Abstract
The spindle-pole body (SPB), the yeast analog of the centrosome, serves as the major microtubule (MT) organizing center in the yeast cell. In addition to this central function, the SPB organizes and concentrates proteins required for proper coordination between the nuclear-division cycle and cytokinesis. For example, the Schizosaccharomyces pombe septation-initiation network (SIN), which is responsible for initiating actomyosin ring constriction and septation, is assembled at the SPB through its two scaffolding components, Sid4 and Cdc11. In an effort to identify novel SIN interactors, we purified Cdc11 and identified by mass spectrometry a previously uncharacterized protein associated with it, Ppc89. Ppc89 localizes constitutively to the SPB and interacts directly with Sid4. A fusion between the N-terminal 300 amino acids of Sid4 and a SPB targeting domain of Ppc89 supplies the essential function of Sid4 in anchoring the SIN. ppc89Δ cells are inviable and exhibit defects in SPB integrity, and hence in spindle formation, chromosome segregation, and SIN localization. Ppc89 overproduction is lethal, resulting primarily in a G2 arrest accompanied by massive enlargement of the SPB and increased SPB MT nucleation. These results suggest a fundamental role for Ppc89 in organization of the S. pombe SPB.
INTRODUCTION
The centrosome is the major microtubule-organizing center (MTOC) in higher eukaryotes (Urbani and Stearns, 1999). Problems in centrosome function such as improper duplication or an inability to anchor MTs lead to defects in chromosome segregation that result in incomplete genome inheritance to the progeny. Furthermore, centrosomal defects resulting in an inability to properly organize MTs cause nuclear positioning, cell polarity and intracellular trafficking defects (Jaspersen and Winey, 2004). Additionally, there is a growing list of regulatory and signaling molecules localized to the centrosome that underscores its key role as a signaling center (Doxsey et al., 2005).
The spindle-pole body (SPB) is the yeast analog of the centrosome; its structure, composition, and organization are well characterized in the budding yeast Saccharomyces cerevisiae (Adams and Kilmartin, 2000). The S. cerevisiae SPB appears as a multilayered cylindrical structure that is always embedded in the nuclear envelope (NE) as visualized by electron microscopy (EM). Each SPB can be divided into three layers: the outer plaque on the cytoplasmic face, the central plaque in the plane of the NE, and the inner plaque on the nuclear side of the NE (Adams and Kilmartin, 2000; Jaspersen and Winey, 2004). The site of new SPB formation, called the half-bridge, is joined to the central plaque (Jaspersen and Winey, 2004).
Far less is known about the SPB in the fission yeast Schizosaccharomyces pombe. The complete composition of the S. pombe SPB has not been determined, and there remains some uncertainty even as to when during the cell cycle the S. pombe SPB duplicates. Although it was first reported to duplicate in late G2 (Ding et al., 1997), a recent study indicates that the SPB duplicates at the G1/S boundary from a half-bridge (Uzawa et al., 2004), much like the S. cerevisiae SPB. Structural studies show that the S. pombe SPB is an amorphous body with an electron-dense central line. It is tethered to the outside of the NE until mitosis, when it embeds in the NE (Ding et al., 1997; Uzawa et al., 2004). Cut11, which localizes to the NE and nuclear pore complexes throughout the cell cycle, is needed for the SPB to embed in the NE and becomes concentrated at the site of SPB insertion during mitosis (West et al., 1998). During meiosis, the S. pombe SPB leads the nucleus in dynamic oscillatory movements termed “horsetail” (reviewed by Sawin, 2005), which are necessary for normal rates of recombination and also serve to initiate spore formation at the end of meiosis (Shimoda, 2004).
Although the full complement of S. pombe SPB components has not been determined, some proteins have been identified based on homology to S. cerevisiae components or through genetic screens. One structural component identified is the coiled-coil protein Pcp1, the homologue of S. cerevisiae Spc110 (Flory et al., 2002). Spc110 binds calmodulin and links the gamma tubulin complex (γ-TuC) to the central plaque of the SPB (Knop and Schiebel, 1997; Sundberg and Davis, 1997). Another structural element is centrin/Cdc31. This protein is a part of the SPB half-bridge and controls SPB duplication (Paoletti et al., 2003). Several proteins have been identified that localize at the SPB between the NE and the SPB. One such component is Sad1. Sad1 contains one transmembrane helix domain and is required for mitotic functions of the SPB (Hagan and Yanagida, 1995). Interacting with Sad1 are Kms1 and Kms2, which contain coil-coiled and transmembrane helices and are not essential for vegetative growth (Niwa et al., 2000; Miki et al., 2004). Kms1 is essential for telomere clustering and SPB function during meiosis (Shimanuki et al., 1997). The less well-characterized kms2 mutant is reported to have mitotic defects (Miki et al., 2004). Another group of SPB proteins has been identified based on their specific requirement during meiosis. Among these proteins are Hrs1/Mcp6, a coiled-coiled protein produced specifically during meiosis (Saito et al., 2005; Tanaka et al., 2005), and Ssm4 (Niccoli et al., 2004), a p150-Glued protein, which are both required for horsetail nuclear movement and recombination of sister chromosomes, and Spo15, a meiotic coiled-coil SPB protein involved in spore membrane formation (Ikemoto et al., 2000).
In addition to the MTOC functions discussed above, the SPB in S. pombe functions as an assembly site for a signal transduction network, called the septation initiation network (SIN) (reviewed by McCollum and Gould, 2001; Simanis, 2003; Krapp et al., 2004b). The SIN is a GTPase-regulated protein kinase pathway that coordinates proper chromosome segregation with mitotic exit and cytokinesis. All components of this pathway localize to the SPB during at least a portion of the cell cycle, and the signal for cytokinesis is thought to emanate from there (Bardin and Amon, 2001; McCollum and Gould, 2001; Simanis, 2003; Krapp et al., 2004b). Components of the SIN, as well as its regulators, are localized to the SPB through their association with its two scaffolding components, Sid4 and Cdc11, which are constitutively localized to the SPB (Chang and Gould, 2000; Krapp et al., 2001, 2004a; Tomlin et al., 2002; Morrell et al., 2004). Sid4 appears to be the SIN component most proximal to the SPB, because its function is required for the SPB localization of all other known SIN components. The C-terminus of Sid4 directs the protein to the SPB, whereas the Sid4 N-terminus binds directly to the C-terminus of Cdc11 as well as to the polo-like kinase Plo1 and the SIN inhibitor Dma1 (Guertin et al., 2002; Tomlin et al., 2002; Morrell et al., 2004). The N-terminus of Cdc11 binds Spg1, Cdc16, Sid2, and Cdk1-Cdc13 (Morrell et al., 2004). However, although Sid4 is stably associated with the SPB, it is not clear how it is integrated into the SPB.
In an effort to reveal additional components, anchors, or regulators of the SIN previously unidentified through genetic screens, we performed a TAP (tandem affinity purification) analysis on Cdc11. Mass spectrometric analysis of proteins copurifying with Cdc11 identified a previously uncharacterized protein, which we named Ppc89, encoded by the SPAC4H3.11c locus. In this work, we characterize the relationship of this protein to the SIN and its role in SPB function.
MATERIALS AND METHODS
Strains, Media, and Genetic Methods
S. pombe strains used in this study (Table 1) were grown in yeast extract (YE) medium or minimal medium with the appropriate supplements (Moreno et al., 1991). Crosses were performed on glutamate medium (minimal medium lacking ammonium chloride and containing 0.01 M glutamate). S. pombe transformations were performed by a lithium-acetate method (Keeney and Boeke, 1994). Expression of genes regulated by the nmt promoter was achieved by growing cells in the presence of 5 μg/ml thiamine (promoter repressed) and then washing them into medium lacking thiamine (promoter induced; Siam et al., 2004).
Table 1.
