Abstract
The reciprocal expression of GATA-1 and GATA-2 during hematopoiesis is an important determinant of red blood cell development. Whereas Gata2 is preferentially transcribed early in hematopoiesis, elevated GATA-1 levels result in GATA-1 occupancy at sites upstream of the Gata2 locus and transcriptional repression. GATA-2 occupies these sites in the transcriptionally active locus, suggesting that a “GATA switch” abrogates GATA-2-mediated positive autoregulation. Chromatin immunoprecipitation (ChIP) coupled with genomic microarray analysis and quantitative ChIP analysis with GATA-1-null cells expressing an estrogen receptor ligand binding domain fusion to GATA-1 revealed additional GATA switches 77 kb upstream of Gata2 and within intron 4 at +9.5 kb. Despite indistinguishable GATA-1 occupancy at −77 kb and +9.5 kb versus other GATA switch sites, GATA-1 functioned uniquely at the different regions. GATA-1 induced histone deacetylation at and near Gata2 but not at the −77 kb region. The −77 kb region, which was DNase I hypersensitive in both active and inactive states, conferred equivalent enhancer activities in GATA-1- and GATA-2-expressing cells. By contrast, the +9.5 kb region exhibited considerably stronger enhancer activity in GATA-2- than in GATA-1-expressing cells, and other GATA switch sites were active only in GATA-1- or GATA-2-expressing cells. Chromosome conformation capture analysis demonstrated higher-order interactions between the −77 kb region and Gata2 in the active and repressed states. These results indicate that dispersed GATA factor complexes function via long-range chromatin interactions and qualitatively distinct activities to regulate Gata2 transcription.
Complex developmental processes, including blood cell development from stem cells or hematopoiesis, are intricately regulated via transcriptional networks. Constituents of such networks include cell type-specific and ubiquitous transcription factors that constitute large families of homologous proteins. For example, two of the six GATA transcription factor family members, GATA-1 and GATA-2, have unique and overlapping biological roles in regulating erythropoiesis (4, 6). Whereas GATA-2 is expressed in multipotent hematopoietic precursors, endothelial cells, mast cells, and neurons (12, 28, 29, 37-39, 42, 57, 67), GATA-1 is expressed predominantly in erythroid, megakaryocytic, and eosinophil cell lineages (15, 38, 58, 68). These unique expression patterns reflect certain distinct biological functions, since GATA-1 is essential for erythropoiesis (46, 51, 62), whereas GATA-2 regulates the development and function of multipotent hematopoietic precursor cells (32, 56, 57).
Despite certain distinct biological functions, GATA-1 and GATA-2 redundantly regulate the development and/or survival of mouse embryonic erythroblasts (16) and share common molecular mechanisms. The C-terminal zinc fingers of both factors bind the (A/T)GATA(A/G) DNA motif (26, 34, 36). The N-terminal zinc fingers bind the coregulator Friend of GATA-1 (FOG-1) (8, 60) and have intrinsic DNA binding activity, with a preference for GATC and AGATCT, respectively (18, 40, 45). The GATA-1 zinc finger region also binds the histone acetyltransferases CBP/p300 (2).
Targeted deletion of Fog1 blocks erythroid maturation similarly to the Gata1 knockout (59, 60). Taken together with data obtained from altered-specificity mutants (8), the results establish FOG-1 as a key GATA-1 coregulator. GATA-1-mediated transcriptional activation often requires FOG-1, whereas GATA-1-mediated repression appears to always be FOG-1 dependent (8). Mutation of four of the nine FOG-1 zinc fingers abrogates GATA-1 binding (5), and the integrity of a single residue, V205, within the N-terminal zinc finger of GATA-1 is required for FOG-1 binding (8, 41). The FOG-1 N terminus binds the nucleosome remodeling and histone deacetylase complex (21), a common mediator of transcriptional repression (65). Three of the nine zinc fingers of FOG-1 bind transforming acidic coiled coil protein 3, which regulates FOG-1 subcellular localization (17, 52).
Although FOG-1 has not been reported to have sequence-specific DNA binding activity or to affect GATA-1 DNA binding activity, FOG-1 facilitates GATA-1 chromatin occupancy (30, 43) and GATA-2 displacement from certain chromatin sites (43). Analysis of chromatin occupancy in erythroid cells expressing endogenous GATA-2 or physiological levels of an estrogen receptor ligand binding domain fusion to GATA-1 (ER-GATA-1) revealed occupancy at identical regions of the β-globin locus (23, 24). Only a small subset of the WGATAR motifs were occupied, suggesting an essential role for chromatin organization in regulating occupancy. FOG-1-dependent occupancy sites include the βmajor promoter (30), which is transcribed in adult erythroid cells. FOG-1 is also required for GATA-1 to bring the far-upstream β-globin locus control region (LCR) into proximity of the βmajor promoter (61), which might reflect a FOG-1 requirement for full GATA-1 occupancy (30, 43) or a previously undescribed function.
Studies on Gata2 and β-globin transcriptional regulation revealed the chromatin occupancy facilitator activity of FOG-1 (30, 43). GATA-1 directly represses Gata2 transcription by assembling complexes on −3.9 and −2.8 kb, and to a lesser extent −1.8 kb, regions relative to the Gata2 1S hematopoietic promoter (19, 35, 43). As GATA-2 occupies these regions in the transcriptionally active state, we proposed that GATA-2 confers positive autoregulation, and displacement of GATA-2 by GATA-1, termed a “GATA switch,” abrogates autoregulation and induces broad histone deacetylation (19). FOG-1 is required for both the GATA switch (43) and post-GATA switch repression (21). GATA switches occur at multiple loci in erythroid cells (1, 43, 44).
The GATA motifs of the −2.8 kb region of the Gata2 chromosomal locus are functional in transfection (35) and mouse transgenesis (27) assays. Gata2 promoter-lacZ transgenes require GATA motifs within the −2.8 kb region for expression in multipotent hematopoietic precursor cells (27). However, bacterial artificial chromosome transgenesis experiments indicate that sequences ∼100 to 150 kb upstream of Gata2 are required for rescue of hematopoiesis in Gata2-null mice (66), and previous studies analyzed only GATA-1 occupancy and histone H3 and H4 acetylation (acH3 and acH4) to ∼40 kb upstream of the 1S promoter (19, 35). We describe far-upstream and intronic GATA factor-dependent enhancer elements at which GATA switches occur and reveal unique mechanistic insights regarding GATA factor function through dispersed complexes within an endogenous chromosomal locus.
MATERIALS AND METHODS
Antibodies.
Anti-diacetylated histone H3 (06-599), anti-tetra-acetylated H4 (06-866), and anti-dimethylated histone H3 at lysine 4 (H3-dimeK4) (07-030) antibodies were obtained from Upstate Biotechnology. Rabbit anti-GATA-1 and -GATA-2 antibodies against N-terminal fragments of the bacterially expressed GATA factors, which were purified to near-homogeneity, were described previously (23). Rabbit anti-CBP polyclonal antibody (SC-369) was obtained from Santa Cruz Biotechnology. Rabbit anti-FOG-1 polyclonal antibody was described previously (43). Preimmune serum served as controls.
Primers.
Primer sequences are available upon request.
Cell culture.
G1E (63) and G1E-ER-GATA-1 stably expressing a GATA-1 fusion to the human estrogen receptor ligand binding domain (20) cells were maintained in Iscove's modified Dulbecco's medium (GIBCO/BRL) containing 2% penicillin-streptomycin (GIBCO/BRL), 2 U/ml erythropoietin, 120 nM monothioglycerol (Sigma), 0.6% conditioned medium from a Kit ligand-producing CHO cell line, and 15% fetal bovine serum (GIBCO/BRL). G1E-ER-GATA-1 cell medium contained 1 μg/ml puromycin. Mouse erythroleukemia (MEL) cells were maintained in Dulbecco's modified Eagle's medium (Biofluids) containing 5% fetal bovine serum (FBS), 5% calf serum, and 1% antibiotic-antimycotic (all components from GIBCO/BRL). NIH 3T3 cells were maintained in Dulbecco's modified Eagle's medium (Biofluids) containing 10% fetal bovine serum and 1% antibiotic-antimycotic.
Quantitative ChIP assay.
