Abstract
The Fanconi anemia (FA) pathway is a DNA damage-activated signaling pathway which regulates cellular resistance to DNA cross-linking agents. Cloned FA genes and proteins cooperate in this pathway, and monoubiquitination of FANCD2 is a critical downstream event. The cell cycle checkpoint kinase ATR is required for the efficient monoubiquitination of FANCD2, while another checkpoint kinase, ATM, directly phosphorylates FANCD2 and controls the ionizing radiation (IR)-inducible intra-S-phase checkpoint. In the present study, we identify two novel DNA damage-inducible phosphorylation sites on FANCD2, threonine 691 and serine 717. ATR phosphorylates FANCD2 on these two sites, thereby promoting FANCD2 monoubiquitination and enhancing cellular resistance to DNA cross-linking agents. Phosphorylation of the sites is required for establishment of the intra-S-phase checkpoint response. IR-inducible phosphorylation of threonine 691 and serine 717 is also dependent on ATM and is more strongly impaired when both ATM and ATR are knocked down. Threonine 691 is phosphorylated during normal S-phase progression in an ATM-dependent manner. These findings further support the functional connection of ATM/ATR kinases and FANCD2 in the DNA damage response and support a role for the FA pathway in the coordination of the S phase of the cell cycle.
Fanconi anemia (FA) is an autosomal recessive, or X-linked, cancer susceptibility disorder characterized by cellular hypersensitivity to DNA cross-linking agents, such as mitomycin C (MMC) (11, 12). The 11 cloned FA genes (A, B, C, D1, D2, E, F, G, L, M, and J) cooperate in a novel DNA damage response pathway, the FA pathway (6). Eight of the FA proteins (A, B, C, E, F, G, L, and M) are subunits of a nuclear E3 ubiquitin ligase required for monoubiquitination of the downstream FANCD2 protein on lysine 561, which is a critical step for the function of the FA pathway (8). The FANCL subunit is the putative catalytic E3 ligase subunit of the complex (19). Monoubiquitinated D2 interacts with FANCD1/BRCA2 (36). The recently cloned FANCJ protein is a helicase (3, 15, 16, 18) which may work in concert with FANCD2-Ub and BRCA2 or may function independently of the FA pathway. Disruption of any of the proteins in the pathway leads to MMC hypersensitivity and chromosome instability.
Increasing evidence supports a role for the FA pathway during normal S-phase progression in mammalian cells. First, the FA complex is associated with chromatin during S phase and interacts with the helicase BLM, suggesting that the complex plays a “sensor” role at the advancing replication fork (21, 22) Second, the newly cloned FANCM subunit of the FA complex appears to promote its chromatin association during S phase (20, 26). Third, the FA complex activates FANCD2 monoubiquitination during S phase, leading to the association of monoubiquitinated FANCD2 with BRCA1 and RAD51 in replication foci (34). Fourth, the FA pathway is required for normal homologous recombination repair, a mode of DNA repair executed in S phase (28). Finally, the FA pathway is regulated, at least in part, by the checkpoint kinases ATM and ATR (1, 35). These kinases are believed to function during DNA replication fork progression in S phase and following DNA damage.
ATR and ATM have different effects on the FA pathway. ATR is required for efficient monoubiquitination of FANCD2 (1). Disruption of ATR, by small interfering RNA (siRNA) or by germ line mutations, as in the related autosomal recessive disease Seckel syndrome, results in decreased MMC-inducible D2 monoubiquitination and in MMC hypersensitivity. The mechanism of this ATR requirement remains unknown. ATR may increase FANCD2 monoubiquitination by (i) directly phosphorylating FANCD2 and increasing its subsequent monoubiquitination by the FA complex, (ii) phosphorylating subunits of the FA complex and increasing ubiquitin E3 ligase activity, or (iii) phosphorylating other upstream members of the FA pathway, such as BRCA1 and CHK1.
In contrast, the checkpoint kinase ATM phosphorylates FANCD2 but is generally not required for the monoubiquitination of FANCD2 (35). ATM-deficient cells have normal to increased FANCD2 monoubiquitination and have relatively normal cellular resistance to MMC, compared to FA cells. Following ionizing radiation (IR), ATM phosphorylates multiple sites on FANCD2, including residues S222, S1401, S1404, and S1418. Interestingly, the IR-inducible phosphorylation of FANCD2 on S222 by ATM activates an intra-S-phase checkpoint response. Mutation of this residue (S222A) disrupts this checkpoint response but has no effect on FANCD2 monoubiquitination by the FA complex.
Phosphorylations have been observed on several other FA core complex proteins, including FANCA (39), FANCG (7, 23), and FANCM (20). To date, there is no evidence that these phosphorylation events contribute appreciably to the assembly of the core complex or to its intrinsic E3 ubiquitin ligase activity.
In an attempt to further understand the differential effects of ATR and ATM on the FA pathway, we identified other DNA damage-inducible FANCD2 phosphorylation sites. Here we demonstrate that two novel DNA damage-induced phosphorylation sites on FANCD2, T691 and S717, are required for normal cellular resistance to MMC and for the establishment of the intra-S-phase checkpoint. Phosphorylation of these sites may also promote the accumulation of monoubiquitinated FANCD2. Both sites are phosphorylated by ATR and ATM in vitro and in vivo, but the relative contributions of these kinases to each site appear to differ. Finally, we generated activation-specific antibodies (anti-pThr and anti-pSer) specific for these sites, which have allowed us to examine the kinetics of FANCD2 phosphorylation after DNA damage or during S-phase progression.
MATERIALS AND METHODS
Cell lines and culture conditions.
PD20 (FA-D2) and HeLa cells were grown in Dulbecco's modified Eagle medium supplemented with 15% fetal calf serum (FCS). Stable FA-D2 transfectants (PD20) expressing wild-type FANCD2, the FANCD2(S222A) mutant, or the FANCD2(K561R) mutant were described previously (35). AT22IJE-T fibroblasts expressing either empty vector or full-length ATM were described previously (44). The Epstein-Barr virus (EBV)-transformed human lymphoblast lines PD20 (wild type), GM1526 (ATM deficient), EUFA1020 (NBS1 deficient), and HSC230 (FANCB deficient) were grown in RPMI plus 15% FCS. Irradiation was delivered using a Gammacell-40 irradiator. UV treatment was done using a Stratalinker (Stratagene) after gentle aspiration of the growth medium.
Plasmids and retroviral infection.
