Abstract
The Drosophila dysfusion basic-helix-loop-helix-PAS transcription factor gene is expressed in specialized fusion cells that reside at the tips of migrating tracheal branches. dysfusion mutants were isolated, and genetic analysis of live embryos revealed that mutant tracheal branches migrate to close proximity but fail to recognize and adhere to each other. Misexpression of dysfusion throughout the trachea further indicated that dysfusion has the ability to both inhibit cell migration and promote ectopic tracheal fusion. Nineteen genes whose expression either increases or decreases in fusion cells during development were analyzed in dysfusion mutant embryos. dysfusion upregulates the levels of four genes, including the shotgun cell adhesion protein gene and the zona pellucida family transmembrane protein gene, CG13196. Misexpression experiments with CG13196 result in ectopic tracheal fusion events, suggesting that it also encodes a cell adhesion protein. Another target gene of dysfusion is members only, which inhibits protein nuclear export and influences tracheal fusion. dysfusion also indirectly downregulates protein levels of Trachealess, an important regulator of tracheal development. These results indicate that fusion cells undergo dynamic changes in gene expression as they switch from migratory to fusion modes and that dysfusion regulates a discrete, but important, set of these genes.
One important aspect of insect tracheal morphogenesis involves the fusion of tracheal branches to form an interconnected oxygen-bearing tubular network (8). Branches contain a specialized fusion cell at their tips (26). These fusion cells extend actin-rich filopodia (24, 31) that guide migration in response to guidance cues, similar to axonal growth cone guidance (6). Thus, tracheal branches from adjacent hemisegments are directed to close proximity by the external guidance cues and then recognize and adhere to each other. After joining, further cellular changes occur, including (i) cytoskeletal reorganization, (ii) the breaking and reassembling of cellular junctions, and (iii) the formation of a continuous lumen (34). The complexity of tracheal branching and fusion suggests that the fusion cell is a complex, dynamic structure with changing patterns of gene expression. One of the major challenges in studying tracheal morphogenesis is to understand how regulatory pathways control and coordinate these changes in gene expression.
The molecular biology, cell biology, and genetics of branch migration and fusion have been productively studied in recent years. The trachea-expressed Breathless (Btl) tyrosine kinase receptor is activated by Branchless and is required for filopodial extension and migration (14, 24, 30). Additional proteins, such as Roundabout (Robo), Robo2, and Slit, are involved in branch-specific pathfinding (6). Analysis of enhancer trap lines first led to the identification of genes prominently expressed in or absent from fusion cells (26), and their regulation was initiated with the discovery of the escargot (esg) gene (27, 31), which encodes a zinc finger transcription factor. esg is expressed specifically in fusion cells during branching, and esg mutants have defects in tracheal fusion. Several genes whose expression is dependent on esg were identified. They included shotgun (shg), whose levels are upregulated in fusion cells, and Drosophila serum response factor (DSRF), which is expressed in adjacent terminal tracheal cells but repressed in fusion cells. Shg, a cadherin cell surface protein, is implicated in adhesive interactions and the formation of new adherens junctions in the fused trachea (31, 33). These results and other discoveries began to dissect the cellular and molecular bases of tracheal branch migration and fusion.
Another transcriptional target of Esg is dysfusion (dys), which encodes a basic-helix-loop-helix-PAS (bHLH-PAS) transcription factor that is expressed specifically in all fusion cells (12). RNA interference (RNAi)-mediated reduction of Drosophila dys function demonstrated that the dorsal branches (DBs), lateral trunk (LT) branches, and ganglionic branches (GBs) failed to fuse, although the cellular and molecular natures of these defects were not assessed. Dys is evolutionarily well conserved with a human ortholog (NXF or LE-PAS) that is prominently expressed in the brain (17, 21) and with a Caenorhabditis elegans ortholog (C15C8.2) that is expressed in the pharynx (23). Particularly noteworthy is the existence of several other Drosophila bHLH-PAS proteins that have important roles in tracheal development. The trachealess (trh) gene plays an early role in initiating tracheagenesis (10, 37) and directly regulates btl (20). The similar (sima) gene encodes the Drosophila ortholog of mammalian hypoxia-inducible factors (19) and functions in trachea to control the transcriptional response to hypoxia (1, 4, 16). All three proteins function as heterodimers with the same bHLH-PAS protein, Tango (Tgo) (29). Interestingly, Trh levels in fusion cells are reduced as Dys appears, and the reduction is dependent on dys function (12). Several questions arise regarding the function of dys. At the cellular level, does Dys function in branch migration, cell recognition/adhesion, or fusion events? What genes are regulated by dys, and do they constitute a functionally discrete subset of esg-regulated genes? Do dys-regulated genes share common biochemical functions? Are Trh protein levels and other genes downregulated by dys via transcriptional, posttranscriptional, or competitive mechanisms? How widespread are transcriptional changes in fusion cells?
In this study, the dynamics of fusion cell gene expression were examined and the functional role of dys investigated in much greater detail than previously reported. dys mutants were isolated, and analysis of their phenotypes showed that branch migration is normal but fusion cells fail to stably adhere, resulting in a failure of tracheal fusion. Misexpression of dys throughout the trachea resulted in severely malformed trachea in which secondary and tertiary branching was stunted and lumen formation inhibited at fusion sites. In addition, misexpression embryos often showed ectopic fusion events. Nineteen trachea-expressed genes were analyzed in wild-type and dys mutant embryos. Levels of four fusion cell-expressed genes declined in dys mutants. Misexpression of dys also resulted in ectopic expression of target genes. Two target genes are implicated in tracheal cell adhesion: shg and CG13196, a zona pellucida (ZP) gene (11). Embryos that misexpress CG13196 showed tracheal abnormalities consistent with it functioning as an adhesion protein gene. In addition, dys can indirectly regulate fusion cell protein levels and subcellular localization. Thus, dys controls multiple aspects of tracheal fusion, by upregulating and downregulating fusion cell constituents by using direct and indirect biochemical mechanisms.
MATERIALS AND METHODS
Drosophila strains.
The Drosophila strain Df(3R)Espl3 [96F1; 97B1] is deleted for the dys gene and was utilized in an ethyl methanesulfonate (EMS) screen to obtain dys mutants. Additional deletion strains used to characterize the EMS-generated mutants were Df(3R)Exel6204 [96F9; 97A6] and Df(3R)ME61 [96F12-14; 97C4-5] (all three deficiency strains are available from the Bloomington Drosophila Stock Center). In addition, the EMS-generated mutants were crossed to gro1, which is uncovered by Df(3R)Espl3. Misexpression experiments utilized btl-Gal4 (Tien Hsu) (28), engrailed (en)-Gal4 (Mark Peifer) (36), and UAS-green fluorescent protein (GFP) transgenic strains. UAS-actin-GFP (Mark Peifer) was used in the live imaging experiments. The following trachea-expressed lacZ enhancer trap and transgenic lines were crossed into a dys2 mutant background: Fusion-4 (members only [mbo]), Fusion-5 (bloated tubules [blot]), Antifusion-1 (CrebA), and Terminal-3 (ribbon) (all from Mark Krasnow) and shg-lacZ (Shigeo Hayashi) (26, 31). All dys mutant strains were balanced over P[w+; Kr-Gal4 UAS-GFP] (3) so that homozygous mutant embryos could be unambiguously identified.
Isolation of EMS-generated dys mutants.
