Abstract
The phytohormone abscisic acid (ABA) plays an important role in modulating plant growth, development, and stress responses. In a genetic screen for mutants with altered drought stress responses, we identified an ABA-overly sensitive mutant, the abo1 mutant, which showed a drought-resistant phenotype. The abo1 mutation enhances ABA-induced stomatal closing and increases ABA sensitivity in inhibiting seedling growth. abo1 mutants are more resistant to oxidative stress than the wild type and show reduced levels of transcripts of several stress- or ABA-responsive genes. Interestingly, the mutation also differentially modulates the development and growth of adjacent guard cells. Map-based cloning identified ABO1 as a new allele of ELO2, which encodes a homolog of Saccharomyces cerevisiae Iki3/Elp1/Tot1 and human IκB kinase-associated protein. Iki3/Elp1/Tot1 is the largest subunit of Elongator, a multifunctional complex with roles in transcription elongation, secretion, and tRNA modification. Ecotopic expression of plant ABO1/ELO2 in a tot1/elp1Δ yeast Elongator mutant complements resistance to zymocin, a yeast killer toxin complex, indicating that ABO1/ELO2 substitutes for the toxin-relevant function of yeast Elongator subunit Tot1/Elp1. Our results uncover crucial roles for ABO1/ELO2 in modulating ABA and drought responses in Arabidopsis thaliana.
Water stress caused by drought and soil salinity is an important environmental factor that limits the productivity and distribution of plants. The cellular and molecular mechanisms of plant responses to water stress have been analyzed extensively (48, 59, 63). Water stress can induce the accumulation of the phytohormone abscisic acid (ABA) (59). ABA plays a vital role in triggering stomatal closure, which reduces transpirational water loss and constitutes an essential part of plant drought tolerance (48, 58, 63). Analysis of Arabidopsis thaliana mutants has defined several ABA response loci that encode proteins such as protein phosphatases and kinases, which greatly affect guard cell movement (10, 45, 58).
Recent studies indicate that transcripts of protein-coding genes are regulated at all steps of RNA metabolism, from transcription initiation to RNA processing (50). A great deal of information about plant transcriptional regulators that bind the promoters to initiate gene transcription in response to water stress has been collected (61). In contrast, much less is known about proteins involved in RNA processing (30). Nevertheless, recent studies point to a central role of RNA processing in regulating ABA sensitivity and osmotic stress responses. The RNA-binding protein FCA was reported to be an ABA receptor, although it appears to function in ABA regulation of flowering rather than in seed dormancy or drought tolerance (44). ABH1, a cap-binding protein, functions in early ABA signaling (20). A recessive mutation in the SAD1 gene encoding an Sm-like snRNP required for mRNA splicing, export, and degradation rendered plants hypersensitive to ABA and drought (56). The Arabidopsis HYL1 gene encodes a nuclear double-stranded RNA-binding protein. A knockout mutation of the HYL1 gene caused abnormal development, increased sensitivity to abscisic acid, and reduced sensitivity to auxin and cytokinin (33). HYL1 controls gene expression likely through microRNA-mediated gene regulation, although the targeted genes related to ABA sensitivity are still unknown (18). AKIP1 isolated from Vicia faba is a single-stranded RNA-binding protein which can bind to a dehydrin mRNA after phosphorylation by an ABA-activated protein kinase (32). The CRYOPHYTE/LOS4 gene encoding a DEAD box RNA helicase is essential for mRNA export, and the cryophyte mutant is hypersensitive to ABA during seed germination (14, 15). The double-stranded RNA-binding protein FRY2/CPL1 negatively regulates ABA and osmotic stress responses possibly through modulating RNA polymerase II activity by dephosphorylating Ser-5 of its C-terminal domain (27, 28, 57). Transcriptional elongation mediated by RNA polymerase II is a pivotal process in gene regulation, is highly regulated in eukaryotes by numerous factors in mRNA biogenesis and maturation, and is an emerging topic of active study in biology (50).
We report here the isolation and characterization of the Arabidopsis ABA-overly sensitive 1 (abo1) mutant and map-based cloning of ABO1. The abo1 mutant was isolated on the basis of its drought-resistant phenotype. The abo1 mutation enhances ABA sensitivity in both seedling growth and stomatal closure. ABO1 is a new allele of ELO2 (38), which encodes a homolog of yeast Iki3/Elp1/Tot1 or human IκB kinase-associated protein (IKAP), collectively the largest subunit of Elongator, a complex with roles in secretion, tRNA modification, and mRNA transcription elongation (8, 11, 13, 19, 25, 39, 43, 51, 60). These findings suggest that Elongator is important for plants to respond to ABA and drought exposure and that ABO1/ELO2 may play a vital role in ABA signal transduction pathways.
MATERIALS AND METHODS
Plant growth conditions.
Plants were grown in 340-ml pots filled with a mixture of peat/forest soil and vermiculite (3:1) in a greenhouse at 22°C, with light intensity of 50 μmol m−2 s−1 and 70% rH under long-day conditions (16-h-light/8-h-dark cycle). Seedlings were germinated and grown on Murashige and Skoog (MS) medium (M5519; Sigma) supplemented with 3% (wt/vol) sucrose and 0.8% agar under the same growth conditions.
Isolation of the abo1 mutant, growth conditions, and genetic analysis.
The isolation of the abo1-1 mutant of Arabidopsis thaliana (Columbia gl1 background) was performed by use of a water loss screening system. To identify mutants, ethyl methyl sulfonate (EMS)-mutagenized M2 seeds were sown on MS medium. Four-day-old seedlings were transferred to soil and grown for 2 weeks with sufficient watering, and then water was withheld. The abo1-1 mutant was identified as a plant surviving the drought treatment, while other plants around it wilted and died. The abo1-1 mutant was backcrossed to the wild-type plant in the original background, and the resulting F1 seedlings as well as F2 progeny from self-fertilized F1 plants were evaluated in a drought stress assay.
Three T-DNA insertion mutants (Columbia background, SALK database accession no. SALK_004690, SALK_011529, and SALK_084199) were obtained from the Arabidopsis Biological Resource Center (Columbus, Ohio). Allelic tests done by crossing the four mutants (the abo1 mutant and the three T-DNA insertion mutants) revealed that they were allelic and so were named abo1-1 (EMS-mutagenized mutant) and abo1-2, abo1-3, and abo1-4 (T-DNA insertion mutants). All subsequent physiological and phenotypic analyses were carried out using the abo1-1 mutant that had been backcrossed to the wild type four times to remove other mutations.
Genetic mapping.
abo1-1 mutant plants (Columbia gl1 background) were crossed to Landsberg wild-type plants. A total of 1,077 abo1-1 mutant plants were selected from the self-fertilized F2 population by the leaf water loss assay combined with the identification of the abnormal stomatal development phenotype. DNA was isolated from individual mutant plants and analyzed for recombination events with simple sequence length polymorphism (SSLP) markers. The following primer pairs for SSLP markers that are polymorphic between Columbia gl1 and Landsberg were developed: for F8L15, forward primer 5′-CGGTAATACCTATGGAGCCGCCG-3′ and reverse primer 5′-GCGCATGGTACCGCTAATGGCAG-3′; for MUA22, forward primer 5′-GGAGAGACTGATGGACGCCATTTG-3′ and reverse primer 5′-GTCCTCATCAAGGGGCTGCAGAGG-3′; for T24H18, forward primer 5′-CAGTGCATGGTTTGCATGGGAA-3′ and reverse primer 5′-TTTACGCAGGACATGTTTCCTCTC-3′; for T6I14, forward primer 5′-AAGGACCAGCGTGGCTCAAG-3′ and reverse primer 5′-AATCACTCACTGCCTCTTTGGAGG-3′; and for MXE10, forward primer 5′-CGTCAGGGTGCTGCTTTTCTC-3′ and reverse primer 5′-GTGCCTGCACATTGATCACCATC-3′.
Stomatal aperture bioassays.
Stomatal closing assays were conducted as described previously (41). Rosette leaves were floated in solutions containing 50 μM CaCl2, 10 mM KCl, 10 mM MES [2-(N-morpholino)ethanesulfonic acid]-Tris, pH 6.15, and exposed to light (150 μmol m−2 s−l) for 2 h. Subsequently, ABA was added to the solution at 0.5 to 10 μM to assay for stomatal closing. After ABA treatment for 2 h, stomatal apertures were measured as described previously (41). Values are means ± standard errors (SE) (n = 30). Significance (P < 0.05) was assessed by Student's t test.
