Abstract
HER4 expression in human breast cancers correlates with a positive prognosis. While heregulin inhibits the growth of HER4-positive breast cancer cells, it does so by undefined mechanisms. We demonstrate that heregulin-induced HER4 activity inhibits cell proliferation and delays G2/M progression of breast cancer cells. While investigating pathways of G2/M delay, we noted that heregulin increased the expression of BRCA1 in a HER4-dependent, HER2-independent manner. Induction of BRCA1 by HER4 occurred independently of the cell cycle. Moreover, BRCA1 expression was elevated in HER4-postive human breast cancer specimens. Heregulin stimulated c-Jun N-terminal kinase (JNK), and pharmacologic inhibition of JNK impaired heregulin-enhanced expression of BRCA1 and mitotic delay; inhibition of Erk1/2 did not. Knockdown of BRCA1 with small interfering RNA in a human breast cancer cell line interfered with HER4-mediated mitotic delay. Heregulin/HER4-dependent mitotic delay was examined further with an isogenic pair of mouse mammary epithelial cells (MECs) derived from mice harboring homozygous LoxP sites flanking exon 11 of BRCA1, such that one cell line expressed BRCA1 while the other cell line, after Cre-mediated excision, did not. BRCA1-positive MECs displayed heregulin-dependent mitotic delay; however, the isogenic BRCA1-negative MECs did not. These results suggest that heregulin-mediated growth inhibition in HER4-postive breast cancer cells requires BRCA1.
Cell proliferation, differentiation, and survival are highly coordinated processes during the development and maturation of the mammary gland, and control of these mechanisms is critical for the prevention of breast cancers (reviewed in reference 39). Aberrant regulation of the HER/ErbB family of receptor tyrosine kinases (RTKs) and their ligands is a common occurrence in many human cancers, including breast cancer (14, 15, 45). This family consists of four related members, HER1/ErbB1/EGFR (epidermal growth factor receptor), HER2/ErbB2/Neu, HER3/ErbB3, and HER4/ErbB4. Each protein is comprised of a large amino-terminal extracellular domain, a transmembrane domain, and a large intracellular domain with a tyrosine-rich carboxy-terminal region and a tyrosine kinase-like sequence (27, 33, 56). The tyrosine kinase activity of the ErbBs is induced upon ligand interaction, leading to receptor dimerization (homo- and heterodimerization) and subsequent receptor transphosphorylation. Although HER2 is the preferential heterodimeric partner, HER2 does not bind any conventional ligand within the two major families of ErbB ligands (EGF-like ligands and heregulin [HRG]/neuregulin-like ligands) and therefore relies on HER1, HER3, or HER4 for activation of its tyrosine kinase activity.
The well-documented growth stimulatory effects of HER1 and HER2 have driven the investigation of ErbB signaling in breast cancer. HER1 is expressed in nearly all human carcinomas, including breast cancers, while nearly 20 to 25% of breast cancers overexpress HER2 and/or exhibit gene amplification at the her2 locus (26, 37, 40, 48). Expression of either HER1 or HER2 in tumor specimens correlates with a shorter survival period, a higher grade of malignancy, and an overall poor prognosis (10, 12-14, 19, 24). Genetically engineered animal models of breast cancer confirm the role of HER1 and HER2 in driving proliferation of the mammary epithelium (reviewed in reference 39). Recent evidence suggests that increased expression of HER3 in breast cancers also correlates with a poor prognosis. HER3 is overexpressed in about 20% of all breast cancers and is frequently coexpressed with HER2 (2, 5, 10, 23, 31, 52, 53). This has generated the hypothesis that HER2/HER3 heterodimers may function to simultaneously drive cellular proliferation and survival in breast cancer cells.
In contrast, there is evidence that HER4 expression correlates with a more differentiated tumor grade, longer survival, and positive prognostic indicators, such as estrogen receptor expression (1, 17, 29, 38, 41, 44). Women whose breast tumors express HER4 exhibit the lowest risk of death due to cancer compared to women whose tumors express HER1, HER2, or HER3. During breast development, HER4 expression and activity (measured by tyrosine phosphorylation) are lowest during phases of epithelial cell proliferation and highest during phases of differentiation (35). Mammary glands from mice that lack HER4 activity, either by Cre-Lox technology, cardiac-specific transgene rescue of a HER4 knockout (with the mammary epithelium therefore remaining ErbB4 deficient), or mammary-specific expression of a dominant-negative HER4 mutant, have lactational defects due to an impaired program of differentiation (16, 20, 43). Decreased bioavailability of HB-EGF (a ligand for both HER1 and HER4) in the mammary epithelium causes a similar lactational deficiency (57). Taken together, the evidence suggests that HER4 signaling may impair cellular proliferation or promote differentiation of breast epithelium. In support of this hypothesis, it was reported that heregulin-dependent growth inhibition occurs in HER4-positive breast cancer-derived cell lines but not in HER4-negative cells (34).
We investigated the mechanism by which heregulin mediates growth inhibition in a HER4-dependent manner, using human breast cancer cell lines and BRCA1-deficient mouse mammary epithelial cells (MECs). The data demonstrate that heregulin induces a cell cycle delay in the early phases of mitosis and increases expression of the tumor suppressor protein BRCA1. Mitotic delay in response to HER4 requires the expression of BRCA1.
MATERIALS AND METHODS
Cell culture and transfections.
SUM102 cells were derived from a microinvasive primary breast tumor, whereas SUM44 cells were derived from a metastatic pleural effusion. SUM44, SUM44-pBABE, SUM44-5R, SUM102, SUM102-pLXSN, SUM102-HER4, SUM102-HER4-pBABE, and SUM102-HER4-5R cells were grown in serum-free, growth factor-defined media as previously described (34). BT-474 and MCF-7 cells were cultured in 5% CO2 in growth medium (Dulbecco's modified Eagle's medium/F12 [Gibco Life Sciences] supplemented with 10% fetal bovine serum [FBS], EGF [10 ng/ml; Invitrogen], and insulin [5 μg/ml; Sigma-Aldrich, St. Louis, MO]) with hydrocortisone (1 μg/ml; Sigma-Aldrich) unless otherwise indicated. SKBr3 cells were cultured in McCoy's 5A medium supplemented with 15% FBS. HeLa cells were transfected with empty pLXSN or pLXSN-HER4 (34) and then selected with 250 μg/ml G418. BT-474 and HeLa cells were transfected with pcDNA3-BRCA1 and pcDNA3-BRCA1S1423A (a gift from Michael Kastan, St. Jude Children's Research Hospital, Memphis, TN) and then selected with 400 μg/ml G418. BRCA1+ and BRCA1− mouse MECs have been described previously (36) and were cultured in Dulbecco's modified Eagle's medium/F12 (Gibco Life Sciences) supplemented with 10% FBS. All cells were grown in a humidified incubator at 37°C with 5% CO2. Where indicated, cells were treated with HRGβ1 (10 ng/ml; a gift from Genentech), the ErbB inhibitor GW572016/lapatinib (5 μM; GlaxoSmithKline, Research Triangle Park, NC), hydroxyurea (10 μM; Sigma-Aldrich), or olomoucine (10 μg/ml; Promega, Madison, WI) according to the manufacturer's directions. Small interfering RNA (siRNA) sequences targeting BRCA1 or nonspecific siRNA sequences were purchased from Santa Cruz Biotechnologies and were transiently transfected into cells in serum-free medium for 4 h, according to the manufacturer's directions, using an siRNA transfection reagent (Santa Cruz Biotechnologies). The medium was changed after 4 h to serum-free medium plus insulin and hydrocortisone in the presence or absence of HRG. Cell lysates were harvested 30 h after transfection.
Quantitative reverse transcription-PCR (RT-PCR).
Gene-specific 5′-3′ oligonucleotides and intervening fluorescent dye-labeled probes for human genes encoding HER4, heregulin, BRCA1, keratin 18, and amphiregulin were designed using Primer Express software (ABI/Perkin-Elmer). The nonextendable probes were synthesized, labeled with 5′ 6-carboxyfluorescein reporter and 3′ 6-carboxytetramethylrhodamine quencher dyes (Integrated DNA Technologies), and purified by high-performance liquid chromatography. Real-time fluorescence quantitative PCR (qPCR) was performed with an ABI PRISM 7900 instrument (PE Bio). mRNA sequences for each gene were transcribed in vitro, using MEGAscript (Ambion), and used as positive controls and absolute quantitation standards for the assays. Amplification of twofold serial dilutions of RNA was used to construct standard linear curves that permitted accurate measurements of 200 to 90 million template copies. Total RNA was isolated from each cell line or from human tumor specimens by using an RNeasy kit (QIAGEN) and was treated with RNase-free DNase (Ambion). Total RNA (10 ng) isolated from each cell line was assayed. De-identified human tumor specimens were obtained from the Tissue Procurement Core Facility, UNC-Lineberger Comprehensive Cancer Center. The tumor identification numbers listed in Table 1 cannot be linked to clinical identifiers for this anonymous donor collection.
TABLE 1.
Quantitation of HER4 and BRCA1 transcripts by real-time RT-PCRa
| Tumor no. | Avg no. of transcripts
|
Presence of HRG | |
|---|---|---|---|
| HER4 | BRCA1 | ||
| 1 | 50 | 817 | − |
| 2 | 330 | 301 | + |
| 3 | 470 | 3,411 | − |
| 4 | 960 | 821 | − |
| 5 | 1,220 | 891 | + |
| 6 | 1,450 | 1,882 | − |
| 7 | 2,710 | 1,087 | + |
| 8 | 8,450 | 1,770 | + |
| 9 | 8,770 | 2,617 | − |
| 10 | 12,180 | 1,603 | − |
| 11 | 16,570 | 2,427 | + |
| 12 | 20,860 | 6,207 | + |
| 13 | 22,960 | 11,126 | + |
| 14 | 25,950 | 2,318 | − |
| 15 | 49,330 | 6,447 | − |
| 16 | 50,910 | 4,583 | + |
| 17 | 57,820 | 4,714 | + |
| 18 | 77,930 | 8,922 | + |
| 19 | 95,860 | 4,934 | + |
Breast cancer specimens were used to measure absolute levels of HER4 and BRCA1 mRNAs by qPCR. Each sample was independently analyzed three times. qPCR was used to detect the presence (+) or absence (−) of HRG in each of the breast tumor specimens.
