Abstract
Silencing at the rDNA, HM loci, and telomeres in Saccharomyces cerevisiae requires histone-modifying enzymes to create chromatin domains that are refractory to recombination and RNA polymerase II transcription machineries. To explore how the silencing factor Sir2 regulates the composition and function of chromatin at the rDNA, the association of histones and RNA polymerase II with the rDNA was measured by chromatin immunoprecipitation. We found that Sir2 regulates not only the levels of K4-methylated histone H3 at the rDNA but also the levels of total histone H3 and RNA polymerase II. Furthermore, our results demonstrate that the ability of Sir2 to limit methylated histones at the rDNA requires its deacetylase activity. In sir2Δ cells, high levels of K4-trimethylated H3 at the rDNA nontranscribed spacer are associated with the expression of transcription units in the nontranscribed spacer by RNA polymerase II and with previously undetected alterations in chromatin structure. Together, these data suggest a model where the deacetylase activity of Sir2 prevents euchromatinization of the rDNA and silences naturally occurring intergenic transcription units whose expression has been associated with disruption of cohesion complexes and repeat amplification at the rDNA.
INTRODUCTION
Modified histones at silent genomic domains contribute to a chromatin environment that is refractory to gene expression and genetic recombination (reviewed in Strahl and Allis, 2000; Turner, 2000; Jenuwein and Allis, 2001). In Saccharomyces cerevisiae, chromatin at the homothallic mating-type loci HML and HMR, telomeres, and the ribosomal DNA locus (rDNA) silences genetic recombination and expression of native and ectopic genes transcribed by RNA polymerase (Pol) II. Silencing at the HM loci and telomeres has been studied extensively, whereas the mechanisms of Pol II silencing at the rDNA are not well characterized (reviewed in Moazed, 2001; Rusche et al., 2003). Increasing our understanding of the factors and mechanisms that regulate silencing at the rDNA will provide insight into the pathways that regulate gene expression and genome stability.
In S. cerevisiae, the rDNA contains ∼150–200 tandem copies of a 9.1-kilobase (kb) repeat, with each repeat containing a Pol I-transcribed 35S rRNA gene and a nontranscribed spacer (NTS) that is subdivided into NTS1 and NTS2 by the Pol III-transcribed 5S rRNA gene (reviewed in Warner, 1999; Figure 1). Despite high levels of transcription by Pol I and Pol III in the rDNA locus, Pol II-transcribed genes integrated into the rDNA are silenced (referred to as rDNA silencing) (Bryk et al., 1997; Fritze et al., 1997; Smith and Boeke, 1997). Additionally, silent chromatin at the rDNA is essential for repression of genetic recombination (Gottlieb and Esposito, 1989; Davis et al., 2000; Kobayashi et al., 2004) and extension of replicative life span (reviewed in Guarente, 2000).
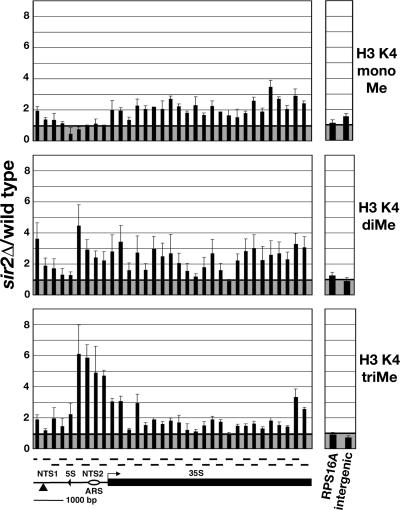
Figure 1.
Altered distribution of K4-methylated histone H3 at the rDNA in sir2Δ cells relative to wild-type cells. Graphical representations of the average ratio of %IP of K4-monomethylated (top; ±SE; n = 2 or 3), K4-dimethylated (middle; ±SE; n = 3), and K4-trimethylated (bottom; ±SE; n = 4) histone H3 in sir2Δ cells (MBY1238) relative to wild-type cells (MBY1198) at the rDNA, the RPS16A gene, and an intergenic region on chr VIII. Data presented were normalized to total histone H3 to correct for reduced levels of H3 in sir2Δ cells at some regions of the rDNA repeat (see text and Supplemental Figure S1). The structure of a 9.1-kb rDNA repeat unit is shown below the bottom panel, indicating the location of the Pol I-transcribed 35S rRNA (35S) gene and the NTS that is divided into NTS1 and NTS2 by the Pol III-transcribed 5S rRNA gene (5S). ARS, replication origin; bent line with arrow, transcription start site of the 35S rRNA gene; ▴, location of a silenced Ty1his3AI element present in one repeat, referred to as rDNA-Ty1 in text. Horizontal lines above the rDNA indicate PCR products generated during the analysis of ChIP experiments by qPCR.
Chromatin-associated proteins and modified histones regulate silencing of Pol II transcription at the rDNA (Bryk et al., 1997; Fritze et al., 1997; Smith and Boeke, 1997; Smith et al., 1999; Straight et al., 1999; Hoppe et al., 2002; Park et al., 2002; Dror and Winston, 2004; Kobayashi et al., 2004; Kuzuhara and Horikoshi, 2004; Machin et al., 2004; Ye et al., 2005). Net1 and the NAD-dependent histone deacetylase Sir2 are members of the RENT complex, which functions in silencing at the rDNA (Shou et al., 1999; Straight et al., 1999). The RENT complex is the functional equivalent of the Sir2–3-4 complex necessary for silencing at the HM loci and telomeres. In addition to being required for the association of Sir2 with the rDNA, Net1 interacts with Pol I and regulates the structure of the nucleolus (Straight et al., 1999; Shou et al., 2001). The histones H3 and H4 are substrates for the deacetylase activity of Sir2. At the HM loci and telomeres, hypoacetylated H3 and H4 promote the interaction of Sir3 and Sir4 with nucleosomes, thereby facilitating the formation and spread of silent chromatin (Braunstein et al., 1993, 1996; Hecht et al., 1995; Wu and Grunstein, 2000; Carmen et al., 2002; Liou et al., 2005). Hypoacetylated histones are present at the rDNA (Bryk et al., 2002; Buck et al., 2002; Sandmeier et al., 2002; Huang and Moazed, 2003), although how they contribute to rDNA silencing remains unclear.
Cells lacking K4-methylated histone H3 exhibit defects in transcriptional silencing at the rDNA and telomeres (Nislow et al., 1997; Briggs et al., 2001; Bryk et al., 2002; Nagy et al., 2002; Mueller et al., 2006). Set1 is the catalytic subunit of the COMPASS complex that is required for mono-, di-, and trimethylation of histone H3 on K4. In set1Δ cells, the lack of K4-methylated histone H3 is associated with the aberrant spreading of Sir proteins from telomeres into adjacent sequences. Reduced concentration of Sir complexes at telomeres in cells lacking Set1 is hypothesized to cause the loss-of-telomeric–silencing defect in set1Δ cells (Ng et al., 2002, 2003a; Meneghini et al., 2003; Martin et al., 2004; Katan-Khaykovich and Struhl, 2005). In contrast, at the rDNA, the levels of the silencing factors Sir2 and Net1 are equivalent in set1Δ and wild-type cells (Bryk et al., 2002). Moreover, in set1Δ cells, the levels of acetylated histone H3 and H4 remain low, indicating that Sir2 maintains its deacetylase function at the rDNA in the absence of K4-methylated H3. Thus, the mechanism by which silencing is lost at the rDNA in set1Δ cells is not equivalent to that at telomeres and has yet to be discovered.
In wild-type cells, silent chromatin at the rDNA contains low levels of acetylated H3 and H4 and low levels of K4-methylated histone H3. In cells lacking Set1 where rDNA silencing is impaired, histones at the rDNA remain hypoacetylated but are unmethylated. In contrast, in cells lacking Sir2, the levels of acetylated histone H3 and H4 are increased at several positions in the rDNA repeat and the level of K4-dimethylated H3 is increased at the NTS (Bryk et al., 2002). Together, these observations indicate that rDNA silencing is controlled by a specific combination of hypoacetylated and hypomethylated histones and that perturbations that alter the profile of modified histones disrupt rDNA silencing. To gain insight into the relationship between Sir2 and K4-methylated histone H3, we used chromatin immunoprecipitation (ChIP) and real-time PCR analysis to generate high-density profiles of K4-methylated histone H3 and Pol II across the rDNA repeat in wild-type and sir2Δ cells. Here, we show that novel changes occur at the rDNA in sir2Δ cells that reveal the central role of deacetylase activity of Sir2 in preventing aberrant Pol II transcription and protecting the structure and function of the unique chromatin domain present at the rDNA in S. cerevisiae. Furthermore, our findings provide mechanistic insight into the recent observation that high levels of transcription in the rDNA NTS are associated with amplification of rDNA repeats (Kobayashi and Ganley, 2005).
MATERIALS AND METHODS
Yeast Strains, Plasmids, and Media
Standard media recipes were used (Rose et al., 1990). YPADT is YPD medium supplemented with adenine sulfate (40 mg/l) and l-tryptophan (20 mg/l). Yeast strains MBY1198 (wild type), MBY1217 (set1Δ), and MBY1238 (sir2Δ) have been described previously (Briggs et al., 2001; Bryk et al., 2002). The XhoI–NotI fragment containing SIR2 or sir2H364Y from plasmids pAR455 (SIR2 HIS3 CEN) or pAR456 (sir2H364Y HIS3 CEN) (Rudner et al., 2005) were subcloned into pRS415 (LEU2 CEN; Stratagene) by using standard techniques. The resulting plasmids, MBB406 (pSIR2 LEU2 CEN) and MBB407 (psir2H364Y LEU2 CEN), were verified by DNA sequencing. pSIR2 and psir2H364Y were transformed into MBY1238 (Bryk et al., 2002). pSIR2 was also transformed into MBY1249 (MATα his3Δ200 ade2Δ::hisG leu2Δ0 trp1Δ63 ura3Δ0 met15Δ0 Ty1his3AI-236 Ty1ade2AI-515 set1Δ1:: TRP1 sir2Δ::KANMX4). The integrity of the pSIR2 and psir2H364Y plasmids was verified in silencing assays and by measuring the level of K9, K14-acetylated histone H3 at the rDNA by ChIP (our unpublished data). ChIP experiments (Supplemental Figure S3) using an anti-Sir2 antibody showed that the level of Sir2 protein at the rDNA in sir2Δ cells containing psir2H364Y was 0.5- to 1.6-fold of wild type, consistent with previously published reports (Tanny et al., 1999; Hoppe et al., 2002). Constructs for NTS-specific RNA probes were made by cloning PCR-generated NTS1 or NTS2 fragments into pSP70 (Promega, Madison, WI) to make MBB419 (pSP70-NTS1) or MBB413 (pSP70-NTS2). In vitro transcription reactions with SP6 or T7 RNA polymerase and linearized plasmid in the presence of [α-32P]CTP were performed to generate strand-specific RNA probes. Yeast strains with wild-type (wt) or temperature-sensitive (ts) Pol II used in temperature-shift experiments were made by genetic crosses: MBY1987 [MATα his3Δ200 leu2Δ0 ura3Δ0 lys2-128δ rpb1Δ187::HIS3 (pRP114 RPB1 LEU2 CEN)]; MBY1988 [his3Δ200 leu2Δ0 ura3-52 lys2-128δ rpb1Δ187::HIS3 sir2Δ::KANMX4) (pRP114 RPB1 LEU2 CEN)]; MBY1989 [MATα his3Δ200 leu2Δ0 ura3Δ0 rpb1Δ187::HIS3 (pRP1-1 rpb1-1 URA3 CEN)]; and MBY1992 [his3Δ200 leu2Δ0 lys2-128δ met15Δ0 trp1Δ63 ura3-52 sir2Δ::KANMX4 rpb1Δ187::HIS3 (pRP1-1 rpb1-1 URA3 CEN)].
