Abstract
FilaminC (FLNc) is the muscle-specific member of a family of actin binding proteins. Although it interacts with many proteins involved in muscular dystrophies, its unique role in muscle is poorly understood. To address this, two models were developed. First, FLNc expression was stably reduced in C2C12 myoblasts by RNA interference. While these cells start differentiation normally, they display defects in differentiation and fusion ability and ultimately form multinucleated “myoballs” rather than maintain elongated morphology. Second, a mouse model carrying a deletion of last 8 exons of Flnc was developed. FLNc-deficient mice die shortly after birth, due to respiratory failure, and have severely reduced birth weights, with fewer muscle fibers and primary myotubes, indicating defects in primary myogenesis. They exhibit variation in fiber size, fibers with centrally located nuclei, and some rounded fibers resembling the in vitro phenotype. The similarity of the phenotype of FLNc-deficient mice to the filamin-interacting TRIO null mice was further confirmed by comparing FLNc-deficient C2C12 cells to TRIO-deficient cells. These data provide the first evidence that FLNc has a crucial role in muscle development and maintenance of muscle structural integrity and suggest the presence of a TRIO-FLNc-dependent pathway in maintaining proper myotube structure.
Filamins are a family of actin-cross-linking proteins first purified by their ability to bind and precipitate actin (11). All three filamins—filamin A, B, and C—consist of an N-terminal actin-binding domain followed by 24 antiparallel β-sheet repeats and dimerize at the C terminus (20). Filamins, through their interactions with more than 30 diverse proteins, are involved in multiple processes, including cell-cell and cell matrix connections, mechanoprotection, and various signaling networks (8). Mutations in filamins A and B display a wide range of phenotypic diversity, further demonstrating the role of filamins in multiple pathways in multiple tissues (8, 18).
FilaminC (FLNc) expression is restricted to striated muscle (13, 24), and it has been shown to interact with many muscle proteins that, when mutated, give rise to muscular dystrophies (6). FLNc localization is abnormal in a number of muscle diseases (3, 19, 24), and a mutation in FLNc has recently been associated with a novel form of myofibrillar myopathy (28).
FLNc localizes both to the Z-disks of the sarcomeres and, in low amounts, to the sarcolemma of the muscle (24, 27). In the sarcomere, it interacts with multiple Z-disk proteins, including myotilin (27) and myozenins (7, 9, 22). At the sarcolemma, FLNc interacts with transmembrane proteins δ- and γ-sarcoglycan (24). Calpain-3, an intracellular cystine protease, cleaves FLNc and abolishes FLNc's binding to the sarcoglycans (10, 23). In patients with mutated γ-sarcoglycan and in Duchenne muscular dystrophy patients, where the membrane localization of sarcoglycans is lost, the amount of FLNc at the sarcolemma greatly increases (24), suggesting a dynamic role for FLNc in muscle. Although FLNc has been shown to interact with many crucial proteins in muscle, its specific functions are not clear. Its localization to the Z-disk in the early stages of myofibril formation has suggested a role in sarcomerogenesis (26). Recently, a dominant form of myofibrillar myopathy with clinical features of a late-onset limb girdle muscular dystrophy has been ascribed to a specific mutation in the FLNC gene (28). This nonsense mutation in the dimerization domain at the very C terminus of FLNc results in formation of massive FLNc aggregates. Abnormal aggregation of FLNc results in altered distribution of myotilin and the dystrophin-sarcoglycan complex and hence gives rise to myofibrillar degeneration (28).
To better understand the role of FLNc in muscle, we used siRNA (short interfering RNA) to “knock down” the expression of FLNc in the mouse myoblast cell line C2C12. Cell lines that do not express FLNc were able to fuse but could not form elongated myotubes. Moreover, these cells showed decreased fusion ability and altered expression of muscle-specific genes, indicating differentiation defects. To validate the importance of FLNc in vivo, we created a Flnc knockout model in mice by deleting the last 8 exons. This model shows that FLNc is critical for mouse viability with homozygous mutant pups dying from respiratory failure shortly after birth. The FLNc-deficient mice have less muscle mass than and a decreased number of primary fibers compared to control littermates, and their muscles exhibit excessive fiber size variation and centrally located nuclei. Similar to the in vitro phenotype, some of the muscle fibers appear shortened and rounded with loss of striated staining of sarcomeric proteins, suggesting disorganization of the muscle structure. Both in vitro and in vivo studies indicate that FLNc is critical for normal myogenesis as well as maintaining the structural integrity of the muscle fibers.
MATERIALS AND METHODS
Generation of siRNA expression constructs and C2C12 knockdown lines.
The DNA sequence for the mouse Flnc mRNA was obtained from the NCBI database using homology to the human Flnc mRNA (NM_001458). The partial mouse Trio mRNA (BC051169) sequence was obtained from the NCBI database (http://www.ncbi.nlm.nih.gov/). Potential sites for siRNA sequences were identified using the Dharmacon siDESIGN Center algorithm. The sequence sites targeted were used to design the hairpin sequence for insertion into the pSirenRetroQ vector (BD Biosciences).
The primers used were as follows (boldface letters indicate the mRNA sequence targeted by the shRNA): Flnc8F, 5′-GATCCGACGGTACCTGCAAAGTCATTCAAGAGATGACTTTGCAGGTACCGTCTTTTTTG-3′; Flnc8R, 5′-AATTCAAAAAAGACGGTACCTGCAAAGTCATCTCTTGAATGACTTTGCAGGTACCGTCG-3′; Trio2F, 5′-GATCCACACCAGAATGATGTTCGATTCAAGAGATCGAACATCATTCTGGTGTTTTTTTG-3′; Trio2R, 5′-AATTCAAAAAAACACCAGAATGATGTTCGATCTCTTGAATCGAACATCATTCTGGTGTG-3′.
Forward and reverse primers for the insert were synthesized, annealed, and ligated to the pSirenRetroQ vector using the EcoRI and BamHI restriction sites according to the manufacturer's protocol. The resulting plasmids were transfected into the 293E (ecotropic) packaging cell line using Lipofectamine 2000 reagent (Invitrogen). The resulting viruses were harvested after 2 days, passed through a 0.45-μm-pore-size filter to avoid any detached cells. C2C12 cells were plated on six-well plates (50,000/well), and a day later, the viruses were mixed with Polybrene and incubated with the cells for 15 min. The plates were centrifuged for 30 min at 37°C, 1,140 × g, and the viral supernatant was then removed. The next day, infected cells were plated in selection medium containing puromycin (2 μg/ml).
C2C12 cells were plated on gelatin-coated plates and grown in high-serum conditions (20% fetal bovine serum, 1% penicillin/streptomycin, Dulbecco's modified Eagle medium, and 2 μg/ml puromycin) as single-cell myoblasts. To induce differentiation, the cells were switched to low-serum conditions (2% horse serum, 1% penicillin/streptomycin in Dulbecco's modified Eagle medium without puromycin) when they reached 100% confluence.
Construction of Flnc targeting construct and screening.
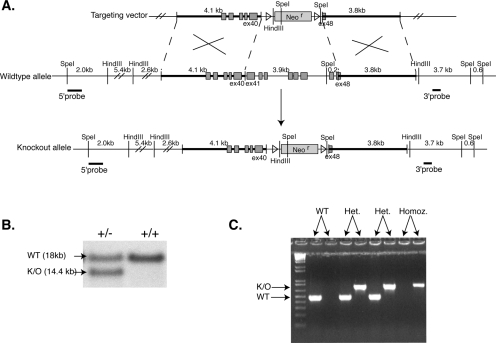
For the construction of the knockout mice, the services of Incyte Genomics and InGenious Targeting, Inc., were used. The sequence of the mouse Flnc exon 40 was obtained through the GenBank expression sequence tag database. The sequence of this expression sequence tag (accession no. AA139804) was used to design a PCR probe, which was used to screen a mouse 129SvJ BAC (bacterial artificial chromosome) library. A BAC containing exons 36 to 48 was obtained and sequenced. The 4.1-kb 5′ arm was cloned into the targeting vector using the SfiI and blunt-end Ecl136II restriction enzymes to digest the isolated BAC. For the 3.7-kb 3′ arm, the BAC was digested using Bsp120II restriction enzyme (see Fig. 4A). The homologous recombination of this targeting vector resulted in the deletion of exon 40 to exon 48 and replaced this region with a neomycin resistance gene. The targeting vector was linearized and electroporated into 129SvJ embryonic stem (ES) cells. The recombinant clones were selected by neomycin resistance. Three different probes were used for Southern blot analysis to confirm the correct targeting of the Flnc gene. The 5′ probe was a 0.7-kb SpeI/Acc65i fragment. Southern blot analysis of the SpeI-digested genomic DNA resulted in the 5′ probe to hybridize to an 18-kb fragment for the wild-type allele and a 14.4-kb fragment for the targeted allele (Fig. 4B). The 3′ probe was a 0.4-kb XbaI fragment, which hybridized to either a 7.8-kb or 7.4-kb fragment in the Southern blot analysis of the SpeI-digested genomic DNA, indicating the wild-type allele or the targeted allele, respectively. Finally, a probe for the neomycin resistance gene was used for Southern blot analysis of HindIII-digested genomic DNA, which hybridized to a 6-kb fragment in the targeted allele. Five successfully targeted clones were isolated. One such clones was chosen, injected into blastocysts, and implanted into the uterus of the pseudopregnant mice. The chimeras were mated to C57BL/6 mice.
