasize the effects of alterations within the AD1 helical domain. Deletion of the LXXLL-containing region (Δ 16-23) largely abrogated binding (3% of wild type, 4.8% of Δ 397-405). Substitution of an alanine residue for either of the conserved leucines at positions 16 or 20 profoundly impaired binding (7.5% and 15%, respectively, of that of Δ 397-405), suggesting a requirement for a bulky, hydrophobic side chain at these positions. Interestingly, while binding was reconstituted by a conservative isoleucine substitution at position 20, an isoleucine substitution at position 16 had little effect on the KIX interaction, suggesting that the leucine residue at position 16, but not that at position 20, must satisfy additional structural requirements beyond that of supplying a bulky aliphatic side chain.
FIG. 3.
The LXXLL motif of E2A-PBX1b is required for interaction with CBP. (A and B) Pull-down experiments using bacterially expressed GST-E2A 1-483, or variants, and in vitro-translated, 35S-labeled CBP 1-688. The images show PhosphorImager-generated autoradiographs; the asterisks and arrowheads to the left indicate the bands related to the CBP fragment and the GST-E2A fusion proteins, respectively. The relative values shown in the graphs represent the signal from the uppermost bands in the autoradiographs normalized according to the corresponding, Coomassie-stained GST-E2A protein. (C) Pull-down experiments using GST-E2A proteins and nuclear extract from leukemia-derived RCH ACV cells. Proteins retained by GST-E2A were immunoblotted with anti-CBP. (D) Mammalian two-hybrid assay. CBP 586-673 (i.e., the KIX domain) fused to a GAL4 DNA binding domain can recruit E2A to GAL4 binding sites on a luciferase reporter plasmid, but this activity is impaired by alterations in E2A that affect KIX binding.
We next investigated the requirement for helicity in the KIX interaction by substituting proline residues for the serine and leucine at positions 17 and 19, respectively. Although the proline substitutions profoundly impaired KIX binding, a similar effect was observed when (helix-permissive) alanines were substituted, indicating specific requirements for the side chains of either S17 or L19. Since L19 is conserved in the LXXLL motif, its side chain is likely to participate in important interactions. Therefore, we evaluated the effect on KIX binding of substitutions involving the serine residue at position 17 in isolation. Substitution of proline was associated with a substantial, deleterious effect on binding (34% relative to that of wild-type E2A 1-483; Fig. 3B) comparable to that associated with the L20A substitution. Substitution of an alanine residue at this position also resulted in reduced binding, although the reduction was of lesser magnitude (83% relative to the wild type). The disproportionate, deleterious effect of the proline, relative to the alanine, substitution on KIX binding supports a requirement for helicity in the AD1 domain.
In order to correlate these results produced using an in vitro-translated fragment of CBP with those obtained using endogenously expressed, full-length CBP, the pull-down experiments were repeated using nuclear extract prepared from the t(1;19)-associated, ALL-derived cell line RCH ACV. Proteins retained by GST-E2A 1-483, or mutants, were analyzed by immunoblotting with an anti-CBP antibody. The results are consistent with those obtained using the smaller, in vitro-translated portion of CBP (Fig. 3C). We used a mammalian two-hybrid assay to determine whether the effects on KIX binding of the various amino acid substitutions or deletions in AD1 of E2A observed in vitro are reproduced in the nuclei of intact, living cells. Cotransfection of 293T cells with E2A 1-483 fused to two tandem copies of the VP16 activation domain and the KIX domain of CBP fused to the GAL4 DNA binding domain resulted in potent transactivation of a GAL4-responsive luciferase reporter (Fig. 3D). Alterations within AD1 of E2A that reduced KIX binding in vitro also impaired the interaction as measured in this assay, suggesting that the structural constraints that affect the interaction in vitro also pertain in vivo.
In aggregate, these results indicate that the structural integrity and helical conformation of the LXXLL motif are required in the direct interaction between E2A proteins and the KIX domain of CBP/p300. The correlation between the results of the binding and cell immortalization assays supports the notion that interaction with CBP/p300 contributes to the oncogenic functions of E2A-PBX1.
A critical leucine within the LXXLL motif is required in E2A-PBX1b oncogenesis.
