Abstract
We have demonstrated that silencing of luteinizing hormone receptor (LHR) gene transcription is mediated via a proximal Sp1 site at its promoter. Trichostatin A (TSA) induced histone acetylation and gene activation in JAR cells that prevailed in the absence of changes in Sp1/Sp3 expression, their binding activity, disassociation of the histone deacetylase/mSin3A complex from the Sp1 site, or demethylation of the promoter. This indicated a different mechanism involved in TSA-induced derepression. The present studies have revealed that phosphatidylinositol 3-kinase/protein kinase Cζ (PI3K/PKCζ)-mediated Sp1 phosphorylation accounts for Sp1 site-dependent LHR gene activation. TSA caused marked phosphorylation of Sp1 at serine 641 in JAR and MCF-7 cells. Blockade of PI3K or PKCζ activity by specific inhibitors, kinase-deficient mutants, or small interfering RNA abolished the effect of TSA on the LHR gene and Sp1 phosphorylation. PKCζ was shown to associate with Sp1, and this association was enhanced by TSA. Sp1 phosphorylation at serine 641 was required for the release of the pRb homologue p107 from the LHR gene promoter, while p107 acted as a repressor of the LHR gene. Inhibition of PKCζ activity blocked the dissociation of p107 from the LHR gene promoter and markedly reduced Sp1 phosphorylation and transcription. These results have demonstrated that phosphorylation of Sp1 by PI3K/PKCζ is critical for TSA-activated LHR gene expression. These studies have revealed a novel mechanism of TSA action through derecruitment of a repressor from the LHR gene promoter in a PI3K/PKCζ-induced Sp1 phosphorylation-dependent manner.
The luteinizing hormone receptor (LHR) is a G-protein-coupled receptor that is essential for sexual development and reproductive function in mammals. The LHR gene is expressed primarily in gonads but is also present in several nongonadal tissues and breast and placenta cancer tissues and cells (21).
Characterization of the mechanism(s) involved in the regulation of LHR gene transcription has revealed that its basal promoter activity is governed by two activating Sp1/Sp3 binding domains and an inhibitory direct-repeat motif recognized by the nuclear orphan receptors EAR2 and EAR3/COUP-TFI (47-49, 56-58). We have further demonstrated that the status of histone modifications and DNA methylation in the LHR gene promoter region operate coordinately to elicit marked silencing or reactivation of this gene's expression in human choriocarcinoma JAR cells and breast tumor MCF-7 cells (59, 60). Maximal derepression of the LHR gene in these cells is achieved upon hyperacetylation of histones H3 and H4 within its promoter and complete demethylation of the CpG island encompassing the promoter (60). These findings have illustrated the critical participation of an epigenetic program in the control of LHR gene transcriptional activity. The proximal Sp1 site of the LHR gene promoter was shown to be an essential mediator of trichostatin A (TSA)-induced LHR gene activation. This site was also identified as an anchor to recruit the histone deacetylase (HDAC)/mSin3A complex to the LHR gene promoter, leading to promoter-localized chromatin condensation through histone hypoacetylation (59). A TSA challenge, however, did not evoke a change in the patterns of DNA binding at this site, the levels of the Sp1 and Sp3 proteins, or release of the HDAC/mSin3A complex from the LHR gene promoter in JAR cells.
Sp1/Sp3 binding site-dependent gene activation via HDAC inhibition has also been observed for other genes in various cell lines treated with a variety of HDAC inhibitors (8, 20, 51, 55). Despite the growing body of evidence in this regard, its mechanism is still far from clear. It has also not yet been defined whether Sp1 and Sp3, which are both ubiquitously expressed and bind to GC-rich sequences with similar activities, contribute equally to HDAC inhibitor-activated gene expression. The lack of change in Sp1/Sp3 binding properties and their protein expression status in most of the cases studied imply that another mechanism(s) may be involved. Posttranslational modifications of Sp1 and Sp3 are known to have a significant impact on Sp1 site-regulated target gene expression in many physiological settings (5, 14, 40). These include target genes involved in cell growth, apoptosis, angiogenesis, and tumorigenesis. In particular, phosphorylation and dephosphorylation of Sp1 in response to diverse exogenous stimuli and environmental cues can alter the transactivation activity of Sp1 or its association with other transcription factors or cofactors, resulting in a change in Sp1 site-controlled gene expression (4, 6, 15, 44). The present studies have demonstrated that the participation of a signal transduction mechanism is essential for derepression of the LHR gene transcription induced by the HDAC inhibitor TSA. Our findings have revealed a novel mechanism of TSA action through derecruitment of a repressor from the LHR gene promoter in a phosphatidylinositol 3-kinase/protein kinase Cζ (PI3K/PKCζ)-induced Sp1 phosphorylation-dependent manner.
MATERIALS AND METHODS
Reagents, expression vectors, and antibodies.
TSA, PD98059, H89, Gö6983, Gö6976, Rottlerin, a myristoylated PKCζ pseudosubstrate peptide inhibitor (PS-PKCζ), wortmannin, and LY294002 were obtained from Calbiochem (San Diego, CA). The human LHR promoter/luciferase reporter gene construct used has been described in our previous studies (57). The PKCζ kinase-inactive mutant construct was generously provided by Alex Toker (Harvard Medical School, Boston, MA). The PI3K p110α dominant negative (DN) mutant was a gift from Bai Lu (National Institute of Child Health and Human Development, NIH). The Flag-Sp1 expression vector was kindly provided by Adrian Black (Roswell Park Cancer Institute, Buffalo, NY). The mutant Flag-Sp1 plasmid, in which serine 641 was mutated to alanine, was constructed on the basis of wild-type (WT) Flag-Sp1 with the QuikChange II site-directed mutagenesis kit (Stratagene, La Jolla, CA) and verified by sequencing analyses. The pCMV-Sp1, pCMV-pRb, and pCMV-p107 expression plasmids were obtained from OriGene (Rockville, MD). The pcDNA1.1-Sp3 expression vector was described previously (29).
The antibodies for various PKC isoforms, Sp3, actin, tubulin, HDAC1, HDAC2, mSin3A, pRb, and p107 were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Whole-cell lysates of HeLa, 3611-RF, Jurkat, and A431 cells obtained from Santa Cruz were used as positive controls to verify the efficiency of the antibodies for individual PKC isoforms (HeLa cell lysates for PKCα, βI, βII, and ɛ; 3611-RF cell lysates for PKCγ and -μ; Jurkat cell lysates for PKCδ, -ζ, and -θ; A431 cell lysates for PKCι). Lung tissue lysates (rat) obtained from Abcam (Cambridge, MA) were utilized as a positive control for PKCη antibody. Antibodies for phospho-PKCζ (Thr 410) and phospho-PKCδ (Thr 505) were provided by Cell Signaling (Danvers, MA). Phosphoserine (p-Ser), phosphothreonine (p-Thr), or phosphotyrosine (p-Tyr) antibodies and Flag antibody were purchased from Sigma (St. Louis, MO). The Sp1 antibody for immunoprecipitation (IP) was also from Sigma. Antibodies for the total and acetylated histones H3 and H4 were obtained from Upstate (Charlottesville, VA).
Cell culture and transfection.
Human choriocarcinoma JAR cells and MCF-7 cells (human mammary gland carcinoma cells) from the American Type Culture Collection (Manassas, VA) were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum and 1% antibiotic-antimycotic solution (Invitrogen, Carlsbad, CA).
Transfections were carried out with Lipofectamine and Plus reagents (Invitrogen) according to the procedure recommended by the manufacture. For reporter gene analyses of cells transfected with expression plasmids, the DNA amount used in each well was adjusted with empty vector plasmid so that every well contained the same amount of DNA. At 24 h posttransfection, cells were treated with the indicated inhibitors for 1 h, followed by TSA treatment for an additional 24 h. Luciferase activities were normalized to light units per microgram of protein and expressed as means ± standard errors. All experiments were performed at least three times in triplicate wells.
RNA isolation, reverse transcription (RT)-PCR, and real-time PCR.
Total RNA from treated or untreated cells was extracted with an RNeasy Kit (QIAGEN, Valencia, CA). RNA was pretreated with amplification grade DNase I (Invitrogen) at room temperature for 15 min before RT-PCR with a QIAGEN One Step RT-PCR kit. The primers for amplification of a 475-bp fragment encoding a C-terminal region of the human LHR gene (nucleotides 1557 to 2031) were 5′ GGAAACCACTCTCTCACAAGT 3′ (forward) and 5′ GGTGGATTGAGAAGGCTTATTTG 3′ (reverse). The primers for amplification of a 620-bp human β-actin gene fragment were 5′ CCTCGCCTTTGCCGATCC 3′ (forward) and 5′ GGATCTTCATGAGGTAGTCAGTC 3′ (reverse).
For quantitative analyses of LHR gene mRNA levels, total RNA was reverse transcribed with a SuperScript III kit (Invitrogen) and subjected to PCR analyses with SYBR green Master Mix and an ABI 7500 sequence detection system (Applied Biosystems, Foster City, CA). The PCR conditions consist of a 10-min hot start at 95°C, followed by 40 cycles of 30 s at 95°C and 1 min at 60°C. The primers used were 5′ ATGGCAATCCTCATCTTCACCG 3′ (forward) and 5′ GTGAATATTGCATACAGAAATG 3′ (reverse). Each sample was run in triplicate, and the results were normalized to the level of β-actin mRNA as described previously (60).
Whole-cell lysate and nuclear protein preparation.
JAR whole-cell lysates were prepared with the M-PER mammalian protein extraction reagent of Pierce (Rockford, IL). The cytosolic and nuclear fractions from treated and untreated cells were isolated with the NucBuster protein extraction kit of Novagen (San Diego, CA) by following the protocol included. All of the preparations were performed in the presence of 1× Complete protease inhibitor cocktail (Roche, Mannheim, Germany) and 1× Halt phosphatase inhibitor cocktail (Pierce).