FIG. 4.
Knockout scheme and genotyping strategy. (A) The last eight exons of the Flnc gene, ∼4.1 kb, are replaced by the neomycin resistance gene flanked by loxP sites. The targeting vector was linearized and electroporated into the 129SvJ ES cell line. ES cells that had successfully recombined out the last 8 exons of the Flnc gene were screened using three different Southern probes. (B) The 5′ probe when hybridized to SpeI-digested genomic DNA was expected to show an 18-kb fragment for the wild-type allele and a 14.4-kb allele for the targeted allele. The successfully targeted cell line shows both alleles, whereas the wild-type cells show only the 18-kb wild-type allele. (C) The mice generated were genotyped using PCR. The primers used to genotype the knockout allele amplified an 800-bp product; the primers for the wild-type (WT) allele gave rise to a 490-bp product.
DNA isolation and analysis of pups.
Small tail clips were taken from pups and incubated at 56°C in 500 μl of SLB buffer (100 mM Tris-HCl, pH 8.5, 5 mM EDTA, pH 8.0, 0.2% sodium dodecyl sulfate [SDS], 200 mM NaCl) with proteinase K overnight. The DNA was precipitated with 500 μl of isopropanol, washed with 70% ethanol, and then resuspended in 200 μl of sterile water. The following primer sets were used for PCRs: knockout allele, 5′-GGATGTAGTCTCCCTTCTCCTTGA-3′ and 5′-CCCGATTCGCAGCGCATCGCCTTC-3′; wild-type allele, 5′-GGATGTAGTCTCCCTTCTCCTTGA-3′ and 5′-CCCGTGACTGCCTACTCTGCCCTTC-3′. An annealing temperature of 59°C and an extension time of 2 min were used. The PCR products were then separated on a 1.2% Seakem agarose gel, stained with ethidium bromide, and visualized under UV light (see Fig. 4C).
Pathological analysis of embryos and neonates.
All animal studies used in the study were reviewed and approved by the Children's Hospital Boston Animal Care and Use Committee (protocol number A04-12-155R). The pups were fixed using Bouin's fixative for 6 days, transferred into 70% ethanol, and routinely embedded in paraffin, and 6-μm sections were taken and stained with hematoxylin and eosin. The sections were visualized on a Nikon Eclipse E1000 microscope, and images were gathered with a Spot insight color camera using Spot software 4.1.1.
RNA isolation and quantitative reverse transcription (RT)-PCR.
RNA was isolated using the QIAGEN RNeasy kit according to the manufacturer's protocol. Briefly, cells were lysed using RLT buffer with β-mercaptoethanol. The resulting lysates were homogenized by being passed through a Qiashredder column (QIAGEN). Frozen tissue samples were first lysed using a Dounce homogenizer in RLT buffer and further homogenized by being passed through a Qiashredder column. The RNA was then bound to the RNeasy column. To eliminate genomic DNA contamination, the samples were incubated with DNase using QIAGEN′s RNase-free DNase kit according to the manufacturer's protocol during RNA isolation.
First-strand cDNA was synthesized from 1 μg of total RNA using the Applied Biosystems TaqMan reverse transcription kit according to the manufacturer's protocol. Quantitative PCR was performed with 20 ng of cDNA using the SYBR green PCR master mix (Applied Biosystems) with the following primers and concentrations: FlncF, 5′-CTTCACGCGGAGCAGTCATACG-3′ (75 mM); FlncR, 5′-GTGGACTCCTCCACTCGGACTTC-3′ (300 mM); TbpF, 5′-CCGTGAATCTTGGCTGTAAACTTG-3′ (600 mM); TbpR, 5′-CAACGCAGTTGTCCGTGGCTCTCT-3′ (300 mM); Caveolin3F, 5′-GGACATTCACTGCAAGGAGATAGAC-3′ (150 mM); Caveolin3R, 5′-CTTCCATACACCGTCGAAGCTGTAG-3′ (300 mM); MyotilinF, 5′-CGGCTGCACGTTCCCACATCACAAG-3′ (300 mM); MyotilinR, 5′-CTCCTGGATGGCATCCTGATCATTG-3′ (150 mM); α7 integrinF, 5′-CATTGACCCCAGAGCCATCGATCTG-3′ (300 mM); α7 integrinR, 5′-CTGTAGCTGCTGGGCACAGCGACATAG-3′ (300 mM); TrioF, 5′-CTTCTGCCAGCCTGAGGGTTCTAG-3′(300 mM); TrioR, 5′-GCCAAGCTCAGCCACTTCACTGTAG-3′ (300 mM); HprtF, 5′-GGTGGAGATGATCTCTCAAC-3′ (300 mM); HprtR, 5′-CCAGGGAAAGCAAAGTTTGCATTG-3′ (75 mM); FlnaF, 5′-GTTGTCATCCAGGACCCTACAG-3′ (75 mM); FlnaR, 5′-GGCTGATAGCTACAGCGATAG-3′ (300 mM); FlnbF, 5′-GACATCGGCATTGAGGTGGAGGATC-3′ (75 mM); FlnbR, 5′-GGCTGCACTGGTTTGTAGACACATC-3′ (300 mM). The PCRs were performed, and the amount of product was measured on ABI Prism 7700 Sequence Detector. Amounts were then normalized to the amount of internal control (Tbp).
Protein isolation and Western blotting.
Total protein lysates were prepared from culture cells using lysis buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1% NP-40, 0.01% SDS, and 1× protease inhibitor cocktail) at 4°C for 15 min. The cellular lysates were scraped off and centrifuged at 16,000 × g for 10 min at 4°C. The supernatant was isolated and stored frozen at −80°C. To obtain protein extracts from tissue, cold pyrophosphate buffer (20 mM Na4P2O7, 20 mM NaH2PO4, 1 mM MgCl2, 1× protease inhibitor cocktail) was added to the tissue sample and homogenized using a Dounce homogenizer. The samples were then centrifuged for 10 min at 16,000 × g at 4°C, and supernatant was saved frozen at −80°C. Protein concentrations were measured using the Lowry assay (Bio-Rad).
Prior to electrophoresis, protein lysates were denatured and reduced using LCS loading buffer and sample reducing buffer (Invitrogen) and incubated at 70°C for 10 min. Proteins were separated by electrophoresis using 4 to 12% Bis-Tris gels with morpholinepropanesulfonic acid (MOPS) SDS running buffer (Invitrogen) according to molecular weight, and following electrophoresis, the proteins were transferred onto nitrocellulose membranes. To inhibit unspecific binding, the membranes were incubated for half an hour at room temperature with blocking solution (1× phosphate-buffered saline [PBS], 0.1% Tween 20, 5% [wt/vol] nonfat dry milk) and then incubated overnight at 4°C with primary antibodies diluted in blocking solution. The membranes were washed twice for 20 min with wash solution (1× PBS, 0.1% Tween 20) and incubated with horseradish peroxidase-conjugated donkey anti-mouse or anti-rabbit immunoglobulin G antibodies (diluted 1 in 10,000 in blocking solution) for 1 h at room temperature. The membranes were then incubated with chemiluminescence substrate (Pierce) for 5 min. The antibody protein complex was visualized with Kodak Biomax MR film developed in a Kodak XOMAT 2000A processor.
Antibodies.
Polyclonal FLNc-specific antibody (FLN2-A2, described by Thompson et al. [24]) diluted 1 in 2,000 (Western) or 1 in 100 (IF-C2C12) was used to detect FLNc. Mouse monoclonal α-actinin (clone EA-53; Sigma) was diluted 1 in 2,000 for Western analysis and 1 in 800 for immunofluorescence, and β-actin (ab6276; abcam) was diluted 1 in 10,000 and used as a loading control in Western blot analysis. Goat polyclonal anti-TRIO (D-20; Santa Cruz Biotechnology, Inc.) was diluted 1 in 50 for immunofluorescence and 1 in 250 for Western blot analysis. Mouse monoclonal antimyogenin (clone F5D; Abcam) and rabbit polyclonal anti-connexin43 (c6219; Sigma) antibodies were diluted 1 in 250 and 1 in 10,000 for Western blot analysis, respectively. Previously described antidystrophin rabbit polyclonal 6-10 (12) and mouse monoclonal anti-slow myosin (NOQ7.5.4D; Sigma) antibodies were diluted 1 in 1,000 for immunofluorescence analysis.
