Abstract
We identified zebra fish forkhead transcription factor l1 (zfoxl1) as a gene strongly expressed in neural tissues such as midbrain, hindbrain, and the otic vesicle at the early embryonic stage. Loss of the function of zfoxl1 effected by morpholino antisense oligonucleotide resulted in defects in midbrain and eye development, and in that of formation of the pectoral fins. Interestingly, ectopic expression of shh in the midbrain and elevated pax2a expression in the optic stalk were observed in foxl1 MO-injected embryos. In contrast, expression of pax6a, which is negatively regulated by shh, was suppressed in the thalamus and pretectum regions, supporting the idea of augmentation of the shh signaling pathway by suppression of foxl1. Expression of zfoxl1-EnR (repressing) rather than zfoxl1-VP16 (activating) resulted in a phenotype similar to that induced by foxl1-mRNA, suggesting that foxl1 may act as a transcriptional repressor of shh in zebra fish embryos. Supporting this notion, foxl1 suppressed isolated 2.7-kb shh promoter activity in PC12 cells, and the minimal region of foxl1 required for its transcriptional repressor activity showed strong homology with the groucho binding motif, which is found in genes encoding various homeodomain proteins. In view of all of our data taken together, we propose zfoxl1 to be a novel regulator of neural development that acts by suppressing shh expression.
Forkhead genes are a family of transcription factors that play important roles during embryonic development (7, 24). The members of this family share a forkhead domain (FHD), which is a highly conserved DNA-binding domain consisting of about 110 amino acids. The Forkhead was named after two spiked-head structures in embryos of the Drosophila fork head mutant, which is defective in formation of the anterior and posterior gut (47). The term “winged helix” is also used to describe the structure of FHD, which consists of a helix-turn-helix core of three α-helices, flanked by two loops or wings. The three-dimensional structure of an FHD bound to a DNA target, determined by X-ray crystallography, was reported earlier (9). As a large number of transcription factors from yeast to humans have been identified as members of this family, the nomenclature of chordate forkhead transcription factors was revised in 2000, and a new family name, Fox, after Forkhead box, was established (18). According to the amino acid similarity of their conserved forkhead domains, these genes are divided into 17 subclasses, or clades (A to Q).
In contrast to most helix-turn-helix proteins, forkhead proteins bind DNA as monomers. Target sequence specificity has been determined for several forkhead genes by selection of binding sites from pools of short, random-sequence oligonucleotides (7). Forkhead proteins have been shown to act mostly as transcriptional activators, but transrepression by forkhead proteins has been reported in some instances (7). In contrast to the high degree of sequence homology within the DNA-binding domain, there is an almost total lack of similarity between activation or repression domains in different forkhead proteins (7). Only short activation motifs have been recognized in certain subfamilies of forkhead proteins. However, reported transactivation and transrepression domains of FOX family members lack typical motifs or features found for other transcription factors, and thus the detailed mechanism as to how FOX proteins act as transcription factors is largely unknown. Recent studies on knockout mice for Fox genes have revealed their vital roles in the development of a diverse range of organs. In addition to the null mutant mice of Fox genes, various mutations in forkhead genes, which are associated with human developmental disorders, including immune, skeletal, circulatory, and craniofacial defects, provide important information for understanding the biological functions of FHD genes (7). There has been a large number of reports indicating roles of FOX genes in the central nervous system (CNS). Several mutations of FOX genes have been detected in patients with ocular diseases (24).
By degenerate PCR using primers for FHD, we isolated the zebra fish homologue of mammalian FoxL1. By using morpholino antisense oligonucleotide (MO)-based knockdown of foxl1 and RNA overexpression experiments, we found important roles of foxl1 in neural development that had not reported before as biological activities of mouse Foxl1. Furthermore, negative regulation of the shh gene by foxl1 was shown, suggesting that the transcriptional repressor activity may be exerted through interaction with groucho. Taken together, our data allow us to propose new mechanisms for regulation of CNS formation by foxl1 in zebra fish, i.e., the negative regulation of shh.
MATERIALS AND METHODS
Maintenance of fish.
Wild-type zebra fish (Danio rerio) were purchased from a local pet shop, bred, and raised for several months in our laboratory. The fish were fed two times daily and maintained under a 14-h day-10-h night cycle, as described previously (49). In some cases, embryos were maintained in 0.2 mM 1-phenyl-2-thiourea to inhibit pigmentation (49).
Cloning, DNA construction, and reverse transcription-PCR (RT-PCR) assay.
Foxl1 was isolated from head cDNA by degenerate PCR using primers designed based on the homologous region of Xenopus and mouse FHD sequences. Messenger RNAs were prepared from the head part of 100 zebra fish embryos at 24 h postfertilization (hpf) and reverse transcribed. The cDNAs were then subjected to PCR amplification using primers designed based on the conserved forkhead region. A total of 50 clones were sequenced and, among them, one of the novel genes was designated foxl1. Full-length foxl1 (AB191484) was isolated by 5′ and 3′ RACE (rapid amplification of cDNA ends) by using a RACE system (Invitrogen). The expressed sequence tag (EST) database was also used to isolate a part of the 5′ side region of foxl1. For mRNA synthesis, full-length foxl1 (1.8 kb) was subcloned into the pCS2+ vector.
A 5′ untranslated region deletion mutant foxl (Δ5′-foxl1) lacking the foxl1 first morpholino target sequence at its 5′ untranslated region was constructed by PCR mutagenesis. Construction of foxl1-VP16, which consisted of the VP16 transcription activation domain (amino acids 414 to 490) fused to the C terminus of FHD (amino acids 43 to 157) of foxl1, and that of foxl1-EnR, created by fusing the engrailed transcription repressor domain (amino acids 1 to 298) to the C terminus of the FHD of foxl1, was done by PCR mutagenesis. pCS2-VP16 and pCS2-EnR were kindly provided Robert Davis and Minoru Watanabe. Δfoxl1, which had the internal FHD (amino acids 46 to 156) deletion, was constructed by deletion of the region between NaeI and DrdI sites.
To confirm the effect of MO, we constructed an enhanced green fluorescent protein (EGFP) plasmid by substituting its 5′ ATG region with a foxl1-derived ATG region that contained the target sequence of the first and second foxl1-MO. The cytomegalovirus (CMV) promoter was localized upstream of the foxl1-derived region.
The Gal4 DNA-binding domain (dbd) fused to the various deletion mutants of foxl1was constructed by PCR mutagenesis.
Semiquantitative RT-PCR to examine the expression pattern of zebra fish foxl1 was done by using total RNA extracted from various zebra fish tissues (23). The primer sequences (upstream and downstream) and length of the amplified fragments were 5′-TTTCGGACTCTGCAGCGGACA-3′ and 5′-AGAGAACTCGGGTGACGAGCTT-3′ (566 bp). The conditions for PCR were 30 cycles of 94°C for 1 min, 60°C for 1 min, and 72°C for 1 min.
Microinjection of morpholino (MO) or RNA into zebra fish embryos.
MO antisense oligonucleotides (GENE-TOOLS, LLC) were designed to recognize 25 bases upstream of the AUG of foxl1. The sequence of the first foxl1-MO was 5′-CATGACTTCGGAGGACAGATTCAGT-3′ (the start codon is underlined). A second MO was designed to bind immediately upstream of the first MO, and its sequence was 5′-CTGGGGAGATCGCCACCGG-3′. The standard control MO available from GENE-TOOLS, which had no effect on embryonic development under our experimental conditions, was used as an injection control. Capped sense RNAs of foxl1 were synthesized by using a Message mMachine in vitro transcription kit (Ambion, Austin, Texas). The amounts of injected RNAs were 50 to 250 pg/embryo.
Whole-mount and section in situ hybridization, light microscopy, Alcian blue staining, and TUNEL assay.
Probes for dlx2, hlx1, otx2, krox20, engrailed2, pax2a, pax6a, shh, ptc1, foxb1.2, and arx genes were cloned by PCR amplification (31), and PCR products were subcloned into the pGEM-T-Easy vector. Zebra fish phox2a in pBKCMV was kindly provided by S. Guo (14). Whole-mount and section in situ hybridization and light microscopy analyses were done as previously described (22). Alcian blue staining was done as previously reported (28). TUNEL (terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling) was done to detect apoptotic cells. Embryos were fixed and stored in methanol. After rehydration, an in situ TUNEL assay was performed by using an in situ apoptosis detection kit (TaKaRa) according to the manufacturer's instructions.
Cell culture, transfection, and reporter assay.
Rat pheochromocytoma (PC12) cells (44) or NIH 3T3 cells (105 cells/well) were plated in wells of 24 plates 1 day before transfection and then transfected with various combinations of plasmid DNAs by using Fugene6 transfection reagent (Roche). The luciferase assay was done as described earlier (37).
RESULTS
Isolation of a novel forkhead transcription factor highly expressed in the nervous system of zebra fish embryos.
