Abstract
Histone deacetylase (HDAC) inhibitors are a promising class of anticancer agents for the treatment of solid and hematological malignancies. The precise mechanism by which HDAC inhibitors mediate their effects on tumor cell growth, differentiation, and/or apoptosis is the subject of intense research. Previously we described a family of multiprotein complexes that contain histone deacetylase 1/2 (HDAC1/2) and the histone demethylase BHC110 (LSD1). Here we show that HDAC inhibitors diminish histone H3 lysine 4 (H3K4) demethylation by BHC110 in vitro. In vivo analysis revealed an increased H3K4 methylation concomitant with inhibition of nucleosomal deacetylation by HDAC inhibitors. Reconstitution of recombinant complexes revealed a functional connection between HDAC1 and BHC110 only when nucleosomal substrates were used. Importantly, while the enzymatic activity of BHC110 is required to achieve optimal deacetylation in vitro, in vivo analysis following ectopic expression of an enzymatically dead mutant of BHC110 (K661A) confirmed the functional cross talk between the demethylase and deacetylase enzymes. Our studies not only reveal an intimate link between the histone demethylase and deacetylase enzymes but also identify histone demethylation as a secondary target of HDAC inhibitors.
In eukaryotes, the nucleosome serves as the in vivo target of multiple chromatin-modifying enzymes (17). Strikingly, tail domains of histones as well as the nucleosome core domain are subject to several posttranslational modifications, including methylation, acetylation, phosphorylation, and ubiquitylation (33). Such histone modifications are hypothesized as “histone code” to modulate many cellular processes by recruiting regulatory transcription complexes and changing gene expression (16).
In particular, histone methylation was considered irreversible. However, recent studies have revealed that histone methylation can be reversed by several histone demethylases, including PAD4/PADI4, BHC110/LSD1, and JmjC domain-containing demethylases (6, 24, 31, 34-36). PAD4/PADI4 has been reported to convert monomethyl arginine to citrulline by demethylimination. The JmjC domain-containing demethylases contain a JmjC domain responsible for their enzymatic activity and demethylate mono-, di- or trimethylated lysines by a hydroxylation-based mechanism (31, 35, 36). BHC110 demethylates mono- and dimethyl histone H3 lysine 4 (H3K4) and belongs to a family of FAD-dependent polyamine oxidases which use molecular oxygen as an electron acceptor to oxidize an amine group (24). In addition, we and others have shown that CoREST, the corepressor of REST (RE1-silencing transcription factor) protein, mediates nucleosomal demethylation by BHC110 (18, 26). Surprisingly, androgen receptor was recently reported to alter BHC110 enzymatic activity, leading to the demethylation of histone H3 lysine 9 (20).
BHC110 has been isolated as a component of many multiprotein complexes (1, 9, 12, 14, 15, 25, 30). One such complex, termed BHC (BRAF-HDAC complex), mediates the repression of neuron-specific genes (1, 12). Importantly, we have shown that such corepressor complexes share two enzymatic core subunits: histone deacetylase (HDAC1/2) and BHC110 (13). Here we provide evidence that the enzymatic activities of the histone demethylase and the deacetylase are intimately linked. Such cross talk between the two enzymes is seen only when nucleosomal substrates are used and is mediated through different domains of the CoREST protein. Importantly, we show that due to such functional connections, HDAC inhibitors diminish histone demethylation. Given that some HDAC inhibitors have reached clinical trials (21), these findings not only reveal a second mechanism of action for HDAC inhibitors but also point to the future potential of histones demethylase inhibitors as therapeutics for cancer.
MATERIALS AND METHODS
Histones, nucleosomes, expression plasmids, and other reagents.
Bulk histones were purchased from Sigma (H9250). Nucleosomes were purified from a HeLa nuclear pellet as previously described (2, 32). Mammalian expression vectors encoding BHC110 and its mutant (K661A) have been described (18). Bacterial expression plasmids encoding FLAG-CoREST and FLAG-ELM2 were previously described (18). Mammalian expression plasmids encoding FLAG-CoREST or FLAG-CoREST with ELM2 or SANT domains deleted (FLAG-ΔELM2, FLAG-ΔSANT2, FLAG-ΔSANT1, and FLAG-ELM2) were generated using pFLAG-CMV2 or p3xFLAG-CMV14 (Sigma). Anti-dimethyl K4 H3 antibodies (12-460), anti-acetyl (K9/K14) H3 antibodies (06-599), and sodium butyrate were purchased from Upstate. Anti-H3 (ab1791) and anti-HDAC2 (34-6400) antibodies were from Abcam Ltd. (Cambridge, United Kingdom) and Zymed laboratories, respectively. Anti-FLAG antibody (F3165) and trichostatin A (TSA; T8552) were from Sigma.
Affinity purification of the BHC110 complex and recombinant proteins.
