Abstract
DNA methylation and histone methylation are two key epigenetic modifications that help govern heterochromatin dynamics. The roles for these chromatin-modifying activities in directing tissue-specific development remain largely unknown. To address this issue, we examined the roles of DNA methyltransferase 1 (Dnmt1) and the H3K9 histone methyltransferase Suv39h1 in zebra fish development. Knockdown of Dnmt1 in zebra fish embryos caused defects in terminal differentiation of the intestine, exocrine pancreas, and retina. Interestingly, not all tissues required Dnmt1, as differentiation of the liver and endocrine pancreas appeared normal. Proper differentiation depended on Dnmt1 catalytic activity, as Dnmt1 morphants could be rescued by active zebra fish or human DNMT1 but not by catalytically inactive derivatives. Dnmt1 morphants exhibited dramatic reductions of both genomic cytosine methylation and genome-wide H3K9 trimethyl levels, leading us to investigate the overlap of in vivo functions of Dnmt1 and Suv39h1. Embryos lacking Suv39h1 had organ-specific terminal differentiation defects that produced largely phenocopies of Dnmt1 morphants but retained wild-type levels of DNA methylation. Remarkably, suv39h1 overexpression rescued markers of terminal differentiation in Dnmt1 morphants. Our results suggest that Dnmt1 activity helps direct histone methylation by Suv39h1 and that, together, Dnmt1 and Suv39h1 help guide the terminal differentiation of particular tissues.
The development of an organism depends upon the concerted spatial and temporal regulation of tissue-specific transcriptional programs. The execution and maintenance of such developmental transcriptional programs are likely to rely on dynamic epigenetic changes in heterochromatin structure during early development (31). Among the key biochemical modifications playing an important role in establishing epigenetic states is methylation on cytosine in DNA (3). A long-standing hypothesis regarding DNA methylation is that de novo methylation is set up in a tissue-specific manner following implantation. This allows genes that are essential for the development of a specific tissue to remain unmethylated and thus transcriptionally competent. Conversely, genes with actions that might oppose the proper development of specific tissues become silenced by methylation. This hypothesis, however, remains controversial. Indeed, a number of studies challenge the hypothesis that DNA methylation plays an important role in directing tissue-specific gene expression during development (56, 57). For example, Walsh and Bestor found that the methylation status of the 5′ regions of a number of tissue-specific genes failed to correlate with their expression in the tissues of fetal and newborn mice (56). In addition, CpG islands are rarely methylated in normal tissues and the vast majority of CpG methylation within the human genome appears to be targeted to transposons, which constitute up to 45% of the human genome. Methylation within transposons appears to limit transposon action, thereby maintaining genome integrity (14).
However, a number of studies support the idea that methylation is correlated with altered expression in some tissue-specific genes (40, 59). For example, the adjacent region of the globin gene was shown to be hypomethylated in erythroid tissues but was completely methylated in nonerythroid tissues (29, 55). Also, the cytosine methylation status of a CpG-rich region within the promoter of maspin (SERPINB5) was shown to be correlated inversely to its expression in various tissues (12). Finally, Song et al. recently identified and confirmed tissue-specific differential methylation of genes that exhibit tissue-specific expression patterns (44). These studies indicate a potential role for methylation in tissue-specific gene regulation. However, it is still unknown whether these specific methylation patterns are established during development to silence specific genes.
Prominent examples of DNA methylation playing a role in development include monoallelic X chromosome inactivation and genomic imprinting, each of which results in permanent and heritable epigenetic states and gene repression (16, 27). Additional evidence for the role of methylation in development comes from studies with mice showing that DNA methylation is essential for embryonic viability. For example, DNMT1 knockout mice show delayed development and fail to survive past midgestation, possibly due to aberrant expression of imprinted genes and chromosomal instability (23). Also, knockout of Dnmt3b is lethal whereas Dnmt3a homozygous mice die at about 4 weeks of age (36). Studies with nonmammalian vertebrates also indicate that DNA methylation is essential for gastrulation. For example, chick embryos treated with 5-azacytidine display perturbed normal cellular migration and embryonic axis patterning (60). In another case, transient knockdown of Dnmt1 during early development in Xenopus laevis led to premature expression of mesodermal marker genes (48). With zebra fish, embryos treated with 5-azacytidine and 5-aza-2′-deoxycytidine just prior to midblastula transition show abnormal patterning of somites, shortened and thickened axial mesoderm, and loss of differentiated notochord and midline muscle (28).
The critical link between DNA methylation and transcriptional silencing is the recruitment of histone modifications that promote silencing. Several studies have suggested that DNA methylation and histone H3K9 methylation are interdependent (10). Studies with Neurospora crassa (51, 52) and Arabidopsis thaliana (18, 19) suggest that H3K9 methylation might control overall levels of DNA methylation. Furthermore, DNA methylation might exert a feedback loop on the levels of global H3K9 methylation, as human cell lines lacking DNMT1 have reduced levels of H3K9 dimethylation and H3K9 trimethylation (6). In Arabidopsis, loss of the DNMT1 ortholog MET1 results in the loss of H3K9 methylation from specific loci, although global H3K9 methylation levels remain unaffected (46). Interestingly, mice deficient in both of the major H3K9 trimethyl (H3K9me3) histone methyltransferases (SUV39H1 and SUV39H2) display severely reduced viability and the surviving animals harbor chromosomal instability (39), phenotypes shared with DNMT1 knockout mice. It is therefore possible that these two processes work in concert to establish and maintain heterochromatin levels during development.
Although the above-mentioned studies support a requirement for DNA and histone methyltransferases in early development, these studies leave a key issue unresolved: whether DNA and histone methyltransferases are required to execute tissue-specific developmental programs. Here, we report that both Dnmt1, the primary maintenance DNA methyltransferase, and Suv39h1, the major H3K9 methyltransferase, direct terminal differentiation within specific organs of zebra fish. We found that zebra fish embryos deficient in either Dnmt1 or Suv39h1 specified the formation of retina, exocrine pancreas, and intestine but failed to complete terminal differentiation within these tissues. In contrast, terminal differentiation of other organs, including endocrine pancreas and liver, appeared unaffected. Phenotypic similarities of Dnmt1 and Suv39h1 knockdown in zebra fish embryos support their cooperative roles in gene regulation during development. We propose that specific DNA and histone methyltransferases work in concert to control the spatial and temporal regulation of transcriptional programs that direct differentiation within specific organs.
MATERIALS AND METHODS
Zebra fish morpholino, mRNA, and plasmid injections.
Zebra fish stocks and embryo culture were maintained as described previously (34). Morpholinos (Mo), mRNA, and plasmids were injected at the 1-cell stage. Morpholino oligonucleotides were ordered from Gene Tools LLC. The morpholino sequences are as follows: for Dnmt1 Mo, 5′-ACAATGAGGTCTTGGTAGGCATTTC-3′; for Suv39h1 Mo, 5′-TAATAATCTGACCTTGTGTTTTTTG-3′; and for control Mo, 5′-CCTCTTACCTCAGTTACAATTTATA-3′. Morpholinos were dissolved in 1× Danieau buffer. Zebra fish dnmt1 (GenBank accession number NM_131189) and zebra fish suv39h1 (GenBank accession number BC076417) were cloned in pcDNA3.1/nV5-DEST vector (Invitrogen) for injections of plasmids. For mRNA injections, zebra fish dnmt1, zebra fish suv39h1, and human DNMT1 (GenBank accession number NM_001379) were cloned into pCRII-TOPO vector (Invitrogen). mRNA was transcribed using an mMessage machine kit (Ambion).
Whole-mount in situ hybridizations and histological analyses.
