Abstract
Regulation of gene transcription in vascular smooth muscle cells (VSMCs) by serum response factor (SRF) plays a crucial role in vascular development and in the pathophysiology of vascular diseases. Nevertheless, the regulation of specific genes by SRF in vascular diseases is poorly understood. Therefore, we investigated the regulation of smooth muscle myosin light chain kinase (smMLCK) by using spontaneously hypertensive rats (SHR) as an experimental model. We found that smMLCK expression in blood vessels increases during the development of hypertension and is always greater in blood vessels from SHR compared with normotensive rats. Analysis of the DNA sequences of the promoters isolated from SHR and normotensive rats revealed that SHR contain a 12-base pair insertion adjacent to the CArG box. This insertion increases SRF binding to the CArG box and positively regulates SRF-dependent promoter activity. The increase in smMLCK expression was blocked by dominant-negative SRF, dominant-negative Ras, or antisense oligonucleotides to ERK. In vivo, inhibiting MEK decreased smMLCK expression and blood pressure in SHR partly by decreasing SRF binding to the smMLCK promoter. These data provide novel insight into the regulation of smMLCK expression at the molecular level and demonstrate the importance of SRF in regulating smMLCK promoter activity in SHR.
INTRODUCTION
Several recent studies have demonstrated the importance of serum response factor (SRF) in the regulation of gene transcription in vascular smooth muscle cells (VSMCs) (Wang and Olson, 2004). SRF binds to a CArG [CC(A/T)6GG] box DNA sequence within the regulatory regions of smooth muscle genes. The CArG element was first identified in the promoter of c-fos, which is downstream of Ras (Treisman, 1992). Ras, through a cascade of events that includes MEK activation and the phosphorylation and activation of ERK, phosphorylates ternary complex factors (TCFs), one of the ETS-domain transcription factors. Phosphorylation of TCFs facilitates their association with SRF, and, in turn, stabilizes the interaction of SRF with CArG boxes in the promoter regions of target genes (Buchwalter et al., 2004). The importance of SRF in regulating gene expression in VSMCs is underscored by the presence of two or more CArG boxes in nearly all smooth muscle genes (Miano, 2003) and its well-defined role in VSMC differentiation (Owens et al., 2004).
Nevertheless, Ras–MEK–ERK–SRF signaling can have pleiotropic effects in VSMCs. ERK activation by platelet-derived growth factor (PDGF), an antagonist of smooth muscle differentiation (Holycross et al., 1992), reduced the expression of smooth muscle cell marker genes such as SM22α, SM α-actin, and SM-MHC (Wang et al., 2004; Hayashi et al., 1999). In contrast, ERK activation by angiotensin II, a potent vasocontractor and a trophic factor in the development of many vascular diseases (Touyz, 2003), increased the expression of SM-α-actin gene via stimulation of SRF binding to the CArG element (Yoshida et al., 2004). Therefore, the mechanisms by which SRF regulates smooth muscle-specific genes and their roles in vascular diseases have not been fully defined.
One mechanism by which SRF could target smooth muscles is by regulating genes unique to VSMCs. That is, the product of a specific gene activated by SRF could have downstream effects on smooth muscles that result in hypertension or other vascular disease. Smooth muscle myosin light chain kinase (smMLCK) is a candidate for such a role, because it is the key regulator of smooth muscle contraction. Smooth muscle contraction requires ATP hydrolysis by actin and myosin II. The actin–myosin II interaction in VSMCs is regulated by the phosphorylation and dephosphorylation of the 20-kDa regulatory light chain of myosin-II (MLC20) (de Lanerolle and Paul, 1991). MLC20 phosphorylation (MLC-P) is catalyzed by smMLCK in the presence of Ca2+/calmodulin. MLC20 dephosphorylation is catalyzed by myosin phosphatase 1 (MPase1) (Uehata et al., 1997). MLC20 phosphorylation and dephosphorylation are required for smooth muscle contraction and relaxation, and the balance between the activities of smMLCK and MPase 1 determines the intracellular level of MLC-P. Other experiments have implicated MLC-P in cell motility (Wilson et al., 1991; Klemke et al., 1997), cytokinesis (Yamakita et al., 1994), the timing of mitosis (Fishkind et al., 1991), and VSMC apoptosis (Fazal et al., 2005).
Despite the pivotal role of smMLCK in VSMCs, the genetic and molecular mechanisms of smMLCK gene expression have not been elucidated, and the regulation of the smMLCK promoter by genetic and regulatory factors remains largely unknown. This is partly because of the complexity of the MYLK gene that encodes smMLCK. In humans, a single functional MYLK gene is located on chromosome 3qcen–q21 spanning >270 kilobases and containing at least 34 exons that encode three proteins (Potier et al., 1995): nonmuscle MLCK (220 kDa), smMLCK (130 kDa), and telokin (20 kDa). Nonmuscle MLCK is the biggest product of the gene, containing the entire coding region of smMLCK and a large extension at the amino terminus (Gallagher et al., 1997). Telokin contains only the carboxy-terminal 154 codons of nonmuscle and smMLCK (Gallagher and Herring, 1991).
