Abstract
The androgen receptor (AR) is a hormone-dependent transcription factor critically involved in human prostate carcinogenesis. Optimal transcriptional control of androgen-responsive genes by AR may require complex interaction among multiple coregulatory proteins. We have previously shown that the AR coregulator TIP60 can interact with human PIRH2 (hPIRH2). In this study, we uncover important new functional role(s) for hPIRH2 in AR signaling: (i) hPIRH2 interacts with AR and enhances AR-mediated transcription with a dynamic pattern of recruitment to androgen response elements in the prostate-specific antigen (PSA) gene; (ii) hPIRH2 interacts with the AR corepressor HDAC1, leading to reduced HDAC1 protein levels and inhibition of transcriptional repression; (iii) hPIRH2 is required for optimal PSA expression; and (iv) hPIRH2 is involved in prostate cancer cell proliferation. In addition, overexpression of hPIRH2 protein was detected in 73 of 82 (89%) resected prostate cancers, with a strong correlation between increased hPIRH2 expression and aggressive disease, as signified by high Gleason sum scores and the presence of metastatic disease (P = <0.0001 and 0.0004, respectively). Collectively, our data establish hPIRH2 as a key modulator of AR function, opening a new direction for targeted therapy in aggressive human prostate cancer.
The androgen receptor (AR) is an intracellular mediator of androgen signaling required for prostate development, differentiation, and carcinogenesis. Prostate cancer (the most prevalent male malignancy) is androgen dependent (23), and AR-inhibitory hormone manipulation(s) reduces tumor proliferation, even in metastatic disease. However, hormone manipulation ultimately fails: tumors relapse, producing androgen-independent cancer (AIPC), for which novel treatments are required.
Like other steroid receptors, AR can interact with many proteins, including chaperone, scaffolding, or cytoskeleton proteins involved in AR folding, transformation, and stability (3, 13, 17, 39); signaling proteins involved in AR phosphorylation and activation (43, 44, 49); and coregulatory proteins that modulate AR transcriptional activities (32). These transcriptional coregulators can be grouped into corepressors such as histone deacetylase 1 (HDAC1) and Mdm2 (6, 11, 12, 27) or coactivators that can repress or enhance AR transcriptional activities, respectively (19). Coactivators include histone acetyltransferase proteins such as p300/CBP and TIP60 (10, 11, 42); ATP-dependent SWI/SNF remodeling factors BRG1 and hBRM (31); p160 proteins, including SRC-1 and SRC-3/AIB1 (38, 47); other modifying factors, such as CARM1/PRMT1, PIAS1, and E6-AP (4, 24, 37, 47, 50); and factors not known to contain modifying activities, such as FHL2 and the TRAP complex (35, 48). Some of these coactivators are overexpressed in prostate cancers (14, 16, 20, 26, 28, 29). Importantly, inhibition of p300 and ARA54 reduces the proliferation of prostate cancer cells (7, 8, 34).
We and others have begun to characterize human PIRH2 (hPIRH2) that interacts directly with AR, TIP60, and p53 (1, 25, 30). PIRH2 can act as an ubiquitin ligase for p53, resulting in reduced p53 protein levels, while hPIRH2 itself is ubiquitylated and targeted for proteasome-mediated destruction (25, 30).
We assessed here the effects of hPIRH2 overexpression or depletion on AR signaling and show that hPIRH2 has an important role in regulating AR transcriptional activities by interacting directly with both AR and the AR corepressor HDAC1. In addition, we demonstrate abnormal expression of hPIRH2 in human prostate cancer.
MATERIALS AND METHODS
Constructs, cell culture, and proliferation assays.
hAR, hPIRH2-MYC, CMV-p300, pBJ5-HDAC1, and luciferase reporter assays have been described (2, 11, 30). Reporter gene assays in which small interfering RNA (siRNA) was introduced were performed as described previously (21). Cell lines were cultured as described previously (2). WST-1 proliferation assays were performed at 48 h posttransfection as recommended by Roche.
Antibodies, immunoprecipitation, ChIP, nickel capture, and immunohistochemistry.
hPIRH2 antibodies were BL588 (Bethylabs) and Ab3886 (Abcam). Others used were MYC 9B11 (Cell Signaling), HDAC1 (Upstate), ubiquitin (Santa Cruz), α-tubulin (Sigma), and AR C-19 (Santa Cruz). Chromatin immunoprecipitations (ChIPs) were performed as described previously (30, 45). Real-time PCR was performed on inputs and recovered material (Applied Biosystems) with the oligonucleotide pairs AREI (5′-CCTAGATGAAGTCTCCATGAGCTACA-3′ and 5′-GGGAGGGAGAGCTAGCACTTG-3′) and AREIII (5′-GCCTGGATCTGAGAGAGATATCATC-3′ and 5′-ACACCTTTTTTTTCTGGATTG-3′), using SYBR Green I (Sigma Aldrich). Immunofluorescence was performed as described previously (30). Nickel capture to recover His-tagged HDAC1 or His-ubiquitin conjugates was performed as described previously (12). Green fluorescent protein (GFP)-tagged hPIRH2 was used because hPIRH2-MYC also contains a His tag. Immunohistochemistry was performed on 4-μm sections of untreated patient samples retrieved by transurethral resection (16). Slides were scored by two independent observers, shielded from clinical data. Immunoreactivity was negative, weak, medium, or strong (scored as 0 to 3). Correlation with clinical parameters was confirmed by using nonparametric Kruskal-Wallis and Mann-Whitney U tests (18, 29). hPIRH2 Ab3886 produced the same staining pattern as hPIRH2 BL588 (not shown).
RNA interference (RNAi) and real-time PCR.
siRNAs si1 (5′-UCAACUAGAUCGCUUUAAADTDT-3′) and si2 (5′-AAGCUGGAGGACGUAGAAUDTDT-3′ [nonsilencing] and 5′-UUCUCCGAACGUGUCACGUDTDT-3′) (QIAGEN and Eurogentec) and HDAC1 sequence as described previously (21) were transfected with RNAiFect (QIAGEN). Quantitative real-time PCR was performed on cDNA by using oligonucleotide sequences corresponding to prostate-specific antigen (PSA) (5′-ATGTGGGTCCCGGTTGTCT-3′ and 5′-AGCGCCAATCCACGTCA-3′), GAPDH (glyceraldehyde-3-phosphate dehydrogenase; 5′-CGACCACTTTGTCAAGCTCA-3′ and 5′-GGGTCTTACTCCTTGGAGGC-3′), and SYBR Green I.
