Abstract
Genomes contain tandem repeat blocks that are at risk of expansion or contraction. The mechanisms of destabilization of the human minisatellite CEB1 (arrays of 36- to 43-bp repeats) were investigated in a previously developed model system, in which CEB1-0.6 (14 repeats) and CEB1-1.8 (42 repeats) alleles were inserted into the genome of Saccharomyces cerevisiae. As in human cells, CEB1 is stable in mitotically growing yeast cells but is frequently rearranged in the absence of the Rad27/hFEN1 protein involved in Okazaki fragments maturation. To gain insight into this mode of destabilization, the CEB1-1.8 and CEB1-0.6 human alleles and 47 rearrangements derived from a CEB1-1.8 progenitor in rad27Δ cells were sequenced. A high degree of polymorphism of CEB1 internal repeats was observed, attesting to a large variety of homology-driven rearrangements. Simple deletion, double deletion, and highly complex events were observed. Pedigree analysis showed that all rearrangements, even the most complex, occurred in a single generation and were inherited equally by mother and daughter cells. Finally, the rearrangement frequency was found to increase with array size, and partial complementation of the rad27Δ mutation by hFEN1 demonstrated that the production of novel CEB1 alleles is Rad52 and Rad51 dependent. Instability can be explained by an accumulation of unresolved flap structures during replication, leading to the formation of recombinogenic lesions and faulty repair, best understood by homology-dependent synthesis-strand displacement and annealing.
All prokaryotic and eukaryotic genomes contain repeated elements that are dispersed or clustered in tandem arrays. Tandem repeats are classified by the length of the repeat unit. Arrays of short repeats (1 to 9 bp) are termed microsatellites, and repeats of 10 to 100 bp characterize minisatellites. The determination of numerous genome sequences has allowed the chromosomal distribution of repetitive elements to be described previously (6, 18, 77). Microsatellites and minisatellites are spread along the chromosomes and are located in both intergenic and coding sequences; in humans, the expansion of intragenic triplet repeats is associated with human neurodegenerative disorders such as ataxias, Huntington's chorea, and fragile X syndrome (22). Minisatellite repeat expansions are also implicated in human diseases, including epilepsy associated with the EPM1 gene (45), diabetes associated with the insulin gene (5), and cancer related to the HRAS1 minisatellite locus (44). Recently, systematic annotations of the Saccharomyces cerevisiae genome indicated that minisatellite sequences are common (63, 78). In addition to the previously identified minisatellites located in subtelomeric regions (Y′ elements) and elsewhere (24, 28), several arrays were found in coding regions, and interestingly, most minisatellite-containing genes encode cell wall proteins (63, 78). Since the repeats are in frame, they are functionally significant. The variation in the number of repeats in the FLO1 gene was found to correlate with variations in cell adhesion and flocculation (78). This example illustrates that the addition or deletion of minisatellite repeat units in individuals of a population is a potent mechanism for creating functional diversity and a source of molecular evolution for rapid adaptation to environmental change.
The mechanism(s) of microsatellite expansion or contraction have been extensively studied in vivo, in both prokaryotes and eukaryotes, and in vitro (61, 64). A prominent contributor to size variation seems to be replication slippage, but slippage associated with homologous recombination or unequal exchange can also alter the size of arrays (19, 64). In contrast, the mechanism(s) of minisatellite destabilization is less understood. This was studied in the human germ line because minisatellite sequences are relatively stable in somatic cells, but some loci such as MS32, CEB1, and B6.7 are significantly destabilized in the female and/or male germ line (8, 9, 32, 74). For example, CEB1, the subject of the present study, exhibits male-specific mutation rates as high as 20% in sperm, but CEB1 size variation in lymphocytes is much lower (1.8 × 10−4) (7, 8). The determination of the internal structure of minisatellites that underwent rearrangement in the human germ line revealed a diversity of novel structures; interallelic conversion events and, in some cases, polarity in the location of exchanges within the arrays were also observed, suggesting the involvement of meiotic recombination in repeat instability (7, 9, 10, 32, 35, 37, 74). The location of highly unstable minisatellite chromosomal regions that are relatively active in meiotic recombination further sustained this mechanistic link between homologous recombination and size variation (9, 34). The instability of human minisatellites has been modeled in S. cerevisiae, and most of the features of human MS32, MS205, CEB1, and HRAS1 minisatellite instability can be reproduced during yeast sporulation (2, 15, 27, 31). Further, physical and mutant analyses of CEB1 alleles inserted in yeast chromosomes have demonstrated that their destabilization is passively initiated at nearby Spo11-dependent DNA double-strand break (DSB) sites, which are natural initiation sites of homologous recombination in meiosis (15). Similarly, the insertion of the HRAS1 minisatellite into the yeast genome at the HIS4 promoter creates a meiotic recombination hot spot characterized by meiosis-specific DSBs flanking the minisatellite insertion (31). Analyses of the internal structures of variant alleles isolated from single sperm or yeast ascospores have revealed the formation of a large variety of rearranged alleles, including those produced by simple deletions and duplications as well as complex events, raising the possibility that different DNA-dependent processes may be at work. The mechanisms invoked thus far include intra- and interallelic gene conversions that allow local transfer of information between repeats, single-strand annealing (SSA), which causes the internal deletion of contiguous repeats within an array, and various forms of synthesis-dependent strand annealing (SDSA) mechanisms, which provoke a large variety of deletion and duplication events. Further, mismatch repair and loop processing processes also contribute to the formation of the final recombinant products (7, 9, 15, 31). The extreme complexity of some rearrangements (15, 74) and ambiguities in determining their internal structures are also consistent with rare mutagenic events and the participation of the nonhomologous end-joining pathway.
The means by which minisatellite sequences are destabilized and rearranged in somatic cells also remain to be elucidated. In human blood cells and yeast cells, the frequency of minisatellite size variation is very low. In humans, analysis of the internal structure of variant CEB1 and MS32 alleles revealed that rearrangements frequently resulted in simple intra-allelic events (8, 36) that could be explained by SSA if the initiating DSB lesion occurred within the array. Repeat duplication and other complex events can be explained by the SDSA model (59, 64). This mechanism was also proposed to explain expansions and contractions of a 36-bp minisatellite by gene conversion in yeast mitotic cells (60). The absence of the Spo11 nuclease in somatic cells implies that the putative DSB lesions may have arisen by the action of another nuclease or from other spontaneous lesions occurring within or near the array. The strong link between replication and the maintenance of genome stability (42) suggests that stochastic replication defects are likely to contribute to repeat instability. Several studies in S. cerevisiae demonstrated that mutations in genes encoding proteins involved in replication can destabilize minisatellites. Most noteworthy are mutations affecting DNA2, POL3, and RAD27 (41, 50, 51). The strongest effect is seen upon inactivation of the Rad27 protein, homologue of the mammalian hFEN1, or Flap endonuclease I (41, 50, 51). In this study, we found that the CEB1-1.8 allele in homozygous rad27Δ diploid cells was destabilized 170-fold over the wild-type level, reaching a rate of 13.9% per cell/generation at 37°C, a restrictive growth temperature for the rad27Δ mutant (50; present results).
The in vitro biochemical activities of the evolutionarily conserved Rad27/hFEN1 proteins have been extensively studied previously (4, 26, 48, 67). Rad27/hFEN1 is a structure-specific nuclease that has a 5′-3′ flap endonuclease activity required for the maturation of Okazaki fragments during lagging strand replication. Thus, in the absence of Rad27, minisatellite destabilization may be initiated by the accumulation of unprocessed or aberrantly processed 5′ flap structures. The synthetic lethality of the rad27Δ mutation in combination with null mutations in any of the genes of the Rad52 recombinational repair pathway (16, 49, 57, 72, 75, 76) suggested that this pathway is required for the subsequent formation of rearrangements. Unfortunately, it is not possible to determine the role of Rad52 family proteins in minisatellite rearrangements because the rad27Δ rad52Δ mutant is inviable. Similarly, the inviability of rad27Δ rad51Δ mutants prevents testing the contribution of the SSA pathway, which is Rad52 dependent but Rad51 independent (30, 70, 73).
In the present report, we have explored the mechanism(s) of CEB1 destabilization in the absence of Rad27. First, we have analyzed the nature of the rearrangements by sequencing CEB1 progenitor alleles and 47 rearrangements. Previously, the internal structures of CEB1 arrays and other minisatellites were determined by the minisatellite variant repeat mapping by PCR technique (MVR-PCR) (33), which took advantage of preexisting sequence variations among repeat units within tandem arrays (38). In the case of CEB1, analysis of three polymorphic sites located within the repeats served to characterize rearrangements, but in several instances, complete array structures could not be determined (7, 8, 15, 50). The sequence information reported here definitively describes these rearrangements, and comparisons with the progenitor CEB1-1.8 allele allows their origins to be traced. The rearrangements are extremely diverse and arise by simple deletion, double deletion, and complex events. Second, considering the complexity of many of the rearrangements led us to ask whether they result from independent rearrangements accumulating over successive generations. To test this hypothesis, we performed pedigree analyses and found that the rearrangements arise in a single generation. Finally, to determine the role of the Rad52 pathway in the production of CEB1 rearrangements, we partially complemented the rad27Δ mutant with the human hFEN1 cDNA (23, 25) by constructing viable rad27Δ hFEN1 rad52Δ (or rad51Δ) CEB1-3.0 haploid strains. The elimination of Rad52 or Rad51 activity suppresses CEB1 rearrangements. We discuss the mechanisms of minisatellite size variation in light of our findings.
MATERIALS AND METHODS
Media and yeast strains.
Strains were grown in standard media including yeast extract-peptone-dextrose (YPD) and synthetic medium (yeast nitrogen base-ammonium sulfate-dextrose) supplemented when necessary with appropriate nutrients (3). G418-resistant colonies were selected on YPD plates containing Geneticin (200 μg/ml). Yeast cells were sporulated in 1% K acetate as described previously (3). The relevant genotypes and sources of haploid and diploid S. cerevisiae strains (S288C background) in this study are indicated in Table 1. The rad27::HIS3 complete deletion was introduced by crosses with the isogenic strains FW2612 (MATα rad27::HIS3) or FW2617 (MATa rad27::HIS3) (75). The rad27::KanMX4-disrupted strain was constructed as follows. The rad27::KanMX4 cassette and flanking regions were amplified from the Y04963 BY strain of the EUROSCARF deletant collection with primers 25183 and 24817, described by Tishkoff et al. (75), and introduced into ORD6767-13C, and transformants were verified by Southern analysis (ORT5027-1). The rad51::LEU2 disruption of ORD6781-35A originated from ORT2615 (68). The rad52::LEU2 disruption in ORD6771-21A and ORD6780-22A originated from ORT2915.4 (68). The human CEB1-0.6 (14 repeats) and CEB1-1.8 (42 repeats) alleles (7) are inserted in the 5′ intergenic region of the ARG4 locus (15). The other CEB1 alleles (hereafter named CEB1-X) range in size from 5 to 65 repeats and were derived from the CEB1-1.8 allele in ORD6708. The CEB1-2.2 (49 repeats)/CEB1-0.6 strain ORD6708-8 is a rearranged derivative of ORD6708; further destabilization of the CEB1-2.2 allele gave rise to ORD6708-823, which contains the CEB1-3.0 expansion (65 repeats). Sporulation of this strain yielded the haploid segregant ORD6708-823-7A, subsequently used to introduce, by genetic crosses, the CEB1-3.0 allele (Table 1).
TABLE 1.
