Abstract
CLOCK and BMAL1 are bHLH-PAS-containing transcription factors that bind to E-box elements and are indispensable for expression of core circadian clock components such as the Per and Cry genes. A key step in expression is the heterodimerization of CLOCK and BMAL1 and their accumulation in the nucleus with an approximately 24-h periodicity. We show here that nucleocytoplasmic shuttling of BMAL1 is essential for transactivation and for degradation of the CLOCK/BMAL1 heterodimer. Using serial deletions and point mutants, we identified a functional nuclear localization signal and Crm1-dependent nuclear export signals in BMAL1. Transient-transfection experiments revealed that heterodimerization of CLOCK and BMAL1 accelerates their turnover, as well as E-box-dependent clock gene transcription. Moreover, in embryonic mouse fibroblasts, robust transcription of Per2 is tightly associated with massive degradation of the CLOCK/BMAL1 heterodimer. CRY proteins suppressed this process during the transcription-negative phase and led to nuclear accumulation of the CLOCK/BMAL1 heterodimer. Thus, these findings suggest that the decrease of BMAL1 abundance during the circadian cycle reflects robust transcriptional activation of clock genes rather than inhibition of BMAL1 synthesis.
Almost all organisms from bacteria to mammals have developed circadian physiology and behavior to adapt to the environmental changes created by the rotation of our planet. Such daily rhythms are controlled by genetically determined and self-sustaining circadian clocks that are composed of networks of transcription-translation feedback loops involving sets of clock genes (15, 30, 37). In mammals, a network of feedback loops functions robustly not only in the master circadian pacemaker, the suprachiasmatic nucleus (SCN) of the hypothalamus (26), but also in most tissues (including liver, heart, lung, and muscle) and even in immortalized cell lines (1, 50, 51). These widely dispersed circadian systems are primarily synchronized by the SCN to coordinate circadian timing in vivo (29, 36).
One of the main questions in clock biology is how the time-keeping system is controlled at the molecular level. Intensive studies in the mouse using genetic and molecular approaches have largely clarified the structure of the central feedback loop (3, 30). Two bHLH-PAS-containing transcription factors, CLOCK and BMAL1, form heterodimers that bind to E-box enhancer elements in the promoters of target genes driving the transcription of three period genes (Per1, Per2, and Per3) and two cryptochrome genes (Cry1 and Cry2) (11, 14, 17, 18). After the PER and CRY proteins have been translated in the cytoplasm, they form heterocomplexes that translocate into the nucleus and inhibit their own transcription. CRY plays a crucial role in this negative feedback process by interacting directly with the CLOCK/BMAL1 heterodimers (12, 22).
Analysis of Bmal1 defective mice has revealed the indispensable role of BMAL1 as the mainspring of the molecular clockwork; thus, targeted disruption of Bmal1 results in complete loss of both circadian behavior and expression of the core clock regulators, Per1 and Per2, in the SCN (5). This strongly supports the notion that the expression of Per1 and Per2 is tightly coupled to the transcriptional activity of BMAL1. A recent study, using transient-transfection assays in HEK293 cells and Bmal1-deficient fibroblasts, indicated that the nuclear accumulation and degradation of the CLOCK proteins, and their phosphorylation, are largely dependent on BMAL1, although the precise mechanisms involved remained to be elucidated (20). On the other hand, previous studies of Cry1/Cry2 double mutant mice revealed sustained high-level expression of both Per1 and Per2 in the liver and mid-to-high levels in the SCN (28, 45, 46). In these mutants, however, Bmal1 transcription was maintained at moderately low levels that were comparable to the trough of Bmal1 transcript levels in the wild type, whereas both mutant and wild-type mice exhibited arrhythmic expression of similar levels of the Clock genes (38). Thus, it is tempting to postulate that moderately low levels of Bmal1 RNA can lead to sufficient BMAL1 synthesis to permit robust transcription of Per1 and Per2 in the absence of the transcriptional inhibition normally exerted by the CRY proteins.
In contrast to this notion, the level of BMAL1 and even of CLOCK in the liver of the Cry-deficient mice was significantly lower than in wild-type animals, especially at the time when nuclear accumulation of CRY peaked in the wild type (e.g., CT 18) (23). Further analysis of subcellular fractions showed that the dramatic decrease in both proteins in the mutant hepatocytes was due to a dearth of the proteins in the nucleus rather than in the cytoplasm. More surprising was the finding that the nuclear accumulation and/or abundance of CLOCK and BMAL1 reach a minimum at the time when maximal transcriptional enhancement of Per1 and Per2 was anticipated, both in vitro and in vivo (20, 23). These paradoxical results led us to dissect the molecular mechanisms underlying the regulation of the transactivation and inhibition of the CLOCK/BMAL1 heterodimer responsible for driving clock gene transcription.
In the present study, we demonstrate that BMAL1 has a functional nuclear localization signal (NLS) and nuclear export signals (NES) in its N-terminal and PAS domains, respectively, and shuttles between the cytoplasm and the nucleus to permit CLOCK to accumulate in the nucleus. This process appears to be crucial for proteolysis of the CLOCK/BMAL1 heterodimer, as well as for the transcription of its target genes. These findings have prompted us to propose a new model of circadian regulation of clock gene transcription. In this model, shuttling of BMAL1 dynamically controls transactivation of the CLOCK/BMAL1 heterodimer, which is tightly coupled to its own degradation via ubiquitin-dependent or -independent pathways.
MATERIALS AND METHODS
Plasmid constructs and primers.
cDNAs encoding wild-type mBmal1 (GenBank accession number NM007489) and mClock (GenBank accession number NM007715) were kindly provided by S. M. Reppert (University of Massachusetts). The deletion mutants were generated with these cDNAs as templates, by a PCR-based method as described previously (39). The resulting PCR products were cloned into pcDNA 3.1-DEST and epitope-tagged pcDNA3.1-DEST by Gateway cloning (Invitrogen). Plasmids encoding NLS or NES mutants of Bmal1 were generated by QuikChange site-directed mutagenesis as detailed by the manufacturer (Stratagene) using the wild-type cDNA as a template. The PCR primers used for the site-directed mutagenesis were as follows: for the NLS1 mutant, F1 (5′-GGTGTGGACTGCAATCGCGCGGCGAAAGGCAGTGCC ACTGAC-3′); for the NLS2 mutant, F1 (5′-GTCAGTGGCACTGCCTTTCGCCGCGCGATTGCAGTCCACACC-3′); for the NES1 mutant, F1 (5′-CCAGGAAGTTAGATAAAGCCACCGTGCTAAGGATGGCT-3′) and F2 (5′-CCAGGAAGTTAGATAAAGCCACCGTGGCAAGGATGGCTGTTCAG-3′); for the NES2 mutant, F1 (5′-GATGACGAAGCGAAACACCTAATTCTCAGGGCAGCAGATGGA-3′) and F2 (5′-GATGACGAAGCGAAACACGCAATTCTCAGGGCAGCAGATGGA-3′); and for the NES3 mutant, F1 (5′-ACAGCTATTTTGGCGTATGCACCACAGGAACTTCTAGGTACA-3′) and F2 (5′-GGCGACAGCTATTTTGGCGTATGCACCACAGGAAGCTCTAGGTAC-3′). All constructs were verified by sequencing.
Cell culture and transfection.
Wild-type and mutant mouse embryo fibroblasts (MEFs), HeLa cells, and NIH 3T3 cells were cultured in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (Invitrogen) under 9% CO2 at 37°C. One day after seeding, cells were transfected by using LipofectAMIN Plus (Invitrogen). For luciferase reporter assays, the mouse Per1-promoter (−1803 to + 40) was cloned into pGL3-Basic Vector (Promega) as a fusion with firefly luciferase, and the product was transfected into NIH 3T3 cells, together with the plasmids carrying various clock genes. At 36 h after transfection, cells were harvested, and the luciferase activity was determined with a luminometer (Turner Designs).
Antibodies.
Polyclonal antibodies against mouse BMAL1 and BRG1 were generated in rabbits by injection with recombinant GST-BMAL1(480-625) and GST-BRG1(65-382), respectively. The rabbit antisera were affinity purified on CNBr-activated Sepharose columns with covalently bound antigen. Antibodies specific for CLOCK and CRY1 were purchased from Santa Cruz. Antibodies 9E10 and F7 were used to detect c-Myc and hemagglutinin (HA) epitopes, respectively. Donkey secondary antibodies conjugated to horseradish peroxidase, fluorescein isothiocyanate (FITC), and TRITC (tetramethyl rhodamine isothiocyanate) were purchased from Jackson Immunoresearch. Other antibodies were from Sigma.
Immunoprecipitation, immunoblotting, and immunofluorescence.