The retroviral expression vectors pMMP-puro, pMMP-puro FANCD2wt, pMMP-puro FANCD2(S222A), and pMMP-puro FANCD2(K561R) (35) were described previously. pMMP-puro EGFP-FANCD2wt was constructed by adding the enhanced green fluorescent protein (EGFP) cDNA sequence (from pEGFP-N1 [Clontech]) to the N terminus of the FANCD2 cDNA sequence. The T691A, S717A, and T691-S717A FANCD2 cDNAs were generated using a QuikChange site-directed mutagenesis kit (Stratagene). All cDNA inserts were verified by DNA sequencing. Production of pMMP retroviral supernatants and infection of fibroblasts were done as previously described (27). After 48 h, cells were trypsinized and transferred to medium containing 1 μg/ml puromycin. Dead cells were removed, and surviving cells were grown under continuous selection in puromycin. MMC sensitivity assays were performed as previously described (35).
Generation of phosphospecific antibodies.
Phosphospecific antibodies for human FANCD2 were generated by immunization of rabbits with keyhole limpet hemocyanin (KLH)-conjugated peptides (Zymed Laboratories [South San Francisco, CA]). The KLH-conjugated phosphopeptide EILGD[PO3]SQHAD was used to generate anti-pS222, and the KLH-conjugated phosphopeptide LEEYD[PO3]TQDG was used to generate anti-pT691. KLH-conjugated GGPVT[PO3]SQESG was used to generate anti-pS717. Antibodies were affinity purified using the corresponding phosphorylated peptide-conjugated gels and unphosphorylated peptide-conjugated gels.
Immunoblotting.
Cells were lysed with 1× sample buffer (50 mM Tris-HCl [pH 6.8], 0.6% 2-mercaptoethanol, and 2% sodium dodecyl sulfate), boiled for 10 min, and separated on 3 to 8% Tris-acetate gradient gels (Invitrogen). After electrophoresis, proteins were transferred to nitrocellulose membranes using a submerged transfer apparatus (Bio-Rad). Membranes were then probed with the appropriate antibodies. Antibodies included anti-ATM (1:500; Novus), anti-ATR (1:500; Santa Cruz [N19]), anti-pS222 FANCD2 (1:400), anti-pT691 FANCD2 (1:500), anti-pS717 FANCD2 (1:200), and anti-FANCD2 (FI17) (1:1,000; Santa Cruz).
In vitro kinase assay.
The in vitro kinase assay was done as described previously (1).
siRNA and transfection.
Expression of targeted genes was knocked down by transient expression of siRNA directed against ATM (5′-AACATACTACTCAAAGACATT-3′) (1), ATR (5′-AACCTCCGTGATGTTGCTTGA-3′), and LacZ (5′-AACGTACGCGGAATACTTCGA-3′). Cells were plated at least 16 h prior to transfection in six-well plates to yield ∼60% confluence at the time of transfection in 1 ml of medium devoid of antibiotics. siRNA oligonucleotides were diluted in 100 μl Opti-MEM I to yield a final concentration of 100 nM siRNA and were mixed with 6 μl of Lipofectamine 2000 (Invitrogen) diluted in 100 μl of OptiMEM I. Complex formation was allowed to proceed for 20 min at room temperature prior to dropwise addition to cells. siRNA complexes were incubated with cells for 6 h, at which time the medium was changed and Lipofectamine washed off. A second transfection of the same siRNA was carried out 24 h following the first transfection. Cells were treated with IR 48 h later.
RDS assay.
The radioresistant DNA synthesis (RDS) assay was performed as described previously (35) with minor modifications. Briefly, cells were plated in 60-mm-diameter plates, incubated with 15 nCi of [14C]thymidine for 24 h, and cultured in the absence of labeling medium for 24 h. The cells were then treated in triplicate with 10 Gy of IR. At 30 min following DNA damage, cells were incubated with 3.0-μCi/ml [3H]thymidine for 15 min. Cells were fixed, loaded onto glass fiber filters, and counted as previously described (25).
Cell cycle synchronization.
Cells were synchronized as described previously (34) with the exception of the AT22IJE-T+vector and AT22IJE-T+ATM cell lines, where 10 mM thymidine was used instead of 2 mM thymidine.
Flow cytometry.
Flow cytometry was done as previously described (1).
RESULTS
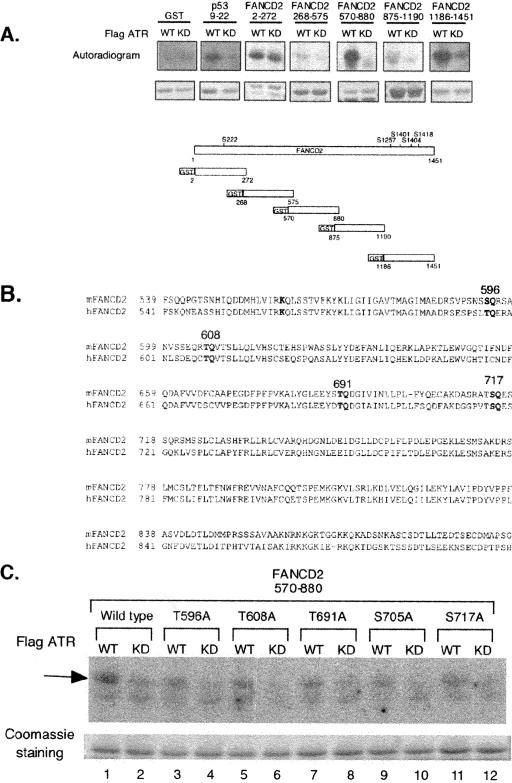
Initially, we examined the primary sequence of FANCD2 for additional ATR/ATM phosphorylation sites (Fig. 1). ATM and ATR phosphorylate the sequences SQ and TQ (13). We generated five partially overlapping glutathione S-transferase (GST) fusion proteins spanning the 1,451 amino acids of FANCD2 and subjected these polypeptides to in vitro kinase reactions. As expected, two of the fusion proteins (GST-2-272, containing Ser222, and GST-1186-1451, containing Ser1401, Ser1404, and Ser1418) were readily phosphorylated by ATR in vitro (Fig. 1A), consistent with our previous report of ATM-dependent phosphorylation of FANCD2. In addition, another GST fusion protein, containing amino acids 570 to 880, was readily phosphorylated by ATR in vitro (Fig. 1A). The phosphorylation of this fragment of FANCD2 was particularly interesting, since it contains amino acid residues near K561, the site of FANCD2 monoubiquitination. This fragment of ATR contains four conserved sites which are potential sites of ATR phosphorylation (S596, T608, T691, and S717) (Fig. 1B). Mutation of each of these residues to alanine, one at a time, decreased ATR-dependent phosphorylation in vitro (Fig. 1C), suggesting that any of these sites may be an ATR phosphorylation site in vivo.