The goal of this study was to investigate the regulation of smMLCK gene expression. Therefore, as an initial step in determining the potential role of altered smMLCK gene expression in essential hypertension, we studied the promoter of this gene in spontaneously hypertensive rats (SHR) and normotensive rats. Our studies demonstrated the presence of an insertion mutation in the smMLCK promoter in SHR not found in normotensive rats. Of interest, this mutation seems to mediate increased binding and promoter responsiveness to SRF. Other in vitro studies established that SRF regulation of smMLCK expression is downstream of Ras activation and requires ERK activation. In vivo, we found that blocking MEK, a downstream effector of Ras, decreased smMLCK expression and prevented the development of hypertension in SHR.
MATERIALS AND METHODS
Isolation and Sequence Analysis of Intron 14
The structure of rat MYLK gene was analyzed using National Center for Biotechnology Information PubGene (ID 288057) and EMBL-Ensembl (ENSRNOT00000036465). The rat MYLK is a predicted sequence based on the mouse MYLK sequence, and no supporting clones are available in the Rat Genome Clone Registry. Therefore, to isolate intron 14 of the rat MYLK, we designed two primers based on EMBL-Ensembl sequences and performed PCR amplification by using genomic DNA as a template. Genomic DNA was extracted from the spleens of SHR, Sprague Dawley (SD), and Wistar-Kyoto (WKY) rats by using a DNeasy tissue kit according to the manufacturer’s instructions (QIAGEN, Valencia, CA). PCR amplification was then carried out on genomic DNA using 5 μM of each primer (Figure 3B). The 5′ primer corresponded to nucleotides 18–38 and the 3′ primer corresponded to nucleotides 462–482 in intron 14 of the MYLK gene (Figure 3B). PCR conditions were 1 min at 94°C, 1 min at 50°C, and 1 min at 72°C for 35 cycles with 0.4 U of Taq polymerase (Bioline, Valley Park, MO). The PCR products were cloned into a Topo vector (Invitrogen, Carlsbad, CA) and subjected to DNA sequencing by using an ABI Prism 3100 genetic analyzer (Applied Biosystems, Foster City, CA) in the Research Resource Center at University of Illinois at Chicago (Chicago, IL). To verify zygosity of the insertion mutation found in the intron 14 of MYLK from SHR and SHR-stroke-prone (SHR-SP), both + and − strands of DNA were sequenced using two sequencing primers that have opposite orientations. Polymorphisms were determined by comparing their DNA sequences among strains (SHR, SHR-SP, SD, and WKY rats) and by comparing them with the sequences published in EMBL-Ensembl. Four animals per strain were used for DNA sequencing, and each DNA sequencing was repeated at least twice to reduce sequencing errors.
Figure 3.
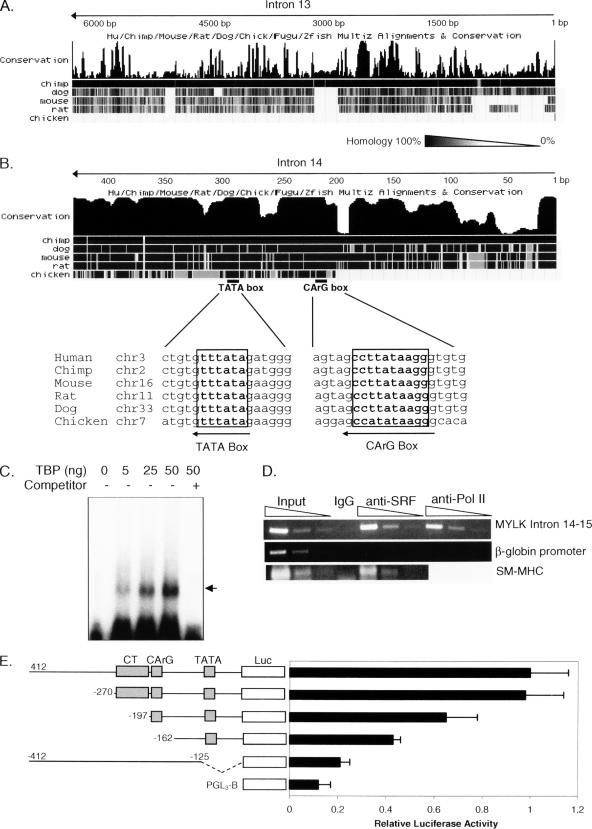
Isolation and analysis of intron 14 of the MYLK gene. (A) Exon–intron structure of the rat smMLCK gene. The translation start site of the smMLCK in exon 15 is shown (ATG). (B) DNA sequences of intron 14 from SHR and WKY rats. Intron 14 was isolated from genomic DNA from SHR and WKY rats using PCR. DNA sequences of the 5′ and 3′ primers are shown (italics, underlined). The transcription start site (+1) of smMLCK was identified using primer extension and 5′ RACE (Supplemental Figure 1). Analysis of transcription elements using the Transcription Element Search System identified >55 elements, including SRF (CArG box) and TBP (TATA box) binding sites. Comparison of these sequences revealed the presence of a 12-base pair insertion (Insertion) in the SHR sequence not found in the WKY sequence. WKY also contains four different single nucleotide polymorphisms (bold, caps) compared with SHR and other rats.