Real-time PCR quantitative chromatin immunoprecipitation (ChIP) analysis was conducted as described previously (22). Chromatin fragments averaged ∼400 bp. Before β-estradiol treatment, cells were grown for at least 24 h in medium containing 15% charcoal-stripped FBS to eliminate steroids. Cells were cultured in medium containing 15% FBS with or without 1 μM β-estradiol (Sigma) for 48 h. Immunoprecipitated DNA was analyzed by real-time PCR (ABI Prism 7000, PE Applied Biosystems). Primers were designed by PRIMER EXPRESS 1.0 (PE Applied Biosystems) to amplify 50- to 150-bp amplicons and were based on sequence from GenBank accession no. AB009272 and sequences in Ensembl (www.ensembl.org/Mus_musculus/geneview?gene=ENSMUSG00000015053). Samples from three or more independent experiments were analyzed. Product was measured by SYBR green fluorescence in 15-μl reaction volumes. The amount of product was determined relative to a standard curve of input chromatin. Dissociation curves confirmed the homogeneity of PCR products.
RNA isolation and RT-PCR.
RNA was prepared from the same cultures used for ChIP. Total RNA was purified with TRIzol (GIBCO/BRL). Before cDNA synthesis, RNA (1.2 μg) was treated with RQ1 DNase (Promega) for 30 min at 37°C. cDNA was prepared by annealing RNA (1 μg) with 250 ng of a 5:1 mixture of random and oligo(dT) primers preheated at 68°C for 10 min. This was followed by incubation with reverse transcriptase (Superscript II; GIBCO/BRL) combined with 10 mM dithiothreitol (DTT), RNasin (Promega), and 0.5 mM deoxynucleoside triphosphates at 42°C for 1 h. Reactions were diluted to a final volume of 150 μl and heat inactivated at 98°C for 5 min. Reactions (15 μl) contained 2.0 μl of cDNA, 7.5 μl SYBR Green (Applied Biosystems), and the appropriate primers. Product accumulation was monitored by using SYBR Green fluorescence. Control reactions lacking reverse transcriptase (RT) yielded very little to no signal. Relative expression levels were determined from a standard curve of serial dilutions of untreated G1E-ER-GATA cDNA samples. Forward and reverse primers for real-time RT-PCR (5′-3′): Gapdh, GAAGGTACGGAGTCAACGGATTT and GAATTTGACCATGGGTGGAAT; Gata2-exon3/4, GCAGAGAAGCAAGGCTCGC and CAGTTGACACACTCCCGGC; Rpn1-exon 4/5, CTGACAGTGGGATCTCCTCCAT and CAATCTCATCCCGGTAATAGACATC; Rab7-exon3/4, GGTGATGGTGGACGACAGACT and GCCACACCAAGAGACTGGAAC.
Transient transfection and luciferase assays.
DNA (4 μg) was diluted in 500 μl of Opti-MEM1 reduced-serum medium per well. DMRIE-C (4 μl) (Invitrogen) was diluted in 500 μl Opti-MEM1 reduced-serum medium per well. The DNA solution was combined with the DMRIE-C solution and incubated at room temperature for 30 min. G1E and MEL cells were isolated by centrifugation at 240 × g for 8 min at 4°C. Cells were diluted to 5 × 106 per ml with Opti-MEM1 reduced-serum medium. One million cells were added per well and incubated for 4.5 h at 37°C. Iscove's modified Dulbecco's medium supplemented with 15% FBS (G1E) or Dulbecco's modified Eagle's medium supplemented with 7.5% FBS and 7.5% calf serum (MEL) was added (2 ml). Cells were incubated for 48 h at 37°C, isolated by centrifugation at 240 × g at 4°C, and washed with phosphate-buffered saline. Lysates were analyzed for luciferase activity and protein concentration, and luciferase activity was normalized by the protein concentration of the lysate.
Microarray amplicon generation and ChIP-chip analysis.
The generation of amplicons from the individual ChIPs was adapted from published protocols (25, 47). Briefly, G1E-ER-GATA cells were grown in charcoal-stripped FBS-containing medium, as described above, before treatment with 1 μM β-estradiol for 48 h. Cells were cross-linked with 1% formaldehyde for 10 min. DNA isolated from ChIP was blunted with T4 DNA polymerase. Two unidirectional linkers, oligoJW102 (GCGGTGACCCGGGAGATCTGAATTC) and oligoJW103 (GAATTCAGATC) were annealed and ligated to the blunted DNA samples. Amplicons were created by PCR; each sample consisted of 5 μl of 10× Taq polymerase buffer, 1 μl of 40 mM deoxynucleoside triphosphates, 3 μl of 25 mM MgCl2, 6.5 μl of 5 M betaine, 1 μl of oligoJW102 (20 μM), 1 μl of Taq (Promega, M1861), and 25 μl of the blunt-ended and ligated chromatin. PCR was run with one cycle at 95°C for 2 min, 55°C for 2 min, and 72°C for 5 min. Twenty-six cycles were then run at 95°C for 0.5 min, 55°C for 0.5 min, and 72°C for 1 min. Finally the products were extended at 72°C for 4 min and then held at 4°C until purified using the QIAquick PCR purification kit according to the manufacturer's instructions. DNA was reamplified (2 μl) for 20 cycles under the conditions above and purified. Samples were quantitated and analyzed via hybridization by Nimblegen. A tiled microarray containing a portion of chromosome 6 (88,423,000 to 88,643,000) containing the Gata2 locus (88,528,257 to 88,541,604) was generated by Nimblegen. The microarray was constructed using 50-bp probes with a 3-bp interval between the 5′ end of a probe and the 5′ end of the next probe on the microarray.
3C assay.
Chromosome conformation capture (3C) analysis was conducted as described previously, except that NcoI was used to digest chromatin (61). The following primers were used to analyze ligation products: −109 kb, TAGGATTGATTGAGCTGTCAGCAGAGCTGG; −77 kb, GCTGGGATTACACCACACTCAACCAGAAAG; −23 kb, AGCCTACACCTCCACATTATAGTCCATCAC; −2.8 kb, TCAGTGTCAACCAAAGCAATAGCTCCGCAG; 1S, AAAGAGGGAAATTACCTGCTCCGGGCGAG; +9.5 kb, TGCCGGTCCGGAAACAGATACACGAAGTTT; +25 kb ACTTCTCCAATGGCTACCTGCATTATCCGC; α-tubulin, TGCATGGTGGCTCTTCTTAGCTCTGGATAG and AGAATTCCAGACCAACCTGGTACCCTACT; β-globin HS3, AGTTCTGCAGATCAGTGCCCAACAGTTCAG; βmajor and βminor, TCAGGATCCACATGCAGCTTGTCACAGTG.
DNase I hypersensitive site analysis.
G1E-ER-GATA-1 cells were treated with 1 μM tamoxifen or left untreated for 23 h. Cells (2 × 107 per condition) were collected by centrifugation at 240 × g for 10 min at 4°C. Cells were washed with ice-cold phosphate-buffered saline and resuspended in 1.5 volumes of lysis buffer (10 mM Tris-HCl, 10 mM NaCl, 3 mM MgCl2, 0.2% NP-40, 5 mM DTT, 0.5 mM phenylmethylsulfonyl fluoride, and 20 μg/ml leupeptin, pH 7.5) and incubated for 5 min on ice. Nuclei were collected by centrifugation at 500 × g for 5 min at 4°C, resuspended in 1.5 volumes of nuclei wash buffer (10 mM Tris-HCl, 10 mM NaCl, 3 mM MgCl2, 5 mM DTT, 0.5 mM phenylmethylsulfonyl fluoride, and 20 μg/ml leupeptin, pH 7.5), and collected by centrifugation. Nuclei were resuspended in 1 ml nuclei wash buffer and were divided into 200-μl aliquots, and DNase I digestion was initiated by addition of 0, 5, 7.5, or 15 U DNase I (Worthington). Reactions were incubated for 8 min at 37°C and terminated by addition of 0.5 ml SDS stop buffer (10 mM Tris-HCl, 25 mM EDTA, 1% sodium dodecyl sulfate, pH 8.0) with vigorous mixing. Proteinase K (Promega) was added to a final concentration of 0.4 mg/ml, and reactions were incubated for at least 15 h at 37°C. Genomic DNA (20 μg) was digested with 5 U/μg EcoRI in a 100-μl volume for 15 h at 37°C. Genomic DNA fragments were used as size markers. The fragments were generated by sequential digestion of genomic DNA with BanI followed by XhoI, XbaI, or BanI alone. Equal amounts of DNA (20 μg or 4 μg for markers) were resolved on a 1.0% agarose gel and analyzed by Southern blotting with a high-specific-activity random-primed 32P-labeled probe. After high-stringency washing, radioactivity was measured by PhosphorImager analysis with ImageQuant software (Molecular Dynamics). The probe spanning the −74.2 kb to −77.9 kb region of Gata2 was generated by PCR amplification of plasmid DNA containing a portion of the upstream sequence of the Gata2 locus using the primers (5′ to 3′) GCACCCATGCCTGTTTGTGCCCTTTG and GGTGATCTGAGAAATCAGTGTCATAAAGCCAGAC.