The striking effect of alterations affecting single amino acids (i.e., L20A or S17P) in our tissue culture-based proliferation/immortalization assay (Fig. 1B) suggested that oncogenesis by E2A-PBX1b may be exquisitely dependent on the integrity of the LXXLL motif in AD1. We evaluated this possibility using a whole-animal model. Irradiated mice in which hematopoiesis is reconstituted with bone marrow cells that have been retrovirally transduced with retroviruses conferring expression of E2A-PBX1 develop a lethal, transplantable myeloproliferative disease (19, 38). Therefore, sublethally irradiated mice were subjected to tail vein injection of bone marrow cells immediately after retroviral transduction with either naturally occurring E2A-PBX1b or the engineered, L20A mutant. Although amino acid substitutions of L16 and L20 of E2A appear to have equivalent, deleterious effects on the KIX interaction (Fig. 3A), we chose to evaluate the effects of only the L20A substitution in this whole-animal experiment. This decision was based on consideration of the three-dimensional structure of the complex between the KIX domain of CBP and the core activation domain of c-Myb, as determined recently by nuclear magnetic resonance spectroscopy (42). The KIX-interactive domain of c-Myb contains an LXXLL motif (underlined in 296-KELELLLM) that, as noted recently by others, shows considerable similarity to the LXXLL motif in E2A (14-KELSDLLD) (32). In the c-Myb/KIX complex, the side chain of the leucine residue at position L302 is inserted into a deep hydrophobic pocket in the KIX domain and plays a major role in stabilizing the complex. Since protein-protein interactions involving deep hydrophobic pockets may be especially susceptible to disruption by small molecules, we wished to evaluate the possibility of a critical requirement in leukemia induction for the corresponding amino acid in E2A-PBX1, specifically the leucine at position 20.
The efficacy of retroviral transduction was monitored using a colony-forming assay with drug selection; the mice that received E2A-PBX1b and the L20A variant were injected with equivalent numbers of colony-forming cells (1,155 and 901, respectively). Expression of both recombinant oncoproteins was documented by immunoblotting lysates from an aliquot of the transduced bone marrow cells (data not shown).
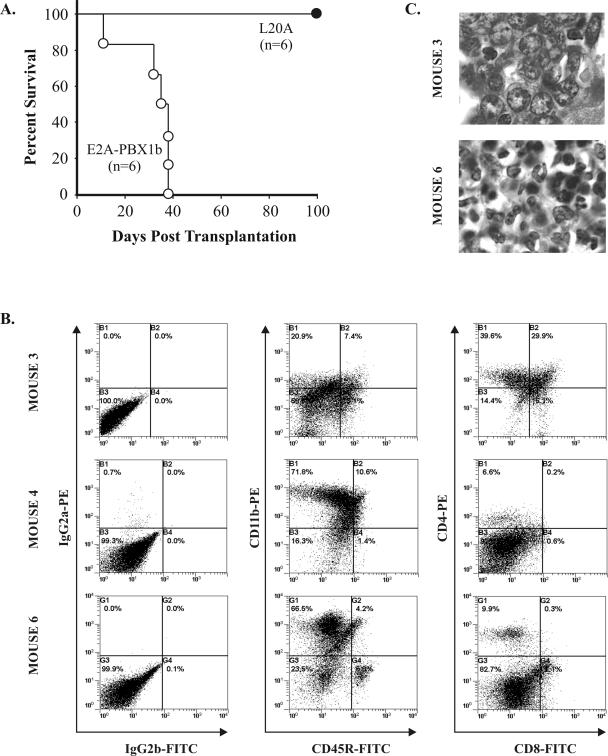
The six animals reconstituted with E2A-PBX1b-transduced marrow rapidly developed a wasting illness; all were consequently euthanized by 38 days (mean survival, 32 days, standard deviation, 11 days) subsequent to transplantation (Fig. 4A). At autopsy, these mice manifested enlarged spleens and livers. The immunophenotype of the splenic cell population was determined by flow cytometry using five animals. In four of these, the predominant cell population showed an immunophenotype characteristic of myeloid progenitors: CD11b positive, Gr-1 negative, CD45R weak expression or negative, CD4/CD8 negative (Fig. 4B and Table 1). The fifth animal (designated mouse 3) developed a disease with a predominantly lymphoid immunophenotype, including coexpression of the T-subset markers CD4 and CD8 in a substantial proportion of cells, characteristic of T-lymphoblastic leukemia/lymphoma. Histopathological examination confirmed expansion of the spleen and infiltration of the liver and other tissues with abnormal cells of evident hematopoietic origin. In the four animals with immunophenotypically myeloid disease, many cells were associated with morphological features characteristic of myeloid differentiation, including the presence of myelocytes and metamyelocytes (Fig. 4C). In contrast, no morphological evidence of myeloid differentiation was apparent in the proliferative cells in mouse 3. Instead, the cytoplasm was less abundant and the nuclei were larger and more round in contour and contained chromatin that appeared more dispersed and, generally, nucleoli that were more prominent. These morphological features are characteristic of progenitors that are more primitive or lymphoid.
