Abstract
Histone acetylation provides a switch between transcriptionally repressive and permissive chromatin. By regulating the chromatin structure at specific promoters, histone acetyltransferases (HATs) carry out important functions during differentiation and development of higher eukaryotes. HAT complexes are present in organisms as diverse as Saccharomyces cerevisiae, humans, and flies. For example, the well-studied yeast SAGA is related to three mammalian complexes. We previously identified Drosophila melanogaster orthologues of yeast SAGA components Ada2, Ada3, Spt3, and Tra1 and demonstrated that they associate with dGcn5 in a high-molecular-weight complex. To better understand the function of Drosophila SAGA (dSAGA), we sought to affinity purify and characterize this complex in more detail. A proteomic approach led to the identification of an orthologue of the yeast protein Ada1 and the novel protein encoded by CG4448, referred to as WDA (will decrease acetylation). Embryos lacking both alleles of the wda gene exhibited reduced levels of histone H3 acetylation and could not develop into adult flies. Our results point to a critical function of dSAGA and histone acetylation during Drosophila development.
The packaging of DNA into chromatin has dramatic consequences on enzymatic processes that require DNA: replication, recombination, repair, and transcription. Eukaryotic cells can overcome the repressive effects of chromatin by covalently modifying the core histone tails (30). The posttranslational modifications of histone tails include acetylation, methylation and ubiquitylation of lysine residues, phosphorylation of serines and threonines, and methylation of arginine residues (16).
The histone code hypothesis states that the combination of different marks on single histones, single nucleosomes, and nucleosomal domains may determine the recruitment of specific factors and result in unique downstream events (53).
The steady-state balance of histone acetylation in the nucleus is maintained by the opposing actions of histone acetyltransferases (HATs) and deacetylases (7, 38). The Saccharomyces cerevisiae transcriptional coactivator protein Gcn5 is a HAT that serves as the catalytic subunit of the multiprotein HAT complexes SAGA, SLIK, ADA, and A2 (22, 48, 51). The 1.8-MDa SAGA complex is comprised of several groups of proteins. The first group consists of the Ada family of proteins (which includes Gcn5, also named Ada4). Ada genes were isolated as mutations that allow yeast growth when GAL4-VP16 is overexpressed (22). The second group contains Spt proteins, isolated as suppressors of transcription initiation defects caused by promoter insertions of the Ty transposon (22, 52). The TATA-binding protein-associated factors (TAFs) present in SAGA seem to carry out a structural function (23, 57). In addition, SAGA has the ATM/phosphatidylinositol 3-kinase-related protein Tra1, the ubiquitin protease Ubp8, and the most recently identified Sgf proteins (24, 32, 46).
Gcn5 is conserved from yeast to humans. In fact, mammals carry two paralogues, Gcn5 and PCAF, which are highly similar in primary structure and in vitro functions (6, 56). Although orthologues for yeast Tra1 and the TAFs are present in higher eukaryotes, only a few of the yeast Ada and Spt proteins from SAGA share amino acid identity with proteins from higher eukaryotes. Three SAGA-related PCAF/Gcn5-containing complexes have been isolated from mammalian cells: PCAF, STAGA, and TFTC. These complexes can activate transcription in vitro from chromatin templates (4, 35, 39).
Studies in multicellular organisms provided evidence of the fundamental roles of Gcn5 during development. Experiments done with mice revealed that Gcn5 is essential for viability, but PCAF is not. Gcn5 knockout mice fail to develop particular mesodermal lineages and die between 10 and 11 days of embryogenesis (59). In addition, mutations in plant ada2b and gcn5 affect development and gene expression (55). Flies homozygous for a dGcn5 mutant allele display defects in metamorphosis and proliferation of imaginal discs. Furthermore, dGcn5 mutants show significantly reduced levels of acetylated histone H3 on polytene chromosomes (8).
The relevance of Gcn5-containing complexes in development is further supported by evidence that links the mammalian STAGA complex to neurodegenerative disorders. Spinocerebellar ataxia type 7 is caused by polyglutamine expansions in the ataxin-7 protein, a recently characterized subunit of STAGA. Incorporation of the mutated version of ataxin-7 into STAGA dramatically reduces the stability of the complex and its ability to acetylate histone H3, leading to retinal degeneration in mice (40). Similar mutations in the yeast homologue Sgf73 negatively affect the HAT activity of the SAGA and SLIK complexes (37).
We previously fractionated Drosophila melanogaster cells and demonstrated the association of dGcn5, dAda3, dAda2B, dSpt3, and dTra1 in a high-molecular-weight HAT complex similar to yeast SAGA (31). This Drosophila HAT complex will be referred to as dSAGA. To address the subunit composition and function of dSAGA, we affinity purified this complex and characterized it in more detail. MudPIT analysis of affinity purified dSAGA revealed two new subunits. These uncharacterized proteins in dSAGA are encoded by the genes CG31865/CG31866 and CG4448. By sequence homology, we determined that the CG31865/CG31866 gene product is an orthologue of the yeast and human Ada1 proteins. We named the CG4448 gene product WDA (will decrease acetylation). WDA contains six WD repeats, which are necessary for the incorporation of WDA into dSAGA. Chromatographic fractionation confirmed that WDA is part of the 1.5-MDa dSAGA complex. We generated deletions in the wda gene and observed that the mutant animals died during second larval instar. The homozygous mutant embryos showed reduced levels of histone H3 acetylation, predominantly in the central nervous system. Ubiquitous expression of WDA from a transgene restored viability and histone H3 acetylation.
MATERIALS AND METHODS
Plasmids used in this study.
The plasmids generated in this study are described in Table 1. pRmHa3-CHA2FL2 and pUAST were previously described (3, 25). The cDNAs for wda, dAda1, and wdaΔC were isolated by reverse transcription-PCR. For reverse transcription, total RNA was isolated from 12- to 18-h Oregon R embryos using Trizol (Invitrogen). Single-stranded cDNA molecules were generated using the SuperScript first-strand synthesis system (Invitrogen) and subsequently amplified by PCR using Pfu turbo (Stratagene).
TABLE 1.
Plasmids generated in this study
| Construct name | Vector | Insert (accession no.) |
|---|---|---|
| pRmHa3-wda-HA2FL2 | pRmHa3-CHA2FL2 | cDNA for CG4448 (NM_142879) |
| pRmHa3-dAda1-HA2FL2 | pRmHa3-CHA2FL2 | cDNA for CG31866 (NM_164980) |
| pRmHa3-wdaΔC-HA2FL2 | pRmHa3-CHA2FL2 | cDNA coding for amino acids 1 to 393 of WDA (NP_651136) |
| pQE12-wdaΔC | pQE12 | cDNA coding for amino acids 1 to 393 of WDA |
| pUAST-wda | pUAST | cDNA for CG4448 (NM_142879) |
Transfections and generation of stable lines.
S2 cells were transfected with Effectene (QIAGEN) according to the manufacturer's instructions. Stable cell lines were established by cotransfection of the pRmha3-derived constructs and pCoHygro (Invitrogen), followed by a 2- to 3-week selection in media containing 0.25 mg/ml hygromycin (Invitrogen).
Preparation and fractionation of nuclear extracts.
Nuclear extracts were prepared as described previously (49). Affinity purifications were carried out using nuclear extracts prepared from 8 liters of cells, grown to a density of 3 × 106 cells/ml. CuSO4 was not added to the medium to keep low levels of expression of the tagged proteins.
To obtain protein fractions enriched in HATs, nuclear extracts were incubated with Ni-nitrilotriacetic acid-agarose beads, as previously described. Anion-exchange (MonoQ) and gel filtration (Superose 6) chromatography fractionations were performed as described previously (25).
Generation of antibodies to WDA.
A DNA fragment coding for the N-terminal part of WDA (amino acids 1 to 393) was amplified by PCR using pRmHa3-wda-HA2FL2 as a template and inserted into pQE12 (QIAGEN). Expression and purification of the recombinant His-tagged protein were performed as described previously (25). Rats and rabbits were immunized with the recombinant soluble WDA fragment (Pocono Rabbit Farm and Laboratory, Inc.).
Coimmunoprecipitations, Western blots, and HAT assays.
Coimmunoprecipitations using anti-FLAG antibodies or polyclonal antibodies were performed as described previously (25). Antibodies directed to dGcn5 (rabbit, 1:3,000), dAda2B (guinea pig, 1:1,000), dSpt3 (rabbit, 1:1,000), dAda2A (rabbit, 1:1,000), Atac1 (rat, 1:1,000), and WDA (rat, 1:500) were used in Western blots (25, 31) as well as antibodies against FLAG (1:5,000; Sigma), acetylated H3 K9 (1:500; Upstate), and H3 (1:500; Abcam).