Cell growth and TUNEL assays.
Cells were plated in 96-well plates at a density of 0.5 × 104 cells per well and grown in serum-free medium plus insulin and hydrocortisone, as described above, with or without recombinant heregulin β1 for 48 h. Relative cell numbers were analyzed using a 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assay (Promega, Madison, WI) according to the manufacturer's directions. For terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) analysis (for in situ detection of apoptotic cells), cells were cultured with or without HRG for 48 h or in the presence of paclitaxel (Taxol; 50 nM) for the final 16 h of culture. Cells were fixed in methanol and then analyzed using a DeadEnd colorimetric TUNEL analysis kit (Promega, Madison, WI).
Immunoprecipitation and Western analysis.
Whole-cell extracts were prepared by washing cells with cold phosphate-buffered saline and lysing them in cold NLB (20 mM HEPES [pH 7.3], 50 mM sodium fluoride, 10% glycerol, 1% Triton X-100, 5 mM EDTA, 0.5 M NaCl, 1 mM Na3VO4, aprotinin [6 μg/ml], and leupeptin [10 μg/ml]). Receptor proteins were precipitated with a purchased antibody against HER2 (clone 9G6.10; Neomarkers, Inc.) and with antibodies against HER1 (V22), HER3 (1511), and HER4 (132), each produced in our lab and described previously (34). Immunocomplexes were pulled down with protein A/G- or protein A-agarose (Santa Cruz) for 3 h at 4°C, washed three times with NLB, and analyzed by immunoblotting using an antiphosphotyrosine antibody (PY20-HRP; Santa Cruz Biotechnologies), as previously described (34). Whole-cell lysates were analyzed directly by immunoblotting using antibodies against the following proteins: cyclin D1, cyclin A, cyclin B1, total cdc2, and BRCA, obtained from Santa Cruz Biotechnologies; HER4 (clone HFR) and STAT5A, obtained from Neomarkers Lab Vision; and phosphoserine pRb, phosphotyrosine 15 cdc2, phospho-serine 10 histone H3, phospho- and total JNK, phospho- and total p44/42, and phospho- and total p38, obtained from Cell Signaling.
Cell cycle analysis.
Cells growing in serum-free medium supplemented with insulin and hydrocortisone were harvested by trypsinization, fixed in ice-cold methanol, labeled with 50 μg/ml of propidium iodide (Sigma-Aldrich), and treated with RNase A. A total of 10,000 stained nuclei per sample was analyzed in a FACSCalibur flow cytometer (Becton Dickinson). Where indicated, fixed cells were stained with anti-phospho-Ser 10 histone H3 antibody (Cell Signaling), washed, and incubated with a fluorescein isothiocyanate (FITC)-conjugated anti-rabbit secondary antibody (Molecular Probes, Invitrogen) prior to fluorescence-activated cell sorting (FACS) analysis. Where indicated, cells were labeled with bromodeoxyuridine (BrdU; 10 mg/ml) for 6 h prior to cell fixation. DNA in BrdU-labeled cells was denatured in 2 M HCl for 30 min, and then cells were stained with FITC-conjugated anti-BrdU antibody (Becton Dickinson) prior to analysis. DNA histograms were modeled using Modfit-LT software (Verity).
In vitro cdc2 kinase activity.
Total cdc2 was immunoprecipitated from whole-cell extracts, washed in ice-cold phosphate-buffered saline, equilibrated in ice-cold kinase buffer (50 mM HEPES, pH 7.5, 10 mM MgCl2, 2.5 mM EGTA, 1 mM dithiothreitol, 0.1 mM NaF, 0.1 mM Na3VO4, 1 μM ATP) for 20 min, and resuspended in 20 μl of kinase buffer. cdc2 immunoprecipitates were assayed for kinase activity against 0.1 μg histone H1 (Roche Applied Science). Kinase reactions were performed in a final volume of 30 μl in the presence of 0.5 μM biotinylated ATP (Promega, Madison, WI) for 45 min at 30°C. The entire reaction volume was electrophoresed in an 8% polyacrylamide gel at 60 mA for 3 h at 4°C, transferred to a polyvinylidene difluoride membrane, and then analyzed by immunoblotting using a horseradish peroxidase-conjugated anti-biotin antibody (Santa Cruz Biotechnologies).
RESULTS
Heregulin decreases growth of HER4-positive breast cancer cells.
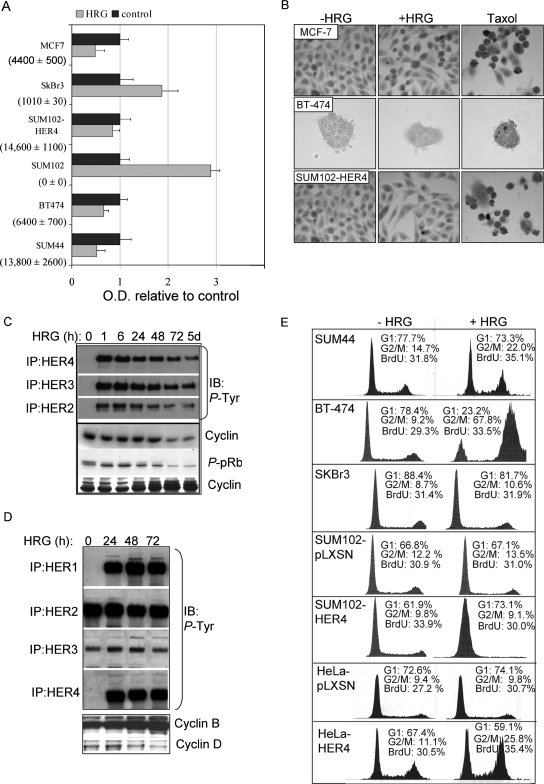
A panel of human breast cancer cells was examined by real-time quantitative RT-PCR to measure the absolute level of HER4 mRNA in each cell line. Three lines (SUM44, BT-474, and MCF-7) expressed >4,000 copies of HER4 mRNA (per 10 ng total RNA), while two lines (SUM102 and SkBR-3) expressed <1,000 copies (Fig. 1A). In fact, SUM102 cells contained no detectable HER4 transcripts, consistent with previous results (34). Western analysis of SUM102 and SKBr3 cells did not reveal any HER4 protein expression (data not shown). SUM102-HER4 cells, stably transfected with pLXSN-HER4 encoding full-length human HER4 (Jma/Cyt-1 isoform), expressed >14,000 HER4 transcripts.
FIG. 1.
HRG-mediated growth inhibition requires HER4 expression. (A) Absolute levels of HER4 mRNA transcripts were measured by quantitative real-time RT-PCR and are shown for each cell line in parentheses (average number of transcripts ± standard deviation [SD]; n = 3). Equal numbers of cells from each cell line were plated and cultured for 48 h with or without HRG; relative cell numbers were determined by using the MTS assay, which converts cell density to optical density (OD). The values represent the average OD ± SD, represented as a ratio to the OD produced by cells grown in the absence of HRG. Each experiment was performed in triplicate and repeated three times. (B) In situ TUNEL analysis of HRG-treated MCF-7, BT-474, and SUM44 cells treated with or without HRG for 0 to 48 h. (C and D) Western analysis of whole-cell extracts or HER1, HER2, HER3, and HER4 immunoprecipitates from SUM44 cells (C) or BT-474 cells (D), treated with HRG for the indicated time courses, to detect the expression of cyclin D, cyclin B, or phospho-pRb or phosphotyrosine residues. (E) Cell cycle analysis of cells cultured in serum-free medium with or without HRG for 72 h. The percentages of the cell population in the G1 and G2/M phases of the cell cycle are shown. In addition, cells were labeled with BrdU for 6 h, stained with an FITC-conjugated anti-BrdU antibody, and analyzed by flow cytometry. The percentages of BrdU-positive cells are indicated. A total of >10,000 nuclei per sample were counted (n = 3).
It was previously reported that HRG-mediated growth inhibition requires HER4 expression. To confirm and extend these findings, cells were plated in equal numbers (0.5 × 106 cells), and relative cell numbers were determined after 48 h of incubation with or without HRG (Fig. 1A). HER4-positive cells grown in the presence of HRG displayed a decreased cell number compared to cells grown in the absence of HRG. In contrast, HER4-negative cells displayed a greater number of cells after 48 h in the presence of HRG than did untreated control cells. These results are consistent with previous findings suggesting that HRG-mediated growth inhibition is a HER4-dependent process. Although some reports suggest that HER4 expression may stimulate the growth of various cell types, it is possible that the effect of HER4 on growth may be modulated by variables including the splice variant of HER4 (there are four naturally occurring splice variants), the cell type, the complement of ErbB receptors within the cell, and the cell type. There are four HER4 isoforms, which are thought to result from alternative splicing. HER4-Jma harbors an ADAM17/TACE cleavage site in the extracellular juxtamembrane domain that is absent from HER4-JMb. HER4-Cyt1 harbors a 16-amino-acid sequence in the cytoplasmic domain that binds to phosphatidylinositol 3-kinase and the transcription factor YAP. This region is absent from HER4-Cyt2. We have determined that SUM44, BT-474, and SUM102-HER4 cells express the Jma-Cyt1 isoform of HER4.
Heregulin results in accumulation of HER4-positive cells in G2 or M phase.