FIG. 4.
A critical leucine side chain in the LXXLL motif is required in oncogenesis by E2A-PBX1b. (A) Extended survival of mice transplanted with bone marrow expressing the L20A mutant of E2A-PBX1b. (B) Immunophenotypes of cells obtained from the enlarged spleens of representative mice transplanted with bone marrow cells expressing unmodified E2A-PBX1b. The cells from mouse 3 express markers (i.e., CD4 and CD8) consistent with T-lineage progenitors, whereas those from mice 4 and 6 express a myeloid (i.e., CD11b) marker. (C) Photomicrographs of liver infiltrates. The cells from mouse 6 manifest nuclear features (i.e., smaller size, darker chromatin, elongated or horseshoe-shaped nuclei) consistent with granulocytic differentiation, whereas those from mouse 3 appear lymphoid or more primitive.
TABLE 1.
Immunophenotypes of splenic cells from mice transplanted with E2A-PBX1b-transduced bone marrow
| Mouse | % of positive cells
|
|||||
|---|---|---|---|---|---|---|
| CD11b | CD45R | CD4/ CD8+/+ | CD4/ CD8+/− | CD4/ CD8−/+ | CD4/ CD8−/− | |
| 1 | NDa | ND | ND | ND | ND | ND |
| 2 | 73 | 29 | 2 | 11 | 1 | 86 |
| 3 | 28 | 21 | 30 | 40 | 16 | 14 |
| 4 | 82 | 12 | 0 | 7 | 0 | 93 |
| 5 | 86 | 8 | 1 | 4 | 17 | 78 |
| 6 | 71 | 11 | 0 | 10 | 7 | 83 |
ND, not done.
Most importantly, all of the six mice transplanted with bone marrow transduced with the L20A mutant remained alive and apparently healthy at 100 days after bone marrow transplantation. Two of these animals died subsequently, at 117 and 125 days. Necropsies uncovered abdominal tumors in both mice that involved the retroperitoneal soft tissues. On microscopic examination, the tumor cells appeared myeloid and infiltrated the bone marrow and liver. The spleens were curiously small (weighing 0.15 g each) and appeared uninvolved by the myeloproliferative process histologically. The other four animals remained alive and apparently well at 334 days posttransplantation. Therefore, using this whole-animal model, leukemogenesis by E2A-PBX1b appears exquisitely dependent on the hydrophobic, KIX-interactive leucine side chain at position 20. The evident importance of this residue demands elucidation as to the mechanism of its contribution.
Functional mapping of E2A binding requirements on the KIX domain.
Of the relatively large number of proteins known to interact with the KIX domain of CBP/p300, the interactions with the CREB protein and c-Myb are especially well characterized. Nuclear magnetic resonance solution structures have been generated for the KIX domain in complex with the phosphorylated kinase-inducible activation domain (pKID) of CREB or the core transcriptional activation domain of c-Myb (36, 42). In both structures, the transcriptional activation domains make multiple hydrophobic interactions with a shallow, hydrophobic groove formed by the α1 and α3 helices of KIX. The interaction with c-Myb, but not pKID, relies on penetration of a critical leucine (L302) side chain into a deep hydrophobic pocket in KIX. The KIX interactions with c-Myb and pKID are disrupted by amino acid substitutions in KIX (Y650A, A654Q, and Y658A, or “YAY”) that affect the hydrophobic pocket and shallow hydrophobic groove without, according to structure prediction analyses, affecting KIX secondary structure (23). In an initial effort to explore experimentally the possibility that E2A might have KIX binding requirements similar to those of c-Myb, the effects of the YAY substitutions on E2A and c-Myb binding were compared.