RESULTS
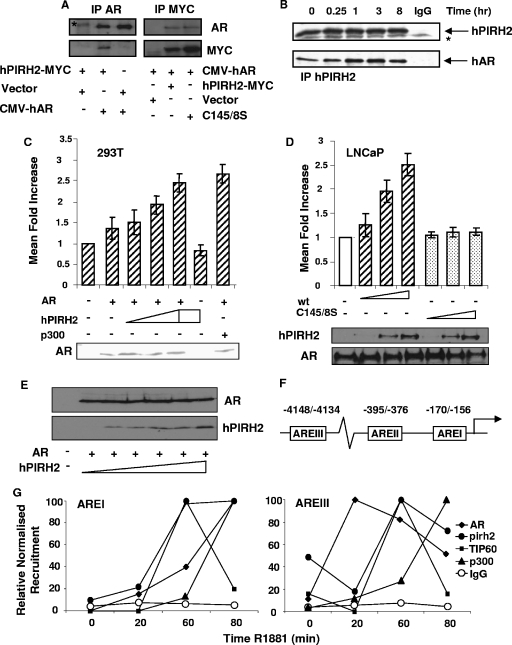
Murine Pirh2 was reported to interact with the hAR N terminus in yeast and in vitro (1). This interaction was tested in human cells; overexpressed hPIRH2-MYC and hAR were specifically coimmunoprecipitated from 293T cells (Fig. 1A). Endogenous hPIRH2 and hAR were also coimmunoprecipitated from LNCaP prostate cancer cells that had been cultured in steroid depleted medium (SDM). However, this interaction was further enhanced in the presence of synthetic androgen R1881 (Fig. 1B).
FIG. 1.
hPIRH2 interacts with AR in human cells and increases AR-mediated transcription. (A) 293T cells were transfected with hPIRH2-MYC and either empty vector or full-length hAR (CMV-hAR) as shown. Immunoprecipitation was performed with AR C-19 antibody, and recovered material was probed with AR and MYC antibodies (left panels). 293T cells were also transfected with full-length hAR and the indicated vectors encoding MYC-tagged hPIRH2. Proteins were immunoprecipitated with MYC antibody and then probed for AR and MYC (right panels). (B) LNCaP cells cultured in SDM were stimulated with 10 nM R1881 for specified times. Proteins were immunoprecipitated with hPIRH2 BL588 and then probed for hPIRH2 and AR. Immunoglobulin G (IgG) corresponds to nonimmune immunoglobulins. The asterisk represents a nonspecific band. (C) 293T cells were transfected with a PSA-driven luciferase reporter gene, CMV-hAR, and increasing amounts of hPIRH2-MYC or CMV-driven p300. Empty vector was used to equalize the DNA quantities transfected. Cells were starved in SDM and then treated with 10 nM R1881 or vehicle. No stimulation was observed in the absence of androgens in any scenario (not shown). Reporter assays were performed in triplicate on three occasions, and values were adjusted to constitutive β-galactosidase activity. Error bars represent the standard deviation. Western blotting shows similar AR levels, where transfected. (D) Reporter gene assays were performed in LNCaP cells as in panel C except that CMV-hAR was omitted. Western blotting shows relative hPIRH2 levels and similar AR levels. (E) 293T cells were transfected with AR and increasing amounts of hPIRH2. Equal quantities of lysates were probed with the indicated antibodies, as shown. (F) Diagram shows ARE positions within human PSA gene. (G) LNCaP cells were used for ChIP with the specified antibodies against endogenous proteins. Inputs and recovered material were analyzed by real-time PCR to calculate the recruitment changes. The maximal observed recruitment was assigned as 100%. AREI and AREIII profiles are shown in the left and right panels, respectively.
That R1881 enhanced hPIRH2-hAR interactions prompted assessment of hPIRH2 to influence AR transcriptional activity. AR-deficient 293T cells were transfected with a PSA promoter-driven AR-responsive luciferase reporter gene. In the presence of androgens, hPIRH2 cotransfection produced a dose-dependent enhancement of reporter gene activity to levels similar to those of the previously confirmed AR coactivator p300 (Fig. 1C). This did not occur in the absence of either cotransfected hAR (Fig. 1C) or androgens (not shown), and Western blotting demonstrated no significant alterations in AR protein levels upon hPIRH2 transfection (Fig. 1C, lower panel). AR-positive LNCaP cells were used in similar reporter gene assays, and the hPIRH2 RING domain double mutant that lacks ubiquitin ligase activity (hPIRH2C145/8S) was included (25, 30). hPIRH2 cotransfection again led to increased reporter gene activity, whereas hPIRH2C145/8S expression did not (Fig. 1D), despite retaining hAR interactions (Fig. 1A). Again, AR levels were largely unaltered upon hPIRH2 transfection, and wild-type hPIRH2 was expressed at similar levels to mutant hPIRH2 (Fig. 1D, lower panel). In addition, overexpression of increasing quantities of hPIRH2 did not alter AR steady-state protein levels in transfected 293T cells (Fig. 1E). These novel data are the first to suggest that hPIRH2 is an AR coregulator.
To confirm a transcriptional role for hPIRH2, recruitment of endogenous hPIRH2 and functionally related proteins to well-characterized androgen response elements (AREI and AREIII) within the PSA gene was examined in LNCaP cells (Fig. 1F and G). ChIP with hPIRH2 antibody at 20, 60, or 80 min after R1881 stimulation, combined with real-time PCR, demonstrated that hPIRH2 is recruited to both AREI and AREIII in response to androgens (Fig. 1G). The analysis revealed distinct hPIRH2 recruitment profiles to the two AREs. hPIRH2 was increasingly recruited to AREI at 20, 60, and 80 min, coinciding with AR and p300 recruitment (Fig. 1G, left graph). Association of the AR coactivator TIP60 was not detected until 60 min after ligand exposure but was then largely lost after 80 min (Fig. 1G, left graph). Examination of AREIII revealed initial loss of hPIRH2 and TIP60 upon ligand exposure (Fig. 1G, right graph), whereas AR recruitment was robustly detected at this time (Fig. 1G, right graph). After 60 min, hPIRH2 and TIP60 recruitment simultaneously peaked to levels above those prior to ligand exposure, and AR recruitment could still be detected (Fig. 1G, right graph). At 80 min TIP60, hPIRH2 and AR began to dissociate (Fig. 1G, right graph), demonstrating a cyclical, coordinated recruitment profile of hPIRH2 and TIP60 to AREIII. Further analyses showed that hPIRH2 and TIP60 were not recruited to non-ARE-containing regions between AREI and AREIII (not shown). p300 exhibited a noncyclical similar recruitment profile to AREIII and AREI, and in no scenario did hPIRH2 and p300 exhibit robust corecruitment, demonstrating a specific association between hPIRH2 and TIP60. In keeping with these data, hPIRH2-TIP60 protein-protein interactions have been previously described (30).