HAT reactions were performed as described previously (15) using either purified HAT complexes or complexes immobilized to protein A-Sepharose beads (Amersham).
Affinity purifications and mass spectrometry.
TAP-dGcn5 purification and MudPIT analysis were previously described (25). Anti-FLAG purifications were performed as described previously (25) using the stable lines that express dAda1-HA2FL2 and WDA-HA2FL2. MudPIT analysis of the affinity-purified complexes was carried out as previously described (32).
Generation of deletions in the wda gene.
All fly stocks used in this study were kindly provided by the Bloomington Drosophila Stock Center at Indiana University.
To generate deletions that disrupt the wda gene, we mobilized the P-element KG03744, located ∼300 bp upstream of the wda gene, and screened for imprecise excisions. The P-element was mobilized by crossing the stock y1 w67c23; ry506 P{SUPor-P}KG03744 (Bloomington stock number 13345) to y1 w*; ry506 Sb1 P{Δ2-3}99B/TM6 flies (Bloomington stock number 3664). Males containing the P-element KG03744 over the transposase gene Δ2-3 were crossed to y1 w1118; D3, gl3/TM3, Sb1, Ser1 virgins. The P-element excisions were screened by the loss of eye color (due to loss of the white gene in the P-element). To determine whether any of the hop-out events was imprecise, we isolated DNA from 15 flies corresponding to each excision event using DNAzol (Invitrogen) according to the manufacturer's protocol. To screen for deletions, PCRs were performed using primers designed in such a way that they would amplify an ∼3-kb fragment if the excision was precise or DNA molecules of smaller size if excisions were imprecise. The three independent deletion lines obtained will be referred to as wda4, wda8, and wda11.
Lethality test of wda mutants.
Flies wda11/TM3, Sb1, Ser1 were mated to the stock w1118; DrMio/TM3, P{GAL4-twi.G}2.3, P{UAS-2xEGFP}AH2.3, Sb1, Ser1 (Bloomington stock number 6663). The progeny with genotype wda11/TM3, P{GAL4-twi.G}2.3, P{UAS-2xEGFP}AH2.3, Sb1, Ser1 was propagated for further studies.
Embryos from a wda11/TM3, P{GAL4-twi.G}2.3, P{UAS-2xEGFP}AH2.3, Sb1, Ser1 population were collected on apple juice plates and aged to generate a collection of embryos at stages 12 to 15. The homozygous mutant embryos wda11/wda11, identified by their lack of fluorescence, and the heterozygous embryos wda11/TM3, P{GAL4-twi.G}2.3, P{UAS-2xEGFP}AH2.3, Sb1, Ser1, showing intermediate fluorescence, were transferred to fresh apple plates to determine the stage at which they died. The heterozygous embryos were used as controls.
Generation of transgenic flies.
The cDNA for wda was cloned into pUAST (3). Injections were carried out by Genetic Services, Inc. Determination of the chromosomal locations of the transgene was carried out using standard genetic crosses. A transgene (UAS-wda) in chromosome 3 was recombined onto the chromosome carrying the deletion of wda to generate the stock wda11, UAS-wda/TM3, Sb1, Ser1. In addition, the tubulin-GAL4 driver from the stock y1 w*; P{tubP-GAL4}LL7/TM3, Sb1 (Bloomington stock number 5138) was recombined onto the chromosome carrying wda11 to obtain wda11, P{tubP-GAL4}LL7/TM3, Sb1, Ser1. In these flies, the yeast GAL4 protein is expressed in the same ubiquitous pattern as tubulin. To determine whether the UAS-wda transgene could rescue the lethality of wda11/wda11 animals, flies with the genotype wda11, UAS-wda/TM3, Sb1, Ser1 were mated to wda11, P{tubP-GAL4}LL7/TM3, Sb1, Ser1 flies, and the bristle phenotype (Sb1 or Sb+) of the progeny was determined in 200 adult flies.
Immunohistochemistry.
Embryos from the stock wda11/TM3, P{GAL4-twi.G}2.3, P{UAS-2xEGFP}AH2.3, Sb1, Ser1 were collected on apple juice plates at 0- to 4-h intervals at 25°C and aged for 14 h at 25°C to obtain a population of embryos at stages 16 to 17. Embryos derived from the cross of wda11, UAS-wda/TM3, P{GAL4-twi.G}2.3, P{UAS-2xEGFP}AH2.3, Sb1, Ser1 to wda11, P{tubP-GAL4}LL7/TM3, P{GAL4-twi.G}2.3, P{UAS-2xEGFP}AH2.3, Sb1, Ser1 were collected under the same conditions. Embryos were dechorionated for 3 min in 50% bleach and fixed in a 1:1 mixture of 4% formaldehyde and heptane. Staining was performed as described previously (29). The primary antibodies used were anti-acetylated H3 K9 (rabbit, 1:100; Upstate), anti-tetra-acetylated H4 (rabbit, 1:100; Upstate) and anti-green fluorescent protein (anti-GFP, mouse, 1:100; Roche). The anti-GFP antibodies were included in the staining reaction to distinguish homozygous mutant embryos (GFP negative) from balancer-containing embryos (GFP positive). The secondary antibodies used were Alexa Fluor 488 goat anti-rabbit (1:250; Molecular Probes) and Alexa Fluor 660 goat anti-mouse (1:250; Molecular Probes).
Determination of bulk levels of H3 acetylation in the embryos.
Embryos from the stock wda11/TM3, P{GAL4-twi.G}2.3, P{UAS-2xEGFP}AH2.3, Sb1, Ser1 were collected on apple juice plates and aged to generate a collection at stages 16 and 17. Embryos derived from the cross of wda11, UAS-wda/TM3, P{GAL4-twi.G}2.3, P{UAS-2xEGFP}AH2.3, Sb1, Ser1 to wda11, P{tubP-GAL4}LL7/TM3, P{GAL4-twi.G}2.3, P{UAS-2xEGFP}AH2.3, Sb1, Ser1 were collected under the same conditions. Embryos were dechorionated for 3 min in 50% bleach, and GFP-negative embryos (wda11/wda11 or wda11, UAS-wda/wda11, tubP-GAL4) were homogenized in sodium dodecyl sulfate-containing loading buffer (1 μl buffer per embryo). Extracts prepared from stage 16 to 17 Oregon R embryos were used as controls. Five microliters of extract was analyzed by Western blotting.
GFP-sorting of homozygous mutant embryos.
To isolate large amounts of homozygous mutant embryos, stage 16 to 17 embryos from the stock wda11/TM3, P{GAL4-twi.G}2.3, P{UAS-2xEGFP}AH2.3, Sb1, Ser1 were dechorionated and sorted by their lack of fluorescence using the COPAS PLUS Drosophila embryo sorting instrument (Union Biometrica) (17). After sorting, embryo lysates were prepared in the presence of 50 mM Tris-Cl, pH 8, 300 mM NaCl, 1% NP-40, 1 mM phenylmethylsulfonyl fluoride, 1 μg/ml pepstatin A, and 1 μg/ml leupeptin.
RESULTS
The products of the genes CG4448 and CG31865/CG31866 associate with TAP-dGcn5.
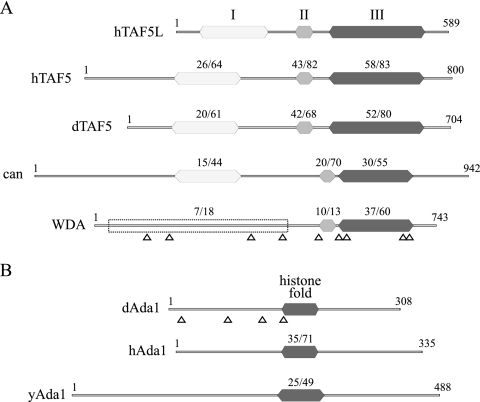
In a previous report, we affinity purified dGcn5-containing complexes and identified dGcn5-associated proteins by multidimensional protein identification technology (MudPIT) (25). In addition to known dGcn5-interacting proteins, such as dAda3, dAda2A, and dAda2B, the MudPIT analysis revealed several peptides encoded by the transcripts CG4448 and CG31865/CG31866. CG4448 and CG31865/CG31866 were identified by 9 and 4 different peptides, respectively (Fig. 1A and B).
FIG. 1.