Stomatal development assays.
Epidermal strips from rosette leaves of 3-week-old seedlings of wild-type and mutant plants were examined for stomata under a light microscope (B5-223 IEP; Motic China Group Co., Ltd.). For scanning electron microscopy, rosette leaves of 3-week-old seedlings of wild-type and mutant plants were fixed as described previously (5), and stomata were observed under a scanning electron microscope (S-570; Hitachi, Japan).
Water loss measurements and determination of ABA content.
Rosette leaves of mutant and wild-type plants growing under normal conditions for 3 weeks were detached and weighed immediately on a piece of weighting paper and then placed on a laboratory bench (40% rH) and weighed at designated time intervals. Three replicates were done for each line. The percentage of loss of fresh weight was calculated on the basis of the initial weight of the plants. ABA contents were determined as described previously (6).
pABO1-GUS chimeric construct and histochemical analysis.
A promoter fragment of 2,173 bp of the ABO1 gene, defined as pABO1, which contains the promoter region of ABO1, its partial coding region, and the partial coding region of ABO1's upstream gene (MSH12.16), was amplified from Columbia gl1 genomic DNA by PCR with the primer pair 5′-CACCGGAAGGAAGAGAGCTGAAGGGC-3′ (to add CACC at the 5′ end) and 5′-GAGGGTCAGAGGGATTCAGAAGG-3′. The amplified fragment was cloned into a Gateway Technology system for cloning and expression (Invitrogen), resulting in a transcriptional fusion of the ABO1 promoter and its partial coding region with the GUS coding region. The ABO1 promoter-GUS fusion construct was introduced into Agrobacterium tumefaciens and transferred into plants. Thirty T2 transgenic lines were subjected to β-glucuronidase (GUS) assays. GUS staining was performed as described previously (47).
RNA gel blot analysis.
Seedlings grown on MS medium for 3 weeks were transferred to a solution containing 100 μM ABA or no ABA (for control) for 3 h or 5 h. Total RNA was isolated and analyzed as previously described (6). An RD29A fragment (967 bp) was amplified by PCR with forward primer 5′-GACGAGTCAGGAGCTGAGCTG-3′ and reverse primer 5′-CGATGCTGCCTTCTCGGTAGAG-3′. A fragment (552 bp) of the RD29B gene was amplified by using forward primer 5′-CCGACGGGAACTCATGATCAGTTC-3′ and reverse primer 5′-CACTTCCACCTCCTTTGTAGCCG-3′. A COR47 fragment (413 bp) was amplified by PCR with forward primer 5′-GAAGCTCCCAGGACACCACGAC-3′ and reverse primer 5′-CAGCGAATGTCCCACTCCCAC-3′. An ABI1 fragment (517 bp) was amplified by PCR with forward primer 5′-CGCAGGTCCTTTCAGGCCATTC-3′ and reverse primer 5′-GCCATGGCCGTCGTAAACAC-3′. These gene fragments were used as probes. A tubulin gene was used as a loading control.
Elongator cross-complementation studies with yeast and plant.
For phenotypic complementation assays, the yeast tot1/elp1Δ mutant LFY3, deleted for the Elongator subunit 1 gene (TOT1/ELP1) (21), was transformed with pDJ98, a vector for galactose-regulated expression of the plant Elongator subunit 1, ABO1/ELO2 (38). pDJ98 construction involved PaeI and SacI restriction of pMD18.T, Klenow fill-in of the resulting 4-kb ABO1/ELO2 cDNA, and subcloning into SmaI-restricted pBluescript (Stratagene) to yield pDJ79. Following pDJ79 restriction by SalI, the ABO1/ELO2 cDNA was cloned into SalI-cut pYEX-GAL, a pYEX-BX (Clontech) expression vector derivative with its CUP1 promoter replaced by the GAL1 promoter. The latter process involved BamHI/HindIII replacement of the GAL1 promoter fragment from plasmid pRB1438 (kindly provided by Mike Stark, University of Dundee, United Kingdom). Collectively, 2μm multicopy vector pDJ98 allows galactose-driven expression of ABO1/ELO2, its maintenance is selectable by URA3, and its copy number is amplifiable by use of leu2d, a transcriptionally compromised marker (52).
Sensitivity tests towards endogenous expression of the lethal γ-toxin subunit of zymocin involved LFY3 cells transformed with pDJ98, empty 2μm vector pYEX-GAL, and pFF14, a 2μm vector carrying the yeast TOT1/ELP1 gene (11). Following subsequent transformations with pHMS14 (a GAL1-γ-toxin expression vector) or pHMS22 (an empty GAL1-promoter control) (11), 10-fold serial dilutions of these yeast tester strains were grown on 2% (vol/vol) glucose synthetic complete medium (46) lacking tryptophan, leucine, and histidine. The response towards γ-toxin expression was monitored after the strains were shifted onto 2% (vol/vol) galactose medium. Growth proceeded for 3 to 4 days at 30°C. Testing the effects of chemical stress towards growth performance of the LFY3 transformants involved addition of 5 to 10 mM caffeine (Sigma) to complete yeast extract-peptone-dextrose (YPD) media (46) and cultivation at 30°C. Thermosensitivity was tested between 30°C and 39°C.
RESULTS
Identification of the abo1 mutant.
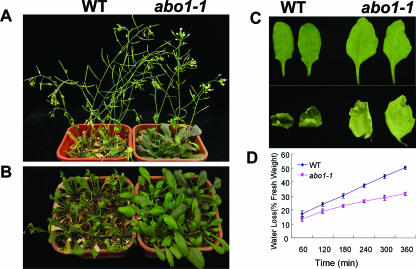
Water was withheld from an EMS-mutagenized population of Arabidopsis (Columbia gl1 background) seedlings grown in a growth room for 2 weeks to screen for mutants with increased drought sensitivity or resistance. In order to recover drought-sensitive mutants, we collected 10 EMS-mutagenized M1 plants as a small pool. Approximately 108 M2 seedlings from each pool were planted in 12 pots filled with soil. We screened approximately 57,000 plants from 534 pools. Putative mutants with drought response phenotypes were scored by the degree of leaf wilting compared with that of neighboring plants. The abo1-1 mutant was identified by its less severe leaf wilting and enhanced drought resistance. Figure 1A shows wild-type and abo1-1 plants grown for 3 weeks and then treated with drought by withholding water for 11 days. abo1-1 leaves were still turgid, whereas wild-type leaves showed serious wilting. After an additional 5 days, we rewatered the treated plants and found that 100% of abo1-1 plants survived, while all wild-type plants died. We also treated 2-week-old plants in soil by withholding water and obtained a similar result (Fig. 1B), indicating that the abo1-1 plant is more drought resistant in two different growth stages. The drought-resistant phenotype of the abo1 mutant was further evaluated by measuring water loss from detached leaves. As shown in Fig. 1C and D, detached leaves of the abo1-1 mutant lost water more slowly than did those of the wild-type plants.
FIG. 1.
abo1-1 mutant plants are resistant to drought stress. Reduced wilting was observed for abo1 mutant plants during water stress treatment. Wild-type (WT) and abo1-1 plants were grown with sufficient water for (A) 3 or (B) 2 weeks, and then water was withheld for 11 days. (C) Water loss in detached leaves from 3-week-old plants placed in a room (40% rH) for 14 h. (D) Comparison of rates of water loss from detached leaves of the wild-type and abo1-1 plants. Water loss is expressed as the proportion of initial fresh weight. Values are means ± SE from 15 leaves for each of three independent experiments.
In order to determine whether the abo1-1 mutant phenotype is caused by mutation in a single nuclear gene, we crossed the abo1-1 plant with the wild-type plant, and all resulting F1 plants showed the wild-type phenotype under drought stress. An F2 progeny from self-fertilized F1 segregated in an ∼3:1 ratio (198 to 67) between wild-type and abo1-1 mutant phenotypes. These results indicate that abo1-1 has a recessive mutation in a single nuclear gene.
Stomatal closure and seedling growth of the abo1-1 mutant are hypersensitive to ABA.