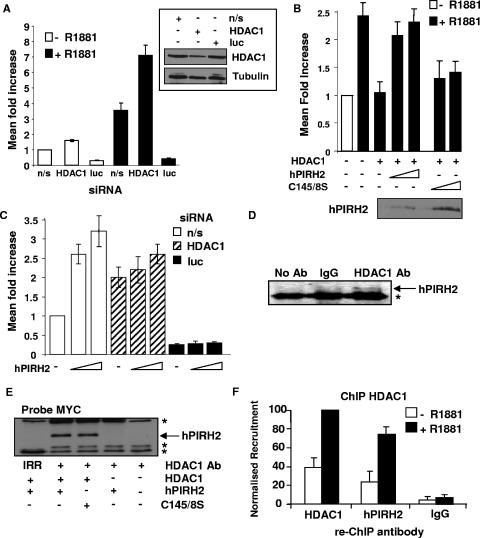
To examine a possible mechanism by which hPIRH2 might increase AR-mediated transcription, hPIRH2 was assessed for its ability to overcome the effects of the potent AR corepressor and histone deacetylase, HDAC1 (11). First, the effects of HDAC1 on the PSA reporter gene were determined in LNCaP cells. Prior transfection of HDAC1 siRNA led to a potent derepression of the subsequently transfected PSA reporter gene, whereas Western blotting showed HDAC1 knockdown in these cells (Fig. 2A). In agreement with previous findings (11, 12), reporter gene assays in LNCaP cells using the PSA-driven luciferase reporter gene demonstrated a strong repression of AR-mediated transcription by HDAC1, in the presence of synthetic androgens (Fig. 2B). Strikingly, cotransfection of wild-type hPIRH2 abrogated this HDAC1-mediated AR repression (Fig. 2B). Interestingly, introduction of the hPIRH2C145/8S mutant, which previously did not increase AR activity, did not substantially overcome the repressive effects of HDAC1 (Fig. 2B). To specifically test whether hPIRH2 enhances AR by HDAC1 downregulation, reporter gene assays were performed in LNCaP cells transfected with the HDAC1 siRNA. As shown in Fig. 2C, HDAC1 depletion reduced the capacity for hPIRH2 to stimulate AR-mediated transcription. siRNA directed against luciferase acted as an internal control to confirm gene silencing was effective at the time of measuring reporter gene activity (Fig. 2C). Considering that hPIRH2 was able to overcome the repressive effects of HDAC1, protein-protein interaction between these coregulators was tested. Untransfected LNCaP cell lysates were subject to immunoprecipitation using the HDAC1 antibody or other control antisera. Only in the presence of HDAC1 antibody was hPIRH2 specifically recovered, demonstrating that endogenous HDAC1 and hPIRH2 complex in LNCaP cells (Fig. 2D). Because the hPIRH2C145/8S mutant could not overcome the repressive effects of HDAC1 on AR, hPIRH2C145/8S was compared against wild-type hPIRH2 for its ability to interact with HDAC1 in transfected 293T cells (Fig. 2E). Immunoprecipitation with HDAC1 antibody specifically recovered both wild-type hPIRH2 and hPIRH2C145/8S, showing that both interact with HDAC1. Immunoprecipitation of transfected cell lysate with an irrelevant antibody did not recover hPIRH2 (Fig. 2E). Having confirmed that hPIRH2 and HDAC1 can interact in human cells, their ability to co-occupy the androgen-responsive PSA promoter was next tested. In keeping with a role of the hPIRH2-HDAC1 interaction with modulation of AR-mediated transcription, ChIP with HDAC1 antibody, followed by re-ChIP with hPIRH2 antibody, revealed that the two factors were simultaneously present at AREI 80 min after R1881 stimulation (Fig. 2F). Re-ChIP performed with HDAC1 or nonspecific antibodies acted as controls (Fig. 2F).
FIG. 2.
hPIRH2 can derepress HDAC1-mediated effects on AR and interacts with HDAC1. (A) LNCaP cells were transfected with the indicated siRNAs and then 24 h later with the PSA reporter gene. Cells were starved for 48 h, stimulated with R1881 or vehicle overnight, and then harvested. (B) LNCaP cells were transfected with the indicated vectors and used for reporter gene assays as in Fig. 1D. (C) LNCaP cells were transfected with the indicated siRNAs and then subjected to reporter gene assays as described in Fig. 1D. (D) LNCaP cell lysates were immunoprecipitated with either HDAC1 antibody, nonimmune immunoglobulins (IgG), or in the absence of antibody (No Ab) as indicated. Material was probed with hPIRH2 BL588. (E) 293T cells were transfected with the indicated vectors. Cell lysates were then used for immunoprecipitation with HDAC1 antibody or irrelevant FRS2 antibody (IRR), where indicated. Material was probed with MYC antibody directed against hPIRH2. (F) LNCaP cells were starved in SDM and then stimulated with 10 nM R1881 for 80 min where indicated. Cells were subjected to ChIP first with HDAC1 antibody, and then the material was subjected to re-ChIP with either HDAC1, hPIRH2 BL588, or nonimmune immunoglobulins (IgG). Recovered material was analyzed by real-time PCR as in Fig. 1G using AREI-specific oligonucleotides.
In order to delineate the mechanism by which hPIRH2 might derepress HDAC1, an alternative reporter gene assay was used that lacks AR and AREs. In this assay, hPIRH2 was tested for its ability to modulate the transcriptional activity of either the heterologous GAL4-DBD or a GAL4-DBD-HDAC1 chimera. 293T cells were transfected with plasmids encoding these factors and a GAL4-responsive luciferase reporter gene (Fig. 3A). As expected, the GAL4-DBD-HDAC1 chimera had substantially lower transcriptional activity than the GAL4-DBD (Fig. 3A). Transfection of hPIRH2 or the hPIRH2C145/8S mutant did not have a notable impact on the GAL4-DBD (Fig. 3A). However, in the presence of GAL4-DBD-HDAC1, hPIRH2 transfection led to a substantial increase in reporter gene expression, whereas transfection of the hPIRH2C145/8S mutant did not have a noticeable impact on reporter gene expression (Fig. 3A). The hPIRH2C145/8S mutant lacks ubiquitin ligase activity, which raised the possibility that hPIRH2 represses HDAC1 in a ubiquitin-dependent manner. To establish whether this might be the case, the previous reporter gene assay was repeated in the presence of overexpressed ubiquitin. As expected, transfection of hPIRH2 again led to derepression of HDAC1. However, cotransfection of ubiquitin slightly enhanced this effect (Fig. 3B). More notably, cotransfection of a mutant form of ubiquitin (7KR, in which all lysine residues are mutated to arginine residues) that is incapable of being incorporated into polyubiquitin chains almost completely abrogated the derepression of HDAC1 by hPIRH2 (Fig. 3B).