Aliquots of 2 × 105 NIH 3T3 cells were plated in 35-mm plates 24 h prior to transfection and then incubated with 2.5 μg (total) of DNA. Immunoprecipitation was performed 30 h after transfection. Cells were harvested with 300 μl of radioimmunoprecipitation assay buffer (50 mM HEPES [pH 7.4], 150 mM NaCl, 1% NP-40, 1 mM EDTA, 1 mM EGTA, 1 mM phenylmethylsulfonyl fluoride, 0.5% sodium deoxycholate, 1 mM NaF, 1 mM Na3VO4, 1× protease inhibitor cocktail [Roche]) and then were centrifuged at maximum speed for 20 min at 4°C. The supernatants were transferred to fresh tubes, and 2 μg of anti-Myc or anti-BMAL1 antibodies was added, followed by incubation at 4°C. After 1.5 h, 40 μl of 50% protein-A agarose was added, and incubation continued for 1 h at 4°C. After the supernatants were discarded carefully, 20 μl of 2× sodium dodecyl sulfate sample buffer was added, and the samples were boiled for 10 min, separated on a 6% sodium dodecyl sulfate-polyacrylamide gel, and transferred to polyvinylidene difluoride membranes. Immunoblotting was performed with appropriate primary antibodies, and horseradish peroxidase-conjugated secondary antibodies for enhanced chemiluminescence detection.
For immunofluorescence analysis, cells grown on 0.1% gelatin-coated coverslips were fixed with 3.75% paraformaldehyde in phosphate-buffered saline and blocked with 10% goat serum. They were then incubated with anti-BMAL1 or anti-Myc antibodies and stained with FITC-conjugated anti-rabbit immunoglobulin G (IgG) or TRITC-conjugated anti-mouse IgG. Nuclei were visualized with DAPI (4′,6′-diamidino-2-phenylindole; Molecular Probes).
Heterokaryon assays.
Heterokaryon assays were performed as previously described (7, 48). NIH 3T3 cells were transfected with Myc-tagged Bmal1 constructs (wild type or NESmutBMAL1) and/or Brg1. After 24 h, NIH 3T3 cells were exposed to leptomycin B (LMB; 10 ng/ml), a specific inhibitor of Crm1-dependent nuclear export, for 5 h. They were then mixed with equal numbers of LMB-treated (nontransfected) HeLa cells and cocultured for 6 h in the presence of LMB (10 ng/ml) and cycloheximide (CHX; 50 μg/ml). The CHX concentration was increased to 100 μg/ml for the last 30 min of culture before cell fusion. After fusion using 50% polyethylene glycol 3350 for 2 min, the cells were washed with PBS and incubated in cell culture medium containing CHX (100 μg/ml) for another 4 h. BMAL1 and BRG1 were visualized by immunofluorescence staining as described above.
Real-time reverse transcription-PCR.
First-strand cDNAs were synthesized from the total RNA of MEFs by reverse transcription. Real-time PCR of Bmal1 and Clock mRNA was normalized to GAPDH (glyceraldehyde-3-phosphate dehydrogenase) as an endogenous reference and was performed with a 7300 Real-Time PCR system (ABI). The primers for GAPDH were 5′-CATGGCCTTCCGTGTTCCTA-3′ and 5′-CCTGCTTCACCACCTTCTTGA-3′. The primers for BMAL1 were 5′-CCAAGAAAGTATGGACACAGACAAA-3′ and 5′-GCATTCTTGATCCTTCCTTGGT-3′, and the primders for Clock were 5′-AGGCACAGACATTATCCG-3′ and 5′-ACCGTCTCATCAAGGGAC-3′. PCR was carried out with 2× Dynamo SYBR Green qPCR Master Mix (Finnzymes) and 40 ng of template cDNA after initial incubation with uracil-N-glycosylase at 50°C for 2 min. Denaturation was at 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 30 s, annealing at 56°C for 30 s and extension at 72°C for 60 s. All reactions were performed in triplicate. Relative expression levels were calculated after correction for the expression of GAPDH.
Real-time monitoring of bioluminescence.
Real-time monitoring was performed as previously described (44). Wild-type and Cry1/Cry2-deficient MEFs were plated at 1.0 × 106 cells per dish in 35-mm dishes 24 h before transfection. Cells were transfected with 0.5 μg of the full-length Per2 promoter fused with destabilized firefly luciferase (dsLuc). When the cells reached confluence after transfection, the medium was replaced with DMEM supplemented with 1% fetal bovine serum, and the cells were maintained for another 2 days. At zero time, they were treated with 1 μM dexamethasone (Sigma), and after 2 h their medium was replaced with 2 ml of culture medium (DMEM supplemented with 10% fetal bovine serum) supplemented with 50 mM HEPES (pH 7.2) and 0.1 mM luciferin (Sigma). Bioluminescence was measured with a dish-type luminometer (AB-2500; ATTO).
RESULTS
Identification of the essential domain for nuclear localization of BMAL1.
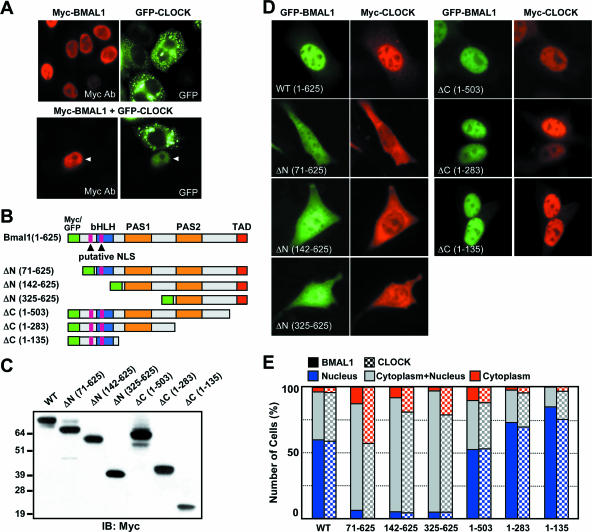
To explore the molecular mechanisms underlying transactivation of the CLOCK/BMAL1 heterodimer, we examined the subcellular distribution of BMAL1 and CLOCK when they were expressed singly or together in NIH 3T3 fibroblasts (Fig. 1A). We tagged expression constructs encoding the full-length sequences of both proteins with Myc or green fluorescent protein (GFP) at their N termini. When singly expressed, their cellular locations were in striking contrast, regardless of the epitope type: BMAL1 was located in the nucleus and CLOCK was in the cytoplasm; on the other hand, when they were coexpressed they were both found in the nucleus (Fig. 1A and data not shown). These observations are consistent with the previous finding that circadian oscillations of the nuclear accumulation of CLOCK depend on BMAL1 (20).
FIG. 1.
Subcellular distribution of truncated BMAL1 mutants and their effects on nuclear translocation of CLOCK. (A) Myc-tagged BMAL1 and GPF-CLOCK were introduced alone (upper panels) or together (lower panel) into HeLa cells. Arrowhead indicates the nucleus of a cell expressing both proteins. (B) Schematic diagram of the GFP-or Myc-tagged BMAL1 constructs. The putative NLS, bHLH, PAS, and TADs are indicated as purple, blue, yellow, and red boxes, respectively. (C) Putative molecular sizes of truncated BMAL1 mutants confirmed by immunoblot analysis with anti-Myc antibody (9E10). (D) Representative fluorescence images demonstrating the subcellular localization of each construct and coexpressed wild-type CLOCK. The GFP-fused BMAL1 constructs and Myc-tagged wild-type CLOCK were coexpressed in NIH 3T3 cells, which were stained with 9E10 to visualize the distribution of wild-type CLOCK. (E) Quantitation of the subcellular distribution of BMAL1 (solid bars) and CLOCK (checked bars). The subcellular localization was categorized as nuclear (blue), cytoplasmic and nuclear (gray), and cytoplasmic (red). The data were obtained from two independent experiments, and in each experiment more than 100 cells were counted.
To investigate the domains responsible for nuclear translocation of BMAL1, we performed a computer-based sequence analysis and found two putative N-terminal NLSs (Fig. 1B). To determine whether these NLS motifs were functional and whether BMAL1 possessed other motifs affecting its cellular distribution, we constructed a series of plasmids expressing BMAL1 deletions N-terminally tagged with Myc or GFP (Fig. 1B). Western blot analysis demonstrated that Myc-tagged full-length BMAL1 and its various mutants were expressed in NIH 3T3 cells and that they were of the expected sizes (Fig. 1C). The cellular locations of the GFP-tagged full-length BMAL1 and mutants were analyzed when coexpressed with Myc-tagged full-length CLOCK (Fig. 1D). The subcellular distributions of both BMAL1 and CLOCK were classified into one of three categories: cytoplasm dominant, nucleus dominant, or equal distribution in cytoplasm and nucleus (Fig. 1E). Full-length BMAL1 (1-625) and each of its C-terminal deletion mutants (1-503, 1-283, and 1-135) were predominantly localized in the nucleus of the transfected cells. Intriguingly, the large C-terminal deletion mutants (1-283 and 1-135) were more concentrated in the nucleus than the full-length protein. In contrast, all of the N-terminal deletion mutants (71-625, 142-625, and 325-625) were diffusely distributed throughout the cell. The findings were the same whether or not CLOCK was coexpressed (data not shown). The overall distribution patterns of CLOCK largely depended on those of BMAL1, although CLOCK showed greater cytoplasmic localization than the coexpressed BMAL1 proteins lacking N-terminal regions (Fig. 1C and D). These findings suggest that the N-terminal region (i.e., from positions 1 to 70) containing the putative NLS is essential for nuclear localization of CLOCK, as well as of BMAL1.