BHC110 and its mutant complexes were purified from 150- to 200-mg nuclear extracts isolated from stable cell lines using anti-FLAG M2 affinity resin as previously described (18). Baculoviral recombinant proteins (FLAG-BHC110, FLAG-HDAC1, and FLAG-CoREST-His6) were purified from Sf21 insect cells infected by recombinant viruses using anti-FLAG M2 affinity resin (Sigma) as previously described (18). Bacterial recombinant proteins (FLAG-CoREST and FLAG-ELM2) were purified from BL21 cells. BHC110-associated proteins were identified and described previously (12, 13, 18). The amount of BHC110 in complexes was determined by silver staining, and amounts of recombinant proteins were determined by colloidal staining compared with known amounts of bovine serum albumin.
Demethylation and deacetylation assay.
Demethylation and deacetylation assays were performed as previously described (18).
Coimmunoprecipitation.
HEK-293 cells were transiently transfected with mammalian expression plasmids encoding FLAG-CoREST and its mutants (FLAG-ΔELM2, FLAG-ΔSANT2, FLAG-ΔSANT1, and FLAG-ELM2) and harvested 36 h after transfection. Whole-cell extracts were prepared using a lysis buffer (20 mM Tris [pH 8.0], 137 mM NaCl, 1.5 mM MgCl, 1% Triton X-100, 10% glycerol, 0.2 mM phenylmethylsulfonyl fluoride [PMSF], and 1 mM dithiothreitol [DTT]) and immunoprecipitated with anti-FLAG M2 resin. The beads were washed extensively with a wash buffer (20 mM Tris-HCl [pH 8.0], 250 mM KCl, 0.2 mM EDTA, 0.1% NP-40, 10% glycerol, 0.2 mM PMSF, and 1 mM DTT) and eluted with phosphate-buffered saline containing 0.5 μg/ml FLAG peptide. The eluates were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), followed by Western blotting using anti-BHC110, anti-HDAC2, and anti-FLAG antibodies.
Quantitative chromatin immunoprecipitation.
HEK 293 cells were transiently transfected with plasmids expressing BHC110 or its mutant (K661A) for 14 to 16 h using Fugene (Roche). For trichostatin A treatment, HEK 293 cells were treated with 100 ng/ml trichostatin A for 14 to 16 h. Dimethyl K4 H3 and acetyl H3 levels on synapsin promoters were measured by quantitative chromatin immunoprecipitation assay as previously described (12) and expressed as the increase (-fold) over the control. Quantitative PCR was performed with a Finnzymes DyNAmo HS SYBR Green qPCR kit using Opticon2 (MJ Research). Data are presented as means and standard errors of the means.
RESULTS
BHC110-containing complexes display enhanced deacetylation.
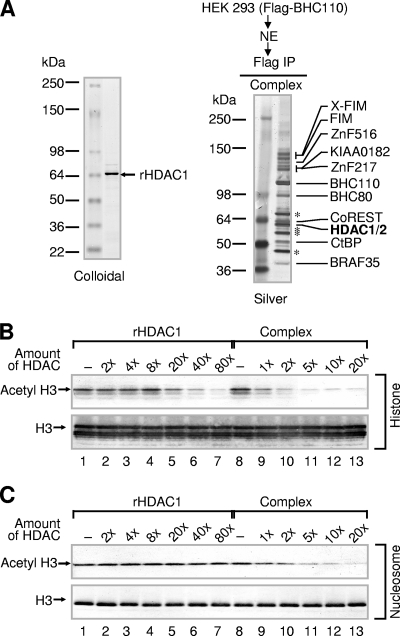
To further define the enzymatic activities of BHC110-containing complexes and to assess whether the enzymes BHC110 and HDAC1/2 are functionally linked, we compared the activity of recombinant HDAC1 to that of HDAC1 in association with BHC110-containing complexes (Fig. 1A). While recombinant HDAC1 displayed HDAC activity toward histone substrates, it was completely inert when nucleosomes were used as substrates (Fig. 1B and C; analysis was performed using antibodies against acetylated lysine 9 and 14 of histone H3, since antibodies against individual acetylated lysines displayed reduced sensitivity and could not be used reliably). In contrast, a BHC110-containing complex(es) not only displayed greater HDAC activity (nearly 20-fold) for histones than recombinant HDAC1 but also deacetylated nucleosomes readily (Fig. 1B and C). These results indicate that additional components of BHC110-containing complexes are required for enhanced activity of HDAC1 on core histones and for mediating nucleosomal deacetylation by HDAC1.
FIG. 1.
Recombinant HDAC1 fails to deacetylate nucleosomes. (A) Analysis of recombinant HDAC1 (rHDAC1) isolated from Sf21 insect cells by colloidal staining (left) and analysis of the BHC110 complex isolated from nuclear extracts (NE) by silver staining (right). (B) Comparison of histone deacetylation activities between recombinant HDAC1 and the BHC110 complex. Bulk histones were used as substrates, and reaction mixtures were analyzed by SDS-PAGE, followed by Western blotting. (C) Comparison of nucleosomal deacetylation activities between recombinant HDAC1 and the BHC110 complex. Nucleosomes were used as substrates, and reaction mixtures were analyzed by SDS-PAGE, followed by Western blotting. “1×” corresponds to approximately 5 ng of recombinant HDAC1 or HDAC1/2 in the complex. “Acetyl H3” represents acetyl K9/K14 H3.