FIG. 3.
hPIRH2 reduces the repressive activity of HDAC1 on transcription, causes HDAC1 ubiquitination, and reduces HDAC1 levels. (A) 293T cells were transfected with a GAL4-responsive luciferase reporter vector, either GAL4-DBD or HDAC1-DBD, and the indicated vectors as in Fig. 1C and then used in reporter gene assays. (B) 293T cells were transfected as in panel A except that CMV-driven ubiquitin or a mutant form of ubiquitin in which all lysine residues are mutated to arginine (7KR) were included. Reporter assays were performed in triplicate, on three occasions, and values were adjusted to constitutive β-galactosidase activity. Error bars represent the standard deviation. (C) 293T cells were transfected with His-tagged HDAC1 and either empty vector or EGFP-hPIRH2 and then treated with 5 μM MG-132. Denatured cell lysates were subjected to nickel capture assay and then recovered material was probed with ubiquitin (left panel) and HDAC1 (right panel) antibodies. (D) 293T cells were transfected with the indicated vectors, and cell lysates were subjected to nickel capture assay as in panel C. Recovered material was probed with HDAC1 antibody. (E) LNCaP cells were transfected with either nonsilencing (n/s) siRNA or hPIRH2 siRNA. Equal quantities of cell lysates were probed for endogenous hPIRH2 or HDAC1 as indicated (right panels).
To examine the possibility that hPIRH2 might ubiquitinate HDAC1 or that hPIRH2 is somehow involved in regulation of HDAC1 stability, 293T cells were transfected with His-tagged HDAC1 and then subjected to nickel capture under denaturing conditions. Recovered material was probed with ubiquitin antibody to detect ubiquitin-conjugated HDAC1. In the presence of the proteasomal inhibitor MG-132, several slowly migrating HDAC1 species were detected that are likely to be ubiquitin-conjugated HDAC1. However, cotransfection of hPIRH2 produced a noticeable increase in high-molecular-weight ubiquitin-conjugated HDAC1, as detected with the ubiquitin antibody (Fig. 3C). When the purified material was probed with HDAC1 antibody, this confirmed that HDAC1 had been recovered, and an additional higher-molecular-weight HDAC1 species appeared upon cotransfection of hPIRH2. This additional HDAC1 species may represent ubiquitin-conjugated HDAC1 (Fig. 3C, right panel). In an attempt to enhance the observed hPIRH2-mediated HDAC1 ubiquitination, His-tagged ubiquitin was overexpressed with hPIRH2 or Mdm2, which has previously been shown to ubiquitinate HDAC1 (12). Nickel purified, ubiquitin-tagged proteins were analyzed by Western blotting with HDAC1 antibody (Fig. 3D). High-molecular-weight, ubiquitin-conjugated HDAC1 could be detected upon His-ubiquitin expression, and these species were enriched upon hPIRH2 transfection, suggesting that hPIRH2 does indeed ubiquitinate HDAC1 (Fig. 3D). Mdm2 transfection had a similar effect (Fig. 3D). Transfection of ubiquitin ligase-deficient hPIRH2C145/8S abolished HDAC1 ubiquitination, suggesting that the ubiquitin ligase activity of hPIRH2 is required for HDAC1 ubiquitination and that hPIRH2C145/8S may act as a dominant-negative form (Fig. 3D, compare lanes 2 and 6). In order to determine whether hPIRH2 might regulate HDAC1 steady-state protein levels, hPIRH2 siRNA was transfected into LNCaP cells in the absence of proteasomal inhibition. Detection of hPIRH2 by Western blotting demonstrated hPIRH2 knockdown, while analysis of HDAC1 levels demonstrated that HDAC1 protein levels increased in response to hPIRH2 depletion (Fig. 3E, right panels).
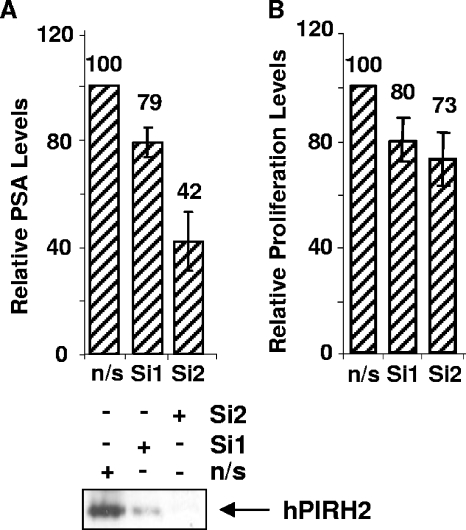
The data suggest that hPIRH2 may have an important role in AR-mediated transcription. The androgen-responsive LNCaP cell line is known to be at least partly dependent upon androgens for the proliferation and expression of androgen-responsive genes such as PSA. In order to examine whether hPIRH2 might be able to influence gene expression and proliferation in LNCaP cells, a gene-silencing approach was undertaken. Transfection of two siRNAs against hPIRH2 produced 21 to 58% decreases in PSA transcript levels compared to nonsilencing siRNA, as assessed by quantitative PCR (Fig. 4A). Western blotting confirmed hPIRH2 knockdown (Fig. 4A, lower panel). In addition, hPIRH2 silencing resulted in a 20 to 27% decreased proliferation of LNCaP cells compared to nonsilencing transfectants (Fig. 4B).
FIG. 4.
hPIRH2 controls PSA expression and LNCaP cell proliferation. (A) Nonsilencing siRNA (n/s) or siRNA against hPIRH2 (Si1 and Si2) was transfected into LNCaP cells. After 48 h, PSA and GAPDH transcript levels were measured by quantitative PCR. PSA levels were normalized to GAPDH levels (upper panel). Equal quantities of transfected cell lysates were also used for Western blotting (lower panel). (B) LNCaP cells were transfected as in panel A, and then proliferation was measured by using a WST-1 assay at 48 h posttransfection. Proliferation of nonsilencing siRNA-transfected cells was assigned as 100%. All error bars represent standard deviations.