In order to determine the cause of the drought-resistant phenotype of the abo1-1 mutant, we performed experiments to determine whether this phenotype is related to increased sensitivity of stomatal closure to ABA. abo1-1 and wild-type plants were exposed to high humidity for 12 h to open stomata fully. Epidermal peels from these plants were used to analyze stomatal responses to ABA. In response to ABA treatment at different concentrations, the closure of preopened abo1-1 stomata was greatly enhanced compared to that of the wild type (Fig. 2A and B). Exposure to 0.5 μM ABA enhanced stomatal closure to the same degree in abo1-1 plants as 10 μM ABA did in the wild type (Fig. 2B). Stomata in abo1-1 plants were almost completely closed with 1 μM ABA treatment for 2 h, but the stomata did not completely close even with 20 μM ABA in wild-type plants for the same treatment time (data not shown). By contrast, darkness-induced stomatal closure in abo1-1 plants was similar to that of the wild type (Fig. 2C and D). These results indicate that the abo1-1 mutation specifically enhances the sensitivity of stomatal closure to ABA.
FIG. 2.
Stomatal closure in abo1-1 plants is hypersensitive to ABA (A and B) but not to darkness (C and D). Data represent the means ± SE from 30 stomata measured for each data point, from three independent experiments. WT, wild type. Bar, 10 μm.
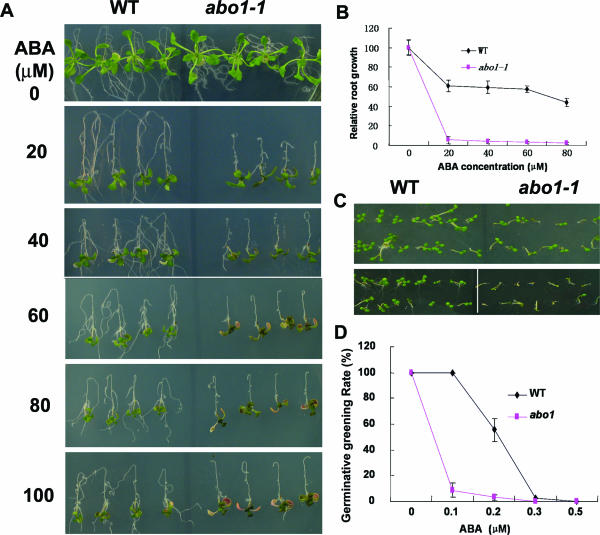
We also analyzed the ABA sensitivities of abo1-1 plants in seed germination and seedling growth. Seedlings grown for 5 days on MS medium were transferred to MS media containing different concentrations of ABA. One week later, the plants were scored for visible changes. Without ABA, the root growth of abo1-1 plants was slower than that of wild-type plants but they had similar levels of shoot growth. In the presence of different concentrations of ABA, both root and shoot growth of abo1-1 plants were more inhibited than that of the wild type (Fig. 3A and B). At higher concentrations of ABA (more than 70 μM), abo1-1 leaves showed more anthocyanin pigments, and some cotyledons became chlorotic, which was rarely seen in wild-type plants even at 100 μM ABA. However, supplementation of different concentrations of ABA on agar plates did not produce a difference in abo1 seed germination compared to that of the wild type (Fig. 3C). The postgermination growth of abo1-1 seedlings was more impaired than that of the wild type, as indicated by the ratio of seedlings with green cotyledons to the total number of seedlings after germination for 1 week (Fig. 3D). In order to determine whether abo1 mutants are specifically sensitive to ABA, we also tested the effects of other phytohormones, including methyl jasmonate, salicylic acid, indole-3-acetic acid, gibberellins (GA3), and ethylene, on seedling growth but failed to find any difference between the abo1 mutant and the wild type (data not shown).
FIG. 3.
Seedling growth of abo1-1 plants is hypersensitive to ABA. (A) Comparison of abo1-1 and wild-type (WT) seedlings grown on MS medium for 5 days and then transferred to MS medium containing different concentrations of ABA. The pictures were taken 7 days after transfer. (B) Comparison of relative root lengths between abo1-1 and wild-type plants. Values are means ± SE (n = 30). (C) Seed germination of abo1-1 and wild-type plants. Wild-type and abo1-1 seeds were planted on MS agar medium (upper rows) or MS medium containing 0.1 μM ABA (lower rows). Plates were transferred to a growth chamber after 3-day stratification. The pictures were taken 7 days after transfer. (D) Seedling greening rates of abo1-1 and wild-type plants. Ratio of green seedlings to total seedlings was determined after plates for seed germination were cultured for 7 days. Values are means ± SE from three independent experiments, with 100 seeds used per genotype per data point for each experiment.
The abo1 mutation enhances oxidative stress tolerance.
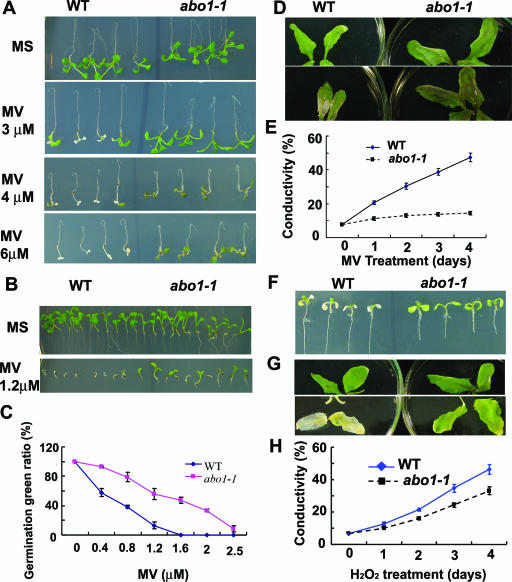
ABA can induce the expression of genes encoding antioxidant enzymes (17, 62). ABA also induces the production of reactive oxygen species that serve as a second messenger in ABA signaling in guard cells (24, 42). In order to test whether the ABA sensitivity of abo1-1 seedlings might be connected with reactive oxygen species, we analyzed the response of abo1-1 mutant plants to oxidative stress. Methyl viologen (MV) (paraquat) inhibits electron transport in the reduction of NADP to NADPH during photosynthesis and thus enhances H2O2 production in chloroplasts under light (53). Seedlings grown on MS medium for 5 days were transferred to new MS medium supplemented with different concentrations of MV. The abo1-1 seedlings grew well on MS medium supplemented with 3 μM MV, while more than 50% of wild-type seedlings were killed at this concentration (Fig. 4A) after a 5-day treatment. Exposure to 4 μM MV completely killed all wild-type seedlings, but all abo1 mutant seedlings still survived even at 6 μM MV. Seed germination was inhibited to similar extents for both abo1-1 and wild-type plants by various concentrations of MV. After radicles emerged, the young seedlings were more sensitive to MV. Figure 4B shows that exposure to 1.2 μM MV for 10 days prevented the cotyledon greening of more than 90% of the wild-type seedlings, whereas only 40% of the abo1-1 cotyledons were affected. Dose-response analysis further indicates that abo1-1 mutant seedlings are less sensitive to MV than wild-type seedlings (Fig. 4C).
FIG. 4.
Seed germination and seedling growth of abo1 mutant plants are resistant to oxidative stress. (A) Five-day-old seedlings of abo1-1 and wild-type (WT) plants were transferred to MS medium or MS medium containing different concentrations of MV. The pictures were taken after 5 days. (B) Germination comparison of wild-type and abo1-1 plants in the presence of 1.2 μM MV. The pictures were taken after 10 days. (C) Effects of different concentrations of MV on seedling greening rates of abo1-1 and wild-type plants grown on MS medium containing different concentrations of MV after 10 days. (D) Comparison of MV effects on abo1 mutant and wild-type leaves. (G) Mature leaves were floated on 1 mM MV for 5 days. (E) Ion leakage analysis of leaves treated with 1 mM MV at different times. (F) Five-day-old seedlings of abo1-1 and wild-type plants were transferred to MS medium containing 10 μM Rose Bengal. Mature leaves were treated with 10 mM H2O2 for 5 days. The pictures were taken after 3 days. (H) Ion leakage analysis of H2O2 effects on abo1 mutant and wild-type leaves.
The leaves taken from plants grown in soil were treated with 1 μM MV. After 5 days, the abo1-1 leaves showed enhanced anthocyanin pigmentation but without any apparent chlorotic symptom. In contrast, wild-type leaves were severely damaged and displayed chlorotic spots or bleaching (Fig. 4D). Electrolyte leakage (conductivity) is often used as an indicator of tissue damage. The abo1-1 leaves treated with 1 μM MV showed substantially less ion leakage than the wild-type leaves (Fig. 4E). Rose Bengal is another reagent that generates H2O2 when exposed to the light. Exposure to 10 μM Rose Bengal for 3 days severely damaged the cotyledons of wild-type seedlings, compared with no chlorotic symptom observed for abo1 mutant cotyledons (Fig. 4F). We also found that wild-type leaves were more damaged than abo1-1 leaves by treatment with 10 mM H2O2 for 5 days (Fig. 4G). This was supported by ion leakage measurements at different time points (Fig. 4H). These results show that abo1-1 mutant plants are more resistant to oxidative stress and suggest a possible connection between the ABA and oxidative stress response phenotypes.
