Abstract
RANKL, a protein essential for osteoclast development and survival, is stimulated by parathyroid hormone (PTH) via a PTH receptor 1/cyclic AMP (cAMP)/protein kinase A (PKA)/CREB cascade, exclusively in osteoblastic cells. We report that a bacterial artificial chromosome-based transcriptional reporter construct containing 120 kb of RANKL 5′-flanking region was stimulated by dibutyryl-cAMP in stromal/osteoblastic cells, but not other cell types. Full cAMP responsiveness was dependent upon a conserved 715-bp region located 76 kb upstream from the transcription start site, which we identified by sequential deletion analysis and by comparison of human and mouse genomic sequences in silico. This region contained conserved consensus sequences which bound CREB and the osteoblast-specific transcription factor Runx2, and when mutated blunted cAMP responsiveness. Overexpression of Runx2 potentiated cAMP responsiveness of the endogenous RANKL gene in a cell-type-specific manner. Lastly, PTH responsiveness of the endogenous RANKL gene was abrogated in mice from which we deleted this conserved upstream region. Thus, PTH responsiveness of the RANKL gene is determined by a distant regulatory region that responds to cAMP in a cell-type-specific manner and Runx2 may contribute to such cell-type specificity.
Calcium homeostasis is essential for life in terrestrial vertebrates, and parathyroid hormone (PTH) has evolved as the principle hormone responsible for maintaining the extracellular levels of calcium within a physiologic range (57). When calcium is needed, PTH stimulates intestinal calcium absorption indirectly by increasing the synthesis of 1,25-dihydroxyvitamin D3 [1,25(OH)2D3] in the kidney and also lowers the amount of urinary calcium excreted. In addition, PTH mobilizes calcium from bone by stimulating the production of osteoclasts—the highly specialized cells that are responsible for bone resorption.
It has been established during the last few years that osteoclasts are derived from hematopoietic precursors in a process that is strictly dependent on the interaction of the receptor activator of NF-κB ligand (RANKL), a membrane-bound member of the tumor necrosis factor (TNF) family of cytokines (Tnfsf11) produced by bone marrow stromal cells of the mesenchymal lineage, with RANK, which is expressed on the surface of the hematopoietic precursors (3, 61). Since the stromal cells that produce RANKL are apparently of the osteoblast lineage, they are commonly referred to as stromal/osteoblastic cells (38, 64). Furthermore, extensive evidence suggests that the main mechanism by which PTH stimulates osteoclastogenesis is increased expression of RANKL (15, 29, 41, 65). Importantly, increased RANKL expression is associated with the osteoclastogenic effects of all other agents that increase bone resorption, including 1,25(OH)2D3 and cytokines (32, 69). Albeit, PTH may also stimulate osteoclast formation by inhibiting expression of osteoprotegerin (OPG), a soluble decoy receptor for RANKL (58).
Besides stromal/osteoblastic cells, high levels of RANKL expression are restricted to lymphocyte-containing tissues and mammary epithelium (1, 32, 69). Consistent with an important biologic role of RANKL in these three tissues, RANKL-deficient mice are devoid of osteoclasts and lymph nodes and also exhibit disrupted mammary gland development (12, 31). Importantly, in all three tissues, biologically relevant levels of RANKL are apparently achieved only after stimulation of its production by the appropriate stimuli: namely, other osteoclastogenic cytokines or hormones in stromal/osteoblastic cells (53, 69), T-cell receptor (TCR) activation in lymphocytes (30), or pregnancy hormones in mammary epithelium (12). In line with in vivo evidence that RANKL becomes biologically relevant only upon stimulation of its basal expression, osteoclastogenesis in cocultures of hematopoietic precursors and stromal/osteoblastic cells occurs only when the latter cell type is stimulated with hormones or cytokines (32, 34, 48, 69). Heretofore, the molecular basis of the tissue-specific expression of RANKL and its regulation by calciotropic hormones remained unknown.
In studies leading to the work presented here, we and others have established that stimulation of RANKL by PTH in stromal/osteoblastic cells is mediated by the classical PTH receptor 1 (PTHR1) and that it requires cyclic AMP (cAMP) generation, downstream activation of protein kinase A (PKA), and PKA-induced phosphorylation and activation of the cAMP response element (CRE)-binding protein (CREB) (15, 29, 36). Notably, activation of PKA and CREB is required for full induction of RANKL by not only PTH, but also by 1,25(OH)2D3 and the gp130 cytokine oncostatin M (OSM) (15), indicating that PKA and CREB are shared components of at least three osteoclastogenic signaling pathways. CREB is broadly expressed and activated by a wide range of extracellular signals, but heretofore the mechanism whereby CREB activation contributes to RANKL expression specifically in stromal/osteoblastic cells remained unknown.
Previous attempts to identify mechanisms that directly control RANKL gene transcription have yielded inconsistent results. Indeed, while we and others have shown that promoter-reporter constructs containing up to 7 kb of the murine RANKL 5′-flanking region do not respond to hormones such as 1,25(OH)2D3 (11, 23, 55), Kitazawa et al. have found that constructs containing approximately 1 kb of 5′-flanking region are stimulated two- to threefold by this hormone (25, 26). Although differences in experimental conditions may play a role in this discrepancy, the low level of reporter gene stimulation relative to stimulation of the endogenous gene clearly suggests the existence of important regulatory regions outside the proximal 5′-flanking region.
The goal of the work presented herein was to explore mechanisms that control the tissue-specific expression of RANKL upon activation of the PKA-CREB pathway. We report that the tissue specificity of RANKL regulation by PTH is not determined by the presence or absence of the PTH receptor, nor is it determined by the activation of the PKA-CREB cascade or the methylation of a CpG island in the first exon of the RANKL gene. Instead, the restricted expression of the RANKL gene in response to activation of PKA is due to a distant transcriptional enhancer that resides 76 kb upstream from the first exon of RANKL. This enhancer responds to CREB activation in a cell-type-specific manner and is required for full PTH responsiveness of the endogenous RANKL gene. Moreover, the osteoblast-specific transcription factor Runx2 likely contributes to this cell-type specificity.
MATERIALS AND METHODS
Materials and cell lines.
Bovine PTH(1-34) (NLE8,18, TYR34) and dibutyryl-cAMP (db-cAMP) were purchased from Sigma (St. Louis, MO). Murine OSM was purchased from R&D Systems (Minneapolis, MN). 1,25-(OH)2 vitamin D3 [1,25(OH)2D3] was purchased from Biomol (Plymouth Meeting, PA). The Hepa (CRL-2026) and RAW264.7 (71-TIB) cell lines were obtained from the American Type Culture Collection (Rockville, MD). UAMS-32 and UAMS-32P cells have been described previously (15, 53). To create a version of UAMS-32P cells suitable for stable transfection with reporter constructs harboring a neomycin phosphotransferase (neo) coding sequence, a second version was generated by infecting UAMS-32 cells with a retroviral vector expressing human PTHR1 as previously described (15), except that the neo sequence in the retroviral vector pLEN was replaced with a blasticidin S-deaminase coding sequence to generate a vector designated pLEB. All cells were maintained in alpha minimal essential medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum and 1% each penicillin, streptomycin, and glutamine.
Animal studies.
Four-month-old female C57BL/6J mice were injected intraperitoneally with phosphate-buffered saline (PBS) or db-cAMP (100 mg/kg) in a total volume of 100 μl. Weanling mice were placed on a normal or calcium-deficient diet for 7 days as previously described (54). All studies involving mice were approved by the Institutional Animal Care and Use Committees of the University of Arkansas for Medical Sciences and the Central Arkansas Veterans Healthcare System.
Reporter constructs.
A firefly luciferase reporter vector, designated pINTluc, that allows efficient stable transfection of reporter constructs was derived from pIRES2-EGFP (Clontech, Palo Alto, CA). Briefly, the enhanced green fluorescent protein (EGFP) coding sequence of pIRES2-EGFP was replaced with a PCR product consisting of the luciferase coding sequence of pGL3-Basic (Promega, Madison WI). The cytomegalovirus promoter in the resulting plasmid was removed by digestion with AseI and NheI followed by filling-in and blunt-end ligation. The SV40 promoter upstream from the neomycin phosphotransferase gene was then replaced with a minimal herpesvirus thymidine kinase promoter to yield plasmid pIRESluc. Lastly, the internal ribosome entry site (IRES) was replaced with a synthetic intron obtained from phRLnull (Promega). Sequences of all PCR-amplified fragments and cloning junctions for this and subsequent constructs were verified by automated sequencing. The complete nucleotide sequences of pIRESluc and pINTluc are available upon request.
Nucleotide locations for the following constructs are all relative to the transcription start site of the respective gene and are given in base pairs unless otherwise indicated. For the bacterial artificial chromosome (BAC)-based reporter constructs, the relative positions of particular regions were determined using the University of California—Santa Cruz genome browser and the August 2005 assembly of the mouse genome. The RANKL-2kb reporter construct was created by excising a fragment spanning −2014 to +27 from a BAC clone containing the murine RANKL gene (BAC-18). BAC-18 was obtained from Genome Systems, Inc., St. Louis, MO, by screening a mouse embryonic stem (ES) cell BAC library with a 400-bp fragment of the proximal murine RANKL promoter. The 2-kb fragment was inserted into the SacI and HindIII sites of pINTluc. The −97 construct was created by PCR amplification of the region from −97 to +27 of the murine RANKL gene and insertion into the HindIII and SmaI sites of pINTluc. Construct 7B-97 was created by inserting a filled-in 8.4-kb ApaI-AgeI fragment from BAC-18, spanning kb −81.5 −73.1, into the filled HindIII site of the −97 construct. Construct CNS1-A-97 was created by inserting a 707-bp PCR fragment, spanning kb −76.9 to −76.2, into the filled HindIII site of the −97 construct. Construct CNS1-B-97 was created by inserting a 1,965-bp PCR fragment, spanning kb −76.9 to −74.9 kb, into the filled HindIII site of the −97 construct. Constructs Delta 1 through Delta 5 were created by PCR amplification of subregions of CNS1-A followed by insertion into the SacI site of the −97 construct. Sequences of primers used to amplify the subregions are presented in Table S1 in the supplemental material. CNS1-A-97 constructs containing point mutations in various potential cis-acting elements were generated by site-directed mutagenesis, as previously described (55), using the primers indicated in Table S1 in the supplemental material. The IL-6-1.4kb construct was created by inserting a filled EcoRI fragment spanning positions −1379 to +45 of the murine interleukin-6 (IL-6) gene into the SmaI site of pINTluc. The 3×CRE construct was created by inserting a 106-bp NheI (filled)-HindIII fragment from pCRE-SEAP (Clontech) into the SacI (filled) and HindIII sites of the −97 construct.
