Abstract
Prostate cancers (PCas) become resistant to hormone withdrawal through increased androgen receptor (AR) signaling. Here we show increased AR-mediated transcription efficiency in PCa cells that have acquired the ability to grow in low concentrations of androgen. Compared to androgen-dependent PCa cells, these cells showed increased activity of transiently transfected reporters and increased mRNA synthesis relative to levels of AR occupancy of the prostate-specific antigen (PSA) gene. The locus also displayed up to 10-fold-higher levels of histone H3-K9/K14 acetylation and H3-K4 methylation across the entire body of the gene. Although similar increased mRNA expression and locus-wide histone acetylation were also observed at another kallikrein locus (KLK2), at a third AR target locus (TMPRSS2) increased gene expression and locus-wide histone acetylation were not seen in the absence of ligand. Androgen-independent PCa cells have thus evolved three distinctive alterations in AR-mediated transcription. First, increased RNA polymerase initiation and processivity contributed to increased gene expression. Second, AR signaling was more sensitive to ligand. Third, locus-wide chromatin remodeling conducive to the increased gene expression in the absence of ligand was apparent and depended on sustained AR activity. Therefore, increased AR ligand sensitivity as well as locus-specific chromatin alterations contribute to basal gene expression of a subpopulation of specific AR target genes in androgen-independent PCa cells. These features contribute to the androgen-independent phenotype of these cells.
The molecular processes that mediate transcription orchestrate cell proliferation, differentiation, and disease progression. Central to this regulation is the dynamic organization and modification of nucleosomes, the basic repeating unit of chromatin that is comprised of 146 bp of DNA wrapped around histone octamers. Accessibility to transcription factors and activation of genes largely depend on diverse posttranslational modifications of amino termini (36, 47) and the more recently implicated globular domains of histones (6). These modifications include acetylation, phosphorylation, and methylation, which covalently add acetyl, phospho, and methyl groups, respectively, to specific residues of core histones. The well-characterized acetylation and methylation of lysines in histones H3 and H4 are highly correlated with transcriptional activation. Acetylation, catalyzed by histone acetyltransferases such as p300/CBP, is reversed by the activity of histone deacetylases, which mediate transcriptional repression (21). Methylation at histone H3 (K4) is catalyzed by specific methyltransferases often found in large complexes such as ALL-1 (32). This process is reversed by the action of a recently identified lysine-specific histone demethylase, LSD1 (39, 40). The complex interactions between the different histone tail modifications have led to the “histone code hypothesis,” which suggests that specific histone modifications affect and interact with other histone modifications, thus serving as markers for the recruitment of associated factors for the regulation of chromatin function (41). This system regulates DNA synthesis, genome stability, and the efficiency of gene expression in patterns that are clonally maintained (17). It is therefore expected that clonally derived cancer cells will have distinguishing patterns of locus-specific histone modifications related to the cancer phenotype. In the case of prostate cancer (PCa), gene expression at androgen receptor (AR) target loci is of particular interest as a focal point for examining such patterns of histone modification as they may relate to disease initiation and progression.
AR signaling serves as a prototypical model of transcriptional regulation and is critical in all phases of PCa development, including the evolution of resistance to androgen ablation therapies. Resistance to androgen ablation is associated with an altered capacity of the AR to remain functional in the androgen-depleted environment (reviewed in references 1, 4, 7, and 12). Growth of androgen-independent PCa cells remains AR dependent, as disruption of the AR by a specific antibody or ribozyme inhibits proliferation (46). Furthermore, aberrant AR activity in androgen-independent PCa includes AR hypersensitivity to androgens and insensitivity to antagonists, possibly due to increased AR expression and/or the impact of other molecular factors in tumors. It was conclusively demonstrated that increased AR expression is necessary and sufficient to convert androgen-sensitive PCa to an androgen-independent state (5). Moreover, specific expression in mouse prostate epithelial cells of a trans-AR gene containing a gain-of-function mutation (with increased basal activity and response to coregulators) resulted in PCa development in 100% of the animals (16), proving that aberrant AR signaling was sufficient to cause PCa.
An important component of the AR-mediated phenotype observed in androgen-independent prostate cancer is the increased sensitivity to castrate levels of androgen that leads to androgen-independent AR target gene expression and tumor recurrence (5). Because it was recently shown that global histone acetylation and dimethylation patterns predicted the risk of PCa recurrence (38), we hypothesized that part of the increased sensitivity of the AR to low levels or absence of androgen may reside in target gene histone alterations that allow AR-mediated transcription to occur at higher-than-normal efficiency.
In our recent work (18-20, 25, 26) we have made extensive use of the LNCaP/C4-2B cell culture model of PCa progression. LNCaP cells endogenously express the AR and are dependent on the natural AR ligand dihydrotestosterone (DHT) for growth and prostate-specific antigen (PSA) expression (45). C4-2B is an androgen-independent subline of LNCaP that still expresses a functional AR (9). It was obtained by passage, growth, and isolation of LNCaP from bone metastasis in castrated athymic mice (42). In our most recently published study (19) we have compared AR activity in LNCaP and C4-2B cell lines using PSA expression as a target gene readout. We observed very little PSA expression in LNCaP cells in the absence of androgens, while substantial expression was observed in C4-2B cells under the same condition. The addition of DHT to the culture medium increased the expression of PSA in both cell types. In that study chromatin immunoprecipitation (ChIP) analysis unexpectedly revealed that AR was not significantly detectable at any site within the PSA gene locus in the absence of DHT in either cell type. We concluded that androgen-independent expression of PSA in C4-2B cells did not rely on the direct occupancy of the AR at the PSA locus but based on results obtained with small interfering RNA (siRNA)-mediated AR knockdown was nevertheless affected indirectly via an unknown AR-dependent mechanism(s).
Therefore, in the present study we set out to investigate mechanisms involved in AR-mediated gene expression in androgen-independent PCa cells and report data consistent with three novel concepts that occur during the progression of PCa to androgen independence. First, increased RNA polymerase initiation and processivity contribute to increased gene expression. Second, AR-mediated transcription is sensitive to lower ligand concentrations. Third, significant androgen-independent chromatin alterations at certain AR target loci depend on sustained AR signaling.
MATERIALS AND METHODS
Cell culture and materials.
Human prostate cancer LNCaP and PC-3 cells, obtained from the American Type Culture Collection (ATCC; Manassas, VA), and C4-2B cells, obtained from ViroMed Laboratories (Minneapolis, MN), were maintained in RPMI 1640 supplemented with 5% (vol/vol) fetal bovine serum (FBS). CV-1 cells obtained from the ATCC were grown in Dulbecco modified Eagle medium (DMEM) with 5% FBS. 5α-DHT, dexamethasone (Dex), and MG132 were purchased from Sigma Chemical Co. (St. Louis, MO). Antibodies were anti-AR (N20), anti-polymerase II (N20), antiactin (Santa Cruz Biotechnology, Santa Cruz, CA), anti-acetylated H3 (anti-AcH3)-K9/K14, anti-dimethyl-H3-K4 (Upstate Biotechnology, Inc., Lake Placid, NY), anti-trimethyl-H3-K4, and anti-histone H3 (Abcam, Cambridge, MA). The probasin-luc (ARR3-thymidine kinase-luciferase) and MMTV-luc (mouse mammary tumor virus-luciferase) expression vectors have been used previously in our studies (20). PSA-luciferase (PGL3-PSA5.85) was a generous gift from Hong-Wu Cheng, University of California at Davis, and was constructed by inserting 5.85 kb of the entire PSA upstream sequence into pGL3 basic vector (29).
Transient-transfection and luciferase assays.