A pull-down assay was devised in which GST-CBP 586-673 (containing the KIX domain) was evaluated for its ability to interact with 35S-labeled, in vitro-translated c-Myb (residues 211 to 360, including the activation domain) or E2A-PBX1a. Both E2A-PBX1a and c-Myb were pulled down by GST-CBP 586-673 of the wild-type sequence (Fig. 5A and B, respectively). However, the YAY amino acid substitutions within the KIX domain affected the c-Myb and E2A interactions differently, as reflected in the pull-down experiments. Whereas the substitutions completely abrogated retention of c-Myb, the quantity of retained E2A-PBX1b was reduced more modestly, to 16% or 29% of the quantity retained by GST-CBP 586-673 of the wild-type sequence, in two independent experiments (Fig. 5 and data not shown). Thus, the E2A interaction appeared less sensitive than the c-Myb interaction to disruption of the hydrophobic pocket and groove of KIX.
FIG. 5.
Functional mapping of E2A binding requirements on the KIX domain. (A) Pull-down experiment using bacterially expressed GST-CBP 586-673 and in vitro-translated, 35S-labeled E2A-PBX1a. CBP amino acids 586-673 encompass the KIX domain and seven additional C-terminal residues. The YAY (i.e., Y650A, A654Q, and Y658A) amino acid substitutions target the binding surface on the KIX domain used by c-Myb. Radiolabeled E2A-PBX1a is indicated by a small arrow at right. (B) Pull-down experiment using 35S-labeled c-Myb (amino acids 211 to 360). (C) Mammalian two-hybrid assay. CBP 586-673 (i.e., the KIX domain) fused to a GAL4 DNA binding domain can recruit either E2A or c-Myb to GAL4 binding sites on a luciferase reporter plasmid. The YAY amino acid substitutions have a greater, deleterious effect on c-Myb binding.
We used our mammalian two-hybrid assay to verify this apparent difference in the KIX-binding properties of E2A and c-Myb in the context of live, intact cells. Cotransfection of 293T cells with a portion of c-Myb fused to tandem VP16 activation domains and the KIX domain fused to the DNA-binding domain of GAL4 activated a luciferase reporter in a KIX-dependent manner (Fig. 5C). Incorporation of the YAY amino acid substitutions in the GAL4-KIX protein essentially abrogated this signal. Cotransfection of a VP16-E2A fusion protein similarly resulted in strong, KIX-dependent transactivation of the reporter. However, in contrast to c-Myb, the YAY mutations resulted in only a modest reduction in transactivation. Thus, despite circumstantial evidence suggesting that the LXXLL-containing activation domain of E2A may interact with KIX in a manner similar to that of c-Myb, these results warn that the interactions may differ in functionally important respects.
DISCUSSION
First described for transcriptional coactivators that interact with nuclear receptors in a ligand-dependent manner, the LXXLL motif has since been identified within a considerable number of protein-protein interaction domains involved in transcriptional regulatory functions beyond those associated with hormone signaling (reviewed in reference 32) (17). The sequence GTDKELSDLLDFS, which includes the LXXLL motif (underlined) and immediately adjacent residues, is strictly conserved in the mammalian E-proteins HEB, E2-2, and E12/E47, and the integrity of the LXXLL elements is required for the transcriptional function of AD1 (28, 40). Beyond this transcriptional role, our results are the first to argue that oncogenesis by E2A-PBX1b is critically dependent on a functional LXXLL motif.
Several observations support the notion that the LXXLL motif of E2A-PBX1 contributes to oncogenesis through recruitment of CBP/p300. Earlier studies have indicated that E12/E47 can interact with CBP/p300, in vitro or in vivo; we mapped the CBP/p300-interactive domain in E2A-PBX1 to a restricted portion of AD1 that includes the LXXLL motif (5, 8, 13, 33, 37). A recent proteomic search for proteins that interact in an LXXLL motif-dependent manner with the AD1 portion of the E-protein HEB identified CBP/p300 as a major binding partner (40). Finally, the data presented in Fig. 1 and 3 of our current study indicate that single amino acid substitutions that abrogate the ability of E2A-PBX1b to immortalize primary hematopoietic cells in culture (i.e., L20A and S17A) also disrupt the interaction with CBP.