To isolate forkhead transcription factors that may play a role in neurogenesis in zebra fish, we performed degenerate PCR by using primers designed to bind to the conserved region of the FHD of mouse Foxe3 and Xenopus FoxE4 forkhead genes, both of which are involved in eye formation (6, 20). We isolated a total of 50 clones as described in Materials and Methods; among these, 9 clones appeared to have conserved motifs of FHD. Only one of the nine clone showed an exact match with a known fox gene, axial (foxa2), whereas the other eight clones showed incomplete similarity with previously deposited sequences. We cloned the full length of one of those genes, clone1, by using 3′ and 5′ RACE and EST sequences in a database. A region in the N-terminal half of clone1 (amino acids 50 to 149) contained conserved motifs of the FHD and showed more than 85% homology at the amino acid level with that region of mouse Foxl1 (fkh6) and human FOXL1 (FREAC7; Fig. 1A). However, we could not find any significant homology in regions other than FHD of clone1 with any other genes or motifs by database searching. We designated clone1 foxl1 according to the suggestion of the Fox Nomenclature Committee (www.biology.pomona.edu/fox.html). Recently, a zebra fish EST clone that was isolated by the NIH-MGC project was deposited as “similar to forkhead box L1” (zgc:63595). The sequences of that clone and ours were nearly identical except for the presence of a three-amino-acid gap in the gene at around amino acid position 235 of our clone. We suspect that the difference was caused by a polymorphism, since different sources of zebra fish were used.
FIG. 1.
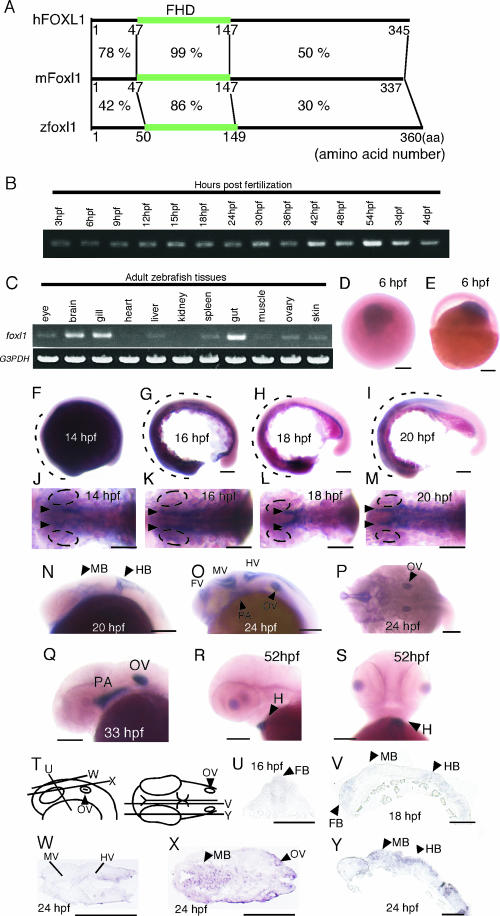
Expression of foxl1 during zebra fish embryonic development. (A) Schematic showing the degree of sequence similarity at the amino acid level of the whole region among zebra fish foxl1, mouse Foxl1, and human FOXL1. Numbers indicate level of amino acids sequence similarity. (B and C) The expression of foxl1 in various stages of zebra fish embryonic developments (B) and in tissues from adult zebra fish (C) was examined by semiquantitative RT-PCR. G3PDH was used as a control. (D to Y) The expression of foxl1 mRNA in zebra fish embryos at various developmental stages. (D to S) Whole-mount in situ hybridization patterns of foxl1 expression during zebra fish embryo development. View from the animal pole (D) and lateral view with the animal pole at top (E) of embryos, lateral views with anterior to left of embryos (F to I, N, O, Q, and R), dorsal views with anterior to left of embryos (J to M and P), and a ventral view with anterior at the top of an embryo (S) are shown. (T to Y) Section in situ hybridization of foxl1 expression during embryonic development. Schematic representation of the lateral view (left) and dorsal view (right) of the embryos indicating the cutting position for sections from panels U to Y is given (T). Cross-section with the anterior at the top (U), sagittal sections with anterior to the left (V and Y), and horizontal sections with anterior to the left (W and X) are shown. Broken lines in panels F to I indicate area corresponding to foxl1 expression domains. Abbreviations: FB, forebrain; MB, midbrain; HB, hindbrain; FV, forebrain ventricle; MV, midbrain ventricle; HV, hindbrain ventricle; OV, otic vesicle; PA, pharyngeal arch; H, heart. Ovals in panels J to M indicate eye field. Scale bars, 100 μm.
Radiation hybrid mapping using the LN54 panel (17), which was kindly provided by the Ottawa Health Research Institute, assigned zebra fish foxl1 to linkage group 18; and the site1 protease gene (38), which is located around 2 Mb downstream of human FOXL1 and mouse Foxl1, was found approximately 3 Mb upstream of zfoxl1 by comparative analysis. Mouse Foxl1 maps to chromosome 8, and human FOXL1 maps to 16q24.
Results of RT-PCR for foxl1 using cDNA from embryos of various stages from 3 hpf to 4 days postfertilization (dpf) showed that foxl1 was expressed throughout the various developmental stages (Fig. 1B). We next performed RT-PCR using total RNAs isolated from various tissues of the adult zebra fish (Fig. 1C). The foxl1 transcripts were strongly expressed in the brain, gill, and gut and weakly in the eye, liver, spleen, ovary, and skin in adult zebra fish.
Whole-mount and section in situ hybridization using digoxigenin-labeled RNA as probes showed that the expression of foxl1 was first observed when gastrulation started in the dorsal part of the animal pole region (Fig. 1D and E). During somite stages, the expression of foxl1 was detected in the lateral mesoderm as two stripes (Fig. 1J to M, arrowheads) and in a part of the forebrain, midbrain, and hindbrain (Fig. 1F to I, broken lines, and Fig. 1N, U, V, X and Y, arrowheads). At 24 hpf, foxl1 expression was observed to be strong in the forebrain ventricle, midbrain ventricle, and hindbrain ventricle (Fig. 1O, P, and W). Strong expression of foxl1 was also detected in the otic vesicles (Fig. 1O, P, and X, arrowheads), and weak expression was detected in the pharyngeal arch primordia (Fig. 1O, arrowhead). In addition to the brain, foxl1 was also expressed in the neural retina (Fig. 1Y). At 33 hpf, the expression in the brain was diminished; in contrast, the expression in the pharyngeal arch primordia and otic vesicles became stronger (Fig. 1Q). At 52 hpf, strong expression of foxl1 was observed in the heart, but no signals were observed in other tissues (Fig. 1R and S, arrowheads).
Foxl1 antisense morpholinos perturbed development of the CNS.
To examine the functions of zfoxl1 during zebra fish embryonic development, we used morpholino antisense oligonucleotides (MO) to knock down the function of foxl1. A MO was designed to bind to the translation initiation site of foxl1 (foxl1 first MO), and a second MO was designed to bind to a 5′ upstream region of the target site of the first MO. We examined the effects of foxl1-MOs in terms of their ability to suppress the expression of EGFP from a fusion gene consisting of the MO target region including the first ATG of foxl1 followed by an EGFP-coding region (pCS2-foxl1/EGFP). Since the plasmid contained the target region of the foxl1 MOs, the expression of EGFP should be abolished by the coinjection with the MO. pCS2-foxl1/EGFP was injected into fertilized eggs with or without foxl1 MO, and the expression of EGFP driven by the CMV promoter was examined by fluorescence microscopy at 24 hpf. In nearly 100% of the eggs, the expression of EGFP was abolished by either the first or the second foxl1 MO (data not shown).
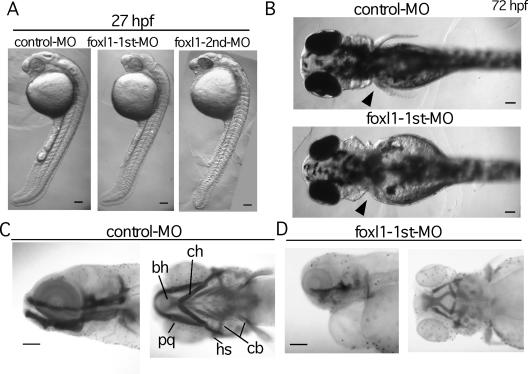
Then the MOs were injected into zebra fish embryos at the one- or two-cell stage, and standard control MO available from the manufacturer was used as a negative control. Until 16 hpf, no apparent defect in the morphology of zebra fish embryos injected with foxl1 MO was observed (data not shown). At 27 hpf, although development of the whole body showed no significant defects and the body size of foxl1 MO-injected embryos was nearly the same as that of the control embryos, the eye and head regions revealed severe degeneration (Fig. 2A). Neural defects were the most striking phenotype of the foxl1 MO-injected embryos; however, in addition to them, more than 40% of the embryos had small or no pectoral fins at 72 hpf (Fig. 2B and Table 1). Since foxl1 was expressed in the primordia of pharyngeal arches, we next examined the craniofacial cartilage structure of foxl1 MO-injected embryos by Alcian blue staining. In the control MO-injected embryos at 5 dpf, individual cartilaginous elements of the pharyngeal skeleton were observed in both lateral and ventral views (Fig. 2C). In contrast, in the foxl1 first MO-injected embryos, most elements of the pharyngeal skeleton were absent or had been displaced (Fig. 2D).