Primer Extension and Rapid Amplification of 5′ cDNA Ends (5′ RACE)
The primer extension analysis was carried out as described previously (Boorstein and Craig, 1989). Briefly, 5 pmol of gene-specific primer 1 or primer 2 (Supplemental Figure 1A) was labeled at the 5′ end by using T4 polynucleotide kinase (MBI Fermentas, Hanover, MD) and [γ-32P]ATP (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom) at 37°C for 30 min. Total RNA (20 μg) extracted from A7r5 cells was denatured at 85°C for 4 min. Annealing of the labeled primer and total RNA was accomplished at 50°C for 1 h. Reverse transcription reaction was performed according to the manufacturer’s instructions using SuperScript II reverse transcriptase (Invitrogen, Carlsbad, CA). The reaction mixtures were denatured at 70°C for 10 min and loaded on 5% or 8% 7 M urea-polyacrylamide gels. The size of the primer extension products was determined by comparison with a 100-base pair or 10-base pair DNA ladder (Supplemental Figure 1, B and C). 5′ RACE was performed on 5 μg of total RNA isolated from A7r5 cells by using the GeneRacer kit according to the manufacturer’s instructions (Invirogen). A GeneRacer 5′ nested primer and the gene-specific primer 1 were used to amplify the 5′ ends of smMLCK mRNA. The size of 5′ RACE product was determined by comparing it with size markers (Supplemental Figure 1D) after electrophoresis on a 2% agarose gel. This product was cloned into a Topo vector (Invitrogen) and subjected to DNA sequencing.
Cell Culture, Transfection, and Infection
VSMCs were explanted from aortas of 4- to 7-wk-old SHR or WKY rats as described previously (Ross, 1971) and cultured in DMEM with 10% fetal bovine serum. To investigate GTPase signaling, VSMCs were transfected with control oligonucleotides (scrambled sequence) or antisense oligonucleotides (Robinson et al., 1996) to ERK (5 μM each; Calbiochem, San Diego, CA) by using LipofectAMINE. Other VSMCs were transfected with 0.5 μg of N17Ras, N19Rho, or empty vector (pcDNA 3.1) by using FuGENE 6 transfection reagent (Roche, Basel, Switzerland). VSMCs from WKY rats grown in six-well plates (90–100% confluent) were infected for 2 h with either an adenovirus that expresses a short form of SRF (AdSRF-S) (Davis et al., 2002) or a control virus that expresses green fluorescent protein (GFP) (AdGFP). The viruses were washed out, and the cells were grown as described above for 48 h. VSMCs were harvested, and cell lysates were subjected to Western blot analyses.
Reporter Activity Assays
A 465- and a 453-base pair fragment of intron 14 from SHR and WKY rats, respectively, was amplified by PCR and cloned into a pGL3-Basic firefly luciferase vector. Progressive deletion fragments of the regions from −412 base pairs to −162 base pairs of the intron were also obtained by PCR and cloned into the pGL3-Basic vectors. To create a mutated construct, the 12-base pair sequence found in SHR was then inserted into intron 14 from WKY rats by using a QuikChange XL site-directed mutagenesis kit (Stratagene, La Jolla, CA). The integrity of the constructs was confirmed by DNA sequencing. A 100:1 ratio of smMLCK-firefly luciferase vectors to cytomegalovirus-Renilla luciferase vectors (Promega, Madison, WI) was used to transfect COS-7 cells. Firefly and Renilla luciferase activities were measured using a dual-luciferase assay system (Promega), and the ratio of firefly:Renilla luciferase activities was calculated to correct for differences in transfection efficiency. When intron14/firefly luciferase was cotransfected with SRF (Chen and Schwartz, 1996) and N17Ras or N19Rho expression vectors, the luciferase activity was normalized to protein concentration in the cell extract.
Electromobility Gel Shift Assay
Nuclear extracts were prepared as described previously (Dignam et al., 1983) from Cos-7 cells expressing SRF (Chen and Schwartz, 1996) or pcDNA 3.1 plasmids. Recombinant TATA-binding protein (TBP) was expressed and purified as described previously (Peterson et al., 1990). Intron 14 (Figure 4C) or the oligonucleotides representing defined regions of the SHR or WKY smMLCK promoters (Figure 6A) were 5′-end–labeled with T4 polynucleotide kinase (Invitrogen) and [γ-32P]ATP. The binding reaction was carried out in a total volume of 10 μl containing ∼10,000 cpm (1–5 ng) of the labeled DNA, 2 μg of the nuclear extract, and 1 μg of poly(dI-dC) at room temperature for 30 min. For antibody supershift assays, 1.2 μg of antibody to SRF (Santa Cruz Biotechnology, Santa Cruz, CA) was added, and the reaction mixtures were incubated at 4°C for an additional hour with gentle shaking. The reaction mixtures were then loaded on 4% native polyacrylamide gels, and electrophoresis was carried out at 350 V for 30 min. Radioactive bands were visualized by autoradiography.
