Abstract
The human β-globin genes are expressed in a developmental stage-specific manner in erythroid cells. Gene-proximal cis-regulatory DNA elements and interacting proteins restrict the expression of the genes to the embryonic, fetal, or adult stage of erythropoiesis. In addition, the relative order of the genes with respect to the locus control region contributes to the temporal regulation of the genes. We have previously shown that transcription factors TFII-I and USF interact with the β-globin promoter in erythroid cells. Herein we demonstrate that reducing the activity of USF decreased β-globin gene expression, while diminishing TFII-I activity increased β-globin gene expression in erythroid cell lines. Furthermore, a reduction of USF activity resulted in a significant decrease in acetylated H3, RNA polymerase II, and cofactor recruitment to the locus control region and to the adult β-globin gene. The data suggest that TFII-I and USF regulate chromatin structure accessibility and recruitment of transcription complexes in the β-globin gene locus and play important roles in restricting β-globin gene expression to the adult stage of erythropoiesis.
The mammalian β-globin genes are expressed exclusively in erythroid cells and organized along the chromosome according to the timing of expression during development (5, 39). The embryonic genes are located at the 5′ end of the β-globin gene locus, and the adult genes are located at the 3′ end. Tissue- and developmental stage-specific expression of the genes is regulated by complex mechanisms involving gene-proximal and -distal DNA regulatory elements (40). High-level transcription of β-globin genes is mediated by the locus control region (LCR), a powerful regulatory DNA element located far upstream of the ɛ-globin gene (19, 15). The LCR is composed of several DNase I-hypersensitive sites (HS) that function together to regulate chromatin structure and globin gene expression (2, 7, 16, 28, 29, 33). The arrangement of the genes with respect to the LCR is important for correct developmental expression of the β-globin genes (11, 32, 34). Inversion of the genes relative to the LCR activates the adult β-globin gene in embryonic cells and represses ɛ-globin gene expression throughout development (42).
Recent models propose that macromolecular protein complexes involved in modulating chromatin structure and transcription of the β-globin genes are first recruited to the LCR (23, 43). These activities are used to establish stage-specific accessible chromatin domains, and transcription complexes are subsequently transferred from the LCR to the globin genes. Stage-specific modification of chromatin structure would provide a means to restrict the transfer of macromolecular complexes from the LCR to individual globin genes (18).
The activation of gene expression is controlled at different levels, including positioning genes to transcriptionally active domains in the nucleus, increasing chromatin accessibility, recruitment of transcription complexes, transcription elongation, and posttranscriptional mechanisms (21, 31). It is becoming increasingly clear that one role DNA binding proteins play is to recruit coactivators that modify histones or mobilize nucleosomes at regulatory sites. It is believed that the modification and mobilization of nucleosomes render DNA accessible for interactions with transcription complexes. Recent reports support the view that chromatin-remodeling activities act locally at promoter and perhaps enhancer regions to allow the subsequent assembly of transcription or enhanceosome complexes (27). These activities, which are often part of large protein complexes, must be recruited to the site of action by proteins that recognize specific DNA motifs. EKLF represents a well-described example of a protein that recruits chromatin-remodeling activities to regulatory sites in the β-globin gene locus. EKLF interacts with LCR elements HS2 and HS3, as well as with the adult β-globin gene, and recruits chromatin-remodeling activities (1, 3). The ablation of EKLF in mice leads to a reduction of DNase I HS formation in HS3 and in the β-globin gene promoter (17). EKLF is particularly important for β-globin gene expression and does not significantly affect the expression of other genes in the locus (17).
TFII-I and USF are ubiquitously expressed helix-loop-helix proteins that are involved in the transcriptional regulation of genes containing E-box or initiator sequences. USF1 and USF2 are highly related proteins that interact with DNA either as homo- or heterodimers (25). They often bind E-box motifs in the vicinity, sometimes downstream, of transcription start sites and have been shown to aid in the formation of transcription complexes (6, 12). Interestingly, a recent genomic survey revealed that many Drosophila genes harbor E-box elements located about 60 bp downstream of the transcription start site (20). Likewise, TFII-I has been implicated in the recruitment of transcription complexes to promoters containing an initiator sequence overlapping the transcription start site (38). USF and TFII-I can act as activators or repressors of transcription, depending on interacting proteins and the sequence context of the promoter region (26, 30, 36, 37, 46).
A recent report demonstrated that USF is a critical component of the boundary function of the chicken HS4 (cHS4) insulator (47). cHS4 establishes a boundary between open and closed chromatin in the chicken β-globin gene locus. It was shown that USF interacts with cHS4 in vivo and recruits histone-modifying activities that may be involved in separating inactive from active chromatin domains. Thus, part of the in vivo activating function of USF may be due to the recruitment of chromatin-modifying activities to DNA regulatory elements. USF has also been shown to interact with a conserved E-box located in LCR element HS2 (4, 14). A mutation of this E-box impairs the enhancer function of HS2.
We have previously shown that TFII-I and USF interact with the core promoter of the β-globin gene in vitro and in vivo (22). The interaction of TFII-I was more pronounced in erythroid cells in which the β-globin gene is repressed. Here we demonstrate that reducing TFII-I activity by RNA interference (RNAi) or the expression of a dominant-negative mutant leads to the derepression of the β-globin gene in erythroid cells. Moreover, we show that TFII-I is complexed with histone deacetylase 3 (HDAC3) and that both proteins together interact with the β-globin core promoter. A reduction of USF activity by expression of a dominant-negative protein causes a decrease of RNA polymerase (Pol) II recruitment to the globin gene locus and the repression of β-globin gene transcription. The results support the hypothesis that antagonistic activities exerted by TFII-I and USF during embryonic and adult erythroid development contribute to the stage-specific expression of the β-globin gene.
MATERIALS AND METHODS
Construction of protein expression vectors.
The plasmid pCMV/A-USF was obtained from Charles Vinson (NIH) and has been described previously (35). The A-USF open reading frame, including an N-terminal hemagglutinin (HA) tag, was amplified by PCR. The reaction was carried out using Elongase (Invitrogen), 0.4 mM forward primer (5′-GCGCCTTAAGCTCCACCATGGCGTATCC-3′) containing an AflII restriction site (underlined), 0.4 mM reverse primer (5′-GCGCTCTAGAAGAAGCTTTTAGTTGCTGTCATTC-3′) containing a XbaI restriction site (underlined), 1.5 mM Mg2+, and 5 ng pCMV/A-USF as a template in a final volume of 50 μl. PCR conditions were as follows: 94°C for 30 s, 35 cycles of 94°C for 30 s, 61°C for 30 s, and 68°C for 30 s, with a final extension at 68°C for 2 min. The amplified product and pcDNA4/TO (Invitrogen) were digested with AflII and XbaI and subsequently ligated to create pTO/A-USF.
pET11d/USF1 was digested with BamHI and XbaI to isolate the USF1 open reading frame containing an N-terminal Flag tag. The fragment was subcloned into pcDNA4/TO and then properly oriented by digestion with PmeI and ligation back into pcDNA4/TO to create pTO/USF1.
pET11d/p70 and pET11d/TFII-I were digested with NcoI and SpeI to isolate the open reading frames. The fragments were subcloned into the pLitmus 29 vector (New England Biolabs). A subsequent digest with BamHI and SpeI isolated the open reading frames, which were subcloned into the pcDNA 3.1(+) vector (Invitrogen). The open reading frames were properly oriented by digestion with PmeI and ligation back into pcDNA 3.1(+). The open reading frames were isolated from pcDNA 3.1(+) by digestion with AflII and XbaI and subcloned into pcDNA4/TO, thus creating pTO/p70 and pTO/TFII-I. pTO/HA-p70, expressing an HA-tagged p70 mutant and containing a neomycin resistance gene, was created by amplifying the p70 open reading frame by PCR using pTO/p70 as a template and the forward primer containing a BamHI site (5′-GCGCGGATCCCATGGCCCAAGTTGCAATGTCC-3′) and the reverse primer containing an XbaI site (5′-GCGCTCTAGAGTTCAGGTTTTTTAACAACGAAC-3′). The PCR product and pTO/A-USF were digested with BamHI and XbaI and subsequently ligated, creating pcDNA4/TO/HA-p70. This construct was digested with XbaI and BstZ171 to remove the Zeocin resistance gene. pCMV-566 (obtained from Charles Vinson) was digested with PciI, filled in using a Klenow reaction, and then digested with XbaI to isolate a fragment containing the neomycin resistance gene. These two fragments were then ligated to create pTO/HA-p70. pTO, the empty control vector, was created by digesting pcDNA4/TO with XbaI and BstZ171. pCMV-566 was digested with PciI, filled in using a Klenow reaction, and digested with XbaI. The fragments were then ligated.
