Abstract
Hypoxia and DNA damage stabilize the p53 protein, but the subsequent effect that each stress has on transcriptional regulation of known p53 target genes is variable. We have used chromatin immunoprecipitation followed by CpG island (CGI) microarray hybridization to identify promoters bound by p53 under both DNA-damaging and non-DNA-damaging conditions in HCT116 cells. Using gene-specific PCR analysis, we have verified an association with CGIs of the highest enrichment (>2.5-fold) (REV3L, XPMC2H, HNRPUL1, TOR1AIP1, glutathione peroxidase 1, and SCFD2), with CGIs of intermediate enrichment (>2.2-fold) (COX7A2L, SYVN1, and JAG2), and with CGIs of low enrichment (>2.0-fold) (MYC and PCNA). We found little difference in promoter binding when p53 is stabilized by these two distinctly different stresses. However, expression of these genes varies a great deal: while a few genes exhibit classical induction with adriamycin, the majority of the genes are unchanged or are mildly repressed by either hypoxia or adriamycin. Further analysis using p53 mutated in the core DNA binding domain revealed that the interaction of p53 with CGIs may be occurring through both sequence-dependent and -independent mechanisms. Taken together, these experiments describe the identification of novel p53 target genes and the subsequent discovery of distinctly different expression phenomena for p53 target genes under different stress scenarios.
The tumor microenvironment is a major factor influencing tumor growth, metastatic potential, and response to chemotherapeutic drugs and radiation therapy (7). The hypoxia-inducible factor (HIF) family of transcription factors regulates the expression of key enzymes in anaerobic glycolysis and proangiogenic factors (i.e., vascular endothelial growth factor), aiding cells in adapting to a low-oxygen environment (15). Thus, a tumor adapts to hypoxia first by sustaining itself via anaerobic metabolism and then by stimulating angiogenesis to acquire more nutrients and oxygen. However, the resulting vasculature tends to have aberrant structures with irregular blood flow and leaky walls, paradoxically creating regions of transient hypoxia and reoxygenation (76). Reoxygenation after hypoxia causes DNA damage, exacerbating genomic instability (25). The porous structure of the tumor vasculature may also provide an escape route for metastasizing cells. Repeated cycles of hypoxia, vascular formation, and reoxygenation select for cells that are more likely to survive in a restrictive environment. The consequences of this process are cancer cells that are more aggressive, more likely to metastasize, and more likely to be resistant to radiation and chemotherapy. Intimately tied to this process is the selection for cells that have lost the expression of functional p53 (19).
A variety of cellular stresses, including DNA damage, oncogenic transformation, and hypoxia, stabilize the p53 protein by activating a host of stress-inducible kinases that phosphorylate p53 and MDM2, resulting in increased levels of p53 (17). Stabilized p53 binds palindromic DNA sequences found near the promoters of genes regulating cell cycle arrest (p21/Cip and GADD45A), apoptosis (APAF1, Fas, BAX, PERP, etc.), DNA repair (MSH2 and proliferating cell nuclear antigen [PCNA]), and p53 stability (HDM2) (reviewed in reference 26). While many genes known to be regulated by p53 are induced by the recruitment of coactivator proteins like CBP/p300, a distinct subset of genes (AFP, MAP4, etc.) is repressed primarily through interactions with corepressor complexes (31, 56, 57, 59). These studies demonstrate that p53 can perform different functions on different promoters under stress.
While p53 stabilization is usually thought of as a response to genotoxic stress, stabilization also occurs in the absence of DNA damage, as in the case of hypoxia (2, 19, 20, 23). Although the transcriptional consequences of stabilizing p53 during hypoxia remain to be elucidated, there is evidence that interactions between p53 and the transcription coactivator p300 are reduced compared to DNA-damaging stresses, while interactions between p53 and the corepressor mSin3a are retained. Accordingly, genes normally induced by p53 during DNA damage are unaffected by severe hypoxic stress (22, 43). Since the transcriptional upregulation of these targets is important for initiating the senescent and apoptotic effects of p53 during DNA damage, there may be a distinct set of genes affected during hypoxia that achieve similar results through different mechanisms (22).
Expression microarray analysis has been used to identify direct targets of transcription factors during a wide variety of conditions but is complicated by the fact that a number of genes may be secondary or tertiary targets. The additional process of identifying and testing potential regulatory elements near genes of interest may also further hamper the process. Chromatin immunoprecipitation (IP) (ChIP) coupled with DNA microarray analysis (ChIP-chip) is a powerful approach to identify the direct targets of transcription factor action. CpG island (CGI) arrays are a useful and economical microarray platform for ChIP-chip studies. CGIs are evolutionarily conserved elements corresponding to the promoters and regulatory regions of more than 50% of the genes in the human genome (6). CGI arrays have been used in a number of studies to identify regulatory elements bound directly by transcription factors, including E2F, MYC, and Suz12 (40, 53, 60, 82). To date, there have been several publications describing global ChIP-based approaches for identifying new p53-regulated genes, but none have used CGI arrays to do so (9, 27, 36, 81). The reasons for using other approaches may be based on the finding that only 12% of the identified p53 binding sites (p53BS) on chromosomes 21 and 22 were located within 1 kb of a CpG island (9). Nevertheless, we proceeded to utilize CGI arrays for three reasons. First, while chromosome 22 contains many CpG islands, chromosome 21 has relatively few (11, 12). The proximity of p53BS to CGIs might be more accurately determined within a broader survey of CGIs in the human genome. Second, if the estimated proximity of p53BS to CGIs is correct, even that number represents a large number of potentially new targets. Third, given that CpG islands are classically thought to regulate so-called housekeeping genes, we would be sampling a cohort of genes whose regulation by p53 could have profound effects on cellular function (6).
In this study, we describe the use of ChIP coupled with CGI microarray analysis to identify promoter regions bound by p53 during hypoxia and DNA damage. After confirming some of the targets by ChIP-PCR, we then analyzed the expression of a number of these targets. We have identified a number of previously unknown target promoters that are bound by p53 regardless of the stress. Although p53 binds to promoters in vivo during both hypoxia and DNA damage, the corresponding effect on gene expression is highly dependent on the specific stress and promoter. Mutation of p53 in the core DNA binding domain identified sequence-independent binding to some CGI loci. Using REV3L as a model, a luciferase reporter assay and a electrophoretic mobility shift assay (EMSA) confirmed the direct regulation of REV3L expression through a functional p53 response element. These studies confirm the complex nature of transcriptional regulation by p53 under both hypoxic and DNA-damaging stresses and highlight the complex nature of transcriptional regulation in general.
MATERIALS AND METHODS
Cell lines, culture conditions, and reporter assays.
HCT116 p53+/+, p53−/−, and p21−/− colon carcinoma cells were maintained in McCoy's medium supplemented with 10% fetal bovine serum. H1299 lung carcinoma cells stably transfected with HRE-p53 constructs have been described previously (22, 24). H1299-derived cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. All transfections were carried out using the Lipofectamine Plus system (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Luciferase activity was measured using the Dual Luciferase assay system (Promega, Madison, WI) according to the manufacturer's instructions.
Plasmid constructs.
A total of 1.8 kb of human genomic sequence spanning 1.2 kb of the REV3L promoter and 600 bp of the native transcript was amplified by PCR from human genomic DNA using Pfx DNA polymerase (Invitrogen). The 5′ and 3′ amplification primers included NheI and XhoI restriction sites, respectively. The amplified product was ligated into the NheI and XhoI sites of pGL3 basic (Promega), and the sequence of the construct was confirmed by dideoxy sequencing. The reporter plasmids pG13luc and pM15luc have been described previously (14) and were obtained from Bert Vogelstein (Johns Hopkins University). The pCEP4-p53 expression vector was obtained from Laura Attardi (Stanford University) with permission from Jennifer Pietenpol (Vanderbilt University).
Hypoxia treatment.
Cells were plated in glass dishes, and treatment was carried out in a hypoxia chamber (<0.02% O2; Sheldon Corp., Cornelius, OR) or in a mixed-gas incubator (2% O2).
RNA isolation and Northern hybridization.
Total RNA was isolated from 106 to 107 cells grown in monolayers with TRIzol reagent (Invitrogen) according to the manufacturer's protocol. Northern hybridization assays were performed using 5 μg of total RNA. Radiolabeled probes were synthesized by random priming (GE Health Care, Piskataway, NJ) from DNA fragments obtained by PCR amplification of human cDNA. Primers are available upon request.
Precipitation of [3H]thymidine-labeled DNA with TCA.
Cells were plated at 1 × 104 cells/well. One hour prior to harvesting, cells were labeled with 1 μCi/ml [3H]thymidine. Cells were lysed in a solution containing 25 mM Tris-HCl (pH 8.0), 25 mM EDTA, and 0.5% sodium dodecyl sulfate. Fifty percent trichloroacetic acid (TCA) was then added to a final concentration of 12%, and samples were incubated on ice for 20 min. Precipitated nucleic acids were collected on Whatman GF/C glass fiber filters (2.4-cm diameter). The filters were washed three times with ice-cold 5% TCA-20 mM sodium pyrophosphate and once with 70% ethanol. The amount of radioactivity was counted after the addition of Ecolume scintillant. Each experiment was carried out in triplicate.
Chromatin immunoprecipitation.
ChIP assays were performed as described previously by Krieg et al. (44), with the following modifications. HCT116 cells were exposed to the respective stress for approximately 12 h prior to formaldehyde fixation (variations are indicated for the respective experiments). For cells exposed to hypoxia (0.02% O2), cells were fixed within the chamber to avoid reoxygenation. For experiments conducted with H1299 cells stably transfected with murine p53 under the control of five copies of the hypoxia response element, cells were incubated for 12 h in a mixed-gas incubator equilibrated to 2% O2. H1299-derived cells were removed from the incubator immediately prior to formaldehyde fixation. Fixed and lysed cells were sonicated with eight 10-s bursts using a Sonix Vibra-cell 130 sonicator set to 90% power and equipped with a 3-mm tip. Diluted sonicates were measured for protein content using bicinchoninic acid reagent with bovine serum albumin as a standard (Pierce, Rockford, IL). After preclearing, approximately 200 μg of protein extract was incubated with 4 μg DO-1 anti-p53 antibody (Santa Cruz Biotechnology) or nonspecific mouse immunoglobulin G (IgG) overnight prior to the addition of a protein A-Sepharose slurry (Sigma, St. Louis, MO). Five percent of the sample (approximately 10 μg of DNA) from each immunoprecipitation was reserved for input controls. Immunoprecipitated complexes were washed, eluted, and de-cross-linked as previously described (44). DNA was purified with QIAquick PCR purification columns according to the manufacturer's instructions (QIAGEN Sciences, MA). A total of 1 to 2.5% of each IP was assayed by PCR using primers specific for a region of interest. For semiquantitative PCR, the sample signal was calculated by comparison to titrated inputs, separated on an agarose gel, stained with ethidium bromide, imaged using a GelDoc apparatus (Bio-Rad, Hercules, CA), and quantified with Imagequant (GE Health Care, Piscataway, NJ). Primers were designed with Primer3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) using default settings altered in the following manner: the optimal melting temperature was 63°C (range of 60 to 65°), and there was one GC clamp. Primer sequences are available upon request.
CGI array hybridization and analysis.
