Abstract
The orphan nuclear receptor liver receptor homolog 1 (LRH-1) has been reported to play an important role in bile acid biosynthesis and reverse cholesterol transport. Here, we show that LRH-1 is a key player in the control of the hepatic acute-phase response. Ectopic expression of LRH-1 with adenovirus resulted in strong inhibition of both interleukin-6 (IL-6)- and IL-1β-stimulated haptoglobin, serum amyloid A, and fibrinogen β gene expression in hepatocytes. Furthermore, induction of the hepatic inflammatory response was significantly exacerbated in HepG2 cells expressing short hairpin RNA targeting LRH-1 expression. Moreover, transient-transfection experiments and electrophoretic mobility shift and chromatin immunoprecipitation assays revealed that LRH-1 regulates this cytokine-elicited inflammatory response by, at least in part, antagonizing the CCAAT/enhancer binding protein β signaling pathway. Finally, we show, by using LRH-1 heterozygous mice, that LRH-1 is involved in the control of the inflammatory response at the hepatic level in vivo. Taken together, our results outline an unexpected role for LRH-1 in the modulation of the hepatic acute-phase response.
Nuclear receptors (NRs) are ligand-activated transcription factors which have central roles in nearly every aspect of development and adult physiology (22, 34). NRs have been shown to be key players in the control of the inflammatory response (53). Among them, peroxisome proliferator-activated receptors (PPAR) and liver X receptors (LXR) have been reported to be molecular links between lipid metabolism and the inflammatory response (10, 27). They exert their anti-inflammatory activities in various tissues, including the liver and macrophages, by antagonizing cytokine-mediated gene expression via transcriptional cross talk with NF-κB, AP-1, CCAAT/enhancer binding protein (C/EBP), and signal transducers and activators of transcription (STAT) signaling pathways (12, 23). The liver is the primary site of the acute-phase response (APR), which is the host reaction in response to trauma, infection, tissue damage, or acute inflammation (3). This APR plays an important role in limiting tissue injury and the innate immune response. A major component of this APR is the altered hepatic synthesis of a wide spectrum of proteins involved in coagulation, lipid metabolism, and the complement system (20). Cytokines, more specifically, interleukin-6 (IL-6) and IL-1β, are the physiological mediators of this APR. They are mainly produced by monocytes and macrophages at inflammatory sites. This cytokine-induced APR is characterized by changes in the concentrations of plasma proteins known as acute-phase proteins (APPs) that are largely produced by hepatocytes. These may be positive APPs (up-regulated) or negative APPs (down-regulated). Negative APPs include carrier proteins such as albumin, whereas positive APPs play a direct role in the inflammatory process (5, 20). C-reactive protein (CRP), serum amyloid A (SAA), fibrinogen β (FGB), haptoglobin (HP), and plasminogen activator inhibitor 1 (PAI-1, SERPINE1) are the best defined. APPs have been divided into two major classes according to their responsiveness to cytokines; class I APPs are responsive to IL-1β and other cytokines, whereas class II APPs are solely responsive to IL-6 and related cytokines. APP expression is regulated at the transcriptional level by three important transcription factors, NF-κB, STAT3, and C/EBPs. Chronic changes in the levels of these APPs are considered to be detrimental rather than beneficial to the host. Interestingly, many APPs are well-characterized markers of chronic inflammatory diseases such as atherosclerosis (8). Among them, CRP has been shown in certain studies to be a more powerful predictor of cardiovascular risk than traditional risk factors such as low-density lipoprotein (LDL). Indeed, CRP levels predict cardiovascular risk independently of LDL or high-density lipoprotein (HDL) cholesterol levels (47).
The orphan NR liver receptor homolog 1 (LRH-1; NR5A2) is the mammalian homolog of the Drosophila fushi tarazu F1 receptor (FTZ-F1; NR5A3) and, like FTZ-F1, binds its cognate target sequence (5′-[Py]CAAGG[Py]C[Pu]-3′) as a monomer (21). It is highly expressed in the liver, intestine, ovary, and pancreas (21, 48). Recently, several groups reported independently the crystal structure of the LRH-1 ligand-binding domain and identified phospholipids buried in the hydrophobic cavity, raising the possibility that phospholipids are natural ligands for LRH-1 (30, 32, 39, 54). LRH-1 plays an important role in the embryonic development via the regulation of key transcription factors such as Oct4 and hepatic nuclear factors HNF-3β, HNF-4α, and HNF-1α (25, 40). Recently, LRH-1 was shown to regulate expression of aromatase (CYP19) in ovarian and adipose tissues (11, 40) and adiponectin in adipocytes (26). Furthermore, LRH-1 has recently been reported to be involved in intestinal crypt cell renewal by coactivating β-catenin on the cyclin D1 promoter (6). In addition, LRH-1 is believed to be a key player in cholesterol homeostasis (15). LRH-1 has been shown to play a pivotal role in the transcriptional regulation of CYP7A1, the rate-limiting enzyme of the bile acid biosynthetic pathway (37), and CYP8B1, the oxysterol 12α-hydroxylase required for cholic acid production (7). Moreover, LRH-1 has been reported to regulate expression of APO AI (13), ABCG5/ABCG8 (18), CETP (33), SR-BI (48), and the carboxyl ester lipase (16), thereby implicating this receptor in HDL remodeling and cholesterol transport.
Since NRs have been shown to be at the crossroads between lipid metabolism and the inflammatory response, we investigated the potential involvement of LRH-1 in the hepatic APR. Overexpression studies and gene-silencing experiments indicate that LRH-1 modulates the cytokine-stimulated APR in vitro but also in vivo, outlining an unexpected role for LRH-1 in hepatic inflammation control.
MATERIALS AND METHODS
Cell culture.
HepG2 cells (American Type Culture Collection, Manassas, VA) were maintained in basic Eagle's medium supplemented with 2 mM glutamine, 1% nonessential amino acids, and 10% (vol/vol) fetal calf serum (FCS) in an atmosphere of 5% CO2 at 37°C. Cynomolgus primary hepatocytes were provided by Biopredic (Rennes, France) and cultured in William's E medium supplemented with 1% FCS, 10 nM dexamethasone, 100 U/ml penicillin, 100 μg/ml streptomycin, and insulin-transferrin-selenium G.
Plasmids.
Plasmid pSG5-LRH-1 has been previously described (13). Plasmid pSG5 was purchased from Stratagene (La Jolla, CA). The pCEBP, pNF-κB, and pSTAT3 reporter vectors were provided by Panomics. The human HP promoter construct (−460 to +1) was obtained by PCR amplification with human genomic DNA (Clontech) as the template. The resulting PCR product was inserted as a KpnI/HindIII fragment into the pGL3 basic vector (Promega), yielding HP-Luc. The mutation of the C/EBP binding site within the human HP promoter was obtained by site-directed mutagenesis (Stratagene, La Jolla, CA) with the oligonucleotides 5′-CAAGTGTGAAGCGATATCCTCAACTCTTAACAG-3′ and 5′-CTGTTAAGAGTTGAGGATATCGCTTCACACTTG-3′. All constructs were verified by DNA sequence analysis.
Transient-transfection assays.