The inserts of all generated plasmids were sequenced to verify that no mutations were introduced and ensure that coding sequences were in the correct reading frames.
Cell culture and transfections.
The K562 cells used in this study were obtained from Ann Dean (NIH) and grown in RPMI 1640 medium supplemented with 15% fetal bovine serum and 5% penicillin-streptomycin. Murine erythroleukemia (MEL) cells were grown in RPMI containing 10% fetal bovine serum and 5% antibiotic-antimycotic. Cells were grown in 5% CO2 at 37°C and maintained at a density between 1 × 105 cells/ml and 5 × 105 cells/ml.
Stable MEL cell lines were created following the protocol for FuGENE 6 transfection reagent (Roche Molecular Biochemicals). Stable pTO/A-USF-pcDNA6/TR (Invitrogen) and pTO-pcDNA6/TR MEL cell lines were created by cotransfecting pTO/A-USF/TO/A-USF or pTO (control vector) with pcDNA6/TR. Stable pTO/p70-pcDNA6/TR, pTO/TFII-I-pcDNA6/TR, and pTO-pcDNA6/TR (control vector) MEL cell lines were created by transfecting stable MEL pcDNA6/TR cells (previously transfected with 3 μg of pcDNA6/TR) with 1 μg of pTO/p70, pTO/TFII-I, or pTO. Stable pTO/USF1-pcDNA6/TR and pTO-pcDNA6/TR (control vector) MEL cell lines were created by transfecting MEL pcDNA6/TR cells with either pTO/USF1 or pTO. Cells were allowed to recover for 48 h before selection with 5 μg/ml blasticidin and 300 μg/ml Zeocin.
For K562 transfections, 106 cells were resuspended in 100 μl of cell line Nucleofector solution V (Amaxa GmbH) and nucleofected with 1 μg of pTO/HA-p70/neo or pTO and 3 μg of pcDNA6/TR, using the protocol for K562 cells. Cells were allowed to recover for 3 days before selection with 4 μg/ml blasticidin and 300 μg/ml G418.
To induce protein expression, stable MEL and K562 cell lines were treated with 1 μg/ml doxycycline (MEL cells for 36 h and K562 cells for 12 h) at a cell concentration of approximately 2 × 105 cells/ml.
Murine embryonic stem cells were cultured and differentiated into erythroid cells as previously described by Levings et al. (24).
Chromatin immunoprecipitation, double-chromatin immunoprecipitation, and coimmunoprecipitation.
Chromatin immunoprecipitation (ChIP) was performed as previously described by Leach et al. (22). Double ChIP was carried out following the same ChIP procedure with the addition of dialysis of the elutant from the first immunoprecipitation against 200 ml of dilution buffer with complete mini-protease inhibitors (Roche) using Slide-A-Lyse mini-dialysis units (Pierce) at 4°C with constant stirring for 2 h. Samples were removed from dialysis units, and 500 μl dilution buffer was added to each. The second immunoprecipitation was performed, again following the previously referenced ChIP protocol.
The immunoprecipitation of TFII-I from K562 cells was carried out following the ChIP protocol as previously described by Leach et al. (22), with the addition of protease inhibitors in all buffers for coimmunoprecipitation assay. Immunoprecipitation of USF1 was performed using an extract from 1 × 108 K562 or MEL cells lysed with 1 ml of NP-40 buffer (150 mM NaCl, 1% NP-40, 50 mM Tris, pH 8.0). Extract was precleared with 100 μl of anti-rabbit immunoglobulin G (IgG) beads (eBioscience) for 30 min. Precipitation was carried out by incubating extract with 10 μg of antibody for 2.5 h. Complexes were captured by incubating extracts with 100 μl of anti-rabbit IgG beads for 2 h. All incubations were performed on a spinning wheel at 4°C. Samples were washed three times with NP-40 buffer and eluted with Laemmli buffer (Bio-Rad) at 95°C for 10 min. The eluted samples were loaded onto an either 7.5% or 10% Ready gel (Bio-Rad) and analyzed by Western blotting.
Antibodies used in the experiments are as follows: acetyl-histone H3 06-599 and RNA polymerase II clone CTD4H8 05-623 (both purchased from Upstate Biotechnology, Inc.); USF1 (M-86) sc-8983, USF2 (N-18) sc-861, and p300 (N-15) sc-584 (all purchased from Santa Cruz Biotechnology); IgG C6409 (anti-chicken Iga; Sigma); anti-TFII-I (a gift from R. G. Roeder, Rockefeller University); and anti-HDAC3 (a gift from E. Seto, University of South Florida).
RNA interference.
siGENOME SMARTpool reagents for human TFII-I (catalog no. M-013638-0), HDAC3 (catalog no. M-003496-00), USF2 (catalog no. M-003618-00), siCONTROL nontargeting pool (catalog no. D-001206-13-05), and siGLO RISC-free (catalog no. D-00160-01-05) were obtained from Dharmacon. A total of 106 K562 cells were nucleofected with 0.5 μg of short interfering RNA (siRNA) or mock transfected following the protocol for K562 cells (Amaxa GmbH). RNA and protein from cells were collected after 48, 72, and 96 h. Transfection efficiency was determined by first fixing and then DAPI (4′,6′-diamidino-2-phenylindole) staining K562 siGLO cells. Pictures were taken using an Axioplan 2 microscope (Zeiss) under 40× power.
Protein isolation and Western blotting.
Western blotting experiments were performed essentially as previously described by Leach et al. (22). A total of 20 μg of protein, unless otherwise noted, was run on either a 7.5% or a 10% Ready gel (Bio-Rad). The detection of proteins on membranes was performed with an ECL Plus system according to the manufacturer's instructions (Amersham Pharmacia Biotech). The primary antibodies used were the same as those used for ChIP, in addition to anti-HA tag (a gift from Michael Kilberg, University of Florida) and anti-TFII-I (a gift from Ananda Roy, Tufts University). The secondary antibodies used were goat anti-rabbit IgG-horseradish peroxidase (Kirkegaard & Perry Laboratories and Santa Cruz Biotechnology; sc-2004), goat anti-mouse IgG-horseradish peroxidase sc-2005 (Santa Cruz Biotechnology), and anti-rabbit IgG TrueBlot (eBiosciences) for USF1 coimmunoprecipitation. The concentration of antibodies used followed the manufacturers' guidelines.
RNA isolation, real-time PCR, and PCR.
RNA was isolated using the guanidine thiocyanate method (10) and reverse transcribed as described previously (7). RNA for RNAi experiments was isolated using the Aurum total RNA kit (Bio-Rad) and reverse transcribed as described above. Quantitative PCR (qPCR) was performed using the Opticon 2 (MJ Research). The reactions were carried out using either DyNAmo HS SYBR green master mix (MJ Research) or iQ SYBR green super mix (Bio-Rad Laboratories). Real-time PCR conditions were as follows: 95°C for 15 min, 40 cycles of 94°C for 10 s, 59°C for 20 s, and 72°C for 30 s (the plate was read after each extension step), final extension at 72°C for 5 min, a melting curve from 55°C to 95°C (reading every 0.5°C), and reannealing at 72°C for 5 min. The 20-μl reaction mixtures contained 20 ng of cDNA from dominant-negative and overexpression assays, a 1:40 dilution of cDNA from RNAi assays, or 2 μl of DNA from ChIP assays, 0.3 μM of forward and reverse primer, and 10 μl of the 2× SYBR green master mix. All primers for expression analysis span introns to ensure exclusive amplification of cDNA. Amplicons ranged in size from 110 to 120 bp. All reactions were carried out in duplicate with a no-template control. Standard curves were generated using 10-fold serial dilutions of cDNA or input DNA for ChIP assay from the appropriate cell line. Final quantification analysis was performed using the relative standard curve method, and for reverse transcription (RT)-qPCR, results were reported as the relative expression of the control after normalization of the transcript to the endogenous control, which was either β-actin (data not shown) or GAPDH.