S. pombe strains used in this study
| Strain | Genotypea | Source | |
|---|---|---|---|
| KGY246 | h− | ade6-M210 ura4-D18 leu1-32 | Lab stock |
| KGY247 | h+ | ade6-M210 ura4-D18 leu1-32 | Lab stock |
| KGY248 | h− | ade6-M216 ura4-D18 leu1-32 | Lab stock |
| KGY249 | h+ | ade6-M216 ura4-D18 leu1-32 | Lab stock |
| KGY262 | h+ | ppc89-CFP::KanR sid4-YFP::KanR ade6-M210 ura4-D18 leu1-32 | This studyb |
| KGY774 | h+/h− | ppc89Δ::ura4+/ppc89+ade6-M210/ade6-M216 ura4-D18/ura4-D18 leu1-32/leu1-32 | See text |
| KGY794 | h+ | ppc89-YFP::KanRsid4-CFP::KanRade6-M21aura4-D18 leu1-32 | This studyb |
| KGY1234 | h− | sid4-SA1 ade6-M210 ura4-D18 leu1-32 | Balasubramanian et al. (1998) |
| KGY1341 | h+ | sid4-myc::KanR ade6-M210 ura4-D18 leu1-32 | Chang and Gould (2000) |
| KGY1358 | h+/h− | sid4Δ::ura4+/sid4+ade6-M210/ade6-M216 ura4-D18/ura4-D18 leu1-32/leu1-32 | Chang and Gould (2000) |
| KGY3271 | h+ | sid4-YFP::KanR cdc11-CFP::KanR ade6-M21X ura4-D18 leu1-32 | This studyb |
| KGY3275 | h− | ppc89-YFP::KanR cdc11-CFP::KanR ade6-M210 ura4-D18 leu1-32 | This studyb |
| KGY3526 | h− | sid4-CFP::KanR cdc11-YFP::KanR ade6-M21X ura4-D18 leu1-32 | This studyb |
| KGY3888 | h− | cdc11-TAP::KanRade6-M210 ura4-D18 leu1-32 | See text |
| KGY4288 | h− | ppc89-HA::KanRade6-M210 ura4-D18 leu1-32 | See text |
| KGY4293 | h− | ppc89-TAP::KanRade6-M210 ura4-D18 leu1-32 | See text |
| KGY4294 | h+ | ppc89-GFP::KanRade6-M210 ura4-D18 leu1-32 | See text |
| KGY4297 | h+ | ppc89-HA::KanRsid4-myc::KanRade6-M21X ura4-D18 leu1-32 | This studyb |
| KGY4318 | h+ | ppc89-CFP::KanRcdc11-YFP::KanRade6-M210 ura4-D18 leu1-32 | This studyb |
| KGY4325 | h+ | ppc89-GFP::KanRsid4-SA1 ade6-M21X ura4-D18 leu1-32 | This studyb |
| KGY4334 | h− | sid4-YFP::KanRade6-M210 ura4-D18 leu1-32 | See text |
| KGY4439 | h− | sid4-CFP::KanRade6-M210 ura4-D18 leu1-32 | See text |
| KGY4608 | h− | sid4-YFP-CFP::KanRade6-M210 ura4-D18 leu1-32 | See text |
| KGY5185 | h+ | leu1-32::pJK148-nmt81-ppc89+ppc89Δ::ura4+ade6-M21X ura4-D18 | See text |
| KGY5328 | h− | leu1-32::pJK148-nmt81-ppc89+ade6-M210 ura4-D18 | See text |
| KGY5331 | h− | sid4-GFP::KanRleu1-32::pJK148-nmt1-ppc89+ade6-M210 ura4-D18 | This studyb |
| KGY5332 | h− | ppc89-GFP::KanRleu1-32::pJK148-nmt1-ppc89+ade6-M210 ura4-D18 | This studyb |
| KGY5333 | h− | leu1-32::pJK148-nmt1-ppc89+ade6-M210 ura4-D18 | See text |
| KGY5458 | h+ | cdc11-GFP::KanRleu1-32::pJK148-nmt1-ppc89+ade6-M210 ura4-D18 | This studyb |
| KGY5459 | h+ | alp6-GFP::KanRleu1-32::pJK148-nmt1-ppc89+ade6-M210 ura4-D18 | This studyb |
| KGY5537 | h− | pcp1-YFP::KanRleu1-32::pJK148-nmt1-ppc89+ade6-M210 ura4-D18 | This studyb |
| KGY5576 | h+ | sid4-CFP::KanRnmt81-GFP-atb2+::KanRleu1-32::pJK148-nmt1-ppc89+ade6-M210 ura4-D18 | This studyb |
| KGY5647 | h− | cdc11-GFP::KanRleu1-32::pJK148-nmt81-ppc89+ppc89Δ::ura4+ade6-M21X ura4-D18 | This studyb |
| KGY5648 | h+ | sid4-GFP::KanRleu1-32::pJK148-nmt81-ppc89+ppc89Δ::ura4+ade6-M21X ura4-D18 | This studyb |
| KGY5649 | h+ | alp6-GFP::KanRleu1-32::pJK148-nmt81-ppc89+ppc89Δ::ura4+ade6-M21X ura4-D18 | This studyb |
| KGY5650 | h+ | pcp1-GFP::KanRleu1-32::pJK148-nmt81-ppc89+ppc89Δ::ura4+ade6-M21X ura4-D18 | This studyb |
| KGY5762 | h+ | cut11-GFP::ura4+leu1-32::pJK148-nmt81-ppc89+ppc89Δ::ura4+ade6-M21X ura4-D18 | This studyb |
a Some ade6 alleles could be either 210 or 216, and 21X denotes this uncertainty.
b Constructed by crosses among other strains listed here or between such strains and strains described by West et al. (1998), Chang and Gould (2000), Flory et al. (2002), Tomlin et al. (2002), Sawin et al. (2004), and Venkatram et al. (2004).
All of the indicated tagged alleles of ppc89+, cdc11+, sid4+, alp6+, cut11+, and pcp1+ were tagged at the 3′ end of their chromosomal loci with sequences encoding 13 copies of the Myc epitope, 3 copies of the HA epitope, enhanced green fluorescent protein (eGFP), cyan fluorescent protein (CFP), the yellow fluorescent protein (YFP), the tandem affinity purification (TAP) tag (Tasto et al., 2001), or a YFP-CFP double cassette (Tomlin and Gould, unpublished results) by a PCR-mediated strategy as described previously (Bähler et al., 1998). Proper integration of these epitope cassettes was confirmed by PCR.
Deletion of the complete ppc89+ open reading frame (ORF) was achieved by PCR-based one-step homologous recombination as described by Bähler et al. (1998), using ura4+ as a selectable marker. The amplified fragment was transformed into a h−/h+ ade6-M210/ade6-M216 ura4-D18/ura4-D18 leu1-32/leu1-32 diploid strain, and stable integrants were selected. Deletion of one copy of ppc89+ in strain KGY774 was confirmed by PCR. Sporulation of KGY774 and tetrad dissection revealed that ppc89+ was essential for vegetative growth. sid4+ deletion was performed previously (Chang and Gould, 2000).
The ppc89+ ORF was amplified from S. pombe genomic DNA by PCR and cloned into pSK+. The pSKppc89 clone was sequenced in its entirety to confirm that no mutations had been introduced into the ORF. To clone ppc89+ and pieces thereof under control of the nmt promoters in the pREP1, pREP81, and pREP81GFP vectors (Basi et al., 1993; Maundrell, 1993; Drummond and Hagan, 1998), NdeI and BamHI sites were added to the 5′ and 3′ ends, respectively, of oligonucleotides used for PCR amplification. The NdeI site in each oligonucleotide contributed the initiating methionine codon. Stop codons were incorporated into the oligonucleotides just upstream of the BamHI sites. Similarly, the N-terminus of sid4+ was amplified by PCR using a 5′AseI and 3′NdeI site to allow an N-terminal fusion with the C-terminal sequences of ppc89+ in the pREP vectors. To facilitate depletion and overproduction of Ppc89, the nmt81-ppc89 and nmt1-ppc89 portions of the pREP plasmids were excised with PstI and BamHI and cloned into pJK148 (Keeney and Boeke, 1994). The resultant plasmids were linearized within the leu1+ gene by digestion with Eco47III and integrated into the leu1 locus of KGY246 to create strains KGY5328 and KGY5333, respectively. To generate a conditional ppc89+ shutoff strain, KGY5328 was crossed to the heterozygous ppc89+ deletion strain (KGY774) and allowed to sporulate on glutamate medium. Random spore analysis was then performed, selecting for haploid cells that were Leu+, Ura+, Ade−, and dead in the presence of thiamine.
Fluorescence Microscopy and FRET Analysis
Strains producing YFP- and CFP-tagged proteins were grown in YE medium and visualized live. Images were acquired digitally on a Carl Zeiss MicroImaging Axiovert II inverted microscope (Thornwood, NY) equipped with a Plan Apo 100/1.40 lens, a piezo-electric Z-axis stepper objective motor (Physik Instrumente, Auburn, MA), an UltraView LCI real-time scanning-head confocal (Perkin Elmer-Cetus, Wellesley, MA), a 488-nm argon ion laser (for YFP excitation), and a 442-nm helium cadmium laser (for CFP excitation). Images were captured on an Orca-ER charge-coupled-device camera (Hamamatsu, Bridgewater, NJ). Z-series optical sections for each filter set were captured at 0.5-μm intervals using Ultra-View software (Perkin Elmer-Cetus). Subsequently, the images were deconvolved, merged, and rendered into a single plane using Volocity 3.5.1 software (Improvision, Lexington, MA).
Cells expressing GFP fused at the N-terminus of full-length Ppc89 or Ppc89 fragments from the nmt81 promoter were grown in the absence of thiamine and fixed in methanol or visualized live. To visualize MTs, cells were fixed in methanol and stained with TAT-1 antibodies to α-tubulin (Woods et al., 1989). Microscopy was performed on a Zeiss Axioskop II equipped with a z-focus motor drive and GFP and DAPI filter sets (ChromaTechnology, Rockingham, VT). Images were captured with an Orca II charge-coupled-device camera (Hamamatsu) and processed and analyzed with Open-Lab 4.0.3 software (Improvision).
For fluorescence resonance energy transfer (FRET), cells were imaged on a DeltaVision microscope (Applied Precision, Issaquah, WA) and analyzed as described by Muller et al. (2005) (http://depts.washington.edu/∼yeastrc/). Briefly, 100 images were captured for each strain. Exposure times were 0.4 s with 2 × 2 binning and a final image size of 512 × 512. The order of image acquisition was YFP, FRET, CFP, and DIC. Images were analyzed with the SoftWoRx program from Applied Precision. For each strain, the tagged protein replaced the wild-type protein and was expressed under the control of the native promoter. FRETR = FRET channel ÷ SpilloverTotal (Muller et al., 2005). The SpilloverYFP factor was determined from the Sid4-YFP strain KGY4334 and was 0.233 ± 0.040 (n = 85). The SpilloverCFP factor was determined from the Sid4-CFP strain KGY4439 and was 0.486 ± 0.060. SpilloverTotal = (CFPchannel × SpilloverCFP) + (YFPchannel × SpilloverYFP). All fluorescence channels were background-subtracted.