RESULTS AND DISCUSSION
Identification of a conserved far-upstream region of GATA factor occupancy at the endogenous Gata2 locus.
We generated tiled microarrays containing the murine Gata2 locus and ∼100 kb upstream and downstream for ChIP-chip analysis to comprehensively analyze ER-GATA-1 occupancy at the endogenous Gata2 locus in GATA-1-null G1E cells stably expressing ER-GATA-1 (G1E-ER-GATA-1). Cross-linked ER-GATA-1-chromatin complexes were immunoprecipitated with anti-GATA-1 antibody. DNA immunoprecipitated with preimmune or anti-GATA-1 antibodies was hybridized to the Gata2 locus microarray using established ChIP-chip methodology (25).
ER-GATA-1 occupied the −3.9 to −2.8 kb region (Fig. 1), as expected from our previous quantitative ChIP analyses (19, 35). ER-GATA-1 also occupied a highly conserved region 77 kb upstream of the 1S promoter (Fig. 1). Occupancy was not detected, however, at other highly conserved regions, both upstream and downstream of Gata2 (Fig. 1). Based on the simplicity of the WGATAR motif, 264 motifs reside within the ∼220-kb tiled sequence. GATA-1 can also bind simple derivatives of this motif on naked DNA templates; 382, 465, and 688 nGATAR, WGATAn, and nGATAn motifs, respectively, reside within the tiled region. Thus, the exquisite specificity of GATA motif occupancy at a small subset of the regions containing GATA motifs is analogous to results from studies of ER-GATA-1 and endogenous GATA-2 occupancy at the β-globin locus (23, 24).
FIG. 1.
ChIP-chip analysis reveals ER-GATA-1 occupancy far upstream of Gata2. Gata2 locus organization is illustrated at the top. Open and filled boxes depict noncoding and coding exons, respectively. Arrows pointing down depict GATA switch sites and the −77 kb region. The VISTA plot depicts percent sequence identity between mouse and human (M/H) or mouse and dog (M/D) as a function of genomic coordinates. The first three ChIP-chip profiles depict results comparing chromatin immunoprecipitation with anti-GATA-1 and preimmune antibodies. The bottom three ChIP-chip profiles compare signals obtained upon immunoprecipitation with anti-GATA-1 antibody with that of input chromatin. Note that no qualitative differences were detected in the six ChIP-chip profiles. These results are representative of those obtained from three independent ChIP-chip experiments. The “peaks” delineated with Signal Map software represent the highest signal/background ratios obtained using default software settings.
To validate the ChIP-chip results, quantitative ChIP was conducted with real-time PCR. ER-GATA-1 occupancy was measured at 34 amplicons spanning all conserved (mouse to human) WGATAR, WGATA, GATAR, and GATA motifs of the Gata2 locus within the region analyzed by ChIP-chip in β-estradiol-treated G1E-ER-GATA-1 cells (Fig. 2A). Occupancy was detected at the −77, −3.9, and −2.8 kb regions, confirming the ChIP-chip results, and lower signals were detected at the −1.8 and +9.5 kb (intron 4) regions (Fig. 2B).
FIG. 2.
Quantitative ChIP analysis of ER-GATA-1, GATA-2, and FOG-1 occupancy at the endogenous Gata2 locus. (A) Gata2 locus organization. Open and filled boxes depict noncoding and coding exons, respectively. Arrows pointing down depict GATA switch sites and the −77 kb region. The VISTA plot depicts sequence identity between mouse and human using the mouse sequence as a reference. Coordinate 1 reflects the first nucleotide of the Gata2 1S exon. (B to E) Quantitative ChIP analysis of ER-GATA-1 (B), GATA-2 (C), FOG-1 (β-estradiol-treated cells) (D), and FOG-1 (untreated cells) (E) chromatin occupancy at regions containing conserved nGATAn sites in untreated (transcriptionally active) or β-estradiol-treated (48 h) (transcriptionally repressed) G1E-ER-GATA-1 cells (mean ± standard error, two to three independent experiments). Asterisks indicate three nonconserved GATA sites residing within chromatin highly enriched in histone H3 and H4 acetylation and H3-dimeK4.
Endogenous GATA-2 is expressed in G1E cells lacking activated ER-GATA-1 (19, 63). Our previous studies at the β-globin and Gata2 loci demonstrated that ER-GATA-1, GATA-1, and GATA-2 occupy the same chromatin sites (19, 23, 35, 43). However, although GATA-2 occupancy at the −1.8 kb region of the Gata2 locus is comparable in magnitude to that of the −2.8 kb region, ER-GATA-1 (and GATA-1) occupancy at the −1.8 kb region is considerably lower than at the −2.8 kb region. Endogenous GATA-2 occupied the −77, −3.9, −2.8, and −1.8 kb regions, and a lower signal was detected at the +9.5 kb region (Fig. 2C). Endogenous FOG-1 colocalized at these regions in untreated and β-estradiol-treated G1E-ER-GATA-1 cells, with the signals being highest in β-estradiol-treated cells (Fig. 2D). Thus, ER-GATA-1, GATA-2, and FOG-1 occupy similar regions at the endogenous Gata2 locus, suggesting that ER-GATA-1 and GATA-2 reside at these regions in complexes containing FOG-1.
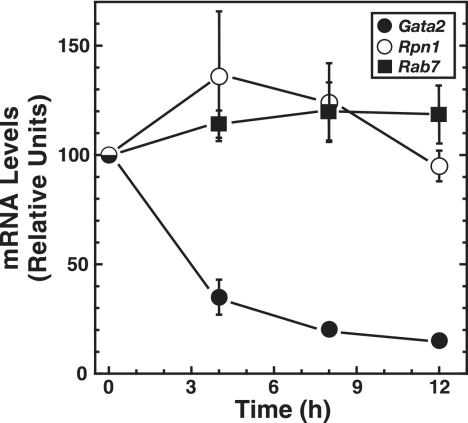
The −77 kb region resides ∼33 kb downstream of the Rpn1 promoter, and Rab7 is ∼39 kb upstream of Rpn1. Conceivably, the GATA switch at the −77 kb region might regulate Rpn1 and Rab7 transcription rather than that of Gata2. A rigorous Affymetrix microarray analysis of ER-GATA-1-mediated gene expression changes showed that Rpn1 decreased slightly (∼2-fold) by 14 h of β-estradiol treatment and did not change further by 30 h (64). Rab7 expression was constant for up to 30 h. To ensure that these microarray results are relevant to our culture conditions, real-time RT-PCR was used to quantitate Rab7 and Rpn1 mRNA levels. Rab7 and Rpn1 expression was constant under conditions in which ER-GATA-1 activation strongly repressed Gata2 transcription (Fig. 3). These results are inconsistent with a role for the −77 kb GATA factor complexes in regulating Rab7 and Rpn1.
FIG. 3.
ER-GATA-1-mediated Gata2 repression is not associated with altered transcription of nearest-neighbor genes. Quantitative real-time RT-PCR was used to measure murine Gata2, Rpn1, and Rab7 mRNA levels in untreated or β-estradiol-treated (for 4, 8, and 12 h) G1E-ER-GATA-1 cells. The graph depicts relative mRNA levels normalized by the level of Gapdh mRNA (mean ± standard error, three independent experiments).
ER-GATA-1 concomitantly occupies the −77 kb region and downstream GATA switch sites but instigates deacetylation in a spatially restricted manner.