Histone demethylase and deacetylase activities are linked.
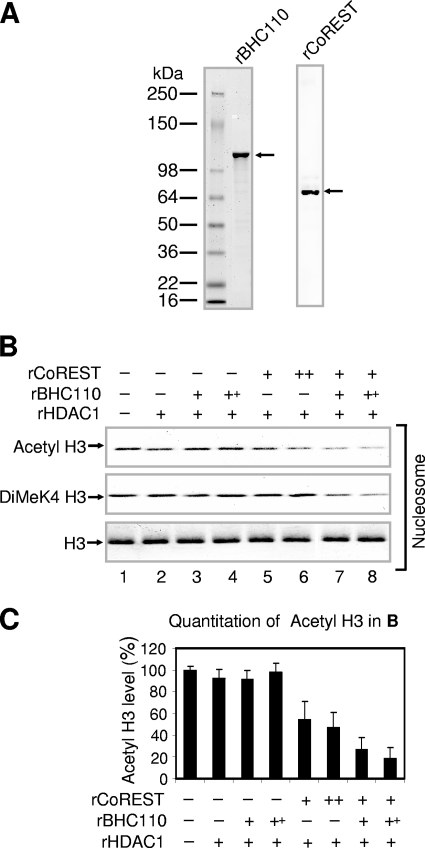
Since nucleosomes are the in vivo substrates of BHC110 complexes at specific promoters, we wished to uncover the factor(s) that mediates the deacetylation of nucleosomes. CoREST was analyzed since it contains two tandem SANT domains which in other corepressors were shown to modulate HDAC activity (11, 37). BHC110 and CoREST were produced in insect cells and purified to homogeneity (Fig. 2A). Analysis of nucleosomal deacetylation using recombinant HDAC1 and CoREST indicated that CoREST was required for mediating nucleosomal deacetylation (Fig. 2B and C). Surprisingly, addition of increasing concentrations of BHC110 further enhanced deacetylation of nucleosomal histone H3, suggesting a role for the demethylase activity in promoting deacetylation by HDAC1 (Fig. 2B and C).
FIG. 2.
Demethylation of nucleosomes enhances deacetylation. (A) Analysis of recombinant BHC110 (rBHC110) and rCoREST isolated from Sf21 insect cells by colloidal staining. (B) Effect of recombinant BHC110 and CoREST on nucleosomal deacetylation. The amounts of recombinant proteins used were approximately as follows: BHC110, 800 ng (+) and 1200 ng (++); HDAC1, 400 ng (+); CoREST, 400 ng (+) and 800 ng (++). Nucleosomes were used as substrates, and reaction mixtures were analyzed by SDS-PAGE, followed by Western blotting. (C) Quantitation of acetyl H3 levels shown in panel B. The acetyl H3 levels in the absence of recombinant proteins (−) were set as 100%. Data are means plus standard deviations from three experiments.
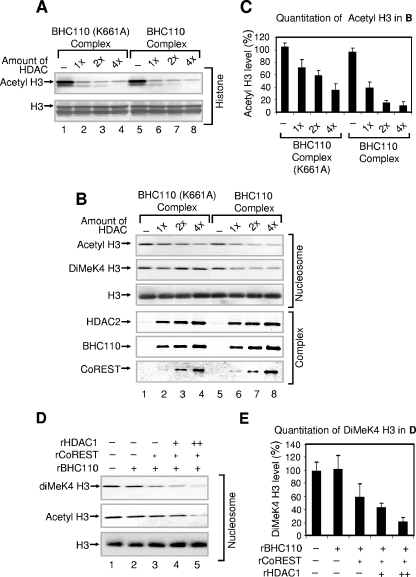
To assess whether the increased deacetylation of nucleosomal histone H3 might require the enzymatic activity of BHC110, we compared the wild-type BHC110 complexes to that of mutant BHC110 (K661A) complex unable to demethylate histone H3 lysine 4 (3) (Fig. 3A and B). This conserved lysine in the monoamine oxidase crystal structure was shown to direct essential interactions with the flavin cofactor through water molecules (3, 4). While the two BHC110-containing complexes display identical subunit compositions and similar deacetylation activities for histone H3 (Fig. 3A and data not shown), the mutant BHC110 (K661A) complex displayed decreased activity in deacetylating nucleosomal histone H3 (Fig. 3B and C). These results demonstrate a role for BHC110 enzymatic activity in regulating deacetylation by HDAC1.
FIG. 3.