FIG. 2.
Abrogation of foxl1 function by MO antisense oligonucleotide. (A to D) Control or foxl1 MO was injected into fertilized eggs at the one- or two-cell stage, and embryos were examined at various stages. Lateral views (anterior at the top) of embryos at 27 hpf (A) and dorsal views (anterior to the left) of embryos at 72 hpf (B) injected with control MO or foxl1 MO. The arrowheads in panel B indicate the position of pectoral fin. (C and D) Craniofacial development in foxl1 MO-injected embryos. Alcian blue-stained control (C) and foxl1 (D) MO-injected embryos at 5 dpf are shown. Lateral views with anterior to the left (left panels) and ventral views with anterior to the left (right panels) are seen. Abbreviations: bh, basihyal; cb, ceratobranchial; ch, ceratohyal; hs, hyosymplectic; pq, palatoquadrate. Scale bars, 100 μm.
TABLE 1.
Abnormal phenotypes caused by foxl1 MO injection
| Material injected (amt [ng]) | No. of embryos examined | No. (%) of embryos with normal or abnormal phenotypes at 27 hpf
|
||||
|---|---|---|---|---|---|---|
| Normal | Abnormal
|
Lethal | Severe | |||
| Brain | Small fina | |||||
| Control MO (10) | 96 | 82 (85) | 0 (0) | 0 (0) | 14 (15) | 0 (0) |
| foxl1 first MO (10) | 256 | 91 (36) | 138 (54) | 37/81a (46) | 11 (4) | 16 (6) |
| foxl1 second MO (10) | 273 | 59 (22) | 164 (60) | NEb | 38 (14) | 12 (4) |
Fin abnormality was examined in 96 embryos with control MO and in 81 embryos with foxl1 first MO.
NE, not examined.
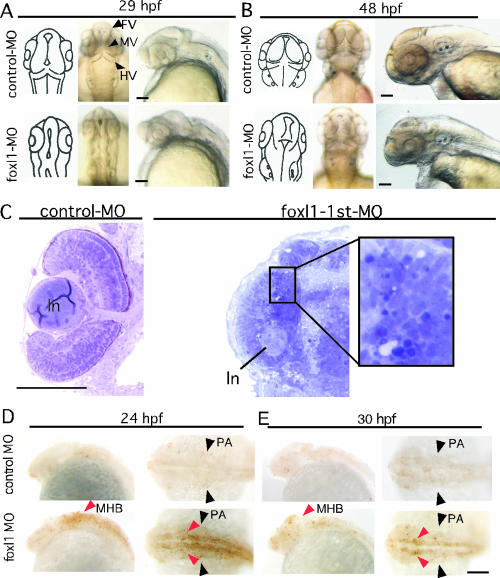
Therefore, we examined the morphological development of the brain and eye of the foxl1 MO-injected embryos in more detail. By 29 hpf, in the control MO-injected embryos, the brain had morphologically developed, and three ventricles—the forebrain, midbrain, and hindbrain ventricles—were evident (Fig. 3A, upper panels, arrowheads). At that time, the lens and retina of the eye could be clearly observed. In contrast, embryos injected with foxl1 MO showed small eyes and brain, and development of the MV and HV had hardly occurred (Fig. 3A, lower panels). At 48 hpf, the sizes of both the eyes and head were still smaller than in the control (Fig. 3B, lower panels), and the structure of the tectum and cerebellum had degenerated (Fig. 3B, dorsal view). Embryos with a mild phenotype survived at least 5 days, but their brain size was smaller than that of the control embryos (data not shown). Plastic sections made through the tectal region (54-hpf embryos) showed clear three-layer formation in the retina of the control embryos (Fig. 3C, left panel). In contrast, no layer formation of the retina, and some dark-stained cells, which were surmised to be apoptotic cells, were observed in the foxl1 MO-injected embryos (Fig. 3C, right panel). We further examined whether the apoptosis occurred by using the TUNEL in situ staining method (Fig. 3D and E). We found that foxl1 MO-injected embryos exhibited an increased number of apoptotic cells in their brain, especially in the midbrain and hindbrain (Fig. 3D and E, lower panels, red arrowheads). However, no signals were observed in the pectoral fin buds (Fig. 3D and E, black arrowheads), indicating that different mechanisms were operative in brain and fin bud defects caused by downregulation of foxl1 by MO.
FIG. 3.
Abnormal development of brain in foxl1 MO-injected embryos. (A and B) Detailed structure of head portion of zebra fish embryos injected with control or foxl1 MO. Left panels show schematic representation of dorsal views of the brain structure. Middle panels are dorsal views with anterior at the top, and right panels are lateral views with anterior to the left. (C) Absence of layer formation of neural retina in foxl1 MO-injected embryos. Plastic sections of the eye region of 54-hpf zebra fish embryos injected with control (left panel) or foxl1 MO (right panel) are shown. ln, lens. (D and E) Apoptotic cells in foxl1 MO-injected embryos. Embryos injected with control (upper panels) or foxl1 (lower panels) MOs were harvested at the indicated stages. Whole-mount TUNEL in situ staining was done. Lateral views with anterior to the left and dorsal views with anterior to the left are seen. Red arrowheads indicate an increased number of apoptotic cells in MHB. Black arrowheads indicate pharyngeal arch (PA). Scale bars, 100 μm.
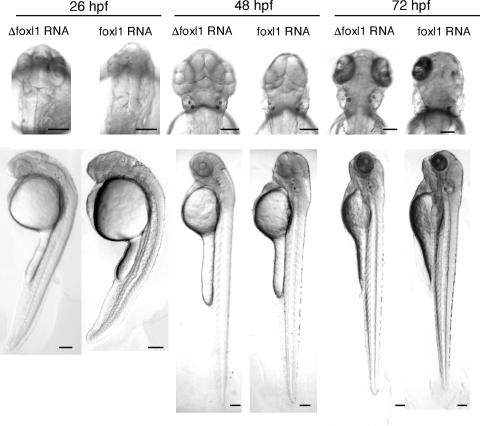
To confirm the specificity of these effects, we used a second MO synthesized against a different region of the foxl1 mRNA. The second MO had similar effects on zebra fish embryo development, i.e., small eyes and degenerated brain (Table 1), and had a similar dose requirement (data not shown). We next examined whether the phenotype induced by the foxl1 MO injection could be reversed by coexpression of foxl1 or not. We constructed a 5′ untranslated region deletion mutant foxl1 (Δ5′-foxl1) which lacks target sequence of the foxl1 first MO in the 5′ untranslated sequence and coinjected the Δ5′-foxl1 mRNA along with the foxl1 first MO. The expression of the Δ5′-foxl1 did not reverse the neural phenotype (data not shown), allowing us to surmise that the ectopic expression of foxl1 itself causes severe neural defects (Fig. 6).
FIG. 6.
Overexpression of mRNA encoding foxl1. Poly(A) mRNA of foxl1 or Δfoxl1, which lacks the foxl1 DNA-binding domain, was injected into fertilized eggs at the one- or two-cell stage, and embryos were harvested at the indicated time points. Dorsal views of the head region with anterior at the top are shown in the upper panels, and lateral views of whole embryos with anterior at the top are shown in the lower panels. Scale bars, 100 μm.
Expression of shh and its related genes were perturbed in foxl1 MO-injected zebra fish embryos.
In order to define these phenotypes in detail, we injected embryos with foxl1-MO and examined them for alterations in the expression of various marker genes of neural tissues.