We demonstrated previously that ER-GATA-1 displaces GATA-2 at the −2.8 kb region concomitant with transcriptional repression, as measured by loss of primary Gata2 transcripts (19), mRNA, and protein. Furthermore, GATA switches at the −3.9 and −2.8 kb regions exhibited indistinguishable kinetics (35). Whereas GATA-2 occupancy at the −1.8 kb region rapidly declined upon ER-GATA-1 activation, little to no ER-GATA-1 could be cross-linked to this region, unlike the −3.9 and −2.8 kb regions.
It is instructive to compare how ER-GATA-1 instigates Gata2 repression versus βmajor activation. At the β-globin locus, ER-GATA-1 occupies the DNase I hypersensitive sites (HSs) of the LCR prior to the βmajor promoter. Moreover, low-level ER-GATA-1 activity is sufficient for occupancy at the HSs but not the promoter (23). We tested whether ER-GATA-1 occupies the −77 kb region prior to the −3.9, −2.8, and +9.5 kb regions, analogous to the sequential occupancy at the β-globin locus, or if occupancy at the −77, −3.9, −2.8, and +9.5 kb regions occurs concomitantly (Fig. 4A). Graded ER-GATA-1 activation with increasing concentrations of β-estradiol resulted in concentration-dependent occupancy at the −77 kb region, which differed only slightly from the −3.9, −2.8, and +9.5 kb regions (Fig. 4B). Thus, by contrast to the β-globin locus, comparable ER-GATA-1 activity is required for occupancy at the multiple GATA switch sites of the Gata2 locus.
FIG. 4.
Concomitant GATA switches at multiple upstream regions and intron 4 of the Gata2 locus. (A) Sequential (1) and concomitant (2) GATA switch models. (B) Quantitative ChIP analysis of ER-GATA-1 and GATA-2 occupancy at −77, −3.9, −2.8, and +9.5 kb (intron 4) regions in untreated and β-estradiol-treated (10, 40, or 200 nM) G1E-ER-GATA-1 cells (mean ± standard error, four independent experiments).
Since ER-GATA-1 occupancy at the −77 kb region was indistinguishable vis-à-vis ER-GATA-1 activity requirements from that at other GATA switch sites of the Gata2 locus, we predicted that ER-GATA-1 occupancy instigates identical molecular events at the distinct occupancy sites. However, ER-GATA-1 can mediate either activation or repression at different loci (8), raising the possibility that ER-GATA-1 functions differently at distinct sites within a chromosomal locus. To determine whether ER-GATA-1 regulates the histone modification pattern similarly at the distinct sites, quantitative ChIP was used to comprehensively define specific epigenetic components of the Gata2 histone modification pattern in both the transcriptionally active and repressed states.
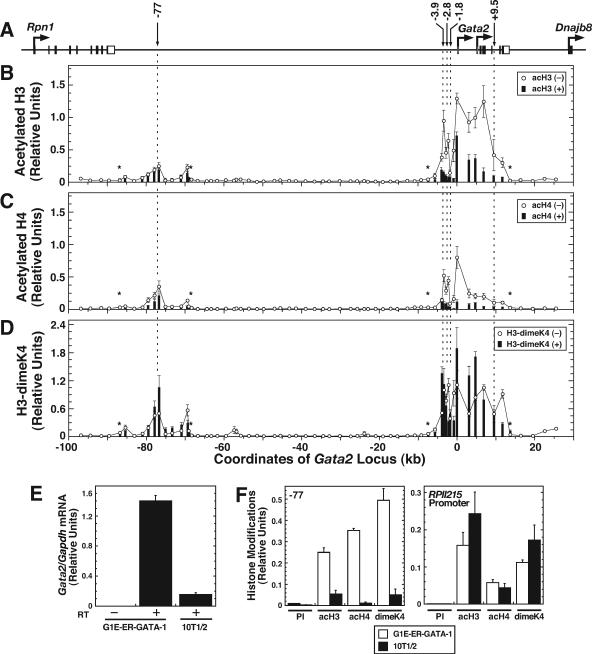
As described previously (19), ER-GATA-1 activation decreased acH3 and acH4 at the Gata2 1S promoter, open reading frame, and extending ∼4 kb upstream of the promoter (Fig. 5A to C). By contrast, enriched acH3 and acH4 at the −77 kb region was insensitive to ER-GATA-1 activation. H3-dimeK4 was constant at most sites but increased twofold or less at the 1G promoter (Fig. 5D). Intriguingly, ER-GATA-1 occupancy exerts spatially restricted deacetylation at GATA switch sites near the 1S promoter but not at the −77 kb region. Treatment of uninduced G1E-ER-GATA-1 cells for 2 h with the transcriptional elongation inhibitor 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole, which abrogated RNA polymerase II accumulation within Gata2 exon 3, did not significantly reduce acH3 and acH4 at the −2.8 kb, −77 kb, and exon 3 regions (data not shown). This result suggests that ER-GATA-1-dependent deacetylation at and near Gata2 is not a consequence of reduced transcriptional elongation.
FIG. 5.
ER-GATA-1 induces deacetylation at and near the Gata2 promoter and open reading frame but not at the −77 kb region. (A) Gata2 locus organization. Open and filled boxes depict noncoding and coding exons, respectively. Arrows pointing down depict GATA switch sites. The value 1 reflects the first nucleotide of the Gata2 1S exon. (B to D) Quantitative ChIP analysis of acH3 (B), acH4 (C), and H3-dimeK4 (D) across the locus with an average distance of 2 kb between primer sets in untreated and β-estradiol-treated (48 h) G1E-ER-GATA-1 cells (mean ± standard error, two to five independent experiments). Asterisks indicate the limit of signal detection in the treated condition. DNA immunoprecipitated with preimmune (PI) antibody was analyzed with all primer sets, and the average signal from all experiments is 0.0035. (E) RT-PCR analysis of Gata2 mRNA in untreated G1E-ER-GATA-1 and 10T1/2 cells. Gata2 mRNA levels were normalized by the levels of Gapdh mRNA. (F) Quantitative ChIP analysis of acH3, acH4, and H3-dimeK4 at the −77 kb region (left) and the constitutively active RPII215 promoter (right) in untreated G1E-ER-GATA-1 and 10T1/2 cells.
The ER-GATA-1 insensitivity of histone acetylation at the −77 kb region implied that mechanisms responsible for establishing and/or maintaining acetylation at this region differ from those operational at GATA switch sites near the 1S promoter. Histone acetylation at the GATA switch sites near the promoter, the 1S and 1G promoters, and the open reading frame is highly enriched in erythroid, but not nonerythroid, cells. To determine whether the −77 kb region shares this cell type-specific epigenetic regulation, we compared acH3, acH4, and H3-dimeK4 at the −77 kb region in untreated G1E-ER-GATA-1 cells and murine 10T1/2 embryonic fibroblast cells (48). Gata2 mRNA transcripts were detected at a low level in 10T1/2 cells (Fig. 5E). By contrast to the high enrichments of acH3, acH4, and H3-dimeK4, relative to signals obtained with a preimmune antibody, in G1E-ER-GATA-1 cells, little to no enrichments were detected at the −77 kb region in 10T1/2 cells (Fig. 5F, left). Comparable high enrichments of the epigenetic marks were detected at the RPII215 promoter in both cell lines (Fig. 5F, right). Thus, mechanisms that establish and/or maintain enriched acH3, acH4, and H3-dimeK4 at the −77 kb region are erythroid cell specific. Based on the ER-GATA-1 insensitivity of acH3 and acH4 at the −77 kb region, clearly other erythroid-cell-specific factors establish and/or maintain epigenetic marks at this region.
GATA factor occupancy sites in chromatin are commonly DNase I HSs, and HSs often demarcate functional complexes. We tested whether the −77 kb GATA switch site corresponds to an HS. High-resolution Southern blot analysis indicated the presence of an HS at the −77 kb region (−77.1 to −77.2) in both the active and repressed locus, with the signals being slightly higher at the repressed locus (Fig. 6).
FIG. 6.
DNase I hypersensitivity at the −77 region. Nuclei were isolated from untreated or tamoxifen-treated (23 h) G1E-ER-GATA-1 cells and digested with increasing concentrations of DNase I. After cleavage of purified genomic DNA with BanI, fragments were analyzed by Southern blotting. The PhosphorImager scan shown is representative of three independent DNase I digests and analyses. The diagram on the right illustrates the Gata2 locus coordinates of fragments determined with genomic molecular size markers. 0, no-DNase I control.
Distinct functional properties of the GATA switch sites.