Figure 4.
Analysis of introns 13 and 14. (A and B) Homology analysis of the introns 13 (A) or 14 (B) among species. Darkness gradients from black to white represent homology from 100 to 0%. Note that the TATA box and the CArG box are conserved in all the species. (C) TBP binds to intron 14 of the MYLK gene. Purified TBP was incubated with 32P-labeled intron 14 from WKY and analyzed using an EMSA. TBP induced a concentration-dependent appearance of a slower migrating band (arrow). A 10-fold excess of cold, nonradioactive competitor eliminated this band. (D) ChIP assays were performed with antibodies to SRF or RNA polymerase II. Nonspecific IgG was used as a negative control (IgG). Input chromatin (0.2, 0.02, and 0.002%) and immunoprecipitated DNA (4, 0.4, and 0.04%) were amplified with primers specific for the |gb-globin promoter, SM-MHC promoter or intron 14 of MYLK gene. SRF and RNA polymerase II are not present in the β-globin promoter. However, SRF bound to the SM-MHC promoter and to intron 14 of the MYCK gene in VSMC from WKY rats. RNA polymerase II also bound to the intron 14. (E) Serial deletion of intron 14 isolated from WKY rats. Deletion fragments were isolated using PCR and cloned into the pGL3-Basic luciferase reporter vector. The numbers indicate the location of the first nucleotide in each fragment, whereas the black bars indicate relative luciferase activities. CT, CArG, and TATA represent CT repeats, CArG box, and TATA box, respectively. The means ± SE from four experiments are shown.
Figure 6.
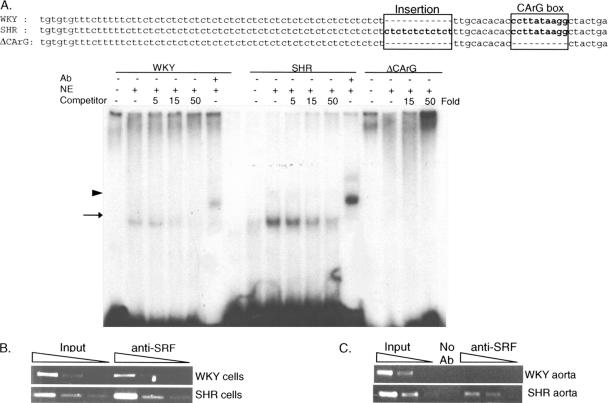
Increased SRF binding to the CArG box in the SHR promoter. (A) Two oligonucleotides representing regions of the smMLCK promoters containing the CT repeats and CArG box were synthesized. The only difference between them was the presence of a 12-base pair insertion in the SHR oligonucleotides. A WKY oligonucleotide without the CArG box was also synthesized and used as a negative control (ΔCArG). Nuclear extracts were isolated from cells expressing SRF (NE). For competition titration assays, three different amounts of the cold competitors (5-, 15-, or 50-fold to 32P-labeled probe) were incubated with the nuclear extracts and 32P-labeled WKY or SHR oligonucleotides. Autoradiography demonstrated the presence of slower migrating bands (arrow) that decreased in intensity as the concentration of the competitors was increased. These bands were always more intense when incubated with the oligonucleotides representing the SHR promoter, and SRF antibodies supershifted this band (arrowhead). (B) In vitro ChIP assays. Cross-linked chromatin from VSMCs of SHR or WKY rats was immunoprecipitated with antibodies to SRF. Input chromatin (0.1, 0.01, and 0.001%) and immunoprecipitated DNA (2, 0.2, and 0.02%) were amplified with primers specific for intron 14. (C) In vivo ChIP assays. Cross-linked chromatin was isolated from aortas removed from SHR or WKY rats. Input chromatin (1, 0.1, and 0.01%) and immunoprecipitated DNA (20, 2, and 0.2%) were amplified with the same primers used for the in vitro ChIP assays. Comparing the relative abundance of the signals confirmed increased binding of SRF to the intron in SHR compared with the intron in WKY rats.
Chromatin Immunoprecipitation (ChIP) Assays
ChIP assays were performed as described previously (Hofmann et al., 2004), with minor modifications. Briefly, VSMCs from WKY rats were fixed directly with formaldehyde. Cross-linked chromatin was immunoprecipitated with antibodies to SRF (Upstate Biotechnology, Lake Placid, NY) or RNA polymerase II (Santa Cruz Biotechnology). The precipitated chromatin DNA was then purified and subjected to PCR analysis. The primers for the β-globin promoter and the smooth muscle myosin heavy chain (SM-MHC) promoter were designed as described previously (Manabe and Owens, 2001). The smMLCK promoter-specific primers were 5′-TCAGGAACCGGGTTGGCGAATGCA-3′ and 5′-TGCATTCGCCAACCCGGTTCCTGA-3′. The PCR products were separated on agarose gels, visualized with ethidium bromide, and the staining intensities of the bands were quantified using a densitometer. The relative enrichment of intron 14 was determined by calculating the ratio of intron 14 present in the immunoprecipitates compared with intron 14 in the input chromatin. In vivo ChIP assays were performed as described by McDonald et al. (2006) with minor modification. Briefly, aortas were quickly dissected and cleaned in ice-cold phosphate-buffered saline (PBS) to remove blood, connective tissue, and endothelial cells. The cleaned aortas were snap frozen in liquid nitrogen and stored at −80°C. The aortas were transferred directly to 1% formaldehyde at 37°C for 10 min and washed four times with ice-cold PBS. The tissues were homogenized in a lysis buffer by using a PowerGen 700D homogenizer (Fisher Scientific, Pittsburgh, PA) and taken through the rest of the ChIP procedure.
