Abstract
The signaling cascades activated by insulin and IGF-1 contribute to the control of multiple cellular functions, including glucose metabolism and cell proliferation. In most cases these effects are mediated, at least in part, by insulin receptor substrates (IRS), one of which is insulin receptor substrate 1 (IRS-1). R-Ras is a member of the Ras family of GTPases and is involved in a variety of biological processes, including integrin activation, cell migration, and control of cell proliferation. Here we demonstrate that both R-Ras and BCAR3, a regulator of R-Ras activity that has been implicated in breast cancer, regulate the level of IRS-1 protein in estrogen-dependent MCF-7 and ZR75 breast cancer cells. In particular, expression of a constitutively activated R-Ras mutant, R-Ras38V, or of BCAR3 accelerates the degradation of IRS-1, leading to the impairment of signaling through insulin but not epidermal growth factor receptors. Moreover, knockdown of endogenous R-Ras levels in MCF-7 cells inhibits IRS-1 degradation induced by estrogen signaling blockade but not by long-term insulin treatment. Consistent with these results, both R-Ras38V expression and estrogen signaling blockade lead to the degradation of IRS-1, at least in part, through calpain activity. These findings show that R-Ras activity mediates inhibition of insulin signaling associated with suppression of estrogen action, implicating this GTPase in a growth-inhibitory mechanism associated with antiestrogen treatment of breast cancer.
Estrogen and insulin/IGF-1 have potent positive effects upon the proliferation of mammary epithelial cells and estrogen-dependent breast cancer cells. While the former functions mainly through cytoplasmic estrogen receptor activation, the latter functions through cell surface insulin or IGF-1 receptors and subsequent tyrosine phosphorylation of a family of insulin receptor substrates. One of these substrates, insulin receptor substrate 1 (IRS-1), has been implicated in mediating the mitogenic activities of both insulin and IGF-1 (32). The proper regulation of insulin/IGF-1 signaling may be of particular importance in breast cancer etiology, since elevated levels of both IGF-1 and IRS-1 have been associated with breast cancer (8, 24).
Recent studies have demonstrated cooperative cross talk between estrogen and insulin/IGF-1 signaling pathways. For example, estrogen enhances insulin signaling by increasing IGF-1 (29), IGF-1 receptor (27), and IRS-1 (12, 14, 25) levels in cells through elevated transcription. Moreover, inhibition of estrogen signaling by loss of estrogen receptors (19) or by exposure to antiestrogens decreases IRS-1 levels through decreased IRS-1 synthesis (4, 25) and increased degradation (15). The latter phenomenon raises the possibility that at least part of the growth inhibitory effects of estrogen antagonists used in breast cancer therapy may be through concomitant suppression of insulin/IGF-1 signaling.
The R-Ras GTPase is a close relative of the Ras proto-oncogenes, displaying both common and distinct activities (28). For example, Ras proteins and R-Ras have very similar effector-binding domains; thus, the active GTP-bound forms of both proteins bind and activate phosphatidylinositol (PI) 3-kinase (PI3K) (13). However, for reasons that are not clear, R-Ras is not an effective activator of other known Ras targets, such as Raf kinases and Ral guanine nucleotide exchange factors (GEFs). Consistent with these observations, R-Ras can promote cell division, but its activity is not as potent as that of Ras in the cell systems studied to date (5, 26). In contrast, R-Ras can promote inside/out integrin signaling (34), while Ras cannot, although the effector R-Ras uses to promote this specific function is not known. R-Ras has also been implicated in axon guidance as a mediator of Eph receptor signaling (9). In particular, an R-Ras-specific GTPase-activating protein, plexin-B1, inhibits R-Ras activity (21). Studies with R-Ras knockout mice have implied that this GTPase functions in vascular regeneration. R-Ras has also been shown to be overexpressed in gastric carcinoma (16).
R-Ras and Ras proteins are also activated by both common and distinct guanine nucleotide exchange factors. For example Ras-GRF1 activates both Ras and R-Ras, but Sos activates only Ras (7). In contrast C3G and BCAR3 activate R-Ras but not Ras proteins (6, 7, 20). BCAR3 is of particular interest here because its overexpression has been shown to promote estrogen-independent proliferation of MCF-7 and ZR75 cells (30), possibly through activation of R-Ras (33).
Here we show that both BCAR3 and its target in cells, R-Ras, promote the degradation of IRS-1 in estrogen-dependent breast cancer cells. Moreover, we show that R-Ras participates in cross talk between estrogen and insulin/IGF-1 signaling by mediating, at least in part, the downregulation of IRS-1 levels that occurs in response to estrogen antagonist treatment of these cells.
MATERIALS AND METHODS
Reagents.
Antibodies against IRS-1, phospho-AKT(serS473), phosphorylated extracellular signal-regulated kinase 1 (ERK1)/ERK2, and phosphorylated-Jun N-terminal kinase (thr183/tyr185) were purchased from Cell Signaling Technology (Beverly, MA). Ral A polyclonal antibody was from BD Biosciences-Pharmingen (San Diego, CA). Antibodies against ERK1, Ral A, R-Ras, hemagglutinin, Myc (9E10), and total phosphotyrosine (4G10) were purchased from Santa Cruz Biotechnologies (Santa Cruz, CA), The inhibitors MG132, lactacystin, ALLN, calpeptin, and LY294002 were from Calbiochem (La Jolla, CA). Wortmannin was purchased from Sigma. All tissue culture reagents and Oligofectamine were from Invitrogen Life Technologies (Carlsbad, CA).
Recombinant plasmids and transfection technology.