PCR for semiquantitative ChIP samples was performed using the following conditions: 2 μl DNA, 1 μM each forward and reverse primer, and 8 μl of 2.5× master mix (Eppendorf) in a 20-μl reaction mixture. PCR cycle parameters were as follows: 95°C for 5 min, 30 cycles of 95°C for 95 s, 60°C for 95 s, and 72°C for 95 s, and a final extension at 72°C for 2 min.
Real-time RT primers used were as follows: mouse βmaj-globin, upstream (US) 5′-CTGTGGGGAAAGGTGAACTC-3′ and downstream (DS) 5′-GCAGAGGCAGAGGATAGGTC-3′; mouse β-actin, US, 5′-ACTGCTCTGGCTCCTAGCAC-3′, and DS, 5′-ACATCTGCTGGAAGGTGGAC-3′; mouse GAPDH, US, 5′-CCAAGGTCATCCATGACAACT-3′, and DS, 5′-ATCACGCCACAGCTTTCC-3′; human β-globin, US, 5′-GCACGTGGATCCTGAGAACT-3′, and DS, 5′-GCCACCACTTTCTGA TAGGC-3′; human ɛ-globin, US, 5′-CTGAGTGAGCTGCACTGTGA-3′, and DS, 5′-TGCACTTCAGGGGTGAACTC-3′; human β-actin, US, 5′-CCAACCGCGAGAAG ATGAC-3′, and DS, 5′-ACGATGCCAGTGGTACGG-3′; and human GAPDH US, 5′-GAAGGTGAAGGTCGGAGTCA-3′, and DS, 5′-GAGGTCAATGAAGGGGTCAT-3′. Primers for ChIP and double-ChIP samples used were as follows: mouse βmaj-globin promoter, US, 5′-AAGCCTGATTCCGTAGAGCCACAC-3′, and DS, 5′-CCCACAGGCAAGAGACAGCAGC-3′; mouse HS2, US, 5′-TGCAGTACCACTG TCCAAGG-3′, and DS, 5′-ATCTGGCCACACACCCTAAG-3′; mouse GAPDH promoter, US, 5′-GATGATGGAGGACGTGATGG-3′, and DS, 5′-GGCTGCAGGAGAAGAAAATG-3′; human ɛ-globin, US, 5′-GCTCCTT TATATGAGGCTTTCTTGG-3′, and DS, 5′-AATGCACCATGATGCCAGG-3′; human β-globin, US, 5′-TATCTTAGAG GGAGGGCTGAGGGTTTG-3′, and DS, 5′-CCAACTTCATCCACGTTCACCTTGC-3′ and human HS2/-3 linker, US, 5′-TGGGGACTCG AAAATCAAAG-3′, and DS, 5′-AGTAAGAAGCAAGGGCCACA-3′. Primers used for pTO/TFII-I RT-PCR were as follows: US, 5′-CCCGGAGTTCTTGTATGTGG-3′, and DS (bovine growth hormone), 5′-TAGAAGGCACAGTCGAGG-3′.
P values were determined using the Microsoft Excel t test function. Data with a P value smaller than 0.05 are considered statistically significant.
RESULTS
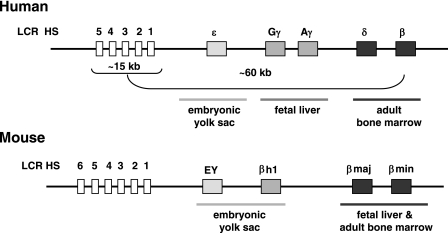
We previously demonstrated that the helix-loop-helix proteins USF and TFII-I interact with sequences located at or downstream of the transcription start site of the adult β-globin gene (22). The interaction of TFII-I was more pronounced in cells not expressing the β-globin gene, suggesting that it may be involved in repressing β-globin gene expression. In contrast, USF1 and USF2 efficiently interact with the β-globin gene in cells that express the gene. In this study, we examined the consequence of reducing or increasing the activities of TFII-I and USF on β-globin gene expression in human K562 erythroleukemia cells, in which the β-globin gene is repressed, and in MEL cells, which express the adult β-globin gene. Figure 1 shows an outline of the organization and expression of the β-globin genes in human and mouse loci.
FIG. 1.
Schematic of the organizational structure of the human and murine β-globin gene loci. The human β-globin locus depicted on top consists of five genes which are expressed in a developmental stage-specific manner in erythroid cells as outlined. The expression of the genes is regulated by a locus control region composed of five DNase I HS and located about 15 to 27 kbp upstream of the embryonic ɛ-globin gene. The murine β-globin gene locus, which is depicted on the bottom, consists of four genes which are expressed either in erythroid cells of the embryonic yolk sac (EY or βH1) or in definitive erythroid cells derived from fetal liver or bone marrow hematopoiesis (βmaj and βmin). The murine LCR also contains multiple HS required for high-level globin gene expression.
Repression of β-globin gene transcription by TFII-I and HDAC3.
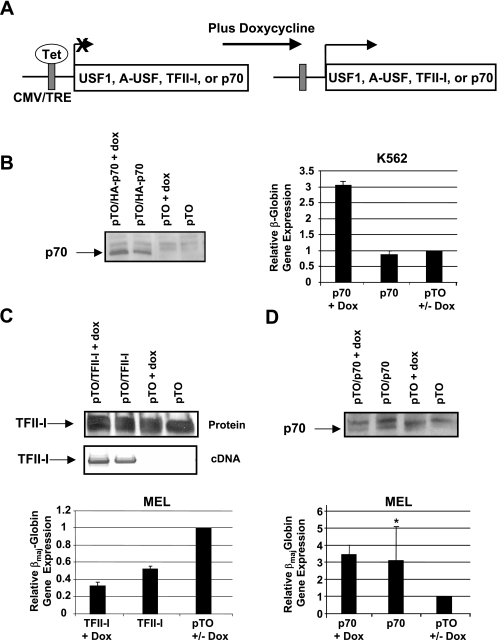
In this study, we overexpressed wild-type or mutant TFII-I and USF1 proteins in erythroid cell lines. The effect of expressing these proteins on β-globin gene expression was monitored by quantitative RT-PCR. We related β-globin gene expression to the expression of two internal control genes, β-actin (data not shown) and GAPDH, and obtained similar results. We first used a dominant-negative mutant of TFII-I (p70) that lacks the C-terminal acidic activation domain as described previously (9). p70 forms dimers with TFII-I but does not allow for interactions with coregulators. The sequence encoding full-length wild-type TFII-I or p70 was cloned into an expression vector and placed under the control of the cytomegalovirus (CMV) promoter and the Tet operator. We generated stable K562 or MEL cell lines with a Tet repressor expression construct. These cells were then stably transfected with constructs expressing either wild-type TFII-I or the p70 mutant. The expression of these proteins was induced by incubating the cells with doxycycline (Fig. 2A). The expression of TFII-I and p70 was confirmed by RT-PCR and/or Western blotting (Fig. 2B to D). Overexpression of TFII-I was difficult to assess from the Western blotting experiment, but the message was detected by RT-PCR using primers specific for TFII-I expressed from the transfected plasmid (Fig. 2C). The expression of p70 in K562 cells led to a threefold increase in β-globin gene expression. The p70 mutant protein is expressed in transfected cells even in the absence of doxycycline, demonstrating that there is leaky expression from the CMV/tetracycline-responsive element promoter. Overexpression of TFII-I in MEL cells decreased βmaj-globin gene expression by about 70%, whereas the expression of the dominant-negative mutant (p70) increased βmaj-globin gene expression three- to fourfold (Fig. 2D). We observed higher expression of p70 and TFII-I in the absence of doxycycline in MEL cells relative to p70 expression in K562 cells; however, the expression of the proteins in the untreated MEL cells had effects on globin gene expression similar to those in Dox-treated cells. The MEL cells used in these experiments express the βmaj-globin gene in the absence of dimethyl sulfoxide (DMSO) induction. Incubating the cells in the presence of 3% DMSO leads to a 5- to 10-fold increase in βmaj-globin gene expression (data not shown). It is interesting that the DMSO-mediated increase is similar to that with the up-regulation of βmaj-globin gene expression by p70. Taken together, the data demonstrate that TFII-I functions as a repressor of β-globin gene expression in both MEL and human K562 cells.