HepG2 cells, plated in 24-well plates at 50 to 60% confluence in basic Eagle's medium supplemented with 10% FCS, were transiently transfected with reporter and receptor expression plasmids with Fugene 6 reagent (Roche Molecular Biochemical, Indianapolis, IN) as indicated in the figure legends. The pSEAP2 expression plasmid (Clontech) was cotransfected to assess transfection efficiency. At 48 h posttransfection, cells were collected and assayed for luciferase and alkaline phosphatase activities. All experiments were repeated at least three times.
Stable cell line generation.
HepG2 cells stably expressing short hairpin RNA (shRNA) targeting LRH-1 expression or LacZ as a control were engineered with the BLOCK-iT inducible H1 RNA interference entry vector kit (Invitrogen, Carlsbad, CA). Briefly, 50-nucleotide DNA oligonucleotides encoding the shRNA targeting human LRH-1 or LacZ were ligated into the pENTR/H1/T0 vector, yielding pshLRH-1 and pshLacZ, respectively. The sequence targeting LRH-1 (GenBank accession number AB019246) corresponds to the coding region (nucleotides 1438 to 1458) relative to the first nucleotide of the start codon. HepG2 cells (40% confluence) were then transfected with pshLRH-1 or pshLacZ by using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA) and following the manufacturer's instructions. At 24 h posttransfection, cells were refed with fresh medium containing zeocin for stable cell line generation. After 10 days of culture, several zeocin-resistant colonies were identified and then expanded and each clone was characterized regarding LRH-1 gene knockdown by real-time quantitative PCR (RT-QPCR).
RNA analysis.
Total RNA was extracted with TRIZOL (Invitrogen) by following the manufacturer's instructions. The RNA was treated with DNase I (Ambion Inc., Austin, Texas) at 37°C for 30 min, followed by inactivation at 75°C for 5 min. RT-QPCR assays were performed with an Applied Biosystems 7900 sequence detector. Total RNA (1 μg) was reverse transcribed with random hexamers with a TaqMan reverse transcription reagent kit (Applied Biosystems) by following the manufacturer's protocol. RNA expression levels were determined by SYBR green assays as previously described (13). Cyclophilin transcript was used as an internal control to normalize the variations in RNA amounts. Gene expression levels are expressed relative to cyclophilin mRNA levels. All of the primers used in this study are available upon request.
Adenovirus generation.
Recombinant adenovirus expressing green fluorescent protein (Ad-GFP) or LRH-1 (Ad-LRH-1) was obtained by homologous recombination in Escherichia coli after insertion of the cDNAs into pAdCMV2. Viral stocks were created as previously described (13). Viral titers were determined by a plaque assay on 293 cells and expressed as numbers of PFU per milliliter. Cells were infected, in most of the experiments, at a multiplicity of infection (MOI) of 50 viral particles per cell by adding virus stocks directly to the HepG2 culture medium.
Electrophoretic mobility shift assay.
Double-stranded oligonucleotides (5′-ATCAAGTGTGAAGCAAGAGCTCA-3′) were end labeled with [γ-32P]ATP by using T4 polynucleotide kinase according to standard procedures. Nuclear extracts were prepared as previously described (14), and protein concentration was determined with the bicinchoninic acid assay kit (Bio-Rad). Five micrograms of extract was incubated with 100,000 cpm of labeled probe for 20 min at room temperature in 20 μl of buffer containing 10 mM Tris (pH 7.5), 50 mM NaCl, 1 mM dithiothreitol, 1 mM EDTA, 5% glycerol, 0.3 μg bovine serum albumin (BSA), and 2 μg of poly(dI-dC). DNA-protein complexes were analyzed by electrophoresis in a 5% nondenaturing polyacrylamide gel with 0.5× Tris-borate-EDTA buffer. The gel was then dried and exposed at −80°C for autoradiography.
Chromatin immunoprecipitation (ChIP) assays.
ChIP assays were performed essentially as previously described (13). Briefly, 3 × 106 HepG2 cells were cross-linked for 30 min at 4°C by adding an 11% formaldehyde-containing solution. Cross-linking was stopped by adding glycine to a final concentration of 125 mM for 5 min. Cells were then rinsed with phosphate-buffered saline (PBS), harvested, and centrifuged at 600 × g for 5 min at 4°C. Pellets were resuspended in lysis buffer (50 mM HEPES-KOH at pH 8.0, 1 mM EDTA, 0.5 mM EGTA, 140 mM NaCl, 10% glycerol, 0.5% NP-40, 0.25% Triton, 1 mM phenylmethylsulfonyl fluoride [PMSF], leupeptin-pepstatin A-aprotinin at 5 μg/ml each) and rotated for 10 min at 4°C. The nuclei were collected by centrifugation, resuspended in 10 ml wash buffer (10 mM Tris-HCl at pH 8, 1 mM EDTA, 0.5 mM EGTA, 200 mM NaCl, 1 mM PMSF, leupeptin-pepstatin A-aprotinin at 5 μg/ml each), and rotated again. Washed nuclei were centrifuged and resuspended in 1× radioimmunoprecipitation assay buffer (10 mM Tris-HCl at pH 8, 1 mM EDTA, 0.5 mM EGTA, 140 mM NaCl, 1% Triton, 0.1% sodium dodecyl sulfate, 0.1% Na-deoxycholate, 1 mM PMSF, leupeptin-pepstatin A-aprotinin at 5 μg/ml each) and subsequently sonicated, leading to the generation of DNA fragment sizes of 0.3 to 1.5 kb. Samples were cleared by centrifugation at 16,000 × g for 10 min at 4°C. Ten percent of the cleared supernatant was used as the input, and the remaining volume was immunoprecipitated with a C/EBPβ monoclonal antibody (sc-7962 at 5 μg/ml; Santa Cruz). After extensive washings, proteins were digested by adding proteinase K (100 μg/ml) and placed at 55°C for 3 h, followed by 6 h at 65°C to reverse cross-links. DNA was extracted by a standard procedure, and pellets were resuspended in Tris-EDTA buffer. The human proximal HP promoter region was PCR amplified with the oligonucleotides HPCHIP_F (5′-GCCTGGGCAACAGGAGTGAAA.-3′) and HPCHIP_R (5′-CTTGGTTGGTCTTGCCTCTGG-3′). PCRs were analyzed by electrophoresis on a 1% agarose gel.
LPS challenge.
Experimental protocols were approved by the GlaxoSmithKline Institutional Animal Care and Use Committee. Female wild-type and LRH-1+/− mice (25) (eight per group) received an intraperitoneal injection of a lipopolysaccharide (LPS) solution (40 μg per mouse) or saline. At 2.5 h postinjection, animals were sacrificed, blood was recovered for serum preparation, and the liver was quickly removed, frozen in liquid nitrogen, and used for RNA extraction. Acute-phase protein levels in plasma were measured by enzyme-linked immunosorbent assays (ELISAs) (CRP, Kamiya; HP, ICL; SAA, Phase).
Statistical analysis.
Results are shown as means ± the standard errors of the means. Statistical significance was determined with the Student t test. Differences with P < 0.05 were considered to be statistically significant.