Wild-type (WT) and activated R-Ras38V, both with Myc tags in their N termini, were cloned into pcDNA3 (Invitrogen Life Technologies). pcDNA3Myc-R-Ras38V64E was generated by single mutation in amino acid 64 from D to E in the R-Ras38V background. Mutation of the proline-rich region of R-Ras38V was obtained by inducing double mutations of P202 and P203 to A202 and A203 to generate pcDNA3Myc-R-Ras38V-202A/203A. Constructs were stably transfected into MCF-7 cells by calcium phosphate precipitation and selected with G418. Multiple individual positive clones were screened for by Western blotting. R-Ras small interfering RNA (siRNA), 203AAG AUC UGC AGU GUG GAU GGC224, was purchased from Dharmacon Research, Inc. MCF-7 cells (0.5 × 106) were plated in a 60-mm dish overnight and transiently transfected with 25 nM siRNA or control laminin siRNA by use of Oligofectamine (Invitrogen). After 48 h, cells were used for analysis. To generate stable MCF-7 cells with knockdown of R-Ras, oligonucleotide 205G AUC UGC AGU GUG GAU GGC224 was cloned into pSUPER.retro vector (OligoEngine, Washington) to create R-Ras short hairpin RNA (shRNA). R-Ras shRNA-expressing and control empty plasmids were transfected into MCF-7 cells by use of calcium phosphate, and multiple stable clones were obtained after the selection with puromycin. An R-Ras mutant resistant to shRNA was generated by site-directed mutagenesis and transfected into R-Ras shRNA cells. Cells were selected for neomycin resistance. The WT sequence was 205G AUC UGC AGU GUG GAU GGC224, and that for the resistant R-Ras clone was as follows (mutations are indicated in boldface): 205G AUA UGT AGC GUC GAC GGC224.
Cell culture and immunoblot analysis.
MCF-7 or ZR75 cells were maintained in minimal essential alpha medium supplemented with 5% fetal calf serum (HyClone). 3T3L1 cells were grown in Dulbecco's minimal essential medium (DMEM) containing 10% calf serum, while INS-1 cells were grown in DMEM plus 10% fetal calf serum. Cells were plated in 60-mm dishes with a density of 0.5 × 106 cells/dish and serum starved in regular medium or E2-free medium (charcoal-stripped serum and dye-free DMEM) containing ICI 182,780 (10 μM) for 24 h and then treated for defined time periods with the following inhibitors: MG132 (25 μM), lactacystin (25 μM), ALLN (5 μM), calpeptin (10 μM), or LY294002 (25 μM); wortmannin (100 nM); or growth factors insulin (1 μM) or epidermal growth factor (EGF) (10 μM). Control cells were treated with the same amount of vehicle. After treatment or stimulation, cells were lysed in 50 mM Tris-HCl (pH 7.5), 1% NP-40, 75 mM NaCl, 10 mM sodium orthovanadate, 1 mM phenylmethylsulfonyl fluoride, 20 μM leupeptin, and 1 μM beta-glycerolphosphate. Protein concentration was determined by use of a Micro bicinchoninic acid protein assay reagent kit (Pierce, Illinois). The results were visualized by enhanced chemiluminescence (PerkinElmer, Massachusetts). Digital images of blots were produced by densitometric scans of autoradiographs and quantified using NIH Image 1.63 software, working within the linear range of the readouts.
Reverse transcription-PCR.
Total RNA was isolated using TRIzol reagent (Invitrogen Life Technologies). The total RNA (0.5 μg) was used for reverse transcription to make cDNA in a 25-μl reaction mixture containing 4 U RNasin RNase inhibitor (Promega), 1 mM deoxynucleoside triphosphate, 50 mM Tris-HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, 0.1 μg of oligodeoxythymidine triphosphate 12-18, 4 mM dithiothreitol, and 50 units of Moloney murine leukemia virus reverse transcriptase (Invitrogen) at 37°C for 1 h. Reactions were stopped by heat inactivation at 95°C for 10 min. Subsequently, 2.5 μl of reverse transcription reaction product was used for PCR in a 50-μl reaction mixture in the presence of 50 pmol of sense primer (CTTCTGTCAGGTGTCATCC) and antisense primer (CTCTGCAGCAATGCCTGTTC) for IRS-1 (291-bp fragment), 1× ThermoPol reaction buffer, 200 μM deoxynucleoside triphosphates, and one unit of Vent DNA polymerase for 25 cycles in an Eppendorf Mastercycler gradient machine (Eppendorf, Westbury, NY). The PCR product was run in a 1.5% agarose gel and photographed. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control, where a 358-bp fragment was generated using the following pair of primers: CTACTGGCGCTGCCAAGGCTG (sense) and GCCATGAGGTCCACCACCCTG (antisense).
Turnover experiments.
MCF-7 cells (5 × 106) overexpressing either pcDNA3Myc-R-Ras38V or empty pcDNA3 were plated into 100-mm dishes. After 24 h, cells were incubated in the presence of 10 μm cycloheximide for various amounts of time. Cell extracts were prepared, and the levels of IRS-1 or control proteins (Ral A and AKT) were determined by immunoblot analysis.
RESULTS
Expression of activated R-Ras decreases insulin but not EGF activation of ERK in MCF-7 cells.
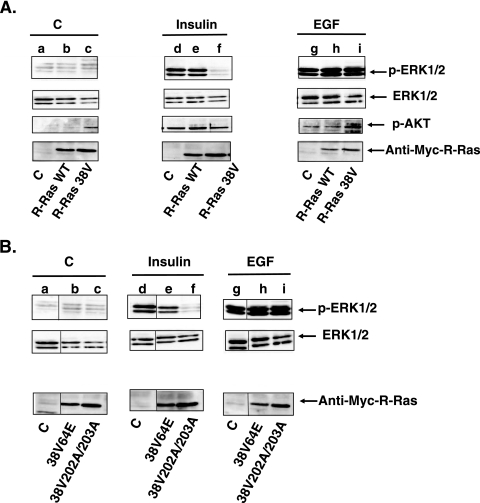
Recently, we studied the consequence of expressing a constitutively activated form of the R-Ras GTPase, R-RasG38V, on the proliferation of MCF-7 breast cancer cells (33). In the process of characterizing these cells, we examined whether R-Ras activity influences well-characterized signaling transduction pathways induced by growth factors such as EGF and insulin. To this end, we compared the abilities of insulin and EGF to stimulate phosphorylation of ERK mitogen-activated protein kinase in MCF-7 cells stably expressing either empty vector, wild-type R-Ras, or activated R-Ras (Fig. 1A, top row). We found that expression of activated R-Ras38V (lanes c and f) but not of WT R-Ras (lanes b and e) or of empty vector (lanes a and d) dramatically suppressed (∼80%) insulin activation of ERK. Strikingly, expression of activated R-Ras38V had no detectable effect on the activation of ERK by EGF (lanes g to i). Total ERK levels were comparable in all cell lines (Fig. 1A, second row).