Electron Microscopy
Aliquots of cells expressing Ppc89-GFP were prepared for EM as described previously (Giddings et al., 2001). Briefly, cells were harvested by vacuum filtration onto 0.45-μm Millipore filters and cryofixed by high-pressure freezing in a HPM-010 (BAL-TEC/RMC, Tucson, AZ). Frozen samples were freeze-substituted in 0.25% glutaraldehyde and 0.1% uranyl acetate in acetone at −80°C, infiltrated with liquid Lowicryl HM20 (Electron Microscopy Sciences, Fort Washington, PA) at −20°C, and polymerized under UV at −45°C. For immunolabeling, 60-nm-thick sections were retrieved on formvar-coated nickel slot grids, and then floated on a series of drops containing 1) blocking solution of 1% nonfat dry milk in phosphate-buffered saline plus 0.1% Tween 20 (PBST), 2) primary antibody (affinity-purified rabbit anti-GFP) diluted 1:150 in blocking solution, and 3) goat anti-rabbit-IgG secondary antibody conjugated to 15-nm colloidal gold (Ted Pella, Redding, CA) diluted 1:20 in blocking solution. The labeled grids were rinsed in PBST followed by distilled water. Sections were stained with 2% uranyl acetate and lead citrate and then viewed in a Philips (Mahwah, NJ) CM10 transmission electron microscope operating at 80 kV. Images were recorded with a Gatan (Pleasanton, CA) BioScan digital camera or on Kodak (Eastman Kodak, Rochester, NY) 4489 electron microscope film.
Protein Methods
Total cell extracts of S. pombe were prepared in NP-40 buffer (Gould et al., 1991), and immunoprecipitations (McDonald et al., 1999) were carried out using 5 μg of either 12CA5 (anti-HA) or 9E10 (anti-Myc) monoclonal antibodies (both from Vanderbilt Molecular Recognition Shared Resource). After 1 h of incubation, the immunoprecipitates were washed six times in NP-40 buffer and then resuspended in 2× SDS-PAGE sample buffer (McDonald et al., 1999).
For immunoblotting, proteins were resolved by 10% SDS-PAGE and transferred by electroblotting to PVDF membrane (Immobilon P; Millipore, Bedford, MA). Antibodies 12CA5 and 9E10 were used at 2 μg/ml in Tris-buffered saline to detect epitope-tagged proteins. These antibodies were then detected using horseradish-peroxidase–conjugated goat anti-mouse-IgG secondary antibodies (0.8 mg/ml; Jackson ImmunoResearch Laboratories, West Grove, PA) at a dilution of 1:50,000. Immunoblots were visualized using ECL reagents (Amersham Pharmacia Biotech, Piscataway, NJ).
Maltose-binding protein (MBP) and MBP fused to full-length Ppc89 were produced in Escherichia coli using plasmid pMAL-c2 (New England Biolabs, Beverly, MA) and purified on amylose beads (New England Biolabs) per the manufacturer’s instructions. Briefly, the DNA sequence encoding Ppc89 was amplified by PCR from wild-type genomic DNA using primers that introduced EcoRI and BamHI sites at the 5′ and 3′ ends, respectively. The product was cut with these enzymes and cloned into similarly cut pMAL-c2 to create an MBP-ppc89 fusion. To create pSK(+)sid4 (284–660; pKG3159), DNA was amplified by PCR from wild-type genomic DNA using primers that introduced EcoRI and BamHI sites at the 5′ and 3′ ends, respectively. The product cut with these restriction enzymes and cloned into similarly cut pSK(+). pSK(+) containing full-length Prp19 (pKG1781) was constructed previously (Ohi and Gould, 2002). Both pKG1781 and pKG3159 were translated in vitro in the presence of [35S]-trans label (ICN Pharmaceuticals, Irvine, CA) with the use of the TNT-coupled reticulocyte-lysate system (Promega, Madison, WI). After the transcription/translation reaction was allowed to proceed for 90 min at 30°C, 1 ml of binding buffer (20 mM Tris-HCl, pH 7.0, 150 mM NaCl, 2 mM EDTA, 0.1% NP-40) was added to the lysates. The lysates were then clarified by centrifugation at 14,000 rpm for 15 min. Purified MBP or MBP-Ppc89 bound to amylose beads was mixed with 35S-labeled Sid4 fragment or full-length Prp19 in binding buffer and incubated for 1 h at 4°C. The beads were washed five times in binding buffer, and the proteins were resolved by 10% SDS-PAGE, treated with Amplify (Amersham Pharmacia Biotech), and exposed to film.
Two-Hybrid Analyses
The yeast two-hybrid system used in this study was described previously (James et al., 1996). Various portions of ppc89+ or sid4+ were amplified by PCR from genomic DNA and cloned into the bait plasmid pGBT9 and/or the prey plasmid pGAD424 (Clontech, Palo Alto, CA). To test for protein interactions, both bait and prey plasmids were cotransformed into S. cerevisiae strain PJ69-4A. Leu+ and Trp+ transformants were selected and then scored for positive interactions by streaking onto synthetic dextrose plates lacking adenine and histidine.
RESULTS
Identification of Ppc89
To identify previously unrecognized SIN components, interactors, or SPB anchors, sequences encoding a TAP cassette were introduced at the 3′ end of the cdc11+ ORF, creating strain KGY3888 (see Materials and Methods). After TAP, the eluate was digested with proteases and subjected to two-dimensional liquid chromatography and tandem mass spectrometry as described (Link et al., 1999; MacCoss et al., 2002). Proteins within the mixture were then identified using the SEQUEST algorithm (Eng et al., 1994). Besides Cdc11, Sid4, and known contaminants of TAP complexes (Gould et al., 2004), the protein identified by the next greatest sequence coverage in this purification was encoded by a previously uncharacterized ORF, SPAC4H3.11c. The predicted molecular mass of this protein is 89 kDa. We named this protein Pombe Pole Component 89 (Ppc89) based on its molecular mass and localization (see below). Ppc89 is predicted to contain two blocks of coiled-coils in its central region as well a third coiled-coil region at its C-terminus (schematized in Figures 2A and 4A). Blast searches failed to reveal obvious homologues of this protein in any other organism for which a genome sequence is available. We determined that ppc89+ is an essential gene by tetrad analysis of diploid cells in which one copy of ppc89+ had been replaced with ura4+ (see Materials and Methods).
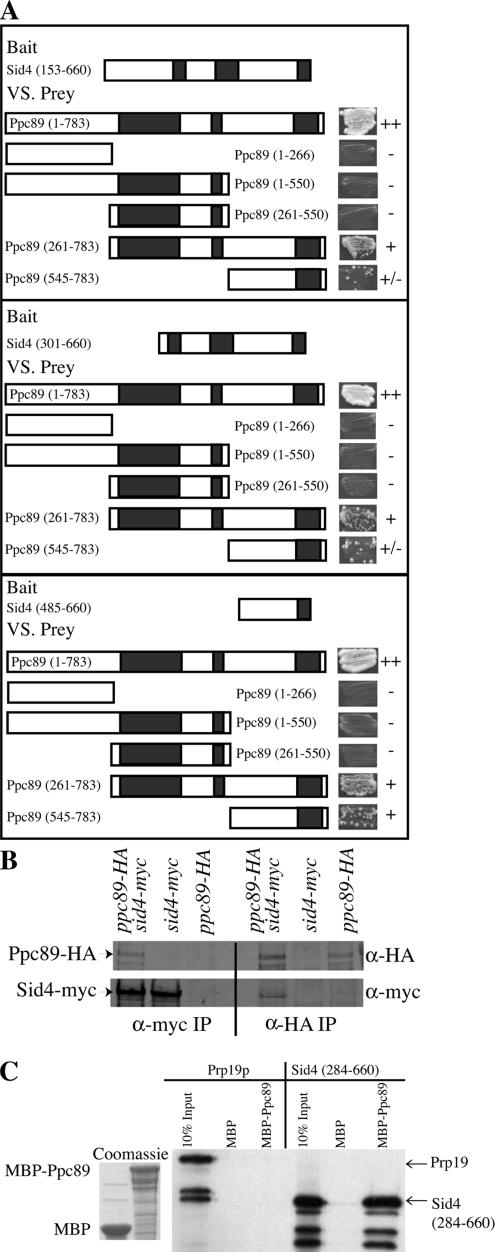
Figure 2.
Ppc89 and Sid4 interact directly through their C-termini. (A) S. cerevisiae strain PJ69-4A was cotransformed with plasmids expressing the indicated regions of Sid4 and Ppc89. Black boxes indicate regions of predicted coiled-coil. Transformants carrying both plasmids were scored for growth on −Ade −His plates. Pluses indicate strong growth, plus/minus indicates little growth, and minuses indicate no growth. (B) Lysates from ppc89-HA3 (KGY4288), sid4-myc13 (KGY1341), or ppc89-HA3 sid4-myc13 (KGY4297) strains were immunoprecipitated using anti-myc (left panel) or anti-HA (right panel) antibodies. After SDS-PAGE, blots were probed with anti-HA (top panel) or anti-myc (bottom panel) antibodies. (C) MBP and MBP-Ppc89 were produced in E. coli, and the soluble portion was bound to amylose beads. The amounts of recombinant proteins added to the reactions are shown by Coomassie staining in the left panel. Prp19 and Sid4 (284–660) were expressed using an in vitro reticulocyte lysate system in the presence of [35S]methionine, and 10% of the reaction was used in the input lane. After incubation with proteins bound to beads and washing of the beads, samples were separated by SDS-PAGE, and proteins were detected by autoradiography (right panel).