Cell growth is a balance between cell cycle progression and cell death. However, using in situ TUNEL analysis to detect apoptotic cells, we did not observe increased apoptosis in HRG-treated breast cancer cells at any time point observed, regardless of HER4 expression (Fig. 1B). As a positive control, 50 nM paclitaxel induced apoptotic cell death in each cell line examined. Next, we examined the effects of HRG on the cell cycle of SUM44 cells, using Western analysis to detect molecular markers of each cell cycle phase (Fig. 1C). HRG resulted in prolonged tyrosine phosphorylation of HER2, -3, and -4 in SUM44 cells over 5 days of treatment with HRG (Fig. 1C). SUM44 cells do not express HER1 at the RNA or protein level (34). Decreased expression of cyclin D1 was observed within 24 to 72 h of HRG treatment, suggesting a decreased percentage of cells in G1/S phase, a supposition reinforced by decreased phosphorylation of the retinoblastoma gene product, pRb. By 24 h, increased protein expression of cyclin B1 was observed in HRG-treated cells, and it continued to increase throughout the 5-day period. It is known that cyclin B1 expression begins to increase in late S phase and remains elevated until its degradation in mitosis (anaphase) (reviewed in reference 30). Therefore, our data suggest that HRG-dependent HER4 signaling may increase in the proportions of cells in either G2 or M phase.
We examined the effects of HRG on the expression of cyclin D and cyclin B in another HER4-positive cell line, BT-474. BT-474 cells differ from SUM44 cells in that BT-474 cells express HER1 and overexpress HER2. Vast overexpression of HER2 in BT-474 cells may explain the high basal levels of HER2 and HER3 tyrosine phosphorylation observed (Fig. 1D). HRG induced the tyrosine phosphorylation of HER4 and HER1 (presumably via heterodimerization) through 72 h. Similar to the case in SUM44 cells, the cyclin B level increased and remained elevated in HRG-treated BT-474 cells, while cyclin D expression decreased.
Cell cycle analysis of propidium iodide-stained cells demonstrated that HRG increased the proportion of 4N DNA (4N)-containing cells among SUM44 and BT-474 cells (Fig. 1E). However, HRG did not significantly alter the number of cells undergoing DNA synthesis, as determined by analyzing the percentage of cells that incorporated BrdU over a 6-hour labeling period (>10,000 cells; n = 3) (Fig. 1E). This confirms that HRG delays progression through G2 or M in these two HER4-positive cell lines. The response of BT-474 cells to HRG was particularly pronounced. HeLa cells stably transfected with HER4 also displayed an increase in the G2/M population compared to that in HeLa cells expressing the empty pLXSN vector. SkBR-3 and SUM102-pLXSN cells did not display any HRG-dependent alterations of the 4N-containing population of cells, consistent with the observation that HRG did not inhibit the growth of these HER4-negative cell lines (33). However, while HRG-mediated growth inhibition of SUM102-HER4 cells was observed (Fig. 1A), HRG increased the proportion of 2N- rather than 4N-containing cells in this particular cell line. Therefore, HRG-mediated growth inhibition in SUM102-HER4 cells occurs by a different mechanism than that in SUM44 or BT-474 cells.
Mitotic delay in response to heregulin.
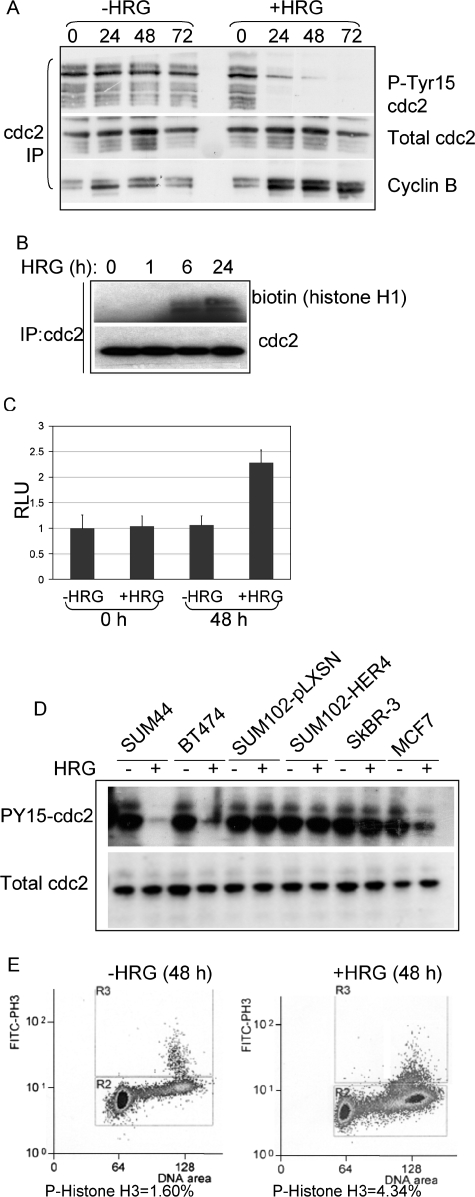
Because cells in the G2 phase of the cell cycle and cells undergoing mitosis each contain 4N DNA, we were unable to determine in which phase HRG-treated SUM44 and BT-474 cells were delayed. Therefore, we examined the activity of cdc2, a kinase that is acti vated at the G2/M transition. In this process, cdc2 interacts with its catalytic partner, cyclin B1, becomes dephosphor ylated at tyrosine 15, and becomes kinase active. Total cdc2 was immunoprecipitated from whole-cell extracts of SUM44 cells cultured in the presence or absence of HRG for 72 h. Immunoprecipitates were analyzed by Western analysis, revealing an HRG-dependent decrease in inhibitory Tyr15 phosphorylation of cdc2 and increased cyclin B coprecipitation with cdc2 (Fig. 2A). In contrast, SUM44 cells grown without HRG did not display any alterations in the level of cdc2 tyrosine 15 phosphorylation or in cdc2-cyclin B coprecipitation (Fig. 2A). Immunoprecipitates of cdc2 were used for an in vitro kinase assay, with a histone H1 peptide as the substrate. Although equal amounts of cdc2 were immunoprecipitated from SUM44 cells cultured with and without HRG at each time point, we found enhanced cdc2 kinase activity in cells treated with HRG for at least 24 h (Fig. 2B). Similarly, a quantitative, luciferase-based assay detecting in vitro cdc2 kinase activity demonstrated increased kinase activity in cdc2 immunoprecipitates from SUM44 cells treated for 48 h with HRG (Fig. 2C).
FIG. 2.
HRG delays progression through mitosis. (A) Western analysis of cdc2 immunoprecipitates from whole-cell extracts of SUM44 cells cultured with or without HRG for 0 to 72 h, detecting total cdc2, phosphotyrosine 15 cdc2, or coprecipitation of cyclin B. (B) In vitro kinase activity of cdc2 immunoprecipitates from SUM44 cells cultured in the presence of HRG for 0 to 24 h. cdc2 immunoprecipitates were divided. One-half was analyzed by Western analysis for total levels of cdc2; the other half was used in the cdc2 kinase assay, directing phosphorylation against a fragment of histone H1. (C) Quantification of a luciferase-linked in vitro cdc2 kinase assay (Promega), with luciferase activity measured in relative light units (RLU). Cells were cultured for 0 or 48 h with or without HRG, and cdc2 was immunoprecipitated from 500 μg whole-cell extract to be used directly in each reaction. Results are presented as the average relative light units per sample ± SD (n = 3). (D) Western analysis to detect phosphotyrosine 15 cdc2 and total cdc2 in cdc2 immunoprecipitates from a series of cell lines cultured with or without HRG for 48 h. (E) FACS analysis of BT-474 cells treated with or without HRG for 0 or 48 h. Methanol-fixed cells were stained with an antibody against phospho-histone H3 (and an FITC-labeled secondary antibody) and with propidium iodide. A total of >20,000 cells were analyzed by flow cytometry. The percentage of cells that were positive for FITC is shown below each panel.
To determine if HRG increased cdc2 activity in other HER4-positive breast cancer cell lines, we examined phospho-tyrosine 15 cdc2 expression as a surrogate marker of cdc2 activity. HER4-positive SUM44, BT-474, and MCF-7 cells each displayed decreased cdc2 Tyr15 phosphorylation, while HER4-negative cells (SUM102 and SkBR-3) did not (Fig. 2D). Interestingly, SUM102-HER4 cells did not display HRG-dependent alterations in phospho-cdc2 levels, consistent with flow cytometry results showing no HRG-dependent accumulation of SUM102-HER4 cells with 4N DNA (Fig. 1).
Phosphorylation of histone H3 at serine 10 occurs specifically during mitosis. We observed a substantial increase in Ser10 phosphorylation of histone H3 in SUM44 cells treated with HRG for 48 h by FACS analysis of cells stained with an antibody against phospho-Ser 10 histone H3 (Fig. 2E). This suggests that HRG-treated SUM44 cells may accumulate in early mitosis.
Increased expression of BRCA1 in heregulin-treated cells requires HER4 but not HER2.
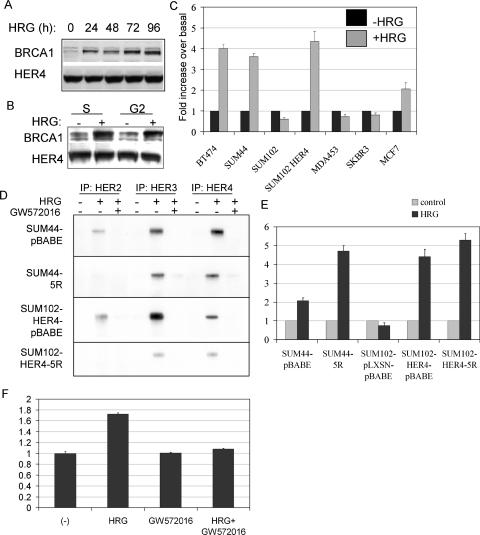
We examined the expression of several regulators of progression through the G2 and M phases in HRG-treated cells. While the expression of many proteins was unaltered by HRG (CHK1, CHK2, p53, and AuroraA) (data not shown), we found increased expression of BRCA1 within 24 h in HRG-treated BT-474 cells; elevated BRCA1 expression was sustained through 96 h (Fig. 3A). Because BRCA1 mRNA levels are regulated in a cell cycle-dependent manner, we determined if changes in BRCA1 expression were secondary to the HRG-induced cell cycle changes or if they occurred independently of the cell cycle. We treated BT-474 cells with or without HRG in the presence of hydroxyurea (to synchronize cells in S phase) or olomoucine (to synchronize cells at the G2/M boundary). We found increased expression of BRCA1 in HRG-treated synchronized cells compared to that in synchronized cells that were not treated with HRG (Fig. 3B), suggesting that BRCA1 induction by HRG occurred independently of the cell cycle.