Immunoprecipitated DNA was blunt ended, ligated onto amplification linkers, and amplified by PCR as described previously by Oberley et al. (61). Ten nanograms of input was amplified in parallel reactions for hybridization controls. Two micrograms of amplified DNA corresponding to DO-1-immunoprecipitated DNA, input chromatin, and IgG control served as templates for random primer labeling using deoxynucleoside triphosphates (Invitrogen) spiked with amino-allyl dUTP (Sigma, St. Louis, MO). Inputs were performed in duplicate and pooled to provide the same control for both the IgG and the DO-1 immunoprecipitations. Amino-allyl-labeled amplicons corresponding to the DO-1 and IgG samples were incubated with Cy5 Mono-reactive NHS ester dye packs (GE Health Care, Piskataway, NJ), while the corresponding input samples were labeled with Cy3 Mono-reactive NHS ester dye (GE Health Care, Piskataway, NJ). Hybridization and wash conditions were described previously by Oberley et al. (61). Hybridized microarrays were scanned using an Axon 2000 scanner. Scanned images were visually inspected for defects prior to analysis with Gene Pix 4.0 software. Damaged spots were discarded. Data were loaded into the Stanford Microarray Database for image normalization and calculation of the log (base 2) of the normalized ratio of Cy5 to Cy3 based on the mean. Normalized ratios were exported to Excel (Microsoft, Redmond, WA). Values for IgG hybridizations were subtracted from the corresponding values obtained from the respective DO-1 hybridizations. The data presented are the mean enrichments for four separate ChIP experiments: two from hypoxia-treated HCT116 p53+/+ cells and two from adriamycin-treated cells. CGI clone identification numbers corresponding to spots enriched greater than twofold were identified by searching the CpG Island Microarray Bioinformatics Database (http://derlab.med.utoronto.ca/CpGIslands/) for the identities and genomic locations of corresponding CGIs (28). Loci were cross-referenced to the University Health Network Human CpG Island Microarray Database at the University of Toronto (http://data.microarrays.ca/cpg/). Clones with no corresponding sequences were discarded.
Identification of putative p53 response elements.
CGI sequences corresponding to enriched microarray spots were downloaded from the CpG Island Microarray Bioinformatics Database. One kilobase of sequence flanking either side of the CGI was searched for p53 response element (p53RE) sequences using MOTIF (http://motif.genome.jp/) and MatInspector (http://www.genomatix.de/) (8). For MOTIF searches, stringency was reduced to identify sites with a cutoff score of 75% similarity to the consensus. For MatInspector, the core similarity was set to 75%, while matrix similarity was reduced to “optimized-0.1.” All sites recognized as p53REs (including half-sites) were mapped to the sequence used to search for sites using Jellyfish (LabVelocity, Los Angeles, CA). For ChIP-PCR, primers were designed to flank putative elements identified by both search engines. Similarity to the consensus sequence (WWWCRRGYYY-N0-13-WWWCRRGYYY) (13), clustering of half-site sequences, and proximity to CGI sequence were all used as criteria for primer design.
qRT-PCR.
HCT116 cells (3 × 105 cells) were treated with severe hypoxia (0.02% O2) or 0.3 μg/ml adriamycin. One microgram of total RNA was reverse transcribed with MMLV reverse transcriptase (Invitrogen) with 5 μM random primers according to the manufacturer's instructions. Approximately 0.5% of each reverse transcription reaction mixture was added to reactions containing the following in a total volume of 10 μl: 5 μl 2× SYBR green master mix (ABI, Foster City, CA) and 50 nM forward and reverse primers specific for the genes of interest. Detection and data analysis were carried out with the ABI PRISM 7900 sequence detection system using 18S rRNA as an internal control. PCR primers were obtained from the Primer Bank Database (http://pga.mgh.harvard.edu/primerbank/). Primers were tested against pooled cDNA samples and analyzed by agarose electrophoresis to verify the formation of a single band after 40 rounds of PCR. Primer sequences are available upon request.
EMSAs.
EMSAs were performed as described previously by Johnson et al. (37), with the following modifications. Protein extracts were prepared from HCT116 p53+/+ cells treated with 0.3 μM adriamycin for 6 h. Thirty micrograms of protein extracts was incubated with 4.5 μg salmon sperm DNA in a solution containing 50 mM Tris (pH 8.0), 50 mM KCl, 7.5% glycerol, 0.5 mM EDTA, 0.5% Triton X-100, and 1 mM dithiothreitol for 20 min on ice. Pantropic antibody PAb 421 (0.3 μg; Cell Signaling) was added as required. A total of 40,000 cpm (approximately 15 fmol) of labeled oligonucleotide probe corresponding to a p53 response elements from the promoters of CDKN1A (p21), REV3L, SYVN1, and HNRPUL1 were added, and the reaction mixtures were incubated at room temperature for 20 min. Mutated competitor oligonucleotides contained alterations in the core sequence (CxxG was altered to TXXT). Sequences used for EMSA are as follows (putative binding elements are underlined): TGGCCATCAGGAACATGTCCCAACATGTTGAGCTCTGGCA (CDKN1A [see reference 1]), TCACCGGCAGCTGACAAGTCCTTACATGTACTTGCGCGTGCCCGGGACGCAGC (REV3L), GGCGTGGGTCGTACCAAGTGCGTCCTGCCCTGGCTCGCACTGCGCCCTACG (SYVN1), CAGAATTACACAGACACAAGCATATTTACAAACATGCTCAGGCAGATACCCGCCACGAATAG (HNRPUL1 RE1), and GACACAGACCTAGAGTCAAGTGACACAAATACTCAAGAGACATGTAGAGTACAACAGGTAC (HNRPUL1 RE2).
RESULTS
p53 binds the p21 and HDM2 promoters during both hypoxia and DNA damage.
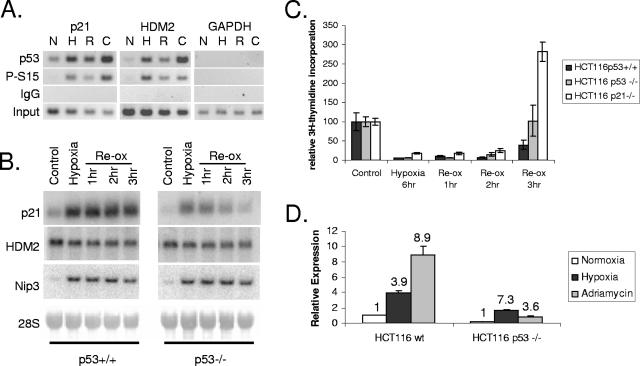
While hypoxia has been shown to stabilize p53, there have been few comprehensive studies regarding the ability of p53 to bind DNA and regulate transcription compared to DNA-damaging conditions. In order to determine the ability of p53 to bind known cognate response elements during hypoxic stress, chromatin immunoprecipitation of p53 was performed with cellular extracts from HCT116 cells exposed to hypoxia for 12 h. With an antibody against unmodified p53, substantial enrichment of the region spanning the p21 and HDM2 p53REs was observed during hypoxia, similar to what was observed after 12 h of treatment with 6 μM camptothecin (a topoisomerase poison known to induce p53) but more than what was observed for the corresponding normoxic control (Fig. 1A). The strength of stress-specific binding was particularly evident when an antibody against phosphorylated serine 15 was used, where no promoter enrichment was observed without stress.
FIG. 1.
p53 binds to known promoters under both hypoxic and genotoxic stresses. (A) Chromatin immunoprecipitation of p21 and HDM2 promoters. HCT116 cells were exposed to 12 h of hypoxia (0.02% O2) (H), 3 h of reoxygenation (R) following hypoxia, 12 h of camptothecin treatment (C), or normoxic control conditions (N) and processed for ChIP using DO-1 (p53) or an antibody against p53 phosphorylated on serine 15 (S15P). DNA was interrogated with primers specific for the p21 and HDM2 p53REs. GAPDH (glyceraldehyde-3-phosphate dehydrogenase) served as a negative control. (B) Northern blot analysis of p21 and HDM2 expression during hypoxia and reoxygenation. p53+/+ and p53−/− HCT116 cells were exposed to 12 h of hypoxia (H), followed by 1, 2, or 3 h of reoxygenation (Re-ox)-induced DNA stress. RNA was detected using probes specific for both p21 and HDM2. Nip3L served as a control for hypoxia, while 18S served as a loading control. (C) p53+/+, p53−/−, and p21−/− HCT116 cells were exposed to hypoxia for 6 h and then reoxygenated for the times indicated. Reentry into S phase is shown as a function of DNA synthesis determined by thymidine incorporation. (D) Quantitative real-time PCR analysis of p21 expression in both p53+/+ and p53−/− HCT116 cells. Cells were exposed to either hypoxia (black) or 0.3 μg/ml adriamycin (gray) for 24 h. Expression was normalized to the normoxic control in the p53+/+ cells (white). Numbers above bars refer to induction (n-fold) within the same cell type.
Binding of p53 to the p21 promoter corresponded to activation during DNA damage but not during hypoxia. Under hypoxia, both p53+/+ and p53−/− HCT116 cells upregulated expression of the p21 gene. Although the overall magnitude of induction under hypoxia was approximately the same, the presence of p53 maintained a higher overall level of p21 expression during both normoxic and hypoxic conditions (Fig. 1B and 1D). Under conditions of reoxygenation-induced DNA damage, the presence of p53 maintains p21 induction for at least 3 h after reoxygenation (Fig. 1B) and leads to reoxygenation-induced G1 arrest (Fig. 1C). The absence of p53 results in the rapid diminution of p21 expression and a loss of p21 arrest. Treatment of HCT116 cells with 0.3 μM adriamycin, a DNA-intercalating drug known to induce p53 function, strongly induced p21 expression in a p53-dependent manner (Fig. 1D). Although p53 binding does not contribute to the hypoxic regulation of p21, there is a rapid change in the transcriptional activity of p53 upon reoxygenation. In contrast, the HDM2 oncogene showed no dependence on p53 for expression under hypoxia- or reoxygenation-dependent stress (Fig. 1B). Thus, p53 can bind to the same targets under hypoxia as it does during DNA damage, but the transcriptional effects vary widely with the promoter.
p53 binds to the same CpG islands during hypoxia and DNA damage.
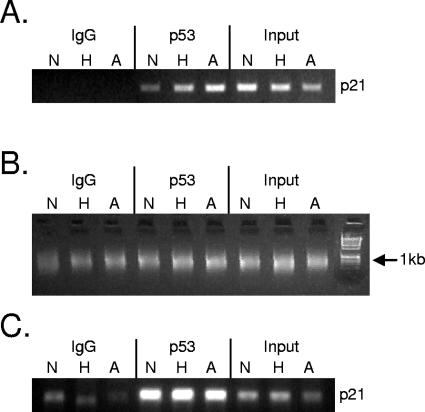
Since p53 bound to two well-characterized promoters irrespective of the specific stress, we investigated whether this effect was a global phenomenon. To address this question, DNA from two independent ChIP experiments was amplified and hybridized to CpG island microarrays using the protocol described previously by Oberley et al. (61). Both experiments consisted of HCT116 p53+/+ cells treated with severe hypoxia, adriamycin, or no stress for 12 h and processed for ChIP with DO-1 antibody (described in Materials and Methods). Both immunoprecipitated chromatin and input DNAs were ligated to linker primers and amplified by PCR. Chromatin was checked before and after amplification by PCR with primers specific for the 5′ p53RE in the p21 promoter (Fig. 2A and C). Before amplification, there was a distinct difference between the unstressed control and stressed cells (Fig. 2A). After ligation of linkers and PCR amplification, chromatin from all IP conditions was approximately 600 bp in length (Fig. 2B). After the amplification process, there was still a robust difference between the no-antibody control and the IP samples (Fig. 2C), but the stress-specific enrichment evident in Fig. 2A was lost. This is likely due to nonlinear amplification of the immunoprecipitated DNA during PCR. Amplicons derived from immunoprecipitated chromatin were labeled with Cy5 dye, mixed with Cy3-labeled amplicons derived from input DNA, and hybridized to an array containing approximately 12,000 CpG islands (Sanger 12k) (described in Materials and Methods). Spots enriched for p53 binding by more than twofold were selected for further analysis (approximately 150 loci). Chromosomal loci corresponding to enriched spots were identified from the CpG Island Microarray Bioinformatics Database (28) and the University Health Network Human CpG Island Microarray Database (see Materials and Methods). Repetitive loci, duplicated sequences, and loci farther than 10 kb from identified genes were removed from the list. The top 44 remaining loci are presented in Table 1. Genes located near these loci perform a broad range of functions including DNA synthesis and repair (REV3L [48] and PCNA [73]), transcription (XPMC2H [55], HNRPUL1 [45], BANP [67], ATF7 [64], ZNF215 [3], and MYC [21]), mitochondrial function (CA5A [58] and COX7A2L [79]), and mitotic spindle assembly (CDCA8) (16). One target has direct involvement in the secretory functions of the cell (COH1) (41), while another target has a direct link to the unfolded protein response (UPR) (SYVN1) (75). CGI ChIP-chip to identify p53 targets genes therefore identifies a broad range of potential target genes that may mediate direct effects on p53 function.