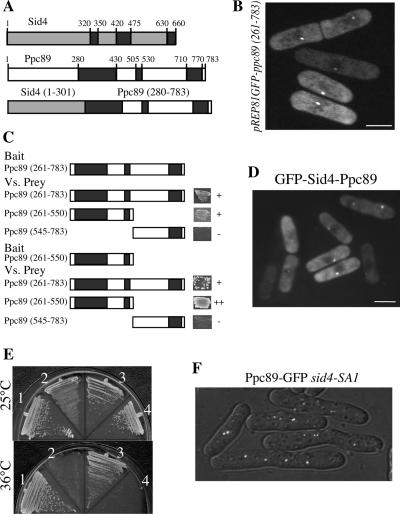
Figure 4.
The essential function of the Sid4 C-terminus in SIN signaling is SPB targeting. (A) Schematic representation of Sid4, Ppc89, and a Sid4-Ppc89 fusion. Predicted coiled-coil domains are indicated by black boxes. Numbers indicate amino acids. (B) Wild-type cells (KGY246) expressing GFP-ppc89 (261–783) from the pREP81 vector were grown in the absence of thiamine for 18 h at 32°C. An image of live cells was captured. (C) Bait and prey vectors expressing the indicated regions of Ppc89 were cotransformed into S. cerevisiae strain PJ69-4A. Transformants carrying both plasmids were scored for growth on a medium selecting for a two-hybrid interaction, as indicated to the right of the schematics. Black boxes indicate regions of predicted coiled coil. (D) sid4-SA1 cells (KGY1234) were transformed with pREP81-GFPsid4-ppc89, and live-cell images were captured after 24 h of growth in the absence of thiamine at 25°C. (E) Strain KGY1234 was transformed with pREP81sid4+ (1), pREP81ppc89+ (2), pREP81sid4-ppc89 (3), and pREP81 (4). Transformants were incubated on medium lacking thiamine at 25°C (top panel) or 36°C (bottom panel) for 4 d. (F) ppc89-GFP sid4-SA1 cells (KGY4325) were incubated at the restrictive temperature of 36°C for 4 h, and live-cell images were captured. Bars, 5 μm.
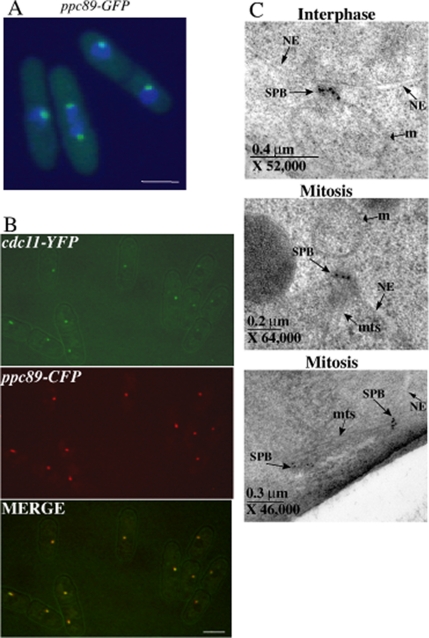
To determine the intracellular localization of Ppc89 and investigate other properties of this protein, sequences encoding eGFP, Myc13, HA3, YFP, CFP, or the TAP tag were appended to the 3′end of the chromosomal ppc89+ ORF so that normal control of fusion-protein expression was maintained. The resultant strains were wild type in morphology and growth rate, indicating that the tags did not disrupt Ppc89 function. Pcp89-GFP was detected as one or two dots adjacent to the nucleus throughout the cell cycle (Figure 1A), a staining pattern indicative of SPB localization. To confirm that these dots corresponded to SPBs, we examined the localization of Pcp89-CFP in cells also producing Cdc11-YFP. Cdc11 is known to reside at SPBs throughout the cell cycle (Krapp et al., 2001; Tomlin et al., 2002). The individual and merged images indicate that Pcp89 and Cdc11 colocalize to SPBs (Figure 1B), an expected result given that the two proteins copurified. Ppc89 was also found to localize to SPBs through immuno-EM. ppc89-GFP cells in exponential phase were fixed by high-pressure freezing and stained with antibodies to GFP and then with colloidal-gold–labeled secondary antibodies. In interphase cells, gold particles were localized in the central region of the SPB that lay adjacent to the NE (Figure 1C and Supplementary Figure 1). In mitotic cells, gold particles appeared in the central region of the SPB in the same plane as the NE or slightly toward the cytoplasmic side (Figure 1C, Supplementary Figure 1, and unpublished observations). A reciprocal TAP experiment was also performed on a ppc89-TAP strain (KGY4293), and mass-spectrometric analysis revealed the presence of Ppc89, Sid4, and Cdc11 (unpublished observations). Taken together, these results suggest that Ppc89 is closely associated with the SIN scaffolding proteins Cdc11 and Sid4 at the SPB.
Figure 1.
Ppc89 localizes to the SPB throughout the cell cycle. (A) ppc89-GFP cells (KGY 4294) were fixed in ethanol and stained with DAPI to visualize DNA. GFP is shown in green; DAPI is shown in blue. (B) Live ppc89-CFP cdc11-YFP cells (KGY 4318) were imaged, and the images were then merged. Bars, 5 μm. (C) Immunoelectron micrographs of ppc89-GFP cells (KGY4294). Top, gold particles labeling Ppc89-GFP in an interphase cell in which the SPB lies just outside the nuclear envelope; middle and bottom, mitotic cells showing SPBs labeled with gold particles. NE, nuclear envelope; m, mitochondrion; mts, microtubules.
Ppc89 Interacts with Sid4
To understand the basis of the copurification of Ppc89 with Cdc11 and Sid4, we tested whether it bound to either or both of these proteins directly. First, directed two-hybrid analysis was performed. Although no positive interactions were detected between fragments of Cdc11 and Ppc89, positive interactions were detected between Sid4 and Ppc89 (Figure 2A). The regions within the two proteins responsible for their interaction were narrowed to their C-terminal regions containing predicted coiled-coils (Figure 2A). As expected from the above results, Sid4 and Ppc89 were able to coimmunoprecipitate from a sid4-myc13 ppc89-HA3 strain (but not from singly tagged strains) when either an anti-Myc or an anti-HA antibody was used for precipitation (Figure 2B). Moreover, MBP-Ppc89 produced and purified from Escherichia coli, but not MBP alone, was able to interact with the C-terminus of Sid4 (residues 284–660) that was produced in a cell-free coupled transcription/translation system from rabbit reticulocytes, whereas neither MPB nor MBP-Ppc89 interacted with a functionally unrelated control protein, Prp19 (Figure 2C). These results indicate that there is a direct association between Sid4 and Ppc89 mediated by their respective C-termini.
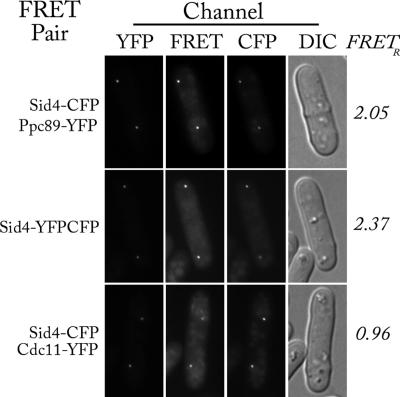
Evidence for the close proximity of the C-termini of Ppc89 and Sid4 in vivo was provided by FRET. The C-termini of the two proteins were tagged with YFP and CFP, respectively, and examined by fluorescence microscopy (Figure 3, top row). The extent of energy transfer was determined using the FRETR metric (see Materials and Methods), which measures the relative increase of fluorescence in the FRET channel compared with a baseline calculated from the fluorescence in the CFP and YFP channels. In the absence of energy transfer, FRETR has a value of 1. FRETR for the Ppc89-YFP and Sid4-CFP pair was 2.05 (Table 2, row 1). There was no significant change when the tags were reversed (Table 2, row 2). This high FRET was comparable to that of a positive control, Sid4-YFP-CFP, which had a FRETR of 2.37 (Figure 3, middle row; Table 2, row 3). Neither Ppc89 nor Sid4 was near enough to Cdc11 for FRET (Figure 3, bottom row; Table 2, lines 4–7). From these values, we conclude that the C-termini of Ppc89 and Sid4 are probably within 100 Å of each other in the cell.
Figure 3.
The close proximity of the C-termini of Sid4 and Ppc89 leads to FRET. Images were captured in four channels: YFP (500-nm excitation, 545-nm emission), FRET (440-nm excitation, 545-nm emission), CFP (440-nm excitation, 480-nm emission), and DIC. For each strain, the images have the same minimum and maximum pixel intensity, allowing visual comparison of the images. FRETR was calculated as described in Materials and Methods. Strains were KGY794, KGY4608, and KGY3526.
Table 2.
Summary of FRET results
| Row | Strain | CFP donor | YFP acceptor | FRETR | N |
|---|---|---|---|---|---|
| 1 | KGY794 | Sid4-CFP | Ppc89-YFP | 2.05 ± 0.12 | 126 |
| 2 | KGY262 | Ppc89-CFP | Sid4-YFP | 1.86 ± 0.16 | 58 |
| 3 | KGY4608 | Sid4-YFP-CFP | Sid4-YFP-CFP | 2.37 ± 0.20 | 90 |
| 4 | KGY4318 | Ppc89-CFP | Cdc11-YFP | 0.96 ± 0.09 | 113 |
| 5 | KGY3275 | Cdc11-CFP | Ppc89-YFP | 1.02 ± 0.08 | 104 |
| 6 | KGY3526 | Sid4-CFP | Cdc11-YFP | 0.96 ± 0.10 | 88 |
| 7 | KGY3271 | Cdc11-CFP | Sid4-YFP | 0.95 ± 0.09 | 81 |
The C-terminus of Ppc89 is an SPB-binding Module
The overall organization of predicted coiled-coil domains within Ppc89 is very similar to that of Sid4 (Figure 4A). Sid4 dimerizes through its central coiled-coil domain (Chang and Gould, 2000) and binds to the SPB through its very C-terminal region (Tomlin et al., 2002). Furthermore, Sid4 SPB binding is enhanced by its ability to dimerize; residues 300–560 very effectively localize GFP to the SPB in cells that also express full-length Sid4 (Tomlin et al., 2002). To determine if similar regions of Ppc89 have similar functions, we generated a GFP fusion to residues 261–783 of Ppc89 and produced it in wild-type cells. As expected, this fusion protein localized correctly to SPBs (Figure 4B). To determine if Ppc89, like Sid4, is able to self-associate, two-hybrid analysis was performed. As with Sid4, Ppc89 interacted robustly with itself, and this activity could be narrowed to the central coiled-coil region (Figure 4C).