Regulation of gene expression by ABO1.
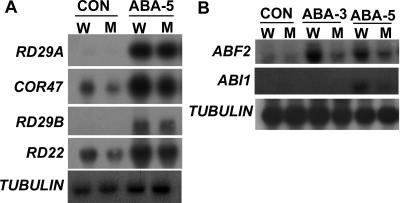
To ascertain whether the ABA response defects of the abo1 mutation are related to an altered expression of ABA- and stress-responsive genes, we conducted Northern blot analysis of different stress-inducible genes. We used the abo1-2 mutant for Northern blot analysis because it is more likely a null allele (see below). The expression of RD29A and COR47 genes is partly ABA independent, while RD22 and RD29B are dependent on ABA (58). Under our conditions, both COR47 and RD22 are expressed at a low level in wild-type and mutant untreated control samples, but the expression levels of both genes are lower in the mutant than in the wild type (Fig. 5A). In response to 20 μM ABA treatment for 5 h, the expression of all four genes is induced to higher levels in the wild type than in the mutant, although the expression difference is rather small for RD29A and RD29B (Fig. 5A).
FIG. 5.
Expression of ABA- and stress-responsive genes in abo1-2 (M) and wild-type (W) seedlings. Three-week-old seedlings growing on agar plates were treated with 20 μM ABA for 3 h (ABA-3) or 5 h (ABA-5). A tubulin gene was used as a loading control. Con, seedlings without any treatment.
The transcription factor ABF2/AREB1 (26) positively regulates the expression of ABA-responsive genes, whereas the protein phosphatase 2C ABI1 negatively regulates ABA responses (16, 35). We found that the expression of ABF2/AREB1 is more induced in the wild type than in the abo1-2 mutant upon ABA treatment (Fig. 5B). The induction of the ABI1 transcript by ABA is lower in the abo1 mutant than in the wild type at 5 h of treatment (Fig. 5B).
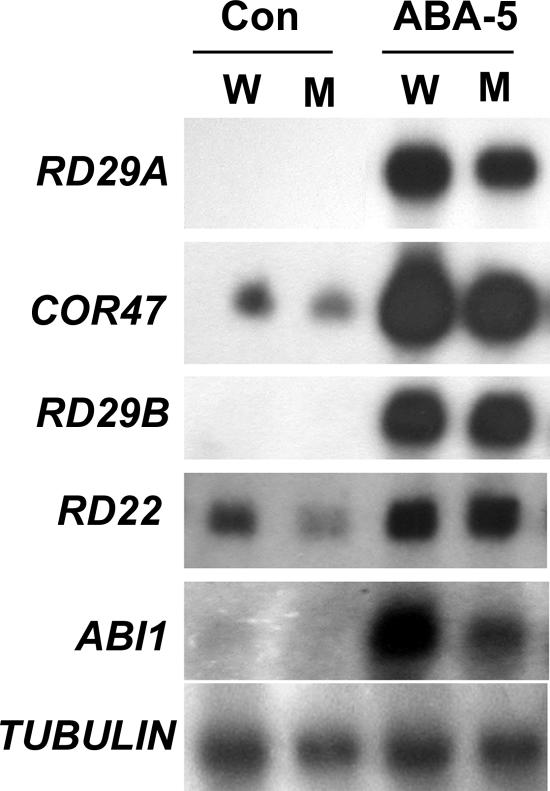
We further compared the expression levels of some of these genes in response to a higher ABA concentration (100 μM). The transcripts of RD29A, COR47, and ABI1 are induced to lower levels in the abo1 mutant than in the wild type, whereas the expression levels of RD29B and RD22 are similar between the abo1 mutant and the wild type (Fig. 6). These results suggest that ABA-responsive genes are less sensitive to ABA induction in the abo1 mutant but that high concentrations of ABA may compensate for the difference for some of the genes.
FIG. 6.
Expression of ABA- and stress-responsive genes in abo1-2 (M) and wild-type (W) seedlings treated with 100 μM ABA for 5 h (ABA-5). A tubulin gene was used as a loading control. Con, seedlings without any treatment.
ABO1 appears to differentially regulate the development and growth of two adjacent pairs of guard cells.
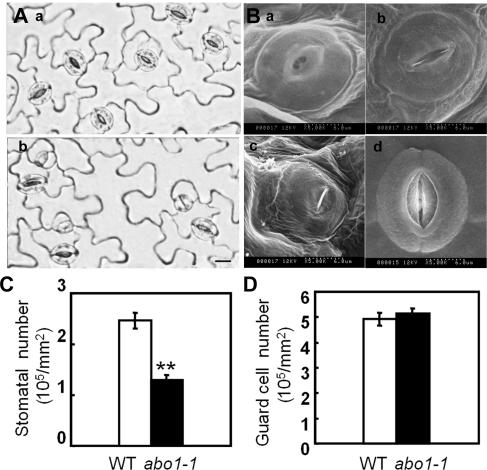
Drought resistance could be contributed by both ABA sensitivity of stomatal movement and stoma number. A stoma is formed through one or more asymmetric cell divisions followed by the symmetric division of the guard mother cell (36). During our experiments on stomatal movement, we noticed that the number of stomata with pores that could be observed under a light microscope in the abo1-1 mutant was only about half of that in the wild type (Fig. 7A and C). Some pairs of guard cells did not form normal stomata or formed stomata with very small pores (Fig. 7B). However, the total number of guard cells that form stomata with or without pores in the abo1-1 plant is almost the same as that in the wild-type plant (Fig. 7D). The abo1 mutation appears to affect only the development and growth of guard cells and their adjacent pavement cells and not the division and differentiation of their precursor cells. These results suggest that, in addition to increased ABA sensitivity of fully developed stomata, the drought-resistant phenotype of the abo1-1 mutant may be caused in part by the reduced number of stomata on the leaf surface.
FIG. 7.
Comparison of stomatal morphologies of abo1-1 and wild-type plants. (A) Light microscopy of abaxial epidermises from mature wild-type (a) and abo1-1 (b) leaves. In wild-type plants, only normally developed guard cells with formal stomata are observed. In contrast, in the abo1-1 mutant, only one pair of guard cells forms normal stoma among two pairs of adjacent stomata. Bar, 10 μm. (B) Scanning electron microscopy of abo1-1 (abnormal) (a, b, and c) and wild-type (d) stomata. (C and D) Quantitative analyses of (C) the numbers of stomata and (D) the pairs of guard cells in abaxial epidermises of abo1-1 and wild-type (WT) plants. Values are means ± SE (n = 30). **, P < 0.01.
Map-based cloning of ABO1.
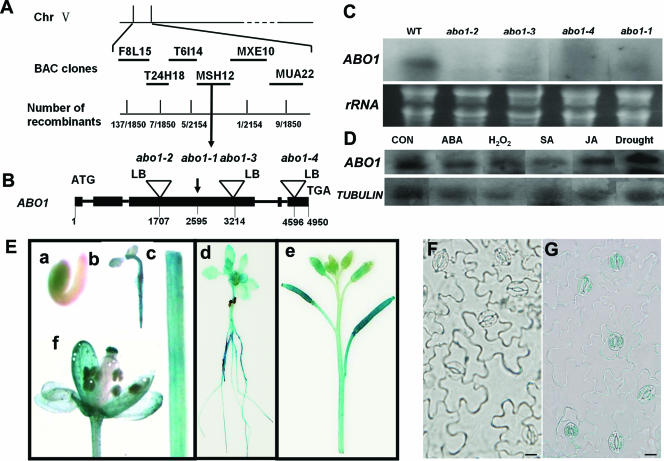
We used a map-based cloning strategy to identify the mutation responsible for the abo1 mutant phenotype. The abo1-1 mutant in the Columbia background was crossed with wild-type plants in the Landsberg background. The phenotypes of the resultant F2 seedlings were determined, and DNA was extracted from each plant and examined with SSLP markers designed according to information from Cereon Genomics (http://www.Arabidopsis.org). Initial mapping located abo1 to the top of chromosome V. Further mapping positioned ABO1 to within six bacterial artificial chromosome clones: F8L15, T24H18, T6I14, MSH12, MXE10, and MUA22. Continued mapping delimited ABO1 to a region within the bacterial artificial chromosome clones T6I14, MSH12, and MXE10 (Fig. 8A). Several predicted open reading frames were amplified by PCR and sequenced. A single nucleotide mutation from G2595 (counting from the first putative ATG of the genomic sequence) to A was found in abo1-1 in the third predicted exon of the putative gene At5g13680 (MSH12.15). The mutation was predicted to change amino acid Trp746 (encoded by TGG) to a stop codon (encoded by TAG) and to result in a C-terminal truncation mutant lacking almost half of the polypeptide sequence of the ABO1 protein (Fig. 8B).