FIG. 1.
ATR phosphorylates FANCD2 on T691 and S717 in vitro and in vivo. (A) Schematic showing GST fusion proteins containing variable regions of human FANCD2. The fusion proteins were incubated with immune complexes containing either wild-type ATR or a kinase-inactive form of ATR, and in vitro kinase reactions was performed. Three fragments of FANCD2 were phosphorylated by ATR in vitro (2-272, 570-880, and 1186-1451). (B) Sequence of the 570-880 region of FANCD2, with the five putative ATR phosphorylation sites indicated. (C) In vitro ATR kinase reactions, using GST fusion proteins with the indicated mutations.
We used mass spectrometry to examine tryptic fragments of FANCD2, isolated from IR-treated cells (35). Mass spectrometry revealed the presence of tryptic fragments containing either phospho-T691 or phospho-S717, indicating that these sites are phosphorylated in vivo (data not shown). We therefore raised rabbit antisera against FANCD2 peptides containing these phosphorylated residues. Specifically, we raised rabbit polyclonal antisera directed against LEEXDpT(691)QDG and against GGPVTpS(717)QESG of FANCD2. These antisera were compared to the anti-FANCD2 [anti-pS(222) antibody generated in our previous study (35)].
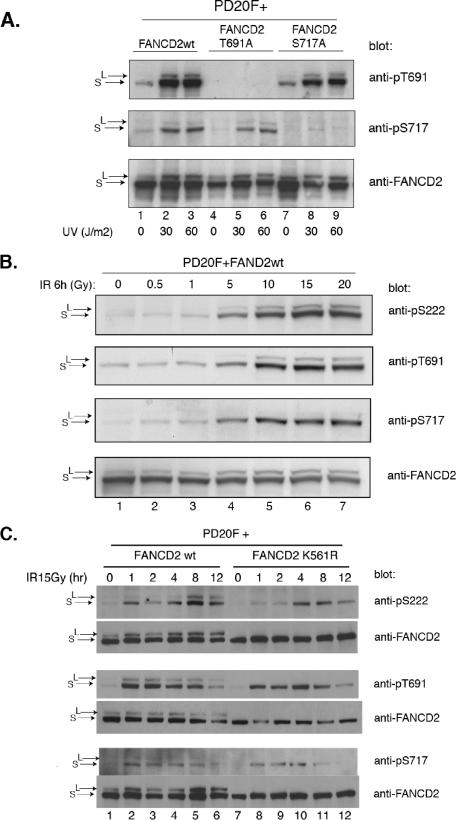
We tested the specificity of these new antisera (Fig. 2). FA-D2 cells (PD20 fibroblasts), which lack expression of endogenous FANCD2 protein, were stably transfected with the cDNA encoding either wild-type FANCD2, FANCD2(T691A), or FANCD2(S717A). The transfected cells were exposed to UV light as a source of DNA damage (Fig. 2A). Previous studies have indicated that UV light is a potent stimulator of FANCD2 monoubiquitination (8) and of ATR activity. The wild-type FANCD2 protein was phosphorylated on both T691 and S717 following DNA damage. Phosphorylation at these sites was observed on both the unubiquitinated (S) and the monoubiquitinated (L) isoforms of FANCD2 (Fig. 2A, lanes 2 and 3). The T691A mutant protein was phosphorylated on one site (S717), and the S717A mutant protein was phosphorylated only on the other site (T691). These results indicate that (i) the antisera are specific for the specific phosphopeptides for which they were raised and (ii) the sites are phosphorylated in vivo independently.
FIG. 2.
In vivo phosphorylation of FANCD2 on T691 and S717 following DNA damage. (A) FA-D2 cells were stably transfected with the cDNAs encoding either wild-type FANCD2, FANCD2(T691A), or FANCD2(S717A). Cells were treated with UV irradiation or left untreated, as indicated, and proteins from whole-cell extracts were isolated, electrophoresed, and immunoblotted with the indicated antisera. (B) FA-D2 cells, stably transfected with wild-type FANCD2, were exposed to IR, as indicated, and total cellular proteins were immunoblotted with the indicated antisera. The anti-pS222 antiserum has been described (35). (C) FA-D2 cells were stably transfected with either wild-type FANCD2 or the K561R mutant. Cells were irradiated, and cell lysates were probed by immunoblotting with the indicated anti-FANCD2 antiserum.
We next tested dose-dependent phosphorylation of FANCD2 following DNA damage. IR activated the dose-dependent phosphorylation of T691, S717, and S222 in the 0- to 20-Gy radiation range (Fig. 2B). The monoubiquitinated (L) isoform of FANCD2 was only weakly phosphorylated on S717. Phosphorylation of these three sites was detected at IR doses as low as 5 Gy.
We also tested the kinetics of phosphorylation at these three FANCD2 residues. We transfected FA-D2 cells with the cDNA encoding either wild-type FANCD2 or the FANCD2(K561R) mutant, which cannot be monoubiquitinated (Fig. 2C). Following IR, FANCD2 and monoubiquitinated FANCD2 were rapidly phosphorylated on both T691 and S717, within only 1 h of IR exposure. In contrast, the S222 site was only weakly phosphorylated at early time points (1 h and 2 h after IR exposure) and strongly phosphorylated later, after 4 to 8 h of IR exposure. Phosphorylation of FANCD2 occurred at all three sites, even in the absence of FANCD2 monoubiquitination (Fig. 2C, K561R mutant), indicating that monoubiquitination is not required for phosphorylation. Previous studies have indicated that the FANCD2 L isoform is localized to the chromatin (24). Taken together, these results suggest that FANCD2 can be phosphorylated even before chromatin association of the monoubiquitinated (L) isoform.
Phosphorylation of FANCD2 on T691 and S717 promotes the monoubiquitination of FANCD2.