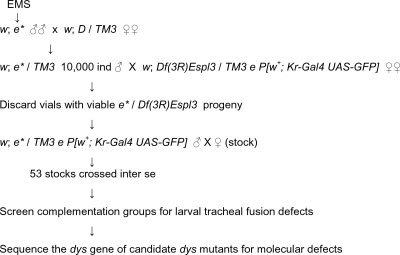
Mutations in dys were identified in a two-step procedure in which EMS mutations were screened over Df(3R)Espl3 for lethality and then examined for tracheal fusion defects. Isogenic w; es males were starved overnight and then fed a 1% sucrose solution with 25 mM EMS (Sigma) for 12 h. The mutagenized (*) males were then mated en masse to w; D/TM3 females. Male progeny (10,000) of the genotype w; es */TM3 were individually crossed to Df(3R)Espl3/TM3 e P[w+; Kr-Gal4 UAS-GFP] females. Males were discarded, and lack of complementation to Df(3R)Espl3 was assayed by scoring the vials for the absence of Sb+ flies. Mutant stocks (es */TM3 e P[w+; Kr-Gal4 UAS-GFP]) were generated from 53 vials with no Sb+ flies. Stocks were crossed inter se for complementation and representatives of each complementation group crossed to Df(3R)Exel6204 and Df(3R)ME61 for further genetic mapping. Members of each complementation group were assayed for tracheal fusion defects resembling the dys RNAi fusion defect (12).
Phenotypic analysis of larvae.
Second-instar larvae homozygously mutant for dys were placed onto grape juice agar plates and chilled at 4°C for 30 min. The larvae were then transferred to a slide and covered in 70% glycerol, and their trachea were examined by bright-field microscopy.
DNA sequence analysis of dys mutants.
Sequence analysis of the dys gene in dys mutant strains was carried out on genomic DNA isolated from homozygous dys EMS mutant embryos. The mutant embryos, balanced over P[w+; Kr-Gal4 UAS-GFP], were examined under a GFP stereoscope and selected manually by their absence of GFP fluorescence. The genomic DNA used as a template for PCRs encompassed the coding sequence and splice sites for all 10 dys exons.
Immunostaining and in situ hybridization of embryos.
Whole-mount embryos were immunostained and hybridized by in situ hybridization using standard techniques (13, 22). The following primary antibodies were used for immunostaining: anti-DSRF mouse monoclonal antibody (MAb) (Active Motif), rat anti-Dys, rabbit anti-Dys, MAb 2A12, rabbit anti-diphospho-extracellular signal-regulated kinase (Sigma E7028), anti-Tgo MAb, anti-β-galactosidase MAb, rabbit anti-β-galactosidase, and rat anti-Trh. The following secondary antibodies were used: Alexafluor 546-labeled anti-mouse immunoglobulin M (IgM), Alexafluor 488-labeled anti-rat IgG, and Cy5-labeled anti-rabbit IgG. Plasmids containing apontic (apt), btl, center divider (cdi), CG13196, CG15252, dys, esg, headcase (hdc), knirps (kni), knirps-related (knrl), trh, and ventral veinless (vvl) cDNA clones (all from Origene except dys) were used to generate RNA probes for in situ hybridization. Embryos were examined using a Zeiss 510 confocal microscope.
Generation of UAS-dys transgenes.
UAS-dys and UAS-Δb-dys transgenes were generated for misexpression experiments. A cDNA fragment containing a copy of the dys coding region was cloned into the EcoRI site of pUAST (2) to create UAS-dys. The UAS-Δb-dys transgene contained a copy of the dys coding sequence with a deletion of the entire Dys basic region (ANKSTKGASKMRR) cloned into the XbaI site of pUAST. Two UAS-dys lines were analyzed and showed similar tracheal defects. Seven UAS-dys-Δb lines were analyzed; all had the same wild-type tracheal morphology upon misexpression.
Time-lapse observations of tracheal branch migration and fusion.
Imaging of tracheal migration and fusion was carried out by examining wild-type and dys mutant P[btl-Gal4] P[UAS-actin-GFP] embryos, in which actin-GFP was expressed in the trachea (24). The dys mutant chromosome was balanced over TM3 P[Kr-Gal4] P[UAS-GFP]. Homozygous dys mutants were identified because of cortical actin-GFP in the trachea, due to UAS-actin-GFP expression, whereas nonmutant embryos with one or two copies of TM3 P[Kr-Gal4] P[UAS-GFP] showed cytoplasmic GFP in the trachea, due to the presence of cytoplasmic GFP driven by btl-Gal4 acting on P[UAS-GFP]. Embryos were collected at room temperature and dechorionated. They were then mounted on a glass coverslip and immersed in halocarbon oil 700 (Sigma) on slides containing an oxygen-permeable membrane. GFP fluorescent images were captured on a Zeiss LSM-510 confocal microscope with a 60× oil immersion lens for 2 h (wild-type embryos) and 4 h (dys mutant embryos). Typically, projections consisted of ten 2-μm slices captured along the z axis every 2 min for wild-type embryos and every 4 min for dys mutant embryos. Movies were assembled from projections by using Image J software.
RESULTS
Isolation of dys mutant alleles.
The strategy for isolating dys mutants was based on RNAi results. Previously, it was shown that injection of dys double-stranded RNA into Drosophila embryos resulted in (i) a loss of detectable Dys protein, (ii) embryonic and larval tracheal fusion defects, and (iii) a high degree of larval lethality (12). These results provided the basis for an EMS screen for dys mutants (Fig. 1), based on dys being a vital locus with easily detectable tracheal defects. In our screen, mutagenized males were crossed to a deletion strain, Df(3R)Espl3, that removes dys, and stocks that failed to complement the deficiency strain were screened for tracheal defects. Representative mutant larvae from each complementation group were analyzed for tracheal defects by examination with phase-contrast microscopy. Mutant larvae from group 3 (Table 1), which contains nine different mutants, had tracheal fusion defects (Fig. 2A to F) identical to those observed in the dys RNAi embryos (12). Identical, severe tracheal fusion phenotypes were observed in homozygous mutants of all nine dys alleles, when examined as either second-instar larvae or stage 17 embryos. Embryos heterozygous for combinations of two different dys alleles and dys/Df(3R)Espl3 mutants all showed similar severe phenotypes identical to those of homozygous dys mutants. Thus, all mutants appear to have a severe, genetically null dys phenotype. dys mutant strains are lethal, with animals dying as second- or third-instar larvae.
FIG. 1.
Isolation of EMS-mutagenized dys alleles.
TABLE 1.
Mutants isolated in the EMS mutagenesis
| Complementation groupa | Mutant(s) |
|---|---|
| Df(3R)Exel6204 | |
| 1 | 1, 13 |
| 2 (gro) | 2, 3, 16, 17, 18, 26, 27, 28, 31, 33, 43, 45, 59 |
| 3 (dys) | 4, 6, 14, 35, 42, 51, 53, 54, 57 |
| 4 | 5, 15, 20, 34, 39, 40, 52, 55 |
| 5 | 7, 12, 60 |
| 6 | 8, 11, 19, 23, 32, 46, 56 |
| 7 | 9, 10, 49 |
| 8 | 38, 44, 47, 48 |
| 9 | 41 |
| 10 | 58 |
| Df(3R)Espl3 | |
| 11 | 21 |
| 12 | 24 |
| 13 | 30 |
| 14 | 37 |
| 15 | 50 |
Fifty-three mutants failed to complement Df(3R)Espl3 [96F1; 97B1]. These mutants were crossed inter se and placed into 15 complementation groups numbered 1 to 15. Ten of those complementation groups also failed to complement Df(3R)Exel6204 [96F9; 97A6], and five lie outside that chromosomal interval. The group 3 mutants were named dys to dys in ascending order. Complementation group 2 contains alleles of groucho (gro).