CV-1 cells were seeded in 96-well plates at 7 × 103 cells/well in phenol-red free DMEM containing 5% charcoal/dextran-stripped FBS (CSS). One day later cells were transfected with MMTV-luc reporter (100 ng/well) along with AR (10 ng/well) or glucocorticoid receptor (GR) (10 ng/well) using Lipofectamine 2000 (Invitrogen Corp., Carlsbad, CA) according to the manufacturer's protocol. The next day cells were exposed to 5 μM MG132 or dimethyl sulfoxide (DMSO) vehicle for 2 h, after which the media were changed to introduce 10 nM DHT or 100 nM Dex or ethanol (EtOH) vehicle for a subsequent 24-hour treatment. Cells were lysed using Passive Lysis Buffer (Promega, Madison, WI), and luciferase activity determinations in cell lysates were conducted as previously described (20).
LNCaP (3 × 105 cells/well) and C4-2B (1.5 × 105 cells/well) cells were plated in 12-well plates and grown in phenol red-free RPMI 1640 containing 5% CSS for 2 days. Cells were then transfected with an AR-responsive firefly luciferase reporter plasmid, PSA-luc, probasin-luc, or MMTV-luc (1 μg/well) along with pRL-SV40 Renilla luciferase plasmid (Promega) (10 ng/well) using Lipofectamine 2000 (Invitrogen Corp.) according to the manufacturer's protocol. After transfection, cells were grown in phenol red-free RPMI 1640 containing 5% CSS with different concentrations of DHT as indicated for 24 h. Dual luciferase activity determinations in cell lysates were conducted according to the manufacturer's protocol using the dual luciferase reporter assay system (Promega).
Histone isolation and immunoblotting.
Total histone isolation and immunoblotting were performed as previously described using the indicated antibodies (18, 26).
Real-time RT-PCR.
After the indicated treatments, total RNA from cells or tissues was extracted using the SV Total RNA Isolation System (Promega). A two-step reverse transcription-PCR (RT-PCR) method was employed using the TaqMan Gold RT-PCR kit (Applied Biosystems, Branchburg, NJ) and Opticon (MJ Research, Inc., Waltham, MA). The primers and probes of real-time PCR for PSA pre-mRNA, PSA mature mRNA, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA expression were as previously described (20). The primers and probes of real-time PCR for p16, KLK2, and TMPRSS2 mRNA expression were as follows: p16, 5′-CTGCCCAACGCACCGA-3′ (forward), 5′-CGCTGCCCATCATCATGAC-3′ (reverse), 5′-6-FAM-TGGATCGGCCTCCGACCGTAACT-BHQ-1-3′ (probe); KLK2, GCTGCCCATTGCCTAAAGAAG (forward), TGGGAAGCTGTGGCTGACA (reverse), 5′-6-FAM-CGGCACAACCTGTTTGAGCCTGAAGA-BHQ-1-3′ (probe); TMPRSS2, 5′-CCTGCAAGGACATGGGTATA-3′ (forward), CCGGCACTTGTGTTCAGTTTC-3′ (reverse), 5′-6-FAM-TAGCCAAGGAATAGTGGATGACAGCGGA-BHQ-1-3′ (probe). Triplicate PCRs were conducted. GAPDH mRNA expression was analyzed for each sample in parallel.
ChIP assays.
LNCaP (6 × 106 cells/150-mm dish), C4-2B (3 × 106 cells/150-mm dish), and PC-3 (3 × 106 cells/150-mm dish) cells were cultured in phenol red-free RPMI 1640 supplemented with 5% CSS for 3 days. After the indicated treatments, ChIP assays were conducted as described previously (20). DNA samples from ChIP preparations were analyzed by real-time PCR using AmpliTaq Gold PCR Master Mix (Applied Biosystems) and Opticon (MJ Research, Inc.). The primers and probes (synthesized by Biosearch Technologies, Novato, CA) for the PSA, p16, KLK2, and TMPRSS2 loci are listed in Tables S1 to S4, respectively, in the supplemental material. Duplicate immunoprecipitations (IPs) for each antibody and duplicate real-time PCRs for each IP sample were performed. Input values were obtained from samples treated in the same way as the experimental ones, except that no IP steps were performed. The results are given as percentages of input.
siRNA transfection.
C4-2B cells (1.5 × 106 cells/150-mm dish) were grown in phenol red-free RPMI 1640 containing 5% CSS for 2 days. Cells were transfected with an AR siRNA duplex (sense, 5′-ACG UUU ACU UAU CUU AUG CTT-3′; antisense, 5′-GCA UAA GAU AAG UAA ACG UTT-3′) directed against the coding region of AR mRNA. Mock transfection (without siRNA) and a nonspecific (NS) siRNA duplex (sense, 5′-AAU UUU ACU CGC UCG AUU UTT-3′; antisense, 5′-AAA UCG AGC GAG UAA AAU UTT-3′) were used as controls. All siRNA duplexes were transfected at a final concentration of 100 nM using Oligofectamine reagent (Invitrogen Corp.) according to the manufacturer's instructions. After transfection, cells were grown in phenol red-free RPMI 1640 containing 5% CSS for 4 days and then ChIP assays were conducted as described above. AR protein levels of 2% sonicated input from ChIP experiments were measured by immunoblotting.
CWR22 prostate cancer xenograft model and tissue ChIP.
The CWR22 human prostate tumors were provided by C. W. Gregory of the University of North Carolina (Chapel Hill, NC) and maintained as xenografts. The CWR22 androgen-dependent and androgen-independent prostate tumors were generated in nude mice (obtained from Charles River Laboratories, Inc., Wilmington, MA) as described previously (24) with the following modifications. The tumors were injected subcutaneously (s.c.) as 1 million dissociated cells in Matrigel (BD Biosciences, Bedford, MA) into nude mice containing 12.5-mg sustained-release testosterone pellets (implanted s.c. 2 days prior to tumor transplantation). Tumor growth was determined weekly by caliper measurement. Tumor volume was calculated with the following formula: 0.52 × L × W2, where L is length and W is width. After 3 weeks, when tumors had grown to a volume size of approximately 0.5 cm3, mice were castrated and randomized into two study groups. In group 1 (n = 5), s.c. 12.5-mg testosterone pellets were left to maintain consistent serum levels of testosterone. In group 2 (n = 4), s.c. 12.5-mg testosterone pellets were removed. Once tumors reached a volume of about 3 cm3, mice were sacrificed by cervical dislocation. Fresh tumors were harvested and excised into several pieces (∼0.2 cm3). Tumors for the study of RNA were frozen in liquid nitrogen. Tumors for ChIP assay were cut into small pieces and immediately fixed with 1% formaldehyde at room temperature for 10 min. The cross-linking reaction was terminated by adding glycine at a final concentration of 0.125 M. Tissues were washed with cold 1× phosphate-buffered saline and homogenized. Cell pellets were resuspended in lysis buffer (1% sodium dodecyl sulfate, 10 nM EDTA, 50 nM Tris-HCl, pH 8.0, 2× protease inhibitors). After sonication, the resulting soluble chromatins were stored at −80°C. ChIP assays and quantitative real-time PCR were performed later as described above. Blood was collected, and serum testosterone levels were determined by a highly specific radioimmunoassay (15).
RESULTS
Unliganded AR occupancy at the PSA enhancer is associated with high PSA expression in androgen-independent PCa cells.