FIG. 1.
Expression of R-Ras38V downregulates insulin but not EGF signaling in MCF-7 cells. (A) MCF-7 lines expressing empty vector (C), Myc-tagged WT R-Ras (R-Ras WT), or Myc-tagged R-Ras38V were serum starved overnight and then stimulated with either buffer, insulin (1 μM), or EGF (10 nM) for 10 min, and cell lysates were assayed for p-ERK (top row), total ERK (second row), p-AKT (third row), or Myc-R-Ras (fourth row). (B) Cell lines expressing effector mutations in R-Ras were treated as described for panel A. The results are representative of experiments performed at least in triplicate with at least two individual clones for each cell line.
R-Ras38V also perturbed insulin but not EGF activation of AKT (Fig. 1A, third row). These experiments were more complex because R-Ras38V elevates basal AKT activity in unstimulated cells (33). Nevertheless, the data show that insulin further activated AKT in R-Ras38V cells to levels only ∼50% of that found with insulin-stimulated control cells expressing empty vector or WT R-Ras (compare lanes c and f to a and d and to b and e). Thus, R-Ras activity has a specific inhibitory effect on insulin signaling in MCF-7 cells.
To begin to understand how R-Ras influences insulin signaling, we tested mutant forms of activated R-Ras that are known to suppress interaction with specific proteins. One mutation, R-Ras38V64E, is in the effector-binding domain that suppresses R-Ras interaction with PI 3-kinase (and potentially with unidentified effectors) (13). The other mutation, R-Ras38V202A/203A, destroys a putative SH3-binding site in R-Ras that has been reported to bind to the adaptor Nck (31). Insulin activation of ERK was no longer inhibited in cells stably expressing R-Ras38V64E (Fig. 1B, top row, compare lanes a and d with b and e). In contrast, insulin activation of ERK continued to be suppressed in cells expressing R-Ras38V202A/203A (compare lanes a and d with c and f). Thus, R-Ras interaction with PI 3-kinase or another R-Ras effector that has effector-binding properties similar to those of PI 3-kinase is involved in the suppression of insulin signaling. In contrast, R-Ras interaction with Nck is not involved.
Expression of activated R-Ras suppresses the level of IRS-1 in MCF-7 and ZR75 cells.
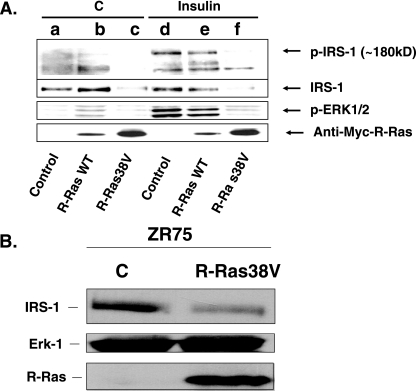
Since activated R-Ras specifically suppressed insulin signaling, we investigated where in the insulin signaling cascade this effect arose. To this end, MCF-7 cells expressing empty vector (control), wild-type R-Ras, or activated R-Ras (R-Ras38V) were left untreated or were treated with insulin, and cell lysates were probed with antiphosphotyrosine antibodies (Fig. 2A, top row). Strikingly, insulin enhancement of the tyrosine phosphorylation of a 180-kDa protein, i.e., approximately of the size of IRS-1, was reduced in lysates from cells expressing activated R-Ras (lane f) compared to cells expressing empty vector (lane d) or wild-type R-Ras (lane e). To directly assess the effect of activated R-Ras expression on IRS-1 that could account for this defect, immunoblotting of cell lysates was performed using IRS-1-specific antibodies (Fig. 2A, second row). This experiment clearly shows that expression of activated R-Ras, but not of wild-type R-Ras or empty vector, led to an ∼4-fold decrease in expression of IRS-1 protein in MCF-7 cells (compare lanes a, b, d, and e to c and f). This effect apparently caused the defect in insulin stimulation of IRS-1 tyrosine phosphorylation (first row) and insulin activation of ERK (third row) in cells expressing activated R-Ras38V (fourth row). This effect was specific, since R-Ras38V expression did not reduce the expression of Ral A or Jun N-terminal protein kinase activity in these cells (data not shown).
FIG. 2.
Expression of R-Ras38V causes reduced expression of IRS-1 in MCF-7 and ZR75 cells. (A) MCF-7 cells expressing empty vector control, WT R-Ras, or R-Ras38V were treated with buffer (buffer C or insulin) for 10 min, and then lysates were probed for phosphotyrosine (top row), anti-IRS-1 (second row), phospho-ERK (third row), or Myc-R-Ras (fourth row). The results are representative of experiments performed at least in triplicate with at least two individual clones for each cell line. kD, kDa. (B) ZR75 cells stably expressing empty vector (lane C) or activated R-Ras (R-Ras38V) were generated and assayed for IRS-1 levels by immunoblotting.
This effect was not limited to MCF-7 cells, as activated R-Ras had a similar effect on IRS-1 levels in another estrogen-dependent breast cancer cell line, ZR75 (Fig. 2B). Since regulation of IRS-1 levels is clearly important for metabolic diseases such as diabetes, we tested the effects of activated R-Ras expression in cell types used in studies on glucose metabolism, such as 3T3 L1 fibroblasts that can differentiate into adipocytes and ins-1 cells derived from pancreatic islets. In neither case did expression of R-Ras38V alter IRS-1 l levels (data not shown). Thus, the mechanism by which R-Ras promotes IRS degradation may be active in a subset of cell types, including those derived from mammary epithelium.