Oligonucleotides for ChIP
For analysis of histone H3 at HMR and TEL-VIR by real-time PCR, oligonucleotides were designed to amplify ∼250- to 300-base pair products (sequences available upon request). The primers used to analyze the rDNA (Huang and Moazed, 2003), RPS16A, and the intergenic region on chr VIII (Ng et al., 2003c) have been described previously. For analysis of the promoter and 5′ end of the rDNA-Ty1 element by quantitative radioactive PCR, the primers annealed to the Ty1A region and downstream of the 5S rRNA gene (Bryk et al., 2002).
Chromatin Immunoprecipitation
Cells were grown and lysates prepared as described previously (Strahl-Bolsinger et al., 1997; Bryk et al., 2002) with the following modifications. Strains containing pSIR2 or psir2H364Y were grown in synthetic complete media lacking leucine with 2% glucose. Chromatin solutions were sonicated 12 times for 20 s each at 4°C by using a Branson Sonifier 250 at power setting 1.5 and 100% duty cycle to shear the chromatin to an average length of <500 base pairs. For each ChIP, 200 μl of sheared chromatin was incubated in a volume of 500 μl with specific antibodies for ∼12 h at 4°C. The antisera used were anti-histone H3, 5 μl (ab1791; Abcam, Cambridge, MA); anti-K4monoMeH3, 6 μl (ab8895; Abcam); anti-K4diMeH3, 15 μl (07-030; Upstate Biotechnology, Lake Placid, NY); anti-K4triMeH3, 5 μl (ab8580; Abcam); anti-Sir2, 1 μl (gift from Danesh Moazed, Department of Cell Biology, Harvard Medical School, Boston, MA); and anti-RNA Pol II carboxy-terminal domain (CTD) (4H8), 10 μl (ab5408; Abcam). The specificity of the antisera for K4-methylated H3 was verified by peptide blots by using unmodified and K4-methylated H3 peptides (our unpublished data).
Analysis of ChIPs
Quantitative radioactive PCRs were performed as described previously (Bryk et al., 2002) to determine the level of K4-di- and -trimethylated H3 at the rDNA-Ty1 element in wild-type (MBY1198), sir2Δ (MBY1238), and set1Δ (MBY1217) cells. ChIP experiments shown in Figures 1, 2, 4, and 5 and Supplemental Figures S1–S3 were analyzed by quantitative real-time PCR (qPCR). Duplicate reactions containing 1 μl of input DNA (1:100) or immunoprecipitated DNA (1:10) were amplified in 20 μl containing 1.25 μM of each oligonucleotide and 1× SYBR Green Dynamo Hot Start PCR mix (Finnzymes, Espoo, Finland) by using an iCycler Iq real-time PCR machine (Bio-Rad, Hercules, CA). The PCR parameters were 1 cycle of 95°C, 15 min; 40 cycles of 95°C for 30 s, 60°C for 30 s, 72°C for 30 s; 1 cycle at 95°C, 1 min; 1 cycle at 55°C, 1 min; and 80 cycles starting from 55°C with an increasing gradient of 0.5°C for 10 s each cycle to acquire a melting curve for each primer pair. The threshold cycle value was detected at the 72°C elongation step. Percent age of immunoprecipitation (%IP) was calculated by dividing the signal from IP DNA by the signal from input DNA. The %IP values from wild-type and sir2Δ cells analyzed by ChIP for K4-monomethylated H3, K4-dimethylated H3, and K4-trimethylated H3 were corrected for background by subtracting the %IP value obtained for set1Δ cells. For the 33 pairs of rDNA oligonucleotides, the range of average %IP values obtained from set1Δ cells were 0.031-0.128 for K4-monomethylated H3, 0.086-0.384 for K4-dimethylated H3, and 0.012-0.207 for K4-trimethylated H3. Graphs showing %IP data obtained for K4-methylated forms of histone H3 at the rDNA and two control loci before normalization to total histone H3 levels are shown in Supplemental Figure S2. The %IP values of K4-di- and -trimethylated H3 from cells containing pSIR2 or psir2H364Y were corrected for background by subtracting the %IP values obtained from set1Δ sir2Δ cells (MBY1249) containing pSIR2. The %IP values from wild-type and sir2Δ cells analyzed by ChIP for RNA Pol II and total histone H3 were corrected for background by subtracting the %IP value from a no-antibody control.
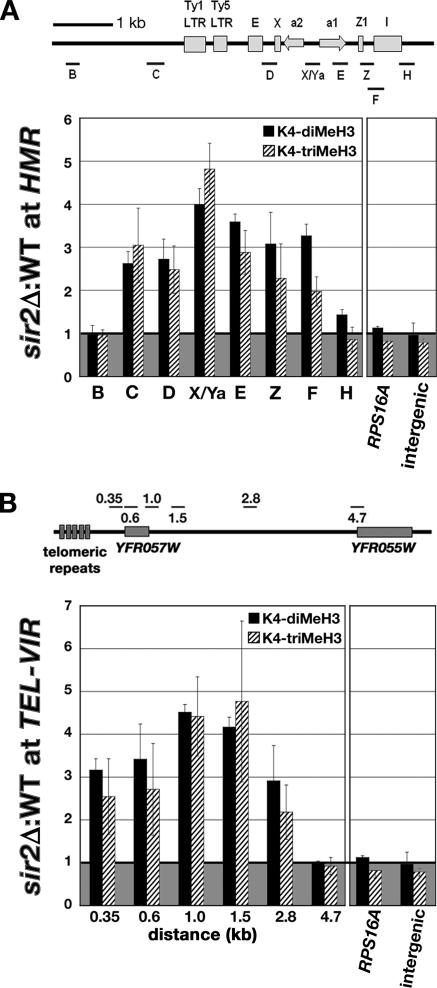
Figure 2.
The levels of K4 di- and trimethylated histone H3 were increased at HMR and TEL-VIR in sir2Δ cells. (A) Diagram of HMR located on chr III with the positions of silencers (E and I), cis-sequences (X and Z1), Ty long terminal repeats (LTR), and the genes HMRa1 and HMRa2. Short lines labeled B, C, D, X/Ya, E, Z, F, and H indicate the location of the PCR products used in ChIP analysis. The graph shows the average ratio of %IP of K4-dimethylated H3 (black bars) and K4-trimethylated H3 (hatched bars) at HMR, the RPS16A gene, and an intergenic region on chr VIII in sir2Δ cells relative to wild-type cells. (B) Diagram of TEL-VIR (right telomere on chr VI) with the positions of ORFs (YFR057W and YFR055W) and telomeric repeats indicated. Short lines labeled 0.35, 0.6, 1.0, 1.5, 2.8, and 4.7 indicate regions analyzed by qPCR. The numbers indicate the distance (in kb) from the right end of the PCR product to the right end of chr VI. Graph shows the average ratio of %IP of K4-di- and -trimethylated H3 at TEL-VIR in sir2Δ cells relative to wild-type cells. For both panels, the average ratio of % IP ± SE from sir2Δ cells to wild-type cells from three independent experiments is plotted.
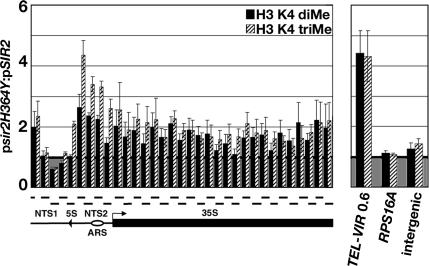
Figure 4.
Deacetylase activity of Sir2 is required to exclude K4-di- and -trimethylated H3 from the rDNA. Graph showing the average ratio of %IP of K4-dimethylated H3 (H3 K4 diMe, black bars) and K4-trimethylated H3 (H3 K4 triMe, hatched bars) across rDNA repeat (left) and control loci (right) in psir2H364Y/pSIR2 cells. The labels for the representation of rDNA repeat are defined in legend to Figure 1. Average ratio of %IP ± SE for three independent experiments is shown.
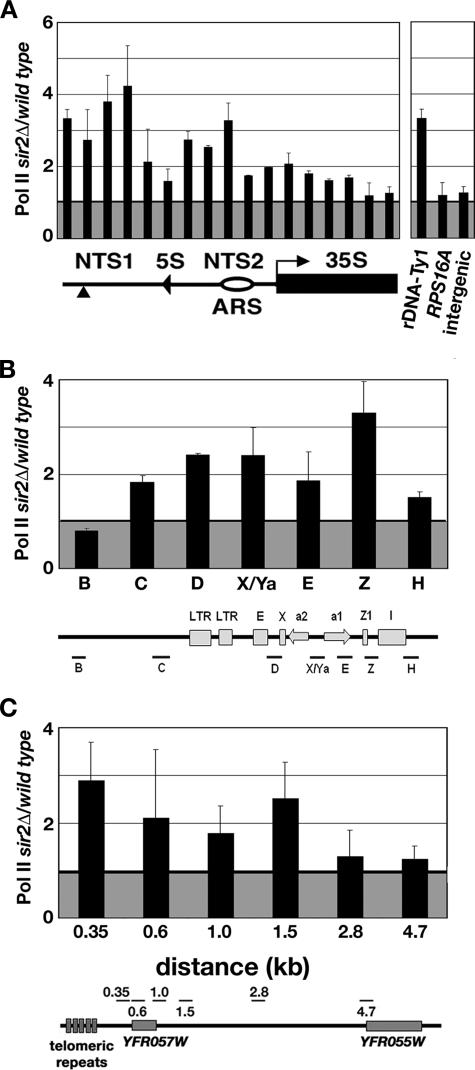
Figure 5.