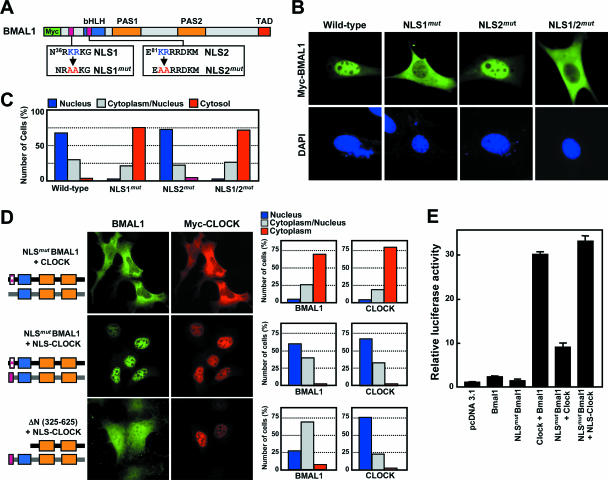
To confirm the functionality of the two putative NLS motifs (i.e., NLS1 and NLS2), we made constructs expressing Myc-tagged full-length BMAL1 with alanine substituted for adjacent basic residues in NLS1 and/or NLS2 (Fig. 2A). Mutation of NLS1 abolished nuclear accumulation, whereas mutation of NLS2 did not (Fig. 2B and C). It is therefore most likely that NLS1 (referred to as NLS hereafter) is the functional NLS. We next examined the role of the NLS in nuclear translocation of CLOCK and transcriptional activation of the CLOCK/BMAL1 heterodimer. As shown in Fig. 2D and E, the NLS mutant of BMAL1 failed to induce either nuclear import of CLOCK or Per1 promoter-dependent luciferase activity in NIH 3T3 cells when coexpressed with Myc-tagged full-length CLOCK and wild-type CLOCK, respectively. However, these defects were fully overcome by coexpression of NLS-CLOCK containing the wild-type NLS sequence. However, NLS-CLOCK was unable to overcome the malfunction of a truncated BMAL1 (142-625) lacking the N-terminal bHLH domain as well as the NLS motif. These findings provide good evidence that the N-terminal NLS of BMAL1 is not only sufficient for its translocation to the nucleus but is also crucial for transactivation of the CLOCK/BMAL1 heterodimer.
FIG. 2.
Identification of the BMAL1 NLS and its role in transactivation of the CLOCK/BMAL1 heterodimer. (A) Schematic diagram of the putative NLS motifs and substitution of the consensus basic amino acid residues RK by AA in the two motifs (indicated as NLS1mut and NLS2mut). (B) Representative fluorescence images of BMAL1 carrying intact or mutant NLS motifs in NIH 3T3 cells. (C) Quantitation of the subcellular distribution of the NLS mutants of BMAL1. Subcellular localization is categorized as nuclear (blue), cytoplasmic and nuclear (gray), and cytoplasmic (red). More than 200 cells were evaluated in each experiment. (D) The addition of a functional NLS to CLOCK induces nuclear translocation of CLOCK regardless of any defects in the coexpressed BMAL1. The cells were transiently cotransfected with constructs encoding either NLS1mut or ΔN(325-625) BMAL1 and Myc-tagged CLOCK (upper panels) or NLS-CLOCK, which was fused with the active NLS1 of BMAL1 at its N terminus (middle and lower panels). Quantitative data on the subcellular localization of the two proteins are given on the right as depicted in panel C. (E) Role of the NLS in transactivation of the CLOCK/BMAL1 heterodimer. Cells were transfected with constructs encoding a Per1-luciferase reporter and the indicated transcriptional activators. Transcription of the reporter gene is expressed as luciferase activity relative to the response in the absence of activator. Each value is the mean ± the standard error of the mean (SEM) of three independent experiments, and each experiment was performed using three replicates.
BMAL1 shuttles between cytoplasm and nucleus using the two NES sequences in its PAS domains.
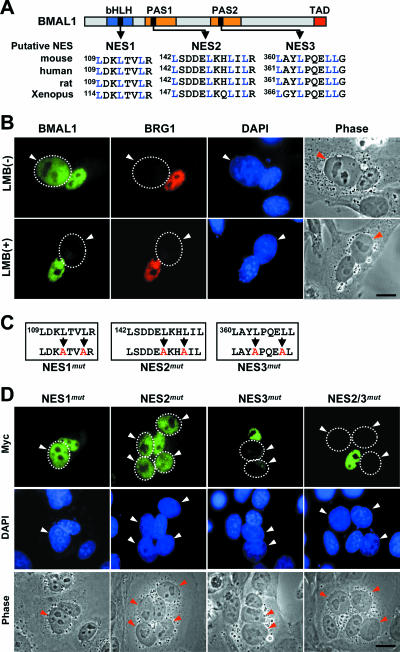
As described above, BMAL1 protein has intrinsic nuclear import ability, but nevertheless some exogenously expressed full-length BMAL1 remains in the cytoplasm. The fact that a larger percentage of the C-terminal deletion proteins lacking the PAS domains (1-283 and 1-135) were found in the nucleus (75.3 and 88.5%, respectively) than in the case of the full-length protein (62.4%) (see Fig. 1E) suggested that additional domains, probably in the C-terminal region (i.e., from positions 135 to 503) were involved in retaining BMAL1 in the cytoplasm. Comparison of the amino acid sequences of mammalian BMAL1s revealed three conserved putative NES containing leucine-rich sequences (25) in the bHLH and two PAS domains (designated NES1, NES2, and NES3, respectively) (Fig. 3A). We therefore carried out a heterokaryon assay (see Materials and Methods) to test whether nuclear export activity plays a role in regulating the localization of BMAL1. For this experiment, NIH 3T3 cells cotransfected with plasmids encoding BMAL1-GFP and the nuclear protein BRG1 (31) were cultured for 24 h and fused with HeLa cells in the presence of the protein synthesis inhibitor, CHX, and further incubated for 2 h. Nuclear staining with DAPI was used to distinguish between human nuclei (diffuse nuclear staining) and mouse nuclei (speckled nuclear staining). The GFP-BMAL1 signal initially present in the mouse nuclei was subsequently equally strong in adjacent human nuclei, whereas BRG1 protein was retained in the mouse nuclei (Fig. 3B). Clearly, BMAL1 but not BRG1 was exported out of the NIH 3T3 nuclei, and some of it was imported into the HeLa nuclei of the same heterokaryons. This nucleocytoplasmic shuttling of BMAL1 was completely blocked by the fungal antibiotic, LMB, which inhibits nuclear export of proteins by binding to the nuclear export receptor, Crm1 (21, 42). These data demonstrate that BMAL1 shuttles between nucleus and cytoplasm via a Crm1-dependent nuclear export pathway.
FIG. 3.
Nucleocytoplasmic shuttling of BMAL1 and identification of the BMAL1 NES. (A) Schematic positions of the three putative NES of BMAL1 and comparison of the amino acid sequences of these motifs in mouse, human, rat, and Xenopus laevis. Conserved leucine residues are shown in blue. (B) Crm1-dependent nucleocytoplasmic shuttling of BMAL1. Heterokaryon assays were performed with NIH 3T3 cells as donors and HeLa cells as acceptors. Donor cells transiently cotransfected with GFP-BMAL1 and BRG1, a nonshuttling nuclear protein, were seeded, with HeLa cells, onto coverslips. After incubation with CHX and LMB, the cells were fused with polyethylene glycol and further cultured in the presence of CHX alone (upper panels) or both drugs (lower panels). Transfected donor nuclei were identified by immunofluorescence staining with anti-BRG1 and TRITC-conjugated secondary antibody. Acceptor nuclei (HeLa nuclei) are indicated by the dotted circles and arrowheads. (C) Site-directed mutations of the three putative NES. The BMAL1 variants are designated NES1mut, NES2mut, and NES3mut, respectively. The alanine residues substituted in place of leucines in each NES domain are shown in red. (D) Effects of the NES mutations on BMAL1 shuttling. NIH 3T3 cells were transfected with GFP-fused NES1mut, NES2mut, NES3mut, or NES2/3mut, and nucleocytoplasmic shuttling was evaluated by using the heterokaryon assay as described in panel B. Phase-contrast images of the cells confirming the cytoplasmic continuity of the heterokaryons. Scale bar, 25 μm.
To determine which putative NES motifs are functional for shuttling, we constructed full-length GFP-BMAL1 mutants containing alanine instead of leucine residues in each individual putative NES separately or in both NES2 and NES3 (Fig. 3C). The resulting constructs (i.e., NES1mut, NES2mut, NES3mut, and NES2/3mut) were tested for shuttling activity (Fig. 3D). Site-directed mutation of NES2 and NES3 but not NES1 substantially reduced BMAL1 shuttling, and the NES3 mutation had the most effect. Moreover, translocation of BMAL1 from mouse nuclei to human nuclei was completely inhibited in the NES2 and NES3 double mutant. Thus, we conclude that both NES2 and NES3 are responsible for the nuclear export of BMAL1.
Nucleocytoplasmic shuttling of BMAL1 is essential for target gene transcription and nuclear accumulation of CLOCK.