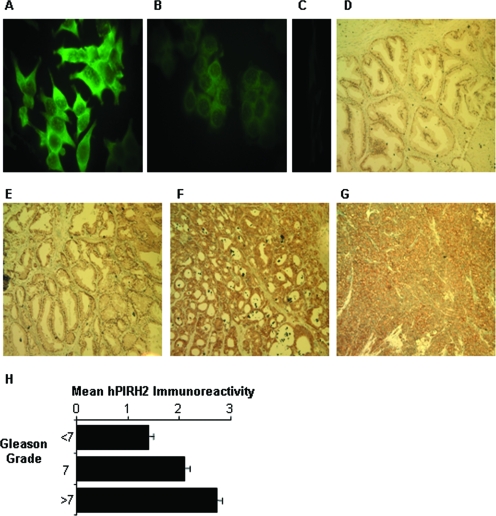
Considering the involvement of hPIRH2 in prostate cancer cell proliferation, hPIRH2, like other AR coregulators, might be expected to be aberrantly expressed in prostate tumors. BL588 hPIRH2 antibody has previously been used for immunohistochemistry (9). In LNCaP cells, this antibody produced a diffuse nuclear/cytoplasmic immunofluorescence staining pattern (Fig. 5A to C) as observed previously for GFP-tagged hPIRH2 (30). To examine expression in human prostate tumors, paraffin-embedded prostate tissue samples, retrieved by transurethral resection, were assayed by immunohistochemistry. Samples were obtained from untreated patients at the time of diagnosis. Some patients did go on to develop AIPC posttreatment. hPIRH2 staining in tissues was detected in both cytoplasmic and nuclear compartments, and the intensity was categorized as negative (score of 0), weak (score of 1), medium (score of 2), or strong (score of 3). hPIRH2 expression in the benign sample group was weak or absent (mean staining score of 0.9, Fig. 5D). In lower-grade cancers (Gleason sum score of <7) staining was weak or medium (mean score of 1.4; Fig. 5E). In Gleason sum score 7 samples, hPIRH2 exhibited mainly medium expression, indicating that with a mean staining score of 2.1, hPIRH2 staining was significantly stronger in Gleason sum score 7 samples than in low-grade cancers (P = 0.0078 [Kruskal-Wallis]) or benign tissue (P = 0.0001 [Kruskal-Wallis]) (Fig. 5F). Strikingly, most high-grade tumors (above Gleason sum score 7) exhibited strong expression, while others exhibited medium staining (mean staining score of 2.79; Fig. 5G). hPIRH2 expression in these tumors was statistically significantly stronger than in Gleason sum score 7 tumors (P = 0.0005 [Kruskal-Wallis]), low Gleason sum score tumors (P = 0.0001, Kruskal-Wallis) or benign tissues (P = 0.0001, Kruskal-Wallis). This is summarized in Fig. 5H and Table 1.
FIG. 5.
Expression patterns of hPIRH2. (A) LNCaP cells were fixed and subject to immunofluorescence with hPIRH2 BL588 antibody. (B) MCF-7 breast cancer cells were treated as in panel A. (C) Negative control immunofluorescence experiment with no primary antibody. (D to G) Immunohistochemical staining performed on human transurethral resection samples of either benign prostatic hyperplasia (D) or prostate cancers of increasing Gleason grade (E to G) with hPIRH2 BL588 antibody. (H) Mean hPIRH2 immunoreactivity in prostate cancers. Error bars represent the standard deviations.
TABLE 1.
PIRH2 expression in human prostate tumors
| Clinical parameter | No. of tumors with a staining intensity score of:
|
P | |||
|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | ||
| Gleason score (n = 82) | 0.0001 | ||||
| 4-6 | 0 | 9 | 6 | 0 | |
| 7 | 0 | 0 | 9 | 1 | |
| 8-10 | 0 | 0 | 12 | 45 | |
| Bone metastasis (n = 73) | 0.0004 | ||||
| No | 0 | 8 | 17 | 15 | |
| Yes | 0 | 0 | 8 | 25 | |
No correlation was observed between hPIRH2 immunostaining and PSA levels, patient survival, or patients that developed AIPC versus androgen-responsive cancers. Interestingly, a correlation was observed between hPIRH2 staining score and the presence of bone metastases (mean staining score of 2.76) or absence of bone metastases (mean staining score of 2.18) at diagnosis (P = 0.0004 [Mann-Whitney U test]) (Table 1). The data show that hPIRH2 is overexpressed in prostate cancers with increasing Gleason sum scores, and strong expression correlates with the presence of bone metastases.
DISCUSSION
Here we show that the E3 ubiquitin ligase hPIRH2 can directly interact with components of the AR signaling pathway, resulting in increased AR-mediated transcription. Although conserved RING domain residues of hPIRH2 are not essential for these interactions, the RING domain double mutant hPIRH2C145/8S does not increase AR-mediated transcription, suggesting hPIRH2 ubiquitin ligase activity is required for increased AR transcriptional activity. Our data suggest that hPIRH2 can ubiquitylate HDAC1 in cells and that hPIRH2 knockdown stabilizes HDAC1. The finding that hPIRH2 can reduce HDAC1 levels and overcome the transcriptional repression of AR by HDAC1 affords a mechanism through which hPIRH2 ubiquitin ligase activity can increase AR-mediated transcription. Indeed, HDAC1 silencing reduced the capacity of hPIRH2 to enhance AR transcriptional activity. Previous experiments suggesting that hPIRH2 could not increase AR activity were performed in cell lines different from those used here (1). Their HDAC1 status is unknown. Our findings are reminiscent of derepressive factors, such as SENP1, that can increase AR-mediated transcription through the modulation of HDAC1 function (5) and suggest that hPIRH2 could increase AR-mediated transcription via a derepressive mechanism. Aside from the hPIRH2-HDAC1 interaction, hPIRH2 may also regulate AR directly via their protein-protein interactions.