A luciferase reporter construct, designated BAC1, was generated from a BAC clone containing the entire murine RANKL gene as well as 120 kb of 5′-flanking region and 14 kb of 3′-flanking region (BAC-18) via recombinogenic targeting in Escherichia coli by the method of Lee et al. (35), also known as recombineering (5). Briefly, BAC-18 DNA was introduced into E. coli strain DY380 by electroporation, and the integrity of the BAC DNA was verified by restriction enzyme digestion and pulsed-field gel electrophoresis. A targeting construct was generated by PCR amplification of an IRES-luciferase-tk-neo cassette from pIRESluc using primers with 50 bp of homology to the 5′ and 3′ ends of the RANKL 3′-untranslated region (UTR). This cassette also contains a prokaryotic transcriptional promoter upstream from the neo/kan sequence. The amplified DNA was digested with DpnI and gel purified to eliminate template DNA. Induction of recombinogenic proteins and electroporation with the targeting construct were performed as described by Lee et al. (35). Recombinants were selected by plating the electroporated cells on LB plates containing 20 μg/ml kanamycin and 20 μg/ml chloramphenicol. Bacterial colonies were screened for correct targeting by generating a liquid culture from each and using 2 μl of culture as a template for a PCR utilizing primers flanking the recombination site. The primers were designed such that a product of 1,353 bp or 4,600 bp would be generated from wild-type (WT) or correctly recombined BACs, respectively. Correct junctions were verified by sequence analysis of BAC Maxiprep DNA. The sequences of primers used to generate the targeting construct for BAC1 are presented in Table S1 in the supplemental material. The sequences of the primers used in screening for homologous recombinants are also presented in Table S1 in the supplemental material. The generation of the BAC-6 reporter construct was similar to that of BAC1, except the 50-bp homology arms of the targeting construct matched the 50 bp of sequence immediately downstream of the start codon in exon 1 of the RANKL gene and the 50 bp of vector sequence immediately downstream of the 3′ end of the murine genomic DNA insert contained in BAC-18, respectively. In addition, the selection cassette used for BAC-6 was generated from pINTluc, resulting in a construct in which the intron-luciferase-tk-neo cassette replaced all sequences downstream from the start codon located in exon 1 of the murine RANKL gene contained in BAC-18. All remaining BAC-based reporters were derived from BAC1 via recombineering using cassettes containing a blasticidin resistance cassette amplified from pEF-BSD (Invitrogen) with primers homologous to 50-bp regions flanking the region to be deleted. All primers used for BAC construct generation and screening are presented in Table S1 in the supplemental material.
Transfections and retroviral infections.
Luciferase reporter constructs were introduced into cells by stable transfection using Lipofectamine (Invitrogen). For each construct, 5 × 105 cells were plated in a 6-cm dish and transfected 16 h later with 5 μg of linearized DNA. BAC-based reporters were linearized via NotI sites located within the BAC vector. Two days after transfection, cells were collected by trypsinization and replated into three 10-cm dishes. Twenty-four hours later, G418 was added to a concentration of 0.15 mg/ml for UAMS-32P or Hepa cells and 0.35 mg/ml for RAW264.7 cells. After 9 to 12 days, colonies from the three plates were harvested by trypsinization and pooled. For each construct, only pools containing 40 or more colonies were used. Stably transfected cell pools for each construct were plated at a density of 2 × 104 to 4 × 104 cells/well in 48-well plates, grown to confluence, and treated with various stimulating agents for 4 to 24 h. Cells were then washed twice with phosphate-buffered saline and incubated in lysis buffer (25 mM glycyl-glycine, pH 7.8, 1% Triton X-100, 15 mM MgSO4, 4 mM EGTA) for 15 min and then kept at −20°C for 30 min. Cells were thawed and centrifuged at 500 × g in a plate holder to pellet debris. Firefly luciferase activity and protein concentration of the cleared lysates were determined as previously described (54). Full-length murine Runx2 (type II) or Runx2 DNA binding domain (DBD)-encoding cDNAs were inserted into the filled BamHI site of pLEB to generate pRunx2-LEB and pDBD-LEB, respectively. Retroviral particles were generated from these constructs and transductions were performed as previously described (53).
Gel mobility shift assays.
Nuclear extract preparation and gel mobility shift assays were performed as previously described (55). The following double-stranded oligonucleotides were used in this study (only the top strands are shown): CNS1-OSE2 (5′-AGAATATCACCACATCAAACAC-3), mutant CNS1-OSE2 (5′-AGAATATCAGAACATCAAACAC-3), proximal CRE (5′-AGGAAGTTTGATGTCACTGTATGT-3), mutant proximal CRE (5′-AGGAAGTTTGAATTCACTGTATGT-3′) distal CRE (5′-ACGTTCCCAGTTGGCGTCAGGGAAGCTA-3′), and mutant distal CRE (5′-ACGTTCCCAGTTGAATTCAGGGAAGCTA-3′). Anti-CREB (sc-186X) and anti-Runx2 (sc-10758X) antibodies used for supershift assays were obtained from Santa Cruz Biotechnology (Santa Cruz, CA).
ChIP assays.
Chromatin immunoprecipitation (ChIP) assays were performed as previously described (24). Briefly, UAMS-32P cells were grown to confluence in 10-cm dishes and treated with vehicle or 1.5 mM db-cAMP for 6 h. Cells were then washed twice with PBS and cross-linked by adding a solution of 1.5% formaldehyde in PBS. Cells were extracted in a solution containing 10 mM EDTA, 0.5 mM EGTA, 10 mM HEPES, pH 6.5, and 0.25% Triton X-100 followed by a solution containing 1 mM EDTA, 0.5 mM EDTA, 10 mM HEPES, pH 6.5, and 200 mM NaCl. Nuclear pellets were then resuspended in a solution containing 10 mM EDTA, 50 mM Tris-HCl, pH 8.1, 0.5% Empigen BB, and 1% sodium dodecyl sulfate and sonicated three times for 10 s (each) at room temperature. The sonicated chromatin was centrifuged and diluted 2.5-fold in ChIP buffer (20 mM Tris-HCl, pH 8.1, 150 mM NaCl, 2 mM EDTA, and 1% Triton X-100) and precleared by a 3-h incubation at 4°C with DNA- and bovine serum albumin (BSA)-treated Zysorbin (Zymed, San Francisco, CA). Cleared chromatin was incubated overnight at 4°C with either anti-CREB antibody (Upstate, Charlottesville, VA), anti-Runx2 antibody (Santa Cruz Biotechnology, Santa Cruz, CA), or rabbit immunogobulin G (IgG), and the immunoprecipitates were collected by incubation for 2 h with Zysorbin. Precipitates were washed and then eluted in a solution containing 1% sodium dodecyl sulfate and 0.1 M NaHCO3 followed by overnight incubation at 65°C to reverse cross-links. DNA was purified using QIAquick PCR purification kits (QIAGEN, Valencia, CA) and subjected to PCR amplification using the following primer sets: CNS1-A (5′-GGTCAAGAGGGGCCTGACTT-3′ and 5′-GCAGTGTGTAAACAAAGAGA-3′), the 3′ end of CNS1-B (5′-CCAAAGAGAGCTGAGGTAAA-3′ and 5′-CAGCTTTTCCCCAGCTCAGT-3′) and the transcriptional start site (TSS; (5′-CAGAAACCAACCACTGGACCCAA-3′ and 5′-CAGGAACATGGAGCGGGAGG-3′). PCR products were resolved on a 2% agarose gel and visualized by ethidium bromide staining.
RNA analysis.
Total RNA was purified from cell cultures and tissues using Ultraspec reagent (Biotecx Laboratories, Houston, TX), according to the manufacturer's directions. RNase protection probes for murine IL-6, RANKL, and ribosomal protein L32 were purchased from Pharmingen (San Diego, CA) and used according to the manufacturer's directions. Taqman quantitative reverse transcription-PCR (RT-PCR) was performed as previously described (54) using the following primer probe sets from Applied Biosystems (Foster City, CA): RANKL (Mm0041908-m1); IL-6 (Mm00446190-m1); c-fos (Mm00487425-m1); osteocalcin (forward, 5′-GCTGCGCTCTGTCTCTCTGA-3′; reverse, 5′-TGCTTGGACATGAAGGCTTTG-3′; probe, 5′-6-carboxyfluorescein [FAM]-AAGCCCAGCGGCC-NFQ-3′ [NFQ, nonfluorescent quencher]), bone sialoprotein (forward, 5′-TTTTGCTCAGCATTTTGGGAAT-3′; reverse, 5′-TGCTTTGATTCTTCGATGGAAATT-3′; probe, FAM-5′-CTGTGCTTTCTCGATGAA-3′-NFQ), and ribosomal protein S2 (forward, 5′-CCCAGGATGGCGACGAT-3′; reverse, 5′-CCGAATGCTGTAATGGCGTAT-3′; probe, FAM-5′-TCCAGAGCAGGATCC-3′-NFQ).
Southern blot analysis.
Genomic DNA from tissues or cell lines was digested with PvuII and HindIII and then divided into four aliquots which were incubated with buffer alone, SmaI, HpaII, or MspI. Digested DNA was fractionated on a 0.8% agarose gel, denatured with NaOH, and transferred to Hybond-N membranes (Amersham, Arlington Heights, IL) according to the manufacturer's instructions. Membranes were hybridized to random-primed, [32P]dCTP-labeled DNA probes using QuikHyb (Stratagene, La Jolla, CA) and washed according to the manufacturer's instructions.
Sequence comparisons.
Sequence comparisons of mouse and human DNA sequences were performed using the web-based zPicture software (56) available at the Lawrence Livermore National Laboratory (www.decode.org). Genomic DNA sequences of the CNS1-A region from different species were obtained by BLAST search of the respective species genomes at the Ensembl genome website (www.ensembl.org) and aligned using the ClustalW multiple sequence alignment program (19).
Generation of mutant mice.
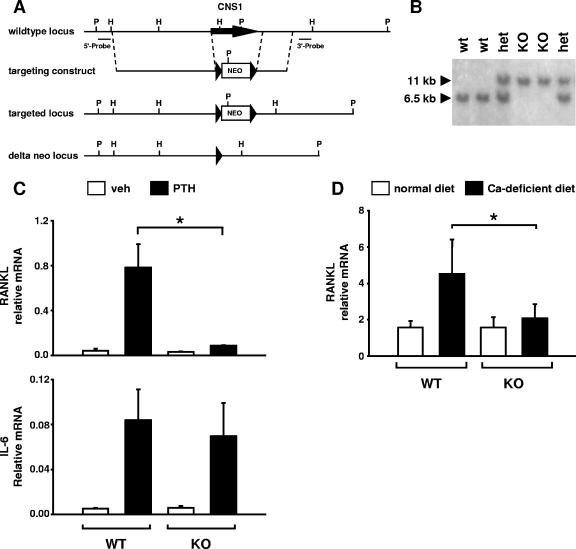
CNS1-deficient mice were created using a targeting vector consisting of a neo cassette flanked by a 4.5-kb left homology arm and a 1.4-kb right homology arm (see Fig. 9A). Both homology arms were obtained from an 8.4-kb ApaI-AgeI fragment subcloned from BAC-18. The left homology arm extended from the ApaI site to an NheI site immediately upstream from the CNS1 region. The right homology arm extended from an HpaI site downstream from the CNS1 homology region to the AgeI site. In addition, the Neo cassette was flanked by loxP recognition sequences. Linearized targeting vector was used to electroporate R1 ES cells (47), and correctly targeted ES cell clones were identified by Southern blotting with probes flanking the homology arms (Fig. 9A). Three independent, correctly targeted ES cell clones were injected into C57BL/6 blastocyts to create chimeric mice. Chimeric mice from two of the three ES cell clones demonstrated germ line transmission. The Neo cassette was removed by crossing the chimeric mice with transgenic mice expressing the Cre recombinase in germ cells (33). Primer sequences used for genotyping CNS1-deficient mice are available upon request.
FIG. 9.
PTH stimulation of RANKL expression is blunted in mice lacking CNS1. (A) The targeting construct used to delete the CNS1 region (indicated by the arrow) from the mouse genome is shown. Germ line-transmitting chimeras were crossed with EIIα-CRE mice to delete the neo selection cassette from the germ line, leaving a single 34-bp loxP site (arrowhead) in place of CNS1. P, PstI; H, HindIII. (B) Southern blot analysis used for genotyping CNS1 KO mice. het, heterozygous mice. (C) Quantitative RT-PCR analysis of bone marrow cultures from homozygous CNS1 KO mice and wild-type littermates. Cells were treated with vehicle or 10−7 M PTH for 24 h, after which RANKL and IL-6 mRNAs were quantified and normalized to ribosomal protein S2. Values shown are means from duplicate cultures of two to three mice per genotype. (D) Quantitative RT-PCR analysis of L5 vertebral RNA from CNS1 homozygous KO mice or wild-type littermates fed a normal or calcium-deficient diet for 7 days. Values are means ± SD of four to five mice per group. *, P < 0.05 by analysis of variance.
Statistics.
Data were analyzed using SigmaStat (SPSS Science, Chicago, IL). All values are reported as the mean ± standard deviation (SD). Differences between group means were evaluated with Student's t test or analysis of variance.