RESULTS
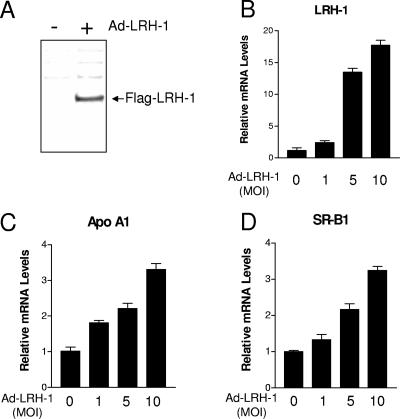
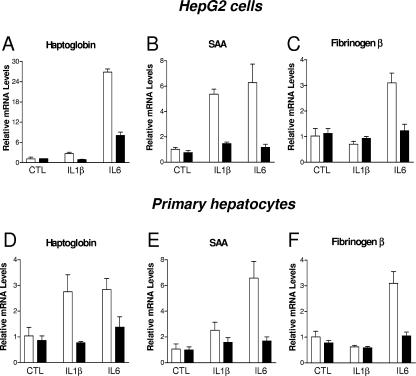
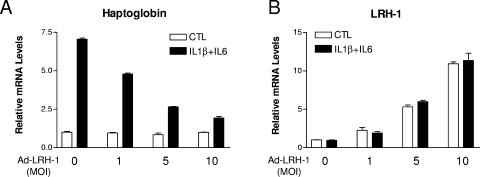
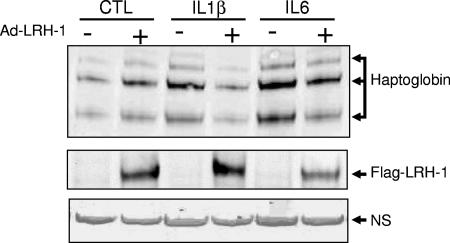
In order to investigate a potential involvement of LRH-1 in the control of the hepatic APR, we generated an adenovirus encoding FLAG-tagged human LRH-1 (Ad-LRH-1). Infection of HepG2 cells, a human hepatic cell line, with Ad-LRH-1 led to robust expression of LRH-1 at both the mRNA and protein levels (Fig. 1A and B). Overexpression of the FLAG-tagged LRH-1 protein resulted in dose-dependent induction of APOA1 and SRB1, two previously characterized LRH-1 target genes (13, 48), thereby confirming the functionality of the LRH-1 protein produced (Fig. 1C and D). Next, HepG2 cells were infected with Ad-LRH-1 or Ad-GFP for 24 h. Cells were subsequently stimulated for 6 h with IL-1β or IL-6, two key mediators of the APR (3). Type 1 and type 2 APR gene expression levels were monitored by RT-QPCR (Fig. 2A to D). Ectopic expression of LRH-1 in HepG2 cells did not significantly affect basal SAA, HP, or FGB gene expression levels. As expected, IL-1β and IL-6 treatment resulted in significant induction of HP, SAA, and FGB gene expression, which was blunted by LRH-1 overexpression (Fig. 2A to C). Similar results were obtained with primary cynomolgus hepatocytes (Fig. 2D to F). In addition, LRH-1 prevented cytokine-mediated CRP gene induction in primary hepatocytes (data not shown). Furthermore, LRH-1 inhibited IL-1β- and IL-6-mediated gene induction in a dose-dependent manner (Fig. 3A). This inhibition also occurs at the protein level, as demonstrated by analysis of HP secretion into the medium by Western blot analysis (Fig. 4). Of note, treatment with IL-1β and IL-6 did not affect LRH-1 mRNA expression in HepG2 cells (Fig. 3B). Taken together, these results suggest that LRH-1 negatively regulates the hepatic inflammatory response elicited by IL-1β or IL-6 in vitro.
FIG. 1.
LRH-1 overexpression in human hepatic cells. (A) Western blot analysis of LRH-1 in HepG2 cells infected with Ad-LRH-1 or Ad-GFP (MOI = 10). (B to D) HepG2 cells cultured in 24-well plates were infected for 24 h with Ad-LRH-1 or Ad-GFP at various MOIs (1, 5, 10). At the end of the treatment period, total RNA was extracted and gene expression levels were monitored by RT-QPCR.
FIG. 2.
Ectopic expression of LRH-1 inhibits the inflammatory response elicited by IL-1β and IL-6 in hepatic cells. HepG2 cells (A to C) and primary cynomolgus hepatocytes (D to F) cultured in 24-well plates were infected for 24 h with Ad-LRH-1 (black bars) or Ad-GFP (white bars) at an MOI of 10 or 50, respectively. Cells were then stimulated with IL-1β (10 ng/ml), IL-6 (10 ng/ml), or the vehicle (PBS-0.1% BSA) for 6 h. At the end of the treatment period, total RNA was extracted and gene expression levels were monitored by RT-QPCR. LRH-1 overexpression resulted in 10- and 20-fold induction of LRH-1 expression in HepG2 cells and in primary hepatocytes, respectively.
FIG. 3.
Ectopic expression of LRH-1 inhibits IL-1β- and IL-6-mediated HP induction in a dose-dependent manner. HepG2 cells (A and B) were infected for 24 h with increasing MOIs of Ad-LRH-1 or Ad-GFP. Cells were then stimulated with IL-1β (10 ng/ml), IL-6 (10 ng/ml), or the vehicle (PBS-0.1% BSA) for 6 h. At the end of the treatment period, HP and LRH-1 mRNA levels were measured by RT-QPCR.
FIG. 4.
Ectopic expression of LRH-1 inhibits IL-1β- and IL-6-mediated HP secretion in HepG2 cells. (Top) Western blot analysis of HP secretion in HepG2 cells infected with Ad-LRH-1 (MOI = 10) or Ad-GFP stimulated with IL-1β, IL-6, or the vehicle (PBS-0.1% BSA) for 6 h. (Middle) Intracellular LRH-1 detection by Western blot assay in HepG2 cells infected with Ad-LRH-1 (MOI = 10) or Ad-GFP stimulated with IL-1β, IL-6, or the vehicle (PBS-0.1% BSA) for 6 h. (Bottom) Nonspecific (NS) band used as a loading control.
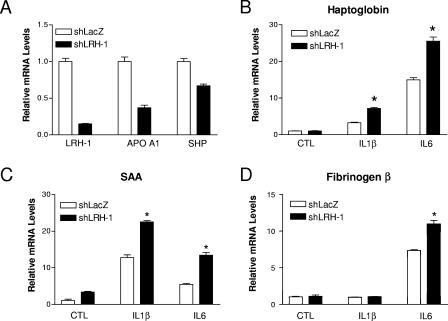
To further explore the anti-inflammatory properties of LRH-1, gene-silencing experiments were carried out with HepG2 cells. To knock down LRH-1 expression, HepG2 cells stably expressing shRNA targeting LRH-1 expression or lacZ as a control were engineered. In those cells, LRH-1 mRNA expression was dramatically reduced (−80%) compared to shLacZ-expressing HepG2 cells, as demonstrated by RT-QPCR (Fig. 5A). In addition, LRH-1 knockdown led to transcriptional inhibition of the expression of both SHP and APOAI (Fig. 5A), two well-established LRH-1 target genes, in hepatic cells (13, 24). Stimulation for 6 h with IL-1β or IL-6 resulted in induction of the genes for HP, SAA, and FGB in control cells (Fig. 5B to D), in line with our previous results (Fig. 2). Interestingly, this inflammatory response was significantly more pronounced in shLRH-1-expressing cells, indicating that LRH-1 negatively regulates the transcriptional response elicited by both IL-1β and IL-6 in hepatic cells (Fig. 5B to D).