FIG. 2.
Repression of β-globin gene expression by TFII-I. (A) Diagram outlining the experimental system for expressing wild-type or dominant-negative proteins. Coding sequences for TFII-I, p70, USF1, and A-USF are under the control of the CMV enhancer/promoter. A Tet operator sequence interacts with the Tet repressor, for which an expression construct is cotransfected into these cells. Doxycycline interacts with the Tet repressor and relieves repression. TRE, tetracycline-responsive element. (B) Analysis of TFII-I function in K562 cells. The expression of dominant-negative mutant p70 was induced in stably transfected K562 cells by using 1 μg doxycycline per ml culture medium for 12 h. The expression of p70 was analyzed by Western blotting (left panel). RNA was isolated from transfected cell lines, reverse transcribed, and analyzed by quantitative PCR for expression of the β-globin gene. β-Globin gene expression is presented as the ratio of expression in pTO/p70 cells using GAPDH as the internal reference, relative to expression in cells transfected with the pTO vector, which was set at 1. +, doxycycline-treated cells; +/−, pTO+Dox cells served as control for doxycycline-treated cells, and pTO-Dox served as control for untreated cells. (C and D) Analysis of TFII-I function in MEL cells. The expression of wild-type TFII-I (C) or the dominant-negative mutant p70 (D) was induced in stably transfected MEL cells lines by using 1 μg doxycycline per ml culture medium for 36 h. The expression of TFII-I or p70 was analyzed by Western blotting and/or RT-PCR analysis as indicated. For RT-PCR analysis of TFII-I expression, RNA was isolated from induced MEL cells harboring expression constructs for TFII-I or the empty vector pTO and converted to cDNA. The cDNA was analyzed by PCR with forward primers corresponding to the 3′ region of the p70 and TFII-I-coding region and the reverse bovine growth hormone primer hybridizing to the pTO vector. For gene expression analysis, RNA was isolated from transfected cell lines, reverse transcribed, and analyzed by quantitative PCR for the expression of the βmaj-globin gene. βmaj-Globin expression is presented as the ratio of expression in pTO/p70 or pTO/TFII-I cells and expression in pTO cells using GAPDH as the internal reference. Doxycycline-treated pTO MEL cells were used as controls for all studies in which protein expression was induced by doxycycline. Data marked with an asterisk have a P value above 0.05. Error bars indicate standard deviations.
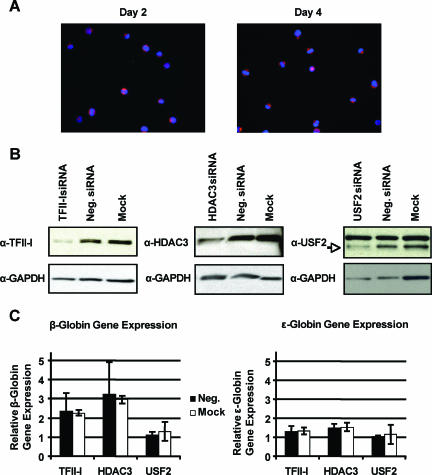
Previous studies have shown that TFII-I interacts with histone deacetylase 3 (46). We reduced the expression of TFII-I, HDAC3, and USF2 by RNA interference in K562 cells (Fig. 3B and C). We transfected K562 cells with siGENEOME SMARTpool reagents (Dharmacon) using a Nucleofector and observed close to 100% transfection efficiency in these cells (Fig. 3A). Western blotting experiments revealed specific reductions in the expression of TFII-I, HDAC3, and USF2 compared to that in cells transfected with a nontargeting siRNA or mock-transfected cells (Fig. 3B). Reducing the activity of either TFII-I or HDAC3 resulted in a significant increase in β-globin gene transcription (Fig. 3C). The increase of β-globin gene expression in cells transfected with TFII-I-directed siRNA was about 2.5-fold. Cells transfected with siRNA directed against HDAC3 revealed a threefold increase in β-globin gene expression in these cells. The expression of ɛ-globin was not affected, demonstrating that TFII-I and HDAC3 specifically act on the β-globin gene. A reduction of USF2 expression had no effect on the expression of either the ɛ-globin or the β-globin gene in K562 cells.
FIG. 3.
TFII-I and HDAC3 are repressors of β-globin gene expression in K562 cells. (A) To determine transfection efficiency, K562 cells were nucleofected with 0.5 μg of siGLO RISC-free. Cells were collected on days 2 and 4, fixed, and stained with DAPI for nuclear visualization; siGLO is seen in red. (B) Knockdown of TFII-I, HDAC3, and USF2 proteins in K562 cells. K562 cells were nucleofected with 0.5 μg of siGENOME SMARTpool targeting either TFII-I, HDAC3, USF2, or 0.5 μg of siCONTROL nontargeting pool (Neg.) or mock transfected. A total of 20 μg of protein was run on gels from cells collected on day 2 for TFII-I and HDAC3 Western blots, and 10 μg of protein was loaded from cells collected on day 3 for the USF2 Western blot. Blots were probed with anti (α)-TFII-I (upper left panel), α-HDAC3 (upper middle panel), and α-USF2 (upper right panel). Blots were stripped and reprobed with α-GAPDH for loading control (bottom panels). (C) Relative β- and ɛ-globin expression of TFII-I, HDAC3, or USF2 knockdown K562 cells. On day 4, RNA was collected from K562 cells transfected as described above, reverse transcribed, and analyzed by real-time PCR. Expression is set relative to either nontargeting siRNA (Neg.) or mock-transfected cells, with GAPDH as the internal reference. Error bars indicate standard deviations.
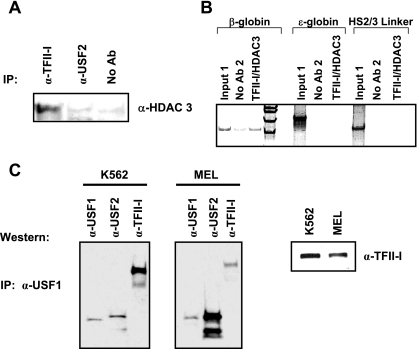
We next analyzed interactions between HDAC3 and TFII-I in K562 cells by coimmunoprecipitation (Fig. 4A). The data show that TFII-I but not USF2 interacts with HDAC3. We examined whether TFII-I and HDAC3 simultaneously interact with the β-globin gene promoter in K562 cells. Using double-chromatin immunoprecipitation, we first precipitated cross-linked and sheared chromatin with TFII-I antibodies. The precipitate was subsequently immunoprecipitated with HDAC3 antibodies. The DNA purified from this precipitate was analyzed by PCR for the presence of the β-globin gene promoter. The results show that HDAC3 interacts with the β-globin promoter in TFII-I-selected chromatin fragments (Fig. 4B). We did not detect interactions of TFII-I or HDAC3 with the embryonic ɛ-globin gene promoter or with a region between LCR HS2 and -3 (Fig. 4B). Together, the data suggest that TFII-I recruits HDAC3 and possibly other coregulators to the β-globin gene to repress transcription. Previous studies have shown that TFII-I interacts with USF. We analyzed protein-protein interactions involving USF1 by coimmunoprecipitation in K562 and MEL cells (Fig. 4C). The results demonstrate that USF1 and USF2 interact efficiently in MEL cells and less efficiently in K562 cells. We observed the same results in experiments using USF2 antibodies for immunoprecipitation and USF1 antibodies for Western blotting (data not shown). USF1 interacts strongly with TFII-I in K562 cells but not as strongly in MEL cells. TFII-I, USF1, and USF2 are expressed at similar levels in K562 and MEL cells as previously shown by Leach et al. (22).