The R-Ras GEF, BCAR3, also reduces IRS-1 protein levels.
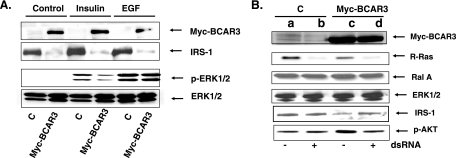
To test whether a protein known to function through R-Ras also leads to suppression of IRS-1 expression, we studied MCF-7 cells stably expressing the R-Ras GEF, BCAR3 (also referred to as AND34 [3]) (Fig. 3A, top row). Expression of Myc-tagged BCAR3 downregulated IRS-1 protein expression by ∼80% (Fig. 3A, second row). As expected, this was accompanied by the reduction of ERK activation (∼60%) (Fig. 3A, third row) in response to insulin but not EGF stimulation.
FIG. 3.
Expression of BCAR3 causes reduced expression of IRS-1 in MCF-7 cells through R-Ras. (A) MCF-7 cells expressing Myc-BCAR3 or empty vector were treated with buffer (control), insulin, or EGF for 10 min. Cell lysates were probed for Myc-BCAR3, IRS-1, p-ERK, or total ERK. (B) Cells shown in panel A were transfected with control siRNA (− dsRNA) or R-Ras specific siRNA (+ dsRNA), and 24 h later the lysates of cells were probed for Myc-BCAR3 (top row), R-Ras (second row), Ral A (third row), ERK (fourth row), IRS-1 (fifth row), or p-AKT (bottom row). The results are representative of experiments performed at least in duplicate with at least two individual clones for each cell line.
To confirm that R-Ras is involved in the ability of BCAR3 to suppress IRS-1 levels, siRNA against R-Ras was transfected into MCF-7 cells expressing empty vector or Myc-BCAR3. Knockdown of R-Ras expression was observed (Fig. 3B, second row), while no effect on the related Ral GTPase or ERK kinase was observed (third and fourth rows). While knockdown of R-Ras had no effect on baseline levels of IRS-1 (Fig. 3B, second row, compare lanes a and b) it almost completely (∼80%) reversed BCAR3-induced downregulation of IRS-1 (Fig. 3B, fourth row, compare lanes c and d). Consistent with this finding, knockdown of R-Ras expression also blocked the ability of BCAR3 to activate AKT, a kinase previously shown to be activated by the expression of either BCAR3 or activated R-Ras (sixth row).
Downregulation of IRS-1 levels by R-Ras is mediated by calpain-induced degradation.
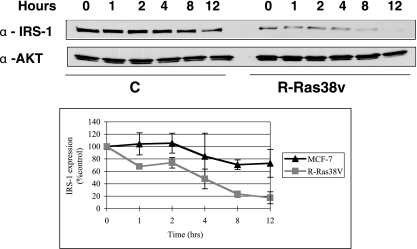
To begin to determine how R-Ras38V downregulates IRS-1 protein levels, turnover experiments were performed on MCF-7 cells expressing either control vector or a vector expressing activated R-Ras38V. In the presence of cycloheximide to prevent new protein synthesis, IRS-1 expression was stable for at least 12 h. In contrast, in R-Ras38V cells IRS-1 levels fell to ∼20% of control levels by 8 h, indicating that R-Ras38V enhances the degradation of IRS-1 (Fig. 4).
FIG. 4.
R-Ras38V increases turnover of IRS-1 in MCF-7 cells. MCF-7 cells expressing either empty vector (pcDNA3) or activated R-Ras (R-Ras38V) were incubated in the presence of cycloheximide for various amounts of time. Cell extracts were prepared, and the levels of IRS-1 or control proteins (Ral A and AKT) were determined by immunoblot analysis. Data from three experiments were pooled and expressed in the graph as the means ± standard deviations.
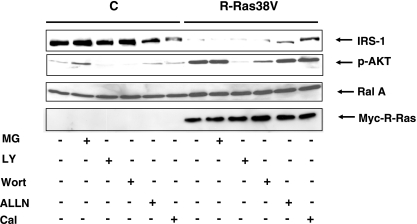
Since mutation of the PI3K-binding site of R-Ras38V reversed R-Ras38V-induced downregulation of IRS-1 protein expression and since PI3K has been implicated in IRS-1 degradation through proteosome activation (11), we examined the effect of LY294002, a specific PI3K inhibitor, on IRS-1 levels in R-Ras38V-expressing MCF-7 cells. However, treatment of LY294002 did not restore IRS-1 levels in R-Ras38V-expressing cells to the levels present in control cells (Fig. 4, top lane). Similar negative results were obtained with another PI3K inhibitor, wortmannin. LY294002 and wortmannin did inhibit PI3K in these cells, as evidenced by the abolishment of p-AKT in treated cells (Fig. 4, second and third lanes). Moreover, treatment of cells with the proteosome inhibitor MG132 also failed to restore IRS-1 to normal levels in R-Ras38V cells. In contrast, exposure of cells to ALLN or calpeptin, two inhibitors of calpain, another mediator of IRS-1 stability, did partially restore IRS-1 levels in R-Ras38V cells (Fig. 5, fourth and fifth lanes). Overall, these findings indicate that elevated R-Ras activity promotes the degradation of IRS-1 at least in part through a calpain-mediated pathway.
FIG. 5.
R-Ras induces enhanced turnover of IRS-1 through a calpain-mediated pathway. Control (C) and R-Ras38V-expressing cells were treated for 24 h with the proteosome inhibitor MG132 (MG), with either of the PI 3-kinase inhibitors LY294002 (LY) or wortmannin (Wort), or with either of the calpain inhibitors ALLN or calpeptin (Cal). Cell lysates were then prepared and probed for IRS-1 (top row) p-AKT (second row), Ral A (third row), or Myc-R-Ras (bottom row). The results are representative of at least three independent experiments and performed on at least two independent R-Ras38V cell lines.