Small interfering RNA (siRNA) analyses.
siRNAs designed to knock down the endogenous expression of Sp1, Sp3, PKCζ, or PKCδ and siRNA for the negative control (NTC) were obtained from Ambion (Austin, TX). Transfections of siRNAs were performed by use of siPORTNeoFX reagent according to the procedures recommended by the manufacture (Ambion). Briefly, the siPORTNeoFX reagent was diluted in Opti-MEM I reduced-serum medium (Invitrogen) and incubated at room temperature for 10 min. This was then mixed with the indicated siRNA (30 nM) diluted in Opti-MEM I reduced-serum medium, followed by incubation at room temperature for 10 min. The complexes formed were dispensed into 24-well or 6-well cell culture plates and overlaid with cells at a density of 1 × 105 cells/ml. At 24 h posttransfection, cells were replaced with fresh medium and grown for an additional 48 h before harvest. For reporter gene analyses of LHR gene promoter activity in the presence of siRNAs of Sp1, Sp3, PKCζ, PKCδ, or NTC, the promoter/luciferase construct was introduced by Lipofectamine and Plus reagents at 24 h posttransfection with siRNA, and cells were terminated 48 h later. TSA was added to the cells 24 h prior to termination.
IP analyses.
For each IP assay, 250 μg nuclear proteins or 1 mg whole-cell lysates in 1 ml IP buffer (20 mM Tris HCl [pH 8.0], 100 mM NaCl, 1 mM EDTA, 0.5% NP-40, 1× protease inhibitor cocktail, 1× phosphatase inhibitor cocktail) was used. The proteins were initially precleared with 0.5 μg normal rabbit or mouse immunoglobulin G (IgG) and 40 μl protein A/G PLUS agarose beads (Santa Cruz) at 4°C for 20 min. The precleared supernatant was incubated with 1 μg of the specific antibody of interest at 4°C overnight with gentle shaking. This was followed by addition of 36 μl protein A/G agarose beads and incubation at 4°C for another 1 h. The protein-antibody-agarose complex was recovered by brief centrifugation and washed three times with 1 ml IP buffer for 5 min each time at 4°C. The complex was resuspended in 30 μl 2× sodium dodecyl sulfate protein sample buffer containing 2.5% β-mercaptoethanol and boiled for 3 min before being subjected to Western blot analyses.
DNA affinity precipitation assays (DAPA).
DAPA were performed as described previously (59). Briefly, 50 μg nuclear extracts was incubated with 0.4 μg biotin-labeled probe harboring the sequences of the proximal Sp1 binding site of the LHR gene promoter. The reaction was carried out on ice for 45 min, which was followed by addition of 30 μl of tetrameric avidin resin (Promega, Madison, WI) with another incubation at 4°C for 1 h. The DNA-protein complex was harvested by brief centrifugation and analyzed by immunoblotting analyses with the antibodies indicated.
Chromatin IP (ChIP).
ChIP experiments were performed with the ChIP assay kit from Upstate Biotechnology (Lake Placid, NY). Briefly, 2 × 107 treated or untreated cells were fixed with 1% formaldehyde at 37°C for 10 min and lysed and sheared by sonication. The soluble chromatin fraction was subjected to IP analyses with the antibodies of interest as indicated. The precipitated complexes were washed sequentially with low-salt, high-salt, LiCl, and Tris-EDTA buffers and extracted twice with freshly prepared 1% sodium dodecyl sulfate-0.1 M NaHCO3. The cross-linking between DNA and proteins was reversed, and DNA was purified by ethanol precipitation. The ChIP-precipitated DNA and input DNA were analyzed by real-time PCR with SYBR green Master Mix in an ABI 7500 sequence detection system. The primers for the LHR gene promoter region were 5′ ACTGGGCACTGTCGCAGGTC 3′ (forward) and 5′ CATGGCCGGCGAACTGGGCT 3′ (reverse), as described previously (60). The primers for the human β-actin promoter were 5′ GGCCAACGCCAA AACTCTCCCTC 3′ (forward) and 5′ GTCTCGGCGGTGGT GGCGCGT 3′. The amplified 236-bp DNA fragment was cloned into the pCRZeroBlunt vector (Invitrogen), and the standard curve was created by a 10-fold dilution of the cloned plasmid DNA for quantitative analyses of the binding of the indicated transcription factors to the β-actin promoter.
RESULTS
PKCζ has a critical role during TSA-induced LHR gene activation.
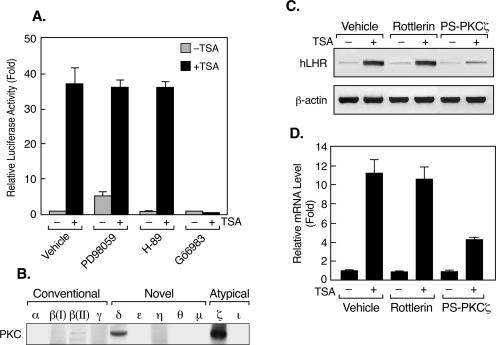
Recent studies have demonstrated that treatment with an HDAC inhibitor (e.g., sodium butyrate [NaB], trapoxin, or apicidin) caused increased activities of mitogen-activated protein kinases (MAPK) and protein kinase C (PKC) in several of the cell lines studied (28, 39, 52). These findings indicated that these signal transduction pathways might contribute to the stimulatory changes induced by the HDAC inhibitors in the transcription of several genes. To investigate the putative participation of a protein kinase(s) in TSA-activated LHR gene expression, reporter gene analyses of LHR gene promoter activity were carried out initially with JAR cells that were pretreated with inhibitors of MAPK, PKA, or PKC prior to incubation with TSA (Fig. 1A). The PKC inhibition by Gö6983 (a broad-spectrum PKC inhibitor) diminished TSA-induced activation of LHR gene promoter activity. In contrast, neither inhibition of MAPK by PD98059 nor that of PKA by H89 had an impact on LHR gene promoter activity. This indicated the participation of a PKC family member(s) in the TSA-mediated activation of LHR gene expression.
FIG. 1.
The PKC pathway is involved in TSA-induced LHR gene activation. (A) Reporter gene analyses of LHR gene promoter activity in JAR cells. Cells were pretreated with PD98059 (10 μM), H-89 (5 μM), or Gö6983 (2 μM) for 1 h prior to incubation with or without TSA (100 ng/ml) for 24 h. Relative promoter activities are indicated as n-fold induction over the activity in the absence of treatment (onefold). (B) Western blot analyses of the endogenous expression of PKC isoforms in JAR cells. (C and D) RT-PCR (C) and real-time PCR (D) analyses of human LHR (hLHR) gene expression in JAR cells that were treated with Rottlerin (5 μM) or PS-PKCζ (50 μΜ) for 1 h, followed by incubation with or without TSA (100 ng/ml) for 24 h. The relative mRNA levels from the quantitative analyses are indicated as n-fold induction over the level in the absence of treatment (onefold).
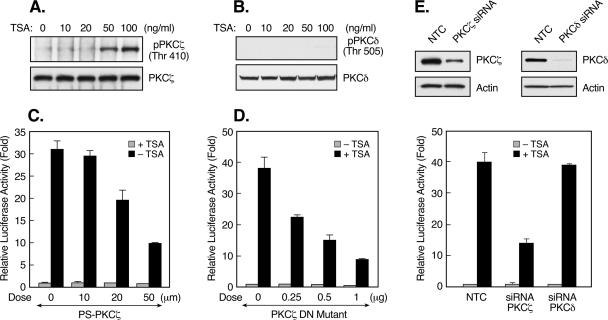
PKC is a family of serine/threonine kinases that includes at least 11 isozymes (19, 46). These PKC isoforms are subgrouped into three categories based on their regulatory properties, namely, conventional PKCs [α, β(I), β(II), and γ], novel PKCs (δ, ɛ, η, θ, and μ), and atypical PKCs (ζ, ι/λ [human/mouse]). Western blot analyses of endogenous PKC isoenzymes in JAR cells revealed predominant expression of PKCδ and -ζ, whereas the levels of other forms of PKC were undetectable, except for PKCβ(II), which displayed a weak signal (Fig. 1B). The efficiency of the PKC antibodies used was confirmed by Western blot analyses of lysates from cell lines expressing known PKC subtypes (see Materials and Methods; data not shown). RT-PCR analyses revealed that inhibition of PKCδ activity by Rottlerin did not affect TSA-mediated induction at the LHR gene mRNA level (Fig. 1C and D). In addition, Gö6976, an inhibitor of conventional PKC, did not have any effect on LHR gene expression endogenously or in reporter gene analyses (data not shown). These results were consistent with nonexpression or minimal expression of the conventional PKC isotypes in these cells (Fig. 1B). However, this induction was largely abolished upon blockade of the activity of the atypical PKC, PKCζ, by PS-PKCζ (Fig. 1C and D). Furthermore, TSA dose dependently induced the activation of PKCζ, as determined by the phosphorylation of this enzyme at threonine 410 (Thr 410) (Fig. 2A), which is recognized to be critical for its activity (2, 42). In contrast, phosphorylation of PKCδ at Thr 505 (control) was not observed (Fig. 2B). Moreover, the critical participation of PKCζ in the up-regulation of LHR gene expression by TSA was confirmed by reporter gene analyses in which both the PKCζ inhibitor and a kinase-inactive mutant form of PKCζ (the DN mutant PKCζ DN) abrogated TSA-activated LHR gene promoter activity in a dose-dependent manner (Fig. 2C and D). The specific role of PKCζ was further demonstrated by siRNA analyses in which TSA-induced LHR gene promoter activity was significantly abolished upon siRNA suppression of PKCζ expression. In contrast, no such effect was observed after knockdown of the expression of PKCδ (Fig. 2E). Collectively, it has been demonstrated that PKCζ is critical for TSA-induced LHR gene activation, since inhibition of its activity markedly diminished the inducible effect of TSA on LHR gene expression.