Although ER-GATA-1 replaces GATA-2 at the −77 kb region, at GATA switch sites near the 1S promoter, and at the +9.5 kb region, the spatially restricted deacetylation suggests that ER-GATA-1 complexes at these regions are intrinsically different. To further compare functional properties of the distinct GATA switch sites, we measured their activities in transient-transfection assays in G1E cells expressing endogenous GATA-2 and MEL cells expressing endogenous GATA-1. Previously, we demonstrated that the −1.8 and −2.8 kb regions activated the Gata2 1S promoter when fused to a luciferase reporter gene in a transient transfection assay in G1E, but not in MEL, cells (35). By contrast, the −3.9 kb region activated the 1S promoter in MEL, but not in G1E, cells. Although caveats exist upon extrapolating transient-transfection data to physiological mechanisms, these results suggest that the activities of complexes assembled at the −1.8 and −2.8 kb regions share more in common with each other than with that of the complex at the −3.9 kb region.
The −77 kb region contains TGATAA, GGATAC, and TGATAGATAG motifs (Fig. 7A), which could potentially mediate the GATA factor occupancy described in Fig. 1 and 2. The −77 kb region, with or without mutations of these motifs, was cloned upstream of the Gata2 1S promoter fused to a luciferase reporter gene. The −77 kb region stimulated reporter activity in both G1E and MEL cells (Fig. 7B). Mutation of the TGATAA motif abrogated enhancer activity, whereas mutations of the GGATAC and TGATAGATAG motifs had no effect. Thus, a single GATA motif within the −77 kb region confers enhancer activity in GATA-1- and GATA-2-expressing cells; the additional GATA motifs had no apparent functional role. Furthermore, the results indicate that GATA-2 and GATA-1 function redundantly through this isolated element in the transfection assay. As described previously (35), the −3.9 kb region conferred enhancer activity in MEL, but not G1E, cells (Fig. 7B). The enhancer activity of the −77 kb region in both G1E and MEL cells also differs from the −2.8 and −1.8 kb regions analyzed previously (35) (Fig. 7C). Unlike other GATA switch sites, the +9.5 kb region exhibited very strong enhancer activity in G1E cells (30-fold), stronger than any other enhancers studied in this system. The +9.5 kb region also conferred enhancer activity in MEL cells, comparable to that of the −77 kb region, but the magnitude of this activity was ∼5-fold less than in G1E cells.
FIG. 7.
Cell type-specific enhancer activity of the −77 kb region. (A) Sequence of the −77 and +9.5 kb regions analyzed in the transient-transfection assay. Conserved GATA motifs within the −77 and +9.5 kb fragments are highlighted. (B) G1E and MEL cells were transiently transfected with reporter plasmids derived from the pGL3 luciferase vector containing the Gata2 1S promoter cloned upstream of luciferase (1SLuc): the GATA motifs were intact in the (−77)1SLuc, (−3.9)1SLuc, and (+9.5)1SLuc plasmids, whereas the GATA motifs were mutated in the (−77mt1)1SLuc, (−77mt2)1SLuc, (−77mt3)1SLuc, and (+9.5mt)1SLuc plasmids. Mutations: −77mt1, TTATCA to TATGAA; −77mt2, GGATAC to ACGCGT; −77mt3, CTATCTATCA to TAGCTAGCGT; +9.5mt, CTATCC to ACGCGT, AGATAA to GACTTC, and GTATCT to GCTAGC. The plots depict luciferase activities of the cell lysates normalized by the protein concentrations of the lysates. The activity of the 1SLuc construct was designated 1.0 (mean ± standard error, 3 to 12 independent experiments). In each experiment, transfections were performed in triplicate. RLU, relative luciferase units. (C) Summary of enhancer activities of the GATA switch sites. +, enhancer activity; +++, strong enhancer activity (∼30-fold); −, no enhancer activity. Parentheses indicate results from our previously published study (35).
The transient-transfection results allow one to segregate the GATA switch sites at the Gata2 locus into four categories: (i) comparable activity in G1E and MEL cells (−77 kb); (ii) activity exclusively in G1E cells (−2.8 and −1.8 kb); (iii) activity exclusively in MEL cells (−3.9 kb); (iv) activity in both G1E and MEL cells, but greater activity in G1E cells (+9.5 kb). It is attractive to propose that such distinct functions reflect the unique composition of nucleoprotein complexes at these GATA switch sites. Proteomics analysis provided evidence that MEL cell GATA-1 in solution assembles at least four multiprotein complexes (49). Considering the high sequence similarity between the zinc finger region and certain additional sequences of GATA-1 and GATA-2, presumably, GATA-2 also assembles diverse complexes, but this has not been reported.
Based on the differential ER-GATA-1 sensitivities of acH3 and acH4 and differential enhancer activities of the −77 kb region versus the downstream GATA switch sites, we tested whether the composition of ER-GATA-1-dependent nucleoprotein complexes assembled at these regions differ. Previously, we showed that ER-GATA-1 decreased CBP/p300 occupancy at the −1.8 kb region, which correlated with loss of DNase I hypersensitivity at this region (19, 35). CBP/p300 occupancy at the −3.9 kb region increased slightly upon ER-GATA-1 activation after 24 h. Quantitative ChIP was used to determine whether CBP/p300 occupies the −77 kb region prior to and after ER-GATA-1 activation in G1E-ER-GATA-1 cells. CBP/p300 occupied the −77 kb region at a higher level than at the −3.9, −1.8, and +9.5 GATA switch sites in untreated cells (Fig. 8). Few to no signals were detected at a −46 kb amplicon, which lacks known regulatory sequences. ER-GATA-1 activation for 48 h increased occupancy at the −3.9 and +9.5 kb GATA switch sites (twofold or less). Occupancy at the −77 region was relatively constant, whereas occupancy at the −1.8 kb region decreased ∼2-fold. As the ER-GATA-1-dependent changes in CBP/p300 occupancy (Fig. 8) and histone acetylation (Fig. 5) do not correlate precisely, it is highly unlikely that CBP/p300 is the sole coregulator mediating establishment of the histone acetylation pattern of the Gata2 locus. Importantly, the differential CBP/p300 recruitment to the −77, −3.9, and +9.5 kb regions versus the −1.8 kb region further highlights intrinsic qualitative differences in the GATA switch site activities.
FIG. 8.
Differential CBP/p300 occupancy at the −77, −3.9, and +9.5 versus the −1.8 kb GATA switch site. CBP/p300 occupancy was measured in untreated and β-estradiol-treated (48 h) G1E-ER-GATA-1 cells by quantitative ChIP analysis at the −77, −3.9, −1.8, and +9.5 kb GATA switch sites and the −46 kb region, which lacks known regulatory sequences (mean ± standard error, three independent experiments).
Gata2 higher-order chromatin conformation: proximity of the −77 kb region to Gata2.
Since ER-GATA-1 activation increases the proximity of the β-globin LCR to the downstream adult β-globin genes in a FOG-1-dependent manner, which correlates with ER-GATA-1 occupancy of both regions (61), the dispersed GATA factor complexes at the Gata2 locus might regulate higher-order chromatin structure. ER-GATA-1 occupancy at the −77 kb region and the additional GATA switch sites might facilitate interactions between these regions. Alternatively, the prior presence of GATA-2 and FOG-1 at these sites might establish a higher-order structure that persists after the GATA switches. Last, since looping is commonly associated with transcriptional activation (7, 13, 50, 55, 61), ER-GATA-1 might disrupt a GATA-2-stabilized loop.
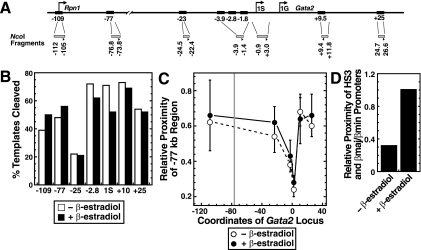
To distinguish among the potential mechanisms described above, 3C analysis (9-11) was conducted to measure the proximity of the −77 kb region relative to the −2.8 and +9.5 kb GATA switch sites, the 1S promoter, and −23 and +25 kb regions lacking known regulatory elements (Fig. 9A). Real-time PCR analysis of NcoI cleavage efficiencies revealed similar efficiencies at the various sites, except the −25 kb site, in which cleavage was lower (Fig. 9B). A decrease in the relative proximity of −77 versus −2.8 and the 1S promoter was detected in all experiments (Fig. 9C). A declining 3C signal as a function of distance is diagnostic of a scenario whereby upstream and downstream regions are not in close proximity (10). By contrast, analysis of the relative proximity of the −77 and +9.5 kb regions yielded a reproducible upward shift in the curve, consistent with the −77 kb region residing closer to the +9.5 kb region relative to other regions tested. The differential ligation of NcoI fragments cannot be explained by variable cleavage (Fig. 9B). Importantly, these results provide evidence for the existence of a chromatin loop in which the upstream −77 kb region communicates with Gata2.