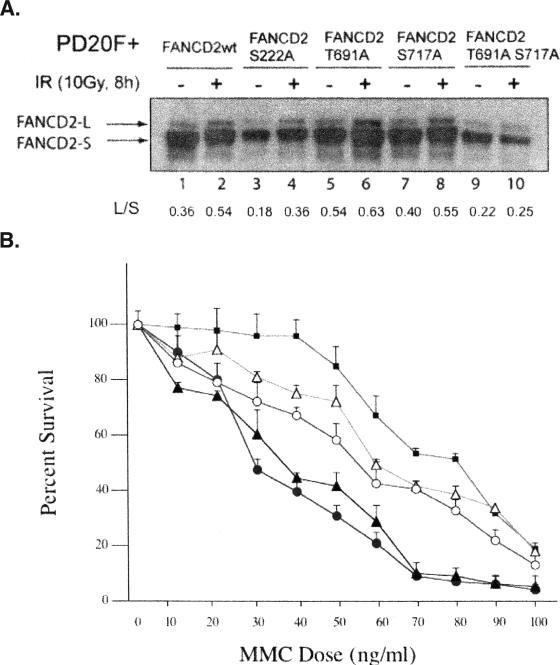
We next determined the effect of mutating these ATR phosphorylation sites on FANCD2 monoubiquitination and function (Fig. 3). FA-D2 fibroblasts were transduced with cDNAs encoding either wild-type FANCD2, single point mutants, or the double point mutant [FANCD2(T691A,S717A)]. All mutant FANCD2 polypeptides underwent some monoubiquitination (Fig. 3A, lanes 3 to 10), demonstrating that phosphorylation of these specific residues is not absolutely required for FANCD2 monoubiquitination. Interestingly, the FA-D2 cells expressing the double point mutant protein had slightly decreased FANCD2 monoubiquitination (Fig. 3A, lanes 9 and 10) and remained hypersensitive to MMC (Fig. 3B). Cells expressing the single point mutants had normal IR-inducible FANCD2 monoubiquitination but exhibited an intermediate level of MMC resistance (Fig. 3B). Taken together, these results demonstrate that the ATR-dependent phosphorylation of FANCD2 on T691 and S717 enhances FANCD2 monoubiquitination but is not essential for this posttranslational modification. This provides a possible molecular explanation for the decreased DNA damage-inducible FANCD2 monoubiquitination and increased MMC sensitivity of ATR-deficient (Seckel syndrome) cells (1).
FIG. 3.
Phosphorylation of T691 and S717 is required for optimal FANCD2 monoubiquitination and downstream functional activity. (A) FA-D2 fibroblasts were stably transduced with the cDNAs encoding the indicated wild-type (wt) or mutant FANCD2 proteins. Cells were treated as indicated, and whole-cell extracts were immunoblotted with anti-FANCD2 antiserum. The autoradiograph was scanned by densitometry, and the ratio of FANCD2-L (monoubiquitinated FANCD2 isoform) to FANCD2-S (unubiquitinated isoform) was calculated (L/S). (B) Cells were examined by the MMC cytotoxicity assay. The results are shown for vector alone (closed triangles), FANCD2 (closed squares), FANCD2(T691A,S717A) (closed circles), FANCD2(T691A) (open triangles), and FANCD2(S717A) (open circles).
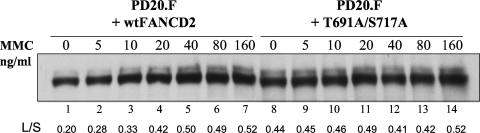
To further confirm that the double mutant of FANCD2 [FANCD2(T691A,S717A)] was deficient in monoubiquitination, we performed an MMC dose response study (Fig. 4). PD20(F) transfectants, expressing relatively equal levels of either wild-type FANCD2 or FANCD2(T691A,S717A), were compared. Again, the PD20 cells expressing the double mutant protein exhibited decreased monoubiquitination compared to those expressing the wild-type FANCD2 protein. Specifically, the double mutant protein exhibited an L/S ratio (i.e., ratio of FANCD2-L to FANCD2-S) of 0.44 in the absence of MMC damage, and this ratio increased to only 0.52 (18% increase) at the highest dose of MMC (Fig. 4, lanes 8 to 14). In contrast, the wild-type FANCD2 protein increased from 0.20 to 0.52 in this MMC dose range (160% increase) (Fig. 4, lanes 1 to 7). Taken together, these results suggest that the MMC hypersensitivity of the PD20 cells expressing the double mutant protein results, at least in part, from a failure to optimally monoubiquitinate FANCD2 after DNA damage.
FIG. 4.
FANCD2(T691A,S717A) fails to upregulate its monoubiquitination after MMC cross-linker damage. FA-D2 fibroblasts were stably transduced with the cDNAs encoding wild-type (wt) or mutant FANCD2 proteins. Cells were treated with MMC, as indicated, and whole-cell extracts were immunoblotted with anti-FANCD2 antiserum. The upper band is monoubiquitinated FANCD2 (FANCD2-L), and the lower band is unubiquitinated FANCD2 (FANCD2-S). The ratio of FANCD2-L to FANCD2-S (L/S) was calculated by densitometry.
It is also interesting that FANCD2(T691A,S717A) paradoxically exhibited elevated monoubiquitination in the absence of DNA damage (Fig. 4, compare lanes 1 and 8). This elevated monoubiquitination probably results from the elevated endogenous DNA damage in cells expressing the mutant polypeptide (lane 8), compared to cells functionally complemented with the wild-type FANCD2 protein (lane 1).
FANCD2 is phosphorylated on T691 and S717 in vivo by ATR and ATM.
We next evaluated the relative contributions of ATR and ATM to the in vivo phosphorylation of FANCD2, by using an siRNA strategy (Fig. 5A). In HeLa cells, siRNA knockdown of ATM reduced phosphorylation of S222 and T691 in response to IR more strongly than siRNA knockdown of ATR. Simultaneous knockdown of ATM and ATR reduced FANCD2 phosphorylation on S717 (Fig. 5A, lane 8), although single knockdown of ATM or ATR did not reduce the phosphorylation of S717 significantly.
FIG. 5.
FANCD2 is phosphorylated on S222, T691, and S717 by both ATR and ATM. (A) HeLa cells stably transfected with the cDNA encoding wild-type EGFP-tagged FANCD2 were exposed to siRNA specific for LacZ, ATM, ATR, or both ATM and ATR. After 48 h, whole-cell lysates were prepared and total cellular protein was analyzed by immunoblotting with antisera specific for the indicated proteins. (B) FA-D2 transfectants were subjected to the RDS assay.
The phosphorylation of FANCD2 on these sites by ATM suggested that these phosphorylated residues might play a functional role in the intra-S-phase checkpoint. The intra-S-phase checkpoint is evaluated by the RDS assay (25, 42). We have previously shown that knockdown of ATM or disruption of FANCD2 on Ser222 (S222A mutant) results in loss of this checkpoint response (35). Interestingly, FA-D2 cells transfected with either FANCD2(T691A) or FANCD2(S717A) failed to activate the intra-S-phase checkpoint (Fig. 5B). That is, these cells, expressing the mutant FANCD2 protein, continued to synthesize DNA, even after sustaining damage from IR. Taken together, these results indicate that ATM-dependent phosphorylation of FANCD2 on T691 and ATM/ATR dependent phosphorylation of S717 contribute to this checkpoint, further underscoring the functional importance of these phosphorylation events for S-phase events.