FIG. 2.
Representative ChIP experiment used for CGI microarray hybridization. (A) Representative ChIP experiment used for CGI array experiments. HCT116 p53+/+ cells were exposed to hypoxia (H), 0.3 μg/ml adriamycin (A), or control conditions (N) for 12 h. PCR against the p21 p53RE was used to verify specific enrichment. (B) Verification of DNA amplification after ligation-mediated PCR (LM-PCR). Approximately 10% of amplified DNA was electrophoresed on a 1.5% agarose gel alongside a 1-kb DNA ladder. All amplicons are approximately 600 bp in length. (C) PCR for p21 p53RE after amplification. Ten nanograms of LM-PCR amplicon was added to a PCR using primers specific for the p21 p53RE. For all figures, IgG refers to samples derived for the IP-negative control, p53 refers to DNA immunoprecipitated with DO-1 antibody, and input refers to the DNA derived from samples prior to immunoprecipitation.
TABLE 1.
Representative list of p53 targets identified by CGI screena
| Gene | Description | Chromosomal coordinateb | Positionc | Enrichment (fold) |
|---|---|---|---|---|
| REV3L | REV3-like catalytic subunit of DNA polymerase ζ (yeast) | 6 | P | 6.55 |
| XPMC2H | Prevents mitotic catastrophe 2 homolog (Xenopus laevis) | 9 | P | 4.82 |
| ADAMTS13 | Von Willebrand factor-cleaving protease | U | ||
| HNRPUL1 | Heterogeneous nuclear ribonucleoprotein U-like 1 | 19 | P | 3.37 |
| LOC400341 | Hypothetical gene supported by GenBank accession no. NM_173613 (pseudo) | 15 | P | 2.69 |
| ARHGAP11A | Rho GTPase-activating protein 11A | P | ||
| TOR1AIP1 | Torsin A-interacting protein 1 | 1 | I-1 | 2.65 |
| LOC440392 | LOC440392 | 16 | U | 2.64 |
| BANP | BTG3-associated nuclear protein | U | ||
| CA5A | Carbonic anhydrase VA, mitochondrial | U | ||
| GPX1* | Glutathione peroxidase 1 | 3 | P | 2.62 |
| SCFD2 | Sec1 family domain-containing 2 | 4 | P | 2.59 |
| COH1 | Vacuolar protein-sorting 13B (yeast), Cohen syndrome protein 1 | 8 | P | 2.47 |
| ZNF611 | Zinc finger protein 611 | 19 | I-1 | 2.46 |
| COX7A2L | Cytochrome c oxidase subunit VIIa polypeptide 2-like | 2 | P | 2.41 |
| TMEM33 | Transmembrane protein 33, FLJ10525 | 4 | P | 2.39 |
| SYVN1 | Synovial apoptosis inhibitor 1 | 11 | P | 2.39 |
| CTDSPL2 | CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like 2 | 15 | P | 2.36 |
| JAG2* | Jagged 2 | 14 | P | 2.33 |
| NUDT14 | Nudix (nucleoside diphosphate-linked moiety X)-type motif 14 | D | ||
| ATF7 | Activating transcription factor 7 | 12 | P | 2.33 |
| HIST1H2BJ | Histone 1, H2bj | 6 | P | 2.33 |
| UBE1C | Ubiquitin-activating enzyme E1C (UBA3 homolog, yeast) | 3 | P | 2.32 |
| ARL6IP5 | ADP-ribosylation-like factor 6-interacting protein 5 | U | ||
| DNMT2 | DNA (cytosine-5) methyltransferase 2 | 10 | P | 2.31 |
| PTP4A2 | Protein tyrosine phosphatase type IVA, member 2 | 1 | I-1 | 2.24 |
| TORC3 | Transducer of regulated CREB protein 3 | 15 | I-1 | 2.24 |
| ZNF215 | Zinc finger protein 215 | 11 | P | 2.23 |
| KBTBD7 | Kelch repeat and BTB (POZ) domain-containing 7 | 13 | P | 2.23 |
| CDCA8 | Cell division cycle-associated 8 | 1 | P | 2.23 |
| FLJ20508 | Chromosome 1 open reading frame 109 | P | ||
| MGA | MAX gene associated | 15 | I-1 | 2.22 |
| LOC342460 | Novel protein similar to heparan sulfate (glucosamine) 3-O-sulfotransferases | 16 | U | 2.20 |
| GPR89 | G-protein-coupled receptor 89 | 1 | P | 2.20 |
| TAF1L | TAF1-like RNA polymerase II, TATA box binding protein-associated factor, 210 kDa | 9 | U | 2.19 |
| RPH3AL | Rabphilin 3A-like (without C2 domains) | 17 | I-5 | 2.19 |
| EIF3S9 | Eukaryotic translation initiation factor 3, subunit 9 eta, 116 kDa | 7 | I-1 | 2.18 |
| MRPL27 | Mitochondrial ribosomal protein L27 | 17 | P | 2.18 |
| EME1 | Essential meiotic endonuclease 1 homolog 1 (Schizosaccharomyces pombe) | P | ||
| PRO1855 | Hypothetical protein PRO1855 | U | ||
| ARL17P1 | ADP-ribosylation factor-like 17 pseudogene 1 | 17 | I-1 | 2.18 |
| ZNF498 | Zinc finger protein 498 | 7 | P | 2.16 |
| ASCC3 | Activating signal cointegrator 1 complex subunit 3 | 6 | E-1/I-1 | 2.16 |
| KIAA1539 | KIAA1539 | 9 | P | 2.15 |
| MYC* | v-myc myelocytomatosis viral oncogene homolog (avian) | 8 | I-2 | 2.14 |
| HIST1H3A | Histone 1, H3a | 6 | E-1 | 2.14 |
| HIST1H1A | Histone 1, H1a | U | ||
| HIST1H4A | Histone 1, H4a | U | ||
| HIST1H4B | Histone 1, H4b | D | ||
| ZFP37 | Zinc finger protein 37 homolog (mouse) | 9 | P | 2.14 |
| GNB2L1 | Guanine nucleotide binding protein (G protein), beta polypeptide 2 like 1 | 5 | P | 2.14 |
| TRIM41 | Tripartite motif-containing 41 | D | ||
| HNRPR | Heterogeneous nuclear ribonucleoprotein R | 1 | P | 2.13 |
| GGH | Gamma-glutamyl hydrolase (conjugase, folylpolygammaglutamyl hydrolase) | 8 | P | 2.12 |
| LOC441193 | Similar to zinc finger protein 469 | 7 | U | 2.12 |
| CDA08 | T-cell-immunomodulatory protein precursor (CDA08) | 16 | I-6 | 2.11 |
| PCNA* | Proliferating cell nuclear antigen | 20 | P | 2.11 |
| CDS2 | CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 2 | U | ||
| C20orf30 | Chromosome 20 open reading frame 30 | U |
Targets are listed in descending order of microarray enrichment. Genes located within 10 kb of a CpG island are shown with the corresponding chromosome, approximate position of the CGI relative to each gene, and average enrichment during stress.
Chromosomal coordinates for each CGI are listed in Fig. S1 of the supplemental material.
The general position of the CpG island relative to each gene is described with a single-letter code: P, promoter; E, exon; I, intron; U, upstream; D, downstream. Numbers define the identity of the respective intron or exon.
CGI ChIP-chip identified known and novel p53 target genes.
As expected, the ChIP-chip screen identified some known p53 target genes: glutathione peroxidase 1 (GPX1) (74), PCNA (72), jagged 2 (JAG2) (70), and c-myc (MYC) (30). GPX1, PCNA, and MYC have p53 response elements that have been described previously (72, 74, 78). The promoters for both GPX1 and PCNA have p53REs within the CGIs identified from the array. Interestingly, three CGIs have been mapped to the PCNA locus (6 kb upstream, 500 bp upstream, and 6 kb downstream) (28), but only one was enriched for p53 binding (2.11-fold enrichment) and contains a previously reported p53RE at −278 bp (72). The MYC locus has several predicted p53REs located throughout the exon and intron structure of the human gene (78). Our CGI screen identified one of these elements located 457 bp into the second intron (+3,391 bp downstream of the primary transcription start site). This location is approximately 400 bp from the 3′ end of the CGI spanning exon 2. A number of p53 target genes that were represented by the CGI library did not appear in the analyzed data, including APAF1, BCL2, GADD45A, TNFRSF10B (Killer/DR5), and PA26 (Table 2). Upon further examination of the sequence surrounding these genes, it was readily apparent that most of the p53 elements described for these genes resided outside of the resolution of the ChIP assay. Three notable exceptions for this phenomenon were DR5/Killer, APAF1, and BCL2. The p53REs for the DR5 and APAF1 loci are located approximately 400 bp away from their respective CGIs. While this is within the theoretical limit of the ChIP-chip experiment, they exhibited weak enrichment (1.93- and 1.4-fold, respectively). A substantial number of targets (CDKN1A [p21/Cip], BAX, BIRC5 [survivin], etc.) were not represented on the Sanger 12k microarray and were not identified in the ChIP-chip screen (Table 2).
TABLE 2.
List of known p53 targets on the CGI 12,000-gene arraya
| p53 target | Position
|
Enrichment (fold) | ||
|---|---|---|---|---|
| p53RE location | CGI 5′ end | CGI 3′ end | ||
| APAF-1 | −765 | −1,204 | −1,045 | 1.54 |
| −604 | ||||
| BCL2 | −526 | −945 | −483 | 1.40 |
| GADD45A | +1,518 (intron 3) | −384 | 429 | 1.36 |
| GPX-1 | +61 | −30 | +278 | 2.62 |
| Killer/DR5 | +245 (intron 1) | 546 | 1,383 | 1.93 |
| HDM2 | +727 (intron 1) | −877 | −475 | 0.83 |
| MMP2 | −1,649 | 209 | 1,041 | 1.4 |
| MYC | +957 | −2,086 | −1,815 | 1.46 |
| +3,391 | 1,154 | 1,513 | 1.13 | |
| 2,130 | 3,008 | 2.14 | ||
| PCNA | −278 | −6,292 | −6,112 | 0.61 |
| −583 | −104 | 2.11 | ||
| 6,682 | 7,093 | 1.68 | ||
| Serpine 1 | −1,458 | 9,903 | 10,498 | 1.28 |
| Sestrin-1 (PA26) | +86,087 (intron 2) | −1,984 | −1,332 | 1.07 |
| Sestrin-3 | −1,502 | −1,592 | 1.22 | |
Each of the known p53 targets found on the CGI 12,000-gene array is listed alphabetically. The positions of p53REs (if mapped) and CpG islands relative to the transcription start site are tabulated. CGIs with more than one representation are listed in order. The known p53 targets not found on the CGI 12,000-gene array are as follows: p21, BAX, PERP, insulin-like growth factor binding protein 3, EGFR, actin 2, survivin, MAP4, and Cdc25C.
p53 response elements are located near CpG islands.