While abrogation of demethylation by K661A mutation decreases deacetylation activity, nucleosomal deacetylation enhances demethylation. (A) Effect of the K661A mutation on histone deacetylation. Bulk histones were used as substrates. (B) Effect of the K661A mutation on nucleosomal deacetylation. Nucleosomes were used as substrates. (C) Quantitation of acetyl H3 levels shown in panel B. The acetyl H3 levels in the absence of the complex (−) were set as 100%. Data are means plus standard deviations from three experiments. “1×” corresponds to approximately 50 ng of HDAC1/2 in the complex. (D) Effect of recombinant HDAC1 and CoREST on nucleosomal demethylation. The amounts of recombinant proteins used were approximately as follows: BHC110, 800 ng (+); HDAC1, 400 ng (+) and 800 ng (++); CoREST, 200 ng (+). (E) Quantitation of dimethyl H3 levels shown in panel D. The dimethyl K4 H3 levels in the absence of recombinant proteins (−) were set as 100%. Data are means plus standard deviations from three experiments. For panels A, B, and D, reaction mixtures were analyzed by SDS-PAGE, followed by Western blotting.
To determine whether the functional association of demethylase and deacetylase enzymes also impacts demethylation of nucleosomes by the deacetylase, we examined the effect of recombinant HDAC1 on nucleosomal demethylation. While CoREST is an essential component of nucleosomal demethylation by BHC110 (18), addition of HDAC1 further augmented BHC110-mediated demethylation (Fig. 3D and E). These results demonstrate that the histone deacetylase and histone demethylase enzymes impact each other's activities on nucleosomal substrates.
HDAC inhibitors diminish H3 K4 demethylation.
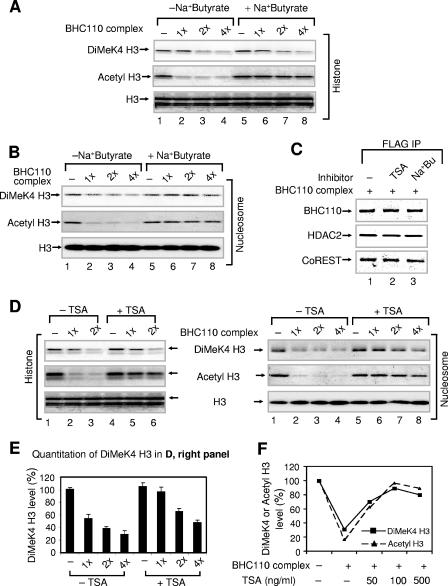
To assess whether the enhancement of demethylation by HDAC1 is due to its catalytic activity, we used HDAC inhibitors to silence histone deacetylation. Inhibition of deacetylase activity by the HDAC inhibitors sodium butyrate and TSA decreased nucleosomal demethylation (Fig. 4A to E). Importantly, the effect of HDAC inhibitors is observed only when nucleosomes are used as substrates for the deacetylation reactions (Fig. 4A, B, and D). Moreover, treatment of the BHC110 complex with increasing concentrations of TSA resulted in similar magnitudes of inhibition of HDAC and histone demethylase activities, attesting to the functional interconnection of the two enzymes (Fig. 4F).
FIG. 4.
Inhibition of deacetylation reduces nucleosomal demethylation. (A) Effect of sodium (Na+) butyrate (100 mM) on histone demethylation. Bulk histones were used as substrates. (B) Effect of sodium butyrate (100 mM) on nucleosomal demethylation. Nucleosomes were used as substrates. “1×” corresponds to approximately 50 ng of HDAC1/2 in the complex. (C) Immunoprecipitation of the BHC110 complexes in the absence or presence of trichostatin A or sodium (Na+) butyrate. The BHC110 complexes were incubated without or with TSA (500 ng/ml) and sodium butyrate (Na+Bu, 100 mM), immunoprecipitated, and analyzed by Western blotting. (D) Effect of TSA (500 ng/ml) on histone (left) and nucleosomal demethylation (right). Bulk histones and nucleosomes, respectively, were used as substrates. (E) Quantitation of dimethyl K4 H3 levels shown in panel D, right panel. The dimethyl K4 H3 levels in the absence of the complex (−) were set as 100%. Data are means plus standard deviations from three different experiments. (F) Effect of different concentrations of trichostatin A on nucleosomal demethylation/deacetylation. Data are the averages of two different points. “1×” corresponds to approximately 50 ng of HDAC1/2 in the complex. In panels A, B, and D, reaction mixtures were analyzed by SDS-PAGE, followed by Western blotting.
To assess whether inhibition of histone deacetylation by HDAC inhibitors impacts the subunit composition of BHC110 complex, Flag-BHC110 affinity eluate was analyzed in the absence or presence of trichostatin A or sodium (Na+) butyrate. As shown in Fig. 4C, Western blot analysis using antibodies to three major subunits (BHC110, HDAC2, and CoREST) of the complex demonstrated that the addition of HDAC inhibitors had no effect on the composition of the complex. Taken together, our data point to an intimate link between the BHC110 and HDAC1 enzymatic activities on nucleosomal substrates and identify histone H3 K4 demethylation as a secondary target of HDAC inhibitors.
The ELM2 domain of CoREST mediates nucleosomal deacetylation.