The similarity in architecture between Sid4 and Ppc89 and their similar localization prompted us to generate a fusion between the N-terminus of Sid4 and the C-terminal self-interaction and SPB-targeting domains of Ppc89 (Figure 4A). We reasoned that if the only region of Sid4 important for SIN signaling was its N-terminal 300 amino acids, then this fusion protein should be able to rescue sid4 mutant alleles. Consistent with this possibility, the Sid4-Ppc89 fusion protein localized to SPBs (Figure 4D) and rescued sid4Δ::ura4+ segregants from strain KGY1358 (unpublished observation) and the temperature-sensitive lethality of sid4-SA1 cells (Figure 4E). Full-length ppc89+, the C-terminal portion of Ppc89 used in the fusion protein, and any sid4 construct lacking its C-terminal coiled-coil region, such as Sid4 (1–300), were not able to rescue growth of sid4-SA1 cells at 36°C (Figure 4E; unpublished observations; Chang and Gould, 2000; Krapp et al., 2001; Tomlin et al., 2002). These data indicate that the first 300 amino acids of Sid4 are the only ones critical for SIN signaling and that the remainder of the molecule is involved in SPB targeting and oligomerization, functions that can be performed by a similar region of Ppc89. These results also suggest that Ppc89 might anchor Sid4 to the SPB rather than Sid4 affecting Ppc89 localization. Consistent with this possibility, Ppc89-GFP was still at SPBs in cells lacking sid4 function (Figure 4F), in contrast to all known SIN components, which require Sid4 for their localization (Krapp et al., 2001; Tomlin et al., 2002; Morrell et al., 2004).
Ppc89 Is Required For SPB Integrity and Function
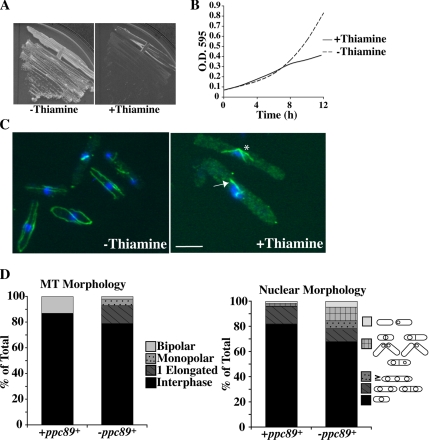
To further elucidate the role of Ppc89, we created a strain containing ppc89Δ::ura4+ and a copy of ppc89+ under control of the low- strength, thiamine-repressible nmt81 promoter integrated at the leu1 locus (see Materials and Methods). These cells were viable in the absence of thiamine, when ppc89+ was expressed, but failed to grow when the promoter was repressed (Figure 5, A and B). Samples taken 8 h after thiamine addition were stained with DAPI to visualize the nuclei and anti-α-tubulin antibodies to visualize MTs (Figure 5, C and D). (Beyond this point, the majority of cells lysed, and no further assessment of phenotype could be performed.) The majority of cells (68%) were found to contain a single nucleus, although a minor population (6%; Figure 5D, right panel) contained multiple nuclei, indicative of a SIN phenotype. A significant percentage of the cells (15%; Figure 5D, right panel) displayed a “cut” phenotype in which the septum cleaved through an undivided chromatin mass. By immunofluorescence, the majority (79%; Figure 5D, left panel) of Ppc89-depleted cells appeared to contain a normal interphase array of MTs. Another substantial population (14%; Figure 5D, left panel) arrested with a single elongated MT bundle. Only 7% of the cells appeared to be in mitosis, as judged by the presence of a spindle, and most of these had monopolar rather than bipolar spindles. (The population of cut cells was not represented in cells stained for MTs, probably because cell-wall digestion frequently destroys them.) Taken together, these data suggest that cells lacking Ppc89 have defects in organizing SPB MTs, including the mitotic spindle, and that, as a consequence, chromosome segregation fails in those cells that enter mitosis.
Figure 5.
Ppc89 depletion is lethal. (A) Cells containing a single copy of ppc89+ under control of the nmt81 promoter (KGY5185) were incubated on minimal medium lacking thiamine (promoter induced) or containing thiamine (promoter repressed). (B) Strain KGY5185 was grown in minimal medium in the absence of thiamine, and thiamine was added to half the culture at time 0. (C) Cells from the 8-h time point in B were fixed with ethanol and stained with DAPI (blue) and antibodies to α-tubulin (green). Arrow indicates single elongated MT bundle; asterisk indicates a monopolar spindle. Bar, 5 μm. (D) Quantification of MT morphologies from (C). Right, nuclear morphologies; left, MT morphologies. n = 300 cells in each case.
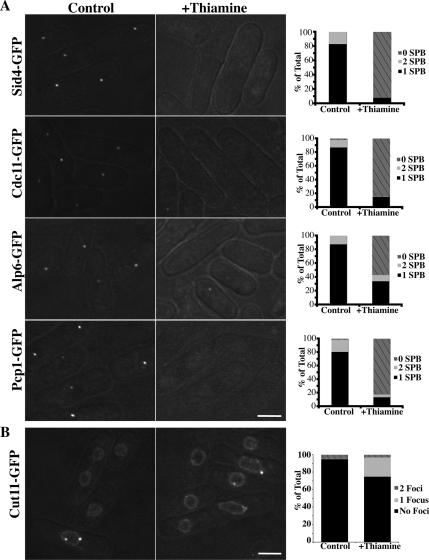
To examine the effects of Ppc89 loss on SIN-component localization, it was depleted from strains producing GFP-tagged Sid4 or Cdc11. In both cases, GFP signals were absent from SPBs (Figure 6A, top two rows). This result is consistent with the data implicating Ppc89 as an anchor for Sid4 at the SPB (Figure 4). In similar experiments, the other SPB components Alp6, a γ-tubulin complex (γ-TuC) component (Vardy and Toda, 2000), and Pcp1 (see Introduction) were examined. Again, the GFP signals at SPBs were absent or significantly reduced after 8 h of ppc89+ repression (Figure 6A, bottom two rows). These data indicate that Ppc89 is required to recruit or maintain a variety of proteins at the SPB and suggest that Ppc89 might be required for SPB integrity.
Figure 6.
Ppc89 depletion results in the loss of multiple factors from the SPB but not in NE fragmentation. (A) Strains producing the indicated GFP-tagged SPB component and carrying a single, integrated copy of ppc89+ under control of the thiamine-repressible nmt81 promoter (strains KGY5647, KGY5648, KGY5649, and KGY5650) were grown for 16 h at 32°C in the absence of thiamine. Each culture was then split into two portions, thiamine was added to one, incubation was continued for 8 h at 32°C, and live-cell images were captured. The numbers of SPBs visualized per cell were scored (graphs on right). (B) As in A, except using strain KGY5762 (cut11-GFP nmt81-ppc89+). The numbers of foci of Cut11-GFP were quantified (graph on right). Bars, 5 μm.
We next asked whether Ppc89 depletion also affected the integrity of the NE by repressing ppc89+ expression in a strain producing GFP-tagged Cut11, which normally decorates the NE and nuclear pores as well as concentrating at the site of SPB insertion into the NE in mitosis (West et al., 1998). In cells depleted of Ppc89, the NE appeared intact (Figure 6B). However, 36% of the cells had a misshapen nucleus (vs. 2% in control cells), whereas 22% (vs. 0.5% in control cells) displayed a single Cut11-GFP focus (Figure 6B). Although we do not yet understand the reason for the altered NE morphology, these results indicate that NE fragmentation does not account for the observed loss of SPB components in cells lacking Ppc89.
Ppc89 Overproduction Is Lethal and Results in SPB Enlargement
To further examine the role of Ppc89 at the SPB, ppc89+ was placed under the control of the strong, thiamine-regulatable nmt1 promoter. Overproduction of Ppc89 was lethal (Figure 7A), and FACS analysis revealed that the cells arrested with a 2N content of DNA (unpublished observation). The nmt1-ppc89 construct was then integrated into the leu1 locus to facilitate further analysis. To explore the basis of the lethality, Ppc89-overproducing cells were stained with DAPI and antibodies to the known SPB component Sad1 (Hagan and Yanagida, 1995; Figure 7B). The majority of Ppc89-overproducing cells arrested with a single nucleus, but a significant population (22%) displayed a “cut” phenotype. Interestingly, most cells overproducing Ppc89 contained a single, enlarged focus of Sad1 staining. Ppc89 was also overproduced in strains expressing GFP- or YFP-tagged Sid4, Cdc11, Pcp1, Alp6, or Ppc89 itself. In all cases, single, enlarged GFP or YFP foci were detected that trailed out to give a comet-like appearance (Figure 7C). Immunoblot analysis indicated that the overall levels of these proteins did not change with Ppc89 overproduction (unpublished observation). Taken together, these data suggest that Ppc89 overproduction causes a cell cycle arrest, predominantly in G2/M phase, accompanied by the enlargement of a single SPB.
Figure 7.