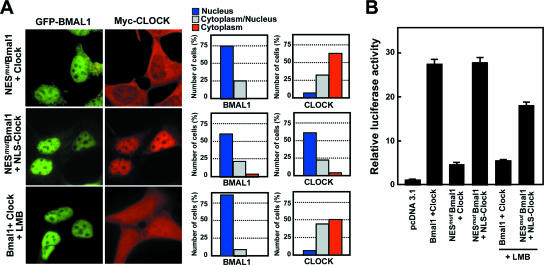
To clarify the role of nucleocytoplasmic shuttling of BMAL1, we introduced NES2/3mut (referred to as NESmutBMAL1) and wild-type CLOCK into the same cells and assessed the effects of the NES mutation on BMAL1-mediated nuclear accumulation of CLOCK and transcriptional activity (Fig. 4). Unexpectedly, in spite of the predominant nuclear localization of the NES mutant, CLOCK was found largely in the cytoplasm (Fig. 4A). This effect did not seem to be caused by disruption of the interaction between CLOCK and BMAL1 because CLOCK coprecipitated to a similar extent with wild-type BMAL1 and the NES mutant (see Fig. 6B). Moreover, when wild-type BMAL1 and CLOCK were coexpressed, treatment with LMB led to the presence of a considerable amount of CLOCK in the cytoplasm, confirming that BMAL1 shuttling is an important step in the nuclear accumulation of CLOCK.
FIG. 4.
BMAL1 shuttling mediates nuclear accumulation of CLOCK and transactivation of the CLOCK/BMAL1 heterodimer. (A) Representative fluorescence images demonstrating that nuclear accumulation of CLOCK is abolished by disruption of BMAL1 shuttling. BMAL1 carrying intact or mutant NES was coexpressed in NIH 3T3 cells with either Myc-tagged wild-type CLOCK (upper and lower panels) or NLS-CLOCK (middle panels). Cells expressing wild-type BMAL1 and CLOCK treated with LMB (10 ng/ml) for 5 h (lower panels). Quantitative data on the subcellular locations of the two proteins are presented on the right. (B) Role of NES in the transactivation of the CLOCK/BMAL1 heterodimer. Cells transfected with constructs encoding the Per1-luciferase reporter and the indicated transcriptional activators were incubated for 5 h in the presence or absence of LMB (10 ng/ml). Transcription of the reporter gene is expressed as luciferase activity relative to the response in the absence of activator. The data are shown as means ± the SEM of independent experiments (n = 3 to 5).
FIG. 6.
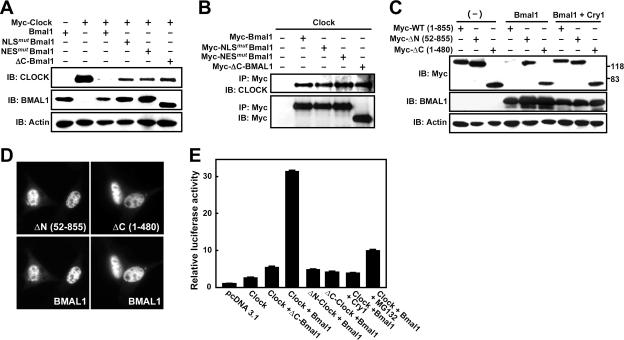
BMAL1-dependent proteolysis of CLOCK is closely related to its transcriptional activity. (A) Intact BMAL1 but not functionally defective BMAL1 induces drastic decreases in CLOCK levels. NIH 3T3 cells were transfected with Myc-Clock alone or together with wild-type Bmal1 or Bmal1 mutants and then analyzed by immunoblotting with the indicated antibodies. (B) Confirmation of protein-protein interactions between CLOCK and BMAL1 mutants. Cells were cotransfected with wild-type CLOCK and Myc-tagged Bmal1 constructs with various defects and then immunoprecipitated with anti-Myc antibodies. The immunoprecipitates were analyzed by immunoblotting with anti-CLOCK or anti-Myc. (C) BMAL1-dependent CLOCK degradation is prevented when CLOCK has functional defects or is coexpressed with CRY1. Cells were transfected with Myc-tagged Clock constructs encoding wild-type or transcriptionally inactive mutants [ΔN (52-855) lacking the DNA-binding domain and ΔC (1-480) lacking TAD] alone or together with Bmal1 and/or Cry1 and then analyzed by immunoblotting with the indicated antibodies. (D) The CLOCK mutants lacking DNA-binding domain or TAD colocalize with BMAL1 in the nucleus. Localization of the Myc-tagged CLOCK mutants and BMAL1 was detected with anti-Myc and anti-BMAL1 antibodies and visualized with TRITC and FITC, respectively. (E) Relative Per1 promoter-dependent luciferase reporter activity induced by combined transfection with the various forms of Clock and Bmal1. The rightmost bar is the result from cells transfected with wild-type Clock and Bmal1 and exposed to 50 μM MG132 for 5 h. Relative luciferase reporter activity is expressed as fold of the control. Each value is the mean ± the SEM of three replicates from a single assay. Similar results were obtained from replicated experiments.
Consistent with this, both the NES mutation of BMAL1 and LMB treatment markedly reduced CLOCK/BMAL1-dependent reporter gene expression (Fig. 4B). However, the use of an NLS-tagged CLOCK with its own nuclear import ability fully compensated for the inhibitory effect of the BMAL1 NES mutation on transcriptional activity but had only a moderate effect on LMB-mediated inhibition of transcription. This could be due to unexpected LMB effects such as defects in the transcriptional machinery, since LMB treatment blocks shuttling not only of BMAL1 but also of a number of nuclear proteins (4, 8, 19). Thus, these findings indicate that nucleocytoplasmic shuttling of BMALl is essential for nuclear accumulation of CLOCK and that this in turn leads to transactivation of the CLOCK/BMAL1 heterodimer.
CLOCK accelerates the turnover of BMAL1 via ubiquitin-dependent proteolysis.
It is understandable that mutation of the NLS of BMAL1 should diminish its transcriptional activity because the mutant form is no longer able to bring about nuclear translocation of the BMAL1/CLOCK complexes required for E-box-dependent clock gene transcription. However, it is somewhat puzzling that the BMAL1 NES mutant failed to accumulate CLOCK in the nucleus despite having intact nuclear import ability. A possible explanation is that nuclear translocation is coupled with proteolytic destruction of CLOCK and that therefore continuous transport of CLOCK to the nucleus is dependent on nucleocytoplasmic shuttling of BMAL1. Subcellular fractionation assays yielded results (Fig. 5A) consistent with this hypothesis; when CLOCK was coexpressed with BMAL1, the amount of exogenously expressed CLOCK in the cytoplasm dropped dramatically without any increase in the amount in the nucleus. In addition, previous findings have suggested that many unstable transcription factors, such as Jun, Fos, Myc, and p53, are rapidly degraded by the ubiquitin-mediated proteasome pathway in a process dependent on their own activation (27, 33, 34).
FIG. 5.
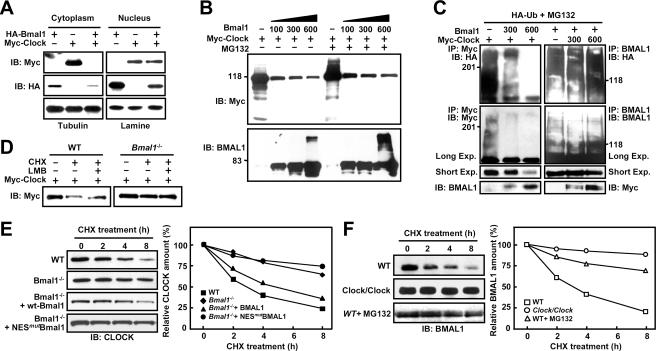
CLOCK and BMAL1 reduce each other's stability. (A) Subcellular fractionation of exogenous CLOCK and BMAL1 in NIH 3T3 cells transfected with HA-tagged Bmal1 and/or Myc-tagged Clock. (B) Effect of the proteasome inhibitor MG132 on the stability of CLOCK and BMAL1. Cells were transfected with a fixed amount of Clock DNA (600 ng) and the indicated amounts of Bmal1 DNA. At 36 h after transfection, they were incubated for 5 h with or without 50 μM MG132. (C) Ubiquitination of CLOCK and BMAL1. Cells were cotransfected with fixed amounts of Myc-Clock (600 ng) and increasing amounts of Bmal1 and vice versa in the presence of HA-ubiquitin. Cells were incubated with 50 μM MG132 for 5 h, and corresponding cell extracts were subjected to immunoprecipitation with anti-Myc or anti-BMAL1 antibodies. Immunoprecipitated proteins were then analyzed by immunoblotting with anti-HA antibodies. (D) Role of BMAL1 shuttling in the decreased CLOCK stability. Wild-type and BMAL1-deficient MEFs transfected with Myc-Clock (600 ng) were treated with CHX (30 μg/ml) alone or with LMB (10 ng/ml) for 5 h and then subjected to immunoblotting with anti-Myc antibodies. (E) Stability of endogenous CLOCK in wild-type and BMAL1-deficient MEFs. BMAL1-deficient cells transfected with wild-type Bmal1 or NESmut Bmal1 were exposed to CHX (30 μg/ml) for the indicated times, and CLOCK levels were assessed by immunoblotting at the indicated times after drug treatment. (F) Stability of endogenous BMAL1 in wild-type and Clock/Clock mutant MEFs. BMAL1 levels were assessed by the same experimental procedure as the CLOCK analysis, except that one set of Clock mutant cells were incubated with MG132 (50 μM) together with CHX (30 μg/ml) until cells were harvested at the indicated time points.
