Abstract
Eukaryotic cilia are assembled via intraflagellar transport (IFT) in which large protein particles are motored along ciliary microtubules. The IFT particles are composed of at least 17 polypeptides that are thought to contain binding sites for various cargos that need to be transported from their site of synthesis in the cell body to the site of assembly in the cilium. We show here that the IFT20 subunit of the particle is localized to the Golgi complex in addition to the basal body and cilia where all previous IFT particle proteins had been found. In living cells, fluorescently tagged IFT20 is highly dynamic and moves between the Golgi complex and the cilium as well as along ciliary microtubules. Strong knock down of IFT20 in mammalian cells blocks ciliary assembly but does not affect Golgi structure. Moderate knockdown does not block cilia assembly but reduces the amount of polycystin-2 that is localized to the cilia. This work suggests that IFT20 functions in the delivery of ciliary membrane proteins from the Golgi complex to the cilium.
INTRODUCTION
Cilia are usually thought of as motile organelles, but many eukaryotes including vertebrates make wide use of cilia for sensory perception. For example, in the vertebrate retina, the photosensitive outer segments of photoreceptor rod and cone cells are developmentally derived from cilia and the first steps of visual cascade occur within the cilium. Likewise, in the nose, olfactory receptors responsible for detecting odors are localized in cilia that project from the cells of the olfactory epithelium. In addition to these two classic sensory cilia, most vertebrate cells have a solitary nonmotile primary cilium projecting from their surfaces (Wheatley, 1995), which also are thought to be sensory organelles (Pazour and Witman, 2003). Defects in primary cilia lead to developmental defects (Nonaka et al., 1998; Marszalek et al., 1999; Takeda et al., 1999; Murcia et al., 2000), polycystic kidney disease (Moyer et al., 1994; Pazour et al., 2000; Lin et al., 2003), pancreatic and liver cysts, hydrocephaly, and skeletal abnormalities (Moyer et al., 1994), illustrating the importance that these organelles play in vertebrates.
It is likely that primary cilia use receptors localized in their membranes to detect extracellular signals. The ciliary membrane is continuous with the plasma membrane of the cell, but appears to be a separate domain with a unique complement of proteins localized to it (Bloodgood, 1990). A number of transmembrane receptors and channels have been localized to the mammalian primary cilium, including the SSTR3 isoform of the somatostatin receptor (Handel et al., 1999), the 5HT6 isoform of the serotonin receptor (Brailov et al., 2000), smoothened, a transmembrane receptor in the hedgehog pathway (Corbit et al., 2005), the PDGFRα isoform of the platelet-derived growth factor receptor (Schneider et al., 2005), and the polycystins, products of the human autosomal dominant polycystic kidney disease genes (Pazour et al., 2002b; Yoder et al., 2002). Analysis of the ciliary membrane has lagged behind the characterization of the ciliary axoneme despite the fact that this membrane is vitally important for the sensory functions carried out by cilia. Work in several systems has firmly established that the axonemal cytoskeleton of eukaryotic cilia and flagella is assembled via a process called intraflagellar transport (IFT; reviewed in Rosenbaum and Witman, 2002; Scholey, 2003), but the role of IFT in movement of membrane proteins is not well characterized. During IFT, large protein complexes are transported along the ciliary microtubules under the ciliary membrane. These large protein complexes, called IFT particles, are composed of at least 17 polypeptides organized in two complexes named A and B (Piperno and Mead, 1997; Cole et al., 1998). Electronmicroscopic analysis of IFT particles in Chlamydomonas flagella showed that they are localized close to both the microtubule axoneme and the flagellar membrane (Kozminski et al., 1993; Pazour et al., 1998) and probably interact with both structures. It is thought that the association with the microtubules is via molecular motors, but the nature of the connection to the flagellar membrane is not known because none of the known IFT particle proteins have any predicted transmembrane domains (Cole, 2003). In Caenorhabditis elegans, movement of membrane channels has been observed in cilia, and the rates are comparable to those of IFT, suggesting that IFT transports ciliary proteins within the ciliary membrane (Qin et al., 2005), and in Chlamydomonas, movement of a membrane-associated kinase into the cilium requires IFT (Pan and Snell, 2003).
All of the IFT proteins characterized to date have been found localized to cilia and to the peri-basal body region within the cell body (Rosenbaum and Witman, 2002). It is thought that the peri-basal body-localized protein represents a cytoplasmic pool from which the IFT proteins organize into particles before moving into the cilium. We here describe the localization of IFT20 to the Golgi complex in mammalian cells. If the Golgi complex is separated from the cilium, then a thin thread of IFT20 is often seen leading from the Golgi stack to the base of the cilium. We further demonstrate that GFP-tagged IFT20 is actively transported in the cell body and along the cilium in mammalian cells, which is the first direct demonstration of IFT in mammals. Using RNA interference (RNAi), we reduced the level of IFT20 in ciliated mammalian cells. Strong reduction of IFT20 levels prevents ciliary assembly, whereas weaker reduction does not block ciliary assembly but reduces the amount of polycystin-2 that gets localized to the ciliary membrane. This suggests that IFT20 plays a role in trafficking of ciliary membrane proteins into the cilium.
MATERIALS AND METHODS
In Vivo Expression of RNAi
The human H1 promoter was amplified from genomic DNA using the primers described in Brummelkamp et al. (2002) and used to replace the CMV promoter, GFP coding region, and SV40 polyadenylation signal of pEGFP-N1 (Clontech, Palo Alto, CA). This plasmid, called pGP676.13, is essentially equivalent to pSUPER (Brummelkamp et al., 2002) except that it carries the neomycin resistance gene.
Complementary oligonucleotides corresponding to the coding region of human IFT20 (gatccccGGAAGAGTGCAAAGACTTTatcaagagtAAAGTCTTTGCACTCTTCCtttttggaaa, agcttttccaaaaaGGAAGAGTGCAAAGACTTTactcttgatAAAGTCTTTGCACTCTTCCggg), mouse IFT20 (gatccccGGAGGAGTGCAAGGACTTTatcaagagtAAAGTCCTTGCACTCCTCCtttttggaaa, agcttttccaaaaaGGAGGAGTGCAAGGACTTTactcttgatAAAGTCCTTGCACTCCTCCggg 3′), rat IFT20 (gatccccGGAAGAGTGCAAGGACTTTatcaagagtAAAGTCCTTGCACTCTTCCtttttggaaa, agcttttccaaaaaGGAAGAGTGCAAGGACTTTactcttgatAAAGTCCTTGCACTCTTCCggg), and rat IFT88 (gatccccCCAACGACCTGGAGATTAAatcaagagtTTAATCTCCAGGTCGTTGGtttttggaaa, agcttttccaaaaaCCAACGACCTGGAGATTAAactcttgatTTAATCTCCAGGTCGTTGGggg) were annealed and cloned into BglII, HindIII digested pGP676.13 to produce pGP677.2, pGP678.12, pJAF43.1, and pJAF135.45, respectively.
Tagged Proteins
To construct a GFP-tagged IFT20, we PCR amplified the open reading frame of IFT20 (accession number AAA81518) from EST clone AA023464 and cloned the product into pEGFP-N1 to produce pJAF2.13. This fuses GFP to the C-terminal end of IFT20. GFP and GST-tagged IFT20 was constructed by amplifying the GST open reading frame from pGEX-6p1 (Amersham Pharmacia Biotechnology, Piscataway, NJ), and cloning this between the IFT20 and GFP open reading frames of pJAF2.13 to produce pJAF25.7. Flag-tagged IFT20 was constructed in p3XFLAG-myc-CMV-26 (Sigma, St. Louis, MO).
Mammalian Cell Culture
IMCD3, NRK, and hTERT-RPE cells (Clontech) were grown in 45% DMEM (high glucose for IMCD3 and NRK, low glucose for hTERT-RPE), 45% F12, 10% fetal calf serum, with penicillin and streptomycin at 37°C in 5% CO2. LLC-PK1 cells were cultured similarly except that they were grown in 90% DMEM (high glucose) rather than a DMEM/F12 mix.
Cells were transfected using Lipofectamine Plus reagent (Invitrogen, Carlsbad, CA) or by electroporation (Bio-Rad, Richmond, CA). Stable cell lines were selected by supplementing the medium with 400 μg/ml G418 (Sigma) or 1 μg/ml puromycin (Sigma).