Whole-mount in situ hybridizations were carried out as described previously (34) by use of digoxigenin-labeled riboprobes for suv39h1, insulin, trypsin, gata6, hnf1β, foxa3, otx5, otx2, irbp, fabp10, and fabp2. For histological analyses, embryos were fixed in 10% neutral buffered formalin, rinsed in phosphate-buffered saline, and embedded in paraffin (for gut sections) or glycol methacrylate (Polysciences) (for eye sections). Five or six micron sections were cut using a Leica microtome and stained in hematoxylin and eosin (for gut sections) or toluidine blue (for eye sections). Sections were analyzed using a Zeiss Axiovert100 microscope, and pictures were taken using an Olympus Magnafire color camera.
Western blotting and antibodies.
Embryos were collected at 96 hours postfertilization (hpf), homogenized in either sodium dodecyl sulfate lysis buffer (63 mM Tris-HCl, pH 6.8, 10% glycerol, 5% β-mercaptoethanol, 3.5% sodium dodecyl sulfate, protease inhibitors) or zebra fish immunoprecipitation buffer (250 mM sucrose, 4 mM EDTA, 100 mM NaCl, 10 mM Tris, pH 7.4, protease inhibitors), and rotated at 4 degrees for 10 min, and supernatant was collected after centrifugation. The protein was separated by denaturing sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to a polyvinylidene difluoride membrane, and probed with the corresponding antibody. Zebra fish Dnmt1 antibody was raised in rabbit against the C terminus of the protein (Covance). Other antibodies used were obtained from the following vendors: human DNMT1 from Imgenex, H3K9me3 from Abcam, H3 from Abcam, V5 from Invitrogen, and vinculin from Sigma.
Genomic DNA isolation and global DNA methylation assay (liquid chromatography-mass spectrometry).
Embryos were collected at 72 hpf, and genomic DNA was isolated using a Puregene DNA isolation kit (Gentra Systems). The global DNA methylation assay was performed using liquid chromatography-mass spectrometry as described previously (45).
RT-PCR and microarray analysis.
Reverse transcriptase PCR (RT-PCR) was performed as described previously (34) by using the following primers: for amplification of zebra fish dnmt1, forward primer 5′-ATGCCTACCAAGACCTCATT-3′ and reverse primer 5′-TTAGTCAGAGAGCTCCATTT-3′; for human DNMT1, forward primer 5′-ATGCCGGCGCGTACC-3′ and reverse primer 5′-CTAGTCCTTAGCAGCTTCCTCC-3′; and for zebra fish suv39h1, forward primer 5′-ATGGCGCGAGATCTAAAAGA-3′ and reverse primer 5′-TTAAAAGAGATATTTCCGGCAG-3′. For checking splice blocking of suv39h1, forward primer 5′-ATTGCAGGGTGCCTTGTAA-3′ and reverse primer 5′-AAGCAGAGCTGCCTTCAT-3′ were used. β-actin primers were used as described previously (34).
Statistical analyses.
Results of statistical analyses are provided in Table 1. P values were calculated using Student's t test. P values represent results for morphants compared against those for control-injected embryos, while P values for the rescue experiments represent comparisons of results between the morphants.
TABLE 1.
Summary of Dnmt1 and Suv39h1 morphant phenotypes
| Marker | % Embryos (n) that stained positive for markera
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Control Mo | Dnmt1 Mo | Dnmt1 Mo + dnmt1WT | Dnmt1 Mo + DNMT1WT | Dnmt1 Mo + dnmt1C1109S | Dnmt1 Mo + dnmt1C1226S | Suv39h1 Mo | Suv39h1 Mo + suv39h1WT | Dnmt1 Mo + suv39h1WT | Dnmt1 Mo + suv39h1H323K | |
| otx5 | 100 (25) | 100 (15) | NA | NA | NA | NA | 100 (26) | NA | NA | NA |
| otx2 | 100 (22) | 100 (22) | NA | NA | NA | NA | 100 (23) | NA | NA | NA |
| irbp | 100 (33) | 6 (48)* | 75 (40)* | 71 (42)* | 7 (56) | 36 (33) | 5 (38)* | 81 (63)* | 80 (94)* | 23 (39) |
| foxa3 | 100 (32) | 98 (49) | NA | NA | NA | NA | 100 (24) | NA | NA | NA |
| gata6 | 100 (42) | 95 (22) | NA | NA | NA | NA | 100 (37) | NA | NA | NA |
| hnf1β | 100 (21) | 100 (18) | NA | NA | NA | NA | 100 (28) | NA | NA | NA |
| fabp2 | 100 (84) | 3 (96)* | 83 (65)* | 99 (57)* | 26 (27) | 23 (66) | 25 (57)* | 87 (85)* | 75 (118)† | 37 (51) |
| insulin | 100 (69) | 100 (57) | NA | NA | NA | NA | 100 (37) | NA | NA | NA |
| trypsin | 99 (84) | 18 (62)* | 72 (43)* | 71 (42)§ | 10 (30) | 23 (47) | 22 (37)* | 73 (49)* | 85 (94)* | 28 (39) |
| fabp10 | 100 (23) | 89 (19) | NA | NA | NA | NA | 76 (21) | NA | NA | NA |
Values shown indicate the percentage of n embryos staining positively for each indicated marker, where n is the total number of embryos used in two or three different replicates of an experiment. *, P < 0.0001; †, P < 0.0005; §, P = 0.0002. All other P values were nonsignificant. The confidence level was set to 95% (i.e., P < 0.05). dnmt1WT, wild-type Dnmt1; suv39h1WT, wild-type Suv39h1; NA, not applicable.
RESULTS
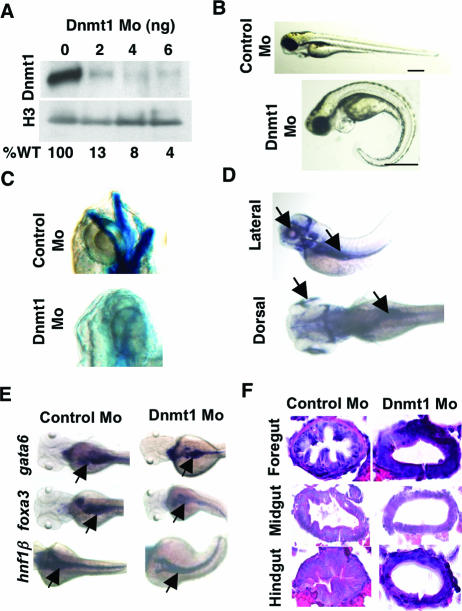
Zebra fish have emerged as a powerful genetic model system for identifying and mapping signaling pathways critical to embryological development (1, 4, 17, 21, 22, 25, 38, 50, 54). In addition, zebra fish harbor orthologs of each of the mammalian DNA methyltransferases, including the founding member, DNMT1 (28, 30, 43). By convention, we present zebra fish genes in lowercase and human genes in uppercase. To evaluate the role of the Dnmt1 protein in zebra fish embryonic development, we designed and injected a translation-blocking antisense morpholino, which efficiently reduced Dnmt1 protein levels (Fig. 1A). At an injection amount of 4 ng, approximately 40% of the embryos died within 24 hpf, an effect that was reminiscent of DNMT1 knockout in mice (23). With the remaining viable embryos, we determined global DNA methylation levels by using a mass spectrometry-based assay (45) and observed an approximate 40% decrease in the global levels of cytosine DNA methylation, with mdC/dG contents of 7.15% ± 0.06% (average ± standard deviation [SD]) and 4.48% ± 0.35% (average ± SD) for the wild type and Dnmt1 morphants, respectively. The viable morphants retained an overall normal appearance but displayed developmental defects, including curled tails, pericardial edema (Fig. 1B), and the absence of the P1 (mandibular) and P2 (hyoid) segments in the jaw (Fig. 1C).