Conserved domains in WDA and dAda1. (A) The amino acid sequences of three conserved regions in human TAF5 (hTAF5), Drosophila TAF5 (dTAF5), cannonball (can), and two conserved regions in WDA were compared to that of human TAF5-like (hTAF5L). Region I (white) consists of a WD40-associated domain. Region II (light gray) contains a noncanonical WD repeat. Region III (dark gray) includes five consensus WD repeats. The percent identity/similarity relative to hTAF5L is shown in each conserved domain. Different 150-amino-acid stretches contained in the empty box were compared to region I of hTAF5L, and the average value of identity/similarity is shown. (B) The amino acid sequence of the conserved histone fold in Drosophila Ada1 (dAda1) was compared to that of human Ada1 (hAda1) and yeast Ada1 (yAda1). The percent identity/similarity relative to dAda1 is shown. Arrowheads point to the different peptides for WDA and dAda1 obtained by MudPIT.
CG4448 encodes a 743-amino-acid polypeptide with a predicted molecular mass of 84 kDa (NP_651136). Analysis of its primary structure revealed that this protein belongs to the superfamily of WD40-repeat proteins (Pfam accession number PF00400). We have named this protein WDA because of the phenotype of a mutant, which is described below. WD repeats have a length of 44 to 60 amino acids and contain a conserved core region flanked by the dipeptides GH and WD (50). The original member of this family is the β subunit of trimeric G proteins that couple receptors for extracellular signals to intracellular enzymes (21). The crystal structure of this protein, which contains seven WD repeats, revealed a β-propeller structure with seven blades made of four-stranded β-twisted sheets. WD-repeat proteins are present in all eukaryotes, with over 100 members in humans. These proteins are involved in a wide variety of cellular functions, including cell division, vesicle transport, RNA synthesis and processing, chromatin assembly, cell cycle regulation, and cytoskeletal assembly. In addition, mutations in several WD-repeat proteins have been implicated in human diseases (34).
A BLAST search was performed to recover potential orthologues of WDA in other organisms. We identified several WD-repeat-containing proteins from vertebrates that share some amino acid sequence with WDA. One of the proteins in this group is human TAF5-like/PAF65β (hTAF5L), a component of the STAGA, PCAF, and TFTC HAT complexes (9). Pairwise alignments between the different vertebrate proteins identified by BLAST indicated that they are at least 65% identical to each other in their entire sequence, while WDA has less than 30% identity to any of the vertebrate proteins. Other proteins identified by BLAST include human and Drosophila TAF5 (hTAF5 and dTAF5) and cannonball (can), a fly tissue-specific TAF required for male gametogenesis (26).
We next compared the domain structure of WDA to that of hTAF5L, hTAF5, dTAF5, and can (Fig. 1A). All five polypeptides carry a noncanonical WD repeat (region II) (Fig. 1A), followed by 5 conserved WD repeats (region III) (Fig. 1A). Moreover, regions II and III in hTAF5L are more closely related to TAF5 than WDA. In addition, all TAF5L and TAF5 proteins, but not WDA, share a conserved N-terminal region, known as WD40-associated domain (region I, Pfam accession number PF04494). While it is a WD40-repeat-containing protein, the lack of the WD40-associated domain suggests that WDA cannot be included in either the TAF5 or TAF5-like families of proteins. This is supported by BLAST searches using only the N terminus of WDA, which do not detect proteins from other species with significant homology.
CG31865 and CG31866 are two independent transcription units located in region 33B5 of chromosome 2 and separated by an ∼3-kb stretch of DNA that harbors the genes CG18789 and CG31864. The amino acid sequences for the two predicted proteins (NP_723707 and NP_723703) are identical except for a single substitution at position 97. In addition, the predicted 5′ (90 bp) and 3′ (70 bp) untranslated regions of their RNA transcripts are identical except for one nucleotide (10, 11). The genomic organization of CG31865 and CG31866 suggests that these two genes arose by a gene duplication event.
Further analysis of the sequence of the CG31865/CG31866 gene product revealed a histone fold motif located in the center of the polypeptide chain (Fig. 1B). Histone fold motifs, present in the globular domains of the core histones, are found in subunits of the general transcription factor TFIID and other transcription factors. In fact, 9 of the 14 TAFs in yeast TFIID contain this motif (18). In addition, four of these TAFs (TAF4, 6, 9, and 12) can be reconstituted in vitro into an octamer-like structure (47). The yeast SAGA subunits Spt3, Spt7, and Ada1 also carry histone folds (2, 19, 20). Blast searches enabled us to identify human and yeast Ada1 as orthologues of CG31865/CG31866. The polypeptides encoded by CG31865/CG31866 and hAda1 are identical in 26% of their amino acid sequences. Thus, we will refer to CG31865/CG31866 as Drosophila Ada1 (dAda1) (Fig. 1B).
WDA and dAda1 associate with dSAGA subunits.
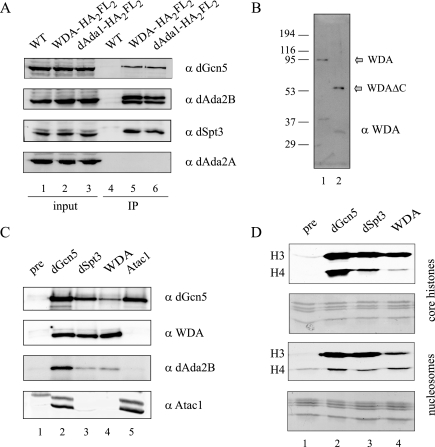
To confirm interactions between WDA, dAda1, and dGcn5, we generated the plasmids pRmHa3-wda-HA2FL2 and pRmHa3-dAda1-HA2FL2, which express C-terminally FLAG-tagged WDA and dAda1, respectively. We prepared whole-cell extracts from S2 cells transiently transfected with these plasmids and used anti-FLAG antibodies in coimmunoprecipitation experiments. Western blots demonstrated that WDA and dAda1 associated with dGcn5 (Fig. 2A, top panel). In addition, tagged WDA and dAda1 interacted with the dSAGA-specific subunits dSpt3 and dAda2B (Fig. 2A, dAda2B and dSpt3 blots) but not dAda2A, which is not in dSAGA but is in the ATAC histone acetyltransferase complex (Fig. 2A, bottom panel) (25). From this experiment, we conclude that dAda1 and WDA are specific components of dSAGA but not ATAC.
FIG. 2.
WDA and dAda1 specifically associate with dSAGA subunits. (A) S2 cells were transfected with pRmHa3-wda-HA2FL2 or pRmHa3-dAda1-HA2FL2 plasmids. After a 1-day induction, whole-cell extracts were prepared and incubated with M2-agarose beads. Untransfected S2 cells (WT) were used as negative controls. The immunoprecipitated material was analyzed by Western blotting using antibodies against dGcn5 (rabbit), dSpt3 (rabbit), dAda2B (guinea pig), and dAda2A (rabbit). Lanes 1 to 3 correspond to 40 μg of whole-cell extract (2% input). Lanes 4 to 6 correspond to the immunoprecipitated material (IP). (B) Ten microliters of a HAT-enriched extract (lane 1) and 20 pg of recombinant WDAΔC (lanes 2) were analyzed by Western blotting using antibodies against WDA (rat). (C) Nuclear extracts were immunoprecipitated using antibodies against dGcn5, dSpt3, WDA, Atac1, or a preimmune bleed (pre). The immunocomplexes were analyzed by Western blotting using antibodies directed to dGcn5 (rabbit), WDA (rat), dAda2B (guinea pig), and Atac1 (rat). (D) Nuclear extracts were immunoprecipitated using antibodies against dGcn5, dSpt3, WDA, or a preimmune bleed (pre). The HAT activity on the beads was assayed using core histones (top panels) or nucleosomes (bottom panels) as substrates. For each substrate, the fluorography showing the labeled histones is located above the corresponding Coomassie gel. The migrations of histones H3 and H4 are shown. α, anti.
To further investigate the association of WDA with dSAGA, we generated polyclonal antibodies against the N-terminal nonconserved portion of WDA, which lacks the WD repeats. We used these antibodies in Western blots but were unable to detect the endogenous protein in nuclear extracts from S2 cells (data not shown). However, these antisera recognized WDA (predicted molecular mass of ∼84 kDa) present in a Ni-agarose HAT complex-enriched fraction (Fig. 2B, lane 1). In addition, the serum detected a truncated recombinant protein purified from E. coli, with a molecular mass of ∼50 kDa (Fig. 2B, lane 2). An identical blot probed with a preimmune bleed from the same animal did not contain signals for endogenous or recombinant WDA (data not shown), which led us to conclude that the antiserum specifically recognized WDA, but was not sensitive enough to detect it in nuclear extracts.