FIG. 2.
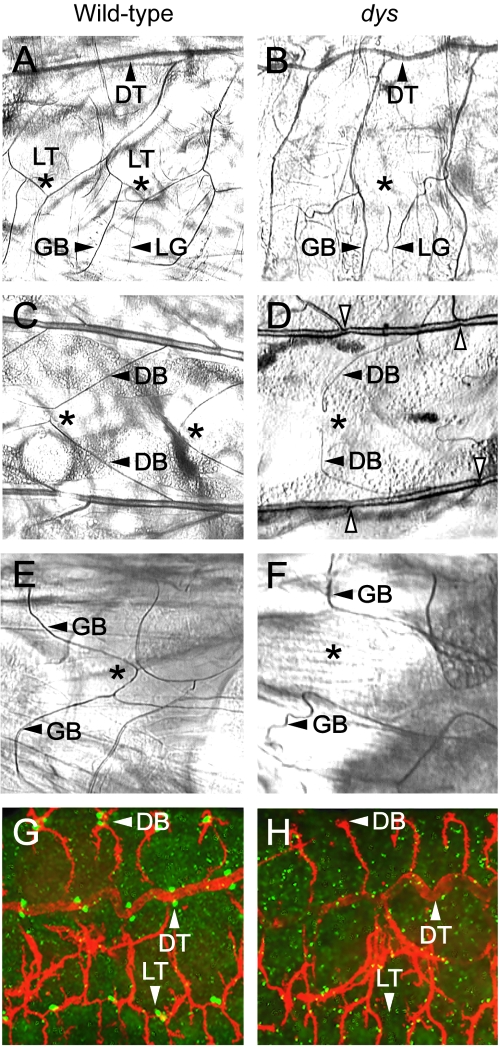
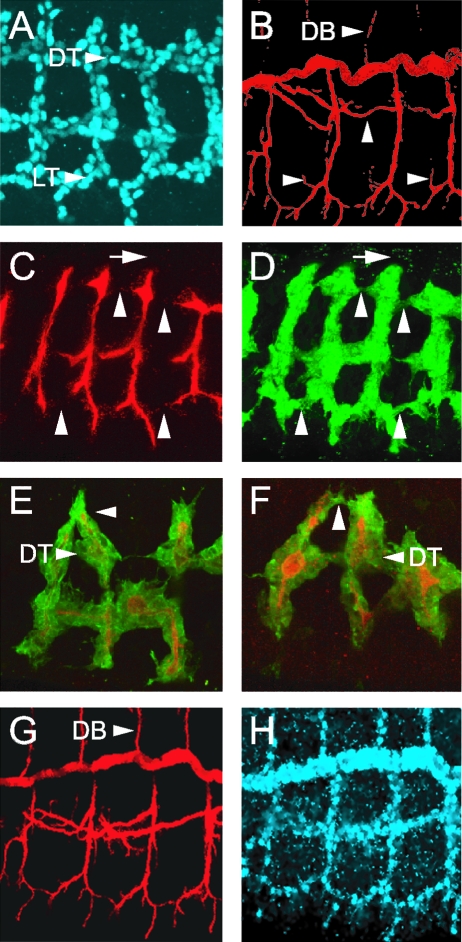
dys mutant embryos show tracheal fusion defects. Anterior is to the left in all panels. (A to F) Trachea from wild-type and dys2/dys3 mutant second-instar larvae were examined by phase microscopy. (A and B) Sagittal view. (A) The LT is fused (*) in a wild-type larva. The DT, GB, and lateral branch G (LG) are shown. (B) A dys mutant larva shows an absence of LT fusion (*). The GB and LG branch still persist and project ventrally. The DT is fused. (C and D) Dorsal view. (C) Wild-type larva showing fusion (*) of the DBs. (D) dys mutant larva showing a lack of DB fusion (*), although branches (arrowheads) are close together. Constrictions are seen in dys mutant DTs at fusion sites (open arrowheads). (E and F) Ventral view. (E) Wild-type larva showing fusion (*) of GBs. (F) dys mutant larva showing a lack of GB fusion (*). (G and H) Sagittal view. Confocal microscopic analysis of whole-mount embryos. (G) Stage 16 wild-type embryo stained with MAb 2A12 (red) and anti-Dys (green), showing fusion (arrowheads) of DBs, DTs, and LTs. Mab2A12 stains the tracheal lumen, and anti-Dys stains the tracheal fusion cells. (H) Stage 16 dys mutant embryo stained with MAb 2A12 (red) and anti-Dys (green), showing an absence of fusion of DB and LT branches. The DT is fused. Note the absence of anti-Dys reactivity.
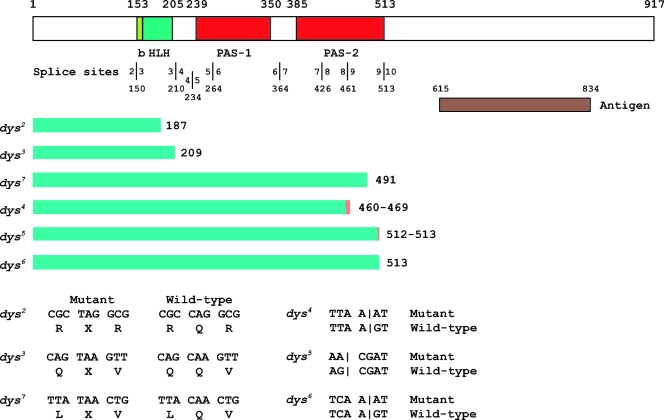
DNA sequence analysis of dys exons and intron/exon boundaries derived from homozygous mutant embryo genomic DNA was used to confirm that complementation group 3 mutants corresponded to the dys gene. The results are summarized in Fig. 3. Three alleles have premature stop codons: dys2 and dys3 have termination codons in the HLH region, and dys7 has a termination codon in the PAS-2 domain. Three alleles, dys4, dys5, and dys6, have mutations that likely affect splicing and would result in proteins truncated within or at the end of the PAS-2 domain. The remaining three alleles show no obvious mutation in the coding region or splice sites. All six dys alleles with recognizable mutations are predicted to produce truncated proteins that lack the C-terminal region, which was used as an antigen to generate Dys antibody (Fig. 3). Consistent with the sequencing data, no Dys immunoreactivity was detected in homozygous mutant embryos from any of these strains (data not shown). Wild-type levels of dys RNA were present in the two dys mutants assayed, dys2 and dys3, suggesting that dys expression is not maintained in tracheal cells by positive autoregulation (data not shown).
FIG. 3.