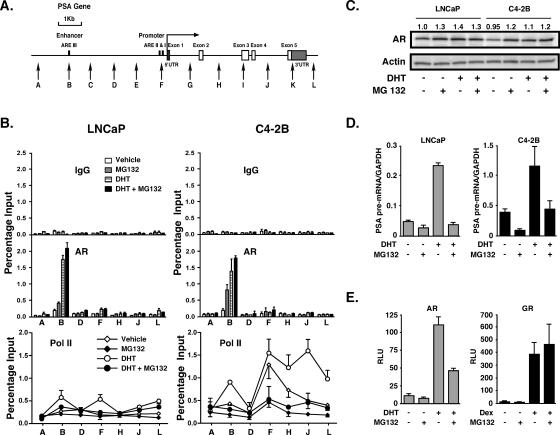
In the present study we have addressed the novel concept that an AR target locus may become more sensitive to the transcription factor by extending our analyses across the entire PSA gene (Fig. 1A). Although AR did not significantly localize to any site at this locus in C4-2B cells in the absence of DHT, it could be detected at the PSA enhancer if the cells were pretreated with the potent 26S proteasome inhibitor MG132 (Fig. 1B) (27). MG132 disrupts proteasome involvement in the cyclical recruitment of transcription factors to steroid receptor-mediated transcription sites (10, 22). The proteasome-mediated turnover of the estrogen receptor (ER) on responsive promoters was previously shown to be an integral feature of ER signaling (33). Therefore, we surmised that inhibition of the proteasome may cause accumulation of AR at the PSA enhancer even if AR occupies the site transiently and that this could result in enhanced detection sensitivity. Significant AR levels accumulated at the PSA enhancer after MG132 treatment in the absence of DHT, more so in C4-2B than in LNCaP cells. However, this did not lead to substantial RNA polymerase II recruitment to any site at the locus (Fig. 1B), consistent with proposals that proteasome-mediated effects on RNA polymerase II resolve stalled polymerase complexes during transcription (14).
FIG. 1.
Unliganded and liganded AR occupancy at the PSA enhancer. A. Schematic representation of the PSA gene. Vertical arrows show real-time PCR targeted regions. UTR, untranslated region. B. LNCaP and C4-2B cells were incubated in phenol red-free RPMI 1640 containing 5% CSS for 3 days and then pretreated with 5 μM MG132 or DMSO vehicle for 2 h, followed by 10 nM DHT or EtOH vehicle for 2 h. AR and RNA polymerase II (Pol II) occupancies at the indicated sites of the PSA gene were examined by ChIP analyses. The results are representative of two independent experiments, and values are presented as percentage input and represent mean values ± standard deviations. IgG, immunoglobulin G. C. LNCaP and C4-2B cells were treated as described for panel B. Immunoblot analyses of AR and actin protein levels from whole-cell extracts were conducted. The number above each lane of AR expression indicates the relative intensity of each band normalized to actin expression. The data are representative of two independent experiments. D. LNCaP and C4-2B cells were treated as described for panel B. PSA pre-mRNA levels were examined by real-time RT-PCR. The results are shown as PSA pre-mRNA/GAPDH ratios. Values are presented as the means ± standard deviations of triplicate determinations. E. CV-1 cells were incubated in phenol-red free DMEM containing 5% CSS for 1 day prior to transfection with MMTV-luc reporter (100 ng/well) along with AR (10 ng/well) or GR (10 ng/well) in 96-well plates. Cells were exposed to 5 μM MG132 or DMSO vehicle for 2 h, followed by 10 nM DHT or 100 nM Dex treatment for 24 h. Data are measured in relative light units (RLU) and are representative of results obtained from three independent experiments. Values are means ± standard deviations of triplicate wells.
Another insight was revealed by the polymerase occupancy data obtained from C4-2B cells not treated with MG132 (Fig. 1B). Although the polymerase occupancy levels were similar between cells treated and cells not treated with DHT at the promoter (site F), the levels were significantly higher through the body of the genes in cells treated with DHT. Polymerase II occupancy at the transcription start site in the absence of DHT (vehicle) in C4-2B cells is in line with significant PSA expression under these conditions, whereas its increased presence through the body of the gene after DHT treatment is consistent with an increase in polymerase processivity (30). However, the fact that the polymerase occupancy levels were higher at the enhancer, promoter, and gene body in C4-2B cells than in LNCaP cells may be due to increases in both processivity and transcription initiation events in C4-2B cells. Exactly how this is achieved is unclear, but it seems to be unrelated to AR occupancy at the enhancer, which was similar in LNCaP and C4-2B cells.
The increased AR accumulation at the PSA enhancer in C4-2B compared to LNCaP cells after MG132 treatment indicates that AR turnover, mediated by proteasomal action, may occur at higher rates in the absence of ligand in C4-2B than in LNCaP cells. The MG132 treatment marginally stabilized the AR protein by 20 to 30% (Fig. 1C). As expected, AR occupancy levels increased to similar levels as a result of 2-h DHT treatment in both cell types. Therefore, AR does in fact localize to the PSA enhancer in C4-2B cells in the absence of ligand, if only transiently and at a relatively low level, and can thus mediate PSA expression in the absence of DHT. These data also suggest that very low steady-state levels of AR occupancy can mediate substantial PSA expression in the absence of DHT specifically in C4-2B cells (Fig. 1D). PSA pre-mRNA expression was 4- to 11-fold higher in C4-2B cells under all conditions (Fig. 1D). Therefore, in androgen-independent C4-2B cells AR-mediated transcription of PSA was markedly more efficient than in LNCaP cells as defined by mRNA synthetic rates relative to AR occupancy levels at the PSA enhancer. As expected, MG132 treatment inhibited PSA expression in both cell types in the presence or absence of DHT. Also, AR-mediated regulation of an ectopically expressed luciferase reporter in CV-1 cells was inhibited by MG132 treatment (Fig. 1E). It is understood that these inhibitory effects result from sequestration of the AR at AR binding sites in response to proteasome inhibition. It is known that MG132 does not inhibit GR activity (11), a phenomenon that we have used in this experiment to demonstrate that the inhibition of AR-mediated transcription by MG132 was not due to cytotoxic effects of the drug.
DHT-stimulated, non-chromatin-integrated, ARE-luciferase reporters are expressed at higher levels and are more sensitive to DHT concentrations in androgen-independent cells.
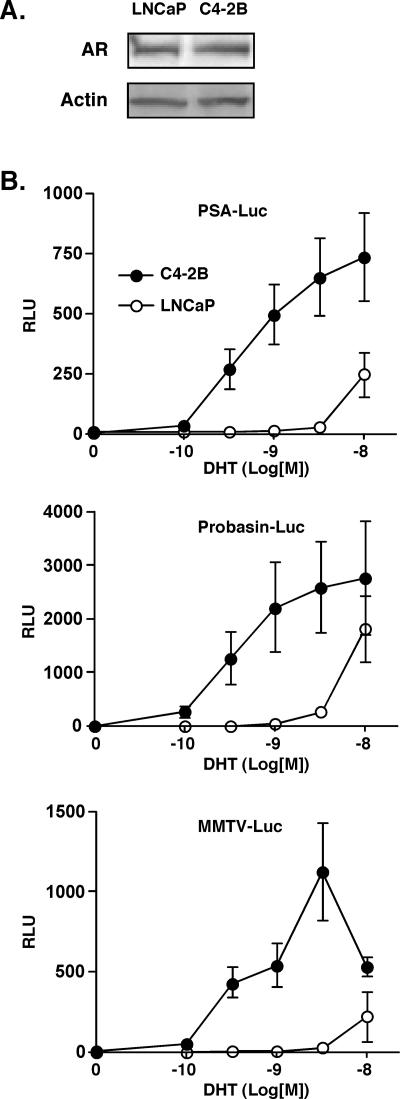
In order to determine AR activity on non-chromatin-integrated promoters, we transiently transfected and measured expression of androgen response element (ARE)-driven luciferase reporters in C4-2B and LNCaP cells (Fig. 2). With the use of any one of three reporter constructs (PSA-, probasin-, or MMTV-luciferase), the C4-2B cells were able to respond to DHT at concentrations lower by several orders of magnitude and with severalfold-higher expression levels. The results could not be explained by differences in transfection efficiency or AR steady-state levels (Fig. 2A). The data indicate that an AR ligand-sensitive phenotype exists in C4-2B cells, unrelated to integrated chromatin architecture of the target locus. This is probably due to altered AR activation mechanisms (4, 8, 12). Similar hypersensitivity to DHT was observed when endogenous PSA expression was measured (19). However, a significant difference between basal expression levels (no added DHT) of the transient reporter and the endogenous PSA was apparent; whereas luciferase expression levels under these conditions were very low and similar in C4-2B and LNCaP cells (Fig. 2), endogenous PSA expression values in the absence of DHT were more than 10-fold higher in C4-2B than in LNCaP cells (Fig. 1D). We conclude that although a chromatin-independent, hypersensitive AR phenotype exists in C4-2B cells, efficient transcription in the absence of DHT depends on chromatin-integrated genes.