FIG. 3.
HRG results in increased expression of BRCA1 in HER4-positive cells. (A) Western analysis of whole-cell extracts from BT-474 cells cultured with HRG for 0 to 96 h, detecting BRCA1 and HER4. (B) BT-474 cells were cultured with or without HRG for 48 h in the presence of hydroxyurea (to synchronize cells in S phase) or olomoucine (a cdc2/cdk1 inhibitor that synchronizes cells in late G2). Whole-cell lysates were analyzed by Western analysis to detect BRCA1 or HER4. (C) Expression of BRCA1 mRNA was measured using real-time quantitative RT-PCR. Each cell line was grown in serum-free medium plus insulin and hydrocortisone, with or without HRG, for 24 h. The BRCA1 transcript level in total RNA (10 ng) was assessed by qPCR as described in Materials and Methods. Comparison to a BRCA1 standard allowed quantitation of transcript numbers. The average number (± SD) is represented as a ratio to that in untreated controls, assigned the value of 1 (n = 3). (D) Western analysis of HER2, HER3, and HER4 immunoprecipitates from cells cultured with or without HRG and with or without the ErbB-specific small-molecule inhibitor GW572016. 5R cells express an intracellular antibody that eliminates HER2 surface expression. Immunoprecipitates were analyzed by Western blotting to detect phosphotyrosine residues. (E) Expression of BRCA1 mRNA was measured by real-time quantitative RT-PCR. Each cell line was grown in serum-free medium plus insulin and hydrocortisone, with or without HRG, for 24 h. Total RNA (10 ng) was analyzed as noted above (n = 3). The number of BRCA1 transcripts in each untreated cell line was given a value of 1, and the number of BRCA1 transcripts in the HRG-treated cell line (24 h of HRG treatment) is given relative to that in the control. Experiments were repeated three times, with each sample analyzed in triplicate. (F) Expression of BRCA1 mRNA was measured by real-time quantitative RT-PCR (as described above) in SUM44 cells cultured with or without HRG (24 h) in the presence or absence of the ErbB tyrosine kinase inhibitor GW572016 (1 μM). Experiments were analyzed in triplicate and repeated three times.
To determine if HRG altered the mRNA expression of BRCA1, real-time RT-PCR was used to quantitate BRCA1 transcripts in HRG-treated cells. HRG increased the number of BRCA1 transcripts >3-fold in HER4-positive SUM44, BT-474, MCF-7, and SUM102-HER4 cells (Fig. 3C). In contrast, SUM102-pLXSN, MDA-MB-453, and SkBR-3 cells, which are HER4 negative, did not demonstrate HRG-enhanced BRCA1 expression. This suggests that HRG-mediated BRCA1 induction requires HER4. However, there was no correlation between basal levels of BRCA1 expression and HER4 expression in untreated cells (data not shown).
Since HRG stimulates the tyrosine kinase activities of both HER2 and HER4, we next examined the requirement for HER2 in HRG-mediated induction of BRCA1. HER2 signaling was impaired by the elimination of HER2 cell surface expression. This was accomplished by sequestration of HER2 in the endoplasmic reticulum, using a single-chain anti-HER2 antibody containing an endoplasmic reticulum targeting sequence known as scFv-5R (34). This protein is referred to herein as 5R, and it was retrovirally expressed in SUM44 and SUM102-HER4 cells. The expression of 5R in each of these cells has been described previously (34), and it completely abolished HRG-dependent HER2 tyrosine phosphor ylation in both SUM44-5R cells and SUM102-HER4-5R cells (Fig. 3D) compared to that in SUM44-pBABE and SUM102-HER4-pBABE cells, respectively. The expression of 5R did not eliminate HRG-dependent tyrosine phosphor ylation of HER4 or HER3 in SUM44-5R or SUM102-HER4-5R cells (Fig. 3D). Previous reports demonstrated that SUM44-pBABE and SUM44-5R cells maintain HRG-dependent growth inhibition (34).
The expression of 5R and elimination of HER2 signaling did not block the HRG-dependent induction of BRCA1 mRNA in SUM44 cells (Fig. 3E). In contrast, SUM44-5R cells displayed a heightened induction of BRCA1 expression in response to HRG compared to SUM44-pBABE cells. Furthermore, in SUM102-HER4 cells, which had acquired an antiproliferative response to HRG by virtue of HER4 expression, sequestration of HER2 did not abolish HRG-mediated induction of BRCA1. Thus, HER2 is not necessary for the HRG-dependent increase in BRCA1 expression in cells with either endogenous (SUM44) or exogenously expressed (SUM102-HER4) HER4. Although increased BRCA1 induction was seen in HRG-treated SUM44- 5R cells compared to that in SUM44-pBABE cells, we did not see exaggerated HRG-mediated growth inhibition in SUM44-5R cells compared to that in SUM44-pBABE cells (data not shown), consistent with previous observations that HRG-mediated growth inhibition is similar in SUM44-5R and SUM44-pBABE cells (34).
We treated SUM44 cells with the pan-ErbB kinase inhibitor GW572016 (1 μM) to determine if ErbB kinase activity is required for the induction of BRCA1 mRNA in these HER4-positive, HER1-negative cells. In the absence of HRG, the expression of BRCA1 was similar in cells treated with or without GW572016 (Fig. 3F). While HRG induced BRCA1 transcript levels in the absence of GW572016, the HRG-mediated increase in BRCA1 was not observed in cells cultured in the presence of GW572016. Since HER2 expression is not required for HRG-inducible BRCA1 expression and since these cells do not express HER1, this suggests that HRG-induced HER4 kinase activity is required for BRCA1 induction.
Clinical correlation of HER4 and BRCA1 expression.
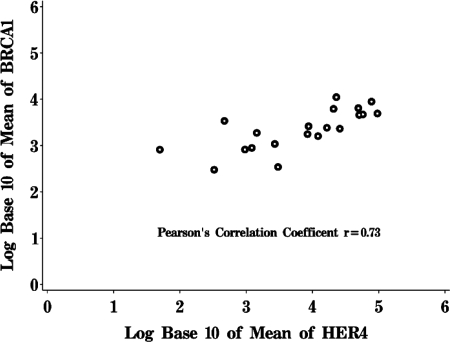
To examine the relationship between HER4 and BRCA1 expression further, we measured mRNA expression of HER4 and BRCA1 in human breast cancer specimens. HER4 mRNA expression was determined using real-time quantitative RT-PCR, measuring the absolute number of HER4 transcripts in 10 ng of tumor total RNA (Table 1). We assayed 19 human breast cancer samples, finding that HER4 mRNA expression could be detected at various levels. We also measured the BRCA1 transcript number in each tumor, using quantitative real-time RT-PCR. Interestingly, a correlation was observed between the numbers of HER4 and BRCA1 transcripts (Fig. 4) (Pearson's correlation coefficient = 0.73). Transcripts encoding HRG were detected in some, but not all, of the breast tumor specimens (Table 1). As controls, we quantified the mRNAs encoding amphiregulin and keratin 18. Unlike that of BRCA1, expression of amphiregulin did not correlate positively or negatively with HER4 expression in these tumor samples, nor did gene expression of the epithelial structural protein keratin 18 (data not shown). The consent forms from these anonymous donors do not allow linkage to clinical data or clinical outcomes. Nonetheless, these results are consistent with our findings for breast cancer cell lines that HER4 enhances the expression of BRCA1 mRNA.
FIG. 4.
HER4-positive breast tumors express greater amounts of BRCA1 than HER4-negative breast tumors. RNAs extracted from frozen breast cancer specimens were used to measure the absolute levels of HER4 and BRCA1 mRNAs by quantitative real-time RT-PCR. Each sample was analyzed independently three times. The figure shows a Pearson's correlation plot, comparing absolute levels of BRCA1 mRNA to those of HER4.
Heregulin-induced BRCA1 expression requires JNK activity.
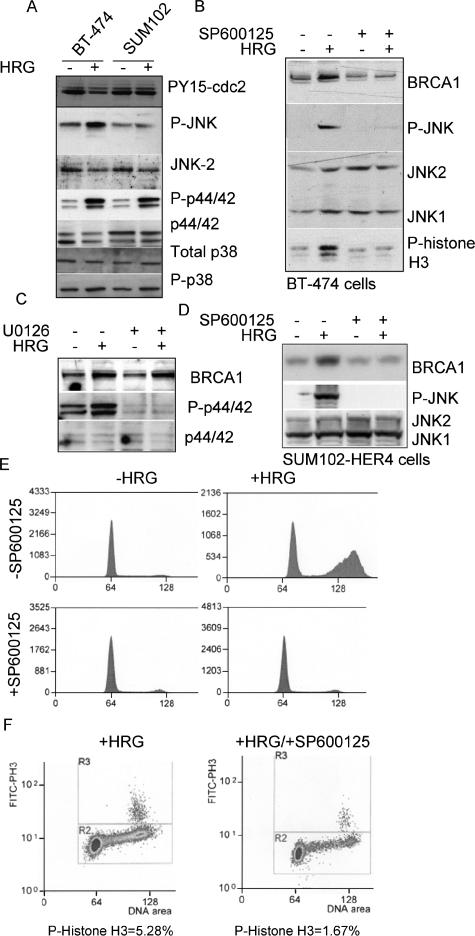
To determine which signaling pathways may be involved in HRG-mediated induction of BRCA1, we examined the activities of JNK, p44/42, and p38 in BT-474 cells (HER4 positive) and in SUM102 cells (HER4 negative). In BT-474 cells and SUM102 cells, HRG-dependent phosphorylation of p44/42 and, to a lesser extent, p38 was observed; however, HRG induced an increase in phosphorylation of JNK in HRG-treated BT-474 cells only (Fig. 5A). Therefore, we treated BT-474 cells with SP600125, a pharmacologic inhibitor of JNK1 and JNK2 activity. SP600125 inhibited HRG-induced JNK phosphorylation without affecting the expression of total JNK1 or JNK2 (Fig. 5B). Moreover, BT-474 cells cultured in the presence of SP600125 did not display the HRG- mediated increase in BRCA1 expression (Fig. 5B) or increased phospho-histone H3 expression. In contrast, inhibition of p44/42 activity using the small-molecule inhibitor U0126 did not affect the HRG-dependent increase in BRCA1 protein expression (Fig. 5C).