FIG. 5.
shRNA-mediated LRH-1 knockdown in HepG2 cells results in an exacerbated inflammatory response. (A) LRH-1, APO AI, and SHP mRNA levels were determined by RT-QPCR in HepG2 cells stably transfected with a vector expressing shRNA targeting LRH-1 or LacZ expression. (B to D) shLRH-1 or shLacZ HepG2 cells were stimulated by IL-1β (10 ng/ml), IL-6 (10 ng/ml), or the vehicle (PBS-0.1% BSA) for 6 h. At the end of the treatment period, HP, SAA, and FBG gene expression levels were measured by RT-QPCR. *, P < 0.05 (shLacZ versus shLRH-1).
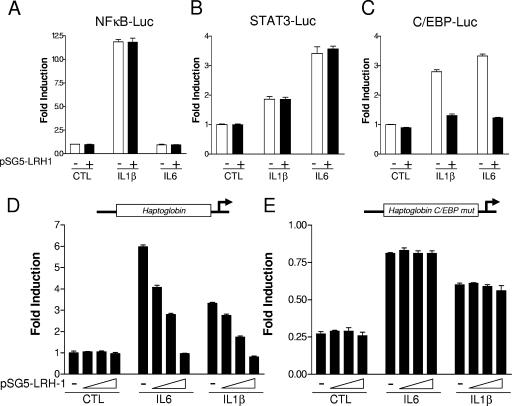
A large number of studies have led to the identification of several signaling pathways controlling the transcription of the APR genes, including those for NF-κB, STAT3, and C/EBP (13, 19, 24, 42, 44). To gain some insight into the molecular mechanism by which LRH-1 exerts these anti-inflammatory properties, the influence of LRH-1 overexpression was tested on minimal promoters driven by multiple copies of NF-κB, STAT3, or C/EBP binding sites (Fig. 6A to C). As expected, IL-1β treatment led to the transcriptional activation of NF-κB, C/EBP, and to a lesser extent STAT3 promoter reporter vectors. By contrast, IL-6 stimulation induced both STAT3 and C/EBP but not NF-κB promoter activation (Fig. 6A to C). Cotransfection of LRH-1 did not affect the basal promoter activity of those reporter vectors, while it completely blocked IL-1β-mediated but also IL-6-mediated C/EBP transcriptional activation (Fig. 6C). By contrast, LRH-1 failed to negatively interfere with the transcriptional activation of both STAT3 and NF-κB (Fig. 6A and B). These results indicate that LRH-1 antagonizes cytokine-induced C/EBP but not STAT3 or NF-κB transcriptional activation. Interestingly, the SAA, HP, and FBG promoters have been shown to contain functional C/EBP binding motifs (4, 19, 38, 46). To further characterize the transcriptional cross talk between LRH-1 and the C/EBP signaling pathway, a 0.45-kb promoter fragment of the human HP gene was cloned upstream of the luciferase reporter gene. Cotransfection of increasing amounts of an LRH-1-expressing vector did not significantly affect the basal human HP promoter activity (Fig. 6D). As expected, treatment with both IL-6 and IL-1β resulted in strong induction of reporter activity (six- and threefold, respectively). This promoter induction was inhibited by LRH-1 cotransfection in a dose-dependent manner (Fig. 6D). To determine whether LRH-1 inhibits cytokine-induced HP promoter activity by antagonizing C/EBP signaling, a promoter construct bearing a mutation for the C/EBP binding site located in the proximal promoter was generated by site-directed mutagenesis. This mutated construct displayed a significantly lower basal activity (−75%) compared to the wild-type promoter (Fig. 6E), in agreement with published reports (4). Interestingly, this promoter construct was still responsive to both IL-1β and IL-6, but to a lesser extent compared to the wild-type construct. Cotransfection of increasing amounts of LRH-1 failed to inhibit this cytokine-induced promoter activity, suggesting that LRH-1 inhibits HP promoter activation by negatively interfering with the C/EBP signaling pathway (Fig. 6E). As a control, LRH-1 cotransfection did not modify the basal activity of the mutated promoter (Fig. 6B). Taken together, these results suggest that LRH-1 antagonizes C/EBP transcriptional activation, leading to HP promoter inhibition.
FIG. 6.
LRH-1 negatively regulates HP gene expression at the transcriptional level, at least in part, by antagonizing the C/EBP signaling pathway. HepG2 cells were transfected with the NF-κB (A), STAT3 (B), or C/EBP (C) reporter construct and pSG5-LRH-1 (200 ng) or the empty vector (pSG5). At 24 h posttransfection, cells were stimulated with IL-6 (10 ng/ml), IL-1β (10 ng/ml), or the vehicle (PBS-0.1% BSA) for 6 h. Luciferase activity was then determined. HepG2 cells were transfected with the wild-type human HP promoter (D) or a construct mutated for the C/EBP binding site (E) and increasing concentrations of pSG5-LRH-1 (0, 50, 100, and 200 ng) or the empty vector (pSG5). At 24 h posttransfection, cells were stimulated with IL-6 (10 ng/ml), IL-1β (10 ng/ml), or the vehicle (PBS-0.1% BSA) for 6 h. Luciferase activity was then determined.
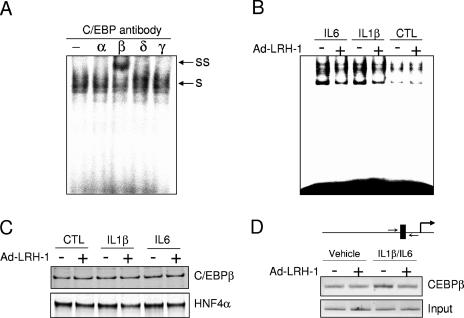
To study the molecular basis of the transcriptional cross talk between LRH-1 and C/EBP signaling, electrophoretic mobility shift assays were performed with nuclear extracts from HepG2 cells. Supershift experiments demonstrated that C/EBPβ was the main C/EBP isoform binding to the HP promoter, in agreement with other reports (45) (Fig. 7A). Ad-GFP-infected HepG2 cells, in the absence of cytokine treatment, display a weak C/EBP DNA-binding activity which was not affected by LRH-1 ectopic expression (Fig. 7B). As expected, IL-1β and IL-6 treatment resulted in an increase in C/EBP DNA-binding capacity in ad-GFP-infected cells, which is consistent with the higher transcriptional activity (Fig. 6C). LRH-1 overexpression sharply reduced this DNA-binding activity. Importantly, nuclear C/EBPβ levels (and HNF4α as a loading control) were not modified by LRH-1 overexpression or by cytokine treatments (Fig. 7C). In order to demonstrate that LRH-1 inhibits cytokine-induced C/EBPβ DNA binding in vivo, ChIP assays were performed. The IL-1β-IL-6 combination induced a significant increase in C/EBPβ occupancy on the HP promoter. This increase was completely abrogated by LRH-1 overexpression, in line with our previous electrophoretic mobility shift assay results (Fig. 7B). Interestingly, under resting conditions, C/EBPβ recruitment to the HP proximal promoter was not affected by LRH-1 ectopic expression. Taken together, these results suggest that LRH-1 negatively interferes with the C/EBP pathway by, at least in part, preventing its DNA binding.
