Abstract
Deficiency in either the interferon consensus sequence binding protein (ICSBP) or neurofibromin 1 (Nf1) increases the proliferative response of myeloid progenitor cell to hematopoietic cytokines. Consistent with this, we previously demonstrated that ICSBP activates transcription of the gene encoding Nf1 (the NF1 gene). In the studies presented here, we determine that ICSBP tyrosine phosphorylation is necessary for the activation of NF1 transcription. Since ICSBP is tyrosine phosphorylated in response to hematopoietic cytokines, these studies identify a novel pathway by which cytokine-induced posttranslational modification of ICSBP results in NF1 transcription. Nf1 subsequently inactivates cytokine-activated Ras, thereby creating a negative feedback mechanism for cytokine-induced proliferation. In these studies, we also determine that ICSBP is a substrate for SHP2 protein tyrosine phosphatase (SHP2-PTP). We find that wild-type SHP2-PTP dephosphorylates ICSBP only in undifferentiated myeloid cells. In contrast, a leukemia-associated, constitutively activated mutant form of SHP2-PTP dephosphorylates ICSBP in both myeloid progenitors and differentiating myeloid cells. Activated SHP2-PTP mutants thereby inhibit ICSBP-dependent NF1 transcription, impairing this negative feedback mechanism on cytokine-activated Ras. Therefore, these studies suggest that leukemia-associated ICSBP deficiency cooperates with leukemia-associated activating mutants of SHP2-PTP to contribute to the proliferative phenotype in myeloid malignancies.
Neurofibromin 1 (Nf1) is a 2,818-amino-acid protein encoded by the NF1 gene (1). The Nf1 protein includes a Ras-GAP domain which plays an important role in regulating the proliferation of myeloid cells in response to hematopoietic cytokines. Specifically, Nf1 Ras-GAP activity antagonizes granulocyte-macrophage colony-stimulating factor (GM-CSF)-, stem cell factor (SCF)-, or macrophage colony-stimulating factor (M-CSF)-induced Ras activation in myeloid cells (2, 4, 34). Therefore, Nf1-deficient myeloid cells exhibit increased proliferation in response at low doses of these hematopoietic cytokines (referred to as cytokine “hypersensitivity”). In humans, congenital Nf1 deficiency is associated with neurofibromatosis (8) and with a myeloproliferative/myelodysplastic disorder referred to as juvenile myelomonocytic leukemia (JMMoL) or juvenile chronic myeloid leukemia (18). Acquired Nf1 deficiency has also been documented for myeloid cells from human subjects with acute myeloid leukemia (AML) and adult myelodysplastic syndrome (MDS) (21). Therefore, although Nf1 expression is not restricted to hematopoietic cells, Nf1 deficiency is implicated in the pathogenesis of several malignant myeloid disorders. Consistent with this, NF1 deficiency results a myeloproliferative disorder in murine models (5, 33).
In previous investigations, we found that cytokine-induced differentiation of either myeloid leukemia cell lines or murine myeloid progenitor cells increases NF1 transcription and Nf1 expression. Cytokines implicated in this effect include M-CSF and gamma interferon (IFN-γ) (34). We also found that cytokine-induced NF1 gene transcription requires the interferon consensus sequence binding protein (ICSBP, or interferon regulatory factor 8 [IRF8]) (34). We determined that ICSBP activates NF1 transcription via a proximal promoter cis element which is similar to the previously described “PRDI” consensus sequence for interferon regulatory factor (IRF) protein-DNA binding (the PRDI consensus is TCACTT; the NF1 cis element is CCACTTCC) (29, 34). However, previous studies determined that ICSBP represses artificial promoter constructs with multiple copies of the PRDI element, while we found that ICSBP activates this similar NF1 cis element (29, 34).
Like Nf1 deficiency, ICSBP deficiency has been described for myeloid cells from human subjects with AML and MDS (25). ICSBP-deficient mice also develop a myeloproliferative disorder, and myeloid cells from these animals exhibit hypersensitivity to the same hematopoietic cytokines as Nf1-deficient cells do (14, 26, 34). Additionally, we found that proliferative abnormalities in myeloid progenitor cells from ICSBP-deficient mice can be rescued by expression of the Nf1 GAP-related domain (GRD) (34).
ICSBP is expressed exclusively in myeloid and B cells (23). Consistent with this, most previously identified ICSBP target genes encode proteins involved in the inflammatory response. For example, ICSBP activates transcription of the genes encoding gp91PHOX (the CYBB gene), p67PHOX (the NCF2 gene), Toll-like receptor 4, and interleukin-12 (IL-12) in phagocytic cells (11, 16, 25, 31). Transcriptional activation of the CYBB and NCF2 genes requires interaction of ICSBP, interferon regulatory factor 1 (IRF1), and the ets protein PU.1, with positive cis elements in these genes (11, 16). This interaction requires ICSBP tyrosine phosphorylation, which occurs during differentiation of myeloid leukemia cell lines or myeloid progenitor cells (16). Therefore, differentiation-stage-specific CYBB and NCF2 transcription is regulated by cytokine-induced posttranslational modification of ICSBP. In undifferentiated myeloid progenitor cells, ICSBP is a substrate for the SHP1 protein tyrosine phosphatase (SHP1-PTP) (16). During myeloid differentiation, SHP1-PTP activity decreases and the activity of Jak2 (and possibly that of other unidentified kinases) increases, resulting in ICSBP tyrosine phosphorylation (15, 16).
SHP1-PTP is a hematopoiesis-specific PTP with structural similarity to the ubiquitously expressed SHP2-PTP (24). SHP1 and SHP2, which can be located in either the nucleus or the cytoplasm (7, 16), contain conserved Src homology 2 (SH2) and single PTP domains. In the inactive form, protein folding renders the PTP domains in these proteins inaccessible to the substrate (12). Activation of SHP1 and SHP2 involves the interaction of their SH2 domains with phosphorylated tyrosine residues from substrates or other proteins. This interaction induces a conformational change which unmasks the PTP domain, activating the phosphatase (12). Few substrates have been identified for either PTP, and the extent of similarity in terms of substrate specificity between SHP1 and SHP2 is not known.
Although SHP1 gene mutations have not been described in association with any human disorder, recent studies identified acquired mutations in the SHP2 gene in some human myeloid malignancies. These mutations were found in myeloid cells from human subjects with AML, MDS, and JMMoL in blast crisis (3, 30). Modeling studies suggest that many of these leukemia-associated SHP2 mutations result in conformational changes which unmask the PTP domain (20, 30). The conclusions of these modeling studies have been specifically confirmed by biochemical analysis of some of these mutants (17, 30). Therefore, these mutations induce constitutive SHP2-PTP activation, which might be anticipated to influence regulation of functional activity, substrate specificity, or both. Consistent with this, functional studies of these leukemia-associated SHP2 mutants indicate that expression in myeloid cells confers hypersensitivity to a number of hematopoietic cytokines, including GM-CSF and IL-3 (6, 22, 28).
Based on this previous work, our current studies investigate the impact of ICSBP tyrosine phosphorylation on cytokine-induced NF1 gene transcription in differentiating myeloid cells. These studies also investigate the effect of constitutively activated, leukemia-associated SHP2 mutants on ICSBP regulation of NF1 transcription. Additionally, we investigate whether activating SHP2 mutants result in cytokine hypersensitivity by impairing Nf1 expression.
MATERIALS AND METHODS
Plasmids and PCR mutagenesis. (i) Protein expression vectors.
The ICSBP cDNA was obtained from Ben Zion-Levi (Technion, Haifa, Israel), and the full-length cDNA was generated by PCR and subcloned into the mammalian expression vector pcDNA as described previously (11). Generation of specific ICSBP tyrosine mutants by PCR site-directed mutagenesis has been previously described (16). An ICSBP mutant with conserved IRF domain tyrosine residues (Y92/95F ICSBP) changed to phenylalanine was used in the current studies. The cDNAs for wild-type and dominant negative (C463S) SHP2 protein tyrosine phosphatase were obtained from Stuart Frank (University of Alabama, Birmingham, AL) (10). A leukemia-associated, activating SHP2 mutant (E76K) (3) was generated by site-directed mutagenesis with the Clontech QuikChange protocol as described previously (11). Mutant clones were sequenced on both strands to verify that only intended mutations had been introduced. Wild-type SHP2 and E76K SHP2 were subcloned into the pMSCVneo vector for retroviral production (Stratagene, La Jolla, CA). These cDNAs were also subcloned into the pcDNAamp vector for in vitro protein translation. Wild-type SHP1 and SHP2 were subcloned into the pGEX1 vector (Amersham, Piscataway, NJ) for expression in Escherichia coli as a fusion protein with glutathione S-transferase (GST). The cDNAs for the wild-type Nf1-GRD and a GAP mutant form of the GRD were a kind gift from D. Wade Clapp (Indiana University, Indianapolis, IN).
(ii) Reporter constructs.
The proximal 973 bp of the human NF1 5′ flank was a kind gift from Meena Upadhyaya (Institute of Medical Genetics, Cardiff, United Kingdom). The promoter fragment was subcloned into the pCATE reporter vector (Promega, Madison, WI). A 337-bp NF1 promoter truncation mutant was generated by PCR and subcloned into the pCATE vector as described previously (34). PCR products were sequenced to ensure that no unintended mutations had been introduced. An artificial promoter construct was generated with four copies of the ICSBP-binding cis element from the NF1 promoter (bp −320 to −337) linked to a minimal promoter and a CAT reporter (the p-TATACAT vector). This construct is referred to as p-nf1TATACAT.
Oligonucleotides.
Oligonucleotides were custom synthesized by MWG Biotech (Piedmont, NC). Oligonucleotides representing sequences in the NF1 promoter were generated to analyze chromatin coimmunoprecipitating with anti-ICSBP antibody (F, 5′-CAGAGGAAAAGCTGGGCTTAAAT-3′; R, 5′-AGATCTGTCCTCCCCGCGGCCGGGG-3′). These oligonucleotide sequences flank the previously identified ICSBP-binding cis element in this promoter. Double-stranded oligonucleotides used in electrophoretic mobility shift assays (EMSA) represented the bp −320 to −337 NF1 promoter sequence (NF1-337; 5′-GGATCCTCACTCCGGTGGC-3′). This sequence has homology to the IRF-binding PRDI consensus (5′-TCACTT-3′), which is underlined. Subcloning of this oligonucleotide into a plasmid vector to generate radiolabeled EMSA probes has been described previously (11).
Myeloid cell line culturing.
The human myelomonocytic cell line U937 (19) was obtained from Andrew Kraft (Hollings Cancer Center, Medical University of South Carolina, Charleston, SC). Cells were maintained and differentiated as described previously (11). For differentiation experiments, U937 cells were treated for 48 h with 400 U per ml human recombinant IFN-γ (Roche, Indianapolis, IN) (11).
Transfections and reporter gene assays. (i) Transfections for promoter analysis.
U937 cells were cultured and transfected as previously described (11, 23, 34). Cells (32 × 106 per sample) were transfected with a vector to express wild-type ICSBP (ICSBP/pcDNAamp), ICSBP with a mutation of conserved IRF domain tyrosines (Y92/95F ICSBP/pcDNAamp), or an empty vector control; wild-type SHP2 (SHP2/pSX), SHP2 with a leukemia-associated activating mutation (E76K SHP2/pSX), or an empty vector control; the pCATE reporter vector with the proximal 337 bp of the human NF1 promoter (337Nf1pCATE) or an empty pCATE vector control; and p-CMVβgal (as a control for transfection efficiency).
In other experiments, U937 cells were transfected with vectors to overexpress wild-type ICSBP or a vector control; wild-type SHP2 (SHP2/pSX), SHP2 with a leukemia-associated activating mutation (E76K SHP2/pSX), dominant negative SHP2 (C463S SHP2), or an empty vector control; the minimal promoter/reporter vector p-TATACAT with four copies of the bp −320 to −337 NF1 sequence (nf1TATACAT); and p-CMVβgal. Transfectants were harvested 48 h after transfection with and without incubation with recombinant human IFN-γ (400 U/ml). Lysates were analyzed for CAT and β-galactosidase activity as described previously (34).
(ii) Transfections for stable pools overexpressing SHP2.
U937 cells were transfected with an empty expression vector or a vector to overexpress wild-type or E76K SHP2 as described above. After 24 h, the media were supplemented with G418 to 1 mg/ml, and stable transfectant pools were selected over 14 days, as previously described (11, 16). Stable transfectant pools were selected instead of clones to compensate for possible integration site effects. All experiments were repeated with at least two independent transfectant pools for each construct.
Murine bone marrow culturing and transduction.
Bone marrow mononuclear cells were obtained from the femurs of wild-type C57BL/6 mice. Sca1+ cells were separated using the Miltenyi magnetic bead system according to the manufacturer's instructions (Miltenyi Biotechnology, Auburn, CA). Bipotential myeloid progenitor cells were cultured (at a concentration of 2 × 105 cells per ml) for 48 h in Dulbecco minimal essential (DME) medium supplemented with 10% fetal calf serum, 1% penicillin-streptomycin, 10 ng/ml murine GM-CSF (R&D Systems Inc., Minneapolis, MN), 10 ng/ml SCF (R&D Systems Inc.), and 5 ng/ml murine recombinant IL-3 (R&D Systems Inc.). After retroviral transduction, cells were either maintained with GM-CSF plus SCF plus IL-3 (myeloid progenitor cells) or switched to DME medium supplemented with 10% fetal calf serum, 1% penicillin-streptomycin, and 10 ng/ml murine recombinant M-CSF (R&D Systems Inc.) for 96 h (monocyte differentiation), with or without 400 U/ml recombinant murine IFN-γ (Roche, Indianapolis, Indiana) for the last 48 h. Cells were harvested and cell lysates were used in Western blot experiments as described below. Aliquots of the cells were used in proliferation assays as described below.
Retroviral transduction of murine bone marrow myeloid cells.
High-titer murine stem cell retroviral supernatants were produced using the pMSCVneo vector (for wild-type or E76K SHP2) or pMSCVpuro vector (for Nf1-GAP GRD or GAP mutant Nf1-GRD) in the PT67 packaging cell line per the manufacturer's instructions (Stratagene). Filtered retroviral supernatants were used immediately or stored at −80°C. Transductions of murine bone marrow myeloid progenitor cells were performed as previously described (34). Briefly, cells were harvested, and 4.0 × 106 cells were plated in 3 ml of DME medium supplemented with 10% fetal calf serum, 10 ng/ml GM-CSF, 10 ng/ml SCF, and 5 ng/ml IL-3. An equal volume of retroviral supernatant was added to each dish, and Polybrene was added to a final concentration of 6 μg/ml. Cells were incubated for 8 h at 37°C in 5% CO2 and then diluted threefold with medium supplemented as described above. Cells were incubated overnight, and the procedure was repeated the next day. The day after transduction, G418 was added to 250 ng/ml. In some experiments, puromycin was added to 1.2 ng/ml. Cells were selected in antibiotic-supplemented media for 48 h and then treated with cytokines as indicated. Each experiment was repeated at least three times. Expression of transduced proteins was independently verified for each experiment by Western blotting. Aliquots of transduced cells were used in proliferation assays.
Isolation of nuclear proteins and electrophoretic mobility shift assays.
Nuclear extract proteins were isolated from U937 cells by the method of Dignam et al. with protease inhibitors (as described previously) (9). In some experiments, U937 cells were differentiated with 400 U/ml of IFN-γ before nuclear protein isolation. For other experiments, total cell lysates were generated from murine bone marrow-derived myeloid progenitor cells from wild-type or ICSBP−/− mice. These cells were lysed in hypotonic buffer (buffer C) and dialyzed against nuclear extract dialysis buffer (buffer D) according to the Dignam protocol cited above.
Oligonucleotide probes were prepared and EMSA and antibody supershift assays were performed as described previously (34). Antibodies to ICSBP (goat polyclonal) and control glutathione S-transferase antibody were obtained from Santa Cruz Biotech (Santa Cruz, CA).
Chromatin immunoprecipitation.