FIG. 8.
Positional cloning and expression pattern of the ABO1 gene. (A) Positional cloning of the ABO1 gene. Chr, chromosome; BAC, bacterial artificial chromosome. (B) Structure of the ABO1 gene, showing the different mutant alleles. LB, T-DNA left border. (C) Expression of the ABO1 gene in different mutant alleles. An rRNA gene was used as a loading control. WT, wild type. (D) ABO1 expression was not induced under different treatment conditions. A tubulin gene was used as a control. Con, seedlings without any treatment; SA, salicylic acid; JA, jasmonic acid. (E) ABO1 promoter-GUS analysis in Arabidopsis transgenic seedlings. (a) One-day-old seedling, (b) 4-day-old seedling, (c) stem, (d) 10-day-old seedling, (e) siliques, and (f) flower. (F) Guard cells on an abaxial epidermis from a nontransgenic plant, used as a negative control. (G) ABO1 promoter-GUS analysis of guard cells on an abaxial epidermis from a transgenic plant. Bar, 10 μm.
We searched the SALK collection database and obtained three independent insertion mutants (SALK_004690, SALK_011529, and SALK_084199) from the Arabidopsis Stock Center (http://www.Arabidopsis.org). Sequencing analysis confirmed the insertion sites provided by the Salk Institute Genome Analysis Laboratory. T-DNA insertions of SALK_004690, SALK_011529, and SALK_084199 were detected before nucleotides 1707, 3214, and 4596 (counting from the first putative ATG of the genomic sequence), respectively. We renamed SALK_004690, SALK_011529, and SALK_084199, respectively, abo1-2, abo1-3, and abo1-4 (Fig. 8B). Northern blot analysis using a probe amplified from nucleotides 1916 to 3037 of ABO1 (counting from the first putative ATG of the genomic sequence) revealed that the expression of the ABO1 gene was detected in the wild type but not in the mutant alleles abo1-1, abo1-2, abo1-3, and abo1-4 (Fig. 8C). We crossed abo1-1 with abo1-2, abo1-3, and abo1-4, and all of the resulting F1 plants showed the abo1-1 drought resistance phenotype. All three T-DNA insertion mutants have, per leaf area, about half the number of guard cells with pores that can be seen under a light microscope compared to the wild type. We also checked the ABA sensitivities of abo1-2, abo1-3, and abo1-4 and found that all of the mutants showed a phenotype similar to that of abo1-1. Taken together, these data demonstrate that ABO1 is the gene mutated in the different abo1 alleles.
ABO1/ELO2 encodes a homolog of yeast Elongator subunit Elp1/Iki3/Tot1.
ABO1 is a new allele of ELO2, which is predicted to encode a protein of 1,319 amino acids with significant similarity to the largest subunit of the yeast Elongator complex, Elp1/Iki3/Tot1 (11, 38). ABO1 shares 50% identity and 68% similarity in the entire amino acid sequence with a putative rice IKI3 (GenBank accession no. NP_910712), 26% identity and 45% similarity in the entire amino acid sequence with the human homolog of yeast Iki3, IKAP (GenBank accession no. AAG43369), and 27% identity and 44% similarity in the entire amino acid sequence with the fission yeast Iki3 (GenBank accession no. NP_595335).
In order to analyze the ABO1/ELO2 gene expression pattern, transgenic Arabidopsis plants expressing an ABO1/ELO2 promoter-ABO1/ELO2 partial coding region-GUS reporter gene were analyzed for GUS activity. The GUS gene was expressed in roots, hypocotyls, stems, leaves, flowers, and siliques (Fig. 8E, panels b to f) but was not detected at the earlier stage of seed germination, which is consistent with the lack of mutant effect on ABA inhibition of seed germination (Fig. 8E, panel a). Consistent with the role of ABO1/ELO2 in guard cells, GUS activity was detected in guard cells of isolated epidermal peels (Fig. 8F and G). It appears that GUS staining is greater in one of the two adjacent pairs of stomata which are usually formed in Arabidopsis. However, because of the low GUS expression and development sequence of the guard cells, it is difficult to differentiate the GUS expression levels of later-formed guard cells from those of earlier-formed guard cells. The GUS expression pattern is consistent with our observed mutant phenotypes. Nevertheless, the GUS expression pattern may not fully reflect the expression pattern of the endogenous ABO1 gene since the ABO1 promoter fragment used for the GUS experiment does not include potential regulatory sequences that may be present in the introns or other parts of the gene.
To determine whether ABO1/ELO2 gene expression is induced by stress, we performed Northern blot experiments using total RNA extracted from 2-week-old seedlings treated with different stresses. The ABO1/ELO2 transcript was not induced by drought or H2O2 or by treatment with exogenous hormones such as ABA, salicylic acid, or jasmonic acid (Fig. 8D), which indicates that ABO1/ELO2 is not a stress-inducible gene.
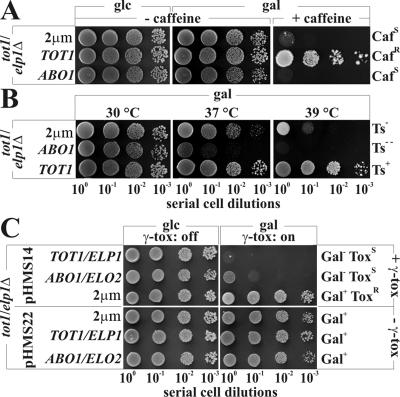
In order to study the plant ABO1/ELO2 gene, we asked whether its expression complements a yeast tot1/elp1Δ mutant lacking the Elongator subunit 1 gene homologous to ABO1/ELO2. In yeast, loss of Elongator elicits sensitivity to thermal and chemical stress and causes resistance to cell death by the zymocin toxin complex (11, 22). Intriguingly, while caffeine sensitivity of the tot1/elp1Δ strain remained unaltered at 30°C under galactose induction of the ABO1/ELO2 gene (Fig. 9A), cell viability of ABO1/ELO2 expressors became progressively compromised at 37°C and ceased completely at 39°C (Fig. 9B) in the absence of caffeine. This effect is not linked to thermosensitivity of the Elongator mutant (11), as tot1/elp1Δ cells carrying empty vector proved to be significantly more thermotolerant than ABO1/ELO2 expressors. As expected, the wild-type yeast gene TOT1/ELP1 efficiently complemented tot1/elp1Δ cells to restore wild-type thermotolerance at 39°C and caffeine resistance (Fig. 9A and B). Hence, under conditions known to interfere with Elongator deficits, ABO1/ELO2 expression is not able to improve cell viability. Contrast this with complementation of γ-toxin resistance due to galactose induction of the ABO1/ELO2 gene (Fig. 9C). Based on normal performance in the presence of pHMS22, a vector devoid of the zymocin subunit gene, we consider the growth arrest of ABO1/ELO2 expressors by the γ-toxin (Fig. 9C) to be fully ascribable to ABO1/ELO2 gene expression. Hence, the ability of ABO1/ELO2 to confer γ-toxin sensitivity in tot1/elp1Δ, an otherwise γ-toxin-resistant Elongator mutant (11), strongly indicates that the plant gene ABO1/ELO2 substitutes for this zymocin-relevant function of the yeast Elongator subunit 1. Whether the ABO1/ELO2 protein assembles into a chimeric Elongator complex is not yet known but appears likely since, in yeast, zymocin toxicity requires a structurally integrated six-subunit complex (9, 12).
FIG. 9.