RESULTS
cAMP control of RANKL expression is tissue specific.
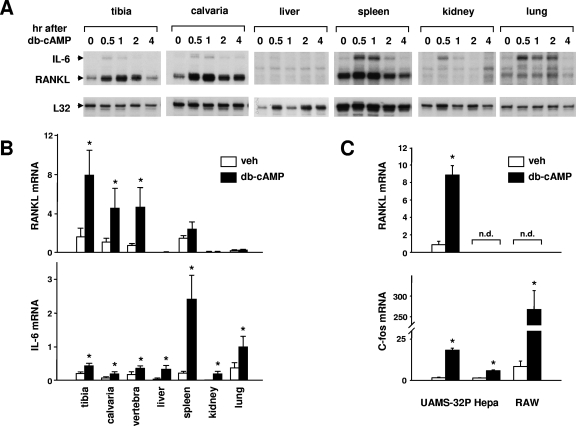
As a prelude to our search for the molecular mechanism responsible for PTH-controlled RANKL expression in a PKA-CREB-dependent manner, we administered db-cAMP, a cell-permeable analog of cAMP, to adult C57BL/6 mice and compared the levels of expression of RANKL mRNA in bone and other tissues (Fig. 1A). The choice of db-cAMP, as opposed to PTH, was based on the reasoning that the former stimulus would allow us to determine the existence of a post-receptor cell-type-specific mechanism(s)—the objective of our work here—as opposed to the mere presence or absence of PTHR1. RANKL mRNA was analyzed at various time intervals following db-cAMP administration by RNase protection assays. db-cAMP significantly stimulated RANKL mRNA in tibia and calvaria, but not in liver, spleen, kidney, or lung (Fig. 1A). In contrast, and consistent with previous studies demonstrating that IL-6 is stimulated via a cAMP-PKA-CREB pathway in many different cell types (18, 20, 46, 70), db-cAMP transiently stimulated IL-6 mRNA in all tissues examined. A quantitative assessment of the regulation of RANKL and IL-6 expression by cAMP was accomplished using real-time RT-PCR analysis of mRNA isolated from the indicated tissues 1 h after the injection of db-cAMP (Fig. 1B). Again, db-cAMP elevated RANKL mRNA in bone (tibia, calvaria, and vertebra) but not liver, spleen, kidney, or lung. In contrast, IL-6 mRNA was stimulated in all tissues examined. In full agreement with the specificity exhibited following db-cAMP administration to mice, 24 h of exposure to db-cAMP stimulated RANKL mRNA in a stromal/osteoblastic cell line (UAMS-32P), but not in a hepatocyte (Hepa) or monocyte (RAW 264.7) cell line (Fig. 1C). Shorter exposure (30 min or 4 h) of the cells to db-cAMP also stimulated RANKL mRNA in UAMS-32P cells but not in Hepa or RAW264.7 cells (data not shown). db-cAMP stimulation of c-fos mRNA demonstrated that each cell line was responsive to cAMP elevation (Fig. 1C).
FIG. 1.
Tissue-specific stimulation of RANKL. (A) RNase protection assay of RNA from mice injected with db-cAMP (100 mg/kg body weight). RNA was isolated from the indicated tissues at the indicated time points and analyzed with probes for IL-6, RANKL, and ribosomal protein L32. Each lane contains RNA from a single mouse, and the RNA analyzed in lane 0 is from an uninjected mouse. (B) Quantitative RT-PCR analysis of RNA from mice injected with PBS (white bars) or db-cAMP at 100 mg/kg body weight (black bars). The indicated tissues were harvested 1 h after injection, and total RNA was prepared. RANKL or IL-6 mRNAs were quantified using Taqman RT-PCR, and the values were normalized to ribosomal protein S2. Each bar represents the mean ± SD of three animals. veh, vehicle. (C) Quantitative RT-PCR analysis of RNA from cell lines treated with vehicle or 1.5 mM db-cAMP for 24 h. Values for RANKL and c-fos were normalized to ribosomal protein S2. Each bar represents the mean ± SD of triplicate wells. *, P < 0.05 by Student's t test versus vehicle. n.d., not detectable.
Previous studies have shown that loss of RANKL expression after serial passage of a murine stromal cell line, ST2, was associated with hypermethylation in a CpG island encompassing the first exon of the RANKL gene (27) and that tissue-specific methylation of such CpG islands in other genes may underlie their tissue-specific expression (4). Therefore, one potential explanation for the cell-type-specific expression of RANKL could be that important regulatory regions in or near the first exon are hypermethylated in nonexpressing cells. To explore this possibility, we examined the methylation status of the CpG island encompassing exon 1 using genomic DNA isolated from cell lines and tissues that either express or do not express RANKL. Southern blot analysis with methylation-sensitive restriction enzymes revealed that this region was hypermethylated in the nonexpressing cell line Hepa and partially methylated in UAMS-32 cells (see Fig. S1 in the supplemental material). In contrast, the CpG island was completely unmethylated in calvaria cells, as well as kidney and liver (see Fig. S1 in the supplemental material), yet only the calvaria cells express RANKL (Fig. 1A and B).
The results presented so far indicated that the tissue- and cell-type-specific expression of RANKL cannot be accounted for by limited activation of signaling pathways or by hypermethylation of the CpG island encompassing the first exon of the RANKL gene. We have shown previously that PTH stimulates RANKL gene transcription as measured by nuclear run-on assay (15). Therefore, we postulated that the cell-type-specific control of RANKL expression by the PTH-cAMP pathway may result from cell-type-specific control of gene transcription. To address this possibility, we next attempted to identify regions of the RANKL gene that mediate the response to cAMP with the eventual goal of identifying trans-acting factors responsible for cell-type-specific control of this gene.
The region conferring cAMP responsiveness is distant from the transcription start site.
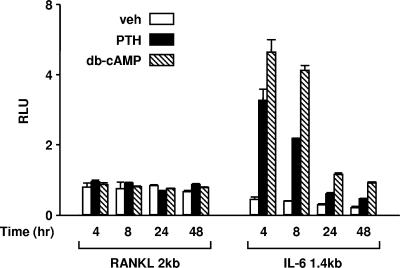
To identify cis-acting sequences that mediate the response to PTH, we initially used a luciferase reporter construct containing 2 kb of the murine RANKL 5′-flanking region (Fig. 2). However, neither PTH nor db-cAMP stimulated the activity of this promoter construct when the cells were treated between 4 and 24 h. However, an IL-6 promoter in the same vector was stimulated by these agents at each time point. These results suggested that cis-acting elements required for cAMP responsiveness of the RANKL gene reside outside the proximal 2-kb 5′-flanking region.
FIG. 2.
The RANKL proximal 2-kb promoter is unresponsive to PTH. UAMS-32P cells were stably transfected with luciferase reporter constructs containing either the region from −2014 to +27 of the murine RANKL promoter (RANKL 2kb) or the region from −1379 to +45 of the murine IL-6 promoter (IL-6 1.4kb). Pools of transfected cells were treated with vehicle (veh; white bars), 10−7 M PTH (black bars), or 1.5 mM db-cAMP (striped bars) for the indicated times before determination of relative light units (RLU). Each value represents the mean ± SD of triplicate wells.
In earlier work, we had also shown that reporter constructs containing up to 7 kb of 5′-flanking region were not regulated by gp130 cytokines or 1,25(OH)2D3 (55), raising the possibility that important control elements of the RANKL gene may be located in regions very distant from the transcription start site. Therefore, we generated a reporter construct using a BAC containing the entire murine RANKL gene as well as 120 kb of 5′-flanking region and 14 kb of 3′-flanking region. This 160-kb fragment does not encode any known transcripts other than RANKL, as determined using the Ensembl genome browser (www.ensembl.org). The reporter construct, designated BAC1 (Fig. 3A), was generated by inserting a cassette, containing the encephalomyocarditis virus (ECMV) internal ribosome entry site followed by a firefly luciferase cDNA and a minimal thymidine kinase promoter-neomycin phosphostransferase gene (IRES-luc-tk-neo), into exon 5 of the BAC clone via homologous recombination in E. coli, a process also known as recombineering (5). The targeting was such that the IRES-luc-tk-neo cassette replaced only the RANKL 3′-UTR contained in exon 5.
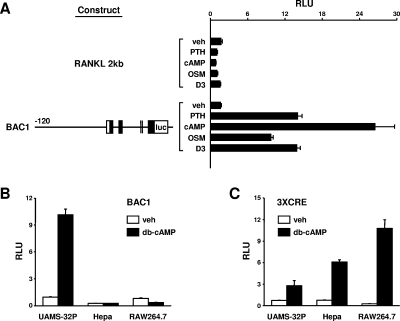
FIG. 3.
A BAC-based RANKL transcriptional reporter construct is responsive to PTH. (A) UAMS-32P cells were stably transfected with the RANKL 2-kb reporter construct (RANKL 2kb) or a reporter construct derived from a BAC containing the entire murine RANKL gene as well as 120 kb of 5′-flanking region and 14 kb of 3′-flanking region (BAC1). The BAC1 construct is depicted graphically with the relative positions of exons indicated by boxes and the RANKL coding region indicated by filled boxes. The sizes of the introns and exons are not drawn to scale. Pools of transfected cells were treated with vehicle (veh), 10−7 M PTH, 1.5 mM db-cAMP, 25 ng/ml OSM, or 10−8 M 1,25(OH)2D3 for 24 h, and relative light units (RLU) were determined. (B) UAMS-32P, Hepa, or RAW264.7 cells stably transfected with the BAC1 reporter construct were treated with 1.5 mM db-cAMP for 24 h, and RLU were determined. (C) UAMS-32P, Hepa, or RAW264.7 cells stably transfected with a reporter construct containing three cAMP response elements upstream from a minimal promoter (3×CRE), were treated with db-cAMP as described for panel B. The values shown in each panel are the mean ± SD of triplicate wells.
The BAC1 reporter construct was used to stably transfect UAMS-32P cells, which were then tested for responsiveness to PTH and db-cAMP, as well as other hormones and cytokines known to stimulate RANKL. In contrast to the 2-kb RANKL promoter construct, BAC1 demonstrated robust responsiveness to PTH or db-cAMP, as well as to OSM and 1,25(OH)2D3 (Fig. 3A). The reproducibility of stable transfection with the BAC-based reporter was verified by multiple transfections with the same DNA preparation as well as independent preparations (see Fig. S2 in the supplemental material). The integrity of the integrated DNA construct was verified by Southern blot analysis with probes derived from the extreme 5′ and 3′ ends of the construct (see Fig. S3 in the supplemental material).
To determine whether the responsiveness of the BAC1 construct was cell type specific, UAMS-32P, Hepa, or RAW264.7 cells stably transfected with BAC1 were treated with vehicle or db-cAMP (Fig. 3B). db-cAMP was used, rather than PTH, since the presence of PTHR1 on Hepa or RAW264.7 cells has not been determined. While activity of the BAC1 construct was stimulated in UAMS-32P cells, it was unresponsive in both Hepa and RAW264.7 cells. However, db-cAMP stimulated a reporter construct containing three copies of the somatostatin cAMP response element (CRE) inserted upstream from a RANKL minimal promoter in each of the cell lines, demonstrating that cAMP activation of CREB did occur in each of them (Fig. 3C). These results indicate that a reporter construct containing extensive 5′- and 3′-flanking region, but not a reporter containing only the proximal 2-kb 5′-flanking region, contains sequences that confer hormonal control of RANKL gene expression in a cell-type-specific manner.