FIG. 9.
Close proximity of the −77 and +9.5 kb GATA switch sites at the endogenous Gata2 locus. (A) The diagram depicts the 3C strategy. NcoI fragments and primers are depicted as shaded rectangles and triangles, respectively. (B) The graph depicts NcoI cleavage efficiencies at the indicated sites determined via real-time PCR. (C) The graph depicts 3C results from four independent experiments (mean ± standard error) measuring the proximity of an NcoI fragment containing the −77 kb region relative to fragments containing the −2.8 and +9.5 kb GATA switch sites, the 1S promoter, −23 kb and +25 kb regions lacking known regulatory elements, or the Rpn1 promoter in untreated or β-estradiol-treated G1E-ER-GATA-1 cells. The vertical gray bar indicates the position of the −77 kb region. Solid line, β-estradiol treated; dotted line, no β-estradiol. (D) The graph depicts a 3C analysis of the relative proximity of β-globin HS3 and the βmajor/βminor promoters (mean, two independent experiments).
The Gata2 higher-order chromatin conformation did not reproducibly change upon ER-GATA-1-induced transcriptional repression. As described previously (61), ER-GATA-1 activation increased the proximity of β-globin LCR HS3 relative to the βmajor promoter (Fig. 9D). As ER-GATA-1 activates the β-globin locus and represses Gata2, the differential influence of ER-GATA-1 on the conformation of the two loci correlates with opposite transcriptional outputs.
Although only a limited number of loci have been studied by 3C, loci can adopt static higher-order structures in both transcriptionally active and inactive states (53), or such structures can be dynamically regulated (13, 61). Ifng adopts a common higher-order chromatin conformation in neutral, Th1, and Th2 T-cells, but secondary stimulation of the cells is associated with remodeling of the common conformation (14). Of course, an apparent static structure might be highly dynamic and/or subject to more subtle structural transitions not detectable by 3C. The static higher-order chromatin scenario is conceptually analogous to the Drosophila melanogaster hsp26 locus, a considerably simpler scenario in which a positioned nucleosome at the promoter serves an architectural function to bring regulatory elements in close proximity, thereby creating a permissive chromatin structure (33, 54). Thus, permissive structures involving a single nucleosome or a broad segment of a locus facilitate transcriptional activation, but establishment of the structure is insufficient to induce preinitiation complex assembly.
It is attractive to propose that the ER-GATA-1 insensitivity of the higher-order chromatin interaction is explained by the presence of GATA-2 and FOG-1 at the GATA switch sites in untreated G1E-ER-GATA-1 cells. Either GATA-2 or ER-GATA-1, in conjunction with FOG-1, would therefore establish the higher-order chromatin conformation. However, since GATA-2 and ER-GATA-1 reside at the GATA switch sites near the 1S promoter and the −77 kb region exhibited a closer apparent proximity to the +9.5 kb GATA switch site than to those near the promoter, the mere presence of GATA factor-FOG-1 complexes appears insufficient to establish a higher-order chromatin interaction, based on what can be measured via 3C analysis.
Chromatin domain regulation via qualitatively distinct activities of dispersed GATA factor complexes.
Herein, we described the identification of a far upstream GATA switch site at the endogenous Gata2 locus. The ER-GATA-1-insensitivity of the upstream Rpn1 and Rab7 genes, the capacity of the −77 kb region to enhance Gata2 1S promoter activity, and the proximity of the −77 kb region to Gata2 provide strong evidence that the −77 kb region regulates Gata2 transcription. Intriguingly, measurements of the ER-GATA-1 sensitivity of histone acetylation, enhancer activities, and nucleoprotein structure indicate that the GATA switch sites of the Gata2 locus have important intrinsic differences, which might reflect qualitatively distinct functions. The multiple lines of evidence implicating the −77 kb region in Gata2 transcriptional regulation, combined with the bacterial artificial chromosome transgenesis data discussed earlier (66), strongly support a model in which Gata2 transcriptional regulation requires long-range chromatin interactions.
Although long-range regulation conferred by dispersed GATA factor complexes appears to be common to both β-globin locus and Gata2 transcriptional regulation (3), our studies have identified important distinctions. ER-GATA-1 occupies upstream and downstream regions of the Gata2 locus concomitantly (Fig. 4), whereas occupancy at upstream and downstream regions of the β-globin chromatin domain occurs sequentially (23). Furthermore, by contrast to the β-globin locus in which ER-GATA-1 solely enhances CBP/p300 occupancy (31), ER-GATA-1 has variable effects on CBP/p300 occupancy at the Gata2 locus: little to no effect at the −77 kb region, increased occupancy at the −3.9 and +9.5 kb regions, and decreased occupancy at the −1.8 kb region (Fig. 8). It will be particularly instructive to extend the comparison of ER-GATA-1-mediated β-globin activation with Gata2 repression to include a larger cohort of GATA factor target genes and to consider the impact of higher-order chromatin structure transitions in the context of the three-dimensional space of the nucleus.
Acknowledgments
We thank Peggy Farnham for helpful discussions on ChIP-chip methodology. We thank Jing Wu for 10T1/2 ChIP samples. We thank Kirby Johnson, Hogune Im, and Jing Wu for critical comments.
This work was funded by NIH grants DK50107 and DK68634. Saumen Pal is a postdoctoral fellow of the American Heart Association.
REFERENCES
- 1.Anguita, E., J. Hughes, C. Heyworth, G. A. Blobel, W. G. Wood, and D. R. Higgs. 2004. Globin gene activation during hematopoiesis is driven by protein complexes nucleated by GATA-1 and GATA-2. EMBO J. 23:2841-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blobel, G. A., T. Nakajima, R. Eckner, M. Montminy, and S. H. Orkin. 1998. CREB-binding protein cooperates with transcription factor GATA-1 and is required for erythroid differentiation. Proc. Natl. Acad. Sci. USA 95:2061-2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bresnick, E. H., K. D. Johnson, S.-I. Kim, and H. Im. 2006. Establishment and regulation of chromatin domains: mechanistic insights from studies of hemoglobin synthesis. Prog. Nucleic Acids Res. Mol. Biol. 81:435-471. [DOI] [PubMed]
- 4.Bresnick, E. H., M. L. Martowicz, S. Pal, and K. D. Johnson. 2005. Developmental control via GATA factor interplay at chromatin domains. J. Cell. Physiol. 205:1-9. [DOI] [PubMed] [Google Scholar]
- 5.Cantor, A. B., S. G. Katz, and S. H. Orkin. 2002. Distinct domains of the GATA-1 cofactor FOG-1 differentially influence erythroid versus megakaryocytic maturation. Mol. Cell. Biol. 22:4268-4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cantor, A. B., and S. H. Orkin. 2002. Transcriptional regulation of erythropoiesis: an affair involving multiple partners. Oncogene 21:3368-3376. [DOI] [PubMed] [Google Scholar]
- 7.Carter, D., L. Chakalova, C. S. Osborne, Y. F. Dai, and P. Fraser. 2002. Long-range chromatin regulatory interactions in vivo. Nat. Genet. 32:623-626. [DOI] [PubMed] [Google Scholar]