We next wished to establish associations between endogenous WDA and dSAGA subunits, such as dGcn5 and dSpt3. Coimmunoprecipitation experiments showed that antibodies directed to dGcn5 and dSpt3 pull down WDA from nuclear extracts (Fig. 2C, lanes 2 and 3, WDA blot). In addition, when we used the WDA serum for the reciprocal immunoprecipitation reaction, the dSAGA subunits dGcn5 and dAda2B were brought down but not the ATAC complex subunit Atac1 (Fig. 2C, lane 4). dGcn5 antibodies again confirmed its presence in both dSAGA and ATAC (Fig. 2C, lane 2). Conversely, a preimmune bleed did not precipitate dGcn5, WDA, dAda2B, or Atac1, although it pulled down a nonspecific band that cross-reacts with the Atac1 serum (Fig. 2C, lane 1). Another way to confirm the association of WDA with the dSAGA HAT complex is by testing the HAT activity of the immunoprecipitated complexes. Antibodies against WDA immunoprecipitated a HAT pattern identical to that of dSpt3 and dGcn5 when either core histones (Fig. 2D, top panels) or nucleosomes (Fig. 2D, lower panels) were used as substrates. This pattern, consisting of a strong H3 signal and weaker H4 is characteristic of yeast and Drosophila SAGA complexes (32).
WDA is a stable subunit of the 1.5-MDa dSAGA complex.
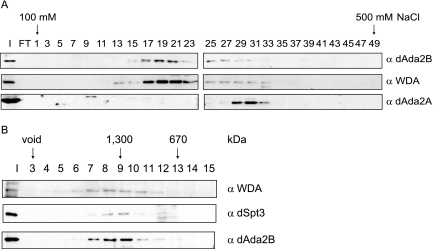
Since our observations provided evidence for a stable association of WDA with dSAGA, we wanted to determine if WDA cofractionated with dSAGA components by anion-exchange chromatography. To this end, we applied a Ni-agarose HAT-enriched fraction to a Mono-Q column and compared the elution profiles of WDA, dAda2B, and dAda2A by Western blotting. This analysis showed that WDA and the dSAGA subunit dAda2B elute in the same range of the salt gradient (Fig. 3A), corresponding to the elution profile of dSAGA. The WDA peak did not overlap with the ATAC complex subunit dAda2A, which elutes from the column at a higher salt (Fig. 3A, bottom panel) (25).
FIG. 3.
WDA is a subunit of the 1.5-MDa dSAGA complex. (A) A HAT-enriched extract was applied to a Mono Q column. The bound proteins were resolved with a linear gradient of 100 to 500 mM NaCl, and odd fractions were analyzed by Western blotting using antibodies against dAda2B (guinea pig), dSpt3 (rabbit), and WDA (rat). I, 10 μl of input; FT, 10 μl of Mono Q flowthrough. (B) Fractions 17 to 23 were pooled, concentrated, and loaded onto a Superose 6 column. The eluted fractions were analyzed for the presence of WDA, dAda2B, and Spt3 by Western blotting. The migration of the molecular mass standards is shown. I, 10 μl input; α, anti.
To determine the apparent molecular mass of the WDA-containing dSAGA complex, we pooled the fractions where WDA was present (fractions 17 to 23), concentrated them, applied them to a Superose 6 gel filtration column, and performed Western analysis on the eluted fractions. Figure 3B points out that WDA, dSpt3, and dAda2B associate in a complex with an apparent molecular mass of ∼1.5 MDa.
Affinity purification of dSAGA.
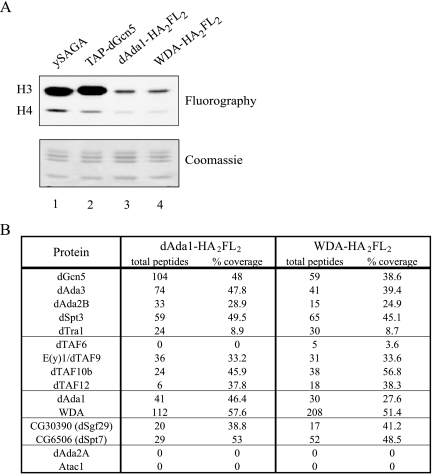
Coimmunoprecipitations indicated that dAda1 and WDA are components of dSAGA. Nuclear extract fractionation provided additional evidence that WDA is a subunit of dSAGA. To gain insight into the subunit composition of the dSAGA complex, we wished to obtain mass spectrometry data from complexes affinity purified from tagged dSAGA-specific subunits. To this end, we established stable cell lines that express C-terminally FLAG-tagged WDA and dAda1 and prepared nuclear extracts from these cell lines. The extracts were subjected to anti-FLAG affinity purification, and the purified material was analyzed for nucleosomal HAT activity to confirm the success of the dSAGA purifications (Fig. 4A). The affinity-purified complexes (lanes 3 and 4) generated an acetylation pattern identical to the H3 pattern of yeast SAGA (lane 1) on nucleosomes. To determine the protein composition in dSAGA and to identify new subunits, the purified complexes were subjected to MudPIT (Fig. 4B). MudPIT analyses turned out to be very informative not only because they confirmed previous findings but because they provided evidence for new subunits in the complex. First, we noticed that the previously identified dSAGA components dGcn5, dAda3, dAda2B, dSpt3 and dTra1 copurified with dAda1 and WDA. Second, we identified peptides for the SAGA-specific TAFs, such as dTAF9 and dTAF12 (23). Thus, it appears that yeast and fly SAGA have a subset of TAFs in common. One possible explanation for not obtaining peptides for TAF6 in the dAda1-FLAG purification could be partial disruption of the interaction of TAF6 with dSAGA due to the tags fused to the C terminus of dAda1. Third, we could confirm that dAda1 and WDA are present in the same complex, since they copurify with each other. Fourth, results from this MudPIT experiment point to the presence of other uncharacterized proteins in dSAGA, a number of which are conserved through evolution. For instance, the proteins CG30390 and CG6506 are orthologues of yeast Sgf29 and Spt7, both of which are subunits of ySAGA (22, 43). Another potential dSAGA subunit revealed by MudPIT is an orthologue of yeast Ubp8 (data not shown). We are currently tagging these new putative dSAGA subunits to confirm their interaction with dGcn5. Fifth, MudPIT confirmed that WDA and dAda1 specifically associate with dSAGA and not the dGcn5-containing complex ATAC, since the purifications did not yield peptides for the ATAC-specific proteins dAda2A and Atac1 (25). Finally, the fact that we could not identify peptides for the TATA-binding protein or TDIID-exclusive TAFs such as TAF1 in our purifications supports our hypothesis that WDA is not a TAF (data not shown).
FIG. 4.
Affinity purification and MudPIT analysis of dSAGA. (A) The nucleosomal HAT activity of affinity-purified dSAGA complexes (lanes 2 and 3) was compared to the HAT pattern of yeast SAGA (ySAGA) and dGcn5-containing complexes purified from cells expressing TAP-dGcn5 (TAP-dGcn5). The fluorography is shown above the Coomassie gel. (B) MudPIT analysis of dSAGA, purified from cells expressing tagged dAda1 or WDA. The table shows the number of nonredundant spectra for each protein (total peptides) and the amino acid sequence coverage (% coverage).
The WD repeats in WDA are required for its incorporation into dSAGA.
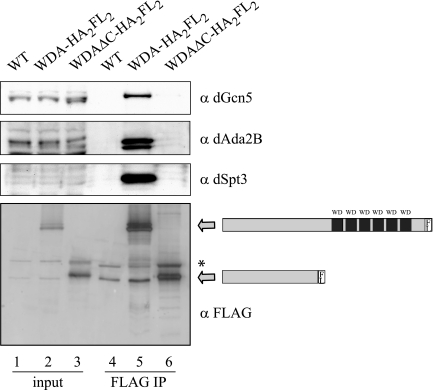
WD repeats are present in a very diverse group of eukaryotic proteins with different cellular functions. WD repeats can create surfaces for protein-protein interactions (50), a feature that may be exploited by WDA. Therefore, we wished to establish the function of the WD repeats in WDA. To achieve this, we expressed a C-terminally FLAG-tagged truncated form of WDA, lacking all the WD repeats, in S2 cells. This protein, referred to as WDAΔC-HA2FL2, was stable and could be detected on Western blots using an anti-FLAG antibody (Fig. 5, lane 3, bottom panel). The anti-FLAG beads could bring down the truncated polypeptide (Fig. 5, lane 6, bottom panel). However, the dSAGA subunits dGcn5, dAda2B, and dSpt3 could not be detected in the immunoprecipitated material (Fig. 5, lane 6). When an identical immunoprecipitation was performed with full-length tagged WDA, the FLAG antibody coimmunoprecipitated not only the tagged protein but also all the dSAGA subunits tested (Fig. 5, lane 5). This experiment confirms a crucial role of the WD repeats in the association of WDA with dSAGA.