U937 cells were cultured with or without IFN-γ for 48 h as described previously (11, 16). Cells for chromatin immunoprecipitation were incubated with formaldehyde and lysed, and lysates were sonicated to generate chromatin fragments with an average size of 2.0 kb (32). Lysates underwent two rounds of immunoprecipitation with either antiserum to ICSBP or preimmune serum as described previously (34). Antibody to ICSBP and control preimmune serum were a kind gift from Stephanie Vogel (University of Maryland, Baltimore, MD). Coprecipitated chromatin was analyzed by PCR for ICSBP antibody-specific coprecipitation of the NF1 gene. For these experiments, input chromatin was a positive control, and chromatin precipitated by preimmune serum was a negative control. PCR products were analyzed by acrylamide gel electrophoresis. The identity of the PCR product was verified by subcloning into a plasmid vector and subsequent dideoxy sequencing.
Immunoprecipitation and Western blotting procedures. (i) Western blots of lysate proteins from murine bone marrow cells.
Murine bone marrow cells were lysed by boiling in 2× sodium dodecyl sulfate (SDS) sample buffer. Lysate proteins (50 μg) were separated by SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to nitrocellulose according to standard techniques. Western blots were serially probed with antibodies to Nf1, SHP2, ICSBP, total extracellular signal-regulated kinase (ERK), phosphorylated ERK, and alpha tubulin (to control for loading). Antibody to Nf1 (rabbit polyclonal raised to the C terminus) was obtained from Santa Cruz Biotech. Antibody to ICSBP (goat polyclonal) was also obtained from Santa Cruz Biotech (same as the antibody used for EMSA).
(ii) Immunoprecipitation and Western blotting.
U937 or murine bone marrow-derived myeloid cell stable transfectants were lysed under denaturing conditions by boiling in denaturing lysis buffer (50 mM Tris [pH 7.5]). Radioimmunoprecipitation assay buffer (10 mM Tris [pH 7.4], 150 mM NaCl, 1% NP-40, 5 mM EDTA) was added to the lysates, and proteins were immunoprecipitated with antibody to phosphotyrosine (clone 4G10; Upstate Biotechnology, Charlottesville, VA), antibody to ICSBP (rabbit polyclonal antiserum from S. Vogel, described above), or preimmune serum, as previously described (11, 16). Precipitated proteins were separated by SDS-PAGE and transferred to nitrocellulose as described above. Western blots were serially probed with antibodies to phosphotyrosine or ICSBP (goat polyclonal from Santa Cruz Biotech).
In vitro protein translation and phosphatase assay.
In vitro-transcribed ICSBP and E76K SHP2 mRNAs were generated from linearized template DNA using the Riboprobe system according to the manufacturer's instructions (Promega). In vitro-translated proteins were generated in rabbit reticulocyte lysate according to the manufacturer's instructions (Promega). Control (unprogrammed) lysates were generated by similar reactions in the absence of input RNA. For these experiments, ICSBP (but not E76K SHP2) was radiolabeled by including [35S]methionine in the translation reaction mixture.
For dephosphorylation assays, in vitro-translated ICSBP (10 μl) was incubated for 30 min at 30°C with either in vitro-translated E76K SHP2 or control lysate (1 μl). The ICSBP tyrosine phosphorylation state was determined by immunoprecipitating the reaction with antiphosphotyrosine or an irrelevant control antibody under denaturing conditions as previously described (16). Precipitated proteins were separated by SDS-PAGE, and autoradiography was then performed to detect coprecipitating in vitro-translated ICSBP.
GST fusion protein pull-down assays.
JM109 E. coli cells transformed with SHP1 or SHP2 in the pGEX vector or control empty vector were grown to log phase, supplemented to 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and incubated for 3 h at 37°C with shaking. The cells were harvested, resuspended in HN buffer (20 mM HEPES [pH 7.4], 0.1 M NaCl, 2 mM MgCl2, 0.1 mM EDTA, 0.5% Nonidet P-40, 0.1% Triton X-100, 2 mM phenylmethylsulfonyl fluoride), and sonicated on ice. Debris was removed by centrifugation, and the lysate was incubated for 30 min at 4°C with glutathione-agarose beads (Sigma Biochemical, St. Louis, MO) and washed extensively with HN buffer. The beads were preincubated for 30 min at 4°C with 5 μl of control rabbit reticulocyte lysate and then for 1 h with 20 μl of [35S]methionine-labeled in vitro-translated protein and washed extensively with HN buffer. Proteins were eluted with SDS-PAGE sample buffer and separated on 10% SDS-PAGE gels, and an autoradiograph was made.
Proliferation assays.
Murine bone marrow myeloid progenitor cells from wild-type or ICSBP−/− C57BL/6 mice were transduced with various combinations of retroviral vectors to express wild-type SHP2, E76K SHP2, ICSBP, the Nf1-GRD, a GAP mutant form of the Nf1-GRD, or an empty vector control. Cells were selected for 96 h in G418 (250 ng/ml) and/or puromycin (1.2 ng/ml) prior to proliferation assays. Cells were harvested and plated at 105 cells per 200 μl in a 96-well dish in DME medium supplemented with antibiotics and IL-3 but without GM-CSF. After 48 h of deprivation, medium was supplemented with a serial dilution of GM-CSF (10-fold from 10 ng/ml to 0.01 ng/ml or no cytokine). Cells were stimulated with GM-CSF for 24 h at 37°C in 5% CO2. In other experiments, deprived cells were cultured for 72 h at 37°C in 5% CO2 in a serial dilution of M-CSF (10-fold from 10 ng/ml to 0.01 ng/ml or no cytokine). For all proliferation assays, [3H]thymidine was added for the last 8 h of culturing. Cells were harvested with an automated cell harvester, and 3H incorporated into DNA was counted by scintillation counting according to standard techniques.
For all proliferation assays, Western blotting was performed on cells from the same transduction procedure and selected and cultured under the same cytokine conditions (GM-CSF at 10 ng/ml plus IL-3 at 10 ng/ml or differentiation with 10 ng/ml of M-CSF). This blotting was performed to verify the comparability of the expression levels of wild-type and mutant proteins and to compare expression levels between experiments. Representative blots are shown for each experiment. Antibodies used for these blots include antibodies to SHP2, Nf1 (rabbit polyclonal raised to the C terminus), ICSBP (goat polyclonal), and control antibodies from Santa Cruz Biotech (described above).
Statistical analysis. (i) Reporter gene assays.
For reporter gene assays, groups of transfectants were initially analyzed by the technique of analysis of variance between groups. Conditions with reporter gene expression levels significantly different from those of the others were identified by calculating the F value and determining the P value for the null hypothesis (i.e., no difference between conditions). For individual pairs of transfectants, paired Student's t tests were used to determine the significance of differences (P value) between individual data sets. Calculations were performed using SigmaPlot and SigmaStat software (Systat Software Inc, Richmond, CA).
(ii) Proliferation assays.
The significance of differences in proliferation under various conditions was determined using a paired Student's t test as described above and as previously reported (34).
RESULTS
ICSBP tyrosine phosphorylation is necessary for IFN-γ-induced NF1 transcription in U937 myeloid leukemia cells.
Previously, we determined that IFN-γ-induced differentiation of U937 myeloid leukemia cells increases Nf1 mRNA abundance and ICSBP-induced NF1 promoter activity (34). In other previous studies, we identified ICSBP tyrosine residues necessary for the activation of CYBB and NCF2 transcription during IFN-γ-induced U937 differentiation (tyrosine [Y] residues 92 and 95 in the IRF domain) (11, 16). We found that phosphorylation of these tyrosine residues is essential for ICSBP to participate in a multiprotein transcriptional activation complex which binds the cis elements in the CYBB and NCF2 genes. In contrast, we found that wild-type and tyrosine mutant ICSBP equivalently repress an artificial promoter construct with multiple copies of the PRDI consensus sequence in U937 cell transfection experiments (not shown) (16, 29, 34). Wild-type and Y92/95F ICSBP proteins were equivalently stable in these studies (11, 16).
Therefore, in the studies reported here, we investigated whether or not mutation of the same tyrosine residues prevents ICSBP-induced NF1 promoter activation. For these studies, U937 cells were transfected with an NF1 promoter/reporter construct with the proximal 337 bp of the NF1 5′ flank (337Nf1pCATE) or an empty vector control (34). Cells were cotransfected with a vector to express wild-type ICSBP, tyrosine mutant ICSBP (Y92/95F ICSBP), or empty expression vector control (pcDNA). Tyrosine phosphorylation of endogenous ICSBP is minimal in undifferentiated U937 cells but increases during IFN-γ differentiation (16, 29). We previously found that tyrosine phosphorylation of overexpressed ICSBP is slightly increased in comparison to that of the endogenous protein in undifferentiated U937 cells (16). However, tyrosine phosphorylation of overexpressed ICSBP is also significantly increased by IFN-γ differentiation of U937 cells (11, 16). Therefore, transfectants were assayed with and without 48 h of IFN-γ differentiation (11, 16, 34).
In these studies, we find that overexpression of wild-type ICSBP significantly increases reporter activity in comparison to Y92/95F ICSBP overexpression or transfection with an empty vector control without (F = 11.04; P = 0.004; n = 6) or with (F = 43.9; P = 0.0001; n = 6) IFN-γ (Fig. 1A). Consistent with our previous studies, wild-type ICSBP overexpression significantly increased NF1 promoter activity in comparison to transfectants with empty, control pcDNA expression vector without (P = 0.014; n = 6) and with (P = 0.0005; n = 6) IFN-γ. In contrast, expression of Y92/95F ICSBP did not significantly alter reporter expression from the 337-bp NF1 promoter fragment in comparison to transfectants with empty, control pcDNA expression vector without (P = 0.84; n = 6) or with (P = 0.78; n = 6) IFN-γ treatment. Reporter expression from empty, control reporter vector was insignificant and was not influenced by ICSBP overexpression or IFN-γ treatment and therefore was subtracted as background from NF1 promoter construct activity. Therefore, the same tyrosine residues that are essential for ICSBP-induced CYBB and NCF2 transcription are also involved in NF1 transcription.
FIG. 1.
Specific ICSBP tyrosine residues are necessary for induction of NF1 transcription. (A) Specific ICSBP tyrosine residues are essential for NF1 promoter activation. U937 cells were cotransfected with a reporter construct containing the proximal 337 bp of the NF1 promoter or an empty reporter vector control and a vector to overexpress wild-type or Y92/95F ICSBP or the empty vector control. Reporter gene activity was assayed after 48 h of incubation with (+ IFNg) or without (no IFNg) IFN-γ differentiation. Wild-type ICSBP induced NF1 promoter activity in U937 cells with and without IFN-γ treatment, consistent with our previous results (34). In contrast, ICSBP with mutation of conserved tyrosine residues in the IRF domain did not significantly increase NF1 promoter activity in these transfectants with and without IFN-γ differentiation. Neither expression of wild-type or Y92/95F ICSBP nor IFN-γ treatment altered the activity of the empty reporter vector control, which was subtracted as background. Bars which are significantly different from others under the same cytokine condition are indicated with asterisks. (B) Specific ICSBP tyrosine residues are essential for activation of the ICSBP-binding-positive cis element in the NF1 promoter. U937 cells were cotransfected with an artificial promoter construct containing four copies of the positive cis element from the NF1 promoter linked to a minimal promoter (p-nf1TATACAT) or a minimal promoter vector control (p-TATACAT) and a vector to overexpress wild-type or Y92/95F ICSBP or the empty vector control. Reporter gene activity was assayed after 48 h of incubation with (+ IFNg) or without (no IFNg) IFN-γ differentiation. Wild-type ICSBP increased reporter activity from the NF1 cis element-containing construct in U937 cells with and without IFN-γ treatment. In contrast, ICSBP with mutation of conserved tyrosine residues in the IRF domain did not significantly increase activity of the NF1 cis element in these transfectants with and without IFN-γ differentiation. Neither wild-type nor Y92/95F ICSBP had a significant impact on the control minimal promoter reporter vector. Bars which are significantly different from others under the same cytokine condition are indicated with asterisks. (C) Wild-type ICSBP and Y92/95F ICSBP are equivalently expressed in U937 transfectants. U937 cells were transfected with a vector to overexpress wild-type or Y92/95F ICSBP or the empty vector control. Lysate proteins from transfectants were analyzed with and without IFN-γ differentiation by Western blotting (WB). Blots were serially probed with an anti-ICSBP antibody (goat polyclonal; see Materials and Methods) or an antibody to alpha tubulin as a loading control. Levels of overexpression of wild-type ICSBP and the tyrosine mutant form of ICSBP were equivalent for these transfectants, consistent with our previous results.
In previous studies, we identified a positive cis element in the NF1 promoter which is activated by ICSBP (−327 to −331 bp relative to the ATG) (34). Therefore, we investigated the role of ICSBP tyrosine phosphorylation in the activation of this specific NF1 cis element. For these studies, U937 cells were transfected with a reporter construct containing a minimal promoter linked to four copies of the ICSBP-binding NF1 cis element (p-nf1TATACAT) or empty control vector (p-TATACAT). Cells were cotransfected with vectors to express wild-type ICSBP, Y92/95F ICSBP, or an empty vector control, and transfectants were analyzed with and without IFN-γ differentiation.
In these studies, we find that ICSBP overexpression significantly increases reporter activity in comparison to that seen for cells transfected with Y92/95F ICSBP expression vector or an empty vector control without (F = 5.62; P = 0.01; n = 9) or with (F = 11.83; P = 0.0006; n = 9) IFN-γ (Fig. 1B). We find that activity of the NF1 cis element is significantly increased by ICSBP overexpression in comparison to transfection with an empty vector control without (P = 0.016; n = 9) or with (P = 0.001; n = 8) IFN-γ differentiation. In contrast, the effect of Y92/95F ICSBP expression is not significantly different from that of transfection with empty control vector on the activity of the NF1 cis element without (P = 0.86; n = 4) or with (P = 0.98; n = 4) IFN-γ treatment of the transfectants. Neither wild-type nor Y92/95F ICSBP impacts reporter expression from empty, minimal promoter p-TATACAT control with or without IFN-γ. These results suggest that the effect of ICSBP tyrosine phosphorylation on NF1 transcription occurs via the previously identified cis element.
To reconfirm our previous results that the expression levels of wild-type ICSBP and Y92/95F ICSBP are equivalent in these studies, Western blotting was performed. Total cell lysates of U937 transfectants were separated by SDS-PAGE, and Western blots were probed for total ICSBP protein (Fig. 1C). Equal loading was determined by reprobing the blot with an antibody to alpha tubulin. A blot from a representative transfection experiment is shown.
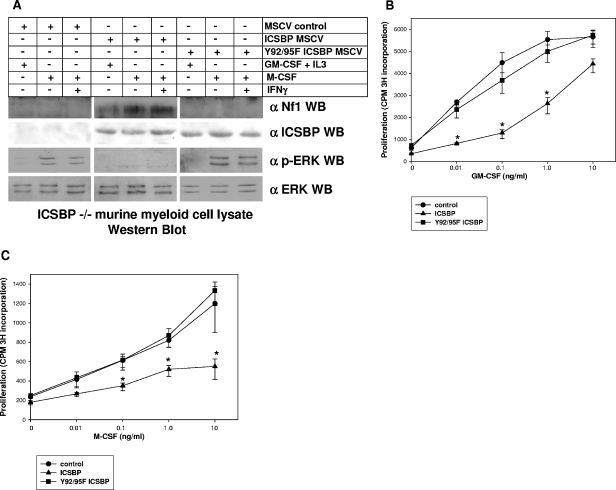
ICSBP tyrosine phosphorylation is necssary for Nf1 expression and cytokine hypersensitivity of differentiating murine bone marrow myeloid cells.