NBS1 is not required for DNA damage-inducible phosphorylation of FANCD2 on T691.
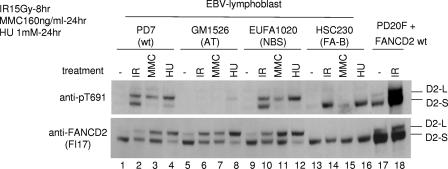
In response to IR, ATM activates multiple cellular substrates, including p53 (2, 4), NBS1 (9, 17, 37, 43), and BRCA1 (5). Many of these checkpoint proteins, including NBS1 and BRCA1, colocalize following IR and cooperate in the IR-activated S-phase checkpoint (38). Furthermore, recent studies have suggested that NBS1 is required for the efficient activation of ATM by IR (41). For instance, NBS1-deficient cells exhibit decreased ATM-dependent phosphorylation of SMC1 (14, 40). We therefore tested the possible requirement of NBS1 in the ATM-dependent phosphorylation of FANCD2 on Thr691 (Fig. 6). Human lymphoblast lines that were either wild type, ATM deficient, NBS1 deficient, or FANCB deficient were exposed to genotoxic stress (IR, MMC, or hydroxyurea [HU] replication arrest), and phosphorylation of FANCD2 on T691 was analyzed. While ATM-deficient cells had decreased phosphorylation of FANCD2 on T691, NBS1-deficient cells and FANCB-deficient cells had normal levels of FANCD2 phosphorylation at this residue. Interestingly, IR activated the phosphorylation of T691 on both the unmodified and the monoubiquitinated isoform of FANCD2 (Fig. 6, lanes 2 and 10), but MMC or HU exposure resulted in a selective increase in phosphorylation of the monoubiquitinated isoform, FANCD2-L. These results suggest that following MMC or HU exposure, the phosphorylated form of FANCD2 is primarily localized to the chromatin, where the monoubiquitinated isoform accumulates. Monoubiquitination may further promote T691 phosphorylation or may prevent or delay dephosphorylation of this site by some unknown phosphatase.
FIG. 6.
NBS1 is not required for DNA damage-inducible phosphorylation of FANCD2 on T691. The indicated EBV-transformed human lymphoblast lines were exposed to either IR, MMC, or HU, as indicated. PD20 (FA-D2) fibroblasts, expressing wild-type FANCD2, were also analyzed. Proteins from whole-cell extracts were immunoblotted with antisera specific for either pT691 or unphosphorylated FANCD2.
ATM-dependent phosphorylation of T691 during normal S-phase progression.
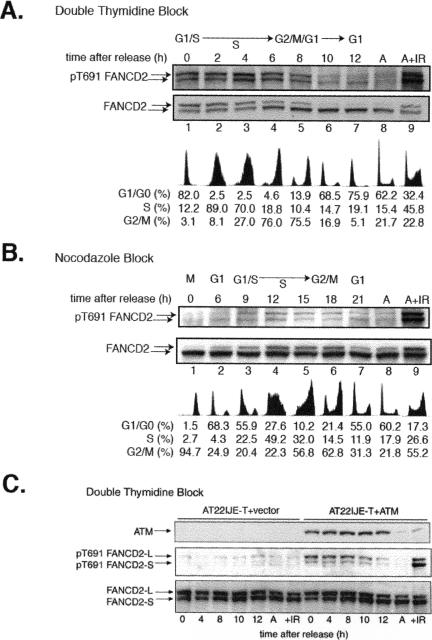
FANCD2 monoubiquitination is also activated during normal S-phase progression, suggesting that FANCD2 phosphorylation may occur in S phase (34). Furthermore, ATM has recently been shown to be activated during normal S-phase progression. We therefore synchronized HeLa cells (by double thymidine block and by nocodazole block) and examined FANCD2 phosphorylation on T691 (Fig. 7). Interestingly, T691 was phosphorylated during normal S-phase progression (Fig. 7A and B).
FIG. 7.
ATM phosphorylates FANCD2 on T691 during normal S-phase progression. (A) HeLa cells were synchronized by double thymidine block and released into S phase. Whole-cell lysates were prepared from the synchronized cell populations at the indicated times, and FANCD2 was immunoblotted with either anti-pT691 or anti-FANCD2. Asynchronous cells (A), treated with IR or untreated, were also evaluated. DNA flow histograms corresponding to the synchronized cells are also shown. (B) HeLa cells were synchronized by the nocodazole block method. (C) ATM-deficient cells (vector control) and the isogenic ATM-corrected control (AT22IJE-T+ATM) were synchronized by double thymidine block and evaluated as described above.
Phosphorylation of T691 occurred throughout S phase (Fig. 7A, lanes 1 to 4). Deubiquitination of FANCD2 began 6 h after release from G1/S block (lane 4), and dephosphorylation of FANCD2 on T691 occurred after the onset of deubiquitination. Similar results were obtained when cells were synchronized by nocodazole block (Fig. 7B). Deubiquitination of FANCD2 began in late S phase (lane 7), and dephosphorylation of FANCD2 on T691 occurred thereafter.
To further evaluate the role of the ATM kinase in the S-phase-specific phosphorylation of FANCD2, we evaluated synchronized cultures of ATM-deficient or ATM-corrected fibroblasts (Fig. 7C). In ATM-deficient cells, S-phase-specific phosphorylation on T691 of FANCD2 was impaired. Correction with the ATM kinase restored S-phase-specific phosphorylation of FANCD2. Taken together, these results indicate that (i) ATM is the prominent cellular kinase which phosphorylates FANCD2 on T691 during S-phase progression and (ii) this phosphorylation of FANCD2 on T691 correlates with the monoubiquitination state of FANCD2 in S phase.
DISCUSSION
The purpose of our study was to identify additional ATR- and ATM-dependent phosphorylation sites on FANCD2 which contribute to DNA damage-dependent activation of the FA pathway. We identified two new DNA damage-inducible phosphorylation sites (T691 and S717) which stimulate the pathway. Mutant forms of FANCD2 lacking these phosphorylation sites (i.e., the T691A-S717 A double mutant) have a slight decrease in DNA damage-inducible FANCD2 monoubiquitination and fail to functionally complement the MMC hypersensitivity of transfected FA-D2 cells.