Immunofluorescence Microscopy
Cells for immunofluorescence microscopy were grown on acid-washed glass coverslips. The cells were either fixed for 30 min in 2% paraformaldehyde in PHEM (0.05 M Pipes, 0.025 M HEPES, 0.01 M EGTA, 0.01 M MgCl2, pH 7.2), followed by a 2-min extraction with 0.1% Triton X-100 in PHEM, or pre-extracted with PHEM containing 0.1% Triton X-100 for 30 s followed by 10 min in cold methanol. After two brief washes in phosphate-buffered saline (PBS), the cells were blocked with 1% bovine serum albumin (BSA) in TBST (0.01 M Tris, pH 7.5, 0.166 M NaCl, 0.05% Tween 20) for 1 h and then incubated with the primary antibodies either overnight at 4°C or for 2 h at room temperature. The cells were then washed four times with 1% BSA/TBST over ∼30 min. The cells were then incubated with 1:2000 dilutions of Alexa 350–, 488–, 594–, 633–, or 680–conjugated anti-mouse IgG or anti-rabbit IgG (Molecular Probes, Eugene, OR) secondary antibodies for 1 h and washed four times with 1% BSA/TBST over ∼30 min followed by a brief wash with PBS. If the cells were being labeled with a lectin or streptavidin, this was added with the secondary antibody. The cells were then mounted with ProLong Antifade (Molecular Probes) and visualized by fluorescence microscopy.
Primary antibodies used included anti-tubulins (611β1, B-5–1-2, GTU-88, Sigma), anti-Flag (Sigma), anti-golgin-97 (CDF4, Molecular Probes), anti-TGN-38 (2F7.1; Affinity Bioreagents, Golden, CO), anti-golgin-96/GM130 (gift of M. Fitzler), anti-giantin (gift of M. Fitzler), anti-protein disulfide isomerase (RL77, Affinity Bioreagents), anti-MmIFT20, anti-MmIFT52, anti-MmIFT57, anti-MmIFT88 (Pazour et al., 2002a), anti-MmIFT140 (gift of B. Walker), and scleroderma patient sera 5051 (gift of S. Doxsey). Antibodies to polycystin-2 were generated by injecting maltose fusion proteins containing portions of mouse polycystin-2 equivalent to the B9 and C2 fragments (Cai et al., 1999). Alexa 488–conjugated Helix pomatia agglutinin was from Molecular Probes.
Images were acquired by an Orca ER camera on a Zeiss Axiovert 200M microscope equipped with a Zeiss 100× plan-Apochromat 1.4 NA objective (Thornwood, NY). Images were captured by Openlab (Improvision, Lexington, MA) and adjusted for contrast in Adobe Photoshop (San Jose, CA). If comparisons are to be made between images, the photos were taken with identical conditions and manipulated equally. For the quantification of polycystin-2 in the cilia, the length, area, and average fluorescence intensity of the cilia were measured using the measurement tools of Openlab. Data were subjected to one-way analysis of variance, followed by post hoc analysis with a Bonferroni Dunn test (SuperANOVA, Abacus Concepts, Berkley, CA). The amount of IFT20 remaining at the centrosomes was similarly quantitated from slides stained with antibodies to IFT20 and centrosomal markers.
Live Cell Imaging
LLC-PK1 cells expressing IFT20-GFP (pJAF2.13) were seeded in glass-bottom dishes. After the cells had reached ∼50% confluence the medium was changed to a HEPES-containing, serum-free, phenol red–free version of the growth medium. After at least 48 h in this medium, the cells were visualized with the Zeiss Axoivert as described above using a stage and objective heater to maintain the cells at 37°C. Cells with green cilia were identified and then photographed 34 times with 0.2 s. exposures at 0.75-s intervals. The images were adjusted for contrast and assembled into a movie in Metamorph (Universal Imaging, West Chester, PA). Kymographs were produced in Metamorph using a 10-pixel-wide selection.
To visualize movement of IFT20 between the cell body and ciliary compartments, IMCD3 cells expressing IFT20-GFP (pJAF2.13) were grown as described above and imaged using a custom-built high-speed microscope and camera system (Rizzuto et al., 1998). Forty Z-stacks (5 planes per stack, 200 nm apart, 10-ms exposure) were acquired at 150-ms intervals. The 3-D data were deconvolved (Carrington et al., 1995), projected to 2-D by summing the five planes and then converted to a movie displayed at six frames per second, which is close to real time (6.6 frames per second).
Protein Analysis
IMCD3 cells that had been transfected with pJAF2.13 (MmIFT20-GFP) or pJAF25.7 (MmIFT20-GFP-GST) were lysed in HMDEK (Cole et al., 1998) containing 1% Triton X-100. After passage through a 22-gauge needle, the extract was clarified by 18,000 × g centrifugation, and the supernatant was incubated with glutathione-conjugated Sepharose (Amersham Pharmacia Biotechnology) for 30 min and washed four times with extraction buffer, and the bound proteins eluted with 100 mM glutathione. The eluted proteins were separated by SDS-PAGE and transferred to membrane for Western blot analysis as described in Pazour et al. (1998). Quantification of Western blot intensity was preformed with a Fuji Film LAS3000 imager (Tokyo, Japan).
RESULTS
IFT20 Is a Golgi-associated Protein
The IFT20 subunit of the Chlamydomonas IFT particle was previously cloned using peptide sequence derived from purified protein (GenBank AAM75748; Pazour et al., 2002a), but was not analyzed in detail. Homologues of IFT20 appear to be present in all ciliated organisms including C. elegans (GenBank AAL77186), Drosophila melanogaster (GenBank NP_724409), Xenopus laevis (GenBank AAL07274), mice (GenBank AAL99202), and humans (GenBank AAH38094), but have not been found in nonciliated organisms such as Arabidopsis or Saccharomyces, suggesting that IFT20 is specifically involved in ciliary processes. Proteins in the IFT20 family are typically around 15 kDa in size and do not share significant homology to other proteins. The IFT20 family does not contain any conserved motifs except for a coiled-coil domain that makes up the C-terminal ∼75% of the peptide. The gene encoding IFT20 appears to be single copy in both mice and humans. However, both species have two other apparently unexpressed regions in their genomes with similarity to IFT20 that are likely to be pseudogenes.
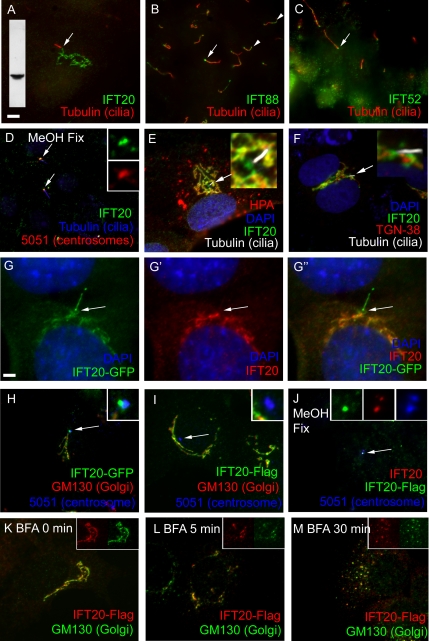
Even though the IFT particle proteins were originally purified from flagella, immunofluorescence microscopy showed that the majority of IFT172, IFT139, IFT81, IFT57 (Cole et al., 1998), and IFT52 (Deane et al., 2001) is found in the cell body at the base of the flagella with only a small amount in the flagella themselves. To determine if a similar distribution occurs in mammalian primary cilia, antibodies to the mouse homologues of IFT20, IFT88/polaris, and IFT52/ngd5 were used to stain cultured mammalian cells fixed in paraformaldehyde (Figure 1). The antibodies to IFT88 and IFT52 (Figure 1, B and C) labeled the bases of the primary cilia and showed weak punctate staining of the cilia similar to what has been observed in Chlamydomonas. Unlike what was observed in Chlamydomonas, these antibodies often labeled the tips of the primary cilia as well. In contrast, cells fixed the same way and stained with IFT20 antibodies showed label on a much larger reticulated structure that appeared similar to the Golgi complex (Figure 1A). In some instances the major focus of IFT20 staining was some distance from the cilium. When this occurred, a thread of staining could usually be seen leading from the major focus of staining to, but not quite touching, the base of the cilium. Cells fixed with paraformaldehyde did not show labeling of the cilia or ciliary basal bodies but this is likely a fixation artifact as the ciliary basal body pool is stained in cells fixed with methanol (Figure 1D) and GFP-tagged IFT20 is found in both of these pools (Figure 1, G and H).
Figure 1.