FIG. 2.
DHT-sensitive AR phenotype in androgen-independent PCa cells. A. LNCaP and C4-2B cells were transiently transfected with PSA-luc reporter plasmid along with Renilla luciferase plasmid as described for panel B. Immunoblot analyses of AR and actin protein levels from whole-cell extracts were conducted. B. LNCaP and C4-2B cells were transiently transfected with one of three AR-responsive firefly luciferase reporter plasmids, PSA-luc, probasin-luc, or MMTV-luc (1 μg/well), along with the pRL-SV40 Renilla luciferase plasmid (10 ng/well) in 12-well plates and incubated with different concentrations of DHT as indicated for 24 h. Dual luciferase assays were conducted. The results were normalized for the internal Renilla control of each well and expressed as the mean relative light units (RLU) ± standard deviation from quadruplicate samples.
The PSA locus is hyperacetylated and hypermethylated in androgen-independent cells.
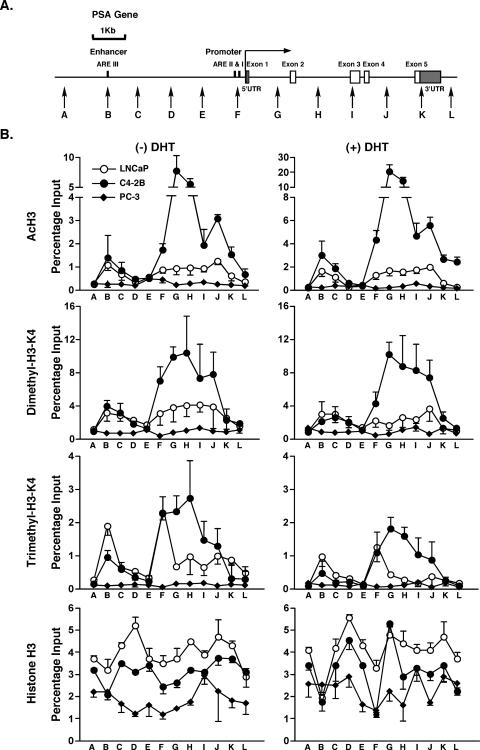
To test the hypothesis that the chromatin structure at the PSA locus may be conducive to AR-mediated transcription at very low levels of AR occupancy and in the absence of added ligand, we examined histone modifications across the entire PSA locus (Fig. 3A). Histone H3 acetylation levels at residues K9 and K14 were markedly higher in C4-2B cells than in either LNCaP or AR-negative PCa cells (PC-3), even in the absence of DHT treatment (Fig. 3B). Very high levels of histone H3 acetylation were apparent at the transcription start site and also most dramatically throughout the body of the gene in C4-2B cells, returning to basal levels only downstream of the 3′ end of the gene. Although the highest levels of acetylation were observed in introns 1 (site G) and 2 (site H), the high levels of acetylation across the entire body of the gene are in contrast to acetylation islands at the transcription start sites of many actively transcribed genes as revealed in genome-wide screens (28, 34, 35). Furthermore, both dimethylated and trimethylated H3-K4 levels in C4-2B cells were significantly increased across the body of the gene (Fig. 3B). The current data suggest that the situation in specifically activated genes, such as PSA in PCa cells, represents a special condition of sustained “open” chromatin structures across entire loci that facilitate hypersensitivity to the AR-mediated transcription. In this regard the PSA locus is similar to the locus control region of the active ɛ-globin gene where histone H3 and H4 acetylation and H3-K4 methylation were found to be continuous over a 17-kb region during active gene expression (23). Similarly, a nearly continuous H3-K4 methylated 60-kb region stretches from HOXA1 to HOXA7 in human lung fibroblasts (2). Loci such as these have apparently evolved to allow sustained expression of their component genes.
FIG. 3.
Histone modification analyses at the PSA locus. A. Schematic representation of the PSA gene. Vertical arrows show real-time PCR-targeted regions. UTR, untranslated region. B. LNCaP, C4-2B, and PC-3 cells were incubated in phenol red-free RPMI 1640 containing 5% CSS for 3 days and then treated with 10 nM DHT or EtOH vehicle for 4 h. AcH3, dimethylated H3-K4, trimethylated H3-K4, and unmodified histone H3 were examined by ChIP analyses at each of the PSA target regions. Duplicate IPs for each antibody and duplicate real-time PCR for each IP sample were done. Values are presented as percentages of input and represent mean values ± standard deviations.
DHT treatment increased the levels of histone H3 acetylation in C4-2B and LNCaP cells but not in AR-negative PC-3 cells (Fig. 3B). Di- and trimethyl-H3 levels at residue K4 decreased after DHT treatment at most sites, as we have reported previously, which is possibly due to epitope masking (26). Total histone H3 density was similar at most sites in all cells, indicating that the observed hyperacetylation and hypermethylation were not because of discrepancies in histone densities. However, at both the enhancer and promoter (sites B and F, respectively), H3 density decreased somewhat after DHT treatment, possibly due to nucleosome displacement at these sites by the docking of transcription initiation factors. Similar dips in levels of histones H3.3 and H3 were seen at active promoters of Drosophila melanogaster and interpreted as areas depleted of nucleosomes (31).
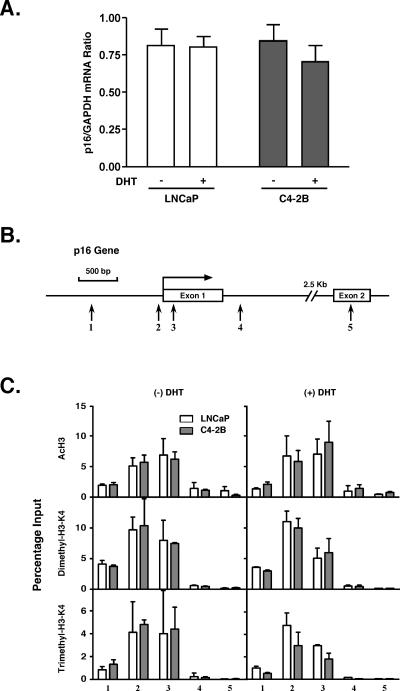
At the non-AR-target p16 locus the levels of above-mentioned histone modifications were similar in LNCaP and C4-2B cells, as was p16 mRNA expression (Fig. 4). Furthermore, the total genome-wide levels of acetylated and methylated histone H3 were similar in the three cell types regardless of DHT exposure (see Fig. S1 in the supplemental material). These results indicate that the dramatic alterations of histones in C4-2B cells are PSA locus specific, which strongly implicates AR activity in the process.