Ppc89 overproduction results in abnormal SPB morphology. (A) Wild-type cells (KGY246) were transformed with either pREP1 vector alone (left sectors) or pREP1-ppc89 (right sectors), and transformants were incubated on minimal medium containing thiamine (promoter repressed) or lacking thiamine (promoter induced). (B) sid4-GFP nmt1-ppc89+ cells (KGY5331) were grown in the presence or absence of thiamine for 24 h at 32°C, fixed with methanol, and stained with DAPI and antibodies to Sad1. Left, quantification of the numbers of nuclei per cell; right, quantification of the number of SPBs per cell, based on Sad1 staining. n = 300 cells in each case. (C) Strains KGY5331, KGY5332, KGY5458, KGY5459, and KGY5537, which produce the indicated GFP- or YFP-tagged SPB component and carry a single, integrated copy of ppc89+ under control of the nmt1 promoter, were grown to midexponential phase at 32°C in medium containing thiamine. Cells were then grown in medium lacking thiamine for 24 h at 32°C, and live-cell images were captured. (D) nmt81-GFP-atb2 sid4-CFP nmt1-ppc89+ (KGY5576) cells were grown and imaged as in C. Bars, 5 μm.
To examine effects on the MT cytoskeleton, we also overproduced Ppc89 in cells expressing both the GFP-tagged α-tubulin gene, atb2+, under control of the nmt81 promoter (Sawin et al., 2004) and Sid4-CFP. Many cells (24%) were observed with greater-than-normal numbers of cytoplasmic MTs emanating from the SPBs (Figure 7D). There were also many cells (53%) with a single, extremely bright MT (or bundle of MTs) that extended from the edge of the nucleus (Figure 7D).
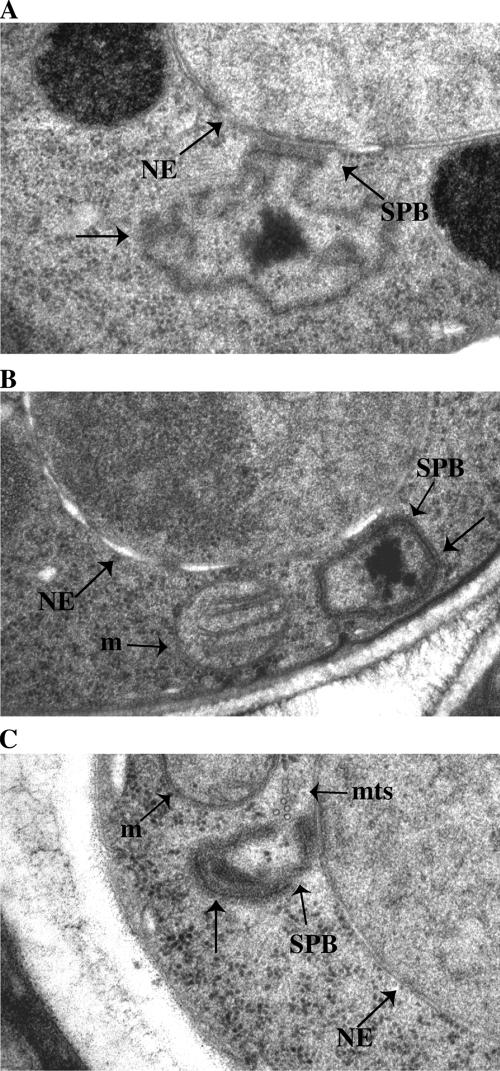
To examine the effect of Ppc89 overproduction on SPB morphology in greater detail, cells were viewed by EM. The majority of cells examined contained an elaborate extension of what appeared to be a reasonably normal SPB-like structure, although it extended away from the NE (Figure 8, A–C). In some sections, a dark line was visible through this structure, as in normal SPBs (Ding et al., 1997; Uzawa et al., 2004). In cells with this additional SPB-associated material, the SPB itself was not embedded within the NE, and the darkly staining material associated with the SPB on the nuclear side of the NE appeared normal. This extension of SPB material into the cytoplasm is consistent with the localization of various SPB proteins as observed by light microscopy in cells overproducing Ppc89 (Figure 7C). Also consistent with the light-microscopy data, nuclear MTs were not detected adjacent to the enlarged SPBs, but cytoplasmic MTs frequently were (Figure 8C and unpublished observations). Interestingly, in Ppc89-overproducing cells, there were no detectable electron-dense centers or irregularities in the surfaces of the SPBs, such as those typically observed between duplicated but unseparated SPBs (Ding et al., 1997; Uzawa et al., 2004; Figure 8, A–C). Therefore, these enlarged SPBs are unlikely to be duplicated.
Figure 8.
Representative electron micrographs of SPBs in cells overproducing Ppc89. Strain KGY5331 was grown as in Figure 7B. Note elaborate cytoplasmic SPB projections (unlabeled arrows). NE, nuclear envelope; m, mitochondria; mts, microtubules.
DISCUSSION
The SPB in yeast functions not only to nucleate and organize MTs but also as a signaling center for coordinating cell cycle events. Although several S. pombe SPB components have been identified, we do not know in detail how they interact with one another to form a functional SPB that organizes signaling modules. One such gap in knowledge is how the SIN signaling complex is tethered to the SPB. It is known that the scaffolding protein Sid4 is the most upstream component of the SIN and a stable SPB component, but it is not known how this protein is integrated into the SPB. In this study, we have identified and characterized a novel component of the S. pombe SPB, Ppc89, which binds directly to Sid4 and appears be the link between the SIN and the SPB. It is also important more globally for the organization of the SPB.
Ppc89 and the SIN
Ppc89 was identified in a TAP analysis of Cdc11, indicating that it associated with one or more SIN components. Through two-hybrid analyses and in vitro binding experiments, Ppc89 was determined to bind Sid4 directly, an association mediated by their respective C-termini and one that can be observed by FRET in vivo. These data indicate that Ppc89 and Sid4 interact on the outer surface of the SPB, where Sid4 (unpublished observation) and Sid2 (Sparks et al., 1999) have been shown to localize by immuno-EM.
In contrast to all known SIN components and regulators (reviewed by (Balasubramanian et al., 2004; Krapp et al., 2004b; Wolfe and Gould, 2005), Ppc89-GFP remains SPB-associated in sid4-SA1 cells, suggesting that Ppc89 links the SIN to the SPB through its association with Sid4. Indeed, immunolocalization of Ppc89-GFP by EM indicated that it localizes more centrally within the SPB than SIN components. Further evidence in support of this relative arrangement of proteins is the finding that the Sid4 N-terminus fused to the Ppc89 coiled-coil domains is fully functional for SIN signaling. This observation not only suggests a SIN tethering role for Ppc89 but indicates that the N-terminal 300 amino acids of Sid4 is solely responsible for its essential function in the SIN. This conclusion is consistent with the evidence that Sid4 residues 1–300 contain the docking sites for the checkpoint protein Dma1p (Guertin et al., 2002), the mitotic kinase Plo1 (Morrell et al., 2004), and also Cdc11, which in turn links to all other SIN components and Cdk1-cyclin B (Krapp et al., 2004a; Morrell et al., 2004). These data also indicate that the central and C-terminal coiled-coil domains of Sid4 serve solely as a SPB targeting module. Other evidence that Ppc89 tethers Sid4 to the SPB is that both Sid4 and Cdc11 were lost from SPBs in the absence of Ppc89 and that some cells with multiple nuclei and no septa, indicative of SIN defects, were observed in Ppc89-depleted cells.
Ppc89 and SPB Functions
Clearly, however, the major defect of cells lacking Ppc89 is not SIN mis-regulation. Depletion and overexpression experiments showed that Ppc89 is necessary for assembling and/or maintaining a variety of proteins at the SPB that perform different functions. For example, a number of MT defects are caused by Ppc89 overproduction or depletion. This is probably due, at least in part, to changes in the amount of SPB-localized γ-TuC, as measured in this study by the localization of Alp6, a core γ-TuC element. The γ-TuC localizes to the outer and inner faces of the S. pombe SPB, as well as to non-SPB interphase microtubule-organizing centers (iMTOCs) and is responsible for nucleating and anchoring MTs (Hagan and Petersen, 2000). The MT defects we observed in Ppc89-depletion and overproduction studies are consistent with defects in SPB MTOC functions. In Ppc89-overproducing cells, a greater number of MTs emanated from the enlarged SPBs, as would be expected if the γ-TuC had been preferentially recruited to SPBs rather than iMTOCs. Reciprocally, most cells depleted of Ppc89 contained a seemingly normal interphase array of MTs that could be generated from non-SPB MTOCs. However, because these non-SPB MTOCs do not contribute to spindle assembly, it is not surprising that bipolar-spindle formation and proper chromosome segregation were disrupted in Ppc89-depleted cells.
Ppc89 and SPB Structure
The aberrant phenotypes arising from overproduction or depletion of Ppc89 differ from those of any previously identified S. pombe SPB component. For example, Pcp1 overproduction induces formation of multiple foci throughout the cytoplasm and nucleus but not enlarged SPBs (Flory et al., 2002). Loss of other central SPB components with essential roles during vegetative growth, such as centrin/Cdc31, leads to formation of monopolar spindles in a high percentage of cells (Paoletti et al., 2003). Because the SPBs can still nucleate MTs in these mutants, it appears that the SPBs remain largely intact in the absence of these proteins. In contrast, our evidence indicates that Ppc89 integrates the assembly of a large number of SPB proteins, including Pcp1, into a well-organized structure. When excess Pcp89 is supplied, at least some portion of the SPB expands. It is interesting that the Ppc89-induced SPB structures extend primarily away from the NE, suggesting that some SPB-NE-tethering factors become limiting. Sad1 appears not to be such a factor, because it appears to be enriched in the nontethered structures. Reciprocally, when Ppc89 is depleted, the SPB apparently disassembles, because we have yet to identify a single SPB marker that remains concentrated as a spot in such cells (this study and unpublished observation). However, until EM studies are performed, the exact defect in SPB structure in the absence of Ppc89 cannot be known. Given that promoter shutoff experiments lead to heterogeneity in the extent of Ppc89 depletion at any given time point, it will be advantageous to perform EM studies on a tight conditional mutant. Such a temperature-sensitive strain is currently being constructed for these purposes.