Association of Pol II with silent loci is increased in sir2Δ cells. Graphs show the average ratio of %IP of Pol II at the (A) rDNA (left), the promoter of the rDNA-Ty1 element and control loci (right); (B) HMR; and (C) TEL-VIR in sir2Δ cells relative to wild-type cells. The representation of the rDNA in A indicates the portion of the rDNA repeat analyzed. Labels for the representations of the rDNA, HMR, and TEL-VIR are in the legends to Figures 1 and 2. The average ratio (±range) of Pol II measured in sir2Δ:wild-type cells from two independent experiments is plotted.
Northern Analysis
Total RNA was isolated from yeast cells as described previously (Bryk et al., 1997). Northern analyses were performed as described previously (Swanson et al., 1991). Strand-specific 32P-labeled RNA probes were used to detect NTS transcripts and PYK1 RNA. An ACT1 (+564 to +1200) probe and 35S rRNA probe were made by PCR amplification of yeast genomic DNA and then purified from an agarose gel and labeled with [α-32P]dATP by random priming (Ausubel et al., 1988). Growth conditions for the Pol II shut-off experiments (Figure 7) were as described previously (Herrick et al., 1990). Briefly, for each strain (MBY1987, MBY1988, MBY1989, and MBY1992), a 50-ml culture in YPADT was grown at 24°C to 1–2 × 107 cells/ml before being split into two 25-ml cultures. Twenty-five milliliters of fresh YPADT at 24°C was added to one culture, and growth was continued at 24°C for 30 min. To the other culture, 25 ml of YPADT at 48°C was added, and the culture was incubated at 36°C for 30 min. For time-course experiments, the volumes of the initial culture and fresh YPADT added were increased so that RNA was isolated from 50 ml of culture at each time point (0, 15, 30, and 60 min). Northern blots were quantified on a GE Healthcare (Little Chalfont, Buckinghamshire, United Kingdom) Storm 860 PhosphorImager by using ImageQuant software.
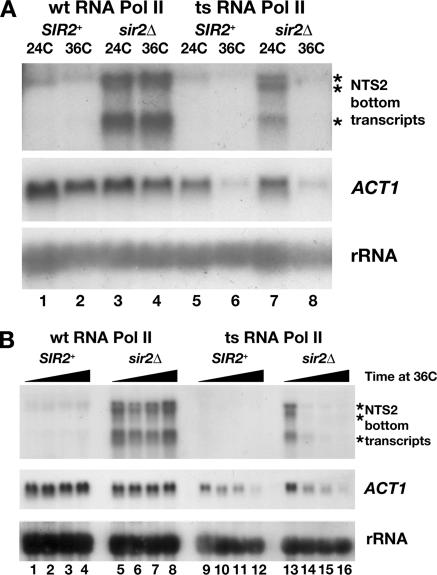
Figure 7.
NTS2 transcripts are made by RNA polymerase II. (A) Cultures of SIR2+ and sir2Δ cells containing a wild-type allele of RPB1 (wt RNA Pol II) or a ts allele rpb1-1 (ts RNA Pol II) were grown at 24°C and either maintained at 24°C or shifted to 36°C for 30 min (see Materials and Methods). Northern analysis of total RNA shows that NTS2 and ACT1 transcript levels are stable at 24 and 36°C in cells with wt RNA Pol II (lanes 1–4, top and middle) but are depleted after 30 min at 36°C in cells with ts RNA Pol II (lanes 5–8). The levels of Pol I transcribed rRNA are stable at 24 and 36°C (lanes 1–8, bottom). (B) Time-course analysis reveals that NTS2 transcripts are depleted 15 min after inactivation of Pol II. Cultures of cells grown at 24C were shifted to 36°C and aliquots were removed at 0, 15, 30, and 60 min. Total RNA was analyzed as described in A. Between 15 and 30 min after the shift to 36°C, the NTS2 transcripts were depleted significantly in cells containing the ts RNA Pol II but continue to be made in cells with the wt RNA Pol II (compare lanes 13–16 to 5–8, top). By 60 min, the level of ACT1 RNA was reduced significantly in cells with ts RNA Pol II, whereas rRNA levels remain stable (compare lanes 1–8 to 9–16, middle and bottom). Asterisks indicate positions of NTS2 transcripts.
Primer Extension
Total RNA (30–50 μg) from MBY1198, MBY1217, and MBY1238 was annealed to 1 pmol of 32P-labeled oligonucleotide (OM454; 5′-GTTGGTTTTGGTTTCGGTTG-3′) at 58°C for 20 min. Extension reactions were performed using the Primer Extension System-AMV Reverse Transcriptase kit (Promega) according to the manufacturer’s protocol. Sequencing reactions with oligonucleotide OM454 and double-stranded DNA template were performed using Sequenase Version 2.0 (USB, Cleveland, OH). Primer extension products and sequencing ladders were separated on 8 M urea, 8% polyacrylamide gels in 1× Tris borate-EDTA. Dried gels were visualized using a GE Healthcare Storm 860 PhosphorImager with ImageQuant software and autoradiography.
Analysis of Chromatin Structure with Micrococcal Nuclease
Yeast cells MBY1198, MBY1217, and MBY1238 were grown in 100 ml of YPADT medium to ∼1.2 × 107/ml and harvested by centrifugation. Preparation of spheroplasts and digestion with micrococcal nuclease (MNase; Worthington, Biochemicals, Freehold, NJ) was performed as described previously with slight modifications (Kent and Mellor, 1995). Spheroplasts (∼1.7 × 108) were incubated with MNase (0, 2.15, 4.3, 6.45, or 8.6 U/ml) for 4 min at 37°C and then the DNA was purified. Purified DNA that had not been treated with MNase was digested with 0.86 U of MNase for 2 min at 37°C to control for MNase sequence preferences. All DNAs were digested to completion with EcoRI or PvuII, separated on 1.5% agarose gels, transferred to nylon membranes, and analyzed by indirect end labeling. Blots of DNA digested with EcoRI were hybridized with a 32P-labeled probe that anneals to +2229 to +2496 of the rDNA locus (beginning of NTS1 is +1), and blots with DNA digested with PvuII were hybridized with a 32P-labeled probe that anneals to +1015 to +1268 of the rDNA. Positions of MNase cleavages and nucleosomes were calculated using restriction fragments as a reference.
RESULTS
Sir2 Excludes K4-Methylated Histone H3 from Silent Chromatin
Cells lacking Sir2 have high levels of acetylated histones at the HM loci and telomeres (Braunstein et al., 1993, 1996) and the rDNA (Bryk et al., 2002; Buck et al., 2002; Sandmeier et al., 2002; Huang and Moazed, 2003). We observed that K4-dimethylated histone H3 was also increased at the rDNA NTS in sir2Δ cells (Bryk et al., 2002), suggesting that Sir2 regulates the levels of K4-methylated histone H3 at the rDNA. Specific antisera against three forms of K4-methylated H3, K4-mono-, di-, and -trimethylated histone H3 have been used in numerous studies investigating the association of K4-methylated H3 with genes that are actively transcribed by RNA Pol II. These studies, in S. cerevisiae, have shown that K4-dimethylated H3 is enriched at euchromatin and that K4-trimethylated H3 is present at high levels at genes transcribed by Pol II (Bernstein et al., 2002; Santos-Rosa et al., 2002, 2004; Krogan et al., 2003; Ng et al., 2003b, c). Therefore, high levels of K4-methylated H3 at the rDNA NTS were unexpected due to the lack of Pol II-transcribed genes at the rDNA.
We examined the effect of Sir2 on the association of each of the three forms of K4-methylated H3 across the rDNA repeat. ChIPs using antisera against histone H3 as well as K4-mono-, di-, or -trimethylated histone H3 were analyzed by qPCR by using 33 primer pairs that amplify 260- to 280-base pair regions at intervals of ∼300 base pairs across the 9.1-kb rDNA repeat and two sets of primers that amplify control loci. Samples from cells lacking Set1 that are devoid of K4-mono-, di-, and -trimethylated histone H3 were included to provide a measurement of background that was subtracted from the signal obtained from wild-type and sir2Δ cells (see Materials and Methods). To determine whether Sir2 affected the level of total histone H3 at the rDNA, ChIP experiments were performed with antisera that recognize the C-terminal tail of histone H3. The interaction of the anti-histone H3 antisera with histone H3 is independent of modifications present at the N terminus of H3. The results of these experiments revealed that the level of total histone H3 was significantly lower at several positions in the rDNA repeat in sir2Δ cells compared with wild-type cells (Supplemental Figure S1). To control for differences in the amount of total histone H3 at the rDNA, in Figure 1, we have normalized data examining the levels of K4-methylated histones at the rDNA to the level of total histone H3 in wild-type and sir2Δ cells.
The results of ChIP experiments using antisera specific for K4-mono, di-, and -trimethylated H3 showed significant changes in the profile of K4-methylated histone H3 at the rDNA in sir2Δ cells compared with wild-type cells (Figure 1 and Supplemental Figure S2). We observed that K4-methylated histone H3 was excluded from the rDNA repeat in a Sir2-dependent manner. The profile of each form of K4-methylated H3 across the rDNA repeat in sir2Δ cells was distinct. In cells lacking Sir2, the level of K4-monomethylated histone H3 was increased over the 35S rRNA gene and the NTS1 portion of the NTS (Figure 1, top; and Supplemental Figure S2, top). The level of K4-dimethylated H3 was higher across most of the rDNA repeat, including regions in NTS1, NTS2, and the 35S rRNA gene (Figure 1, middle; and Supplemental Figure S2, middle). Strikingly, the level of K4-trimethylated histone H3 was increased primarily at the NTS2 region of the rDNA with levels that were approximately five- to sixfold higher in sir2Δ cells than wild-type cells (Figure 1, bottom and Supplemental Figure S2, bottom). Sir2 also affected the level of K4-trimethylated H3 at the beginning and end of the Pol I-transcribed 35S rRNA gene. The levels of K4-methylated H3 were similar in wild-type and sir2Δ cells at genomic loci that were analyzed as controls, RPS16A, a highly expressed Pol II-transcribed gene, and an intergenic region on chr VIII that is devoid of known Pol II-transcribed open reading frames (ORFs), indicating that Sir2 does not regulate K4-methylated H3 at these regions (Figure 1, right and Supplemental Figure S1, right). In summary, our data indicate that in cells lacking Sir2, the rDNA is exposed to the Set1-containing complex COMPASS that is required for the methylation of histone H3 on K4. Moreover, because the K4-trimethylated form of histone H3 has been found exclusively at genes that are actively transcribed by Pol II, our results suggest that Pol II may also be present at the rDNA NTS.