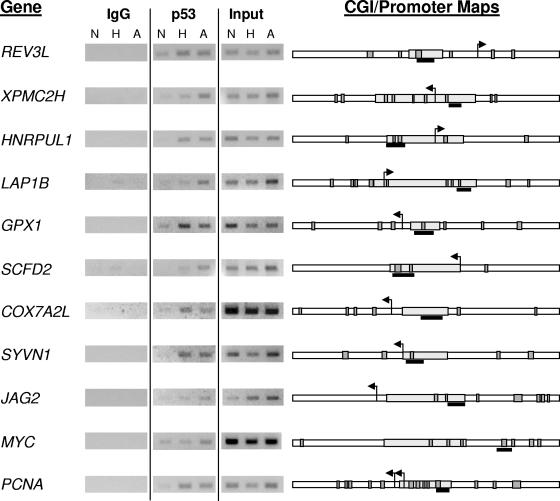
Most reported interactions of p53 with DNA occur through the binding of the central DNA binding domain to the consensus palindromic sequence RRRCWWGYYY (13). This “half-site” sequence is separated from another half-site by 0 to 13 bp in most cases. However, a number of known p53 targets are regulated through a broad range of degenerate sequences (66), making the identification of functional p53REs potentially problematic. In order to individually confirm p53 association with selected promoters, we first identified potential p53REs in the vicinity of identified CGIs. Since the average length of the amplicons used for our ChIP-chip experiments was approximately 600 bp (Fig. 2B), we felt that it was unlikely that we could detect p53-DNA interactions more than 1 kb away from a CpG island sequence. We used two different search engines (MOTIF and MatInspector) (see Materials and Methods) to identify potential p53REs within 1 kb of either side of the CGIs. The stringency of the search parameters was relaxed to account for degeneracy in the p53RE sequences. Only elements identified by both of the search engines were mapped to promoters. PCR primer pairs were designed to span putative regulatory elements near CGIs identified from the array, with preference given to elements located within the CpG island or in clusters. Eleven of the genes screened for p53 binding with ChIP-PCR are shown in Fig. 3. As might be expected from the CGI microarray, despite some variation in p53 recruitment to the various regulatory elements during hypoxia or adriamycin treatment, there was more robust association of p53 during either stress than without stress (normoxia). The majority of the p53REs identified have so far been located near the transcription start site of the gene and within a CGI sequence. One notable exception is the p53 response element located within intron 2 of MYC. The identification of MYC as a target of p53 repression has been reported previously, with the latest report showing direct in vivo evidence of p53 binding in a mouse system (30, 54). We have individually confirmed the association of p53 with a number of targets identified with a global ChIP-chip screen. These results confirm our initial hypothesis that p53 binding to promoters is a result of stress-induced protein stability and is independent of the specific stress.
FIG. 3.
Individual validation of p53 association. ChIP experiments used to verify p53 binding are independent replicates of experiments used for the CGI array experiments. HCT116 cells were exposed to hypoxia (H), adriamycin (A), and control (N) conditions. p53 IPs, no-antibody controls, and inputs are labeled accordingly. CGI sequences are mapped to the right of the ChIP experiments. CGI sequences (gray rectangles) are shown with 1 kb of flanking sequence on either side (white rectangles). Putative p53REs mapped to sequences are shown as dark gray boxes, with the amplicon verifying ChIP enrichment shown as a black bar.
p53 target genes are differentially regulated by stress.
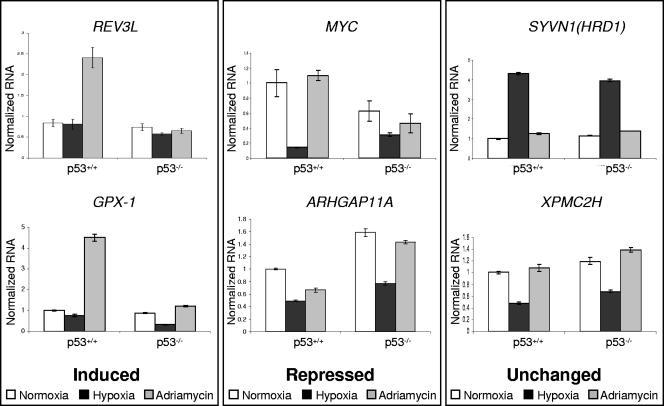
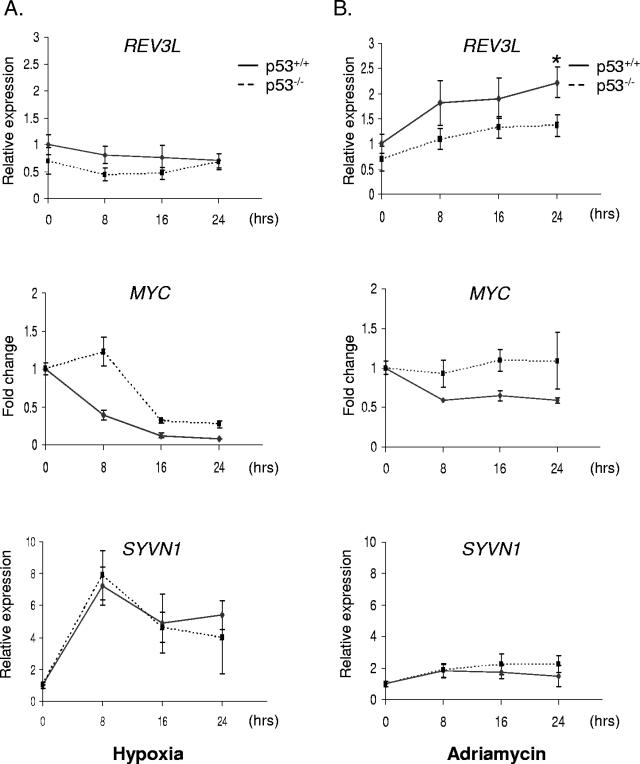
The binding of p53 to target promoters can result in either transcriptional activation or repression, depending on the promoter and the nature of the stimulus. We therefore screened for p53-dependent expression of a number of the targets from the array. RNA was harvested from p53+/+ and p53−/− HCT116 cells after exposure to 0.02% O2 or 0.3 μg/ml adriamycin and assayed by qRT-PCR. We found that REV3L, the human homolog of the catalytic subunit of DNA polymerase ζ (18), is induced by DNA damage (Fig. 4 and 5B). While there is some induction of REV3L by adriamycin in the absence of p53 (Fig. 5B), the expression of p53 in HCT116 cells resulted in a significant (P = 0.03) increase in p53-dependent expression by 24 h of treatment. Variations in the kinetics of REV3L induction may represent subtle differences in the culture conditions between experiments. Consistent with prior publications (74), GPX1 was induced approximately fourfold after 24 h of adriamycin treatment (Fig. 5). GPX1 is a selenoenzyme that uses glutathione as a substrate to convert reactive peroxides into nonreactive alcohols, protecting cells against damage by reactive oxygen species. There was little effect of hypoxia treatment on the expression of REV3L or GPX1 (Fig. 4 and 5A). These results are consistent with previous publications showing that p53 preferentially interacts with the p300 coactivator during DNA damage but not during hypoxic stress (22, 43).
FIG. 4.
Expression analysis of p53 target promoters. p53+/+ and p53−/− HCT116 cells were exposed to hypoxia (black) or adriamycin (gray) for 24 h. Normalized RNA levels of REV3L, GPX1, MYC, ARHGAP11A, SYVN1, and XPMC2H are shown. Analyses are arranged in columns according to whether genes are induced, repressed, or unaffected by the p53 status. Data are representative of multiple similar experiments analyzed in triplicate.
FIG. 5.
Kinetic analysis of p53 regulation. p53+/+ (solid line) and p53−/− (dashed line) HCT116 cells were exposed to (A) hypoxia or (B) 0.3 μg/ml adriamycin. RNA was harvested at 0, 8, 16, and 24 h and analyzed with primers specific for REV3L, MYC, and SYVN1. Data represent the averages of three independent replicates ± standard errors of the means. The asterisk above the 24-h time point for REV3L expression during adriamycin treatment denotes a statistical difference (P = 0.03) in expression as determined by Student's t test.
We also found that the proliferative oncogene MYC was repressed in the presence of p53 (Fig. 4). While MYC was also repressed by hypoxia in the p53−/− cells, this effect exhibited different kinetics (Fig. 5A). In the p53+/+ cell line, there was a 50% decrease in expression within 8 h of hypoxia. In the absence of p53, MYC expression decreased after 16 h of hypoxic exposure. This difference in kinetics highlights a clear mechanistic role for p53 in reducing proliferative gene expression in response to replication stress. While there appears to be a general trend of MYC repression by p53 in response to DNA damage (Fig. 5B), the difference between wild-type and p53 null cells was more difficult to discern. This may be a result of asynchronous growth of the p53−/− cells during adriamycin treatment. These results were consistent with the findings described previously by Ho et al. (30), who showed that repression is the result of p53 recruitment of corepressors to the MYC promoter. A few other CGI targets also appear to be repressed by p53 in a manner dependent and independent of stress: ARHGAP11A, a member of the Rho GTPase-activated protein family with unknown function, is repressed by p53 under normoxic conditions and is further repressed about twofold under DNA-damaging stress (Fig. 5). TOR1AIP1 (LAP1B), a nuclear membrane protein with a potential role in cell cycle regulation (87), is generally repressed by p53 expression but has no discernible dependence on stress (data not shown).
A large number of genes have surprisingly little if any dependence on p53 for expression, even though p53 binds near the promoters of these genes. In particular, the stress response protein SYVN (HRD1) (38) shows strong induction under hypoxia without any dependence on p53 (Fig. 4 and 5A). After 8 h of exposure to hypoxia, SYVN1 is induced approximately eightfold and continues to be expressed at high levels throughout the remainder of the experiment (Fig. 5A). The kinetics of induction are virtually identical in both cell lines, even though p53 has robust stress-dependent binding near the promoter of the gene. More telling is the distinct lack of expression during genotoxic stress (Fig. 4). With adriamycin treatment, SYVN1 expression is identical in both wild-type and knockout cell lines. Another locus that ranks highly as a p53 target and that has been confirmed by PCR (Fig. 3) is the promoter of XPMC2H (REXO4), a homolog of Saccharomyces cerevisiae RNA exonuclease 4 and a potential transcription coregulator (55). Under hypoxic conditions, XPMC2H is down-regulated in both p53+/+ and p53−/− HCT116 cells, but there does not appear to be a difference in expression between cell lines. There is no difference in the expression of the gene in the normoxic controls or the cells treated with adriamycin (Fig. 4). A number of other genes display p53-independent expression during both hypoxia and adriamycin treatment (data not shown). This list of genes includes COX7A2L, a nuclear-encoded mitochondrial protein that may regulate cytochrome c oxidase activity (79); LOC400341, a hypothetical gene that has a CGI with an identical sequence to the one near ARHGAP11A; and BANP, a component of the nuclear scaffold that binds BTG3 and may enhance p53-dependent transcription (67). Two genes, ADAMTS13 and JAG2, are located near confirmed p53 targets but appear to be silenced in HCT116 cells, as expression could not be detected with qRT-PCR. JAG2, a member of the delta family of signaling molecules, is regulated by the p53 family of transcription factors (70). qRT-PCR of JAG2 using cDNA from H1299 cells expressing a tetracycline-inducible p53 construct confirmed this result. These analyses demonstrate that promoters bound by p53 can be regulated in very different ways depending on the specific stress, promoter, or cell type.