It is important to note that phosphorylation of T691 and S717 is not absolutely required for FANCD2 monoubiquitination. Indeed, the double point mutant of FANCD2 exhibits decreased but not absent monoubiquitination. The decrease in damage-inducible monoubiquitination of FANCD2(T691A,S717A) compared to the wild-type protein correlates with the failure of the mutant to correct the MMC hypersensitivity of the FA-D2 cells. Interestingly, a defect in damage-inducible monoubiquitination of FANCD2 also correlates with the MMC hypersensitivity and chromosome instability of some human tumor cell lines (A. D'Andrea, unpublished observation). It will therefore be interesting to determine whether these same tumor lines have decreased FANCD2 phosphorylation, perhaps due to defective upstream ATR or ATM activity.
Recent studies indicate that DNA damage activates the phosphorylation of other FA proteins as well. For instance, we have recently shown that after DNA damage, FANCE is phosphorylated in vivo on two conserved sites (A. D'Andrea, unpublished observation). FANCE phosphorylation promotes FANCE degradation, another necessary precondition for DNA damage-inducible FANCD2 monoubiquitination.
The mechanism by which ATR-dependent phosphorylation of these two residues promotes FANCD2 monoubiquitination is unknown, and several models are possible. First, phosphorylation may promote the binding of FANCD2 to the FA complex, the putative monoubiquitin ligase of D2. Recent studies suggest that the primary subunit of the FA complex which binds FANCD2 is FANCE (10, 30), so perhaps phosphorylation of FANCD2 enhances its interaction with FANCE. Alternatively, phosphorylation of FANCD2 may enhance its interaction with the FANCL subunit (19). Along these lines, FANCL contains WD40 domains, which are possible sites of binding of phosphorylated polypeptides. Second, phosphorylation of FANCD2 may result in its translocation to a cellular compartment in which monoubiquitination occurs. Third, phosphorylation of FANCD2 may stabilize the monoubiquitinated state of FANCD2, perhaps by increasing its affinity for chromatin or by preventing its interaction with the deubiquitinating enzyme USP1 (29). Regardless of the mechanism, phosphorylation of FANCD2 by ATR at these sites appears to slightly enhance FANCD2 monoubiquitination and its downstream function in DNA repair.
Recent studies suggest that the ATR substrate CHK1 may also play a role in FANCD2 monoubiquitination (T. Taniguchi and A. D. D'Andrea, unpublished observation). Knockdown of CHK1 activity, either by siRNA or by specific small-molecule kinase inhibitors of CHK1, resulted in decreased FANCD2 monoubiquitination, even in the setting of increased FANCD2 phosphorylation on T691 and S717. Other recent work confirmed that ATR promotes increased phosphorylation of its substrates in the setting of CHK1 inhibition (33). The mechanism by which CHK1 promotes FANCD2 monoubiquitination is unknown. CHK1 may phosphorylate FANCD2 on another site or may phosphorylate and activate subunits of the FA ubiquitin ligase complex. The direct phosphorylation of FANCD2 by ATR is necessary for the efficient damage-inducible monoubiquitination of FANCD2; CHK1 phosphorylation events may indirectly promote FANCD2 monoubiquitination as well.
Additional roles of FANCD2 phosphorylation on T691 and S717 by ATR and ATM are also possible, and these roles may be independent of FANCD2 monoubiquitination. Phosphorylation may enhance the activity of FANCD2 in homologous recombination repair (28). Phosphorylation may also enhance the formation of FANCD2/FANCE/BRCA2 complexes (36). Regardless of the mechanism, our study demonstrates that failure of phosphorylation at these sites disrupts the downstream function of the FA pathway in the maintenance of MMC resistance.
The two newly identified phosphorylation sites (T691 and S717) add to a growing list of DNA damage-inducible phosphorylation sites on FANCD2 (S222, T691, S717, S1401, S1404, and S1418). Interestingly, these sites differ with respect to several characteristics. First, the kinetics of phosphorylation after DNA damage differ significantly. For example, T691 and S717 are phosphorylated early after IR exposure, while S222 is phosphorylated later. Second, phosphorylation of T691 and S717 enhances K561 monoubiquitination and MMC resistance, but phosphorylation of the other sites has no obvious effect on these processes. Third, phosphorylation of S222, T691, and S717 strongly activates the intra-S-phase checkpoint response, consistent with their regulated phosphorylation during the S phase of the cell cycle. Fourth, some sites are predominantly phosphorylated by ATM while other sites are phosphorylated equally well by ATM and ATR.
The contribution of FANCD2 phosphorylation on S222, T691, and S717 to the intra-S-phase checkpoint is particularly important. The molecular basis of this checkpoint remains unknown. These phosphorylation sites may contribute to the assembly of FANCD2 with other ATM substrates in an S-phase checkpoint complex. Such a complex may play a role in blocking the progression of DNA replication or in the firing of new DNA replication origins. Further studies will be required to determine whether loss of FANCD2 phosphorylation leads to impaired protein complex assembly or defective phosphorylation of other downstream players in the S-phase checkpoint.
The new anti-phospho-Ser and anti-phospho-Thr antibodies for FANCD2 will have considerable utility in future studies. For instance, the antisera can be used to examine the state of phosphorylation of FANCD2 in cells from different FA complementation groups and in various human tumor cells. Absence of FANCD2 phosphorylation may correlate with specific defects in the FA pathway and may be useful in predicting the sensitivity of tumors to different types of radiation or chemotherapy. Also, the antibodies can be used to probe the phosphorylation state of FANCD2 in different subcellular compartments and thereby allow a better understanding of the sequence of signaling events in the FA pathway.
Finally, there is increasing evidence that the checkpoint kinases ATR and ATM function during normal S-phase progression. Gautier and colleagues (31, 32) have demonstrated a requirement for ATM in normal DNA replication fork progression in Xenopus extracts. How the FA pathway contributes to replication fork progression during S phase remains a key unanswered question in the FA field.
Acknowledgments
We thank T. Huang and R. Kennedy for helpful discussions. We thank Y. Shiloh for the AT fibroblast line (AT22IJE-T) and ATM-corrected cells.
This work was supported by NIH grants RO1HL52725, RO1 DK43889, P0150654, P50 CA105009-01, and PO1HL54785 (A.D.D.).