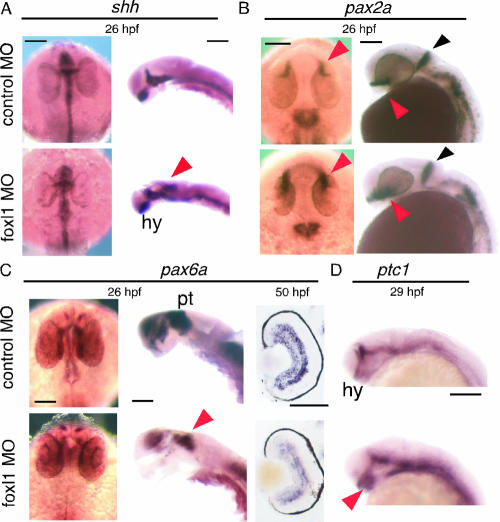
We first examined the shh expression in the foxl1 MO-injected embryos. shh plays a crucial role in the specification of the floor plate and ventral brain identity (11). Interestingly, ectopic shh expression in the posterior part of the midbrain was observed at 24 to 26 hpf (Fig. 4A, arrowhead), and this region corresponded to that showing foxl1 expression around 24 hpf (Fig. 1O and P), suggesting the possibility that foxl1 functions as a negative regulator of shh expression in this region. Since we could not find ectopic expression of shh in other regions of embryos at various other time points (data not shown), the ectopic expression of shh in the midbrain may be a region-specific phenomenon. However, the expression of shh in hypothalamus was expanded (Fig. 4A, hy). The pax2a gene encodes a paired-box transcription factor that is one of the earliest genes to be specifically activated in development of the midbrain, and it is required for the development and organizer activity of this territory (36). pax2a expression in the optic stalk was elevated in foxl1 MO-injected embryos compared to that of control embryos at 26 hpf (Fig. 4B, red arrowheads). In contrast, the expression of pax2a, which is expressed in the anterior edge of the midbrain-hindbrain boundary (MHB; Fig. 4B, black arrowheads), was similar between foxl1 MO- and control MO-injected embryos, but the shape of the expression domain was slightly different. pax6a is widely expressed in dorsal parts of the fore- and midbrain (16) and known to be negatively regulated by shh (26). pax6a was strongly expressed in the thalamus and pretectum of the control, but the expression in the thalamus of the foxl1-MO-injected embryo was strongly reduced (Fig. 4C, left and middle panels, red arrowhead). Section in situ hybridization of pax6a showed that pax6a was expressed in the retina derived from foxl1 MO-injected embryo with a similar pattern to that of the control MO-injected embryos; however, the intensity of the signal was far weaker than that of the control (Fig. 4C, right panels). The activity of Hh proteins is thought to be mediated by their interaction with a large multipass transmembrane protein encoded by the patched (ptc) gene, and overexpression of shh RNA induced ectopic ptc expression (25). We then tested the profile of expression of ptc1. In foxl1 MO-injected embryos, the ptc1 expression was elevated in the hypothalamus (Fig. 4D, red arrowhead).
FIG. 4.
Expression pattern of shh and its related genes in foxl1 MO-injected embryos. Expression of shh, pax2a, pax6a, and ptc1 was examined by whole-mount in situ analysis. Embryos injected with control MO or foxl1 MO were harvested at the indicated stages, and whole-mount in situ hybridization was done. Expression patterns of shh, pax2a, pax6a (dorsal view with anterior at the top and lateral view with dorsal to the right), and ptc1 (lateral view with anterior to the left) are shown. Cross sections of retina are shown in the right panels of pax6a. Embryos with eyes removed are shown in the right panels of shh and the middle panels of pax6a. The results depicted were obtained with antisense probes. Sense probe control experiments were done for all of the probes, and no significant signals were detected (data not shown). The colored arrowheads are as defined in the text. Abbreviations: hy, hypothalamus; pt, pretectum. Scale bars, 100 μm.
Various neural marker genes were abnormal in their expression in foxl1 MO-injected embryos.
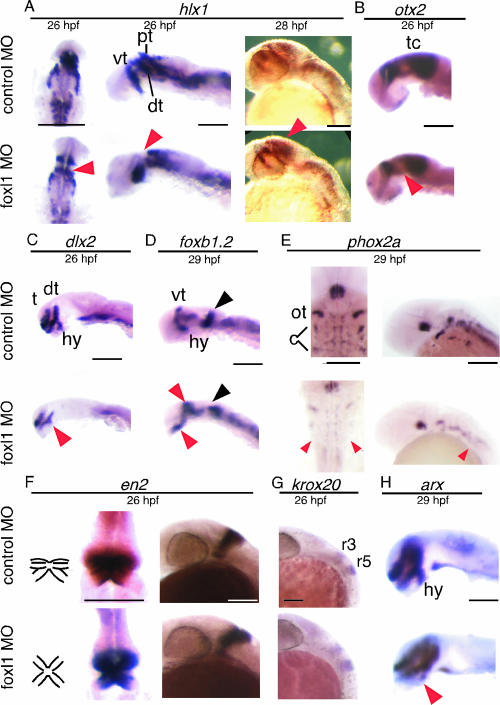
hlx1 is expressed in the presumptive rostral brain in the late gastrula, and then its expression changes dynamically (12). At 26 hpf, we clearly observed three distinct regions of hlx1 expression in the brain of control MO-injected embryos (Fig. 5A, upper left and middle panels). On the other hand, in the foxl1 MO-injected embryos, hlx1 expression in the ventral thalamus was observed, but expression in dorsal thalamus and the pretectum seemed to be fused, or either one was absent (Fig. 5A, lower left and middle panels, arrowheads). Both control and experimental embryos at 28 hpf showed a similar expression pattern in the thalamus (Fig. 5A, right panels), but the expression in the pretectal region was rather enhanced in the foxl1 MO-injected embryos (Fig. 5A, lower right panel, red arrowhead). otx2 is known for its specific function as a transcriptional regulator during forebrain and midbrain development in vertebrates (5). At 26 hpf, otx2 expression was observed in the dorsal thalamus and tectum regions of the control embryos (Fig. 5B, upper panel). In the foxl1 MO-injected embryos, its expression was similar to that in the tectum region of the control MO-injected embryos but was slightly weaker in the dorsal thalamus (Fig. 5B, lower panel, red arrowhead). dlx2 is a member of the distal-less family of genes that may specify positional information in the head (2). dlx2 was clearly expressed as two longitudinal stripes and one positioned perpendicular to them in control MO-injected embryos at 26 hpf (Fig. 5C, upper panel). In the foxl1 MO-injected embryos, the expression of dlx2 was weak in all of the regions and missing from the hypothalamus (hy, Fig. 5C, lower panel, red arrowhead). We next examined markers expressed in the tegmentum. The expression of foxb1.2 (33) was detected in the diencephalon (hypothalamus and ventral thalamus) and at the MHB at 29 hpf. The foxb1.2 expression in the hypothalamus and ventral thalamus (tegmentum) was elevated in foxl1 MO-injected embryos (Fig. 5D, red arrowheads). In addition, the expression pattern of foxb1.2 in MHB was severely perturbed (Fig. 5D, black arrowhead). phox2a is expressed in neural progenitors that give rise to the locus coeruleus and cranial motor and sensory neurons (14). Although the foxl1 MO-injected embryos showed normal phox2a expression in ocular and trochlear motor progenitors or neurons, the expression in cranial sensory progenitors or neurons of the tegmentum was absent (Fig. 5E, red arrowheads). krox20 marks rhombomere 3 and 5 of the hindbrain (34). The expression of krox20 and engrailed homeodomain transcription factor 2 (en2) was indistinguishable between foxl1 MO- and control MO-injected embryos in lateral views at 26 hpf (Fig. 5F and G); however, dorsal views showed abnormal formation of MHB region (Fig. 5F, left diagrams, middle panels). arx (for aristaless related homeobox gene) was reported to regulate forebrain development in Xenopus system (39). arx was expressed in the telencephalon, diencephalon (ventral thalamus and hypothalamus), and floor plate in zebra fish (29). The expression of arx of the floor plate was indistinguishable between foxl1 MO- and control MO-injected embryos. However, a reduction of arx expression was observed in the hypothalamus (Fig. 5H, red arrowhead). The incidence in abnormal gene expression in foxl1 MO-injected embryos described above is summarized in Table 2.
FIG. 5.
Expression pattern of various neuronal markers in foxl1 MO-injected embryos. Expression of various neuronal markers was examined by whole-mount in situ analysis. Embryos injected with control MO or foxl1 MO were harvested at the indicated stages, and whole-mount in situ hybridization was done. The expression patterns of hlx1, phox2a, and en2 (dorsal view with anterior at the top and a lateral view with dorsal to the right) and of otx2, dlx2, foxb1.2, krox20, and arx (lateral view with anterior to the left) are shown. Embryos with eyes removed are shown in the left and middle panels of hlx1, in the middle panels of en2, and in the panels for the otx2, dlx2, foxb1.2, and Arx samples. The results depicted were obtained with antisense probes. Sense probe control experiments were done for all of the probes, and no significant signals were detected (data not shown). The colored arrowheads are as defined in the text. Abbreviations: pt, pretectum; dt, dorsal thalamus; vt, ventral thalamus; tc, tectum; t, telencephalon; hy, hypothalamus; ot, ocular and trochlear motor progenitors or neurons; c, cranial sensory progenitors or neurons; r, rhombomere. Scale bars, 100 μm.
TABLE 2.
Incidence of abnormal expression of various neural markers in foxl1 MO-injected embryosa
| Material injected | No. (%) of embryos with abnormal expression pattern of various neuronal markers
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| shh | pax2a | pax6a | ptc1 | hlx1 | dlx2 | foxb1.2 | phox2a | en2 | arx | |
| Control MO | 0/20 (0) | 0/13 (0) | 0/14 (0) | 0/10 (0) | 0/15 (0) | 0/21 (0) | 0/13 (0) | 0/20 (0) | 0/10 (0) | 0/11 (0) |
| foxl1-MO | 18/34 (53) | 24/29 (83) | 26/28 (93) | 19/31 (61) | 23/24 (83) | 21/29 (72) | 19/24 (79) | 13/19 (68) | 20/21 (95) | 20/24 (83) |
An abnormal expression pattern was evaluated for ectopic expression in midbrain (shh), elevated expression in optic stalks (pax2a), reduced expression in pretectum (pax6a), elevated expression in hypothalamus (ptc1), fused or absent expression in dorsal thalamus and pretectum (hlx1), reduced expression in hypothalamus (dlx2), elevated expression in hypothalamus and ventral thalamus and abnormal shape of MHB (foxb1.2), reduced expression in cranial sensory progenitors or neurons in tegmentum (phox2a), abnormal shape in MHB (en2), and reduced expression in hypothalamus (arx).