Distribution of IFT20 in cells. (A–C) Distribution of IFT particle proteins in mammalian cells. Mouse IMCD3 cells were paraformaldehyde fixed and stained with antibodies to MmIFT20, MmIFT88/polaris, and MmIFT52/ngd5 (green) along with an antibody to acetylated tubulin (red) to mark the position of the primary cilium. Basal body regions are marked with arrows, and distal tips are marked with arrowheads. Inset in A shows a Western blot of IMCD3 protein extract probed with the IFT20 antibody used to label this cell. Size bar is 5 μm and is relevant for all images except G. (D) Mouse IMCD3 cells pre-extracted with Triton X-100, fixed with methanol, and stained with antibodies to IFT20 (green), acetylated tubulin (blue), and autoimmune sera 5051 (red), which labels centrosomes. Insets are 4× enlargements of the green and red channels near the basal body region of the upper cilium. Note that the pre-extraction with Triton X-100 followed by methanol fix removes most of the Golgi-associated IFT20 but reveals the basal body pool. (E) RPE cell stained with anti-IFT20 (green), anti-acetylated tubulin (white), and Helix pomatia agglutinin (red). Note the extensive colocalization of IFT20 and HPA (yellow-orange) and the patch of green at the base of the cilium where HPA is not found. HPA is also found in cytoplasmic vesicles that are not stained with IFT20 antibodies. (F) NRK cells stained with anti-IFT20 and anti-RnTGN-38. Note that this Golgi marker labeled structures very near to IFT20, but there is not extensive overlap. Insets in E and F show 4× enlargements of the regions around the cilia. (G–G″) An IMCD3 cell expressing GFP-tagged IFT20 (G, green) was fixed and stained with an antibody against IFT20 (G′, red). The overlay (G″) shows that the basal body (arrow) and cilia label with GFP but not with IFT20 antibodies, whereas the internal membranes label with both. Size bar, 2.5 μm. (H) IMCD3 cells expressing IFT20-GFP (green) were fixed with paraformaldehyde and stained with the GM130 Golgi marker (red) and the 5051 centrosomal marker (blue). Inset is a 4× enlargement of the centrosome. (I) IMCD3 cells expressing IFT20-Flag (green) were fixed with paraformaldehyde and stained with the GM130 Golgi marker (red) and the 5051 centrosomal marker (blue). Inset is a 4× enlargement of the centrosome. (J) IMCD3 cells expressing IFT20-Flag were pre-extracted with Triton X-100, fixed with methanol, and stained with antibodies to IFT20 (red), Flag (green), and 5051 (centrosomal marker, blue). Inset is a 4× enlargement of the green, red, and blue channels of centrosome. (K–M) IMCD3 cells expressing IFT20-Flag were exposed to brefeldin A for 0, 5, and 30 min, fixed with paraformaldehyde, and stained with antibodies to Flag (red) and GM130 (Golgi, green). Insets are the red and green channels from a portion of the image to illustrate extensive colocalization of the two markers. Similar results were obtained with endogenous IFT20 and IFT20-GFP (unpublished data).
To test the idea that IFT20 is a Golgi-associated protein, cells were labeled with IFT20 and the Golgi markers H. pomatia agglutinin (HPA), giantin, golgin-96/GM130, golgin-97, and trans-Golgi network-38 (TGN-38), as well as markers for endoplasmic reticulum (ER) and mitochondria. All of the Golgi markers showed roughly similar labeling patterns as IFT20 (Figure 1, E and F), whereas no significant colocalization was seen between the mitochondrial or ER markers and IFT20 (unpublished data), indicating that IFT20 is associated with the Golgi complex. Of the Golgi markers, the most extensive colocalization with IFT20 was seen with HPA (Figure 1E), giantin, and golgin-96/GM130 (unpublished data). Golgin-97 and TGN-38 labeled structures near, but not overlapping those labeled by IFT20 (Figure 1F). HPA recognizes terminal α-N-acetylgalactosaminyl residues and is a marker for the cis-Golgi cisternae (Sharon, 1983; Molecular Probes data sheet). Golgin-96/GM130 is thought to be a marker for the cis-Golgi cisternae (Nakamura et al., 1995), whereas giantin is thought to label the cis and medial cisternae (Linstedt and Hauri, 1993; M. Fritzler, personal communication). Golgin-97 and TGN-38 are markers for the trans-Golgi network (M. Fritzler, personal communication; Luzio et al., 1990). This pattern of colocalization suggests that IFT20 is associated with the cis and medial cisternae of the Golgi complex but not extensively with the trans-Golgi network. Interestingly, we often observed lobes of IFT20 staining near the base of the cilium that did not label with the cis or medial Golgi markers (green at the base of the cilium in Figure 1E). These lobes do not appear to be part of the TGN either, because we did not observe colocalization of TGN markers and IFT20 at the base of the cilia (Figure 1F). It is not known whether these lobes represent a tightly packed group of small vesicles or a larger structure.
To rule out the possibility that the IFT20 staining of the Golgi complex is an antibody artifact, GFP-tagged and Flag epitope–tagged IFT20 constructs were prepared and transfected into mouse kidney cells. The majority of the IFT20-GFP and IFT20-Flag localized to Golgi complex (Figure 1, G, H, and I) as had been observed with native protein detected by our IFT20 antibody (Figure 1, A, E, and F). In highly expressing cells, both tagged proteins also showed faint diffuse distribution in the cytoplasm, probably as a result of the overexpression. In addition, IFT20-GFP localized to the basal body and to cilia (Figure 1, G and H). When the cells were fixed with paraformaldehyde, the IFT20-GFP associated with the Golgi complex stained with IFT20 antibodies (Figure 1G). However, the IFT20-GFP around the basal body and in the cilium did not stain with the antibody (Figure 1G″). This suggests that the IFT20 epitope is blocked at the basal body and cilium when the cells are fixed by paraformaldehyde, probably as a result of being sequestered in a large complex. Consistent with this idea, cells expressing IFT20-Flag stained with Flag antibodies do not show labeling of basal bodies and cilia when fixed by paraformaldehyde (Figure 1I) but do show staining of the basal body region when fixed by methanol (Figure 1J) and also show staining of cilia when the cells express large amounts of IFT20-Flag (unpublished data).
To further confirm that IFT20 is a Golgi-associated protein, we examined the fate of IFT20 upon treatment with brefeldin-A. Brefeldin-A rapidly blocks traffic from the ER to the Golgi apparatus and causes the Golgi complex to fragment into small punctate structures within 30 min of application (Klausner et al., 1992). Within 5 min of addition of brefeldin-A (Figure 1L), IFT20 started to disperse from it normal compact peri-nuclear structure (Figure 1K). By 30 min, the dispersal was complete, and all of the IFT20 was now found in small puncta distributed throughout the cytoplasm (Figure 1M). Importantly, the cis-Golgi marker, golgin-96/GM130, showed the same kinetics of dispersion and showed extensive colocalization with IFT20 providing further evidence that IFT20 is associated with the Golgi complex. In Gonium, a green alga (Haller and Fabry, 1998), and in sea urchin embryos (Stephens, 2001), brefeldin-A inhibits reciliation of cells that have been experimentally de-ciliated but does not affect pre-existing cilia. We are unable to experimentally de-ciliate our cells to assay brefeldin-A effects on ciliary assembly but treatment with brefeldin-A for up to 16 h did not appear to affect cilia that were already formed (unpublished data).
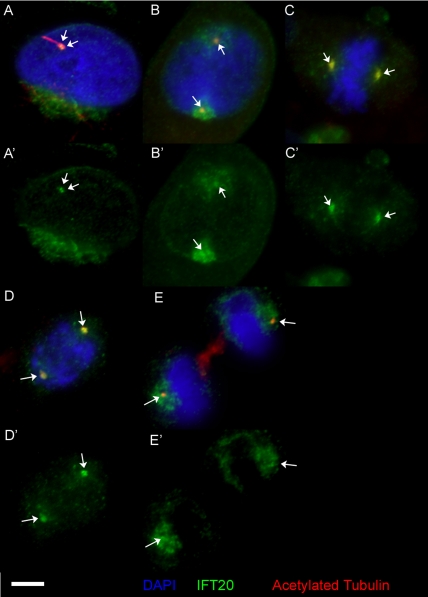
IFT20 Remains Associated with the Centrosomes during the Cell Cycle
To examine the fate of IFT20 during the cell cycle, actively growing cells were fixed with methanol and labeled with antibodies to IFT20, acetylated tubulin, the 5051 scleroderma sera, and DAPI (Figure 2). During interphase, IFT20 antibodies stain one or two small spots that colocalize with staining by acetylated tubulin antibodies (Figure 2A). These spots also label with scleroderma patient serum 5051, which marks these spots as centrosomes (unpublished data). The IFT20 antibody also stains the Golgi complex, although the preservation of this structure is not good under these fix conditions. When the cell enters mitosis, the IFT20 in the Golgi is dispersed into smaller structures that coalesce around the centrosomes. The centrosomes label with IFT20 throughout the cell cycle (Figure 2, B–E).
Figure 2.
IFT20 associates with centrosomes during the cell cycle. (A–E) Actively growing mammalian RPE cells were fixed with methanol and labeled with anti-IFT20 (green, shown alone in A′–E′) and anti-acetylated tubulin (red) antibodies along with DAPI (blue). The position of the centrioles/basal bodies (as determined by 5051 staining, unpublished data) is marked with arrows. Note that a bright focus of IFT20 is associated with the centrosomes at all stages of the cell cycle. Size bar is 5 μm and is relevant for all images.