Tissue isolation and immunohistochemistry.
Mouse pups were sacrificed by decapitation and deskinned, and diaphragm and limb muscles were isolated. The tissues were embedded in OCT (TissueTek), snap-frozen in isopentane cooled in liquid nitrogen, and stored at −80°C. The frozen muscle samples were cut into 10-μm sections using a Microm HM 505E cryostat, placed on Superfrost-plus slides (VWR), and subsequently fixed in methanol for 3 min. The slides were incubated in a blocking solution (10% horse serum, 1× PBS, and 0.1% Triton X-100) at room temperature for at least 1 h to block nonspecific antibody binding. Primary antibodies were incubated with the slides overnight at 4°C and washed three times for 10 min in wash buffer (1× PBS, 0.1% Triton X-100). The primary antibody-protein complexes were detected using secondary antibodies, fluorescein isothiocyanate-conjugated donkey anti-rabbit and Texas Red-conjugated donkey anti-mouse antibodies, diluted 1 in 100 in blocking solution. Slides were incubated at room temperature for 2 h with the secondary antibodies. Slides were then washed three times for 10 min each with the wash solution (1× PBS, 0.1% Triton X-100). Coverslips were mounted using Vectashield mounting medium with 4′,6′-diamidino-2-phenylindole (DAPI; Vector Laboratories Inc.). Samples were visualized under a Nikon Eclipse E1000 fluorescent microscope, and images were obtained using Hamamatsu ORCA-ER digital camera and OpenLab 3.1.5 software.
Fiber number quantification.
Lower right hind limbs of the pups were isolated, embedded in OCT, and snap-frozen in isopentane chilled in liquid nitrogen. Ten-micrometer cross-sections through the hind limb were obtained and stained using dystrophin and slow myosin antibodies as described above. The percentage of primary fibers was calculated as the number of slow myosin-positive fibers divided by the total number of fibers (dystrophin positive) as described in reference (5).
Electron microscopy.
Whole limbs of mutant and wild-type embryonic day 18.5 (e18.5) mice were skinned and fixed overnight at 4°C in 100 mM cacodylate buffer with 1.25% formaldehyde, 2.5% glutaraldehyde, and 0.03% picric acid. Muscles were then postfixed in 1% osmium tetroxide-1.5% potassium ferrocyanide, stained with 1% uranyl acetate in maleate buffer (pH 5.2), and dehydrated in graded ethanol. Following dehydration, samples were incubated in propylene oxide and then infiltrated with a 1:1 mixture of Epon (Marivac Ltd.) and propylene oxide overnight before being embedded in fresh Epon. Eighty- to ninety-nanometer longitudinal muscle sections were mounted on Formvar/carbon-coated copper grids and stained with 2% uranyl acetate in acetone and then in 0.2% lead citrate. Sections were viewed with a JEOL 1200EX transmission electron microscope at 80 kV.
RESULTS
Cells with reduced Flnc mRNA start to differentiate normally but form multinucleated “myoballs.”
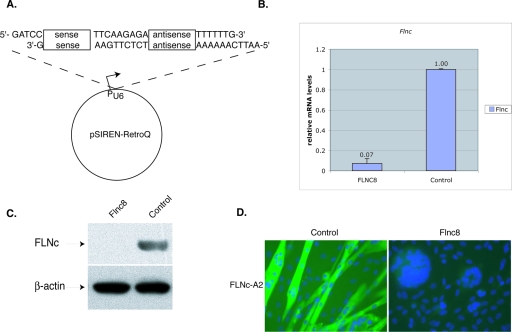
To study the function of FLNc in muscle development, RNA interference technology was used to specifically and stably knock down the expression of FLNc in C2C12 mouse myoblasts. The cells were infected with retroviral vectors containing either Flnc target sequences or a control sequence with no homology to known genes (Fig. 1A). The amount of reduced RNA expression was assessed by quantitative RT-PCR. The results were normalized to the expression of the endogenous TATA binding protein (Tbp) to control for cell number and loading. The amount of Flnc expression in selected lines was calculated as a percentage of the control cell line. One cell line, designated Flnc8, had Flnc mRNA levels reduced down to 7% (Fig. 1B). Both Western blot analysis (Fig. 1C) and immunohistochemistry using FLNc-specific antibodies on Flnc8 cells (Fig. 1D) confirmed near complete suppression of FLNc protein expression.
FIG. 1.
Reduction of Flnc mRNA by siRNA in the C2C12 cell line. (A) To knockdown FLNc expression, 19-bp target sequences were chosen. These sequences were used to design an inverted repeat insert and were cloned into pSirenRetroQ vector (Clontech). When the human U6 promoter is activated, this vector expresses a short hairpin RNA (shRNA). This plasmid was stably integrated into C2C12 cells using retroviral delivery. (B) The amount of Flnc knockdown was determined using quantitative RT-PCR. All of the amounts were first normalized to the endogenous Tbp (TATA binding protein) transcript levels and then to the control cell line, which expressed a shRNA that has no homology to any known mammalian sequences. The cells were allowed to differentiate for >3 days. Results shown are averages of those from five independent experiments, and the error bars represent the standard errors. The Flnc8 line shows a decrease of 93% in Flnc transcripts. (C) Ten micrograms of total protein isolated from the C2C12 cell lines was loaded onto 4 to 12% Bis-Tris gels (Invitrogen), and Western analysis was performed using FLNc-specific polyclonal antibodies. Antibodies against β-actin were used to control for even loading. The Flnc8 line has virtually no FLNc expressed. (D) Immunohistochemical analysis of FLNc expression with Flnc-A2 polyclonal antibodies (green). The Flnc8 line has virtually no FLNc protein detected, whereas in the control cell line, FLNc antibodies stain the fused fibers. Nuclei are labeled with DAPI (blue). Magnification, ×40.
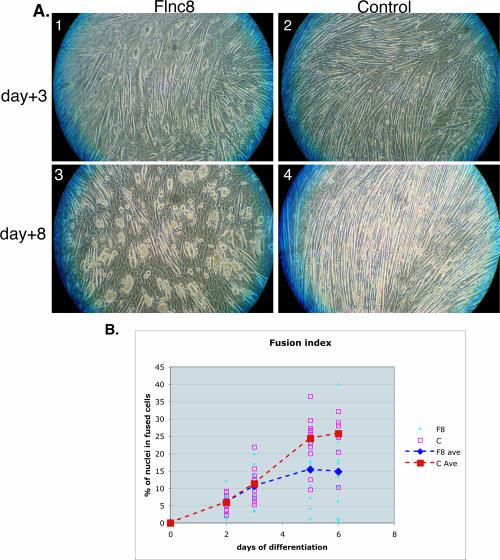
In high-serum conditions, C2C12 cells grow as single myoblasts. When placed under low-serum conditions, these cells differentiate by fusing to form myotubes. FLNc knockdown C2C12 cells, when switched to low-serum media, start differentiating normally, fuse, and start forming myotubes (Fig. 2A, top panels). However, after 3 days of differentiation, myotubes collapse to form multinucleated “myoballs” (Fig. 2A, bottom panels). These myoballs are still attached to the plate and twitch, indicating that the actin myosin structure is at least partially intact. The severity of the phenotype inhibits the study of the internal structures using immunohistochemical assays. The formation of myoballs is not specific to the type of extracellular matrix used for plating and differentiation of the cells. Various matrices, including collagen I, collagen IV, fibronectin, gelatin, and laminin, were tested as substrates, and the phenotype observed was the same (data not shown).
FIG. 2.
Cell morphology and fusion changes in cells lacking FLNc. (A) Switching the Flnc8 cell line to low-serum media induces differentiation and fusion normally (panel 1). After 3 days of low-serum conditions, the tubes collapse and form multinucleated balls (panel 3). The control line shows elongated multinucleated fibers throughout differentiation (panels 2 and 4). (B) The amount of fusion is determined by the number of nuclei in multinucleated myotubes divided by the total number of nuclei. At least 10 randomly chosen fields were counted. The average fusion index of Flnc8 (dashed blue line) is comparable to the control (dashed red line) until day 3, after which time the fusion index of Flnc8 is significantly lower (P < 0.01).