FIG. 4.
Characterization of TFII-I- and USF-interacting proteins in K562 and MEL cells. (A) Coimmunoprecipitation experiment demonstrating interactions between TFII-I and HDAC3. K562 cells were lysed and subjected to immunoprecipitation (IP) with antibodies directed at either TFII-I or USF2. The precipitate was loaded onto a 7.5% denaturing polyacrylamide gel, and the gel was subjected to Western blotting with antibodies against HDAC3. The lane labeled no Ab represents the no-antibody control. (B) Double-ChIP experiment demonstrating simultaneous associations of TFII-I and HDAC3 with the β-globin gene promoter in K562 cells. K562 cells were subjected to ChIP using antibodies against TFII-I. The precipitate was subsequently immunoprecipitated with antibodies against HDAC3. The DNA in the second precipitate was purified and analyzed by PCR using primers specific for the human ɛ- or β-globin genes or for a region between LCR HS2 and -3. The lanes labeled no Ab2 represent samples from the no-antibody control experiments. (C) Coimmunoprecipitation of USF1, USF2, and TFII-I in K562 and MEL cells. K562 or MEL cell extract was precleared with anti (α)-rabbit IgG beads and precipitated with α-USF1, and complexes were captured by incubation with anti-rabbit IgG beads. Complexes were eluted off the beads with Laemmli buffer and incubation at 95°C for 10 min and loaded onto 10% Ready gels (Bio-Rad). After transfer, the membrane was cut into strips to probe with antibodies against either USF1, USF2, or TFII-I. The strips were then reassembled for phosphorimaging. The right panel is total extract from K562 or MEL cells run on a gel and subjected to Western blotting using an antibody against TFII-I. IP, immunoprecipitation.
The data strongly suggest that a protein complex containing the DNA binding protein TFII-I, the coregulator HDAC3, and possibly USF1 interacts with the β-globin gene promoter in embryonic erythroid cells and represses transcription by rendering the chromatin structure inaccessible for the transcription complex.
Activation of β-globin gene expression by USF.
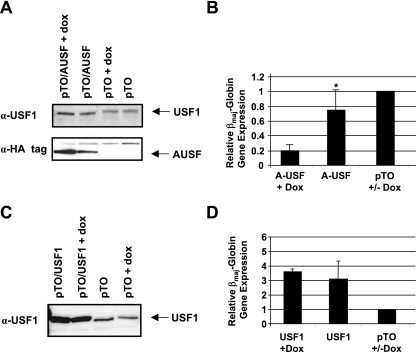
Many studies have shown that USF proteins can function as either activators or repressors of transcription. We previously demonstrated that USF1 and USF2 interact prominently with the adult β-globin gene promoter in MEL cells (22). In addition, in vitro transcription of the β-globin gene was impaired in promoter constructs containing mutations in the USF binding site. To analyze the role of USF in a cellular context, we expressed a dominant-negative mutant of USF, A-USF, in MEL cells. The expression construct was kindly provided by Charles Vinson (NIH). A-USF lacks the basic DNA binding region, which has been replaced by an acidic domain, and represses both USF1 and USF2 activity (35). We generated an A-USF expression construct in which the coding region is under control of the CMV promoter and the Tet operator. This construct was cotransfected into MEL cells with a Tet repressor expression plasmid. A-USF was expressed as an HA-tagged protein, and the induction of A-USF expression by doxycycline was monitored by Western blotting experiments using antibodies against the HA tag (Fig. 5A). In these experiments, we again observed some expression of A-USF in the absence of doxycycline, demonstrating that the Tet repressor does not effectively repress the expression of A-USF. However, after the induction with doxycycline, we detected a higher level of expression of A-USF. The expression of A-USF led to a 5-fold decrease in βmaj-globin gene expression in induced MEL cells transfected with pTO/A-USF and only a 1.5-fold decrease in uninduced cells transfected with pTO/A-USF (Fig. 5B). We also overexpressed USF1 in MEL cells and found that it increased βmaj-globin gene expression three- to fourfold (Fig. 5C and D). The inducible system again revealed some leakiness, as we detected a significant increase in USF1 expression in both the presence and the absence of doxycycline (Fig. 5C). Nevertheless, the combined data demonstrate that USF is an activator of β-globin gene expression in MEL cells.
FIG. 5.
USF activates β-globin gene expression in MEL cells. Stable MEL cell lines were created containing either A-USF (pTO/A-USF) or USF1 (pTO/USF1) and the Tet repressor (pTR). The expression of A-USF and USF1 was induced by incubating the cells with doxycycline and analyzed by Western-blotting experiments (A and C). βmaj-Globin gene expression in transfected cell lines was analyzed by quantitative RT-PCR, using the expression of GAPDH as an internal reference, and compared to the expression in cells transfected with the empty vector (pTO), whose expression level was set to 1. (A and B) Western blot analysis of A-USF expression (A) and analysis of β-globin gene expression (B) in MEL cells transfected with pTO/A-USF or pTO in the presence or absence of doxycycline as indicated. A-USF was detected using an antibody against the HA tag (α-HA), and USF1 was detected with a USF1-specific antibody. Error bars indicate standard deviations. (C and D) Western blot analysis of USF1 expression (C) and analysis of βmaj-globin gene expression (D) in MEL cells transfected with pTO/USF1 or pTO. USF1 was detected with a USF1-specific antibody. Data marked with an asterisk had P values above 0.05. + and +/− are as defined for Fig. 2.
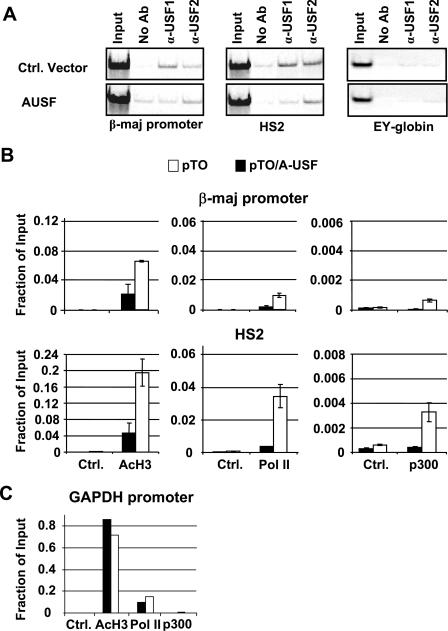
To analyze whether USF is involved in the recruitment of transcription complexes to the globin gene locus, we performed ChIP experiments in MEL cells transfected either with the A-USF expression construct or an empty control vector (pTO) (Fig. 6). We first determined whether the expression of A-USF affects the interaction of USF1 and USF2 with the β-globin gene locus. We and others previously demonstrated that USF interacts with LCR element HS2 and the β-globin gene promoter (4, 14, 22). The expression of A-USF inhibits the binding of USF1 to both HS2 and the βmaj-globin gene promoter. There is less of an effect on the interaction pattern of USF2. This result suggests that A-USF preferentially inhibits USF1/USF2 heterodimers and USF1/USF1 homodimers. We next analyzed the interaction of RNA Pol II and the coregulator p300 as well as the association of acetylated histone H3 (AcH3) with LCR element HS2 and the βmaj promoter by ChIP. The results show that the interactions of essentially all of the factors with the β-globin gene locus were reduced in cells expressing A-USF compared to that in cells harboring the control vector (Fig. 6B). A reduction in factor binding was not only restricted to the β-globin promoter region but also occurred at LCR element HS2, which contains several E-box elements. The decrease in factor binding was accompanied by reductions in the association of AcH3 with the globin locus. There was very little change in factor association with the control GAPDH promoter (Fig. 6C). Overall, the results suggest that USF proteins not only regulate the recruitment of transcription complexes but also play roles in establishing accessible chromatin domains in the globin gene locus.
FIG. 6.