This correlation between the effects of amino acid substitutions on oncogenesis and CBP/p300 recruitment by E2A-PBX1 is not perfect. Whereas the S17P and S17A substitutions differ strikingly from one another in their effect on myeloid cell immortalization by E2A-PBX1 (i.e., S17P abrogates immortalization whereas S17A has no apparent effect; Fig. 1B), the effects of these substitutions on CBP recruitment appear somewhat more modest and relative (Fig. 3B). Such considerations raise the possibility that the LXXLL motif of E2A-PBX1 might interact with additional proteins in mediating oncogenesis. As large proteins with multiple well-characterized domains that mediate interactions with numerous proteins, CBP/p300 may function to coordinate and integrate transcriptional regulation downstream of multiple signaling pathways (15). The KIX domain itself may function as a nexus for interactions involving several transcription factors. For example, core activation domains from the mixed lineage leukemia and c-Jun oncoproteins interact with a surface on the KIX domain that is distinct from, but contiguous with, the hydrophobic groove occupied by pKID or c-Myb such that simultaneous, cooperative interactions with KIX can occur between factors that make use of the adjacent surfaces (9, 14, 16). Therefore, the detailed elucidation of the E2A-KIX interaction, including that at the structural level, is likely to uncover the identity of additional protein partners that participate in higher-order complexes with CBP/p300 and E2A-derived proteins and thereby modulate transcriptional or oncogenic activities.
The LXXLL motif overlaps with the previously described LDFS motif; the leucine at position 20 is both the last residue in LXXLL and the first in LDFS (28). Therefore, our results demonstrating that L20A mutants are defective in both KIX binding and leukemia induction support the importance of both motifs, although our demonstration that mutations that affect amino acids within the LXXLL motif but N terminal to the LDFS motif (i.e., S17P) are similarly deficient shows that a helical LDFS domain is not sufficient for these functions. In fact, it now appears likely that the LXXLL and LDFS motifs contribute to a single functional domain.
The interaction between E2A-PBX1 and CBP/p300 represents a potential drug target. Identifying small-molecule inhibitors of protein-protein interactions has proven challenging, in part because of the large interaction surfaces involved (2). Having demonstrated that a helical transition involving the LXXLL motif of AD1 is required in the interaction between E2A-PBX1 and CBP and in cellular immortalization by E2A-PBX1, it seems reasonable to suggest that compounds that favor alternative, perhaps nonhelical conformations for the LXXLL-containing portion of E2A-PBX1 may be effective antagonists of oncogenesis. Furthermore, our findings argue that oncogenic recruitment of CBP/p300 requires the hydrophobic side chain of L20 of E2A-PBX1. On the basis of comparison with c-Myb, we speculate that L20 may stabilize the association with CBP/p300 by inserting its side chain into the hydrophobic pocket of KIX. The potential involvement of a small binding pocket suggests that the E2A-KIX interaction may be susceptible to inhibition by small molecules that can be accommodated within the pocket. However, our results indicating differential susceptibilities of c-Myb and E2A binding to amino acid substitutions in the α3 helix of KIX intended to affect the hydrophobic pocket and groove (i.e., Y650, A654, and Y658) suggest alternative modes of KIX recognition by these activation domains and underscore the need to characterize the E2A-KIX interaction further at a biophysical and structural level.
Despite the wealth of data pertaining to the function of both E proteins and CBP/p300, the mechanism by which the interaction between E2A-PBX1 and CBP/p300 might contribute to leukemia induction is not clear. Given the function of CBP/p300 as a transcriptional coactivator and the well-documented involvement of PBX1 and its HOX cofactors in hematopoiesis and leukemia, oncogenesis could result from deregulation of PBX/HOX target genes (1). In the context of E2A 1-483, which includes both AD1 and AD2, we showed recently that deletion of the entire LXXLL and contiguous LDFS domains results in only a modest reduction of transcriptional activity to 68% of that of the wild-type sequence (5). Nonetheless, even a modest reduction could be sufficient to impair oncogenesis by this mechanism, thus accounting for prolonged survival of mice transplanted with L20A mutant-expressing bone marrow. The possibility that E2A-PBX1 might exert deleterious effects on a tumor-suppressive function of wild-type E proteins or CBP/p300 through a dominant inhibitory mechanism has also been suggested previously (6). As a relevant example, the chimeric transcription factor AML1-ETO, implicated in cases of acute myelogenous leukemia, can silence transcriptional induction by E proteins through binding to the LXXLL motif, thereby preventing recruitment of CBP/300 (40).