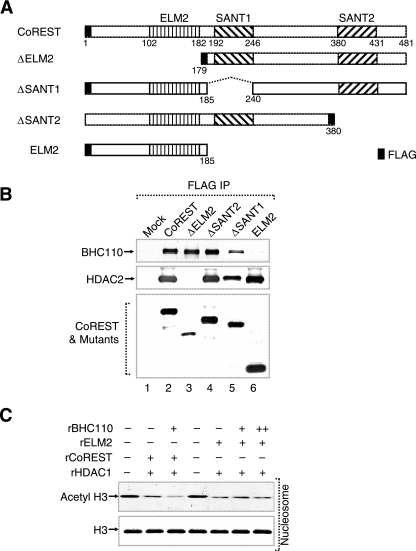
We recently showed that the SANT domains of CoREST are required for nucleosomal demethylation (18). Since CoREST was also required for nucleosomal deacetylation by HDAC1, we mapped the domain in CoREST mediating HDAC1 association (Fig. 5A). These experiments revealed that in contrast to BHC110, which is associated through CoREST SANT domains, HDAC1 association requires the ELM2 domain of CoREST (Fig. 5B). The ELM2 domain was previously shown to associate with HDAC1 in other transcription factors (7, 27). Importantly, not only is the ELM2 domain of CoREST sufficient for HDAC1 association, but also it could confer nucleosomal deacetylation to recombinant HDAC1 (Fig. 5B and C). However, the ELM2 domain alone in the absence of CoREST SANT domains is unable to recruit BHC110 and consequently confer the enhanced deacetylation mediated through BHC110 (Fig. 5B and C). Collectively, these results indicate that different domains of CoREST are required for mediating deacetylation and demethylation and that the cross talk between the two enzymes requires both the ELM2 and the SANT domains of CoREST.
FIG. 5.
The ELM2 domain of CoREST confers nucleosomal deacetylation. (A) Map of five mammalian expression constructs for CoREST and its mutants containing FLAG tag (ΔELM2, ΔSANT2, ΔSANT1, and ELM2). All CoREST constructs lack 30 amino acids from position 31 to 60. (B) Western blot analysis of BHC110 and HDAC2 coimmunoprecipitated with CoREST and its mutants. HEK-293 cells were transfected with CoREST and its mutants, and whole-cell extracts were immunoprecipitated with anti-FLAG M2 resin. The eluates were analyzed by Western blotting using anti-BHC110, anti-HDAC2, and anti-FLAG antibodies. (C) Effect of ELM2 domain on nucleosomal deacetylation. The amounts of recombinant proteins used were approximately as follows: rBHC110, 800 ng (+) and 1600 ng (++); rHDAC1, 400 ng (+); rCoREST, 400 ng (+); rELM2, 150 ng (+). Nucleosomes were used as substrates, and reaction mixtures were analyzed by SDS-PAGE, followed by Western blotting.
The demethylase and the deacetylase enzymes are linked in vivo.
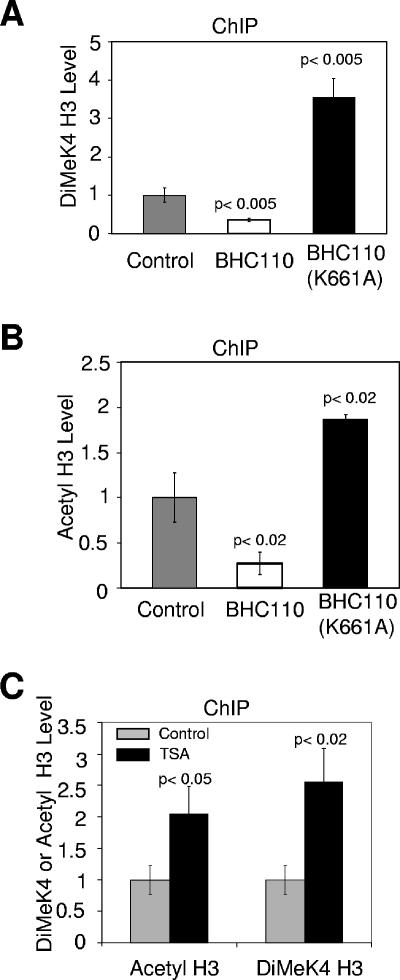
To extend our in vitro observations and to confirm the close functional association of BHC110 and HDAC1/2 in vivo, we examined the levels of dimethyl lysine 4 and acetyl histone H3 (K9 and K14) following ectopic expression of either wild-type or mutant (K661A) BHC110. Consistent with our in vitro findings, while expression of wild-type BHC110 resulted in lower levels of histone H3 methylation and acetylation on the synapsin promoter, the addition of mutant BHC110 (K661A) enhanced the levels of both histone marks (Fig. 6A and B). Moreover, treatment of HEK293 cells with TSA resulted in increased levels of acetylated and methylated nucleosomes in vivo (Fig. 6C). These results extend our functional reconstitution experiments and are consistent with a functional link between the activities of BHC110 and HDAC1/2 in vivo.