Overexpression of foxl1 resulted in failure to form anterior neural structures.
We next examined the effects of overexpressed foxl1 by microinjecting of synthetic capped RNA of foxl1 into one- to two-cell stage zebra fish embryos. We also synthesized RNA for foxl1 lacking FHD (Δ foxl1) as a control. We first examined the effects of RNA injection of Δ foxl1 and found that the injection had no obvious effect on zebra fish development compared to no injected control embryos (Fig. 6 and data not shown). We then compared the effect of the overexpression of foxl1 RNA on zebra fish embryo development. In a large majority of embryos (ca. 30%), the injection of foxl1 RNA resulted in the severe malformation of neural tissues, especially the anterior structures (Fig. 6). The embryos with severe neural phenotypes showed no eyes or expansion of the jaw, whereas the posterior regions of the brain developed normally (Fig. 6). In some cases, the loss of either or both anterior and posterior structures was observed (data not shown). At a lower dose of foxl1 RNA (56 pg), most abnormalities were observed at frequencies similar to those seen with the higher dose of RNA injection (243 pg), but the severe and lethal phenotypes were observed at lower frequencies (data not shown). Embryos with milder phenotypes had smaller eyes, and the brain ventricles (MV and HV) and the MHB failed to develop properly.
Overexpression of foxl1 RNA affected the expression of shh and other genes in the early gastrulation stage.
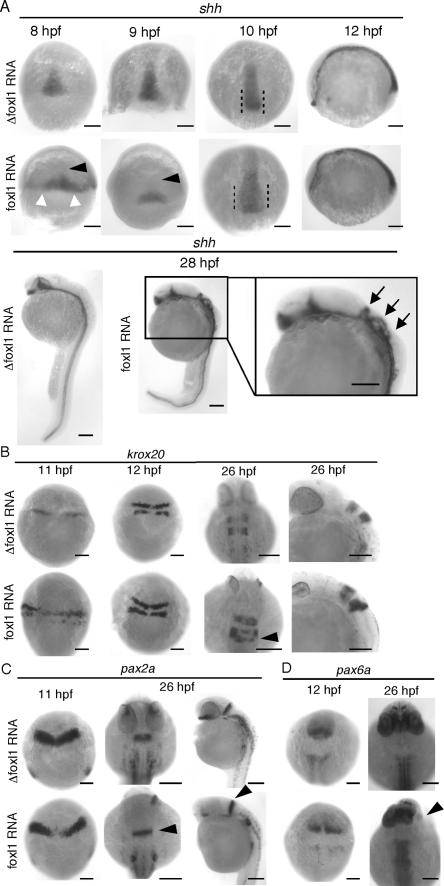
Since ectopic expression of shh was observed with the embryos injected with foxl1 MO, release of the shh gene from transcriptional repression in foxl1 MO-injected embryos was hypothesized. We then performed whole-mount in situ hybridization to examine modification of the pattern and/or strength of shh expression in foxl1 RNA-injected embryos. Interestingly, at the late gastrulation stage, the area of the expression of shh in foxl1 RNA-injected embryos expanded laterally (Fig. 7A, 8 hpf, white arrowheads) but was absent from the region on the animal pole side (Fig. 7A, 8 and 9 hpf, black arrowheads). At 10 hpf, shh expression spread into the animal pole, but the area was still broader laterally than that of Δfoxl1 RNA-injected embryos. At a later stage (28 hpf), the expression of shh in Δfoxl1 RNA-injected embryos was clearly observed only in the neural plate, whereas that in foxl1 RNA-injected embryos expanded dorsally (Fig. 7A, arrows). We then examined the expression of krox20. In 11-hpf embryos, the expression area of krox20 was stronger and expanded laterally in the foxl1 RNA-injected embryos (Fig. 7B). Like the expression of krox20, the expression of pax2a in the foxl1 RNA-injected embryos also expanded laterally at 11 hpf, and at 26 hpf, even in the embryos lacking eyes, the expression of pax2a in the MHB was clearly observed (Fig. 7C, black arrowheads). pax6a marks appropriate positions but was absent from the region that normally produces the eye in the foxl1 RNA-injected embryos at 26 hpf (Fig. 7D, lower right panel, arrowhead).
FIG. 7.
Expression of various markers in early embryonic development of foxl1-overexpressing embryos. Whole-mount in situ hybridization of foxl1 mRNA- or Δfoxl1, which lacks the DNA-binding domain, mRNA-injected embryos. (A) Expression of shh with dorsal views with anterior at the top of 8-, 9-, and 10-hpf embryos and a lateral view with anterior to the left of a 12-hpf embryo are shown in the upper and middle panels. The lower panels are lateral views with dorsal to the right of 28-hpf embryos. White arrowheads show enhanced expression of shh, and the black arrowheads (8 and 9 hpf) indicate the lack of shh expression. Arrows in magnified panel of 28-hpf embryos indicate expanded expression of shh into the dorsal side. (B) The expression pattern of krox20 at 11 and 12 hpf (dorsal views with anterior at the top) and 26 hpf (dorsal views with anterior at the top and a lateral view with dorsal to the right). The arrowhead indicates krox20 expression in rhombomere 5. (C) Expression pattern of pax2a at 11 hpf (dorsal views with anterior at the top) and 26 hpf (dorsal views with anterior at the top and a lateral view with dorsal to the right). Arrowheads indicate pax2a expression in MHB. (D) Expression of pax6a in 12- to 26-hpf embryos are shown by dorsal views with anterior at the top. The arrowheads indicate the lack of pax6a expression.
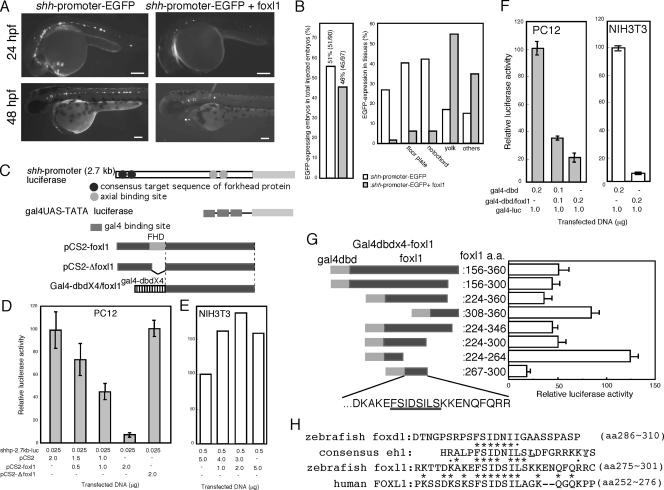
FIG. 8.
Effects of foxl1 on shh promoter activities. (A) shh-promoter-EGFP plasmid was injected into fertilized zebra fish eggs with or without plasmid encoding foxl1. The expression of EGFP at 24 or 48 hpf was examined under a fluorescence microscope. (B) Quantification of suppressive effects of foxl1 for the shh promoter. The left panel shows the total number of EGFP-positive embryos among injected embryos. The right panel shows the population of shh-promoter-dependent EGFP expression in several tissues in the presence or absence of foxl1. EGFP-expression was examined in 26- to 31-hpf embryos under a fluorescence microscope. White and gray bars indicate that shh-promoter-EGFP was injected with control or foxl1 plasmids, respectively. (C) Schematic drawing of shh-promoter-luciferase plasmids and gal4-UAS-luciferase plasmid and various mutants of foxl1. (D and E) Effects of foxl1 on shh promoter activity. Various combinations of the indicated plasmids were used for transfection of PC12 cells (D) or NIH 3T3 cells (E), and luciferase activity driven by the shh promoter was examined after a 2-day culture period (n = 3). In panel E, we repeated this experiment three times independently, and similar results were obtained. (F) Luciferase activities were analyzed by using the gal4-UAS system 2 days after transfection (n = 3). (G) Various regions of foxl1 C-terminal portion were fused to gal4-dbd. PC12 cells were cotransfected with these mutants and gal4-UAS-luciferase, and luciferase activities were determined after 2-day culture period (n = 3). (H) Comparison of the eh1-like sequence among zebra fish foxl1, human FOXL1, and zebra fish foxd1.
Foxl1 suppressed isolated shh-promoter activity in PC12 cells but not in fibroblasts.