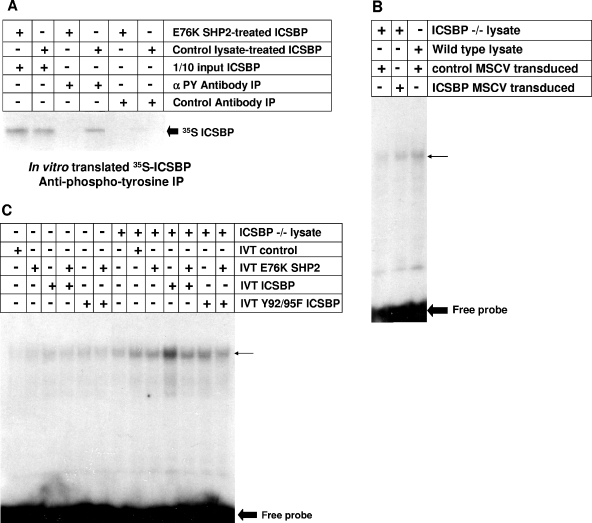
We also investigated the impact of ICSBP tyrosine phosphorylation on Nf1 expression in myeloid progenitor cells from ICSBP-deficient mice, a nontransformed model of myeloid differentiation. We previously found that Nf1 expression is minimal in cultured, wild-type murine bone marrow myeloid progenitor cells but increases during ex vivo M-CSF differentiation of these cells (34). Consistent with this, we found that both ERK phosphorylation (activation) and proliferation decrease after 48 h of M-CSF differentiation. We also found that ICSBP−/− myeloid progenitors express less Nf1 than wild-type progenitors and that Nf1 expression is not induced during ex vivo differentiation of ICSBP-deficient cells (34). Consistent with this, ERK activation and cell proliferation are sustained in ICSBP-deficient, M-CSF-differentiating cells. Additionally, ICSBP expression in ICSBP-deficient cells rescues Nf1 expression, decreases ERK activation, and abolishes GM-CSF and M-CSF hypersensitivity (34). Cytokine hypersensitivity of ICSBP−/− myeloid progenitor cells is also reversed by expression of the Nf1-GRD (34). In previous studies, we also found that M-CSF differentiation increases tyrosine phosphorylation of endogenous ICSBP in wild-type murine bone marrow myeloid progenitor cells and of overexpressed ICSBP in ICSBP−/− cells (16, 34).
Therefore, to investigate the role of ICSBP tyrosine phosphorylation in cytokine-dependent Nf1 expression, cultured ICSBP−/− myeloid progenitor cells were transduced with a murine retroviral stem cell vector to express wild-type ICSBP, Y92/95F ICSBP, or empty control vector (murine stem cell retroviral vector). In control experiments, we found that wild-type ICSBP and tyrosine mutant ICSBP were equivalently expressed in the ICSBP−/− cells (Fig. 2A). Myeloid progenitor cells were maintained with GM-CSF plus SCF plus IL-3 or differentiated to monocytes with M-CSF or M-CSF plus IFN-γ as described previously (34). Nf1 protein expression and ERK activation in the cultured, transduced cells were determined by Western blotting of total cell lysates (Fig. 2A).
FIG. 2.
Specific ICSBP tyrosine residues are necessary for cytokine-induced Nf1 expression in murine bone marrow myeloid cells. (A) Expression of wild-type ICSBP, but not Y92/95F ICSBP, rescues Nf1 protein expression and M-CSF-induced down-regulation of ERK in ICSBP−/− murine bone marrow myeloid progenitor cells. Bone marrow myeloid progenitor cells from ICSBP−/− mice (cultured with GM-CSF, IL-3, and SCF) were transduced with a murine stem cell retroviral vector (MSCV) to express wild-type or Y92/95F ICSBP or an empty vector control. Cell lysates were analyzed by Western blotting (WB) for expression of Nf1, ICSBP, and phospho-ERK during M-CSF differentiation. Wild-type ICSBP, but not Y92/95F ICSBP, rescues Nf1 expression in ICSBP−/− murine myeloid cells. Consistent with this, activated ERK increases during M-CSF differentiation in ICSBP−/− cells transduced with Y92/95F ICSBP or an empty vector control but not with wild-type ICSBP. No ICSBP expression is detected for ICSBP−/− myeloid progenitors transduced with an empty control vector, consistent with expectations. In contrast, ICSBP is approximately equivalently expressed in ICSBP−/− cells transduced with a vector to express wild-type or Y92/95F ICSBP. Antibody to Nf1 used in these experiments is a rabbit polyclonal raised to the C terminus, and antibody to ICSBP is rabbit polyclonal (see Materials and Methods). (B) Expression of wild-type ICSBP, but not Y92/95F ICSBP, rescues GM-CSF hypersensitivity of ICSBP−/− murine bone marrow myeloid progenitor cells. Bone marrow myeloid progenitor cells from ICSBP−/− mice (cultured with GM-CSF, IL-3, and SCF) were transduced with a murine stem cell retroviral vector to express wild-type or Y92/95F ICSBP or an empty vector control (see above). An aliquot of cells was deprived of cytokines for 24 h and then stimulated for 24 h with a dose titration of GM-CSF as indicated. Cells were pulsed with [3H]thymidine for the final 8 h of GM-CSF stimulation, and proliferation was determined by 3H incorporation into DNA. Reconstitution with wild-type ICSBP decreases GM-CSF hypersensitivity, consistent with our previous results (34). In contrast, ICSBP−/− cells overexpressing Y92/95F ICSBP have a GM-CSF dose response which is not significantly different from the dose response of ICSBP-deficient cells transduced with empty vector. (C) Expression of wild-type ICSBP, but not Y92/95F ICSBP, rescues prolonged proliferation of ICSBP−/− murine bone marrow myeloid progenitor cells during M-CSF differentiation. Bone marrow myeloid progenitor cells from ICSBP−/− mice were transduced with a murine stem cell retroviral vector to express wild-type or Y92/95F ICSBP or an empty vector control (see above). An aliquot of cells was deprived of cytokines for 24 h and then differentiated for 72 h with an M-CSF dose titration as indicated. Cells were pulsed with [3H]thymidine for the final 8 h of M-CSF stimulation, and proliferation was determined by 3H incorporation into DNA. Reconstitution with wild-type ICSBP abolishes prolonged proliferation of ICSBP−/− cells during M-CSF-induced differentiation, consistent with our previous results (34). In contrast, ICSBP−/− cells overexpressing Y92/95F ICSBP have a proliferative response to M-CSF which is not significantly different from that of ICSBP-deficient cells transduced with empty vector.
Consistent with our previous results (34), Nf1 is not detected in blots of lysates from control vector-transduced ICSBP−/− myeloid progenitor cells, with or without differentiation. Also consistent with our previous studies, transduction with an ICSBP expression vector induces Nf1 expression in cells treated with GM-CSF plus SCF plus IL-3; the expression level increases during ex vivo M-CSF differentiation. In contrast, expression of Y92/95F ICSBP in ICSBP−/− cells does not rescue Nf1 expression, with or without M-CSF differentiation (Fig. 2A). Consistent with these results, ERK activation (phosphorylation) increases during M-CSF differentiation of ICSBP-deficient myeloid progenitors transduced with an empty vector control or Y92/95F ICSBP expression vector but not with a vector to express wild-type ICSBP (Fig. 2A).
To correlate this result with a physiologic consequence, we determined the impact of expressing wild-type or Y92/95F ICSBP on GM-CSF or M-CSF hypersensitivity of ICSBP−/− myeloid progenitors. For these studies, an aliquot of cultured ICSBP−/− myeloid progenitor cells, transduced with the retroviral vectors described above, was evaluated for proliferative response to a dose titration of GM-CSF (Fig. 2B). Consistent with our previous results, ICSBP reconstitution significantly decreases the sensitivity of ICSBP−/− cells to GM-CSF-induced proliferation (P < 0.02 at 0.01, 0.1, and 1.0 ng/ml of GM-CSF; n = 4). In contrast, expression of Y92/95F ICSBP does not significantly alter the proliferative response of ICSBP−/− cells to this cytokine (P > 0.2; n = 4). Therefore, ICSBP overexpression increases Nf1 expression and decreases GM-CSF hypersensitivity in myeloid progenitor cells in a tyrosine phosphorylation-dependent manner.
In our previous studies, we also found that ICSBP−/− myeloid progenitor cells exhibit a prolonged proliferative response during ex vivo M-CSF differentiation (34). In contrast, wild-type myeloid progenitor cells or ICSBP−/− cells transduced with an ICSBP expression vector undergo proliferation arrest after 48 to 72 h of M-CSF differentiation. Therefore, we tested an aliquot of ICSBP−/− cells expressing either wild-type ICSBP or Y92/95F ICSBP (or transduced with empty control vector) for proliferation in response to an M-CSF dose titration (Fig. 2C). We found that levels of proliferation at 72 h are not significantly different in control vector and Y92/95F ICSBP-transduced cells (P > 0.5; n = 3). In contrast, expression of wild-type ICSBP significantly decreases proliferation of ICSBP−/− cells at 72 h in comparison to empty vector control-transduced cells (P < 0.01; n = 3). Therefore, both M-CSF-induced Nf1 expression and M-CSF hypersensitivity are influenced by phosphorylation of specific ICSBP tyrosine residues.
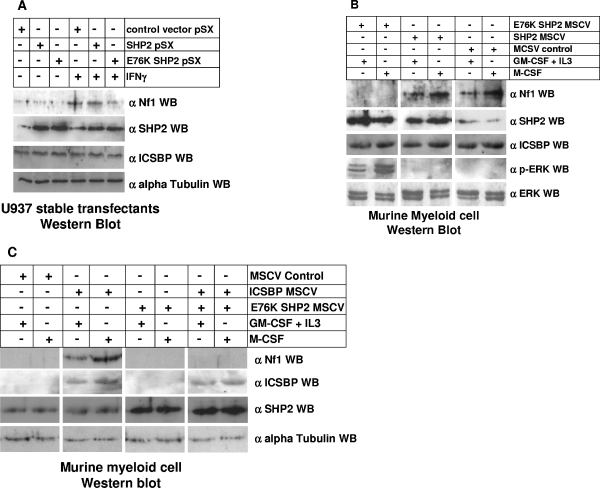
Expression of a leukemia-associated, constitutively activated SHP2 mutant inhibits cytokine-induced Nf1 expression in U937 myeloid leukemia cells.
The above-described results suggest that impairment of cytokine-induced ICSBP tyrosine phosphorylation might decrease Nf1 expression. Therefore, mutations in signaling pathways that regulate ICSBP tyrosine phosphorylation may lead to Nf1 deficiency. In previous studies, we found that ICSBP is a substrate for SHP1-PTP in undifferentiated myeloid cells (16). Although leukemia-associated, activating mutations in the SHP1 gene in have not been described, such mutations have been described in the gene encoding the closely related SHP2-PTP. Therefore, we investigated the impact of one of these mutations (that for E76K SHP2) on Nf1 expression (30). This is one of the most commonly described SHP2 mutant forms found in AML, MDS, and JMMoL cells (3, 19, 30). This mutant form has also been shown to induce GM-CSF hypersensitivity in murine myeloid cell transduction experiments (6, 22, 28).
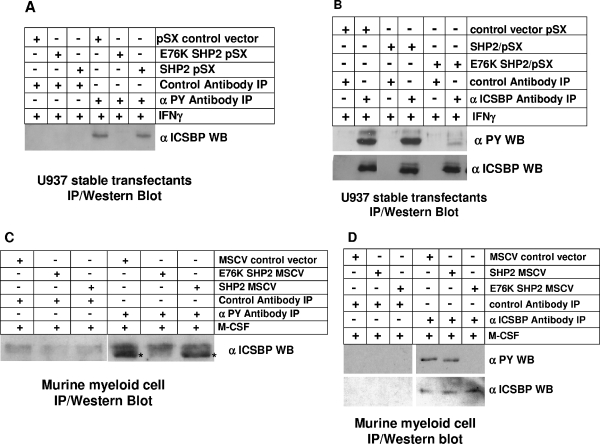
In initial studies, we determined the impact of expressing E76K SHP2 on Nf1 expression in U937 myeloid leukemia cells. For these studies, stable U937 transfectants overexpressing wild-type SHP2, E76K SHP2, or an empty vector control were generated. Stable transfectant pools were selected instead of clones to avoid potential integration site effects, and at least three independent transfectant pools were analyzed for each construct. Lysate proteins from transfected cells with and without IFN-γ differentiation were analyzed for Nf1 expression by Western blotting (Fig. 3A). We found that overexpression of wild-type SHP2 does not significantly decrease Nf1 expression in U937 cells either with or without IFN-γ differentiation. In contrast, expression of E76K SHP2 slightly decreases Nf1 expression in undifferentiated U937 cells and blocks IFN-γ-induced Nf1 expression during differentiation. These blots were also analyzed to verify equivalent levels of overexpression of wild-type SHP2 and E76K SHP2. The blots were stripped and reprobed with an anti-ICSBP antibody to verify that total ICSBP protein abundance is not altered for any of these transfectants (Fig. 3A).
FIG. 3.
Overexpression of E76K SHP2 (constitutively activated mutant), but not wild-type SHP2, decreases IFN-γ-induced Nf1 expression in myeloid cells. (A) Overexpression of E76K SHP2 inhibits IFN-γ-induced Nf1 expression in U937 cells. U937 cells were stably transfected with a vector to overexpress wild-type SHP2, E76K SHP2 (a leukemia-associated, constitutively activated mutant), or an empty vector control and analyzed for expression of Nf1, SHP2, and ICSBP during IFN-γ differentiation by Western blotting. Cells transfected with empty vector demonstrate IFN-γ-induced Nf1 expression, consistent with our previous studies (34). This level is not significantly different from that seen for cells stably overexpressing wild-type SHP2. In contrast, stable overexpression of E76K SHP2 in U937 cells inhibits IFN-γ-induced Nf1 expression. The levels of overexpression of wild-type and activated mutant SHP2 are approximately equivalent in these stable transfectant cells, and overexpression of these proteins does not alter ICSBP protein abundance relative to that seen for empty vector control transfectants. Antibody to Nf1 used in these experiments is a rabbit polyclonal raised to the C terminus, and antibody to ICSBP is rabbit polyclonal (see Materials and Methods). Blots were probed with an antibody to alpha tubulin as a loading control. (B) Expression of E76K SHP2 inhibits Nf1 expression in ex vivo-differentiating murine bone marrow-derived myeloid progenitor cells. Wild-type bone marrow myeloid progenitor cells (cultured with GM-CSF, IL-3, and SCF) were transduced with a murine stem cell retroviral vector (MSCV) to express wild-type SHP2, E76K SHP2, or an empty vector control. Cells were maintained in this medium or were ex vivo differentiated with M-CSF. Cell lysate proteins were analyzed for expression of Nf1, phospho-ERK, SHP2 and ICSBP by Western blotting. Expression of E76K SHP2 decreases Nf1 expression in myeloid progenitors and inhibits M-CSF-induced expression. In contrast, overexpression of wild-type SHP2 does not alter Nf1 expression in comparison to the level seen for cells transduced with the empty control vector. Consistent with this, phospho-ERK expression is increased in cells expressing E76K SHP2 in comparison to the level seen for cells transduced with a vector to express wild-type SHP2 or an empty vector control. Overexpression levels of wild-type SHP2 and activated mutant SHP2 were approximately equivalent in these transduced myeloid progenitor cells, and overexpression of these proteins did not alter ICSBP protein abundance relative to that seen for empty vector control-transduced cells. Antibody to Nf1 used in these experiments is a rabbit polyclonal raised to the C terminus, and antibody to ICSBP is rabbit polyclonal (see Materials and Methods). Blots were probed with an antibody to total ERK as a loading control. (C) ICSBP does not rescue Nf1 expression in ICSBP−/− murine myeloid cells coexpressing E76K SHP2. ICSBP−/− bone marrow myeloid progenitor cells (cultured with GM-CSF, IL-3, and SCF) were transduced with a murine stem cell retroviral vector (MSCV) to express E76K SHP2 or an empty vector control and with a vector to coexpress ICSBP or an empty vector control. Cells were maintained in this medium or were ex vivo differentiated with M-CSF. Cell lysate proteins were analyzed for expression of Nf1, SHP2, and ICSBP by Western blotting. Consistent with previous results, Nf1 expression is rescued in ICSBP−/− cells transduced with vectors to express ICSBP and an empty vector control. In contrast, cotransduction with vectors to express ICSBP and E76K SHP2 does not rescue Nf1 expression in ICSBP−/− cells. ICSBP is not expressed in these cells in the absence of transduction with an ICSBP expression vector, and this expression is not altered by cooverexpression of SHP2. Antibody to Nf1 used in these experiments is a rabbit polyclonal raised to the C terminus, and antibody to ICSBP is rabbit polyclonal (see Materials and Methods). Blots were probed with an antibody to alpha tubulin as a loading control. WB, Western blot.
Expression of a leukemia-associated, constitutively activated SHP2 mutant inhibits Nf1 expression in murine bone marrow myeloid cells.