IFT20 Interacts with IFT Complex B Proteins
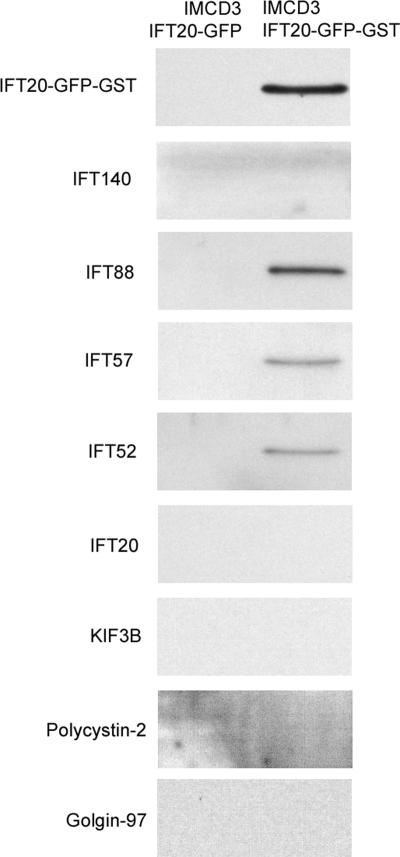
Chlamydomonas IFT20 was purified from the flagellum as part of a large complex of IFT proteins (Cole et al., 1998). However, the observation that IFT20 is found in a different cellular compartment than most IFT particle proteins in mammalian cells called into question the identification of IFT20 as a subunit of the IFT particle. To more directly test the interaction between IFT20 and other IFT particle proteins, we generated an expression construct of IFT20 fused to GFP and glutathione S-transferase (GST) and transfected this into cultured mouse kidney cells. IFT20 fused to GFP was used as a control. Extracts of these cells were incubated with glutathione beads, and the bound proteins eluted with glutathione. IFT52, IFT57, and IFT88 were eluted from the experimental beads but not the control beads, whereas only very small amounts (if any) of IFT140 bound under these conditions (Figure 3). Previous work showed that IFT20 interacts with the KIF3B subunit of the anterograde IFT motor in yeast two-hybrid analysis and that IFT20 and KIF3B can be coimmunoprecipitated with antibodies to other IFT complex B subunits (Baker et al., 2003). We did not observe any KIF3B in our GST pulldowns, nor did we detect the ciliary membrane protein polycystin-2 or the Golgi- associated protein golgin-97, indicating that the binding conditions are stringent and that the observed interaction between IFT20 and the complex B proteins is specific. In Chlamydomonas, the IFT particle purifies as two subcomplexes called A and B, and it was observed that immunoprecipitation using a complex B antibody preferentially immunoprecipitates complex B proteins relative to complex A proteins, and vice versa (Cole et al., 1998). Our results using the complex B subunit IFT20 as bait are consistent with this as IFT52, IFT57, and IFT88 are part of complex B, whereas IFT140 is a subunit of complex A. No endogenous IFT20 bound to the beads, suggesting that this subunit is not present in more than one copy per complex. These results confirm that mouse IFT20 is a subunit of IFT complex B.
Figure 3.
Mammalian IFT20 associates with complex B proteins. Lysates from mouse IMCD3 cells expressing either IFT20-GFP (pJAF2.13) or IFT20-GST-GFP (pJAF25.7) were incubated with glutathione beads and washed, and the bound proteins were eluted with glutathione. The eluted proteins were analyzed by Western blotting with antibodies to the proteins marked on the left side of the images. In this image, IFT20-GFP-GST was detected with the IFT20 antibody and was confirmed with a GFP antibody (unpublished data).
IFT20 Trafficking in the Cell Body and Cilium
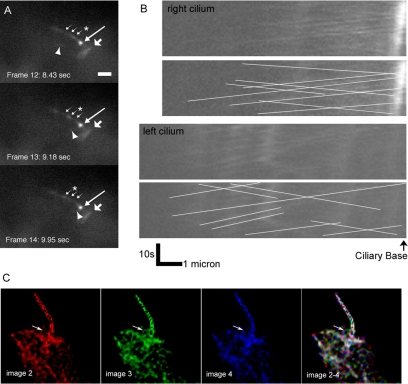
To observe the dynamics of IFT20 in live cells, IMCD3 and LLC-PK1 kidney cell lines expressing IFT20-GFP were generated and examined by fluorescence microscopy similar to technique widely used to study IFT in the sensory cilia of C. elegans (Orozco et al., 1999). In both cell lines, IFT20-GFP was localized to the Golgi complex, the region surrounding the basal body at the base of the cilia, and to the cilia themselves. The IFT20-GFP was brighter in the IMCD3 cells, but LLC-PK1 cells are better for imaging cilia in a single plane because they tend to assemble longer cilia that often lay horizontal on the surface of the cell, whereas the cilia on IMCD3 cells usually project vertically from surface of the cells. In initial experiments, time series of single-plane fluorescence images were captured of IFT20-GFP in LLC-PK1 cells. These images were adjusted for contrast and assembled into a movie (Online Supplementary Data, LLC_IFT20-GFP.mov). Large numbers of particles could be observed moving anterograde from the cell body to the flagellar tip. Retrograde particles were observed less frequently. In Chlamydomonas, the retrograde particles appear smaller (Kozminski et al., 1993), and so it is possible that most of the retrograde particles in the mammalian cilia are below our detection limit. The outward-moving particles traveled at ∼0.6 μm/s, whereas the inward-moving particles traveled at ∼0.7 μm/s. These speeds are slower than in Chlamydomonas (2.5 μm/s anterograde and 4 μm/s retrograde; Kozminski et al., 1993) but similar to what was observed in the middle segment of C. elegans sensory cilia (0.52–0.72 μm/s anterograde, 1.09–1.17 μm/s retrograde; Snow et al., 2004).
Examination of single-plane images taken of the cell bodies indicated that the IFT20-GFP within the Golgi apparatus was quite dynamic and possibly moved between the Golgi complex and the cilium. To better examine this, a time series of rapid Z-stacks was captured of ciliated IFT20-GFP IMCD3 cells. The Z-stacks at each time point were deconvolved, reduced to a single plane by summing the individual planes, and assembled into a movie (Online Supplementary Data, IMCD3_IFT20-GFP.mov and Figure 4C). As was observed in the movies made from single-plane images, IFT20-GFP was clearly moving along the cilium. The IFT20-GFP in the Golgi complex and at the base of the cilium was also highly dynamic and moved from the cell body into the cilium. Figure 4C shows three sequential frames from the movie that were pseudocolored red, green, or blue, and the three images were combined. If the IFT20-GFP did not move during the time interval, the GFP particles would be white in the composite image. Discrete red, green, and blue particles indicate that the IFT20-GFP moved between the images. The arrow illustrates an IFT20-GFP structure that moves from the cell body and into the cilium near the beginning of the movie.
Figure 4.
IFT20 moves in cell bodies and cilia. (A and B) Time-lapse fluorescence images of cells LLC-PK1 cells expressing IFT20-GFP were taken every 0.75 s for 26 s. These were adjusted for contrast and assembled into a movie that plays at ∼5× normal speed. (A) To illustrate the two types of movements observed, three frames (12–14) are shown. The longest arrow marks the basal body. The broad arrow marks IFT20 in the Golgi complex. The three small arrows mark the position of a particle moving from the basal body to the ciliary tip (anterograde movement). The position of the particle in each frame is marked by an asterisk (*). The arrowhead marks the position of a focus of IFT20-GFP that is moving rapidly in the cytoplasm. In the movie, this focus appears near the basal body, moves out into the cell, and then returns back to the basal body. Size bar, 2 μm long. (B) Kymographs showing IFT20-GFP movement in the two cilia in the movie. The top panel of each pair shows the raw kymographs, whereas lines have been drawn on the bottom panel to mark the moving particles. Anterograde particles show upward sloping lines, whereas retrograde particles show downward sloping lines. (C) Five image Z-stacks were taken of IMCD3 cells expressing IFT20-GFP every 150 ms. The Z-stacks were deconvolved, reduced to a single plane by summing, and assembled into a movie. In C three sequential frames from the movie were pseudocolored red, green, or blue (images 2, 3, and 4), and the three images were combined (images 2–4). The discrete colors in the combined image indicate that the IFT20-GFP is highly dynamic. The arrow in image 3 points to a tubular structure moving into the cilium.