To understand how Sir2 affected the distribution of K4-methylated histone H3 at other silent loci, we performed ChIP experiments using wild-type, sir2Δ, and set1Δ cells to obtain profiles of K4-methylated H3 at the HMR locus and near the telomere on the right arm of chr VI (TEL-VIR). Experiments measuring histone H3 at HMR and TEL-VIR revealed that the average ratio of histone H3 in sir2Δ to wild-type cells was between 0.9 and 1.2, indicating that the level of histone H3 was similar in wild-type and sir2Δ cells (our unpublished data). In contrast to the rDNA where we observed distinct profiles of K4-methylated H3, the levels of K4-di- and -trimethylated H3 were increased similarly across ∼4 kb of the HMR locus and up to 2.8 kb from the right end of chr VI (Figure 2, A and B). At HMR, the highest levels of K4 di- and -trimethylated histone H3 were present at a region containing the Pol II-transcribed a1 and a2 genes (Figure 2A, primer pair X/Ya). The region of HMR where we detect changes in the association of K4-methylated histone H3 falls within the boundaries of the silent domain that were mapped in previous studies (Donze et al., 1999; Donze and Kamakaka, 2001). At TEL-VIR, the highest levels of K4 di- and -trimethylated histone H3 were present 1.0–1.5 kb from the end of the chromosome. This region of chr VI contains the promoter and 5′ end of the Pol II-transcribed gene YFR057W, whose expression has been shown to be increased in cells lacking Sir2 or Ubp10 (Wyrick et al., 1999; Emre et al., 2005). In contrast, we did not observe an effect of Sir2 on the levels of K4-methylated H3 at YFR055W a Pol II-transcribed gene located 4.7 kb from the end of chr VI. In summary, in cells lacking Sir2, the rDNA, HMR, and TEL-VIR are exposed to factors that are required for the methylation of histone H3 on K4.
K4-Methylated H3 at the Promoter of the rDNA-Ty1 Element Is Regulated by Sir2
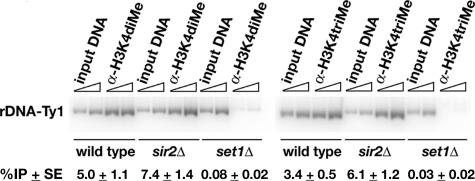
Several of the strains used in this study contain a single copy of a genetically marked Ty1 element located in NTS1 in one of the repeats of the rDNA array (see Figure 1; Bryk et al., 1997). In previous work, we showed that expression of the rDNA-Ty1 element is increased in sir2Δ cells and that the promoter of the rDNA-Ty1 element contains low levels of K4-dimethylated H3 in wild-type cells (Bryk et al., 1997, 2002). It is unlikely that the rDNA-Ty1 element is responsible for the increased levels of K4-methylated H3 observed in sir2Δ cells because it is present in only one of the ∼150–200 rDNA repeats. However, we wanted to determine whether Sir2 affected the levels of K4-methylated H3 at the promoter and 5′ end of the Pol II-transcribed rDNA-Ty1 element, which would be consistent with its increased expression in sir2Δ cells. Using ChIP and radioactive PCR, we compared the levels of K4-di- and -trimethylated H3 at the rDNA-Ty1 element by using primers that amplify a 540-base pair fragment containing the promoter and 5′ end of the rDNA-Ty1 element (Figure 3). We found that the levels of K4-dimethylated H3 and total histone H3 present at the rDNA-Ty1 element were not significantly different in sir2Δ and wild-type cells (Figure 3; our unpublished data). In contrast, the level of K4-trimethylated H3 at the rDNA-Ty1 element was 1.8-fold higher in sir2Δ cells than wild-type cells, consistent with increased expression of the rDNA-Ty1 element in sir2Δ cells (Bryk et al., 1997).
Figure 3.
sir2Δ cells have higher levels of K4-trimethylated H3 at the rDNA-Ty1 element. ChIP experiments were analyzed by quantitative radioactive PCR to measure K4-di- and -trimethylated H3 at the promoter region of the silent Ty1his3AI element in the rDNA in wild-type and sir2Δ cells. Immunoprecipitations using set1Δ cells were performed to provide a measure of background for the ChIP experiments. The average %IP ± SE of K4-dimethylated H3 at the rDNA Ty1 promoter region from three independent experiments in wild-type cells was 5.0 ± 1.1, in sir2Δ cells is 7.4 ± 1.4, and in set1Δ cells is 0.08 ± 0.02. The average %IP ± SE of K4-trimethylated H3 at the rDNA-Ty1 promoter region from four independent experiments in wild-type cells was 3.4 ± 0.5, in sir2Δ cells is 6.1 ± 1.2, and in set1Δ cells is 0.03 ± 0.02. Slanted triangles indicate a twofold increase in the amount of template DNA used in the PCR reactions.
Deacetylase Activity of Sir2 Excludes K4-Di- and -Trimethylated H3 from the rDNA
One model to account for increased levels of K4 methylated histone H3 at the rDNA is that the physical presence of Sir2 prevents the association of factors required for the methylation of histone H3. In this steric model, in the absence of Sir2, rDNA chromatin would be more accessible, allowing higher levels of K4-methylated histone H3. An alternative model posits that the deacetylase activity of Sir2 contributes to a hypoacetylated chromatin domain at the rDNA that hinders the methylation of histone H3.
To distinguish between these two modes of regulation by Sir2, we used a catalytically inactive allele of SIR2, sir2H364Y, which encodes a mutant Sir2 protein that retains the ability to bind to the rDNA but lacks histone deacetylase activity (Tanny et al., 1999; Hoppe et al., 2002). ChIP experiments were performed using antisera against K4-di- and -trimethylated H3 and cross-linked chromatin from sir2Δ cells containing a plasmid with either a wild-type copy of SIR2 (pSIR2) or the mutant allele of SIR2 (psir2H364Y). Signal obtained from set1Δ sir2Δ double mutant cells containing the pSIR2 plasmid was subtracted as background in the analysis of these ChIP experiments (see Materials and Methods). The data revealed that the levels of K4-di- and -trimethylated H3 were higher at the rDNA in cells expressing sir2H364Y compared with cells with pSIR2 (Figure 4). Our results indicate that the deacetylase activity of Sir2 is required to maintain low levels of K4-methylated H3 at the rDNA. In addition, whereas the mutant Sir2 protein did not affect the levels of methylated H3 at RPS16A or the intergenic region, the levels of K4-di- and -trimethylated histone H3 were increased significantly at a region 0.6 kb from the right end of chr VI (Figure 4, right). Together, our results indicate that the deacetylase activity of Sir2 is required to maintain low levels of K4-di- and -trimethylated H3 at the rDNA and TEL-VIR.
Sir2 Excludes RNA Pol II from Silent Chromatin
In sir2Δ cells, the average %IP value of K4-trimethylated H3 of 21–22% measured at NTS2 was similar to the value of 23% measured at the Pol II-transcribed RPS16A gene (Supplemental Figure S2), suggesting that Pol II might be present at higher levels at the rDNA in sir2Δ cells. This idea is supported by several lines of evidence. First, in S. cerevisiae, K4-trimethylated histone H3 has been shown to be associated with genes transcribed by RNA Pol II (Bernstein et al., 2002; Santos-Rosa et al., 2002, 2004). In addition, at HMR and TEL-VIR in sir2Δ cells, we observed the highest levels of K4-trimethylated H3 at regions that contain one or more Pol II-transcribed genes (Figure 2), whose expression is increased in sir2Δ cells (Wyrick et al., 1999; Rusche et al., 2003; Emre et al., 2005). To determine whether the higher level of K4-trimethylated H3 observed over the rDNA NTS region in sir2Δ cells was associated with increased levels of Pol II, we performed ChIP experiments by using antisera that recognize the CTD of Pol II. We found that the association of Pol II with the NTS1 and NTS2 regions of the rDNA, the 5′ end of the rDNA-Ty1 element HMR, and TEL-VIR was higher in sir2Δ cells than in wild-type cells (Figure 5). In contrast, Pol II levels at RPS16A and the intergenic region were equivalent in sir2Δ and wild-type cells (Figure 5A, right). Previous work has shown that Pol II was increased at HMR and TEL-VIR in silencing-defective cells (Chen and Widom, 2005). Our data reveal that Pol II was also increased at the rDNA in sir2Δ cells. Considering our observations of increased levels of K4-trimethylated H3 and Pol II at the rDNA, we wanted to determine whether Pol II transcription was occurring in the NTS in sir2Δ cells.
Sir2 Regulates Expression of Endogenous Transcription Units in the rDNA NTS
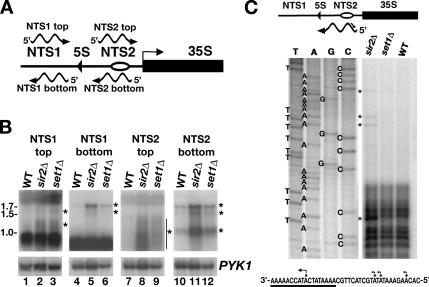
We detected high levels of Pol II at NTS1 and NTS2 consistent with Pol II transcription in the NTS region. A recent report has shown that bidirectional transcripts can be detected from NTS1 in sir2Δ cells (Kobayashi and Ganley, 2005). The observation of high levels of K4-trimethylated H3 in NTS2 suggested to us that Pol II transcription might be occurring in NTS2 as well. Northern analyses were performed using strand-specific probes to detect transcripts with the same polarity as the NTS1 top, NTS1 bottom, NTS2 top, or NTS2 bottom strand to determine whether transcription was occurring in the rDNA NTS region in sir2Δ cells or set1Δ cells, which both exhibit defects in rDNA silencing. To control for equivalent loading of RNA, we used a PYK1-specific RNA probe. NTS-specific transcripts, whose levels were increased significantly in sir2Δ and set1Δ cells, were identified using Northern blot experiments (Figure 6). Hybridization with a probe that detects transcripts with the same polarity as the top strand of NTS1 revealed transcripts in RNA from sir2Δ and set1Δ cells, similar to those previously identified by (Ganley et al., 2005) (Figure 6B, wild type, lane 1; sir2Δ, lane 2; and set1Δ, lane 3). Likewise, a transcript of ∼1.7 kb with the same polarity as the bottom strand of NTS1 was detected in RNA from sir2Δ (lane 5) and set1Δ cells (lane 6). On longer exposure of this blot, a faint 1.5-kb transcript could also be detected in RNA from sir2Δ and set1Δ cells. In lanes 7–9, by using a probe with the same polarity as the top strand of NTS2, we detected a faint smear in RNA from sir2Δ and set1Δ cells that was likely to represent transcripts that ranged in length from ∼0.8 to 1.5 kb. In lanes 10–12, by using a probe to detect RNA with the same polarity as the bottom strand of NTS2, we identified three transcripts of ∼1.7, 1.5, and 1.0 kb in RNA from sir2Δ and set1Δ cells. Although these 1.7-, 1.5-, and 1.0-kb transcripts could be detected at low levels in RNA from wild-type cells (lane 10), they were enriched significantly in RNA from sir2Δ (lane 11) and set1Δ cells (lane 12). Thus, these data show that, in addition to transcripts from NTS1 that have been characterized previously, transcripts can be detected from NTS2.