FIG. 7.
LRH-1 inhibits IL-1β- or IL-6-stimulated C/EBPβ DNA-binding activity. (A) C/EBP supershift assay with nuclear extracts derived from cytokine-stimulated HepG2 cells (s, shifted complex; ss, supershift). (B and C) HepG2 cells were infected for 24 h with Ad-LRH-1 or Ad-GFP at an MOI of 10. Cells were then stimulated with IL-1β (10 ng/ml), IL-6 (10 ng/ml), or the vehicle (PBS-0.1% BSA) for 6 h. Nuclear extracts were then prepared, and C/EBP DNA-binding activity was assessed by electrophoretic mobility shift assay (B). C/EBPβ and HNF4α (as a loading control) protein levels were quantified by Western blot analysis (C). (D) HepG2 cells infected with Ad-LRH-1 or Ad-GFP (MOI = 10), stimulated with IL-1β (10 ng/ml) and IL-6 (10 ng/ml) for 6 h, were subjected to a ChIP assay with a C/EBPβ monoclonal antibody as described in Materials and Methods. In vivo HP promoter occupancy was assessed by amplifying the proximal promoter region by PCR.
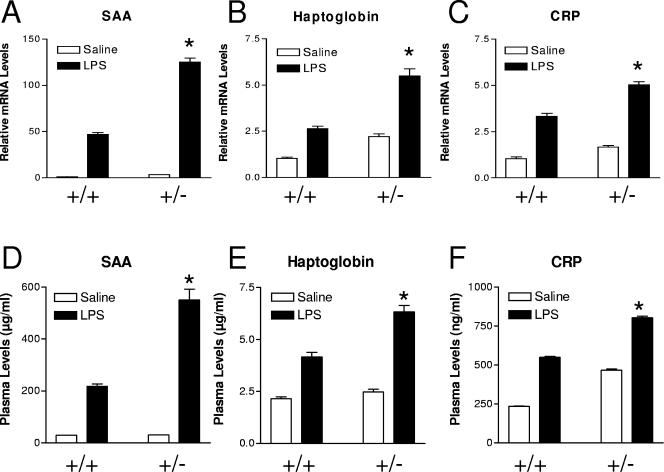
Finally, to further examine the biological relevance of LRH-1 anti-inflammatory properties in vivo, we studied the response to an LPS challenge in wild-type and LRH-1 heterozygous mice. Female mice (eight per group) received an intraperitoneal injection of an LPS solution (40 μg per animal) or saline. At 2.5 h postinjection, animals were sacrificed and the APR was studied at the protein and gene expression levels (Fig. 8). LPS treatment resulted in strong induction of acute-phase gene expression, as measured by RT-QPCR (Fig. 8A to C). This inflammatory response was much more pronounced in LRH-1 heterozygous mice (Fig. 8A to C). Quantification of plasma APP levels by ELISA confirmed the gene expression data, i.e., an exacerbated inflammatory response in LRH-1+/− mice (Fig. 8D to F). Interestingly, basal CRP and gene expression levels were higher in LRH-1 heterozygous than in wild-type mice. As a control, hepatic LRH-1 expression was monitored by RT-QPCR. In wild-type and LRH-1+/− mice, LRH-1 expression was not modified by LPS challenge (data not shown). Collectively, these results demonstrate that LRH-1 is a negative regulator of the hepatic APR in vitro and in vivo.
FIG. 8.
Exacerbated APR to LPS challenge in LRH-1 heterozygous mice. Wild-type and LRH-1+/− mice (eight per group) received an intraperitoneal injection of an LPS solution (40 μg per animal) or saline. At 2.5 h postinjection, animals were sacrificed and plasma acute-phase proteins were measured by ELISAs (A to C). Liver gene expression was also assessed by RT-QPCR (D to F). *, P < 0.05 (wild type plus LPS versus heterozygous plus LPS).
DISCUSSION
LRH-1 has been shown to be an important regulator of cholesterol homeostasis (15) and to be involved in intestinal crypt cell renewal (6). In the present study, we investigated the functional implication of this orphan NR in the control of the hepatic APR. LRH-1 overexpression resulted in inhibition of IL-1- and IL-6-mediated FBG, SAA, CRP, and HP gene expression in hepatocytes. It has also been shown that partial LRH-1 deficiency leads to an exacerbated inflammatory response in vitro and in vivo (Fig. 5 and 8), indicating that LRH-1 is a physiological modulator of the hepatic APR. Several members of the NR superfamily have been shown to play a role in the control of the inflammatory response (23). For instance, LXR and PPARα have been shown to be molecular links between lipid metabolism and inflammation by negatively interfering with several transcription factors, such as C/EBPs, AP-1, and NF-κB (10, 27, 29, 35). In this study, we identified a set of APR genes down-regulated by LRH-1 in hepatic cells. Interestingly, all of those genes contain functional C/EBP binding sites within their promoter regions and we show that LRH-1 was capable of blocking the C/EBP-dependent activation of an isolated C/EBP response element, suggesting that LRH-1 blocks C/EBP signaling. This phenomenon seemed to be specific for C/EBP since transient-transfection experiments revealed that LRH-1 was unable to antagonize cytokine-stimulated NF-κB- and STAT3-driven transcription (Fig. 6A and B), in contrast to other NRs, such as LXR and PPARγ (27). These results suggest that LRH-1 likely inhibits a distinct pattern of inflammatory gene expression compared to LXR, for instance. This is consistent with the concept that every NR may differentially impact the development of the inflammatory response (23). Further studies requiring microarray analysis should help us to better understand the role of LRH-1 during the APR. Interestingly, Schoonjans and coworkers reported an inverse correlation between LRH-1 and tumor necrosis factor alpha (TNF-α) gene expression in the colon (49). In their study, C57BL6 mice received an intraperitoneal injection of LPS (100 μg/mouse). LRH-1 and TNF-α mRNA levels were quantified in colon biopsies at 6 or 24 h postinjection. This high dose of LPS resulted in a significant reduction of LRH-1 expression (−50%) after 24 h with a parallel induction of TNF-α. Furthermore, LRH-1 haploinsufficiency resulted in modulation of the inflammatory response in the intestine (49), in line with the results reported here.