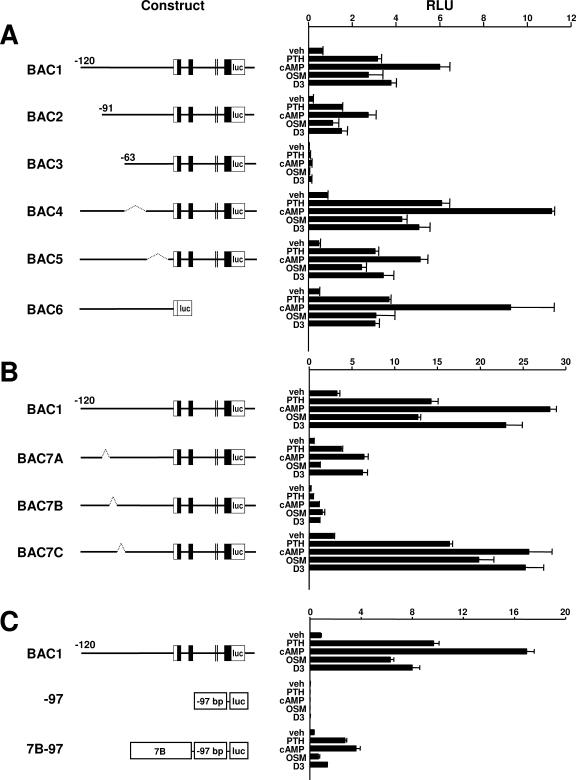
To localize the responsive region, we then sequentially deleted segments of approximately 30 kb from the 5′-flanking region contained in the BAC1 reporter construct again via the recombineering technique (Fig. 4A). In addition, we deleted the entire sequence downstream from the translation start codon located in exon 1 (construct BAC6 in Fig. 4A). Deletion of the region between −63 and −91 kb significantly reduced both basal activity of the reporter construct as well as its responsiveness to PTH, db-cAMP, OSM, and 1,25(OH)2D3. However, deletion of the other 30-kb segments did not dramatically alter responsiveness. To further refine the responsive region, we deleted sequential fragments of approximately 10 kb from within the region from −63 to −91 kb of the BAC1 construct (constructs BAC7A, BAC7B, and BAC7C in Fig. 4B). Deletion of the distal 10 kb selectively reduced the responsiveness to OSM, whereas deletion of the central 10-kb region reduced responsiveness to all stimuli, but had the least affect on OSM responsiveness (Fig. 4B). Deletion of the proximal 10-kb region did not significantly alter the responsiveness to any of the agents. These results suggest that an OSM response element(s) resides within the region deleted in BAC7A and that elements controlling the overall responsiveness of the gene reside within the region deleted in construct BAC7B. In studies leading to the work presented here, we had also shown that the cAMP-CREB pathway is a central regulator of RANKL gene expression in response to a variety of stimuli (15). Thus, the region deleted in BAC7B may also contain important cAMP response elements. Consistent with this hypothesis, insertion of an 8.4-kb fragment containing the majority of this region upstream from an unresponsive minimal RANKL promoter conferred significant responsiveness to PTH, db-cAMP, and 1,25(OH)2D3, but not OSM (Fig. 4C). Since the primary objective of the present study was to identify regions conferring responsiveness to cAMP in a cell-type-specific manner, we subsequently focused our attention on this 8.4-kb region, located approximately 73 kb upstream from exon 1 of the RANKL gene.
FIG. 4.
Localization of hormone- and cytokine-responsive regions. The reporter constructs diagrammed to the left of each panel were used to stably transfect UAMS-32P cells. The sizes of the introns and exons are not drawn to scale. Pools of colonies for each construct were treated in triplicate with vehicle (veh), PTH (10−7 M), db-cAMP (1.5 mM), OSM (25 ng/ml), or 1,25(OH)2D3 (10−8 M) for 24 h, and relative light units (RLU) were determined. Values represent the mean ± SD of triplicate wells. (A) BAC2 through BAC5 were derived from BAC1 via deletion of the following regions: BAC2, −120 kb to −91 kb; BAC3, −120 kb to −63 kb; BAC4, −63 kb to −33 kb; and BAC5, −33 kb to −2 kb. The dotted lines in BAC4 and BAC5 indicate deleted regions. BAC6 was derived from BAC-18 (see Materials and Methods) by replacing all sequences downstream from the start codon in the first exon of the RANKL gene with the luciferase coding sequence. (B) BAC7A, BAC7B, and BAC7C were derived from BAC1 via deletion of the following regions: BAC7A, −91 kb to −82 kb; BAC7B, −82 kb to −73 kb; and BAC7C, −73 kb to −63 kb. The dotted lines indicate deleted regions. (C) The −97 construct contains a RANKL minimal promoter (the first 97 bp of the RANKL 5′-flanking region) inserted upstream from a luciferase coding sequence. The 7B-97 construct was derived from the −97 construct by inserting a fragment, corresponding to the region deleted from construct BAC7B, upstream from the RANKL minimal promoter.
A 715-bp CNS is required for full responsiveness to cAMP.
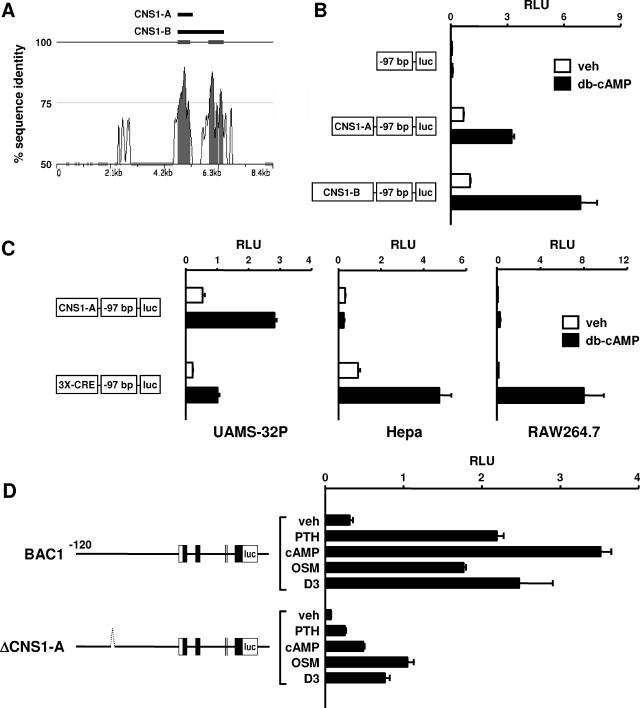
Important regulatory regions are often highly conserved between species (49, 66, 67). Based on this, we used zPicture software (56) to compare human and mouse genomic DNA sequences within the 8.4-kb region that conferred cAMP responsiveness to the minimal RANKL promoter. This comparison identified two highly conserved regions of approximately 700 bp and 900 bp separated by approximately 350 bp in the mouse genome (Fig. 5A). The homology in each region was greater than 70% for more than 100 bp, which conforms to the definition of a conserved noncoding sequence (CNS) (40). Therefore, a 715-bp DNA fragment containing the upstream region was designated CNS1-A and a 1,975-bp fragment containing both the upstream and downstream regions was designated CNS1-B. The full nucleotide sequences of murine CNS1-A and CNS1-B are presented in Fig. S4 in the supplemental material. The 3′ ends of CNS1-A and CNS1-B are located 76.2 kb and 74.9 kb, respectively, upstream from exon 1 of the murine RANKL gene.
FIG. 5.
Identification and analysis of CNS1. (A) A zPicture plot of sequence conservation between mouse (bottom) and human (top) genomic DNA within the 7B region is shown. The percentage of sequence identity using a sliding 100-bp window is indicated by the vertical axis. Portions of the plot filled in gray indicate regions of DNA with at least 100 bp of 70% or more sequence identity. Filled bars above the plot depict the relative positions of fragments CNS1-A and CNS1-B used in promoter studies. (B) Luciferase reporter assay of UAMS-32P cells stably transfected with a minimal RANKL promoter-reporter construct (−97) or the same construct containing either CNS1-A (CNS1-A-97) or CNS1-B (CNS1-B-97). Cells were treated with vehicle (veh) or 1.5 mM db-cAMP for 24 h prior to relative light unit (RLU) determination. (C) Luciferase reporter assay of UAMS-32P, Hepa, or RAW264.7 cells stably transfected with either CNS1-A-97 or a reporter construct containing the same minimal promoter (−97) with three cAMP response elements from the somatostatin promoter inserted upstream (3×CRE-97). Cells were treated with vehicle or db-cAMP for 24 h and analyzed as for panel B. (D) Luciferase reporter assay of UAMS-32P cells stably transfected with either BAC1 or a reporter construct derived from BAC1 in which the 715 bp corresponding to CNS1-A was deleted (ΔCNS1-A). The dotted line in ΔCNS1-A indicates the deleted region but is not drawn to scale. Cells were treated with vehicle, 10−7 M PTH, 1.5 mM db-cAMP, 25 ng/ml OSM, or 10−8 M 1,25(OH)2D3 for 24, and RLU were determined. Values in all bar graphs represent the mean ± SD of triplicate wells.
To determine whether CNS1-A or CNS1-B contained sequences capable of conferring responsiveness to cAMP, each fragment was inserted upstream from a minimal RANKL promoter and used to stably transfect UAMS-32P cells. Both fragments conferred significant cAMP responsiveness to the otherwise unresponsive minimal RANKL promoter (Fig. 5B). Since the construct containing CNS1-B was only approximately 30% more responsive than the construct containing CNS1-A, we concluded that sequences within CNS1-A were sufficient for a significant cAMP response and focused further analysis on this region.
The cell type specificity of the cAMP responsiveness conferred by CNS1-A was determined by comparing its activity in either RANKL-expressing (UAMS-32P) or -nonexpressing (Hepa and RAW264.7) cell lines. The reporter construct containing CNS1-A was stimulated by db-cAMP exclusively in UAMS-32P cells, while a reporter containing three consensus CREs derived from the somatostatin promoter was stimulated in each cell type (Fig. 5C). These results demonstrate that sequences contained within CNS1-A confer responsiveness to cAMP in a cell-type-specific manner.
To determine whether the CNS1-A region was required for cAMP responsiveness of the full-length BAC1 reporter construct, CNS1-A was deleted from the BAC1 construct using recombineering techniques to generate a construct designated ΔCNS1-A. Similar to what was observed with the BAC7B construct, deletion of CNS1-A significantly blunted the responses to PTH, db-cAMP, and 1,25(OH)2D3, and, to a lesser extent, OSM (Fig. 5D), indicating that sequences within the 715-bp CNS1-A fragment play an important role in the responsiveness of the BAC1 construct to the PTH-cAMP pathway.
CNS1-A contains functional CREB and Runx2 binding sites.
In the experiments summarized in Fig. 6, we attempted to localize the cAMP-responsive region as a prelude to identification of possible trans-acting factors. To this end, we generated a series of deletion mutants from the reporter construct harboring CNS1-A upstream from a minimal RANKL promoter and tested for responsiveness to db-cAMP. As shown in Fig. 6A, a 120-bp fragment located near the center of CNS1-A was both necessary and sufficient to confer cAMP responsiveness to the minimal promoter. Visual inspection of this 120-bp region led to the identification of two potential CREs, each with a single nucleotide difference from the consensus CRE sequence (Fig. 6B). In addition, a potential binding site for the osteoblast-specific transcription factor Runx2, designated OSE2 (7), was also observed. Strikingly, each of these sequences was completely conserved among five different mammalian species, with the exception of single base pair in the distal CRE of the chimp sequence (Fig. 6B).
FIG. 6.
Localization of cAMP response elements within CNS1-A. (A) Luciferase reporter assay of UAMS-32P cells stably transfected with the CNS1-A-97 reporter construct or reporter constructs derived from CNS1-A in which different regions of CNS1-A were deleted (Delta 1 through Delta 5). Open boxes indicate the region of CNS1-A retained in each construct. veh, vehicle. (B) Sequence comparison of a portion of DNA contained in the region of CNS1-A retained in the Delta 5 construct shown in panel A. Sequences for rat, dog, human, and chimp CNS1-A were obtained by BLAST search of Ensembl genome databases with mouse CNS1-A. For each search, only a single high-homology region was obtained, and in each case the region was located a similar distance upstream from the RANKL gene. Potential binding sites for Runx2 (OSE2) and CREB (dCRE and pCRE) are highlighted in gray. Nucleotides differing from consensus binding sites are shown in bold. (C) Luciferase reporter assay of UAMS-32P cells stably transfected with CNS1-A-97 or CNS1-A-97 containing point mutations within the indicated consensus sites (mutation indicated by an X over the site). In all reporter assays, cells were treated with vehicle or 1.5 mM db-cAMP for 24 h and relative light units (RLU) were determined.