To further test this hypothesis, we examined the effect of the proteasome inhibitor, MG132, on BMAL1-induced CLOCK degradation. The results of this experiment are shown in Fig. 5B. In the absence of the 26S proteasome inhibitor, CLOCK protein levels decreased markedly with increasing amounts of coexpressed BMAL1. Treatment with MG132 resulted in the accumulation of slowly migrating bands in the cells transfected with Clock alone but did not dramatically alter BMAL1-induced CLOCK degradation. Interestingly, under this condition, the levels of both the naked and the slowly migrating BMAL1s were significantly increased by MG132. We next tested whether ubiquitin bound covalently to these proteins. HA-tagged ubiquitin was cotransfected with a fixed amount of CLOCK and increasing amounts of BMAL1, and vice versa, and ubiquitination of the two proteins was visualized with anti-HA antibodies in immunoprecipitates with anti-Myc or anti-BMAL1 antibodies (Fig. 5C). Consistent with the results from Western blot analysis, CLOCK on its own was strongly ubiquitinated, and coexpression of BMAL1 abolished its ubiquitination, whereas the ubiquitination of BMAL1 increased in parallel with the amount of coexpressed CLOCK. Since endogenous CLOCK did not reveal detectable ubiquitination in the cells, regardless of exogenous expression of BMAL1 (data not shown), the massive ubiquitination of CLOCK without coexpression of BMAL1 appears to be caused by the overexpression of CLOCK itself. Thus, these observations indicate that the ubiquitin-proteasome pathway is implicated in CLOCK-dependent BMAL1proteolysis but not in CLOCK degradation induced by BMAL1.
To further examine whether endogenous BMAL1 reduces the stability of CLOCK and, if so, whether shuttling of BMAL1 is involved in this effect, we compared the stability of CLOCK in BMAL1-deficient MEFs and in wild-type MEFs (Fig. 5D). After blocking de novo protein synthesis with CHX for 5 h, the wild-type fibroblasts had severely reduced CLOCK levels, whereas the BMAL1-deficient fibroblasts did not. Under the same conditions, LMB inhibited CLOCK turnover in the wild-type cells, but no similar effect was evident in the BMAL1-deficient cells. Moreover, assessment of protein half-life using CHX treatment revealed that endogenous CLOCK levels in the wild-type fibroblasts fell rapidly, with an estimated half-life of ∼3 h (Fig. 5E), whereas in the BMAL1-deficent fibroblasts its half-life was extended to ∼12 h. The latter half-life was reduced to ∼5 h by introducing wild-type BMAL1 but not the BMAL1 NES mutant, implying that endogenous BMAL1 affects the stability of CLOCK and that its shuttling activity is crucial for CLOCK turnover. We also compared BMAL1 stability in Clock mutant fibroblasts and wild-type cells. Turnover of BMAL1 was delayed (>12 h) in the Clock mutant and had a half-life similar to that of CLOCK (∼ 4 h) (Fig. 5F). In addition, MG132 efficiently inhibited the rapid degradation of BMAL1 in wild-type cells. Thus, it is likely that CLOCK indeed plays an essential role in proteasome-dependent BMAL1 degradation in vivo.
BMAL1-induced CLOCK proteolysis is coupled with transactivation of the CLOCK/BMAL1 heterodimer.
Recently, it was discovered that the transcriptional activation domains (TADs) of most of the unstable transcription factors overlap functionally with the protein degradation domains (27). As shown above, nuclear translocation is essential for transactivation of the CLOCK/BMAL1 heterodimer. To examine whether the transcription factor activity of BMAL1 is directly connected to CLOCK degradation, we compared CLOCK levels in NIH 3T3 cells coexpressing full-length BMAL1 or BMAL1 variants with specific defects (Fig. 6A). As expected, coexpression with wild-type BMAL1 abolished detectable CLOCK accumulation, whereas neither the NLS mutant nor the NES mutant promoted CLOCK degradation. Moreover, a C-terminally truncated BMAL1 mutant (mutant 1-503) lacking a TAD (positions 588 to 625) failed to induce proteolysis of CLOCK (Fig. 6A) and gene expression from the Per1 promoter (Fig. 6E), although it was mainly located in the nucleus together with CLOCK and had an intact NLS and NES (Fig. 1D and E). All of these BMAL1 mutants were coimmunoprecipitated with CLOCK, confirming that the inhibition of BMAL1-induced CLOCK degradation was not due to the inhibition of heterodimerization (Fig. 6B). The functional defects revealed by these observations on the BMAL1 mutants converge on loss of transcription-enhancing activity, even though the NLS and NES mutations abolish nucleocytoplasmic shuttling. Thus, it appears that BMAL1 promotes CLOCK degradation as a consequence of transactivation rather than nuclear translocation or heterodimerization per se.
To determine whether the transcriptional activity of CLOCK is also required for BMAL1-induced CLOCK degradation, we generated two Myc-tagged CLOCK mutants (52-855 and 1-480), which lack DNA binding or transcription activation domains, and coexpressed them with full-length BMAL1 in the presence or absence of exogenous CRY1. Immunoblotting analysis showed that BMAL1 failed to abolish the accumulation of these CLOCK mutants (Fig. 6C), despite the results from immunofluorescence analysis demonstrating that the mutant proteins were predominantly localized in the nucleus together with BMAL1 (Fig. 6D). Moreover, coexpression of CRY1, a strong negative regulator of CLOCK/BMAL1-dependent transcription, prevented BMAL1-induced degradation of wild-type CLOCK (Fig. 6C). To evaluate the correlation between the transcription enhancing activity of CLOCK and its proteolysis, we performed a transcriptional activity assay in NIH 3T3 cells using a luciferase reporter fused to the Per1 promoter (Fig. 6E). Introduction of CLOCK on its own slightly increased Per1 promoter-dependent gene expression, whereas the combined expression of wild-type CLOCK and BMAL1 caused a marked increase that was abolished by CRY1, as described previously (22, 52). In sharp contrast, when either component of the CLOCK/BMAL1 heterodimer was replaced with a mutant lacking an essential domain for transactivation, the dimeric transcription factor no longer elicited robust induction of reporter gene expression. Interestingly, treatment with MG132, a potent inhibitor of the 26S proteasome, also diminished CLOCK/BMAL1-mediated transcription. Taken together, these findings indicate that transcriptional activation by CLOCK/BMAL1 is tightly coupled to degradation of the two proteins and also implies that this degradation is a prerequisite for E-box-controlled clock gene transcription rather than a necessary consequence of it.
Low BMAL1 abundance indicates high transcriptional activity during the circadian cycle.
We have shown that CRY1 blocks the proteolysis of CLOCK and BMAL1, as well as their transcriptional activity as heterodimers. Lee et al. (23) found that, in the liver, the nuclear accumulation and/or abundance of CLOCK and BMAL1 unexpectedly reached their lowest levels between CT 6 and 9, the peak time for Per1 and Per2 gene transcription, and attained their highest levels between CT 18 and 21 in parallel with maximal CRY levels. Moreover, the levels of both proteins were marginal in Cry-deficient mice and showed no obvious oscillation. These observations led us to speculate that CRYs play an important role in the circadian accumulation of CLOCK and BMAL1. To test this possibility, we monitored the real-time circadian rhythmicity of clock gene expression in wild-type MEFs and Cry-deficient fibroblasts synchronized by treatment with dexamethasone (Dex), a strong inducer of circadian gene expression in culture models (2, 44). For this experiment, we fused the full-length Per2 promoter (1.6 kb) to a firefly luciferase (dsLuc) reporter gene carrying a rapid degradation signal (PEST sequence [32]) at its C terminus to reduce the reporter half-life (<0.5 h) and transiently transfected this reporter into wild-type and Cry-deficient cells. Real-time luciferase assays (see Materials and Methods) revealed that Dex induced robust oscillations of bioluminescence with an approximately 24-h periodicity in the wild-type cells but not in the Cry-deficient cells (Fig. 7A).
FIG. 7.