FIG. 1.
Dnmt1 knockdown in zebra fish. (A) Western analysis of Dnmt1 protein levels in embryos injected with a Dnmt1-directed morpholino (0, 2, 4, or 8 ng). Levels of histone H3 served to normalize Dnmt1 expression levels, as indicated by percent wild-type (WT) levels. (B) Nomarski images of 96-hpf embryos injected with either control or Dnmt1 morpholino. Bar = 0.5 mm. (C) Alcian blue staining showing loss of branchial arches P1 (mandibular) and P2 (hyoid) in Dnmt1 morphants. (D) Whole-mount in situ hybridization of 72-hpf wild-type embryos with dnmt1 shows dnmt1 expression in the retina and gut (indicated by arrows). (E) Early gut tube specification was assessed in control or Dnmt1 morphant embryos (96 hpf) by whole-mount in situ hybridization for expression of gata6, foxa3, and hnf1β (indicated by arrows). (F) Histological sectioning shows loss of columnar cells throughout the intestine of Dnmt1 morphants.
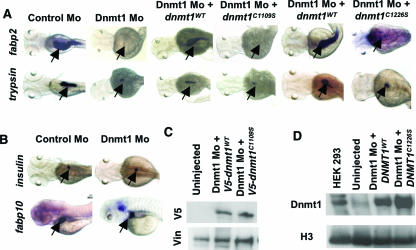
At 72 hpf, dnmt1 is expressed in developing gut, eye, and hindbrain (Fig. 1D) (53). This led us to examine possible defects in these organs. We observed near-normal expression levels of markers of the primordial gut foxa3 (35), gata6 (37), and hnf1β (15) within Dnmt1 morphant embryos (Fig. 1E), thereby confirming formation and specification of the early gut tube. However, cross-sections of the morphant embryo guts revealed failed epithelial differentiation, as evidenced by lack of established columnar cells throughout the intestine (Fig. 1F). This was paralleled by absence of intestinal fatty acid binding protein (fabp2) expression, a marker of terminally differentiated enterocytes (33), in 97% of Dnmt1 morphants (Fig. 2A). The penetrance statistics of all morphant phenotypes, as well as all other statistical analyses, are provided in Table 1. To determine whether lack of intestinal differentiation resulted from specific knockdown of Dnmt1 activity, we coinjected a plasmid encoding V5-tagged zebra fish Dnmt1 or mRNA encoding human DNMT1 along with the Dnmt1 morpholino and confirmed expression by Western analysis (Fig. 2C and D). These coinjections effectively rescued fabp2 expression to near-normal levels in more than 80% of the embryos (Fig. 2A and Table 1). In contrast, coinjection of a catalytically inactive derivative of zebra fish (Dnmt1C1109S) or human (DNMT1C1226S) origin failed to rescue fabp2 expression (Fig. 2A and Table 1). Taken together, these data indicate that zebra fish intestinal differentiation requires the catalytic activity of Dnmt1 and that human DNMT1 activity can substitute for that of zebra fish Dnmt1.
FIG. 2.
Intestinal and pancreatic differentiation defects in Dnmt1 morphants. (A) Whole-mount in situ hybridization assessing levels of fabp2 and trypsin (arrows) in Dnmt1 morphants shows failed differentiation of intestinal epithelial cells and exocrine pancreas. Injection of a plasmid encoding V5-tagged, wild-type Dnmt1 (dnmt1WT) or human DNMT1WT RNA rescues fabp2 and trypsin expression, but use of V5-tagged, catalytically inactive Dnmt1 (dnmt1C1109S) or human DNMT1C1226S does not. (B) Whole-mount in situ hybridization assessing levels of insulin and fabp10 (arrows) in Dnmt1 morphants. Levels of insulin and fabp10 are unchanged, indicating apparent normal development of the endocrine pancreas and liver, respectively. (C) Western analysis confirming equal expression of wild-type or catalytically inactive Dnmt1 within the injected embryos. Vinculin (Vin) served as a loading control. (D) Western analysis showing equal expression of DNMT1WT and DNMT1C1226S RNA in injected embryos. HEK 293 extract was used as a positive control, and histone H3 was used as a loading control.
Organ-specific defects extended to the pancreas, which failed to undergo exocrine differentiation, as indicated by lack of trypsin production (2) (Fig. 2A and Table 1). Remarkably, differentiation of endocrine pancreas and liver appeared unaffected in Dnmt1 morphants, as assessed by the expression levels of insulin (Fig. 2B) (2) and liver fatty acid binding protein (fabp10) (Fig. 2B) (5), respectively. Again, injection of wild-type but not catalytically inactive Dnmt1 (or human DNMT1) restored trypsin expression in 72% and 71% of the morphant embryos, respectively (Fig. 2A and Table 1). As the exocrine pancreas (2, 7, 58), endocrine pancreas, and liver (8) emerge from the same primordial precursors, these findings imply highly specific, temporally regulated roles for Dnmt1 within the developing gut.
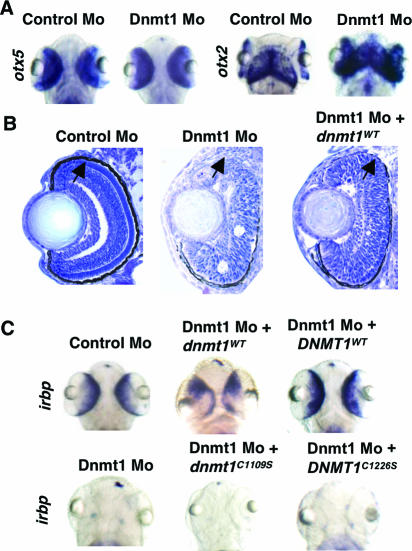
Considering that dnmt1 is strongly expressed within the developing eye (Fig. 1D), it was surprising that the morphological appearance of the eye remained relatively normal in Dnmt1 morphants (Fig. 3A and C). Indeed, markers of retinal development otx2 (32) and otx5 (13) were similarly expressed in both control and Dnmt1 morphants at 96 hpf (Fig. 3A). However, examination of cross-sections of the Dnmt1 morphant eyes revealed a profound disorganization of all retinal layers (Fig. 3B) as well as failed establishment of the dorsal retinal pigmented epithelium (RPE). In keeping with this, expression of interphotoreceptor retinoic binding protein (irbp), which marks terminal differentiation of the RPE and photoreceptor cell layers (49), was lost from the eyes of Dnmt1 morphants (Fig. 3C and Table 1). Consistent with observations of the intestine and exocrine pancreas, irbp expression and organization of retinal cell layers were restored in greater than 70% of the Dnmt1 morphants upon coinjection of wild-type but not catalytically inactive zebra fish Dnmt1 or human DNMT1 (Fig. 3B and C and Table 1).
FIG. 3.
Retinal differentiation defects in Dnmt1 morphants. (A) Control or Dnmt1 morphant embryos (96 hpf) were subjected to whole-mount in situ hybridization for otx5 and otx2. (B) TOPRO-1 staining of histological cross-sections within the Dnmt1 morphant retinas. Arrows indicate the dorsal loss of RPE. Injection of a plasmid encoding V5-tagged, wild-type Dnmt1 (dnmt1WT) rescues the RPE and retinal layering. (C) Whole-mount in situ hybridization of irbp expression within the retina of Dnmt1 morphants. Injection of a plasmid encoding V5-tagged, wild-type Dnmt1 (dnmt1WT) rescues irbp expression, but use of V5-tagged, catalytically inactive Dnmt1 (dnmt1C1109S) does not.