Molecular defects of dys mutants. At the top is a schematic of the Dys protein. The basic-region DNA binding domain (b), HLH dimerization domain, and PAS-1 and PAS-2 domains are labeled below the schematic. Above the schematic are the amino acid positions of domain boundaries. The locations of splice sites are shown below the domain names, and each exon-intron boundary is indicated by a vertical line with the corresponding exon indicated on each side, along with the amino acid residue corresponding to the splice site below. The anti-Dys antibody was raised against animals injected with a bacterial fusion protein antigen containing Dys protein residues 615 to 834. The conceptual protein fragment predicted for each dys mutant gene is indicated by a green bar, followed by the size in amino acids. The minimum extent of the predicted protein is shown in green, and the orange bar indicates translated intronic sequences. dys2, dys3, and dys7 have premature termination codons that should result in a truncated protein. dys4, dys5, and dys6 have splice site mutations. dys4 and dys5 are followed by two numbers; the first indicates the end of the exonic sequence and the second the location of the in-frame termination codon in the adjacent intron. The sequence alteration for each mutant is shown at the bottom. Vertical lines for dys4, dys5, and dys6 indicate locations of splice sites.
dys mutants show tracheal adhesion and fusion defects.
dys mutants were initially examined for larval defects in tracheal fusion (Fig. 2A to F). The GBs, DBs, and LT branches in dys mutant embryos showed a complete absence of fusion in all branches examined. The dorsal trunk (DT) (Fig. 2B) was fused and relatively normal in appearance, although some DT fusion sites were constricted (Fig. 2D). These results were identical to those observed with the dys RNAi experiments (12). Penetrance and expressivity were 100% for dys2/dys3 mutant larvae, as well as for other dys alleles and allelic combinations analyzed. In each case, the mutant tracheal branches had migrated near the point of fusion but failed to fuse. These results were confirmed by staining stage 16 embryos with MAb 2A12, which stains the tracheal lumen (Fig. 2G and H). Mutant embryos showed an absence of fusion by stage 16.
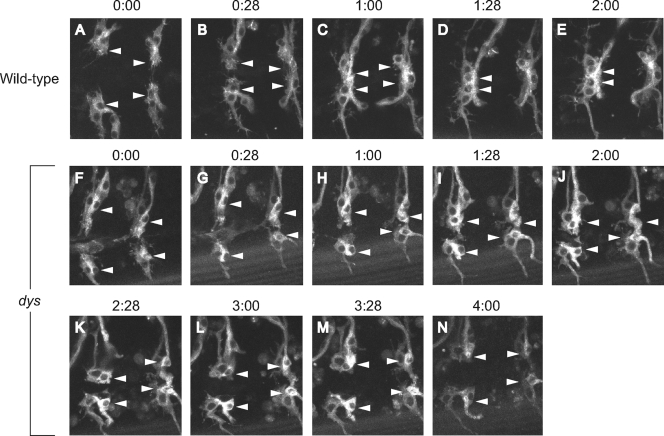
More-detailed analyses of dys mutant embryos were carried out by employing time-lapse microscopy of tracheal migration and fusion (Fig. 4 and data not shown). Wild-type and dys mutant embryos containing btl-Gal4 UAS-actin-GFP were visualized for GFP fluorescence by using confocal microscopy and compared over 4 h of development with respect to migration, filopodial dynamics, and fusion. The btl-Gal4 transgene expresses Gal4 in most tracheal cells, and UAS-actin-GFP allows visualization of actin-rich structures, including tracheal filopodia (24). We focused on the DB, which migrates over large distances along the internal surface of the dorsal epidermis to meet and fuse at the dorsal midline with its partner from the adjacent hemisegment. In both wild-type and dys mutant embryos, the migratory processes, including filopodial extension, were similar. However, in wild-type embryos, the filopodia from the fusion cells touched and adhered, and the fusion cells joined together (Fig. 4A to E). The fusion event was accompanied by intense actin accumulation at the fusion junction. In mutant embryos (Fig. 4F to N), the two fusion cells migrated close to each other, and filopodia made contact. However, the cells failed to adhere and remained separated. In summary, the time-lapse movies indicated that long-range migration in dys mutant embryos is sufficiently normal that fusion cells contact each other via their filopodia. However, the branches fail to stably adhere and fuse.
FIG. 4.
Live imaging of wild-type and dys mutant tracheae. DB migration and fusion were examined by live confocal imaging of tracheal GFP fluorescence. Shown are individual images from a wild-type btl-Gal4 UAS-actin-GFP embryo (A to E) imaged over 2 h and a dys2/dys3 btl-Gal4 UAS-actin-GFP mutant embryo (F to N) imaged over 4 h. Filopodia from tip/fusion cells (arrowheads) can be observed in both migrating wild-type (A and B) and dys mutant (F to N) trachea. Note the accumulation of actin-GFP at sites of fusion in wild-type embryos (C to E). In the dys mutant embryo, the fusion cells come into close range (F to N) but fail to form stable contacts.
Misexpression of dys results in ectopic fusion and inhibition of tracheal migration and lumen formation.
Additional insight into the function of dys was obtained by misexpressing the gene in trachea by using UAS-dys transgenic fly strains crossed to btl-Gal4. This results in dys being expressed at high levels in most tracheal cells (Fig. 5A). Staining of w− control embryos with MAb 2A12 revealed the characteristic and extensive secondary and tertiary branches and branch fusion (Fig. 5B). However, MAb 2A12 staining and examination of GFP in btl-Gal4 UAS-dys UAS-GFP embryos indicated a striking reduction in DB, DT, GB, and visceral branch branching (Fig. 5C and D). While both DT and LT branches were sometimes fused, the connections were abnormal and were characterized by reduced GFP-positive cytoplasmic material and an absence of MAb 2A12 luminal staining. The dys misexpression embryos also resulted in the formation of ectopic tracheal fusion events, connecting tracheal branches that normally never join (Fig. 5E and F). These ectopic fusion events rarely showed the presence of MAb 2A12 luminal staining at the fusion sites. The dys misexpression experiments provided evidence that dys can promote tracheal fusion events, consistent with the dys mutant studies. However, the misexpression experiments also provided evidence that tracheal migration can be inhibited by dys.
FIG. 5.
Misexpression of dys results in tracheal fusion, migration, and lumen formation defects. All embryos are stage 16, and sagittal views are shown. Embryos were stained with combinations of MAb 2A12 (red), anti-Dys (blue), and anti-GFP (green). (A) btl-Gal4 UAS-dys embryo showing the strong misexpression of Dys protein throughout the trachea. The DT and LT are indicated. (B) w− control embryo. Note the extensive branching (arrowheads) illustrated by MAb 2A12 staining, including DBs. (C and D) btl-Gal4 UAS-dys UAS-GFP embryo. (C) MAb 2A12 staining reveals that the trachea shows a severe reduction in secondary and tertiary branching, as well as breaks in lumen formation at normal sites of fusion (arrowheads). Both DBs and GBs are severely stunted (arrows in C and D show lack of DB extension; compare to panels B and G). (D) Staining for cytoplasmic GFP shows that both the DT and LT are often joined at fusion sites, but (C) show an absence of MAb 2A12 luminal staining. (E and F) Two btl-Gal4 UAS-dys UAS-actin-GFP embryos. Arrowheads point to sites of ectopic fusion. While the cytoplasmic GFP staining is continuous, MAb 2A12 luminal staining is absent at fusion sites. The DT is indicated. (G and H) btl-Gal4 UAS-dys-Δb embryo. The trachea has a wild-type pattern of branching.