- 8.Crispino, J. D., M. B. Lodish, J. P. MacKay, and S. H. Orkin. 1999. Use of altered specificity mutants to probe a specific protein-protein interaction in differentiation: the GATA-1:FOG complex. Mol. Cell 3:219-228. [DOI] [PubMed] [Google Scholar]
- 9.Cullen, K. E., M. P. Kladde, and M. A. Seyfred. 1993. Interaction between transcriptional regulatory elements of prolactin chromatin. Science 261:203-206. [DOI] [PubMed] [Google Scholar]
- 10.Dekker, J. 2006. The three ‘c’s of chromosome conformation capture: controls, controls, controls. Nat. Methods 3:17-21. [DOI] [PubMed] [Google Scholar]
- 11.Dekker, J., K. Rippe, M. Dekker, and N. Kleckner. 2002. Capturing chromosome conformation. Science 295:1306-1311. [DOI] [PubMed] [Google Scholar]
- 12.Dorfman, D. M., D. B. Wilson, G. A. Bruns, and S. H. Orkin. 1992. Human transcription factor GATA-2. Evidence for regulation of preproendothelin-1 gene expression in endothelial cells. J. Biol. Chem. 267:1279-1285. [PubMed] [Google Scholar]
- 13.Drissen, R., R. J. Palstra, N. Gillemans, E. Splinter, F. Grosveld, S. Philipsen, and W. de Laat. 2004. The active spatial organization of the beta-globin locus requires the transcription factor EKLF. Genes Dev. 18:2485-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eivazova, E. R., and T. M. Aune. 2004. Dynamic alterations in the conformation of the Ifng gene region during T helper cell differentiation. Proc. Natl. Acad. Sci. USA 101:251-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Evans, T., and G. Felsenfeld. 1989. The erythroid-specific transcription factor Eryf1: a new finger protein. Cell 58:877-885. [DOI] [PubMed] [Google Scholar]
- 16.Fujiwara, Y., A. N. Chang, A. M. Williams, and S. H. Orkin. 2004. Functional overlap of GATA-1 and GATA-2 in primitive hematopoietic development. Blood 103:583-585. [DOI] [PubMed] [Google Scholar]
- 17.Garriga-Canut, M., and S. H. Orkin. 2004. Transforming acidic coiled-coil protein 3 (TACC3) controls friend of GATA-1 (FOG-1) subcellular localization and regulates the association between GATA-1 and FOG-1 during hematopoiesis. J. Biol. Chem. 279:23597-23605. [DOI] [PubMed] [Google Scholar]
- 18.Ghirlando, R., and C. D. Trainor. 2003. Determinants of GATA-1 binding to DNA: the role of non-finger residues. J. Biol. Chem. 278:45620-45628. [DOI] [PubMed] [Google Scholar]
- 19.Grass, J. A., M. E. Boyer, S. Paul, J. Wu, M. J. Weiss, and E. H. Bresnick. 2003. GATA-1-dependent transcriptional repression of GATA-2 via disruption of positive autoregulation and domain-wide chromatin remodeling. Proc. Natl. Acad. Sci. USA 100:8811-8816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gregory, T., C. Yu, A. Ma, S. H. Orkin, G. A. Blobel, and M. J. Weiss. 1999. GATA-1 and erythropoietin cooperate to promote erythroid cell survival by regulating bcl-xl expression. Blood 94:87-96. [PubMed] [Google Scholar]
- 21.Hong, W., M. Nakazawa, Y. Y. Chen, R. Kori, C. R. Vakoc, C. Rakowski, and G. A. Blobel. 2005. FOG-1 recruits the NuRD repressor complex to mediate transcriptional repression by GATA-1. EMBO J. 24:67-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Im, H., J. A. Grass, K. D. Johnson, M. E. Boyer, J. Wu, and E. H. Bresnick. 2004. Measurement of protein-DNA interactions in vivo by chromatin immunoprecipitation. Methods Mol. Biol. 284:129-146. [DOI] [PubMed] [Google Scholar]
- 23.Im, H., J. A. Grass, K. D. Johnson, S.-I. Kim, M. E. Boyer, A. N. Imbalzano, J. J. Bieker, and E. H. Bresnick. 2005. Chromatin domain activation via GATA-1 utilization of a small subset of dispersed GATA motifs within a broad chromosomal region. Proc. Natl. Acad. Sci. USA 102:17065-17070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson, K. D., J. D. Grass, M. E. Boyer, C. M. Kiekhaefer, G. A. Blobel, M. J. Weiss, and E. H. Bresnick. 2002. Cooperative activities of hematopoietic regulators recruit RNA polymerase II to a tissue-specific chromatin domain. Proc. Natl. Acad. Sci. USA 99:11760-11765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirmizis, A., S. M. Bartley, A. Kuzmichev, R. Margueron, D. Reinberg, R. Green, and P. J. Farnham. 2004. Silencing of human polycomb target genes is associated with methylation of histone H3 Lys 27. Genes Dev. 18:1592-1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ko, L. J., and J. D. Engel. 1993. DNA-binding specificities of the GATA transcription factor family. Mol. Cell. Biol. 13:4011-4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kobayashi-Osaki, M., O. Ohneda, N. Suzuki, N. Minegishi, T. Yokomizo, S. Takahashi, K.-C. Lim, J. D. Engel, and M. Yamamoto. 2005. GATA motifs regulate early hematopoietic lineage-specific expression of the Gata2 gene. Mol. Cell. Biol. 25:7005-7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee, M. E., D. H. Temizer, J. A. Clifford, and T. Quertermous. 1991. Cloning of the GATA binding protein that regulates endothelin-1 gene expression in endothelial cells. J. Biol. Chem. 266:16188-16192. [PubMed] [Google Scholar]
- 29.Leonard, M., M. Brice, J. D. Engel, and T. Papayannopoulou. 1993. Dynamics of GATA transcription factor expression during erythroid differentiation. Blood 82:1071-1079. [PubMed] [Google Scholar]
- 30.Letting, D. L., Y. Y. Chen, C. Rakowski, S. Reedy, and G. A. Blobel. 2004. Context-dependent regulation of GATA-1 by friend of GATA-1. Proc. Natl. Acad. Sci. USA 101:476-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Letting, D. L., C. Rakowski, M. J. Weiss, and G. A. Blobel. 2003. Formation of a tissue-specific histone acetylation pattern by the hematopoietic transcription factor GATA-1. Mol. Cell. Biol. 23:1334-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ling, K. W., K. Ottersbach, J. P. van Hamburg, A. Oziemlak, F. Y. Tsai, S. H. Orkin, R. Ploemacher, R. W. Hendriks, and E. Dzierzak. 2004. GATA-2 plays two functionally distinct roles during the ontogeny of hematopoietic stem cells. J. Exp. Med. 200:871-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu, Q., L. L. Wallrath, and S. C. Elgin. 1995. The role of a positioned nucleosome at the Drosophila melanogaster hsp26 promoter. EMBO J. 14:4738-4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin, D. I., and S. H. Orkin. 1990. Transcriptional activation and DNA binding by the erythroid factor GF-1/NF-E1/Eryf 1. Genes Dev. 4:1886-1898. [DOI] [PubMed] [Google Scholar]
- 35.Martowicz, M. L., J. A. Grass, M. E. Boyer, H. Guend, and E. H. Bresnick. 2005. Dynamic GATA factor interplay at a multi-component regulatory region of the GATA-2 locus. J. Biol. Chem. 280:1724-1732. [DOI] [PubMed] [Google Scholar]
- 36.Merika, M., and S. H. Orkin. 1993. DNA-binding specificity of GATA family transcription factors. Mol. Cell. Biol. 13:3999-4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Minegishi, N., J. Ohta, H. Yamagiwa, N. Suzuki, S. Kawauchi, Y. Zhou, S. Takahashi, N. Hayashi, J. D. Engel, and M. Yamamoto. 1999. The mouse GATA-2 gene is expressed in the para-aortic splanchnopleura and aorta-gonads and mesonephros region. Blood 93:4196-4207. [PubMed] [Google Scholar]
- 38.Mouthon, M. A., O. Bernard, M. T. Mitjavila, P. H. Romeo, W. Vainchenker, and D. Mathieu-Mahul. 1993. Expression of Tal-1 and GATA-binding proteins during human hematopoiesis. Blood 81:647-655. [PubMed] [Google Scholar]
- 39.Nardelli, J., D. Thiesson, Y. Fujiwara, F.-Y. Tsai, and S. H. Orkin. 1999. Expression and genetic interaction of transcription factors GATA-2 and GATA-3 during development of the mouse central nervous system. Dev. Biol. 210:305-321. [DOI] [PubMed] [Google Scholar]