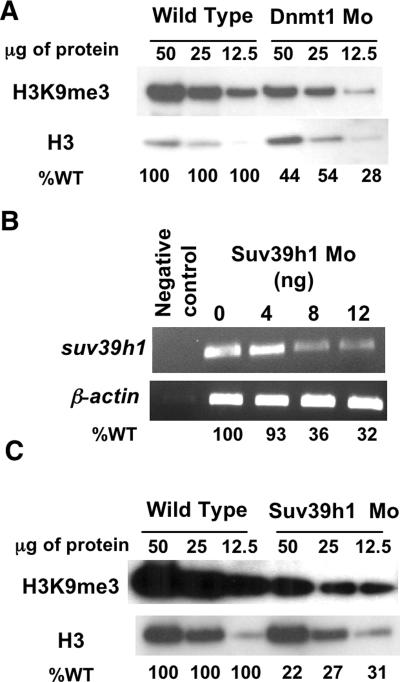
A number of investigations support functional and physical interactions between DNA methyltransferases and H3K9 methyltransferases (9, 10, 14, 42). Consistent with these studies, we observed a significant reduction in the global H3K9me3 levels in Dnmt1 morphants (Fig. 4A). Since SUV39H1 proteins establish H3K9 methylation in heterochromatic regions (39, 41), we next investigated the in vivo functions of Suv39h1 in zebra fish embryonic development. Using database search tools, we identified a zebra fish Suv39h1 ortholog that shares 63% identity with both human and murine SUV39H1 and contains all of the essential functional domains (data not shown). We also determined its developmental expression profile by whole-mount in situ hybridization and RT-PCR (Fig. 5), which demonstrated that suv39h1 expression in zebra fish overlapped with the tissue expression profile of dnmt1.
FIG. 4.
Reduced levels of H3K9me3 in Dnmt1 and Suv39h1 morphant embryos. (A) Western analysis of H3K9me3 levels in embryos injected with a Dnmt1-directed morpholino. The amount of protein loaded in each lane is indicated at top. Analysis of histone H3 loading served to normalize levels of H3K9me3 for morphants relative to levels for the wild type (WT). Percent control levels are indicated at bottom. (B) cDNA was prepared at 96 hpf from embryos injected with increasing amounts of Suv39h1 morpholino (0, 4, 8, and 12 ng); RT-PCR using primers flanking the exon 2/intron 2 boundary of suv39h1 (against which the morpholino was targeted) showed that the level of spliced suv39h1 transcripts was reduced with increasing amounts of Suv39h1 splice-blocking morpholino. β-actin was used as a loading control. Percent control levels are indicated at bottom. (C) H3K9me3 levels in Suv39h1 morphants.
FIG. 5.
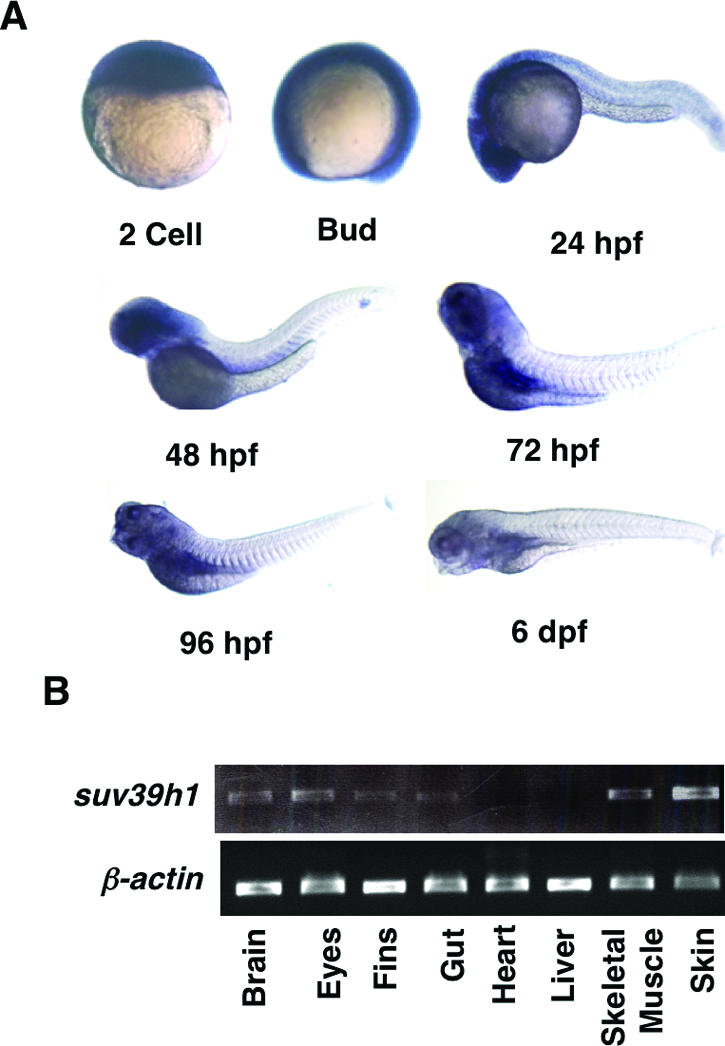
Expression pattern of suv39h1 during embryonic development. (A) Whole-mount in situ hybridization for suv39h1 shows expression of suv39h1 during early embryonic development. suv39h1 expression is ubiquitous until early somatic stages, after which it becomes restricted to the head of the embryo. After 48 hpf, the expression reduces drastically. Very low levels of expression can be seen in the pancreas starting at 72 hpf. dpf, days postfertilization. (B) RT-PCR analysis shows suv39h1 expression in adult tissues of brain, eyes, fins, gut, skeletal muscle, and skin.
To compare the consequences of Suv39h1 loss with those of Dnmt1 loss, we knocked down suv39h1 mRNA transcript levels by using a splice-blocking morpholino directed against the exon 2/intron 2 boundary and verified incomplete splicing by RT-PCR (Fig. 4B). To confirm knockdown of Suv39h1, we analyzed H3K9me3 levels in Suv39h1 morphants by Western blotting and noticed a significant reduction, a reduction that was greater than that observed for Dnmt1 morphants (Fig. 4A and C). Since knockout of H3K9 methyltransferase leads to reduction in global DNA methylation levels in Neurospora crassa (51, 52) and Arabidopsis thaliana (18, 19), we analyzed global levels of 5-methyl-2′-deoxycytidine in genomic DNA from 72-hpf Suv39h1 morphant embryos. We were unable to detect any change in global cytosine methylation levels within the Suv39h1 morphant embryos (for wild-type embryos, the mdC/dG ratio was 7.15% ± 0.06% [average ± SD], whereas for Suv39h1 morphants, it was 7.21% ± 0.46% [average ± SD]), despite robust reductions in the H3K9me3 levels. Although this method does not assess locus-specific DNA methylation changes, these data show that in zebra fish significant reduction in H3K9 methylation does not appreciably reduce DNA methylation.