Interactions of USF, RNA Pol II, p300, and modified histones with the β-globin gene locus in MEL cells expressing dominant-negative A-USF. (A) pTO/A-USF, pTR (A-USF), and pTO, pTR (control [Ctrl.]) MEL cells were induced with doxycycline. Cells were subjected to ChIP, immunoprecipitating with anti (α)-USF1, α-USF2, or no antibody (No Ab). Semiquantitative PCR was performed on samples, including input, using primers specific for the β-major promoter, HS2, or the ɛγ promoter. (B and C) MEL cells transfected with the A-USF expression vector (pTO/A-USF) or empty vector pTO were subjected to ChIP analysis with antibodies against AcH3 and RNA Pol II, or p300 (B). DNA was purified from the immunoprecipitate and analyzed by qPCR with primers specific for the βmaj-globin promoter and LCR element HS2. The error bars represent the results from two independent experiments. Ctrl represents either no antibody or nonspecific IgG antibody. (C) Results from pTO control cells are shown as white boxes. Results from pTO/A-USF cells are represented by black boxes. Samples described in the legend for panel B were analyzed with primers specific to the GAPDH promoter. All quantitative data were subjected to statistical analysis, and the P values were all below 0.05.
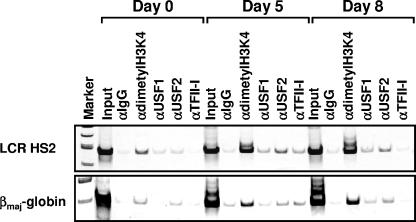
We previously analyzed the association of RNA polymerase II and transcription factors with the murine β-globin gene locus during the erythroid differentiation of murine embryonic stem cells (24). Differentiation is induced by erythropoietin and is accompanied by the expression of the embryonic globin genes at day 5 and the activation of the adult globin genes at day 8. We utilized this differentiation system to examine the interaction patterns of USF and TFII-I with locus control region element HS2 and the βmaj-globin gene promoter by ChIP assay (Fig. 7). As a control, we examined the association of histone H3 dimethylated at lysine 4 (H3K4 dimethyl), which is a mark for transcriptionally competent chromatin. Dimethylated H3K4 is detectable in LCR HS2 at all stages but reduced at the β-globin promoter at day 0, which is consistent with our previous findings (24). USF2 is detectable at the LCR and the βmaj-globin gene promoter in undifferentiated cells at day 0 and continues to interact with these elements throughout differentiation. TFII-I is detectable at HS2 and the β-globin promoter at day 5, while USF1 is found at only HS2 at this stage. At day 8, USF1 interacts with the β-globin promoter and HS2. Interestingly, TFII-I is no longer detectable at HS2 or the β-globin promoter at day 8, when β-globin gene expression is activated. These results are consistent with the conclusion that TFII-I is a repressor of β-globin gene expression, while USF is an activator. Furthermore, these results, together with those of other interaction studies, strongly suggest that USF and TFII-I directly act on the globin gene locus.
FIG. 7.
Interaction of USF and TFII-I with the β-globin gene locus during erythroid differentiation of murine embryonic stem cells. Murine embryonic stem cells were cultured and induced to differentiate as previously described by Levings et al. (24). At day 5, after the addition of erythropoietin, cells were collected and subjected to ChIP analysis using antibodies against dimethylated histone H3K4 (α-dimethyl H3K4), USF1, USF2, and TFII-I. IgG antibodies were used as negative controls in these experiments. The DNA was isolated from the precipitate and analyzed by PCR using sets of primers specific for LCR element HS2 and the βmaj-globin gene promoter.
DISCUSSION
The mechanisms governing the correct expression of β-globin genes during development involve DNA binding proteins that interact with specific regulatory elements in the globin locus in a developmental stage-specific manner and aid in the modification of chromatin structure and/or in the recruitment of transcription complexes (40). Relative gene order and activity of stage-specific DNA binding proteins regulate chromatin accessibility and ultimately the recruitment of active transcription complexes to the genes.
Among the proteins known to mediate stage-specific expression of globin genes is EKLF, which activates the adult β-globin gene but not the embryonic globin genes, and NF-Y, which is involved in the stage-specific expression of the γ-globin genes (13). EKLF recruits ATP-dependent chromatin-remodeling activities to the β-globin gene promoter (1). It is becoming increasingly clear that the local remodeling of nucleosomes in the promoter region is an important step in the activation of gene transcription (27).
Our previous studies demonstrated that β-globin promoter activity is regulated by DNA sequences located downstream of the transcription start site (22). One of these elements is an E-box motif located 60 bp downstream of the transcription initiation site. This element interacts with USF proteins in vitro and is precipitated by USF antibodies in ChIP experiments in cells expressing the β-globin gene. Another element located at and downstream of the transcription start site is composed of overlapping initiator and E-box elements and interacts with TFII-I and USF proteins. Other studies have shown that USF interacts with a functionally important E-box motif in LCR element HS2 (14). In this study, we have analyzed the contribution of helix-loop-helix proteins TFII-I and USF to β-globin gene expression in K562 and MEL cells. The human erythroleukemia cell line K562 exhibits an embryonic-cell-like phenotype and predominantly expresses the ɛ- and γ-globin genes. In contrast, MEL cells express the adult βmaj- and βmin-globin genes but not the embryonic yolk and βh1-globin genes. The results of the present study show that the inhibition of either TFII-I or HDAC3 activity upregulates β-globin gene expression in K562 cells. Furthermore, we show that TFII-I and HDAC3 interact with each other and the β-globin gene promoter in K562 cells. These results confirm and extend previous studies documenting interactions between TFII-I and HDAC3 (44, 46). Our data demonstrate that TFII-I functions as a repressor of β-globin gene expression by recruiting histone deacetylase activity to the promoter. This is consistent with observations that acetylation of the human β-globin promoter region is reduced in erythroid cells with an embryonic phenotype (45). It is likely that the ubiquitously expressed transcription factor TFII-I functions in conjunction with stage- and erythroid-cell-specific activities to repress the expression of the β-globin gene. Repressor complexes have previously been characterized that mediate stage-specific repression of the ɛ-globin gene in adult cells (41). Alternatively, activities that counteract the establishment of inaccessible chromatin mediated by TFII-I and HDAC3 are present in adult erythroid cells but may not be expressed or active at the embryonic stage.
The reversal of gene order in the globin locus with respect to the LCR leads to the expression of the adult β-globin gene in embryonic cells (42). This result shows that proximity to the LCR can override activities involved in repressing β-globin gene expression at the embryonic stage. In the normal configuration, an open and accessible chromatin domain extends over the LCR and the embryonic genes, while TFII-I and other proteins recruit histone-modifying activities to the β-globin gene to establish an inaccessible chromatin domain. In the “genes-inverted” orientation, the β-globin gene becomes part of an environment in embryonic cells that is dominated by activities recruited to and possibly spreading from the LCR. These activities override the stage-specific silencing of the β-globin gene. It is possible that in the wild-type configuration, stage-specific insulators between the embryonic and adult domains prevent the spread of chromatin opening into the region containing the adult genes. Inversion of the genes with respect to the LCR would place the same insulators between the LCR and the embryonic genes. This may explain why the embryonic ɛ-globin gene is not expressed in a transgenic β-globin locus in which the genes are inverted relative to the LCR (42).
USF proteins have previously been shown to interact with HS2 of the β-globin locus control region, which contains conserved E-box motifs (4, 14). In addition, we have shown that USF also interacts with the β-globin gene promoter (22). Mutations in the downstream promoter E-box motif decreased in vitro transcription efficiency of the β-globin gene. Our findings of reduced β-globin gene transcription in MEL cells expressing a dominant-negative mutant are consistent with the in vitro data. However, the ChIP analysis clearly shows that a reduction of USF activity not only affects the recruitment of Pol II to the globin locus but also reduces the modification of histone tails that are associated with accessible chromatin domains. The Felsenfeld group recently showed that USF interacts with a chromatin boundary element in the chicken β-globin gene locus and recruits the coregulators p300 and PCAF, which harbor histone acetyltransferase activity (47). It was discussed that the chromatin-modifying activities recruited by USF could establish an open, accessible chromatin region, counteracting the spread of heterochromatin. USF could play a similar role in establishing accessible chromatin in the LCR and the adult β-globin gene promoter. In fact, our data show that the association of p300 with LCR element HS2 and the βmaj-globin promoter, both of which associate with USF (22, 45), is reduced in cells expressing A-USF. In addition, we have previously shown that RNA polymerase II dissociates from the LCR and globin genes during replication in synchronized K562 cells (45). In these studies, we found that USF interacts with the LCR and globin genes before RNA polymerase II reassociates with the β-globin gene locus. This result supports the hypothesis that USF recruits chromatin-modifying activities that confer or maintain an open chromatin conformation in specific regions of the β-globin gene locus. The ablation of the genes encoding either USF1 or USF2 does not significantly affect the function of the hematopoietic system (8). The ablation of both genes leads to an embryonic lethal phenotype, suggesting that the proteins can compensate for one another during embryonic development. The dominant-negative mutant protein that we used in this study inactivates USF1 and, to a somewhat lesser extent, USF2, which could explain the more dramatic effect on β-globin gene expression.