FIG. 2.
PKCζ is critical for the LHR gene activation induced by TSA. (A) Levels of phospho-PKCζ (Thr 410) in JAR cells treated with increasing doses of TSA. Whole-cell lysates were immunoprecipitated with PKCζ antibody, followed by Western blotting with phospho-PKCζ (Thr 410) antibody. (B) Western blot analyses of phospho-PKCδ (Thr 505) in JAR cells treated with the indicated doses of TSA. The total PKCζ and PKCδ levels in these cells are also shown. (C and D) Reporter gene analyses of LHR gene promoter activity in JAR cells in the presence of increasing doses of a PKCζ inhibitor (C) or of a PKCζ DN mutant construct (D). (E, top) JAR cells were transfected with PKCζ or PKCδ siRNA or a negative control siRNA (NTC), followed by Western blot analyses of the expression of PKCζ or PKCδ protein. The expression of actin in these cells is also shown as a control. (E, bottom) Reporter gene analyses of LHR gene promoter activity in JAR cells cotransfected with PKCζ, PKCδ siRNA, or NTC. Cells were treated with or without TSA for 24 h. Relative luciferase activities are indicated as n-fold induction of enzyme activity in the presence of TSA over enzyme activity in the absence of TSA.
TSA treatment elicited marked Sp1 phosphorylation at serine 641 dependent on the activity of PKCζ.
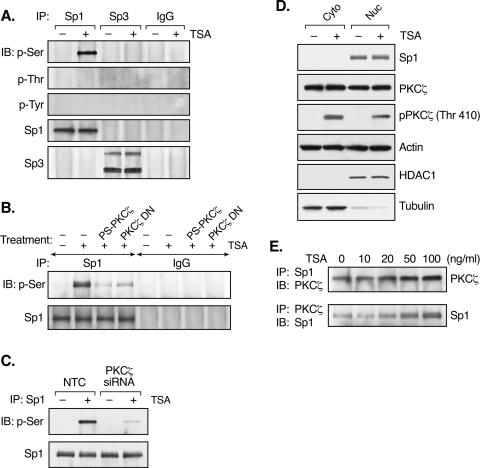
The dependence on the proximal Sp1/Sp3 binding site in the process of TSA-activated LHR gene expression led to studies to determine whether PKCζ exerts its impact through Sp1, Sp3, or both. IP of nuclear extracts from TSA-treated or untreated JAR cells by an Sp1 or Sp3 antibody was performed, followed by Western blot analyses with p-Ser, p-Thr, or p-Tyr antibodies. TSA treatment caused marked Sp1 phosphorylation at serine residues, whereas no phosphorylation of Sp1 at threonine or tyrosine residues was detected (Fig. 3A). In contrast, no phosphorylation of Sp3 was observed in the absence or presence of TSA. Furthermore, the TSA-induced serine phosphorylation of Sp1 was PKCζ dependent since inhibition of the activity of this kinase by its inhibitor, its DN mutant form, and its siRNA significantly reduced Sp1 phosphorylation (Fig. 3B and C). The TSA-induced Sp1 phosphorylation at a serine residue(s) was also in agreement with a previous report showing that PKCζ-elicited Sp1 phosphorylation occurred solely at a serine residue(s) (37).
FIG. 3.
TSA causes marked Sp1 phosphorylation dependent on the activity of PKCζ. (A) Nuclear extracts isolated from TSA-treated (100 ng/ml, 24 h) or untreated JAR cells was immunoprecipitated (IP) with Sp1 or Sp3 antibody or normal rabbit IgG, followed by immunoblotting (IB) with p-Ser, p-Thr, or p-Tyr antibodies. Total Sp1 and Sp3 precipitates were also assayed with Sp1 or Sp3 antibody. (B) JAR cells pretreated with the PKCζ inhibitor PS-PKCζ or overexpressed of the PKCζ DN mutant form were incubated with TSA for 24 h. Nuclear extracts were immunoprecipitated with Sp1 antibody or normal IgG and then immunoblotted with p-Ser antibody. Immunoblotting with Sp1 antibody is also shown. (C) JAR cells transfected with PKCζ or NTC siRNA were treated with or without TSA for 24 h. Whole-cell lysates were immunoprecipitated with Sp1 antibody and then immunoblotted with p-Ser antibody. The total amount of Sp1 immunoprecipitated is also shown. (D) Cytosolic (Cyto) and nuclear (Nuc) fractions prepared from TSA-treated or untreated JAR cells were subject to immunoblotting analyses with antibodies to Sp1, PKCζ, phospho-PKCζ (Thr 410), and actin. HDAC1 and tubulin, used as nuclear and cytosolic markers, were also assessed by immunoblotting. (E) Nuclear extracts isolated from TSA-treated JAR cells were immunoprecipitated with Sp1 antibody and then immunoblotted with PKCζ antibody and vice versa.
PKCζ is distributed evenly in both the nuclear and cytosolic fractions of cells in the presence or absence of TSA (Fig. 3D). The activated form of PKCζ (phosphorylated at Thr 410; Fig. 2A) not only existed in the cytosol but was also present in the nucleus, strengthening the concept that Sp1 could be a nuclear target of this kinase. Coimmunoprecipitation studies demonstrated that endogenous PKCζ was in the immunocomplex precipitated by an Sp1 antibody and, similarly, endogenous Sp1 in the PKCζ immunocomplex (Fig. 3E). These observations have confirmed that Sp1and PKCζ coexisted in the same protein complex. Moreover, TSA treatment caused a significant increase in the association between Sp1 and PKCζ in a TSA dose-dependent manner.
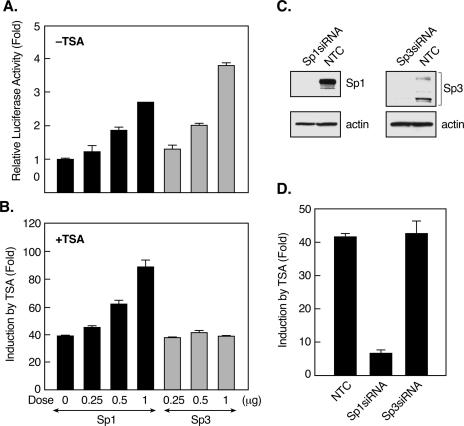
The differential effects of TSA on the phosphorylation of Sp1 and Sp3 raised the possibility that Sp1 and Sp3 act differently during TSA-activated LHR gene expression. In agreement with our previous findings (25), overexpression of Sp1 or Sp3 in JAR cells increased basal LHR gene promoter activity (Fig. 4A). On the other hand, Sp1 augmented the effect of TSA on LHR transcriptional activation, whereas no enhancement was observed with Sp3 (Fig. 4B). Furthermore, transfection of cells with Sp1 siRNA, which caused complete knockdown of Sp1 protein expression, markedly prevented TSA-induced LHR gene promoter activity (Fig. 4C and D). In contrast, suppression of Sp3 protein expression by its siRNA had no effect on the TSA response of the LHR gene. Taken together, these results have illustrated that TSA treatment elicited marked Sp1 but not Sp3 phosphorylation and that Sp1, but not Sp3, has a significant role in TSA-induced LHR gene activation. Coincident with the requirement of PKCζ activation for the effect of TSA on LHR gene expression, TSA-induced Sp1 phosphorylation was dependent on the activity of PKCζ.
FIG. 4.
Sp1, but not Sp3, is required for TSA-activated LHR gene expression. (A and B) Reporter gene analyses of LHR gene promoter activity in JAR cells cotransfected with increasing doses of Sp1or Sp3 expression plasmid. Cells were treated with (B) or without (A) TSA for 24 h. (C) JAR cells were transfected with Sp1 or Sp3 siRNA or negative control siRNA (NTC), followed by immunoblotting analyses of the Sp1 or Sp3 protein level. The level of actin in these cells is shown as a control. (D) Reporter gene analyses of LHR gene promoter activity in JAR cells cotransfected with Sp1, Sp3 siRNA, or NTC. Cells were treated with or without TSA for 24 h. Relative luciferase activities are indicated as n-fold induction of enzyme activity in the presence of TSA over enzyme activity in the absence of TSA.
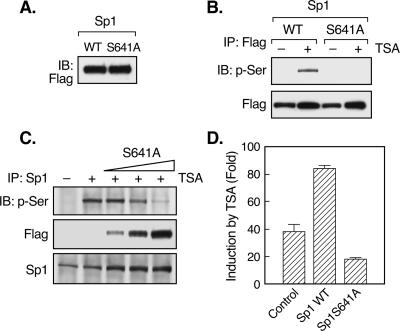
Computational analyses of the putative phosphorylation site(s) of Sp1 by PKCζ with the Scansite program (http://scansite.mit.edu/) at low stringency revealed that serine 641 and threonine 681 in the Sp1 DNA binding domain were probably the candidate sites. This prediction is consistent with in vitro phosphorylation studies of a series of truncated forms of the Sp1 protein, which demonstrated that PKCζ interacted with and phosphorylated the DNA binding domain of Sp1, whereas no interaction or phosphorylation was detected for the rest of the Sp1 molecule (34). To delineate the putative phosphorylation site(s) of Sp1 targeted by PKCζ during TSA-activated LHR gene expression, serine 641 of WT Flag-Sp1 was mutated to alanine (S641A mutant) and expressed in JAR cells (Fig. 5A). Consistent with our previous results, TSA treatment caused significant phosphorylation of Sp1 precipitated with a Flag antibody (Fig. 5B). However, no phosphorylation of the S641A mutant Sp1 protein was detected. These results indicated that Ser 641 is the target site associated with the TSA-induced responses. In addition, overexpression of the Sp1 S641A mutant protein dose dependently reduced the phosphorylation level of Sp1 in TSA-treated cells (Fig. 5C). Functionally, the Sp1 S641 mutant antagonized TSA-activated LHR gene promoter activity, in contrast to the augmentation effect caused by WT Flag-Sp1 (Fig. 5D). Taken together, these results have demonstrated that phosphorylation of Sp1 at serine 641 has a critical role in TSA-induced LHR gene activation.