Since in vivo examination of foxl1 activity by EnR and VP16 (see the supplemental material) did not give conclusive results to judge foxl1 transcriptional activity, we then examined the effects of foxl1 on the cis-regulatory region of the shh promoter by using transfected cell cultures. According to a previous report (30), we cloned the shh 5′ promoter region by PCR and fused it upstream of EGFP (shh-promoter-EGFP). We first confirmed the appropriate expression of the shh-promoter-EGFP in the ventral brain, notochord, and the floor plate of 24- and 48-hpf embryos (Fig. 8A, left panels), which showed a pattern similar to that of endogenous shh (data not shown). Then, foxl1 plasmids were coinjected with shh-promoter-EGFP plasmids into embryos. As shown in Fig. 8A, the expression of shh-promoter-EGFP was strongly suppressed by the coexpression of foxl1. Total number of EGFP-expressing embryos only slightly reduced by the coexpression of foxl1 (Fig. 8B, left panel). However, when we quantify the extent of reduced shh-promoter-EGFP expression in various tissues, we found that the specific repressive activity of foxl1 in the ventral brain, floor plate, and notochord (Fig. 8B, right panel). We then constructed shh promoter-luciferase plasmid (shh-promoter-luc) to analyze the effects of foxl1 on luciferase activities in transfected PC12 cells. Coexpression of foxl1 dose dependently suppressed shh-promoter induced luciferase activity in the PC12 cells (Fig. 8D). Since Δfoxl1 did not have any effect on the luciferase activity (Fig. 8D), the essential role of the forkhead DNA-binding domain for repression of shh promoter activity by foxl1 was strongly suggested.
We next examined the cell type specificity of foxl1 by conducting luciferase assays. In mouse fibroblastic NIH 3T3 cells, surprisingly, foxl1 did not show any suppressive effects on the luciferase activity induced by the shh promoter (Fig. 8E). We performed the same experiments with HeLa cells and obtained the same results (data not shown), strongly suggesting that foxl1 does not exert its activity in nonneural cells.
foxl1 exerted its transcriptional suppressor activity through its putative groucho binding domain.
Next, we sought to define the region of zfoxl1 responsible for the observed transcriptional suppressor activity by performing deletion analysis of foxl1 by using the gal4-UAS system. Since the FHD is localized near the N terminus of foxl1, we first examined whether the side of foxl1 C-terminal to its FHD contained transcriptional repressor activity. The C terminus of the dbd was fused to the N terminus of the C-terminal half of foxl1 (gal4-dbd/foxl1, Fig. 8C), and PC12 cells were transfected with it, along with a luciferase reporter gene driven by a tandem repeat of gal4 binding domain (gal4UAS-TATA-luciferase, Fig. 8C). gal4-dbd/foxl1 strongly suppressed gal4-UAS-luciferase activity (Fig. 8F) and, interestingly, this suppression was observed in not only PC12 cells but also in NIH 3T3 cells (Fig. 8F), suggesting that this C-terminal transcriptional-suppressing region acts regardless of the cell type. To further delineate the transcriptional repression domain, we made several deletion constructs of gal4-dbd/foxl1 (Fig. 8G) and then examined the transrepression activity in PC12 cells. As shown in Fig. 8G, the region between amino acids 267 to 300 appeared to be essential and sufficient for the activity. We then compared this sequence with other sequences previously reported to function as transrepressor regions of several transcriptional factors. We found that a part of the conserved transcription repressor sequence of the HOX transcription factor, called the engrailed eh1 region (40) and known as a groucho binding domain, was included in our sequence (Fig. 8G, black underlined amino acid sequence, and Fig. 8H). Previous comparative studies on various classes of homeoproteins (n = 30) showed different values of conservation of each amino acid (40), and we found that the eh1-like sequence contained in foxl1 corresponded to the most conserved part of the engrailed eh1 region. We then searched a database to find whether other FHD protein contained this sequence and found the motif in human FOXL1 (Fig. 8H) but not in mouse Foxl1 (not shown). Further database searching revealed that foxd family members (foxd1, d2, d3, d4, and d5) contained a sequence that partially matched the eh1 consensus (Fig. 8H).
DISCUSSION
Knockdown of foxl1 in zebra fish embryos resulted in a phenotype similar to that of shh-overexpressing zebra fish embryos.
In the present study, we reported on a novel forkhead transcription factor in zebra fish that plays important roles in the development of the anterior region of the brain. Foxl1 MO-injected embryos had defects in neural development and formation of pharyngeal arches, but other types of organogenesis except for the fin development seemed rather normal. The most striking structural abnormalities of foxl1 MO-injected embryos were the small eyes and morphological defects in their anterior segment of the brain. The phenotype induced by foxl1 MO injection showed significant similarity with the phenotypes of shh-overexpressing zebra fish embryos (4, 15, 26). Previous reports on overexpression of shh described malformation of the anterior brain, especially of the third ventricle, and indicated that eye development was strongly perturbed (4, 15, 26). The lens was frequently reduced in size or sometimes even absent from the eyes of shh-injected embryos (4). The distribution of pax2 transcripts was earlier reported to be widened in the area of the optic stalk of shh-overexpressing embryos (15, 26). Although we could observe ectopic expression of shh only in the midbrain of the foxl1 MO-injected embryos, enhanced expression of pax2a in optic stalks suggests that enhancement of shh expression, though undetectable, occurred in the eye. In accordance with this possibility, no lens was found in some cases of foxl1 MO-injected embryos. However, retinal pigment epithelium was present, unlike in the case of the shh-overexpressing embryos. The eyeless cave-dwelling form of Astyanax mexicanus has a phenotype of eye degeneration (50). Expansion of shh and twhh genes was observed along the anterior embryonic midline in cave fishes, and hyperactivation of downstream genes such as pax2a was reported (51), supporting the idea of the involvement of shh in foxl1 MO-injected embryos.
shh is thought to play important roles in pectoral fin development (32). Since zp50 and axial expressing domains were unaltered in their anteroposterior extent, it was anticipated that shh overexpression modifies primarily dorsoventral patterning rather than disrupting the formation of the anterior brain per se (15). We examined zp50 expression in foxl1 MO-injected embryos and found it to be indistinguishable between control and foxl1 MO-injected embryos (data not shown), supporting the idea that suppression of foxl1 expression did not affect anteroposterior formation.
Systemic administration of retinoic acid to zebra fish embryos resulted in the induction of ectopic expression of shh and abnormal pectoral fin bud morphology (1). We could not observe the expression of foxl1 in the fin bud. Furthermore, the ectopic expression of shh in fin bud was also undetectable when zfoxl1 was knocked down by MO, but we could not exclude the possibility that a lower-than-detectable level of foxl1 and shh caused malformation of the fin in the foxl1 MO-injected embryos.
Suppressive effects of foxl1 on shh promoter activity.
As expected from the findings of ectopic expression of shh by foxl1 knockdown by MO injection, we observed suppression of activity of the zebra fish shh promoter by foxl1 in both in vivo and in vitro experiments. Previous reports on deletion analysis of the zebra fish shh promoter identified an important region of the promoter for shh expression that contained two retinoic acid response elements (RAREs) and two axial (HNF3β) binding sites (8, 30). HNF3β is one of the FOX proteins, and targeted mutation of HNF3β in the mouse results in impaired notochord development and loss of shh expression (3, 48). We cloned the full-length zebra fish axial and examined the possible competition for axial binding sites in the shh promoter between axial and foxl1 in NIH 3T3 cells and PC12 cells by transient-transfection analysis; however, we could not observe enhancement of transcriptional activity of the isolated shh promoter by coexpression of axial in these cell lines under our experimental conditions (data not shown). In addition, since we could not exclude the possibility that overexpressed foxl1 had reduced sequence specificity for targets, we do not know whether foxl1 acts through axial binding sites or not.
Tissue-specific activity of zebra fish foxl1 may be exerted through its forkhead domain.
We found that foxl1 had transcriptional repressor activity in PC12 cells but not in NIH 3T3 or HeLa cells. Experiments using gal4-dbd showed that the C-terminal-region-dependent transcriptional suppression was not tissue specific. Taken together, our data suggest that FHD and/or the N-terminal region is responsible for tissue-specific repression activity of foxl1. LIN-31, a FOX protein in Caenorhabditis elegans, is thought to act as either a repressor or an activator depending on its state of phosphorylation (43). As in the case of LIN-31, we cannot exclude that foxl1 acts in both positive and negative ways. This notion is supported by the dramatic phenotype induced by the foxl1-VP16 construct in zebra fish embryos. However, there is a possibility that foxl1-VP16 acts as a dominant negative to wild-type foxl1, leading to the developmental defects.
When we compared the endogenous expression of foxl1 and shh, there was some complementary expression of shh and foxl1. In the embryos at 24 hpf, foxl1 was expressed in the ventricles of the forebrain, midbrain, and hindbrain and in the pharyngeal arch and otic vesicle. At this stage, shh was not expressed in these sites, and the expression was observed in the hypothalamus and floor plate (16), where foxl1 expression was not observed. However, ectopic expression or enhanced expression of shh was not observed in all of the expression domains of foxl1. The shh gene was ectopically expressed in the midbrain in foxl1 MO-injected embryos, and this region correlated with the area of foxl1 expression. However, the only other perturbation of shh expression in foxl1 MO-injected embryos was expansion of the shh expression domain observed in the hyopthalamus. As a reason why shh was not induced in other regions normally expressing the foxl1, such as the hindbrain and forebrain, the lack of a positive regulator of the shh promoter may be a possible reason. axial, a positive regulator of shh promoter (8), is expressed in domains overlapping with shh in the embryonic shield, notochord, and floor plate (21, 41). However, no clear expression of it was observed in midbrain and hindbrain. In a previous report on shh overexpression, widespread ectopic expression of shh was not detected (4), which was surmised to indicate that the injected shh RNA had been degraded by the stage at which the embryos were fixed. Similar reasoning may be applied to explain our observations on foxl1 MO-injected embryos.