FIG. 6.
Deacetylation and demethylation are functionally cooperative in vivo. (A) Analysis of dimethyl K4 H3 levels on the synapsin promoter by quantitative chromatin immunoprecipitation (ChIP) assay after transient transfection of HEK 293 cells with plasmids encoding BHC110 or its mutant (K661A). (B) Analysis of acetyl H3 levels on synapsin promoter by quantitative ChIP assay after transient transfection of HEK 293 cells with plasmids encoding BHC110 or its mutant (K661A). (C) Analysis of dimethyl K4/acetyl H3 levels on synapsin promoter by quantitative ChIP assay after trichostatin A treatment (100 ng/ml) of HEK 293 cells.
DISCUSSION
Our present study reveals an intimate physical and functional link between nucleosomal demethylation and deacetylation through the action of HDAC1, BHC110, and CoREST. We show that both enzymes require CoREST for their activities on nucleosomal substrates. While the enzymatic activity of BHC110 is required for optimal activity of histone deacetylase, inhibition of histone deacetylation diminishes nucleosomal demethylation. These results are consistent with previous findings that BHC110/CoREST preferentially demethylates the hypoacetylated nucleosomes and that acetylation of K9 in H3 peptide decreases binding of recombinant BHC110 (10, 26).
Since both dimethyl H3K4 and acetyl H3K9/K14 marks are often associated with gene activation in mammalian cells, the repressive function of the BHC110 complex may be enhanced by removing both marks through a synergistic interplay between histone demethylase and acetylase enzymes. Similar cross talks between histone modifications have previously been reported to modulate gene expression in eukaryotic cells. Rad6-mediated ubiquitination of H2B lysine 123 is required for H3K4 methylation by COMPASS and H3K79 methylation by Dot1, leading to telomeric gene silencing in Saccharomyces cerevisiae (8, 23, 29). In contrast, phosphorylation of histone H3 serine 10 by Snf1 unidirectionally facilitates acetylation of H3K14 by Gcn5 to activate gene transcription (19).
Importantly, the two enzymes in the BHC110 complex are linked when nucleosomes but not core histones are used as substrates, reflecting differences in how histone tails are packaged in nucleosomes. These results are consistent with our previous observations that inhibition of histone deacetylation by HDAC inhibitors in experiments using the three-subunit complex (BHC110/CoREST/HDAC1) had no significant effect on core histone demethylation (18). Moreover, we show that while recombinant HDAC1 fails to deacetylate nucleosomal substrates, addition of the ELM2 domain of CoREST is sufficient to confer nucleosomal deacetylation in recombinant HDAC1. The ELM2 domain is also required for the association of HDAC1 with CoREST (Fig. 5B and C). Previously, it has been shown that BHC110 interacts with the SANT domains of CoREST and requires either of these domains for its enzymatic activity for nucleosomes (18). Therefore, although both histone deacetylase and demethylase require CoREST for their nucleosomal activities, they may have different strategies in unmasking the nucleosomal histone tails. Since the minimal complex of the two enzymes and CoREST is sufficient to display enzymatic activities found in the native complex, we speculate that the other components of the complex may have other roles, such as recruitment of the corepressor to specific genomic loci or regulation of the complex's enzymatic activities.
How does CoREST mediate the synergy between the two enzymes and confer nucleosomal deacetylation and demethylation? A likely scenario for the action of CoREST entails its role as a bridge between the two enzymes and the nucleosome. Each enzyme is associated with a different domain of CoREST and through this interaction could influence the activity of the other enzyme. Moreover, such interactions with CoREST may increase nucleosomal accessibility of BHC110 and HDAC1 by inducing conformational changes in both enzymes. Structural studies will be required to unravel the exact mechanism by which the two enzymes are coupled.
Histone deacetylase inhibitors are in phase I clinical trials and show promise as a new generation of anticancer therapeutics (21). Although HDAC inhibitors can cause growth arrest of cancer cells by increasing the expression of tumor suppressor proteins such as p21 (5, 22, 28), the downstream effects of these inhibitors are not entirely clear. In this study, we found that HDAC inhibitors diminish H3 K4 demethylation due to the functional link between HDAC activity and BHC110 activity, providing another mechanism of action for HDAC inhibitors. Whether inhibition of histone demethylation is an important component of the antitumor activity of these drugs is not known. Future studies aimed at identifying in vivo inhibitors of H3K4 demethylation should be able to address these questions and may reveal a new class of anticancer agents.
Acknowledgments
We thank Gail Mandel for a CoREST cDNA construct.
R.S. was supported by NIH grant R01 GM 61204.