To determine whether the consensus sites for CREB or Runx2 were important for the cAMP responsiveness conferred by the CNS1-A region, each site was mutated in the context of the CNS1-A-minimal promoter-reporter construct. The potential binding sites were mutated to sequences known to disrupt respective transcription factor protein binding (13, 55) and stably transfected into UAMS-32P. Mutation of either potential CRE site blunted, while mutation of both sites together abolished, the response to db-cAMP (Fig. 6C). In addition, mutation of the potential Runx2 binding site also blunted the response to db-cAMP (Fig. 6C).
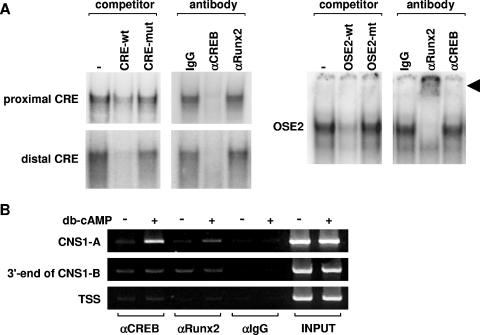
Furthermore, we determined whether CREB or Runx2 could bind to the CRE or OSE2 sequences of the RANKL CNS1-A, respectively, using nuclear extracts from UAMS-32P cells and gel mobility shift assays. Complexes formed by both the proximal and distal CRE probes were competed by the respective unlabeled wild-type, but not mutant, probes (Fig. 7A). Moreover, an antibody directed against CREB, but not an antibody directed against Runx2, blocked formation of the protein-DNA complex formed by either the proximal or distal CRE probes (Fig. 7A). Similarly, the complex formed by the OSE2 sequence was competed by wild-type, but not mutant, unlabeled probe and an antibody to Runx2, but not CREB, supershifted the complex (Fig. 7A). ChIP assays were used to confirm binding of CREB and Runx2 to the CNS1-A region within intact cells (Fig. 7B). Treatment with db-cAMP induced binding of both CREB and Runx2 at the CNS1-A region, but not at the 3′ end of the CNS1-B region or at the transcription start site of the RANKL gene.
FIG. 7.
CREB and Runx2 bind to CNS1-A. (A) Gel mobility shift assays using probes for proximal and distal CREs or OSE2 present in CNS1-A. Probes are indicated to the left of each panel, and cold competitors (200-fold excess) or antibodies are indicated above each panel. Nuclear extracts were from UAMS-32P cells. A supershifted complex of the OSE2 probe and anti-Runx2 antibody is indicated by the arrowhead. wt, wild type; mut, mutant. (B) UAMS-32P cells were treated with either vehicle or 1.5 mM db-cAMP for 6 h and then subjected to ChIP analysis using antibodies to CREB, Runx2, or control IgG (αCREB, αRunx2, and αIgG, respectively). The immunoprecipitated DNA was isolated and then amplified using a primer set centered on the OSE2 and CRE sites within CNS1A (30 cycles), a primer set encompassing the 3′ end of CNS1-B (30 cycles), or a primer set centered on the TSS (32 cycles). PCR analyses were all performed within the linear range of amplification.
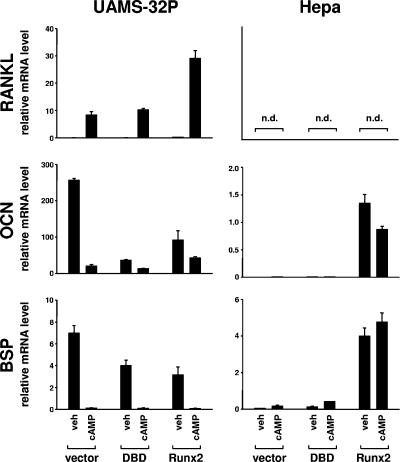
The close proximity of the OSE2 and CRE sites, together with the reduction in cAMP responsiveness observed with mutation of the OSE2 site, raised the possibility that Runx2 may contribute to the cell-type-specific responsiveness of RANKL to the PTH-cAMP pathway. We therefore overexpressed murine Runx2 in UAMS-32P and Hepa cells using retroviral transduction and determined the effect on the cAMP responsiveness of the endogenous RANKL gene. In parallel, we transduced cells with a construct expressing only the DBD of Runx2, which has been shown to act in a dominant-negative manner on some Runx2 target genes (8). Overexpression of Runx2 in UAMS-32P cells resulted in a threefold-higher stimulation of RANKL mRNA by db-cAMP compared to that in vector-transduced cells (Fig. 8). Expression of osteocalcin and bone sialoprotein was suppressed by overexpression of Runx2 in UAMS-32P cells, a phenomenon which has been observed previously in osteoblastic cells in vitro (22, 63) and, in the case of osteocalcin, in vivo (39). RANKL mRNA levels were undetectable in Hepa cells and were not altered by overexpression of Runx2 (Fig. 8). Nonetheless, overexpression of Runx2 in Hepa cells induced expression of osteocalcin and bone sialoprotein, thereby demonstrating that Runx2 levels in the transduced cells were sufficient for stimulation of known Runx2 target genes (Fig. 8). In full agreement with earlier work from our laboratory (15), expression of the Runx2-DBD in UAMS-32P cells had no effect on the stimulation of RANKL by PTH but did suppress osteocalcin and bone sialoprotein mRNA levels (Fig. 8). Together the results of the comparison of UAMS-32P and Hepa cells overexpressing Runx2 demonstrate that (i) increased Runx2 levels potentiate PTH stimulation of RANKL in stromal/osteoblastic cells and (ii) elevation of Runx2 levels alone is not sufficient to confer RANKL expression in cell types that do not normally express this cytokine.
FIG. 8.
Overexpression of Runx2 elevates endogenous RANKL mRNA. Shown are the results of quantitative RT-PCR analysis of UAMS-32P cells (left panels) or Hepa cells (right panels) transduced with an empty retroviral expression vector (vector) or the same vector expressing the DNA-binding domain of Runx2 (DBD) or full-length Runx2 (Runx2). Cell were treated with vehicle (veh) or 1.5 mM db-cAMP for 24 h, and RANKL, osteocalcin (OCN), and bone sialoprotein (BSP) mRNAs were quantified and normalized to ribosomal protein S2 mRNA. n.d., not detectable.
Generation of CNS1-KO mice: demonstration of the critical role of this enhancer in PTH stimulation of the endogenous RANKL gene.
Finally, to establish unequivocally the significance of CNS1 for PTH regulation of the endogenous RANKL gene, we designed a targeting construct to delete a 2,372-bp region containing CNS1 from the mouse genome (Fig. 9A) and used it to generate mice deficient in this distant transcriptional enhancer (knockout [KO] mice). The region deleted corresponds to CNS1-B in Fig. 5A. Electroporation of murine ES cells with the targeting construct and screening with probes flanking both ends of the homology arms resulted in the identification of multiple clones that had undergone homologous recombination replacing CNS1 with a neomycin-phosphotransferase expression cassette flanked by loxP recognition sites. Three clones were used to create chimeric mice, and two of these demonstrated germ line transmission of the mutant allele. Mice generated from both ES cell clones displayed identical phenotypes. To avoid potential contributions of the floxed neo selection cassette to RANKL gene expression, mice heterozygous for the mutant allele were crossed with transgenic mice expressing Cre recombinase in germ line cells. This resulted in heterozygous mice harboring one wild-type allele and one allele in which the 2,372 bp encompassing CNS1 was replaced with a single 34-bp loxP recognition site, indicated by a 6.5-kb band and an 11-kb band, respectively, on Southern blots using the 5′-flanking probe and genomic DNA digested with PstI (Fig. 9B).
Mice homozygous for the CNS1 deletion (KO) were obtained at the expected Mendelian frequency from mating of heterozygous mice and did not display the obvious osteopetrotic characteristics observed in RANKL-deficient mice. Specifically, previous radiographic analysis of RANKL KO mice revealed a lack of bone marrow cavity formation, abnormally shaped long bones, and a lack of tooth eruption (31), all due to a lack of osteoclastic bone resorption. Therefore, we compared wild-type and CNS1 KO mice radiographically at 5 months of age. The radiographs demonstrated the presence of bone marrow cavities in long bones, normally shaped long bones, and erupted teeth in the CNS1 KO mice, indicating that they are not osteopetrotic (see Fig. S5 in the supplemental material).
To determine whether loss of CNS1 altered the response of the endogenous RANKL gene to PTH, bone marrow cells from WT and KO mice were cultured in vitro under conditions that promote osteoblast differentiation and PTH receptor expression as we have previously described (54). In these experiments, cultures were treated with vehicle or PTH for 24 h and RANKL mRNA was quantified using quantitative RT-PCR. As shown in Fig. 9C, PTH stimulation of RANKL mRNA was significantly blunted in KO cells. In contrast, PTH stimulation of IL-6 mRNA was indistinguishable in cells from the KO mice as compared to cells from the WT control (Fig. 9C, bottom panel). Next we determined whether loss of CNS1 altered the response of the RANKL gene in vivo to elevation of endogenous PTH via a calcium-deficient diet. Seven days of calcium-deficient diet elevated PTH levels to similar extents in both WT and KO mice (WT normal diet, 9.8 ± 5.8 pg/ml, and calcium-deficient diet, 70.2 ± 58.7 pg/ml; KO normal diet, 8.1 ± 8.2 pg/ml, and calcium-deficient diet, 86.6 ± 26.6 pg/ml). However, whereas RANKL mRNA was significantly elevated in bone from WT mice receiving the calcium-deficient diet, RANKL mRNA was not elevated in bone from KO mice receiving the calcium-deficient diet (Fig. 9D).
DISCUSSION
The essential role of RANKL in the birth and survival of osteoclasts, and thereby in bone resorption—a process critical for mineral and skeletal homeostasis—has been extensively documented during the last few years (3, 61). Moreover, RANKL has attracted extensive attention, both as the most proximal pathogenetic factor of numerous diseases associated with increased bone resorption and as the prime target for the development of rational and effective pharmacotherapeutics for bone and mineral disorders. However, the molecular mechanism(s) by which this gene is controlled in health and disease remains poorly, if at all, understood.
The evidence presented herein demonstrates for the first time that an enhancer sequence residing 76 kb upstream from the transcription start site of the RANKL gene, designated CNS1, is largely responsible for the cell-type-specific response of the RANKL gene to PTH, the principle hormone that controls calcium metabolism. Moreover, we show that deletion of the CNS1 enhancer from the mouse genome abrogates PTH responsiveness of the RANKL gene. Remarkably, the proximal portion of this enhancer region also contributes to transcriptional regulation of RANKL by 1,25(OH)2D3 (24a).
Skeletal development and tooth eruption were grossly normal in mice lacking the CNS1 enhancer. PTH-knockout mice are not osteopetrotic (45) and do not demonstrate a significant decrease in RANKL mRNA expression in bone (44). Thus, we were not necessarily expecting an osteopetrotic phenotype in mice lacking the CNS1 region. PTH is only one of many factors known to control RANKL expression, and it is unlikely that the CNS1 enhancer is essential for control of RANKL by all of these factors. Consistent with this notion, deletion analysis of our reporter constructs indicated that the response elements for gp130 cytokines such as OSM are located upstream from the CNS1 enhancer and that deletion of the CNS1 enhancer only mildly affected the response of the reporter construct to OSM (Fig. 5D). Thus, while response of the RANKL gene to PTH is significantly blunted in the CNS1 KO mice, it is likely that responsiveness to gp130 cytokines, as well as responsiveness to other known and unknown factors, remains intact or only mildly affected in the CNS1 KO animals.