Binding of p53 to CGI targets is partially dependent on a functional DNA binding domain.
Aside from its well-described function as a transcription factor, numerous reports in the literature have also described p53 as a structural factor that mediates recognition and repair of DNA damage (4, 71). In contrast to the “classical” model of p53 binding DNA through contacts of the core DNA binding domain with palindromic sequences, p53 can also bind certain DNA structures through the C-terminal domain (47). This structural mode of binding by p53 may recruit repair proteins to DNA structures like sites of recombination, mispaired bases, or double-strand breaks (71). Since several CGIs do not have distinct transcriptional phenomena associated with p53 binding, it is possible that some of the CGI sequences identified from our screen represent this alternative mode of binding. Additionally, there is a growing body of evidence demonstrating the association of transcription factors with promoters through binding to more general transcription factors like AP-1 and NF-Y (33-35, 80). Mutations in the core DNA binding domain would be expected to abolish classical interactions with DNA while preserving these other modes of binding.
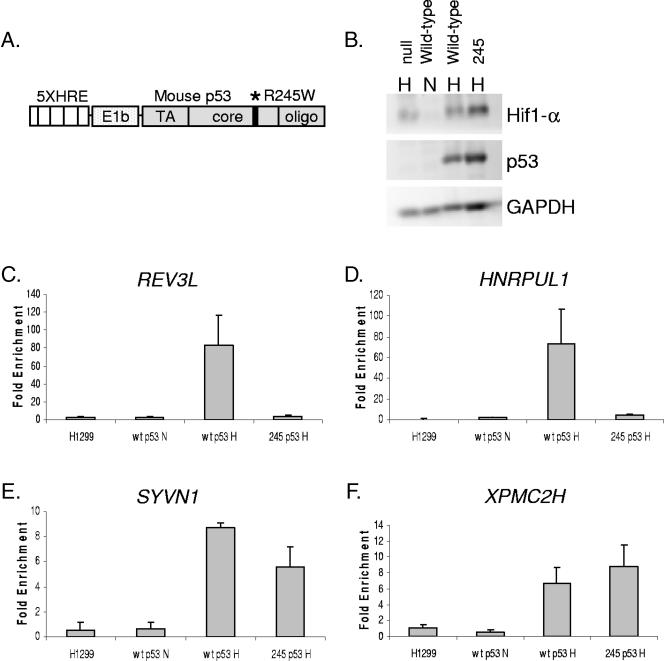
In order to distinguish between classical and other modes of binding, we made use of a series of cell lines derived from H1299 lung carcinoma cells. These isogenic lines express various forms of murine p53 under the control of five copies of the hypoxia response element, allowing the expression of p53 only during HIF stabilization (Fig. 6A) (22). H1299 cells expressing either wild-type p53 (p53wt) or p53 mutated at the core DNA binding residue 245 (p53245) were exposed to hypoxia for 12 h to allow full expression of p53 (Fig. 6B). Cells were fixed with formaldehyde and processed for ChIP using CM5 anti-p53 antibody. Using primers specific for the REV3L, HNRPUL1, XPMC2H, and SYVN1 p53 binding regions, p53 association was quantified with semiquantitative PCR against titrated input DNA (Fig. 6C to F). For all four of the regions screened, there was a robust increase in the association of p53wt when it was expressed during hypoxia (Fig. 6, wtp53N versus wtp53H), reflecting the massive increase in p53 protein expression. In contrast, there is a distinct difference in p53245 association: while the association with REV3L and HNRPUL1 decreases to near background levels when arginine 245 is mutated, there is only a slight reduction in p53 interactions with the SYVN1 binding region (Fig. 6E) and none with XPMC2H (Fig. 6F). Therefore, binding of p53 to these two nonresponsive CGIs is likely the result of p53 associating with DNA independently of the core DNA binding domain.
FIG. 6.
Binding to CGI targets is mediated primarily through the core DNA binding domain. (A) Diagram of the HRE-p53 expression construct. The location of the R245W core DNA binding domain mutation (mouse p53245 [mp53245]) is indicated with an asterisk. (B) Western blot demonstrating hypoxic expression of p53. H1299 cells stably transfected with either HRE-mp53wt or HRE-mp53245 were incubated in 2% O2 for 12 h, harvested, and analyzed for p53 expression by Western blot using CM5 polyclonal antibody. (C to F) H1299 lung carcinoma cells stably expressing either HRE-mp53wt or HRE-mp53245 were exposed to 2% O2 for 12 h, fixed in formaldehyde, and processed for ChIP using CM5 antibody. Data represent the ratio of CM5 to IgG signals (Fold Enrichment), as calculated by semiquantitative PCR against the REV3L (C), HNRPUL1 (D), SYVN1 (E), and XPMC2H (F) p53 binding sites.
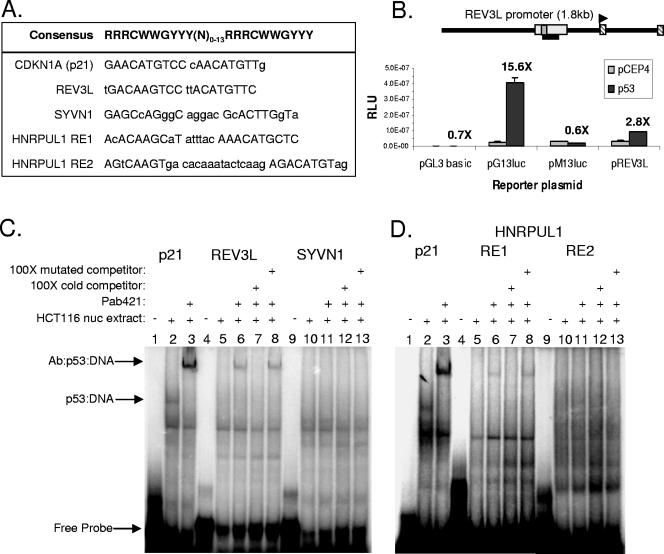
To further confirm the nature of the p53-DNA interaction with these various CGI sequences, the p53REs located within the REV3L, HNRPUL1, and SYVN1 promoters were analyzed by EMSA (Fig. 7). Within the ChIP amplicon for REV3L, there is one p53 response element (Fig. 7A). Located 433 bp upstream of the transcription start site, this element deviates from the consensus sequence by 3 bp. This element formed a complex in vitro with p53 when supplemented with PAb 421 (Fig. 7C, lane 6). This binding could be competed away with an excess of unlabeled probe but not a probe containing mutations in the core CXXG sequence of the binding site (Fig. 7C, lanes 7 and 8, respectively), indicating that p53 binds the REV3L promoter through a classical p53-p53RE interaction. In a luciferase reporter assay, the REV3L promoter is induced by the expression of p53 by approximately threefold (Fig. 8B), recapitulating the native expression pattern of the gene in HCT116 cells and verifying a p53-dependent activation mechanism. Showing similar binding characteristics to REV3L was one of the p53 elements from the HNRPUL1 promoter. The region shown to be occupied by p53 (Fig. 3 and 6D) contains two potential p53 response elements located 361 bp (RE1) and 286 bp (RE2) upstream of the transcription start site (Fig. 7A). RE1 is bound by p53 in EMSA experiments, while RE2 is not (Fig. 7D, lanes 3 to 8 versus lanes 9 to 13). Binding to RE1 is specific for p53, as the shifted band appears only with PAb 421 and can be competed away with the unlabeled element but not a mutated form. RE1 deviates from the consensus by only 2 bp, but the two half-sites are separated by 6 bp, likely reducing the affinity of p53 compared to the p21 probe (Fig. 7C and D). We could not detect in vitro binding to the sequence most likely to be a functional p53 element in the SYVN1 promoter (Fig. 7A and C, lanes 9 to 13). This p53RE, located 69 bp upstream of the transcription start site, deviates from the consensus by 6 bp. The two half-sites are separated by 13 bp, making it unlikely that p53 binds through a conventional mechanism. All of these results are consistent with those shown in Fig. 6, where we could differentiate native promoter associations that were dependent on a functional DNA binding domain (REV3L and HNRPUL1) from those that were not (SYVN1). These results suggest that the association of p53 with CGIs represents multiple DNA binding modes.
FIG. 7.
p53 binds the REV3L and HNRPUL1 p53 response elements in vitro. (A) p53 response elements corresponding to their respective promoters are shown, with deviations from the consensus displayed in lowercase type. (B) H1299 cells were transfected with 1 ng of pCEP4 or pCEP4-p53; 400 ng of pGL3 basic, pG13luc (positive control), pM13luc (negative control), or pREV3luc (promoter diagram shown above the graph); and 0.5 ng of pRLSV40. Inductions (n-fold) are shown above the respective constructs. Results represent averages of triplicate experiments. (C and D) EMSA of putative p53 response elements. RLU, relative light units. (C) EMSA of REV3L and SYVN1 p53REs. HCT116 whole-cell extract was incubated with labeled oligonucleotides corresponding to the p21, REV3L, and SYVN1 p53REs, respectively (lanes 2, 5, and 10). Reaction mixtures were incubated with PAb 421 (Ab) specific for p53 to generate supershifted bands (lanes 3, 6 to 8, and 11 to 13). One hundred-fold excess unlabeled competitor and mutated probes were used to probe binding specificities (lanes 7 and 12 and lanes 8 and 13, respectively). Lanes 1, 4, and 9 contain free probe but no extract. Arrows denote the migration of free probe, p53-probe complexes (visible only with p21), and antibody supershift of the p53-DNA complexes. nuc, nuclear. (D) EMSA to identify the functional p53 response element in the HNRPUL1 promoter. Lanes correspond to the same conditions described in C, with the respective probes shown above each set of experiments.
FIG. 8.
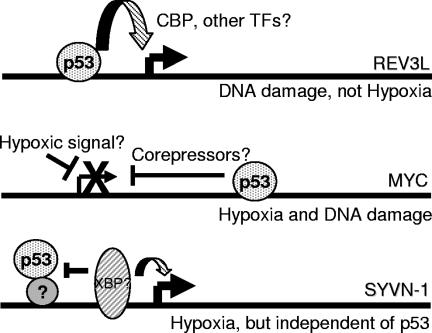
Model for p53 action on CGI targets. Under stress, p53 is stabilized and binds to response element sequences located near a number of promoters. During DNA damage, p53 induces the expression of the REV3L gene, perhaps through an increased association with transcriptional coactivators. In contrast, stress-specific signals during hypoxia may cooperate with p53 bound to the second intron of the MYC gene to recruit corepressors. Either of these conditions may depend on the actions of accessory transcription factors. Other genes like SYVN1 and a number of other genes identified on the CGI array may have promoter environments that permit indirect p53 binding but lack the necessary factors required for p53 to contribute to either an activating or a repressive effect. TFs, transcription factors.
DISCUSSION
CGIs to identify p53 targets.