FIG. 5.
Ser 641 of Sp1 is the target phosphorylation site during TSA-induced LHR gene activation. (A) WT or serine 641-mutated (to alanine, S641A) Flag-Sp1 plasmid was transfected into JAR cells. The expression of WT and mutant Sp1 proteins was assayed by immunoblotting (IB) analyses with Flag antibody. (B) JAR cells transfected with WT or S641A mutant Flag-Sp1 were treated with or without TSA for 24 h. Whole-cell lysates were immunoprecipitated with Flag antibody, followed by immunoblotting with p-Ser antibody. Total immunoprecipitated Flag-Sp1 proteins are also shown. (C) JAR cells were transfected with increasing doses of Flag-Sp1 S641A mutant construct, followed by treatment with TSA for 24 h. Whole-cell lysates were immunoprecipitated with Sp1 antibody, followed by immunoblotting with p-Ser, Flag, or Sp1 antibody. (D) Reporter gene analyses of LHR gene promoter activity in JAR cells transfected with a WT or S641A mutant Sp1 construct. Relative luciferase activity is shown as n-fold induction of enzyme activity in the presence of TSA over that in the absence of TSA.
Participation of PI3K during TSA-induced LHR gene activation.
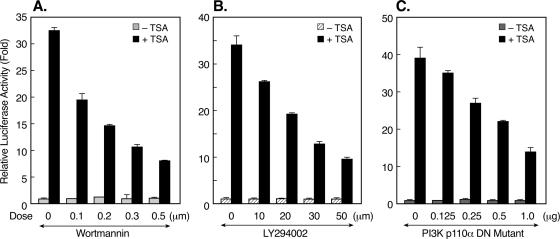
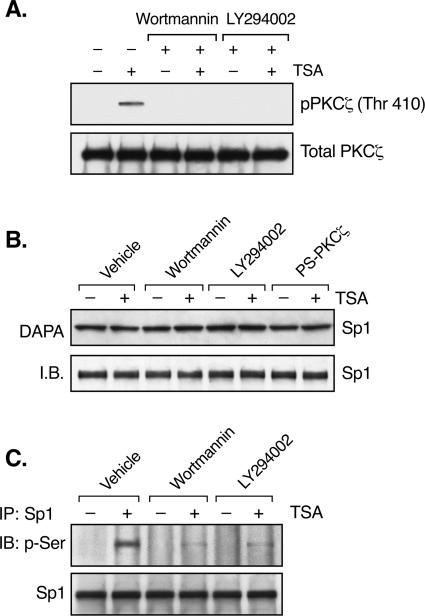
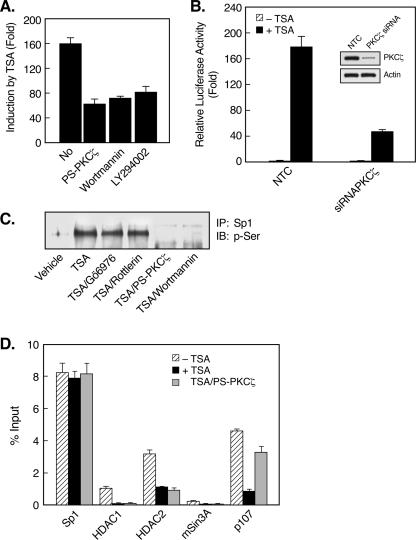
The activation of PKCζ by TSA indicated the participation of another kinase(s) as an upstream effector of PKCζ during TSA-induced LHR gene activation. Recent studies have implied that PKCζ is a downstream target for PI3K in diverse physiological settings (13, 43, 45). Also, PI3K has been shown to be involved in the regulation of the gene for p21 by apicidin, an HDAC inhibitor (32). To address whether PI3K contributes to TSA-activated LHR gene expression, blockade of PI3K activity by the inhibitor wortmannin or LY294002 was tested in reporter gene analyses of LHR gene promoter activity. Both wortmannin and LY294002 caused a dose-dependent reduction of promoter activity in the presence of TSA (Fig. 6A and B). This reduction was specific to the TSA-mediated effect since no change in LHR gene basal promoter activity was observed. We further confirmed the effect of PI3K on LHR gene promoter activity with a kinase-inactive mutant form of the PI3K p110α subunit. Similarly, the DN mutant PI3K p110α construct resulted in a significant reduction in the LHR gene promoter activity induced by TSA (Fig. 6C). Moreover, the PKCζ phosphorylation induced by TSA was abolished upon treatment with wortmannin (0.5 μM) or LY294002 (50 μM) (Fig. 7A). Coincident with our previous studies showing a lack of effect of TSA on the binding activity or level of the Sp1 protein (59), no changes in these parameters were observed in the presence of wortmannin or LY294002 or the PS-PKCζ mutant (Fig. 7B). However, inhibition of PI3K markedly suppressed TSA-induced Sp1 phosphorylation (Fig. 7C), demonstrating an important role for PI3K in TSA-activated LHR gene expression. These results, in combination with the observation that no nuclear location of PI3K was detected in the absence or presence of TSA (data not shown), have demonstrated that PI3K acts upstream of PKCζ, which is present in both the nuclear and cytoplasmic compartments, and that the activity of this kinase is necessary for PKCζ-elicited phosphorylation of Sp1.
FIG. 6.
Participation of PI3K in TSA-induced LHR gene activation. (A and B) Reporter gene analyses of LHR gene promoter activity in JAR cells that were pretreated with wortmannin or LY294002 for 1 h, followed by incubation with or without TSA (100 ng/ml) for 24 h. (C) Reporter gene analyses of LHR gene promoter activity in JAR cells in the presence of a DN PI3K p110α mutant construct. Cells were treated with or without TSA for 24 h prior to analyses. Relative luciferase activities are indicated as n-fold induction over enzyme activity in the absence of drug treatment or the PI3K p110α DN mutant form.
FIG. 7.
PI3K activity is required for PKCζ activation and Sp1 phosphorylation. (A) JAR cells were pretreated with or without wortmannin (0.5 μM) or LY294002 (50 μM) for 1 h, followed by incubation with or without TSA (100 ng/ml) for 24 h. Whole-cell lysates were immunoprecipitated with PKCζ antibody, followed by immunoblotting (IB) with phospho-PKCζ antibody (Thr 410). The total amounts of PKCζ precipitated from these cells are also shown as a control. (B) DAPA of Sp1 binding activity in JAR cells. Nuclear extracts isolated from JAR cells treated with or without TSA in the presence of wortmannin, LY294002, or PS-PKCζ were incubated with the biotin-labeled proximal Sp1 site of the LHR gene promoter, followed by immunoblotting with Sp1 antibody. The level of Sp1 protein is also shown. (C) IP of the nuclear extracts from panel A by Sp1 antibody was followed by immunoblotting with an antibody recognizing p-Ser and with an Sp1 antibody, respectively.
Release of p107 from the TSA-activated LHR gene promoter in a PKCζ-dependent manner.
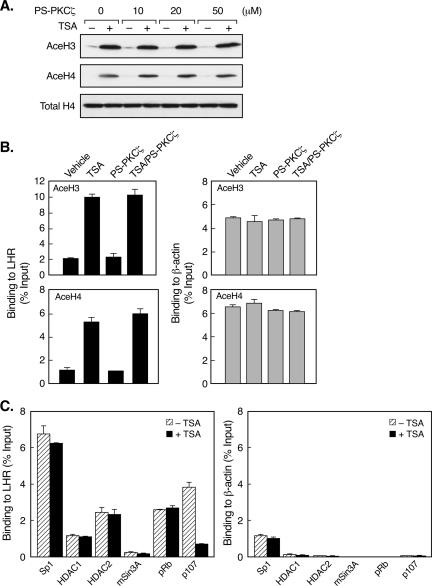
Promoter-specific histone hyperacetylation resulting from HDAC inhibition has been recognized as crucial for the activation of target genes. To further elucidate the mechanism underlying the relevance of PKCζ in TSA-induced LHR gene expression, the putative impact of this kinase on histone acetylation status in the LHR gene promoter region was next investigated. Blockade of PKCζ activity did not affect the global histone H3 and H4 hyperacetylation in TSA-treated cells (Fig. 8A). Also, as demonstrated by quantitative ChIP analyses, the presence of acetylated H3 or H4 at the LHR gene promoter which was evoked by TSA treatment remained unchanged in the presence of the PKCζ inhibitor (Fig. 8B, left). These results thus indicate a lack of effect of PKCζ on LHR gene promoter-associated histone acetylation status.
FIG. 8.