FIG. 4.
p16 gene expression and histone modifications. A. LNCaP and C4-2B cells were incubated in phenol red-free RPMI 1640 containing 5% CSS for 3 days and treated with 10 nM DHT and EtOH vehicle for 18 h. p16 and GAPDH mRNA levels were measured by real-time RT-PCR. The p16 expression levels are shown as p16/GAPDH mRNA ratios. Values are presented as the means ± standard deviations of triplicate determinations. B. Schematic representation of the p16 gene. Vertical arrows show real-time PCR-targeted regions. C. ChIP DNA samples, obtained from the experiment depicted in Fig. 3B, were analyzed for AcH3, dimethylated H3-K4, and trimethylated H3-K4 histones at the p16 locus.
In vitro electromobility shift assays using an androgen response element containing double-stranded DNA oligonucleotide as a probe revealed no differences in activity from whole-cell extracts of LNCaP and C4-2B cells (see Fig. S2 in the supplemental material), indicating that the cellular milieu was not different between the two cell types in allowing AR to interact with its cognate DNA response element.
Overall the results indicate that the hypersensitization of the PSA locus by significant histone H3 acetylation and methylation that encourage an “open” chromatin structure is a likely mechanism behind the elevated androgen-independent PSA expression observed in C4-2B cells.
Gene expression and histone H3 acetylation at two other AR target loci.
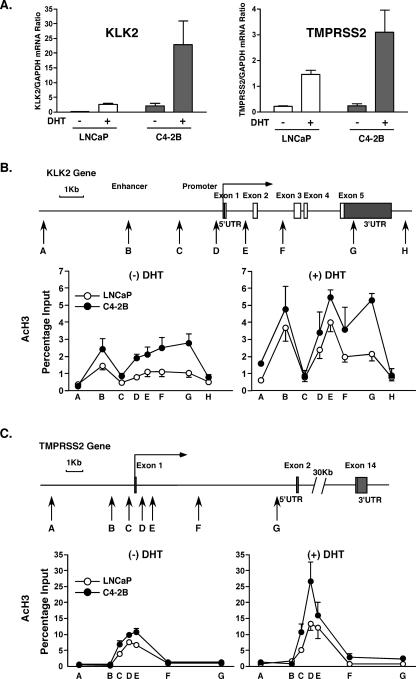
In order to determine the generality of AR-mediated gene overexpression and locus-wide histone hyperacetylation found at the PSA locus, we analyzed two additional AR target loci, tissue kallikrein 2 (KLK2) and TMPRSS2. KLK2 is 18.5 kb downstream from the PSA locus (also known as KLK3) and similarly belongs to a family of 15 tissue kallikrein genes on chromosome 19 (3). KLK2 is a distinct locus from PSA, and histone H3 acetylation increases observed at the PSA locus do not continuously extend to the KLK2 locus (data not shown). TMPRSS2 is an AR target gene on chromosome 21, and its promoter (and androgen dependence) is often translocated to ETS transcription factor genes in PCa (43, 44). This translocation has not happened in LNCaP cells (44) and presumably also not in C4-2B cells, since in both cell lines TMPRSS2 expression was significantly stimulated by DHT (Fig. 5A). In the absence of DHT, KLK2 was significantly more expressed in C4-2B than in LNCaP, whereas expression levels of TMPRSS2 were similar in the two cell lines under the same nonligand conditions. The expression of both genes in both cell lines was significantly induced by the addition of DHT to the medium. In the absence of DHT, the KLK2 locus had an enhanced histone H3 acetylation pattern across the entire gene body in C4-2B compared to LNCaP cells (Fig. 5B). At the TMPRSS2 locus only the promoter and 5′ end were acetylated and the levels were similar in the two cell lines in the absence of DHT (Fig. 5C). At both loci, and in both cell lines, the levels of acetylation increased after DHT treatment. Thus, KLK2 behaved with greater similarity to PSA than does TMPRSS2.
FIG. 5.
mRNA expression and histone H3 acetylation at KLK2 and TMPRSS2 loci. A. LNCaP and C4-2B cells were incubated in phenol red-free RPMI 1640 containing 5% CSS for 3 days and treated with 10 nM DHT and EtOH vehicle for 16 h. KLK2, TMPRSS2, and GAPDH mRNA levels were measured by real-time RT-PCR. The KLK2 and TMPRSS2 expression levels are shown as KLK2/GAPDH and TMPRSS2/GAPDH mRNA ratios. Values are presented as the means ± standard deviations of triplicate determinations. B. Schematic representation of the KLK2 gene. Vertical arrows show real-time PCR targeted regions. LNCaP and C4-2B cells were incubated in phenol red-free RPMI 1640 containing 5% CSS for 3 days and then treated with 10 nM DHT or EtOH vehicle for 4 h. AcH3 was examined by ChIP analyses at each of the KLK2 target regions. C. Schematic representation of the TMPRSS2 gene. Vertical arrows show real-time PCR targeted regions. ChIP DNA samples, obtained from the experiment described for panel B, were analyzed for AcH3 at the TMPRSS2 locus. UTR, untranslated region.
Increased histone acetylation is associated with the development of androgen independence in vivo.
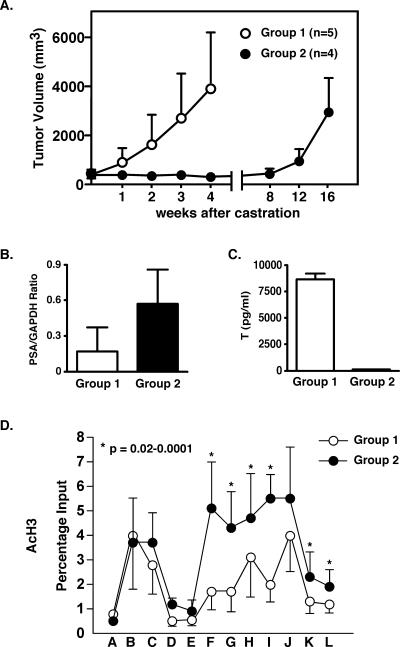
To determine whether the data linking histone modification with PCa progression reported thus far are confined to the LNCaP/C4-2B cell culture model, we opted to investigate CWR22 mouse xenografts, a powerful model system to study clinically relevant PCa progression to androgen independence in a nontissue culture system. CWR22 tumors express AR and secrete PSA and whereas they are initially androgen dependent, will progress to androgen independence following castration while retaining a transcriptionally active AR in the recurrent tumors (24). Therefore, the CWR22 human PCa xenograft model mimics clinical PCa progression and provides an excellent system for studying histone alterations at the PSA locus in the context of failed androgen ablation therapy and subsequent tumor relapse. In the present study this process lasted 12 to 16 weeks following castration (Fig. 6A).
FIG. 6.
PSA mRNA expression and histone modification in the CWR22 mouse model. A. Tumor volumes were measured at the time of castration and subsequent weeks thereafter. Group 1 mice (containing s.c. 12.5-mg testosterone pellets and bearing androgen-dependent tumors) were sacrificed 4 weeks after castration. Group 2 mice (without s.c. testosterone pellets and bearing androgen-independent tumors) were sacrificed 16 weeks after castration. B. Total RNA from the frozen tumor of each mouse was extracted. The PSA and GAPDH mRNA levels were measured by real-time RT-PCR. The PSA expression values are shown as PSA/GAPDH mRNA ratios. Columns show the mean of each group; error bars show the standard deviations. The two-sided P value between the two groups was 0.054 as calculated using the Student t test. C. Blood from each mouse was collected, and serum testosterone levels were measured by radioimmunoassay. Columns show the mean of each group; error bars show the standard deviation. D. ChIP assays were conducted as described in Materials and Methods. AcH3 histone was examined at the PSA locus. Values are presented as percentages of input and represent mean values ± standard deviations of each group. The two-sided P value between two groups was calculated using the Student t test.