Cross-complementation studies of a yeast tot1/elp1Δ Elongator mutant by expression of plant ABO1/ELO2. (A) Caffeine sensitivity. LFY3 (tot1/elp1Δ) cells transformed with vector pFF14 (TOT1/ELP1) or pDJ98 (ABO1/ELO2) or empty vector (2μm) were serially diluted and spotted onto ABO1/ELO2-repressing (glucose [glc]) or -inducing (galactose [gal]) YPD medium in the absence (− caffeine) or presence (+ caffeine) of 7.5 mM caffeine. Growth proceeded for 2 to 3 days at 30°C. Caffeine-sensitive (CafS) and -resistant (CafR) drug responses are indicated. (B) Thermosensitivity. LFY3 (tot1/elp1Δ) transformants (transformed as described for panel A) were replica plated on ABO1/ELO2-inducing galactose YPD medium and grown for 3 days at 30°C, 37°C, and 39°C. Thermotolerant (Ts+) responses are distinguished from sensitive (Ts−) and hypersensitive (Ts−−) responses. (C) Resistance to zymocin γ-toxin (γ-tox) subunit. LFY3 (tot1/elp1Δ) cells (transformed as described for panel A) were transformed with GAL1-γ-toxin expression vector (pHMS14) (top panels) or empty GAL1 control (pHMS22) (bottom panels). Upon replica spotting from glucose-repressing to galactose-inducing synthetic complete medium, cell death by γ-toxin was monitored (γ-tox: on). pHMS22 transformants carrying no γ-toxin (− γ-tox) served as galactose-utilizing (Gal+) controls. Growth on galactose in the presence of γ-toxin (+ γ-tox) reflects resistance towards γ-toxin (ToxR), and failure to do so equals sensitivity (ToxS).
DISCUSSION
Several genetic loci important in ABA signaling have been identified previously based on seed germination sensitivity to ABA (10) and abnormal bioluminescence emitted from plants carrying the RD29A luciferase gene under ABA treatment in Arabidopsis (7). We have succeeded in isolating mutations through directly screening the drought-resistant or leaf-wilting plants growing on soil under drought stress conditions (6). The abo1 mutations isolated in this study greatly increased drought tolerance. abo1 mutants show ABA hypersensitivity in the inhibition of seedling growth and the promotion of stomatal closing. Furthermore, the abo1 mutant is more resistant to oxidative stress, which may be related to its ABA hypersensitivity and increased drought tolerance. Interestingly, mutations in ABO1/ELO2 also influence the development of guard cells, resulting in stomata reduced to half the number in the wild type. These results indicate that ABO1/ELO2 represents a unique mechanism for modulating drought tolerance in Arabidopsis.
Studies with yeast and animals have demonstrated the importance of Elongator, a histone acetyltransferase complex, in controlling gene expression at the elongating stage of transcription and potentially mRNA processing (13, 25). In yeast, holo-Elongator comprises two subcomplexes: core-Elongator, consisting of subunits Elp1-3/Tot1-3, and the smaller Elp4-6/Tot5-7 module (11, 29, 55). Elp1/Tot1 is the largest subunit with homology to the human IKAP, which can cause a severe neurodegenerative disorder called familial dysautonomia when mutated (1). In Arabidopsis, putative homologs of all six yeast Elongator subunits were predicted based on comparative genomics (37). Three Arabidopsis loci homologous to the yeast Elongator gene ELP1, ELP3, and ELP4 subunits and responsible for phenotypes of the elongata2 (elo2), elo3, and elo1 mutants have been cloned recently (38).
Our findings that induction of ABO1/ELO2 from the GAL1 promoter restored sensitivity of a tot1/elp1Δ Elongator mutant towards growth inhibition by the zymocin γ-toxin subunit suggest functional Elongator conservation. In line with previous reports that zymocin-induced cell death requires holo-Elongator (9, 12), complementation by ABO1/ELO2 implies that tot1/elp1Δ cells expressing the plant gene are likely to assemble an Elongator chimera that accommodates ABO1/ELO2 and that supports Elongator function in yeast. Based on differential phenotypic displays, i.e., complemented zymocin resistance, unaltered caffeine sensitivity, and enhancement of thermosensitivity, cross-complementation by ABO1/ELO2, however, is considered to be partial. Although the capability of ABO1/ELO2 to rescue caffeine sensitivity of tot1/elp1Δ cells by upregulating ABO1/ELO2 expression was not investigated, we cannot exclude the possibility that full Elongator competence may require excess ABO1/ELO2 levels in yeast. In support, complementation of the γ-toxin phenotype required plasmid-coupled ABO1/ELO2 expression under the control of leu2d that amplifies plasmid copy number (52). Alternatively, differential phenotypes may indicate that some Elongator functions are conserved from yeast to plants while others are not. Our observation that ABO1/ELO2 induction enhanced thermosensitivity points to proliferation-relevant aspects that may distinguish plant from yeast Elongator. Congruently, the mechanism by which Elongator affects cell proliferation reportedly differs between yeast and plants (38) and hELP3, human Elongator subunit 3, hardly replaced the yeast homolog (31). Whether Elongator's roles in transcription (13, 39), secretion (43), or tRNA modification (19) are hijacked by zymocin is under study. Recent reports that zymocin particularly targets Elongator-dependent tRNA species support the latter option (23, 34) and provide a strong case to study whether Elongator's novel role in tRNA modification is conserved among yeast, plants, and mammals.
Recent studies identified several Arabidopsis mutations that affect ABA signaling by controlling RNA metabolism (30). Among them, an ABH1 mutation encodes an mRNA cap-binding protein which, together with CBP20, forms a heterodimeric nuclear cap-binding complex (20, 40), and a SAD1 mutation encodes a multifunctional Sm-like protein which is a component of snRNPs functioning in pre-RNA splicing and mRNA transport and degradation (56). Mutations in either ABH1 or SAD1 or CBP20 render plants hypersensitive to ABA in seed germination and seedling growth (20, 40, 56). Interestingly, mutations in ABO1/ELO2 lead to similar ABA sensitivity of seedling growth but cause no clear change in response to other plant hormones. All of these phenotypes are also observed with ABH1 and SAD1 mutants. Because all three genes (SAD1, ABO1/ELO2, and ABH1) are involved in different stages during mRNA processing, together these studies suggest that the RNA-processing machinery or part of it is intimately involved in early ABA signaling for stress tolerance (20, 30, 56).
Our Northern blotting results indicated that mutations in ABO1/ELO2 affect the expression of some stress-inducible genes. Although abo1 mutants are hypersensitive to ABA in both stomatal closing and seedling growth, the expression levels of ABA-responsive genes did not show hypersensitivity to exogenous ABA in the abo1 mutant. In fact, upon 20 μM ABA treatment the expression levels of stress-responsive marker genes RD29A, RD29B, RD22, and COR47 as well as ABF2/AREB1 and ABI1 are lower in the abo1 mutant. ABO1/ELO2 is a single-copy nonessential gene in Arabidopsis. As discussed above, one of Elongator's functions is to facilitate mRNA transcription elongation by RNA polymerase II (13, 25, 39). The mutations in ABO1/ELO2 would decrease the transcript levels of its target mRNAs. It has also been shown that both ABH1 and SAD1 have minor effects on global gene expression and that only a limited number of genes are altered by the abh1 or sad1 mutations (20, 56). At present, the molecular mechanism underlying the differential gene expression effects of the mRNA processing-related mutations is not known. However, based on the recent report that ABA can affect RNA processing by binding to the RNA-binding protein FCA (44), it is possible that ABA may directly modulate these RNA processing steps by binding to one or more of the RNA processing factors.
Another interesting phenotype of the abo1 mutant is a reduced stoma number. In abo1 mutants, the total number of stomata observed on the leaf surface was half of the number in the wild type, but the total numbers of guard cells were similar. The abundance of stomata is an important factor influencing water use efficiency. Several genes affecting guard cell patterning and development have been isolated recently (2-4, 36, 49, 54). Mutations in SDD, YODA, and TMM genes showed increased stoma number in mutant plants, which indicates that these genes are negative regulators in guard cell formation. Results suggest that SDD, YODA, and TMM function upstream and FLP, CDKB1, and FAMA function downstream in the guard cell development signaling pathway (3, 4). The guard cell phenotypes caused by ABO1/ELO2 mutations are different from those of the guard cell-related mutants in the literature. ABO1/ELO2 affects only the growth and development but not the division and differentiation of the pairs of guard cells originating from satellite meristemoid mother cells. Another important difference is that the abo1 mutations impair not only stomatal development but also stomatal sensitivity to ABA, whereas other stomatal developmental mutants do not have defects in ABA sensitivity (2-4, 36, 49, 54). Because ABO1/ELO2 is suggested to influence the mRNA elongation process, ABO1/ELO2 may not directly participate in controlling the growth and development of guard cells. Instead, ABO1/ELO2 may regulate another gene(s) that plays a role in guard cell growth and development. The fact that abo1 mutants are hypersensitive to ABA-induced stomatal closure suggests that some genes responsible for stomatal closure could also be involved in regulating guard cell growth. Our results suggest that the growth and development of pairs of guard cells originating from meristemoid mother cells and satellite meristemoid mother cells are modulated by different genes, possibly through different signaling pathways.