R-Ras functions in the degradation of IRS-1 mediated by estrogen antagonism.
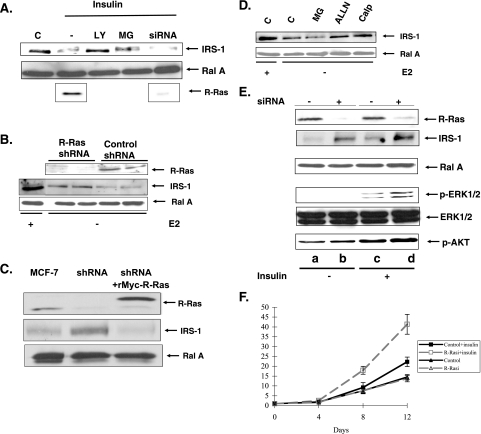
A variety of extracellular signals have been reported to promote the degradation of IRS-1 in breast cancer cells. One is long-term exposure to insulin. Insulin signals through PI 3-kinase to promote the phosphorylation IRS-1 and subsequent proteolysis through the proteosome. However, the results described above showed that R-Ras does not promote degradation of IRS-1 through PI 3-kinase or the proteosome. Consistent with these findings, depletion of R-Ras through siRNA transfection did not prevent degradation of IRS-1 induced by long-term insulin stimulation (Fig. 6A). In contrast, either the PI3K inhibitor LY294002 or the proteosome inhibitor MG132 did restore or at least partially restore IRS-1 levels under these conditions. Thus, R-Ras does not participate in IRS-1 downregulation induced by long-term insulin treatment.
FIG. 6.
R-Ras participates in the downregulation of IRS-1 levels induced by estrogen depletion but not by long-term insulin stimulation. (A) MCF-7 cells were exposed to buffer (lane C) or insulin for 24 h. Cells were exposed to LY294002 (LY), MG132 (MG), or R-Ras siRNA 24 h before insulin addition, and then lysates were probed for levels of IRS-1 (top row), Ral A (second row), or R-Ras (bottom row). (B) MCF-7 cells stably expressing R-Ras shRNA or empty shRNA vector were exposed to complete medium (+) or medium depleted of estrogen and containing ICI 183,780 (−) for 24 h. Twenty-four h later, cell lysates were probed for levels of R-Ras, IRS-1, or Ral A. (C) MCF-7 cells, MCF-7 cells expressing R-Ras shRNA, or MCF-7 cells expressing R-Ras shRNA plus an shRNA-resistant Myc-R-Ras (rMyc-R-Ras) were grown in the absence of E2, and levels of R-Ras, IRS-1, and Ral A were probed as described above. (D) MCF-7 cells were exposed to complete medium (+) or medium depleted of E2 in the presence of ICI 183,780 for 24 h (−). Then, cells were exposed to either buffer, MG132 (MG), ALLN, or calpeptin (Calp) for another 24 h. Cells lysates were then probed for either IRS-1 or Ral A. (E) Cells stably expressing either control vector or vector expressing R-Ras shRNA were exposed to media lacking estrogen and containing ICI 183.780 for 24 h then treated with either buffer or insulin (10 μM) overnight. Cell lysates were then probed for either R-Ras, Ral, IRS-1, phospho-ERK, total ERK, or phospho-AKT. The results are representative of at least three independent experiments. (F) MCF-7 cells and MCF-7 cells stably expressing R-Ras shRNA were plated at 104 cells per dish in estrogen-depleted media. After 24 h, 100 nM insulin along with ICI 182,780 was added to some dishes every 48 h. Cell numbers were counted at the indicated times. The data represent the averages of data from experiments performed in triplicate (± standard deviations). Darker lines, control cells (MCF-7); lighter lines, shRNA-expressing cells (R-Rasi); triangles, no insulin; squares, insulin.
IRS-1 levels in breast cancer cells have also been shown to be regulated by estrogen, where estrogen signaling promotes IRS-1 expression (14, 19) and blockage of estrogen signaling promotes IRS-1 down-regulation (4, 15, 27). To determine whether R-Ras contributes to estrogen antagonism-induced downregulation of IRS-1, the levels of IRS-1 protein were assessed for untreated MCF-7 cells and for MCF-7 cells transfected with siRNA against R-Ras either under normal growing conditions or in medium lacking estrogen and containing the estrogen antagonist ICI 183,780. Figure 6B shows that IRS-1 levels, suppressed by estrogen inhibition, were partially restored to normal (∼70%) by R-Ras depletion.
Importantly, the siRNA effect was specific because IRS-1 levels were not restored by R-Ras shRNA expression in cells expressing a form of R-Ras that is resistant to siRNA treatment, rMyc-R-Ras (Fig. 6C). Moreover, consistent with these and previous results described above, the calpain inhibitors ALLN and calpeptin but not the proteosome inhibitor MG132 partially restored IRS-1 levels under these antiestrogen conditions (Fig. 6D). Thus, R-Ras function is required for IRS-1 downregulation that occurs in response to blockade of estrogen signaling.
Knockdown of R-Ras by RNA interference enhances MCF-7 cell response to insulin stimulation.
The sensitivities to insulin of control MCF-7 cells and of MCF-7 cells with reduced expression of R-Ras were compared under conditions where estrogen signaling was blocked by the addition of the estrogen antagonist ICI183,780. In these studies, control and R-Ras-specific shRNA-expressing cells were stimulated with control buffer or insulin. Consistent with previously described results (Fig. 6B), knockdown of R-Ras (Fig. 6E, top row) increased IRS-1 protein expression compared to what was seen for control cells expressing control vector (second row) in these estrogen-inhibited cells. Again, knockdown of R-Ras had no effect on the expression of a close family member, the Ral A protein (third row). Consistent with these findings, insulin activation of both ERK (fourth and fifth rows) and AKT (bottom row) was magnified ∼2-fold in cells expressing low levels of R-Ras. Moreover, insulin stimulation of MCF-7 cell proliferation under these conditions was enhanced in cells lacking R-Ras ∼2-fold after 12 days (Fig. 6F). Thus, R-Ras normally functions to suppress insulin signaling upon blockade of estrogen-signaling MCF-7 cells, and decreased R-Ras function sensitizes breast cancer cells to insulin's growth-promoting signals under these antiestrogen conditions.