Immunocytochemistry
A7r5 immortalized rat aorta smooth muscle cells that have been studied extensively (Wang et al., 2004), or VSMCs isolated from SHR were plated on fibronectin-coated coverslips (BD Biosciences, Palo Alto, CA). Cells were serum starved overnight and treated with 20 μM U0126 or dimethyl sulfoxide (DMSO) for 24 h. After fixation with 4% paraformaldehyde in PBS for 20 min, cells were permeabilized with 0.1% Triton/PBS for 5 min. After blocking with 1% bovine serum albumin/phosphate-buffered saline for 30 min, cells were incubated overnight with primary anti-SRF antibodies (Upstate Biotechnology) diluted 1:20 in blocking solution. Cells were then washed with PBS and incubated with biotinylated anti-mouse antibodies (Vector Laboratories, Burlingame, CA) diluted 1:300 in PBS for 1 h. Cells were washed again with PBS and incubated with Alexa Fluor 488-labeled streptavidin (Invitrogen) diluted 1:1000 in PBS for 45 min. Subsequently, coverslips were rinsed with PBS, mounted in VECTASHIELD (Vector Laboratories) and analyzed using a Zeiss 510 laser-scanning confocal microscope (Carl Zeiss, Thornwood, NY).
Northern Blot Analyses
Total RNA was extracted from aortas of 12-wk-old SHR and WKY rats by using a RNeasy Fibrous Tissue Mini kit (QIAGEN). Purified RNA (2 μg) was separated on 1% agarose-formaldehyde denaturing gels and transferred to nylon membranes. A 1790-base pair fragment of the smMLCK cDNA (amino acids 426-1022 based on rabbit smMLCK cDNA) labeled with [γ-32P]dCTP was used to probe the blots. Hybridization was carried out at 68°C, overnight. The blots were washed two times with 2× SSC containing 0.1% SDS and two times with 0.5× SSC containing 0.1% SDS. The blots were then subjected to autoradiography, and the amount of mRNA in each band was quantified densitometrically.
Animals and Procedures
All animal procedures were performed with prior approval of the Institutional Animal Care Committee and in accordance with National Institutes of Health guidelines. Systolic blood pressure (SBP) was measured using tail-cuff sphygmomanometry (IITC Incorporated/Life Sciences Institute, Woodland Hills, CA). Drugs were delivered via an osmotic pump (DURECT, Cupertino, CA) implanted subcutaneously in the shoulder. The pumps (capacity of 2 ml and a delivery rate of 2.45 μl/h) were filled with 13.5 mM U0126 or 27 mM ML-7 (BIOMOL Research Laboratories, Plymouth Meeting, PA) dissolved in 50% DMSO or vehicle alone. Three weeks later, rats were killed, and the aortas were removed. Vessels were immediately placed in ice-cold acetone containing 10% trichloroacetic acid (TCA) and 10 mM dithiothreitol (DTT). Endothelial cells were removed by gently rubbing the inside of the vessels with a cell lifter, and connective tissue was removed by dissection. The cleaned vessels were then frozen on dry ice and stored at −80°C for biochemical analysis.
Immunoblot Analyses
Cells or pieces of blood vessels placed in acetone containing 10% TCA and 10 mM DTT were washed three times with ether and extracted in a buffer containing 9 M urea, 10 mM DTT, and 20 mM Tris, pH 8.0. Protein concentrations were measured using the Bradford Protein Assay (Bio-Rad, Hercules, CA), and equivalent amounts of protein were subjected to SDS-PAGE or urea-glycerol PAGE. Proteins separated by SDS-PAGE were transferred to nitrocellulose and probed with an antibody to smMLCK (de Lanerolle et al., 1981), the broad specificity C4 antibody to actin (Lessard, 1988), antibodies to Rho A or phosphorylated MPase1 (Santa Cruz Biotechnology), or an antibody to phosphorylated ERK1/2 (Cell Signaling Technology, Beverly, MA). Urea-glycerol PAGE was used to separate the unphosphorylated and phosphorylated forms of MLC20, transferred to nitrocellulose, probed with an antibody to MLC20 (Fazal et al., 2005), and the stoichiometry of phosphorylation (mol PO4/mol MLC20) was calculated as described previously (Obara et al., 1989). All immunoreactive bands were visualized using enhanced chemiluminescence (GE Healthcare) and quantified densitometrically.