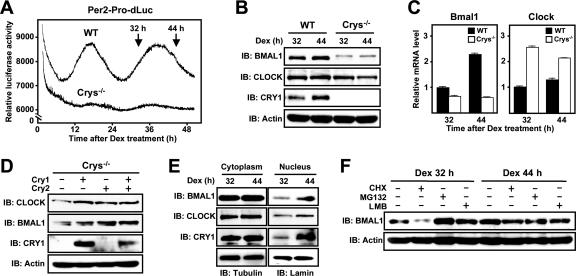
Circadian regulation of the abundance and the stability of CLOCK and BMAL1 in vivo. (A) Real-time measurement of Per2 promoter-controlled destabilized luciferase (Per2-Pro-dsLuc) activity in live cells. Wild-type MEFs (WT) and Cry-deficient MEFs (Crys−/−) carrying Per2-Pro-dsLuc with the Per2 promoter were synchronized by treatment with 1 μM Dex, and their luciferase activities were monitored in real time (see Materials and Methods). (B) Changes in the levels of BMAL1, CLOCK, and CRY1 during the circadian cycle. Each protein was analyzed at the times corresponding to the maximal and minimal transcription of Per2-Pro-dsLuc, respectively (32 and 44 h). (C) Relative mRNA levels of Bmal1 and Clock at the same circadian time points (32 h and 44 h) in wild-type and Cry-deficient cells. mRNA levels in the wild-type (▪) and Cry-deficient cells (□) were quantified by real-time PCR (see Materials and Methods). The data are presented as means ± the SEM (n = 3). (D) Transient expression of CRY1 and CRY2 in Cry-deficient cells increases endogenous CLOCK and BMAL1 levels. The cellular levels of the indicated proteins were analyzed by immunoblotting at 24 h after transfection with Cry-expressing plasmids. (E) Nuclear accumulation of CRY1 is correlated with accumulation of BMAL1 and CLOCK during the circadian cycle. The levels of each protein in nuclear and cytoplasmic fractions of wild-type MEFs were assessed by immunoblotting at 32 and 44 h after Dex treatment. (F) Massive degradation of BMAL1 via the ubiquitin-proteasome pathway during the transcriptionally active phase. Wild-type cells were treated with CHX (30 μg/ml), MG132 (50 μM), and LMB (10 ng/ml) 4 h before the indicated times and were subjected to immunoblotting with the indicated antibodies.
Next, we analyzed the levels of CLOCK and BMAL1 protein and RNA in these MEFs at the times when they appear to have maximal and minimal Per2 transcriptional activity (Dex 32 h and Dex 44 h, respectively) (Fig. 7B and C). BMAL1 abundance in total lysates of the wild-type cells was lower at the transcription maximum than at the transcription minimum, thus showing a pattern similar to the CRY1 protein profile; on the other hand, there was no significant change in the abundance of CLOCK protein. In sharp contrast, in the Cry-deficient fibroblasts, BMAL1 protein abundance was extremely low, and BMAL1 was hyperphosphorylated at both time points despite the presence of mRNA levels comparable to those in the wild-type cells. In the same cells, CLOCK protein levels were also low relative to the Clock transcript levels. Moreover, the introduction of CRY1 and/or CRY2 into the Cry-deficient cells led to a significant increase in both BMAL1 and CLOCK levels (Fig. 7D). These findings provide strong evidence that CRY proteins are involved in stabilizing BMAL1 and CLOCK in vivo.
To further dissect the molecular events leading to circadian oscillations of CLOCK and BMAL1, we examined their subcellular distribution in the synchronized MEFs at the same two time points. Fractionation experiments demonstrated that the decrease in BMAL1 levels at the peak of Per2 transcription (Dex 32 h) was mainly due to reduced nuclear accumulation rather than cytosolic accumulation (Fig. 7E). Similarly, the CLOCK protein was also lower in the nucleus at the Dex 32-h point, whereas the level of the protein in total lysates was little changed. These reductions in nuclear CLOCK and BMAL1 were tightly correlated with the nuclear profile of CRY1. Furthermore, treatment with CHX during the transcriptionally active phase elicited a drastic decrease in BMAL1 level, and treatment with either MG132 or LMB led to protein levels that were even higher than the peak levels in the untreated cells (Fig. 7F). All of these drug effects, however, were blunted at the phase of minimal transcription when CRY proteins accumulate in the nucleus (Fig. 7F). Thus, it appears that the nucleocytoplasmic shuttling activity of BMAL1 facilitates its own proteasomal degradation and that the CRYs inhibit this process in the nucleus, causing nuclear accumulation of the heterodimeric factor at the same time as abolishing its transcriptional activation.
DISCUSSION
The heterodimeric transcription factor CLOCK/BMAL1 plays an indispensable role in generating daily rhythms in mammals by occupying the positive limb of the transcription-translation feedback loop of the molecular clock. Throughout the circadian cycle, the mRNA and protein profiles of BMAL1 display robust rhythmic changes without time delays, whereas CLOCK is constitutively expressed. A recent study suggested that BMAL1 mediates the nuclear translocation of CLOCK in a circadian manner and that this BMAL1-dependent translocation provides an additional clock regulatory mechanism by generating periodic availability of the heterodimeric transactivation complex (20). In addition, it has been demonstrated that the absolute molar concentration of BMAL1 is much lower than that of CLOCK in vivo (23). On the basis of these findings, it is widely accepted that BMAL1 abundance is rate limiting for the transcriptional activation of CLOCK/BMAL1 heterodimers.
Ironically, however, BMAL1 protein levels reach a trough when BMAL1 is displaying the greatest transcriptional activity and peak during the transcription inhibition phase in diverse tissues and cells, including the SCN (23, 41, 43) (Fig. 7). Furthermore, the decline in BMAL1 at the transcriptional peaks is not due to a reduction of the protein in the cytosol but in the nucleus (Fig. 7). To explain these paradoxical results, we have dissected the molecular events underlying transcriptional activation of CLOCK/BMAL1. Our findings lead us to propose the model of the molecular clock shown in Fig. 8, which we believe accounts for most previous findings.
FIG. 8.
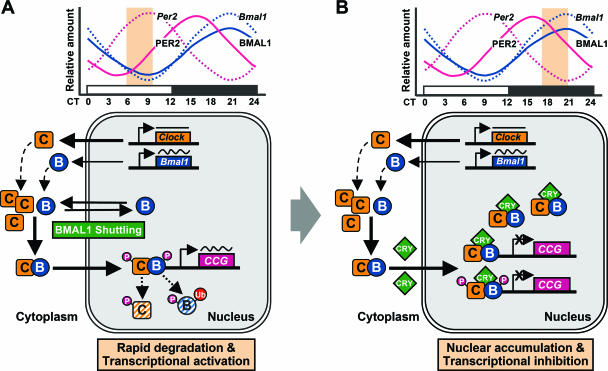
Model of the circadian regulation of the degradation of CLOCK/BMAL1 coupled with its functional activation. (A) BMAL1 shuttling promotes the rapid degradation and functional activation of CLOCK/BMAL1 heterodimers during the transcriptionally active stage. (B) CRY proteins impede the rapid degradation and functional activation of CLOCK/BMAL1 heterodimers in the transcriptional inhibition stage. The upper panels are schematic diagrams of the temporal regulation of Per2 (pink) and Bmal1 (blue) transcripts (dotted line) and proteins (solid line). Yellow-shaded boxes indicate the periods of transcriptional activation (A) and inhibition (B), respectively. The lower panels show the molecular events regulating the transcriptional activity and stability of CLOCK/BMAL1 during the indicated circadian phases. C (yellow box), CLOCK; B (blue circle), BMAL1; CRY (green diamond), cryptochrome; P (pink circle), phosphate; Ub (red circle), ubiquitin; CCG (pink box), core clock genes.
During the transcriptional activation stage, BMAL1 shuttling promotes nuclear translocation of CLOCK and E-box-dependent clock gene transcription, coupled with rapid proteolysis of both BMAL1 and CLOCK via ubiquitin-dependent and -independent pathways, respectively (Fig. 8A). Thus, transcription-coupled protein degradation seems to be sufficient to lead to the BMAL1 protein minimum as well as nuclear depletion of CLOCK/BMAL1 despite the massive de nova synthesis of BMAL1 required for transcriptional activation at the peak of target gene transcription. Indeed, treatment of wild-type MEFs with MG132 at this time dramatically increased BMAL1 levels, whereas CHX treatment reduced them (Fig. 7F). In contrast, at the transcriptional minimum, the increased levels of the CRY proteins facilitate the binding of CRY to the CLOCK/BMAL1 complex, thereby stabilizing this complex, as well as suppressing its transcriptional activity (Fig. 8B). This notion is supported by the fact that neither MG132 nor CHX caused a significant change of BMAL1 abundance in the cells at the phase of minimum transcription (Fig. 7F). Cry-deficient cells, however, had low protein levels relative to their mRNA levels, and this effect was reversed by the exogenous expression of CRY1 and/or CRY2 (Fig. 7B and D). Therefore, the increased CRY levels should permit nuclear accumulation of CLOCK/BMAL1 without massive protein synthesis.
Our model emphasizes that transactivation of CLOCK/BMAL1 is tightly coupled with its degradation. This scenario is consistent with the recently proposed “black widow model” (27). According to this model, most unstable transcription factors, including Jun, Fos, Myc, p53, and HIF1-α, possess TADs that overlap functionally with their degradation signals and control transcription of downstream genes by mechanisms involving the ubiquitin-proteasome system. Processes that limit the activity, location, and abundance of transcription activators might play an important role in keeping their functions tightly in check, and proteolysis could be a major mechanism regulating transcriptional activity. Our results demonstrate that the active complex of BMAL1 and CLOCK promotes rapid proteolysis of both its component proteins in an activation-dependent manner, like a “one shot, one kill” mechanism, although only BMAL1 degradation is mediated by the ubiquitin-proteasome pathway. It might be, therefore, that the rapid and controlled destruction of CLOCK/BMAL1 allows tight control of periodicity by ensuring that activation of the target gene is linked to ongoing synthesis of its transcriptional regulator.