The biochemical mechanism of the dys misexpression phenotypes (and thus likely to be relevant to the dys mutant phenotypes) was assessed using a mutant version of dys that lacks the putative DNA binding basic region (Δb-dys) in the dys misexpression assay. Embryos that were btl-Gal4 UAS-Δb-dys showed trachea that were wild type in appearance (Fig. 5G and H). The Δb-dys gene was misexpressed at high levels as revealed by anti-Dys immunostaining of misexpression embryos (Fig. 5H), and it was able to dimerize with Tgo and translocate to the nucleus as revealed by staining en-Gal4 UAS-Δb-dys embryos with anti-Tgo (data not shown). Thus, the inability of Δb-dys to generate misexpression phenotypes is likely due to its inability to bind DNA. While the relationship between the dys misexpression and mutant phenotypes is uncertain, these results indicated that the ability of dys to inhibit cell migration and promote tracheal fusion requires the ability to bind DNA.
Regulation of fusion cell gene expression by Dys.
Since Dys is a transcription factor expressed in tracheal fusion cells, important insight into its biological function can be revealed by identifying transcriptional target genes. We examined 19 trachea-expressed genes in both wild-type and dys mutant embryos for potential regulation by dys. The genes fall into three categories: (i) those prominently expressed in fusion cells, (ii) those expressed in terminal cells but not fusion cells, and (iii) those expressed in tracheal cells but downregulated in fusion cells.
(i) Fusion cell-expressed genes.
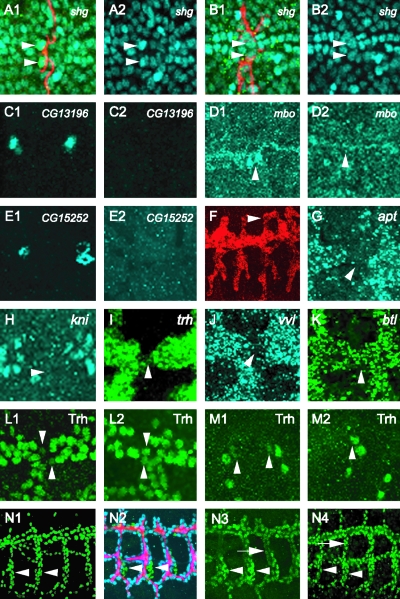
Previous work identified a number of fusion cell-expressed genes that are regulated by esg (27), including blot, dys, hdc, mbo, and shg. Another trachea-expressed gene, cdi, is not regulated by esg. Expression of these genes, and also esg, CG13196 (a ZP family gene expressed in fusion cells) (11), and CG15252 (which encodes a protein of unknown function) (32), was examined in dys mutant embryos. Four genes, i.e., shg, CG13196, CG15252, and mbo (Fig. 6A to E) were found to be regulated by dys, whereas the other five, i.e., blot, cdi, dys, esg, and hdc, were not regulated by dys (data not shown). The expression of shg is upregulated by dys from a basal level, whereas dys is required for the initiation of CG13196, CG15252, and mbo expression. shg expression is enhanced in both DB and DT fusion cells, but dys (as well as esg [31]) controls expression in only DBs and not DTs. shg and CG13196 are membrane proteins that may control fusion cell recognition and adhesion events, whereas mbo attenuates protein nuclear export. Levels of shg and mbo were assayed by examining shg-lacZ and mbo/Fusion-4-lacZ lines, indicating that the control by dys is likely to be at the transcriptional level.
FIG. 6.
Regulation of gene expression in tracheal fusion cells. Each panel consists of a projection of three consecutive 1-μm confocal slices. (A to D) shg upregulation in DBs is dependent on dys function. Stage 16 shg-lacZ embryos were stained with anti-β-galactosidase (A1 and B2; blue), MAb 2A12 (A1 and B1; red), and anti-Dys (not shown). The use of anti-Dys staining allows for selection of homozygous mutant embryos, since they do not possess antibody-reactive protein. Fusion cells (arrowheads) express a higher level of β-galactosidase than surrounding tracheal cells in wild-type DB (A1 and A2). This upregulation was not observed in dys mutant DB fusion cells (B1 and B2, arrowheads). (C1 to M2) Embryos were hybridized to gene-specific probes and a dys RNA probe and immunostained with anti-Dys. dys RNA hybridization revealed fusion cells, and anti-Dys allowed identification of dys mutant embryos. (C1 and C2) CG13196 fusion cell expression is dependent on dys. Stage 15 wild-type DT fusion cells express CG13196 (C1), but dys mutants (C2) do not. (D1 and D2) mbo expression is regulated by dys. Fusion-4 embryos, which contain an mbo-lacZ enhancer trap gene, were immunostained with anti-β-galactosidase. Stage 15 wild-type DT (D1) fusion cells (arrowheads) show high levels of lacZ expression, whereas β-galactosidase expression is absent in dys mutant DT (D2). (E1 and E2) CG15252 fusion cell expression is dependent on dys. Stage 15 wild-type DT fusion cells (E1) express CG15252, but dys mutants (E2) do not. (F) Misexpression of CG13196 in btl-Gal4 UAS-CG13196 embryos results in ectopic tracheal branch fusion (arrowhead). (G to K) Stage 15 embryos were hybridized to apt (G), kni (H), trh (I), vvl (J), and btl (K) probes as well as to a dys probe to mark fusion cells (not shown). RNA levels of all five genes are downregulated in fusion cells but are not regulated by dys (not shown). (L1 to M2) dys downregulates Trh protein levels. Trh protein levels are reduced in wild-type (L1) DT fusion cells (arrowheads) and (M1) DB fusion cells (arrowheads). In dys mutant embryos, Trh proteins levels increased to levels comparable to those in other tracheal cells in both DTs (L2) and DBs (M2). (N1 to N4) Embryos stained with ant-Trh (green). (N1) w− control embryo. Trh is localized to nuclei at uniform levels throughout the trachea. Cells of the spiracular branch (arrowheads) show the same levels of Trh as other tracheal cells. (N2 and N3) btl-Gal4 UAS-dys embryo. (N2) Embryo stained with MAb 2A12 (red), anti-Dys (blue), and anti-Trh (green). btl-Gal4 UAS-dys is expressed at high levels in all tracheal cells except the spiracular branch cells (green; arrowheads). (N3) Trh levels are reduced in dys-positive cells (arrow) compared to spiracular branch cells (arrowheads). (N4) btl-Gal4 UAS-dys-Δb embryo. Trh levels are uniform throughout the trachea, as indicated by comparable Trh levels in dys-positive cells (arrow) compared to spiracular branch cells (arrowheads).
The shg gene encodes a cadherin cell adhesion protein implicated in tracheal morphogenesis and branch fusion (31, 33). CG13196 encodes a ZP protein of unknown function. An attempt to abolish CG13196 function by using RNAi showed only weak effects on tracheal fusion (data not shown). In contrast, misexpression of CG13196 in btl-Gal4 UAS-CG13196 embryos revealed sites of ectopic fusion (Fig. 6F), suggesting that CG13196 encodes an adhesion protein. This suggests that Dys controls expression of a group of two or more proteins involved in tracheal recognition/adhesion events.
(ii) Terminal cell-expressed genes.
While screening for genes expressed in tracheal cells, Samakovlis et al. (26) identified a class of trachea-expressed genes that were expressed in terminal cells, which in the case of DBs are adjacent to fusion cells. Analysis of two terminal-class genes, including DSRF, in esg mutants indicated that esg represses expression of both genes in fusion cells. We investigated whether terminal genes, including DSRF and ribbon, were negatively regulated by dys in fusion cells. In both cases, expression was unaffected in fusion cells (data not shown). While this analysis is limited to these two genes, it indicates that the negative regulation of DSRF in fusion cells by esg is not mediated by dys, and transcriptional repression in fusion cells may not be a common role of dys.