REFERENCES
- 1.Andreassen, P. R., A. D. D'Andrea, and T. Taniguchi. 2004. ATR couples FANCD2 monoubiquitination to the DNA-damage response. Genes Dev. 18:1958-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banin, S., L. Moyal, S. Shieh, Y. Taya, C. Anderson, L. Chessa, N. Smorodinsky, C. Prives, Y. Reiss, Y. Shiloh, and Y. Ziv. 1998. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science 281:1674-1677. [DOI] [PubMed] [Google Scholar]
- 3.Bridge, W. L., C. J. Vandenberg, R. J. Franklin, and K. Hiom. 2005. The BRIP1 helicase functions independently of BRCA1 in the Fanconi anemia pathway for DNA crosslink repair. Nat. Genet. 37:953-957. [DOI] [PubMed] [Google Scholar]
- 4.Canman, C. E., D. S. Lim, K. A. Cimprich, Y. Taya, K. Tamai, K. Sakaguchi, E. Appella, M. B. Kastan, and J. D. Siliciano. 1998. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science 281:1677-1679. [DOI] [PubMed] [Google Scholar]
- 5.Cortez, D., Y. Wang, J. Qin, and S. J. Elledge. 1999. Requirement of ATM-dependent phosphorylation of brca1 in the DNA damage response to double-strand breaks. Science 286:1162-1166. [DOI] [PubMed] [Google Scholar]
- 6.D'Andrea, A. D., and M. Grompe. 2003. The Fanconi anaemia/BRCA pathway. Nat. Rev. Cancer 3:23-34. [DOI] [PubMed] [Google Scholar]
- 7.Futaki, M., S. Watanabe, S. Kajigaya, and J. M. Liu. 2001. Fanconi anemia protein, FANCG, is a phosphoprotein and is upregulated with FANCA after TNF-alpha treatment. Biochem. Biophys. Res. Commun. 281:347-351. [DOI] [PubMed] [Google Scholar]
- 8.Garcia-Higuera, I., T. Taniguchi, S. Ganesan, M. S. Meyn, C. Timmers, J. Hejna, M. Grompe, and A. D. D'Andrea. 2001. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol. Cell 7:249-262. [DOI] [PubMed] [Google Scholar]
- 9.Gatei, M., S. P. Scott, I. Filippovitch, N. Soronika, M. F. Lavin, B. Weber, and K. K. Khanna. 2000. Role for ATM in DNA damage-induced phosphorylation of BRCA1. Cancer Res. 60:3299-3304. [PubMed] [Google Scholar]
- 10.Gordon, S. M., and M. Buchwald. 2003. Fanconi anemia protein complex: mapping protein interactions in the yeast 2- and 3-hybrid systems. Blood 102:136-141. [DOI] [PubMed] [Google Scholar]
- 11.Joenje, H., and K. J. Patel. 2001. The emerging genetic and molecular basis of Fanconi anaemia. Nat. Rev. Genet. 2:446-457. [DOI] [PubMed] [Google Scholar]
- 12.Kennedy, R. D., and A. D. D'Andrea. 2005. The Fanconi anemia/BRCA pathway: new faces in the crowd. Genes Dev. 19:2925-2940. [DOI] [PubMed] [Google Scholar]
- 13.Kim, S. T., D. S. Lim, C. E. Canman, and M. B. Kastan. 1999. Substrate specificities and identification of putative substrates of ATM kinase family members. J. Biol. Chem. 274:37538-37543. [DOI] [PubMed] [Google Scholar]
- 14.Kim, S. T., B. Xu, and M. B. Kastan. 2002. Involvement of the cohesin protein, Smc1, in Atm-dependent and independent responses to DNA damage. Genes Dev. 16:560-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levitus, M., Q. Waisfisz, B. C. Godthelp, Y. de Vries, S. Hussain, W. W. Wiegant, E. Elghalbzouri-Maghrani, J. Steltenpool, M. A. Rooimans, G. Pals, F. Arwert, C. G. Mathew, M. Z. Zdzienicka, K. Hiom, J. P. De Winter, and H. Joenje. 2005. The DNA helicase BRIP1 is defective in Fanconi anemia complementation group Nat. Genet. 37:934-935. [DOI] [PubMed] [Google Scholar]
- 16.Levran, O., C. Attwooll, R. T. Henry, K. L. Milton, K. Neveling, P. Rio, S. D. Batish, R. Kalb, E. Velleuer, S. Barral, J. Ott, J. Petrini, D. Schindler, H. Hanenberg, and A. D. Auerbach. 2005. The BRCA1-interacting helicase BRIP1 is deficient in Fanconi anemia. Nat. Genet. 37:931-933 [DOI] [PubMed] [Google Scholar]
- 17.Lim, D. S., S. T. Kim, B. Xu, R. S. Maser, J. Lin, J. H. Petrini, and M. B. Kastan. 2000. ATM phosphorylates p95/nbs1 in an S-phase checkpoint pathway. Nature 404:613-617. [DOI] [PubMed] [Google Scholar]
- 18.Litman, R., M. Peng, Z. Jin, F. Zhang, J. Zhang, S. Powell, P. R. Andreassen, and S. B. Cantor. 2005. BACH1 is critical for homologous recombination and appears to be the Fanconi anemia gene product FANCJ. Cancer Cell 8:255-265. [DOI] [PubMed] [Google Scholar]
- 19.Meetei, A. R., J. P. de Winter, A. L. Medhurst, M. Wallisch, Q. Waisfisz, H. J. van de Vrugt, A. B. Oostra, Z. Yan, C. Ling, C. E. Bishop, M. E. Hoatlin, H. Joenje, and W. Wang. 2003. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat. Genet. 35:165-170. [DOI] [PubMed] [Google Scholar]
- 20.Meetei, A. R., A. L. Medhurst, C. Ling, Y. Xue, T. R. Singh, P. Bier, J. Steltenpool, S. Stone, I. Dokal, C. G. Mathew, M. Hoatlin, H. Joenje, J. P. de Winter, and W. Wang. 2005. A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat. Genet. 37:958-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meetei, A. R., S. Sechi, M. Wallisch, D. Yang, M. K. Young, H. Joenje, M. E. Hoatlin, and W. Wang. 2003. A multiprotein nuclear complex connects Fanconi anemia and Bloom syndrome. Mol. Cell. Biol. 23:3417-3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mi, J., and G. M. Kupfer. 2004. The Fanconi anemia core complex associates with chromatin during S phase. Blood 105:759-766. [DOI] [PubMed] [Google Scholar]