The erythroleukemia cell lines used in this study do not accurately reflect the identities of erythroid cells that differentiate during human or mouse development, and it will be important to determine the functions of USF and TFII-I during the development and differentiation of erythroid cells in an animal model. We are currently in the process of generating transgenic mice expressing dominant-negative forms of USF and TFII-I exclusively in erythroid cells. Our examination of the interaction pattern of USF and TFII-I with the β-globin gene locus during erythroid differentiation of murine embryonic stem cells demonstrates that TFII-I dissociates and USF associates with the β-globin promoter when it is activated.
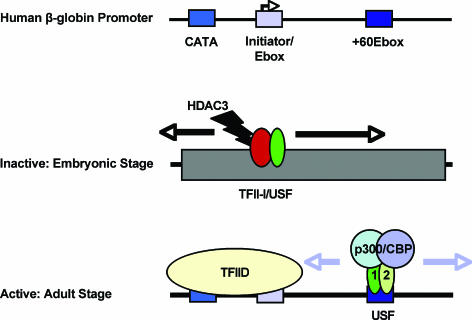
In summary, our data strongly suggest that the ubiquitously expressed helix-loop-helix proteins TFII-I and USF participate in restricting β-globin gene expression to the adult stage of erythropoiesis (Fig. 8). We propose that the primary function of these proteins is to recruit protein complexes that regulate chromatin accessibility in the β-globin gene locus. Tissue- and stage-specific regulation of the β-globin gene locus involves complex mechanisms, and to understand this process, it is important to determine the function of not only erythroid-cell-specific activities but also ubiquitously acting factors that function together with cell-type-specific proteins.
FIG. 8.
Model for β-globin gene regulation by helix-loop-helix proteins USF and TFII-I. The β-globin promoter consists of a TATA-like sequence (CATA) located 25 bp upstream of the transcription start site, an initiator with an overlapping E-box, and a downstream E-box element located at +60. We propose that a protein complex containing TFII-I, HDAC3, and possibly other proteins interacts with the β-globin promoter in embryonic and fetal erythroid cells. The modification of histones by HDAC3 confers or maintains inaccessibility of the globin gene to the transcription complex. In adult cells, USF interacts with the β-globin promoter and recruits coactivator complexes that modify the chromatin structure to increase accessibility for transcription complexes.
Acknowledgments
We thank Takeesha Roland and Jian Tang for expert technical assistance and our colleagues in the laboratory and Tom Yang (UF) for stimulating discussions. We thank Rolf Renne (UF) for allowing us to use the Nucleofector. We thank Charles Vinson (NIH) for the A-USF expression construct, Robert G. Roeder for USF and TFII-I expression constructs and TFII-I antibodies, Michael Kilberg (UF) for anti-HA antibodies, Ed Seto (University of South Florida) for HDAC3 antibodies, Ananda Roy for TFII-I antibodies, and Ann Dean (NIH) for the K562 cell line.
This work was supported by alumni fellowships (UF) to V.J.C.-D. and K.F.V. and grants from the NIH to J.B. (DK58209 and DK52356).
REFERENCES
- 1.Armstrong, J. A., J. J. Bieker, and B. M. Emerson. 1998. A SWI/SNF related chromatin remodeling complex, E-RC1, is required for tissue-specific transcriptional regulation by EKLF in vitro. Cell 95:93-104. [DOI] [PubMed] [Google Scholar]
- 2.Bender, M. A., J. N. Roach, J. Hallow, J. Close, R. Alami, E. E. Bouhassira, M. Groudine, and S. N. Fiering. 2001. Targeted deletion of 5′HS1 and 5′HS4 of the beta-globin locus control region reveals additive activity of the DNase I hypersensitive sites. Blood 98:2022-2027. [DOI] [PubMed] [Google Scholar]
- 3.Bieker, J. J. 2001. Kruppel-like factors: three fingers in many pies. J. Biol. Chem. 276:34355-34358. [DOI] [PubMed] [Google Scholar]
- 4.Bresnick, E. H., and G. Felsenfeld. 1993. Evidence that the transcription factor USF is a component of the human beta-globin locus control region heteromeric protein complex. J. Biol. Chem. 268:18824-18834. [PubMed] [Google Scholar]
- 5.Bulger, M., and M. Groudine. 1999. Looping versus linking: toward a model for long-distance gene activation. Genes Dev. 13:2465-2477. [DOI] [PubMed] [Google Scholar]
- 6.Bungert, J., I. Kober, F. Düring, and K. H. Seifart. 1992. Transcription factor eUSF is an essential component of isolated transcription complexes on the duck histone H5 gene and it mediates the interaction of TFII-D with a TATA-deficient promoter. J. Mol. Biol. 223:885-898. [DOI] [PubMed] [Google Scholar]
- 7.Bungert, J., U. Dave, K.-C. Lim, K. H. Lieuw, J. A. Shavit, Q. Liu, and J. D. Engel. 1995. Synergistic regulation of human β-globin gene switching by locus control region elements HS3 and HS4. Genes Dev. 9:3083-3096. [DOI] [PubMed] [Google Scholar]
- 8.Casado, M., V. S. Vallet, M. Kahn, and S. Vaulont. 1999. Essential role in vivo of upstream stimulatory factors for a normal dietary response of the fatty acid synthase gene in the liver. J. Biol. Chem. 274:2009-2013. [DOI] [PubMed] [Google Scholar]
- 9.Cheriyath, V., C. D. Novina, and A. L. Roy. 1998. TFII-I regulates Vβ promoter activity through an initiator element. Mol. Cell. Biol. 18:4444-4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 11.Dillon, N., T. Trimborn, J. Strouboulis, P. Fraser, and F. Grosveld. 1997. The effect of distance on long-range chromatin interactions. Mol. Cell 1:131-139. [DOI] [PubMed] [Google Scholar]
- 12.Du, H., A. L. Roy, and R. G. Roeder. 1993. Human transcription factor USF stimulates transcription through the initiator elements of the HIV-1 and AdML promoters. EMBO J. 12:501-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duan, Z., G. Stamatoyannopoulos, and Q. Li. 2001. Role of NF-Y in in vivo regulation of the γ-globin gene. Mol. Cell. Biol. 21:3083-3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elnitski, L., W. Miller, and R. Hardison. 1997. Conserved E-boxes function as part of the enhancer in hypersensitive site 2 of the beta-globin locus control region. Role of basic helix-loop-helix proteins. J. Biol. Chem. 272:369-378. [DOI] [PubMed] [Google Scholar]
- 15.Engel, J. D., and K. Tanimoto. 2000. Looping, linking and chromatin activity: new insights into β-globin locus regulation. Cell 100:499-502. [DOI] [PubMed] [Google Scholar]
- 16.Fiering, S., E. Epner, K. Robinson, Y. Zhuang, A. Telling, M. Hu, D. I. K. Martin, T. Enver, T. J. Ley, and M. Groudine. 1995. Targeted deletion of 5′HS2 of the murine β-globin LCR reveals that it is not essential for proper regulation of the β-like globin locus. Genes Dev. 9:2203-2213. [DOI] [PubMed] [Google Scholar]
- 17.Gillemans, N., R. Tewari, F. Lindeboom, R. Rottier, T. de Wit, M. Wijgerde, F. Grosveld, and S. Philipsen. 1998. Altered DNA-binding specificity mutants of EKLF and Sp1 show that EKLF is an activator of the beta-globin locus control region in vivo. Genes Dev. 12:2863-2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gribnau, J., K. Diderich, S. Pruzina, R. Calzolari, and P. Fraser. 2000. Intergenic transcription and developmental remodeling of chromatin structure in the human β-globin locus. Mol. Cell 5:377-386. [DOI] [PubMed] [Google Scholar]