Strong Reduction of IFT20 in RPE Cells Blocks Ciliary Assembly
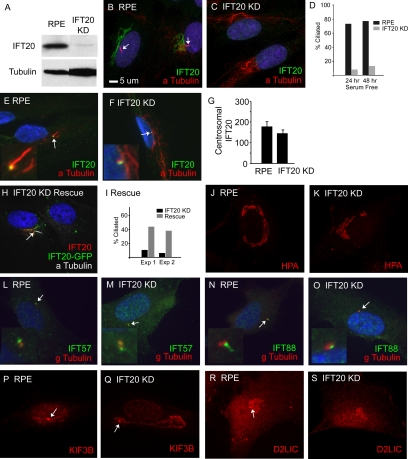
To disrupt the function of IFT20 in cells, we used the in vivo expression of a short hairpin RNA (shRNA) molecule (Brummelkamp et al., 2002) corresponding to the IFT20 message, to reduce the amount of IFT20 produced. This construct was transfected into human RPE cells, and a subclone that showed a strong reduction in IFT20 staining was selected for more analysis (Figure 5). IFT20 was almost undetectable by Western blotting (Figure 5A) or immunofluorescence (Figure 5, B and C) in the cells transfected with the IFT20 knockdown construct. The knockdown of IFT20 greatly reduced the percentage of cells that were ciliated after 24 or 48 h of serum starvation (Figure 5D). Serum starvation causes cells to enter G0, which are often more highly ciliated than mitotically active cells (Tucker and Pardee, 1979). The ciliary assembly phenotype could be rescued by transfection with a mouse IFT20-GFP expression construct (Figure 5, H and I), indicating that the lack of cilia was due to the expression of the hairpin RNA directed against the IFT20 message in these human cells. Mouse IFT20 differs from human IFT20 by one nucleotide in the target sequence. Even though the cells lack cilia and the overall level of knock down is high, the amount of IFT20 at the centrosomes is not greatly reduced in the knockdown cell line (Figure 5, E, F, and G). The structure of the Golgi apparatus did not appear to be significantly affected by reducing the level of IFT20 (Figure 5, J and K). Consistent with the relatively minor reduction of IFT20 at the centrosomes (Figure 5G), the lack of IFT20 did not prevent IFT57 or IFT88 from accumulating at the basal body (Figure 5, L–O) although small reductions as seen with pericentrin (Jurczyk et al., 2004) were not ruled out. The reduction in IFT20 levels does not appear to affect Golgi-localized kinesin-2 (Allan et al., 2002; KIF3B, Figure 5, P and Q) or dynein 2 (Grissom et al., 2002; D2LIC, Figure 5, R and S). These results indicate that IFT20 is required for ciliary assembly, but is not necessary for the integrity of the Golgi complex or the localization of the two IFT motors to the Golgi complex.
Figure 5.
Strong IFT20 knockdown blocks ciliary assembly. (A) Western blot of RPE and IFT20 KD cells probed with the IFT20 antibody. The IFT20KD cells were generated by transfection of RPE cells with pGP677.2 followed by dilution cloning to purify a cell line with a strong reduction in the amount of IFT20. Western blot with an anti-tubulin antibody as a loading control shows that more protein was loaded from the knockdown cells than the control cells. (B and C) RPE and IFT20KD cells labeled with anti-MmIFT20 (green), anti-acetylated tubulin (red), and DAPI (blue). Cilia are marked with arrows. Note the absence of IFT20 staining and lack of cilia in C. (D) The IFT20KD cells are less ciliated than control cells. Percent ciliation was determined by counting the number of cilia found on 100 cells that had been stained with anti-acetylated tubulin antibodies. (E and F) RPE and IFT20KD cells were pre-extracted with Triton X-100, methanol fixed, and then stained with antibodies to IFT20 (green), acetylated tubulin (red), and DAPI (blue). Note that IFT20 remains at the centriole/basal body (arrows) in both cell lines. Insets are 4× enlargements of the basal body regions. (G) The amount of IFT20 remaining at the centrosome was quantified on 25 cells from each population. Error bars, SEM. (H and I) Knockdown cells were electroporated with a construct that expresses mouse IFT20 tagged with GFP. This restores cilia assembly (arrow in H, graph in I). The percent of ciliated cells were determined after two separate transfections (Exp 1 and Exp 2). (J and K) RPE and IFT20KD cells stained with HPA, a lectin that labels the cis and medial cisternae of the Golgi complex. Note that the Golgi structure appears normal in the knockdown cells. (L–O) RPE and IFT20KD cells stained with anti-IFT57 (green; L and M) and anti-IFT88 (green; N and O), along with DAPI (blue) and anti-gamma tubulin (red). Insets are 4× enlargements of the basal body regions. Note that a spot of IFT57 or IFT88 is found near one of the two centrioles in both control and knockdown cells. (P–S) RPE and IFT20KD cells stained with anti-KIF3B (P and Q) and anti D2LIC (R and S). Note that the Golgi staining appears similar in knockdown and control cells. Basal bodies are marked with arrows. Size bar in B is 5 μm and applies to all images.
The localization of IFT20 to the centrosomes during the cell cycle (Figure 2) suggested that it might play a role in mitosis. Flow cytometric measurement of DNA content of cycling cells or cells blocked in G2 by nocodazole did not show any differences between control and IFT20 knockdown cells, indicating that either IFT20 is not required for the mitosis or that the amount of IFT20 that remained at the centrosomes was sufficient for any function that IFT20 may have in mitosis.
Moderate Reduction of IFT20 Levels Reduces Ciliary Polycystin-2
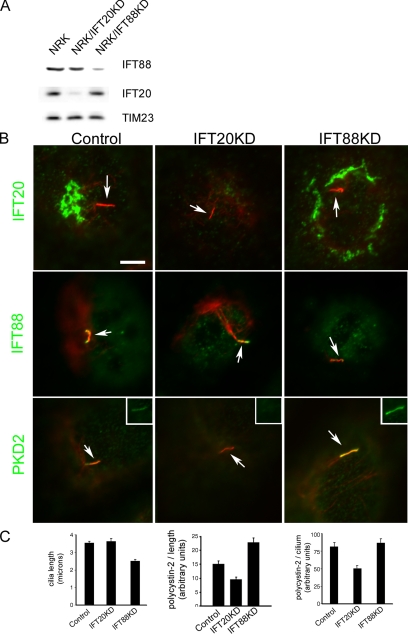
Ciliary membrane proteins are synthesized on the rough ER and processed through the Golgi complex (Bloodgood, 1990). Because IFT20 is associated with the Golgi complex, it is possible that IFT20 plays a role in the trafficking or sorting of ciliary membrane proteins. To begin to test this idea, we examined how a moderate reduction in the amounts of IFT20 or IFT88 affected the ciliary levels of polycystin-2. Polycystin-2 is a transmembrane protein in the TRP superfamily of cation channels (Mochizuki et al., 1996) and is localized to cilia (Pazour et al., 2002b). Rat NRK cells were chosen for this analysis because they are highly ciliated and have higher amounts of polycystin-2 in their cilia than IMCD3 or RPE cells. shRNA constructs targeting rat IFT20 and IFT88 were transfected into NRK cells. After stable integration and drug selection, lines showing reductions in IFT20 and IFT88 were selected for further analysis (Figure 6). The IFT20 knockdown line retained 14% of the normal amount of the IFT20, whereas the IFT88 knockdown line retained 46% of the normal amount of IFT88. Knocking down IFT20 or IFT88 did not appear to greatly affect the levels of the other protein (Figure 6A). Both cell lines retained the ability to assemble cilia (Figure 6, B and C). Average ciliary length on the IFT20 knockdown line was similar to the control cells (3.6 vs. 3.5 μm), whereas the IFT88 cells had shorter cilia (2.5 μm; Figure 6C).
Figure 6.
Moderate IFT20 knockdown reduces ciliary polycystin-2 levels. Rat kidney NRK cells lines with reduced IFT20 or IFT88 were generated by expression of shRNA constructs. (A) Western blot analysis showing the reduction of IFT20 and IFT88 in the two cell lines. TIM23 was used as a loading control. (B) Immunofluorescence images showing that cilia (arrows) are still formed in these cells (acetylated tubulin, red). IFT20 (top row) and IFT88 (middle row) are reduced in the respective knockdown line but not greatly altered in the reciprocal line. Ciliary polycystin-2 (PKD2, green) is reduced in the IFT20 knockdown line (bottom middle) compared with the control cells (bottom left) and the IFT88 knockdown line (bottom right). The insets in the bottom row show ciliary polycystin-2 staining without acetylated tubulin staining. (C) Bar graphs show average (n = 50 per condition) ciliary length, polycystin-2 per unit length, and polycystin-2 per cilium. To obtain polycystin-2 per unit length, the amount of polycystin-2 in each cilium was divided by its length. Error bars, SEM.