DISCUSSION
The experiments described in this paper reveal that the R-Ras GTPase participates in the downregulation of insulin/IGF-1 signaling that occurs in estrogen-dependent breast cancer cells in response to a blockade of estrogen signaling. This conclusion is based on the following observations. First, overexpression of a constitutively activated R-Ras allele induced the downregulation of IRS-1 levels in estrogen-dependent MCF-7 and ZR75 cells. As expected, this effect was associated with a loss of insulin but not EGF signaling in these cells. A similar phenomenon was also observed when BCAR3, a GEF for R-Ras, was overexpressed in MCF-7 cells, and this effect was blunted when R-Ras levels were reduced in cells by siRNA treatment. Finally, downregulation of IRS-1 levels induced by antiestrogen treatment, but not by long-term insulin treatment, was suppressed, and insulin stimulation of ERK, AKT, and cell proliferation were all enhanced when R-Ras levels were reduced in MCF-7 cells. This action of R-Ras appears to be cell type specific, since activated R-Ras downregulated insulin signaling in two estrogen-dependent breast cancer cell lines but not in fibroblasts or pancreatic cells.
IRS-1 levels are known to be regulated by protein turnover. The most thorough investigations have been in the context of studies of long-term insulin/IGF-1 signaling, where treatment with these ligands in multiple cell types, including MCF-7, promotes the proteolysis of IRS-1 through PI 3-kinase-dependent ubiquitination and subsequent engagement of the proteosome-mediated degradation pathway (11). The present study shows that although R-Ras can induce the breakdown of IRS-1in MCF-7 cells, R-Ras is not involved in this insulin/IGF-1-induced process, since neither PI 3-kinase nor proteosome inhibitors prevented IRS-1 downregulation by R-Ras. Moreover, suppression of R-Ras levels in MCF-7 cells also failed to prevent insulin-induced IRS-1 turnover.
Recent studies have indicated that estrogen also regulates IRS-1 levels in breast cancer cells. For example, estrogen stimulation of cells increases IRS-1 gene expression (12, 14, 25), while blockers of estrogen signaling downregulate IRS-1 gene expression and enhance its breakdown (4, 15, 25). The experiments described here show that R-Ras contributes to negative cross talk between estrogen and insulin signaling, since R-Ras depletion by siRNA treatment of MCF-7 cells suppressed IRS-1 degradation. Additional support for this notion came from inhibitor studies showing that IRS-1-induced downregulation by activated R-Ras and by an estrogen antagonist were both blocked by the calpain inhibitors ALLN and calpeptin.
How R-Ras connects to calpain-mediated protein degradation remains to be determined; however, mutant analysis of R-Ras at sites known to interact with other proteins yielded some hints. For example, R-Ras has been reported to interact with the Nck adaptor protein (31), but a mutation blocking that interaction failed to suppress R-Ras effects on IRS-1 levels in MCF-7 cells, indicating that this interacting protein is not involved. R-Ras is also known to bind to and activate PI 3-kinase (13). In fact, a point mutation in the effector domain that blocks PI 3-kinase activation, V64E, also blocks R-Ras effects on IRS-1. However, R-Ras activation of PI 3-kinase is not involved, because incubation with a PI 3-kinase inhibitor did not reverse the effects of R-Ras on IRS-1 levels. This finding implies that to mediate IRS-1 downregulation, amino acid 64 in the effector domain of R-Ras interacts with an additional R-Ras effector that remains to be identified. Interestingly, integrin activation by R-Ras has also been shown to be blocked by the V64E mutation but not by inhibitors of PI3K (18). Thus, it remains possible that these two functions share a common as-yet-unidentified R-Ras effector. R-Ras has also been shown to promote focal adhesions through Cas phosphorylation and cell migration in breast cancer cells, but these phenomena were sensitive to PI 3-kinase inhibition, which suggests that these activities do not share a mechanism used by R-Ras to downregulate IRS-1 (10).
How does this newly detected activity of BCAR3 and R-Ras relate to previous studies showing that they both promote estrogen-independent proliferation in MCF-7 cells? We showed previously that R-Ras activation of PI 3-kinase is involved in promoting estrogen-independent proliferation, since expression of a dominant negative version of the PI 3-kinase target, AKT, blocked R-Ras effects on proliferation (33). However, PI 3-kinase-induced AKT activation was found to be insufficient to promote estrogen-independent proliferation, suggesting that another PI 3-kinase-induced pathway is also involved. In fact, a more recent study showed that BCAR3 promotes estrogen-independent proliferation, at least in part, through PI 3-kinase-induced Rac activation (33). Thus, it appears that R-Ras-induced downregulation of IRS-1 and enhancement of estrogen independent proliferation function through different effector pathways emanating from R-Ras. In support of the idea that these two functions of R-Ras are independent is the finding that the prevention of R-Ras-induced downregulation of IRS-1 by overexpression of IRS-1 in MCF-7 cells did not block estrogen-independent proliferation by activated R-Ras (data not shown). Thus, excessive R-Ras and a loss of R-Ras function can both potentially promote cell proliferation but under different cellular conditions and through the use of different R-Ras effectors.