Figure 6.
Endogenous transcription units in the rDNA NTS are expressed in rDNA silencing-defective mutants. (A) Schematic of rDNA NTS showing the polarity of transcripts detected by probes used in Figure 6B. Other labels, as in Figure 1. (B) Representative Northern blots of total RNA from wild-type, sir2Δ, and set1Δ cells hybridized to strand-specific probes that are complementary to transcripts with the same polarity as the NTS1 top (lanes 1–3), NTS1 bottom (lanes 4–6), NTS2 top (lanes 7–9), and NTS2 bottom (lanes 10–12) strands. For each blot, PYK1 RNA levels were used to normalize for the amount of RNA analyzed. Numbers on left, transcript length in kb. Asterisks or line with asterisks, position of NTS-specific transcripts. (C) Primer extension analysis to map the 5′ ends of the NTS2 bottom-strand transcripts. Schematic at top represents an NTS2 transcript and the oligonucleotide (OM454, line with arrowhead) that was used to map the 5′ ends (not to scale). DNA sequence ladder and primer extension reactions revealed one major 5′ end within the consensus TSS (underlined in DNA sequence representation of the bottom strand of NTS2 at base of C) and three minor start sites upstream of the TSS. Asterisks and/or bent arrows indicate 5′ ends.
The sequence of the rDNA NTS2 region was scanned by eye for the presence of Pol II-specific regulatory sequences. We identified a consensus Pol II transcription start site (TSS) sequence, 5′-A-(Arich)5-N-Py-A-(A/T)-N-N-(Arich)6-3′, where A is the TSS (Zhang and Dietrich, 2005), between the rDNA autonomously replicating sequence (ARS) in NTS2 and beginning of the 35S rRNA gene (Figure 6C, bottom). This sequence was recognized previously as a conserved sequence element (rCNS6) in the rDNA NTS of several yeasts related to S. cerevisiae (Ganley et al., 2005). No sequences with an exact match to a consensus TATA box sequence, 5′-TATA(A/T)A(A/T)-3′, were found upstream of the TSS sequence in NTS2. After narrowing down the endpoints of the transcripts by using Northern analysis with oligonucleotide probes that spanned the rDNA NTS (our unpublished data), we performed primer-extension reactions to map the 5′ ends of the transcripts with the same polarity as the bottom strand of NTS2 (hereafter referred to as NTS2 transcripts). Total RNA from wild-type, sir2Δ, and set1Δ cells was subject to reverse transcription with a primer that was extended toward the predicted 5′ end of the NTS2 transcripts (see schematic at top of Figure 6C). In RNA from sir2Δ cells, the major 5′ end of the NTS2 transcripts was detected within the TSS consensus sequence at the position corresponding to the A (Figure 6C), indicating that the major transcription start site for the NTS2 transcripts was at the same base that was identified for >200 Pol II-transcribed genes in S. cerevisiae (Zhang and Dietrich, 2005). Minor 5′ ends were detected upstream of the major TSS. Although longer exposures of the primer-extension gels revealed the presence of these bands in the set1Δ lane, they were barely or not at all detectable in the wild-type lane. Based on our Northern and 5′-end mapping data, we conclude that the NTS2 transcripts initiate ∼300 base pairs upstream of the 35S rRNA gene and extend across NTS2 and the 5S rRNA gene into NTS1. Consistent with this conclusion, we were able to detect the 1.7- and 1.5-kb transcripts in RNA from sir2Δ cells using a probe to the 5S rRNA gene (our unpublished data).
To determine whether the NTS2 transcripts were made by Pol II, we used wild-type and sir2Δ strains lacking the genomic RPB1 gene that encodes the largest subunit of Pol II with either the wt RPB1 gene or a ts mutant allele, rpb1-1 provided from a plasmid (see Materials and Methods). Strains with rpb1-1 make functional Pol II at 24°C; however, upon shift of these cells to 36°C, Pol II becomes nonfunctional. RNA from cultures grown continuously at 24°C and from cultures shifted from 24 to 36°C for 30 min was hybridized with probes specific for the NTS2 transcripts, Pol II-transcribed ACT1 RNA, and Pol I-transcribed rRNA (Figure 7A). In wild-type and sir2Δ cells with RPB1 (wt RNA Pol II), ACT1 and rRNA transcript levels were maintained in cells shifted to 36°C (Figure 7A, lanes 1–4, middle and bottom), as were the NTS2 transcripts in total RNA from sir2Δ cells (Figure 7A, lanes 3 and 4, top). However, in cells with rpb1-1 (ts RNA Pol II, Figure 7A, lanes 5–8), ACT1 RNA was present in cultures grown at 24°C but disappeared after 30 min at 36°C (compares lanes 5 and 6 or 7 and 8). Likewise, the NTS2 transcripts present in the sir2Δ cells were gone after 30 min at 36°C (compare lanes 7 and 8). The level of transcript from the Pol I-transcribed 35S rRNA gene was stable at 36°C. These data suggest that the NTS2 transcripts were made by Pol II and not by Pol I. In addition, it is unlikely that the NTS2 transcripts were made by Pol III, as numerous polyT>5 stretches, which act as terminators for Pol III (Allison and Hall, 1985; Braglia et al., 2005), are present in the rDNA NTS.
The stability of ACT1 and NTS2 transcripts in RPB1 and rpb1-1 cells was measured over time at 36°C. Cultures grown at 24°C were shifted to 36°C, and aliquots of culture were removed for isolation of total RNA at 0, 15, 30 and 60 min after the shift to 36°C. A representative Northern blot in Figure 7B shows that although the levels of NTS2 transcripts were maintained in sir2Δ cells with wt RNA Pol II (lanes 5–8), the NTS2 transcripts were depleted in the rpb1-1 cells 15–30 min after the shift to 36°C (lanes 13–16). Likewise, ACT1 transcript levels were decreased over the time course at 36°C in rpb1-1 cells. Together, these data indicate that the NTS2 transcripts were made by Pol II. In further support of this conclusion, we found that the NTS2 and ACT1 transcripts were enriched in RNA immunoprecipitations using antisera against the trimethylguanosine cap present at the 5′ end of Pol II-transcribed RNAs (our unpublished data).
Nucleosome Positioning at NTS2 Is Altered in Cells with rDNA-silencing Defects
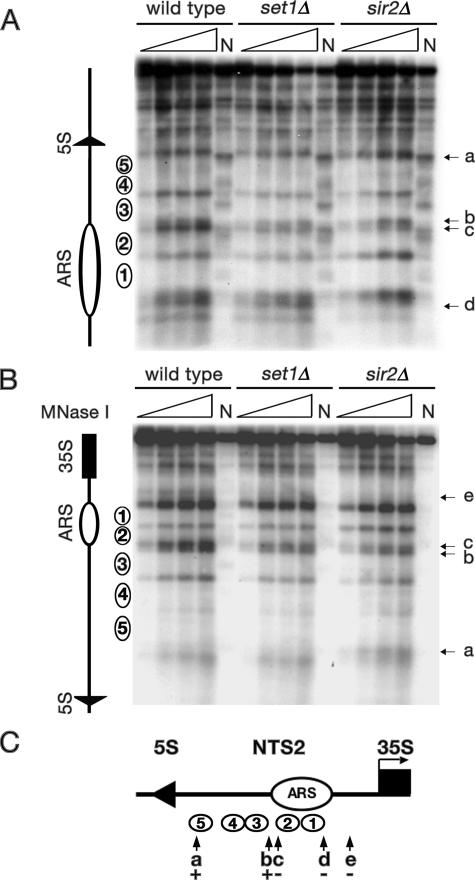
Previous studies revealed that cells lacking Sir2 have altered chromatin structure in the rDNA (Fritze et al., 1997; Cioci et al., 2002); however, these studies did not focus on NTS2 where we predict changes in nucleosome positioning might occur because of increased transcription of the NTS2 by Pol II. To determine whether nucleosome positioning in the NTS2 region of the rDNA was altered in cells lacking Sir2 or Set1, we performed indirect end-labeling analysis of MNase-digested chromatin. The rDNA repeats in S. cerevisiae exist in two configurations that are correlated with their accessibility to a cross-linking reagent psoralen and Pol I transcription activity (Dammann et al., 1993). Open rDNA repeats are accessible to psoralen and transcribed by Pol I, whereas closed repeats are inaccessible to psoralen and not transcribed. It is important to consider that similar to the ChIP analyses, MNase experiments at the rDNA evaluate populations of cells, with each cell containing 150–200 rDNA repeats that are in different conformations with variable MNase accessibilities. It is possible that there are stronger effects on individual repeats, however, the alterations we observed represent the average accessibility of all the rDNA repeats. Thus, the ability to see even modest alterations in MNase cleavage over 150–200 repeats is significant.
Five positioned nucleosomes have been mapped in the NTS2 region of the rDNA (Figure 8, open circles labeled 1–5; Vogelauer et al., 1998). We detected alterations in MNase accessibility at several positions in the NTS2 region in sir2Δ cells and set1Δ cells (Figure 8, A–C). Between the ARS and the transcription start site of the 35S rRNA gene, in the region containing the TSS for the NTS2 transcripts, an MNase cleavage site was clearly missing in chromatin from sir2Δ cells and set1Δ cells that was present in wild-type chromatin (Figure 8, B and C, arrow marked e). This change may reflect protection by Pol II or its associated factors in sir2Δ and set1Δ cells. Other changes in MNase accessibility were detected in sir2Δ cells only, including a new cleavage site in NTS2 upstream of the Pol III-transcribed 5S gene (Figure 8, A–C, arrow marked a) and a missing cleavage site near the ARS (Figure 8, A and C, arrow marked d). In addition, we detected subtle changes in two MNase cleavage sites between nucleosomes 2 and 3 (Figure 8, A–C, arrows marked b and c). Our results indicate that rDNA chromatin structure was altered near the TSS sequence in the NTS2 region of the rDNA repeat in sir2Δ cells and set1Δ cells. These changes in chromatin structure are consistent with the observation that sir2Δ and set1Δ cells, which have profoundly different types of modified histones at the rDNA, both have defects in silencing of Pol II-transcribed genes at the rDNA.
Figure 8.
MNase accessibility of chromatin in the NTS2 region of the rDNA is altered in set1Δ and sir2Δ cells. DNA purified after MNase treatment of spheroplasts from wild-type, set1Δ, or sir2Δ cells was digested with EcoRI (A) or PvuII (B). Schematic of NTS2 with five positioned nucleosomes (numbered ovals) identified here and in a previous study (Vogelauer et al., 1998) is shown on the left of each panel and in C. Triangles, increasing amounts of MNase; N, naked DNA; arrows with lowercase letters, altered MNase accessibility (see text); other labels as in Figure 1. (C) The NTS2 region of the rDNA. Numbered circles, positioned nucleosomes; + and −, extent of MNase digestion in sir2Δ cells relative to wild-type cells.