We also investigated the impact of overexpressing wild-type or E76K SHP2 on Nf1 expression during ex vivo differentiation of murine bone marrow myeloid progenitor cells. For these studies, wild-type bone marrow myeloid progenitors were cultured with GM-CSF plus SCF plus IL-3 and transduced with a vector to express SHP2, E76K SHP2, or an empty vector control. Transduced cells were either maintained with GM-CSF plus SCF plus IL-3 or differentiated with M-CSF as described above. Western blotting was performed on lysates from transduced cells, with and without M-CSF differentiation. In initial experiments, blots were probed with antibodies to Nf1, phospho-ERK, and total ERK as described above (Fig. 3B). Blots were also probed with an anti-SHP2 antibody to verify equivalent overexpression of wild-type SHP2 and E76K SHP2 and with an anti-ICSBP antibody to verify that ICSBP protein abundance is not altered by transduction with any of these vectors.
We found that overexpression of wild-type SHP2 does not significantly impact Nf1 protein abundance in myeloid progenitors or ex vivo M-CSF-differentiated cells. In contrast, expression of E76K SHP2 decreases Nf1 expression in myeloid progenitor cells and prevents induction of Nf1 expression during M-CSF-induced differentiation (Fig. 3B). Consistent with this, expression of E76K SHP2, but not wild-type SHP2, results in sustained ERK activation during M-CSF differentiation.
We found that Nf1 expression in differentiating ICSBP−/− murine myeloid progenitor cells is rescued by wild-type but not Y92/95F ICSBP (see above). Therefore, we investigated the ability of ICSBP to rescue Nf1 expression in ICSBP−/− myeloid cells cotransduced with a vector to express E76K SHP2. For these studies, ICSBP−/− murine myeloid progenitors were cultured with GM-CSF plus IL-3 plus SCF and transduced with vectors to express ICSBP and E76K SHP2 (or an empty vector control). Cells were transduced with vectors to express each protein alone and to coexpress ICSBP with E76K SHP2. Cells were either maintained with GM-CSF plus SCF plus IL-3 or differentiated with M-CSF as described above. Western blots of cell lysates were serially probed with antibodies to Nf1, SHP2, ICSBP, and alpha tubulin (as a loading control). We found that coexpression of E76K SHP2 prevents ICSBP from rescuing Nf1 expression in ICSBP-deficient cells (Fig. 3C). However, overexpression of ICSBP was not influenced by cooverexpression of E76K SHP2.
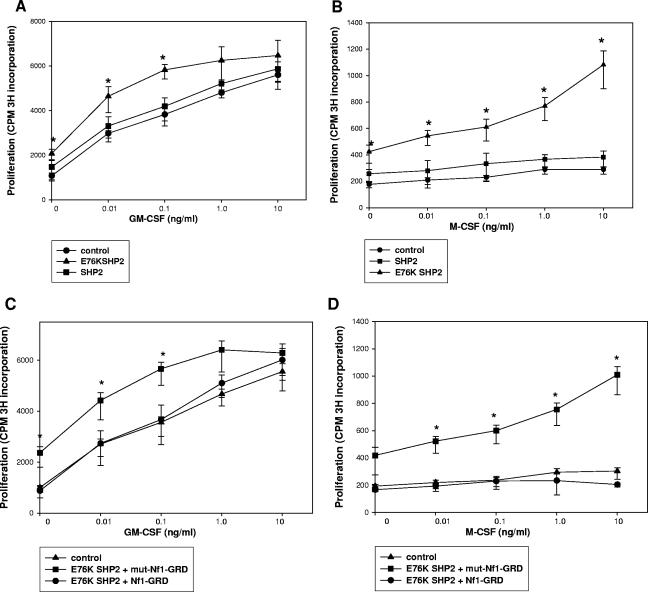
Expression of a leukemia-associated, constitutively activated SHP2 mutant induces cytokine hypersensitivity in murine bone marrow myeloid cells.
These studies suggest that expression of leukemia-associated, activated SHP2 mutants blocks cytokine-induced Nf1 expression during myeloid differentiation. To determine the physiologic significance of this, we initially investigated the impact of wild-type SHP2 and E76K SHP2 on cytokine hypersensitivity in murine bone marrow myeloid cells. For these experiments, an aliquot of the cultured, transduced wild-type murine bone marrow myeloid cells which are described above was also evaluated for proliferation in response to a GM-CSF dose titration (Fig. 4A). We found that overexpression of wild-type SHP2 does not significantly alter the proliferative response to a dose titration of GM-CSF in comparison to vector control-transduced cells (P > 0.5; n = 3). In contrast, low GM-CSF doses induce more proliferation in cells overexpressing E76K SHP2, consistent with the previous results of other investigators (6, 22, 28). This increase in proliferation is significant at the lowest GM-CSF doses (P < 0.002 for 0, 0.01, and 0.1 ng/ml GM-CSF; n = 3) and approaches statistical significance at higher doses.
FIG. 4.
E76K SHP2 expression induces cytokine hypersensitivity in murine bone marrow myeloid progenitor cells which is reversed by expression of the Nf1-GRD. (A) E76K SHP2 expression in murine bone marrow myeloid progenitor cells induces GM-CSF hypersensitivity. Wild-type bone marrow myeloid progenitor cells (cultured with GM-CSF, IL-3, and SCF) were transduced with a murine stem cell retroviral vector to express wild-type SHP2, E76K SHP2, or an empty vector control (Fig. 3B). An aliquot of cells was deprived of cytokines for 24 h and then stimulated for 24 h with a GM-CSF dose titration as indicated. Cells were pulsed with [3H]thymidine for the final 8 h of GM-CSF stimulation, and proliferation was determined by 3H incorporation into DNA. The GM-CSF proliferative response of SHP2-overexpressing cells is not significantly different from that of control vector-transduced cells. In contrast, E76K SHP2 overexpression resulted in an increased proliferative response to GM-CSF in these cells. (B) E76K SHP2 expression in murine bone marrow myeloid progenitor cells results in prolonged proliferation in response to M-CSF differentiation. Wild-type bone marrow myeloid progenitor cells (cultured with GM-CSF, IL-3, and SCF) were transduced with a murine stem cell retroviral vector to express wild-type SHP2, E76K SHP2, or an empty vector control (Fig. 3B). An aliquot of cells was deprived of cytokines for 24 h and then differentiated for 72 h with a dose titration of M-CSF as indicated. Cells were pulsed with [3H]thymidine for the final 8 h of M-CSF stimulation, and proliferation was determined by 3H incorporation into DNA. At 72 h, the M-CSF proliferative response of SHP2-overexpressing cells is not significantly different from that of control vector-transduced cells. In contrast, E76K SHP2 overexpression results in an increased and prolonged proliferative response to M-CSF in these cells. (C) GM-CSF hypersensitivity in E76K SHP2-overexpressing murine bone marrow myeloid progenitor cells is abolished by expression of the Nf1-GRD. Wild-type bone marrow myeloid progenitor cells (cultured with GM-CSF, IL-3, and SCF) were transduced with a murine stem cell retroviral vector to express E76K SHP2 and the Nf1-GRD, E76K SHP2 and a GAP mutant Nf1-GRD, or an empty vector control. Cells were deprived of cytokines for 24 h and then stimulated for 24 h with a GM-CSF dose titration as indicated. Cells were pulsed with [3H]thymidine for the final 8 h of GM-CSF stimulation, and proliferation was determined by 3H incorporation into DNA. GM-CSF hypersensitivity in E76K SHP2-overexpressing cells is abolished by coexpression of the Nf1-GRD. In contrast, GM-CSF hypersensitivity in E76K SHP2 cells is not significantly altered by coexpression of a GAP mutant form of the Nf1-GRD. (D) The prolonged proliferative response to M-CSF in E76K SHP2-overexpressing murine bone marrow myeloid progenitor cells is abolished by expression of the Nf1-GRD. Wild-type bone marrow myeloid progenitor cells (cultured with GM-CSF, IL-3, and SCF) were transduced with a murine stem cell retroviral vector to express E76K SHP2 and the Nf1-GRD, E76K SHP2 and a GAP mutant Nf1-GRD, or an empty vector control. Cells were deprived of cytokines for 24 h and then differentiated for 72 h with a dose titration of M-CSF, as indicated. Cells were pulsed with [3H]thymidine for the final 8 h of M-CSF stimulation, and proliferation was determined by 3H incorporation into DNA. Prolonged M-CSF-induced proliferation in E76K SHP2-overexpressing cells is abolished by coexpression of the Nf1-GRD. In contrast, prolonged M-CSF-induced proliferation in E76K SHP2 cells was not significantly altered by coexpression of a GAP mutant form of the Nf1-GRD.
Based on these results, we also tested the impact of expressing this leukemia-associated SHP2 mutant on prolonged proliferation of murine bone marrow myeloid cells during M-CSF differentiation. For these studies, an aliquot of transduced cells was treated with a dose titration of M-CSF. Proliferation was determined after 72 h, as described above (Fig. 4B). Consistent with previous results, we found there is little proliferation of empty vector-transduced cells at this time point. Similarly, M-CSF-differentiating cells transduced with the wild-type SHP2 expression vector did not proliferate significantly after 72 h. However, expression of E76K SHP2 in these cells results in sustained proliferation after 72 h of M-CSF stimulation. The increase in proliferation is statistically significant (P < 0.002; n = 3) at all M-CSF doses.
ICSBP deficiency, Nf1 deficiency, and E76K SHP2 expression all result in GM-CSF hypersensitivity and a sustained proliferative response to M-CSF. We were interested in determining the contribution of impaired Nf1 expression to cytokine hypersensitivity in E76K SHP2-expressing cells. Therefore, we investigated the impact of cooverexpressing E76K SHP2 and the Nf1-GRD on the proliferative response of cultured murine myeloid progenitor cells to GM-CSF and M-CSF. For these experiments, wild-type murine myeloid progenitor cells were cultured as described above and transduced with a retroviral vector to express E76K SHP2, with and without coexpression of the Nf1-GRD or a GAP mutant form of the Nf1-GRD (mut-Nf1-GRD). Proliferation in response to a dose titration of GM-CSF was determined as described above (Fig. 4C). We found that cooverexpression of a GAP mutant form of the Nf1-GRD does not significantly alter the proliferative response of E76K SHP2-expressing cells to GM-CSF (P > 0.5; n = 3). In contrast, coexpression of the wild-type Nf1-GRD and E76K SHP2 significantly decreases GM-CSF-induced proliferation of the transduced cells at all cytokine doses (P < 0.002; n = 3).
We also studied whether expression of the Nf1-GRD in E76K SHP2-expressing cells blocks sustained proliferation in response to M-CSF treatment (Fig. 4D). We found that expression of the wild-type Nf1-GRD abolishes the sustained proliferative response to M-CSF in murine myeloid cells expressing E76K SHP2. Indeed, M-CSF-induced proliferation in cells expressing Nf1-GRD plus E76K SHP2 is not significantly different from that in control, empty vector-transduced cells (P > 0.5; n = 3). In contrast, expression of mut-Nf1-GRD does not alter the M-CSF proliferative response in cells expressing E76K SHP2. These results further suggest the possibility that cytokine-induced Nf1 expression is downstream of SHP2-PTP activity in myeloid cells and indicate the functional consequences to this relationship.
In these studies, overexpression of E76K SHP2 was not altered by transduction of the cells with a vector to express wild-type or mutant forms of the Nf1-GRD (not shown). Because the antibodies available do not recognize the GRD of Nf1, overexpression of these proteins was not confirmed by Western blotting, consistent with previous studies by other investigators (33).
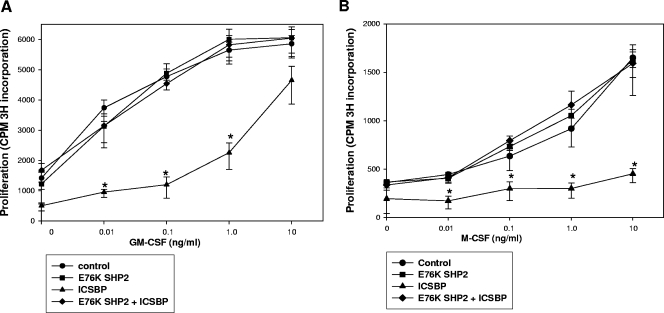
ICSBP expression does not rescue cytokine hypersensitivity in ICSBP−/− murine bone marrow myeloid cells expressing a leukemia-associated, constitutively activated SHP2 mutant.
To determine the impact of ICSBP deficiency on E76K SHP2-induced hypersensitivity, additional experiments were performed. Initially, we determined the impact of E76K SHP2 on proliferation in response to GM-CSF or M-CSF in ICSBP-deficient cells. We also investigated the ability of ICSBP reconstitution to rescue cytokine hypersensitivity in ICSBP−/− cells expressing E76K SHP2. For these experiments, an aliquot of the ICSBP−/− cultured, transduced wild-type murine bone marrow myeloid cells, described above, was evaluated for the proliferative response to a dose titration of GM-CSF (Fig. 5A). We found that GM-CSF-induced proliferation is not significantly different in ICSBP−/− cells transduced with E76K SHP2 or with an empty vector control (P > 0.4; n = 3). In these studies, ICSBP expression rescued GM-CSF hypersensitivity, consistent with our previous results (P < 0.01; n = 3). However, proliferation in response to a dose titration of GM-CSF was not significantly different between ICSBP−/− cells transduced with vectors to express E76K SHP2 plus ICSBP and control vector-transduced cells (P > 0.5; n = 3).
FIG. 5.
Cytokine hypersensitivity of ICSBP−/− murine myeloid cells is not rescued by expression of ICSBP in the presence of coexpression of E76K SHP2. (A) ICSBP expression fails to rescue GM-CSF hypersensitivity in ICSBP−/− murine bone marrow myeloid progenitor cells coexpressing E76K SHP2. ICSBP−/− bone marrow myeloid progenitor cells (cultured with GM-CSF, IL-3, and SCF) were transduced with a murine stem cell retroviral vector to express E76K SHP2 (or an empty vector control) and a vector to express ICSBP (or an empty vector control) (Fig. 3C). An aliquot of cells was deprived of cytokines for 24 h and then stimulated for 24 h with a GM-CSF dose titration as indicated. Cells were pulsed with [3H]thymidine for the final 8 h of GM-CSF stimulation, and proliferation was determined by 3H incorporation into DNA. The GM-CSF proliferative response of E76K SHP2-overexpressing cells or of cells coexpressing E76K SHP2 and ICSBP is not significantly different from that of control vector-transduced cells. In contrast, ICSBP overexpression results in a decreased proliferative response to GM-CSF in these cells. (B) ICSBP expression fails to rescue the prolonged proliferation in response to M-CSF differentiation in ICSBP−/− murine bone marrow myeloid progenitor cells coexpressing E76K SHP2. ICSBP−/− bone marrow myeloid progenitor cells (cultured with GM-CSF, IL-3, and SCF) were transduced with a murine stem cell retroviral vectors to express E76K SHP2 (or an empty vector control) and a vector to express ICSBP (or an empty vector control) (Fig. 3C). An aliquot of cells was deprived of cytokines for 24 h and then differentiated for 72 h with a dose titration of M-CSF as indicated. Cells were pulsed with [3H]thymidine for the final 8 h of M-CSF stimulation, and proliferation was determined by 3H incorporation into DNA. At 72 h, the M-CSF proliferative response of ICSBP-expressing ICSBP−/− cells is minimal, consistent with previous results. In contrast, E76K SHP2 expression or coexpression of E76K SHP2 and ICSBP results in an increased and prolonged proliferative response to M-CSF that is not significantly different from that of cells transduced with empty control vectors.
We also studied an aliquot of these transduced ICSBP−/− bone marrow cells for the proliferative response to a dose titration of M-CSF. We found that sustained proliferation in M-CSF-treated ICSBP−/− cells was not significantly altered by expression of E76K SHP2 (P > 0.2; n = 3) (Fig. 5B). Consistent with our previous studies, we found that the ICSBP expression rescues proliferation arrest in response to M-CSF differentiation in ICSBP−/− myeloid progenitor cells. However, the proliferative response to M-CSF was not significantly different between ICSBP−/− cells transduced with empty vector and cells transduced with E76K SHP2 plus ICSBP (P > 0.6; n = 3) (Fig. 5B). Therefore, these studies are consistent with the impact of E76K SHP2 on tyrosine phosphorylation of ICSBP and the impact of phosphorylation of ICSBP on cytokine hypersensitivity.