Acknowledgments
This work was supported by grants from the National Basic Research Program of China (2003CB114300), the National High Technology Research and Development Program of China (863 project), the National Nature Science Foundation of China (30421002), and the Programme of Introducing Talents of Discipline to Universities (B06003) to Z.G. R.S. and D.J. are grateful for support from a DFG-sponsored grant within SFB648 and acknowledge a donation by the Fonds der Chemischen Industrie, Frankfurt, Germany.
We thank the Arabidopsis Biological Resource Center (Columbus, Ohio) for providing the T-DNA lines, M. J. R. Stark (Dundee, United Kingdom) for yeast strains and plasmids.
REFERENCES
- 1.Anderson, S. L., R. Coli, I. W. Daly, E. A. Kichula, M. J. Rork, S. A. Volpi, J. Ekstein, and B. Y. Rubin. 2001. Familial dysautonomia is caused by mutations of the IKAP gene. Am. J. Hum. Genet. 68:753-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berger, D., and T. Altmann. 2000. A subtilisin-like serine protease involved in the regulation of stomatal density and distribution in Arabidopsis thaliana. Genes Dev. 14:1119-1131. [PMC free article] [PubMed] [Google Scholar]
- 3.Bergmann, D. C., W. Lukowitz, and C. R. Somerville. 2004. Stomatal development and pattern controlled by a MAPKK kinase. Science 304:1494-1497. [DOI] [PubMed] [Google Scholar]
- 4.Boudolf, V., R. Barroco, A. Engler Jde, A. Verkest, T. Beeckman, M. Naudts, D. Inze, and L. De Veylder. 2004. B1-type cyclin-dependent kinases are essential for the formation of stomatal complexes in Arabidopsis thaliana. Plant Cell 16:945-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burton, R. A., D. M. Gibeaut, A. Bacic, K. Findlay, K. Roberts, A. Hamilton, D. C. Baulcombe, and G. B. Fincher. 2000. Virus-induced silencing of a plant cellulose synthase gene. Plant Cell 12:691-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, Z., X. Hong, H. Zhang, Y. Wang, X. Li, J. K. Zhu, and Z. Gong. 2005. Disruption of the cellulose synthase gene, AtCesA8/IRX1, enhances drought and osmotic stress tolerance in Arabidopsis. Plant J. 43:273-283. [DOI] [PubMed] [Google Scholar]
- 7.Chinnusamy, V., B. Stevenson, B. H. Lee, and J. K. Zhu. 2002. Screening for gene regulation mutants by bioluminescence imaging. Sci. STKE 2002:PL10. [DOI] [PubMed] [Google Scholar]
- 8.Cohen, L., W. J. Henzel, and P. A. Baeuerle. 1998. IKAP is a scaffold protein of the IkappaB kinase complex. Nature 395:292-296. [DOI] [PubMed] [Google Scholar]
- 9.Fichtner, L., F. Frohloff, D. Jablonowski, M. J. Stark, and R. Schaffrath. 2002. Protein interactions within Saccharomyces cerevisiae Elongator, a complex essential for Kluyveromyces lactis zymocicity. Mol. Microbiol. 45:817-826. [DOI] [PubMed] [Google Scholar]
- 10.Finkelstein, R. R., S. S. Gampala, and C. D. Rock. 2002. Abscisic acid signaling in seeds and seedlings. Plant Cell 14(Suppl.):S15-S45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frohloff, F., L. Fichtner, D. Jablonowski, K. D. Breunig, and R. Schaffrath. 2001. Saccharomyces cerevisiae Elongator mutations confer resistance to the Kluyveromyces lactis zymocin. EMBO J. 20:1993-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frohloff, F., D. Jablonowski, L. Fichtner, and R. Schaffrath. 2003. Subunit communications crucial for the functional integrity of the yeast RNA polymerase II elongator (gamma-toxin target (TOT)) complex. J. Biol. Chem. 278:956-961. [DOI] [PubMed] [Google Scholar]
- 13.Gilbert, C., A. Kristjuhan, G. S. Winkler, and J. Q. Svejstrup. 2004. Elongator interactions with nascent mRNA revealed by RNA immunoprecipitation. Mol. Cell 14:457-464. [DOI] [PubMed] [Google Scholar]
- 14.Gong, Z., C. H. Dong, H. Lee, J. Zhu, L. Xiong, D. Gong, B. Stevenson, and J. K. Zhu. 2005. A DEAD box RNA helicase is essential for mRNA export and important for development and stress responses in Arabidopsis. Plant Cell 17:256-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gong, Z., H. Lee, L. Xiong, A. Jagendorf, B. Stevenson, and J. K. Zhu. 2002. RNA helicase-like protein as an early regulator of transcription factors for plant chilling and freezing tolerance. Proc. Natl. Acad. Sci. USA 99:11507-11512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gosti, F., N. Beaudoin, C. Serizet, A. A. Webb, N. Vartanian, and J. Giraudat. 1999. ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell 11:1897-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guan, L., and J. G. Scandalios. 1998. Two structurally similar maize cytosolic superoxide dismutase genes, Sod4 and Sod4A, respond differentially to abscisic acid and high osmoticum. Plant Physiol. 117:217-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han, M. H., S. Goud, L. Song, and N. Fedoroff. 2004. The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proc. Natl. Acad. Sci. USA 101:1093-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang, B., M. J. Johansson, and A. S. Bystrom. 2005. An early step in wobble uridine tRNA modification requires the Elongator complex. RNA 11:424-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hugouvieux, V., J. M. Kwak, and J. I. Schroeder. 2001. An mRNA cap binding protein, ABH1, modulates early abscisic acid signal transduction in Arabidopsis. Cell 106:477-487. [DOI] [PubMed] [Google Scholar]
- 21.Jablonowski, D., L. Fichtner, M. J. Stark, and R. Schaffrath. 2004. The yeast elongator histone acetylase requires Sit4-dependent dephosphorylation for toxin-target capacity. Mol. Biol. Cell 15:1459-1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jablonowski, D., F. Frohloff, L. Fichtner, M. J. Stark, and R. Schaffrath. 2001. Kluyveromyces lactis zymocin mode of action is linked to RNA polymerase II function via Elongator. Mol. Microbiol. 42:1095-1105. [DOI] [PubMed] [Google Scholar]
- 23.Jablonowski, D., S. Zink, C. Mehlgarten, G. Daum, and R. Schaffrath. 2006. tRNA wobble uridine methylation by Trm9 identifies Elongator's key role for zymocin-induced cell death in yeast. Mol. Microbiol. 59:677-688. [DOI] [PubMed] [Google Scholar]
- 24.Jiang, M., and J. Zhang. 2001. Effect of abscisic acid on active oxygen species, antioxidative defence system and oxidative damage in leaves of maize seedlings. Plant Cell Physiol. 42:1265-1273. [DOI] [PubMed] [Google Scholar]
- 25.Kim, J. H., W. S. Lane, and D. Reinberg. 2002. Human Elongator facilitates RNA polymerase II transcription through chromatin. Proc. Natl. Acad. Sci. USA 99:1241-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim, S., J. Y. Kang, D. I. Cho, J. H. Park, and S. Y. Kim. 2004. ABF2, an ABRE-binding bZIP factor, is an essential component of glucose signaling and its overexpression affects multiple stress tolerance. Plant J. 40:75-87. [DOI] [PubMed] [Google Scholar]
- 27.Koiwa, H., A. W. Barb, L. Xiong, F. Li, M. G. McCully, B. H. Lee, I. Sokolchik, J. Zhu, Z. Gong, M. Reddy, A. Sharkhuu, Y. Manabe, S. Yokoi, J. K. Zhu, R. A. Bressan, and P. M. Hasegawa. 2002. C-terminal domain phosphatase-like family members (AtCPLs) differentially regulate Arabidopsis thaliana abiotic stress signaling, growth, and development. Proc. Natl. Acad. Sci. USA 99:10893-10898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koiwa, H., S. Hausmann, W. Y. Bang, A. Ueda, N. Kondo, A. Hiraguri, T. Fukuhara, J. D. Bahk, D. J. Yun, R. A. Bressan, P. M. Hasegawa, and S. Shuman. 2004. Arabidopsis C-terminal domain phosphatase-like 1 and 2 are essential Ser-5-specific C-terminal domain phosphatases. Proc. Natl. Acad. Sci. USA 101:14539-14544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krogan, N. J., and J. F. Greenblatt. 2001. Characterization of a six-subunit Holo-Elongator complex required for the regulated expression of a group of genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:8203-8212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuhn, J. M., and J. I. Schroeder. 2003. Impacts of altered RNA metabolism on abscisic acid signaling. Curr. Opin. Plant Biol. 6:463-469. [DOI] [PubMed] [Google Scholar]