Suv39h1 morphants did not show a significantly increased rate of death at 24 hpf (∼10% in Suv39h1 morphants, compared to ∼5% in control-injected embryos). However, they displayed overall morphological defects that were strikingly similar to those of Dnmt1 morphants, including pericardial edema and curled tails (Fig. 6A). However, unlike the jaws of Dnmt1 morphants, the jaws of Suv39h1 morphants developed normally (data not shown), suggesting that there are both unique and overlapping development requirements for Dnmt1 and Suv39h1. We tested this concept by examining expression of terminal differentiation markers of the eye and gut that were absent in Dnmt1 morphants. Similarly to Dnmt1 morphants, Suv39h1 morphants lacked expression of irbp in the retina (Fig. 6B and Table 1). This defect was rescued when embryos were coinjected with the Suv39h1 morpholino and a plasmid containing V5-tagged, wild-type zebra fish Suv39h1, thereby confirming specific knockdown of suv39h1 (Fig. 6B and Table 1). Similarities between Dnmt1 and Suv39h1 morphants extended to the gut: Suv39h1 morphants expressed the primordial gut tube markers foxa3, gata6, and hnf1β (Table 1 and data not shown) but did not express fabp2 within the intestinal epithelial cell layer (Fig. 6B). As with irbp, coinjection of suv39h1 rescued fabp2 expression (Fig. 6B and Table 1). Dnmt1 and Suv39h1 morphants also shared loss of trypsin and retention of insulin and fabp10 (Fig. 6B and Table 1). The above-described findings indicate that Dnmt1 and Suv39h1 cooperate to promote tissue-specific differentiation.
FIG. 6.
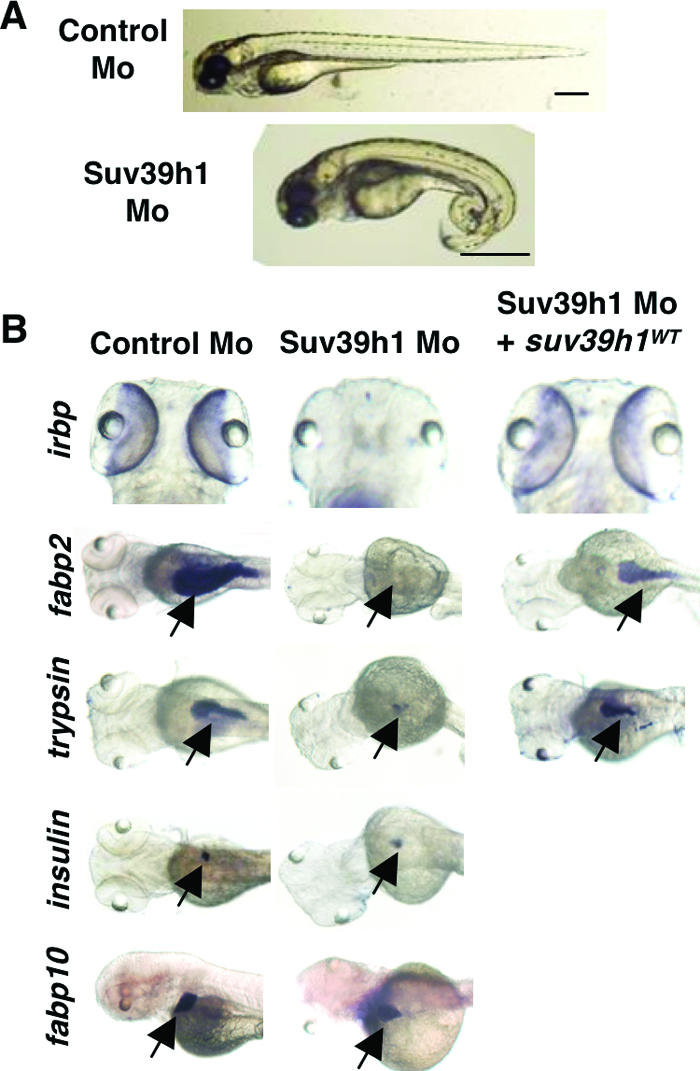
Suv39h1 morphants produce phenocopies of Dnmt1 morphants. (A) Nomarski image of 96-hpf embryos injected with either control morpholino or Suv39h1 morpholino. Bar = 0.5 mm. (B) Whole-mount in situ hybridization for irbp, fabp2, trypsin, insulin, and fabp10 (arrows) in an Suv39h1 morphant in comparison to a control embryo indicates failed terminal differentiation of retina, exocrine pancreas, and intestine. irbp, fabp2, and trypsin (arrows) expression levels were rescued in Suv39h1 morphants by coinjection of V5-tagged, wild-type suv39h1 (suv39h1WT). As seen with Dnmt1 morphants, Suv39h1 morphants continue to express insulin and fabp.
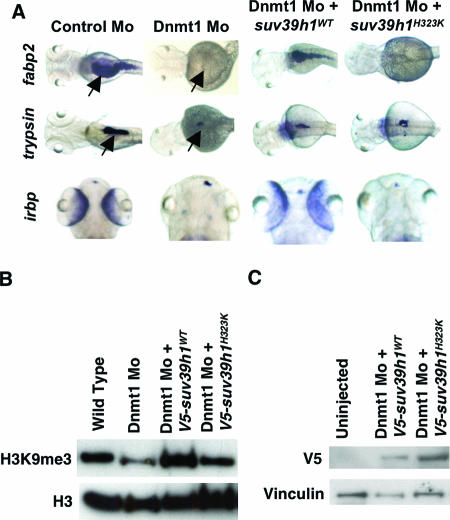
Reduction of global H3K9me3 and CpG methylation levels in Dnmt1 morphants and no change in DNA methylation levels in Suv39h1 morphants suggest that H3K9me3 levels might be dependent upon CpG methylation levels. Also, phenotypic similarities between Dnmt1 and Suv39h1 morphants suggest that maintaining H3K9me3 levels might be important for terminal differentiation programs. Hence, we tested whether increasing H3K9me3 levels in Dnmt1 morphants by overexpression of suv39h1 would rescue the terminal differentiation defects. Strikingly, injection of suv39h1 mRNA (100 pg) or a plasmid containing V5-tagged, wild-type zebra fish Suv39h1 (12 pg) rescued the expression of fabp2, trypsin, and irbp in Dnmt1 morphants (Fig. 7A). In contrast, injection of a catalytically inactive Suv39h1 derivative (suv39h1H323K) failed to rescue the phenotypic defects (Fig. 7A). Furthermore, overexpression of wild-type suv39h1, but not of suv39h1H323K, increased H3K9me3 levels in Dnmt1 morphants (Fig. 7B). As a control, expression of wild-type suv39h1 and suv39h1H323K constructs was confirmed by Western blotting using V5 antibody (Fig. 7C). Taken together, these data suggest that reduced levels of H3K9 methylation account for the terminal differentiation defects present in Dnmt1 and Suv39h1 morphants.
FIG. 7.
Suv39h1 overexpression rescues Dnmt1 morphants. (A) Whole-mount in situ hybridization for irbp, fabp2, and trypsin (arrows) in embryos injected with Dnmt1 morpholino alone or coinjected with plasmid encoding V5-tagged, wild-type Suv39h1 (suv39h1WT) or catalytic mutant Suv39h1 (suv39h1H323K) indicates that Dnmt1 morphants can be rescued by overexpression of wild-type Suv39h1 but not catalytically inactive Suv39h1. (B) Western analysis showing expression of V5-tagged suv39h1WT and suv39h1H323K in embryos injected with these constructs along with Dnmt1 morpholino. Vinculin is used as a loading control. (C) Western analysis of H3K9me3 levels in embryos injected with Dnmt1 morpholino alone or coinjected with suv39h1WT or suv39h1H323K. Analysis of histone H3 loading served to normalize levels of H3K9me3.
DISCUSSION
Our data support an overlapping requirement for Dnmt1 and Suv39h1 in directing tissue-specific, terminal differentiation within particular organs during zebra fish development. These findings suggest a model wherein DNA and histone methyltransferases do not simply contribute to transcription in general but rather help execute specific developmental programs.