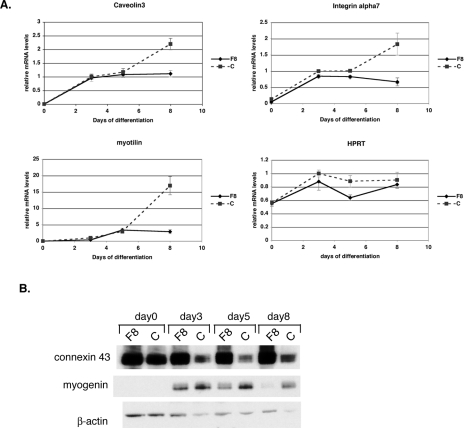
Fusion index of Flnc8 cultures, measured as a percentage of nuclei in multinucleated fibers, is similar to the control cells until day 3, after which they fuse at a slower rate (Fig. 2B). Finally, at day 5, the amount of fusion in Flnc8 cultures is significantly lower than in control cells (P < 0.05). As measured by quantitative RT-PCR, muscle protein expression in the control C2C12 line starts at low levels at the time differentiation is induced (day 0) and then significantly increases as differentiation continues (Fig. 3A). In contrast to the decrease in cell fusion, the expression of muscle-specific genes measured (caveolin3, myotilin, α7 integrin, β1D integrin, desmin, sarcoglycans, obscurin, myozenin-1) in the Flnc8 cell line remain at the same level as the control line until day 5. However, after day 5, there is no further increase in the expression of muscle markers in FLNc knockdown cells, while in control cells, their expression continues to increase (Fig. 3A and data not shown). Relative expression of the housekeeping gene Hprt shows no significant difference in the Flnc8 line, indicating that the decrease in transcription is specific to muscle genes (Fig. 3A). At day 5, FLNc knockdown cells have a lower fusion index than controls, while transcriptionally, they retain the same amount of gene expression, suggesting that failure of the cells to fuse is a primary consequence of FLNc loss rather than of secondary gene expression. Furthermore, the dramatic stall in transcription of muscle genes seen after day 5 does not appear to be a consequence of decreased fusion but is more likely to be due to a block in further differentiation. To check if the decreased amount of fusion is due to apoptosis in FLNc-deficient myotubes, a terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assay was performed on the FLNc-deficient versus control cells, and no significant difference in apoptosis was observed (data not shown).
FIG. 3.
Myogenic differentiation defects in cells lacking FLNc. (A) Quantitative RT-PCR was used to measure the amount of various muscle transcripts throughout cell differentiation. On day 0, cells were switched to the low-serum condition. All amounts were normalized to an internal control (Tbp) and then to the amount of transcription in control cells at day 3. The experiment was performed in triplicate, and the error bars represent the standard errors. The three muscle genes shown here, caveolin3, myotilin, and α7 integrin, have a very low expression on day 0 both for control (C) and Flnc8 (F8) cell lines. The expressions of all three genes increases in both cell lines until day 5, after which the transcript level in the control line (dashed line) continues to increase, whereas the Flnc8 (solid line) does not show any change. Constitutively expressed Hprt does not show a significant difference in expression between the control and the Flnc knockdown line, indicating that the stall in transcription is specific to muscle genes. (B) Ten micrograms of total protein isolated from the C2C12 cell lines throughout myogenic differentiation was loaded onto 4 to 12% Bis-Tris gels (Invitrogen), and Western analysis was performed using connexin43- and myogenin-specific antibodies. Antibodies against β-actin were used to control for even loading.
Myogenin is a transcription factor that regulates muscle differentiation (15). In control cells, myogenin is turned on at the onset of differentiation (Fig. 3B). In FLNc-deficient cells, myogenin is initially turned on, but as differentiation progresses, it is no longer expressed. Connexin43 is a gap junction protein known to be down regulated in C2C12 cells at the onset of muscle differentiation (17). While, in control cells, expression of connexin43 decreases as muscle differentiation proceeds, in FLNc-deficient cells, its expression remains elevated (Fig. 3B). This suggests that FLNc expression is critical for the decrease in connexin43 expression that occurs in normal muscle differentiation. Moreover, changes seen in myogenin and connexin43 expression appear around day 3 of differentiation, earlier than the muscle-specific transcriptional changes, indicating primary consequences of FLNc loss. The decrease in fusion, block in expression of muscle-specific genes, decrease in myogenin, and failure to repress connexin43 expression all indicate defects in myogenesis due to loss of FLNc protein in the C2C12 model system.
Flnc Δ41-48−/− mice are neonatally lethal.
To study the effect of FLNc loss in vivo, a FLNc-deficient mouse model was created. Since we expected a very severe phenotype in mice, we elected to create a partial knockout and delete the last 8 exons of the Flnc gene. FLNc protein has an actin-binding domain at its N terminus and 24 filamin-like repeats interspersed with a FLNc unique domain and a hinge domain. Exons 41 to 48 include the last five repeats of the FLNc protein and the hinge region but exclude the FLNc unique domain (see Fig. 5A). The deletion includes the region that interacts with the sarcoglycans (24) and myozenin (7, 22) and allows dimerization (16) as well as the hinge region that gets cleaved by calpain-3 (10, 23). Similar truncation mutations in FLNa have caused loss-of-function or partial-loss-of-function phenotypes in human patients, suggesting that this knockout scheme would generate, at minimum, a partial-loss-of-function model (8).
FIG. 5.
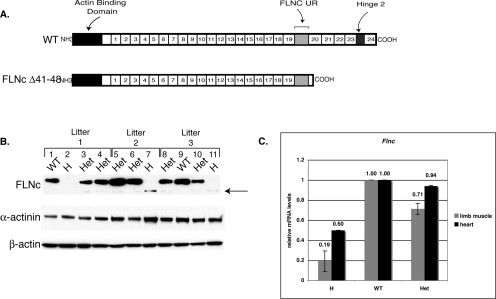
Truncated Flnc is expressed at low levels in the knockout mice. (A) The knockout is expected to have a truncated protein lacking the last four repeats and the hinge 2 region, but the FLNc unique region against which the FLNc antibodies are made is retained. (B) To determine the amount of truncated protein present, 15 μg of total lysates obtained from limbs of the pups was separated by SDS gel electrophoresis. The Western blot analysis was performed using Flnc-A2 antibodies raised against the unique region. Homozygotes (H) showed a faint band (arrow), whereas the heterozygotes (Het) showed both the wild-type (WT) and truncated alleles. Comparison of the bands in the heterozygotes indicates that the truncated protein is made at very low levels. Western blot analysis with β-actin and sarcomeric α-actinin was performed as a loading control. (C) The amount of Flnc transcript was determined using quantitative RT-PCR. Total RNA was isolated from limb muscles and hearts of homozygous (n = 3 and n = 2), heterozygous (n = 3 and n = 2), and wild-type (n = 1) littermates. The amount of mRNA was first normalized to endogenous Tbp expression in the control and then to the amount of Flnc transcript present in the wild-type animal. The error bars represent the standard errors of the means.
A targeting vector with a deletion of a 4.5-kb region containing the genomic sequences between exons 41 to the stop codon in exon 48 was created (Fig. 4A). Correct targeting of the region in ES cells was assessed by Southern analysis using 5′, 3′, and neo probes (Fig. 4B and data not shown). One of five ES clones found to carry the correct insert was injected into blastocysts and implanted into the uterus of the pseudopregnant mice. The resulting chimeras were mated to C57BL/6 mice, and the heterozygotes were identified by PCR (Fig. 4C).
To see if truncated protein is expressed, Western blot analysis was performed on muscle protein lysates obtained from limbs. Previously described FLNc-A2 antibodies were raised against a peptide at the unique region therefore able to recognize truncated protein (Fig. 5A) (24). Western blot analysis shows that truncated protein is made at very low levels (Fig. 5B). The levels of truncated protein vary between litters (lanes 2, 7, and 11) as well as within a litter (compare lanes 3 and 4). To determine if the expression of truncated FLNc was reduced at the mRNA or protein level, mRNA amounts of Flnc were measured using quantitative RT-PCR. Primers were designed against exon 40 retained in the knockout allele. Total RNA was isolated from limb muscles as well as from the heart, and results were normalized to the expression of endogenous TATA binding protein (Tbp) to control for equal loading. The amount of Flnc expression was calculated as a percentage of that of wild-type littermates. In limb muscles, Flnc transcript levels are reduced to 20% of the wild-type levels, indicating that truncated FLNc is unstable at the mRNA level (Fig. 5C). Truncated Flnc mRNA levels are also reduced in the heart tissue but to a lesser extent (Fig. 5C).