Expression of a leukemia-associated, constitutively activated SHP2 mutant inhibits ICSBP-dependent, cytokine-induced NF1 transcription.
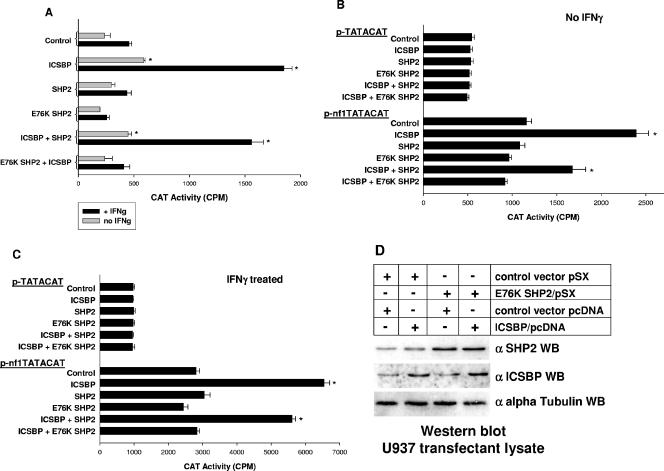
Based on the data described above, we investigated whether activated SHP2 inhibits cytokine-induced NF1 transcription. In our initial investigations, we determined the effect of overexpressing wild-type or E76K SHP2 on the activity of the proximal 337 bp of the NF1 promoter. For these studies, we used the same constructs employed in our studies of ICSBP tyrosine phosphorylation described above. U937 myeloid cells overexpressing wild-type or E76K SHP2 (or transfected with the pSX empty vector control) were cotransfected with a 337-bp NF1 promoter/reporter construct (337Nf1pCATE) or an empty pCATE vector control and a vector to express ICSBP or an empty expression vector control (pcDNA).
In these studies, we found that overexpression of ICSBP or ICSBP plus SHP2 significantly increased activity from the NF1 promoter (for the entire group, F = 13.4; P = 0.0001; n = 3; for the group without ICSBP or ICSBP plus SHP2, F = 1.2; P = 0.3) (Fig. 6A). In undifferentiated U937 transfectants, we found that overexpression of either wild-type or E76K SHP2 alone does not significantly decrease NF1 promoter activity, although repression by E76K SHP2 approaches significance (comparison between control and SHP2, P > 0.51; n = 3; between control and E76K SHP2, P < 0.04; n = 3). Although cooverexpression of SHP2 slightly decreases transcriptional activation of the NF1 promoter by overexpressed ICSBP, this decrease does not reach statistical significance (comparison between ICSBP and ICSBP plus SHP2, P = 0.05; n = 3). In contrast, cooverexpression of ICSBP and E76K SHP2 significantly decreases the impact of ICSBP overexpression on NF1 promoter activity (P < 0.004; n = 3). In these experiments, reporter activity from the empty control vector is not altered by overexpression of any of these proteins, and this activity was subtracted as background.
FIG. 6.
Activated SHP2 influences ICSBP-induced NF1 transcription in U937 myeloid leukemia cells. (A) E76K SHP2 inhibits ICSBP-induced NF1 promoter activity in IFN-γ-differentiated U937 myeloid leukemia cells. U937 cells were cotransfected with a reporter construct containing the proximal 337 bp of the NF1 promoter or the empty reporter vector control and a vector to overexpress various combinations of ICSBP, wild-type or E76K SHP2, or an empty vector control. Reporter gene activity was assayed after 48 h of incubation with (+ IFNg) or without (no IFNg) IFN-γ differentiation. Overexpression of wild-type SHP2 does not significantly decrease NF1 promoter activity with or without cooverexpression of ICSBP or with or without IFN-γ differentiation. In contrast, although E76K SHP2 alone does not significantly decrease NF1 promoter activity in undifferentiated U937 cells, expression of this mutant significantly decreases promoter activity in IFN-γ-treated transfectants. Additionally, E76K SHP2 significantly decreases ICSBP-induced NF1 promoter activity in both undifferentiated and IFN-γ-treated U937 cells. Bars which are significantly different from others under the same cytokine condition are indicated with asterisks. (B) E76K SHP2 inhibits ICSBP-induced activation of the ICSBP-binding-positive cis element in the NF1 promoter in undifferentiated U937 myeloid leukemia cells. U937 cells were cotransfected with an artificial promoter construct containing four copies of the ICSBP-binding-positive cis element from the NF1 promoter linked to a minimal promoter (p-nf1TATACAT) or the empty minimal promoter vector control (p-TATACAT) and a vector to overexpress various combinations of ICSBP, wild-type or E76K SHP2, or an empty vector control. Reporter gene activity was assayed after 48 h of incubation. Neither wild-type nor E76K SHP2 overexpression alone significantly inhibited reporter activity from the NF1 cis element-containing construct. Cooverexpression of ICSBP and wild-type SHP2 slightly decreases ICSBP-induced reporter activity. In contrast, cooverexpression of E76K SHP2 completely blocks ICSBP-induced activity of the NF1 cis element. None of the overexpressed proteins have a significant impact on the control minimal promoter reporter vector. Bars which are significantly different from others under the same cytokine condition are indicated with asterisks. (C) E76K SHP2 inhibits ICSBP-induced activation of the ICSBP-binding-positive cis element in the NF1 promoter in IFN-γ-differentiated U937 myeloid leukemia cells. U937 cells were cotransfected with an artificial promoter construct containing four copies of the ICSBP-binding-positive cis element from the NF1 promoter linked to a minimal promoter (p-nf1TATACAT) or the empty minimal promoter vector control (p-TATACAT) and a vector to overexpress various combinations of ICSBP, wild-type or E76K SHP2, or an empty vector control. Reporter gene activity was assayed after 48 h of incubation with IFN-γ differentiation. Similar to the results for undifferentiated transfectants, neither wild-type nor E76K SHP2 overexpression alters reporter expression from the NF1 cis element-containing construct. Additionally, cooverexpressed wild-type SHP2 does not significantly decrease the impact of overexpressed ICSBP on reporter activity from this construct. In contrast, E76K SHP2 significantly decreases ICSBP-induced activation of the NF1 cis element in differentiated U937 cells. None of the overexpressed proteins have a significant impact on the control minimal promoter reporter vector. Bars which are significantly different from others under the same cytokine condition are indicated with asterisks. (D) Cooverexpression of E76K SHP2 does not alter ICSBP overexpression in U937 transfectants. U937 cells stably transfected with a vector to express E76K SHP2 or the empty vector control were cotransfected with a vector to overexpress ICSBP (or an empty vector control). Cell lysates from transfected cells were analyzed by Western blotting (WB) for SHP2 and ICSBP expression. ICSBP and SHP2 are overexpressed in the cells transfected with these expression vectors, as anticipated. Overexpression of one protein does not significantly impact the overexpression of the other. Antibody to ICSBP is rabbit polyclonal (see Materials and Methods). Blots were probed with antibody to alpha tubulin as a loading control.
Since IFN-γ differentiation increases the impact of ICSBP on NF1 promoter activity in U937 cells (34), we performed the same experiments with IFN-γ-treated transfectants (Fig. 6A). As for the studies done in the absence of IFN-γ differentiation, NF1 promoter activity levels are significantly different in transfectants overexpressing ICSBP with and without wild-type SHP2 (for the entire group, F = 122.3; P = 0.0001; n = 3; for the group without ICSBP or ICSBP plus SHP2, F = 1.0; P = 0.5). Consistent with our previous studies, ICSBP overexpression significantly increases NF1 promoter activity in comparison to control vector transfectants (P < 0.001; n = 3). Similar what is seen for to undifferentiated transfectants, overexpression of SHP2 alone does not significantly decrease NF1 promoter activity in IFN-γ-treated transfectants (P = 0.5; n = 3). In contrast, expression of E76K SHP2 alone significantly decreases NF1 promoter activity in differentiated transfectants (P < 0.01; n = 3). We also found that NF1 promoter activity in transfectants cooverexpressing ICSBP and wild-type SHP2 is not significantly different from that in transfectants overexpressing ICSBP alone (P = 0.1; n = 3). In contrast, cooverexpression of ICSBP and E76K SHP2 significantly decreases the impact of ICSBP on NF1 promoter activity in IFN-γ-treated transfectants (P < 0.001; n = 3). Indeed, NF1 reporter activity is not significantly different between IFN-γ-treated transfectants overexpressing ICSBP plus E76K SHP2 and control vector transfectants (P > 0.5; n = 3). Therefore, these results indicate that E76K SHP2 expression inhibits ICSBP-induced NF1 transcription in cytokine-treated U937 cells.
We next determined the impact of wild-type or E76K SHP2 on the ICSBP-binding NF1 promoter cis element. For these studies, U937 cells overexpressing wild-type or E76K SHP2 (or transfected with empty pSX control vector) were transfected with the minimal promoter construct containing multiple copies of the NF1 promoter cis element linked to a reporter gene (p-nf1TATACAT), described above. Control cells were transfected with an empty vector containing the minimal promoter and reporter (p-TATACAT). Cells were cotransfected with a vector to express ICSBP or an empty vector control (pcDNA). Reporter activity was assayed as previously described.
In a result similar to that obtained with the experiments described above, NF1 cis element activity is significantly increased in cells overexpressing ICSBP, with or without wild-type SHP2 (for the entire group, F = 18.8; P < 0.0001; n = 3; for the group without ICSBP or ICSBP plus SHP2, F = 0.9; P = 0.6). In these studies, ICSBP overexpression significantly increases reporter expression from the NF1 cis element-containing construct (P < 0.001; n = 3) (Fig. 6B). ICSBP-induced reporter activity is slightly decreased by cooverexpression of wild-type SHP2 (P = 0.02; n = 3) but significantly decreased by cooverexpression of E76K SHP2 (P < 0.001; n = 3). Expression of the minimal promoter/reporter construct (p-TATACAT) is not altered by overexpression of any of these proteins in U937 transfectants.
We also determined the impact of IFN-γ differentiation on the ability of overexpressed wild-type or E76K SHP2 to influence ICSBP activation of the NF1 cis element in U937 transfectants. These experiments were performed as described above, and reporter activity was assayed after 48 h of IFN-γ differentiation (Fig. 6C). As in our previous studies, ICSBP overexpression, with or without wild-type SHP2 overexpression, significantly increases the activity of the minimal promoter construct with multiple copies of the NF1 cis element (for the entire group, F = 171.1; P < 0.0001; n = 3; for the group without ICSBP or ICSBP plus SHP2, F = 1.3; P = 0.3). In contrast, cooverexpression of E76K SHP2 abolishes the ICSBP-induced increase in reporter activity (comparison to control vector transfectants, P > 0.8; n = 3). These studies indicate that activated SHP2 significantly abolishes ICSBP-induced NF1 transcription in cytokine-treated cells via the previously identified ICSBP-binding NF1 promoter cis element.
In these studies, expression of E76K SHP2 decreases the impact of ICSBP overexpression on these reporter constructs. We hypothesize that this is due to an effect of E76K SHP2 on the phosphorylation state of ICSBP. However, another possible explanation might be that expression of E76K SHP2 decreases ICSBP overexpression. To investigate the latter possibility, Western blotting of lysate proteins from U937 transfections, with and without IFN-γ differentiation, was performed. These studies showed that E76K SHP2 expression does not impact overexpression of ICSBP in U937 cells (Fig. 6D). The role of E76K SHP2 on ICSBP tyrosine phosphorylation is investigated below.
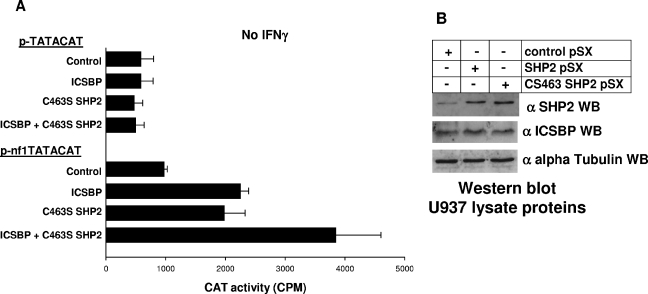
The studies described above suggest that activated, mutant SHP2 inhibits ICSBP-dependent activation of NF1 transcription. However, these studies do not indicate the impact of endogenous, wild-type SHP2 on ICSBP functional activity. Therefore, we investigated the role of SHP2 in regulating ICSBP-induced NF1 transcription in undifferentiated myeloid cells. For these experiments, U937 stable transfectant pools were generated with a vector to express a previously described dominant negative form of SHP2 (C463S SHP2) (10). In these studies, U937 cells were transfected with p-nf1TATACAT or control p-TATACAT reporter constructs, described above. Cells were cotransfected with a vector to express C463S SHP2 or empty control vector (pSX) and a vector to express ICSBP or an empty vector control (pcDNA). Reporter gene transcription was assayed as described above (Fig. 7A).
FIG. 7.
Endogenous SHP2 activity influences the impact of ICSBP on NF1 promoter activity in U937 myeloid leukemia cells. (A) C463S SHP2 (dominant negative mutant) activates the positive cis element in the NF1 promoter and increases ICSBP-induced activation in U937 myeloid leukemia cells. U937 cells were cotransfected with an artificial promoter construct containing four copies of the ICSBP-binding-positive cis element from the NF1 promoter linked to a minimal promoter (p-nf1TATACAT) or the empty minimal promoter vector control (p-TATACAT) and a vector to overexpress various combinations of ICSBP, C463S SHP2, or an empty vector control. Reporter gene activity was assayed after 48 h of incubation. C463S SHP2 expression alone significantly increases reporter expression from the NF1 cis element-containing construct. Additionally, cooverexpression of C463S SHP2 significantly increases the impact of ICSBP overexpression on this cis element in undifferentiated transfectants. Neither overexpressed protein has a significant impact on the control minimal promoter reporter vector. Bars which are significantly different from others under the same cytokine condition are indicated with asterisks. (B) Wild-type SHP2 and CS463 SHP2 (a dominant negative) are equivalently overexpressed in U937 transfectants. Stable U937 transfectants were generated with wild-type SHP2 (see above), a dominant negative mutant form of SHP2 (CS463), or the empty control vector. Cell lysates were analyzed by Western blotting (WB) for expression of SHP2 and ICSBP. Both forms of SHP2 are equivalently overexpressed in these U937 transfectants. Overexpression of wild-type SHP2 and CS463 SHP2 does not influence expression of ICSBP. Antibody to ICSBP used in these experiments was a goat polyclonal (see Materials and Methods). Blots were probed with antibody to alpha tubulin as a loading control.
We found that expression of the dominant negative form of SHP2 increases reporter expression from the NF1 cis element-containing construct to an extent similar to that of the overexpression of ICSBP (P = 0.5; n = 3). Additionally, cooverexpression of ICSBP and C463S SHP2 significantly increases reporter expression from the p-nf1TATACAT construct in comparison to overexpression of ICSBP or C463S SHP2 alone (P < 0.01; n = 3). Therefore, inhibition of SHP2-PTP activity in undifferentiated myeloid cells increases the impact of ICSBP on the NF1 cis element.
In control experiments, we found that wild-type and CS463 SHP2 are equivalently overexpressed in these U937 transfectants by Western blotting of cell lysate proteins (Fig. 7B). We also found that expression of this dominant negative SHP2 does not influence ICSBP expression in U937 transfection experiments.
Expression of a leukemia-associated, constitutively activated SHP2 mutant inhibits binding of endogenous ICSBP to the NF1 cis element.