The fact that R-Ras activity suppresses insulin signaling in MCF-7 cells may have clinical significance. First, epidemiological studies have shown a link between insulin and IGF-1 signaling (both of which function through IRS-1) and breast cancer (8). High IGF-1 plasma levels are correlated with breast cancer risk, and this correlation has been linked to the association between obesity and breast cancer (2). In addition, elevated levels of IGF-1 receptors and IRS-1 have been found in breast cancer tissue (22, 24). Moreover, some of the effects of estrogen on mammary cell proliferation may be mediated by enhancement of insulin/IGF-1 signaling through estrogen-induced increase in the levels and activities of multiple components of the insulin/IGF-1 signaling cascade, including IGF-1, IGF-1R, and IRS-1 (27). Moreover, blocking IGF-1R function not only blocks IGF-1 signaling but also blocks estrogen-induced gene activation and cell growth (1, 17).
That R-Ras participates in the downregulation of IRS-1 levels specifically in response to inhibition of estrogen signaling in MCF-7 cells is of particular interest, because there is a growing appreciation that some of the growth-inhibitory effects of antiestrogens in breast cancer may be due to their negative effects on insulin and/or IGF-1 signaling (23). Studies with cell culture systems (15) and with whole animals (4) showed that antiestrogens, including ICI 182,780 and tamoxifen, suppress insulin/IGF-1 signaling by suppressing the expression or activity of the IGF-1 receptor and IRS-1. Thus, the present study implies that a completely functional R-Ras signaling system in estrogen-dependent breast cancer cells may be required for the full potency of antiestrogens to be achieved.
Acknowledgments
This work was supported by grants to L.A.F. (PHS grant CA47391 from the NCI) and to Y.Y. from the Canadian Institute of Health Research Postdoctoral Fellowship and from the Susan G. Komen Breast Cancer Foundation. Support was also from the GRASP Digestive Disease Center (P30-DK34928 GRASP).
REFERENCES
- 1.Arteaga, C. L., E. Coronado, and C. K. Osborne. 1988. Blockade of the epidermal growth factor receptor inhibits transforming growth factor alpha-induced but not estrogen-induced growth of hormone-dependent human breast cancer. Mol. Endocrinol. 2:1064-1069. [DOI] [PubMed] [Google Scholar]
- 2.Boyd, D. B. 2003. Insulin and cancer. Integr. Cancer Ther. 2:315-329. [DOI] [PubMed] [Google Scholar]
- 3.Cai, D., L. K. Clayton, A. Smolyar, and A. Lerner. 1999. AND-34, a novel p130Cas-binding thymic stromal cell protein regulated by adhesion and inflammatory cytokines. J. Immunol. 163:2104-2112. [PubMed] [Google Scholar]
- 4.Chan, T. W., M. Pollak, and H. Huynh. 2001. Inhibition of insulin-like growth factor signaling pathways in mammary gland by pure antiestrogen ICI 182,780. Clin. Cancer Res. 7:2545-2554. [PubMed] [Google Scholar]
- 5.Cox, A. D., T. R. Brtva, D. G. Lowe, and C. J. Der. 1994. R-Ras induces malignant, but not morphologic, transformation of NIH3T3 cells. Oncogene 9:3281-3288. [PubMed] [Google Scholar]
- 6.Gotoh, T., D. Cai, X. Tian, L. A. Feig, and A. Lerner. 2000. p130Cas regulates the activity of AND-34, a novel Ral, Rap1 and R-Ras guanine nucleotide exchange factor. J. Biol. Chem. 275:30118-30123. [DOI] [PubMed] [Google Scholar]
- 7.Gotoh, T., Y. Niino, M. Tokuda, O. Hatase, S. Nakamura, M. Matsuda, and S. Hattori. 1997. Activation of R-Ras by Ras-guanine nucleotide-releasing factor. J. Biol. Chem. 272:18602-18607. [DOI] [PubMed] [Google Scholar]
- 8.Hankinson, S. E., W. C. Willett, G. A. Colditz, D. J. Hunter, D. S. Michaud, B. Deroo, B. Rosner, F. E. Speizer, and M. Pollak. 1998. Circulating concentrations of insulin-like growth factor-I and risk of breast cancer. Lancet 351:1393-1396. [DOI] [PubMed] [Google Scholar]
- 9.Kruger, R. P., J. Aurandt, and K. L. Guan. 2005. Semaphorins command cells to move. Nat. Rev. Mol. Cell Biol. 6:789-800. [DOI] [PubMed] [Google Scholar]
- 10.Kwong, L., M. A. Wozniak, A. S. Collins, S. D. Wilson, and P. J. Keely. 2003. R-Ras promotes focal adhesion formation through focal adhesion kinase and p130 Cas by a novel mechanism that differs from integrins. Mol. Cell. Biol. 23:933-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee, A. V., J. L. Gooch, S. Oesterreich, R. L. Guler, and D. Yee. 2000. Insulin-like growth factor I-induced degradation of insulin receptor substrate 1 is mediated by the 26S proteasome and blocked by phosphatidylinositol 3′-kinase inhibition. Mol. Cell. Biol. 20:1489-1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee, A. V., J. G. Jackson, J. L. Gooch, S. G. Hilsenbeck, E. Coronado-Heinsohn, C. K. Osborne, and D. Yee. 1999. Enhancement of insulin-like growth factor signaling in human breast cancer: estrogen regulation of insulin receptor substrate-1 expression in vitro and in vivo. Mol. Endocrinol. 13:787-796. [DOI] [PubMed] [Google Scholar]
- 13.Marte, B. M., P. Rodriguez-Viciana, S. Wennstrom, P. H. Warne, and J. Downward. 1997. R-Ras can activate the phosphoinositide 3-kinase but not the MAP kinase arm of the Ras effector pathways. Curr. Biol. 7:63-70. [DOI] [PubMed] [Google Scholar]