When heterozygous Flnc Δ41-48 mice were mated to each other, no homozygous pups were found by 3 weeks of age, indicating that the homozygotes were dying either in utero or shortly after birth (Table 1). Timed matings were performed, and pups were harvested at various stages of development. Homozygous mice were observed from e12.5 until birth, indicating that Flnc Δ41-48−/− mice were able to develop to term (Table 1). All newborn (P0) homozygous mice were found dead at birth (Fig. 6A and Table 1). To further diagnose the time of death of the homozygous mutants, Cesarean sections were performed on the pregnant mice at e18.5, and the embryos were removed, cleaned, and resuscitated. The homozygous mice were able to take several short breaths but died shortly after birth, whereas the heterozygous and wild-type mice survived (Table 1). Moreover, the homozygotes were less active and, even when stimulated to move, they responded very little compared to the wild-type and heterozygote littermates (see Movie S1 in the supplemental material).
TABLE 1.
Genotyping results of matings between heterozygotesa
| Age | No. of surviving mice
|
||
|---|---|---|---|
| Homozygote | Heterozygote | Wild type | |
| >3 wk | 0 | 112 | 52 |
| P0 | 22 (0/22) | 62 (61/62) | 28 (28/28) |
| e18.5 | 34 (0/22) | 99 (64/69) | 32 (16/16) |
| e17.5 | 10 | 23 | 11 |
| e16.5 | 14 | 26 | 16 |
| e14.5 | 3 | 4 | 3 |
| e12.5 | 3 | 3 | 2 |
The genotyping results of the litters in matings between heterozygotes. No homozygotes were found among animals more than 3 weeks old. To determine the time of death of the pups, timed matings were performed. The numbers in parentheses indicate ratios of pups that were found alive (P0) or survived after resuscitation (e18.5). Although heterozygote mice appear to be overrepresented in e18.5 pups, it is still within a range consistent with Mendelian distribution of a single locus (P > 0.025).
FIG. 6.
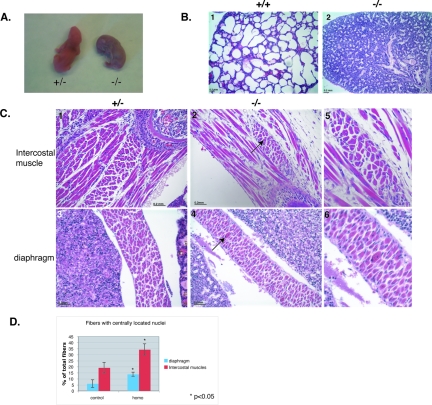
Flnc knockout mice die at birth, and muscles are myopathic. (A) All homozygous mice died within 10 min of birth and turned blue, whereas the control mice pinked up and continued breathing. (B) Hematoxylin (blue) and eosin (red) staining of the paraffin-embedded sections of mouse pups taken at ×10 magnification. The lungs of the homozygous mutants were not inflated (panel 2) compared to those of the wild-type littermates (panel 1). (C) Hematoxylin (blue, nuclei) and eosin (red, cytoplasm) staining of homozygous and control mice taken at ×20 magnification. The knockout mice have fewer muscle fibers and exhibit fibrosis. Muscles involved in respiration, such as intercostals, are severely decreased and show infiltration of connective tissue in homozygotes (panel 2) compared to the control littermates (panel 1). Other muscles, such as the diaphragm, show less drastic differences (panel 4 compared to panel 5) but still show centrally located nuclei (panel 5, arrow). Panels 3 and 6 show the magnified images of the centrally located nuclei in these muscles. (D) The percentage of the fibers with centrally located nuclei was counted in control animals (n = 2) and homozygous mutant animals (n = 3) in three different intercostal muscles and two random fields in the diaphragm. The percentage of fibers with centrally located nuclei was significantly increased in homozygous mutant mice (P < 0.05). The error bars represent the standard errors.
Analysis of the lungs of the newborns demonstrated that the lungs of the homozygotes were not inflated (Fig. 6B, panel 2) compared to the controls (panel 1), indicating a respiratory problem likely leading to the death of the neonates.
Flnc Δ41-48−/− mice show defects in skeletal muscle.
FLNc is expressed predominantly in the cardiac and skeletal muscle (24, 26, 29). To see how much these tissues are affected by the loss of FLNc, we harvested neonates and examined muscles microscopically. While the hearts of the homozygotes appeared normal and unaffected, skeletal muscles were found to have severe abnormalities compared to the littermate controls. Muscle fibers showed centrally located nuclei (Fig. 6C, panels 2 and 3) and infiltration of connective tissue. Some muscles responsible for respiration, such as intercostal muscles, were especially affected (Fig. 6C, panel 2). The diaphragm was less severely affected, but fibers with centrally located nuclei were still observed (Fig. 6C, panels 5 and 6). In fact, the percentage of fibers with centrally located nuclei was significantly increased both in intercostal and diaphragm muscles (Fig. 6D).
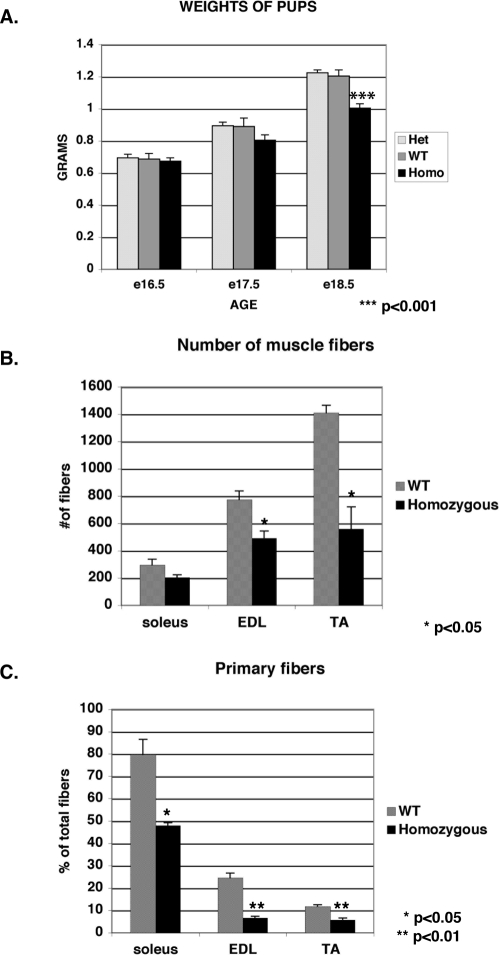
Many muscle groups showed decreased muscle mass, including trunk, limb, intercostal, tongue, extraocular, and facial muscles. The decrease in muscle mass correlated with the lower overall weight of the pups (Fig. 7A). Homozygotes were comparable to wild-type pups in weight at e16.5 but show a significantly lower weight just before birth (P < 0.001). To determine if the decrease in muscle mass was due to lower number of fibers or decrease in fiber mass, the lower hind limbs of pups at e18.5 were isolated, and the number of muscle fibers were determined in three hind-limb muscles: soleus, tibialis anterior (TA), and extensor digitorum longus (EDL). In all three muscle groups, the number of muscle fibers was reduced compared to the wild-type littermates, suggesting that the decrease in muscle mass and weight is due to the lower number of muscle fibers (Fig. 7B). To determine if this significant decrease in fiber number was due to increased apoptosis, TUNEL assays were performed on these lower hind limb muscles. No significant increase in the number of fibers with apoptotic nuclei was seen in the FLNc-deficient mice, indicating that the lower fiber number was due to problems in fiber differentiation as opposed to fiber wasting (data not shown).
FIG. 7.
The FLNc-deficient mice exhibit decreased muscle mass and defects in primary myogenesis. (A) The homozygotes are comparable in weight at embryonic day 16.5 but fall behind as the animals increase in weight until birth. The homozygotes weigh significantly less then the wild-type (WT) and heterozygous counterparts at embryonic day 18.5 (P < 0.001). (B) To determine if the decrease in muscle mass was due to decreased number of fibers, the numbers of fibers were counted in three different lower hind limb muscles (soleus, EDL, TA) at embryonic day 18.5. The number of muscle fibers was significantly reduced in the homozygotes (n = 3) compared to the wild-type littermates (n = 3) in the EDL and TA muscles. (C) The percentage of primary fibers was also significantly reduced for all three muscles in homozygotes. The error bars indicate the standard errors of the means.
Myogenesis occurs in two phases during embryogenesis (4). Primary myogenesis starts at e12 and is followed by secondary myogenesis, starting around e15 (4). To determine whether the decrease in fiber number was due to selective loss of either primary or secondary fibers, the percentage of primary fibers was determined in the three hind- limb muscles. The number and the percentage of primary fibers in these muscles were significantly reduced compared to the wild-type littermates, indicating a defect in primary myogenesis in these mice (Fig. 7C).