Earlier studies of Dnmt1 and Suv39h1/Suv39h2 knockout in other organisms have established a more general role for these enzymes in maintaining normal gene expression or chromosomal stability (23, 39). For example, examination of DNMT1−/− mouse embryos at embryonic day 10.5 revealed that they more closely resembled embryos of wild-type mice at embryonic day 9.5, thereby suggesting developmental delays (23). In addition, two embryonic organs, the heart and the brain, showed increased cell death and decreased cell proliferation. Follow-up studies demonstrated that fibroblasts derived from DNMT1 conditional knockout mice displayed robust activation of p53 (20). Similarly, transient knockout of Dnmt1 in Xenopus at the blastula stage resulted in severe p53-dependent apoptosis of cells undergoing early tissue specification (47, 48). Furthermore, combined knockout of SUV39H1 and SUV39H2 in mice caused early embryonic lethality along with genomic instability (39). Although each of the previous studies revealed a clear requirement for DNA and histone methylation in maintaining cell homeostasis during early development, none addressed the potential for control of tissue-specific, terminal differentiation programs by these enzymes.
Consistent with studies of mice and Xenopus, we noticed ∼40% lethality during gastrulation in Dnmt1 morpholino-injected embryos (23, 48). However, unlike previous studies, we investigated the terminal differentiation states of multiple organs in the Dnmt1 and Suv39h1 morphant zebra fish surviving at 72 and 96 hpf. This analysis revealed organ-specific developmental defects rather than general developmental defects: differentiation of the retina, exocrine pancreas, and intestine were defective, whereas differentiation of the liver and endocrine pancreas appeared normal. As injection of antisense morpholino constructs comes with inherent variability of target knockdown, it is possible that the difference between survival and lethality in these embryos resulted directly from differences in extent of Dnmt1 knockdown in each population. However, it is important to note that the embryos displaying tissue-specific differentiation defects survived with significant loss of Dnmt1 protein (Fig. 1A) as well as substantial reductions in global levels of 5-methyl-cytidine. Consequently, not only do these findings support a role for Dnmt1 and Suv39h1 during early development, but they also provide evidence for tissue-specific functions of these enzymes. Further studies are needed to define the specific targets of Dnmt1 and Suv39h1 that control terminal differentiation in that markers such as fabp2 and irbp are unlikely direct targets of Dnmt1 and Suv39h1.
Our data are consistent with a model wherein DNA methylation and histone methylation act in concert to regulate chromatin states of one or more key tissue-specific regulators of terminal differentiation. It is possible that maternal contributions of Dnmt1 and Suv39h1 may have protected early embryo viability, organ specification, and differentiation. However, this would not appear to account for the selective, terminal differentiation defects seen with some tissues but not others. Likewise, although increased genetic instability may account for 40% lethality upon knockdown of Dnmt1, it would not readily account for selective tissue differentiation abnormalities. Indeed, the expression domains of dnmt1 and suv39h1 appear to support tissue-specific functions in that each is expressed highly in retina and gut, the very tissues showing terminal differentiation defects. Given that knockdown by morpholino injection is incomplete, our data may imply that the surviving morphant embryos retain residual amounts of Dnmt1 activity that are sufficient for the terminal differentiation of some tissues, such as liver. Alternatively, our findings might predict that DNA methyltransferases other than Dnmt1 may be required for terminal differentiation of those organs which remain unaffected by Dnmt1 and Suv39h1 knockdown. Indeed, zebra fish bear additional DNA and histone methyltransferases that are conserved in all vertebrates, raising the possibility that additional enzymes may likewise direct tissue-specific differentiation (43). In support of this, recent evidence indicates that methylation of a developmentally regulated gene (ntl) in zebra fish depends selectively upon Dnmt7 (43).
Human DNMT1 and SUV39H1 interact both in vitro and in vivo (11), and both of these proteins have been implicated in repression of genes and formation of heterochromatin (10). For example, DNMT1 and SUV39H1 have been shown to act together to repress transcription of the estrogen receptor alpha promoter in breast cancer (26). However, the evidence for their concerted roles in early development has been lacking. We noted that Dnmt1 morphant embryos showed a dramatic reduction in H3K9 trimethyl levels, indicating a dependence of H3K9 methylation on cytosine methylation in zebra fish. Indeed, phenotypes of Dnmt1 and Suv39h1 knockdown were strikingly similar within the gut and eyes, thereby confirming an overlapping role for each in intestinal and retinal tissue development. Notably, Suv39h1 morphants showed no decrease in global DNA methylation levels. These findings align well with observations of human cells wherein a DNMT1−/− colon carcinoma cell line displayed substantial decreases in H3K9 trimethylation (6). Taken together, these observations imply that Dnmt1 activity lies upstream of Suv39h1, a idea confirmed by our finding that injection of suv39h1 mRNA into Dnmt1 morphants rescued gut and retinal differentiation defects. Importantly, these data indicate that vertebrates are distinct from Neurospora and Arabidopsis, in which DNA methylation clearly relies on histone methylation (19, 51). In Neurospora, mutation in H3K9 methyltransferase abolished almost all DNA methylation, both at symmetric and at nonsymmetric loci (51). In Arabidopsis, mutations in KRYPTONITE histone H3K9 methyltransferase led to loss of CpNpG methylation but not CpG methylation (19). Interestingly, only plants bear orthologs of the DNA methyltransferase CHROMOMETHYLASE3, which utilizes its chromodomains to recognize histone methylation at two important locations along the H3 tail (H3K9 and H3K27), thereby targeting DNA methylation to histone-methylated regions (24). Thus, it appears that the dependency relationships between histone and DNA methylation differ between plants and vertebrates.
In conclusion, our evidence supports the notion that Dnmt1, the primary maintenance DNA methyltransferase, and Suv39h1, the major H3K9 methyltransferase, help direct terminal differentiation within specific organs of the developing zebra fish. These findings offer support for a model wherein organ-specific terminal differentiation requires the coordinated efforts of specific DNA and histone methyltransferases.
Acknowledgments
This work was supported by grants from the National Cancer Institute (to D.A.J.), the Howard Hughes Medical Institute (to B.R.C.), the Huntsman Cancer Foundation (to D.A.J. and B.R.C.), the Ralph Wilson Medical Research Foundation, the Roswell Park Alliance Foundation, and Phi Beta Psi (to A.R.K.).
We thank the DNA peptide resource, the DNA sequencing resource, the microarray resource, and the centralized zebra fish animal resource at the University of Utah.