- 23.Mi, J., F. Qiao, J. B. Wilson, A. A. High, M. J. Schroeder, P. T. Stukenberg, A. Moss, J. Shabanowitz, D. F. Hunt, N. J. Jones, and G. M. Kupfer. 2004. FANCG is phosphorylated at serines 383 and 387 during mitosis. Mol. Cell. Biol. 24:8576-8585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Montes de Oca, R., P. R. Andreassen, S. P. Margossian, R. C. Gregory, T. Taniguchi, X. Wang, S. Houghtaling, M. Grompe, and A. D. D'Andrea. 2005. Regulated interaction of the Fanconi anemia protein, FANCD2, with chromatin. Blood 105:1003-1009. [DOI] [PubMed] [Google Scholar]
- 25.Morgan, S. E., C. Lovly, T. K. Pandita, Y. Shiloh, and M. B. Kastan. 1997. Fragments of ATM which have dominant-negative or complementing activity. Mol. Cell. Biol. 17:2020-2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mosedale, G., W. Niedzwiedz, A. Alpi, F. Perrina, J. B. Pereira-Leal, M. Johnson, F. Langevin, P. Pace, and K. J. Patel. 2005. The vertebrate Hef ortholog is a component of the Fanconi anemia tumor-suppressor pathway. Nat. Struct. Mol. Biol. 12:763-771. [DOI] [PubMed] [Google Scholar]
- 27.Naf, D., G. M. Kupfer, A. Suliman, K. Lambert, and A. D. D'Andrea. 1998. Functional activity of the Fanconi anemia protein FAA requires FAC binding and nuclear localization. Mol. Cell. Biol. 18:5952-5960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakanishi, K., Y. G. Yang, A. J. Pierce, T. Taniguchi, M. Digweed, A. D. D'Andrea, Z. Q. Wang, and M. Jasin. 2005. Human Fanconi anemia monoubiquitination pathway promotes homologous DNA repair. Proc. Natl. Acad. Sci. USA 102:1110-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nijman, S. M., T. T. Huang, A. M. Dirac, T. R. Brummelkamp, R. M. Kerkhoven, A. D. D'Andrea, and R. Bernards. 2005. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol. Cell 17:331-339. [DOI] [PubMed] [Google Scholar]
- 30.Pace, P., M. Johnson, W. M. Tan, G. Mosedale, C. Sng, M. Hoatlin, J. de Winter, H. Joenje, F. Gergely, and K. J. Patel. 2002. FANCE: the link between Fanconi anaemia complex assembly and activity. EMBO J. 21:3414-3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shechter, D., V. Costanzo, and J. Gautier. 2004. ATR and ATM regulate the timing of DNA replication origin firing. Nat. Cell Biol. 6:648-655. [DOI] [PubMed] [Google Scholar]
- 32.Shechter, D., and J. Gautier. 2005. ATM and ATR check in on origins: a dynamic model for origin selection and activation. Cell Cycle 4:235-238. [PubMed] [Google Scholar]
- 33.Syljuasen, R. G., C. S. Sorensen, L. T. Hansen, K. Fugger, C. Lundin, F. Johansson, T. Helleday, M. Sehested, J. Lukas, and J. Bartek. 2005. Inhibition of human Chk1 causes increased initiation of DNA replication, phosphorylation of ATR targets, and DNA breakage. Mol. Cell. Biol. 25:3553-3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taniguchi, T., I. Garcia-Higuera, P. R. Andreassen, R. C. Gregory, M. Grompe, and A. D. D'Andrea. 2002. S-phase-specific interaction of the Fanconi anemia protein, FANCD2, with BRCA1 and RAD51. Blood 100:2414-2420. [DOI] [PubMed] [Google Scholar]
- 35.Taniguchi, T., I. Garcia-Higuera, B. Xu, P. R. Andreassen, R. C. Gregory, S. T. Kim, W. S. Lane, M. B. Kastan, and A. D. D'Andrea. 2002. Convergence of the Fanconi anemia and ataxia telangiectasia signaling pathways. Cell 109:459-472. [DOI] [PubMed] [Google Scholar]
- 36.Wang, X., P. R. Andreassen, and A. D. D'Andrea. 2004. Functional interaction of monoubiquitinated FANCD2 and BRCA2/FANCD1 in chromatin. Mol. Cell. Biol. 24:5850-5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu, X., V. Ranganathan, D. S. Weisman, W. F. Heine, D. N. Ciccone, T. B. O'Neill, K. E. Crick, K. A. Pierce, W. S. Lane, G. Rathbun, D. M. Livingston, and D. T. Weaver. 2000. ATM phosphorylation of Nijmegen breakage syndrome protein is required in a DNA damage response. Nature 405:477-482. [DOI] [PubMed] [Google Scholar]
- 38.Xu, B., S. Kim, and M. B. Kastan. 2001. Involvement of Brca1 in S-phase and G2-phase checkpoints after ionizing irradiation. Mol. Cell. Biol. 21:3445-3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamashita, T., G. M. Kupfer, D. Naf, A. Suliman, H. Joenje, S. Asano, and A. D. D'Andrea. 1998. The Fanconi anemia pathway requires FAA phosphorylation and FAA/FAC nuclear accumulation. Proc. Natl. Acad. Sci. USA 95:13085-13090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yazdi, P. T., Y. Wang, S. Zhao, N. Patel, E. Y. Lee, and J. Qin. 2002. SMC1 is a downstream effector in the ATM/NBS1 branch of the human S-phase checkpoint. Genes Dev. 16:571-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.You, Z., C. Chahwan, J. Bailis, T. Hunter, and P. Russell. 2005. ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol. Cell. Biol. 25:5363-5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zampetti-Bosseler, F., and D. Scott. 1981. Cell death, chromosome damage and mitotic delay in normal human, ataxia telangiectasia and retinoblastoma fibroblasts after X-irradiation. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 39:547-558. [DOI] [PubMed] [Google Scholar]
- 43.Zhao, S., Y.-C. Weng, S.-S. F. Yuan, Y.-T. Lin, H.-C. Hsu, S.-C. J. Lin, E. Gerbino, M.-H. Song, M. Z. Zdzienicka, R. A. Gatti, J. W. Shay, Y. Ziv, Y. Shiloh, and E. Y.-H. P. Lee. 2000. Functional link between ataxia-telangiectasia and Nijmegen breakage syndrome gene products. Nature 405:473-477. [DOI] [PubMed] [Google Scholar]
- 44.Ziv, Y., A. Bar-Shira, I. Pecker, P. Russell, T. J. Jorgensen, I. Tsarfati, and Y. Shiloh. 1997. Recombinant ATM protein complements the cellular A-T phenotype. Oncogene 15:159-167. [DOI] [PubMed] [Google Scholar]