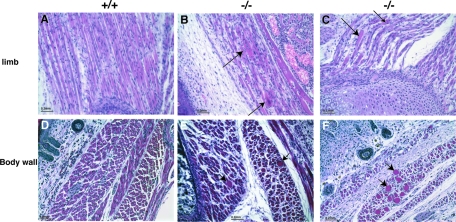
A comparison of cross-sections showed that the Flnc knockout muscles had great variation in fiber diameter and large fibers with centrally located nuclei (Fig. 8, panels E and F). Moreover, rounded fibers with centrally located nuclei were observed (Fig. 8, panels B and C). These fibers resembled the in vitro phenotype seen in FLNc knockdown C2C12 line when examined longitudinally (Fig. 8, panels B and C and 9A).
FIG. 8.
Abnormal muscle fiber pathology in Flnc knockout mice. Hematoxylin (blue, nuclei) and eosin (pink, cytoplasm) staining of muscles was visualized at ×20 magnification. Longitudinal sections show rounded fibers with centrally located nuclei (panels B and C, arrows). Cross-sections show vast variations in fiber size with giant fibers (panels E and F, arrows).
Since FLNc has been shown to interact with a variety of muscle proteins, the effect of loss of FLNc on the expression and specific localization of these proteins was tested. Immunofluorescence analysis demonstrated that the membrane staining of dystrophin, δ-sarcoglycan, and caveolin-3 was unchanged in the knockout mice (data not shown). Most fibers showed the striated staining of many sarcomeric proteins, such as titin, obscurin, actinins, myozenin-1, myotilin, and myosin (data not shown). However, sarcomeric staining in the rounded fibers was disorganized, indicating that the internal structure is disturbed (Fig. 9B). In fact, some fibers show diffuse staining of α-actinin in the some areas (Fig. 9B, panel 1), whereas the rest of the fiber retains some striated α-actinin staining (Fig. 9B, panels 1 and 2).
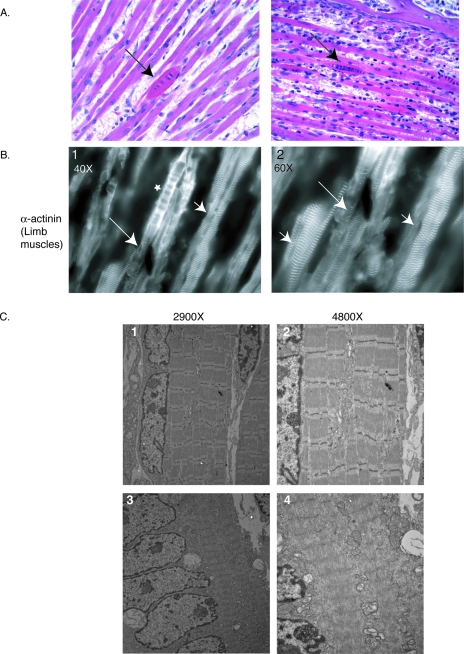
FIG. 9.
Rounded fibers in FLNc-deficient mice. (A) Hematoxylin and eosin staining of facial muscles showing rounded fibers (arrows) among unaffected fibers in mutant mice. (B) ×40 and ×60 magnifications of the α-actinin staining in limb muscles from a homozygous mouse show expected striated staining in many fibers (panels G and H, arrowheads) as well as diffuse staining in rounded fibers (panel G, marked by a star). The specific fiber in this field is half disorganized, with areas of striation still retained (panels G and H, marked by arrows). (C) Electron micrographs of muscles from forelimbs of mutant mice. Most of the muscle fibers exhibit normal sarcomeres (top panels). The rounded fibers with centrally located nuclei show extensive thick and thin filament crossover and loss of distinct Z-lines (bottom panels).
When examined ultrastructurally, most of the fibers in mutant mice exhibit normal sarcomeric structure (Fig. 9C, top panels). The few rounded and centrally nucleated fibers, however, show sarcomeres with thin and thick filaments overlapping without distinct Z-lines (Fig. 9C, bottom panels).
Trio-deficient mice and myoblasts exhibit similar phenotypes.
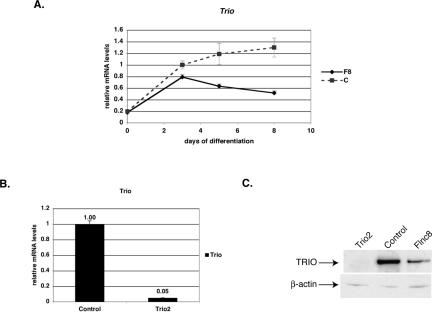
TRIO, a Dbl homology guanine nucleotide exchange factor family protein, when deleted in mice, gives rise to a late embryonic lethality with abnormalities in neural development as well as problems in late myogenesis and accumulation of spherical myotubes (14). Similar to FLNc-deficient mice, some muscle fibers of Trio knockout mice exhibit shortened and round morphology. TRIO is a guanine nucleotide exchange factor involved in activating the Rho family of small GTPases and is involved in axon guidance and cell migration (1). TRIO has been shown to interact with FLNa and be involved in actin remodeling in membrane ruffle formations (2). To determine if the level of TRIO was changed in FLNc-deficient cells, the amount of Trio transcript was quantitated and found to be highly decreased in FLNc-deficient cells (Fig. 10A). Moreover, unlike other muscle transcripts where the transcription levels were comparable to the control cells until 5 days of differentiation, the Trio levels were lower even at 3 days after differentiation and then showed a decline (Fig. 10A).
FIG. 10.
Trio expression is lowered in FLNc-deficient cells. (A) Quantitative RT-PCR analysis of Trio transcript levels in FLNc-deficient cells (F8) versus control cells throughout myogenic differentiation. On day 0, cells were switched to low-serum conditions. All amounts were normalized to an internal control (Tbp) and then to the amount of transcription in control cells at day 3. The data shown are the averages of results from three experiments, and error bars represent standard errors. (B) The amount of Trio knockdown was determined using quantitative RT-PCR. All of the amounts were first normalized to the endogenous Tbp (TATA binding protein) transcript levels and then to the control cell line. The cells were allowed to differentiate for 8 days. Experiments were performed in triplicate, and the error bars represent standard errors. The Trio2 line shows a decrease of 95% in Trio transcripts. (C) Ten micrograms of total protein isolated from the C2C12 cell lines was loaded onto 3 to 8% Tris-acetate gels (Invitrogen), and Western analysis was performed using anti-TRIO antibodies. Antibodies against β-actin were used to control for even loading. The Trio2 line has virtually no TRIO protein expressed.
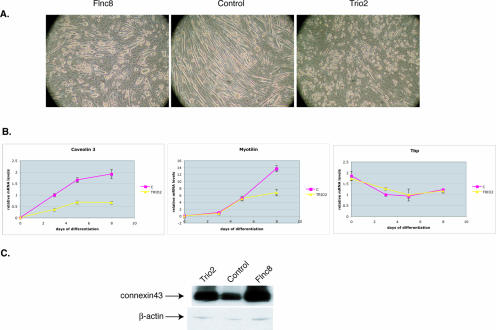
To see if loss of TRIO in C2C12 cells would result in a similar fiber phenotype, a TRIO-deficient C2C12 cell line was created using short interfering RNAs. Quantitative PCR analysis revealed that the Trio mRNA levels were reduced to 5% of that of the control cell line (Fig. 10B). Western blot analysis of the total protein lysate obtained from Trio-deficient cells confirmed the virtual loss of TRIO protein expression (Fig. 10C). These cells were able to grow normally in the single-cell stage, but when switched to low-serum conditions, they formed multinucleated “myoballs,” similar to the morphology exhibited by FLNc-deficient C2C12 cells (Fig. 11A). Similar to FLNc-deficient cells, the transcription of muscle-specific genes was decreased in TRIO-deficient cells (Fig. 11B) and expression of the gap junction protein connexin43 remained elevated (Fig. 11C). These results suggest that TRIO-deficient cells are similar not only in the morphological phenotype to the FLNc-deficient cells but also in differentiation phenotype, further suggesting the involvement of TRIO and FLNc in a common pathway in skeletal muscle.
FIG. 11.
Phenotype of TRIO-deficient cells. (A) TRIO-deficient cells when switched to low-serum media formed multinucleated myoballs similar to FLNc-deficient cells. (B) Quantitative RT-PCR analysis of muscle-specific transcripts caveolin3 and myotilin in TRIO-deficient cells (Trio2) versus the control cell line (C). Constitutively expressed Tbp does not show a significant difference in expression between the control and the Trio knockdown line, indicating that the change in transcription is specific to muscle genes. Day 0 is when the cells were switched to the low-serum condition. All amounts were normalized to an internal control (Hprt) and to the amount of transcription in control cells at day 3. The experiment was performed in triplicate, and error bars represent standard errors. (C) Connexin43 expression is increased in Trio-deficient cells similar to the FLNc-deficient cells. Ten micrograms of total protein, isolated from the C2C12 cell lines, was loaded onto 4 to 12% Bis-Tris gels (Invitrogen), and Western analysis was performed using connexin43-specific antibodies. Antibodies against β-actin were used to control for even loading.