Statistical Analysis
Results are expressed as mean ± SE. The data were analyzed using an unmatched Student’s t test in most experiments. In Figure 5, B and C, a one-way ANOVA (SigmaStat; Systat Software, Point Richmond, CA) followed by unmatched Student’s t test was used to evaluate the data. p < 0.05 was considered statistically significant.
Figure 5.
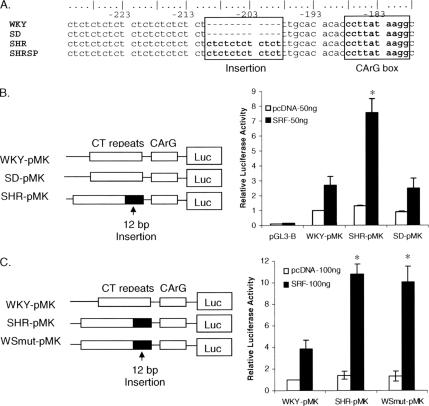
Increased responsiveness to SRF in the intron 14 from SHR. (A) Analysis of intron 14 in other normotensive and hypertensive rat strains. Stroke-prone SHR (SHRSP) also contains the 12-base pair insertion, whereas normotensive SD and WKY rats do not. (B) Activity of intron 14 from WKY (WKY-pMK) rats, SD (SD-pMK) rats, and SHR (SHR-pMK) was analyzed using luciferase reporter gene assays. Although coexpression with 50 ng of the SRF plasmid increased the activities of all three promoters, it increased SHR promoter activity much more than the activities of the WKY and SD promoters (7.2-, 2.7-, and 2.1-fold compared with control cells transfected with 50 ng of pcDNA 3.1, respectively). The means ± SE are shown. *p < 0.01 compared with WKY-pMK plus SRF. (C) The role of the 12-base pair insert was investigated by extending the CT repeats in the WKY promoter by 12 base pairs (WSmut-pMK) as shown schematically. Luciferase activity assays on cells transfected with WKY-pMK, SHR-pMK, or WSmut-pMK, with or without SRF, showed that the 12-base pair sequence increased the activity of the WKY promoter to the same level as the SHR promoter. This experiment was repeated three times. *p < 0.01 compared with the activity of WKY-pMK plus SRF.
RESULTS
SBP, smMLCK Expression, and MLC-P in SHR
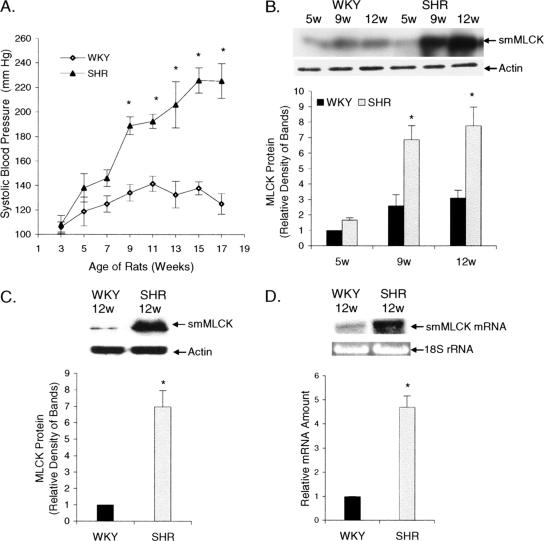
SHR develop hypertension as they grow older without any provocative dietary or environmental stimuli (Okamoto and Aoki, 1963) (Figure 1A). However, changes in vascular contractile proteins during the development of hypertension in SHR have not been elucidated. Therefore, we isolated aortas and small mesenteric arteries of SHR and normotensive WKY rats and analyzed the changes in smMLCK expression. Western blot analyses showed that the amount of smMLCK protein increased in SHR with age and that the level of smMLCK protein was always higher in aortas of SHR compared with WKY rats (Figure 1B). Analysis of resistant arteries also showed that smMLCK protein expression is greater in small mesenteric arteries of 12-wk-old SHR compared with the age-matched WKY rats (Figure 1C). Northern blot analyses performed on total RNA extracted from aortas of 12-wk-old SHR or WKY rats showed that smMLCK mRNA levels were also higher in the aortas from SHR compared with the WKY rats (Figure 1D). Thus, smMLCK expression in blood vessels increases during the development of hypertension in SHR, and smMLCK expression is regulated at the mRNA level.
Figure 1.
Increases in blood pressure and smMLCK expression in SHR. (A) SBP was measured every 2 wk in SHR and WKY rats. Values are mean ± SE, n = 6, *SHR versus WKY, p < 0.001. (B and C) Western blot analyses were performed on aortas (B) or small mesenteric arteries (C) removed from SHR and WKY rats of different ages by using antibodies to smMLCK or actin (loading control). Ratios of smMLCK to actin were calculated, and the mean ± SE are shown. (D) Northern blot analysis showing the level of smMLCK mRNA in aortas from 12-wk-old SHR and age-matched WKY rats. 18S rRNA was used as a loading control. *SHR versus WKY, p < 0.01. n = 4 (B–D).