The C/EBPs belong to a family of leucine zipper transcription factors involved in the control of inflammatory and native immunity functions (42). All of the genes inhibited by LRH-1 in response to cytokine stimulation contain functional C/EBP DNA-binding sites within their promoter regions. Site-directed mutagenesis and transfection experiments revealed that LRH-1 negatively regulates APR gene expression by antagonizing the C/EBP signaling pathway. Interestingly, the C/EBP-mutated reporter construct was responsive to treatment with both IL-1β and IL-6, but to a lesser extent compared to the wild-type promoter. However, LRH-1 cotransfection failed to inhibit this transcriptional activation, suggesting that LRH-1 does not interfere with other transcription factors, such as STAT3, involved in HP promoter regulation (43). LRH-1 inhibits C/EBP transcriptional activity by, at least in part, reducing C/EBP DNA binding in vitro and in vivo, as demonstrated by electrophoretic mobility shift and ChIP assays (Fig. 7). Supershift experiments suggest that C/EBPβ is the main C/EBP involved in this transcriptional cross talk; however, LRH-1 overexpression did not affect total or nuclear C/EBPβ levels (Fig. 7). Preliminary coimmunoprecipitation experiments failed to demonstrate a direct protein-protein interaction between LRH-1 and C/EBPβ in hepatic cells (data not shown). Interestingly, C/EBPβ is known to undergo a number of posttranslational modifications, such as phosphorylation and SUMOylation, that modulate its transcriptional activity (50). For instance, C/EBPβ phosphorylation by protein kinase C enhances its transcriptional activity (51). More recently, Kim and coworkers identified a conserved inhibitory domain within C/EBP that is a target for SUMOylation (28). Attachment of SUMO-1 to a lysine residue identified in this region decreases the transcriptional inhibitory function of this domain (28). Interestingly, Pascual and colleagues recently reported a SUMOylation-dependent pathway involved in PPARγ-mediated transrepression of the inflammatory response genes (41). Whether or not this mechanism is also operative for other NRs implicated in transcriptional cross talk has not been determined. Similarly to PPARγ, LRH-1 has been shown to be SUMOylated (9, 31). Further studies are required to determine whether LRH-1 antagonizes C/EBPβ transcriptional activity via a SUMOylation-dependent pathway.
LRH-1 was originally identified as a key player in cholesterol homeostasis (15). LRH-1 is involved in the control of bile acid biosynthesis via regulation of the expression of the genes for both CYP7A1 and CY8B1 (7, 37). Moreover, LRH-1 has also been reported to regulate the expression of APOAI (13), CETP (33), and SR-BI (48), thereby implicating this receptor in HDL remodeling and cholesterol transport. HDL per se is known to have anti-inflammatory properties (2). For instance, HDL is known to modulate LDL-induced monocyte chemotactic activity in the human artery wall (36). Furthermore, HDL plays an important role as an antioxidant by inhibiting phospholipid oxidation and by decreasing the activity of minimally modified LDL (55). However, during the APR or another chronic systemic inflammatory response, HDL can assume proinflammatory and proatherogenic characteristics (17, 52). HDL isolated from patients with coronary artery disease increased LDL-induced recruitment of monocytes to endothelial and smooth-muscle cells (1). SAA, an APP that is dramatically induced in plasma during the APR, is able to displace APOA1 from HDL in vitro (52). In addition, because SAA binds to proteoglycans, SAA-containing HDL may be retained by the vascular matrix, leading to the formation of proatherogenic particles that do not participate in reverse cholesterol transport anymore. By negatively regulating the expression of APP that are well-established markers of coronary artery disease and by controlling HDL biosynthesis and remodeling, LRH-1 may quantitatively affect and qualitatively modulate HDL levels, ultimately resulting in a slowdown of the development of atherosclerosis. Further studies using transgenic animals should help to address this question.
In conclusion, our results outline an unexpected role for LRH-1 in the control of the hepatic APR and suggest that LRH-1 could be a novel molecular link between cholesterol homeostasis and inflammation. On the basis of these observations, LRH-1 appears to be an interesting target for the treatment of dyslipidemia and atherosclerosis.
Acknowledgments
We thank John Bisi (GSK, gene interference) for providing FLAG-tagged Ad-LRH-1, Stéphane Huet (GSK, CVU CEDD) for critical reading of the manuscript, and Beverly H. Koller and Anne M. Latour (Department of Medicine, University of Chapel Hill, Chapel Hill, NC) for LRH-1 heterozygous mice.
REFERENCES
- 1.Ansell, B. J., M. Navab, S. Hama, N. Kamranpour, G. Fonarow, G. Hough, S. Rahmani, R. Mottahedeh, R. Dave, S. T. Reddy, and A. M. Fogelman. 2003. Inflammatory/antiinflammatory properties of high-density lipoprotein distinguish patients from control subjects better than high-density lipoprotein cholesterol levels and are favorably affected by simvastatin treatment. Circulation 108:2751-2756. [DOI] [PubMed] [Google Scholar]
- 2.Ansell, B. J., M. Navab, K. E. Watson, G. C. Fonarow, and A. M. Fogelman. 2004. Anti-inflammatory properties of HDL. Rev. Endocr. Metab. Disord. 5:351-358. [DOI] [PubMed] [Google Scholar]
- 3.Baumann, H., and J. Gauldie. 1994. The acute phase response. Immunol. Today 15:74-80. [DOI] [PubMed] [Google Scholar]
- 4.Baumann, H., K. K. Morella, G. P. Jahreis, and S. Marinkovic. 1990. Distinct regulation of the interleukin-1 and interleukin-6 response elements of the rat haptoglobin gene in rat and human hepatoma cells. Mol. Cell. Biol. 10:5967-5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Black, P. H. 2003. The inflammatory response is an integral part of the stress response: implications for atherosclerosis, insulin resistance, type II diabetes and metabolic syndrome X. Brain Behav. Immun. 17:350-364. [DOI] [PubMed] [Google Scholar]
- 6.Botrugno, O. A., E. Fayard, J. S. Annicotte, C. Haby, T. Brennan, O. Wendling, T. Tanaka, T. Kodama, W. Thomas, J. Auwerx, and K. Schoonjans. 2004. Synergy between LRH-1 and beta-catenin induces G1 cyclin-mediated cell proliferation. Mol. Cell 15:499-509. [DOI] [PubMed] [Google Scholar]