DISCUSSION
It has been previously demonstrated that FLNc interacts with proteins important in normal muscle development, and a mutation in FLNC in humans results in myofibrillar myopathy. However, still little is known about the function of FLNc in muscle. This study shows that FLNc has a role in two distinct aspects of muscle physiology: muscle differentiation and maintenance of elongated muscle fiber structure.
Two approaches were undertaken to study the effects of loss of filaminC in muscle. First, a C2C12 cell line with virtually no FLNc protein expression was created. FLNc loss in vitro also results in a severe morphological phenotype. The cells start differentiation normally, but as differentiation progresses, fusion slows and expression of transcriptional markers of differentiation prematurely plateau, indicating a block of the later stages of differentiation. Furthermore, myotubes do not maintain an elongated tube morphology and collapse to form large multinucleated “balls.” Similar to this phenotype, in FLNc-deficient mice harboring a deletion of the last eight exons of the Flnc gene, few short, rounded fibers with centrally located nuclei and disorganized sarcomeric structure are observed. These mice were born alive but appear to have difficulty breathing, are not able to move as much as the control littermates, and quickly die due to respiratory failure. The histopathological analysis of these mice revealed decreased muscle mass, increased numbers of fibers with centrally located nuclei, and variability in myofiber size.
This study suggests that the role of FLNc in muscle has two components. First, loss of FLNc results in defects in primary myogenesis evidenced by a decrease in the overall number of muscle fibers and specifically by a decrease in the ratio of primary muscle fibers to secondary muscle fibers in the knockout mice. This is further supported by the differentiation defects seen in the knockdown C2C12 cell line. Another critical role for FLNc in muscle development is in the maintenance of elongated muscle morphology. Both in vivo and in vitro, loss of FLNc expression results in the formation of rounded muscle fibers. In vivo, most muscles show a normal sarcomeric structure and normal staining of sarcomeric proteins, indicating that loss of FLNc does not affect sarcomerogenesis. In contrast, FLNc plays an essential role in maintaining muscle cell structure.
This is consistent with our understanding of the function of other members of the filamin family. Filamins are actin-binding proteins, first isolated by their ability to cross-link and precipitate purified actin (21). The filamins are implicated in multiple pathways, such as cell motility and signaling, and mutations in filamin A and B have been shown to cause neuronal migration defects as well as abnormal development of the brain, bone, and cardiovascular system (8). It has been shown that overexpression of specific isoforms of FLNb results in increased fusion in C2C12 (25). This paper shows that loss of FLNc in C2C12 cells decreases fusion and the Flnc-deficient mouse exhibits less muscle, reinforcing the hypothesis that filamins are involved in differentiation of the muscle cells. Moreover van der Flier et al. had seen differences in the morphology of the C2C12 myotubes: cells overexpressing the FLNb isoform exhibited thinner myotubes (25). The opposite phenotype is seen in cells lacking FLNc, where there are rounded myofibers in the mouse and myoballs in the C2C12 lines.
FLNc has been shown to interact with many muscle proteins involved in muscular dystrophies. No localization differences were seen in the interacting proteins analyzed, including myotilin, sarcoglycans, dystrophin, and myozenin, in fibers that had not collapsed. This would indicate that FLNc does not have a role in the expression and localization of these proteins. Rather, FLNc is likely involved in integrating signals from these proteins. A possible role for FLNc in signaling is not surprising when compared to the functions of FLNa and FLNb, which have been shown to be involved in many cellular signaling processes (20).
The aberrant fiber morphology found in FLNc-deficient mice resembles the muscle morphology of TRIO-deficient mice. TRIO, a Dbl homology guanine nucleotide exchange factor family protein, when deleted in mice, gives rise to a late embryonic lethality with abnormalities in neural development as well as problems in late myogenesis and accumulation of spherical myotubes without any cardiac defects (14). TRIO activates multiple Rho GTPases and is involved in axon guidance and cell migration (1). TRIO has been shown to interact with FLNa and be involved in actin remodeling in membrane ruffle formations (2). To further reinforce the similarities between TRIO- and FLNc-deficient muscle, a C2C12 model for the loss of TRIO was created. These cells, when directed to differentiate, formed round multinucleated myotubes similar to FLNc-deficient cells. They also showed similar decreases in expression of muscle-specific genes as well as changes in differentiation markers. The similarity between the skeletal muscle defect seen in TRIO-deficient mice and C2C12 cells and the Flnc-deficient mice and C2C12 cells would suggest that this phenotype of rounded fibers is a result of the perturbation of a pathway common to TRIO and filaminC. Conversely, it would also imply that the skeletal muscle defects in Trio−/− mice are due to perturbation of TRIO-FLNc pathway and that the neuronal defects are due to perturbation of the TRIO-FLNa pathway.
The mice described in this paper have the last eight exons of the Flnc gene deleted. Both the heterozygotes and homozygotes express the truncated protein but at very low levels. The heterozygotes do not exhibit any abnormal phenotype related to the presence of this truncated form, indicating that this deletion is a hypomorphic or null mutation. Therefore, one would expect a full knockout to have either an earlier embryonic lethality or a similar neonatal lethality phenotype. The mutation seen in some myofibrillar myopathy patients is a nonsense mutation in the last β-sheet repeat, involved in proper dimerization of FLNc. The FLNc in these patients forms large aggregates and hence results in muscle degeneration. We believe this is a gain-of-function mutation in which aggregation of FLNc due to the perturbed dimerization results in accumulation of a number of muscle proteins and therefore gives rise to disease phenotype. The specific nature of this mutation demonstrates the toxicity of the FLNc aggregates; however, it does not explain the function of FLNc in muscle.
Both the in vivo and the in vitro phenotypes of cells lacking FLNc indicate that FLNc is essential in muscle development and viability. FLNc has been shown to interact with numerous muscle proteins that are crucial for muscle membrane integrity and when mutated give rise to muscular dystrophies, thus suggesting the FLNC gene as a strong candidate for muscular dystrophies with unknown causes. Loss-of-function mutations in FLNC would likely result in a very severe congenital or even an embryonic lethality phenotype. Different mutations in nonmuscle filamins have been found to selectively perturb specific systems that result in different phenotypes. Similarly, the dimerization domain mutation in FLNC gives rise to a late-onset myofibrillar myopathy, and the loss-of-function mutation in mice give rise to severe neonatal lethality. FLNc is also expressed to a lesser extent in the cardiac muscle. The neonatal mice do not show a cardiac defect but this does not eliminate the possibility of later onset cardiac phenotypes. We therefore hypothesize that various different mutations in FLNC in humans would give rise to phenotypes ranging from severe congenital myopathies to late-onset myofibrillar myopathies to cardiac diseases.
FilaminC is an actin binding protein mainly expressed in skeletal muscle and, to a lesser extent, in cardiac muscle and has been shown to interact with crucial muscle proteins and mislocalized in a number of muscle diseases. Since the only mutation in humans that has been observed so far has been a gain-of-function mutation, it has not been clear previously how important a role FLNc plays in muscle physiology. This is the first comprehensive study revealing information regarding the normal function of FLNc in muscle. The two approaches taken in this study demonstrate that FLNc has an important role in primary myogenesis and muscle differentiation and is essential for the viability of mice. Moreover, the presence of abnormally rounded fibers in both in vivo and in vitro models indicates a novel role for FLNc in the maintenance of myofiber structure.
Supplementary Material
Acknowledgments
We thank Forrester J. Liddle and the members of the Kunkel lab for useful discussion of the manuscript. We also thank Roderick Bronson and Hart Lidov for technical and analytical help in pathological analysis of the mice.