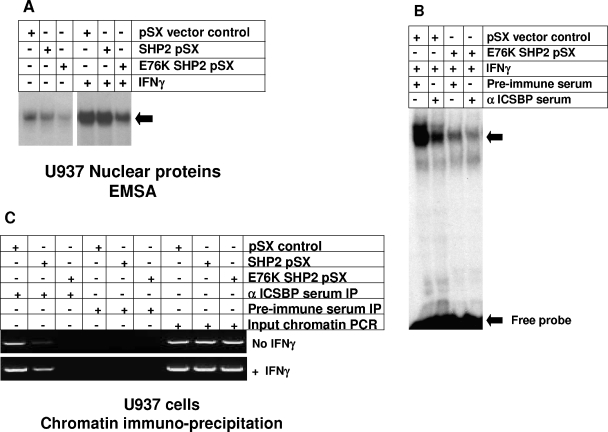
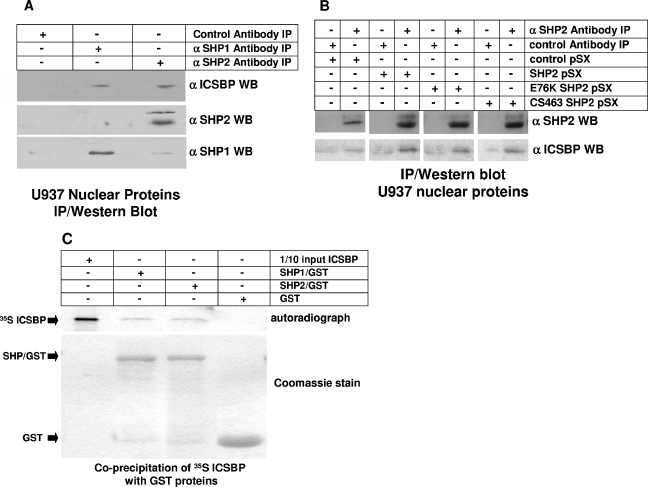
To identify the mechanism by which expression of an activated SHP2 mutant abolishes ICSBP-induced NF1 transcription, we investigated the impact of this mutant on ICSBP binding to the NF1 promoter. In previous studies, we identified a specific protein complex that interacts with the positive cis element at −320 to −336 bp of the NF1 promoter (34). We found that binding of this complex is increased by IFN-γ differentiation of U937 myeloid leukemia cells (34). We determined that ICSBP is a component of this complex by EMSA and chromatin immunoprecipitation (34). Therefore, we performed initial experiments to determine if stable expression of either wild-type or E76K SHP2 alters binding of the ICSBP-containing complex to −320 to −336 bp of the NF1 promoter. The U937 stable transfectants overexpressing wild-type or E76K SHP2 or an empty vector control are described above. Nuclear proteins were extracted from these transfectants with and without IFN-γ differentiation.
By using EMSA with nuclear proteins from untreated cells, we found that overexpression of wild-type SHP2 slightly decreases the binding of the previously described complex in comparison to that seen with nuclear proteins from control vector-transduced cells (Fig. 8A). However, binding of the complex significantly decreases in EMSA with nuclear proteins from untreated U937 cells expressing E76K SHP2. EMSA with nuclear proteins from control, IFN-γ-differentiated U937 cells demonstrates an increase in complex binding to the −320- to −336-bp NF1 probe, consistent with previous results (Fig. 8A). EMSA with nuclear proteins from IFN-γ-treated, wild-type SHP2-overexpressing U937 transfectants demonstrates protein complex binding that is not different from that seen for IFN-γ-treated vector control cells. In contrast, stable expression of E76K SHP2 significantly inhibits IFN-γ induction of protein complex binding to the −320- to −336-bp NF1 probe.
FIG. 8.
SHP2-PTP activity influences ICSBP interaction with the positive cis element in the NF1 promoter in U937 myeloid leukemia cells. (A) E76K SHP2 expression decreases in vitro interaction of a specific protein complex with the NF1 cis element in assays with U937 nuclear proteins. EMSA were performed with a probe representing the ICSBP-binding NF1 cis element and nuclear proteins isolated from U937 cells stably overexpressing wild-type or E76K SHP2 or transduced with an empty vector control, as indicated. Nuclear proteins were isolated from U937 cells with and without IFN-γ differentiation. Consistent with our previous studies, IFN-γ treatment increases binding of a specific protein complex to this probe in control vector assays (34). In assays with nuclear proteins from undifferentiated U937 cells, overexpression of wild-type SHP2 slightly decreases protein complex binding to the NF1 probe, and binding is almost completely abolished by E76K SHP2 expression. In contrast, only E76K SHP2 overexpression demonstrably decreases complex binding in assays with nuclear proteins from IFN-γ-differentiated U937 stable transfectants. (B) ICSBP is a component of the in vitro complex which binds to the NF1 cis element with and without E76K expression. EMSA were performed with a probe representing the ICSBP-binding NF1 cis element and nuclear proteins isolated from U937 cells stably overexpressing E76K SHP2 or transduced with an empty vector control as indicated. Nuclear proteins were isolated from IFN-γ-differentiated U937 cells. Binding assays were performed with preincubation with anti-ICSBP antibody or preimmune serum as indicated. Binding of the specific complex is disrupted by the ICSBP-specific antibody (goat polyclonal; see Materials and Methods), but not by an irrelevant control antibody, consistent with our previous results with nontransfected U937 cells (34). (C) Both wild-type SHP2 and E76K SHP2 decrease in vivo interaction of ICSBP with the NF1 cis element in undifferentiated U937 myeloid leukemia cells, but only E76K SHP2 significantly decreases ICSBP binding in IFN-γ-differentiated U937 cells. In vivo interaction of ICSBP with the positive NF1 cis element in U937 cells stably overexpressing wild-type or E76K SHP2 or transduced with the empty control vector was analyzed by chromatin immunoprecipitation. Chromatin coprecipitating with an anti-ICSBP antibody (rabbit polyclonal antiserum; see Materials and Methods) was PCR amplified with primers flanking the ICSBP-binding NF1 cis element. Chromatin coprecipitating with preimmune serum was a negative control in these studies. Total input chromatin (nonprecipitated) was a positive control. Overexpression of wild-type SHP2 decreases and overexpression of E76K SHP2 abolishes in vivo binding of ICSBP to the NF1 cis element in undifferentiated U937 cells. However, in IFN-γ-differentiated cells, only E76K SHP2 overexpression demonstrates a significant effect on ICSBP binding to this cis element.
To verify that ICSBP is a component of the protein complex binding the −320- to −336-bp NF1 probe in these experiments, additional EMSA were performed (Fig. 8B). Nuclear proteins from IFN-γ-treated U937 stable transfectants were preincubated with antibody to ICSBP or preimmune serum. EMSA were performed to determine whether an ICSBP cross-immunoreactive protein is a component of the complex binding the −320- to −336-bp probe. Consistent with previous studies, we found that ICSBP antibody disrupts protein complex binding to the −320- to −336-bp NF1 probe in EMSA with nuclear proteins from IFN-γ-differentiated control U937 cells. In EMSA with nuclear proteins from IFN-γ-treated U937 stable transfectants expressing E76K SHP2, anti-ICSBP antibody slightly disrupts this complex. These studies suggest that ICSBP is a component of the protein complex binding to this NF1 cis element in these transfectant cells and that binding is decreased by E76K SHP2 expression.
We also used chromatin immunoprecipitation to determine if overexpression of wild-type or E76K SHP2 influences ICSBP binding to the positive cis element in the NF1 promoter. We previously used this technique to demonstrate that IFN-γ differentiation of U937 cells increases ICSBP binding to this region of the NF1 promoter (34). U937 cells stably transfected with wild-type or E76K SHP2 expression vector or an empty vector control were assayed for in vivo ICSBP binding to the NF1 cis element. DNA-protein cross-links were generated in vivo, and chromatin coprecipitation with anti-ICSBP antibody (or control preimmune serum) was analyzed by PCR for the presence of the NF1 cis element. Stable transfectant cells were analyzed with and without IFN-γ differentiation.
In these studies, we found that overexpression of wild-type SHP2 slightly decreases in vivo ICSBP binding to the NF1 cis element in differentiated and undifferentiated U937 cells (Fig. 8C). In contrast, overexpression of E76K SHP2 abolishes in vivo ICSBP binding to the NF1 cis element in this assay with and without IFN-γ differentiation. Consistent with the EMSA results, these studies suggest that E76K SHP2 expression impairs ICSBP binding to the positive NF1 cis element. This is consistent with the results of the functional studies described above.
Expression of a leukemia-associated, constitutively activated SHP2 mutant inhibits cytokine-induced tyrosine phosphorylation of ICSBP.
ICSBP tyrosine phosphorylation occurs during cytokine-induced myeloid differentiation (16, 34). In the current studies, we found that ICSBP tyrosine phosphorylation is necessary for Nf1 expression during cytokine-induced differentiation of both U937 myeloid leukemia cells and murine bone marrow myeloid progenitor cells. We also found that expression of a constitutively active form of SHP2 inhibits ICSBP-induced Nf1 expression in these two models of myeloid differentiation. Therefore, we next investigated whether expression of this leukemia-associated SHP2 mutant impairs cytokine-induced ICSBP tyrosine phosphorylation.
We first determined the impact of overexpressing either wild-type or E76K SHP2 on ICSBP tyrosine phosphorylation in differentiating U937 cells. For these studies, stable U937 transfectants (see above) were differentiated with IFN-γ under conditions previously determined to result in tyrosine phosphorylation of endogenous ICSBP. Wild-type SHP2 and E76K SHP2 were shown to be equivalently overexpressed in these transfectants, as discussed above (Fig. 3A). Nuclear proteins were isolated and immunoprecipitation was performed under denaturing conditions with antibody to phosphotyrosine or an irrelevant control antibody. Immunoprecipitated proteins were analyzed by probing Western blots with an anti-ICSBP antibody (Fig. 9A). In parallel experiments, nuclear proteins were also immunoprecipitated with antibody to ICSBP or an irrelevant control antibody. Immunoprecipitated proteins were analyzed by Western blots serially probed with antiphosphotyrosine antibody and anti-ICSBP antibody (as a loading control) (Fig. 9B).
FIG. 9.
E76K SHP2 expression significantly decreases ICSBP tyrosine phosphorylation in cytokine-differentiated myeloid cells. (A) E76K SHP2 expression decreases ICSBP tyrosine phosphorylation in IFN-γ-differentiated U937 myeloid leukemia cells. U937 cells were stably transfected with vectors to overexpress wild-type or E76K SHP2 or an empty vector control. Cells were differentiated with IFN-γ to induce ICSBP tyrosine phosphorylation. Nuclear proteins were immunoprecipitated under denaturing conditions with an antibody to phosphotyrosine residues or an irrelevant control antibody, as indicated. Immunoprecipitated proteins were separated by SDS-PAGE, and Western blots were probed with an anti-ICSBP antibody (goat polyclonal; see Materials and Methods). IFN-γ-induced ICSBP tyrosine phosphorylation is abolished by E76K SHP2 expression in U937 cells. In contrast, overexpression of wild-type SHP2 decreases ICSBP tyrosine phosphorylation only slightly under these conditions. (B) E76K SHP2 expression decreases ICSBP tyrosine phosphorylation in IFN-γ-differentiated U937 myeloid leukemia cells without altering ICSBP protein abundance. U937 cells were stably transfected with vectors to overexpress wild-type or E76K SHP2 or an empty vector control. Cells were differentiated with IFN-γ to induce ICSBP tyrosine phosphorylation. Nuclear proteins were immunoprecipitated under denaturing conditions with an antibody to ICSBP (rabbit polyclonal antiserum) or preimmune serum as indicated. Immunoprecipitated proteins were separated by SDS-PAGE, and Western blots were serially probed with an antiphosphotyrosine antibody and an anti-ICSBP antibody (goat polyclonal; see Materials and Methods). IFN-γ-induced ICSBP tyrosine phosphorylation is abolished by E76K SHP2 expression in U937 cells. In contrast, overexpression of wild-type SHP2 decreases ICSBP tyrosine phosphorylation only slightly under these conditions. Total ICSBP protein is not altered by expression of wild-type or E76K SHP2. (C) E76K SHP2 expression decreases ICSBP tyrosine phosphorylation in M-CSF-differentiated murine bone marrow myeloid progenitor cells. Wild-type bone marrow myeloid progenitor cells (cultured with GM-CSF, IL-3, and SCF) were transduced with a murine stem cell retroviral vector (MSCV) to express wild-type SHP2, E76K SHP2, or an empty vector control. Cells were maintained in this medium or ex vivo differentiated with M-CSF. Cell lysate proteins were immunoprecipitated under denaturing conditions with an antibody to phosphotyrosine residues or an irrelevant control antibody as indicated. Immunoprecipitated proteins were separated by SDS-PAGE and analyzed by use of Western blots probed with an anti-ICSBP antibody (goat polyclonal; see Materials and Methods). Although ICSBP tyrosine phosphorylation in M-CSF-differentiated cells is slightly decreased by wild-type SHP2 overexpression, it is abolished by expression of E76K SHP2. The asterisk indicates the specific immunoreactive band. (D) E76K SHP2 expression decreases ICSBP tyrosine phosphorylation in M-CSF-differentiated murine bone marrow myeloid progenitor cells but does not alter ICSBP protein abundance. Wild-type bone marrow myeloid progenitor cells (cultured with GM-CSF, IL-3, and SCF) were transduced with a murine stem cell retroviral vector (MSCV) to express wild-type SHP2, E76K SHP2, or an empty vector control. Cells were maintained in this medium or ex vivo differentiated with M-CSF. Cell lysate proteins were immunoprecipitated under denaturing conditions with an antibody to ICSBP (rabbit polyclonal) or preimmune serum as indicated. Immunoprecipitated proteins were separated by SDS-PAGE and analyzed by use of Western blots serially probed with antiphosphotyrosine and anti-ICSBP (goat polyclonal) antibodies. ICSBP tyrosine phosphorylation in M-CSF-differentiated cells is abolished by expression of E76K SHP2. Total ICSBP protein is not altered by expression of wild-type or E76K SHP2. IP, immunoprecipitation; WB, Western blot.
Consistent with previous studies, we found that tyrosine-phosphorylated ICSBP is abundant in IFN-γ-treated U937 cells. Additionally, we found that wild-type SHP2 overexpression does not significantly decrease ICSBP tyrosine phosphorylation under these conditions. In contrast, expression of E76K SHP2 inhibits IFN-γ-induced ICSBP tyrosine phosphorylation. Abundance of total ICSBP protein is not altered by overexpression of either wild-type or E76K SHP2 in these U937 stable transfectants, as also demonstrated in Fig. 3A. These results suggest that expression of leukemia-associated, activated SHP2 blocks differentiation-induced ICSBP tyrosine phosphorylation in this myeloid leukemia cell line.
We next investigated the impact of overexpressing wild-type or E76K SHP2 on tyrosine phosphorylation of endogenous ICSBP in ex vivo-differentiating murine bone marrow myeloid cells. In our previous studies, we found that endogenous ICSBP was minimally tyrosine phosphorylated in murine bone marrow myeloid progenitor cells cultured with GM-CSF plus IL-3 (34). We also found that ICSBP tyrosine phosphorylation increased during ex vivo M-CSF differentiation of these cells (34). Therefore, we determined the tyrosine phosphorylation state of ICSBP in M-CSF-differentiated murine bone marrow-derived myeloid cells transduced either with wild-type or E76K SHP2 or with empty retroviral vector control. Cell lysates were immunoprecipitated under denaturing conditions with antibody to phosphotyrosine or with an irrelevant control antibody. Immunoprecipitates were analyzed by use of Western blots probed with anti-ICSBP antibody (Fig. 9C). Cell lysates were also immunoprecipitated with an antibody to ICSBP or with an irrelevant control antibody. Immunoprecipitates were analyzed by Western blotting with antiphosphotyrosine antibody and anti-ICSBP antibody as described above for U937 immunoprecipitates (Fig. 9D).
In these studies, we found that overexpression of wild-type SHP2 only minimally decreases ICSBP tyrosine phosphorylation in comparison to what is seen for vector control-transduced cells. In contrast, expression of E76K SHP2 inhibits M-CSF-induced tyrosine phosphorylation of ICSBP under these conditions. Overexpression of wild-type and E76K SHP2 does not significantly alter the abundance of total ICSBP protein in comparison to control vector-transduced cells, as was also found for the results shown in Fig. 3B. Wild-type SHP2 and E76K SHP2 were equivalently overexpressed in these experiments, consistent with data shown in Fig. 3C (not shown). These studies suggest that the leukemia-associated form of SHP2 inhibits ICSBP tyrosine phosphorylation in response to cytokine-induced differentiation in nontransformed myeloid progenitor cells.
ICSBP interacts with SHP2.