REFERENCES
- 1.Amatruda, J. F., J. L. Shepard, H. M. Stern, and L. I. Zon. 2002. Zebrafish as a cancer model system. Cancer Cell 1:229-231. [DOI] [PubMed] [Google Scholar]
- 2.Biemar, F., F. Argenton, R. Schmidtke, S. Epperlein, B. Peers, and W. Driever. 2001. Pancreas development in zebrafish: early dispersed appearance of endocrine hormone expressing cells and their convergence to form the definitive islet. Dev. Biol. 230:189-203. [DOI] [PubMed] [Google Scholar]
- 3.Bird, A. P. 1996. The relationship of DNA methylation to cancer. Cancer Surv. 28:87-101. [PubMed] [Google Scholar]
- 4.Davidson, A. J., P. Ernst, Y. Wang, M. P. Dekens, P. D. Kingsley, J. Palis, S. J. Korsmeyer, G. Q. Daley, and L. I. Zon. 2003. cdx4 mutants fail to specify blood progenitors and can be rescued by multiple hox genes. Nature 425:300-306. [DOI] [PubMed] [Google Scholar]
- 5.Denovan-Wright, E. M., M. Pierce, M. K. Sharma, and J. M. Wright. 2000. cDNA sequence and tissue-specific expression of a basic liver-type fatty acid binding protein in adult zebrafish (Danio rerio). Biochim. Biophys. Acta 1492:227-232. [DOI] [PubMed] [Google Scholar]
- 6.Espada, J., E. Ballestar, M. F. Fraga, A. Villar-Garea, A. Juarranz, J. C. Stockert, K. D. Robertson, F. Fuks, and M. Esteller. 2004. Human DNA methyltransferase 1 is required for maintenance of the histone H3 modification pattern. J. Biol. Chem. 279:37175-37184. [DOI] [PubMed] [Google Scholar]
- 7.Field, H. A., P. D. Dong, D. Beis, and D. Y. Stainier. 2003. Formation of the digestive system in zebrafish. II. Pancreas morphogenesis. Dev. Biol. 261:197-208. [DOI] [PubMed] [Google Scholar]
- 8.Field, H. A., E. A. Ober, T. Roeser, and D. Y. Stainier. 2003. Formation of the digestive system in zebrafish. I. Liver morphogenesis. Dev. Biol. 253:279-290. [DOI] [PubMed] [Google Scholar]
- 9.Freitag, M., and E. U. Selker. 2005. Controlling DNA methylation: many roads to one modification. Curr. Opin. Genet. Dev. 15:191-199. [DOI] [PubMed] [Google Scholar]
- 10.Fuks, F. 2005. DNA methylation and histone modifications: teaming up to silence genes. Curr. Opin. Genet. Dev. 15:490-495. [DOI] [PubMed] [Google Scholar]
- 11.Fuks, F., P. J. Hurd, R. Deplus, and T. Kouzarides. 2003. The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res. 31:2305-2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Futscher, B. W., M. M. Oshiro, R. J. Wozniak, N. Holtan, C. L. Hanigan, H. Duan, and F. E. Domann. 2002. Role for DNA methylation in the control of cell type specific maspin expression. Nat. Genet. 31:175-179. [DOI] [PubMed] [Google Scholar]
- 13.Gamse, J. T., Y. C. Shen, C. Thisse, B. Thisse, P. A. Raymond, M. E. Halpern, and J. O. Liang. 2002. Otx5 regulates genes that show circadian expression in the zebrafish pineal complex. Nat. Genet. 30:117-121. [DOI] [PubMed] [Google Scholar]
- 14.Goll, M. G., and T. H. Bestor. 2005. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 74:481-514. [DOI] [PubMed] [Google Scholar]
- 15.Gong, H. Y., C. J. Lin, M. H. Chen, M. C. Hu, G. H. Lin, Y. Zhou, L. I. Zon, and J. L. Wu. 2004. Two distinct teleost hepatocyte nuclear factor 1 genes, hnf1alpha/tcf1 and hnf1beta/tcf2, abundantly expressed in liver, pancreas, gut and kidney of zebrafish. Gene 338:35-46. [DOI] [PubMed] [Google Scholar]
- 16.Heard, E. 2005. Delving into the diversity of facultative heterochromatin: the epigenetics of the inactive X chromosome. Curr. Opin. Genet. Dev. 15:482-489. [DOI] [PubMed] [Google Scholar]
- 17.Hsu, K., J. P. Kanki, and A. T. Look. 2001. Zebrafish myelopoiesis and blood cell development. Curr. Opin. Hematol. 8:245-251. [DOI] [PubMed] [Google Scholar]
- 18.Jackson, J. P., L. Johnson, Z. Jasencakova, X. Zhang, L. PerezBurgos, P. B. Singh, X. Cheng, I. Schubert, T. Jenuwein, and S. E. Jacobsen. 2004. Dimethylation of histone H3 lysine 9 is a critical mark for DNA methylation and gene silencing in Arabidopsis thaliana. Chromosoma 112:308-315. [DOI] [PubMed] [Google Scholar]
- 19.Jackson, J. P., A. M. Lindroth, X. Cao, and S. E. Jacobsen. 2002. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416:556-560. [DOI] [PubMed] [Google Scholar]
- 20.Jackson-Grusby, L., C. Beard, R. Possemato, M. Tudor, D. Fambrough, G. Csankovszki, J. Dausman, P. Lee, C. Wilson, E. Lander, and R. Jaenisch. 2001. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat. Genet. 27:31-39. [DOI] [PubMed] [Google Scholar]
- 21.Langenau, D. M., T. Palomero, J. P. Kanki, A. A. Ferrando, Y. Zhou, L. I. Zon, and A. T. Look. 2002. Molecular cloning and developmental expression of Tlx (Hox11) genes in zebrafish (Danio rerio). Mech. Dev. 117:243-248. [DOI] [PubMed] [Google Scholar]
- 22.Langenau, D. M., D. Traver, A. A. Ferrando, J. L. Kutok, J. C. Aster, J. P. Kanki, S. Lin, E. Prochownik, N. S. Trede, L. I. Zon, and A. T. Look. 2003. Myc-induced T cell leukemia in transgenic zebrafish. Science 299:887-890. [DOI] [PubMed] [Google Scholar]
- 23.Li, E., T. H. Bestor, and R. Jaenisch. 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69:915-926. [DOI] [PubMed] [Google Scholar]
- 24.Lindroth, A. M., D. Shultis, Z. Jasencakova, J. Fuchs, L. Johnson, D. Schubert, D. Patnaik, S. Pradhan, J. Goodrich, I. Schubert, T. Jenuwein, S. Khorasanizadeh, and S. E. Jacobsen. 2004. Dual histone H3 methylation marks at lysines 9 and 27 required for interaction with CHROMOMETHYLASE3. EMBO J. 23:4286-4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, T. X., N. G. Howlett, M. Deng, D. M. Langenau, K. Hsu, J. Rhodes, J. P. Kanki, A. D. D'Andrea, and A. T. Look. 2003. Knockdown of zebrafish Fancd2 causes developmental abnormalities via p53-dependent apoptosis. Dev. Cell 5:903-914. [DOI] [PubMed] [Google Scholar]
- 26.Macaluso, M., C. Cinti, G. Russo, A. Russo, and A. Giordano. 2003. pRb2/p130-E2F4/5-HDAC1-SUV39H1-p300 and pRb2/p130-E2F4/5-HDAC1-SUV39H1-DNMT1 multimolecular complexes mediate the transcription of estrogen receptor-alpha in breast cancer. Oncogene 22:3511-3517. [DOI] [PubMed] [Google Scholar]
- 27.Martienssen, R., Z. Lippman, B. May, M. Ronemus, and M. Vaughn. 2004. Transposons, tandem repeats, and the silencing of imprinted genes. Cold Spring Harbor Symp. Quant. Biol. 69:371-379. [DOI] [PubMed] [Google Scholar]
- 28.Martin, C. C., L. Laforest, M. A. Akimenko, and M. Ekker. 1999. A role for DNA methylation in gastrulation and somite patterning. Dev. Biol. 206:189-205. [DOI] [PubMed] [Google Scholar]
- 29.McGhee, J. D., and G. D. Ginder. 1979. Specific DNA methylation sites in the vicinity of the chicken beta-globin genes. Nature 280:419-420. [DOI] [PubMed] [Google Scholar]