To determine how the reductions in IFT88 and IFT20 affected ciliary polycystin-2 levels, we quantified the amount of polycystin-2 on cilia from control, IFT20, and IFT88 knockdown cells. When examined either as total amount of polycystin-2 per cilium or as amount of polycystin-2 per micron of cilium length, the IFT20 knockdown cilia had significantly less (p < 0.001) polycystin-2 in them (Figure 6C). The cilia from IFT88 knockdown cells showed about the same amount of polycystin-2 in them as the control cells, but this was more concentrated because of the shorter length of the knockdown cilia. The observation that reduction in IFT20 also reduces the amount of polycystin-2 on cilia results support a role for IFT20 in the trafficking of polycystin-2 into cilia.
DISCUSSION
In this work, we show that the intraflagellar transport protein IFT20 is a Golgi-associated protein required for ciliary assembly and the localization of polycystin-2 to cilia. Before this study, all of the IFT-particle proteins examined had been found concentrated around the bases of cilia and, to a lesser extent, in the cilia themselves (Rosenbaum and Witman, 2002). It is thought that the proteins at the base of the cilium constitute a cytoplasmic pool from which the IFT particles organize before entering the cilium to carry their cargos to the site of assembly at the distal tip. In addition to its Golgi localization, IFT20 is also found in the peri-basal body pool at the base of the cilium along with the other IFT proteins. All the data indicates that IFT20 is an essential component of the classic IFT complex. IFT20 cosediments with other IFT particle proteins as a 17S complex when isolated from Chlamydomonas (Cole et al., 1998) and mouse (Pazour et al., 2002a) and can interact with other IFT complex B proteins in a GST-IFT20 pulldown experiment (Figure 3) and immunoprecipitation (Baker et al., 2003). Furthermore, it is transported along ciliary microtubules at rates that are consistent with IFT (Figure 4). Like other IFT-particle proteins, it also appears to be required for ciliary assembly as demonstrated by the reduced number of cilia observed on IFT20 knockdown cells (Figure 5). These cells showed a large reduction in the total amount of IFT20 (Figure 5A), but the amount that remained was concentrated at the basal bodies/centrioles such that this pool was not greatly affected (Figure 5G). The observation that ciliary assembly was affected even though the centrosomal pool was not greatly depleted indicates that IFT20 plays additional roles beyond the classic IFT system.
The localization of IFT20 to the Golgi complex suggests that the additional role of IFT20 is related to this organelle. Kinesin-2, the anterograde IFT motor, and dynein 2, the retrograde IFT motor are both localized to the Golgi complex in addition to the cilium and basal body regions. Disruption of kinesin-2’s function by expression of a dominant negative fragment of a motor subunit dramatically reduced HPA lectin staining of the Golgi cisternae, probably due to reduced ER-to-Golgi transport (Le Bot et al., 1998). The exact role of dynein-2 in Golgi function is not clear; however, injection of antibodies against this protein caused dispersion of the cisternae (Vaisberg et al., 1996). IFT20 does not appear to play a role in either of these processes as Golgi localization and structure, and HPA staining are not altered when the levels of IFT20 are strongly reduced (Figure 5, J and K). However, the amount of polycystin-2 that gets trafficked to the cilium is reduced. Polycystin-2 is a transmembrane protein that is highly abundant in the ER (Cai et al., 1999) and is processed through the Golgi complex before being inserted into the ciliary membrane.
Very little is known about the mechanism by which membrane proteins are sorted and delivered to the ciliary domain of the plasma membrane; however, the limited data available suggest that there is an active and highly directed mechanism for placement of proteins on the ciliary membrane. In algae (Ochromonas and Chlamydomonas) the secretion of the mastigonemes, which are ciliary membrane proteins, appears to be polarized and occur only at the anterior end of the cell where the cilia are found (Bouck, 1971; Nakamura et al., 1996). In Ochromonas, the mastigoneme-containing vesicles could be observed fusing with the plasma membrane just outside of the cilium (Bouck, 1971). It is thought that the fusion site is outside of the cilium because vesicles are too large to fit through the constriction or “flagellar pore” at the base of the cilium (see Rosenbaum and Witman, 2002). A similar situation has been observed with opsin in vertebrate photoreceptors (Papermaster et al., 1985). Opsin is a seven-transmembrane protein that is concentrated in the membranes of the photoreceptor outer segment, which is a modified cilium. IFT plays a role in assembly and maintenance of the outer segment (Marszalek et al., 2000; Pazour et al., 2002a), and it is likely that the mechanism of membrane protein transport into this organelle is evolutionarily conserved. In the photoreceptor cell body, opsin-containing vesicles appear to be trafficked directly from the Golgi complex to the base of the cilium where they fuse with the plasma membrane. In frogs, the opsin docking sites have a ninefold symmetry (Papermaster et al., 1985) that is similar to the distribution of the basal body pool of IFT52 in Chlamydomonas (Deane et al., 2001). Once the ciliary membrane protein–containing vesicle fuse with the plasma membrane, the ciliary membrane protein must be directed into the cilium and not allowed to diffuse away into the rest of the plasma membrane. In photoreceptors, this process is highly efficient, and opsin is not found on the inner segment membrane except in disease states. The mechanism by which this is accomplished is unknown; however, a dynein light chain (Tai et al., 1998) and a small G-protein (Deretic et al., 2005) have been shown to bind to the tail of opsin and are proposed to direct opsin-containing vesicles from the Golgi to the base of the cilium. IFT20 may be acting in a similar manner, although our inability to enrich polycystin-2 in IFT20-GST pulldowns indicates that IFT20 does not bind to polycystin-2 with high affinity and suggests that the interaction may be less direct.
In C. elegans, an AP1μ1 clathrin subunit encoded by the UNC-101 gene is required for proper localization of membrane proteins to the cilia. This protein appears to be acting at the level of sorting or packaging at the trans-Golgi network because unc-101 mutant animals mostly lacked dendritic vesicles containing ciliary membrane proteins (Dwyer et al., 2001). Mammalian epithelial cells have two AP1m isoforms, AP1m1 is thought to be involved in sorting between the trans-Golgi network and endosomes, whereas AP1m2 appears to be involved in sorting proteins for the basal lateral membrane (Folsch et al., 2001). It is not known if either of these proteins are involved in sorting polycystin-2 to cilia; however, it is unlikely that the AP1m2 subunit is critical, because LLC-PK1 cells lack this protein (Folsch et al., 2001) and still sort polycystin-2 to cilia (Geng et al., 2006). Also in C. elegans, KLP-6 kinesin, a kinesin-3 subfamily member, is required for proper localization of polycystin-2 to cilia. Animals with mutations in klp-6 show accumulations of polycystin-2 in the dendrites, at the transition zone and in some cilia, whereas other cilia show decreased amounts of polycystin-2. It is not clear at what step KLP-6 is functioning, but because the protein was not observed moving along cilia or dendrites, it was proposed that it may be a linker between polycystin-2 and microtubules that ensures the proper distribution of polycystin-2 (Peden and Barr, 2005). The role of mammalian kinesin-3 family members in localization or transport of polycystin-2 has not been examined.
One possible function of IFT20 is to mark vesicles that contain proteins destined for the ciliary membrane. IFT20 could associate with these vesicles when the proteins are sorted in the Golgi complex. After delivery of the ciliary-destined vesicles to the base of the cilium, IFT20 could initiate assembly of the remainder of the IFT particle and associated motors on the surface of the vesicle. The IFT particle could then couple fusion of the vesicle at the base of the cilium and transport of the membrane proteins into the cilium. Recently, it has been hypothesized that IFT evolved from the clathrin/COPI-coated vesicle transport system of eukaryotic cells (Jekely and Arendt, 2006). This model is based on the observation that even though components of the clathrin/COPI and IFT systems do not have any significant primary sequence similarity, many of the subunits of these systems have an unusual arrangement of N-terminal WD-40 repeats and C-terminal TPR domains that suggest an ancient common ancestor. In this model, the clathrin/COPI transport system duplicated in the proto-eukaryotic cell and evolved to transport selected proteins from the Golgi complex to a specialized plasma membrane domain. This specialized domain evolved into the modern cilium with coevolution of the duplicated clathrin/COPI system into the modern IFT system. Although this model is highly speculative, and will require substantial structural biology to back up the claims of a common ancestor between clathrin/COPI components and IFT subunits, it is plausible and intriguing. In this model, IFT20 could be playing the role of an adaptin to couple membranous cargo proteins to the particle. However, regardless of the evolutionary origin of the IFT system, the localization of IFT20 to the Golgi complex as well as to the basal body region and cilium indicates that it is in a unique position to couple the directed movement of proteins destined for the ciliary membrane through the endomembrane system, to the base of the cilium, and then into the cilium itself.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. George B. Witman for his support of this work and his critical insight; Drs. Jovenal San Agustin, Bethany Walker, Steve Doxsey and Marvin Fitzler for reagents; and Dr. Julie Jonassen for assistance with statistics. We also thank our colleagues for helpful discussion and critical comments on the manuscript. This work was supported by grants from the National Institutes of Health (GM-60992 to G.J.P.) and a Worcester Foundation for Biomedical Research Foundation Scholar Award (to G.J.P.).