Residual PTH responsiveness was observed in each of the reporter constructs lacking the CNS1-A region as well as in cells from CNS1 KO mice in vitro. This residual responsiveness may result from additional response elements located outside the CNS1-A region as well as from posttranscriptional regulation of the mRNA. We have reported previously that PTH prolongs that half-life of RANKL mRNA (15), and it is unknown at present whether replacement of the 3′-UTR in the reporter constructs with the IRES-luciferase cassette abrogates this regulation. Moderate PTH elevation in vivo, via use of the calcium-deficient diet, did not significantly stimulate RANKL mRNA in CNS1 KO mice, suggesting that there is no residual PTH stimulation in vivo. One explanation for the difference between the in vivo and in vitro results may be that basal levels of RANKL mRNA are higher in vivo due to local and systemic factors that contribute to tonic RANKL expression but are not present in the culture medium of our in vitro studies. This higher basal level in vivo may mask any low-level residual stimulation conferred by either additional response elements outside the CNS1-A region or posttranscriptional regulation.
We found in earlier studies that CREB is required for full stimulation of RANKL by PTH (15). Thus, the identification in the present work of two CRE-like sequences in the PTH-responsive region of the RANKL gene was not unexpected. However, two important findings indicate that activation of CREB alone is not sufficient for RANKL expression. First, db-cAMP failed to stimulate either the endogenous RANKL gene or RANKL transcriptional reporter constructs in liver and monocyte cell lines, whereas control genes or reporter constructs known to broadly respond to CREB activation were stimulated in these cell lines. Second, and perhaps more striking, injection of db-cAMP into mice elevated RANKL mRNA in bone, but not in several other tissues, even though IL-6 mRNA was elevated in all tissues. Thus, significant RANKL expression was restricted to tissues containing cells of the osteoblastic lineage and/or lymphocytes, even when cAMP signaling pathways were activated in many other tissues. It is likely, therefore, that other transcription factors act in concert with CREB to achieve the cell-type-specific control of RANKL by PTH.
There are several precedents for CREB cooperation with such cell-type-specific transcription factors. Inducible expression of TCR-α depends on an enhancer, located 68 kb downstream from the transcription start site (68), which contains a consensus CRE that functions synergistically with the lymphocyte-enriched transcription factors AML-1, Ets-1, and LEF-1 (42). Liver-specific induction of tyrosine aminotransferase (TAT) is due to an enhancer located 3.6 kb upstream from the transcription start site that consists of a binding site for the liver-enriched HNF4 transcription factor and a CRE (50). Neither the CRE nor the HNF4 site alone confers high promoter activity or stimulation by cAMP, indicating that the sites act synergistically to confer cAMP regulation in a cell-type-specific manner (2). In addition, kidney-specific expression of the urokinase-type plasminogen activator gene (LLC-PK1) also involves a cAMP-responsive enhancer, which is located 3.4 kb upstream from the transcription start-site (59). Induction by cAMP and cell type specificity are mediated by two imperfect CREs adjacent to an imperfect LFB3 binding site.
While some CREs that act in a tissue-restricted manner are located close to the transcription start site of the gene, as in the case of melanocyte-restricted microphthalmia-associated transcription factor (21), the CREs in the TCR-α, TAT, and LLC-PK1 genes, as well as in the RANKL gene, are located more than a kilobase away from the transcription start site. Studies with reporter constructs suggest that consensus CREs become less effective at mediating a response to CREB activation with increasing distance from the transcription start site (62). Thus, the mechanistic basis of the tissue-type-specific responsiveness of some cAMP-responsive genes may indeed depend on the physical distance of the CREs from the transcription start site, so that effective stimulation is possible only when additional transcription factors function together with CREB. Such synergism may involve structural changes in chromatin that allow distal and proximal regions of the promoter to interact (6).
Four distinct observations obtained from the studies described herein provide compelling evidence that the osteoblast-specific transcription factor, Runx2, binds to and participates in the transcriptional activity of the distant RANKL enhancer. First, there is a Runx2 binding site (OSE2) within the RANKL CNS1 region which is highly conserved, in sequence and in its relative position to the CREs among mammalian species. Second, this OSE2 site efficiently bound Runx2 protein in both gel shift and ChIP assays. Third, mutation of this site within the context of a CNS1-containing reporter construct blunted PTH stimulation of the construct. And fourth, overexpression of Runx2 in a stromal/osteoblastic cell line potentiated cAMP stimulation of the endogenous RANKL gene.
These findings are consistent with published studies utilizing Runx2-deficient mice which display reduced RANKL mRNA expression and reduced osteoclast numbers in vivo (16, 28). In addition, primary calvaria cells from these mice express less RANKL mRNA and display a reduced ability to support osteoclast formation in vitro (16). Consistent with these studies, reintroduction of Runx2 via adenoviral transduction partially restored RANKL expression to a cell line derived from Runx2-deficient mice (10). Nonetheless, it has been recently reported that 1,25(OH)2D3 was able to stimulate RANKL mRNA and osteoclast support in additional cell lines derived from calvaria of Runx2-deficient mice (52). Thus, Runx2 is not absolutely required for RANKL expression in stromal/osteoblastic cells and it remains possible that other members of the Runx family, such as Runx1 or Runx3, may substitute for Runx2 in Runx2-deficient cells.
We have reported previously (15, 55), and confirmed in the present study, that expression of the Runx2 DBD, which can act as a dominant negative for some Runx2 target genes, did not alter RANKL expression. We had originally concluded from this observation that Runx2 is unlikely to directly regulate RANKL expression. However, such a conclusion seems at odds with the evidence mentioned above, as well as previous work by others (10, 16, 17). One possible explanation that may help resolve this apparent discrepancy relies on the mechanism by which the DBD acts as a dominant-negative protein. Ducy et al. demonstrated that the DBD bound more avidly than full-length Runx2 to the OSE2 derived from the murine osteocalcin promoter, thereby allowing it to displace endogenous Runx2 from the osteocalcin gene (8). Based on this observation, the DBD should only function as a dominant negative for genes in which it has a higher affinity for the OSE2 site than full-length Runx2. Thus, the DBD may not act as a dominant negative with respect to RANKL expression because it may not have a higher affinity for the OSE2 present in the CNS1 region. Affinity binding studies will ultimately be required to determine whether this is the case.
The observation in the present report that overexpression of Runx2 in a liver cell line was not sufficient for RANKL expression leads us to conclude that, while Runx2 may potentiate RANKL expression, it is not sufficient for RANKL expression. Thus, even if Runx2 or another RUNX family member participates in the stromal/osteoblast-specific expression of RANKL, other factors are required to work with Runx2 and CREB to allow induction of RANKL by PTH in these cells. There are several additional conserved sequences within the 715-bp conserved region of CNS1, and it is likely that one or more of these sequences may be binding sites for such factors. Additional in silico analyses as well as footprinting techniques should help to identify these factors in future studies.
Recent work by the Karsenty laboratory indicates that sympathetic nervous system signaling stimulates bone resorption by stimulating RANKL expression in immature osteoblastic cells via the ATF4 transcription factor, which binds a CRE-like element in the proximal RANKL promoter (9). However, consistent with results presented here, this pathway does not appear to be involved in PTH control of RANKL since PTH stimulation of RANKL mRNA was normal in cells from mice deficient in either β2-adrenergic receptors or ATF4 (9).
The work reported here also strongly suggests that a distant regulatory region upstream from CNS1 controls responsiveness to OSM, presumably via binding of STAT transcription factors (53). Nonetheless, prolactin stimulates RANKL mRNA in mammary epithelial cells via a Jak2/STAT5a pathway that requires a STAT-binding sequence in the proximal murine RANKL promoter, and mutation of this site in the context of a −965-bp promoter reporter construct eliminated the twofold stimulation of the reporter by prolactin (60). This site, therefore, may not be important for STAT regulation in stromal/osteoblastic cells since our reporter construct containing 2 kb of proximal 5′-flanking region was not stimulated by OSM in UAMS-32P cells. In contrast, our BAC-based reporter was significantly stimulated by OSM and deletion of a 10-kb region located approximately 80 kb upstream from exon 1 specifically abolished this stimulation. Furthermore, deletion of the CNS1 region in our studies also blunted a full response to OSM, suggesting that multiple regions located far upstream from exon 1 are required for responsiveness to gp130 cytokines.
Kitazawa et al. have found that reporter constructs containing proximal RANKL promoter sequences are stimulated by 1,25(OH)2D3 in a murine stromal cell line, ST-2 (25). In the case of the murine promoter, this stimulation was abolished by mutation of a putative vitamin D receptor element (VDRE) at −937 bp relative to the transcription start site. However, we and others have not detected significant 1,25(OH)2D3 stimulation of reporter constructs containing up to 7 kb of 5′-flanking region (11, 23, 55). A possible explanation for these discordant findings is that factors required for stimulation of RANKL transcription are more limiting in some cell lines, such as UAMS-32, compared to ST-2 cells. However, we showed that stable integration of the 2-kb RANKL promoter into UAMS-32P cells, a maneuver that should overcome any potential limiting factors, still did not result in stimulation by 1,25(OH)2D3 (Fig. 2). This finding, together with the low level of reporter gene stimulation in these previous studies, suggests that additional regulatory regions exist outside the proximal 5′-flanking region. Consistent with this contention, we observed significant stimulation of our BAC-based reporter constructs by 1,25(OH)2D3. More important, Pike and coworkers, using ChIP/chip analysis, have independently identified a proximal portion of the CNS1 sequence as a region of the RANKL gene required for the ability of 1,25(OH)2D3 to stimulate RANKL transcription (24a). Based on the latter finding, we propose that the entire highly conserved region, which we labeled CNS1-B in our Fig. 5A, be designated the RANKL distal control region, or DCR.
In closing, in the work presented here we have identified a critical enhancer of RANKL gene transcription utilizing a strategy that combined mapping of response elements by deletion analysis together with in silico identification of CNSs within the mapped regions. There are at least 40 additional CNSs within the 160-kb fragment characterized in this study (not shown). Therefore, our combined approach was more efficient than a systematic analysis of all of the CNSs contained within the BAC clone since the majority of these lie within regions that were dispensable for stimulated RANKL BAC reporter activity. While it is clear that conservation of a sequence alone does not guarantee its functional significance (51), it remains possible that CNSs that were dispensable for PTH control of RANKL are nonetheless important for regulation by signals, or for expression in cell types, not analyzed in our study. As expected from our BAC-based mapping results, deletion of CNS1 from the mouse genome blunted PTH stimulation of the RANKL gene in vitro and in vivo. Thus, generation of CNS1 KO mice validated our combined strategy for mapping long-range enhancer regions. It is important to note that germ line deletion of enhancers identified by reporter analyses frequently fails to demonstrate a significant effect on gene expression, possibly due to redundancy in regulatory sequences (14, 37, 43). We expect that CNS1 KO mice will provide a unique and novel tool for determining the significance of PTH control of RANKL expression for both calcium and skeletal homeostasis.
Supplementary Material
Acknowledgments
We thank Isaac P. Foote, Andrea Dickens, and Priscilla Cazer for technical assistance. We also thank Sungtae Kim and J. Wesley Pike for advice, protocols, and reagents to perform the ChIP assays. Finally, we thank Teresita Bellido and Robert Jilka for critical reading of the manuscript.