Engrailed eh1 motif was found in the transrepressor region of foxl1.
We found a conserved motif called engrailed eh1, which is responsible for transcriptional repression of a large number of homeoprotein transcription factors, in the foxl1 C-terminal region (40). Zebra fish foxl1 contains a sequence that has similarity only with the central region of the eh1 domain and, interestingly, comparison of sequences of various homeoproteins indicates the central region to be the most conserved region, with the adjacent region on either side being far less conserved (40). We found exactly the same motif in both zfoxl1 and human FOXL1, whereas the motif in mouse Foxl1 was different. Database searching did not find this conserved motif in any other FHD-containing protein: however, partially matched motifs (FSI, FSIxxI, FSIDxI, FSIxS[I], FSIDS, or FxIxSI) were found in the C-terminal region of human, mouse, chicken, and Xenopus proteins of the FOXD family. Xenopus FoxD1 and FoxD5a are known as transcriptional repressors (27, 42), but the mechanism of their repression is not yet known.
The eh1 motif binds to the groucho corepressor (45). The groucho protein does not bind to DNA directly but is recruited to its targets by protein-protein interactions with a variety of transcriptional repressors (10). Mammalian Foxg1 (BF1) and Foxa2 act as a transcription repressor by forming a complex with groucho family proteins and histone deacetylases (46, 52). Tetrapeptide motifs such as WRPW and WRPY are known to be necessary and sufficient for recruitment of groucho. A conserved sequence of the FoxG1 (BF1) family, YWPMSPFLSLH is also thought to be involved in groucho binding (52). Although the engrailed eh1 motif appeared to bind with groucho, there was no similarity in sequence between eh1 and the WRPW motif.
Biological function of foxl1 among species.
As discussed above, mouse Foxl1 does not have the conserved eh1-like motif. The mouse Foxl1 gene is expressed in the mesenchymal layer of the developing and mature gastrointestinal tract (35). In addition, Foxl1-deficient mice exhibit various defects in the epithelial layer of the gastrointestinal tract and gut-associated lymphoid tissues such as Peyer's patches (13, 19). There are no common features of the expression pattern between mouse Foxl1 and zebra fish foxl1. Furthermore, when we consider the primary structure, except for FHD, there is almost no similarity between mouse Foxl1 and zebra fish foxl1. In addition to these findings, the knockout phenotype of the mouse Foxl1 was quite different from that of zebra fish. Therefore, we wonder whether mouse Foxl1 is a functional counterpart of zfoxl1 or not. There are several cases of FOX proteins whose expression pattern and biological roles are conserved among different species. In the case of axial, in addition to the forkhead domain, several short amino acids stretches are conserved between mouse and zebra fish axial. In the case of FoxG1 (BF1), high homology was observed in the entire region of the molecules of mouse and zebra fish, a finding in contrast to the lack of similarity in primary amino acid sequence between mouse and zebra fish foxl1. We surmise that different evolutionary changes occurred for each FHD protein, leading to the diverse structures and functions of certain FOX proteins.
Supplementary Material
Acknowledgments
We thank C. Neumann (EMBL), Y. Niikura, and R. Kurita for general discussions and Y. Matsumura for secretarial help.
This study was supported by the RIKEN Center for Developmental Biology.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Akimenko, M. A., and M. Ekker. 1995. Anterior duplication of the sonic hedgehog expression pattern in the pectoral fin buds of zebrafish treated with retinoic acid. Dev. Biol. 170:243-247. [DOI] [PubMed] [Google Scholar]
- 2.Akimenko, M. A., M. Ekker, J. Wegner, W. Lin, and M. Westerfield. 1994. Combinatorial expression of three zebrafish genes related to distal-less: part of a homeobox gene code for the head. J. Neurosci. 14:3475-3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ang, S. L., and J. Rossant. 1994. HNF-3 beta is essential for node and notochord formation in mouse development. Cell 78:561-574. [DOI] [PubMed] [Google Scholar]
- 4.Barth, K. A., and S. W. Wilson. 1995. Expression of zebrafish nk2.2 is influenced by sonic hedgehog/vertebrate hedgehog-1 and demarcates a zone of neuronal differentiation in the embryonic forebrain. Development 121:1755-1768. [DOI] [PubMed] [Google Scholar]
- 5.Boncinelli, E., and R. Morgan. 2001. Downstream of Otx2, or how to get a head. Trends Genet. 17:633-636. [DOI] [PubMed] [Google Scholar]
- 6.Brownell, I., M. Dirksen, and M. Jamrich. 2000. Forkhead Foxe3 maps to the dysgenetic lens locus and is critical in lens development and differentiation. Genesis 27:81-93. [DOI] [PubMed] [Google Scholar]
- 7.Carlsson, P., and M. Mahlapuu. 2002. Forkhead transcription factors: key players in development and metabolism. Dev. Biol. 250:1-23. [DOI] [PubMed] [Google Scholar]
- 8.Chang, B.-E., P. Blader, N. Fischer, P. W. Ingham, and U. Strahle. 1997. Axial (HNF3β) and retinoic acid receptors are regulators of the zebrafish sonic hedgehog promoter. EMBO J. 16:3955-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clark, K. L., E. D. Halay, E. Lai, and S. K. Burley. 1993. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature 364:412-420. [DOI] [PubMed] [Google Scholar]
- 10.Courey, A. J., and S. Jia. 2001. Transcriptional repression: the long and the short of it. Genes Dev. 15:2786-2796. [DOI] [PubMed] [Google Scholar]
- 11.Echelard, Y., D. J. Epstein, B. St-Jacques, L. Shen, J. Mohler, J. A. McMahon, and A. P. McMahon. 1993. Sonic hedgehog, a member of a family of putative signaling molecules, is implicated in the regulation of CNS polarity. Cell 75:1417-1430. [DOI] [PubMed] [Google Scholar]
- 12.Fjose, A., J. C. Izpisua-Belmonte, C. Fromental-Ramain, and D. Duboule. 1994. Expression of the zebrafish gene hlx-1 in the prechordal plate and during CNS development. Development 120:71-81. [DOI] [PubMed] [Google Scholar]
- 13.Fukuda, K., H. Yoshida, T. Sato, T. A. Furumoto, Y. Mizutani-Koseki, Y. Suzuki, Y. Saito, T. Takemori, M. Kimura, H. Sato, M. Taniguchi, S. Nishikawa, T. Nakayama, and H. Koseki. 2003. Mesenchymal expression of Foxl1, a winged helix transcriptional factor, regulates generation and maintenance of gut-associated lymphoid organs. Dev. Biol. 255:278-289. [DOI] [PubMed] [Google Scholar]
- 14.Guo, S., J. Brush, H. Teraoka, A. Goddard, S. W. Wilson, M. C. Mullins, and A. Rosenthal. 1999. Development of noradrenergic neurons in the zebrafish hindbrain requires BMP, FGF8, and the homeodomain protein soulless/Phox2a. Neuron 24:555-566. [DOI] [PubMed] [Google Scholar]
- 15.Hauptmann, G., and T. Gerster. 1996. Complex expression of the zp-50 pou gene in the embryonic zebrafish brain is altered by overexpression of sonic hedgehog. Development 122:1769-1780. [DOI] [PubMed] [Google Scholar]
- 16.Hauptmann, G., and T. Gerster. 2000. Regulatory gene expression patterns reveal transverse and longitudinal subdivisions of the embryonic zebrafish forebrain. Mech. Dev. 91:105-118. [DOI] [PubMed] [Google Scholar]
- 17.Hukriede, N. A., L. Joly, M. Tsang, J. Miles, P. Tellis, J. A. Epstein, W. B. Barbazuk, F. N. Li, B. Paw, J. H. Postlethwait, T. J. Hudson, L. I. Zon, J. D. McPherson, M. Chevrette, I. B. Dawid, S. L. Johnson, and S. C. Ekker. 1999. Radiation hybrid mapping of the zebrafish genome. Proc. Natl. Acad. Sci. USA 96:9745-9750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaestner, K. H., W. Knochel, and D. E. Martinez. 2000. Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev. 14:142-146. [PubMed] [Google Scholar]
- 19.Kaestner, K. H., D. G. Silberg, P. G. Traber, and G. Schutz. 1997. The mesenchymal winged helix transcription factor Fkh6 is required for the control of gastrointestinal proliferation and differentiation. Genes Dev. 11:1583-1595. [DOI] [PubMed] [Google Scholar]