FIG. 5.
BRCA1 induction and growth inhibition in response to HRG requires JNK activity. (A) Western analysis of whole-cell extracts of BT-474 or SUM102 cells treated with or without HRG for 48 h. Antibodies used for Western analysis are shown to the right. Results shown are representative of three experiments. (B) Western analysis of BT-474 cells cultured for 48 h with or without HRG and with or without SP600125 (a small-molecule pan-JNK inhibitor). Antibodies used for Western analysis are shown to the right. (C) Western analysis of BT-474 cells cultured for 48 h with or without HRG and with or without U0126 (a small-molecule p44/42 inhibitor). Antibodies used for Western analysis are shown to the right. (D) Western analysis of SUM102-HER4 cells cultured for 48 h with or without HRG and with or without SP600125 (a small-molecule pan-JNK inhibitor). Antibodies used for Western analysis are shown to the right. (E) Cell cycle analysis of propidium iodide-stained BT-474 cells cultured for 48 h with or without HRG and with or without SP600125. (F) FACS analysis of BT-474 cells treated with HRG and with or without SP600125 for 48 h. Methanol-fixed cells were stained with an antibody against phospho-histone H3 (and an FITC-labeled secondary antibody) and with propidium iodide. A total of >20,000 cells were analyzed by flow cytometry. The percentage of cells that were positive for FITC is shown below each panel.
Because BRCA1expression is lowest in the G1 phase of the cell cycle and since JNK inhibition may halt the cells in G1, therefore indirectly inhibiting HRG-induced BRCA1 expression, we examined the HRG-mediated induction of BRCA1 in SUM102-HER4 cells. These cells have increased BRCA1 mRNA expression in the presence of HRG (Fig. 3) but do not undergo a G2/M delay in response to HRG, instead demonstrating an increase in G1 cells (Fig. 1). Treatment of SUM102-HER4 cells with HRG induced the phosphorylation of JNK and increased protein expression of BRCA1 (Fig. 5D). Inhibition of JNK activity with SP600125 in SUM102-HER4 cells also impaired the induction of BRCA1, suggesting that the induction of BRCA1 by HRG is cell cycle independent and that JNK is required for induction of BRCA1 by HRG.
Consistent with these observations, SP600125 impaired the HRG-dependent accumulation of BT-474 cells with 4N DNA (Fig. 5E). The HRG-induced increase in phospho-histone H3-positive cells was also impaired by SP600125 (Fig. 5F). These data suggest that JNK may be required for the HRG-mediated cell cycle delay at the G2/M transition.
Heregulin-mediated growth inhibition requires BRCA1.
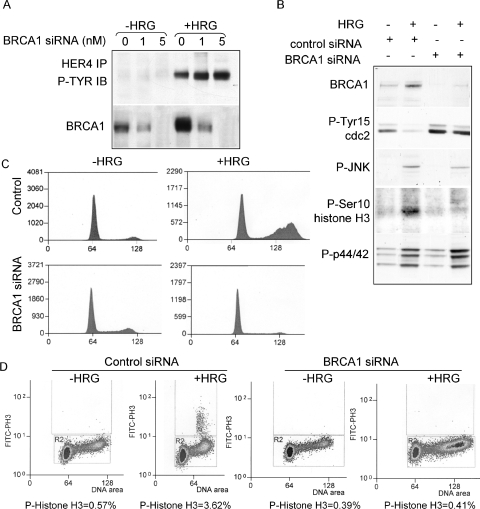
To examine the role of BRCA1 in HRG/HER4-mediated growth inhibition, we inhibited the expression of BRCA1 in BT-474 cells, using transient transfection of siRNA sequences targeting BRCA1. Cells were transfected and then cultured in serum-free medium in the presence or absence of HRG for 30 h. Cells transfected with BRCA1 siRNA sequences demonstrated a dose-dependent decrease in BRCA1 expression without altering HRG-induced tyrosine phosphorylation of HER4 (Fig. 6A). Western analysis demonstrated that the HRG-dependent dephosphorylation of cdc2 at tyrosine 15 was not observed in the absence of BRCA1, nor was the HRG-dependent increase in immunodetection of phosphorylation of histone H3 at Ser10 (Fig. 6B). In contrast, HRG-induced phosphorylation of JNK and p44/42 was unaffected by transfection with BRCA1 siRNA, demonstrating that the effects of BRCA1 inhibition are beyond the initial signaling events. These results further suggest that JNK activation in response to HRG is required upstream of BRCA1 induction, although our results do not rule out other functions of JNK that may lie downstream of BRCA1 induction.
FIG. 6.
BRCA1 expression is required for HRG-mediated growth inhibition in BT-474 cells. (A) Western analysis of BT-474 cells transfected with increasing concentrations of BRCA1 siRNA sequences and treated with or without HRG. Cells were collected at 30 h posttransfection. (Top) Western analysis of HER4 immunoprecipitates from whole-cell extracts to detect phosphotyrosine residues. (Bottom) Western analysis of whole-cell extracts to detect BRCA1. (B) Western analysis of whole-cell lysates from BT-474 cells transfected with 10 nM BRCA1 or control siRNA and treated with or without HRG. Lysates were collected 30 h after transfection. Antibodies used for Western analysis are shown to the left of the panel. (C) Cell cycle analysis of propidium iodide-stained BT-474 cells transfected with 10 nM BRCA1 or control siRNA and treated with or without HRG. Cells were collected after 30 h. A total of >10,000 nuclei were counted per sample. (D) FACS analysis of BT-474 cells transfected with control or BRCA1 siRNA and then treated with HRG and with or without SP600125 for 36 h. Methanol-fixed cells were stained with an antibody against phospho-histone H3 (and an FITC-labeled secondary antibody) and with propidium iodide. A total of >20,000 cells were analyzed by flow cytometry. The percentage of cells that were positive for FITC is shown below each panel.
Transfection of cells with BRCA1 siRNA impaired the HRG-mediated accumulation of BT-474 cells with 4N DNA, as determined by flow cytometric analysis of propidium iodide-stained cells (Fig. 6C). In contrast, BT-474 cells transfected with a nonspecific siRNA sequence displayed an HRG-mediated increase in the 4N DNA-containing cell population. While cells transfected with control siRNA sequences displayed an HRG-induced increase in the population of phospho-histone H3-positive cells compared to that in untreated cells, transfection with BRCA1 siRNA sequences impaired the HRG-mediated induction of phospho-histone H3-positive cells (Fig. 6D). This suggests that the HRG-dependent delay in the G2/M transition is not observed in cells transfected with BRCA1 siRNA.
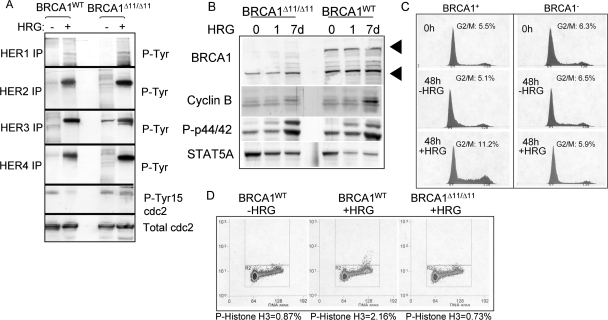
To further test the requirement for BRCA1 in HRG-mediated growth inhibition, we used a genetic model of BRCA1 depletion. Mouse MECs were derived from a mouse model in which exon 11 of the BRCA1 gene is flanked with LoxP sites (36). Exon 11 was excised by retroviral expression of Cre recombinase in culture, producing an isogenic pair of MECs differing only with respect to BRCA1 expression, referred to herein as BRCA1WT and BRCA1Δ11/Δ11 MECs. We cultured these two cell lines in serum-free medium in the presence or absence of HRG for 0 to 48 h, resulting in HRG-dependent tyrosine phosphorylation of HER2, HER3, HER4, and, to a lesser extent, HER1 at 48 h (Fig. 7A). We analyzed whole-cell extracts from BRCA1WT and BRCA1Δ11/Δ11 MECs treated for 48 h with HRG for phosphorylation of cdc2 at tyrosine 15 by Western analysis. While equal amounts of cdc2 were detected regardless of HRG treatment or BRCA1 status, we found an HRG-dependent decrease in cdc2 phosphorylation at Tyr15 in BRCA1WT MECs but not in BRCA1Δ11/Δ11 MECs.
FIG. 7.
Growth of BRCA1-deficient mammary epithelial cells is not inhibited by HRG. (A) Mouse mammary epithelial cells carrying a targeted deletion of exon 11 of the BRCA1 gene (BRCA1−) and their congenic wild-type counterparts (BRCA1+) were cultured for 0 to 7 days with or without HRG. (Top panels) Western analysis of HER4 immunoprecipitates to detect tyrosine phosphorylation. (Bottom panels) Western analysis of whole-cell lysates. Antibodies used for analysis are shown to the left of each panel. (B) Cells were grown with or without HRG for 0, 1, or 7 days in serum-free medium. Cells were collected and analyzed by Western analysis using the antibodies indicated to the left. (C) Analysis of propidium iodide-stained cells by flow cytometry. BRCA1WT and BRCA1Δ11/Δ11 cells were treated with or without HRG for 0 or 48 h prior to analysis. (D) FACS analysis of BRCA1+ and BRCA1− MECs treated with or without HRG for 7 days. Methanol-fixed cells were stained with an antibody against phospho-histone H3 (and an FITC-labeled secondary antibody) and with propidium iodide. A total of >20,000 cells were analyzed by flow cytometry. The percentage of cells that were positive for FITC is shown below each panel.