TSA causes release of p107 from the LHR gene promoter independently of histone acetylation status. (A) Immunoblotting analyses of the levels of acetylated histones H3 (AceH3) and H4 (AceH4) in TSA-treated or untreated JAR cells in the presence of different doses of the PKCζ inhibitor PS-PKCζ. The total H4 protein level is also shown. (B) Quantitative ChIP analyses of the association of acetylated H3 or H4 with the LHR gene promoter (left) and the human β-actin promoter (right) in JAR cells treated with or without TSA, PS-PKCζ, or both. (C) Quantitative ChIP analyses of the association of Sp1, HDAC1, HDAC2, mSin3A, pRb, and p107 with the LHR gene promoter (left) and the human β-actin promoter (right) in TSA-treated or untreated JAR cells. Results are expressed as percentages of the total input DNA.
On the other hand, it was conceivable that PKCζ might exert its action through a change at the protein-protein interaction level. Thus, the effort was directed toward the identification of a protein(s) whose associations with the LHR gene promoter were changed by TSA treatment. Consistent with our previous findings (59), the binding of HDAC1/2 and the corepressor protein mSin3A was unchanged in the absence or presence of TSA (Fig. 8C, left). This was also the case for histone acetyltransferase p300 and CBP corepressor proteins NcoR and SMRT (data not shown). We further tested for retinoblastoma protein pRb and its close homologue p107 since pRb is an HDAC-interacting protein and pRb and p107 have been shown to associate with the Sp1 binding site (9, 18, 36). TSA treatment profoundly decreased the association of p107 with the LHR gene promoter (Fig. 8C, left). In contrast, the binding of pRb was not affected by TSA. The release of p107 from the LHR gene promoter was found to be largely dependent on the activity of PKCζ, inhibition of which substantially prevented TSA-induced p107 dissociation (Fig. 9A). In addition, these observed changes were confirmed to be specifically associated with the LHR gene, whose promoter activity was markedly affected by TSA, whereas no such effects were detected for the β-actin gene, whose expression was not influenced by TSA (Fig. 1, 8, and 9).
FIG. 9.
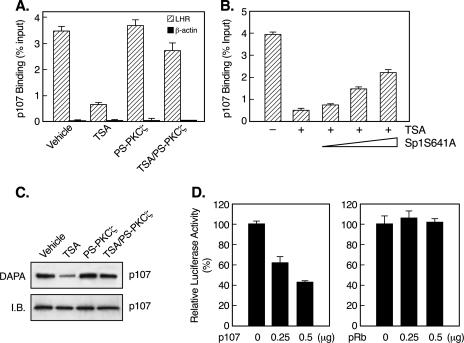
Release of p107 from the LHR gene promoter in a PKCζ-dependent manner. (A) Quantitative ChIP analyses of p107 association with the LHR gene promoter and the human β-actin promoter in JAR cells treated with or without TSA, PS-PKCζ, or both. (B) JAR cells were transfected with increasing doses of an S641A Flag-Sp1 mutant construct, followed by treatment with TSA for 24 h. Quantitative ChIP assays were carried out for analyses of p107 binding to the LHR gene promoter under these conditions. (C) DAPA of p107 association with the proximal Sp1 site of the LHR gene promoter. Nuclear extracts from cells treated as described for panel A were incubated with a biotin-labeled Sp1 probe, followed by immunoblotting analyses with a p107 antibody. The level of p107 protein is also shown (I.B.). (D) Reporter gene analyses of LHR gene promoter activity in JAR cells in the presence of a p107 or pRb expression plasmid. Relative luciferase activities are expressed as percentages of enzyme activity in the absence of the p107 or Rb construct (100%).
The dependency of p107 release on the phosphorylation status of Sp1 was further analyzed by ChIP analyses of JAR cells, where increasing doses of the Flag-Sp1 S641A mutant construct were expressed (Fig. 9B). In agreement with the previous phosphorylation and functional analyses (Fig. 5), this phosphorylation-deficient Sp1 mutant reduced TSA-elicited p107 release from the LHR gene promoter in a dose-dependent manner. Consistent with the results of ChIP assays, DAPA have shown that TSA caused significant detachment of p107 from the proximal Sp1 site of the LHR gene promoter, whereas this effect was counteracted upon the blockade of PKCζ activity (Fig. 9C). Furthermore, functional analyses have demonstrated that p107 acted as a repressor of LHR gene expression since coexpression of p107 but not pRb with the LHR gene promoter repressed promoter activity dose dependently (Fig. 9D).
We had previously reported that TSA caused marked LHR gene derepression in MCF-7 cells (60). The response elicited by TSA in this cell line was more prominent than in JAR cells (160-fold versus 40-fold), and in contrast to JAR cells, the LHR gene promoter is completely demethylated in MCF-7 cells under basal conditions. This probably facilitated the dissociation of the HDAC/mSin3A complex from the LHR gene promoter observed in the presence of TSA, while such a dissociation of the complex did not occurred in JAR cells treated with TSA (59, 60). However, at that point we could not determine whether the properties observed, such as DNA methylation status and release of the HDAC/mSin3A inhibitory complex, in these cell types would solely account for the differences observed in LHR gene promoter activities. Therefore, it was important to learn whether the TSA-induced release of p107 from the LHR promoter observed in JAR cells also occurred in MCF-7 cells. The activation of LHR gene expression by TSA in MCF-7 cells was Sp1 site dependent, and the expression patterns of PKC isoforms in these cells were identical to those observed in JAR cells (data not shown). Reporter gene analyses revealed that the activation of LHR gene promoter activity by TSA was significantly repressed by inhibitors of PI3K (wortmannin, LY294002) and PKCζ (PS-PKCζ) and by PKCζ siRNA (Fig. 10A and B). Similar to JAR cells, TSA treatment resulted in Sp1 serine phosphorylation in MCF-7 cells and this phosphorylation was abolished in the presence of PS-PKCζ or wortmannin but not Gö6976 or Rottlerin (Fig. 10C). Furthermore, besides the dissociation of the HDAC/mSin3A complex, significant release of p107 from the LHR gene promoter was observed in TSA-treated MCF-7 cells (Fig. 10D). In particular, release of p107 required the activity of PKCζ because treatment of cells with PS-PKCζ in combination with TSA prevented p107 dissociation. In contrast, dissociation of the HDAC/mSin3A complex from the LHR gene promoter was not affected by a blockade of PKCζ activity (Fig. 10D). This was consistent with our previous finding demonstrating that recruitment or derecruitment of the HDAC/mSin3A complex is dependent on the methylation status of the promoter (60). Collectively, these results have demonstrated the participation of the PKCζ and PI3K signaling pathways in TSA-induced LHR gene activation in both JAR and MCF-7 cells. We have elucidated a novel mechanism by which PKCζ-dependent Sp1 phosphorylation and release of a repressor protein from the LHR gene promoter are critical for the LHR gene activation elicited by TSA.
FIG. 10.
Activation of LHR gene expression in MCF-7 cells by TSA involves PKCζ/PI3K, Sp1phosphorylation, and p107 release. (A) Reporter gene analyses of LHR gene promoter activity in MCF-7 cells which were treated with or without TSA (500 μg/ml, 24 h) in the presence or absence of PS-PKCζ, wortmannin, or LY294002. Relative luciferase activities are expressed as n-fold induction of enzyme activity in the presence of TSA over that observed in the absence of TSA. (B) Reporter gene analyses of LHR gene promoter activity in MCF-7 cells cotransfected with PKCζ siRNA or a negative control (NTC). Cells were treated with or without TSA for 24 h. The PKCζ and actin protein levels in these cells are also shown. (C) Sp1 phosphorylation status in MCF-7 cells. Nuclear extracts were isolated from untreated MCF-7 cells or cells treated with TSA in the absence or presence of Gö6976, Rottlerin, PS-PKCζ, or wortmannin. Nuclear extracts were immunoprecipitated with Sp1 antibody, followed by immunoblotting (IB) with p-Ser antibody. (D) Quantitative ChIP analyses of the association of indicated transcription factors to the LHR gene promoter in MCF-7 cells. Cells were treated with or without TSA in the presence of PS-PKCζ.
DISCUSSION
These studies have identified a mechanism in which Sp1 phosphorylation and release of the p107 repressor from the LHR gene promoter mediate TSA-evoked LHR gene activation in JAR and MCF-7 cells. PKCζ is responsible for the marked Sp1 phosphorylation induced by TSA, and blockade of its activity largely abrogates Sp1 phosphorylation, p107 dissociation, and the TSA-induced increase in LHR gene expression. Serine 641 of the Sp1 DNA binding domain has been identified as the functional phosphorylation site, and mutation of this site significantly prevented Sp1 phosphorylation, p107 release, and TSA-mediated LHR gene activation. Moreover, PI3K was found to be an upstream effector of PKCζ's action. Inhibition of PI3K activity abolishes TSA-induced PKCζ activation, Sp1 phosphorylation, and the TSA-induced increase in LHR gene expression. Collectively, these findings have demonstrated that Sp1 phosphorylation by a PI3K/PKCζ kinase pathway is a critical component in HDAC inhibitor-elicited derepression of a target gene such as the LHR gene.
Our recent studies revealed that the proximal Sp1/Sp3 binding site is essential for activation of LHR gene expression by the HDAC inhibitor TSA (59). The present observations have demonstrated that although both Sp1and Sp3 function as activators of LHR gene expression under basal conditions, they have different roles in TSA-induced LHR gene activation. Sp1 is a critical mediator of the effect of TSA on the LHR gene, whereas Sp3 is noncontributory (Fig. 4). Consistent with the differences in the functional roles of Sp1 and Sp3 in TSA-mediated LHR gene activation, TSA induced marked Sp1 phosphorylation with no change in Sp3. In cells exposed to exogenous stimuli, Sp1 phosphorylation is recognized as an important factor in Sp1 site-regulated gene expression, particularly for genes involved in viral infection and cell growth (5, 14). Although Sp1 phosphorylation was observed in Hep3B and SL2 cells treated with NaB (an HDAC inhibitor) (12), the functional impact of the signaling pathways and mechanisms involved in the activation of target gene transcription by HDAC inhibitors have not been determined. Our studies have demonstrated the direct relevance of Sp1 phosphorylation in HDAC inhibitor-regulated gene expression and have revealed that phosphorylation of Sp1 was attributed to the activity of the PI3K/PKCζ pathway.