The ability of the p53 tumor suppressor to regulate transcription depends on the specific promoter or stress. This disparity is particularly evident in the case of hypoxia, where p53 can exert its effects on apoptosis without interacting with transcription coactivators (22, 43). In this study, we have undertaken a global screening approach to identify promoters that are bound by p53 under both DNA-damaging and hypoxic stresses. Initially, the goal was to determine the promoter specificity of p53 during different stresses, but our initial analysis revealed that p53 bound the same targets regardless of the stress. Additionally, examining loci bound by p53 under both stresses would be more likely to reveal insight into the mechanism of transcriptional regulation by p53. Using CGI arrays to screen p53 ChIP experiments, we have identified genes that are known p53 targets (GPX1, PCNA, JAG2, and MYC) while also identifying unique targets (REV3L, SYVN1, LAP1B, XPMC2H, HNRPUL1, etc.). Although we have not confirmed associations with every target identified from the array, we have used gene-specific PCR analysis to verify associations with loci that were highly enriched (REV3L and GPX1) and weakly enriched (MYC and PCNA) for p53 binding during stress. Since we also started with DNA that had very specific enrichment for p21, we feel that the data represent a broad sampling of p53 association throughout the human genome.
Although we did not observe enrichment of all of the known p53-dependent targets on the array, most that were not identified were located outside the resolution limit of the ChIP amplicons (Fig. 3). Three loci fell within the theoretical range of the ChIP experiment but were not enriched enough to meet our criteria for inclusion: APAF1, BCL2, and TNFRSF10B (Killer/DR5). These three genes are involved in the apoptotic response of p53 to cellular stress. It is interesting that a recent publication using a functional screen of ChIP-isolated p53 targets also failed to observe the enrichment of these and other apoptotic targets of p53 (27). Microarray analysis of p53-dependent expression in response to adriamycin treatment in HCT116 cells did reveal an induction of APAF1, TNFRSF6 (Fas/Apo1), TNFRSF10B (Killer/DR5), and BBC3 (see Table S2 in the supplemental material). Therefore, the subthreshold enrichment of these targets may be due to technical limitations of the conditions used for amplification of the immunoprecipitated DNA.
p53 targets identified from the CGI array are regulated in a stress-specific manner.
The genes that we identified from this screen could be characterized as induced, repressed, or nonresponsive to p53 binding (modeled in Fig. 8). An induced gene of particular interest was REV3L, the catalytic subunit of DNA polymerase ζ (18). Although p53-dependent induction is not as high as it is for genes like p21, a two- to threefold increase of a key DNA repair enzyme may have profound consequences. DNA polymerase ζ is essential for translesion synthesis, resulting in increased survival after cisplatin treatment or exposure to DNA-alkylating chemicals (86). Translesion synthesis allows replication to progress past DNA lesions when nucleotide excision repair (NER) mechanisms have been overwhelmed (65). Previous work conducted with colon carcinoma cells has described an increase in cisplatin sensitivity after genetic ablation of p53 (49). This was later shown to be due to REV3L expression and activity (50), although a direct link between the two phenomena was not described. Additionally, homozygous deletions of REV3L in mice resulted in embryonic lethality due to an inability to repair genetic lesions that occur during the developmental process (84). Embryonic fibroblasts derived from these mice accumulate gross genetic abnormalities and become quiescent (85). Thus, our data describing a p53-dependent induction of REV3L expression at the transcriptional level provide a direct link between tolerance to DNA damage and p53 activity. It is also interesting that DDB2, the subunit of the NER complex that binds to DNA lesions and recruits the NER machinery, is also regulated by p53 (32). Thus, p53 may influence both aspects of the cellular response to genotoxic DNA lesions.
We have also identified GPX1 as a direct p53 target induced by DNA damage. Prior reports have identified GPX1 as a p53 target gene, with a p53 response element mapped to the promoter (36, 74). Our ChIP-chip screen confirms these prior findings while also demonstrating that GPX1 is not induced in response to hypoxic stress, even though p53 is clearly bound to the promoter (Fig. 3 and 4B). Neither REV3L nor GPX1 was induced by p53 under hypoxia, nor were they significantly repressed. This is consistent with prior observations of p21, HDM2, and GADD45A, despite the fact that p53 is more likely to be associated with corepressor complexes than with coactivators (43). Contributions from other transcription factors may override any repressive effect of hypoxia-stabilized p53. Thus, p53 target genes that mediate cell cycle arrest and apoptosis in response to DNA damage do not necessarily contribute to p53 functions during other stress conditions.
One of the goals of the CGI ChIP-chip screen was to identify p53 target genes regulated by hypoxic stress. MYC displayed a distinct p53-dependent repression in wild-type HCT116 cells that was distinct from the generic decrease that occurs during hypoxia (51). This phenomenon may require cooperation with promoter elements that facilitate the recruitment of corepressor complexes (83). In HCT116 p53+/+ cells, treatment with adriamycin also appeared to repress MYC over time, but due to the heterogeneous response of the isogenic p53−/− cell, this result was not as robust. This evidence is consistent with expression microarray data comparing gene repression phenomena under hypoxia to those under DNA-damaging stresses (see Table S2 in the supplemental material). Until recently, there was little chromatin immunoprecipitation evidence that p53 represses MYC through the recruitment of corepressor complexes to the promoter (29). A bioinformatics approach identified multiple potential p53REs located throughout the MYC locus (78). Our observation that p53 associates with the second intron of MYC does not rule out an association with regions closer to the promoter of the gene (30).
Repression of MYC by p53 during hypoxia likely represents a distinct mechanism of cell cycle arrest in the absence of DNA damage. While the p21 gene is classically thought of as a p53 target, it is regulated independently of p53 by hypoxia (Fig. 1) (42). MYC represses the p21 promoter during proliferative conditions, but HIF can displace MYC during hypoxia to activate the gene (42). Induction of p21 during hypoxia may be enhanced by p53-dependent repression of MYC. Since MYC has a multitude of effectors (21), it is likely that its repression during hypoxia may have many other downstream effects on cell cycle regulation. However, p53 also induces apoptosis under hypoxia (22, 24), implying that there may be yet another cohort of regulated genes responsible for further suppressing tumor formation and progression.
CGI ChIP-chip also revealed the identities of p53 targets that did not have a transcriptional dependence on p53. A particularly notable example is the stress response protein SYVN1 (HRD1), an E3 ubiquitin ligase important for retrograde transport and degradation of unfolded proteins in the endoplasmic reticulum (39). SYVN1 is thought to be regulated by the UPR (38), which can be triggered by a number of stimuli including hypoglycemia, calcium depletion, and severe hypoxia (69). SYVN is clearly induced by the hypoxic conditions used for this study, indicating that the UPR is activated regardless of p53 binding. Our data are also supported by a recent publication describing an association of p53 and NF-Y with a number of promoters regulating the unfolded protein response. Consistent with our results, p53 occupied the SYVN1 promoter without affecting transcription (10).
The inability of p53 to regulate the expression of SYVN1 and other genes is interesting for several reasons. Because studies of transcription factor function have largely been restricted to a select set of promoters identified by changes in gene expression, very little is known about transcription factors binding to regions without a direct transcriptional effect. It is well known that some genes are regulated by combinatorial action; general or tissue-specific factors contribute architectures conducive to the functions of an inducible protein (46, 52, 68). This leaves open the possibility of the inverse situation, where inducible transcription factors bind to any number of promoters lacking the accessory factors necessary for a transcriptional response. This scenario of a nonresponsive promoter environment can be generalized to explain the large number of genes that are not regulated by p53 in response to any stress. Other groups using ChIP-coupled discovery methods have also discovered nonresponsive p53 targets (27, 36, 81). This is consistent with observations from a global analysis of dMyc binding in Drosophila melanogaster, where approximately 25% of the bound sites did not have a corresponding regulation of transcription (62).
CGIs are evolutionarily conserved regulatory sequences characterized by early replication and an open chromatin structure (6). Genome-wide binding by p53 to these sites may be part of a broader mechanism of creating chromosomal architecture conducive to maximizing the stress response (4). It may be that some of these targets are regulated by p53 in other cell types (5) or represent off-target binding to regions more responsive to other p53 family members (63, 70). Additionally, p53 may have a direct role in aspects of DNA recombination and repair (71). Transcription-independent association of p53 with CGIs may therefore represent in vivo examples of interactions with structures indicative of sites of recombination, repetitive sequences, or hairpin structures (77).
The data presented in this study demonstrate the utility of CGI arrays for the discovery of p53 binding sites under different stresses. After confirming specific p53-CGI interactions under both hypoxic and DNA-damaging stresses, we determined the transcriptional consequences of some of these interactions. Using this approach, we have identified novel targets associated with DNA repair, stress response, and cell cycle regulation. We observe that transcriptional regulation of p53 targets is highly dependent on the specific stress and promoter. The actions of p53 can thus be separated into two distinct mechanisms: generic stress-induced protein stability and DNA binding followed by promoter-specific responses to stress-stabilized p53. The mechanism of DNA binding may influence the ability of p53 to regulate a given promoter. These experiments raise several interesting questions regarding the regulation of transcription by p53, with particular emphasis on the functions of p53 during hypoxic stress.
Supplementary Material
Acknowledgments
We thank Lawrence Heisler and Sandy Der at the University of Toronto for providing access to the CpG Island Microarray Database, Bert Vogelstein at Johns Hopkins University for providing the HCT116-derived cell lines and the pG13 and pM15 luciferase vectors, Jennifer Pietenpol at Vanderbilt University for permission to use the pCEP4-p53 expression plasmid, Janoz Demeter of the Stanford Microarray Database for expert help with creating CGI gene list files, and Ali Salim for expert assistance with microarray hybridization techniques. We also thank Denise Chan, Patrick Sutphin, and Sacha Krieg for critical reading of the manuscript.