DISCUSSION
Modified histones play a central role in regulating gene expression and gene silencing in eukaryotes. We have uncovered a functional relationship between Sir2, Pol II, and K4-methylated histone H3 at the rDNA in S. cerevisiae. Cells lacking Sir2 not only have high levels of acetylated histones but also high levels of K4-methylated H3 at the rDNA. Our data suggest that the change, from silent chromatin containing K4-hypomethylated H3 to active chromatin containing high levels of K4-methylated H3, reflects increased transcription by Pol II at the rDNA NTS in cells lacking Sir2.
K4-Methylated Histone H3 and Pol II Are Excluded from Silent Chromatin by Sir2
We have analyzed K4-methylated H3 at silent loci in wild-type and sir2Δ cells by ChIP. Our results show that Sir2 excludes K4 mono-, di- and -trimethylated histone H3 from silent chromatin domains, including HMR, TEL-VIR, and the rDNA (Figures 1–3). Using a catalytically inactive form of Sir2, we have determined that the deacetylase activity of Sir2 is required to exclude K4-di- and -trimethylated H3 from the rDNA (Figure 4). Consistent with the primary role of Sir2 at silent chromatin, no significant difference in the levels of K4-methylated H3 was detected at the euchromatic and non-rDNA intergenic regions in sir2Δ cells.
In S. cerevisiae, high levels of K4-trimethylated histone H3 are associated with actively transcribed genes in euchromatic domains of the genome (Bernstein et al., 2002; Santos-Rosa et al., 2002). These observations are supported by work showing that the Pol II elongation complex Paf1C recruits the histone H3 K4-methylation complex COMPASS to genes being transcribed by Pol II (Krogan et al., 2003; Ng et al., 2003b, c). The results of experiments in Figures 5–7 show that the level and activity of Pol II are increased at the rDNA NTS in sir2Δ cells. Interestingly, in cells lacking Paf1, the levels of K4-methylated H3 at the rDNA NTS are reduced, suggesting that even the low levels of K4-methylated H3 observed at the rDNA are dependent on transcription (Mueller et al., 2006). Finally, our MNase data show that the structure of chromatin in NTS2 is altered in sir2Δ cells. Based on these results, we propose that the deacetylase activity of Sir2 prevents the conversion of silent chromatin at the rDNA, normally inaccessible to Pol II, to active chromatin with the potential to be transcribed. This conclusion is supported by results showing that the level of K4-dimethylated H3, a mark of chromatin with the potential to be transcribed by Pol II, was increased over the rDNA repeat in cells lacking Sir2 and that the highest level of K4-trimethylated H3 was in the NTS, where Pol II transcription was occurring.
The observation of increased K4-methylated H3 and increased transcription at the rDNA in S. cerevisiae cells lacking Sir2 is reminiscent of recent reports regarding the role of modified histones in regulating Pol I transcription and rDNA chromatin structure in higher eukaryotes (reviewed in Grummt and Pikaard, 2003; McStay, 2006). In several Arabidopsis species, disruption of the histone deacetylase (HDAC) activities of HDT1, a plant-specific HDAC, or HDA6, a member of the RPD3 family of HDACs, caused normally silenced rDNA repeats to acquire characteristics of active rDNA repeats, including high levels of Pol I and K4-methylated H3, and low levels of K9-methylated H3 and DNA methylation (Lawrence et al., 2004; Probst et al., 2004; Earley et al., 2006). Unlike the Arabidopsis system, in S. cerevisiae, transcription by Pol I is not increased in cells that lack the histone deacetylase RPD3 (Oakes et al., 1999, 2006). Likewise, no changes in Pol I transcription have been observed in cells lacking Sir2 (Oakes et al., 1999, 2006). Despite the differences, a common thread between the yeast and plant systems is that loss of specific HDACs results in rDNA that is more like euchromatin, and, subsequently, has altered function.
It is important to note that each rDNA repeat in S. cerevisiae contains an ARS; however, only a small fraction (∼20%) of these replication origins fire during S phase to replicate the rDNA locus (Linskens and Huberman, 1988). Molecular imaging experiments have shown that a large proportion of rDNA origins (50%) are activated in cells lacking Sir2 (Pasero et al., 2002). The high level of K4-trimethylated H3 at the rDNA may be associated with increased firing of the rDNA ARS elements in sir2Δ cells. Experiments to address the role of K4-methylated H3 and NTS transcription in origin firing at the rDNA are currently underway.
Additional Factors May Limit K4-Methylated H3 at the rDNA
In cells lacking Sir2, increased levels of K4-trimethylated histone H3 were observed across an ∼4-kb region of HMR and up to 2.8 kb from TEL-VIR, whereas the increase at the rDNA was limited primarily to NTS2 (Figures 1 and 2). The region of HMR where we detect changes in the association of K4-methylated histone H3 falls within the boundaries of the silent domain mapped in previous studies (Donze et al., 1999; Donze and Kamakaka, 2001). From these observations, we conclude that COMPASS has greater access to HMR and TEL-VIR than to the rDNA in sir2Δ cells. Given that Pol II transcription units have been identified in the 35S rRNA gene and NTS1 (see below), it is possible that Sir2-independent mechanisms exist that limit the association of Pol II and K4-methylated H3 with these other regions. One possibility is that transcription by Pol I excludes factors required for methylation of H3. This can be tested by measuring the levels of K4-methylated H3 at the rDNA in cells that lack Pol I and make rRNA from a plasmid-borne 35S rRNA gene under the control of a Pol II promoter, similar to the cells used to demonstrate the requirement for Pol I transcription in rDNA silencing (Buck et al., 2002; Cioci et al., 2003).
In addition, a trans-histone regulatory pathway has been identified where the ubiquitylation of histone H2B by Rad6 is required for the di- and trimethylation of histone H3 on K4 and K79 (Briggs et al., 2002; Sun and Allis, 2002; Shahbazian et al., 2005). In cells lacking Ubp10, an enzyme required for the removal of ubiquitin from H2B at silent domains in S. cerevisiae, the level of K4-trimethylated H3 was increased at the 5S and 35S rRNA gene (Emre et al., 2005). We measured low levels of K4-trimethylated H3 at these regions in sir2Δ cells, suggesting that Ubp10 limits K4-methylated H3 even in the absence of Sir2.
Silencing at the rDNA, HM loci, and telomeres requires the deacetylase activity of Sir2. The known protein targets of Sir2 are lysine residues in histones H3 and H4, including K9 and K14 of H3 and K16 of H4 (reviewed in Rusche et al., 2003). Recent work has demonstrated that O-acetyl-ADP-ribose (AAR), a product of the Sir2 deacetylation reaction, promotes a conformational change in the Sir2-3-4 complex that may contribute to the formation of silent chromatin (Liou et al., 2005). Although we have determined that Sir2 restricts the access of Pol II and the K4-methylation machinery to silent chromatin, our in vivo studies do not separate the contributions of hypoacetylated histone tails and AAR. In vitro studies using purified factors and chromatin templates should distinguish between the functions of these products of Sir2.
Cells lacking Set1 exhibit defects in silencing at the rDNA and telomeres (Nislow et al., 1997; Briggs et al., 2001; Bryk et al., 2002; Nagy et al., 2002; Mueller et al., 2006). Unlike what has been observed at telomeres (Ng et al., 2002, 2003a; Meneghini et al., 2003; Martin et al., 2004; Katan-Khaykovich and Struhl, 2005), silencing factors remain at the rDNA in set1Δ cells (Bryk et al., 2002), and thus the mechanism behind loss of silencing at the rDNA in set1Δ cells is not consistent with the model proposed to explain loss of silencing at telomeres. One model that we are currently testing is that release of Sir proteins from telomeres in set1Δ cells interferes with rDNA silencing. However, at this time, we are unable to determine whether the changes in chromatin accessibility or the loss of rDNA silencing seen in set1Δ cells are direct or indirect.
Other Pol II Transcription Units in the rDNA
In addition to the NTS2 transcripts that we have identified, the rDNA contains other Pol II transcription units. A naturally occurring Pol II-transcribed gene TAR1 has been identified on the antisense strand of the 35S rRNA gene ∼3–4 kb away from NTS2 (Coelho et al., 2002). Although its expression was shown to be responsive to Sir2, we did not observe high levels of K4-trimethylated H3 over the TAR1 ORF. Previous studies have characterized mutants that lack subunits of Pol I and survive by transcribing the 35S rRNA genes with Pol II (Conrad-Webb and Butow, 1995; Vu et al., 1999). However, in RNA from sir2Δ cells, Pol II-derived 35S rRNA transcripts have not been detected (Oakes et al., 1999, 2006). Moreover, we did not detect high levels of K4-trimethylated H3 or Pol II over the 35S rRNA gene, which would be expected if the 35S rRNA gene was being transcribed by Pol II.
Recent work has shown that in sir2Δ cells, bidirectional transcripts from NTS1 displace cohesin complexes from the rDNA leading to high levels of unequal sister chromatid exchange and rDNA repeat amplification (Kobayashi and Ganley, 2005). In addition to the transcripts from NTS1, we have identified transcripts from NTS2. Although transcription and the level of Pol II is increased clearly over both NTS1 and NTS2 in sir2Δ cells, we were surprised that the level of K4-trimethylated H3 over NTS1 was increased only ∼1.5- to 2.0-fold. Despite our efforts to correct for total histone H3, we suspect reduced levels of total H3 at NTS1 (Supplemental Figure S1) may have contributed to the lower level of K4-trimethylated H3 measured at NTS1 in ChIP experiments (Figure 1 and Supplemental Figure S2). Nonetheless, the NTS transcription units display histone marks found on euchromatic genes, suggesting that not only Pol II- but also Pol II-associated factors required for the trimethylation of H3 on K4 have access to the rDNA in sir2Δ cells.
Three possible ORFs of 71, 64, and 55 amino acids were identified in the sequence of the 1.7-kb NTS2 transcript, but no significant matches to known eukaryotic proteins from the nonredundant protein database were identified using BLASTp searches. Likewise, significant conservation of S. cerevisiae NTS DNA sequences was noted in several yeast species related to S. cerevisiae by BLASTn, but not in nonyeast organisms. It is possible that the NTS RNAs are noncoding RNAs. Intergenic transcription has been associated with regulatory pathways involving chromatin and gene expression in several organisms (reviewed in Bernstein and Allis, 2005). Interestingly, intergenic transcripts from the rDNA spacer that are made by Pol I have recently been shown to regulate the heterochromatin structure of rDNA repeats in mouse cells (Mayer et al., 2006). Experiments are currently underway to analyze the Pol II-transcribed NTS transcripts from S. cerevisiae in detail and to determine whether transcription through NTS2 alters rDNA recombination, silencing of Pol II marker genes, and/or DNA replication from the origin present in each rDNA repeat.