The extension of the SPB induced by Ppc89 overproduction bears some similarity to superplaque structures formed by overproduction of S. cerevisiae Spc42. These superplaques are formed when the central plaque of the SPB enlarges laterally and bulges out of the NE (Donaldson and Kilmartin, 1996; Jaspersen et al., 2004). This enlargement, and the central role played by Spc42 in SPB organization (Muller et al., 2005), have led us to consider the possibility that S. pombe Ppc89 is a functional analog of S. cerevisiae Spc42. It should be noted that neither of these proteins has an evident homologue in the other yeast, although both contain extensive regions of predicted coiled-coil. Even if Ppc89 and Spc42 are not strictly functional equivalents, it is likely that Ppc89 function or protein level is regulated to control the extent of SPB growth during the cell cycle. This might occur similarly to the regulation of SPB duplication through Cdc28-mediated phosphorylation of Spc42 (Donaldson and Kilmartin, 1996; Jaspersen et al., 2004). Because Sid4 and Cdc11 bind the mitotic kinases Plo1 and Cdc2-Cdc13, respectively (Morrell et al., 2004), Ppc89 would be proximal to these regulators and a potential target.
The defects associated with Ppc89 depletion and overproduction suggest that, in addition to Sid4, it interacts with at least one other protein to organize SPBs. Preliminary mass spectrometric analysis of a Ppc89 purification indicates that several coil-coil proteins including Pcp1 (Flory et al., 2002), Kms2 (Miki et al., 2004), and Cut12 (Bridge et al., 1998) copurify (unpublished observation). These are strong candidates for additional direct binding partners and studies are underway to test this possibility.
Although our data suggest that Ppc89 anchors the SIN to the SPB, it is not yet clear how Ppc89 is organized within the SPB. Specifically, we do not know if the N-terminus of Ppc89 is buried within the SPB, facing into the cytoplasm to bind additional proteins, or both. The self-interacting central coiled-coil domain could allow dimerization in a head-to-head or head-to-tail configuration, and it could also allow oligomerization. Determining the configuration of Ppc89 at the SPB will help us understand how it organizes S. pombe SPB components into functional units.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Thomas Giddings and Erin White of the University of Colorado Electron Microscopy Service for providing electron micrographs. We thank all members of Gould lab for their advice and helpful discussions. We are also grateful to Rob Carnahan for help with confocal microscopy, Keith Gull for providing TAT-1 antibodies, Iain Hagan for providing anti-Sad1 antibodies, Trisha Davis for the Pcp1-GFP strain, and Dannel McCollum for the Cut11-GFP strain. J.A.R. was supported by National Institutes of Health training grant T32CA09385. This work was supported by the Howard Hughes Medical Institute, of which K.L.G. is an Investigator.
Footnotes
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-01-0039) on June 14, 2006.
REFERENCES
- Adams I. R., Kilmartin J. V. Spindle pole body duplication: a model for centrosome duplication? Trends Cell Biol. 2000;10:329–335. doi: 10.1016/s0962-8924(00)01798-0. [DOI] [PubMed] [Google Scholar]
- Bähler J., Wu J.-Q., Longtine M. S., Shah N. G., McKenzie A., 3rd, Steever A. B., Wach A., Philippsen P., Pringle J. R. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Balasubramanian M. K., Bi E., Glotzer M. Comparative analysis of cytokinesis in budding yeast, fission yeast and animal cells. Curr. Biol. 2004;14:R806–R818. doi: 10.1016/j.cub.2004.09.022. [DOI] [PubMed] [Google Scholar]
- Balasubramanian M. K., McCollum D., Chang L., Wong K. C., Naqvi N. I., He X., Sazer S., Gould K. L. Isolation and characterization of new fission yeast cytokinesis mutants. Genetics. 1998;149:1265–1275. doi: 10.1093/genetics/149.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardin A. J., Amon A. MEN and SIN: what’s the difference? Nat. Rev. Mol. Cell Biol. 2001;2:815–826. doi: 10.1038/35099020. [DOI] [PubMed] [Google Scholar]
- Basi G., Schmid E., Maundrell K. TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene. 1993;123:131–136. doi: 10.1016/0378-1119(93)90552-e. [DOI] [PubMed] [Google Scholar]
- Bridge A. J., Morphew M., Bartlett R., Hagan I. M. The fission yeast SPB component Cut12 links bipolar spindle formation to mitotic control. Genes Dev. 1998;12:927–942. doi: 10.1101/gad.12.7.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L., Gould K. L. Sid4p is required to localize components of the septation initiation pathway to the spindle pole body in fission yeast. Proc. Natl. Acad. Sci. USA. 2000;97:5249–5254. doi: 10.1073/pnas.97.10.5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding R., West R. R., Morphew D. M., Oakley B. R., McIntosh J. R. The spindle pole body of Schizosaccharomyces pombe enters and leaves the nuclear envelope as the cell cycle proceeds. Mol. Biol. Cell. 1997;8:1461–1479. doi: 10.1091/mbc.8.8.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson A. D., Kilmartin J. V. Spc42p: a phosphorylated component of the S. cerevisiae spindle pole body (SPB) with an essential function during SPB duplication. J. Cell Biol. 1996;132:887–901. doi: 10.1083/jcb.132.5.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doxsey S., McCollum D., Theurkauf W. Centrosomes in cellular regulation. Annu. Rev. Cell Dev. Biol. 2005;21:411–434. doi: 10.1146/annurev.cellbio.21.122303.120418. [DOI] [PubMed] [Google Scholar]
- Drummond D. R., Hagan I. M. Mutations in the bimC box of Cut7 indicate divergence of regulation within the bimC family of kinesin related proteins. J. Cell Sci. 1998;111(Pt 7):853–865. doi: 10.1242/jcs.111.7.853. [DOI] [PubMed] [Google Scholar]
- Eng J. K., McCormack A. L., Yates J. R. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Amer. Soc. Mass. Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- Flory M. R., Morphew M., Joseph J. D., Means A. R., Davis T. N. Pcp1p, an Spc110p-related calmodulin target at the centrosome of the fission yeast Schizosaccharomyces pombe. Cell Growth Differ. 2002;13:47–58. [PubMed] [Google Scholar]
- Giddings T. H., Jr, O’Toole E. T., Morphew M., Mastronarde D. N., McIntosh J. R., Winey M. Using rapid freeze and freeze-substitution for the preparation of yeast cells for electron microscopy and three-dimensional analysis. Methods Cell Biol. 2001;67:27–42. doi: 10.1016/s0091-679x(01)67003-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould K. L., Moreno S., Owen D. J., Sazer S., Nurse P. Phosphorylation at Thr167 is required for Schizosaccharomyces pombe p34cdc2 function. EMBO J. 1991;10:3297–3309. doi: 10.1002/j.1460-2075.1991.tb04894.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould K. L., Ren L., Feoktistova A. S., Jennings J. L., Link A. J. Tandem affinity purification and identification of protein complex components. Methods. 2004;33:239–244. doi: 10.1016/j.ymeth.2003.11.019. [DOI] [PubMed] [Google Scholar]
- Guertin D. A., Venkatram S., Gould K. L., McCollum D. Dma1 prevents mitotic exit and cytokinesis by inhibiting the septation initiation network (SIN) Dev. Cell. 2002;3:779–790. doi: 10.1016/s1534-5807(02)00367-2. [DOI] [PubMed] [Google Scholar]
- Hagan I., Yanagida M. The product of the spindle formation gene sad1+ associates with the fission yeast spindle pole body and is essential for viability. J. Cell Biol. 1995;129:1033–1047. doi: 10.1083/jcb.129.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan I. M., Petersen J. The microtubule organizing centers of Schizosaccharomyces pombe. Curr. Top. Dev. Biol. 2000;49:133–159. doi: 10.1016/s0070-2153(99)49007-6. [DOI] [PubMed] [Google Scholar]
- Ikemoto S., Nakamura T., Kubo M., Shimoda C. S. pombe sporulation-specific coiled-coil protein Spo15p is localized to the spindle pole body and essential for its modification. J. Cell Sci. 2000;113:545–554. doi: 10.1242/jcs.113.3.545. [DOI] [PubMed] [Google Scholar]
- James P., Halladay J., Craig E. A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen S. L., Huneycutt B. J., Giddings T. H., Jr, Resing K. A., Ahn N. G., Winey M. Cdc28/Cdk1 regulates spindle pole body duplication through phosphorylation of Spc42 and Mps1. Dev. Cell. 2004;7:263–274. doi: 10.1016/j.devcel.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Jaspersen S. L., Winey M. The budding yeast spindle pole body: structure, duplication, and function. Annu. Rev. Cell Dev. Biol. 2004;20:1–28. doi: 10.1146/annurev.cellbio.20.022003.114106. [DOI] [PubMed] [Google Scholar]