This work was supported by an NIH grant (CA 88480) awarded to A.J.G.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Adimoolam, S., and J. M. Ford. 2002. p53 and DNA damage-inducible expression of the xeroderma pigmentosum group C gene. Proc. Natl. Acad. Sci. USA 99:12985-12990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alarcon, R., C. Koumenis, R. K. Geyer, C. G. Maki, and A. J. Giaccia. 1999. Hypoxia induces p53 accumulation through MDM2 down-regulation and inhibition of E6-mediated degradation. Cancer Res. 59:6046-6051. [PubMed] [Google Scholar]
- 3.Alders, M., A. Ryan, M. Hodges, J. Bliek, A. P. Feinberg, O. Privitera, A. Westerveld, P. F. Little, and M. Mannens. 2000. Disruption of a novel imprinted zinc-finger gene, ZNF215, in Beckwith-Wiedemann syndrome. Am. J. Hum. Genet. 66:1473-1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allison, S. J., and J. Milner. 2004. Remodelling chromatin on a global scale: a novel protective function of p53. Carcinogenesis 25:1551-1557. [DOI] [PubMed] [Google Scholar]
- 5.Amundson, S. A., M. Bittner, Y. Chen, J. Trent, P. Meltzer, and A. J. Fornace, Jr. 1999. Fluorescent cDNA microarray hybridization reveals complexity and heterogeneity of cellular genotoxic stress responses. Oncogene 18:3666-3672. [DOI] [PubMed] [Google Scholar]
- 6.Antequera, F., and A. Bird. 1993. Number of CpG islands and genes in human and mouse. Proc. Natl. Acad. Sci. USA 90:11995-11999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown, J. M., and A. J. Giaccia. 1998. The unique physiology of solid tumors: opportunities (and problems) for cancer therapy. Cancer Res. 58:1408-1416. [PubMed] [Google Scholar]
- 8.Cartharius, K., K. Frech, K. Grote, B. Klocke, M. Haltmeier, A. Klingenhoff, M. Frisch, M. Bayerlein, and T. Werner. 2005. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics 21:2933-2942. [DOI] [PubMed] [Google Scholar]
- 9.Cawley, S., S. Bekiranov, H. H. Ng, P. Kapranov, E. A. Sekinger, D. Kampa, A. Piccolboni, V. Sementchenko, J. Cheng, A. J. Williams, R. Wheeler, B. Wong, J. Drenkow, M. Yamanaka, S. Patel, S. Brubaker, H. Tammana, G. Helt, K. Struhl, and T. R. Gingeras. 2004. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell 116:499-509. [DOI] [PubMed] [Google Scholar]
- 10.Ceribelli, M., M. Alcalay, M. A. Vigano, and R. Mantovani. 2006. Repression of new p53 targets revealed by ChIP on chip experiments. Cell Cycle 5:1102-1110. [DOI] [PubMed] [Google Scholar]
- 11.Craig, J. M., and W. A. Bickmore. 1994. The distribution of CpG islands in mammalian chromosomes. Nat. Genet. 7:376-382. [DOI] [PubMed] [Google Scholar]
- 12.Cross, S. H., V. H. Clark, M. W. Simmen, W. A. Bickmore, H. Maroon, C. F. Langford, N. P. Carter, and A. P. Bird. 2000. CpG island libraries from human chromosomes 18 and 22: landmarks for novel genes. Mamm. Genome 11:373-383. [DOI] [PubMed] [Google Scholar]
- 13.el-Deiry, W. S., S. E. Kern, J. A. Pietenpol, K. W. Kinzler, and B. Vogelstein. 1992. Definition of a consensus binding site for p53. Nat. Genet. 1:45-49. [DOI] [PubMed] [Google Scholar]
- 14.el-Deiry, W. S., T. Tokino, V. E. Velculescu, D. B. Levy, R. Parsons, J. M. Trent, D. Lin, W. E. Mercer, K. W. Kinzler, and B. Vogelstein. 1993. WAF1, a potential mediator of p53 tumor suppression. Cell 75:817-825. [DOI] [PubMed] [Google Scholar]
- 15.Forsythe, J. A., B. H. Jiang, N. V. Iyer, F. Agani, S. W. Leung, R. D. Koos, and G. L. Semenza. 1996. Activation of vascular endothelial growth factor gene transcription by hypoxia-inducible factor 1. Mol. Cell. Biol. 16:4604-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gassmann, R., A. Carvalho, A. J. Henzing, S. Ruchaud, D. F. Hudson, R. Honda, E. A. Nigg, D. L. Gerloff, and W. C. Earnshaw. 2004. Borealin: a novel chromosomal passenger required for stability of the bipolar mitotic spindle. J. Cell Biol. 166:179-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giaccia, A. J., and M. B. Kastan. 1998. The complexity of p53 modulation: emerging patterns from divergent signals. Genes Dev. 12:2973-2983. [DOI] [PubMed] [Google Scholar]
- 18.Gibbs, P. E., W. G. McGregor, V. M. Maher, P. Nisson, and C. W. Lawrence. 1998. A human homolog of the Saccharomyces cerevisiae REV3 gene, which encodes the catalytic subunit of DNA polymerase zeta. Proc. Natl. Acad. Sci. USA 95:6876-6880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Graeber, T. G., C. Osmanian, T. Jacks, D. E. Housman, C. J. Koch, S. W. Lowe, and A. J. Giaccia. 1996. Hypoxia-mediated selection of cells with diminished apoptotic potential in solid tumours. Nature 379:88-91. [DOI] [PubMed] [Google Scholar]
- 20.Graeber, T. G., J. F. Peterson, M. Tsai, K. Monica, A. J. Fornace, Jr., and A. J. Giaccia. 1994. Hypoxia induces accumulation of p53 protein, but activation of a G1-phase checkpoint by low-oxygen conditions is independent of p53 status. Mol. Cell. Biol. 14:6264-6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grandori, C., S. M. Cowley, L. P. James, and R. N. Eisenman. 2000. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu. Rev. Cell Dev. Biol. 16:653-699. [DOI] [PubMed] [Google Scholar]
- 22.Hammond, E. M., D. J. Mandell, A. Salim, A. J. Krieg, T. M. Johnson, H. A. Shirazi, L. D. Attardi, and A. J. Giaccia. 2006. Genome-wide analysis of p53 under hypoxic conditions. Mol. Cell. Biol. 26:3492-3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hammond, E. M., N. C. Denko, M. J. Dorie, R. T. Abraham, and A. J. Giaccia. 2002. Hypoxia links ATR and p53 through replication arrest. Mol. Cell. Biol. 22:1834-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hammond, E. M., and A. J. Giaccia. 2005. The role of p53 in hypoxia-induced apoptosis. Biochem. Biophys. Res. Commun. 331:718-725. [DOI] [PubMed] [Google Scholar]
- 25.Hammond, E. M., S. L. Green, and A. J. Giaccia. 2003. Comparison of hypoxia-induced replication arrest with hydroxyurea and aphidicolin-induced arrest. Mutat. Res. 532:205-213. [DOI] [PubMed] [Google Scholar]
- 26.Harms, K., S. Nozell, and X. Chen. 2004. The common and distinct target genes of the p53 family transcription factors. Cell. Mol. Life Sci. 61:822-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hearnes, J. M., D. J. Mays, K. L. Schavolt, L. Tang, X. Jiang, and J. A. Pietenpol. 2005. Chromatin immunoprecipitation-based screen to identify functional genomic binding sites for sequence-specific transactivators. Mol. Cell. Biol. 25:10148-10158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heisler, L. E., D. Torti, P. C. Boutros, J. Watson, C. Chan, N. Winegarden, M. Takahashi, P. Yau, T. H. Huang, P. J. Farnham, I. Jurisica, J. R. Woodgett, R. Bremner, L. Z. Penn, and S. D. Der. 2005. CpG island microarray probe sequences derived from a physical library are representative of CpG islands annotated on the human genome. Nucleic Acids Res. 33:2952-2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ho, J., and S. Benchimol. 2003. Transcriptional repression mediated by the p53 tumour suppressor. Cell Death Differ. 10:404-408. [DOI] [PubMed] [Google Scholar]
- 30.Ho, J. S., W. Ma, D. Y. Mao, and S. Benchimol. 2005. p53-dependent transcriptional repression of c-myc is required for G1 cell cycle arrest. Mol. Cell. Biol. 25:7423-7431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoffman, W. H., S. Biade, J. T. Zilfou, J. Chen, and M. Murphy. 2002. Transcriptional repression of the anti-apoptotic survivin gene by wild type p53. J. Biol. Chem. 277:3247-3257. [DOI] [PubMed] [Google Scholar]
- 32.Hwang, B. J., J. M. Ford, P. C. Hanawalt, and G. Chu. 1999. Expression of the p48 xeroderma pigmentosum gene is p53-dependent and is involved in global genomic repair. Proc. Natl. Acad. Sci. USA 96:424-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Imbriano, C., A. Gurtner, F. Cocchiarella, S. Di Agostino, V. Basile, M. Gostissa, M. Dobbelstein, G. Del Sal, G. Piaggio, and R. Mantovani. 2005. Direct p53 transcriptional repression: in vivo analysis of CCAAT-containing G2/M promoters. Mol. Cell. Biol. 25:3737-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jakacka, M., M. Ito, F. Martinson, T. Ishikawa, E. J. Lee, and J. L. Jameson. 2002. An estrogen receptor (ER) alpha deoxyribonucleic acid-binding domain knock-in mutation provides evidence for nonclassical ER pathway signaling in vivo. Mol. Endocrinol. 16:2188-2201. [DOI] [PubMed] [Google Scholar]
- 35.Jakacka, M., M. Ito, J. Weiss, P. Y. Chien, B. D. Gehm, and J. L. Jameson. 2001. Estrogen receptor binding to DNA is not required for its activity through the nonclassical AP1 pathway. J. Biol. Chem. 276:13615-13621. [DOI] [PubMed] [Google Scholar]
- 36.Jen, K. Y., and V. G. Cheung. 2005. Identification of novel p53 target genes in ionizing radiation response. Cancer Res. 65:7666-7673. [DOI] [PubMed] [Google Scholar]
- 37.Johnson, T. M., E. M. Hammond, A. Giaccia, and L. D. Attardi. 2005. The p53QS transactivation-deficient mutant shows stress-specific apoptotic activity and induces embryonic lethality. Nat. Genet. 37:145-152. [DOI] [PubMed] [Google Scholar]
- 38.Kaneko, M., M. Ishiguro, Y. Niinuma, M. Uesugi, and Y. Nomura. 2002. Human HRD1 protects against ER stress-induced apoptosis through ER-associated degradation. FEBS Lett. 532:147-152. [DOI] [PubMed] [Google Scholar]
- 39.Kikkert, M., R. Doolman, M. Dai, R. Avner, G. Hassink, S. van Voorden, S. Thanedar, J. Roitelman, V. Chau, and E. Wiertz. 2004. Human HRD1 is an E3 ubiquitin ligase involved in degradation of proteins from the endoplasmic reticulum. J. Biol. Chem. 279:3525-3534. [DOI] [PubMed] [Google Scholar]
- 40.Kirmizis, A., S. M. Bartley, A. Kuzmichev, R. Margueron, D. Reinberg, R. Green, and P. J. Farnham. 2004. Silencing of human polycomb target genes is associated with methylation of histone H3 Lys 27. Genes Dev. 18:1592-1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kolehmainen, J., G. C. Black, A. Saarinen, K. Chandler, J. Clayton-Smith, A. L. Traskelin, R. Perveen, S. Kivitie-Kallio, R. Norio, M. Warburg, J. P. Fryns, A. de la Chapelle, and A. E. Lehesjoki. 2003. Cohen syndrome is caused by mutations in a novel gene, COH1, encoding a transmembrane protein with a presumed role in vesicle-mediated sorting and intracellular protein transport. Am. J. Hum. Genet. 72:1359-1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koshiji, M., Y. Kageyama, E. A. Pete, I. Horikawa, J. C. Barrett, and L. E. Huang. 2004. HIF-1alpha induces cell cycle arrest by functionally counteracting Myc. EMBO J. 23:1949-1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koumenis, C., R. Alarcon, E. Hammond, P. Sutphin, W. Hoffman, M. Murphy, J. Derr, Y. Taya, S. W. Lowe, M. Kastan, and A. Giaccia. 2001. Regulation of p53 by hypoxia: dissociation of transcriptional repression and apoptosis from p53-dependent transactivation. Mol. Cell. Biol. 21:1297-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krieg, A. J., S. A. Krieg, B. S. Ahn, and D. J. Shapiro. 2004. Interplay between estrogen response element sequence and ligands controls in vivo binding of estrogen receptor to regulated genes. J. Biol. Chem. 279:5025-5034. [DOI] [PubMed] [Google Scholar]