FIG. 5.
The WD repeats in WDA are required for the incorporation of WDA into dSAGA. S2 cells were transfected with pRmHa3-wda-HA2FL2 and pRmHa3-wdaΔC-HA2FL2 plasmids. After a 1-day induction, whole-cell extracts were prepared and incubated with M2-agarose beads. Untransfected S2 cells (WT) were used as negative controls. The immunoprecipitated material was analyzed by Western blotting using antibodies against dGcn5 (rabbit), dSpt3 (rabbit), dAda2B (guinea pig), and FLAG. Lanes 1 to 3 correspond to 40 μg of whole-cell extract (2% input). Lanes 4 to 6 correspond to the immunoprecipitated material (FLAG IP). The arrows show the migration of tagged WDA and WDAΔC. The asterisk points to a cross-reacting band recognized by the anti-FLAG antibodies. α, anti.
Using a pull-down assay followed by mass spectrometry analysis, Wysocka et al. identified the human WD-repeat protein WDR5 as an interaction partner of methylated histone H3 on lysine 4 (K4). WDR5, a subunit of a number of methyltransferase complexes, has transactivation properties and is important for the conversion from di- to tri-methylated H3 (58). This observation raises the possibility that WDA, in a similar fashion, could bind to methylated nucleosomes to propagate acetylation by dSAGA through chromatin. To address this question, we performed in vitro binding experiments to test if H3 peptides methylated on K4 could pull down recombinant WDA with stronger affinity than the unmodified H3 peptides. We did not observe that WDA preferentially bound to di-methylated H3 on K4 (data not shown). It would be interesting to see if the related fly protein WDS (will die slowly), encoded by an essential gene and the closest orthologue of WDR5, can bind to methylated histones like its human counterpart does (27, 58).
WDA is encoded by an essential gene.
To ascertain the phenotype of flies devoid of WDA, we generated a wda null allele via imprecise excision of the KG03744 transposon, located ∼300 bp upstream of the wda gene. We analyzed the hop-out events by PCR and sequencing (data not shown) and confirmed imprecise excisions of the P-element in three independent events. The mutant alleles wda4, wda8, and wda11 were missing the predicted TATA box, the translation initiation codon, the first exon, and part of the second exon of the wda gene (Fig. 6A). Due to the absence of transcription and translation regulatory regions, we anticipate all three alleles to be protein null. In addition, all three alleles were lethal when homozygous. wda11 was chosen for further studies because of the removal of a larger portion of the wda gene than of the other two alleles.
FIG. 6.
Deletions in the wda gene are homozygous lethal. (A) Schematic representation of the wda locus, showing the position of the P-element KG03744 (insertion) and the DNA deleted after P-element excision (deletions 4, 8, and 11). Exons are represented by boxes. 5′ and 3′ untranslated regions are open boxes, while translated sequences are filled in with gray. (B) Genetic crosses were carried out with the flies whose genotypes are shown to the left. The bristle phenotype (Sb+ or Sb1) was scored in the progeny (200 flies). The percentage of flies corresponding to each genotype is shown. a, flies with phenotype Sb+; b, flies with phenotype Sb1; c, animals carrying two copies of the balancer chromosome die as embryos.
To determine the lethal phase of wda11 flies, embryos from a wda11/TM3, P{GAL4-twi.G}2.3, P{UAS-2xEGFP}AH2.3, Sb1, Ser1 population were collected on apple juice plates and aged to generate a collection of embryos at stages 12 to 15. The homozygous mutant wda11/wda11 embryos were identified by their lack of fluorescence and separated from the fluorescent, balancer-containing embryos. The majority of these mutant embryos (98%) could hatch into larvae. Mutant first-instar larvae did not present abnormalities in their mouth parts, spiracles, or movement behavior and could develop into second instar. However, second-instar larvae could not develop beyond that developmental stage.
When we examined the genomic sequence of the alleles wda4, wda8, and wda11, we realized that the predicted transcription start for the gene CG13827, adjacent to wda, was missing. To address whether the lethality was due to the loss of wda or CG13827, we attempted to rescue the lethality of wda11 flies using the GAL4/UAS binary system (3). Transgenic flies carrying the cDNA coding for WDA under control of UAS were recombined onto the wda11 chromosome. Simultaneously, we recombined the tubulin-GAL4 driver onto the wda11 chromosome. The genotypes of the resulting stocks were wda11, UAS-wda/TM3, Sb1, Ser1 and wda11, P{tubP-GAL4}LL7/TM3, Sb1, Ser1. These stocks were crossed to each other, and their adult progeny were scored for the absence of the dominant marker Sb1 in the balancer chromosome TM3, Sb1, Ser1 (Fig. 6B). We observed that 39% of the adult progeny did not carry the balancer chromosome, having the genotype wda11, UAS-wda/wda11, P{tubP-GAL4}LL7. The rest of the progeny were Sb1, corresponding to the genotypes wda11, UAS-wda/TM3, Sb1, Ser1 and wda11, P{tubP-GAL4}LL7/TM3, Sb1, Ser1 (both of them carry the balancer chromosome). These results indicated that the wda transgene could rescue the lethality of the wda11 allele, when expressed under control of a ubiquitous promoter, and suggested that the lethality in wda11 flies was due to lack of expression of WDA and not CG13827.
WDA is required for histone H3 acetylation in Drosophila embryos.
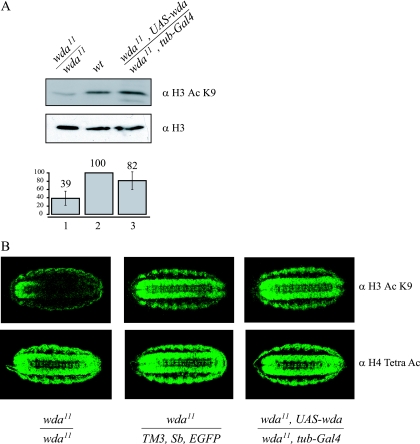
Previous studies demonstrated that inactivation of the dSAGA subunits dGcn5 and dAda2B causes a decrease in H3 acetylation (8, 41, 44). Therefore, we wished to determine whether the novel dSAGA protein WDA is linked to histone H3 acetylation. To determine the overall level of acetylated H3 in embryos, we prepared whole-cell extracts from wild-type embryos at stage 16 to 17, homozygous wda11/wda11 embryos and homozygous mutant embryos expressing WDA from a transgene (wda11, UAS-wda/wda11, P{tubP-GAL4}LL7). We analyzed equivalent amounts of extract by Western blotting, probing with antibodies against H3 or acetylated lysine 9 on histone H3 (ac H3 K9) (Fig. 7A). The blots reveal a significant decrease in histone H3 acetylation on K9 in the mutant embryos (compare lanes 1 and 2, top panel), relative to the total levels of histone H3. When we introduced a transgene carrying the cDNA for wda in the mutant background, acetylated H3 is restored to levels closer to those of the wild type (compare lanes 2 and 3, top panel). The H3 blots confirm equal loading on all three samples (Fig. 7A, middle panel).
FIG. 7.
Deletions in the wda gene result in reduced levels of histone H3 acetylation. (A) Whole-cell extracts from homozygous mutant (lane 1), wild-type (lane 2), and mutant embryos expressing WDA from a transgene (lane 3) were analyzed by Western blotting using antibodies against H3 and acetylated H3 K9. The intensities of the bands on the Western blots from three independent experiments were quantified. The levels of acetylated H3 were normalized to the total amount of H3 in each sample to determine the levels of acetylated H3 relative to the wild type. (B) Embryos were stained with antibodies against acetylated H3 K9 (top panels) or tetra-acetylated H4 (bottom panels). The genotypes of the embryos are indicated. α, anti.
Quantitation of the bands revealed a reduction of over 50% in H3 K9 acetylation in the mutants, which was restored to over 80% of wild-type levels by introduction of a transgene (Fig. 7, bottom panel).
Western analysis revealed a reduction in the global levels of histone H3 K9 acetylation in mutant embryos. We wished to investigate if this reduction could also be observed by immunostaining of embryos at the same stage of development. We collected stage 16 to 17 embryos and stained them with antibodies against ac H3 K9 (see Materials and Methods). We observed a reduction in the staining intensity of the homozygous mutant embryos, compared to control embryos, carrying a wild-type copy of the wda gene (compare top left and middle panels). This reduction in ac H3 K9 signal was clear in the central nervous system of stage 16 embryos (Fig. 7B). Furthermore, when WDA is expressed from a transgene, in the same pattern as tubulin, the acetylation of H3 in the central nervous system is restored (compare top middle and right panels). In contrast, we did not observe differences in the staining pattern when we used the tetra-acetylated H4 antibodies (Fig. 7B, bottom panels). These results are consistent with experiments performed with dAda2b mutant flies. dAda2b mutants are homozygous lethal and display reduced levels of H3 acetylation in stage 16 embryos. In addition, the overall acetylated H3 signal in polytene chromosomes is severely reduced (41, 44). We could not test for acetylation levels of wda mutants on polytene chromosomes, since wda mutants die during second larval instar, while dAda2b mutants die during pupal stage.