In the present experiment we established two groups of mice: group 1 CWR22 tumors were grown under the influence of testosterone, whereas group 2 tumors were grown under total ablation conditions, as evidenced by the absence of measurable testosterone levels in this group (Fig. 6C). Notably, group 1 mice had on average lower steady-state levels of PSA mRNA than did group 2 mice (Fig. 6B). Histone modifications were analyzed by ChIP assays performed on minced tumor material, which was harvested when the tumors reached about 3 cm3. Group 2 mice, with androgen-independent tumors, displayed statistically significant elevated histone H3-K9/K14 acetylation at the transcription start site as well as at most of the sites across the body of the PSA gene (Fig. 6D). Interestingly, this difference was not observed at the PSA enhancer, although substantial acetylation was observed in both groups at this site. Histone H3-K4 di- and trimethylation were somewhat elevated in group 2, but due to the variation between animals statistical significance was not reached (data not shown).
These results show that at the PSA locus the histone H3 acetylation in androgen-independent tumors (i.e., group 2 mice) is increased compared to that in androgen-dependent tumors (group 1 mice). The results from this animal model system are therefore in agreement with those obtained in the LNCaP/C4-2B culture system and prove that the histone modifications at the PSA locus are not an artifact of cell culture, nor are they specific to a given model system.
Histone alterations at the PSA locus in androgen-independent PCa cells are AR dependent.
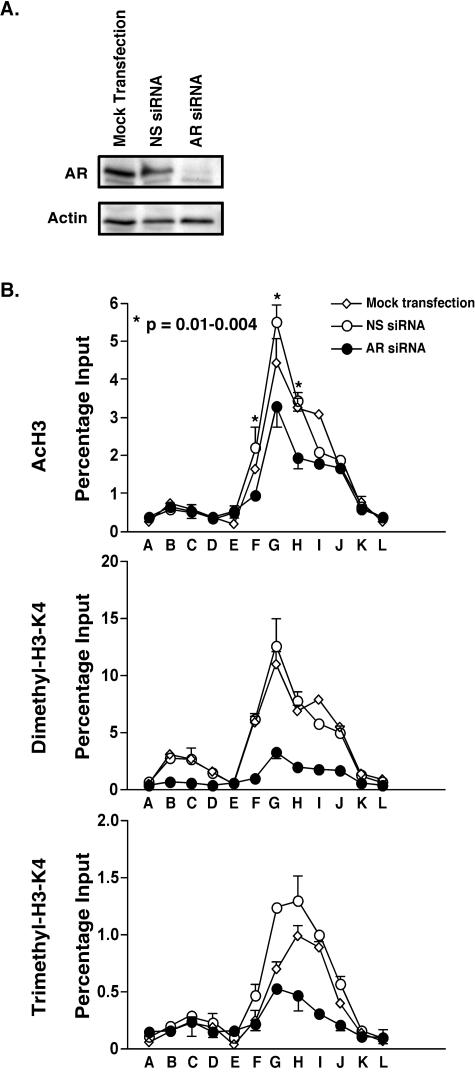
In our previous study we observed that siRNA knockdown of AR using two independent siRNA constructs inhibited PSA expression in C4-2B cells grown in the absence and presence of androgens (19). Recently it was also shown that cell proliferation was attenuated by siRNA-mediated knockdown of the AR (13). In the present experiments we knocked down AR and analyzed histone modifications at the PSA locus. As our work has previously demonstrated, after 4 days of AR knockdown nearly all AR protein was depleted from the cells as shown by immunoblot analysis (Fig. 7A). Histone H3-K9/K14 acetylation and histone H3-K4 methylation (both di- and trimethylation) were reduced at many sites after the AR knockdown (Fig. 7B). With regard to acetylated histones, this was most apparent at sites F to H, but less so at the other sites, whereas both di- and trimethylated levels were quite dramatically reduced by the treatment at most sites. We interpret the difference between the reduction levels in acetylation versus methylation as reflecting a lower turnover rate of acetylation than of methylation marks. It is interesting that the levels of dimethylated sites were more dramatically reduced than the levels of the trimethylated ones, again suggesting a difference in turnover rates. Nevertheless, the fact that the histone alterations were reduced after AR knockdown indicates that the altered modified histone levels seen in C4-2B cells are maintained by sustained AR activity, possibly due to sustained transcription at this locus. Therefore, the AR represents the link between gene expression and histone alterations at the PSA locus.
FIG. 7.
Histone alterations at the PSA locus after siRNA-mediated knockdown of AR. A. C4-2B cells were incubated in phenol red-free RPMI 1640 containing 5% CSS for 2 days and were either mock transfected or transfected with AR siRNA directed against the coding region of the AR gene or with NS siRNA. Four days after transfection, AR and actin protein levels were measured by immunoblotting. B. Four days after siRNA transfection, AcH3, dimethylated H3-K4, and trimethylated H3-K4 were examined by ChIP analyses at the PSA locus as described in the legend to Fig. 3. The two-sided P value between AR siRNA and NS siRNA acetylation data was calculated using the Student t test.
DISCUSSION
Progression of PCa is typified by conversion of cells from androgen dependence to independence as a result of ablation therapy failure. The consensus view is that this is due to the conversion of AR signaling from ligand dependence to independence (most recently reviewed in reference 8). Our present studies revealed that androgen-independent PCa cells became more amenable to AR-mediated gene expression because of increased polymerase engagement at target genes, an AR hypersensitive to ligand, and chromatin alterations of target genes conducive to increased transcription efficiency. This combined phenotype resulted in elevated AR signaling at very low or no androgen levels. We propose that cells containing this phenotype are selected during the transition from androgen-dependent to -independent PCa. This can be appreciated by considering the AR as the “seed” and AR target loci as the “soil” that combine to yield more efficient transcription as a consequence of more effective polymerase engagement. Locus-wide chromatin alterations were observed at some, but not all, AR target loci. Thus, the two kallikrein genes (PSA and KLK2), behaved similarly in that locus-wide histone acetylation in C4-2B cells not exposed to ligand was associated with substantial gene expression at the loci. Such genes belong to a subset of AR target genes, since TMPRSS2 did not behave in this manner and acetylation was confined to the 5′ end of the gene with similar levels of acetylation in the two cell lines in the absence of ligand. We do not know how many genes belong to each of the two groups of AR targets; neither do we know the functional significance of the two groups with respect to PCa progression to androgen independence. It is even possible that simply the size of a particular locus determines the extent of locus-wide acetylation. On the other hand, functional selection of efficient expression of kallikreins may aid tumor growth in a diminished-androgen environment; it is proposed elsewhere that kallikreins are major players in cancer progression (3). Many more AR target genes need to be analyzed before general rules can be established that determine efficiency of expression and associated locus-wide histone modifications in androgen-independent PCa cells. However, the chromatin structure and gene expression behavior at the two kallikrein loci provide a novel insight in AR-mediated gene expression during PCa progression to androgen independence.
It is known that AR expression/activity and its associated signaling partners may be altered during the androgen-independent development of PCa clones (1, 4, 5, 12). However, these proposed mechanisms that focus solely on alterations of the AR itself cannot explain the data presented here or those of our previous work (18-20). Although KLK2 behaved similarly to PSA, here we have concentrated on PSA gene expression since PSA is the best-studied kallikrein gene and was the target of our previous investigations. It is remarkable that the rate of PSA mRNA expression did not correlate with AR occupancy at the PSA enhancer or promoter when LNCaP and C4-2B cells were compared. In the androgen-independent C4-2B cell line PSA mRNA expression in the absence of DHT was nearly an order of magnitude higher than that in LNCaP even though the AR occupancy was barely detectable. When DHT was added to the culture medium, PSA mRNA expression rates increased in both cell lines but the expression in LNCaP reached only a level comparable to that in C4-2B cells not treated with DHT. These results indicate that perhaps the PSA locus itself was more conducive to transcriptional stimulation in C4-2B cells, either by histone modifications or by loss of repressors.