- 19.Grosveld, F., G. B. van Assendelft, D. R. Greaves, and G. Kollias. 1987. Position-independent, high-level expression of the human β-globin gene in transgenic mice. Cell 51:975-985. [DOI] [PubMed] [Google Scholar]
- 20.Hulf, T., P. Bellosta, M. Furrer, D. Steiger, D. Svensson, A. Barbour, and P. Gallant. 2005. Whole-genome analysis reveals a strong positional bias of conserved dMyc-dependent E-boxes. Mol. Cell. Biol. 25:3401-3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kosak, S. T., and M. Groudine. 2004. Gene order and dynamic domains. Science 306:644-647. [DOI] [PubMed] [Google Scholar]
- 22.Leach, K. M., K. F. Vieira, S. H. Kang, A. Aslanian, M. Teichmann, R. G. Roeder, and J. Bungert. 2003. Characterization of the human β-globin downstream promoter region. Nucleic Acids Res. 31:1292-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levings, P. P., and J. Bungert. 2002. The human β-globin locus control region: a center of attraction. Eur. J. Biochem. 269:1589-1599. [DOI] [PubMed] [Google Scholar]
- 24.Levings, P. P., Z. Zhou, K. F. Vieira, V. J. Crusselle-Davis, and J. Bungert. 2006. Recruitment of transcription complexes to the β-globin locus control region and transcription of hypersensitive site 3 prior to erythroid differentiation of murine embryonic stem cells. FEBS J. 273:746-755. [DOI] [PubMed] [Google Scholar]
- 25.Massari, M. E., and C. Murre. 2000. Helix-loop-helix proteins: regulators of transcription in eukaryotic organisms. Mol. Cell. Biol. 20:429-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McMurray, H. R., and D. J. McCance. 2003. Human papillomavirus type 16 E6 activates TERT gene transcription through induction of c-Myc and release of USF-mediated repression. J. Virol. 77:9852-9861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mellor, J. 2005. The dynamics of chromatin remodeling at promoters. Mol. Cell 19:147-157. [DOI] [PubMed] [Google Scholar]
- 28.Milot, E., J. Strouboulis, T. Trimborn, M. Wijgerde, E. de Boer, A. Langeveld, K. Tan-un, W. Vergeer, N. Yannoutsos, F. Grosveld, and P. Fraser. 1996. Heterochromatin effects on the frequency and duration of LCR-mediated gene transcription. Cell 87:105-114. [DOI] [PubMed] [Google Scholar]
- 29.Navas, P. A., K. R. Peterson, Q. Li, E. Skarpidi, A. Rohde, S. E. Shaw, C. H. Clegg, H. Asano, and G. Stamatoyannopoulos. 1998. Developmental specificity of the interaction between the locus control region and embryonic or fetal globin genes in transgenic mice with an HS3 core deletion. Mol. Cell. Biol. 17:4188-4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ogura, Y., M. Azuma, Y. Tsuboi, Y. Kabe, Y. Yamaguchi, T. Wada, H. Watanabe, and H. Handa. 2006. TFII-I down-regulates a subset of estrogen-responsive genes through its interaction with an initiator element and estrogen receptor alpha. Genes Cells 11:373-381. [DOI] [PubMed] [Google Scholar]
- 31.Orphanides, G., and D. Reinberg. 2002. A unified theory of gene expression. Cell 108:439-451. [DOI] [PubMed] [Google Scholar]
- 32.Peterson, K. R., and G. Stamatoyannopoulos. 1993. Role of gene order in developmental control of human gamma- and beta-globin gene expression. Mol. Cell. Biol. 13:4836-4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peterson, K. R., C. H. Clegg, P. A. Navas, E. J. Norton, T. G. Kimbrough, and G. Stamatoyannopoulos. 1996. Effect of deletion of 5′ HS3 or 5′HS2 of the human beta-globin locus control region on the developmental regulation of globin gene expression in beta-globin locus yeast artificial chromosome transgenic mice. Proc. Natl. Acad. Sci. USA 93:6605-6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peterson, K. R. 2003. Hemoglobin switching: new insights. Curr. Opin. Hematol. 10:123-129. [DOI] [PubMed] [Google Scholar]
- 35.Qyang, Y., X. Luo, T. Lu, P. M. Ismail, D. Krylov, C. Vinson, and M. Sawadogo. 1999. Cell-type-dependent activity of the ubiquitous transcription factor USF in cellular proliferation and transcriptional activation. Mol. Cell. Biol. 19:1508-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ribeiro, A., D. Pastier, D. Kardassis, J. Chambaz, and P. Cardot. 1999. Cooperative binding of upstream stimulatory factor and hepatic nuclear factor 4 drives the transcription of the human apolipoprotein A-II gene. J. Biol. Chem. 274:1216-1225. [DOI] [PubMed] [Google Scholar]
- 37.Roy, A. L., H. Du, P. D. Gregor, C. D. Novina, E. Martinez, and R. G. Roeder. 1997. Cloning of an INR- and E-box binding protein, TFII-I, that interacts physically and functionally with USF1. EMBO J. 16:7091-7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roy, A. L., M. Meisterernst, P. Pognonec, and R. G. Roeder. 1991. Cooperative interaction of an initiator-binding transcription initiation factor. Nature 354:245-248. [DOI] [PubMed] [Google Scholar]
- 39.Stamatoyannopoulos, G., and A. Nienhuis. 1994. Hemoglobin switching, p 107-155. In G. Stamatoyannopoulos, A. Nienhuis, P. Majerus, and H. Varmus (ed.), The molecular basis of blood diseases. W. B. Saunders, Philadelphia, Pa.
- 40.Stamatoyannopoulos, G. 2005. Control of globin gene expression during development and erythroid differentiation. Exp. Hematol. 33:259-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanimoto, K., Q. Liu, F. Grosveld, J. Bungert, and J. D. Engel. 2000. Context dependent EKLF responsiveness defines the developmental specificity of the human ɛ-globin gene in erythroid cells of YAC transgenic mice. Genes Dev. 14:2778-2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tanimoto, K., Q. Liu, J. Bungert, and J. D. Engel. 1999. Effects of altered gene order or orientation of the locus control region on human β-globin gene expression in mice. Nature 398:344-348. [DOI] [PubMed] [Google Scholar]
- 43.Tuan, D., S. Kong, and K. Hu. 1992. Transcription of the HS2 enhancer in erythroid cells. Proc. Natl. Acad. Sci. USA 89:11219-11223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tussie-Luna, M. I., D. Bayarsaihan, E. Seto, F. H. Ruddle, and A. L. Roy. 2002. Physical and functional interactions of histone deacetylase 3 with TFII-I family proteins and PIASxβ. Proc. Natl. Acad. Sci. USA 99:12807-12812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vieira, K. F., P. P. Levings, M. A. Hill, V. J. Crusselle, S.-H. L. Kang, J. D. Engel, and J. Bungert. 2004. Recruitment of transcription complexes to the β-globin gene locus in vivo and in vitro. J. Biol. Chem. 279:50350-50357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wen, Y. D., W. D. Cress, A. L. Roy, and E. Seto. 2003. Histone deacetylase 3 binds to and regulates the multifunctional transcription factor TFII-I. J. Biol. Chem. 278:1841-1847. [DOI] [PubMed] [Google Scholar]
- 47.West, A. G., S. Huang, M. Gaszner, M. D. Litt, and G. Felsenfeld. 2004. Recruitment of histone modifications by USF proteins at a vertebrate barrier element. Mol. Cell 16:453-463. [DOI] [PubMed] [Google Scholar]