We demonstrate that hPIRH2 is contained within transcriptional complexes occupying AREs on the PSA gene. Interestingly, hPIRH2 exhibits different recruitment dynamics at AREI and AREIII, implying that it might perform different roles at the two AREs, in response to androgens. Notably, hPIRH2 and TIP60 exhibit cyclical corecruitment to AREIII, suggesting that they are both present in a transcriptional complex, confirming previous hPIRH2-TIP60 protein-protein interactions (30). Cyclical recruitment of transcriptional coregulators, including TIP60, is now an established phenomenon that is intrinsic to transcriptional activation by steroid receptors (33), although little work has been performed to comprehensively assess the recruitment of AR coregulators to androgen-responsive genes (45). We demonstrate by re-ChIP analysis that hPIRH2 and HDAC1 co-occupy AREI in response to androgens. This raises the possibility that hPIRH2 may target HDAC1 for destruction while the two proteins are present in a transcriptional complex and supports the concept that proteolysis of transcriptional regulators is intrinsically linked to active transcription (36, 41). It would seem that hPIRH2 can form transcriptional complexes with corepressors such as HDAC1, as well as coactivators such as TIP60. Although hPIRH2 seems to target HDAC1 proteolysis, the role of hPIRH2 interaction with coactivators such as TIP60 is unknown. We speculate that transcriptional modulation by hPIRH2 involves the use of both coactivators and corepressors. The finding that RNAi-mediated hPIRH2 knockdown reduces PSA expression implies that hPIRH2 does play an important role in transcriptional complexes that regulate gene expression.
hPIRH2 has previously been reported to repress p53 transcriptional activity and ubiquitylate p53, leading to its destruction (25). HDAC1 is known to act in concert with Mdm2 in order to repress p53 transcriptional activity (22). It is difficult to reconcile the role we describe for hPIRH2 in derepression of HDAC1 with a role for hPIRH2 as a p53 corepressor. However, the fact that hPIRH2 can derepress HDAC1 when it is fused the GAL4-DBD suggests that hPIRH2 can derepress HDAC1 not only in the context of AR-mediated transcription. Importantly, the p53 corepressor Mdm2 can also ubiquitylate HDAC1, leading to its destruction (12). In light of these facts, the hPIRH2-HDAC1 interaction in p53 signaling should be examined. The emergence of hPIRH2 in the upregulation of AR activity and the ubiquitylation of p53 bears similarities to E6-AP, a steroid receptor coactivator required for proper development of murine tissues, including the prostate (37, 46).
hPIRH2 knockdown resulted in reduced LNCaP prostate cancer cell proliferation. Although hPIRH2 depletion in LNCaP cells may increase p53 activities, thereby reducing proliferation, we have not observed increased p53 protein levels in this case (data not shown). The mechanism by which hPIRH2 controls proliferation of LNCaP cells is currently unknown, although this could conceivably involve regulation of AR signaling. A recent study showed Mdm2 antisense treatment of prostate cancer cell lines resulted in reduced proliferation and increased sensitivity to chemotherapy (51), in a manner independent of AR or p53 status. This suggests that ubiquitin ligase enzymes do have critical targets, other than p53, involved in proliferation of prostate cancer cells.
hPIRH2 expression, like that of some other AR coregulators (see the introduction), is altered in human prostate cancers. Expression is weak in benign prostate and low-Gleason-grade cancers (with no apparent statistical significant difference between the two) and is increased in higher-grade cancers. Increased expression in Gleason sum score 7 tissues indicates that hPIRH2 overexpression occurs during disease progression. Strong hPIRH2 expression correlated with the presence of bone metastases, likely reflecting a cohort of patients with highly aggressive tumors, although it is unknown whether hPIRH2 might directly participate in metastasis. It is quite possible that hPIRH2 overexpression in prostate tumors impacts upon pathways other that AR signaling (such as p53 signaling) and in this regard the role of hPIRH2 in tumorigenesis should be studied. hPIRH2 was also recently shown to be overexpressed in lung cancer (9). Notably, two hPIRH2-interacting partners, TIP60 and HDAC1, also exhibit aberrant expression in prostate cancer, pointing toward a role for these coregulators in prostate carcinogenesis (15, 40).
The data indicate that prostate cancer cells are sensitive to reduced hPIRH2 activities. Pharmacological inhibition of enzymes such as hPIRH2 is therefore attractive in the search for new treatments for human prostate cancer.
Acknowledgments
This study was funded by MRC PROMPT grant G0100100/64424 and European Union Framework 6 (PRIMA program grant).
REFERENCES
- 1.Beitel, L. K., Y. A. Elhaji, R. Lumbroso, S. S. Wing, V. Panet-Raymond, B. Gottlieb, L. Pinsky, and M. A. Trifiro. 2002. Cloning and characterization of an androgen receptor N-terminal-interacting protein with ubiquitin-protein ligase activity. J. Mol. Endocrinol. 29:41-60. [DOI] [PubMed] [Google Scholar]
- 2.Brady, M. E., D. M. Ozanne, L. Gaughan, I. Waite, S. Cook, D. E. Neal, and C. N. Robson. 1999. Tip60 is a nuclear hormone receptor coactivator. J. Biol. Chem. 274:17599-17604. [DOI] [PubMed] [Google Scholar]
- 3.Brinkmann, A. O., L. J. Blok, P. E. de Ruiter, P. Doesburg, K. Steketee, C. A. Berrevoets, and J. Trapman. 1999. Mechanisms of androgen receptor activation and function. J. Steroid Biochem. Mol. Biol. 69:307-313. [DOI] [PubMed] [Google Scholar]
- 4.Chen, D., H. Ma, H. Hong, S. S. Koh, S. M. Huang, B. T. Schurter, D. W. Aswad, and M. R. Stallcup. 1999. Regulation of transcription by a protein methyltransferase. Science 284:2174-2177. [DOI] [PubMed] [Google Scholar]