- 7.Castillo-Olivares, A., and G. Gil. 2000. α1-Fetoprotein transcription factor is required for the expression of sterol 12α-hydroxylase, the specific enzyme for cholic acid synthesis: potential role in the bile acid-mediated regulation of gene transcription. J. Biol. Chem. 275:17793-17799. [DOI] [PubMed] [Google Scholar]
- 8.Chait, A., C. Y. Han, J. F. Oram, and J. W. Heinecke. 2005. Thematic review series: the immune system and atherogenesis. Lipoprotein-associated inflammatory proteins: markers or mediators of cardiovascular disease? J. Lipid Res. 46:389-403. [DOI] [PubMed] [Google Scholar]
- 9.Chalkiadaki, A., and I. Talianidis. 2005. SUMO-dependent compartmentalization in promyelocytic leukemia protein nuclear bodies prevents the access of LRH-1 to chromatin. Mol. Cell. Biol. 25:5095-5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chinetti, G., J. C. Fruchart, and B. Staels. 2000. Peroxisome proliferator-activated receptors (PPARs): nuclear receptors at the crossroads between lipid metabolism and inflammation. Inflamm. Res. 49:497-505. [DOI] [PubMed] [Google Scholar]
- 11.Clyne, C. D., C. J. Speed, J. Zhou, and E. R. Simpson. 2002. Liver receptor homologue-1 (LRH-1) regulates expression of aromatase in preadipocytes. J. Biol. Chem. 277:20591-20597. [DOI] [PubMed] [Google Scholar]
- 12.Delerive, P., J. C. Fruchart, and B. Staels. 2001. Peroxisome proliferator-activated receptors in inflammation control. J. Endocrinol. 169:453-459. [DOI] [PubMed] [Google Scholar]
- 13.Delerive, P., C. M. Galardi, J. E. Bisi, E. Nicodeme, and B. Goodwin. 2004. Identification of liver receptor homolog-1 as a novel regulator of apolipoprotein AI gene transcription. Mol. Endocrinol. 18:2378-2387. [DOI] [PubMed] [Google Scholar]
- 14.Dignam, J. D., P. L. Martin, B. S. Shastry, and R. G. Roeder. 1983. Eukaryotic gene transcription with purified components. Methods Enzymol. 101:582-598. [DOI] [PubMed] [Google Scholar]
- 15.Fayard, E., J. Auwerx, and K. Schoonjans. 2004. LRH-1: an orphan nuclear receptor involved in development, metabolism and steroidogenesis. Trends Cell Biol. 14:250-260. [DOI] [PubMed] [Google Scholar]
- 16.Fayard, E., K. Schoonjans, J. S. Annicotte, and J. Auwerx. 2003. Liver receptor homolog 1 controls the expression of carboxyl ester lipase. J. Biol. Chem. 278:35725-35731. [DOI] [PubMed] [Google Scholar]
- 17.Fogelman, A. M. 2004. When good cholesterol goes bad. Nat. Med. 10:902-903. [DOI] [PubMed] [Google Scholar]
- 18.Freeman, L. A., A. Kennedy, J. Wu, S. Bark, A. T. Remaley, S. Santamarina-Fojo, and H. B. Brewer, Jr. 2004. The orphan nuclear receptor LRH-1 activates the ABCG5/ABCG8 intergenic promoter. J. Lipid Res. 45:1197-1206. [DOI] [PubMed] [Google Scholar]
- 19.Fuller, G. M., and Z. Zhang. 2001. Transcriptional control mechanism of fibrinogen gene expression. Ann. N. Y. Acad. Sci. 936:469-479. [DOI] [PubMed] [Google Scholar]
- 20.Gabay, C., and I. Kushner. 1999. Acute-phase proteins and other systemic responses to inflammation. N. Engl. J. Med. 340:448-454. [DOI] [PubMed] [Google Scholar]
- 21.Galarneau, L., J. F. Pare, D. Allard, D. Hamel, L. Levesque, J. D. Tugwood, S. Green, and L. Belanger. 1996. The α1-fetoprotein locus is activated by a nuclear receptor of the Drosophila FTZ-F1 family. Mol. Cell. Biol. 16:3853-3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giguere, V. 1990. Structure and function of the nuclear receptor superfamily for steroid, thyroid hormone and retinoic acid. Genet. Eng. 12:183-200. [DOI] [PubMed] [Google Scholar]
- 23.Glass, C. K., and S. Ogawa. 2006. Combinatorial roles of nuclear receptors in inflammation and immunity. Nat. Rev. Immunol. 6:44-55. [DOI] [PubMed] [Google Scholar]
- 24.Goodwin, B., S. A. Jones, R. R. Price, M. A. Watson, D. D. McKee, L. B. Moore, C. Galardi, J. G. Wilson, M. C. Lewis, M. E. Roth, P. R. Maloney, T. M. Willson, and S. A. Kliewer. 2000. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol. Cell 6:517-526. [DOI] [PubMed] [Google Scholar]
- 25.Gu, P., B. Goodwin, A. C. Chung, X. Xu, D. A. Wheeler, R. R. Price, C. Galardi, L. Peng, A. M. Latour, B. H. Koller, J. Gossen, S. A. Kliewer, and A. J. Cooney. 2005. Orphan nuclear receptor LRH-1 is required to maintain Oct4 expression at the epiblast stage of embryonic development. Mol. Cell. Biol. 25:3492-3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iwaki, M., M. Matsuda, N. Maeda, T. Funahashi, Y. Matsuzawa, M. Makishima, and I. Shimomura. 2003. Induction of adiponectin, a fat-derived antidiabetic and antiatherogenic factor, by nuclear receptors. Diabetes 52:1655-1663. [DOI] [PubMed] [Google Scholar]
- 27.Joseph, S. B., A. Castrillo, B. A. Laffitte, D. J. Mangelsdorf, and P. Tontonoz. 2003. Reciprocal regulation of inflammation and lipid metabolism by liver X receptors. Nat. Med. 9:213-219. [DOI] [PubMed] [Google Scholar]
- 28.Kim, J., C. A. Cantwell, P. F. Johnson, C. M. Pfarr, and S. C. Williams. 2002. Transcriptional activity of CCAAT/enhancer-binding proteins is controlled by a conserved inhibitory domain that is a target for sumoylation. J. Biol. Chem. 277:38037-38044. [DOI] [PubMed] [Google Scholar]
- 29.Kleemann, R., P. P. Gervois, L. Verschuren, B. Staels, H. M. Princen, and T. Kooistra. 2003. Fibrates down-regulate IL-1-stimulated C-reactive protein gene expression in hepatocytes by reducing nuclear p50-NFκB-C/EBP-β complex formation. Blood 101:545-551. [DOI] [PubMed] [Google Scholar]
- 30.Krylova, I. N., E. P. Sablin, J. Moore, R. X. Xu, G. M. Waitt, J. A. MacKay, D. Juzumiene, J. M. Bynum, K. Madauss, V. Montana, L. Lebedeva, M. Suzawa, J. D. Williams, S. P. Williams, R. K. Guy, J. W. Thornton, R. J. Fletterick, T. M. Willson, and H. A. Ingraham. 2005. Structural analyses reveal phosphatidyl inositols as ligands for the NR5 orphan receptors SF-1 and LRH-1. Cell 120:343-355. [DOI] [PubMed] [Google Scholar]
- 31.Lee, M. B., L. A. Lebedeva, M. Suzawa, S. A. Wadekar, M. Desclozeaux, and H. A. Ingraham. 2005. The DEAD-box protein DP103 (Ddx20 or Gemin-3) represses orphan nuclear receptor activity via SUMO modification. Mol. Cell. Biol. 25:1879-1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li, Y., M. Choi, G. Cavey, J. Daugherty, K. Suino, A. Kovach, N. C. Bingham, S. A. Kliewer, and H. E. Xu. 2005. Crystallographic identification and functional characterization of phospholipids as ligands for the orphan nuclear receptor steroidogenic factor-1. Mol. Cell 17:491-502. [DOI] [PubMed] [Google Scholar]