Increased MAPK or PKC activity has been detected in cells treated with HDAC inhibitors. Inhibition of the MEK-ERK activity in K562 leukemia cells was shown to abrogate the Gαi2 gene activation induced by NaB and Helminthosporium carbonium toxin, but not TSA (54). Also, blockade of the MEK activity in CHP126 neuroepithelioma cells reduced the activation of the choline acetyltransferase gene by NaB, TSA, or trapoxin (23). On the other hand, the apicidin-mediated p21 gene activation in HeLa cells required an Sp1 binding site and the activity of PKCɛ, while the Sp1 phosphorylation status was not evaluated (32). However, since apicidin caused significant translocation of PKCɛ to the plasma membrane, it is unlikely that PKCɛ would exert an action in the nucleus. Our studies have shown that the atypical PKC isoform PKCζ is required for TSA-induced LHR gene expression, whereas the participation of conventional or novel PKCs, MAPK, and PKA was excluded. The lack of conventional PKC, novel PKC, and MAPK action was further confirmed by the use of their DN or constitutively active mutant forms (data not shown). This evidence indicates that different kinase pathways are utilized and associated with the effects of various HDAC inhibitors and that they are subject to the nature of the inhibitors and/or the genes studied. This notion is also supported by gene expression profiling analyses of T24 bladder and MDA breast carcinoma cells treated with three HDAC inhibitors (26). It was found that the set of genes influenced by the structurally related compounds suberoylanilide hydroxamic acid and TSA was substantially different from those regulated by MS-27-275, a reagent that is chemically unrelated to suberoylanilide hydroxamic acid and TSA.
The array of protein kinases that has been found to mediate Sp1 phosphorylation includes DNA-PK, PKA, PKCζ, casein kinase II, ERK, cyclin-dependent kinase, and others whose identities are not yet known (1, 6, 7, 16, 27, 30, 33). However, PKCζ-induced Sp1 phosphorylation has been exclusively associated with the regulation of growth factor genes (35, 37, 38). The present study has identified the gene for LHR as a novel target of PKCζ and has linked the PKCζ pathway and the upstream activator PI3K to the changes elicited by the HDAC inhibitor. TSA strongly enhances PKCζ activation and its association with Sp1. It has been further demonstrated that serine 641 in the Sp1 DNA binding domain is important for TSA-activated LHR gene expression. Mutation of this site simultaneously diminished the Sp1 phosphorylation and LHR gene induction elicited by TSA treatment, illustrating a causative role for Sp1 phosphorylation in this process. PI3K is responsible for PKCζ activation and Sp1 phosphorylation during TSA-activated LHR gene expression. The current literature on signaling pathways and their integration in the regulation of gene expression by HDAC inhibitors is limited, but some reports have indicated a role for PI3K in this process (28). Specifically, inhibition of PI3K activity has been shown to repress the apicidin-induced phosphorylation of PKCɛ and activation of the p21 gene (32). However, whether the action of TSA impacts the PI3K pathway, directly or indirectly, is not yet clear. PI3K could indirectly affect the activity of PKCζ through phosphatidylinositol 3,4,5-trisphosphate generated by activation of its lipid kinase activity through a known substrate (13). Alternatively, PI3K could influence downstream effectors through its intrinsic protein kinase activity. In this study, activation of PI3K was indicated by AKT phosphorylation in MCF-7 cells (data not shown), in addition to the inhibition of this enzyme's effect by its specific inhibitors wortmannin and LY294002 and by its DN mutant form (see above). However, inhibition of AKT by its DN mutant form did not affect TSA-induced LHR gene activation, implying a lack of participation of AKT in PKCζ activation. It remains to be determined whether PKCζ is a target for PI3K protein kinase activity, as has been recognized for insulin receptor substrate docking protein and more recently for tropomyosin (34, 41). Our preliminary results indicated that treatment of JAR and MCF-7 cells with insulin or platelet-derived growth factor, which are known PI3K agonists, did not mimic the induction of LHR gene expression by TSA (data not shown). This indicates that a mechanism(s) other than a direct stimulatory effect on PI3 kinase activity is probably involved. TSA, alternatively, may influence protein phosphatase activity or its interaction with another protein(s), which in turn could change the phosphorylation status of a target protein. The finding that HDAC inhibitors induced dissociation of protein phosphatase 1 from HDAC6, causing a change in the AKT phosphorylation level in prostate cancer cells, favors such a mode of action (11).
Although phosphorylation of Sp1 by PKCζ increases its DNA binding activity in the upregulation of platelet-derived growth factor B-chain gene expression by nogalamycin (37), activation of vascular endothelial growth factor/vascular permeability factor gene expression by hepatocyte growth factor, which is also dependent on PKCζ/Sp1 phosphorylation, does not involve a change in Sp1 binding activity (38). Sp1 is ubiquitously expressed, and its interaction with a number of transcription factors and cofactors is influenced by its phosphorylation status (14, 31). Modulation of such protein interactions appears to be a critical component of the specificity of Sp1 site-dependent gene regulation. Our studies have shown that TSA causes prominent release of p107 but not pRb from the LHR gene promoter and that p107 acts as a repressor of LHR gene promoter activity. Although pRb and p107 were originally recognized as potent repressors of E2F-regulated gene expression during cell cycle progression (9, 22, 24), they are also involved in nuclear receptor (Nur, SF-1, HNF4, and others)-, TGF-β1-, and Sp1-dependent gene regulation via association with SRC/p160 coactivators, Smad, and Sp1 proteins (3, 10, 50, 53). Similar functional roles of pRb and p107 were observed for several genes, but no redundant or substitute functions of these two proteins have been demonstrated (17). Association of p107 and Sp1 has been observed previously in multiple cell lines (HeLa, COS, 293, NIH 3T3, and mouse L cells), and p107 had an Sp1 site-dependent silencing effect on the simian virus 40 early promoter and the transforming growth factor β1 promoter in Saos-2 cells (18). However, it has not been defined in these studies whether the association of p107 and Sp1 is subject to regulation by the phosphorylation status of Sp1. In our study, the observations that p107 release occurred in a PKCζ-dependent manner and that the Sp1S641A phosphorylation-deficient mutant significantly antagonized this p107 dissociation have indicated a clear involvement of the kinase activity, where Sp1 phosphorylation at serine 641 would elicit a conformational change to disrupt the balance of the p107/Sp1 complex.
Our results and a previous report have shown that HDAC inhibitor-mediated PKC activation (ζ/ɛ) does not affect the histone acetylation level in the target gene promoter regions (32). TSA-induced LHR promoter activity is significantly reduced (60 to 70%) during blockade of Sp1 expression by its siRNA by inhibition of the activities of PKCζ and PI3K. The residual promoter activity (30 to 40%) indicates the presence of a mechanism involving histone acetylation independently of Sp1 phosphorylation or could be related to the residual Sp1 phosphorylation observed during repression of the activities of PKCζ and PI3K (Fig. 3 and 7). On the other hand, since marked TSA-induced histone hyperacetylation in the LHR gene promoter region is concurrent with Sp1 phosphorylation, the possibility cannot be excluded that the changes in chromatin structure might provide a permissive state of the LHR gene promoter favoring the phosphorylation of Sp1 and/or the release of p107. Similarly, dissociation of the HDAC/mSi3A complex from the LHR gene promoter in MCF-7 cells was independent of the effect of PI3K/PKCζ and Sp1 phosphorylation. These findings indicate that DNA methylation and histone acetylation levels in the LHR gene promoter region and dissociation of the HDAC/mSin3A complex from the LHR gene promoter, singly or combined, are not sufficient to elicit derepression of the LHR promoter to attain maximal activation of LHR gene expression. Multiple layers of regulation, including PKCζ/PI3K-induced Sp1 phosphorylation, histone acetylation, DNA demethylation of the LHR gene promoter, and release of the inhibitory HDAC/mSin3A complex and p107, all account for TSA-induced LHR gene activation and derepression (a model is shown in Fig. 11).
FIG. 11.
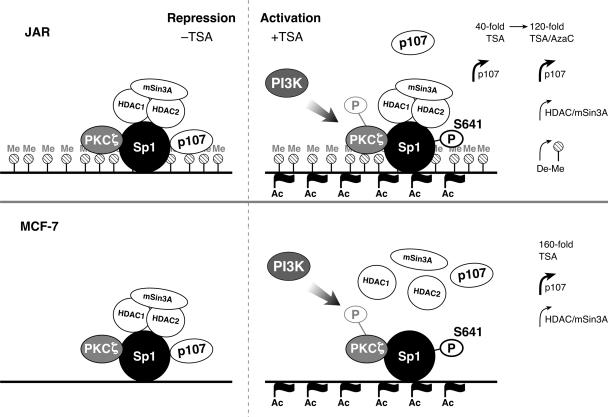
Model of transcriptional derepression and activation of LHR gene expression by TSA. The LHR gene promoter is hypermethylated in JAR cells (top) but demethylated in MCF-7 cells (bottom) (60). In the absence of TSA, the LHR gene promoter is repressed in both cell types and the HDAC/mSin3A complex and p107 are both anchored by Sp1 (left). No Sp1 phosphorylation was observed under these repressive conditions. TSA treatment of JAR cells induced LHR gene promoter-localized histone hyperacetylation without an effect on its DNA methylation status. TSA caused (i) Sp1 phosphorylation dependent on the activity of PKCζ and PI3K and (ii) release of p107 but not of the HDAC/mSin3A complex from the LHR gene promoter. This causes partial LHR gene derepression with a 40-fold induction of its promoter activity (top right). Upon promoter demethylation and HDAC/mSin3A dissociation by TSA-5-azacytidine treatment, maximal derepression of LHR gene expression was observed (60). In MCF-7 cells, TSA induced maximal derepression of LHR gene expression (160-fold). This was caused by histone hyperacetylation and DNA demethylation at its promoter (60) and release of the HDAC/mSin3A complex, more critically, triggered by the dissociation of p107 which resulted from Sp1 phosphorylation by PKCζ and PI3K (bottom right). Me, 13 methylated CpG sites within the promoter of the LHR (60); Ac, acetylated histones; P, phosphorylated; De-Me, promoter DNA demethylation.