In previous studies, we found that ICSBP interacts with SHP1 in vitro and in vivo and is a substrate for SHP1-PTP activity (16). Based on our functional data described above, we investigated whether ICSBP also interacts with SHP2. First, we investigated the interaction of ICSBP with SHP2 in U937 myeloid leukemia cells. For these studies, nuclear proteins were immunoprecipitated with antibody to SHP2. Other investigators previously demonstrated that SHP2 is found in both the nuclei and cytoplasms of U937 cells (7). As a positive control, the lysates were also immunoprecipitated with anti-SHP1 antibody or with an irrelevant antibody as a negative control. Immunoprecipitates were analyzed by probing Western blots with an anti-ICSBP antibody (Fig. 10A). Blots were serially probed with anti-SHP2 and anti-SHP1 antibodies as controls. We found that ICSBP coimmunoprecipitates with SHP1, consistent with previous investigations (16). Additionally, we found a level of coprecipitation of ICSBP with SHP2 which is approximately equivalent to that for coprecipitation with SHP1. These studies suggest that ICSBP interacts either with SHP2 or with an SHP2-interacting protein.
FIG. 10.
ICSBP interacts with SHP2-PTP. (A) ICSBP coimmunoprecipitates with either SHP1 or SHP2 from U937 myeloid leukemia cell lysate proteins. Nuclear proteins from U937 cells were immunoprecipitated under nondenaturing conditions with an anti-SHP1 or -SHP2 antibody or an irrelevant control antibody. Anti-SHP1 was a positive control (16) and an irrelevant antibody a negative control for ICSBP coimmunoprecipitation in these studies. Immunoprecipitating proteins were separated by SDS-PAGE and analyzed by use of Western blots serially probed with antibodies to SHP1, SHP2, and ICSBP (goat polyclonal antibody) as indicated. ICSBP coimmunoprecipitates from U937 lysate proteins with antibody to either SHP1 or SHP2. (B) ICSBP coprecipitates equivalently with overexpressed wild-type SHP2, E76K SHP2, and CS463 SHP2 from U937 myeloid leukemia cell lysates. Nuclear proteins from stable U937 transfectants overexpressing wild-type, E76K, or CS463 SHP2 (or transfected with an empty vector control) were immunoprecipitated under nondenaturing conditions with an anti-SHP2 antibody or an irrelevant control antibody. Immunoprecipitating proteins were separated by SDS-PAGE and analyzed by use of Western blots serially probed with antibodies to SHP2 and ICSBP (goat polyclonal) as indicated. ICSBP coimmunoprecipitates with equivalent efficiencies from nuclear proteins derived from cells overexpressing SHP2, E76K SHP2, or CS463 SHP2. Consistent with the lower SHP2 expression seen for vector control-transduced cells, ICSBP is less abundant in immunoprecipitates from these cells. (C) Recombinant ICSBP interacts with recombinant SHP1 or SHP2. ICSBP was in vitro translated (in the presence of [35S]methionine) in rabbit reticulocyte lysates. SHP1 and SHP2 were expressed in E. coli as fusion proteins with glutathione S-transferase. Interaction between ICSBP and SHP1/GST, SHP2/GST, or control GST was determined by pull-down assay. SHP1/GST is a positive control and GST a negative control for specific ICSBP interaction in this assay (16). Equality of loading was determined by Coomassie staining of the SDS-PAGE gels as shown. We found that ICSBP interacts specifically with either SHP1/GST or SHP2/GST. IP, immunoprecipitation; WB, Western blot.
We were also interested in determining if ICSBP coprecipitates equivalently with wild-type, activated, and dominant negative forms of SHP2. Differences in interaction levels might imply additional mechanisms by which SHP2 mutants influence ICSBP tyrosine phosphorylation and therefore activity. For example, the E76K SHP2 mutant might function as a substrate trap, suggesting that dephosphorylation is only one mechanism by which this activated mutant form impacts ICSBP transcriptional activation function. For these studies, we used the U937 stable transfectants discussed in the sections above. In these transfectants, wild-type SHP2, E76K SHP2, and CS463 SHP2 are equivalently expressed (as is demonstrated in Fig. 3A and 7B). As in the experiment described above, nuclear proteins from U937 stable transfectants were immunoprecipitated under denaturing conditions with an antibody to SHP2. Immunoprecipitates were analyzed by Western blotting for interaction with ICSBP. Blots were also probed with antibody to SHP2 to ensure equivalent levels of immunoprecipitation of the wild-type and mutant forms of this protein (Fig. 10B). We found that ICSBP coimmunoprecipitates equivalently with each of these overexpressed forms of SHP2. Less ICSBP coprecipitates with SHP2 from empty vector-transfected cells, consistent with the decreased expression of SHP2 in these cells. Since overexpression of wild-type SHP2, E76K SHP2, and CS463 SHP2 does not alter abundance of the ICSBP protein, these studies indicate that each of these forms of SHP2 interacts with ICSBP. These studies also suggest that interaction of ICSBP with the activated or dominant negative forms of SHP2 is equivalent to the interaction with wild-type protein in the context of overexpression in U937 cells.
To further investigate whether the interaction between ICSBP and SHP2 is direct or indirect, we performed assays with recombinant proteins. For these studies, SHP2 was expressed in E. coli as a fusion protein with glutathione S-transferase (SHP2/GST). SHP1/GST was a positive control and GST a negative control for specific ICSBP interaction in these studies (16). Equivalent amounts of GST proteins were incubated with in vitro-translated, 35S-labeled ICSBP, and proteins were purified by affinity to a glutathione-linked matrix. Coprecipitating ICSBP was identified by SDS-PAGE followed by autoradiography, as previously described (16). We found that ICSBP coprecipitates with both SHP1/GST and SHP2/GST but not with control GST (Fig. 9C). Loading was evaluated by Coomassie staining of the gel to verify affinity purification of the GST proteins (Fig. 10C).
E76K SHP2 dephosphorylates ICSBP in vitro.
The studies described above suggest that ICSBP interacts with SHP2. However, it is possible that ICSBP is dephosphorylated not by SHP2 but by another protein in a multiprotein complex. Therefore, we determined the ability of recombinant SHP2 to dephosphorylate recombinant ICSBP. For these studies, in vitro-translated, 35S-labeled ICSBP was incubated with in vitro-translated (unlabeled) SHP2. In previous studies, we demonstrated that ICSBP is tyrosine phosphorylated when in vitro translated in rabbit reticulocyte lysate (16). In the current studies, the impact of E76K SHP2 on ICSBP tyrosine phosphorylation was compared to the impact of incubation in control reticulocyte lysate. Reaction mixtures were immunoprecipitated under denaturing conditions with antiphosphotyrosine antibody or an irrelevant control antibody. Tyrosine-phosphorylated ICSBP was identified by autoradiography of immunoprecipitates as previously described (16) (Fig. 11A). We found that incubation with E76K SHP2 decreases ICSBP tyrosine phosphorylation in this assay.
FIG. 11.
ICSBP is dephosphorylated in vitro by E76K SHP2. (A) ICSBP is a substrate for SHP2 in vitro. In vitro-translated, [35S]methionine-labeled ICSBP was incubated with in vitro-translated (unlabeled) activated (E76K) SHP2 or control reticulocyte lysate. Incubation reactions were immunoprecipitated under denaturing conditions with an antiphosphotyrosine or an irrelevant control antibody. Immunoprecipitating proteins were separated by SDS-PAGE, and ICSBP was detected by autoradiography. Tyrosine-phosphorylated ICSBP was detected after incubation with control lysate but not with E76K SHP2. Input protein lanes are a control for loading. MSCV, murine stem cell retroviral vector. (B) ICSBP expression in ICSBP−/− bone marrow murine myeloid progenitor cells reconstitutes the positive NF1 cis element-binding protein complex in EMSA. Wild-type and ICSBP−/− murine bone marrow myeloid progenitor cells (cultured with GM-CSF, IL-3, and SCF) were transduced with a murine retroviral vector to express ICSBP (for ICSBP−/− cells) or with an empty vector control (for both wild-type and ICSBP−/− cells). Cells were ex vivo differentiated with M-CSF, and total cell lysates were isolated. Lysates were used in EMSA with the NF1 cis element probe as indicated. Expression of ICSBP in ICSBP−/− cells substantially reconstituted the protein complex binding the probe representing the positive NF1 cis element. The upper arrow represents the ICSBP-containing complex, and the lower arrow represents free probe. (C) Treatment with E76K SHP2 prevents in vitro-translated (IVT) ICSBP from reconstituting the protein complex that binds the NF1 cis element in EMSA with lysate proteins from ICSBP−/− murine myeloid cells. ICSBP−/− murine bone marrow myeloid progenitor cells (cultured with GM-CSF, IL-3, and SCF) were ex vivo differentiated with M-CSF, and total cell lysates were isolated. Wild-type ICSBP and Y92/95F ICSBP were in vitro translated in reticulocyte lysate. These proteins (or control reticulocyte lysate) were preincubated with in vitro-translated E76K SHP2 (or control lysate). In vitro-translated proteins were used in EMSA with the positive NF1 cis element probe with lysate from M-CSF-differentiated ICSBP−/− murine myeloid cells or buffer only as indicated. In vitro-translated wild-type ICSBP, but not Y92/95F ICSBP, reconstitutes protein complex binding to the NF1 cis element probe in EMSA with ICSBP-deficient lysates. Preincubation of in vitro-translated ICSBP with E76K SHP2, but not control reticulocyte lysate, abolishes the ability of ICSBP to reconstitute this complex. In vitro-translated proteins do not generate a specific protein complex with this probe in the absence of lysate proteins from ICSBP−/− cells. The upper arrow represents the ICSBP-containing complex, and the lower arrow represents free probe.
We next investigated the functional significance of E76K SHP2 dephosphorylation of ICSBP for ICSBP binding to the NF1 cis element. In these studies, we determined the effect of in vitro treatment of ICSBP with recombinant E76K SHP2 on ICSBP interaction with the NF1 cis element probe. The goal of these studies was to determine if in vitro phosphatase treatment prevents ICSBP from reconstituting the protein complex which binds the NF1 cis element in EMSA with lysate proteins from ICSBP−/− murine myeloid cells. In initial experiments, we investigated protein complex binding to the NF1 cis element probe in EMSA with lysate proteins from ex vivo M-CSF-differentiated murine myeloid progenitor cells. We compared levels of complex binding in EMSA with lysates from ICSBP−/− cells transduced with empty vector, ICSBP−/− cells transduced with ICSBP expression vector, and wild-type cells (Fig. 11B). In these studies, we found that protein complex binding to the NF1 cis element probe in EMSA with lysate proteins from M-CSF-differentiated ICSBP−/− cells is increased by transduction with an ICSBP expression vector (Fig. 11B).
Therefore, we determined whether in vitro binding of this protein complex to the NF1 cis element probe is increased by including in vitro-translated ICSBP in the binding reaction. For these studies, lysate proteins from M-CSF-differentiated ICSBP−/− murine myeloid cells were incubated with in vitro-translated wild-type or Y92/95F ICSBP (or control reticulocyte lysate). These proteins were used in EMSA with the NF1 cis element probe (Fig. 11C). We found that protein complex binding to the probe was increased by inclusion of wild-type ICSBP, but not Y92/95F ICSBP, in the binding reaction. Therefore, we preincubated in vitro-translated wild-type or Y92/95F ICSBP (or control lysate) with E76K SHP2 (or control lysate) under the conditions used to obtain the results shown in Fig. 11A. EMSA was performed with the NF1 cis element probe and lysate proteins from M-CSF-differentiated ICSBP−/− cells supplemented with wild-type or Y92/95F ICSBP (or control reticulocyte lysate) which was preincubated with E76K SHP2 (or with control reticulocyte lysate). We found that preincubation with E76K SHP2 prevented wild-type ICSBP from reconstituting the NF1 cis element-binding protein complex in EMSA with lysate proteins from ICSBP−/− cells (Fig. 11C). In contrast, preincubation with control reticulocyte lysate did not have any impact on ICSBP binding to the NF1 cis element probe in these experiments.
None of these in vitro-translated proteins significantly bound the NF1 cis element probe in the absence of lysate proteins from ICSBP−/− cells. This result suggests that ICSBP binds the NF1 cis element as part of a multiprotein complex. For all of these experiments, unprogrammed reticulocyte lysate was used to equalize the total amount of in vitro-translated proteins in binding reactions. In vitro-translated proteins that were not incubated with E76K SHP2 were sham incubated in control reticulocyte lysate.
DISCUSSION
In previous studies, we found that ICSBP activates NF1 transcription in differentiating myeloid cells (34). In the current study, we determine that NF1 transcriptional activation requires cytokine-induced phosphorylation of specific tyrosine residues in the ICSBP-IRF domain. These results therefore identify a novel pathway by which hematopoietic cytokines, such as M-CSF and IFN-γ, induce NF1 transcription in myeloid cells. Our current studies also suggest that ICSBP is a substrate for SHP2-PTP in undifferentiated myeloid cells, thereby identifying SHP2-PTP as a negative regulator of this pathway. However, we find that a previously described, leukemia-associated, constitutively activated SHP2 mutant dephosphorylates ICSBP in both undifferentiated and cytokine-differentiated myeloid cells. Consistent with these results, the current studies and previous work by others indicate that the expression of leukemia-associated, activated SHP2 mutants induces cytokine hypersensitivity in myeloid cells and myeloproliferation in murine models, similar to ICSBP or Nf1 deficiency.
Our results suggest that such leukemia-associated SHP2 mutants may not be susceptible to the (unknown) mechanisms which normally regulate PTP activity during myelopoiesis. In our previous studies, we found that SHP1 protein decreases in the nuclear fraction and increases in the cytoplasmic fraction during IFN-γ differentiation of U937 cells (16). Other investigators found that SHP2 translocates to the nucleus in response to prolactin stimulation in mammary epithelial cells in a Stat5-dependent manner (7). In our studies, IFN-γ treatment of U937 cells did not alter SHP2 protein in the nuclear fraction. However, there are other possible mechanisms for SHP2 regulation. For example, cytokine-induced signaling events might decrease protein interaction with the SHP2 SH2 domains, altering conformation and masking the PTP domain. It is possible that cytokine signaling decreases availability of a nonsubstrate protein which interacts with this domain. Or, if phosphorylated tyrosine residues on the substrate are necessary for SH2 domain interaction, these residues might be unavailable due to cytokine-induced interaction with other proteins. This issue is the subject of ongoing investigations in the lab.
Our studies suggest that leukemia-associated, activating SHP2 mutants decrease the impact of hematopoietic cytokines on ICSBP tyrosine phosphorylation. This prevents cytokine-induced Nf1 expression, resulting in unopposed Ras activation in response to these cytokines. Consistent with this, we found that expression of activated SHP2 prevents ICSBP expression from rescuing GM-CSF and M-CSF hypersensitivity in ICSBP-deficient myeloid cells. Also, cytokine hypersensitivity of ICSBP-deficient myeloid cells is not altered by E76K SHP2 expression These studies suggest that SHP2 and ICSBP influence cytokine-induced proliferation through a common pathway. These studies also identify a mechanism by which leukemia-associated SHP2 mutants impact Nf1 expression, thereby contributing to the proliferative/leukemic phenotype. However, SHP2 is likely to impact multiple substrates relevant to proliferation. Therefore, there are likely to be additional genetic lesions that are complemented by activating SHP2 mutants and that contribute to disease progression in myeloid malignancy.
Previous investigations indicate that ICSBP protein abundance is one factor which influences ICSBP target gene regulation. Other studies indicate that posttranslational modification of ICSBP impacts protein-protein and protein-DNA interactions, also impacting target gene transcription. For example, we found that ICSBP tyrosine phosphorylation is important for the determination of lineage- and differentiation-stage-specific CYBB and NCF2 transcription (16). Similarly, our current studies indicate the importance of cytokine-induced ICSBP tyrosine phosphorylation for NF1 transcriptional regulation. Other investigators found that ICSBP represses expression of an artificial promoter construct with multiple copies of the PRDI consensus sequence in U937 transfections (29). This repression is abolished by ICSBP tyrosine phosphorylation during IFN-γ differentiation. Consistent with these results, we found that the PRDI construct is repressed by both wild-type and tyrosine mutant ICSBP in undifferentiated U937 cells but only by the tyrosine mutant form in IFN-γ-treated transfectants.