WDA is not required for the structural integrity of dSAGA.
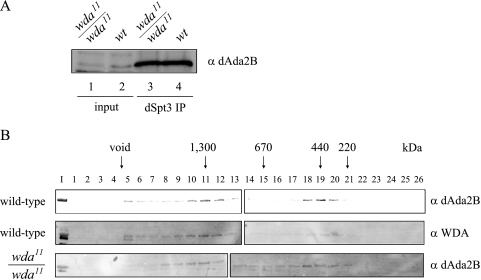
One possible explanation for the defects in H3 acetylation in the wda mutants could be that WDA is an architectural subunit and its removal disrupts the integrity of the complex. To address this possibility, we prepared whole-cell extracts from wild-type and wda11/wda11 embryos. We carried out immunoprecipitations using antibodies directed to dSpt3 and analyzed the immunoprecipitates with antibodies to dAda2B (Fig. 8A). Western blot analysis indicates that dSpt3 antibodies can pull down dAda2B from mutant extracts as efficiently as from wild-type embryos (lanes 3 and 4), suggesting that removal of WDA does not interfere with the stability of the complex. A low-resolution structure of the yeast SAGA complex indicates that Spt3 and Gcn5 are physically separated in different modules within SAGA (57). Since Ada2 and Ada3 potentiate the nucleosomal HAT activity of Gcn5, it is likely that those three proteins reside in the same module within SAGA (1). Our coimmunoprecipitation experiment indicates that dAda2B and dSpt3, likely to be in distinct modules of dSAGA, can still associate in the absence of WDA.
FIG. 8.
WDA is not required for the structural integrity of dSAGA. (A) Three milligrams of whole-cell extracts from wild-type or wda11/wda11 embryos were immunoprecipitated with antibodies directed to dSpt3 and analyzed by Western blotting with dAda2B antibodies. Lanes 1 and 2 correspond to 50 μg of whole-cell extract from mutant and wild-type embryos, respectively. Lanes 3 and 4 show the immunoprecipitated material from mutant and wild-type extracts, respectively. (B) Forty milligrams of whole-cell extracts from wild-type or wda11/wda11 embryos were loaded onto a Superose 6 gel filtration column. The elution profiles were determined by probing the column fractions with anti-WDA and -dAda2B antibodies. Molecular mass standards were run under the same conditions. I, 40 μg of whole-cell extracts; α, anti.
We also tested the integrity of the complex lacking WDA by gel filtration chromatography. To this end, we applied extracts from wild-type or wda11/wda11 embryos to a Superose 6 column and looked for shifts in the elution profiles of wild-type and WDA-lacking dSAGA, which would reflect changes in their apparent molecular weights. Western analysis of the column fractions corresponding to wild-type dSAGA indicated cofractionation of WDA and Ada2B in a 1.5-MDa complex (Fig. 8B, fractions 10 to 12, top and middle panels). In addition, these two proteins are present in a small complex (fractions 18 to 20), which could represent a real dSAGA subcomplex present in embryos or result from degradation of the 1.5-MDa complex during our fractionation procedure. The elution profile of mutant dSAGA, determined with the dAda2B antibodies, is identical to that of wild-type dSAGA (Fig. 8B, bottom panel), except for the fact that WDA antibodies do not detect any signal on Western blots (data not shown). The gel filtration results agree with the coimmunoprecipitation experiment, pointing us to conclude that WDA is not an architectural protein required to hold dSAGA subunits together.
DISCUSSION
Previous studies from our lab have shown that Drosophila orthologues of the yeast proteins Gcn5, Spt3, Ada2, Ada3, and Tra1 cofractionated in anion-exchange and size-exclusion chromatography, suggesting that flies harbor a complex similar to SAGA (dSAGA) (31). This study describes the affinity purification of this complex for the first time. Proteomic approaches combined with biochemical assays allowed us to confirm that dSAGA contains dGcn5, dAda3, dSpt3, dAda2B, dTra1, and TAFs. In addition to the previously characterized subunits, this study uncovered the new components dAda1 and WDA.
Our mass spectrometry study of affinity-purified dSAGA unveiled peptides corresponding to CG31865/CG31866. We utilized BLAST searches and multiple-sequence alignments as tools to determine that CG31865/CG31866 encodes an orthologue of the yeast and human Ada1. Coimmunoprecipitation experiments proved that dAda1 associated with dSAGA. Yeast Ada1 is required for the structural integrity of SAGA and, together with Spt7 and Spt20, is part of a subset of SAGA subunits that show the most severe phenotypes when their genomic copies are deleted (52). For example, ada1Δ cells, unlike spt3Δ or gcn5Δ cells, fail to grow in media containing caffeine or the DNA synthesis inhibitor hydroxyurea. Moreover, ada1Δ is synthetically lethal with mutations in SWI/SNF subunits; however, ada2Δ and ada3Δ are not (52). We suggest that dAda1 may provide a structural role in dSAGA similar to that of its yeast counterpart.
A novel protein identified by MudPIT was WDA. We observed that this WD-repeat-containing protein has a unique N-terminal domain with a different amino acid composition from other WD-containing proteins in the database. The presence of a subunit containing six WD repeats seems to be an evolutionary feature of SAGA-related complexes. Yeast SAGA and human STAGA contain the WD-repeat proteins yTAF5 and hPAF65β/hTAF5L, respectively (23, 35). Temperature-sensitive mutations in yTAF5 impair the association of yTAF5 with SAGA subunits and cause transcription defects. In addition, mutations in yTAF5 affect the overall stability of yAda1 and ySpt7. Interestingly, the mutations in yTAF5 localize to the WD repeats, suggesting an essential role of these repeats in ySAGA structure and function (14).
The WD repeats in WDA are required for the interaction of WDA with dSAGA. Different chromatin-related proteins make protein-protein interactions through their WD repeats. For instance, the transcription corepressors Groucho (Gro) and Tup1 display WD repeats at their C termini. The WD domain of Drosophila Gro is required for direct interactions with the DNA-binding proteins Engrailed and Hairy. In addition, point mutations in the WD domain of yeast Tup1 disrupt its interaction with the repressor α2 (12). Another example of a WD-repeat protein involved in chromatin metabolism is Drosophila p55, a subunit of the remodeling complex NURF, the chromatin assembly factor (CAF1) and an uncharacterized HAT (36). In some cases, several WD-repeat-containing proteins can be found in the same complex, illustrated by the yeast COMPASS complex, involved in histone methylation, that contains Swd1, Swd2, and Swd3, all of which have WD repeats (13).
We generated small deficiencies in the wda gene by imprecise excision of a P-element and observed that this gene was required for viability. The homozygous mutant animals died during second larval instar. A transgene containing the cDNA for wda could rescue the lethality when expressed in a pattern similar to that of tubulin. The recombinant adult flies, homozygous for the deletion but expressing WDA from the transgene, had no obvious morphological abnormalities and were fertile. The fertility of the adults and the fact that UAS-wda cannot be expressed in the germ line indicate that WDA is dispensable for oogenesis (45). In contrast, dGcn5 and dAda2B, both of which are in dSAGA, are required for oogenesis. These observations illustrate how different dSAGA subunits are involved in distinct functions during development. In fact, mutations in different yeast SAGA subunits also display different phenotypes under specific growth conditions (52). Moreover, genome-wide analysis in yeast demonstrated that the set of genes affected by the loss of Spt3 does not completely overlap with the groups of genes regulated by Gcn5 (33).
The immunofluorescent staining of embryos with antibodies directed to acetylated K9 on H3 complemented the analysis of bulk levels of acetylated K9 on H3, suggesting that K9 of H3 is one of the in vivo targets for dSAGA. Our results are consistent with a previous report that describes the requirement of dAda2b for viability and histone acetylation (44). Both dAda2B and WDA contribute to the in vivo H3 HAT activity of dSAGA. However, wda null animals die earlier than dAda2b null strains, which could indicate distinct functions of these two proteins within dSAGA. An alternative explanation for the difference in lethality phases of wda and dAda2b mutants could be a difference in maternal loading and stability of WDA and dAda2B proteins. A higher amount or higher stability of maternal dAda2B could enable development of the mutants until pupal stage.