Abbreviations used:
- HPA
Helix pomatia agglutinin
- IFT
intraflagellar transport
- RPE
retinal pigmented epithelium.
Footnotes
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-02-0133) on June 14, 2006.
REFERENCES
- Allan V. J., Thompson H. M., McNiven M. A. Motoring around the Golgi. Nat. Cell Biol. 2002;4:E236–E242. doi: 10.1038/ncb1002-e236. [DOI] [PubMed] [Google Scholar]
- Baker S. A., Freeman K., Luby-Phelps K., Pazour G. J., Besharse J. C. IFT20 links kinesin II with a mammalian intraflagellar transport complex that is conserved in motile flagella and sensory cilia. J. Biol. Chem. 2003;278:34211–34218. doi: 10.1074/jbc.M300156200. [DOI] [PubMed] [Google Scholar]
- Bloodgood R. A. Ciliary and Flagellar Membranes. New York: Plenum Press; 1990. [Google Scholar]
- Bouck G. B. The structure, origin, isolation and composition of the tubular mastigonemes of the Ochromonas flagellum. J. Cell Biol. 1971;50:362–384. doi: 10.1083/jcb.50.2.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brailov I., Bancila M., Brisorgueil M., Miquel M., Hamon M., Verge D. Localization of 5-HT(6) receptors at the plasma membrane of neuronal cilia in the rat brain. Brain Res. 2000;872:271–275. doi: 10.1016/s0006-8993(00)02519-1. [DOI] [PubMed] [Google Scholar]
- Brummelkamp T. R., Bernards R., Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- Cai Y., Maeda Y., Cedzich A., Torres V. E., Wu G., Hayashi T., Mochizuki T., Park J. H., Witzgall R., Somlo S. Identification and characterization of polycystin-2, the PKD2 gene product. J. Biol. Chem. 1999;274:28557–28565. doi: 10.1074/jbc.274.40.28557. [DOI] [PubMed] [Google Scholar]
- Carrington W. A., Lynch R. M., Moore E. D., Isenberg G., Fogarty K. E., Fay F. S. Superresolution three-dimensional images of fluorescence in cells with minimal light exposure. Science. 1995;268:1483–1487. doi: 10.1126/science.7770772. [DOI] [PubMed] [Google Scholar]
- Cole D. G. The intraflagellar transport machinery of Chlamydomonas reinhardtii. Traffic. 2003;4:435–442. doi: 10.1034/j.1600-0854.2003.t01-1-00103.x. [DOI] [PubMed] [Google Scholar]
- Cole D. G., Diener D. R., Himelblau A. L., Beech P. L., Fuster J. C., Rosenbaum J. L. Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J. Cell Biol. 1998;141:993–1008. doi: 10.1083/jcb.141.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbit K. C., Aanstad P., Singla V., Norman A. R., Stainier D. Y., Reiter J. F. Vertebrate Smoothened functions at the primary cilium. Nature. 2005;437:1018–1021. doi: 10.1038/nature04117. [DOI] [PubMed] [Google Scholar]
- Deane J. A., Cole D. G., Seeley E. S., Diener D. R., Rosenbaum J. L. Localization of intraflagellar transport protein IFT52 identifies basal body transitional fibers as the docking site for IFT particles. Curr. Biol. 2001;11:1586–1590. doi: 10.1016/s0960-9822(01)00484-5. [DOI] [PubMed] [Google Scholar]
- Deretic D., Williams A. H., Ransom N., Morel V., Hargrave P. A., Arendt A. Rhodopsin C terminus, the site of mutations causing retinal disease, regulates trafficking by binding to ADP-ribosylation factor 4 (ARF4) Proc. Natl. Acad. Sci. USA. 2005;102:3301–3306. doi: 10.1073/pnas.0500095102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer N. D., Adler C. E., Crump J. G., L’Etoile N. D., Bargmann C. I. Polarized dendritic transport and the AP-1 mu1 clathrin adaptor UNC-101 localize odorant receptors to olfactory cilia. Neuron. 2001;31:277–287. doi: 10.1016/s0896-6273(01)00361-0. [DOI] [PubMed] [Google Scholar]
- Folsch H., Pypaert M., Schu P., Mellman I. Distribution and function of AP-1 clathrin adaptor complexes in polarized epithelial cells. J. Cell Biol. 2001;152:595–606. doi: 10.1083/jcb.152.3.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng L., Okuhara D., Yu Z., Tian X., Cai Y., Shibazaki S., Somlo S. Polycystin-2 traffics to cilia independently of polycystin-1 by using an N-terminal RVxP motif. J. Cell Sci. 2006;119:1383–1395. doi: 10.1242/jcs.02818. [DOI] [PubMed] [Google Scholar]
- Grissom P. M., Vaisberg E. A., McIntosh J. R. Identification of a novel light intermediate chain (D2LIC) for mammalian cytoplasmic dynein 2. Mol. Biol. Cell. 2002;13:817–829. doi: 10.1091/mbc.01-08-0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller K., Fabry S. Brefeldin A affects synthesis and integrity of a eukaryotic flagellum. Biochem. Biophys. Res. Commun. 1998;242:597–601. doi: 10.1006/bbrc.1997.8015. [DOI] [PubMed] [Google Scholar]
- Handel M., Schulz S., Stanarius A., Schreff M., Erdtmann-Vourliotis M., Schmidt H., Wolf G., Hollt V. Selective targeting of somatostatin receptor 3 to neuronal cilia. Neuroscience. 1999;89:909–926. doi: 10.1016/s0306-4522(98)00354-6. [DOI] [PubMed] [Google Scholar]
- Jekely G., Arendt D. Evolution of intraflagellar transport from coated vesicles and autogenous origin of the eukaryotic cilium. BioEssays. 2006;28:191–198. doi: 10.1002/bies.20369. [DOI] [PubMed] [Google Scholar]
- Jurczyk A., Gromley A., Redick S., Agustin J. S., Witman G., Pazour G. J., Peters D. J., Doxsey S. Pericentrin forms a complex with intraflagellar transport proteins and polycystin-2 and is required for primary cilia assembly. J. Cell Biol. 2004;166:637–643. doi: 10.1083/jcb.200405023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klausner R. D., Donaldson J. G., Lippincott-Schwartz J. Brefeldin A: insights into the control of membrane traffic and organelle structure. J. Cell Biol. 1992;116:1071–1080. doi: 10.1083/jcb.116.5.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminski K. G., Johnson K. A., Forscher P., Rosenbaum J. L. A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc. Nat. Acad. Sci. USA. 1993;90:5519–5523. doi: 10.1073/pnas.90.12.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bot N., Antony C., White J., Karsenti E., Vernos I. Role of xklp3, a subunit of the Xenopus kinesin II heterotrimeric complex, in membrane transport between the endoplasmic reticulum and the golgi apparatus. J. Cell Biol. 1998;143:1559–1573. doi: 10.1083/jcb.143.6.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F., Hiesberger T., Cordes K., Sinclair A. M., Goldstein L. S., Somlo S., Igarashi P. Kidney-specific inactivation of the KIF3A subunit of kinesin-II inhibits renal ciliogenesis and produces polycystic kidney disease. Proc. Natl. Acad. Sci. USA. 2003;100:5286–5291. doi: 10.1073/pnas.0836980100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linstedt A. D., Hauri H. P. Giantin, a novel conserved Golgi membrane protein containing a cytoplasmic domain of at least 350 kDa. Mol. Biol. Cell. 1993;4:679–693. doi: 10.1091/mbc.4.7.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzio J. P., Brake B., Banting G., Howell K. E., Braghetta P., Stanley K. K. Identification, sequencing and expression of an integral membrane protein of the trans-Golgi network (TGN38) Biochem. J. 1990;270:97–102. doi: 10.1042/bj2700097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marszalek J. R., Liu X., Roberts E. A., Chui D., Marth J. D., Williams D. S., Goldstein L. S. B. Genetic evidence for selective transport of opsin and arrestin by kinesin-II in mammalian photoreceptors. Cell. 2000;102:175–187. doi: 10.1016/s0092-8674(00)00023-4. [DOI] [PubMed] [Google Scholar]
- Marszalek J. R., Ruiz-Lozano P., Roberts E., Chien K. R., Goldstein L. S. B. Situs inversus and embryonic ciliary morphogenesis defects in mouse mutants lacking the KIF3A subunit of kinesin-II. Proc. Nat. Acad. Sci. USA. 1999;96:5043–5048. doi: 10.1073/pnas.96.9.5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki T., et al. PKD2, a gene for polycystic kidney disease that encodes an integral membrane protein. Science. 1996;272:1339–1342. doi: 10.1126/science.272.5266.1339. [DOI] [PubMed] [Google Scholar]