Taken together, the present studies have demonstrated that derecruitment of repressor protein p107 from the LHR gene promoter in a PI3K/PKCζ-induced Sp1 phosphorylation-dependent manner is critically important for TSA-induced activation of the LHR gene.
Acknowledgments
We thank Alex Toker (Department of Pathology, Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, MA), Bai Lu (Laboratory of Cellular and Synaptic Neurophysiology, National Institute of Child Health and Human Development, NIH, Bethesda, MD), and Adrian Black (Department of Pharmacology and Therapeutics, Roswell Park Cancer Institute, Buffalo, NY) for kindly providing the dominant negative mutant PKCζ, dominant negative mutant PI3Kp110α, and Flag-Sp1 construct, respectively.
Our research is supported by the Intramural Research Program of the National Institute of Child Health and Human Development of the National Institutes of Health.
REFERENCES
- 1.Armstrong, S. A., D. A. Barry, R. W. Leggett, and C. R. Mueller. 1997. Casein kinase II-mediated phosphorylation of the C terminus of Sp1 decreases its DNA binding activity. J. Biol. Chem. 272:13489-13495. [DOI] [PubMed] [Google Scholar]
- 2.Bandyopadhyay, G., M. L. Standaert, M. P. Sajan, L. M. Karnitz, L. Cong, M. J. Quon, and R. V. Farese. 1999. Dependence of insulin-stimulated glucose transporter 4 translocation on 3-phosphoinositide-dependent protein kinase-1 and its target threonine-410 in the activation loop of protein kinase C-ζ. Mol. Endocrinol. 13:1766-1772. [DOI] [PubMed] [Google Scholar]
- 3.Batsche, E., J. Desroches, S. Bilodeau, Y. Gauthier, and J. Drouin. 2005. Rb enhances p160/SRC coactivator-dependent activity of nuclear receptors and hormone responsiveness. J. Biol. Chem. 280:19746-19756. [DOI] [PubMed] [Google Scholar]
- 4.Benasciutti, E., G. Pages, O. Kenzior, W. Folk, F. Blasi, and M. P. Crippa. 2004. MAPK and JNK transduction pathways can phosphorylate Sp1 to activate the uPA minimal promoter element and endogenous gene transcription. Blood 104:256-262. [DOI] [PubMed] [Google Scholar]
- 5.Black, A. R., J. D. Black, and J. Azizkhan-Clifford. 2001. Sp1 and Krüppel-like factor family of transcription factors in cell growth regulation and cancer. J. Cell. Physiol. 188:143-160. [DOI] [PubMed] [Google Scholar]
- 6.Black, A. R., D. Jensen, S. Y. Lin, and J. C. Azizkhan. 1999. Growth/cell cycle regulation of Sp1 phosphorylation. J. Biol. Chem. 274:1207-1215. [DOI] [PubMed] [Google Scholar]
- 7.Bonello, M. R., and L. M. Khachigian. 2004. Fibroblast growth factor-2 represses platelet-derived growth factor receptor-α (PDGFR-α) transcription via ERK1/2-dependent Sp1 phosphorylation and an atypical cis-acting element in the proximal PDGFR-α promoter. J. Biol. Chem. 279:2377-2382. [DOI] [PubMed] [Google Scholar]
- 8.Camarero, N., A. Nadal, M. J. Barrero, D. Haro, and P. F. Marrero. 2003. Histone deacetylase inhibitors stimulate mitochondrial HMG-CoA synthase gene expression via a promoter proximal Sp1 site. Nucleic Acids Res. 31:1693-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang, Y. C., S. Illenye, and N. H. Heintz. 2001. Cooperation of E2F-p130 and Sp1-pRb complexes in repression of the Chinese hamster dhfr gene. Mol. Cell. Biol. 21:1121-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, C. R., Y. Kang, P. M. Siegel, and J. Massague. 2002. E2F4/5 and p107 as Smad cofactors linking the TGFβ receptor to c-myc repression. Cell 110:19-32. [DOI] [PubMed] [Google Scholar]
- 11.Chen, C. S., S.-C. Weng, P.-H. Tseng, H.-P. Lin, and C.-S. Chen. 2005. Histone acetylation-independent effect of histone deacetylase inhibitors on Akt through the reshuffling of protein phosphatase 1 complexes. J. Biol. Chem. 280:38879-38887. [DOI] [PubMed] [Google Scholar]
- 12.Choi, H. S., J. H. Lee, J. G. Park, and Y. I. Lee. 2002. Trichostatin A, a histone deacetylase inhibitor, activates the IGFBP-3 promoter by upregulating Sp1 activity in hepatoma cells: alteration of the Sp1/Sp3/HDAC1 multiprotein complex. Biochem. Biophys. Res. Commun. 296:1005-1012. [DOI] [PubMed] [Google Scholar]
- 13.Chou, M. M., W. Hou, J. Johnson, L. K. Graham, M. H. Lee, C. S. Chen, A. C. Newton, B. S. Schaffhausen, and A. Toker. 1998. Regulation of protein kinase C zeta by PI 3-kinase and PDK-1. Curr. Biol. 8:1069-1077. [DOI] [PubMed] [Google Scholar]
- 14.Chu, S., and T. J. Ferro. 2005. Sp1: regulation of gene expression by phosphorylation. Gene 348:1-11. [DOI] [PubMed] [Google Scholar]
- 15.Chun, R. F., O. J. Semmes, C. Neuveut, and K. T. Jeang. 1998. Modulation of Sp1 phosphorylation by human immunodeficiency virus type 1 Tat. J. Virol. 72:2615-2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chupreta, S., M. Du, A. Todisco, and J. L. Merchant. 2000. EGF stimulates gastrin promoter through activation of Sp1 kinase activity. Am. J. Physiol. Cell Physiol. 278:C697-C708. [DOI] [PubMed] [Google Scholar]
- 17.Classon, M., B. K. Kennedy, R. Mulloy, and E. Harlow. 2000. Opposing roles of pRB and p107 in adipocyte differentiation. Proc. Natl. Acad. Sci. USA 97:10826-10831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Datta, P., P. Raychaudhuri, and S. Bagchi. 1995. Association of p107 with Sp1: genetically separable regions of p107 are involved in regulation of E2F- and Sp1-dependent transcription. Mol. Cell. Biol. 15:5444-5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dempsey, E. C., A. C. Newton, D. Mochly-Rosen, A. P. Fields, M. E. Reyland, P. A. Insel, and R. O. Messing. 2000. Protein kinase C isozymes and the regulation of diverse cell responses. Am. J. Physiol. Lung Cell Mol. Physiol. 279:L429-L438. [DOI] [PubMed] [Google Scholar]
- 20.Doetzlhofer, A., H. Rotheneder, G. Lagger, M. Koranda, V. Kurtev, G. Brosch, E. Wintersberger, and C. Seiser. 1999. Histone deacetylase 1 can repress transcription by binding to Sp1. Mol. Cell. Biol. 19:5504-5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dufau, M. L. 1998. The luteinizing hormone receptor. Annu. Rev. Physiol. 60:461-496. [DOI] [PubMed] [Google Scholar]
- 22.Dyson, N. 1998. The regulation of E2F by pRB-family proteins. Genes Dev. 12:2245-2262. [DOI] [PubMed] [Google Scholar]
- 23.Espinos, E., and M. J. Weber. 1998. Activation of the MAP kinase cascade by histone deacetylase inhibitors is required for the stimulation of choline acetyltransferase gene promoter. Brain Res. Mol. Brain Res. 56:118-124. [DOI] [PubMed] [Google Scholar]
- 24.Frolov, M. V., and N. J. Dyson. 2004. Molecular mechanisms of E2F-dependent activation and pRB-mediated repression. J. Cell Sci. 117:2173-2181. [DOI] [PubMed] [Google Scholar]
- 25.Geng, Y., C. H. Tsai-Morris, Y. Zhang, and M. L. Dufau. 1999. The human luteinizing hormone receptor gene promoter: activation by Sp1 and Sp3 and inhibitory regulation. Biochem. Biophys. Res. Commun. 263:366-371. [DOI] [PubMed] [Google Scholar]
- 26.Glaser, K. B., M. J. Staver, J. F. Waring, J. Stender, R. G. Ulrich, and S. K. Davidsen. 2003. Gene expression profiling of multiple histone deacetylase (HDAC) inhibitors: defining a common gene set produced by HDAC inhibition in T24 and MDA carcinoma cell lines. Mol. Cancer Ther. 2:151-163. [PubMed] [Google Scholar]
- 27.Haidweger, E., M. Novy, and H. Rotheneder. 2001. Modulation of Sp1 activity by a cyclin A/CDK complex. J. Mol. Biol. 306:201-212. [DOI] [PubMed] [Google Scholar]
- 28.Han, J. W., S. H. Ahn, Y. K. Kim, G. U. Bae, J. W. Yoon, S. Hong, H. Y. Lee, Y. W. Lee, and H. W. Lee. 2001. Activation of p21(WAF1/Cip1) transcription through Sp1 sites by histone deacetylase inhibitor apicidin: involvement of protein kinase C. J. Biol. Chem. 276:42084-42090. [DOI] [PubMed] [Google Scholar]