(iii) Fusion cell-downregulated genes.
We examined expression of a number of genes that are broadly expressed during tracheal development and found that downregulation in fusion cells is common. These genes included six that encode transcription factors: apt, crebA (Antifusion-1), kni, knrl, trh, and vvl (Fig. 6G to J). In addition, expression of the btl receptor, which is a direct transcriptional target of Trh, was downregulated (Fig. 6K). Consistent with a reduction in btl, mitogen-activated protein kinase (MAPK) activation, as assayed by immunostaining for phosphorylation with anti-diphospho-MAPK, was also reduced (data not shown), as noted previously (9). When tested in dys mutants, transcript levels of all eight genes plus the diphospho-MAPK gene remain reduced, indicating that the downregulation is not dys dependent (data not shown).
It was noted previously that in wild-type embryos Trh protein levels were reduced in fusion cells (Fig. 6L1 and M1), and injection of dys RNAi restored Trh protein levels to the levels found in the adjacent tracheal cells (12). Using dys mutant and misexpression embryos, Trh protein levels were examined in fusion cells. In dys mutant embryos, Trh proteins levels were comparable to the levels observed in adjacent cells in DTs (Fig. 6L2) and DBs (Fig. 6M2). Misexpression of dys in btl-Gal4 UAS-dys embryos also showed a reduction in Trh protein levels (Fig. 6N1 to 3). In contrast, trh RNA levels were not affected in either dys mutant or misexpression embryos (data not shown). Misexpression of Δb-dys did not result in a reduction of Trh protein levels (Fig. 6N4), indicating that Trh protein reduction requires Dys DNA binding and transcriptional regulation. Since Trh protein levels, but not RNA levels, are negatively regulated by Dys transcriptional activity, most likely Dys activates transcription of an unknown gene, whose function is to regulate Trh protein levels.
Misexpression of dys results in ectopic expression of Dys target genes.
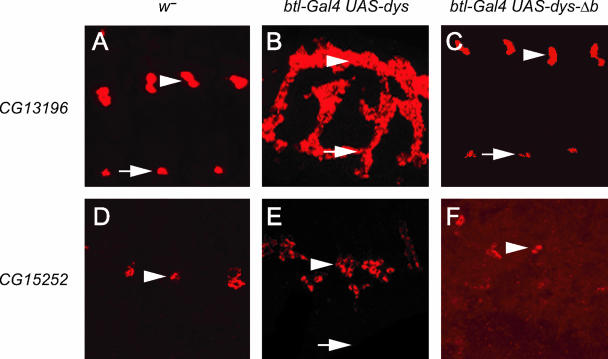
CG13196 is expressed in all tracheal fusion cells (Fig. 7A), and CG15252 is expressed only in DT fusion cells (Fig. 7D). Expression of both genes was assayed in btl-Gal4 UAS-dys embryos. CG13196 was robustly misexpressed throughout the trachea, including all primary branches (Fig. 7B). CG15252 was misexpressed, but the misexpression was restricted mainly to DTs and some cells in the adjacent transverse connective (Fig. 7E). Clearly absent was CG15252 expression in DBs, LTs, and GBs. The esg gene, which is expressed in fusion cells but is not a Dys target gene, was not ectopically expressed in btl-Gal4 UAS-dys embryos (not shown). Thus, ectopic tracheal gene expression by Dys seems specific for Dys target genes and not for all fusion cell-expressed genes. CG13196 and CG15252 expression required Dys DNA binding, since misexpression of dys-Δb failed to activate their transcription (Fig. 7C and F). Neither gene is expressed in nontracheal sites of dys expression, such as leading-edge and anal pad cells, nor does btl-Gal4 UAS-dys central nervous system midline cell expression result in ectopic expression of CG13196 or CG15252 in the central nervous system midline cells. Thus, while Dys:Tgo is able to efficiently activate transcription of fusion cell target genes throughout the trachea, it is unable to activate them indiscriminately in all cell types.
FIG. 7.
Misexpression of dys in trachea results in ectopic expression of dys target genes. All panels show sagittal views of stage 16 embryos; anterior is to the left. Positions of DTs (arrowheads) and LTs (arrows) are indicated. (A to C) w− (A), btl-Gal4 UAS-dys (B), and btl-Gal4 UAS-dys-Δb (C) embryos were hybridized to a CG13196 probe (red). Embryos were also stained with anti-Dys to monitor misexpression (not shown). (A) In w− control embryos, CG13196 is expressed in fusion cells of DTs and LTs. (B) CG13196 expression is found throughout the trachea when dys is ectopically expressed. (C) Only wild-type expression of CG13196 in DTs and LTs is observed when dys-Δb is misexpressed. (D to F) w− (D), btl-Gal4 UAS-dys (E), and btl-Gal4 UAS-dys-Δb (F) embryos were hybridized to a CG15252 probe (red). (D) CG15252 is expressed only in DT fusion cells in w− control embryos. (E) CG15252 expression is ectopically expressed when dys is misexpressed in all tracheal cells, but it is restricted to DT and some nearby tracheal cells. It is completely absent from LTs and GBs. (F) CG15252 expression is restricted to DT fusion cells and is not ectopically expressed in btl-Gal4 UAS-dys-Δb embryos.
DISCUSSION
The isolation of dys mutants allowed detailed phenotypic analysis using time-lapse confocal microscopy of live embryos. Migration and the presence of filopodia during DB branching appeared normal in dys mutant embryos. The two DB fusion cells moved close together, and their filopodia were observed to touch. However, unlike wild-type cells, the dys mutant fusion cells failed to stably adhere. Thus, no fusion occurred, and ultimately the branches retracted from one another. Additional insight into the role of dys emerged from experiments in which dys was misexpressed throughout the trachea. Ectopic fusion events were observed, which is consistent with the dys mutant phenotype and indicates that dys promotes fusion cell recognition/adhesion. In addition, dys misexpression results in a strong reduction in both tracheal branching and formation of MAb 2A12 luminal material at fusion sites. The reduction in branching suggests that another possible function of dys is to inhibit migration in preparation for fusion. The reduction in MAb 2A12 luminal material at sites of normal and ectopic fusion suggests that the fusion process is incomplete. This can be explained for the ectopic fusion results by proposing that Dys activates genes that can mediate fusion but not lumen formation. Since the ectopic cells are not fusion cells, the additional lumen-forming functions would not be present. In contrast, since lumen formation was also defective at normal fusion sites, it is possible that Dys overexpression inhibits lumen formation. In summary, the genetic and misexpression experiments suggest that dys is activated late in the branching process to inhibit migration and promote branch recognition and adhesion. It may also play additional roles after branches join.
Since dys encodes a transcription factor, it is expected that it functions by regulating gene expression. Previous work had identified several genes prominently expressed in fusion cells, as well as additional trachea-expressed genes whose fusion cell expression was low or absent (26, 31). In this paper, we further showed that a number of prominent trachea-expressed genes are also downregulated in fusion cells, indicating that this is a common occurrence. Expression of 19 genes was assayed in dys mutant embryos to identify Dys target genes. RNA levels of four genes (CG13196, CG15252, mbo, and shg) were reduced. In contrast, Trh protein levels, which normally decline in fusion cells, increased in dys mutants. These results were confirmed by dys misexpression experiments, in which CG13196 and CG15252 were increased and Trh protein levels declined. Despite dys expression in all tracheal fusion cells, there exist branch-specific differences in Dys-regulated gene expression. CG13196 is expressed in all fusion cells, and dys is required for its expression. In contrast, shg is upregulated in DB and DT fusion cells, but only DB upregulation requires Dys, an effect also seen for Esg (31). CG15252 is expressed only in DT fusion cells, and this restriction may be due to the positive or negative action of branch-specific transcription factors, such as Spalt major (DT specific, positively acting) (7, 15) or Kni and Knrl (non-DT branches, negatively acting) (5, 18).