- 30.Mhanni, A. A., J. A. Yoder, C. Dubesky, and R. A. McGowan. 2001. Cloning and sequence analysis of a zebrafish cDNA encoding DNA (cytosine-5)-methyltransferase-1. Genesis 30:213-219. [DOI] [PubMed] [Google Scholar]
- 31.Morgan, H. D., F. Santos, K. Green, W. Dean, and W. Reik. 2005. Epigenetic reprogramming in mammals. Hum. Mol. Genet. 14:R47-R58. [DOI] [PubMed] [Google Scholar]
- 32.Mori, H., Y. Miyazaki, T. Morita, H. Nitta, and M. Mishina. 1994. Different spatio-temporal expressions of three otx homeoprotein transcripts during zebrafish embryogenesis. Brain Res. Mol. Brain Res. 27:221-231. [DOI] [PubMed] [Google Scholar]
- 33.Mudumana, S. P., H. Wan, M. Singh, V. Korzh, and Z. Gong. 2004. Expression analyses of zebrafish transferrin, ifabp, and elastaseB mRNAs as differentiation markers for the three major endodermal organs: liver, intestine, and exocrine pancreas. Dev. Dyn. 230:165-173. [DOI] [PubMed] [Google Scholar]
- 34.Nadauld, L. D., I. T. Sandoval, S. Chidester, H. J. Yost, and D. A. Jones. 2004. Adenomatous polyposis coli control of retinoic acid biosynthesis is critical for zebrafish intestinal development and differentiation. J. Biol. Chem. 279:51581-51589. [DOI] [PubMed] [Google Scholar]
- 35.Odenthal, J., and C. Nusslein-Volhard. 1998. fork head domain genes in zebrafish. Dev. Genes Evol. 208:245-258. [DOI] [PubMed] [Google Scholar]
- 36.Okano, M., D. W. Bell, D. A. Haber, and E. Li. 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99:247-257. [DOI] [PubMed] [Google Scholar]
- 37.Pack, M., L. Solnica-Krezel, J. Malicki, S. C. Neuhauss, A. F. Schier, D. L. Stemple, W. Driever, and M. C. Fishman. 1996. Mutations affecting development of zebrafish digestive organs. Development 123:321-328. [DOI] [PubMed] [Google Scholar]
- 38.Peifer, M., and P. Polakis. 2000. Wnt signaling in oncogenesis and embryogenesis—a look outside the nucleus. Science 287:1606-1609. [DOI] [PubMed] [Google Scholar]
- 39.Peters, A. H., D. O'Carroll, H. Scherthan, K. Mechtler, S. Sauer, C. Schofer, K. Weipoltshammer, M. Pagani, M. Lachner, A. Kohlmaier, S. Opravil, M. Doyle, M. Sibilia, and T. Jenuwein. 2001. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107:323-337. [DOI] [PubMed] [Google Scholar]
- 40.Razin, A., and H. Cedar. 1991. DNA methylation and gene expression. Microbiol. Rev. 55:451-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rea, S., F. Eisenhaber, D. O'Carroll, B. D. Strahl, Z. W. Sun, M. Schmid, S. Opravil, K. Mechtler, C. P. Ponting, C. D. Allis, and T. Jenuwein. 2000. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406:593-599. [DOI] [PubMed] [Google Scholar]
- 42.Robertson, K. D. 2005. DNA methylation and human disease. Nat. Rev. Genet. 6:597-610. [DOI] [PubMed] [Google Scholar]
- 43.Shimoda, N., K. Yamakoshi, A. Miyake, and H. Takeda. 2005. Identification of a gene required for de novo DNA methylation of the zebrafish no tail gene. Dev. Dyn. 233:1509-1516. [DOI] [PubMed] [Google Scholar]
- 44.Song, F., J. F. Smith, M. T. Kimura, A. D. Morrow, T. Matsuyama, H. Nagase, and W. A. Held. 2005. Association of tissue-specific differentially methylated regions (TDMs) with differential gene expression. Proc. Natl. Acad. Sci. USA 102:3336-3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Song, L., S. R. James, L. Kazim, and A. R. Karpf. 2005. Specific method for the determination of genomic DNA methylation by liquid chromatography-electrospray ionization tandem mass spectrometry. Anal. Chem. 77:504-510. [DOI] [PubMed] [Google Scholar]
- 46.Soppe, W. J., Z. Jasencakova, A. Houben, T. Kakutani, A. Meister, M. S. Huang, S. E. Jacobsen, I. Schubert, and P. F. Fransz. 2002. DNA methylation controls histone H3 lysine 9 methylation and heterochromatin assembly in Arabidopsis. EMBO J. 21:6549-6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stancheva, I., C. Hensey, and R. R. Meehan. 2001. Loss of the maintenance methyltransferase, xDnmt1, induces apoptosis in Xenopus embryos. EMBO J. 20:1963-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stancheva, I., and R. R. Meehan. 2000. Transient depletion of xDnmt1 leads to premature gene activation in Xenopus embryos. Genes Dev. 14:313-327. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 49.Stenkamp, D. L., L. L. Cunningham, P. A. Raymond, and F. Gonzalez-Fernandez. 1998. Novel expression pattern of interphotoreceptor retinoid-binding protein (IRBP) in the adult and developing zebrafish retina and RPE. Mol. Vis. 4:26. [PubMed] [Google Scholar]
- 50.Stern, H. M., and L. I. Zon. 2003. Cancer genetics and drug discovery in the zebrafish. Nat. Rev. Cancer 3:533-539. [DOI] [PubMed] [Google Scholar]
- 51.Tamaru, H., and E. U. Selker. 2001. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature 414:277-283. [DOI] [PubMed] [Google Scholar]
- 52.Tamaru, H., X. Zhang, D. McMillen, P. B. Singh, J. Nakayama, S. I. Grewal, C. D. Allis, X. Cheng, and E. U. Selker. 2003. Trimethylated lysine 9 of histone H3 is a mark for DNA methylation in Neurospora crassa. Nat. Genet. 34:75-79. [DOI] [PubMed] [Google Scholar]
- 53.Thisse, B., and C. Thisse. 2004. Fast release clones: a high throughput expression analysis. ZFIN direct data submission. The Zebrafish Information Network, Eugene, Oreg. [Online.] http://zfin.org/cgi-bin/webdriver?MIval=aa-pubview2.apg&OID=ZDB-PUB-040907-1.
- 54.Torbenson, M., J. H. Lee, M. Choti, W. Gage, S. C. Abraham, E. Montgomery, J. Boitnott, and T. T. Wu. 2002. Hepatic adenomas: analysis of sex steroid receptor status and the Wnt signaling pathway. Mod. Pathol. 15:189-196. [DOI] [PubMed] [Google Scholar]
- 55.van der Ploeg, L. H., and R. A. Flavell. 1980. DNA methylation in the human gamma delta beta-globin locus in erythroid and nonerythroid tissues. Cell 19:947-958. [DOI] [PubMed] [Google Scholar]
- 56.Walsh, C. P., and T. H. Bestor. 1999. Cytosine methylation and mammalian development. Genes Dev. 13:26-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Warnecke, P. M., and S. J. Clark. 1999. DNA methylation profile of the mouse skeletal α-actin promoter during development and differentiation. Mol. Cell. Biol. 19:164-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yee, N. S., K. Lorent, and M. Pack. 2005. Exocrine pancreas development in zebrafish. Dev. Biol. 284:84-101. [DOI] [PubMed] [Google Scholar]
- 59.Yisraeli, J. A. M. S. 1984. Gene methylation patterns and expression, p. 353-378. In A. Razin, H. Cedar, and A. D. Riggs (ed.), DNA methylation: biochemistry and biological significance. Springer, New York, N.Y.
- 60.Zagris, N., and T. Podimatas. 1994. 5-Azacytidine changes gene expression and causes developmental arrest of early chick embryo. Int. J. Dev. Biol. 38:741-744. [PubMed] [Google Scholar]