One of our animals transplanted with E2A-PBX1b marrow developed a disease with immunophenotypic and morphological features characteristic of T-progenitor lymphoma/leukemia. This observation is novel, since earlier investigators have reported exclusively myeloid proliferations ensuing in mice after adoptive transfer of E2A-PBX1-transduced marrow, although T- or B-lymphoid or myeloid neoplasms can develop in E2A-PBX1 transgenic mice (7, 12, 19, 38). This result requires validation in order to rule out possible E2A-PBX1-independent mechanisms, such as local transcriptional effects related to provirus insertion. Nonetheless, the observation expands the existing evidence indicating that E2A-PBX1 is capable of transforming cells of diverse hematopoietic lineages, despite its strong association with lymphoid disease in humans, and suggests that it may be possible to develop a lymphoid model of E2A-PBX1-induced ALL by use of a retrovirus-based system that is more experimentally tractable than the transgenic approach.
In summary, we have shown that the functional integrity of an LXXLL motif at the extreme N terminus of E2A-PBX1 is required for oncogenesis. Since this motif participates in a direct interaction with the KIX domain of CBP/p300, it is likely that the LXXLL-KIX interaction is important in leukemia induction. This interaction is amenable to detailed characterization, including at the structural level, and could be susceptible to disruption by novel small molecules.
Acknowledgments
This work was supported by a research grant to D.P.L. from the National Cancer Institute of Canada and an operating grant to S.P.S. from the Canadian Institutes of Health Research.
We are grateful to Matthew Gordon for technical assistance with flow cytometry. Circular dichroism experiments were made possible by the Queen’s University Protein Function Discovery Facility.
REFERENCES
- 1.Abramovich, C., and R. K. Humphries. 2005. Hox regulation of normal and leukemic hematopoietic stem cells. Curr. Opin. Hematol. 12:210-216. [DOI] [PubMed] [Google Scholar]
- 2.Arkin, M. R., and J. A. Wells. 2004. Small-molecule inhibitors of protein-protein interactions: progressing towards the dream. Nat. Rev. Drug Discov. 3:301-317. [DOI] [PubMed] [Google Scholar]
- 3.Aronheim, A., R. Shiran, A. Rosen, and M. D. Walker. 1993. The E2A gene product contains two separable and functionally distinct transcription activation domains. Proc. Natl. Acad. Sci. USA 90:8063-8067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bain, G., E. C. Robanus Maandag, D. J. Izon, D. Amsen, A. M. Kruisbeek, B. C. Weintraub, I. Krop, M. S. Schlissel, A. J. Feeney, M. van Roon, M. van der Valk, H. P. J. te Riele, A. Berns, and C. Murre. 1994. E2a proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell 79:885-892. [DOI] [PubMed] [Google Scholar]
- 5.Bayly, R., L. Chuen, R. A. Currie, B. D. Hyndman, R. Casselman, G. A. Blobel, and D. P. LeBrun. 2004. E2A-PBX1 interacts directly with the KIX domain of CBP/p300 in the induction of proliferation in primary hematopoietic cells. J. Biol. Chem. 279:55362-55371. [DOI] [PubMed] [Google Scholar]
- 6.Bayly, R., and D. P. LeBrun. 2000. Role for homodimerization in growth deregulation by E2a fusion proteins. Mol. Cell. Biol. 20:5789-5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bijl, J., M. Sauvageau, A. Thompson, and G. Sauvageau. 2005. High incidence of proviral integrations in the Hoxa locus in a new model of E2a-PBX1-induced B-cell leukemia. Genes Dev. 19:224-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bradney, C., M. Hjelmeland, Y. Komatsu, M. Yoshida, T. P. Yao, and Y. Zhuang. 2003. Regulation of E2A activities by histone acetyltransferases in B lymphocyte development. J. Biol. Chem. 278:2370-2376. [DOI] [PubMed] [Google Scholar]
- 9.Campbell, K. M., and K. J. Lumb. 2002. Structurally distinct modes of recognition of the KIX domain of CBP by Jun and CREB. Biochemistry 41:13956-13964. [DOI] [PubMed] [Google Scholar]