Relevant genotypes of yeast strains used in this study
| Strain | Relevant genotype | Source or reference |
|---|---|---|
| ORD6708 | MATa/MATα CEB1-1.8 ARG4/CEB1-0.6 arg4Bg rad27::HIS3/rad27::HIS3 | 50 |
| ORD6708-Xa | MATa/MATα CEB1-XaARG4/CEB1-0.6 arg4Bg rad27::HIS3/rad27::HIS3 | This study |
| ORD6708-823-7A | MATa CEB1-3.0 ARG4 rad27::HIS3 | This study |
| ORD5330-3C | MATa CEB1-0.6 arg4Bg rad27::HIS3 | This study |
| ORD6713-8D | MATa CEB1-1.8 ARG4 rad27::HIS3 | This study |
| ORD6728-1C | MATaARG4 rad27::HIS3 | This study |
| MGD131-102A | MATaarg4 Δ2060 | 65 |
| ORT2914 | MATα CEB1-1.8 ARG4 | This study |
| ORD6780-11A | MATa CEB1-3.0 ARG4 | This study |
| ORT5019-2 | MATaarg4 Δ2060 upLEU2::pRAD27-hFEN1-URA3b | This study |
| ORD6767-13C | MATα CEB1-1.8 ARG4 upLEU2::pRAD27-hFEN1-URA3b | This study |
| ORT5027-1 | MATα CEB1-1.8 ARG4 rad27::KanMX4 upLEU2::pRAD27-hFEN1-URA3b | This study |
| ORD6778-4B | MATα CEB1-3.0 ARG4 rad27::KanMX4 upLEU2::pRAD27-hFEN1-URA3b | This study |
| ORD6771-21A | MATα CEB1-1.8 ARG4 rad27::KanMX4 upLEU2::pRAD27-hFEN1-URA3brad52::LEU2 | This study |
| ORD6780-22A | MATα CEB1-3.0 ARG4 rad27::KanMX4 upLEU2::pRAD27-hFEN1-URA3brad52::LEU2 | This study |
| ORD6781-35A | MATa CEB1-3.0 ARG4 rad27::KanMX4 upLEU2::pRAD27-hFEN1-URA3brad51::LEU2 | This study |
Strains used for analysis of array size on CEB1 instability in rad27Δ mutant, X represents the variable size of the CEB1 allele with various numbers of repeats (5 to 65).
hFEN1 cDNA, under the promoter of RAD27 gene (pRAD27), is integrated in the 5′ noncoding region of the LEU2 open reading frame (upLEU2). The complete genotype is available on request.
Expression of the human hFEN1 protein.
The human hFEN1 cDNA (1,143 bp) was isolated from the plasmid pMR102194 (25), sequenced and corrected by site-directed mutagenesis (QuikChange site-directed mutagenesis kit; Stratagene) to replace a spurious T mutation (creating a stop codon) at position 1009 by the wild-type C nucleotide. The corrected hFEN1 cDNA was amplified using the Pfu Ultra enzyme (Stratagene) with the oligonucleotides 5′-CGGACACCGGAAGAAAAAATATGGGAATTCAAGGCCTG-3′ and 5′-ACAGCCGCGGTACTTATTTTCCCCTTTTAAAC-3′, which include a SacII site (underlined) and the 3′ end of the RAD27 promoter (in italics). The RAD27 promoter (pRAD27) (coordinates −683 to −1 from the ATG codon of the RAD27 coding region) was amplified from the plasmid pRS314-RAD27 (62) using the Pfu Ultra enzyme (Stratagene) and the oligonucleotides 5′-TATGCCCGGGGGAGGATGCAAATATGGTGATTTG-3′ and 5′-CAGGCCTTGAATTCCCATATTTTTTCTTCCGGTGTCCG-3′, which include an XmaI site (underlined) and the first 18 bases of the human hFEN1 cDNA (in italics). The resulting PCR products (hFEN1 cDNA and pRAD27), which share 38 bp of identity at the 3′ end of pRAD27 and the 5′ end of hFEN1, were pooled and amplified with oligonucleotides containing XmaI and SacII restriction sites. The resulting pRAD27-hFEN1 PCR product was digested with XmaI and SacII and cloned into the integrative plasmid pRS306 containing the URA3 marker to create pJL10. The yeast ADH1 terminator was then inserted at the 3′ end of the hFEN1 cDNA, and finally, the 650-bp fragment containing the 5′ noncoding region of the LEU2 open reading frame (ORF) was inserted at the KpnI/XhoI sites, creating pJL33. This plasmid was linearized with BstEII (partial digestion) and introduced into the 5′ noncoding region of LEU2 (upLEU2) in the yeast strain MGD131-102A by transformation and selection for uracil prototrophy. Correct integration of the plasmid (pRAD27-hFEN1-URA3) was verified by Southern analysis of Ura+ transformants, and the resulting strain ORT5019-2 was used in further crosses. The expression of the human hFEN1 protein was verified in the rad27Δ strain ORT5027-1 by Western analysis using an anti-hFEN1 polyclonal antibody (Santa Cruz).
Identification of CEB1 rearrangements.
To examine the behavior of CEB1 during vegetative growth, strains were inoculated into 5 ml YPD medium at a density of 2 × 105 cells/ml and grown for 7 generations at 30°C or 37°C. Single cells were plated on YPD agar and incubated at 23°C to reduce the occurrence of additional rearrangements during colony growth. Genomic DNA was prepared from cultures inoculated from individual colonies and grown overnight in YPD liquid media at 23°C. Genomic DNA was extracted and digested with AluI, and the resulting fragments were separated by electrophoresis in 0.8% agarose gels and transferred under vacuum (Qbiogene) onto Hybond N+ membranes (Macherey-Nalgen). The membranes were hybridized with a radiolabeled CEB1-0.6 PCR fragment, which also detects the CEB1-1.8 allele or rearranged alleles. The migration of size markers (λ DNA digested by EcoRI-HindIII [Promega] and 1-kb DNA ladder [Gibco]) in parallel lanes allowed the size of rearranged CEB1 alleles generated from CEB1-1.8, CEB1-0.6, or CEB1-X parental alleles to be estimated with a resolution of at least two motifs (approximately 100 bp), except for tracts longer than 2 kb.
Pedigree analysis of CEB1 rearrangements.
Cells grown at 23°C were patched onto YPD, and a micromanipulator was used to deposit 12 individual cells with small buds at known positions (MSM; Singer Instruments). The plate was incubated at 30°C and regularly inspected to separate mother and daughter cells. At each generation, the mother cell was left at the same place to give rise to a colony, while the daughter cell was deposited nearby. This procedure was repeated for one or several generations. The plates were incubated for 4 days at 23°C to obtain colonies for DNA analysis.
Sequencing of CEB1 alleles.
The internal structure of the CEB1-0.6 and CEB1-1.8 alleles and of rearranged alleles was determined by DNA sequencing. CEB1 regions were amplified with primers specific to yeast sequences flanking the human minisatellite, as previously described (50). To minimize the introduction of mutations by the Taq polymerase, the Pfu Ultra enzyme (Stratagene) was used exclusively for PCR. To increase the quantity of amplified DNA, and minimize the presence of stochastic PCR-dependent mutations, 8 amplifications were performed in parallel for each CEB1 allele. These PCR products were pooled, precipitated, and electrophoresed in 0.8% agarose gels to verify their size (as predicted by prior Southern blot analysis) and to purify the CEB1 DNA fragment with the NucleoSpin Extract II kit (Macherey-Nalgen). PCR products smaller than CEB1-0.6 (700 bp) were sequenced using the two flanking primers PSF, 5′-CATCATGACAGATCCGAGCT-3′, and PSR, 5′-CGCAGATCCTCTCCTGTGCC-3′, the Big Dye Terminator version 3.1 kit (Perkin Elmer), and 20% betaine (Sigma). PCR products longer than 700 bp were cloned into pGEM-T Easy (Promega) after the addition of dATP at both 3′ extremities by incubation with recombinant Taq polymerase (Invitrogen), deoxynucleoside triphosphates (dNTPs), and enzyme buffer for 6 min at 72°C. The cloned alleles were sequenced as described above, which generally allowed overlapping sequences of 500 to 600 bp to be read. Alleles longer than 1.2 kb were subcloned. For example, the CEB1-1.8 allele in the vector pGEM-T Easy was subcloned by partial digestion with PstI, which cuts at various sites in the minisatellite but not in the vector. The linearized plasmid was purified on a 0.8% agarose gel and cut at the unique NsiI site in the polylinker, allowing various numbers of contiguous CEB1 motifs to be deleted. Linear plasmids of various sizes were gel purified, and the compatible NsiI and PstI ends were ligated. Five overlapping subclones that covered the entire CEB1-1.8 minisatellite were sequenced.
Structural analysis of CEB1 rearrangements.
The progenitor alleles CEB1-0.6 and CEB1-1.8 and newly rearranged alleles were sequenced and compared to determine the origins of the repeats. When several choices were possible due to repeat redundancy, the most parsimonious interpretation that retained the greatest number of parental repeats and minimized the number of events (deletion, insertion, and duplication) was made. For example, since parental repeats 7 and 31 were identical, the rearranged repeat was assigned a designation of 7 if the neighboring repeats resembled repeats 6 and 8. Numerous polymorphisms among the repeats were exploited to identify junctions. For example, in the rearrangement M12B (see Fig. 3A; also see Fig. S2 in the supplemental material), the 7/2 junction unambiguously associated polymorphisms specific to the beginning of motif 7 with polymorphisms specific to the end of motif 2, and this hybrid sequence was followed by motif 3. The junction regions of each rearrangement are outlined with their sequences in Fig. S2 and S3 in the supplemental material. Motifs for which it was not possible to determine a minimalist origin were designated X.
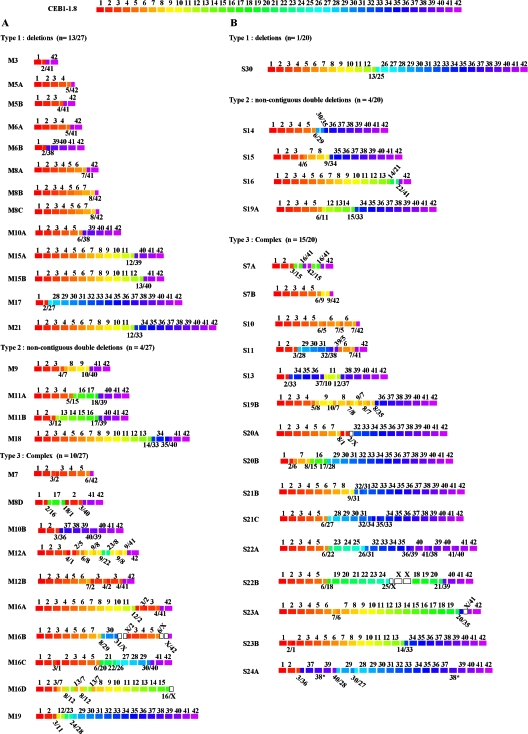
FIG. 3.
Rearrangements of CEB1-1.8 obtained in a rad27Δ diploid (ORD6708) carrying CEB1-1.8 and CEB1-0.6 after growth at 30°C for seven generations (A) and in a rad27Δ haploid (ORD6713-8D) carrying CEB1-1.8 in pedigree experiments (B). Each of the 42 CEB1-1.8 repeats is represented by a colored box and numbered (top). A total of 27 rearrangements obtained after growth for seven generations and a total of 20 rearrangements obtained during pedigree analysis are classified in three categories (types 1 to 3). The name of each rearranged allele is at the left. Hybrid repeats are represented by the two colors corresponding to the fused repeats. A white box indicates a repeat of unknown origin or which cannot be attributed to a specific repeat in the parental CEB1-1.8 allele.
RESULTS
Sequence polymorphism of the CEB1-0.6 and CEB1-1.8 alleles.