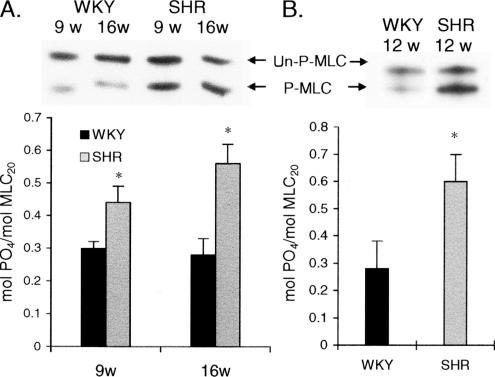
Because the increase in smMLCK expression could directly affect MLC-P, we quantified MLC-P in the blood vessels by using urea-glycerol gel electrophoresis and Western blot analysis. MLC-P was significantly greater in the aortas of 9- or 16-wk-old SHR (0.44 and 0.56 mol PO4/mol MLC20, respectively) compared with age-matched WKY rats (∼0.3 mol PO4/mol MLC20 in 9- and 16-wk-old WKY rats) (Figure 2A). The level of MLC-P in small mesenteric arteries was also higher in 12-wk-old SHR (0.6 mol PO4/mol MLC20) compared with the age-matched WKY rats (0.28 mol PO4/mol MLC20) (Figure 2B). Thus, smMLCK expression and MLC-P increased in the arteries of SHR with age and correlated with the development of hypertension in SHR.
Figure 2.
Increase in MLC-P in SHR. MLC-P was quantified in aortas (A) and in small mesenteric arteries (B) of SHR and WKY rats of differing ages. The stoichiometry of MLC-P (bottom) was calculated after urea-glycerol gel immunoblotting (top). Un-P-MLC and P-MLC indicate un- and monophosphorylated forms of MLC20. Each experiment was repeated at least three times, and mean ± SE are shown in each bar graph. *SHR versus WKY, p < 0.01.
Identification of the smMLCK Promoter
The increase in smMLCK mRNA (Figure 1D) could be due to the changes in transcriptional regulation of smMLCK promoter activity. To investigate this possibility, we analyzed the structure of the rat MYLK gene by using National Center for Biotechnology Information PubGene (ID 288057) and EMBL-Ensembl (ENSRNOT00000036465). This analysis showed that the rat MYLK gene is located on chromosome 11q22 and contains at least 33 exons that encode nonmuscle MLCK, smMLCK, and telokin. This analysis also indicated that the translation start site (ATG) of the rat smMLCK is located in exon 15 (Figure 3A). To identify the transcription start site of the rat smMLCK, we performed primer extension analyses and 5′ RACE. Primer extension analyses were carried out using total RNAs isolated from A7r5 cells and two different antisense oligonucleotides as probes (Supplemental Figure 1A). One probe (primer 1) corresponded to nucleotides 248–280 in exon 15 and was located 22 base pairs upstream of the ATG. Primer extension with primer 1 produced a single fragment of ∼320 nucleotides (Supplemental Figure 1B). This experiment suggested that the transcription start site (+1) resided in the intron proceeding exon 15 (intron 14). Therefore, we designed another probe (primer 2) targeting the nucleotides close to the predicted start site. This probe corresponded to nucleotide 456–475 in intron 14 (Supplemental Figure 1A). Primer extension produced a single fragment of 40 nucleotides (Supplemental Figure 1C), supporting the idea that nucleotide 435 in the intron 14 is the transcription start site. We then performed 5′ RACE to map the transcription start site precisely. 5′ RACE with primer 1 produced a single fragment of ∼340 nucleotides that contained 320 base pairs of cDNA and 23 base pairs of GeneRacer 5′ nested primer (Supplemental Figure 1D). Sequencing the 5′ RACE product confirmed that the +1 nucleotide resided at nucleotide 435 in intron 14 and was located 342 base pairs upstream of the ATG start codon in exon 15 (Supplemental Figure 1A).
We next isolated intron 14 from genomic DNA obtained from SHR and WKY rats by using a PCR method. Analysis of intron 14 for cis-acting transcription elements by using the Transcription Element Search System (http://www.cbil.upenn.edu/tess) showed that the intron 14 contains >55 transcription factor binding sites, including a TATA box and a CArG box and multiple Ras-responsive elements, such as c-Myb, Ap-2αA, ATF-1, and c-Ets-1 (Figure 3B). As an initial step in identifying important regulatory elements, we performed comparative sequence analyses of intron 14 found in various vertebrate species by using the University of California San Diego genome browser (Figure 4, A and B). This analysis was based on the hypothesis that important regulatory elements of a promoter are conserved between species due to functional constraints. It showed that the DNA sequences of intron 14 are highly conserved among humans, chimps, dogs, mice, chickens and rats (Figure 4B). In contrast, intron 13, which does not contain important regulatory elements, is poorly conserved (Figure 4A). Importantly, the TATA and CArG elements of intron 14 are 100% conserved among species.