Whole-cell extracts revealed an HRG-dependent increase in cyclin B expression and increased phosphorylation of histone H3 serine 10, consistent with an increase in the G2/M population of BRCA1WT cells (Fig. 7B). In contrast, HRG failed to enhance cyclin B expression in BRCA1Δ11/Δ11 MECs. The absence of exon 11 of BRCA1 did not affect other aspects of HER4 signaling, such as HRG-dependent phosphorylation of p44/42. Western analysis of STAT5A demonstrated equal loading in each lane of the gel. An HRG-dependent increase in the G2/M population of BRCA1WT cells compared to that of untreated cells was observed (Fig. 7C). In contrast, BRCA1Δ11/Δ11 cells did not have an increased 4N population in response to 48 h of incubation with HRG. The HRG-dependent increase in 4N DNA was also observed in BRCA1WT cells examined 7 days after HRG treatment, but not in BRCA1Δ11/Δ11 cells (data not shown). Phospho-histone H3 was detected at higher levels in HRG-treated BRCA1WT MECs than in untreated BRCA1WT cells (Fig. 7D); however, BRCA1Δ11/Δ11 cells did not display increased phospho-histone H3 staining after being treated with HRG. These results are consistent with the hypothesis that HRG induces a BRCA1-dependent G2/M checkpoint.
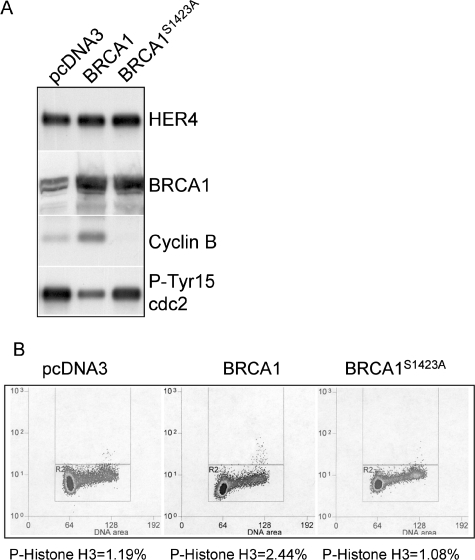
To determine if BRCA1 overexpression is sufficient to induce mitotic delay in the absence of HRG signaling, we generated BT-474 cell lines stably transfected with expression vectors expressing human BRCA1 or BRCA1S1423A, with the latter being deficient in a BRCA1-dependent G2/M checkpoint induced by ionizing radiation (53, 54). Selection-resistant clones were analyzed individually for expression of BRCA1, and more than five clones were pooled together for further analysis. While total HER4 expression was equal across all samples analyzed, Western analysis demonstrated that BRCA1 expression was increased in cells stably transfected with the BRCA1 or BRCA1S1423 expression construct (Fig. 8A). Although the cells were grown under serum-free conditions and not treated with HRG, cells expressing BRCA1 displayed an increase in the level of cyclin B expression compared to that in cells expressing BRCA1S1423A or cells transfected with the empty parental vector pcDNA3. Cells that overexpressed BRCA1 contained less phospho-Tyr 15 cdc2 than did controls. Finally, the percentage of phospho-histone H3-positive cells increased in BT474 cells overexpressing BRCA1 (Fig. 8B) but not in BT-474 cells overexpressing BRCA1S1423A, suggesting that BRCA1 overexpression may be sufficient to delay mitosis in the absence of HRG.
FIG. 8.
Overexpression of BRCA1 is sufficient to induce mitotic delay in the absence of HRG. (A) Western analysis of lysates from BT-474 cells stably transfected with pcDNA3, pcDNA3-BRCA1, or pcDNA3S1423A (defective in G2/M checkpoint induced by ionizing radiation). Cells were cultured in serum-free medium for 16 h prior to analysis. Antibodies used for Western analysis are shown to the right. (B) FACS analysis of BT-474 pcDNA3-, pcDNA3-BRCA1-, or pcDNA3S1423A-expressing cells grown in serum-free medium for 16 h, fixed in methanol, and stained with a phosphoserine 10 histone H3 antibody. The percentage of cells positive for phospho-histone H3 is shown below each panel (n > 20,000 cells).
DISCUSSION
Members of the ErbB/HER family of receptor tyrosine kinases are known to stimulate the proliferation of breast epithelial cells and to play causative roles in the formation and progression of breast cancer. However, recent evidence suggests that HER4 activation prevents the growth of breast epithelial cells while promoting cellular differentiation. Most studies agree that HER4 expression correlates with better outcomes for human breast cancer patients. However, the mechanism by which HER4 promotes a better prognosis or decreases the growth of breast cancer cells is currently unknown. We describe results herein suggesting that HER4 impairs the growth of breast cancer cells through a BRCA1-dependent delay in mitotic progression.
It is known that BRCA1 mRNA expression is regulated in a cell cycle-specific manner, with the lowest levels of BRCA1 expression detected during G1 and with increasing levels detected as cells transition through S phase and G2 (8, 12, 46). This is consistent with the proposed functions of BRCA1 in mediating DNA repair and checkpoint controls in the later phases of the cell cycle. Because BRCA1 expression is influenced by cell cycle dynamics, it is possible that the regulation of BRCA1 expression by HER4 activity might be an indirect reflection of the demonstrated HER4-induced cell cycle delay in late G2 or early M phase. However, our data with SUM102-HER4 cells demonstrated that HRG increased BRCA1 expression >4-fold (Fig. 3), while cells accumulated in G1 (Fig. 1). Furthermore, HRG treatment of synchronized BT-474 cells increased BRCA1 expression regardless of the cell cycle phase in which cells were synchronized. Therefore, it seems as if HER4 increases BRCA1 expression by mechanisms that are independent, at least in part, of cell cycle dynamics.
The mechanism by which HER4 triggers BRCA1 expression is currently unknown. Two likely possibilities are that HER4 stabilizes BRCA1 transcripts and that HER4 induces the transcription of BRCA1. Given that the intracellular domain of HER4 becomes liberated upon ligand binding to form a constitutively active tyrosine kinase that can translocate to the nucleus (25, 47), bind transcription factors such as p53, YAP, and STAT5A (4, 18, 28, 51), and perhaps exhibit transcriptional activity on its own (25), it is interesting to speculate that HER4 may contribute directly to the transcriptional activation of the BRCA1 gene. Alternatively, HER4 could activate gene transcription of BRCA1 through a conventional RTK signaling cascade. The observations that JNK signaling was triggered by HER4 activity and that JNK was required for HRG-induced BRCA1 induction (Fig. 5) suggest that classical RTK signaling pathways, or perhaps tyrosine kinase signaling by the liberated intracellular domain of HER4, may be involved in the regulation of BRCA1, as opposed to direct transcriptional activation by the HER4 intracellular domain. It is clearly possible that both signaling pathways (classical signaling and a novel action of the intracellular fragment of HER4) may simultaneously be involved in BRCA1 induction. Similarly, while Fig. 8 demonstrates that overexpression of BRCA1 produces the same G2/M delay as does HRG-induced HER4 activity, it is possible that maximal growth inhibition via HER4 involves both BRCA1 induction and other HER4-dependent signaling steps.
The observation that molecular signals initiated at or emanating from the cell surface could enhance expression of the BRCA1 gene may hold therapeutic potential, given the importance of BRCA1 as a tumor suppressor. In addition to the numerous inactivating mutations or deletions in the BRCA1 gene detected in hereditary breast cancer families (9, 22), decreased BRCA1 expression is often detected in sporadic cases of breast cancer (42). Our studies showed that human breast cancers expressing higher levels of HER4 mRNA generally displayed greater BRCA1 mRNA levels (Fig. 4), supporting the idea that HER4 expression may confer a better breast cancer prognosis, in part by virtue of elevated BRCA1 expression. Although our translational analysis examined only a small number of tumors (n = 19), we found repeated evidence that HER4-expressing breast cancer-derived cell lines had increased BRCA1 expression at both the protein and mRNA levels when treated with HRG (Fig. 3), while HER4− cell lines did not. HRG-dependent induction of BRCA1 mRNA required HER4 tyrosine kinase activation (Fig. 3F) but was independent of HER2 and, in fact, might be enhanced in the absence of HER2 (Fig. 3D and E). The introduction of HER4 into a HER4-negative breast cancer cell line conferred HRG-mediated BRCA1 induction. Because BRCA1 is a protein with numerous and diverse functions, including control of genomic stability, regulation of cell cycle checkpoints, DNA repair, and regulation of gene transcription, BRCA1 has many pathways by which it can carry out its critically important tumor suppressor activity (58). The interest in this bona fide human tumor suppressor has produced much data and many hypotheses to explain how it works; our findings add to this list the possibility that increased expression of BRCA1 in response to HER4 activation may confer greater control over cellular decisions regarding growth, particularly in the G2/M region of the cell cycle (see below).
Although the accumulation of cells with 4N DNA in response to HRG may have suggested a delay in G2 or at the G2/M transition, our data suggest that HER4-positive breast cancer cells are delayed in early mitosis. Mitotic delay was demonstrated by increased phosphorylation of histone H3, sustained cyclin B expression and cyclin B-cdc2 interaction, and sustained cdc2 activity, each of which initially becomes evident in early mitosis. As cells progress through mitosis and chromosomes align in anaphase, cyclin B dissociates from cdc2, cdc2 becomes inactivated, and cyclin B is targeted for degradation by the anaphase promoting complex. Because we observed sustained cyclin B-cdc2 association and cdc2 activity in response to HRG, it is likely that HRG resulted in a delay at a checkpoint in mitosis known as the spindle assembly checkpoint (SAC). Most of what is known with regard to the SAC has been studied in response to cellular damage rather than in response to a physiologic ligand such as HRG. We examined several SAC proteins in HRG-treated BT-474 cells but did not find any difference in the subcellular localization of BRCA1, cdc20, MAD2, or BubR1 (data not shown), nor did we observe any overall change in the phosphorylation status of these four proteins, as assessed by phosphoserine and phosphothreonine Western analysis of immunoprecipitates from cells treated with or without HRG. This does not rule out HRG-dependent posttranslational modifications of BRCA1. In fact, HRG-dependent BRCA1 phosphorylation has been reported previously (3). The data herein showing that BRCA1S1473A overexpression did not result in a G2/M delay may suggest that HRG-dependent modification by a serine-threonine kinase is involved.