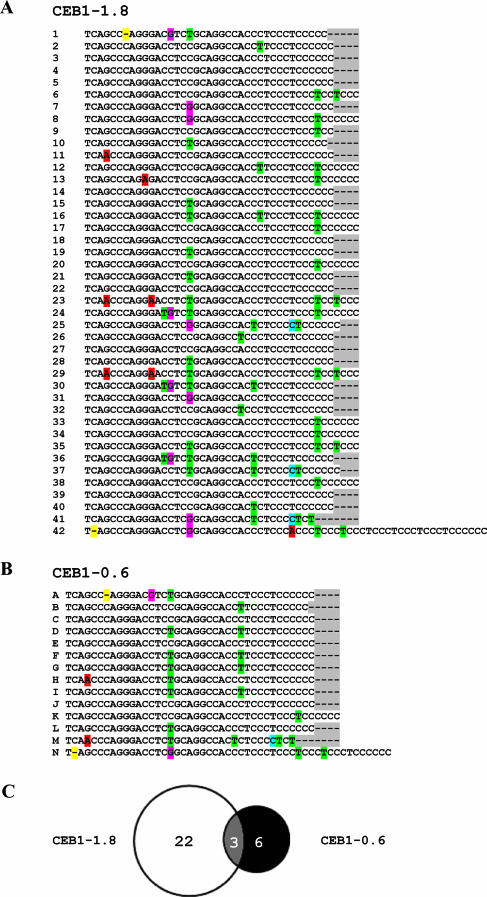
CEB1 rearrangements in human or yeast cells were originally analyzed by the MVR-PCR technique (7-10, 15, 50, 51), which provided a useful overview of minisatellite alleles. To more extensively characterize CEB1 rearrangements, we have made major advances in amplifying and sequencing these GC-rich elements (see Materials and Methods), so we can now routinely and unambiguously determine the sequence of medium-sized arrays (up to 30 repeats). These improvements, combined with limited subcloning, allowed us to sequence the CEB1-1.8 allele, comprising 42 repeats (Fig. 1A) and the CEB1-0.6 allele, with 14 repeats (Fig. 1B). A comparison of these sequences led to several conclusions. First, both alleles were confirmed to consist of tandem repeat motifs; unrelated motifs or repeats were not found. Second, the sizes of the motifs (designated in Fig. 1 by numbers for CEB1-1.8 and letters for CEB1-0.6) vary from 36 to 43 bp. For both alleles, the last motif (which adjoins unique human chromosomal DNA) is slightly longer: 50 bp for the CEB1-0.6 allele (repeat N) and 62 bp for the CEB1-1.8 allele (repeat 42). Although repeat N of CEB1-0.6 is 12 nucleotides shorter than repeat 42 of CEB1-1.8, both alleles share the same terminal sequence composed of several TCCC motifs. Third, as found in previous MVR-PCR analyses, the repeats in each allele are polymorphic; specifically, the 42 CEB1-1.8 repeats include 25 different motifs, and the motifs that are not unique are present in 2 to 8 copies. The most common motif is represented 8 times in successive and dispersed positions (repeats 3, 4, 5, 14, 18, 22, 27, and 39). The extent of CEB1-0.6 polymorphism is similar, as the 14 repeats comprise 9 different motifs, of which the motifs represented by C and D are present in three and four copies, respectively. Repeat C of CEB1-0.6 is identical to repeat 3 of CEB1-1.8, but repeat D is unique to the CEB1-0.6 allele, although the two polymorphisms in this repeat are found individually in other repeats. Figure 1C illustrates the relationship between the CEB1-0.6 and CEB1-1.8 repeats. The sequence data confirm that these alleles are extremely polymorphic. There are 21 and 17 polymorphic sites within the CEB1-1.8 and CEB1-0.6 alleles, respectively. These polymorphisms are mostly single-base substitutions, but they also include two single-base-pair deletions (in repeats 42 and N) and variations in the length of the cytosine tract at the end of each repeat, which explains most of the variation in repeat length. CEB1-0.6 and CEB1-1.8 share only three specific repeat motifs, which are the most common motifs for each allele: the motif of repeats 3, 4, 5, 14, 18, 22, 27, 39, C, E, and J; the motif of repeats 17, 20, 33, 34, 38, and K; and the motif of repeats 15, 19, 21, 28, and L.
FIG. 1.
Sequences of CEB1-1.8 (A) and CEB1-0.6 (B) alleles; polymorphic DNA bases are highlighted. CEB1-1.8 contains 42 repeats (1 to 42), and among these, 25 are unique. CEB1-0.6 contains 14 repeats (A to N), and among these, 9 are unique. The Venn diagram (C) shows the number of repeats shared by the two alleles.
The rearrangement frequency increases with the number of repeats in CEB1 arrays.
We previously examined the mitotic behavior of the CEB1-1.8 and CEB1-0.6 alleles inserted at allelic positions upstream of the ARG4 gene on chromosome VIII (15, 50). Compared to wild-type cells, a rad27Δ/rad27Δ CEB1-0.6/CEB1-1.8 diploid strain (ORD6708) exhibited a strong stimulation (170-fold) of CEB1 size alteration but a sharp difference in the behavior of the two alleles: CEB1-1.8 was rearranged in 41% of cells, while CEB1-0.6 was rearranged in only 2% of cells (50). A similar difference was observed when these diploid cells were grown at permissive (23°C), semipermissive (30°C), and subpermissive (37°C) temperatures (50), as well as in haploid rad27Δ strains (51). The relative stability of the 14-repeat CEB1-0.6 array was surprising, since shorter microsatellite sequences are known to be destabilized in the absence of Rad27 (14, 21, 41, 66). This observation raises the possibility that there is a threshold size for minisatellite instability in rad27Δ cells.
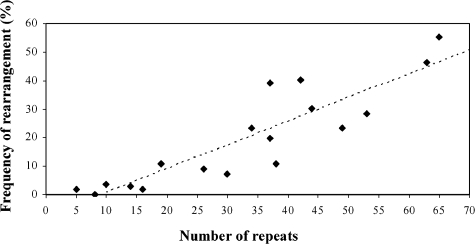
To systematically investigate the relationship between array size and stability, we isolated a series of alleles of various lengths derived from CEB1-1.8 (see Materials and Methods) and measured their rearrangement frequencies. CEB1-0.6 was used as an internal control, and the experiments were performed in rad27Δ/rad27Δ diploid cells carrying the CEB1-0.6 allele on one chromosome and a test allele (CEB1-X) at the same position on the homologous chromosome. The starting lengths of the CEB1-X arrays varied from 5 to 65 repeats. For each strain (rad27Δ/rad27Δ CEB1-0.6/CEB1-X), alterations in the lengths of the CEB1 alleles were determined by Southern analysis, as previously described (see Materials and Methods) (50). For each strain, 56 independent colonies were examined. The overall frequencies of CEB1-X allele rearrangement, including CEB1-0.6, vary by at least 2 orders of magnitude, from less than 2% per cell for one of the shortest alleles (8 repeats) to 55% for the longest allele (65 repeats) (Fig. 2). Rearrangement frequencies increase steadily with allele size (regression coefficient r = 0.79) without an apparent threshold or plateau. This linear relationship indicates that the size of the CEB1 repeat array is the major factor influencing its stability. However, we noted that destabilization frequencies can vary up to twofold for alleles of the same size (for example, in the case of the two CEB1 variants with 37 repeats), implying that factors other than size influence the extent of stabilization. We also analyzed the relationship between CEB1 allele size and the ratio of contractions to expansions. The data in Fig. S1A in the supplemental material confirm a previously reported bias in favor of contractions (50, 51). With the exception of CEB1-0.6, which gives rise to a twofold excess of expansions, this trend is valid with respect to allele size (see Fig. S1B in the supplemental material) and rearrangement frequency (see Fig. S1C in the supplemental material).
FIG. 2.
Relationship between rearrangement frequency (% per cell after seven generations) and the number of CEB1 repeats in rad27Δ diploids (ORD6708-X series) carrying CEB1-X and CEB1-0.6, where CEB1-X indicates an allele of variable length. For the linear regression curve, the regression coefficient is R2 = 0.79. Precise numbers are in Fig. S1 in the supplemental material.
Sequences of the CEB1-1.8 rearrangements.
To characterize the internal structures of CEB1 rearrangements generated in the absence of the Rad27 function and trace their origins, we determined the nucleotide sequence of 27 contractions obtained in a rad27Δ/rad27Δ CEB1-1.8/CEB1-0.6 diploid (ORD6708) grown at 30°C. These variants, identified by Southern analysis of genomic DNA isolated from individual colonies, resulted from shortening of the CEB1-1.8 allele, while the parental CEB1-0.6 fragment remained unchanged. Selective amplification and sequencing of variant alleles (see Materials and Methods) showed that these 27 rearrangements were all unique. Their nucleotide sequences are reported in Fig. S2 in the supplemental material. Sequence analysis confirmed that they contained only motifs and polymorphisms derived from the CEB1-1.8 progenitor array; no CEB1-0.6-specific polymorphisms were present within the altered arrays, nor was there evidence of mutagenic events. These results indicate that all rearrangements resulted from intra-allelic events involving CEB1-1.8.
Comparison with the progenitor CEB1-1.8 allele allowed three classes of rearrangements to be distinguished (Fig. 3A), based on the most parsimonious assessment of the number of events involved (see Materials and Methods). The first class (type 1) comprises 13 variants (48%), schematically illustrated in Fig. 3A. These rearrangements correspond to a simple loss of a variable number (21 to 39) of contiguous repeats to create a hybrid junction repeat. In some cases where two rearrangements were of identical length, sequencing revealed that they were distinct from each other and therefore not clonal. This is illustrated by the M8A, M8B, and M8C rearrangements, which consist of eight repeats (Fig. 3A; see also Fig. S2 in the supplemental material). In Fig. S2 and S3 in the supplemental material, the distance between the reassociated polymorphisms is outlined on the sequence of each rearrangement. This distance is variable and can be as short as 2 nucleotides (S24A). The second class of rearrangement (type 2) includes double deletion alleles and is represented by 4 variants (15%), which in every case contain two junction repeats that define two noncontiguous deletions (Fig. 3A). One example, the M9 rearrangement, arose by a deletion of repeats 5 and 6 and formation of a 4/7 junction repeat plus a deletion of repeats 11 to 39, creating a hybrid 10/40 repeat (see Fig. S2 in the supplemental material). The other rearrangements of this class contain 1 to 4 repeats separating the flanking deletion junctions. The third class of rearrangement (type 3), more complex alleles, comprises 10 variants (37%). These cases are characterized by several novel combinations of repeats and polymorphisms (Fig. 3A). They potentially originated by repeat deletion, repeat addition, and/or fusion of repeats within short stretches of homology, but they are extremely difficult to unambiguously interpret. The difficulty of analyzing these rearrangements is due to partial sequence redundancy among the 42 motifs of the CEB1-1.8 array (see above) and to the multiplicity of events that must be invoked. For example, the M7 rearrangement has only 7 repeats, but it includes a large deletion between repeats 7 to 41 that creates a 6/42 junction repeat and a duplication of repeat 2. Another case, the M8D rearrangement, combines a large deletion of repeats 4 to 39 that creates a 3/40 junction repeat, an insertion of repeats 16, 17, and 18 between repeats 1 and 2, and a duplication of repeats 1 and 2 (see Fig. S2 in the supplemental material). The most extreme case is perhaps the M16B rearrangement, which can be described as an internal loss of 20 repeats that creates an 8/29 junction repeat (Fig. 3A; see also Fig. S2 in the supplemental material) and a duplication of repeats 2, 3, 4, and 5 and part of repeat 6 that is apparently inserted between repeats 31 and 42, creating a deletion between repeats 32 and 41. This insertion is flanked by two junction repeats of uncertain origin, 31/X and X/42, because the polymorphisms of repeat 31/X are found in 8 CEB1-1.8 repeats, and the polymorphisms of the junction repeat X/42 are also frequently represented throughout the array. The complexity of these rearrangements is suggestive of multiple molecular events occurring over short distances that can create mosaics from preexisting sequence.
Pedigree analysis of CEB1-1.8 rearrangements.