There are some other posttranslational modifications that have been postulated to be required for regulating activity and/or stability of CLOCK/BMAL1, although their functional relevance is still elusive. A prominent example is phosphorylation. Biochemical analysis has revealed that hyperphosphorylation of both proteins occurs in the nucleus but not in the cytoplasm (20, 23). This event could play a substantial role in the activation-coupled degradation of them. Indeed, several previous studies have demonstrated that phosphorylation is a prerequisite step for the ubiquitin-dependent proteolysis, including PER2 protein (10, 16, 40). Another possible mechanism implicated in functional regulation of the CLOCK/BMAL1 heterodimer is SUMO modification. Recently, Sassone-Corsi and coworkers have demonstrated that BMAL1 undergoes rhythmic sumoylation in parallel with Per gene transcription, and this process is required for circadian expression of BMAL1 and for its instability (6). Thus, investigation of the relationship between ubiquitination and sumoylation of BMAL1 could provide insight into the precise mechanisms that control CLOCK/BMAL1-mediated circadian gene activation.
The molecular basis of the mammalian circadian clock has been largely uncovered, but there are several important questions remain unanswered. One such question is what is the mechanism that causes the fundamental feedback loop to oscillate stably with an approximately 24-h periodicity? The general delay model assumes that the most important parameter driving this periodicity is a “time-delay event” that regulates the production of functional clock proteins from their mRNAs (24, 35). In mammals, both the negative and the positive elements of the feedback loop, Per1, Per2, and Bmal1, exhibit a robust circadian rhythm with respect to mRNA levels, but the rhythmicity of Bmal1 mRNA is antiphase to the oscillations of the Per genes (9, 49). The time lags between the transcription and translation of the Per genes are around 6 h in rodent tissues and cells (13, 23, 47). On the basis of the empirical data, a working model for the mammalian clock hypothesizes that that the Bmal1 mRNA rhythm drives the BMAL1 protein rhythm with a 4- to 6-h delay, because this would increase the availability of CLOCK/BMAL1 heterodimers at the time they are required to drive the transcription of the Per and Cry genes (38). Unexpectedly, however, there is little delay between the appearance of Bmal1 mRNA and BMAL1 protein (23, 41). Our present findings provide several indications that transcriptional activation of CLOCK/BMAL1 is tightly associated with its degradation. This rapid activation-coupled degradation may alter the apparent BMAL1 profile by masking the absolute amount of newly made BMAL1, thereby filling the temporal gap between synthesis of its RNA and of its protein. Thus, an alternative mechanism explaining how a 24-h time constant is built into the molecular clockwork would include this hidden time delay between the transcription and translation of Bmal1.
Finally, it is important to establish whether the activation-coupled degradation of BMAL1 is essential for the timing keeping system in the SCN. This issue needs to be explored further because we cannot rule out the possibility that there is some functional redundancy within the SCN and not within peripheral clocks; this would allow the SCN to dominate the peripheral oscillators even in the presence of a genetic defect that disrupts normal clock function (29). However, the fact that BMAL1 levels in the SCN of rats also reach a trough at the time predicted for the peak of Per gene transcription (41) supports our contention that the rapid degradation of BMAL1 during the transcriptional active phase may be an intrinsic feature of core clockwork throughout the entire organism and reinforces the idea of a hidden time delay between the transcription and translation of Bmal1.
In conclusion, we have demonstrated that BMAL1 shuttles between the cytoplasm and the nucleus, using its functional NLS and NES, to translocate CLOCK to the nucleus and thereby allow dynamic control of CLOCK/BMAL1 transactivation, which is tightly coupled with its degradation via ubiquitin-dependent or -independent pathways. These findings reveal a new aspect of the molecular clock suggesting that the decrease in BMAL1 abundance during the circadian cycle is due to its robust transcriptional activation rather than inhibition of its synthesis.
Acknowledgments
We thank M. Antoch for generous gifts of the embryonic fibroblasts from clock-defective mice.
This study was supported by the Brain Research Center of the 21st Century Frontier Program in Neuroscience from the Korean Ministry of Science and Technology. K.H.L. was supported by the BK 21 program from the Korean Ministry of Education and Human Resources Development.
REFERENCES
- 1.Balsalobre, A., F. Damiola, and U. Schibler. 1998. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell 93:929-937. [DOI] [PubMed] [Google Scholar]
- 2.Balsalobre, A., S. A. Brown, L. Marcacci, F. Tronche, C. Kellendonk, H. M. Reichardt, G. Schutz, and U. Schibler. 2000. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science 289:2344-2347. [DOI] [PubMed] [Google Scholar]
- 3.Bell-Pedersen, D., V. M. Cassone, D. J. Earnest, S. S. Golden, P. E. Hardin, T. L. Thomas, and M. J. Zoran. 2005. Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat. Rev. Genet. 6:544-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bittinger, M. A., E. McWhinnie, J. Meltzer, V. Iourgenko, B. Latario, X. Liu, C. H. Chen, C. Song, D. Garza, and M. Labow. 2004. Activation of cAMP response element-mediated gene expression by regulated nuclear transport of TORC proteins. Curr. Biol. 14:2156-2161. [DOI] [PubMed] [Google Scholar]
- 5.Bunger, M. K., L. D. Wilsbacher, S. M. Moran, C. Clendenin, L. A. Radcliffe, J. B. Hogenesch, M. C. Simon, J. S. Takahashi, and C. A. Bradfield. 2000. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell 103:1009-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cardone, L., J. Hirayama, F. Giordano, T. Tamaru, J. J. Palvimo, and P. Sassone-Corsi. 2005. Circadian clock control by SUMOylation of BMAL1. Science 309:1390-1394. [DOI] [PubMed] [Google Scholar]
- 7.Chestukhin, A., L. Litovchick, K. Rudich, and J. A. DeCaprio. 2002. Nucleocytoplasmic shuttling of p130/RBL2: novel regulatory mechanism. Mol. Cell. Biol. 22:453-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cushman, I., D. Stenoien, and M. S. Moore. 2004. The dynamic association of RCC1 with chromatin is modulated by Ran-dependent nuclear transport. Mol. Biol. Cell 15:245-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunlap, J. C. 1999. Molecular bases for circadian clocks. Cell 96:271-290. [DOI] [PubMed] [Google Scholar]
- 10.Eide, E. J., M. F. Woolf, H. Kang, P. Woolf, W. Hurst, F. Comacho, E. L. Vielhaber, A. Giovanni, and D. M. Virshrup. 2005. Control of mammalian circadian rhythm by CKIɛ-regulated proteasome-mediated PER2 degradation. Mol. Cell. Biol. 25:2795-2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gekakis, N., D. Staknis, H. B. Nguyen, F. C. Davis, L. D. Wilsbacher, D. P. King, J. S. Takahashi, and C. J. Weitz. 1998. Role of the CLOCK protein in the mammalian circadian mechanism. Science 280:1564-1569. [DOI] [PubMed] [Google Scholar]
- 12.Griffin, E. A., Jr., D. Staknis, and C. J. Weitz. 1999. Light-independent role of CRY1 and CRY2 in the mammalian circadian clock. Science 286:768-771. [DOI] [PubMed] [Google Scholar]
- 13.Hardin, P. E. 2004. Transcription regulation within the circadian clock: the E-box and beyond. J. Biol. Rhythms 19:348-360. [DOI] [PubMed] [Google Scholar]
- 14.Hogenesch, J. B., Y. Z. Gu, S. Jain, and C. A. Bradfield. 1998. The basic-helix-loop-helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proc. Natl. Acad. Sci. USA 95:5474-5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hastings, M. H., A. B. Reddy, and E. S. Maywood. 2003. A clockwork web: circadian timing in brain and periphery, in health and disease. Nat. Rev. Neurosci. 4:649-661. [DOI] [PubMed] [Google Scholar]
- 16.Huang, H., K. M. Regan, F. Wang, D. Wang, D. I. Smith, and J. M. A. van Deursen. 2005. Skp2 inhibits FOXO1 in tumor suppression through ubiquitin-mediated degradation. Proc. Natl. Acad. Sci. USA 102:1649-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin, X., L. P. Shearman, D. R. Weaver, M. J. Zylka, G. J. de Vries, and S. M. Reppert. 1999. A molecular mechanism regulating rhythmic output from the suprachiasmatic circadian clock. Cell 96:57-68. [DOI] [PubMed] [Google Scholar]