All of the dys misexpression defects (and thus probably the mutant defects) require Dys DNA binding, since deletion of the dys basic region, and presumably its ability to bind DNA, abolishes the tracheal defects. Although trh RNA levels decline in fusion cells along with protein levels, the RNA reduction is not dys dependent. Thus, dys likely regulates transcription of a gene that regulates Trh protein levels. Similarly, the requirement of Dys DNA binding to regulate Trh protein levels does not support a model (12) in which Trh levels are reduced as a consequence of Dys competing for their common dimerization partner, Tgo, since this is unlikely to require DNA binding.
The recognition/adhesive properties promoted by dys may be mediated by two Dys target genes, shg and CG13196. Shg is a well-studied adhesion protein, and CG13196 encodes a ZP transmembrane protein, although its function and subcellular localization are unknown. Misexpression of CG13196 results in ectopic fusion events consistent with it playing a role in cell adhesion. Thus, one key role of dys may be to promote tracheal fusion by controlling expression of two or more cell adhesion protein genes. They could work together in the same cellular process or in different aspects of tracheal fusion, lumen formation, or function. The identification of mbo as a transcriptional target of Dys is intriguing, since mbo mutants have a tracheal fusion defect (35) and it attenuates protein nuclear export (25). Although the fusion cell protein cargo regulated by mbo is unknown, it presumably includes proteins that are localized to nuclei in fusion cells.
The two major transcription factors studied to date that control fusion cell transcription and development are esg and dys. esg precedes dys during fusion cell development and controls expression of dys in DBs and GBs but not DTs (12) (the case for LTs is unknown, since fusion cells die in esg mutants [27]). dys itself does not affect esg expression (12). The tracheal fusion phenotypes of both genes are similar. The DT is the one branch that still fuses in both esg and dys mutants, although both show constrictions at the sites of DT fusion (27, 31). Previous work on esg revealed that, genetically, it is required for both activation of fusion cell gene expression and repression of terminal cell gene expression in fusion cells (27, 31). In this study, we found that dys constitutes a transcriptional pathway that carries out a subset of esg functions, focused on upregulating expression of genes involved in cell adhesion and protein localization, although future work may uncover additional target genes. Since a large number of genes are either activated or downregulated in tracheal fusion cells (L. Jiang, unpublished data), it will be important to continue genetic and molecular studies to determine which genes are targets of Esg and Dys and whether their control is direct or indirect.
One model of Drosophila dys function is that dys acts as a developmental timer near the end of tracheal branching to inhibit migration and promote cell adhesion and fusion. The adhesion component works, in part, through activation of shg and (possibly) CG13196. The inhibition of migration has only been postulated from misexpression experiments and needs to be confirmed by alternative approaches. It is also important to note that the switch from migration to fusion can also include changes in gene expression that are independent of dys. For example, it is shown here that RNA levels of btl, a gene required for tracheal migration, are downregulated in fusion cells in a dys-independent mode. dys is expressed in a variety of Drosophila embryonic cell types, including leading edge, brain, gut, and anal pad (12), and the mammalian ortholog is prominently expressed in the brain (17, 21). However, the function of dys in these cell types is unknown, although a potential connection between tracheal fusion cells and both migrating neuronal axon growth cones and leading-edge cells is worth investigating. The role of dys in controlling fusion cell behavior suggests that it is worthwhile to look in tissues that undergo branching morphogenesis, such as the vertebrate lung and vascular system, for regulatory proteins expressed in tip/fusion cells that control the migration, recognition, and fusion properties of their branches.
Acknowledgments
We thank Julie Gates, Shigeo Hayashi, Tien Hsu, Mark Krasnow, Mark Peifer, Tony Perdue, Benny Shilo, Christos Samakovlis, and the Bloomington Fly Stock Center for stocks and reagents and Joseph Kearney for helpful comments on the manuscript. We remember the contributions to tracheal development by the late Peter Kolodziej, who also generously contributed advice and reagents to this project.
This project was funded by a grant from the National Science Foundation (Developmental Mechanisms) to S.T.C.
REFERENCES
- 1.Arquier, N., P. Vigne, E. Duplan, T. Hsu, P. P. Therond, C. Frelin, and G. D'Angelo. 2006. Analysis of the hypoxia-sensing pathway in Drosophila melanogaster. Biochem. J. 393:471-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brand, A. H., and N. Perrimon. 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118:401-415. [DOI] [PubMed] [Google Scholar]
- 3.Casso, D., F. Ramirez-Weber, and T. B. Kornberg. 2000. GFP-tagged balancer chromosomes for Drosophila melanogaster. Mech. Dev. 91:451-454. [DOI] [PubMed] [Google Scholar]
- 4.Centanin, L., P. J. Ratcliffe, and P. Wappner. 2005. Reversion of lethality and growth defects in Fatiga oxygen-sensor mutant flies by loss of hypoxia-inducible factor-alpha/Sima. EMBO Rep. 6:1070-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, C. K., R. P. Kuhnlein, K. G. Eulenberg, S. Vincent, M. Affolter, and R. Schuh. 1998. The transcription factors KNIRPS and KNIRPS RELATED control cell migration and branch morphogenesis during Drosophila tracheal development. Development 125:4959-4968. [DOI] [PubMed] [Google Scholar]
- 6.Englund, C., P. Steneberg, L. Falileeva, N. Xylourgidis, and C. Samakovlis. 2002. Attractive and repulsive functions of Slit are mediated by different receptors in the Drosophila trachea. Development 129:4941-4951. [DOI] [PubMed] [Google Scholar]
- 7.Franch-Marro, X., and J. Casanova. 2002. spalt-induced specification of distinct dorsal and ventral domains is required for Drosophila tracheal patterning. Dev. Biol. 250:374-382. [PubMed] [Google Scholar]
- 8.Ghabrial, A., S. Luschnig, M. M. Metzstein, and M. A. Krasnow. 2003. Branching morphogenesis of the Drosophila tracheal system. Annu. Rev. Cell Dev. Biol. 19:623-647. [DOI] [PubMed] [Google Scholar]
- 9.Ikeya, T., and S. Hayashi. 1999. Interplay of Notch and FGF signaling restricts cell fate and MAPK activation in the Drosophila trachea. Development 126:4455-4463. [DOI] [PubMed] [Google Scholar]