- 29.Hu, Z. Z., L. Zhuang, J. Meng, and M. L. Dufau. 1998. Transcriptional regulation of the generic promoter III of the rat prolactin receptor gene by C/EBPβ and Sp1. J. Biol. Chem. 273:26225-26235. [DOI] [PubMed] [Google Scholar]
- 30.Jackson, S., T. Gottlieb, and K. Hartley. 1993. Phosphorylation of transcription factor Sp1 by the DNA-dependent protein kinase. Adv. Second Messenger Phosphoprot. Res. 28:279-286. [PubMed] [Google Scholar]
- 31.Kaczynski, J., T. Cook, and R. Urrutia. 2003. Sp1 and Krüppel-like transcription factors. Genome Biol. 4:206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim, Y. K., J. W. Han, Y. N. Woo, J. K. Chun, J. Y. Yoo, E. J. Cho, S. Hong, H. Y. Lee, Y. W. Lee, and H. W. Lee. 2003. Expression of p21 (WAF1/Cip1) through Sp1 sites by histone deacetylase inhibitor apicidin requires PI 3-kinase-PKC epsilon signaling pathway. Oncogene 22:6023-6031. [DOI] [PubMed] [Google Scholar]
- 33.Milanini-Mongiat, J., J. Pouyssegur, and G. Pages. 2002. Identification of two Sp1 phosphorylation sites for p42/p44 mitogen-activated protein kinases. Their implication in vascular endothelial growth factor gene transcription. J. Biol. Chem. 277:20631-20639. [DOI] [PubMed] [Google Scholar]
- 34.Naga Prasad, S. V., A. Jayatilleke, A. Madamanchi, and H. A. Rockman. 2005. Protein kinase activity of phosphoinositide 3-kinase regulates beta-adrenergic receptor endocytosis. Nat. Cell Biol. 7:785-796. [DOI] [PubMed] [Google Scholar]
- 35.Pal, S., K. P. Claffey, H. T. Cohen, and D. Mukhopadhyay. 1998. Activation of Sp1-mediated vascular permeability factor/vascular endothelial growth factor transcription requires specific interaction with protein kinase C zeta. J. Biol. Chem. 273:26277-26280. [DOI] [PubMed] [Google Scholar]
- 36.Puri, P. L., S. Iezzi, P. Stiegler, T. T. Chen, R. L. Schiltz, G. E. Muscat, A. Giordano, L. Kedes, J. Y. Wang, and V. Sartorelli. 2001. Class I histone deacetylases sequentially interact with MyoD and pRb during skeletal myogenesis. Mol. Cell 8:885-897. [DOI] [PubMed] [Google Scholar]
- 37.Rafty, L. A., and L. M. Khachigian. 2001. Sp1 phosphorylation regulates inducible expression of platelet-derived growth factor B-chain gene via atypical protein kinase C-ζ. Nucleic Acids Res. 29:1027-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reisinger, K., R. Kaufmann, and J. Gille. 2003. Increased Sp1 phosphorylation as a mechanism of hepatocyte growth factor (HGF/SF)-induced vascular endothelial growth factor (VEGF/VPF) transcription. J. Cell Sci. 116:225-238. [DOI] [PubMed] [Google Scholar]
- 39.Rivero, J. A., and S. E. Adunyah. 1998. Sodium butyrate stimulates PKC activation and induces differential expression of certain PKC isoforms during erythroid differentiation. Biochem. Biophys. Res. Commun. 248:664-668. [DOI] [PubMed] [Google Scholar]
- 40.Samson, S., and N. Wong. 2002. Role of Sp1 in insulin regulation of gene expression. J. Mol. Endocrinol. 29:265-279. [DOI] [PubMed] [Google Scholar]
- 41.Sanchez-Margalet, V., and S. Najib. 2001. Sam68 is a docking protein linking GAP and PI3K in insulin receptor signaling. Mol. Cell. Endocrinol. 183:113-121. [DOI] [PubMed] [Google Scholar]
- 42.Standaert, M. L., G. Bandyopadhyay, L. Perez, D. Price, L. Galloway, A. Poklepovic, M. P. Sajan, V. Cenni, A. Sirri, J. Moscat, A. Toker, and R. V. Farese. 1999. Insulin activates protein kinases C-zeta and C-lambda by an autophosphorylation-dependent mechanism and stimulates their translocation to GLUT4 vesicles and other membrane fractions in rat adipocytes. J. Biol. Chem. 274:25308-25316. [DOI] [PubMed] [Google Scholar]
- 43.Standaert, M. L., L. Galloway, P. Karnam, G. Bandyopadhyay, J. Moscat, and R. V. Farese. 1997. Protein kinase C-zeta as a downstream effector of phosphatidylinositol 3-kinase during insulin stimulation in rat adipocytes. Potential role in glucose transport. J. Biol. Chem. 272:30075-30082. [DOI] [PubMed] [Google Scholar]
- 44.Stark, M., and Y. G. Assaraf. 2006. Loss of Sp1 function via inhibitory phosphorylation in antifolate-resistant human leukemia cells with down-regulation of the reduced folate carrier. Blood 107:708-715. [DOI] [PubMed] [Google Scholar]
- 45.Toker, A. 2000. Protein kinases as mediators of phosphoinositide 3-kinase signaling. Mol. Pharmacol. 57:652-658. [PubMed] [Google Scholar]
- 46.Toker, A. 1998. Signaling through protein kinase C. Front. Biosci. 3:D1134-D1147. [DOI] [PubMed] [Google Scholar]
- 47.Tsai-Morris, C. H., Y. Geng, E. Buczko, and M. L. Dufau. 1998. A novel human luteinizing hormone receptor gene. J. Clin. Endocrinol. Metab. 83:288-291. [DOI] [PubMed] [Google Scholar]
- 48.Tsai-Morris, C. H., Y. Geng, X. Z. Xie, E. Buczko, and M. L. Dufau. 1994. Transcriptional protein binding domains governing basal expression of the rat luteinizing hormone receptor gene. J. Biol. Chem. 269:15868-15875. [PubMed] [Google Scholar]
- 49.Tsai-Morris, C. H., X. Xie, W. Wang, E. Buczko, and M. L. Dufau. 1993. Promoter and regulatory regions of the rat luteinizing hormone receptor gene. J. Biol. Chem. 268:4447-4452. [PubMed] [Google Scholar]
- 50.Udvadia, A., D. Templeton, and J. Horowitz. 1995. Functional interactions between the retinoblastoma (Rb) protein and Sp-family members: superactivation by Rb requires amino acids necessary for growth suppression. Proc. Natl. Acad. Sci. USA 92:3953-3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Walker, G. E., E. M. Wilson, D. Powell, and Y. Oh. 2001. Butyrate, a histone deacetylase inhibitor, activates the human IGF binding protein-3 promoter in breast cancer cells: molecular mechanism involves an Sp1/Sp3 multiprotein complex. Endocrinology 142:3817-3827. [DOI] [PubMed] [Google Scholar]
- 52.Witt, O., K. Sand, and A. Pekrun. 2000. Butyrate-induced erythroid differentiation of human K562 leukemia cells involves inhibition of ERK and activation of p38 MAP kinase pathways. Blood 95:2391-2396. [PubMed] [Google Scholar]
- 53.Yamabe, Y., A. Shimamoto, M. Goto, J. Yokota, M. Sugawara, and Y. Furuichi. 1998. Sp1-mediated transcription of the Werner helicase gene is modulated by Rb and p53. Mol. Cell. Biol. 18:6191-6200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang, J., Y. Kawai, R. W. Hanson, and I. J. Arinze. 2001. Sodium butyrate induces transcription from the Gαi2 gene promoter through multiple Sp1 sites in the promoter and by activating the MEK-ERK signal transduction pathway. J. Biol. Chem. 276:25742-25752. [DOI] [PubMed] [Google Scholar]
- 55.Yokota, T., Y. Matsuzaki, K. Miyazawa, F. Zindy, M. F. Roussel, and T. Sakai. 2004. Histone deacetylase inhibitors activate INK4d gene through Sp1 site in its promoter. Oncogene 23:5340-5349. [DOI] [PubMed] [Google Scholar]
- 56.Zhang, Y., and M. L. Dufau. 2001. EAR2 and EAR3/COUP-TFI regulate transcription of the rat LH receptor. Mol. Endocrinol. 15:1891-1905. [DOI] [PubMed] [Google Scholar]
- 57.Zhang, Y., and M. L. Dufau. 2000. Nuclear orphan receptors regulate transcription of the gene for the human luteinizing hormone receptor. J. Biol. Chem. 275:2763-2770. [DOI] [PubMed] [Google Scholar]
- 58.Zhang, Y., and M. L. Dufau. 2003. Repression of the luteinizing hormone receptor gene promoter by cross talk among EAR3/COUP-TFI, Sp1/Sp3, and TFIIB. Mol. Cell. Biol. 23:6958-6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang, Y., and M. L. Dufau. 2002. Silencing of transcription of the human luteinizing hormone receptor gene by histone deacetylase-mSin3A complex. J. Biol. Chem. 277:33431-33438. [DOI] [PubMed] [Google Scholar]
- 60.Zhang, Y., N. Fatima, and M. L. Dufau. 2005. Coordinated changes in DNA methylation and histone modifications regulate silencing/derepression of luteinizing hormone receptor gene transcription. Mol. Cell. Biol. 25:7929-7939. [DOI] [PMC free article] [PubMed] [Google Scholar]