- 5.Cheng, J., D. Wang, Z. Wang, and E. T. Yeh. 2004. SENP1 enhances androgen receptor-dependent transcription through desumoylation of histone deacetylase 1. Mol. Cell. Biol. 24:6021-6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng, S., S. Brzostek, S. R. Lee, A. N. Hollenberg, and S. P. Balk. 2002. Inhibition of the dihydrotestosterone-activated androgen receptor by nuclear receptor corepressor. Mol. Endocrinol. 16:1492-1501. [DOI] [PubMed] [Google Scholar]
- 7.Debes, J. D., L. J. Schmidt, H. Huang, and D. J. Tindall. 2002. p300 mediates androgen-independent transactivation of the androgen receptor by interleukin 6. Cancer Res. 62:5632-5636. [PubMed] [Google Scholar]
- 8.Debes, J. D., T. J. Sebo, C. M. Lohse, L. M. Murphy, A. L. de Haugen, and D. J. Tindall. 2003. p300 in prostate cancer proliferation and progression. Cancer Res. 63:7638-7640. [PubMed] [Google Scholar]
- 9.Duan, W., L. Gao, L. J. Druhan, W. G. Zhu, C. Morrison, G. A. Otterson, and M. A. Villalona-Calero. 2004. Expression of Pirh2, a newly identified ubiquitin protein ligase, in lung cancer. J. Natl. Cancer Inst. 96:1718-1721. [DOI] [PubMed] [Google Scholar]
- 10.Fu, M., C. Wang, X. Zhang, and R. G. Pestell. 2004. Acetylation of nuclear receptors in cellular growth and apoptosis. Biochem. Pharmacol. 68:1199-1208. [DOI] [PubMed] [Google Scholar]
- 11.Gaughan, L., I. R. Logan, S. Cook, D. E. Neal, and C. N. Robson. 2002. Tip60 and histone deacetylase 1 regulate androgen receptor activity through changes to the acetylation status of the receptor. J. Biol. Chem. 277:25904-25913. [DOI] [PubMed] [Google Scholar]
- 12.Gaughan, L., I. R. Logan, D. E. Neal, and C. N. Robson. 2005. Regulation of androgen receptor and histone deacetylase 1 by Mdm2-mediated ubiquitylation. Nucleic Acids Res. 33:13-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Georget, V., B. Terouanne, J. C. Nicolas, and C. Sultan. 2002. Mechanism of antiandrogen action: key role of hsp90 in conformational change and transcriptional activity of the androgen receptor. Biochemistry 41:11824-11831. [DOI] [PubMed] [Google Scholar]
- 14.Gnanapragasam, V. J., H. Y. Leung, A. S. Pulimood, D. E. Neal, and C. N. Robson. 2001. Expression of RAC 3, a steroid hormone receptor coactivator in prostate cancer. Br. J. Cancer 85:1928-1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Halkidou, K., L. Gaughan, S. Cook, H. Y. Leung, D. E. Neal, and C. N. Robson. 2004. Upregulation and nuclear recruitment of HDAC1 in hormone refractory prostate cancer. Prostate 59:177-189. [DOI] [PubMed] [Google Scholar]
- 16.Halkidou, K., V. J. Gnanapragasam, P. B. Mehta, I. R. Logan, M. E. Brady, S. Cook, H. Y. Leung, D. E. Neal, and C. N. Robson. 2003. Expression of Tip60, an androgen receptor coactivator, and its role in prostate cancer development. Oncogene 22:2466-2477. [DOI] [PubMed] [Google Scholar]
- 17.He, B., S. Bai, A. T. Hnat, R. I. Kalman, J. T. Minges, C. Patterson, and E. M. Wilson. 2004. An androgen receptor NH2-terminal conserved motif interacts with the COOH terminus of the Hsp70-interacting protein (CHIP). J. Biol. Chem. 279:30643-30653. [DOI] [PubMed] [Google Scholar]
- 18.Heer, R., D. Douglas, M. E. Mathers, C. N. Robson, and H. Y. Leung. 2004. Fibroblast growth factor 17 is overexpressed in human prostate cancer. J. Pathol. 204:578-586. [DOI] [PubMed] [Google Scholar]
- 19.Heinlein, C. A., and C. Chang. 2002. Androgen receptor (AR) coregulators: an overview. Endocrinol. Rev. 23:175-200. [DOI] [PubMed] [Google Scholar]
- 20.Hong, H., C. Kao, M. H. Jeng, J. N. Eble, M. O. Koch, T. A. Gardner, S. Zhang, L. Li, C. X. Pan, Z. Hu, G. T. MacLennan, and L. Cheng. 2004. Aberrant expression of CARM1, a transcriptional coactivator of androgen receptor, in the development of prostate carcinoma and androgen-independent status. Cancer 101:83-89. [DOI] [PubMed] [Google Scholar]
- 21.Ishizuka, T., and M. A. Lazar. 2003. The N-CoR/histone deacetylase 3 complex is required for repression by thyroid hormone receptor. Mol. Cell. Biol. 23:5122-5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ito, A., Y. Kawaguchi, C. H. Lai, J. J. Kovacs, Y. Higashimoto, E. Appella, and T. P. Yao. 2002. MDM2-HDAC1-mediated deacetylation of p53 is required for its degradation. EMBO J. 21:6236-6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jemal, A., R. C. Tiwari, T. Murray, A. Ghafoor, A. Samuels, E. Ward, E. J. Feuer, and M. J. Thun. 2004. Cancer statistics, 2004. CA Cancer J. Clin. 54:8-29. [DOI] [PubMed] [Google Scholar]
- 24.Koh, S. S., H. Li, Y. H. Lee, R. B. Widelitz, C. M. Chuong, and M. R. Stallcup. 2002. Synergistic coactivator function by coactivator-associated arginine methyltransferase (CARM) 1 and beta-catenin with two different classes of DNA-binding transcriptional activators. J. Biol. Chem. 277:26031-26035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leng, R. P., Y. Lin, W. Ma, H. Wu, B. Lemmers, S. Chung, J. M. Parant, G. Lozano, R. Hakem, and S. Benchimol. 2003. Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell 112:779-791. [DOI] [PubMed] [Google Scholar]
- 26.Li, P., X. Yu, K. Ge, J. Melamed, R. G. Roeder, and Z. Wang. 2002. Heterogeneous expression and functions of androgen receptor cofactors in primary prostate cancer. Am. J. Pathol. 161:1467-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liao, G., L. Y. Chen, A. Zhang, A. Godavarthy, F. Xia, J. C. Ghosh, H. Li, and J. D. Chen. 2003. Regulation of androgen receptor activity by the nuclear receptor corepressor SMRT. J. Biol. Chem. 278:5052-5061. [DOI] [PubMed] [Google Scholar]
- 28.Linja, M. J., K. P. Porkka, Z. Kang, K. J. Savinainen, O. A. Janne, T. L. Tammela, R. L. Vessella, J. J. Palvimo, and T. Visakorpi. 2004. Expression of androgen receptor coregulators in prostate cancer. Clin. Cancer Res. 10:1032-1040. [DOI] [PubMed] [Google Scholar]