- 45.Kzhyshkowska, J., A. Rusch, H. Wolf, and T. Dobner. 2003. Regulation of transcription by the heterogeneous nuclear ribonucleoprotein E1B-AP5 is mediated by complex formation with the novel bromodomain-containing protein BRD7. Biochem. J. 371:385-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Laganiere, J., G. Deblois, C. Lefebvre, A. R. Bataille, F. Robert, and V. Giguere. 2005. From the cover: location analysis of estrogen receptor alpha target promoters reveals that FOXA1 defines a domain of the estrogen response. Proc. Natl. Acad. Sci. USA 102:11651-11656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee, S., B. Elenbaas, A. Levine, and J. Griffith. 1995. p53 and its 14 kDa C-terminal domain recognize primary DNA damage in the form of insertion/deletion mismatches. Cell 81:1013-1020. [DOI] [PubMed] [Google Scholar]
- 48.Li, Z., H. Zhang, T. P. McManus, J. J. McCormick, C. W. Lawrence, and V. M. Maher. 2002. hREV3 is essential for error-prone translesion synthesis past UV or benzo[a]pyrene diol epoxide-induced DNA lesions in human fibroblasts. Mutat. Res. 510:71-80. [DOI] [PubMed] [Google Scholar]
- 49.Lin, X., K. Ramamurthi, M. Mishima, A. Kondo, R. D. Christen, and S. B. Howell. 2001. p53 modulates the effect of loss of DNA mismatch repair on the sensitivity of human colon cancer cells to the cytotoxic and mutagenic effects of cisplatin. Cancer Res. 61:1508-1516. [PubMed] [Google Scholar]
- 50.Lin, X., J. Trang, T. Okuda, and S. B. Howell. 2006. DNA polymerase zeta accounts for the reduced cytotoxicity and enhanced mutagenicity of cisplatin in human colon carcinoma cells that have lost DNA mismatch repair. Clin. Cancer Res. 12:563-568. [DOI] [PubMed] [Google Scholar]
- 51.Liu, L., and M. C. Simon. 2004. Regulation of transcription and translation by hypoxia. Cancer Biol. Ther. 3:492-497. [DOI] [PubMed] [Google Scholar]
- 52.Lomvardas, S., and D. Thanos. 2002. Modifying gene expression programs by altering core promoter chromatin architecture. Cell 110:261-271. [DOI] [PubMed] [Google Scholar]
- 53.Mao, D. Y., J. D. Watson, P. S. Yan, D. Barsyte-Lovejoy, F. Khosravi, W. W. Wong, P. J. Farnham, T. H. Huang, and L. Z. Penn. 2003. Analysis of Myc bound loci identified by CpG island arrays shows that Max is essential for Myc-dependent repression. Curr. Biol. 13:882-886. [DOI] [PubMed] [Google Scholar]
- 54.Moberg, K. H., W. A. Tyndall, and D. J. Hall. 1992. Wild-type murine p53 represses transcription from the murine c-myc promoter in a human glial cell line. J. Cell Biochem. 49:208-215. [DOI] [PubMed] [Google Scholar]
- 55.Montano, M. M., B. M. Wittmann, and N. R. Bianco. 2000. Identification and characterization of a novel factor that regulates quinone reductase gene transcriptional activity. J. Biol. Chem. 275:34306-34313. [DOI] [PubMed] [Google Scholar]
- 56.Murphy, M., J. Ahn, K. K. Walker, W. H. Hoffman, R. M. Evans, A. J. Levine, and D. L. George. 1999. Transcriptional repression by wild-type p53 utilizes histone deacetylases, mediated by interaction with mSin3a. Genes Dev. 13:2490-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Murphy, M., A. Hinman, and A. J. Levine. 1996. Wild-type p53 negatively regulates the expression of a microtubule-associated protein. Genes Dev. 10:2971-2980. [DOI] [PubMed] [Google Scholar]
- 58.Nagao, Y., J. S. Platero, A. Waheed, and W. S. Sly. 1993. Human mitochondrial carbonic anhydrase: cDNA cloning, expression, subcellular localization, and mapping to chromosome 16. Proc. Natl. Acad. Sci. USA 90:7623-7627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nguyen, T. T., K. Cho, S. A. Stratton, and M. C. Barton. 2005. Transcription factor interactions and chromatin modifications associated with p53-mediated, developmental repression of the alpha-fetoprotein gene. Mol. Cell. Biol. 25:2147-2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Oberley, M. J., D. R. Inman, and P. J. Farnham. 2003. E2F6 negatively regulates BRCA1 in human cancer cells without methylation of histone H3 on lysine 9. J. Biol. Chem. 278:42466-42476. [DOI] [PubMed] [Google Scholar]
- 61.Oberley, M. J., J. Tsao, P. Yau, and P. J. Farnham. 2004. High-throughput screening of chromatin immunoprecipitates using CpG-island microarrays. Methods Enzymol. 376:315-334. [DOI] [PubMed] [Google Scholar]
- 62.Orian, A., B. van Steensel, J. Delrow, H. J. Bussemaker, L. Li, T. Sawado, E. Williams, L. W. Loo, S. M. Cowley, C. Yost, S. Pierce, B. A. Edgar, S. M. Parkhurst, and R. N. Eisenman. 2003. Genomic binding by the Drosophila Myc, Max, Mad/Mnt transcription factor network. Genes Dev. 17:1101-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Osada, M., H. L. Park, Y. Nagakawa, K. Yamashita, A. Fomenkov, M. S. Kim, G. Wu, S. Nomoto, B. Trink, and D. Sidransky. 2005. Differential recognition of response elements determines target gene specificity for p53 and p63. Mol. Cell. Biol. 25:6077-6089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Persengiev, S. P., L. R. Devireddy, and M. R. Green. 2002. Inhibition of apoptosis by ATFx: a novel role for a member of the ATF/CREB family of mammalian bZIP transcription factors. Genes Dev. 16:1806-1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Prakash, S., R. E. Johnson, and L. Prakash. 2005. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem. 74:317-353. [DOI] [PubMed] [Google Scholar]
- 66.Qian, H., T. Wang, L. Naumovski, C. D. Lopez, and R. K. Brachmann. 2002. Groups of p53 target genes involved in specific p53 downstream effects cluster into different classes of DNA binding sites. Oncogene 21:7901-7911. [DOI] [PubMed] [Google Scholar]
- 67.Rampalli, S., L. Pavithra, A. Bhatt, T. K. Kundu, and S. Chattopadhyay. 2005. Tumor suppressor SMAR1 mediates cyclin D1 repression by recruitment of the SIN3/histone deacetylase 1 complex. Mol. Cell. Biol. 25:8415-8429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Robyr, D., A. Gegonne, A. P. Wolffe, and W. Wahli. 2000. Determinants of vitellogenin B1 promoter architecture. HNF3 and estrogen responsive transcription within chromatin. J. Biol. Chem. 275:28291-28300. [DOI] [PubMed] [Google Scholar]
- 69.Romero-Ramirez, L., H. Cao, D. Nelson, E. Hammond, A. H. Lee, H. Yoshida, K. Mori, L. H. Glimcher, N. C. Denko, A. J. Giaccia, Q. T. Le, and A. C. Koong. 2004. XBP1 is essential for survival under hypoxic conditions and is required for tumor growth. Cancer Res. 64:5943-5947. [DOI] [PubMed] [Google Scholar]
- 70.Sasaki, Y., S. Ishida, I. Morimoto, T. Yamashita, T. Kojima, C. Kihara, T. Tanaka, K. Imai, Y. Nakamura, and T. Tokino. 2002. The p53 family member genes are involved in the Notch signal pathway. J. Biol. Chem. 277:719-724. [DOI] [PubMed] [Google Scholar]
- 71.Sengupta, S., and C. C. Harris. 2005. p53: traffic cop at the crossroads of DNA repair and recombination. Nat. Rev. Mol. Cell Biol. 6:44-55. [DOI] [PubMed] [Google Scholar]
- 72.Shan, B., J. Xu, Y. Zhuo, C. A. Morris, and G. F. Morris. 2003. Induction of p53-dependent activation of the human proliferating cell nuclear antigen gene in chromatin by ionizing radiation. J. Biol. Chem. 278:44009-44017. [DOI] [PubMed] [Google Scholar]
- 73.Shivji, K. K., M. K. Kenny, and R. D. Wood. 1992. Proliferating cell nuclear antigen is required for DNA excision repair. Cell 69:367-374. [DOI] [PubMed] [Google Scholar]
- 74.Tan, M., S. Li, M. Swaroop, K. Guan, L. W. Oberley, and Y. Sun. 1999. Transcriptional activation of the human glutathione peroxidase promoter by p53. J. Biol. Chem. 274:12061-12066. [DOI] [PubMed] [Google Scholar]
- 75.Tsuchimochi, K., N. Yagishita, S. Yamasaki, T. Amano, Y. Kato, K. Kawahara, S. Aratani, H. Fujita, F. Ji, A. Sugiura, T. Izumi, A. Sugamiya, I. Maruyama, A. Fukamizu, S. Komiya, K. Nishioka, and T. Nakajima. 2005. Identification of a crucial site for synoviolin expression. Mol. Cell. Biol. 25:7344-7356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vaupel, P. 2004. Tumor microenvironmental physiology and its implications for radiation oncology. Semin. Radiat. Oncol. 14:198-206. [DOI] [PubMed] [Google Scholar]
- 77.Walter, K., G. Warnecke, R. Bowater, W. Deppert, and E. Kim. 2005. Tumor suppressor p53 binds with high affinity to CTG · CAG trinucleotide repeats and induces topological alterations in mismatched duplexes. J. Biol. Chem. 280:42497-42507. [DOI] [PubMed] [Google Scholar]
- 78.Wang, L., Q. Wu, P. Qiu, A. Mirza, M. McGuirk, P. Kirschmeier, J. R. Greene, Y. Wang, C. B. Pickett, and S. Liu. 2001. Analyses of p53 target genes in the human genome by bioinformatic and microarray approaches. J. Biol. Chem. 276:43604-43610. [DOI] [PubMed] [Google Scholar]
- 79.Watanabe, T., S. Inoue, H. Hiroi, A. Orimo, H. Kawashima, and M. Muramatsu. 1998. Isolation of estrogen-responsive genes with a CpG island library. Mol. Cell. Biol. 18:442-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Webb, P., G. N. Lopez, R. M. Uht, and P. J. Kushner. 1995. Tamoxifen activation of the estrogen receptor/AP-1 pathway: potential origin for the cell-specific estrogen-like effects of antiestrogens. Mol. Endocrinol. 9:443-456. [DOI] [PubMed] [Google Scholar]
- 81.Wei, C. L., Q. Wu, V. B. Vega, K. P. Chiu, P. Ng, T. Zhang, A. Shahab, H. C. Yong, Y. Fu, Z. Weng, J. Liu, X. D. Zhao, J. L. Chew, Y. L. Lee, V. A. Kuznetsov, W. K. Sung, L. D. Miller, B. Lim, E. T. Liu, Q. Yu, H. H. Ng, and Y. Ruan. 2006. A global map of p53 transcription-factor binding sites in the human genome. Cell 124:207-219. [DOI] [PubMed] [Google Scholar]
- 82.Weinmann, A. S., P. S. Yan, M. J. Oberley, T. H. Huang, and P. J. Farnham. 2002. Isolating human transcription factor targets by coupling chromatin immunoprecipitation and CpG island microarray analysis. Genes Dev. 16:235-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wilkinson, D. S., S. K. Ogden, S. A. Stratton, J. L. Piechan, T. T. Nguyen, G. A. Smulian, and M. C. Barton. 2005. A direct intersection between p53 and transforming growth factor β pathways targets chromatin modification and transcription repression of the α-fetoprotein gene. Mol. Cell. Biol. 25:1200-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wittschieben, J., M. K. Shivji, E. Lalani, M. A. Jacobs, F. Marini, P. J. Gearhart, I. Rosewell, G. Stamp, and R. D. Wood. 2000. Disruption of the developmentally regulated Rev3l gene causes embryonic lethality. Curr. Biol. 10:1217-1220. [DOI] [PubMed] [Google Scholar]
- 85.Wittschieben, J. P., S. C. Reshmi, S. M. Gollin, and R. D. Wood. 2006. Loss of DNA polymerase zeta causes chromosomal instability in mammalian cells. Cancer Res. 66:134-142. [DOI] [PubMed] [Google Scholar]
- 86.Wu, F., X. Lin, T. Okuda, and S. B. Howell. 2004. DNA polymerase zeta regulates cisplatin cytotoxicity, mutagenicity, and the rate of development of cisplatin resistance. Cancer Res. 64:8029-8035. [DOI] [PubMed] [Google Scholar]
- 87.Yang, L., T. Guan, and L. Gerace. 1997. Integral membrane proteins of the nuclear envelope are dispersed throughout the endoplasmic reticulum during mitosis. J. Cell Biol. 137:1199-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.