This study provides additional evidence that the SAGA complex is conserved throughout evolution. The protein complexes purified from yeast, human, and fly cells display similar features. First, they all have a preference for H3 as a substrate. Second, dSAGA can bind to Ni-agarose in the same way the yeast complex does (22). Third, the complexes purified from yeast and flies are similar in size, having apparent molecular masses between 1.5 and 2 MDa (22). Fourth, like the yeast complex, dSAGA and STAGA, can interact with acidic activators (31, 35, 54). In fact, the conserved proteins Tra1 and TRRAP, subunits of SAGA and STAGA, respectively, directly interact with transcription activators (5, 42). Fifth, both SAGA and STAGA can activate transcription from nucleosomal templates in an acetyl coenzyme A-dependent manner (28, 35).
Although the different SAGA complexes from different organisms share a number of components, they also possess nonconserved subunits. For example, yeast SAGA contains Spt8 and Spt20, while human STAGA contains STAF 46, 55, and 60 and dSAGA has WDA and other nonconserved potential subunits that we are currently investigating. These species-specific subunits may provide unique features for each complex to carry out specialized function.
Acknowledgments
We thank members of the Workman and Abmayr labs for helpful discussions and technical advice. We also thank Jeff Haug and Joshua Wunderlich for the EGFP sorting of Drosophila embryos and Danny Stark and Kiran Kocherlakota for help with confocal microscopy. We thank Joanna Wysocka and David Allis for providing the H3 peptides for the pull-down experiments.
REFERENCES
- 1.Balasubramanian, R., M. G. Pray-Grant, W. Selleck, P. A. Grant, and S. Tan. 2002. Role of the Ada2 and Ada3 transcriptional coactivators in histone acetylation. J. Biol. Chem. 277:7989-7995. [DOI] [PubMed] [Google Scholar]
- 2.Birck, C., O. Poch, C. Romier, M. Ruff, G. Mengus, A. C. Lavigne, I. Davidson, and D. Moras. 1998. Human TAF(II)28 and TAF(II)18 interact through a histone fold encoded by atypical evolutionary conserved motifs also found in the SPT3 family. Cell 94:239-249. [DOI] [PubMed] [Google Scholar]
- 3.Brand, A. H., and N. Perrimon. 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118:401-415. [DOI] [PubMed] [Google Scholar]
- 4.Brand, M., K. Yamamoto, A. Staub, and L. Tora. 1999. Identification of TATA-binding protein-free TAFII-containing complex subunits suggests a role in nucleosome acetylation and signal transduction. J. Biol. Chem. 274:18285-18289. [DOI] [PubMed] [Google Scholar]
- 5.Brown, C. E., L. Howe, K. Sousa, S. C. Alley, M. J. Carrozza, S. Tan, and J. L. Workman. 2001. Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science 292:2333-2337. [DOI] [PubMed] [Google Scholar]
- 6.Candau, R., P. A. Moore, L. Wang, N. Barlev, C. Y. Ying, C. A. Rosen, and S. L. Berger. 1996. Identification of human proteins functionally conserved with the yeast putative adaptors ADA2 and GCN5. Mol. Cell. Biol. 16:593-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Candido, E. P., R. Reeves, and J. R. Davie. 1978. Sodium butyrate inhibits histone deacetylation in cultured cells. Cell 14:105-113. [DOI] [PubMed] [Google Scholar]
- 8.Carre, C., D. Szymczak, J. Pidoux, and C. Antoniewski. 2005. The histone H3 acetylase dGcn5 is a key player in Drosophila melanogaster metamorphosis. Mol. Cell. Biol. 25:8228-8238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carrozza, M. J., R. T. Utley, J. L. Workman, and J. Cote. 2003. The diverse functions of histone acetyltransferase complexes. Trends Genet. 19:321-329. [DOI] [PubMed] [Google Scholar]
- 10.Celniker, S. E., and G. M. Rubin. 2003. The Drosophila melanogaster genome. Annu. Rev. Genomics Hum. Genet. 4:89-117. [DOI] [PubMed] [Google Scholar]
- 11.Celniker, S. E., D. A. Wheeler, B. Kronmiller, J. W. Carlson, A. Halpern, S. Patel, M. Adams, M. Champe, S. P. Dugan, E. Frise, A. Hodgson, R. A. George, R. A. Hoskins, T. Laverty, D. M. Muzny, C. R. Nelson, J. M. Pacleb, S. Park, B. D. Pfeiffer, S. Richards, E. J. Sodergren, R. Svirskas, P. E. Tabor, K. Wan, M. Stapleton, G. G. Sutton, C. Venter, G. Weinstock, S. E. Scherer, E. W. Myers, R. A. Gibbs, and G. M. Rubin. 2002. Finishing a whole-genome shotgun: release 3 of the Drosophila melanogaster euchromatic genome sequence. Genome Biol. 3:RESEARCH0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, G., and A. J. Courey. 2000. Groucho/TLE family proteins and transcriptional repression. Gene 249:1-16. [DOI] [PubMed] [Google Scholar]
- 13.Cheng, H., X. He, and C. Moore. 2004. The essential WD repeat protein Swd2 has dual functions in RNA polymerase II transcription termination and lysine 4 methylation of histone H3. Mol. Cell. Biol. 24:2932-2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Durso, R. J., A. K. Fisher, T. J. Albright-Frey, and J. C. Reese. 2001. Analysis of TAF90 mutants displaying allele-specific and broad defects in transcription. Mol. Cell. Biol. 21:7331-7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eberharter, A., S. John, P. A. Grant, R. T. Utley, and J. L. Workman. 1998. Identification and analysis of yeast nucleosomal histone acetyltransferase complexes. Methods 15:315-321. [DOI] [PubMed] [Google Scholar]
- 16.Fischle, W., Y. Wang, and C. D. Allis. 2003. Histone and chromatin cross-talk. Curr. Opin. Cell Biol. 15:172-183. [DOI] [PubMed] [Google Scholar]
- 17.Furlong, E. E., D. Profitt, and M. P. Scott. 2001. Automated sorting of live transgenic embryos. Nat. Biotechnol. 19:153-156. [DOI] [PubMed] [Google Scholar]
- 18.Gangloff, Y. G., C. Romier, S. Thuault, S. Werten, and I. Davidson. 2001. The histone fold is a key structural motif of transcription factor TFIID. Trends Biochem. Sci. 26:250-257. [DOI] [PubMed] [Google Scholar]
- 19.Gangloff, Y. G., S. L. Sanders, C. Romier, D. Kirschner, P. A. Weil, L. Tora, and I. Davidson. 2001. Histone folds mediate selective heterodimerization of yeast TAF(II)25 with TFIID components yTAF(II)47 and yTAF(II)65 and with SAGA component ySPT7. Mol. Cell. Biol. 21:1841-1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gangloff, Y. G., S. Werten, C. Romier, L. Carre, O. Poch, D. Moras, and I. Davidson. 2000. The human TFIID components TAF(II)135 and TAF(II)20 and the yeast SAGA components ADA1 and TAF(II)68 heterodimerize to form histone-like pairs. Mol. Cell. Biol. 20:340-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcia-Higuera, I., J. Fenoglio, Y. Li, C. Lewis, M. P. Panchenko, O. Reiner, T. F. Smith, and E. J. Neer. 1996. Folding of proteins with WD-repeats: comparison of six members of the WD-repeat superfamily to the G protein beta subunit. Biochemistry 35:13985-13994. [DOI] [PubMed] [Google Scholar]
- 22.Grant, P. A., L. Duggan, J. Côté, S. M. Roberts, J. E. Brownell, R. Candau, R. Ohba, T. Owen-Hughes, C. D. Allis, F. Winston, S. L. Berger, and J. L. Workman. 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 11:1640-1650. [DOI] [PubMed] [Google Scholar]
- 23.Grant, P. A., D. Schieltz, M. G. Pray-Grant, D. J. Steger, J. C. Reese, J. R. Yates III, and J. L. Workman. 1998. A subset of TAFIIs are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell 94:45-53. [DOI] [PubMed] [Google Scholar]
- 24.Grant, P. A., D. Schieltz, M. G. Pray-Grant, J. R. Yates III, and J. L. Workman. 1998. The ATM-related cofactor Tra1 is a component of the purified SAGA complex. Mol. Cell 2:863-867. [DOI] [PubMed] [Google Scholar]