- 33.Luo, Y., C. P. Liang, and A. R. Tall. 2001. The orphan nuclear receptor LRH-1 potentiates the sterol-mediated induction of the human CETP gene by liver X receptor. J. Biol. Chem. 276:24767-24773. [DOI] [PubMed] [Google Scholar]
- 34.Mangelsdorf, D. J., C. Thummel, M. Beato, P. Herrlich, G. Schutz, K. Umesono, B. Blumberg, P. Kastner, M. Mark, P. Chambon, and R. M. Evans. 1995. The nuclear receptor superfamily: the second decade. Cell 83:835-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mouthiers, A., A. Baillet, C. Delomenie, D. Porquet, and N. Mejdoubi-Charef. 2005. Peroxisome proliferator-activated receptor α physically interacts with CCAAT/enhancer binding protein (C/EBPβ) to inhibit C/EBPβ-responsive α1-acid glycoprotein gene expression. Mol. Endocrinol. 19:1135-1146. [DOI] [PubMed] [Google Scholar]
- 36.Navab, M., S. S. Imes, S. Y. Hama, G. P. Hough, L. A. Ross, R. W. Bork, A. J. Valente, J. A. Berliner, D. C. Drinkwater, and H. Laks. 1991. Monocyte transmigration induced by modification of low density lipoprotein in cocultures of human aortic wall cells is due to induction of monocyte chemotactic protein 1 synthesis and is abolished by high density lipoprotein. J. Clin. Investig. 88:2039-2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nitta, M., S. Ku, C. Brown, A. Y. Okamoto, and B. Shan. 1999. CPF: an orphan nuclear receptor that regulates liver-specific expression of the human cholesterol 7α-hydroxylase gene. Proc. Natl. Acad. Sci. USA 96:6660-6665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oliviero, S., and R. Cortese. 1989. The human haptoglobin gene promoter: interleukin-6-responsive elements interact with a DNA-binding protein induced by interleukin-6. EMBO J. 8:1145-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ortlund, E. A., Y. Lee, I. H. Solomon, J. M. Hager, R. Safi, Y. Choi, Z. Guan, A. Tripathy, C. R. Raetz, D. P. McDonnell, D. D. Moore, and M. R. Redinbo. 2005. Modulation of human nuclear receptor LRH-1 activity by phospholipids and SHP. Nat. Struct. Mol. Biol. 12:357-363. [DOI] [PubMed] [Google Scholar]
- 40.Pare, J. F., D. Malenfant, C. Courtemanche, M. Jacob-Wagner, S. Roy, D. Allard, and L. Belanger. 2004. The fetoprotein transcription factor (FTF) gene is essential to embryogenesis and cholesterol homeostasis and is regulated by a DR4 element. J. Biol. Chem. 279:21206-21216. [DOI] [PubMed] [Google Scholar]
- 41.Pascual, G., A. L. Fong, S. Ogawa, A. Gamliel, A. C. Li, V. Perissi, D. W. Rose, T. M. Willson, M. G. Rosenfeld, and C. K. Glass. 2005. A SUMOylation-dependent pathway mediates transrepression of inflammatory response genes by PPAR-γ. Nature 437:759-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poli, V. 1998. The role of C/EBP isoforms in the control of inflammatory and native immunity functions. J. Biol. Chem. 273:29279-29282. [DOI] [PubMed] [Google Scholar]
- 43.Rakemann, T., M. Niehof, S. Kubicka, M. Fischer, M. P. Manns, S. Rose-John, and C. Trautwein. 1999. The designer cytokine hyper-interleukin-6 is a potent activator of STAT3-dependent gene transcription in vivo and in vitro. J. Biol. Chem. 274:1257-1266. [DOI] [PubMed] [Google Scholar]
- 44.Ramadori, G., and T. Armbrust. 2001. Cytokines in the liver. Eur. J. Gastroenterol. Hepatol. 13:777-784. [DOI] [PubMed] [Google Scholar]
- 45.Ramji, D. P., A. Vitelli, F. Tronche, R. Cortese, and G. Ciliberto. 1993. The two C/EBP isoforms, IL-6DBP/NF-IL6 and C/EBPδ/NF-IL6β, are induced by IL-6 to promote acute phase gene transcription via different mechanisms. Nucleic Acids Res. 21:289-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ray, A., M. Hannink, and B. K. Ray. 1995. Concerted participation of NF-κB and C/EBP heteromer in lipopolysaccharide induction of serum amyloid A gene expression in liver. J. Biol. Chem. 270:7365-7374. [DOI] [PubMed] [Google Scholar]
- 47.Ridker, P. M., N. Rifai, L. Rose, J. E. Buring, and N. R. Cook. 2002. Comparison of C-reactive protein and low-density lipoprotein cholesterol levels in the prediction of first cardiovascular events. N. Engl. J. Med. 347:1557-1565. [DOI] [PubMed] [Google Scholar]
- 48.Schoonjans, K., J. S. Annicotte, T. Huby, O. A. Botrugno, E. Fayard, Y. Ueda, J. Chapman, and J. Auwerx. 2002. Liver receptor homolog 1 controls the expression of the scavenger receptor class B type I. EMBO Rep. 3:1181-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schoonjans, K., L. Dubuquoy, J. Mebis, E. Fayard, O. Wendling, C. Haby, K. Geboes, and J. Auwerx. 2005. Liver receptor homolog 1 contributes to intestinal tumor formation through effects on cell cycle and inflammation. Proc. Natl. Acad. Sci. USA 102:2058-2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schrem, H., J. Klempnauer, and J. Borlak. 2004. Liver-enriched transcription factors in liver function and development. Part II. The C/EBPs and D site-binding protein in cell cycle control, carcinogenesis, circadian gene regulation, liver regeneration, apoptosis, and liver-specific gene regulation. Pharmacol. Rev. 56:291-330. [DOI] [PubMed] [Google Scholar]
- 51.Trautwein, C., C. Caelles, P. van der Geer, T. Hunter, M. Karin, and M. Chojkier. 1993. Transactivation by NF-IL6/LAP is enhanced by phosphorylation of its activation domain. Nature 364:544-547. [DOI] [PubMed] [Google Scholar]
- 52.Van Lenten, B. J., A. C. Wagner, D. P. Nayak, S. Hama, M. Navab, and A. M. Fogelman. 2001. High-density lipoprotein loses its anti-inflammatory properties during acute influenza A infection. Circulation 103:2283-2288. [DOI] [PubMed] [Google Scholar]
- 53.Wang, L., X. Zhang, W. L. Farrar, and X. Yang. 2004. Transcriptional crosstalk between nuclear receptors and cytokine signal transduction pathways in immunity. Cell. Mol. Immunol. 1:416-424. [PubMed] [Google Scholar]
- 54.Wang, W., C. Zhang, A. Marimuthu, H. I. Krupka, M. Tabrizizad, R. Shelloe, U. Mehra, K. Eng, H. Nguyen, C. Settachatgul, B. Powell, M. V. Milburn, and B. L. West. 2005. The crystal structures of human steroidogenic factor-1 and liver receptor homologue-1. Proc. Natl. Acad. Sci. USA 102:7505-7510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Watson, A. D., J. A. Berliner, S. Y. Hama, B. N. La Du, K. F. Faull, A. M. Fogelman, and M. Navab. 1995. Protective effect of high density lipoprotein associated paraoxonase. Inhibition of the biological activity of minimally oxidized low density lipoprotein. J. Clin. Investig. 96:2882-2891. [DOI] [PMC free article] [PubMed] [Google Scholar]