L.M.K. is an investigator of the Howard Hughes Medical Institute. Funding for this work was generously provided by the Bernard F. and Alva B. Gimbel Foundation (L.M.K.) and by an NIH Neurological Disorders and Stroke grant (L.M.K.) (P01NS040828). I.D. is supported in part by the Albert J. Ryan Foundation.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Bateman, J., and D. Van Vactor. 2001. The Trio family of guanine-nucleotide-exchange factors: regulators of axon guidance. J. Cell Sci. 114:1973-1980. [DOI] [PubMed] [Google Scholar]
- 2.Bellanger, J. M., C. Astier, C. Sardet, Y. Ohta, T. P. Stossel, and A. Debant. 2000. The Rac1- and RhoG-specific GEF domain of Trio targets filamin to remodel cytoskeletal actin. Nat. Cell Biol. 2:888-892. [DOI] [PubMed] [Google Scholar]
- 3.Bonnemann, C. G., T. G. Thompson, P. F. van der Ven, H. H. Goebel, I. Warlo, B. Vollmers, J. Reimann, J. Herms, M. Gautel, F. Takada, A. H. Beggs, D. O. Furst, L. M. Kunkel, F. Hanefeld, and R. Schroder. 2003. Filamin C accumulation is a strong but nonspecific immunohistochemical marker of core formation in muscle. J. Neurol. Sci. 206:71-78. [DOI] [PubMed] [Google Scholar]
- 4.Buckingham, M., L. Bajard, T. Chang, P. Daubas, J. Hadchouel, S. Meilhac, D. Montarras, D. Rocancourt, and F. Relaix. 2003. The formation of skeletal muscle: from somite to limb. J. Anat. 202:59-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cachaco, A. S., S. M. Chuva de Sousa Lopes, I. Kuikman, F. Bajanca, K. Abe, C. Baudoin, A. Sonnenberg, C. L. Mummery, and S. Thorsteinsdottir. 2003. Knock-in of integrin beta 1D affects primary but not secondary myogenesis in mice. Development 130:1659-1671. [DOI] [PubMed] [Google Scholar]
- 6.Dalkilic, I., and L. M. Kunkel. 2003. Muscular dystrophies: genes to pathogenesis. Curr. Opin. Genet. Dev. 13:231-238. [DOI] [PubMed] [Google Scholar]
- 7.Faulkner, G., A. Pallavicini, A. Comelli, M. Salamon, G. Bortoletto, C. Ievolella, S. Trevisan, S. Kojic, F. Dalla Vecchia, P. Laveder, G. Valle, and G. Lanfranchi. 2000. FATZ, a filamin-, actinin-, and telethonin-binding protein of the Z-disc of skeletal muscle. J. Biol. Chem. 275:41234-41242. [DOI] [PubMed] [Google Scholar]
- 8.Feng, Y., and C. A. Walsh. 2004. The many faces of filamin: a versatile molecular scaffold for cell motility and signalling. Nat. Cell Biol. 6:1034-1038. [DOI] [PubMed] [Google Scholar]
- 9.Frey, N., and E. N. Olson. 2002. Calsarcin-3, a novel skeletal muscle-specific member of the calsarcin family, interacts with multiple Z-disc proteins. J. Biol. Chem. 277:13998-14004. [DOI] [PubMed] [Google Scholar]
- 10.Guyon, J. R., E. Kudryashova, A. Potts, I. Dalkilic, M. A. Brosius, T. G. Thompson, J. S. Beckmann, L. M. Kunkel, and M. J. Spencer. 2003. Calpain 3 cleaves filamin C and regulates its ability to interact with gamma- and delta-sarcoglycans. Muscle Nerve 28:472-483. [DOI] [PubMed] [Google Scholar]
- 11.Hartwig, J. H., and T. P. Stossel. 1975. Isolation and properties of actin, myosin, and a new actin binding protein in rabbit alveolar macrophages. J. Biol. Chem. 250:5696-5705. [PubMed] [Google Scholar]
- 12.Lidov, H. G., T. J. Byers, S. C. Watkins, and L. M. Kunkel. 1990. Localization of dystrophin to postsynaptic regions of central nervous system cortical neurons. Nature 348:725-728. [DOI] [PubMed] [Google Scholar]
- 13.Maestrini, E., C. Patrosso, M. Mancini, S. Rivella, M. Rocchi, M. Repetto, A. Villa, A. Frattini, M. Zoppe, P. Vezzoni, et al. 1993. Mapping of two genes encoding isoforms of the actin binding protein ABP-280, a dystrophin like protein, to Xq28 and to chromosome 7. Hum. Mol. Genet. 2:761-766. [DOI] [PubMed] [Google Scholar]
- 14.O'Brien, S. P., K. Seipel, Q. G. Medley, R. Bronson, R. Segal, and M. Streuli. 2000. Skeletal muscle deformity and neuronal disorder in Trio exchange factor-deficient mouse embryos. Proc. Natl. Acad. Sci. USA 97:12074-12078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parker, M. H., P. Seale, and M. A. Rudnicki. 2003. Looking back to the embryo: defining transcriptional networks in adult myogenesis. Nat. Rev. Genet. 4:497-507. [DOI] [PubMed] [Google Scholar]
- 16.Pudas, R., T. R. Kiema, P. J. Butler, M. Stewart, and J. Ylanne. 2005. Structural basis for vertebrate filamin dimerization. Structure (Cambridge) 13:111-119. [DOI] [PubMed] [Google Scholar]
- 17.Reinecke, H., and C. E. Murry. 2000. Transmural replacement of myocardium after skeletal myoblast grafting into the heart. Too much of a good thing? Cardiovasc. Pathol. 9:337-344. [DOI] [PubMed] [Google Scholar]
- 18.Robertson, S. P. 2005. Filamin A: phenotypic diversity. Curr. Opin. Genet. Dev. 15:301-307. [DOI] [PubMed] [Google Scholar]
- 19.Sewry, C. A., C. Muller, M. Davis, J. S. Dwyer, J. Dove, G. Evans, R. Schroder, D. Furst, T. Helliwell, N. Laing, and R. C. Quinlivan. 2002. The spectrum of pathology in central core disease. Neuromuscul. Disord. 12:930-938. [DOI] [PubMed] [Google Scholar]
- 20.Stossel, T. P., J. Condeelis, L. Cooley, J. H. Hartwig, A. Noegel, M. Schleicher, and S. S. Shapiro. 2001. Filamins as integrators of cell mechanics and signalling. Nat. Rev. Mol. Cell Biol. 2:138-145. [DOI] [PubMed] [Google Scholar]
- 21.Stossel, T. P., and J. H. Hartwig. 1975. Interactions between actin, myosin, and an actin-binding protein from rabbit alveolar macrophages. Alveolar macrophage myosin Mg-2+-adenosine triphosphatase requires a cofactor for activation by actin. J. Biol. Chem. 250:5706-5712. [PubMed] [Google Scholar]
- 22.Takada, F., D. L. Vander Woude, H. Q. Tong, T. G. Thompson, S. C. Watkins, L. M. Kunkel, and A. H. Beggs. 2001. Myozenin: an alpha-actinin- and gamma-filamin-binding protein of skeletal muscle Z lines. Proc. Natl. Acad. Sci. USA 98:1595-1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taveau, M., N. Bourg, G. Sillon, C. Roudaut, M. Bartoli, and I. Richard. 2003. Calpain 3 is activated through autolysis within the active site and lyses sarcomeric and sarcolemmal components. Mol. Cell. Biol. 23:9127-9135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson, T. G., Y. M. Chan, A. A. Hack, M. Brosius, M. Rajala, H. G. Lidov, E. M. McNally, S. Watkins, and L. M. Kunkel. 2000. Filamin 2 (FLN2): a muscle-specific sarcoglycan interacting protein. J. Cell Biol. 148:115-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van der Flier, A., I. Kuikman, D. Kramer, D. Geerts, M. Kreft, T. Takafuta, S. S. Shapiro, and A. Sonnenberg. 2002. Different splice variants of filamin-B affect myogenesis, subcellular distribution, and determine binding to integrin [beta] subunits. J. Cell Biol. 156:361-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van der Ven, P. F., W. M. Obermann, B. Lemke, M. Gautel, K. Weber, and D. O. Furst. 2000. Characterization of muscle filamin isoforms suggests a possible role of gamma-filamin/ABP-L in sarcomeric Z-disc formation. Cell Motil. Cytoskeleton 45:149-162. [DOI] [PubMed] [Google Scholar]
- 27.van der Ven, P. F., S. Wiesner, P. Salmikangas, D. Auerbach, M. Himmel, S. Kempa, K. Hayess, D. Pacholsky, A. Taivainen, R. Schroder, O. Carpen, and D. O. Furst. 2000. Indications for a novel muscular dystrophy pathway. gamma-filamin, the muscle-specific filamin isoform, interacts with myotilin. J. Cell Biol. 151:235-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vorgerd, M., P. F. van der Ven, V. Bruchertseifer, T. Lowe, R. A. Kley, R. Schroder, H. Lochmuller, M. Himmel, K. Koehler, D. O. Furst, and A. Huebner. 2005. A mutation in the dimerization domain of filamin c causes a novel type of autosomal dominant myofibrillar myopathy. Am. J. Hum. Genet. 77:297-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xie, Z., W. Xu, E. W. Davie, and D. W. Chung. 1998. Molecular cloning of human ABPL, an actin-binding protein homologue. Biochem. Biophys. Res. Commun. 251:914-919. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.