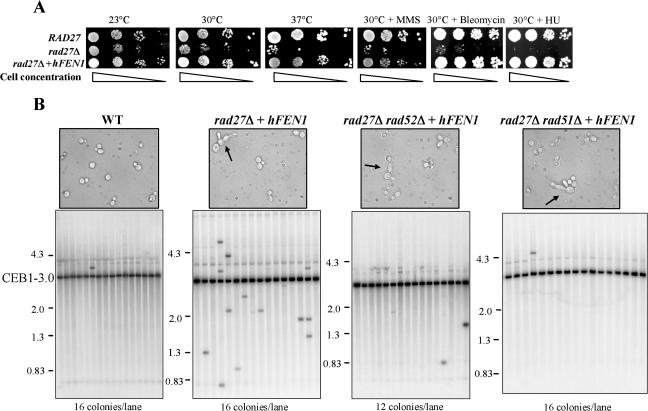
The complexity of the CEB1 rearrangements that arose over seven generations immediately raises the question of whether they arose in a single generation or resulted from alterations that accumulated over several generations. To address this issue, we performed pedigree analysis of haploid rad27Δ CEB1-1.8 cells (ORD6713-8D) to monitor destabilization, as described in Materials and Methods (Fig. 4A). The first parameter measured was cell viability. As controls for the rad27Δ CEB1-1.8 strain, we also examined RAD27 (wild type) strains with or without CEB1-1.8 (ORT2914 and MGD131-102A, respectively) and a rad27Δ strain without CEB1 (ORD6728-1C). As illustrated in Fig. 4B, we found that in the RAD27 background, with or without CEB1-1.8, all cells gave rise to viable progeny (24/24 and 34/34, respectively). In sharp contrast, a significant fraction of rad27Δ and rad27Δ CEB1-1.8 cells did not form colonies: 6/72 (8.3%) and 61/510 (12%), respectively. A similar lethality (6.3%) was observed for rad27Δ cells carrying the shorter CEB1-0.6 allele (ORD5330-3C). These data indicate that the absence of Rad27 results in cell death independently of the presence of the CEB1 minisatellite. In these experiments, we also measured cellular doubling time and observed that it was similar in rad27Δ and wild-type (WT) cells (150 mn and 154 mn, respectively). Therefore, in the absence of Rad27, the high rate of cell death solely explains the overall reduced growth of rad27Δ cells in liquid culture.
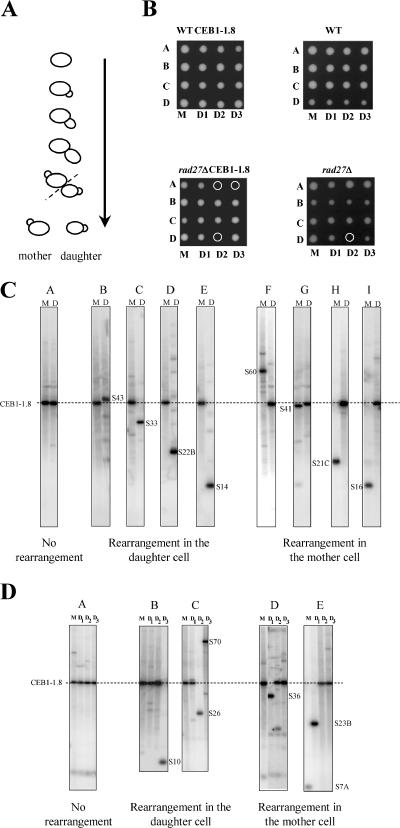
FIG. 4.
Pedigree analysis of rearrangements. (A) Haploid cells were deposited on an agar plate, incubated at 30°C, and regularly inspected to separate mother and daughter cells by micromanipulation after the plate was incubated at 23°C to allow colony formation. (B) Multigeneration pedigree analysis of wild-type (WT) haploids with (ORT2914) or without (MGD131-102A) CEB1-1.8, and of rad27Δ haploids with (ORD6713-8D) or without (ORD6728-1C) CEB1-1.8 for three generations to estimate the lethality rate; four pedigrees (A, B, C, and D) are shown for each strain. The colony formed by the first mother is indicated M, the colony formed by the daughter of M is D1, the daughter of D1 is D2, and so on. Cells which failed to form a colony are indicated by a circle. (C) Southern analysis of single-generation pedigrees of a rad27Δ haploid carrying CEB1-1.8 (ORD6713-8D). DNA extracted from mother (M) and daughter (D) cells was digested by AluI and hybridized with a CEB1 probe. Three cases are presented: no rearrangement (A), only the daughter cell carries a rearrangement (B, C, D, and E), and only the mother call carries a rearrangement (F, G, H, and I). The name of each rearrangement is indicated. (D) Southern analysis of multigeneration pedigrees in a rad27Δ haploid carrying CEB1-1.8 (ORD6713-8D). DNA from mother (M) and daughter (D1, D2, and D3) cells was digested by AluI and hybridized with a CEB1 probe. Three cases are presented: no rearrangement (A), the daughter cell carries the rearrangement (B and C), and the mother cell carries the rearrangement (D and E). The name of the rearrangement is indicated.
The Southern analysis shown in Fig. 4C and D illustrates several examples of single- and multiple-generation pedigrees in which CEB1-1.8 rearrangements occurred. In Fig. 4C, panel A, the CEB1 allele in the mother (M) and daughter (D) colonies is of parental size, indicating that no rearrangement occurred in this lineage. In contrast, the adjacent panels show CEB1 variants. Figure 4C, panels B to E, shows that the mother cell carries a parental CEB1-1.8 allele, whereas daughter cells carry an altered CEB1 allele which is expanded (S43) or contracted (S33, S22B, and S14), indicating that de novo rearrangement occurred in the mother cell and was transmitted to the daughter. In contrast, in panels F to I, CEB1 is altered in the mother cell (contractions and expansions were both observed), while the daughter cell carries an allele of parental size. In these cases, the de novo rearrangement occurred in the mother cell, which transmitted the unaltered chromosome to its daughter. These results demonstrate that CEB1 rearrangements occur in a single generation. Altogether, among 519 pairs of mother and daughter cells, we found 29 rearrangements, indicating a rate of rearrangement per generation of 5.6%, consistent with our previous estimate of 5.9% per generation, which was obtained by analyzing cells after seven generations of growth in liquid media (50).
Next, to determine whether rearrangements could arise in successive generations, we analyzed the frequency and the pattern of CEB1 rearrangements in 72 pedigrees which were extended for 3 to 7 generations without the death of a mother or daughter cell. Of 246 single-cell generations, we observed 15 CEB1 rearrangements, indicating a rate of 6.1% rearrangement per generation. Figure 4D illustrates four examples of multigeneration pedigrees in which a rearrangement occurred (panels B to E). In panel B, lane D3, a large contraction of CEB1-1.8 is visible, and this pedigree indicates that rearrangement occurred in the D2 cell. In contrast, in panel D, a contraction event is visible in the D1 cell, whereas the mother cell (M) and the successive daughters (D2 and D3) exhibit a parental CEB1-1.8 allele. This pedigree indicates that rearrangement occurred and was retained in the D1 cell (the mother of the D2 cell) while the nonrearranged CEB1-1.8 molecule was transmitted to its daughter, D2. Figure 4D also shows two pedigrees in which CEB1 was altered twice and thus in successive generations. As shown in panel C, a contraction occurred in the D1 cell that was inherited by the daughter cell (D2). Then, during the next generation, the rearranged allele underwent expansion and was transmitted to its daughter, D3. And as shown in panel E, a large contraction occurred and was retained in the mother cell (M), while its daughter (D1) inherited the parental CEB1-1.8 allele. This allele was then rearranged in the D1 cell, resulting in a contraction retained in the D1 cell while the D2 cell inherited the original CEB1-1.8 allele, which it transmitted intact to the D3 cell. To estimate the frequency at which rearrangements occurred in each of two successive generations, we first calculated the number of two successive generations among the 72 pedigrees performed, which included a total of 246 single generations (246 − 72 = 174 successive generation doublets). Only one case (Fig. 4D, panel E) with two successive rearrangements of the CEB1-1.8 allele was observed among the 174 successive generation doublets, which provides a frequency of roughly 0.5% (1/174). The case presented in panel C in Fig. 4D cannot be included in this calculation, because the second rearrangement (observed in the D3 cell) occurred in the D2 cell on a previously rearranged CEB1 allele, for which the destabilization frequency per generation has not been determined by pedigree analysis, in contrast to that of the CEB1-1.8 allele (6.1%). Since the expected frequency of two successive but independent CEB1-1.8 rearrangements is 0.37% (6.1% × 6.1%), we conclude that these successive events are likely independent. There is no evidence of recurrent destabilization among generations.
Two additional observations concern the transmission of rearrangements from mother to daughter cells and the ratio of expansions to contractions. The data reported in Table 2 indicate that rearranged alleles were equally inherited by mother and daughter cells (24/44 and 20/44 events, respectively) and, in both situations, most events were contractions (20/24 and 16/20, respectively).
TABLE 2.
Repartition of CEB-1.8 rearrangements in daughter and mother cells in the rad27Δ haploid (ORD6713-8D)
| Rearrangement and generation type | No. of rearrangements in cell type
|
Total no. of rearrangements | |
|---|---|---|---|
| Daughter | Mother | ||
| Contraction | 36 | ||
| Single | 9 | 14 | |
| Multiple | 7 | 6 | |
| Expansion | 8 | ||
| Single | 3 | 3 | |
| Multiple | 1 | 1 | |
| Total | 20 | 24 | 44 |
CEB1-1.8 rearrangements arising in a single generation are diverse and complex.
To further characterize the nature of the CEB1 rearrangements, we determined the sequence of 20 contractions obtained from the pedigree analysis of rad27Δ CEB1-1.8 cells (ORD6713-8D). Several diverse classes of rearrangements were observed in this sample (Fig. 3B; see Fig. S3 in the supplemental matieral). The type 1 class (simple internal deletion) is represented by one rearrangement (S30) resulting from the loss of repeats 14 to 24 and the creation, at the junction, of a 13/25 hybrid repeat. The type 2 class, characterized by a double deletion structure and two hybrid junctions, is represented by 4 cases. The more complex alterations of the type 3 class represent the majority of events (15/20). Again, they are strikingly complex in that they include multiple deletions, duplications, and insertions of repeats, creating novel combinations of preexisting polymorphisms over short distances. The S24A rearrangement, characterized by multiple deletions and the creation of novel junction repeats, has two additional intriguing features. First, it contains an isolated internally modified repeat 38 motif (designated 38*) corresponding to a 4-bp deletion (TCCC) of the triplicate run of TCCC motifs present in the parental repeat. The second intriguing and likely related feature of this S24A rearrangement is the presence of the same modification, embedded within a tract interpreted as repeats 37, 38*, and 39 that is inserted at the beginning of the rearranged array. The repeat 38* modification may have resulted from a short gene conversion event in which part of one of the several repeats which constitutively carries only two TCCC runs at this position was copied; alternatively, it may be due to polymerase slippage. The dual presence of this sequence within the same rearrangement suggests a two-step process in which the primary event (gene conversion or replication slippage) created the modified repeat 38*, which was then duplicated with other flanking repeats at the beginning of the array.
Altogether, the analysis of CEB1 rearrangements obtained by pedigree analysis indicates that (i) they occur in a single generation without recurrence within a cell lineage, (ii) they are as diverse as those isolated after several generations of growth, (iii) the majority are complex, and (iv) complex rearrangements are found in contraction alleles that lost few or many repeats.
The formation of CEB1 rearrangements is Rad52 and Rad51 dependent.