REFERENCES
- 1.Ballas, N., E. Battaglioli, F. Atouf, M. E. Andres, J. Chenoweth, M. E. Anderson, C. Burger, M. Moniwa, J. R. Davie, W. J. Bowers, H. J. Federoff, D. W. Rose, M. G. Rosenfeld, P. Brehm, and G. Mandel. 2001. Regulation of neuronal traits by a novel transcriptional complex. Neuron 31:353-365. [DOI] [PubMed] [Google Scholar]
- 2.Barak, O., M. A. Lazzaro, W. S. Lane, D. W. Speicher, D. J. Picketts, and R. Shiekhattar. 2003. Isolation of human NURF: a regulator of Engrailed gene expression. EMBO J. 22:6089-6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Binda, C., A. Coda, R. Angelini, R. Federico, P. Ascenzi, and A. Mattevi. 1999. A 30-angstrom-long U-shaped catalytic tunnel in the crystal structure of polyamine oxidase. Structure Fold Des. 7:265-276. [DOI] [PubMed] [Google Scholar]
- 4.Binda, C., M. Li, F. Hubalek, N. Restelli, D. E. Edmondson, and A. Mattevi. 2003. Insights into the mode of inhibition of human mitochondrial monoamine oxidase B from high-resolution crystal structures. Proc. Natl. Acad. Sci. USA 100:9750-9755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chopin, V., R. A. Toillon, N. Jouy, and X. Le Bourhis. 2004. P21(WAF1/CIP1) is dispensable for G1 arrest, but indispensable for apoptosis induced by sodium butyrate in MCF-7 breast cancer cells. Oncogene 23:21-29. [DOI] [PubMed] [Google Scholar]
- 6.Cuthbert, G. L., S. Daujat, A. W. Snowden, H. Erdjument-Bromage, T. Hagiwara, M. Yamada, R. Schneider, P. D. Gregory, P. Tempst, A. J. Bannister, and T. Kouzarides. 2004. Histone deimination antagonizes arginine methylation. Cell 118:545-553. [DOI] [PubMed] [Google Scholar]
- 7.Ding, Z., L. L. Gillespie, and G. D. Paterno. 2003. Human MI-ER1 alpha and beta function as transcriptional repressors by recruitment of histone deacetylase 1 to their conserved ELM2 domain. Mol. Cell. Biol. 23:250-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dover, J., J. Schneider, M. A. Tawiah-Boateng, A. Wood, K. Dean, M. Johnston, and A. Shilatifard. 2002. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J. Biol. Chem. 277:28368-28371. [DOI] [PubMed] [Google Scholar]
- 9.Eimer, S., B. Lakowski, R. Donhauser, and R. Baumeister. 2002. Loss of spr-5 bypasses the requirement for the C. elegans presenilin sel-12 by derepressing hop-1. EMBO J. 21:5787-5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forneris, F., C. Binda, M. A. Vanoni, E. Battaglioli, and A. Mattevi. 2005. Hum. histone demethylase LSD1 reads the histone code. J. Biol. Chem. 280:41360-41365. [DOI] [PubMed] [Google Scholar]
- 11.Guenther, M. G., O. Barak, and M. A. Lazar. 2001. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol. Cell. Biol. 21:6091-6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hakimi, M. A., D. A. Bochar, J. Chenoweth, W. S. Lane, G. Mandel, and R. Shiekhattar. 2002. A core-BRAF35 complex containing histone deacetylase mediates repression of neuronal-specific genes. Proc. Natl. Acad. Sci. USA 99:7420-7425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hakimi, M. A., Y. Dong, W. S. Lane, D. W. Speicher, and R. Shiekhattar. 2003. A candidate X-linked mental retardation gene is a component of a new family of histone deacetylase-containing complexes. J. Biol. Chem. 278:7234-7239. [DOI] [PubMed] [Google Scholar]
- 14.Humphrey, G. W., Y. Wang, V. R. Russanova, T. Hirai, J. Qin, Y. Nakatani, and B. H. Howard. 2001. Stable histone deacetylase complexes distinguished by the presence of SANT domain proteins CoREST/kiaa0071 and Mta-L1. J. Biol. Chem. 276:6817-6824. [DOI] [PubMed] [Google Scholar]
- 15.Jarriault, S., and I. Greenwald. 2002. Suppressors of the egg-laying defective phenotype of sel-12 presenilin mutants implicate the CoREST corepressor complex in LIN-12/Notch signaling in C. elegans. Genes Dev. 16:2713-2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 17.Kornberg, R. D., and Y. Lorch. 1999. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 98:285-294. [DOI] [PubMed] [Google Scholar]
- 18.Lee, M. G., C. Wynder, N. Cooch, and R. Shiekhattar. 2005. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature 437:432-435. [DOI] [PubMed] [Google Scholar]