Our comprehensive analyses of histone modifications across the entire PSA locus revealed significant increases in histone H3-K9/K14 acetylation and H3-K4 methylation in androgen-independent cells compared to their androgen-dependent counterparts. These significant alterations of the chromatin structure at the PSA locus suggest an analogy of a “soil” that is more receptive to signaling from the AR “seed.” We have generated evidence that these two aspects are closely linked by analyzing both non-chromatin-integrated and chromatin-integrated AR targets. Our experiments using transiently transfected reporters indicate that the ligand-sensitive AR phenotype in C4-2B cells was chromatin independent. On the other hand, the siRNA knockdown of the AR showed that the chromatin-modified phenotype depended on sustained AR signaling. Furthermore, the significant target gene expression in the absence of added DHT was dependent on both the AR and locus-wide chromatin modifications conducive to gene expression resulting in polymerase initiation at the transcription start site. DHT treatment further increases polymerase processivity, leading to remarkably high levels of gene expression. Thus, to a large extent the “soil” depends on the “seed” but not vice versa. A positive feedback loop may exist between the AR and its target locus with increased AR activity maintaining the altered chromatin state. We tentatively propose that this link may be related to AR and subsequent polymerase engagements of the target locus leading to a “memory” of transcriptional activity that in turn sustains altered patterns of histone modifications. Removal of AR, as accomplished through experimental reduction of AR protein levels, breaks this link and allows the gene locus to return to its prestimulated state. This mechanistic relationship implies that treatment strategies for androgen-independent PCa that currently focus on targeting the AR will be more effective than perhaps previously realized, since targeting the AR will also desensitize AR-regulated gene loci and consequently the entire cancer phenotype. Such treatment strategies are presently being contemplated (37), and our results here support the utility of these approaches to levels not previously appreciated.
Supplementary Material
Acknowledgments
We thank Christopher W. Gregory (University of North Carolina) for supplying us with CWR22 tumors and advice on how to maintain them and Debbie Johnson (USC) for helpful discussions.
This work was supported by grants from the NIH/NCI (R01 CA109147) (to G.A.C.), the United States Department of Defense (W81XWH-04-1-0049 and W81XWH-04-1-0823) (to G.A.C.), and the Prostate Cancer Foundation (to G.A.C.). L.J. was supported by NIH training grant T32CA009320.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Balk, S. 2002. Androgen receptor as a target in androgen-independent prostate cancer. Urology 60:132-138. [DOI] [PubMed] [Google Scholar]
- 2.Bernstein, B. E., M. Kamal, K. Lindblad-Toh, S. Bekiranov, D. K. Bailey, D. J. Huebert, S. McMahon, E. K. Karlsson, E. J. Kulbokas III, T. R. Gingeras, S. L. Schreiber, and E. S. Lander. 2005. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120:169-181. [DOI] [PubMed] [Google Scholar]
- 3.Borgono, C. A., and E. P. Diamandis. 2004. The emerging roles of human tissue kallikreins in cancer. Nat. Rev. Cancer 4:876-890. [DOI] [PubMed] [Google Scholar]
- 4.Buchanan, G., R. A. Irvine, G. A. Coetzee, and W. D. Tilley. 2001. Contribution of the androgen receptor to prostate cancer predisposition and progression. Cancer Metastasis Rev. 20:207-223. [DOI] [PubMed] [Google Scholar]
- 5.Chen, C. D., D. S. Welsbie, C. Tran, S. H. Baek, R. Chen, R. Vessella, M. G. Rosenfeld, and C. L. Sawyers. 2004. Molecular determinants of resistance to antiandrogen therapy. Nat. Med. 10:33-39. [DOI] [PubMed] [Google Scholar]
- 6.Cosgrove, M. S., and C. Wolberger. 2005. How does the histone code work? Biochem. Cell Biol. 83:468-476. [DOI] [PubMed] [Google Scholar]
- 7.Debes, J. D., and D. J. Tindall. 2004. Mechanisms of androgen-refractory prostate cancer. N. Engl. J. Med. 351:1488-1490. [DOI] [PubMed] [Google Scholar]
- 8.Dehm, S. M., and D. J. Tindall. 2005. Regulation of androgen receptor signaling in prostate cancer. Expert Rev. Anticancer Ther. 5:63-74. [DOI] [PubMed] [Google Scholar]
- 9.Denmeade, S. R., L. J. Sokoll, S. Dalrymple, D. M. Rosen, A. M. Gady, D. Bruzek, R. M. Ricklis, and J. T. Isaacs. 2003. Dissociation between androgen responsiveness for malignant growth vs. expression of prostate specific differentiation markers PSA, hK2, and PSMA in human prostate cancer models. Prostate 54:249-257. [DOI] [PubMed] [Google Scholar]
- 10.Dennis, A. P., and B. W. O'Malley. 2005. Rush hour at the promoter: how the ubiquitin-proteasome pathway polices the traffic flow of nuclear receptor-dependent transcription. J. Steroid Biochem. Mol. Biol. 93:139-151. [DOI] [PubMed] [Google Scholar]
- 11.Deroo, B. J., C. Rentsch, S. Sampath, J. Young, D. B. DeFranco, and T. K. Archer. 2002. Proteasomal inhibition enhances glucocorticoid receptor transactivation and alters its subnuclear trafficking. Mol. Cell. Biol. 22:4113-4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feldman, B. J., and D. Feldman. 2001. The development of androgen-independent prostate cancer. Nat. Rev. Cancer 1:34-45. [DOI] [PubMed] [Google Scholar]
- 13.Furutani, T., K. Takeyama, H. Koutoku, S. Ito, N. Taniguchi, E. Suzuki, M. Kudoh, M. Shibasaki, H. Shikama, and S. Kato. 2005. A role of androgen receptor protein in cell growth of an androgen-independent prostate cancer cell line. Biosci. Biotechnol. Biochem. 69:2236-2239. [DOI] [PubMed] [Google Scholar]
- 14.Gillette, T. G., F. Gonzalez, A. Delahodde, S. A. Johnston, and T. Kodadek. 2004. Physical and functional association of RNA polymerase II and the proteasome. Proc. Natl. Acad. Sci. USA 101:5904-5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goebelsmann, U., J. J. Arce, I. H. Thorneycroft, and D. R. Mishell, Jr. 1974. Serum testosterone concentrations in women throughout the menstrual cycle and following HCG administration. Am. J. Obstet. Gynecol. 119:445-452. [DOI] [PubMed] [Google Scholar]
- 16.Han, G., G. Buchanan, M. Ittmann, J. M. Harris, X. Yu, F. J. Demayo, W. Tilley, and N. M. Greenberg. 2005. Mutation of the androgen receptor causes oncogenic transformation of the prostate. Proc. Natl. Acad. Sci. USA 102:1151-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jackson, D. A. 2005. The amazing complexity of transcription factories. Brief. Funct. Genomics Proteomics 4:143-157. [DOI] [PubMed] [Google Scholar]
- 18.Jia, L., C. S. Choong, C. Ricciardelli, J. Kim, W. D. Tilley, and G. A. Coetzee. 2004. Androgen receptor signaling: mechanism of interleukin-6 inhibition. Cancer Res. 64:2619-2626. [DOI] [PubMed] [Google Scholar]