- Keeney J. B., Boeke J. D. Efficient targeted integration at leu1-32 and ura4-294 in Schizosaccharomyces pombe. Genetics. 1994;136:849–856. doi: 10.1093/genetics/136.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M., Schiebel E. Spc98p and Spc97p of the yeast γ-tubulin complex mediate binding to the spindle pole body via their interaction with Spc110p. EMBO J. 1997;16:6985–6995. doi: 10.1093/emboj/16.23.6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapp A., Cano E., Simanis V. Analysis of the S. pombe signalling scaffold protein Cdc11p reveals an essential role for the N-terminal domain in SIN signalling. FEBS Lett. 2004a;565:176–180. doi: 10.1016/j.febslet.2004.03.098. [DOI] [PubMed] [Google Scholar]
- Krapp A., Gulli M. P., Simanis V. SIN and the art of splitting the fission yeast cell. Curr. Biol. 2004b;14:R722–730. doi: 10.1016/j.cub.2004.08.049. [DOI] [PubMed] [Google Scholar]
- Krapp A., Schmidt S., Cano E., Simanis V. S. pombe cdc11p, together with sid4p, provides an anchor for septation initiation network proteins on the spindle pole body. Curr. Biol. 2001;11:1559–1568. doi: 10.1016/s0960-9822(01)00478-x. [DOI] [PubMed] [Google Scholar]
- Link A. J., Eng J., Schieltz D. M., Carmack E., Mize G. J., Morris D. R., Garvik B. M., Yates J. R., 3rd. Direct analysis of protein complexes using mass spectrometry. Nat. Biotechnol. 1999;17:676–682. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- MacCoss M. J., Wu C. C., Yates J. R., 3rd. Probability-based validation of protein identifications using a modified SEQUEST algorithm. Anal. Chem. 2002;74:5593–5599. doi: 10.1021/ac025826t. [DOI] [PubMed] [Google Scholar]
- Maundrell K. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene. 1993;123:127–130. doi: 10.1016/0378-1119(93)90551-d. [DOI] [PubMed] [Google Scholar]
- McCollum D., Gould K. L. Timing is everything: regulation of mitotic exit and cytokinesis by the MEN and SIN. Trends Cell Biol. 2001;11:89–95. doi: 10.1016/s0962-8924(00)01901-2. [DOI] [PubMed] [Google Scholar]
- McDonald W. H., Ohi R., Smelkova N., Frendewey D., Gould K. L. Myb-related fission yeast cdc5p is a component of a 40S snRNP-containing complex and is essential for pre-mRNA splicing. Mol. Cell. Biol. 1999;19:5352–5362. doi: 10.1128/mcb.19.8.5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki F., Kurabayashi A., Tange Y., Okazaki K., Shimanuki M., Niwa O. Two-hybrid search for proteins that interact with Sad1 and Kms1, two membrane-bound components of the spindle pole body in fission yeast. Mol. Genet. Genomics. 2004;270:449–461. doi: 10.1007/s00438-003-0938-8. [DOI] [PubMed] [Google Scholar]
- Moreno S., Klar A., Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Morrell J. L., et al. Sid4p-Cdc11p assembles the septation initiation network and its regulators at the S. pombe SPB. Curr. Biol. 2004;14:579–584. doi: 10.1016/j.cub.2004.03.036. [DOI] [PubMed] [Google Scholar]
- Muller E. G., Snydsman B. E., Novik I., Hailey D. W., Gestaut D. R., Niemann C. A., O’Toole E. T., Giddings T. H., Jr, Sundin B. A., Davis T. N. The organization of the core proteins of the yeast spindle pole body. Mol. Biol. Cell. 2005;16:3341–3352. doi: 10.1091/mbc.E05-03-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niccoli T., Yamashita A., Nurse P., Yamamoto M. The p150-Glued Ssm4p regulates microtubular dynamics and nuclear movement in fission yeast. J. Cell Sci. 2004;117:5543–5556. doi: 10.1242/jcs.01475. [DOI] [PubMed] [Google Scholar]
- Niwa O., Shimanuki M., Miki F. Telomere-led bouquet formation facilitates homologous chromosome pairing and restricts ectopic interaction in fission yeast meiosis. EMBO J. 2000;19:3831–3840. doi: 10.1093/emboj/19.14.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi M. D., Gould K. L. Characterization of interactions among the Cef1p-Prp19p-associated splicing complex. RNA. 2002;8:798–815. doi: 10.1017/s1355838202025050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoletti A., Bordes N., Haddad R., Schwartz C. L., Chang F., Bornens M. Fission yeast cdc31p is a component of the half-bridge and controls SPB duplication. Mol. Biol. Cell. 2003;14:2793–2808. doi: 10.1091/mbc.E02-10-0661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T. T., Tougan T., Okuzaki D., Kasama T., Nojima H. Mcp6, a meiosis-specific coiled-coil protein of Schizosaccharomyces pombe, localizes to the spindle pole body and is required for horsetail movement and recombination. J. Cell Sci. 2005;118:447–459. doi: 10.1242/jcs.01629. [DOI] [PubMed] [Google Scholar]
- Sawin K. E. Meiosis: organizing microtubule organizers. Curr. Biol. 2005;15:R633–R635. doi: 10.1016/j.cub.2005.08.001. [DOI] [PubMed] [Google Scholar]
- Sawin K. E., Lourenco P.C.C., Snaith H. A. Microtubule nucleation at non-spindle pole body microtubule-organizing centers requires fission yeast centrosomin-related protein mod20p. Curr. Biol. 2004;14:763–775. doi: 10.1016/j.cub.2004.03.042. [DOI] [PubMed] [Google Scholar]
- Shimanuki M., Miki F., Ding D. Q., Chikashige Y., Hiraoka Y., Horio T., Niwa O. A novel fission yeast gene, kms1+, is required for the formation of meiotic prophase-specific nuclear architecture. Mol. Gen. Genet. 1997;254:238–249. doi: 10.1007/s004380050412. [DOI] [PubMed] [Google Scholar]
- Shimoda C. Forespore membrane assembly in yeast: coordinating SPBs and membrane trafficking. J. Cell Sci. 2004;117:389–396. doi: 10.1242/jcs.00980. [DOI] [PubMed] [Google Scholar]
- Siam R., Dolan W. P., Forsburg S. L. Choosing and using Schizosaccharomyces pombe plasmids. Methods. 2004;33:189–198. doi: 10.1016/j.ymeth.2003.11.013. [DOI] [PubMed] [Google Scholar]
- Simanis V. Events at the end of mitosis in the budding and fission yeasts. J. Cell Sci. 2003;116:4263–4275. doi: 10.1242/jcs.00807. [DOI] [PubMed] [Google Scholar]
- Sparks C. A., Morphew M., McCollum D. Sid2p, a spindle pole body kinase that regulates the onset of cytokinesis. J. Cell Biol. 1999;146:777–790. doi: 10.1083/jcb.146.4.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundberg H. A., Davis T. N. A mutational analysis identifies three functional regions of the spindle pole component Spc110p in Saccharomyces cerevisiae. Mol. Biol. Cell. 1997;8:2575–2590. doi: 10.1091/mbc.8.12.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Kohda T., Yamashita A., Nonaka N., Yamamoto M. Hrs1p/Mcp6p on the meiotic SPB organizes astral microtubule arrays for oscillatory nuclear movement. Curr. Biol. 2005;15:1479–1486. doi: 10.1016/j.cub.2005.07.058. [DOI] [PubMed] [Google Scholar]
- Tasto J. J., Carnahan R. H., McDonald W. H., Gould K. L. Vectors and gene targeting modules for tandem affinity purification in Schizosaccharomyces pombe. Yeast. 2001;18:657–662. doi: 10.1002/yea.713. [DOI] [PubMed] [Google Scholar]
- Tomlin G. C., Morrell J. L., Gould K. L. The spindle pole body protein Cdc11p links Sid4p to the fission yeast septation initiation network. Mol. Biol. Cell. 2002;13:1203–1214. doi: 10.1091/mbc.01-09-0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbani L., Stearns T. The centrosome. Curr. Biol. 1999;9:R315–R317. doi: 10.1016/s0960-9822(99)80201-2. [DOI] [PubMed] [Google Scholar]
- Uzawa S., Li F., Jin Y., McDonald K. L., Braunfeld M. B., Agard D. A., Cande W. Z. Spindle pole body duplication in fission yeast occurs at the G1/S boundary but maturation is blocked until exit from S by an event downstream of cdc10+ Mol. Biol. Cell. 2004;15:5219–5230. doi: 10.1091/mbc.E04-03-0255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardy L., Toda T. The fission yeast γ-tubulin complex is required in G1 phase and is a component of the spindle assembly checkpoint. EMBO J. 2000;19:6098–6111. doi: 10.1093/emboj/19.22.6098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatram S., Tasto J. J., Feoktistova A., Jennings J. L., Link A. J., Gould K. L. Identification and characterization of two novel proteins affecting fission yeast γ-tubulin complex function. Mol. Biol. Cell. 2004;15:2287–2301. doi: 10.1091/mbc.E03-10-0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West R. R., Vaisberg E. V., Ding R., Nurse P., McIntosh J. R. cut11+: a gene required for cell cycle-dependent spindle pole body anchoring in the nuclear envelope and bipolar spindle formation in Schizosaccharomyces pombe. Mol. Biol. Cell. 1998;9:2839–2855. doi: 10.1091/mbc.9.10.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe B. A., Gould K. L. Split decisions: coordinating cytokinesis in yeast. Trends Cell Biol. 2005;15:10–18. doi: 10.1016/j.tcb.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Woods A., Sherwin T., Sasse R., MacRae T. H., Baines A. J., Gull K. Definition of individual components within the cytoskeleton of Trypanosoma brucei by a library of monoclonal antibodies. J. Cell Sci. 1989;93:491–500. doi: 10.1242/jcs.93.3.491. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.