- 25.Guelman, S., T. Suganuma, L. Florens, S. K. Swanson, C. L. Kiesecker, T. Kusch, S. Anderson, J. R. Yates III, M. P. Washburn, S. M. Abmayr, and J. L. Workman. 2006. Host cell factor and an uncharacterized SANT domain protein are stable components of ATAC, a novel dAda2A/dGcn5-containing histone acetyltransferase complex in Drosophila. Mol. Cell. Biol. 26:871-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hiller, M. A., T. Y. Lin, C. Wood, and M. T. Fuller. 2001. Developmental regulation of transcription by a tissue-specific TAF homolog. Genes Dev. 15:1021-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hollmann, M., E. Simmerl, U. Schafer, and M. A. Schafer. 2002. The essential Drosophila melanogaster gene wds (will die slowly) codes for a WD-repeat protein with seven repeats. Mol. Genet. Genomics 268:425-433. [DOI] [PubMed] [Google Scholar]
- 28.Ikeda, K., D. J. Steger, A. Eberharter, and J. L. Workman. 1999. Activation domain-specific and general transcription stimulation by native histone acetyltransferase complexes. Mol. Cell. Biol. 19:855-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keller, C. A., M. A. Grill, and S. M. Abmayr. 1998. A role for nautilus in the differentiation of muscle precursors. Dev. Biol. 202:157-171. [DOI] [PubMed] [Google Scholar]
- 30.Khorasanizadeh, S. 2004. The nucleosome: from genomic organization to genomic regulation. Cell 116:259-272. [DOI] [PubMed] [Google Scholar]
- 31.Kusch, T., S. Guelman, S. M. Abmayr, and J. L. Workman. 2003. Two Drosophila Ada2 homologues function in different multiprotein complexes. Mol. Cell. Biol. 23:3305-3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, K. K., L. Florens, S. K. Swanson, M. P. Washburn, and J. L. Workman. 2005. The deubiquitylation activity of Ubp8 is dependent upon Sgf11 and its association with the SAGA complex. Mol. Cell. Biol. 25:1173-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee, T. I., H. C. Causton, F. C. Holstege, W. C. Shen, N. Hannett, E. G. Jennings, F. Winston, M. R. Green, and R. A. Young. 2000. Redundant roles for the TFIID and SAGA complexes in global transcription. Nature 405:701-704. [DOI] [PubMed] [Google Scholar]
- 34.Li, D., and R. Roberts. 2001. WD-repeat proteins: structure characteristics, biological function, and their involvement in human diseases. Cell. Mol. Life Sci. 58:2085-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martinez, E., V. B. Palhan, A. Tjernberg, E. S. Lymar, A. M. Gamper, T. K. Kundu, B. T. Chait, and R. G. Roeder. 2001. Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol. Cell. Biol. 21:6782-6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinez-Balbas, M. A., T. Tsukiyama, D. Gdula, and C. Wu. 1998. Drosophila NURF-55, a WD repeat protein involved in histone metabolism. Proc. Natl. Acad. Sci. USA 95:132-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McMahon, S. J., M. G. Pray-Grant, D. Schieltz, J. R. Yates III, and P. A. Grant. 2005. Polyglutamine-expanded spinocerebellar ataxia-7 protein disrupts normal SAGA and SLIK histone acetyltransferase activity. Proc. Natl. Acad. Sci. USA 102:8478-8482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Noland, B. J., J. M. Hardin, and G. R. Shepherd. 1971. Histone acetyltransferase activity in synchronized mammalian cells. Biochim. Biophys. Acta 246:263-268. [DOI] [PubMed] [Google Scholar]
- 39.Ogryzko, V. V., T. Kotani, X. Zhang, R. L. Schlitz, T. Howard, X.-J. Yang, B. H. Howard, J. Qin, and Y. Nakatani. 1998. Histone-like TAFs within the PCAF histone acetylase complex. Cell 94:35-44. [DOI] [PubMed] [Google Scholar]
- 40.Palhan, V. B., S. Chen, G. H. Peng, A. Tjernberg, A. M. Gamper, Y. Fan, B. T. Chait, A. R. La Spada, and R. G. Roeder. 2005. Polyglutamine-expanded ataxin-7 inhibits STAGA histone acetyltransferase activity to produce retinal degeneration. Proc. Natl. Acad. Sci. USA 102:8472-8477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pankotai, T., O. Komonyi, L. Bodai, Z. Ujfaludi, S. Muratoglu, A. Ciurciu, L. Tora, J. Szabad, and I. Boros. 2005. The homologous Drosophila transcriptional adaptors ADA2a and ADA2b are both required for normal development but have different functions. Mol. Cell. Biol. 25:8215-8227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park, J., S. Kunjibettu, S. B. McMahon, and M. D. Cole. 2001. The ATM-related domain of TRRAP is required for histone acetyltransferase recruitment and Myc-dependent oncogenesis. Genes Dev. 15:1619-1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Powell, D. W., C. M. Weaver, J. L. Jennings, K. J. McAfee, Y. He, P. A. Weil, and A. J. Link. 2004. Cluster analysis of mass spectrometry data reveals a novel component of SAGA. Mol. Cell. Biol. 24:7249-7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qi, D., J. Larsson, and M. Mannervik. 2004. Drosophila Ada2b is required for viability and normal histone H3 acetylation. Mol. Cell. Biol. 24:8080-8089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rorth, P. 1998. Gal4 in the Drosophila female germline. Mech. Dev. 78:113-118. [DOI] [PubMed] [Google Scholar]
- 46.Sanders, S. L., J. Jennings, A. Canutescu, A. J. Link, and P. A. Weil. 2002. Proteomics of the eukaryotic transcription machinery: identification of proteins associated with components of yeast TFIID by multidimensional mass spectrometry. Mol. Cell. Biol. 22:4723-4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Selleck, W., R. Howley, Q. Fang, V. Podolny, M. G. Fried, S. Buratowski, and S. Tan. 2001. A histone fold TAF octamer within the yeast TFIID transcriptional coactivator. Nat. Struct. Biol. 8:695-700. [DOI] [PubMed] [Google Scholar]
- 48.Sendra, R., C. Tse, and J. C. Hansen. 2000. The yeast histone acetyltransferase A2 complex, but not free Gcn5p, binds stably to nucleosomal arrays. J. Biol. Chem. 275:24928-24934. [DOI] [PubMed] [Google Scholar]
- 49.Smith, E. R., A. Pannuti, W. Gu, A. Steurnagel, R. G. Cook, C. D. Allis, and J. C. Lucchesi. 2000. The Drosophila MSL complex acetylates histone H4 at lysine 16, a chromatin modification linked to dosage compensation. Mol. Cell. Biol. 20:312-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith, T. F., C. Gaitatzes, K. Saxena, and E. J. Neer. 1999. The WD repeat: a common architecture for diverse functions. Trends Biochem. Sci. 24:181-185. [DOI] [PubMed] [Google Scholar]
- 51.Sterner, D. E., and S. L. Berger. 2000. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 64:435-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sterner, D. E., P. A. Grant, S. M. Roberts, L. J. Duggan, R. Belotserkovskaya, L. A. Pacella, F. Winston, J. L. Workman, and S. L. Berger. 1999. Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol. Cell. Biol. 19:86-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 54.Utley, R. T., K. Ikeda, P. A. Grant, J. Côté, D. J. Steger, A. Eberharter, S. John, and J. L. Workman. 1998. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature 394:498-502. [DOI] [PubMed] [Google Scholar]
- 55.Vlachonasios, K. E., M. F. Thomashow, and S. J. Triezenberg. 2003. Disruption mutations of ADA2b and GCN5 transcriptional adaptor genes dramatically affect Arabidopsis growth, development, and gene expression. Plant Cell. 15:626-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang, L., C. Mizzen, C. Ying, R. Candau, N. Barlev, J. Brownell, C. D. Allis, and S. L. Berger. 1997. Histone acetyltransferase activity is conserved between yeast and human GCN5 and is required for complementation of growth and transcriptional activation. Mol. Cell. Biol. 17:519-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu, P. Y., C. Ruhlmann, F. Winston, and P. Schultz. 2004. Molecular architecture of the S. cerevisiae SAGA complex. Mol. Cell 15:199-208. [DOI] [PubMed] [Google Scholar]
- 58.Wysocka, J., T. Swigut, T. A. Milne, Y. Dou, X. Zhang, A. L. Burlingame, R. G. Roeder, A. H. Brivanlou, and C. D. Allis. 2005. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell 121:859-872. [DOI] [PubMed] [Google Scholar]
- 59.Xu, W., D. G. Edmondson, Y. A. Evrard, M. Wakamiya, R. R. Behringer, and S. Y. Roth. 2000. Loss of Gcn5l2 leads to increased apoptosis and mesodermal defects during mouse development. Nat. Genet. 26:229-232. [DOI] [PubMed] [Google Scholar]