Supplementary Material
ACKNOWLEDGMENTS
We thank Julie Huang for rDNA primer and Kevin Struhl for control primer sequences, Danesh Moazed for Sir2 antisera, Adam Rudner for plasmids, Catalina Alfonso for initiating micrococcal nuclease studies, and Neva Barker for analyzing rDNA sequences. We are grateful to Craig Kaplan, Mike Kladde, and Kelly Williamson for comments on the manuscript. This work was supported by grants from the American Heart Association, Texas Affiliate (0365004Y), the American Cancer Society (RSG-04-049-01-GMC), and the National Institutes of Health (GM-070930) to M.B.
Footnotes
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-03-0205) on June 28, 2006.
REFERENCES
- Allison D. S., Hall B. D. Effects of alterations in the 3′ flanking sequence on in vivo and in vitro expression of the yeast SUP4-o tRNATyr gene. EMBO J. 1985;4:2657–2664. doi: 10.1002/j.1460-2075.1985.tb03984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., Struhl K. Current Protocols in Molecular Biology. New York, NY: Greene Publishing Associates and John Wiley & Sons, Inc; 1988. [Google Scholar]
- Bernstein B. E., Humphrey E. L., Erlich R. L., Schneider R., Bouman P., Liu J. S., Kouzarides T., Schreiber S. L. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc. Natl. Acad. Sci. USA. 2002;99:8695–8700. doi: 10.1073/pnas.082249499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E., Allis C. D. RNA meets chromatin. Genes Dev. 2005;19:1635–1655. doi: 10.1101/gad.1324305. [DOI] [PubMed] [Google Scholar]
- Braglia P., Percudani R., Dieci G. Sequence context effects on oligo(dT) termination signal recognition by Saccharomyces cerevisiae RNA polymerase III. J. Biol. Chem. 2005;280:19551–19562. doi: 10.1074/jbc.M412238200. [DOI] [PubMed] [Google Scholar]
- Braunstein M., Rose A. B., Holmes S. G., Allis C. D., Broach J. R. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 1993;7:592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- Braunstein M., Sobel R. E., Allis C. D., Turner B. M., Broach J. R. Efficient transcriptional silencing in Saccharomyces cerevisiae requires a heterochromatin histone acetylation pattern. Mol. Cell. Biol. 1996;16:4349–4356. doi: 10.1128/mcb.16.8.4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs S. D., Bryk M., Strahl B. D., Cheung W. L., Davie J. K., Dent S. Y., Winston F., Allis C. D. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev. 2001;15:3286–3295. doi: 10.1101/gad.940201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs S. D., Xiao T., Sun Z. W., Caldwell J. A., Shabanowitz J., Hunt D. F., Allis C. D., Strahl B. D. Gene silencing: trans-histone regulatory pathway in chromatin. Nature. 2002;418:498. doi: 10.1038/nature00970. [DOI] [PubMed] [Google Scholar]
- Bryk M., Banerjee M., Murphy M., Knudsen K. E., Garfinkel D. J., Curcio M. J. Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes Dev. 1997;11:255–269. doi: 10.1101/gad.11.2.255. [DOI] [PubMed] [Google Scholar]
- Bryk M., Briggs S. D., Strahl B. D., Curcio M. J., Allis C. D., Winston F. Evidence that Set1, a factor required for methylation of histone H3, regulates rDNA silencing in S. cerevisiae by a Sir2-independent mechanism. Curr. Biol. 2002;12:165–170. doi: 10.1016/s0960-9822(01)00652-2. [DOI] [PubMed] [Google Scholar]
- Buck S. W., Sandmeier J. J., Smith J. S. RNA polymerase I propagates unidirectional spreading of rDNA silent chromatin. Cell. 2002;111:1003–1014. doi: 10.1016/s0092-8674(02)01193-5. [DOI] [PubMed] [Google Scholar]
- Carmen A. A., Milne L., Grunstein M. Acetylation of the yeast histone H4 N terminus regulates its binding to heterochromatin protein SIR3. J. Biol. Chem. 2002;277:4778–4781. doi: 10.1074/jbc.M110532200. [DOI] [PubMed] [Google Scholar]
- Chen L., Widom J. Mechanism of transcriptional silencing in yeast. Cell. 2005;120:37–48. doi: 10.1016/j.cell.2004.11.030. [DOI] [PubMed] [Google Scholar]
- Cioci F., Vogelauer M., Camilloni G. Acetylation and accessibility of rDNA chromatin in Saccharomyces cerevisiae in (Delta)top1 and (Delta)sir2 mutants. J. Mol. Biol. 2002;322:41–52. doi: 10.1016/s0022-2836(02)00749-0. [DOI] [PubMed] [Google Scholar]
- Cioci F., Vu L., Eliason K., Oakes M., Siddiqi I. N., Nomura M. Silencing in yeast rDNA chromatin: reciprocal relationship in gene expression between RNA polymerase I and II. Mol. Cell. 2003;12:135–145. doi: 10.1016/s1097-2765(03)00262-4. [DOI] [PubMed] [Google Scholar]
- Coelho P. S. R., Bryan A. C., Kumar A., Shadel G. S., Snyder M. A novel mitochondrial proteins, Tar1p, is encoded on the antisense strand of the nuclear 25S rDNA. Genes Dev. 2002;16:2755–2760. doi: 10.1101/gad.1035002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad-Webb H., Butow R. A. A polymerase switch in the synthesis of rRNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 1995;15:2420–2428. doi: 10.1128/mcb.15.5.2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammann R., Lucchini R., Koller T., Sogo J. M. Chromatin structures and transcription of rDNA in yeast Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:2331–2338. doi: 10.1093/nar/21.10.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis E. S., Shafer B. K., Strathern J. N. The Saccharomyces cerevisiae RDN1 locus is sequestered from interchromosomal meiotic ectopic recombination in a SIR2-dependent manner. Genetics. 2000;155:1019–1032. doi: 10.1093/genetics/155.3.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donze D., Adams C. R., Rine J., Kamakaka R. T. The boundaries of the silenced HMR domain in Saccharomyces cerevisiae. Genes Dev. 1999;13:698–708. doi: 10.1101/gad.13.6.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donze D., Kamakaka R. T. RNA polymerase III and RNA polymerase II promoter complexes are heterochromatin barriers in Saccharomyces cerevisiae. EMBO J. 2001;20:520–531. doi: 10.1093/emboj/20.3.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dror V., Winston F. The Swi/Snf chromatin remodeling complex is required for ribosomal DNA and telomeric silencing in Saccharomyces cerevisiae. Mol. Cell. Biol. 2004;24:8227–8235. doi: 10.1128/MCB.24.18.8227-8235.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley K., Lawrence R. J., Pontes O., Reuther R., Enciso A. J., Silva M., Neves N., Gross M., Viegas W., Pikaard C. S. Erasure of histone acetylation by Arabidopsis HDA6 mediates large-scale gene silencing in nucleolar dominance. Genes Dev. 2006;20:1283–1293. doi: 10.1101/gad.1417706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emre N. C., et al. Maintenance of low histone ubiquitylation by Ubp10 correlates with telomere-proximal Sir2 association and gene silencing. Mol. Cell. 2005;17:585–594. doi: 10.1016/j.molcel.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Fritze C. E., Verschueren K., Strich R., Easton Esposito R. Direct evidence for SIR2 modulation of chromatin structure in yeast rDNA. EMBO J. 1997;16:6495–6509. doi: 10.1093/emboj/16.21.6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganley A. R., Hayashi K., Horiuchi T., Kobayashi T. Identifying gene-independent noncoding functional elements in the yeast ribosomal DNA by phylogenetic footprinting. Proc. Natl. Acad. Sci. USA. 2005;102:11787–11792. doi: 10.1073/pnas.0504905102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb S., Esposito R. E. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell. 1989;56:771–776. doi: 10.1016/0092-8674(89)90681-8. [DOI] [PubMed] [Google Scholar]
- Grummt I., Pikaard C. S. Epigenetic silencing of RNA polymerase I transcription. Nat. Rev. Mol. Cell Biol. 2003;4:641–649. doi: 10.1038/nrm1171. [DOI] [PubMed] [Google Scholar]
- Guarente L. Sir2 links chromatin silencing, metabolism, and aging. Genes Dev. 2000;14:1021–1026. [PubMed] [Google Scholar]
- Hecht A., Laroche T., Strahl-Bolsinger S., Gasser S. M., Grunstein M. Histone H3 and H4 N–termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell. 1995;80:583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- Herrick D., Parker R., Jacobson A. Identification and comparison of stable and unstable mRNAs in Saccharomyces cerevisiae. Mol. Cell. Biol. 1990;10:2269–2284. doi: 10.1128/mcb.10.5.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe G. J., Tanny J. C., Rudner A. D., Gerber S. A., Danaie S., Gygi S. P., Moazed D. Steps in assembly of silent chromatin in yeast: Sir3-independent binding of a Sir2/Sir4 complex to silencers and role for Sir2-dependent deacetylation. Mol. Cell. Biol. 2002;22:4167–4180. doi: 10.1128/MCB.22.12.4167-4180.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Moazed D. Association of the RENT complex with nontranscribed and coding regions of rDNA and a regional requirement for the replication fork block protein Fob1 in rDNA silencing. Genes Dev. 2003;17:2162–2176. doi: 10.1101/gad.1108403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T., Allis C. D. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Katan-Khaykovich Y., Struhl K. Heterochromatin formation involves changes in histone modifications over multiple cell generations. EMBO J. 2005;24:2138–2149. doi: 10.1038/sj.emboj.7600692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent N. A., Mellor J. Chromatin structure snap-shots: rapid nuclease digestion of chromatin in yeast. Nucleic Acids Res. 1995;23:3786–3787. doi: 10.1093/nar/23.18.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Ganley A. R. Recombination regulation by transcription-induced cohesin dissociation in rDNA repeats. Science. 2005;309:1581–1584. doi: 10.1126/science.1116102. [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Horiuchi T., Tongaonkar P., Vu L., Nomura M. SIR2 regulates recombination between different rDNA repeats, but not recombination within individual rRNA genes in yeast. Cell. 2004;117:441–453. doi: 10.1016/s0092-8674(04)00414-3. [DOI] [PubMed] [Google Scholar]
- Krogan N. J., et al. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol. Cell. 2003;11:721–729. doi: 10.1016/s1097-2765(03)00091-1. [DOI] [PubMed] [Google Scholar]
- Kuzuhara T., Horikoshi M. A nuclear FK506-binding protein is a histone chaperone regulating rDNA silencing. Nat. Struct. Mol. Biol. 2004;11:275–283. doi: 10.1038/nsmb733. [DOI] [PubMed] [Google Scholar]