- 19.Jia, L., and G. A. Coetzee. 2005. Androgen receptor-dependent PSA expression in androgen-independent prostate cancer cells does not involve androgen receptor occupancy of the PSA locus. Cancer Res. 65:8003-8008. [DOI] [PubMed] [Google Scholar]
- 20.Jia, L., J. Kim, H. Shen, P. E. Clark, W. D. Tilley, and G. A. Coetzee. 2003. Androgen receptor activity at the prostate specific antigen locus: steroidal and non-steroidal mechanisms. Mol. Cancer Res. 1:385-392. [PubMed] [Google Scholar]
- 21.Johnstone, R. W. 2002. Histone-deacetylase inhibitors: novel drugs for the treatment of cancer. Nat. Rev. Drug Discov. 1:287-299. [DOI] [PubMed] [Google Scholar]
- 22.Kang, Z., A. Pirskanen, O. A. Janne, and J. J. Palvimo. 2002. Involvement of proteasome in the dynamic assembly of the androgen receptor transcription complex. J. Biol. Chem. 277:48366-48371. [DOI] [PubMed] [Google Scholar]
- 23.Kim, A., and A. Dean. 2004. Developmental stage differences in chromatin subdomains of the beta-globin locus. Proc. Natl. Acad. Sci. USA 101:7028-7033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim, D., C. W. Gregory, F. S. French, G. J. Smith, and J. L. Mohler. 2002. Androgen receptor expression and cellular proliferation during transition from androgen-dependent to recurrent growth after castration in the CWR22 prostate cancer xenograft. Am. J. Pathol. 160:219-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim, J., L. Jia, M. R. Stallcup, and G. A. Coetzee. 2005. The role of protein kinase A pathway and cAMP responsive element-binding protein in androgen receptor-mediated transcription at the prostate-specific antigen locus. J. Mol. Endocrinol. 34:107-118. [DOI] [PubMed] [Google Scholar]
- 26.Kim, J., L. Jia, W. D. Tilley, and G. A. Coetzee. 2003. Dynamic methylation of histone H3 at lysine 4 in transcriptional regulation by the androgen receptor. Nucleic Acids Res. 31:6741-6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee, D. H., and A. L. Goldberg. 1998. Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol. 8:397-403. [DOI] [PubMed] [Google Scholar]
- 28.Liang, G., J. C. Lin, V. Wei, C. Yoo, J. C. Cheng, C. T. Nguyen, D. J. Weisenberger, G. Egger, D. Takai, F. A. Gonzales, and P. A. Jones. 2004. Distinct localization of histone H3 acetylation and H3-K4 methylation to the transcription start sites in the human genome. Proc. Natl. Acad. Sci. USA 101:7357-7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Louie, M. C., H. Q. Yang, A. H. Ma, W. Xu, J. X. Zou, H. J. Kung, and H. W. Chen. 2003. Androgen-induced recruitment of RNA polymerase II to a nuclear receptor-p160 coactivator complex. Proc. Natl. Acad. Sci. USA 100:2226-2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mason, P. B., and K. Struhl. 2005. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol. Cell 17:831-840. [DOI] [PubMed] [Google Scholar]
- 31.Mito, Y., J. G. Henikoff, and S. Henikoff. 2005. Genome-scale profiling of histone H3.3 replacement patterns. Nat. Genet. 37:1090-1097. [DOI] [PubMed] [Google Scholar]
- 32.Nakamura, T., T. Mori, S. Tada, W. Krajewski, T. Rozovskaia, R. Wassell, G. Dubois, A. Mazo, C. M. Croce, and E. Canaani. 2002. ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol. Cell 10:1119-1128. [DOI] [PubMed] [Google Scholar]
- 33.Reid, G., M. R. Hubner, R. Metivier, H. Brand, S. Denger, D. Manu, J. Beaudouin, J. Ellenberg, and F. Gannon. 2003. Cyclic, proteasome-mediated turnover of unliganded and liganded ERalpha on responsive promoters is an integral feature of estrogen signaling. Mol. Cell 11:695-707. [DOI] [PubMed] [Google Scholar]
- 34.Roh, T. Y., S. Cuddapah, and K. Zhao. 2005. Active chromatin domains are defined by acetylation islands revealed by genome-wide mapping. Genes Dev. 19:542-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roh, T. Y., W. C. Ngau, K. Cui, D. Landsman, and K. Zhao. 2004. High-resolution genome-wide mapping of histone modifications. Nat. Biotechnol. 22:1013-1016. [DOI] [PubMed] [Google Scholar]
- 36.Santos-Rosa, H., and C. Caldas. 2005. Chromatin modifier enzymes, the histone code and cancer. Eur. J. Cancer 41:2381-2402. [DOI] [PubMed] [Google Scholar]
- 37.Scher, H. I., G. Buchanan, W. Gerald, L. M. Butler, and W. D. Tilley. 2004. Targeting the androgen receptor: improving outcomes for castration-resistant prostate cancer. Endocr.-Relat. Cancer 11:459-476. [DOI] [PubMed] [Google Scholar]
- 38.Seligson, D. B., S. Horvath, T. Shi, H. Yu, S. Tze, M. Grunstein, and S. K. Kurdistani. 2005. Global histone modification patterns predict risk of prostate cancer recurrence. Nature 435:1262-1266. [DOI] [PubMed] [Google Scholar]
- 39.Shi, Y., F. Lan, C. Matson, P. Mulligan, J. R. Whetstine, P. A. Cole, R. A. Casero, and Y. Shi. 2004. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119:941-953. [DOI] [PubMed] [Google Scholar]
- 40.Shi, Y. J., C. Matson, F. Lan, S. Iwase, T. Baba, and Y. Shi. 2005. Regulation of LSD1 histone demethylase activity by its associated factors. Mol. Cell 19:857-864. [DOI] [PubMed] [Google Scholar]
- 41.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 42.Thalmann, G. N., P. E. Anezinis, S. M. Chang, H. E. Zhau, E. E. Kim, V. L. Hopwood, S. Pathak, A. C. von Eschenbach, and L. W. Chung. 1994. Androgen-independent cancer progression and bone metastasis in the LNCaP model of human prostate cancer. Cancer Res. 54:2577-2581. [PubMed] [Google Scholar]
- 43.Tomlins, S. A., R. Mehra, D. R. Rhodes, L. R. Smith, D. Roulston, B. E. Helgeson, X. Cao, J. T. Wei, M. A. Rubin, R. B. Shah, and A. M. Chinnaiyan. 2006. TMPRSS2:ETV4 gene fusions define a third molecular subtype of prostate cancer. Cancer Res. 66:3396-3400. [DOI] [PubMed] [Google Scholar]
- 44.Tomlins, S. A., D. R. Rhodes, S. Perner, S. M. Dhanasekaran, R. Mehra, X. W. Sun, S. Varambally, X. Cao, J. Tchinda, R. Kuefer, C. Lee, J. E. Montie, R. B. Shah, K. J. Pienta, M. A. Rubin, and A. M. Chinnaiyan. 2005. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 310:644-648. [DOI] [PubMed] [Google Scholar]
- 45.Veldscholte, J., C. A. Berrevoets, C. Ris-Stalpers, G. G. Kuiper, G. Jenster, J. Trapman, A. O. Brinkmann, and E. Mulder. 1992. The androgen receptor in LNCaP cells contains a mutation in the ligand binding domain which affects steroid binding characteristics and response to antiandrogens. J. Steroid Biochem. Mol. Biol. 41:665-669. [DOI] [PubMed] [Google Scholar]
- 46.Zegarra-Moro, O. L., L. J. Schmidt, H. Huang, and D. J. Tindall. 2002. Disruption of androgen receptor function inhibits proliferation of androgen-refractory prostate cancer cells. Cancer Res. 62:1008-1013. [PubMed] [Google Scholar]
- 47.Zhang, Y., and D. Reinberg. 2001. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 15:2343-2360. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.