This work was supported by National Institutes of Health grants R01 AR049794 to C.A.O. and P01 AG13918 to S.C.M. and by a Veterans Affairs Merit Review to S.C.M.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Anderson, D. M., E. Maraskovsky, W. L. Billingsley, W. C. Dougall, M. E. Tometsko, E. R. Roux, M. C. Teepe, R. F. DuBose, D. Cosman, and L. Galibert. 1997. A homologue of the TNF receptor and its ligand enhance T-cell growth and dendritic-cell function. Nature 390:175-179. [DOI] [PubMed] [Google Scholar]
- 2.Boshart, M., F. Weih, A. Schmidt, R. E. Fournier, and G. Schutz. 1990. A cyclic AMP response element mediates repression of tyrosine aminotransferase gene transcription by the tissue-specific extinguisher locus Tse-1. Cell 61:905-916. [DOI] [PubMed] [Google Scholar]
- 3.Boyle, W. J., W. S. Simonet, and D. L. Lacey. 2003. Osteoclast differentiation and activation. Nature 423:337-342. [DOI] [PubMed] [Google Scholar]
- 4.Ching, T. T., A. K. Maunakea, P. Jun, C. Hong, G. Zardo, D. Pinkel, D. G. Albertson, J. Fridlyand, J. H. Mao, K. Shchors, W. A. Weiss, and J. F. Costello. 2005. Epigenome analyses using BAC microarrays identify evolutionary conservation of tissue-specific methylation of SHANK3. Nat. Genet. 37:645-651. [DOI] [PubMed] [Google Scholar]
- 5.Copeland, N. G., N. A. Jenkins, and D. L. Court. 2001. Recombineering: a powerful new tool for mouse functional genomics. Nat. Rev. Genet. 2:769-779. [DOI] [PubMed] [Google Scholar]
- 6.Dekker, J. 2003. A closer look at long-range chromosomal interactions. Trends Biochem. Sci. 28:277-280. [DOI] [PubMed] [Google Scholar]
- 7.Ducy, P., and G. Karsenty. 1995. Two distinct osteoblast-specific cis-acting elements control expression of a mouse osteocalcin gene. Mol. Cell. Biol. 15:1858-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ducy, P., M. Starbuck, M. Priemel, J. Shen, G. Pinero, V. Geoffroy, M. Amling, and G. Karsenty. 1999. A Cbfa1-dependent genetic pathway controls bone formation beyond embryonic development. Genes Dev. 13:1025-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elefteriou, F., J. D. Ahn, S. Takeda, M. Starbuck, X. Yang, X. Liu, H. Kondo, W. G. Richards, T. W. Bannon, M. Noda, K. Clement, C. Vaisse, and G. Karsenty. 2005. Leptin regulation of bone resorption by the sympathetic nervous system and CART. Nature 434:514-520. [DOI] [PubMed] [Google Scholar]
- 10.Enomoto, H., S. Shiojiri, K. Hoshi, T. Furuichi, R. Fukuyama, C. A. Yoshida, N. Kanatani, R. Nakamura, A. Mizuno, A. Zanma, K. Yano, H. Yasuda, K. Higashio, K. Takada, and T. Komori. 2003. Induction of osteoclast differentiation by Runx2 through receptor activator of nuclear factor-kappa B ligand (RANKL) and osteoprotegerin regulation and partial rescue of osteoclastogenesis in Runx2−/− mice by RANKL transgene. J. Biol. Chem. 278:23971-23977. [DOI] [PubMed] [Google Scholar]
- 11.Fan, X., E. M. Roy, T. C. Murphy, M. S. Nanes, S. T. Kim, J. W. Pike, and J. Rubin. 2004. Regulation of RANKL promoter activity is associated with histone remodeling in murine bone stromal cells. J. Cell Biochem. 93:807-818. [DOI] [PubMed] [Google Scholar]
- 12.Fata, J. E., Y. Y. Kong, J. Li, T. Sasaki, J. Irie-Sasaki, R. A. Moorehead, R. Elliott, S. Scully, E. B. Voura, D. L. Lacey, W. J. Boyle, R. Khokha, and J. M. Penninger. 2000. The osteoclast differentiation factor osteoprotegerin-ligand is essential for mammary gland development. Cell 103:41-50. [DOI] [PubMed] [Google Scholar]
- 13.Fax, P., K. S. Lipinski, H. Esche, and D. Brockmann. 2000. cAMP-independent activation of the adenovirus type 12 E2 promoter correlates with the recruitment of CREB-1/ATF-1, E1A(12S), and CBP to the E2-CRE. J. Biol. Chem. 275:8911-8920. [DOI] [PubMed] [Google Scholar]
- 14.Fiering, S., E. Epner, K. Robinson, Y. Zhuang, A. Telling, M. Hu, D. I. Martin, T. Enver, T. J. Ley, and M. Groudine. 1995. Targeted deletion of 5′HS2 of the murine beta-globin LCR reveals that it is not essential for proper regulation of the beta-globin locus. Genes Dev. 9:2203-2213. [DOI] [PubMed] [Google Scholar]
- 15.Fu, Q., R. L. Jilka, S. C. Manolagas, and C. A. O'Brien. 2002. Parathyroid hormone stimulates receptor activator of NFkappa B ligand and inhibits osteoprotegerin expression via protein kinase A activation of cAMP-response element-binding protein. J. Biol. Chem. 277:48868-48875. [DOI] [PubMed] [Google Scholar]
- 16.Gao, Y. H., T. Shinki, T. Yuasa, H. Kataoka-Enomoto, T. Komori, T. Suda, and A. Yamaguchi. 1998. Potential role of Cbfa1, an essential transcriptional factor for osteoblast differentiation, in osteoclastogenesis: regulation of mRNA expression of osteoclast differentiation factor (ODF). Biochem. Biophys. Res. Commun. 252:697-702. [DOI] [PubMed] [Google Scholar]
- 17.Geoffroy, V., M. Kneissel, B. Fournier, A. Boyde, and P. Matthias. 2002. High bone resorption in adult aging transgenic mice overexpressing Cbfa1/Runx2 in cells of the osteoblastic lineage. Mol. Cell. Biol. 22:6222-6233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grassl, C., B. Luckow, D. Schlondorff, and U. Dendorfer. 1999. Transcriptional regulation of the interleukin-6 gene in mesangial cells. J. Am. Soc. Nephrol. 10:1466-1477. [DOI] [PubMed] [Google Scholar]
- 19.Higgins, D. G., J. D. Thompson, and T. J. Gibson. 1996. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 266:383-402. [DOI] [PubMed] [Google Scholar]
- 20.Huang, Y. F., J. R. Harrison, J. A. Lorenzo, and B. E. Kream. 1998. Parathyroid hormone induces interleukin-6 heterogeneous nuclear and messenger RNA expression in murine calvarial organ cultures. Bone 23:327-332. [DOI] [PubMed] [Google Scholar]
- 21.Huber, W. E., E. R. Price, H. R. Widlund, J. Du, I. J. Davis, M. Wegner, and D. E. Fisher. 2003. A tissue-restricted cAMP transcriptional response: SOX10 modulates alpha-melanocyte-stimulating hormone-triggered expression of microphthalmia-associated transcription factor in melanocytes. J. Biol. Chem. 278:45224-45230. [DOI] [PubMed] [Google Scholar]
- 22.Javed, A., G. L. Barnes, B. O. Jasanya, J. L. Stein, L. Gerstenfeld, J. B. Lian, and G. S. Stein. 2001. runt homology domain transcription factors (Runx, Cbfa, and AML) mediate repression of the bone sialoprotein promoter: evidence for promoter context-dependent activity of Cbfa proteins. Mol. Cell. Biol. 21:2891-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kabe, Y., J. Yamada, H. Uga, Y. Yamaguchi, T. Wada, and H. Handa. 2005. NF-Y is essential for the recruitment of RNA polymerase II and inducible transcription of several CCAAT box-containing genes. Mol. Cell. Biol. 25:512-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim, S., N. K. Shevde, and J. W. Pike. 2005. 1,25-Dihydroxyvitamin D3 stimulates cyclic vitamin D receptor/retinoid X receptor DNA-binding, co-activator recruitment, and histone acetylation in intact osteoblasts. J. Bone Miner. Res. 20:305-317. [DOI] [PubMed] [Google Scholar]
- 24a.Kim, S., M. Yamazaki, L. A. Zella, N. K. Shevde, and J. W. Pike. 2006. Activation of receptor activator of NF-κB ligand gene expression by 1,25-dihydroxyvitamin D3 is mediated through multiple long-range enhancers. Mol. Cell. Biol. 26:6469-6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kitazawa, R., and S. Kitazawa. 2002. Vitamin D-3 augments osteoclastogenesis via vitamin D-responsive element of mouse RANKL gene promoter. Biochem. Biophys. Res. Commun. 290:650-655. [DOI] [PubMed] [Google Scholar]
- 26.Kitazawa, S., K. Kajimoto, T. Kondo, and R. Kitazawa. 2003. Vitamin D-3 supports osteoclastogenesis via functional vitamin D response element of human RANKL gene promoter. J. Cell Biochem. 89:771-777. [DOI] [PubMed] [Google Scholar]
- 27.Kitazawa, S., and R. Kitazawa. 2002. Epigenetic control of mouse receptor activator of NF-kappa B ligand gene expression. Biochem. Biophys. Res. Commun. 293:126-131. [DOI] [PubMed] [Google Scholar]
- 28.Komori, T., H. Yagi, S. Nomura, A. Yamaguchi, K. Sasaki, K. Deguchi, Y. Shimizu, R. T. Bronson, Y. H. Gao, M. Inada, M. Sato, R. Okamoto, Y. Kitamura, S. Yoshiki, and T. Kishimoto. 1997. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 89:755-764. [DOI] [PubMed] [Google Scholar]
- 29.Kondo, H., J. Guo, and F. R. Bringhurst. 2002. Cyclic adenosine monophosphate/protein kinase A mediates parathyroid hormone/parathyroid hormone-related protein receptor regulation of osteoclastogenesis and expression of RANKL and osteoprotegerin mRNAs by marrow stromal cells. J. Bone Miner. Res. 17:1667-1679. [DOI] [PubMed] [Google Scholar]
- 30.Kong, Y. Y., U. Feige, I. Sarosi, B. Bolon, A. Tafuri, S. Morony, C. Capparelli, J. Li, R. Elliott, S. McCabe, T. Wong, G. Campagnuolo, E. Moran, E. R. Bogoch, G. Van, L. T. Nguyen, P. S. Ohashi, D. L. Lacey, E. Fish, W. J. Boyle, and J. M. Penninger. 1999. Activated T cells regulate bone loss and joint destruction in adjuvant arthritis through osteoprotegerin ligand. Nature 402:304-309. [DOI] [PubMed] [Google Scholar]
- 31.Kong, Y. Y., H. Yoshida, I. Sarosi, H. L. Tan, E. Timms, C. Capparelli, S. Morony, A. Oliveira-dos-Santos, G. Van, A. Itie, W. Khoo, A. Wakeham, C. R. Dunstan, D. L. Lacey, T. W. Mak, W. J. Boyle, and J. M. Penninger. 1999. OPGL is a key regulator of osteoclastogenesis, lymphocyte development and lymph-node organogenesis. Nature 397:315-323. [DOI] [PubMed] [Google Scholar]
- 32.Lacey, D. L., E. Timms, H. L. Tan, M. J. Kelley, C. R. Dunstan, T. Burgess, R. Elliott, A. Colombero, G. Elliott, S. Scully, H. Hsu, J. Sullivan, N. Hawkins, E. Davy, C. Capparelli, A. Eli, Y. X. Qian, S. Kaufman, I. Sarosi, V. Shalhoub, G. Senaldi, J. Guo, J. Delaney, and W. J. Boyle. 1998. Osteoprotegerin ligand is a cytokine that regulates osteoclast differentiation and activation. Cell 93:165-176. [DOI] [PubMed] [Google Scholar]
- 33.Lakso, M., J. G. Pichel, J. R. Gorman, B. Sauer, Y. Okamoto, E. Lee, F. W. Alt, and H. Westphal. 1996. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc. Natl. Acad. Sci. USA 93:5860-5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lam, J., S. Takeshita, J. E. Barker, O. Kanagawa, F. P. Ross, and S. L. Teitelbaum. 2000. TNF-alpha induces osteoclastogenesis by direct stimulation of macrophages exposed to permissive levels of RANK ligand. J. Clin. Investig. 106:1481-1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee, E. C., D. Yu, J. Martinez de Velasco, L. Tessarollo, D. A. Swing, D. L. Court, N. A. Jenkins, and N. G. Copeland. 2001. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics 73:56-65. [DOI] [PubMed] [Google Scholar]