Despite their complexity, the structure of the rearranged arrays strongly suggests that they arose by a homology-dependent mechanism. Consistent with the known phenotypes of rad27Δ cells, a candidate mechanism to generate such events is the activity of the Rad52 pathway for recombinational repair. The most straightforward genetic assay to establish a role for this pathway in the formation of CEB1 rearrangements is the examination of the frequency of rearrangements in a rad27Δ rad52Δ double mutant. Unfortunately, this is not possible because this combination of mutations confers synthetic lethality at all temperatures (16, 72, 75); similarly, null mutations of the other genes of the Rad52 pathway also confer lethality in the rad27Δ background (16, 49, 57, 76). To circumvent this limitation, we functionally complemented the rad27Δ mutation by expression of the human hFEN1 cDNA (20, 23, 25). To obtain the desired experimental strain (rad27Δ hFEN1 rad52Δ CEB1-1.8), we first constructed a yeast strain expressing the hFEN1 cDNA under the RAD27 promoter (see Materials and Methods). As expected, the expression of hFEN1 significantly improved the growth of rad27Δ cells (ORT5027-1) and restored wild-type levels of resistance to methyl methanesulfonate, bleomycin, and hydroxyurea (Fig. 5A), and it stabilized the CEB1-1.8 allele. At 30°C, in comparison to the high degree of CEB1-1.8 instability observed in rad27Δ cells (41%), the frequency of rearrangement in the rad27Δ hFEN1 CEB1-1.8 strain ORT5027-1 was much lower (0.2%); only a single size alteration was observed among 513 colonies analyzed. This low level is similar to the frequency of 0.3% (5/1,824 events) observed in the RAD27 CEB1-1.8 strain ORT2914. Altogether, these results demonstrate that hFEN1 cDNA under the control of the endogenous RAD27 promoter extensively complements the phenotypic defects of the rad27Δ mutant.
FIG. 5.
Complementation by hFEN1 and instability of the CEB1-3.0 allele in the absence of Rad51/Rad52. (A) Serial dilutions (5,000 to 5 cells) from wild-type (WT) (ORT2914), rad27Δ (ORD6713-8D), and rad27Δ plus hFEN1 (ORT5027-1) haploids were plated on rich media (YPD) at 23°C, 30°C, and 37°C or on rich media with MMS (0.01%), bleomycin (0.2%), or hydroxyurea (10%) at 30°C. (B) Detection by Southern analysis of CEB1-3.0 rearrangements after growth at 37°C in WT (ORD6778-4B), rad27Δ plus hFEN1 (ORT5027-1), rad27Δ rad52Δ plus hFEN1 (ORD6780-22A), and rad27Δ rad51Δ plus hFEN1 (ORD6781-35A) haploids. Each lane contains DNA extracted from pools of 12 or 16 colonies digested by AluI and hybridized with a CEB1 probe. The positions of molecular weight markers (λDNA digested by HindIII and EcoRI) are indicated. The cells were photographed after overnight growth at 37°C; arrows indicate abnormal cell shape due to partial complementation by hFEN1.
One drawback of our hFEN1 complementation is that the frequency of CEB1-1.8 rearrangements per colony is less than 1%, as assayed at the standard temperature of 30°C, which in practice prevents the use of Southern analysis, even of pooled samples, to detect a reduction in destabilization conferred by deletion of the RAD52 gene. To overcome this limitation, we identified conditions in which the complementation of rad27Δ defects by the hFEN1 protein was partial and cells were still viable in the absence of Rad52. First, we assayed a longer CEB1 allele, the CEB1-3.0 variant (65 repeats), which is more unstable than the CEB1-1.8 allele, 7.9% versus 5.6% per generation (Fig. 2). Second, we increased the temperature of the assay from 30°C to 37°C, since in rad27Δ cells, the frequency of CEB1-1.8 rearrangement is ∼2-fold higher at 37°C (13.9% per generation) than at 30°C (5.6% per generation) and ∼4-fold higher than at 23°C (3.2% per generation) (50). Data showing the partial complementation of rad27Δ mutant defects upon expression of hFEN1 cDNA are presented in Fig. 5B. Namely, rad27Δ hFEN1 CEB1-3.0 (ORD6778-4B) cells grown at 37°C exhibit heterogeneous morphology and multiple buds (a phenotype typical of rad27Δ cells), and size variation of the CEB1-3.0 allele was frequently observed. Among 464 rad27Δ hFEN1 CEB1-3.0 colonies analyzed, we observed 21 size variants (4.5%). This is a frequency approximately 20-fold higher (P < 0.001) than for the control RAD27 CEB1-3.0 strain (ORD6780-11A), for which we observed only two rearrangements among 432 colonies (0.25%) (Fig. 5B). We then confirmed the viability of the rad27Δ hFEN1 CEB1-3.0 strain in the absence of Rad52 at 37°C. Upon sporulation of the progeny of the heterozygous diploid strain ORD6780, we easily isolated rad27Δ hFEN1 rad52Δ CEB1-3.0 haploids (e.g., ORD6780-22A) and thus could assay the frequency of CEB1-3.0 alteration after growth at 37°C. Among 348 colonies, we found only two size alterations (Fig. 5B), reflecting a frequency of rearrangement of 0.5%. Compared to the frequency of 4.5% observed in the control rad27Δ hFEN1 CEB1-3.0 strain (ORD6778-4B) also grown at 37°C, we conclude that the absence of Rad52 substantially reduces the frequency of CEB1-3.0 rearrangements. This 10-fold decrease is highly significant (P < 0.001).
Finally, we also assayed the effect of inactivating the RAD51 gene by constructing a rad27Δ hFEN1 rad51Δ CEB1-3.0 strain (ORD6781-35A). This strain was viable at 23, 30, and 37°C, and as for the absence of Rad52, we observed that the absence of Rad51 severely reduced the frequency of CEB1-3.0 rearrangements. Among a total of 384 colonies, we found a single rearrangement (0.25%) (Fig. 5B). Compared to the frequency of 4.5% observed for the rad27Δ hFEN1 CEB1-3.0 strain, the absence of Rad51 reduces CEB1 rearrangements approximately 10-fold (P < 0.001). Altogether, these results demonstrate that the formation of all of the CEB1 rearrangements assayed in the context of partial complementation of the rad27Δ defect by expression of hFEN1 absolutely requires the activity of Rad52 and Rad51, two proteins of recombination and repair pathways.
DISCUSSION
In this study, we have examined the fate of the polymorphic CEB1 minisatellite in the total or partial absence of the Rad27/hFEN1 flap endonuclease. Our main findings are that (i) CEB1 rearrangements, characterized for the first time here by sequencing, are definitively diverse and frequently complex, (ii) the frequency of rearrangement increases with the size of the tandem array, (iii) rearrangements occur in a single generation, and (iv) their production is fully dependent on the activity of the RAD52 recombinational repair pathway.
CEB1 rearrangements are often complex but exhibit features of homology-driven processes.
In the past, the difficulty of accurately amplifying and sequencing long tandem arrays, especially those with GC-rich sequences such as CEB1, limited the high-resolution analysis of repeat rearrangements. At best, the internal structure of the minisatellite array was partially determined by the MVR-PCR method, which exploits sequence variations among repeat units within minisatellite arrays (33, 38). In the case of CEB1, three polymorphic sites within the repeats were utilized (7-10, 15, 50, 51). These sites were sufficient to provide a general overview of parental and rearranged alleles, but only a subset of polymorphic repeats could be visualized, and additional technical issues associated with uncharacterized polymorphisms often interfered with the characterization of some structures. Here, our strategy for sequencing the highly unstable CEB1-1.8 progenitor allele with 42 repeats has greatly improved the resolution of the analysis, since all internal polymorphic markers can be identified (a total of 23 polymorphisms at 21 positions within the repeats) and showed that 25 of the 42 repeats are unique (Fig. 1).
We sequenced 47 CEB1-1.8 contraction alleles ranging in size from 3 to 30 repeats (Fig. 3) and inferred their potential origins by comparison with the progenitor array. The most striking feature of these rearrangements is their extreme diversity; this can be described in several ways. First, most, if not all, possible array lengths were generated. The sizes of all the CEB1-1.8 rearrangements that we have identified so far in rad27Δ or rad27Δ/rad27Δ cells (50; present study) are presented in Fig. S4 in the supplemental material. This distribution ranges from 3 to 100 repeats, with a median length of around 22 repeats. We found that expansions occurred less frequently than contractions and that they represent approximately 25% of all events. CEB1-1.8 expansions are also of various sizes, ranging from 45 to 100 repeats. A second observation describing the diversity of the rearrangements is their internal structure, as characterized by repeat composition and order (Fig. 3). So far, each rearrangement typed by MVR-PCR analysis (50) and sequencing (present study) has been unique, even those of identical length. Third, the rearrangements, whether simple or complex, invariably retain the tandem repetitiveness typical of a minisatellite. The absence of truncated repeats and of internal deletions within the repeats (although several short stretches of identical motifs were noted) and the lack of de novo mutagenesis (with one exception) imply that the rearrangements arose by high-fidelity molecular processes such as homologous recombination and template-directed DNA synthesis. Fourth, all rearrangements retained the first parental repeat (1) and were terminated with a complete or hybrid parental repeat 42 (except the M16D rearranged allele). This feature indicates that the production of the rearrangements either never involved flanking sequences or that it extended transiently outside the repeat array but was resolved without loss or mutagenesis. In conclusion, the sequences of the 47 rearrangements derived from the highly polymorphic CEB1-1.8 allele show that the absence of Rad27 yields an extensive homology-guided reshuffling of the progenitor sequence. With the exception of a single 4-bp deletion within a triplicate tetranucleotide tract, which likely occurred by polymerase slippage during DNA synthesis (as in the case of S24A in Fig. 3B; see also Fig. S3 in the supplemental material), we did not observe a point mutation associated with the rearrangements. It suggests that the production of the rearrangements does not involve error-prone replication or DNA repair pathways dependent on low-fidelity polymerase activities or nonhomologous end-joining mechanisms, although we cannot formally exclude their contribution. All of these features are consistent with the prominent role, demonstrated here, of the homology-dependent Rad52 repair pathway.
CEB1 destabilization in the absence of Rad27/hFEN1 activity likely occurs within the minisatellite array.
Numerous studies have demonstrated that the mammalian hFEN1 (Flap endonuclease) protein and its Rad27 counterpart in S. cerevisiae are structure-specific 5′-3′ exo/endonucleases primarily involved in replication and repair (48). In vitro, the Rad27/hFEN1 protein recognizes various branched DNA structures, particularly the 5′ flaps that are generated by displacement of the RNA primers that initiate lagging-strand synthesis, and it cleaves at the base of the flap. Together with other proteins, notably RNase H1 and the single-stranded DNA binding proteins RPA and Dna2, Rad27/hFEN1 fully removes the RNA primer, thereby generating a nick that is sealed by DNA ligase to form an uninterrupted lagging strand. The inactivation of Rad27/hFEN1 in vivo leads to a constellation of phenotypes consistent with its prominent role in DNA metabolic processes (41, 47, 75). In S. cerevisiae, rad27Δ cells (Fig. 5) resemble other mutants that are defective in DNA replication, and it is widely held that Rad27 is the major structure-specific nuclease required for processing of the >100,000 Okazaki fragments generated during S phase (48). Although the RAD27 gene is not essential in budding or fission yeast, the high frequency (12% per generation) of cell death observed by pedigree analysis (Fig. 4B) at all temperatures (independent of the presence or absence of the CEB1 minisatellite) indicates that alternative flap processing pathways are not functionally equivalent. Alternative modes of Okazaki fragment maturation dependent on Exo1 or Mre11 nuclease activities have been proposed (54), and redundant flap activities have been investigated by various genetic, molecular, and biochemical methods. Another protein that may process flap structures is Dna2, a multifunctional helicase/nuclease (12). Current biochemical and genetic characterization suggest that Dna2 could be involved in the processing of some flaps, with a specific role in processing abnormally long flaps (13, 39, 40). Two other observations supporting a role for Dna2 in Okazaki fragment processing are the synthetic lethality of the dna2-1 rad27Δ double mutant (11) and the suppression of temperature sensitivity that is conferred by the dna2-1 allele in cells that overexpress Rad27 (11). Finally, the low but significant destabilization of the CEB1-1.8 allele in the dna2-1 mutant at subrestrictive temperatures (30 to 32°C) (50) reveals a common phenotype, suggesting roles for Rad27 and Dna2 in similar processes. Indeed, these results suggest that in rad27Δ cells grown at 37°C or below, Dna2 sufficiently processes replicative 5′ flaps to allow proper lagging-strand synthesis in a significant fraction of cells, especially if, in the absence of Rad27, excessive elongation of these flaps is generated due to an enhanced strand displacement activity of DNA polymerase δ. Nonetheless, under these conditions, defects in processing of the 5′ flaps generated during replication may allow them to accumulate and/or mature into abnormal recombinogenic structures, consequently triggering rearrangements of repetitive sequences. Okazaki fragments are currently estimated to be 100 to 150 nucleotides long (1). If they are of similar size in the absence of Rad27, then, on average, every third repeat of a CEB1 allele may be covered by an unprocessed 5′ flap, and several such flaps could be spaced along the CEB1-1.8 allele. A strong implication of this model is that rearrangements are initiated at various positions within the array.