- Moyer J. H., Lee-Tischler M. J., Kwon H.-Y., Schrick J. J., Avner E. D., Sweeney W. E., Godfrey V. L., Cacheiro N. L. A., Wilkinson J. E., Woychik R. P. Candidate gene associated with a mutation causing recessive polycystic kidney disease in mice. Science. 1994;264:1329–1333. doi: 10.1126/science.8191288. [DOI] [PubMed] [Google Scholar]
- Murcia N. S., Richards W. G., Yoder B. K., Mucenski M. L., Dunlap J. R., Woychik R. P. The Oak Ridge Polycystic Kidney (orpk) disease gene is required for left-right axis determination. Development. 2000;127:2347–2355. doi: 10.1242/dev.127.11.2347. [DOI] [PubMed] [Google Scholar]
- Nakamura N., Rabouille C., Watson R., Nilsson T., Hui N., Slusarewicz P., Kreis T. E., Warren G. Characterization of a cis-Golgi matrix protein, GM130. J. Cell Biol. 1995;131:1715–1726. doi: 10.1083/jcb.131.6.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S., Tanaka G., Maeda T., Kamiya R., Matsunaga T., Nikaido O. Assembly and function of Chlamydomonas flagellar mastigonemes as probed with a monoclonal antibody. J. Cell Sci. 1996;109:57–62. doi: 10.1242/jcs.109.1.57. [DOI] [PubMed] [Google Scholar]
- Nonaka S., Tanaka Y., Okada Y., Takada S., Harada A., Kanai Y., Kido M., Hirokawa N. Randomization of left-right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell. 1998;95:829–837. doi: 10.1016/s0092-8674(00)81705-5. [DOI] [PubMed] [Google Scholar]
- Orozco J. T., Wedaman K. P., Signor D., Brown H., Rose L., Scholey J. M. Movement of motor and cargo along cilia. Nature. 1999;398:674. doi: 10.1038/19448. [DOI] [PubMed] [Google Scholar]
- Pan J., Snell W. J. Kinesin II and regulated intraflagellar transport of Chlamydomonas aurora protein kinase. J. Cell Sci. 2003;116:2179–2186. doi: 10.1242/jcs.00438. [DOI] [PubMed] [Google Scholar]
- Papermaster D. S., Schneider B. G., Besharse J. C. Vesicular transport of newly synthesized opsin from the Golgi apparatus toward the rod outer segment. Ultrastructural immunocytochemical and autoradiographic evidence in Xenopus retinas. Invest. Ophthalmol. Vis. Sci. 1985;26:1386–1404. [PubMed] [Google Scholar]
- Pazour G. J., Baker S. A., Deane J. A., Cole D. G., Dickert B. L., Rosenbaum J. L., Witman G. B., Besharse J. C. The intraflagellar transport protein, IFT88, is essential for vertebrate photoreceptor assembly and maintenance. J. Cell Biol. 2002a;157:103–113. doi: 10.1083/jcb.200107108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour G. J., Dickert B. L., Vucica Y., Seeley E. S., Rosenbaum J. L., Witman G. B., Cole D. G. Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene Tg737, are required for assembly of cilia and flagella. J. Cell Biol. 2000;151:709–718. doi: 10.1083/jcb.151.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour G. J., San Agustin J. T., Follit J. A., Rosenbaum J. L., Witman G. B. Polycystin-2 localizes to kidney cilia and the ciliary level is elevated in orpk mice with polycystic kidney disease. Curr. Biol. 2002b;12:R378–R380. doi: 10.1016/s0960-9822(02)00877-1. [DOI] [PubMed] [Google Scholar]
- Pazour G. J., Wilkerson C. G., Witman G. B. A dynein light chain is essential for the retrograde particle movement of intraflagellar transport (IFT) J. Cell Biol. 1998;141:979–992. doi: 10.1083/jcb.141.4.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour G. J., Witman G. B. The vertebrate primary cilium is a sensory organelle. Curr. Opin. Cell Biol. 2003;15:105–110. doi: 10.1016/s0955-0674(02)00012-1. [DOI] [PubMed] [Google Scholar]
- Peden E. M., Barr M. M. The KLP-6 kinesin is required for male mating behaviors and polycystin localization in Caenorhabditis elegans. Curr. Biol. 2005;15:394–404. doi: 10.1016/j.cub.2004.12.073. [DOI] [PubMed] [Google Scholar]
- Piperno G., Mead K. Transport of a novel complex in the cytoplasmic matrix of Chlamydomonas flagella. Proc. Nat. Acad. Sci. USA. 1997;94:4457–4462. doi: 10.1073/pnas.94.9.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H., Burnette D. T., Bae Y. K., Forscher P., Barr M. M., Rosenbaum J. L. Intraflagellar transport is required for the vectorial movement of TRPV channels in the ciliary membrane. Curr. Biol. 2005;15:1695–1699. doi: 10.1016/j.cub.2005.08.047. [DOI] [PubMed] [Google Scholar]
- Rizzuto R., Pinton P., Carrington W., Fay F. S., Fogarty K. E., Lifshitz L. M., Tuft R. A., Pozzan T. Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science. 1998;280:1763–1766. doi: 10.1126/science.280.5370.1763. [DOI] [PubMed] [Google Scholar]
- Rosenbaum J. L., Witman G. B. Intraflagellar transport. Nat. Rev. Mol. Cell Biol. 2002;3:813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- Schneider L., Clement C. A., Teilmann S. C., Pazour G. J., Hoffmann E. K., Satir P., Christensen S. T. PDGFRααsignaling is regulated through the primary cilium in fibroblasts. Curr. Biol. 2005;15:1861–1866. doi: 10.1016/j.cub.2005.09.012. [DOI] [PubMed] [Google Scholar]
- Scholey J. M. Intraflagellar transport. Annu. Rev. Cell Dev. Biol. 2003;19:423–443. doi: 10.1146/annurev.cellbio.19.111401.091318. [DOI] [PubMed] [Google Scholar]
- Sharon N. Lectin receptors as lymphocyte surface markers. Adv. Immunol. 1983;34:213–298. doi: 10.1016/s0065-2776(08)60380-6. [DOI] [PubMed] [Google Scholar]
- Snow J. J., Ou G., Gunnarson A. L., Walker M. R., Zhou H. M., Brust-Mascher I., Scholey J. M. Two anterograde intraflagellar transport motors cooperate to build sensory cilia on C. elegans neurons. Nat. Cell Biol. 2004;6:1109–1113. doi: 10.1038/ncb1186. [DOI] [PubMed] [Google Scholar]
- Stephens R. E. Ciliary protein turnover continues in the presence of inhibitors of golgi function: evidence for membrane protein pools and unconventional intracellular membrane dynamics. J. Exp. Zool. 2001;289:335–349. doi: 10.1002/jez.1015. [DOI] [PubMed] [Google Scholar]
- Tai A. W., Chuang J.-Z., Sung C.-H. Interaction of rhodopsin’s carboxy-terminal cytoplasmic tail with Tctex-1, a cytoplasmic dynein light chain. Mol. Biol. Cell. 1998;9:154a. doi: 10.1016/s0092-8674(00)80800-4. [DOI] [PubMed] [Google Scholar]
- Takeda S., Yonekawa Y., Tanaka Y., Okada Y., Nonaka S., Hirokawa N. Left-right asymmetry and kinesin superfamily protein KIF3A: new insights in determination of laterality and mesoderm induction by kif3A-/- mice analysis. J. Cell Biol. 1999;145:825–836. doi: 10.1083/jcb.145.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker R. W., Pardee A. B. Centriole ciliation is related to quiescence and DNA synthesis in 3T3 cells. Cell. 1979;17:527–535. doi: 10.1016/0092-8674(79)90261-7. [DOI] [PubMed] [Google Scholar]
- Vaisberg E. A., Grissom P. M., McIntosh J. R. Mammalian cells express three distinct dynein heavy chains that are localized to different cytoplasmic organelles. J. Cell Biol. 1996;133:831–842. doi: 10.1083/jcb.133.4.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheatley D. N. Primary cilia in normal and pathological tissues. Pathobiology. 1995;63:222–238. doi: 10.1159/000163955. [DOI] [PubMed] [Google Scholar]
- Yoder B. K., Hou X., Guay-Woodford L. M. The polycystic kidney disease proteins, polycystin-1, polycystin-2, polaris, and cystin, are co-localized in renal cilia. J. Am. Soc. Nephrol. 2002;13:2508–2516. doi: 10.1097/01.asn.0000029587.47950.25. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.