- 20.Kenyon, K. L., S. A. Moody, and M. Jamrich. 1999. A novel fork head gene mediates early steps during Xenopus lens formation. Development 126:5107-5116. [DOI] [PubMed] [Google Scholar]
- 21.Krauss, S., J. P. Concordet, and P. W. Ingham. 1993. A functionally conserved homology of the Drosophila segment polarity gene hh is expressed in tissues with polarizing activity in zebrafish embryos. Cell 75:1431-1444. [DOI] [PubMed] [Google Scholar]
- 22.Kurita, R., H. Sagara, Y. Aoki, B. A. Link, K. Arai, and S. Watanabe. 2003. Suppression of lens growth by aA-crystallin promoter-driven expression of diphtheria toxin results in disruption of retinal cell organization in zebrafish. Dev. Biol. 255:113-127. [DOI] [PubMed] [Google Scholar]
- 23.Kurita, R., Y. Tabata, H. Sagara, K. Arai, and S. Watanabe. 2004. A novel smoothelin-like, actin-binding protein required for choroidal fissure closure in zebrafish. Biochem. Biophys. Res. Commun. 313:1092-1100. [DOI] [PubMed] [Google Scholar]
- 24.Lehmann, O. J., J. C. Sowden, P. Carlsson, T. Jordan, and S. S. Bhattacharya. 2003. Fox's in development and disease. Trends Genet. 19:339-344. [DOI] [PubMed] [Google Scholar]
- 25.Lewis, K. E., J. P. Concordet, and P. W. Ingham. 1999. Characterisation of a second patched gene in the zebrafish Danio rerio and the differential response of patched genes to Hedgehog signalling. Dev. Biol. 208:14-29. [DOI] [PubMed] [Google Scholar]
- 26.Macdonald, R., K. A. Barth, Q. Xu, N. Holder, I. Mikkola, and S. W. Wilson. 1995. Midline signalling is required for Pax gene regulation and patterning of the eyes. Development 121:3267-3278. [DOI] [PubMed] [Google Scholar]
- 27.Mariani, F. V., and R. M. Harland. 1998. XBF-2 is a transcriptional repressor that converts ectoderm into neural tissue. Development 125:5019-5031. [DOI] [PubMed] [Google Scholar]
- 28.Masumoto, J., W. Zhou, F. F. Chen, F. Su, J. Y. Kuwada, E. Hidaka, T. Katsuyama, J. Sagara, S. Taniguchi, P. Ngo-Hazelett, J. H. Postlethwait, G. Nunez, and N. Inohara. 2003. Caspy, a zebrafish caspase, activated by ASC oligomerization is required for pharyngeal arch development. J. Biol. Chem. 278:4268-4276. [DOI] [PubMed] [Google Scholar]
- 29.Miura, H., M. Yanazawa, K. Kato, and K. Kitamura. 1997. Expression of a novel aristaless related homeobox gene “Arx” in the vertebrate telencephalon, diencephalon and floor plate. Mech. Dev. 65:99-109. [DOI] [PubMed] [Google Scholar]
- 30.Muller, F., B. Chang, S. Albert, N. Fischer, L. Tora, and U. Strahle. 1999. Intronic enhancers control expression of zebrafish sonic hedgehog in floor plate and notochord. Development 126:2103-2116. [DOI] [PubMed] [Google Scholar]
- 31.Muto, E., Y. Tabata, T. Taneda, Y. Aoki, A. Muto, K. Arai, and S. Watanabe. 2004. Identification and characterization of Veph, a novel gene encoding a PH domain-containing protein expressed in the developing central nervous system of vertebrates. Biochimie 86:523-531. [DOI] [PubMed] [Google Scholar]
- 32.Neumann, C. J., H. Grandel, W. Gaffield, S. Schulte-Merker, and C. Nusslein-Volhard. 1999. Transient establishment of anteroposterior polarity in the zebrafish pectoral fin bud in the absence of sonic hedgehog activity. Development 126:4817-4826. [DOI] [PubMed] [Google Scholar]
- 33.Odenthal, J., and C. Nusslein-Volhard. 1998. fork head domain genes in zebra fish. Dev. Genes Evol. 208:245-258. [DOI] [PubMed] [Google Scholar]
- 34.Oxtoby, E., and T. Jowett. 1993. Cloning of the zebra fish krox-20 gene (krx-20) and its expression during hindbrain development. Nucleic Acids Res. 21:1087-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perreault, N., J. P. Katz, S. D. Sackett, and K. H. Kaestner. 2001. Foxl1 controls the Wnt/β-catenin pathway by modulating the expression of proteoglycans in the gut. J. Biol. Chem. 276:43328-43333. [DOI] [PubMed] [Google Scholar]
- 36.Picker, A., S. Scholpp, H. Bohli, H. Takeda, and M. Brand. 2002. A novel positive transcriptional feedback loop in midbrain-hindbrain boundary development is revealed through analysis of the zebrafish pax2.1 promoter in transgenic lines. Development 129:3227-3239. [DOI] [PubMed] [Google Scholar]
- 37.Satoh, S., K. Arai, and S. Watanabe. 2004. Identification of a novel splicing form of zebrafish p73 having a strong transcriptional activity. Biochem. Biophys. Res. Commun. 325:835-842. [DOI] [PubMed] [Google Scholar]
- 38.Schlombs, K., T. Wagner, and J. Scheel. 2003. Site-1 protease is required for cartilage development in zebrafish. Proc. Natl. Acad. Sci. USA 100:14024-14029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seufert, D. W., N. L. Prescott, and H. M. El-Hodiri. 2005. Xenopus aristaless-related homeobox (xARX) gene product functions as both a transcriptional activator and repressor in forebrain development. Dev. Dyn. 232:313-324. [DOI] [PubMed] [Google Scholar]
- 40.Smith, S. T., and J. B. Jaynes. 1996. A conserved region of engrailed, shared among all en-, gsc-, Nk1-, Nk2- and msh-class homeoproteins, mediates active transcriptional repression in vivo. Development 122:3141-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Strahle, U., P. Blader, D. Henrique, and P. W. Ingham. 1993. Axial, a zebrafish gene expressed along the developing body axis, shows altered expression in cyclops mutant embryos. Genes Dev. 7:1436-1446. [DOI] [PubMed] [Google Scholar]
- 42.Sullivan, S. A., L. Akers, and S. A. Moody. 2001. foxD5a, a Xenopus winged helix gene, maintains an immature neural ectoderm via transciptional repression that is dependent on the C-terminal domain. Dev. Biol. 232:439-457. [DOI] [PubMed] [Google Scholar]
- 43.Tan, P. B., M. R. lackner, and S. K. Kim. 1998. MAP kinase signaling specificity mediated by the LIN-1 Ets/LIN-31 WH transcription factor complex during Caenorhabditis elegans vulval induction. Cell 93:569-580. [DOI] [PubMed] [Google Scholar]
- 44.Tischler, A. S., and L. A. Greene. 1976. Establishment of a noradrenergic clonal line of rat adrenal pheochromocytoma cells which respond to nerve growth factor. Proc. Natl. Acad. Sci. USA 73:2424-2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tolkunova, E. N., M. Fujioka, M. Kobayashi, D. Deka, and J. B. Jaynes. 1998. Two distinct types of repression domain in engrailed: one interacts with the groucho corepressor and is preferentially active on integrated target genes. Mol. Cell. Biol. 18:2804-2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang, J. C., M. Waltner-Law, K. Yamada, H. Osawa, S. Stifani, and D. K. Granner. 2000. Transducin-like enhancer of split proteins, the human homologs of Drosophila groucho, interact with hepatic nuclear factor 3beta. J. Biol. Chem. 275:18418-18423. [DOI] [PubMed] [Google Scholar]
- 47.Weigel, D., G. Jurgens, F. Kuttner, E. Seifert, and H. Jackle. 1989. The homeotic gene fork head encodes a nuclear protein and is expressed in the terminal regions of the Drosophila embryo. Cell 57:645-658. [DOI] [PubMed] [Google Scholar]
- 48.Weinstein, D. C., A. Ruiz i Altaba, W. S. Chen, P. Hoodless, V. R. Prezioso, T. M. Jessell, and J. E. J. Darnell. 1994. The winged-helix transcription factor HNF-3 beta is required for notochord development in the mouse embryo. Cell 78:575-588. [DOI] [PubMed] [Google Scholar]
- 49.Westerfield, M. 1993. The zebrafish book. University of Oregon Press, Eugene.
- 50.Yamamoto, Y., and W. R. Jeffery. 2000. Central role for the lens in cave fish eye degeneration. Science 289:631-633. [DOI] [PubMed] [Google Scholar]
- 51.Yamamoto, Y., D. W. Stock, and W. R. Jeffery. 2004. Hedgehog signalling controls eye degeneration in blind cavefish. Nature 431:844-847. [DOI] [PubMed] [Google Scholar]
- 52.Yao, J., E. Lai, and S. Stifani. 2001. The winged-helix protein brain factor 1 interacts with groucho and hes proteins to repress transcription. Mol. Cell. Biol. 21:1962-1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.