- 10.Isaac, D. D., and D. J. Andrew. 1996. Tubulogenesis in Drosophila: a requirement for the trachealess gene product. Genes Dev. 10:103-117. [DOI] [PubMed] [Google Scholar]
- 11.Jazwinska, A., and M. Affolter. 2004. A family of genes encoding zona pellucida (ZP) domain proteins is expressed in various epithelial tissues during Drosophila embryogenesis. Gene Expr. Patterns 4:413-421. [DOI] [PubMed] [Google Scholar]
- 12.Jiang, L., and S. T. Crews. 2003. The Drosophila dysfusion basic helix-loop-helix (bHLH)-PAS gene controls tracheal fusion and levels of the trachealess bHLH-PAS protein. Mol. Cell. Biol. 23:5625-5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kearney, J. B., S. R. Wheeler, P. Estes, B. Parente, and S. T. Crews. 2004. Gene expression profiling of the developing Drosophila CNS midline cells. Dev. Biol. 275:473-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klambt, C., L. Glazer, and B.-Z. Shilo. 1992. breathless, a Drosophila FGF receptor homolog, is essential for migration of tracheal and specific midline glial cells. Genes Dev. 6:1668-1678. [DOI] [PubMed] [Google Scholar]
- 15.Kuhnlein, R. P., and R. Schuh. 1996. Dual function of the region-specific homeotic gene spalt during Drosophila tracheal system development. Development 122:2215-2223. [DOI] [PubMed] [Google Scholar]
- 16.Lavista-Llanos, S., L. Centanin, M. Irisarri, D. M. Russo, J. M. Gleadle, S. N. Bocca, M. Muzzopappa, P. J. Ratcliffe, and P. Wappner. 2002. Control of the hypoxic response in Drosophila melanogaster by the basic helix-loop-helix PAS protein similar. Mol. Cell. Biol. 22:6842-6853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moser, M., R. Knoth, C. Bode, and C. Patterson. 2004. LE-PAS, a novel Arnt-dependent HLH-PAS protein, is expressed in limbic tissues and transactivates the CNS midline enhancer element. Brain Res. Mol. Brain Res. 128:141-149. [DOI] [PubMed] [Google Scholar]
- 18.Myat, M. M., H. Lightfoot, P. Wang, and D. J. Andrew. 2005. A molecular link between FGF and Dpp signaling in branch-specific migration of the Drosophila trachea. Dev. Biol. 281:38-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nambu, J. R., W. Chen, S. Hu, and S. T. Crews. 1996. The Drosophila melanogaster similar bHLH-PAS gene encodes a protein related to human hypoxia-inducible factor 1α and Drosophila Single-minded. Gene 172:249-254. [DOI] [PubMed] [Google Scholar]
- 20.Ohshiro, T., and K. Saigo. 1997. Transcriptional regulation of breathless FGF receptor gene by binding of TRACHEALESS/dARNT heterodimers to three central midline elements in Drosophila developing trachea. Development 124:3975-3986. [DOI] [PubMed] [Google Scholar]
- 21.Ooe, N., K. Saito, N. Mikami, I. Nakatuka, and H. Kaneko. 2004. Identification of a novel basic helix-loop-helix-PAS factor, NXF, reveals a Sim2 competitive, positive regulatory role in dendritic-cytoskeleton modulator drebrin gene expression. Mol. Cell. Biol. 24:608-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patel, N. H., P. M. Snow, and C. S. Goodman. 1987. Characterization and cloning of fasciclin III: a glycoprotein expressed on a subset of neurons and axon pathways in Drosophila. Cell 48:975-988. [DOI] [PubMed] [Google Scholar]
- 23.Powell-Coffman, J. A. 2003. bHLH-PAS proteins in C. elegans, p. 51-68. In S. T. Crews (ed.), PAS proteins: regulators and sensors of development and physiology. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 24.Ribeiro, C., A. Ebner, and M. Affolter. 2002. In vivo imaging reveals different cellular functions for FGF and Dpp signaling in tracheal branching morphogenesis. Dev. Cell 2:677-683. [DOI] [PubMed] [Google Scholar]
- 25.Roth, P., N. Xylourgidis, N. Sabri, A. Uv, M. Fornerod, and C. Samakovlis. 2003. The Drosophila nucleoporin DNup88 localizes DNup214 and CRM1 on the nuclear envelope and attenuates NES-mediated nuclear export. J. Cell Biol. 163:701-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Samakovlis, C., N. Hacohen, G. Manning, D. Sutherland, K. Guillemin, and M. A. Krasnow. 1996. Development of the Drosophila tracheal system occurs by a series of morphologically distinct but genetically coupled branching events. Development 122:1395-1407. [DOI] [PubMed] [Google Scholar]
- 27.Samakovlis, C., G. Manning, P. Steneberg, N. Hacohen, R. Cantera, and M. A. Krasnow. 1996. Genetic control of epithelial tube fusion during Drosophila tracheal development. Development 122:3531-3536. [DOI] [PubMed] [Google Scholar]
- 28.Sato, M., and T. B. Kornberg. 2002. FGF is an essential mitogen and chemoattractant for the air sacs of the Drosophila tracheal system. Dev. Cell 3:195-207. [DOI] [PubMed] [Google Scholar]
- 29.Sonnenfeld, M., M. Ward, G. Nystrom, J. Mosher, S. Stahl, and S. Crews. 1997. The Drosophila tango gene encodes a bHLH-PAS protein that is orthologous to mammalian Arnt and controls CNS midline and tracheal development. Development 124:4583-4594. [DOI] [PubMed] [Google Scholar]
- 30.Sutherland, D., C. Samakovlis, and M. A. Krasnow. 1996. branchless encodes a Drosophila FGF homolog that controls tracheal cell migration and the pattern of branching. Cell 87:1091-1101. [DOI] [PubMed] [Google Scholar]
- 31.Tanaka-Matakatsu, M., T. Uemura, H. Oda, M. Takeichi, and S. Hayashi. 1996. Cadherin-mediated cell adhesion and cell motility in Drosophila trachea regulated by the transcription factor Escargot. Development 122:3697-3705. [DOI] [PubMed] [Google Scholar]
- 32.Tomancak, P., A. Beaton, R. Weiszmann, E. Kwan, S. Shu, S. E. Lewis, S. Richards, M. Ashburner, V. Hartenstein, S. E. Celniker, et al. 2002. Systematic determination of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 3:RESEARCH0088. [DOI] [PMC free article] [PubMed]
- 33.Uemura, T., H. Oda, R. Kraut, S. Hayashi, Y. Kotaoka, and M. Takeichi. 1996. Zygotic Drosophila E-cadherin expression is required for processes of dynamic epithelial cell rearrangement in the Drosophila embryo. Genes Dev. 10:659-671. [DOI] [PubMed] [Google Scholar]
- 34.Uv, A., R. Cantera, and C. Samakovlis. 2003. Drosophila tracheal morphogenesis: intricate cellular solutions to basic plumbing problems. Trends Cell Biol. 13:301-309. [DOI] [PubMed] [Google Scholar]
- 35.Uv, A. E., P. Roth, N. Xylourgidis, A. Wickberg, R. Cantera, and C. Samakovlis. 2000. members only encodes a Drosophila nucleoporin required for rel protein import and immune response activation. Genes Dev. 14:1945-1957. [PMC free article] [PubMed] [Google Scholar]
- 36.Ward, M. P., J. T. Mosher, and S. T. Crews. 1998. Regulation of Drosophila bHLH-PAS protein cellular localization during embryogenesis. Development 125:1599-1608. [DOI] [PubMed] [Google Scholar]
- 37.Wilk, R., I. Weizman, L. Glazer, and B.-Z. Shilo. 1996. trachealess encodes a bHLH-PAS protein and is a master regulator gene in the Drosophila tracheal system. Genes Dev. 10:93-102. [DOI] [PubMed] [Google Scholar]