- 19.Lo, W. S., L. Duggan, N. C. Emre, R. Belotserkovskya, W. S. Lane, R. Shiekhattar, and S. L. Berger. 2001. Snf1—a histone kinase that works in concert with the histone acetyltransferase Gcn5 to regulate transcription. Science 293:1142-1146. [DOI] [PubMed] [Google Scholar]
- 20.Metzger, E., M. Wissmann, N. Yin, J. M. Muller, R. Schneider, A. H. Peters, T. Gunther, R. Buettner, and R. Schule. 2005. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 437:436-439. [DOI] [PubMed] [Google Scholar]
- 21.Monneret, C. 2005. Histone deacetylase inhibitors. Eur. J. Med. Chem. 40:1-13. [DOI] [PubMed] [Google Scholar]
- 22.Nakano, K., T. Mizuno, Y. Sowa, T. Orita, T. Yoshino, Y. Okuyama, T. Fujita, N. Ohtani-Fujita, Y. Matsukawa, T. Tokino, H. Yamagishi, T. Oka, H. Nomura, and T. Sakai. 1997. Butyrate activates the WAF1/Cip1 gene promoter through Sp1 sites in a p53-negative human colon cancer cell line. J. Biol. Chem. 272:22199-22206. [DOI] [PubMed] [Google Scholar]
- 23.Ng, H. H., R. M. Xu, Y. Zhang, and K. Struhl. 2002. Ubiquitination of histone H2B by Rad6 is required for efficient Dot1-mediated methylation of histone H3 lysine 79. J. Biol. Chem. 277:34655-34657. [DOI] [PubMed] [Google Scholar]
- 24.Shi, Y., F. Lan, C. Matson, P. Mulligan, J. R. Whetstine, P. A. Cole, and R. A. Casero. 2004. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119:941-953. [DOI] [PubMed] [Google Scholar]
- 25.Shi, Y., J. Sawada, G. Sui, B. Affar el, J. R. Whetstine, F. Lan, H. Ogawa, M. P. Luke, and Y. Nakatani. 2003. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature 422:735-738. [DOI] [PubMed] [Google Scholar]
- 26.Shi, Y. J., C. Matson, F. Lan, S. Iwase, T. Baba, and Y. Shi. 2005. Regulation of LSD1 histone demethylase activity by its associated factors. Mol. Cell 19:857-864. [DOI] [PubMed] [Google Scholar]
- 27.Solari, F., A. Bateman, and J. Ahringer. 1999. The Caenorhabditis elegans genes egl-27 and egr-1 are similar to MTA1, a member of a chromatin regulatory complex, and are redundantly required for embryonic patterning. Development 126:2483-2494. [DOI] [PubMed] [Google Scholar]
- 28.Sowa, Y., T. Orita, S. Minamikawa-Hiranabe, T. Mizuno, H. Nomura, and T. Sakai. 1999. Sp3, but not Sp1, mediates the transcriptional activation of the p21/WAF1/Cip1 gene promoter by histone deacetylase inhibitor. Cancer Res. 59:4266-4270. [PubMed] [Google Scholar]
- 29.Sun, Z. W., and C. D. Allis. 2002. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418:104-108. [DOI] [PubMed] [Google Scholar]
- 30.Tong, J. K., C. A. Hassig, G. R. Schnitzler, R. E. Kingston, and S. L. Schreiber. 1998. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature 395:917-921. [DOI] [PubMed] [Google Scholar]
- 31.Tsukada, Y., J. Fang, H. Erdjument-Bromage, M. E. Warren, C. H. Borchers, P. Tempst, and Y. Zhang. 2006. Histone demethylation by a family of JmjC domain-containing proteins. Nature 439:811-816. [DOI] [PubMed] [Google Scholar]
- 32.Utley, R. T., T. A. Owen-Hughes, L. J. Juan, J. Cote, C. C. Adams, and J. L. Workman. 1996. In vitro analysis of transcription factor binding to nucleosomes and nucleosome disruption/displacement. Methods Enzymol. 274:276-291. [DOI] [PubMed] [Google Scholar]
- 33.Vaquero, A., A. Loyola, and D. Reinberg. 2003. The constantly changing face of chromatin. Sci. Aging Knowledge Environ. 2003:RE4. [DOI] [PubMed]
- 34.Wang, Y., J. Wysocka, J. Sayegh, Y. H. Lee, J. R. Perlin, L. Leonelli, L. S. Sonbuchner, C. H. McDonald, R. G. Cook, Y. Dou, R. G. Roeder, S. Clarke, M. R. Stallcup, C. D. Allis, and S. A. Coonrod. 2004. Human PAD4 regulates histone arginine methylation levels via demethylimination. Science 306:279-283. [DOI] [PubMed] [Google Scholar]
- 35.Whetstine, J. R., A. Nottke, F. Lan, M. Huarte, S. Smolikov, Z. Chen, E. Spooner, E. Li, G. Zhang, M. Colaiacovo, and Y. Shi. 2006. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell 125:467-481. [DOI] [PubMed] [Google Scholar]
- 36.Yamane, K., C. Toumazou, Y. I. Tsukada, H. Erdjument-Bromage, P. Tempst, J. Wong, and Y. Zhang. 2006. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell 125:483-495. [DOI] [PubMed] [Google Scholar]
- 37.Zhang, Y., H. H. Ng, H. Erdjument-Bromage, P. Tempst, A. Bird, and D. Reinberg. 1999. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 13:1924-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]