The view that the key initiation event occurs within the array could account for several other observations and explain the relationship between rearrangement frequency and the total number of repeats. We found that the frequency of destabilization steadily increases over a 27-fold range with the number of repeat units (Fig. 2). This observation indicates that the overall size of the tandem array is the major factor influencing the frequency of destabilization, although some variation among alleles of the same size implies that other factors, such as base composition, secondary structure, or the degree of internal polymorphism, modulate the size parameter. The effect of size may simply reflect the increased probability that more than one recombinogenic flap structure may arise in longer arrays. As discussed below, additional observations concerning the structure of the rearranged alleles further support the view that destabilization of the CEB1 alleles in rad27Δ cells initiates within the array.
The low level of instability of the shortest alleles suggests a threshold situated around 10 repeats. This minimal size (approximately 400 bp) may indicate that a single 5′ flap covering the tandem array is not sufficient to induce a recombinogenic lesion or that, in the absence of Rad27, Okazaki fragments are substantially extended because of the delay in flap processing. Therefore, 5′ flaps may less frequently involve the array. Alternatively, the low frequency of rearrangement for alleles with fewer than 10 repeats, which is similar to the background frequency observed in wild-type cells, raises the possibility that these rearrangements are initiated by a different mechanism. One possible source of destabilization is the stochastic or minisatellite-induced arrest of a replication fork, an event known to induce genome instability (42).
Mechanisms of rearrangement formation.
To examine the mode of rearrangement formation, we performed a pedigree analysis. This study yielded several new insights. First, it allowed us to precisely determine the frequency of CEB1-1.8 rearrangements. Second, we noted that a single rearranged molecule is produced per generation and that it can be retained by either the mother or daughter cell. Finally, we noted that the internal structures of the rearrangements obtained in a single generation are as diverse and complex as those characterized after several generations (and their frequency per generation is also similar). Altogether, these results reveal that the production of rearrangements is asymmetric, consistent with formation during replication.
Assuming that the primary effect of the absence of the Rad27 activity is the accumulation of 5′ flap structures, the rearrangements could arise by several mechanisms. One class of models (50) envisaged that an uncleaved or incompletely cleaved 5′ flap strand containing repeats folds back to reanneal with the template in various registers, thus creating a deletion or a duplication of several contiguous motifs. Two considerations now lead us to disfavor this model. First, it does not easily explain complex rearrangements, which are frequent and occur in a single generation, unless clustered, reiterative, and out-of-register strand interaction and reannealing events are invoked. The second and more compelling reason is that reannealing of the flap is not expected to depend on the Rad52 recombinational repair pathway. Although the annealing activity of Rad52 is compatible with this model (55), the strict dependency on Rad51 activity is inconsistent with the extensively characterized biochemical properties of the eukaryotic Rad51 strand-exchange protein and its orthologue, the RecA protein of Escherichia coli (43) or the human RAD51 protein (71, 79). Alternatively, a more likely model is that an accumulated 5′ flap provokes a secondary recombinogenic lesion such as a single-strand gap or DSB. Thus, rearrangement would result from faulty recombinational repair of this secondary lesion. Rearrangement may occur immediately, if the flap structure triggers a processing nuclease activity, or during the next generation, if, for example, a primary lesion such as a single-stranded DNA nick or gap is converted into a DSB by DNA synthesis in the next round of replication. The formation of a recombinogenic lesion is strongly supported by two genetic observations: first, the demonstration here that the formation of CEB1 rearrangements fully depends on the activity of the Rad52 pathway, which is required for the homology-dependent repair of DSBs or single-strand gaps, and second, that synthetic lethality of the rad27Δ mutation in combination with null mutations of genes in the Rad52 group (16, 49, 57, 72, 75, 76).
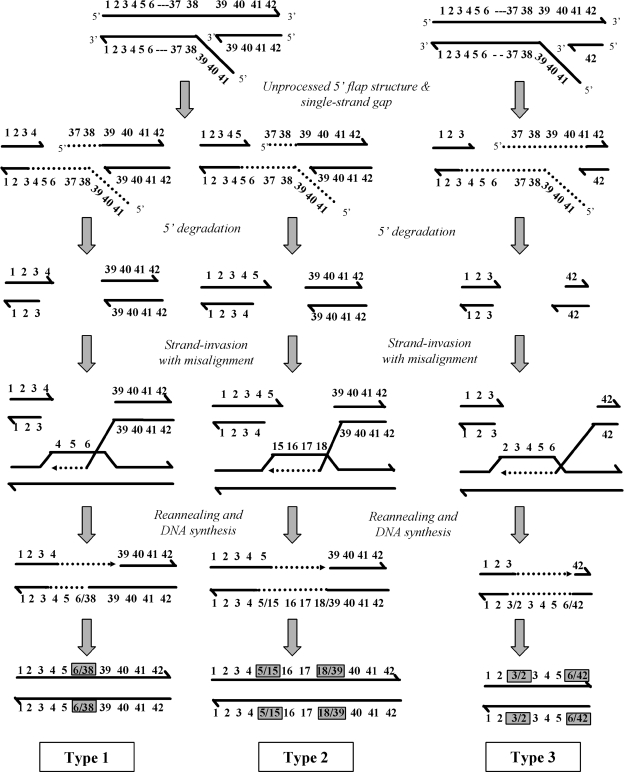
Once a recombinogenic lesion forms, the SDSA model illustrated in Fig. 6 best accounts for the formation of the rearrangements per se and of their diversity. As proposed for the initiation of homologous recombination or the repair of spontaneous or induced DSBs, the processing of an aberrant flap structure into a DSB generates single-stranded molecules. The activity of the Rad51 strand-exchange protein and its cofactors (43, 71, 79) commits the 3′ end to a homology search and catalyzes a canonical but misaligned step of strand invasion using the sister chromatid as a recipient (already replicated upon leading strand synthesis). If the invading strand is not rejected by the presence of mismatches resulting from repeat polymorphism, its 3′ end is extended by DNA synthesis until the newly synthesized strand dissociates from the template and anneals with the complementary strand located on the other side of the DSB in the initiating molecule. Finally, DNA synthesis copies the reannealed strand, thus creating the final rearrangement on the initiating chromosome and leaving the sister chromatid unchanged. Because repeated sequences can align in different registers during strand invasion and the extent of DNA synthesis can vary, the rearrangements can be diverse. As illustrated in Fig. 6, an out-of-register invasion into the array can lead to a type 1 event when DNA synthesis is sufficient to cover the repeats present on the side of the DSB and reannealing occurs in register. Type 2 events occur when strand invasion is out of register and synthesis is insufficient to cover the extremity of the resected DNA, creating a second recombinant junction following out-of-register reannealing (Fig. 6). To explain type 3 complex events, we envisage that multiple rounds of strand invasion, limited DNA synthesis, and destabilization occur and that the accumulation of several flaps along a minisatellite array leads to independent but cumulative events. Precisely, the high density of polymorphisms allows the physical distance between the newly associated polymorphisms to be measured. As shown in Fig. S3 in the supplemental material, this distance can be as short as 2 bp (S24A). Short patch polymorphism reassociation may reflect limited DNA synthesis and/or mismatch correction of heteroduplex DNA formed during the various strand reannealing steps, thus further increasing the diversity of the rearrangements. Finally, small loops on template or donor strands may allow polymerase slippage during DNA synthesis and cause rare internal repeat sequence changes, such as the 4-bp deletion observed in repeat 38 of the S24A rearrangement.
FIG. 6.
Mechanisms of formation of CEB1-1.8 rearrangements in rad27Δ strains according to the SDSA model. The minisatellite repeat units are numbered on each DNA strand (for clarity, not all are shown). Unprocessed 5′ flap structures at extended Okazaki fragments during lagging-strand synthesis can cause replication fork stalling, and a secondary recombinogenic lesion, such as a single-strand gap, can occur in the other DNA strand. After 5′-to-3′ resection (dashed line), the 3′ end invades the sister chromatid in a homology search and can anneal out of register to repeated sequences. After DNA synthesis of a variable extent, the invading strand reanneals to its original complementary strand, and different types of rearrangements are generated according to the register of annealing, the extent of mismatch correction, the extent of DNA synthesis, and the register of reannealing. Three examples are shown: the M10A (type 1), M11A (type 2), and M7 (type 3) rearrangements presented in Fig. 3A.
Previous studies of tandem repeat rearrangements demonstrated an alternative mechanism of DSB repair mediated by the SSA pathway (30, 58, 78). In this case, DNA resection causes complementary single-strand tails to be exposed, and annealing of these tails deletes intervening sequences. Similarly, a DSB located within the minisatellite array can be repaired by SSA of distant repeats, thus generating an internal deletion: i.e., a type 1 event. However, we do not think that the SSA pathway finally contributes to the production of the present type 1 events. First, in contrast to the present results, SSA events are Rad52 dependent but Rad51 independent (30). Second, there is a surprisingly low proportion of type 1 events (17/64) compared to the other two classes. Interestingly, Sugawara et al. recently showed that sequence divergence of 3% of repeated sequences flanking an inducible HO DSB causes a sixfold reduction in repair by SSA compared with repair involving identical sequences (69). It is therefore possible that the low rate of SSA-like events observed here is a consequence of the higher sequence polymorphism of the minisatellite repeats present within the CEB1-1.8 allele (27% divergence for the most divergent motifs: 20 and 41) (Fig. 1A). Another recent study showed that the instability of a natural S. cerevisiae minisatellite (135-bp repeats) present in the FLO1 gene requires Rad52 but not Rad51 (78). The difference with the present results can be explained by the fact that the FLO1 gene contains repeats with long stretches of perfect homology (135 bp), while the maximal length of homology for the CEB1 minisatellite is 43 bp; furthermore, there is no repeat length variation in the FLO1 minisatellite compared to CEB1. In conclusion, the remarkable complexity of many CEB1 rearranged alleles suggests that the processing of the initiating lesion (probably a DSB) likely involves a multistep and reiterated SDSA mechanism that profoundly shuffles repeats to create complex rearrangements in a single “event.”
The heterogeneous features of minisatellite destabilization and rearrangement in yeast and human cells may reflect initiation in or near the array.