- 29.Linja, M. J., K. J. Savinainen, O. R. Saramaki, T. L. Tammela, R. L. Vessella, and T. Visakorpi. 2001. Amplification and overexpression of androgen receptor gene in hormone-refractory prostate cancer. Cancer Res. 61:3550-3555. [PubMed] [Google Scholar]
- 30.Logan, I. R., V. Sapountzi, L. Gaughan, D. E. Neal, and C. N. Robson. 2004. Control of human PIRH2 protein stability: involvement of TIP60 and the proteosome. J. Biol. Chem. 279:11696-11704. [DOI] [PubMed] [Google Scholar]
- 31.Marshall, T. W., K. A. Link, C. E. Petre-Draviam, and K. E. Knudsen. 2003. Differential requirement of SWI/SNF for androgen receptor activity. J. Biol. Chem. 278:30605-30613. [DOI] [PubMed] [Google Scholar]
- 32.McKenna, N. J., and B. W. O'Malley. 2002. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell 108:465-474. [DOI] [PubMed] [Google Scholar]
- 33.Metivier, R., G. Penot, M. R. Hubner, G. Reid, H. Brand, M. Kos, and F. Gannon. 2003. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell 115:751-763. [DOI] [PubMed] [Google Scholar]
- 34.Miyamoto, H., M. Rahman, H. Takatera, H. Y. Kang, S. Yeh, H. C. Chang, K. Nishimura, N. Fujimoto, and C. Chang. 2002. A dominant-negative mutant of androgen receptor coregulator ARA54 inhibits androgen receptor-mediated prostate cancer growth. J. Biol. Chem. 277:4609-4617. [DOI] [PubMed] [Google Scholar]
- 35.Muller, J. M., U. Isele, E. Metzger, A. Rempel, M. Moser, A. Pscherer, T. Breyer, C. Holubarsch, R. Buettner, and R. Schule. 2000. FHL2, a novel tissue-specific coactivator of the androgen receptor. EMBO J. 19:359-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muratani, M., and W. P. Tansey. 2003. How the ubiquitin-proteasome system controls transcription. Nat. Rev. Mol. Cell. Biol. 4:192-201. [DOI] [PubMed] [Google Scholar]
- 37.Nawaz, Z., D. M. Lonard, C. L. Smith, E. Lev-Lehman, S. Y. Tsai, M. J. Tsai, and B. W. O'Malley. 1999. The Angelman syndrome-associated protein, E6-AP, is a coactivator for the nuclear hormone receptor superfamily. Mol. Cell. Biol. 19:1182-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Onate, S. A., S. Y. Tsai, M. J. Tsai, and B. W. O'Malley. 1995. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science 270:1354-1357. [DOI] [PubMed] [Google Scholar]
- 39.Ozanne, D. M., M. E. Brady, S. Cook, L. Gaughan, D. E. Neal, and C. N. Robson. 2000. Androgen receptor nuclear translocation is facilitated by the f-actin cross-linking protein filamin. Mol. Endocrinol. 14:1618-1626. [DOI] [PubMed] [Google Scholar]
- 40.Patra, S. K., A. Patra, and R. Dahiya. 2001. Histone deacetylase and DNA methyltransferase in human prostate cancer. Biochem. Biophys. Res. Commun. 287:705-713. [DOI] [PubMed] [Google Scholar]
- 41.Reid, G., M. R. Hubner, R. Metivier, H. Brand, S. Denger, D. Manu, J. Beaudouin, J. Ellenberg, and F. Gannon. 2003. Cyclic, proteasome-mediated turnover of unliganded and liganded ERα on responsive promoters is an integral feature of estrogen signaling. Mol. Cell 11:695-707. [DOI] [PubMed] [Google Scholar]
- 42.Reutens, A. T., M. Fu, C. Wang, C. Albanese, M. J. McPhaul, Z. Sun, S. P. Balk, O. A. Janne, J. J. Palvimo, and R. G. Pestell. 2001. Cyclin D1 binds the androgen receptor and regulates hormone-dependent signaling in a p300/CBP-associated factor (P/CAF)-dependent manner. Mol. Endocrinol. 15:797-811. [DOI] [PubMed] [Google Scholar]
- 43.Rigas, A. C., D. M. Ozanne, D. E. Neal, and C. N. Robson. 2003. The scaffolding protein RACK1 interacts with androgen receptor and promotes cross-talk through a protein kinase C signaling pathway. J. Biol. Chem. 278:46087-46093. [DOI] [PubMed] [Google Scholar]
- 44.Sadar, M. D. 1999. Androgen-independent induction of prostate-specific antigen gene expression via cross-talk between the androgen receptor and protein kinase A signal transduction pathways. J. Biol. Chem. 274:7777-7783. [DOI] [PubMed] [Google Scholar]
- 45.Shang, Y., M. Myers, and M. Brown. 2002. Formation of the androgen receptor transcription complex. Mol. Cell 9:601-610. [DOI] [PubMed] [Google Scholar]
- 46.Smith, C. L., D. G. DeVera, D. J. Lamb, Z. Nawaz, Y. H. Jiang, A. L. Beaudet, and B. W. O'Malley. 2002. Genetic ablation of the steroid receptor coactivator-ubiquitin ligase, E6-AP, results in tissue-selective steroid hormone resistance and defects in reproduction. Mol. Cell. Biol. 22:525-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tan, J., S. H. Hall, K. G. Hamil, G. Grossman, P. Petrusz, J. Liao, K. Shuai, and F. S. French. 2000. Protein inhibitor of activated STAT-1 (signal transducer and activator of transcription-1) is a nuclear receptor coregulator expressed in human testis. Mol. Endocrinol. 14:14-26. [DOI] [PubMed] [Google Scholar]
- 48.Wang, Q., D. Sharma, Y. Ren, and J. D. Fondell. 2002. A coregulatory role for the TRAP-mediator complex in androgen receptor-mediated gene expression. J. Biol. Chem. 277:42852-42858. [DOI] [PubMed] [Google Scholar]
- 49.Wong, H. Y., J. A. Burghoorn, M. Van Leeuwen, P. E. De Ruiter, E. Schippers, L. J. Blok, K. W. Li, H. L. Dekker, L. De Jong, J. Trapman, J. A. Grootegoed, and A. O. Brinkmann. 2004. Phosphorylation of androgen receptor isoforms. Biochem. J. 383:267-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu, W., H. Cho, and R. M. Evans. 2003. Acetylation and methylation in nuclear receptor gene activation. Methods Enzymol. 364:205-223. [DOI] [PubMed] [Google Scholar]
- 51.Zhang, Z., M. Li, H. Wang, S. Agrawal, and R. Zhang. 2003. Antisense therapy targeting MDM2 oncogene in prostate cancer: effects on proliferation, apoptosis, multiple gene expression, and chemotherapy. Proc. Natl. Acad. Sci. USA 100:11636-11641. [DOI] [PMC free article] [PubMed] [Google Scholar]