- 40.Newton, A., J. P. MacKay, and M. Crossley. 2001. The N-terminal zinc finger of the erythroid transcription factor GATA-1 binds GATC motifs in DNA. J. Biol. Chem. 276:35794-35801. [DOI] [PubMed] [Google Scholar]
- 41.Nichols, K. E., J. D. Crispino, M. Poncz, J. G. White, S. H. Orkin, J. M. Maris, and M. J. Weiss. 2000. Familial dyserythropoietic anaemia and thrombocytopenia due to an inherited mutation in GATA-1. Nat. Genet. 24:266-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Orlic, D., S. Anderson, L. G. Biesecker, B. P. Sorrentino, and D. M. Bodine. 1995. Pluripotent hematopoietic stem cells contain high levels of mRNA for c Kit, GATA-2, p45 NF-E2, and myb and low levels or no RNA for c-fms and the receptors for granulocyte macrophage colony stimulating factor and interleukins 5 and 7. Proc. Natl. Acad. Sci. USA 92:4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pal, S., A. B. Cantor, K. D. Johnson, T. Moran, M. E. Boyer, S. H. Orkin, and E. H. Bresnick. 2004. Coregulator-dependent facilitation of chromatin occupancy by GATA-1. Proc. Natl. Acad. Sci. USA 101:980-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pal, S., M. J. Nemeth, D. M. Bodine, J. L. Miller, J. Svaren, S. L. Thein, P. J. Lowry, and E. H. Bresnick. 2004. Neurokinin-B transcription in erythroid cells: direct activation by the hematopoietic transcription factor GATA-1. J. Biol. Chem. 279:31348-31356. [DOI] [PubMed] [Google Scholar]
- 45.Pedone, P. V., J. G. Omichinski, P. Nony, C. Trainor, A. M. Gronenborn, G. M. Clore, and G. Felsenfeld. 1997. The N-terminal fingers of chicken GATA-2 and GATA-3 are independent sequence-specific DNA binding domains. EMBO J. 16:2874-2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pevny, L., M. C. Simon, E. Robertson, W. H. Klein, S. F. Tsai, V. D'Agati, S. H. Orkin, and F. Costantini. 1991. Erythroid differentiation in chimaeric mice blocked by a targeted mutation in the gene for transcription factor GATA-1. Nature 349:257-260. [DOI] [PubMed] [Google Scholar]
- 47.Ren, B., F. Robert, J. J. Wyrick, O. Aparicio, E. G. Jennings, I. Simon, J. Zeitlinger, J. Schreiber, N. Hannett, E. Kanin, T. L. Volkert, C. J. Wilson, S. P. Bell, and R. A. Young. 2000. Genome-wide location and function of DNA binding proteins. Science 290:2306-2309. [DOI] [PubMed] [Google Scholar]
- 48.Reznikoff, C. A., D. W. Brankow, and C. Heidelberger. 1973. Establishment and characterization of a cloned line of C3H mouse embryo cells sensitive to postconfluence inhibition of division. Cancer Res. 33:3231-3238. [PubMed] [Google Scholar]
- 49.Rodriquez, P., E. Bonte, J. Krijgsveld, K. E. Kolodziej, B. Guyot, A. J. Heck, P. Vyas, E. de Boer, F. Grosveld, and J. Strouboulis. 2005. GATA-1 forms distinct activating and repressive complexes in erythroid cells. EMBO J. 24:2354-2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schleif, R. 1992. DNA looping. Annu. Rev. Biochem. 61:199-223. [DOI] [PubMed] [Google Scholar]
- 51.Simon, M. C., L. Pevny, M. V. Wiles, G. Keller, F. Costantini, and S. H. Orkin. 1992. Rescue of erythroid development in gene targeted GATA-1- mouse embryonic stem cells. Nat. Genet. 1:92-98. [DOI] [PubMed] [Google Scholar]
- 52.Simpson, R. J., S. H. Yi Lee, N. Bartle, E. Y. Sum, J. E. Visvader, J. M. Matthews, J. P. MacKay, and M. Crossley. 2004. A classic zinc finger from friend of GATA mediates an interaction with the coiled-coil of transforming acidic coiled-coil 3. J. Biol. Chem. 279:39789-39797. [DOI] [PubMed] [Google Scholar]
- 53.Spilianakis, C. G., and R. A. Flavell. 2004. Long-range intrachromosomal interactions in the T helper type 2 cytokine locus. Nat. Immunol. 5:1017-1027. [DOI] [PubMed] [Google Scholar]
- 54.Thomas, G. H., and S. C. Elgin. 1988. Protein/DNA architecture of the DNaseI hypersensitive region of the Drosophila hsp26 promoter. EMBO J. 7:2191-2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tolhuis, B., R. J. Palstra, E. Splinter, F. Grosveld, and W. de Laat. 2002. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell 10:1453-1475. [DOI] [PubMed] [Google Scholar]
- 56.Tsai, F.-Y., and S. H. Orkin. 1997. Transcription factor GATA-2 is required for proliferation/survival of early hematopoietic cells and mast cell formation, but not for erythroid and myeloid terminal differentiation. Blood 89:3636-3643. [PubMed] [Google Scholar]
- 57.Tsai, F. Y., G. Keller, F. C. Kuo, M. Weiss, J. Chen, M. Rosenblatt, F. W. Alt, and S. H. Orkin. 1994. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature 371:221-226. [DOI] [PubMed] [Google Scholar]
- 58.Tsai, S. F., D. I. Martin, L. I. Zon, A. D. D'Andrea, G. G. Wong, and S. H. Orkin. 1989. Cloning of cDNA for the major DNA-binding protein of the erythroid lineage through expression in mammalian cells. Nature 339:446-451. [DOI] [PubMed] [Google Scholar]
- 59.Tsang, A. P., Y. Fujiwara, D. B. Hom, and S. H. Orkin. 1998. Failure of megakaryopoiesis and arrested erythropoiesis in mice lacking the GATA-1 transcriptional cofactor FOG. Genes Dev. 12:1176-1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tsang, A. P., J. E. Visvader, C. A. Turner, Y. Fujuwara, C. Yu, M. J. Weiss, M. Crossley, and S. H. Orkin. 1997. FOG, a multitype zinc finger protein as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell 90:109-119. [DOI] [PubMed] [Google Scholar]
- 61.Vakoc, C. R., D. L. Letting, N. Gheldof, T. Sawado, M. A. Bender, M. Groudine, M. J. Weiss, J. Dekker, and G. A. Blobel. 2005. Proximity among distant regulatory elements at the beta globin locus requires GATA-1 and FOG-1. Mol. Cell 17:453-462. [DOI] [PubMed] [Google Scholar]
- 62.Weiss, M. J., G. Keller, and S. H. Orkin. 1994. Novel insights into erythroid development revealed through in vitro differentiation of GATA-1 embryonic stem cells. Genes Dev. 8:1184-1197. [DOI] [PubMed] [Google Scholar]
- 63.Weiss, M. J., C. Yu, and S. H. Orkin. 1997. Erythroid-cell-specific properties of transcription factor GATA-1 revealed by phenotypic rescue of a gene-targeted cell line. Mol. Cell. Biol. 17:1642-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Welch, J. J., J. A. Watts, C. R. Vakoc, Y. Yao, H. Wang, R. C. Hardison, G. A. Blobel, L. A. Chodosh, and M. J. Weiss. 2004. Global regulation of erythroid gene expression by transcription factor GATA-1. Blood 104:3136-3147. [DOI] [PubMed] [Google Scholar]
- 65.Zhang, Y., H. H. Ng, H. Erdjument-Bromage, P. Tempst, A. Bird, and D. Reinberg. 1999. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 13:1924-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou, Y., K. C. Lim, K. Onodera, S. Takahashi, J. Ohta, N. Minegishi, F. Y. Tsai, S. H. Orkin, M. Yamamoto, and J. D. Engel. 1998. Rescue of the embryonic lethal hematopoietic defect reveals a critical role for GATA-2 in urogenital development. EMBO J. 17:6689-6700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zon, L. I., M. F. Gurish, R. L. Stevens, C. Mather, D. S. Reynolds, K. F. Austen, and S. H. Orkin. 1991. GATA-binding transcription factors in mast cells regulate the promoter of the mast cell carboxypeptidase A gene. J. Biol. Chem. 266:22948-22953. [PubMed] [Google Scholar]
- 68.Zon, L. I., Y. Yamaguchi, K. Yee, E. A. Albee, A. Kimura, J. C. Bennett, S. H. Orkin, and S. J. Ackerman. 1993. Expression of mRNA for the GATA-binding proteins in human eosinophils and basophils: potential role in gene transcription. Blood 81:3234-3241. [PubMed] [Google Scholar]