Several features of minisatellite instability in yeast and human cells are convincingly similar, but there are also several differences that remain to be better understood. Two prominent shared features are the relative stability of minisatellites in somatic cells compared to meiotic (germ line) cells and the production of highly diversified rearrangements. In contrast, the effects of minisatellite size, the location of events within arrays and the types of rearrangements can vary. Despite several potential modulating factors, a simple hypothesis is that these differences reflect only the position of the initiating lesion(s), whether within or outside the array. In general, there is no universal relationship between array length and the frequencies of destabilization, with respect to the minisatellite, cell type, or organism under consideration. In the human germ line, the frequency of rearrangement of the CEB1 and B6.7 minisatellites increases with array size (7, 74), but in the case of the MS32 and MS205 minisatellites, destabilization does not correlate with size (37, 52). One interpretation is that the CEB1 and B6.7 minisatellites are destabilized by events that initiate within these arrays but that rearrangement of MS32 and MS205 is provoked by external events. Two other observations are consistent with this interpretation and indicate that, in both cases, destabilization may depend on natural recombination-initiating sites. First, the studies of Monckton et al. (53) have shown that a single-nucleotide polymorphism located near MS32 is associated with a more-than-100-fold variation in the frequency of MS32 destabilization. Second, recent analysis of a meiotic recombinational hotspot in the human genome has identified a highly represented 7-nucleotide sequence (CCTCCCT) (56) that is present intact in several CEB1 repeats and as a version with one substitution in the B6.7 minisatellite, whereas MS32 and MS205 do not contain this sequence or a close match. The third feature that distinguishes these pairs of minisatellites is that MS32 rearrangements are frequently interallelic and polarized with respect to one end of the array (indicative of an initiating element acting in cis to the array). In contrast, CEB1 rearrangements are less polarized and the size effect is largely attributable to an increase in the proportion of intra-allelic events, which again is more compatible with initiation within the array. Thus, consistent with the mode of CEB1 destabilization in yeast meiosis or in the absence of Rad27 in mitotically growing cells, it is likely that germ line destabilization is initiated by events outside the array for MS32 and MS205 but by events within the array for CEB1 and B6.7 and, in all cases, provoked by the Spo11-dependent DSBs that naturally initiate meiotic recombination.
Single-generation rearrangements in rad27Δ-deficient cells could be a source of rapid genome evolution.
More and more precise genome sequence annotation indicates that, like microsatellites, minisatellite repeats are spread along the chromosomes and are located in both intergenic and coding regions (45, 46, 61, 77, 78) and thus makes them interesting structures for exploring the mechanisms underlying genome and gene evolution. The present results, showing that an extreme diversity can be created in a single generation in the absence of Rad27/hFEN1 protein raise the possibility that an inactivation of this gene function in a cell population and the stochastic accumulation of unresolved flaps during replication (in wild-type or mutant cells) could be a source of rapid genome evolution by destabilizing functional repeated sequences. With the Rad27/hFEN1 protein being evolutionarily conserved and the impairment of Rad27 function conferring the strongest mutator phenotype in yeast (29), the situation would be similar to those described for bacteria where mutations of MutS and MutL proteins, involved in mismatch repair and stabilization of microsatellites, play a role in creating genetic diversity (17).
Supplementary Material
Acknowledgments
We thank members of our laboratory and A. El Marjou and J. Buard for technical advice, materials, and helpful discussions; S. Loeillet, H. Debrauwère, C. Soustelle, M. Reagan, M. Hottiger, and D. Tishkoff for providing strains and plasmids; and K. Smith for English correction and helpful discussion.
This study was supported by the Association de la Recherche contre le Cancer (ARC) and IC PIC Paramètres Epigénétiques. C.R. was supported by a graduate student fellowship from the MNERT and the ARC.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Anderson, S., and M. L. DePamphilis. 1979. Metabolism of Okazaki fragments during simian virus 40 DNA replication. J. Biol. Chem. 254:11495-11504. [PubMed] [Google Scholar]
- 2.Appelgren, H., M. Hedenskog, C. Sandstrom, H. Cederberg, and U. Rannug. 1999. Polychlorinated biphenyls induce meiotic length mutations at the human minisatellite MS32 in yeast. Environ. Mol. Mutagen. 34:285-290. [PubMed] [Google Scholar]
- 3.Ausubel, F., R. Brent, R. Kingston, D. Moore, J. Seidman, J. Smith, and K. Struhl. 1987. Current protocols in molecular biology. Wiley, New York, N.Y.
- 4.Bambara, R. A., R. S. Murante, and L. A. Henricksen. 1997. Enzymes and reactions at the eukaryotic DNA replication fork. J. Biol. Chem. 272:4647-4650. [DOI] [PubMed] [Google Scholar]
- 5.Bennett, S. T., A. M. Lucassen, S. C. Gough, E. E. Powell, D. E. Undlien, L. E. Pritchard, M. E. Merriman, Y. Kawaguchi, M. J. Dronsfield, F. Pociot, et al. 1995. Susceptibility to human type 1 diabetes at IDDM2 is determined by tandem repeat variation at the insulin gene minisatellite locus. Nat. Genet. 9:284-292. [DOI] [PubMed] [Google Scholar]
- 6.Benson, G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27:573-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buard, J., A. Bourdet, J. Yardley, Y. Dubrova, and A. J. Jeffreys. 1998. Influences of array size and homogeneity on minisatellite mutation. EMBO J. 17:3495-3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buard, J., A. Collick, J. Brown, and A. J. Jeffreys. 2000. Somatic versus germline mutation processes at minisatellite CEB1 (D2S90) in humans and transgenic mice. Genomics 65:95-103. [DOI] [PubMed] [Google Scholar]
- 9.Buard, J., A. C. Shone, and A. J. Jeffreys. 2000. Meiotic recombination and flanking marker exchange at the highly unstable human minisatellite CEB1 (D2S90). Am. J. Hum. Genet. 67:333-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buard, J., and G. Vergnaud. 1994. Complex recombination events at the hypermutable minisatellite CEB1 (D2S90). EMBO J. 13:3203-3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Budd, M. E., and J. L. Campbell. 1997. A yeast replicative helicase, Dna2 helicase, interacts with yeast FEN-1 nuclease in carrying out its essential function. Mol. Cell. Biol. 17:2136-2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Budd, M. E., W. C. Choe, and J. L. Campbell. 1995. DNA2 encodes a DNA helicase essential for replication of eukaryotic chromosomes. J. Biol. Chem. 270:26766-26769. [DOI] [PubMed] [Google Scholar]
- 13.Budd, M. E., A. H. Tong, P. Polaczek, X. Peng, C. Boone, and J. L. Campbell. 2005. A network of multi-tasking proteins at the DNA replication fork preserves genome stability. PLoS Genet. 1:e61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Callahan, J. L., K. J. Andrews, V. A. Zakian, and C. H. Freudenreich. 2003. Mutations in yeast replication proteins that increase CAG/CTG expansions also increase repeat fragility. Mol. Cell. Biol. 23:7849-7860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Debrauwere, H., J. Buard, J. Tessier, D. Aubert, G. Vergnaud, and A. Nicolas. 1999. Meiotic instability of human minisatellite CEB1 in yeast requires DNA double-strand breaks. Nat. Genet. 23:367-371. [DOI] [PubMed] [Google Scholar]
- 16.Debrauwere, H., S. Loeillet, W. Lin, J. Lopes, and A. Nicolas. 2001. Links between replication and recombination in Saccharomyces cerevisiae: a hypersensitive requirement for homologous recombination in the absence of Rad27 activity. Proc. Natl. Acad. Sci. USA 98:8263-8269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Denamur, E., G. Lecointre, P. Darlu, O. Tenaillon, C. Acquaviva, C. Sayada, I. Sunjevaric, R. Rothstein, J. Elion, F. Taddei, M. Radman, and I. Matic. 2000. Evolutionary implications of the frequent horizontal transfer of mismatch repair genes. Cell 103:711-721. [DOI] [PubMed] [Google Scholar]
- 18.Denoeud, F., G. Vergnaud, and G. Benson. 2003. Predicting human minisatellite polymorphism. Genome Res. 13:856-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ellegren, H. 2004. Microsatellites: simple sequences with complex evolution. Nat. Rev. Genet. 5:435-445. [DOI] [PubMed] [Google Scholar]
- 20.Frank, G., J. Qiu, M. Somsouk, Y. Weng, L. Somsouk, J. P. Nolan, and B. Shen. 1998. Partial functional deficiency of E160D flap endonuclease-1 mutant in vitro and in vivo is due to defective cleavage of DNA substrates. J. Biol. Chem. 273:33064-33072. [DOI] [PubMed] [Google Scholar]
- 21.Freudenreich, C. H., S. M. Kantrow, and V. A. Zakian. 1998. Expansion and length-dependent fragility of CTG repeats in yeast. Science 279:853-856. [DOI] [PubMed] [Google Scholar]
- 22.Gatchel, J. R., and H. Y. Zoghbi. 2005. Diseases of unstable repeat expansion: mechanisms and common principles. Nat. Rev. Genet. 6:743-755. [DOI] [PubMed] [Google Scholar]
- 23.Greene, A. L., J. R. Snipe, D. A. Gordenin, and M. A. Resnick. 1999. Functional analysis of human FEN1 in Saccharomyces cerevisiae and its role in genome stability. Hum. Mol. Genet. 8:2263-2273. [DOI] [PubMed] [Google Scholar]
- 24.Haber, J. E., and E. J. Louis. 1998. Minisatellite origins in yeast and humans. Genomics 48:132-135. [DOI] [PubMed] [Google Scholar]
- 25.Hansen, R. J., E. C. Friedberg, and M. S. Reagan. 2000. Sensitivity of a S. cerevisiae RAD27 deletion mutant to DNA-damaging agents and in vivo complementation by the human FEN-1 gene. Mutat. Res. 461:243-248. [DOI] [PubMed] [Google Scholar]
- 26.Harrington, J. J., and M. R. Lieber. 1994. The characterization of a mammalian DNA structure-specific endonuclease. EMBO J. 13:1235-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He, Q., H. Cederberg, J. A. Armour, C. A. May, and U. Rannug. 1999. Cis-regulation of inter-allelic exchanges in mutation at human minisatellite MS205 in yeast. Gene 232:143-153. [DOI] [PubMed] [Google Scholar]
- 28.Horowitz, H., and J. E. Haber. 1984. Subtelomeric regions of yeast chromosomes contain a 36 base-pair tandemly repeated sequence. Nucleic Acids Res. 12:7105-7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang, M. E., A. G. Rio, A. Nicolas, and R. D. Kolodner. 2003. A genomewide screen in Saccharomyces cerevisiae for genes that suppress the accumulation of mutations. Proc. Natl. Acad. Sci. USA 100:11529-11534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ivanov, E. L., N. Sugawara, J. Fishman-Lobell, and J. E. Haber. 1996. Genetic requirements for the single-strand annealing pathway of double-strand break repair in Saccharomyces cerevisiae. Genetics 142:693-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jauert, P. A., S. N. Edmiston, K. Conway, and D. T. Kirkpatrick. 2002. RAD1 controls the meiotic expansion of the human HRAS1 minisatellite in Saccharomyces cerevisiae. Mol. Cell. Biol. 22:953-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jeffreys, A. J., R. Barber, P. Bois, J. Buard, Y. E. Dubrova, G. Grant, C. R. Hollies, C. A. May, R. Neumann, M. Panayi, A. E. Ritchie, A. C. Shone, E. Signer, J. D. Stead, and K. Tamaki. 1999. Human minisatellites, repeat DNA instability and meiotic recombination. Electrophoresis 20:1665-1675. [DOI] [PubMed] [Google Scholar]