- 10.Carroll, A. J., W. M. Crist, R. T. Parmley, M. Roper, M. D. Cooper, and W. H. Finley. 1984. Pre-B cell leukemia associated with chromosome translocation 1:19. Blood 63:721. [PubMed] [Google Scholar]
- 11.Chang, C. P., W. F. Shen, S. Rozenfeld, H. J. Lawrence, C. Largman, and M. L. Cleary. 1995. Pbx proteins display hexapeptide-dependent cooperative DNA binding with a subset of Hox proteins. Genes Dev. 9:663-674. [DOI] [PubMed] [Google Scholar]
- 12.Dedera, D. A., E. K. Waller, D. P. LeBrun, A. Sen-Majumdar, M. E. Stevens, G. S. Barsh, and M. L. Cleary. 1993. Chimeric homeobox gene E2A-PBX1 induces proliferation, apoptosis, and malignant lymphomas in transgenic mice. Cell 74:833-843. [DOI] [PubMed] [Google Scholar]
- 13.Eckner, R., T. P. Yao, E. Oldread, and D. M. Livingston. 1996. Interaction and functional collaboration of p300/CBP and bHLH proteins in muscle and B-cell differentiation. Genes Dev. 10:2478-2490. [DOI] [PubMed] [Google Scholar]
- 14.Ernst, P., J. Wang, M. Huang, R. H. Goodman, and S. J. Korsmeyer. 2001. MLL and CREB bind cooperatively to the nuclear coactivator CREB-binding protein. Mol. Cell. Biol. 21:2249-2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodman, R. H., and S. Smolik. 2000. CBP/p300 in cell growth, transformation, and development. Genes Dev. 14:1553-1577. [PubMed] [Google Scholar]
- 16.Goto, N. K., T. Zor, M. Martinez-Yamout, H. J. Dyson, and P. E. Wright. 2002. Cooperativity in transcription factor binding to the coactivator CREB-binding protein (CBP). The mixed lineage leukemia protein (MLL) activation domain binds to an allosteric site on the KIX domain. J. Biol. Chem. 277:43168-43174. [DOI] [PubMed] [Google Scholar]
- 17.Heery, D. M., E. Kalkhoven, S. Hoare, and M. G. Parker. 1997. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature 387:733-736. [DOI] [PubMed] [Google Scholar]
- 18.Henthorn, P., M. Kiledjian, and T. Kadesch. 1990. Two distinct transcription factors that bind the immunoglobulin enhancer microE5/kappa 2 motif. Science 247:467-470. [DOI] [PubMed] [Google Scholar]
- 19.Kamps, M. P., and D. Baltimore. 1993. E2A-Pbx1, the t(1;19) translocation protein of human pre-B-cell acute lymphocytic leukemia, causes acute myeloid leukemia in mice. Mol. Cell. Biol. 13:351-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamps, M. P., C. Murre, X. H. Sun, and D. Baltimore. 1990. A new homeobox gene contributes the DNA binding domain of the t(1;19) translocation protein in pre-B ALL. Cell 60:547-555. [DOI] [PubMed] [Google Scholar]
- 21.Kamps, M. P., and D. D. Wright. 1994. Oncoprotein E2A-Pbx1 immortalizes a myeloid progenitor in primary marrow cultures without abrogating its factor-dependence. Oncogene 9:3159-3166. [PubMed] [Google Scholar]
- 22.Kamps, M. P., D. D. Wright, and Q. Lu. 1996. DNA-binding by oncoprotein E2a-Pbx1 is important for blocking differentiation but dispensable for fibroblast transformation. Oncogene 12:19-30. [PubMed] [Google Scholar]
- 23.Kasper, L. H., F. Boussouar, P. A. Ney, C. W. Jackson, J. Rehg, J. M. van Deursen, and P. K. Brindle. 2002. A transcription-factor-binding surface of coactivator p300 is required for haematopoiesis. Nature 419:738-743. [DOI] [PubMed] [Google Scholar]
- 24.LeBrun, D. P. 2003. E2a basic helix-loop-helix transcription factors in human leukemia. Front. Biosci. 8:S206-S222. [DOI] [PubMed] [Google Scholar]
- 25.LeBrun, D. P., and M. L. Cleary. 1994. Fusion with E2A alters the transcriptional properties of the homeodomain protein PBX1 in t(1;19) leukemias. Oncogene 9:1641-1647. [PubMed] [Google Scholar]