- 14.Molloy, C. A., F. E. May, and B. R. Westley. 2000. Insulin receptor substrate-1 expression is regulated by estrogen in the MCF-7 human breast cancer cell line. J. Biol. Chem. 275:12565-12571. [DOI] [PubMed] [Google Scholar]
- 15.Morelli, C., C. Garofalo, M. Bartucci, and E. Surmacz. 2003. Estrogen receptor-alpha regulates the degradation of insulin receptor substrates 1 and 2 in breast cancer cells. Oncogene 22:4007-4016. [DOI] [PubMed] [Google Scholar]
- 16.Nishigaki, M., K. Aoyagi, I. Danjoh, M. Fukaya, K. Yanagihara, H. Sakamoto, T. Yoshida, and H. Sasaki. 2005. Discovery of aberrant expression of R-RAS by cancer-linked DNA hypomethylation in gastric cancer using microarrays. Cancer Res. 65:2115-2124. [DOI] [PubMed] [Google Scholar]
- 17.Nolan, M. K., L. Jankowska, M. Prisco, S. Xu, M. A. Guvakova, and E. Surmacz. 1997. Differential roles of IRS-1 and SHC signaling pathways in breast cancer cells. Int. J. Cancer 72:828-834. [DOI] [PubMed] [Google Scholar]
- 18.Oertli, B., J. Han, B. M. Marte, T. Sethi, J. Downward, M. Ginsberg, and P. E. Hughes. 2000. The effector loop and prenylation site of R-Ras are involved in the regulation of integrin function. Oncogene 19:4961-4969. [DOI] [PubMed] [Google Scholar]
- 19.Oesterreich, S., P. Zhang, R. L. Guler, X. Sun, E. M. Curran, W. V. Welshons, C. K. Osborne, and A. V. Lee. 2001. Re-expression of estrogen receptor alpha in estrogen receptor alpha-negative MCF-7 cells restores both estrogen and insulin-like growth factor-mediated signaling and growth. Cancer Res. 61:5771-5777. [PubMed] [Google Scholar]
- 20.Ohba, Y., N. Mochizuki, S. Yamashita, A. M. Chan, J. W. Schrader, S. Hattori, K. Nagashima, and M. Matsuda. 2000. Regulatory proteins of R-Ras, TC21/R-Ras2, and M-Ras/R-Ras3. J. Biol. Chem. 275:20020-20026. [DOI] [PubMed] [Google Scholar]
- 21.Oinuma, I., Y. Ishikawa, H. Katoh, and M. Negishi. 2004. The Semaphorin 4D receptor Plexin-B1 is a GTPase activating protein for R-Ras. Science 305:862-865. [DOI] [PubMed] [Google Scholar]
- 22.Pandini, G., R. Vigneri, A. Costantino, F. Frasca, A. Ippolito, Y. Fujita-Yamaguchi, K. Siddle, I. D. Goldfine, and A. Belfiore. 1999. Insulin and insulin-like growth factor-I (IGF-I) receptor overexpression in breast cancers leads to insulin/IGF-I hybrid receptor overexpression: evidence for a second mechanism of IGF-I signaling. Clin. Cancer Res. 5:1935-1944. [PubMed] [Google Scholar]
- 23.Pollak, M., J. Costantino, C. Polychronakos, S. A. Blauer, H. Guyda, C. Redmond, B. Fisher, and R. Margolese. 1990. Effect of tamoxifen on serum insulinlike growth factor I levels in stage I breast cancer patients. J. Natl. Cancer Inst. 82:1693-1697. [DOI] [PubMed] [Google Scholar]
- 24.Sachdev, D., and D. Yee. 2001. The IGF system and breast cancer. Endocr.-Relat. Cancer 8:197-209. [DOI] [PubMed] [Google Scholar]
- 25.Salerno, M., D. Sisci, L. Mauro, M. A. Guvakova, S. Ando, and E. Surmacz. 1999. Insulin receptor substrate 1 is a target for the pure antiestrogen ICI 182,780 in breast cancer cells. Int. J. Cancer 81:299-304. [DOI] [PubMed] [Google Scholar]
- 26.Self, A. J., E. Caron, H. F. Paterson, and A. Hall. 2001. Analysis of R-Ras signalling pathways. J. Cell Sci. 114:1357-1366. [DOI] [PubMed] [Google Scholar]
- 27.Stewart, A. J., M. D. Johnson, F. E. May, and B. R. Westley. 1990. Role of insulin-like growth factors and the type I insulin-like growth factor receptor in the estrogen-stimulated proliferation of human breast cancer cells. J. Biol. Chem. 265:21172-21178. [PubMed] [Google Scholar]
- 28.Takai, Y., T. Sasaki, and T. Matozaki. 2001. Small GTP-binding proteins. Physiol. Rev. 81:153-208. [DOI] [PubMed] [Google Scholar]
- 29.Umayahara, Y., R. Kawamori, H. Watada, E. Imano, N. Iwama, T. Morishima, Y. Yamasaki, Y. Kajimoto, and T. Kamada. 1994. Estrogen regulation of the insulin-like growth factor I gene transcription involves an AP-1 enhancer. J. Biol. Chem. 269:16433-16442. [PubMed] [Google Scholar]
- 30.van Agthoven, T., T. L. van Agthoven, A. Dekker, P. J. van der Spek, L. Vreede, and L. C. Dorssers. 1998. Identification of BCAR3 by a random search for genes involved in antiestrogen resistance of human breast cancer cells. EMBO J. 17:2799-2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang, B., J. X. Zou, B. Ek-Rylander, and E. Ruoslahti. 2000. R-Ras contains a proline-rich site that binds to SH3 domains and is required for integrin activation by R-Ras. J. Biol. Chem. 275:5222-5227. [DOI] [PubMed] [Google Scholar]
- 32.Yenush, L., and M. F. White. 1997. The IRS-signalling system during insulin and cytokine action. Bioessays 19:491-500. [DOI] [PubMed] [Google Scholar]
- 33.Yu, Y., and L. A. Feig. 2002. Involvement of R-Ras and Ral GTPases in estrogen-independent proliferation of breast cancer cells. Oncogene 21:7557-7568. [DOI] [PubMed] [Google Scholar]
- 34.Zhang, Z., K. Vuori, H. Wang, J. C. Reed, and E. Ruoslahti. 1996. Integrin activation by R-ras. Cell 85:61-69. [DOI] [PubMed] [Google Scholar]