The role of BRCA1 as a necessary intermediary in HRG- and HER4-dependent growth delay was demonstrated by independent means in assays with human breast cancer cell lines, using RNA interference (Fig. 6), and in genetic experiments using isogenic mouse mammary cell lines that either express BRCA1 or do not. HRG-dependent mitotic delay occurred in BRCA1+ cells and was absent in BRCA1− cells (Fig. 7). Although the precise role of BRCA1 in regulating mitotic progression in response to HER4 activation is still under investigation, it may relate to the ability of BRCA1 to activate the SAC, a checkpoint that ensures the accurate alignment and segregation of chromosomes and prevents cells with misaligned chromosomes from exiting mitosis. The requirement for BRCA1 in the SAC is highlighted by the remarkable number of aneuploid cells derived from mice carrying a homozygous targeted deletion of BRCA1 exon 11, the inability of these cells to inhibit the anaphase promoting complex in the presence of mitotic poisons, and their inability to arrest in mitosis (6, 7, 49, 50, 55).
Evidence from animal models demonstrates that HER4 activity is required for lactational differentiation of the mammary epithelium. Mammary glands from mice that lack HER4 activity as a result of multiple genetic strategies each have lactational defects due to an impaired program of differentiation (16, 20, 43), as measured by a decrease in the expression of milk proteins (β-casein and whey acidic protein) and a decrease in activity of the transcription factor STAT5a, which is required for lactation. Interestingly, the role of BRCA1 in the differentiation of mammary epithelial cells has recently become apparent. BRCA1 expression is spatially and temporally regulated at each distinct stage of mammary gland development (21, 32). Also, mammary glands from mice with a conditional BRCA1 knockout displayed an impaired program of growth and differentiation (55). In cell culture models, the role of BRCA1 in morphological differentiation has been studied in three-dimensional cultures of MCF10A cells. Depletion of BRCA1 using RNA interference revealed that acinus formation, the formation of a single-layered, polarized epithelial structure surrounding a lumen, relied on BRCA1 expression and inversely correlated with increased growth of the MCF10A cells in the absence of BRCA1 expression (11). These studies support a hypothesis currently under investigation that the mechanism by which HER4 regulates mammary differentiation may rely on BRCA1 activity, similar to the BRCA1-dependent mechanism by which HER4 decreases the growth of breast cancer cells, as demonstrated herein.
In summary, our data suggest that HER4-mediated induction of BRCA1 expression may inhibit the growth of breast cancer cells by two mechanisms, by decreasing progression through mitosis and by enhancing cellular differentiation. These data warrant further investigations examining the potential synergy between HER4 and BRCA1 in mitotic delay with regard to tumor formation and progression.
Acknowledgments
We thank Dominic Moore for biostatistical analysis of this work and Michael Kastan for providing BRCA1 expression plasmids.
This work was supported in part by a grant from the Breast Cancer Research Foundation, by National Institute of General Medical Sciences grant GM00678, and by UNC Breast Cancer SPORE grant CA58223.
REFERENCES
- 1.Abd El-Rehim, D. M., S. E. Pinder, C. E. Paish, J. A. Bell, R. S. Rampaul, R. W. Blamey, J. F. Robertson, R. I. Nicholson, and I. O. Ellis. 2004. Expression and co-expression of the members of the epidermal growth factor receptor (EGFR) family in invasive breast carcinoma. Br. J. Cancer 91:1532-1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alimandi, M., A. Romano, M. C. Curia, R. Muraro, P. Fedi, S. A. Aaronson, P. P. Di Fiore, and M. H. Kraus. 1995. Cooperative signaling of ErbB3 and ErbB2 in neoplastic transformation and human mammary carcinomas. Oncogene 10:1813-1821. [PubMed] [Google Scholar]
- 3.Altiok, S., D. Batt, N. Altiok, A. Papautsky, J. Downward, T. M. Roberts, and H. Avraham. 1999. Heregulin induces phosphorylation of BRCA1 through phosphatidylinositol 3-kinase/AKT in breast cancer cells. J. Biol. Chem. 274:32274-32278. [DOI] [PubMed] [Google Scholar]
- 4.Arasada, R. R., and G. Carpenter. 2005. Secretase-dependent tyrosine phosphorylation of Mdm2 by the ErbB-4 intracellular domain fragment. J. Biol. Chem. 280:30783-30787. [DOI] [PubMed] [Google Scholar]
- 5.Bobrow, L. G., R. R. Millis, L. C. Happerfield, and W. J. Gullick. 1997. c-erbB-3 protein expression in ductal carcinoma in situ of the breast. Eur. J. Cancer 33:1846-1850. [DOI] [PubMed] [Google Scholar]
- 6.Brodie, S. G., X. Xu, W. Qiao, W. M. Li, L. Cao, and C. X. Deng. 2001. Multiple genetic changes are associated with mammary tumorigenesis in Brca1 conditional knockout mice. Oncogene 20:7514-7523. [DOI] [PubMed] [Google Scholar]
- 7.Cao, L., W. Li, S. Kim, S. G. Brodie, and C. X. Deng. 2003. Senescence, aging, and malignant transformation mediated by p53 in mice lacking the Brca1 full-length isoform. Genes Dev. 17:201-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, Y., A. A. Farmer, C. F. Chen, D. C. Jones, P. L. Chen, and W. H. Lee. 1996. BRCA1 is a 220-kDa nuclear phosphoprotein that is expressed and phosphorylated in a cell cycle-dependent manner. Cancer Res. 56:3168-3172. [PubMed] [Google Scholar]
- 9.Couch, F. J., M. L. DeShano, M. A. Blackwood, K. Calzone, J. Stopfer, L. Campeau, A. Ganguly, T. Rebbeck, and B. L. Weber. 1997. BRCA1 mutations in women attending clinics that evaluate the risk of breast cancer. N. Engl. J. Med. 336:1409-1415. [DOI] [PubMed] [Google Scholar]
- 10.Eccles, S. A. 2001. The role of c-erbB-2/HER2/neu in breast cancer progression and metastasis. J. Mammary Gland Biol. Neoplasia 6:393-406. [DOI] [PubMed] [Google Scholar]
- 11.Furuta, S., X. Jiang, B. Gu, E. Cheng, P. L. Chen, and W. H. Lee. 2005. Depletion of BRCA1 impairs differentiation but enhances proliferation of mammary epithelial cells. Proc. Natl. Acad. Sci. USA 102:9176-9181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gudas, J. M., T. Li, H. Nguyen, D. Jensen, F. J. Rauscher III, and K. H. Cowan. 1996. Cell cycle regulation of BRCA1 messenger RNA in human breast epithelial cells. Cell Growth Differ. 7:717-723. [PubMed] [Google Scholar]
- 13.Holbro, T., R. R. Beerli, F. Maurer, M. Koziczak, C. F. Barbas III, and N. E. Hynes. 2003. The ErbB2/ErbB3 heterodimer functions as an oncogenic unit: ErbB2 requires ErbB3 to drive breast tumor cell proliferation. Proc. Natl. Acad. Sci. USA 100:8933-8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holbro, T., and N. E. Hynes. 2004. ErbB receptors: directing key signaling networks throughout life. Annu. Rev. Pharmacol. Toxicol. 44:195-217. [DOI] [PubMed] [Google Scholar]
- 15.Hynes, N. E., and H. A. Lane. 2005. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat. Rev. Cancer 5:341-354. [DOI] [PubMed] [Google Scholar]
- 16.Jones, F. E., T. Welte, X. Y. Fu, and D. F. Stern. 1999. ErbB4 signaling in the mammary gland is required for lobuloalveolar development and Stat5 activation during lactation. J. Cell Biol. 147:77-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knowlden, J. M., J. M. Gee, L. T. Seery, L. Farrow, W. J. Gullick, I. O. Ellis, R. W. Blamey, J. F. Robertson, and R. I. Nicholson. 1998. c-erbB3 and c-erbB4 expression is a feature of the endocrine responsive phenotype in clinical breast cancer. Oncogene 17:1949-1957. [DOI] [PubMed] [Google Scholar]
- 18.Komuro, A., M. Nagai, N. E. Navin, and M. Sudol. 2003. WW domain-containing protein YAP associates with ErbB-4 and acts as a co-transcriptional activator for the carboxyl-terminal fragment of ErbB-4 that translocates to the nucleus. J. Biol. Chem. 278:33334-33341. [DOI] [PubMed] [Google Scholar]
- 19.Lohrisch, C., and M. Piccart. 2001. HER2/neu as a predictive factor in breast cancer. Clin. Breast Cancer 2:129-135. [DOI] [PubMed] [Google Scholar]
- 20.Long, W., K. U. Wagner, K. C. Lloyd, N. Binart, J. M. Shillingford, L. Hennighausen, and F. E. Jones. 2003. Impaired differentiation and lactational failure of Erbb4-deficient mammary glands identify ERBB4 as an obligate mediator of STAT5. Development 130:5257-5268. [DOI] [PubMed] [Google Scholar]
- 21.Marquis, S. T., J. V. Rajan, A. Wynshaw-Boris, J. Xu, G. Y. Yin, K. J. Abel, B. L. Weber, and L. A. Chodosh. 1995. The developmental pattern of Brca1 expression implies a role in differentiation of the breast and other tissues. Nat. Genet. 11:17-26. [DOI] [PubMed] [Google Scholar]
- 22.Miki, Y., J. Swensen, D. Shattuck-Eidens, P. A. Futreal, K. Harshman, S. Tavtigian, Q. Liu, C. Cochran, L. M. Bennett, W. Ding, et al. 1994. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 266:66-71. [DOI] [PubMed] [Google Scholar]