We previously identified a mechanism by which ICSBP tyrosine phosphorylation contributes to CYBB and NCF2 transcription during myeloid differentiation (16). We found that phosphorylation of ICSBP tyrosines 92 and 95 is necessary for interaction with PU.1 and IRF1 at composite ets/IRF sequences in these genes (11, 16). In the current studies, we find that phosphorylation of the same tyrosine residues increases cytokine-induced ICSBP interaction with the NF1 cis element. Other investigators found that nonphosphorylated ICSBP binds directly to PRDI sequence DNA binding sites. Tyrosine phosphorylation decreases direct ICSBP binding and increases binding as a heterodimer with IRF1 or IRF2, switching the effect of ICSBP from repression to activation (29). The NF1 cis element is similar to both PRDI and ets/IRF consensus sequences. However, ICSBP does not repress the NF1 cis element in undifferentiated U937 cells, an effect unlike that on the PRDI consensus. Defining the mechanism by which ICSBP tyrosine phosphorylation increases interaction with the NF1 cis element is an area of ongoing investigation in the lab.
Therefore, regulation of ICSBP activity is multifactorial, depending on protein abundance and posttranslational modification. Consistent with this, we find that ICSBP overexpression increases NF1 promoter activity in IFN-γ-treated and untreated U937 cells, although the increase is greater in differentiated transfectants. However, we also find that ICSBP-induced NF1 transcription requires specific tyrosine residues in both undifferentiated and differentiated transfectants. This is consistent with our previous observations that overexpression of ICSBP in U937 cells induces some tyrosine phosphorylation, which is increased by IFN-γ differentiation (16). Our experiments with dominant negative forms of SHP1 (16) and SHP2 suggest that the capacity of the endogenous PTPs to dephosphorylate ICSBP is overwhelmed by ICSBP overexpression in undifferentiated U937 cells. Consistent with this, we find that E76K SHP2 expression has minimal effect on Nf1 protein abundance and NF1 promoter activity in undifferentiated U937 cells but a greater impact in the presence of ICSBP overexpression. ICSBP deficiency is part of the malignant phenotype of U937 cells. Given the minimal tyrosine phosphorylation of endogenous ICSBP in U937 cells, PTP activity may not be rate limiting and additional activity might not influence ICSBP function. However, the impact of SHP2 on endogenous ICSBP is suggested by the ability of a dominant negative SHP2 to increase NF1 promoter activity in the absence of ICSBP overexpression in undifferentiated U937 cells.
In contrast to these results, we find that E76K SHP2 expression decreases Nf1 protein abundance in murine bone marrow myeloid progenitor cells. This difference is likely to be due to differences both in the cytokine conditions of the two models and in the levels of abundance of endogenous ICSBP in normal and transformed cells. Additionally, our studies demonstrate that tyrosine phosphorylation is essential for ICSBP-induced Nf1 expression during M-CSF differentiation of these cells. These studies suggest the importance of ICSBP posttranslational modification in both transformed and nontransformed models of myeloid differentiation. This is consistent with the well-documented role of posttranslational modification in regulating transcription factors that are activated by hematopoietic cytokine receptors (such as Stats).
ICSBP regulates at least three aspects of myeloid differentiation. First, ICSBP regulates transcription of genes involved in the inflammatory response. These genes confer important aspects of the phagocyte phenotype, and their expression is restricted to mature myeloid cells. Second, ICSBP decreases BclXL expression in immature myeloid cells (13). ICSBP repression of BclXL expression is decreased in activated phagocytes, enhancing cell survival. Third, ICSBP activates NF1 gene transcription in response to hematopoietic cytokines (34). Increased Nf1 expression inactivates cytokine-activated Ras, resulting in proliferation arrest after the initial proliferative response. Therefore, ICSBP is involved in regulation of cytokine-induced phenotypic differentiation, apoptosis, and proliferation in myeloid progenitor cells.
In mice, SHP1 deficiency accelerates myeloid differentiation and enhances activation of mature phagocytes. Activation of SHP2 leads to myeloid progenitor expansion in mice. These documented effects of SHP1 and SHP2 are not mutually exclusive. Our studies suggest that either PTP could impact myeloid progenitor hypersensitivity (through NF1 regulation) and differentiation progression (through oxidase gene regulation) via an effect on ICSBP tyrosine phosphorylation. ICSBP expression is frequently decreased in myeloid progenitor cells from subjects with various myeloid malignancies, including MDS (27). Activating SHP2 mutants have been associated with disease progression from myelodysplasia to secondary AML (18, 21). Our current studies suggest that the impact of decreased ICSBP abundance on target gene transcription would be augmented by activated SHP2 mutants which block cytokine-induced ICSBP tyrosine phosphorylation. Combining these two leukemia-associated abnormalities would decrease myeloid-specific gene transcription and impair down-regulation of cytokine activated Ras. The net result would be differentiation blockage, cytokine hypersensitivity, and sustained cellular proliferation, all important components of the malignant myeloid phenotype.
REFERENCES
- 1.Ballester, R., D. Marchuk, M. Boguski, A. Saulino, R. Lecher, M. Wigler, and F. Collins. 1990. The NF1 locus encodes a protein functionally related to mammalian GAP and yeast IRA proteins. Cell 63:851-859. [DOI] [PubMed] [Google Scholar]
- 2.Basu, T. N., D. H. Gutmann, J. A. Fletcher, T. W. Glover, F. S. Colling, and J. Downward. 1992. Aberrant regulation of ras proteins in malignant tumor cells from type 1 neurofibromatosis patients. Nature 356:713-715. [DOI] [PubMed] [Google Scholar]
- 3.Bentires-Ali, M., J. G. Paez, F. S. David, H. Keilhack, B. Halmos, K. Naoki, J. M. Maris, A. Richardson, A. Bardelli, D. J. Sugarbaker, W. G. Richards, J. Du, L. Girard, J. D. Minna, M. L. Loh, D. E. Fisher, V. E. Velculescu, B. Vogelstein, M. Meyerson, W. R. Sellers, and B. G. Neel. 2004. Activating mutations of the Noonan syndrome-associated SHP2/PTPN11 gene in human solid tumors and adult acute myelogenous leukemia. Cancer Res. 64:8816-8820. [DOI] [PubMed] [Google Scholar]
- 4.Bollan, G., D. W. Clapp, S. Shih, F. Adler, Y. Y. Zhang, P. Thompson, B. J. Lange, M. H. Freedman, F. McCormick, T. Jacks, and K. Shannon. 1996. Loss of NF1 results in activation of the Ras signaling pathway and leads to aberrant growth in haematopoietic cells. Nat. Genet. 12:144-148. [DOI] [PubMed] [Google Scholar]
- 5.Brannan, C. I., A. S. Perkins, K. S. Vogel, N. Ratner, M. L. Nordlund, S. W. Reid, A. M. Buchberg, N. A. Jenkins, L. F. Parada, and N. G. Copeland. 1994. Targeted disruption of the neurofibromatosis type-1 gene leads to developmental abnormalities in heart and various neural crest-derived tissues. Genes Dev. 8:1019-1029. [DOI] [PubMed] [Google Scholar]
- 6.Chan, R. J., M. B. Leedy, V. Munugalavadla, C. S. Voorhorst, Y. Li, M. Yu, and R. Kapur. 2005. Human somatic PTPN11 mutations induce hematopoietic cell hypersensitivity to granulocyte-macrophage colony stimulating factor. Blood 105:3737-3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chughtai, N., S. Schimchowitsch, J. J. Lebrun, and S. Ali. 2002. Prolactin induces SHP2 association with Stat5, nuclear translocation and binding to the beta casein gene promoter in mammary cells. J. Biol. Chem. 277:31107-31114. [DOI] [PubMed] [Google Scholar]
- 8.DeClue, J. E., A. G. Papageorge, J. A. Fletcher, S. R. Diehl, N. Ratner, W. C. Vass, and D. R. Lowy. 1992. Abnormal regulation of mammalian p21ras contributes to malignant tumor growth in von Recklinghausen (type 1) neurofibromatosis. Cell 69:265-273. [DOI] [PubMed] [Google Scholar]
- 9.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong, Q., K. A. Siminovitch, L. Fialkow, T. Fukushima, and G. P. Downey. 1999. Negative regulation of myeloid cell proliferation and function by the SH2 domain-containing tyrosine phosphatase-1. J. Immunol. 162:3220-3230. [PubMed] [Google Scholar]
- 11.Eklund, E. A., A. Jalava, and R. Kakar. 1998. PU.1, interferon regulatory factor 1, and interferon consensus sequence-binding protein cooperate to increase gp91phox expression. J. Biol. Chem. 273:13957-13965. [DOI] [PubMed] [Google Scholar]
- 12.Fragale, A., M. Tartaglia, J. Wu, and B. D. Gelb. 2004. Noonan syndrome-associated SHP2/PTPN11 mutants cause EGF-dependent prolonged GAB1 binding and sustained ERK2/MAPK1 activation. Hum. Mutat. 23:267-277. [DOI] [PubMed] [Google Scholar]
- 13.Gabriele, L., J. Phung, J. Fukumoto, D. Segal, I. M. Wang, P. Giannakakou, N. A. Giese, K. Ozato, and H. C. Morse. 1999. Regulation of apoptosis in myeloid cells by interferon consensus sequence-binding protein. J. Exp. Med. 190:411-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holtchke, T., J. Lohler, Y. Kanno, T. Fehr, N. Giese, F. Rosenbauer, J. Lou, K. P. Knobeloch, L. Gabriele, J. F. Waring, M. F. Bachmann, R. M. Zinkernagel, H. C. Morse, K. Ozato, and I. Horak. 1996. Immunodeficiency and chronic myelogenous leukemia-like syndrome in mice with a targeted mutation of the ICSBP gene. Cell 87:307-317. [DOI] [PubMed] [Google Scholar]
- 15.Kakar, R., B. Kautz, and E. A. Eklund. 2005. JAK2 is necessary and sufficient for interferon-gamma-induced transcription of the gene encoding gp91PHOX. J. Leukoc. Biol. 77:120-127. [DOI] [PubMed] [Google Scholar]
- 16.Kautz, B., R. Kakar, E. David, and E. A. Eklund. 2001. SHP1 protein-tyrosine phosphatase inhibits gp91PHOX and p67PHOX expression by inhibiting interaction of PU.1, IRF1, interferon consensus sequence-binding protein, and CREB-binding protein with homologous cis elements in the CYBB and NCF2 genes. J. Biol. Chem. 276:37868-37878. [DOI] [PubMed] [Google Scholar]
- 17.Keilhack, H., F. S. David, M. McGregor, L. C. Cantley, and B. G. Neel. 2005. Diverse biochemical properties of Shp2 mutants. Implications for disease phenotypes. J. Biol. Chem. 280:30984-30993. [DOI] [PubMed] [Google Scholar]
- 18.Largaespada, D. A., C. I. Brannan, N. A. Jenkins, and N. G. Copeland. 1996. Nf1 deficiency causes Ras-mediated granulocyte/macrophage colony stimulating factor hypersensitivity and chronic myeloid leukaemia. Nat. Genet. 12:137-143. [DOI] [PubMed] [Google Scholar]
- 19.Larrick, J. W., S. J. Anderson, and H. S. Koren. 1980. Characterization of a human macrophage-like cell line stimulated in vitro: a model of macrophage functions. J. Immunol. 125:6-14. [PubMed] [Google Scholar]
- 20.Loh, M. L., S. Vattikuti, S. Schubbert, M. G. Reynolds, E. Carlson, K. H. Lieuw, J. W. Cheng, C. M. Lee, D. Stokoe, J. M. Bonifas, N. P. Curtiss, J. Gotlib, S. Meshinchi, M. M. Le Beau, P. D. Emanuel, and K. M. Shannon. 2004. Mutations in PTPN11 implicate the SHP-2 phosphatase in leukemogenesis. Blood 103:2325-2331. [DOI] [PubMed] [Google Scholar]
- 21.Lu, D., R. Nounou, M. Beran, E. Estey, T. Manshouri, H. Kantarjian, M. J. Keating, and M. Albitar. 2003. The prognostic significance of bone marrow levels of neurofibromatosis-1 protein and ras oncogene mutations in patients with acute myeloid leukemia and myelodysplastic syndrome. Cancer 97:441-449. [DOI] [PubMed] [Google Scholar]
- 22.Mohi, M. G., I. R. Williams, C. R. Dearolf, G. Chan, J. L. Kutok, S. Cohen, K. Morgan, C. Boulton, H. Shigematsu, H. Keilhack, K. Akashi, D. G. Gilliland, and B. G. Neel. 2005. Prognostic, therapeutic, and mechanistic implications of a mouse model of leukemia evoked by Shp2 (PTPN11) mutations. Cancer Cell 7:179-191. [DOI] [PubMed] [Google Scholar]
- 23.Nelson, N., M. S. Marks, P. H. Driggers, and K. Ozato. 1993. Interferon consensus sequence-binding protein, a member of the interferon regulatory factor family, suppresses interferon-induced gene transcription. Mol. Cell. Biol. 13:588-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Reilly, A. M., S. Pluskey, S. E. Shoelson, and B. G. Neel. 2000. Activated mutants of SHP-2 preferentially induce elongation of Xenopus animal caps. Mol. Cell. Biol. 20:299-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rehli, M., A. Poltorak, L. Schwarzfischer, S. W. Krause, R. Andreesen, and B. Beutler. 2000. PU.1 and interferon consensus sequence-binding protein regulate the myeloid expression of the human Toll-like receptor 4 gene. J. Biol. Chem. 275:9773-9781. [DOI] [PubMed] [Google Scholar]
- 26.Scheller, M., J. Foerster, C. M. Heyworth, J. F. Waring, J. Lohler, G. L. Gilmore, R. K. Shadduck, T. M. Dexter, and I. Horak. 1999. Altered development and cytokine responses of myeloid progenitors in the absence of transcription factor, interferon consensus sequence binding protein. Blood 94:3764-3771. [PubMed] [Google Scholar]
- 27.Schmidt, M., S. Nagel, J. Proba, C. Theide, M. Ritter, J. F. Waring, F. Rosenbauer, D. Huhn, B. Wittig, I. Horak, and A. Neubauer. 1998. Lack of interferon consensus sequence binding protein (ICSBP) transcripts in human myeloid leukemias. Blood 91:22-29. [PubMed] [Google Scholar]
- 28.Schubbert, S., K. Lieuw, S. L. Rowe, C. M. Lee, X. Li, M. L. Loh, D. W. Clapp, and K. M. Shannon. 2005. Functional analysis of leukemia-associated PTPN11 mutations in primary hematopoietic cells. Blood 106:311-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharf, R., D. Meraro, A. Azriel, A. M. Thornton, K. Ozato, E. F. Petricoin, A. C. Larner, F. Schaper, H. Hauser, and B. Z. Levi. 1997. Phosphorylation events modulate the ability of interferon consensus sequence binding protein to interact with interferon regulatory factors and to bind DNA. J. Biol. Chem. 272:9785-9792. [DOI] [PubMed] [Google Scholar]
- 30.Tartaglia, M., C. M. Neimeyer, A. Fragale, X. Song, J. Buechner, A. Jung, K. Hahlen, H. Hasle, J. D. Licht, and B. D. Gelb. 2003. Somatic mutations in PTPN11 in juvenile myelomonocytic leukemia, myelodysplastic syndromes and acute myeloid leukemia. Nat. Genet. 34:148-150. [DOI] [PubMed] [Google Scholar]
- 31.Wang, I. M., C. Contursi, A. Masumi, X. Ma, G. Trinchieri, and K. Ozato. 2000. An IFN-gamma-inducible transcription factor, IFN consensus sequence binding protein (ICSBP), stimulates IL-12 p40 expression in macrophages. J. Immunol. 165:271-279. [DOI] [PubMed] [Google Scholar]
- 32.Weinmann, A. S., S. M. Bartley, T. Zhang, M. Q. Zhang, and P. J. Farnham. 2001. Use of chromatin immunoprecipitation to clone novel E2F target promoters. Mol. Cell. Biol. 21:6820-6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang, Y., T. A. Vik, J. W. Ryder, E. F. Srour, T. Jacks, K. Shannon, and D. W. Clapp. 1998. Nf1 regulates hematopoietic progenitor cell growth and ras signaling in response to multiple cytokines. J. Exp. Med. 187:1893-1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu, C., G. Saberwal, Y. F. Ly, L. C. Platanias, and E. A. Eklund. 2004. The interferon consensus sequence-binding protein activates transcription of the gene encoding neurofibromin 1. J. Biol. Chem. 279:50874-50885. [DOI] [PubMed] [Google Scholar]