- 31.Li, F., J. Lu, Q. Han, G. Zhang, and B. Huang. 2005. The Elp3 subunit of human Elongator complex is functionally similar to its counterpart in yeast. Mol. Genet. Genomics 273:264-272. [DOI] [PubMed] [Google Scholar]
- 32.Li, J., T. Kinoshita, S. Pandey, C. K. Ng, S. P. Gygi, K. Shimazaki, and S. M. Assmann. 2002. Modulation of an RNA-binding protein by abscisic-acid-activated protein kinase. Nature 418:793-797. [DOI] [PubMed] [Google Scholar]
- 33.Lu, C., and N. Fedoroff. 2000. A mutation in the Arabidopsis HYL1 gene encoding a dsRNA binding protein affects responses to abscisic acid, auxin, and cytokinin. Plant Cell 12:2351-2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu, J., B. Huang, A. Esberg, M. J. Johansson, and A. S. Bystrom. 2005. The Kluyveromyces lactis gamma-toxin targets tRNA anticodons. RNA 11:1648-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Merlot, S., F. Gosti, D. Guerrier, A. Vavasseur, and J. Giraudat. 2001. The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J. 25:295-303. [DOI] [PubMed] [Google Scholar]
- 36.Nadeau, J. A., and F. D. Sack. 2003. Stomatal development: cross talk puts mouths in place. Trends Plant Sci. 8:294-299. [DOI] [PubMed] [Google Scholar]
- 37.Nelissen, H., J. H. Clarke, M. De Block, S. De Block, R. Vanderhaeghen, R. E. Zielinski, T. Dyer, S. Lust, D. Inze, and M. Van Lijsebettens. 2003. DRL1, a homolog of the yeast TOT4/KTI12 protein, has a function in meristem activity and organ growth in plants. Plant Cell 15:639-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nelissen, H., D. Fleury, L. Bruno, P. Robles, L. De Veylder, J. Traas, J. L. Micol, M. Van Montagu, D. Inze, and M. Van Lijsebettens. 2005. The elongata mutants identify a functional Elongator complex in plants with a role in cell proliferation during organ growth. Proc. Natl. Acad. Sci. USA 102:7754-7759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Otero, G., J. Fellows, Y. Li, T. de Bizemont, A. M. Dirac, C. M. Gustafsson, H. Erdjument-Bromage, P. Tempst, and J. Q. Svejstrup. 1999. Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol. Cell 3:109-118. [DOI] [PubMed] [Google Scholar]
- 40.Papp, I., L. A. Mur, A. Dalmadi, S. Dulai, and C. Koncz. 2004. A mutation in the cap binding protein 20 gene confers drought tolerance to Arabidopsis. Plant Mol. Biol. 55:679-686. [DOI] [PubMed] [Google Scholar]
- 41.Pei, Z. M., K. Kuchitsu, J. M. Ward, M. Schwarz, and J. I. Schroeder. 1997. Differential abscisic acid regulation of guard cell slow anion channels in Arabidopsis wild-type and abi1 and abi2 mutants. Plant Cell 9:409-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pei, Z. M., Y. Murata, G. Benning, S. Thomine, B. Klusener, G. J. Allen, E. Grill, and J. I. Schroeder. 2000. Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature 406:731-734. [DOI] [PubMed] [Google Scholar]
- 43.Rahl, P. B., C. Z. Chen, and R. N. Collins. 2005. Elp1p, the yeast homolog of the FD disease syndrome protein, negatively regulates exocytosis independently of transcriptional elongation. Mol. Cell 17:841-853. [DOI] [PubMed] [Google Scholar]
- 44.Razem, F. A., A. El-Kereamy, S. R. Abrams, and R. D. Hill. 2006. The RNA-binding protein FCA is an abscisic acid receptor. Nature 439:290-294. [DOI] [PubMed] [Google Scholar]
- 45.Schroeder, J. I., J. M. Kwak, and G. J. Allen. 2001. Guard cell abscisic acid signalling and engineering drought hardiness in plants. Nature 410:327-330. [DOI] [PubMed] [Google Scholar]
- 46.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 194:3-21. [DOI] [PubMed] [Google Scholar]
- 47.Shi, H., F. J. Quintero, J. M. Pardo, and J. K. Zhu. 2002. The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell 14:465-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shinozaki, K., K. Yamaguchi-Shinozaki, and M. Seki. 2003. Regulatory network of gene expression in the drought and cold stress responses. Curr. Opin. Plant Biol. 6:410-417. [DOI] [PubMed] [Google Scholar]
- 49.Shpak, E. D., J. M. McAbee, L. J. Pillitteri, and K. U. Torii. 2005. Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science 309:290-293. [DOI] [PubMed] [Google Scholar]
- 50.Sims, R. J., III, R. Belotserkovskaya, and D. Reinberg. 2004. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 18:2437-2468. [DOI] [PubMed] [Google Scholar]
- 51.Slaugenhaupt, S. A., and J. F. Gusella. 2002. Familial dysautonomia. Curr. Opin. Genet. Dev. 12:307-311. [DOI] [PubMed] [Google Scholar]
- 52.Spalding, A., and M. F. Tuite. 1989. Host-plasmid interactions in Saccharomyces cerevisiae: effect of host ploidy on plasmid stability and copy number. J. Gen. Microbiol. 135:1037-1045. [DOI] [PubMed] [Google Scholar]
- 53.Suntres, Z. E. 2002. Role of antioxidants in paraquat toxicity. Toxicology 180:65-77. [DOI] [PubMed] [Google Scholar]
- 54.Von Groll, U., D. Berger, and T. Altmann. 2002. The subtilisin-like serine protease SDD1 mediates cell-to-cell signaling during Arabidopsis stomatal development. Plant Cell 14:1527-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Winkler, G. S., T. G. Petrakis, S. Ethelberg, M. Tokunaga, H. Erdjument-Bromage, P. Tempst, and J. Q. Svejstrup. 2001. RNA polymerase II elongator holoenzyme is composed of two discrete subcomplexes. J. Biol. Chem. 276:32743-32749. [DOI] [PubMed] [Google Scholar]
- 56.Xiong, L., Z. Gong, C. D. Rock, S. Subramanian, Y. Guo, W. Xu, D. Galbraith, and J. K. Zhu. 2001. Modulation of abscisic acid signal transduction and biosynthesis by an Sm-like protein in Arabidopsis. Dev. Cell 1:771-781. [DOI] [PubMed] [Google Scholar]
- 57.Xiong, L., H. Lee, M. Ishitani, Y. Tanaka, B. Stevenson, H. Koiwa, R. A. Bressan, P. M. Hasegawa, and J. K. Zhu. 2002. Repression of stress-responsive genes by FIERY2, a novel transcriptional regulator in Arabidopsis. Proc. Natl. Acad. Sci. USA 99:10899-10904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xiong, L., H. Lee, M. Ishitani, and J. K. Zhu. 2002. Regulation of osmotic stress-responsive gene expression by the LOS6/ABA1 locus in Arabidopsis. J. Biol. Chem. 277:8588-8596. [DOI] [PubMed] [Google Scholar]
- 59.Xiong, L., and J. K. Zhu. 2003. Regulation of abscisic acid biosynthesis. Plant Physiol. 133:29-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yajima, H., M. Tokunaga, A. Nakayama-Murayama, and F. Hishinuma. 1997. Characterization of IKI1 and IKI3 genes conferring pGKL killer sensitivity on Saccharomyces cerevisiae. Biosci. Biotechnol. Biochem. 61:704-709. [DOI] [PubMed] [Google Scholar]
- 61.Zhang, J. Z., R. A. Creelman, and J. K. Zhu. 2004. From laboratory to field. Using information from Arabidopsis to engineer salt, cold, and drought tolerance in crops. Plant Physiol. 135:615-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu, D., and J. G. Scandalios. 1994. Differential accumulation of manganese-superoxide dismutase transcripts in maize in response to abscisic acid and high osmoticum. Plant Physiol. 106:173-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhu, J. K. 2002. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53:247-273. [DOI] [PMC free article] [PubMed] [Google Scholar]