- 23.Naidu, R., M. Yadav, S. Nair, and M. K. Kutty. 1998. Expression of c-erbB3 protein in primary breast carcinomas. Br. J. Cancer 78:1385-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Navolanic, P. M., L. S. Steelman, and J. A. McCubrey. 2003. EGFR family signaling and its association with breast cancer development and resistance to chemotherapy. Int. J. Oncol. 22:237-252. [PubMed] [Google Scholar]
- 25.Ni, C.-Y., M. P. Murphy, T. E. Golde, and G. Carpenter. 2001. γ-Secretase cleavage and nuclear localization of ErbB-4 receptor tyrosine kinase. Science 294:2179-2181. [DOI] [PubMed] [Google Scholar]
- 26.Ohgaki, H., et al. 2004. Genetic pathways to glioblastoma: a population-based study. Cancer Res. 64:6892-6899. [DOI] [PubMed] [Google Scholar]
- 27.Olayioye, M. A., R. M. Neve, H. A. Lane, and N. E. Hynes. 2000. The ErbB signaling network: receptor heterodimerization in development and cancer. EMBO J. 19:3159-3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Omerovic, J., E. M. Puggioni, S. Napoletano, V. Visco, R. Fraioli, L. Frati, A. Gulino, and M. Alimandi. 2004. Ligand-regulated association of ErbB-4 to the transcriptional co-activator YAP65 controls transcription at the nuclear level. Exp. Cell Res. 294:469-479. [DOI] [PubMed] [Google Scholar]
- 29.Pawlowski, V., F. Revillion, M. Hebbar, L. Hornez, and J. P. Peyrat. 2000. Prognostic value of the type I growth factor receptors in a large series of human primary breast cancers quantified with a real-time reverse transcription-polymerase chain reaction assay. Clin. Cancer Res. 6:4217-4225. [PubMed] [Google Scholar]
- 30.Porter, L. A., and D. J. Donoghue. 2003. Cyclin B1 and CDK1: nuclear localization and upstream regulators. Prog. Cell Cycle Res. 5:335-347. [PubMed] [Google Scholar]
- 31.Quinn, C. M., J. L. Ostrowski, S. A. Lane, D. P. Loney, J. Teasdale, and F. A. Benson. 1994. c-erbB-3 protein expression in human breast cancer: comparison with other tumour variables and survival. Histopathology 25:247-252. [DOI] [PubMed] [Google Scholar]
- 32.Rajan, J. V., M. Wang, S. T. Marquis, and L. A. Chodosh. 1996. Brca2 is coordinately regulated with Brca1 during proliferation and differentiation in mammary epithelial cells. Proc. Natl. Acad. Sci. USA 93:13078-13083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Riese, D. J., and D. F. Stern. 1998. Specificity within the EGF family/ErbB receptor family signaling network. Bioessays 20:41-48. [DOI] [PubMed] [Google Scholar]
- 34.Sartor, C. I., H. Zhou, E. Kozlowska, K. Guttridge, E. Kawata, L. Caskey, J. Harrelson, N. Hynes, S. Ethier, B. Calvo, and H. S. Earp III. 2001. HER4 mediates ligand-dependent antiproliferative and differentiation responses in human breast cancer cells. Mol. Cell. Biol. 21:4265-4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schroeder, J. A., and D. C. Lee. 1998. Dynamic expression and activation of ERBB receptors in the developing mouse mammary gland. Cell Growth Differ. 9:451-464. [PubMed] [Google Scholar]
- 36.Sgagias, M. K., K. U. Wagner, B. Hamik, S. Stoeger, R. Spieker, L. J. Huber, L. A. Chodosh, and K. H. Cowan. 2004. Brca1-deficient murine mammary epithelial cells have increased sensitivity to CDDP and MMS. Cell Cycle 3:1451-1456. [DOI] [PubMed] [Google Scholar]
- 37.Slamon, D. J., et al. 1987. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 235:177-182. [DOI] [PubMed] [Google Scholar]
- 38.Srinivasan, R., C. E. Gillett, D. M. Barnes, and W. J. Gullick. 2000. Nuclear expression of the c-erbB-4/HER-4 growth factor receptor in invasive breast cancers. Cancer Res. 60:1483-1487. [PubMed] [Google Scholar]
- 39. Stern, D. F. 2003. ErbBs in mammary development. Exp. Cell Res. 284:89-98. [DOI] [PubMed] [Google Scholar]
- 40.Sunpaweravong, P., S. Sunpaweravong, P. Puttawibul, W. Mitarnun, C. Zeng, A. E. Baron, W. Franklin, S. Said, and M. Varella-Garcia. 2005. Epidermal growth factor receptor and cyclin D1 are independently amplified and overexpressed in esophageal squamous cell carcinoma. J. Cancer Res. Clin. Oncol. 131:111-119. [DOI] [PubMed] [Google Scholar]
- 41.Suo, Z., B. Risberg, M. G. Kalsson, et al. 2002. EGFR family expression in breast carcinomas. c-erbB-2 and c-erbB-4 receptors have different effects on survival. J. Pathol. 196:17-25. [DOI] [PubMed] [Google Scholar]
- 42.Thompson, M. E., R. A. Jensen, P. S. Obermiller, D. L. Page, and J. T. Holt. 1995. Decreased expression of BRCA1 accelerates growth and is often present during sporadic breast cancer progression. Nat. Genet. 9:444-450. [DOI] [PubMed] [Google Scholar]
- 43.Tidcombe, H., A. Jackson-Fisher, K. Mathers, D. F. Stern, M. Gassmann, and J. P. Golding. 2003. Neural and mammary gland defects in ErbB4 knockout mice genetically rescued from embryonic lethality. Proc. Natl. Acad. Sci. USA 100:8281-8286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tovey, S. M., C. J. Witton, J. M. Bartlett, P. D. Stanton, J. R. Reeves, and T. G. Cooke. 2004. Outcome and human epidermal growth factor receptor (HER) 1-4 status in invasive breast carcinomas with proliferation indices evaluated by bromodeoxyuridine labelling. Breast Cancer Res. 6:R246-R251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Troyer, K. L., and D. C. Lee. 2001. Regulation of mouse mammary gland development and tumorigenesis by the ERBB signaling network. J. Mammary Gland Biol. Neoplasia 6:7-21. [DOI] [PubMed] [Google Scholar]
- 46.Vaughn, J. P., P. L. Davis, M. D. Jarboe, G. Huper, A. C. Evans, R. W. Wiseman, A. Berchuck, J. D. Iglehart, P. A. Futreal, and J. R. Marks. 1996. BRCA1 expression is induced before DNA synthesis in both normal and tumor-derived breast cells. Cell Growth Differ. 7:711-715. [PubMed] [Google Scholar]
- 47.Vidal, G. A., A. Narres, L. Marrero, and F. E. Jones. 2005. Presenilin-dependent gamma-secretase processing regulates multiple ERBB4/HER4 activities. J. Biol. Chem. 280:19777-19783. [DOI] [PubMed] [Google Scholar]
- 48.Vogt, U., K. Bielawski, C. M. Schlotter, U. Bosse, B. Falkiewicz, and A. J. Podhajska. 1998. Amplification of erbB-4 oncogene occurs less frequently than that of erbB-2 in primary human breast cancer. Gene 223:375-380. [DOI] [PubMed] [Google Scholar]
- 49.Wang, R. H., H. Yu, and C. X. Deng. 2004. A requirement for breast-cancer-associated gene 1 (BRCA1) in the spindle checkpoint. Proc. Natl. Acad. Sci. USA 101:17108-17113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weaver, Z., C. Montagna, X. Xu, T. Howard, M. Gadina, S. G. Brodie, C. X. Deng, and T. Ried. 2002. Mammary tumors in mice conditionally mutant for Brca1 exhibit gross genomic instability and centrosome amplification yet display a recurring distribution of genomic imbalances that is similar to human breast cancer. Oncogene 21:5097-5107. [DOI] [PubMed] [Google Scholar]
- 51.Williams, C. C., J. G. Allison, G. A. Vidal, M. E. Burow, B. S. Beckman, L. Marrero, and F. E. Jones. 2004. The ERBB4/HER4 receptor tyrosine kinase regulates gene expression by functioning as a STAT5A nuclear chaperone. J. Cell Biol. 167:469-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wiseman, S. M., N. Makretsov, T. O. Nielsen, B. Gilks, E. Yorida, M. Cheang, D. Turbin, K. Gelmon, and D. G. Huntsman. 2005. Coexpression of the type 1 growth factor receptor family members HER-1, HER-2, and HER-3 has a synergistic negative prognostic effect on breast carcinoma survival. Cancer 103:1770-1777. [DOI] [PubMed] [Google Scholar]
- 53.Witton, C. J., J. R. Reeves, J. J. Going, T. G. Cooke, and J. M. Bartlett. 2003. Expression of the HER1-4 family of receptor tyrosine kinases in breast cancer. J. Pathol. 200:290-297. [DOI] [PubMed] [Google Scholar]
- 54.Xu, B., S.-T. Kim, and M. B. Kastan. 2001. Involvement of Brca1 in S-phase and G2-phase checkpoints after ionizing irradiation. Mol. Cell. Biol. 21:3445-3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu, X., Z. Weaver, S. P. Linke, C. Li, J. Gotay, X. W. Wang, C. C. Harris, T. Ried, and C. X. Deng. 1999. Centrosome amplification and a defective G2-M cell cycle checkpoint induce genetic instability in BRCA1 exon 11 isoform-deficient cells. Mol. Cell 3:389-395. [DOI] [PubMed] [Google Scholar]
- 56.Yarden, Y., and M. X. Sliwkowski. 2001. Untangling the ErbB signalling network. Nat. Rev. Mol. Cell. Biol. 2:127-137. [DOI] [PubMed] [Google Scholar]
- 57.Yu, W. H., J. F. Woessner, Jr., J. D. McNeish, and I. Stamenkovic. 2002. CD44 anchors the assembly of matrilysin/MMP-7 with heparin-binding epidermal growth factor precursor and ErbB4 and regulates female reproductive organ remodeling. Genes Dev. 16:307-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zheng, L., S. Li, T. G. Boyer, and W. H. Lee. 2000. Lessons learned from BRCA1 and BRCA2. Oncogene 19:6159-6175. [DOI] [PubMed] [Google Scholar]