- Lawrence R. J., Earley K., Pontes O., Silva M., Chen Z. J., Neves N., Viegas W., Pikaard C. S. A concerted DNA methylation/histone methylation switch regulates rRNA gene dosage control and nucleolar dominance. Mol. Cell. 2004;13:599–609. doi: 10.1016/s1097-2765(04)00064-4. [DOI] [PubMed] [Google Scholar]
- Linskens M. H. K., Huberman J. A. Organization of replication of ribosomal DNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 1988;8:4927–4935. doi: 10.1128/mcb.8.11.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou G. G., Tanny J. C., Kruger R. G., Walz T., Moazed D. Assembly of the SIR complex and its regulation by O-acetyl-ADP-ribose, a product of NAD-dependent histone deacetylation. Cell. 2005;121:515–527. doi: 10.1016/j.cell.2005.03.035. [DOI] [PubMed] [Google Scholar]
- Machin F., Paschos K., Torres-Rosell J., Pade C., Aragon L. Condensin regulates rDNA silencing by modulating nucleolar Sir2p. Curr. Biol. 2004;14:125–130. [PubMed] [Google Scholar]
- Martin A. M., Pouchnik D. J., Walker J. L., Wyrick J. J. Redundant roles for histone H3 N-terminal lysine residues in subtelomeric gene repression in Saccharomyces cerevisiae. Genetics. 2004;167:1123–1132. doi: 10.1534/genetics.104.026674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer C., Schmitz K. M., Li J., Grummt I., Santoro R. Intergenic transcripts regulate the epigenetic state of rRNA genes. Mol. Cell. 2006;22:351–361. doi: 10.1016/j.molcel.2006.03.028. [DOI] [PubMed] [Google Scholar]
- McStay B. Nucleolar dominance: a model for rRNA gene silencing. Genes Dev. 2006;20:1207–1214. doi: 10.1101/gad.1436906. [DOI] [PubMed] [Google Scholar]
- Meneghini M. D., Wu M., Madhani H. D. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell. 2003;112:725–736. doi: 10.1016/s0092-8674(03)00123-5. [DOI] [PubMed] [Google Scholar]
- Moazed D. Common themes in mechanisms of gene silencing. Mol. Cell. 2001;8:489–498. doi: 10.1016/s1097-2765(01)00340-9. [DOI] [PubMed] [Google Scholar]
- Mueller J. E., Canze M., Bryk M. The requirement for COMPASS and Paf1 in transcriptional silencing and methylation of histone H3 in Saccharomyces cerevisiae. Genetics. 2006;173:557–567. doi: 10.1534/genetics.106.055400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy P. L., Griesenbeck J., Kornberg R. D., Cleary M. L. A trithorax-group complex purified from Saccharomyces cerevisiae is required for methylation of histone H3. Proc. Natl. Acad. Sci. USA. 2002;99:90–94. doi: 10.1073/pnas.221596698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng H. H., Ciccone D. N., Morshead K. B., Oettinger M. A., Struhl K. Lysine-79 of histone H3 is hypomethylated at silenced loci in yeast and mammalian cells: a potential mechanism for position-effect variegation. Proc. Natl. Acad. Sci. USA. 2003a;100:1820–1825. doi: 10.1073/pnas.0437846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng H. H., Dole S., Struhl K. The Rtf1 component of the Paf1 transcriptional elongation complex is required for ubiquitination of histone H2B. J. Biol. Chem. 2003b;278:33625–33628. doi: 10.1074/jbc.C300270200. [DOI] [PubMed] [Google Scholar]
- Ng H. H., Feng Q., Wang H., Erdjument-Bromage H., Tempst P., Zhang Y., Struhl K. Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes Dev. 2002;16:1518–1527. doi: 10.1101/gad.1001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng H. H., Robert F., Young R. A., Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell. 2003c;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- Nislow C., Ray E., Pillus L. SET1, a yeast member of the trithorax family, functions in transcriptional silencing and diverse cellular processes. Mol. Biol. Cell. 1997;8:2421–2436. doi: 10.1091/mbc.8.12.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes M. L., Siddiqi I., French S. L., Vu L., Sato M., Aris J. P., Beyer A. L., Nomura M. Role of histone deacetylase Rpd3 in regulating rRNA gene transcription and nucleolar structure in yeast. Mol. Cell. Biol. 2006;26:3889–3901. doi: 10.1128/MCB.26.10.3889-3901.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes M., Siddiqi I., Vu L., Aris J., Nomura M. Transcription factor UAF, expansion and contraction of ribosomal DNA (rDNA) repeats, and RNA polymerase switch in transcription of yeast rDNA. Mol. Cell. Biol. 1999;19:8559–8569. doi: 10.1128/mcb.19.12.8559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.-H., Cosgrove M. S., Youngman E., Wolberger C., Boeke J. D. A core nucleosome surface crucial for transcriptional silencing. Nat. Genet. 2002;32:273–279. doi: 10.1038/ng982. [DOI] [PubMed] [Google Scholar]
- Pasero P., Bensimon A., Schwob E. Single-molecule analysis reveals clustering and epigenetic regulation of replication origins at the yeast rDNA locus. Genes Dev. 2002;16:2479–2484. doi: 10.1101/gad.232902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst A. V., et al. Arabidopsis histone deacetylase HDA6 is required for maintenance of transcriptional gene silencing and determines nuclear organization of rDNA repeats. Plant Cell. 2004;16:1021–1034. doi: 10.1105/tpc.018754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M. D., Winston F., Hieter P. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1990. Methods in Yeast Genetics: A Laboratory Course Manual. [Google Scholar]
- Rudner A. D., Hall B. E., Ellenberger T., Moazed D. A nonhistone protein-protein interaction required for assembly of the SIR complex and silent chromatin. Mol. Cell. Biol. 2005;25:4514–4528. doi: 10.1128/MCB.25.11.4514-4528.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusche L. N., Kirchmaier A. L., Rine J. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 2003;72:481–516. doi: 10.1146/annurev.biochem.72.121801.161547. [DOI] [PubMed] [Google Scholar]
- Sandmeier J. J., French S., Osheim Y., Cheung W. L., Gallo C. M., Beyer A. L., Smith J. S. RPD3 is required for the inactivation of yeast ribosomal DNA genes in stationary phase. EMBO J. 2002;21:4959–4968. doi: 10.1093/emboj/cdf498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H., Bannister A. J., Dehe P. M., Geli V., Kouzarides T. Methylation of H3 lysine 4 at euchromatin promotes Sir3p association with heterochromatin. J. Biol. Chem. 2004;279:47506–47512. doi: 10.1074/jbc.M407949200. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H., Schneider R., Bannister A. J., Sherriff J., Bernstein B. E., Emre N. C., Schreiber S. L., Mellor J., Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- Shahbazian M. D., Zhang K., Grunstein M. Histone H2B ubiquitylation controls processive methylation but not monomethylation by Dot1 and Set1. Mol. Cell. 2005;19:271–277. doi: 10.1016/j.molcel.2005.06.010. [DOI] [PubMed] [Google Scholar]
- Shou W., et al. Net1 stimulates RNA polymerase I transcription and regulates nucleolar structure independently of controlling mitotic exit. Mol. Cell. 2001;8:45–55. doi: 10.1016/s1097-2765(01)00291-x. [DOI] [PubMed] [Google Scholar]
- Shou W., Seol J. H., Shevchenko A., Baskerville C., Moazed D., Chen Z. W., Jang J., Charbonneau H., Deshaies R. J. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell. 1999;97:233–244. doi: 10.1016/s0092-8674(00)80733-3. [DOI] [PubMed] [Google Scholar]
- Smith J. S., Boeke J. D. An unusual form of transcriptional silencing in the yeast ribosomal DNA. Genes Dev. 1997;11:241–254. doi: 10.1101/gad.11.2.241. [DOI] [PubMed] [Google Scholar]
- Smith J. S., Caputo E., Boeke J. D. A genetic screen for ribosomal DNA silencing defects identifies multiple DNA replication and chromatin-modulating factors. Mol. Cell. Biol. 1999;19:3184–3197. doi: 10.1128/mcb.19.4.3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl B. D., Allis C. D. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Strahl-Bolsinger S., Hecht A., Luo K., Grunstein M. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- Straight A. F., Shou W., Dowd G. J., Turck C. W., Deshaies R. J., Johnson A. D., Moazed D. Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell. 1999;97:245–256. doi: 10.1016/s0092-8674(00)80734-5. [DOI] [PubMed] [Google Scholar]
- Sun Z. W., Allis C. D. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- Swanson M. S., Malone E. A., Winston F. SPT5, an essential gene important for normal transcription in Saccharomyces cerevisiae, encodes an acidic nuclear protein with a carboxy-terminal repeat. Mol. Cell. Biol. 1991;11:3009–3019. doi: 10.1128/mcb.11.6.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanny J. C., Dowd G. J., Huang J., Hilz H., Moazed D. An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell. 1999;99:735–745. doi: 10.1016/s0092-8674(00)81671-2. [DOI] [PubMed] [Google Scholar]
- Turner B. M. Histone acetylation and an epigenetic code. Bioessays. 2000;22:836–845. doi: 10.1002/1521-1878(200009)22:9<836::AID-BIES9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Vogelauer M., Cioci F., Camilloni G. DNA protein-interactions at the Saccharomyces cerevisiae 35 S rRNA promoter and in its surrounding region. J. Mol. Biol. 1998;275:197–209. doi: 10.1006/jmbi.1997.1451. [DOI] [PubMed] [Google Scholar]
- Vu L., Siddiqi I., Lee B. S., Josaitis C. A., Nomura M. RNA polymerase switch in transcription of yeast rDNA: role of transcription factor UAF (upstream activation factor) in silencing rDNA transcription by RNA polymerase II. Proc. Natl. Acad. Sci. USA. 1999;96:4390–4395. doi: 10.1073/pnas.96.8.4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner J. R. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- Wu J., Grunstein M. 25 years after the nucleosome model: chromatin modifications. Trends Biochem. Sci. 2000;25:619–623. doi: 10.1016/s0968-0004(00)01718-7. [DOI] [PubMed] [Google Scholar]
- Wyrick J. J., Holstege F. C., Jennings E. G., Causton H. C., Shore D., Grunstein M., Lander E. S., Young R. A. Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature. 1999;402:418–421. doi: 10.1038/46567. [DOI] [PubMed] [Google Scholar]
- Ye J., Ai X., Eugeni E. E., Zhang L., Carpenter L. R., Jelinek M. A., Freitas M. A., Parthun M. R. Histone H4 lysine 91 acetylation: a core domain modification associated with chromatin assembly. Mol. Cell. 2005;18:123–130. doi: 10.1016/j.molcel.2005.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Dietrich F. S. Mapping of transcription start sites in Saccharomyces cerevisiae using 5′ SAGE. Nucleic Acids Res. 2005;33:2838–2851. doi: 10.1093/nar/gki583. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.