- 18.Jung, H., Y. Choe, H. Kim, N. Park, G. H. Son, I. Khang, and K. Kim. 2003. Involvement of CLOCK:BMAL1 heterodimer in serum-responsive mPer1 induction. Neuroreport 14:15-19. [DOI] [PubMed] [Google Scholar]
- 19.Knauer, S. K., G. Carra, and R. H. Stauber. 2005. Nuclear export is evolutionarily conserved in CVC paired-like homeobox proteins and influences protein stability, transcriptional activation, and extracellular secretion. Mol. Cell. Biol. 25:2573-2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kondratov, R. V., M. V. Chernov, A. A. Kondratova, V. Y. Gorbacheva, A. V. Gudkov, and M. P. Antoch. 2003. BMAL1-dependent circadian oscillation of nuclear CLOCK: posttranslational events induced by dimerization of transcriptional activators of the mammalian clock system. Genes. Dev. 17:1921-1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kudo, N., N. Matsumori, H. Taoka, D. Fujiwara, E. P. Schreiner, B. Wolff, M. Yoshida, and S. Horinouchi. 1999. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc. Natl. Acad. Sci. USA 96:9112-9117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kume, K., M. J. Zylka, S. Sriram, L. P. Shearman, D. R. Weaver, X. Jin, E. S. Maywood, M. H. Hastings, and S. M. Reppert. 1999. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 98:193-205. [DOI] [PubMed] [Google Scholar]
- 23.Lee, C., J. P. Etchegaray, F. R. Cagampang, A. S. Loudon, and S. M. Reppert. 2001. Posttranslational mechanisms regulate the mammalian circadian clock. Cell 107:855-867. [DOI] [PubMed] [Google Scholar]
- 24.Lema, M. A., D. A. Golombek, and J. Echave. 2000. Delay model of the circadian pacemaker. J. Theor. Biol. 204:565-573. [DOI] [PubMed] [Google Scholar]
- 25.Mattaj, I. W., and L. Englmeier. 1998. Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem. 67:265-306. [DOI] [PubMed] [Google Scholar]
- 26.Moore, R. Y. 1997. Circadian rhythms: basic neurobiology and clinical applications. Annu. Rev. Med. 48:253-266. [DOI] [PubMed] [Google Scholar]
- 27.Muratani, M., and W. P. Tansey. 2003. How the ubiquitin-proteasome system controls transcription. Nat. Rev. Mol. Cell. Biol. 4:192-201. [DOI] [PubMed] [Google Scholar]
- 28.Okamura, H., S. Miyake, Y. Sumi, S. Yamaguchi, A. Yasui, M. Muijtjens, J. H. Hoeijmakers, and G. T. van der Horst. 1999. Photic induction of mPer1 and mPer2 in cry-deficient mice lacking a biological clock. Science 286:2531-2534. [DOI] [PubMed] [Google Scholar]
- 29.Pando, M. P., D. Morse, N. Cermakian, and P. Sassone-Corsi. 2002. Phenotypic rescue of a peripheral clock genetic defect via SCN hierarchical dominance. Cell 110:107-117. [DOI] [PubMed] [Google Scholar]
- 30.Reppert, S. M., and D. R. Weaver. 2002. Coordination of circadian timing in mammals. Nature 418:935-941. [DOI] [PubMed] [Google Scholar]
- 31.Reyes, J. C., C. Muchardt, and M. Yaniv. 1997. Components of the human SWI/SNF complex are enriched in active chromatin and are associated with the nuclear matrix. J. Cell Biol. 137:263-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rogers, S., R. Wells, and M. Rechsteiner. 1986. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science 234:364-368. [DOI] [PubMed] [Google Scholar]
- 33.Salghetti, S. E., M. Muratani, H. Wijnen, B. Futcher, and W. P. Tansey. 2000. Functional overlap of sequences that activate transcription and signal ubiquitin-mediated proteolysis. Proc. Natl. Acad. Sci. USA 97:3118-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salghetti, S. E., A. A. Caudy, J. G. Chenoweth, and W. P. Tansey. 2001. Regulation of transcriptional activation domain function by ubiquitin. Science 293:1651-1653. [DOI] [PubMed] [Google Scholar]
- 35.Scheper, T., D. Klinkenberg, C. Pennartz, and J. van Pelt. 1999. A mathematical model for the intracellular circadian rhythm generator. J. Neurosci. 19:40-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schibler, U., and P. Sassone-Corsi. 2002. A web of circadian pacemakers. Cell 111:919-922. [DOI] [PubMed] [Google Scholar]
- 37.Schibler, U., and F. Naef. 2005. Cellular oscillators: rhythmic gene expression and metabolism. Curr. Opin. Cell Biol. 17:223-229. [DOI] [PubMed] [Google Scholar]
- 38.Shearman, L. P., S. Sriram, D. R. Weaver, E. S. Maywood, I. Chaves, B. Zheng, K. Kume, C. C. Lee, G. T. van der Horst, M. H. Hastings, and S. M. Reppert. 2000. Interacting molecular loops in the mammalian circadian clock. Science 288:1013-1019. [DOI] [PubMed] [Google Scholar]
- 39.Son, G. H., H. Jung, J. Y. Seong, Y. Choe, D. Geum, and K. Kim. 2003. Excision of the first intron from the gonadotropin-releasing hormone (GnRH) transcript serves as a key regulatory step for GnRH biosynthesis. J. Biol. Chem. 278:18037-18044. [DOI] [PubMed] [Google Scholar]
- 40.Srinivas, H., D. M. Juroske, S. Kalyankrishna, D. D. Cody, R. E. Price, X. Xu, R. Narayanan, N. L. Weigel, and J. M. Jurie. 2005. c-Jun N-terminal kinase contributes to aberrant retinoid signaling in lung cancer cells by phosphorylating and inducing proteasomal degradation of retinoic acid receptor α. Mol. Cell. Biol. 25:1054-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tamaru, T., Y. Isojima, T. Yamada, M. Okada, K. Nagai, and K. Takamatsu. 2000. Light and glutamate-induced degradation of the circadian oscillating protein BMAL1 during the mammalian clock resetting. J. Neurosci. 20:7525-7530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tamaru, T., Y. Isojima, G. T. van der Horst, K. Takei, K. Nagai, and K. Takamatsu. 2003. Nucleocytoplasmic shuttling and phosphorylation of BMAL1 are regulated by circadian clock in cultured fibroblasts. Genes Cells 12:973-983. [DOI] [PubMed] [Google Scholar]
- 43.Torres-Farfan, C., M. Seron-Ferre, V. Dinet, and H. W. Korf. 2006. Immuno cytochemical demonstration of day/night changes of clock gene protein levels in the murine adrenal gland: differences between melatonin-proficient (C3H) and melatonin-deficient (C57BL) mice. J. Pineal Res. 40:64-70. [DOI] [PubMed] [Google Scholar]
- 44.Ueda, H. R., W. Chen, A. Adachi, H. Wakamatsu, S. Hayashi, T. Takasugi, M. Nagano, K. Nakahama, Y. Suzuki, S. Sugano, M. Lino, Y. Shigeyoshi, and S. Hashimoto. 2002. A transcription factor response element for gene expression during circadian night. Nature 418:534-539. [DOI] [PubMed] [Google Scholar]
- 45.van der Horst, G. T., M. Muijtjens, K. Kobayashi, R. Takano, S. Kanno, M. Takao, J. de Wit, A. Verkerk, A. P. Eker, D. van Leenen, R. Buijs, D. Bootsma, J. H. Hoeijmakers, and A. Yasui. 1999. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature 398:627-630. [DOI] [PubMed] [Google Scholar]
- 46.Vitaterna, M. H., C. P. Selby, T. Todo, H. Niwa, C. Thompson, E. M. Fruechte, K. Hitomi, R. J. Thresher, T. Ishikawa, J. Miyazaki, J. S. Takahashi, and A. Sancar. 1999. Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc. Natl. Acad. Sci. USA 96:12114-12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yagita, K., F. Tamanini, G. T. van Der Horstqq, and H. Okamura. 2001. Molecular mechanisms of the biological clock in cultured fibroblasts. Science 292:278-281. [DOI] [PubMed] [Google Scholar]
- 48.Yagita, K., F. Tamanini, M. Yasuda, J. H. J. Hoeijmakers, G. T. van der Horst, and H. Okamura. 2002. Nucleocytoplasmic shuttling and mCRY-dependent inhibition of ubiquitylation of the mPER2 clock protein. EMBO J. 21:1301-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamamoto, T., Y. Nakahata, H. Soma, M. Akashi, T. Mamine, and T. Takumi. 2004. Transcriptional oscillation of canonical clock genes in mouse peripheral tissues. BMC Mol. Biol. 5:18-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamazaki, S., R. Numano, M. Abe, A. Hida, R. Takahashi, M. Ueda, G. D. Block, Y. Sakaki, M. Menaker, and H. Tei. 2000. Resetting central and peripheral circadian oscillators in transgenic rats. Science 288:682-685. [DOI] [PubMed] [Google Scholar]
- 51.Yoo, S. H., S. Yamazaki, P. L. Lowrey, K. Shimomura, C. H. Ko, E. D. Buhr, S. M. Siepka, H. K. Hong, W. J. Oh, O. J. Yoo, M. Menaker, and J. S. Takahashi. 2004. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc. Natl. Acad. Sci. USA 101:5339-5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu, H., F. Conte, and C. B. Green. 2003. Nuclear localization and transcriptional repression are confined to separable domains in the circadian protein CRYPTOCHROME. Curr. Biol. 13:1653-1658. [DOI] [PubMed] [Google Scholar]