- 36.Lee, S. K., and J. A. Lorenzo. 2002. Regulation of receptor activator of nuclear factor-kappaB ligand and osteoprotegerin mRNA expression by parathyroid hormone is predominantly mediated by the protein kinase a pathway in murine bone marrow cultures. Bone 31:252-259. [DOI] [PubMed] [Google Scholar]
- 37.Li, S. D., and A. L. Joyner. 2000. Two Pax2/5/8-binding sites in Engrailed2 are required for proper initiation of endogenous mid-hindbrain expression. Mech. Dev. 90:155-165. [DOI] [PubMed] [Google Scholar]
- 38.Liu, B. Y., J. Guo, B. Lanske, P. Divieti, H. M. Kronenberg, and F. R. Bringhurst. 1998. Conditionally immortalized murine bone marrow stromal cells mediate parathyroid hormone-dependent osteoclastogenesis in vitro. Endocrinology 139:1952-1964. [DOI] [PubMed] [Google Scholar]
- 39.Liu, W. G., S. Toyosawa, T. Furuichi, N. Kanatani, C. Yoshida, Y. Liu, M. Himeno, S. Narai, A. Yamaguchi, and T. Komori. 2001. Overexpression of Cbfa1 in osteoblasts inhibits osteoblast maturation and causes osteopenia with multiple fractures. J. Cell Biol. 155:157-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loots, G. G., R. M. Locksley, C. M. Blankespoor, Z. E. Wang, W. Miller, E. M. Rubin, and K. A. Frazer. 2000. Identification of a coordinate regulator of interleukins 4, 13, and 5 by cross-species sequence comparisons. Science 288:136-140. [DOI] [PubMed] [Google Scholar]
- 41.Ma, Y. L., R. L. Cain, D. L. Halladay, X. Yang, Q. Zeng, R. R. Miles, S. Chandrasekhar, T. J. Martin, and J. E. Onyia. 2001. Catabolic effects of continuous human PTH (1-38) in vivo is associated with sustained stimulation of RANKL and inhibition of osteoprotegerin and gene-associated bone formation. Endocrinology 142:4047-4054. [DOI] [PubMed] [Google Scholar]
- 42.Mayall, T. P., P. L. Sheridan, M. R. Montminy, and K. A. Jones. 1997. Distinct roles for P-CREB and LEF-1 in TCR alpha enhancer assembly and activation on chromatin templates in vitro. Genes Dev. 11:887-899. [DOI] [PubMed] [Google Scholar]
- 43.McDevitt, M. A., R. A. Shivdasani, Y. Fujiwara, H. Yang, and S. H. Orkin. 1997. A “knockdown” mutation created by cis-element gene targeting reveals the dependence of erythroid cell maturation on the level of transcription factor GATA-1. Proc. Natl. Acad. Sci. USA 94:6781-6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miao, D., J. Li, Y. Xue, H. Su, A. C. Karaplis, and D. Goltzman. 2004. Parathyroid hormone-related peptide is required for increased trabecular bone volume in parathyroid hormone-null mice. Endocrinology 145:3554-3562. [DOI] [PubMed] [Google Scholar]
- 45.Miao, D. S., B. He, B. Lanske, X. Y. Bai, X. K. Tong, G. N. Hendy, D. Goltzman, and A. C. Karaplis. 2004. Skeletal abnormalities in Pth-null mice are influenced by dietary calcium. Endocrinology 145:2046-2053. [DOI] [PubMed] [Google Scholar]
- 46.Millet, I., T. L. McCarthy, and A. Vignery. 1998. Regulation of interleukin-6 production by prostaglandin E2 in fetal rat osteoblasts: role of protein kinase A signaling pathway. J. Bone Miner. Res. 13:1092-1100. [DOI] [PubMed] [Google Scholar]
- 47.Nagy, A., J. Rossant, R. Nagy, W. Bramow-Newerly, and J. C. Roder. 1993. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc. Natl. Acad. Sci. USA 90:8424-8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakashima, T., Y. Kobayashi, S. Yamasaki, A. Kawakami, K. Eguchi, H. Sasaki, and H. Sakai. 2000. Protein expression and functional difference of membrane-bound and soluble receptor activator of NF-kappa B ligand: modulation of the expression by osteotropic factors and cytokines. Biochem. Biophys. Res. Commun. 275:768-775. [DOI] [PubMed] [Google Scholar]
- 49.Nardone, J., D. U. Lee, K. M. Ansel, and A. Rao. 2004. Bioinformatics for the ‘bench biologist’: how to find regulatory regions in genomic DNA. Nat. Immunol. 5:768-774. [DOI] [PubMed] [Google Scholar]
- 50.Nitsch, D., M. Boshart, and G. Schutz. 1993. Activation of the tyrosine aminotransferase gene is dependent on synergy between liver-specific and hormone-responsive elements. Proc. Natl. Acad. Sci. USA 90:5479-5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nobrega, M. A., Y. Zhu, I. Plajzer-Frick, V. Afzal, and E. M. Rubin. 2004. Megabase deletions of gene deserts result in viable mice. Nature 431:988-993. [DOI] [PubMed] [Google Scholar]
- 52.Notoya, M., E. Otsuka, A. Yamaguchi, and H. Hagiwara. 2004. Runx-2 is not essential for the vitamin D-regulated expression of RANKL and osteoprotegerin in osteoblastic cells. Biochem. Biophys. Res. Commun. 324:655-660. [DOI] [PubMed] [Google Scholar]
- 53.O'Brien, C. A., I. Gubrij, S. C. Lin, R. L. Saylors, and S. C. Manolagas. 1999. STAT3 activation in stromal osteoblastic cells is required for induction of the receptor activator of NF-kappa B ligand and stimulation of osteoclastogenesis by gp130-utilizing cytokines or interleukin-1 but not 1,25-dihydroxyvitamin D-3 or parathyroid hormone. J. Biol. Chem. 274:19301-19308. [DOI] [PubMed] [Google Scholar]
- 54.O'Brien, C. A., R. L. Jilka, Q. Fu, S. Stewart, R. S. Weinstein, and S. C. Manolagas. 2005. IL-6 is not required for parathyroid hormone stimulation of RANKL expression, osteoclast formation, and bone loss in mice. Am. J. Physiol. Endocrinol. Metab. 289:E784-E793. [DOI] [PubMed] [Google Scholar]
- 55.O'Brien, C. A., B. Kern, I. Gubrij, G. Karsenty, and S. C. Manolagas. 2002. Cbfa1 does not regulate RANKL gene activity in stromal/osteoblastic cells. Bone 30:453-462. [DOI] [PubMed] [Google Scholar]
- 56.Ovcharenko, I., G. G. Loots, R. C. Hardison, W. Miller, and L. Stubbs. 2004. zPicture: dynamic alignment and visualization tool for analyzing conservation profiles. Genome Res. 14:472-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Potts, J. T. 2005. Parathyroid hormone: past and present. J. Endocrinol. 187:311-325. [DOI] [PubMed] [Google Scholar]
- 58.Simonet, W. S., D. L. Lacey, C. R. Dunstan, M. Kelley, M. S. Chang, R. Luthy, H. Q. Nguyen, S. Wooden, L. Bennett, T. Boone, G. Shimamoto, M. DeRose, R. Elliott, A. Colombero, H. L. Tan, G. Trail, J. Sullivan, E. Davy, N. Bucay, L. Renshaw-Gegg, T. M. Hughes, D. Hill, W. Pattison, P. Campbell, S. Sander, G. Van, J. Tarpley, P. Derby, R. Lee, and W. J. Boyle. 1997. Osteoprotegerin: a novel secreted protein involved in the regulation of bone density. Cell 89:309-319. [DOI] [PubMed] [Google Scholar]
- 59.Soubt, M. K., R. Marksitzer, P.-A. Menoud, and Y. Nagamine. 1998. Role of tissue-specific transcription factor LFB3 in a cyclic AMP-responsive enhancer of the urokinase-type plasminogen activator gene in LLC-PK1 cells. Mol. Cell. Biol. 18:4698-4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Srivastava, S., M. Matsuda, Z. Hou, J. P. Bailey, R. Kitazawa, M. P. Herbst, and N. D. Horseman. 2003. Receptor activator of NF-kappaB ligand induction via Jak2 and Stat5a in mammary epithelial cells. J. Biol. Chem. 278:46171-46178. [DOI] [PubMed] [Google Scholar]
- 61.Tanaka, S., K. Nakamura, N. Takahashi, and T. Suda. 2005. Role of RANKL in physiological and pathological bone resorption and therapeutics targeting the RANKL-RANK signaling system. Immunol. Rev. 208:30-49. [DOI] [PubMed] [Google Scholar]
- 62.Tinti, C., C. Yang, H. Seo, B. Conti, C. Kim, T. H. Joh, and K. S. Kim. 1997. Structure/function relationship of the cAMP response element in tyrosine hydroxylase gene transcription. J. Biol. Chem. 272:19158-19164. [DOI] [PubMed] [Google Scholar]
- 63.Tsuji, K., Y. Ito, and M. Noda. 1998. Expression of the PEBP2alphaA/AML3/CBFA1 gene is regulated by BMP4/7 heterodimer and its overexpression suppresses type I collagen and osteocalcin gene expression in osteoblastic and nonosteoblastic mesenchymal cells. Bone 22:87-92. [DOI] [PubMed] [Google Scholar]
- 64.Udagawa, N., N. Takahashi, T. Akatsu, T. Sasaki, A. Yamaguchi, H. Kodama, T. J. Martin, and T. Suda. 1989. The bone marrow-derived stromal cell lines MC3T3-G2/PA6 and ST2 support osteoclast-like cell differentiation in cocultures with mouse spleen cells. Endocrinology 125:1805-1813. [DOI] [PubMed] [Google Scholar]
- 65.Udagawa, N., N. Takahashi, E. Jimi, K. Matsuzaki, T. Tsurukai, K. Itoh, N. Nakagawa, H. Yasuda, M. Goto, E. Tsuda, K. Higashio, M. T. Gillespie, T. J. Martin, and T. Suda. 1999. Osteoblasts/stromal cells stimulate osteoclast activation through expression of osteoclast differentiation factor/RANKL but not macrophage colony-stimulating factor: receptor activator of NF-kappa B ligand. Bone 25:517-523. [DOI] [PubMed] [Google Scholar]
- 66.Vavouri, T., and G. Elgar. 2005. Prediction of cis-regulatory elements using binding site matrices—the successes, the failures and the reasons for both. Curr. Opin. Genet. Dev. 15:395-402. [DOI] [PubMed] [Google Scholar]
- 67.Venkatesh, B., and W. H. Yap. 2005. Comparative genomics using fugu: a tool for the identification of conserved vertebrate cis-regulatory elements. Bioessays 27:100-107. [DOI] [PubMed] [Google Scholar]
- 68.Winoto, A., and D. Baltimore. 1989. A novel, inducible and T cell-specific enhancer located at the 3′ end of the T cell receptor alpha locus. EMBO J. 8:729-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yasuda, H., N. Shima, N. Nakagawa, K. Yamaguchi, M. Kinosaki, S. Mochizuki, A. Tomoyasu, K. Yano, M. Goto, A. Murakami, E. Tsuda, T. Morinaga, K. Higashio, N. Udagawa, N. Takahashi, and T. Suda. 1998. Osteoclast differentiation factor is a ligand for osteoprotegerin/osteoclastogenesis-inhibitory factor and is identical to TRANCE/RANKL. Proc. Natl. Acad. Sci. USA 95:3597-3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang, Y., J. X. Lin, and J. Vilcek. 1988. Synthesis of interleukin 6 (interferon-beta 2/B cell stimulatory factor 2) in human fibroblasts is triggered by an increase in intracellular cyclic AMP. J. Biol. Chem. 263:6177-6182. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.