- 26.Lu, Q., P. S. Knoepfler, J. Scheele, D. D. Wright, and M. P. Kamps. 1995. Both Pbx1 and E2A-Pbx1 bind the DNA motif ATCAATCAA cooperatively with the products of multiple murine Hox genes, some of which are themselves oncogenes. Mol. Cell. Biol. 15:3786-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu, Q., D. D. Wright, and M. P. Kamps. 1994. Fusion with E2A converts the Pbx1 homeodomain protein into a constitutive transcriptional activator in human leukemias carrying the t(1;19) translocation. Mol. Cell. Biol. 14:3938-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Massari, M. E., P. A. Jennings, and C. Murre. 1996. The AD1 transactivation domain of E2A contains a highly conserved helix which is required for its activity in both Saccharomyces cerevisiae and mammalian cells. Mol. Cell. Biol. 16:121-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Monica, K., D. P. LeBrun, D. A. Dedera, R. B. Brown, and M. L. Cleary. 1994. Transformation properties of the E2a-Pbx1 chimeric oncoprotein: fusion with E2a is essential, but the Pbx1 homeodomain is dispensable. Mol. Cell. Biol. 14:8304-8314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nourse, J., J. D. Mellentin, N. Galili, J. Wilkinson, E. Stanbridge, S. D. Smith, and M. L. Cleary. 1990. Chromosomal translocation t(1;19) results in synthesis of a homeobox fusion mRNA that codes for a potential chimeric transcription factor. Cell 60:535-545. [DOI] [PubMed] [Google Scholar]
- 31.Phelan, M. L., I. Rambaldi, and M. S. Featherstone. 1995. Cooperative interactions between HOX and PBX proteins mediated by a conserved peptide motif. Mol. Cell. Biol. 15:3989-3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Plevin, M. J., M. M. Mills, and M. Ikura. 2005. The LxxLL motif: a multifunctional binding sequence in transcriptional regulation. Trends Biochem. Sci. 30:66-69. [DOI] [PubMed] [Google Scholar]
- 33.Qiu, Y., A. Sharma, and R. Stein. 1998. p300 mediates transcriptional stimulation by the basic helix-loop-helix activators of the insulin gene. Mol. Cell. Biol. 18:2957-2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quong, M. W., M. E. Massari, R. Zwart, and C. Murre. 1993. A new transcriptional-activation motif restricted to a class of helix-loop-helix proteins is functionally conserved in both yeast and mammalian cells. Mol. Cell. Biol. 13:792-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quong, M. W., W. J. Romanow, and C. Murre. 2002. E protein function in lymphocyte development. Annu. Rev. Immunol. 20:301-322. [DOI] [PubMed] [Google Scholar]
- 36.Radhakrishnan, I., G. C. Perez-Alvarado, D. Parker, H. J. Dyson, M. R. Montminy, and P. E. Wright. 1997. Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: a model for activator:coactivator interactions. Cell 91:741-752. [DOI] [PubMed] [Google Scholar]
- 37.Sartorelli, V., J. Huang, Y. Hamamori, and L. Kedes. 1997. Molecular mechanisms of myogenic coactivation by p300: direct interaction with the activation domain of MyoD and with the MADS box of MEF2C. Mol. Cell. Biol. 17:1010-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thorsteinsdottir, U., J. Krosl, E. Kroon, A. Haman, T. Hoang, and G. Sauvageau. 1999. The oncoprotein E2A-Pbx1a collaborates with Hoxa9 to acutely transform primary bone marrow cells. Mol. Cell. Biol. 19:6355-6366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Dijk, M. A., P. M. Voorhoeve, and C. Murre. 1993. Pbx1 is converted into a transcriptional activator upon acquiring the N-terminal region of E2A in pre-B-cell acute lymphoblastoid leukemia. Proc. Natl. Acad. Sci. USA 90:6061-6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang, J., M. Kalkum, S. Yamamura, B. T. Chait, and R. G. Roeder. 2004. E protein silencing by the leukemogenic AML1-ETO fusion protein. Science 305:1286-1289. [DOI] [PubMed] [Google Scholar]
- 41.Zhuang, Y., P. Soriano, and H. Weintraub. 1994. The helix-loop-helix gene E2A is required for B cell formation. Cell 79:875-884. [DOI] [PubMed] [Google Scholar]
- 42.Zor, T., R. N. De Guzman, H. J. Dyson, and P. E. Wright. 2004. Solution structure of the KIX domain of CBP bound to the transactivation domain of c-Myb. J. Mol. Biol. 337:521-534. [DOI] [PubMed] [Google Scholar]