- 33.Jeffreys, A. J., A. MacLeod, K. Tamaki, D. L. Neil, and D. G. Monckton. 1991. Minisatellite repeat coding as a digital approach to DNA typing. Nature 354:204-209. [DOI] [PubMed] [Google Scholar]
- 34.Jeffreys, A. J., J. Murray, and R. Neumann. 1998. High-resolution mapping of crossovers in human sperm defines a minisatellite-associated recombination hotspot. Mol. Cell 2:267-273. [DOI] [PubMed] [Google Scholar]
- 35.Jeffreys, A. J., D. L. Neil, and R. Neumann. 1998. Repeat instability at human minisatellites arising from meiotic recombination. EMBO J. 17:4147-4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jeffreys, A. J., and R. Neumann. 1997. Somatic mutation processes at a human minisatellite. Hum. Mol. Genet. 6:129-132; 134-6. [DOI] [PubMed] [Google Scholar]
- 37.Jeffreys, A. J., K. Tamaki, A. MacLeod, D. G. Monckton, D. L. Neil, and J. A. Armour. 1994. Complex gene conversion events in germline mutation at human minisatellites. Nat. Genet. 6:136-145. [DOI] [PubMed] [Google Scholar]
- 38.Jeffreys, A. J., V. Wilson, and S. L. Thein. 1985. Hypervariable ‘minisatellite’ regions in human DNA. Nature 314:67-73. [DOI] [PubMed] [Google Scholar]
- 39.Kao, H. I., J. L. Campbell, and R. A. Bambara. 2004. Dna2p helicase/nuclease is a tracking protein, like FEN1, for flap cleavage during Okazaki fragment maturation. J. Biol. Chem. 279:50840-50849. [DOI] [PubMed] [Google Scholar]
- 40.Kim, D. H., K. H. Lee, J. H. Kim, G. H. Ryu, S. H. Bae, B. C. Lee, K. Y. Moon, S. M. Byun, H. S. Koo, and Y. S. Seo. 2005. Enzymatic properties of the Caenorhabditis elegans Dna2 endonuclease/helicase and a species-specific interaction between RPA and Dna2. Nucleic Acids Res. 33:1372-1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kokoska, R. J., L. Stefanovic, H. T. Tran, M. A. Resnick, D. A. Gordenin, and T. D. Petes. 1998. Destabilization of yeast micro- and minisatellite DNA sequences by mutations affecting a nuclease involved in Okazaki fragment processing (rad27) and DNA polymerase delta (pol3-t). Mol. Cell. Biol. 18:2779-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kolodner, R. D., C. D. Putnam, and K. Myung. 2002. Maintenance of genome stability in Saccharomyces cerevisiae. Science 297:552-557. [DOI] [PubMed] [Google Scholar]
- 43.Krogh, B. O., and L. S. Symington. 2004. Recombination proteins in yeast. Annu. Rev. Genet. 38:233-271. [DOI] [PubMed] [Google Scholar]
- 44.Krontiris, T. G., B. Devlin, D. D. Karp, N. J. Robert, and N. Risch. 1993. An association between the risk of cancer and mutations in the HRAS1 minisatellite locus. N. Engl. J. Med. 329:517-523. [DOI] [PubMed] [Google Scholar]
- 45.Lalioti, M. D., H. S. Scott, C. Buresi, C. Rossier, A. Bottani, M. A. Morris, A. Malafosse, and S. E. Antonarakis. 1997. Dodecamer repeat expansion in cystatin B gene in progressive myoclonus epilepsy. Nature 386:847-851. [DOI] [PubMed] [Google Scholar]
- 46.Lander, E. S., L. M. Linton, B. Birren, C. Nusbaum, M. C. Zody, J. Baldwin, K. Devon, K. Dewar, M. Doyle, W. FitzHugh, R. Funke, D. Gage, K. Harris, A. Heaford, J. Howland, L. Kann, J. Lehoczky, R. LeVine, P. McEwan, K. McKernan, J. Meldrim, J. P. Mesirov, C. Miranda, W. Morris, J. Naylor, C. Raymond, M. Rosetti, R. Santos, A. Sheridan, C. Sougnez, N. Stange-Thomann, N. Stojanovic, A. Subramanian, D. Wyman, J. Rogers, J. Sulston, R. Ainscough, S. Beck, D. Bentley, J. Burton, C. Clee, N. Carter, A. Coulson, R. Deadman, P. Deloukas, A. Dunham, I. Dunham, R. Durbin, L. French, D. Grafham, S. Gregory, T. Hubbard, S. Humphray, A. Hunt, M. Jones, C. Lloyd, A. McMurray, L. Matthews, S. Mercer, S. Milne, J. C. Mullikin, A. Mungall, R. Plumb, M. Ross, R. Shownkeen, S. Sims, R. H. Waterston, R. K. Wilson, L. W. Hillier, J. D. McPherson, M. A. Marra, E. R. Mardis, L. A. Fulton, A. T. Chinwalla, K. H. Pepin, W. R. Gish, S. L. Chissoe, M. C. Wendl, K. D. Delehaunty, T. L. Miner, A. Delehaunty, J. B. Kramer, L. L. Cook, R. S. Fulton, D. L. Johnson, P. J. Minx, S. W. Clifton, T. Hawkins, E. Branscomb, P. Predki, P. Richardson, S. Wenning, T. Slezak, N. Doggett, J. F. Cheng, A. Olsen, S. Lucas, C. Elkin, E. Uberbacher, M. Frazier, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409:860-921. [DOI] [PubMed] [Google Scholar]
- 47.Lieber, M. R. 1997. The FEN-1 family of structure-specific nucleases in eukaryotic DNA replication, recombination and repair. Bioessays 19:233-240. [DOI] [PubMed] [Google Scholar]
- 48.Liu, Y., H. I. Kao, and R. A. Bambara. 2004. Flap endonuclease 1: a central component of DNA metabolism. Annu. Rev. Biochem. 73:589-615. [DOI] [PubMed] [Google Scholar]
- 49.Loeillet, S., B. Palancade, M. Cartron, A. Thierry, G. F. Richard, B. Dujon, V. Doye, and A. Nicolas. 2005. Genetic network interactions among replication, repair and nuclear pore deficiencies in yeast. DNA Repair (Amsterdam) 4:459-468. [DOI] [PubMed] [Google Scholar]
- 50.Lopes, J., H. Debrauwere, J. Buard, and A. Nicolas. 2002. Instability of the human minisatellite CEB1 in rad27Delta and dna2-1 replication-deficient yeast cells. EMBO J. 21:3201-3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maleki, S., H. Cederberg, and U. Rannug. 2002. The human minisatellites MS1, MS32, MS205 and CEB1 integrated into the yeast genome exhibit different degrees of mitotic instability but are all stabilised by RAD27. Curr. Genet. 41:333-341. [DOI] [PubMed] [Google Scholar]
- 52.May, C. A., A. J. Jeffreys, and J. A. Armour. 1996. Mutation rate heterogeneity and the generation of allele diversity at the human minisatellite MS205 (D16S309). Hum. Mol. Genet. 5:1823-1833. [DOI] [PubMed] [Google Scholar]
- 53.Monckton, D. G., R. Neumann, T. Guram, N. Fretwell, K. Tamaki, A. MacLeod, and A. J. Jeffreys. 1994. Minisatellite mutation rate variation associated with a flanking DNA sequence polymorphism. Nat. Genet. 8:162-170. [DOI] [PubMed] [Google Scholar]
- 54.Moreau, S., E. A. Morgan, and L. S. Symington. 2001. Overlapping functions of the Saccharomyces cerevisiae Mre11, Exo1 and Rad27 nucleases in DNA metabolism. Genetics 159:1423-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mortensen, U. H., C. Bendixen, I. Sunjevaric, and R. Rothstein. 1996. DNA strand annealing is promoted by the yeast Rad52 protein. Proc. Natl. Acad. Sci. USA 93:10729-10734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Myers, S., L. Bottolo, C. Freeman, G. McVean, and P. Donnelly. 2005. A fine-scale map of recombination rates and hotspots across the human genome. Science 310:321-324. [DOI] [PubMed] [Google Scholar]
- 57.Ooi, S. L., D. D. Shoemaker, and J. D. Boeke. 2003. DNA helicase gene interaction network defined using synthetic lethality analyzed by microarray. Nat. Genet. 35:277-286. [DOI] [PubMed] [Google Scholar]
- 58.Paques, F., and J. E. Haber. 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63:349-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Paques, F., W. Y. Leung, and J. E. Haber. 1998. Expansions and contractions in a tandem repeat induced by double-strand break repair. Mol. Cell. Biol. 18:2045-2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Paques, F., G. F. Richard, and J. E. Haber. 2001. Expansions and contractions in 36-bp minisatellites by gene conversion in yeast. Genetics 158:155-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pearson, C. E., K. N. Edamura, and J. D. Cleary. 2005. Repeat instability: mechanisms of dynamic mutations. Nat. Rev. Genet. 6:729-742. [DOI] [PubMed] [Google Scholar]
- 62.Reagan, M. S., C. Pittenger, W. Siede, and E. C. Friedberg. 1995. Characterization of a mutant strain of Saccharomyces cerevisiae with a deletion of the RAD27 gene, a structural homolog of the RAD2 nucleotide excision repair gene. J. Bacteriol. 177:364-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Richard, G. F., and B. Dujon. 2006. Molecular evolution of minisatellites in hemiascomycetous yeasts. Mol. Biol. Evol. 23:189-202. [DOI] [PubMed] [Google Scholar]
- 64.Richard, G. F., and F. Paques. 2000. Mini- and microsatellite expansions: the recombination connection. EMBO Rep. 1:122-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rocco, V., B. de Massy, and A. Nicolas. 1992. The Saccharomyces cerevisiae ARG4 initiator of meiotic gene conversion and its associated double-strand DNA breaks can be inhibited by transcriptional interference. Proc. Natl. Acad. Sci. USA 89:12068-12072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schweitzer, J. K., and D. M. Livingston. 1998. Expansions of CAG repeat tracts are frequent in a yeast mutant defective in Okazaki fragment maturation. Hum. Mol. Genet. 7:69-74. [DOI] [PubMed] [Google Scholar]
- 67.Shen, B., P. Singh, R. Liu, J. Qiu, L. Zheng, L. D. Finger, and S. Alas. 2005. Multiple but dissectible functions of FEN-1 nucleases in nucleic acid processing, genome stability and diseases. Bioessays 27:717-729. [DOI] [PubMed] [Google Scholar]
- 68.Soustelle, C., M. Vedel, R. Kolodner, and A. Nicolas. 2002. Replication protein A is required for meiotic recombination in Saccharomyces cerevisiae. Genetics 161:535-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sugawara, N., T. Goldfarb, B. Studamire, E. Alani, and J. E. Haber. 2004. Heteroduplex rejection during single-strand annealing requires Sgs1 helicase and mismatch repair proteins Msh2 and Msh6 but not Pms1. Proc. Natl. Acad. Sci. USA 101:9315-9320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sugawara, N., and J. E. Haber. 1992. Characterization of double-strand break-induced recombination: homology requirements and single-stranded DNA formation. Mol. Cell. Biol. 12:563-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sung, P., L. Krejci, S. Van Komen, and M. G. Sehorn. 2003. Rad51 recombinase and recombination mediators. J. Biol. Chem. 278:42729-42732. [DOI] [PubMed] [Google Scholar]
- 72.Symington, L. S. 1998. Homologous recombination is required for the viability of rad27 mutants. Nucleic Acids Res. 26:5589-5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Symington, L. S. 2002. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 66:630-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tamaki, K., C. A. May, Y. E. Dubrova, and A. J. Jeffreys. 1999. Extremely complex repeat shuffling during germline mutation at human minisatellite B6.7. Hum. Mol. Genet. 8:879-888. [DOI] [PubMed] [Google Scholar]
- 75.Tishkoff, D. X., N. Filosi, G. M. Gaida, and R. D. Kolodner. 1997. A novel mutation avoidance mechanism dependent on S. cerevisiae RAD27 is distinct from DNA mismatch repair. Cell 88:253-263. [DOI] [PubMed] [Google Scholar]
- 76.Tong, A. H., M. Evangelista, A. B. Parsons, H. Xu, G. D. Bader, N. Page, M. Robinson, S. Raghibizadeh, C. W. Hogue, H. Bussey, B. Andrews, M. Tyers, and C. Boone. 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294:2364-2368. [DOI] [PubMed] [Google Scholar]
- 77.Vergnaud, G., and F. Denoeud. 2000. Minisatellites: mutability and genome architecture. Genome Res. 10:899-907. [DOI] [PubMed] [Google Scholar]
- 78.Verstrepen, K. J., A. Jansen, F. Lewitter, and G. R. Fink. 2005. Intragenic tandem repeats generate functional variability. Nat. Genet. 37:986-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.West, S. C. 2003. Molecular views of recombination proteins and their control. Nat. Rev. Mol. Cell. Biol. 4:435-445. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.