To determine the functional importance of intron 14, we first performed electrophoretic mobility shift assays (EMSAs) by using 32P-labeled intron 14 from WKY rats and purified TBP. Figure 4C shows that TBP interacts with intron 14 in a concentration-dependent manner. We then used ChIP assays to determine whether SRF binds to the CArG box found in intron 14 of the MYLK gene (Figure 4D). ChIP assays were performed on VSMCs isolated from aortas of WKY rats by using anti-SRF or anti-polymerase II antibodies. The β-globin promoter, which is silent in VSMCs (Manabe and Owens, 2001), was used as a negative control, and the 5′-flanking CArG region of SM-MHC gene was used as a positive control (Manabe and Owens, 2001). Antibodies to SRF or RNA polymerase II failed to precipitate DNA containing the β-globin promoter (Figure 4D). In contrast, these antibodies specifically enriched intron 14 of the MLCK gene. Antibodies to SRF also enriched the SM-MHC gene. Together, the data in Figure 4 demonstrate that intron 14 contains at least part of the smMLCK promoter and that the TATA and CArG boxes are functional and bind TBP and SRF, respectively.
To further investigate the importance of the TATA and the CArG elements in controlling the activity of the smMLCK promoter, we performed luciferase reporter gene activity assays. Serial deletions of intron 14 from WKY rats were cloned into the pGL3-Basic luciferase reporter gene vector (Figure 4E) and used to transfect Cos-7 cells. Deleting nucleotides −412 to −270 decreased promoter activity by only 2%. In contrast, also deleting the CT repeats (−412 to −197) led to a 35% decrease in the promoter activity. Deleting both the CT repeats and the CArG box (−412 to −162) resulted in a 57% decrease in the promoter activity. These data establish the importance of the CArG box and the CT repeats in regulating promoter activity of the smMLCK gene. Next, we investigated the importance of the nucleotides around the TATA box. Deleting nucleotides −125 to +40 almost abolished promoter activity, leaving only 20% of the total activity of the intron 14. Considering that the basal activity of the pGL3-Basic luciferase reporter vector is 12% of the total activity, the data demonstrate that the nucleotides around TATA box are critical for the promoter activity of intron 14.
Increased Responsiveness of the SHR smMLCK Promoter to SRF
Analysis of intron 14 from WKY rats revealed four different single nucleotide polymorphisms compared with intron 14 from SHR and other rat strains (Figure 3B). Comparing the DNA sequences of intron 14 showed that the SHR sequence contains a 12-base pair insertion consisting of six pairs of CT repeats that is not found in normotensive WKY or SD rats (Figures 3B and 5A). This insertion is homozygous and located just 10 base pairs upstream of the CArG box. Further analysis showed that intron 14 from stroke-prone SHR, a closely related genetic strain of SHR (Yamori, 1984), also contain the 12-base pair insertion (Figure 5A). Thus, there is a 12-base pair insertion found in two SHR strains that is absent from two different strains of normotensive rats.
The importance of 12-base pair insertion in regulating the promoter activity of intron 14 was investigated using luciferase reporter gene assays. Intron 14 from SHR, WKY, or SD rats was cloned into the pGL3-Basic luciferase vector (Figure 5B, diagram) and used to cotransfect Cos-7 cells with pcDNA (control) or plasmids expressing SRF. Cotransfection with SRF increased promoter activities of intron 14 from all three rat strains. Importantly, SRF increased the promoter activity of intron 14 from SHR (SHR-pMK) more than those from WKY (WKY-pMK) or SD rats (SD-pMK). Because the 12-base pair insertion is the major difference between introns 14 in SHR and WKY rats, we postulated that adding the 12-base pair sequence to the WKY promoter would stimulate the activity of this promoter to the same level as the SHR promoter. Therefore, we performed site-directed mutagenesis and added the 12-base pair sequence to intron 14 of WKY rats (Figure 5C). Luciferase assays showed that the modified WKY promoter (WSmut-pMK) and the SHR promoter had similar levels of activity in response to SRF (Figure 5C). Thus, the 12-base pair insertion is sufficient to increase SRF-mediated smMLCK promoter activity.
Increased SRF Binding to Intron 14 from SHR, In Vitro and In Vivo
We then investigated how the 12-base pair insert up-regulates SRF-induced smMLCK promoter activity. Because CT or GA dinucleotide repeats are involved in local changes in DNA structure (Lu et al., 2003) and chromatin configuration (Granok et al., 1995), we hypothesized that the insertion of 12 CT repeats in the SHR promoter could facilitate the binding of SRF to the CArG box. This possibility was investigated in two ways. First, we synthesized two different oligonucleotides, representing intron 14 from SHR and WKY rats, containing the CT repeats and CArG box (Figure 5A). The only difference between these two oligonucleotides was the 12-base pair CT repeat found in SHR. An oligonucleotide that has the same sequence as the WKY oligonucleotide but does not contain the CArG box was used as a negative control (ΔCArG). An EMSA was carried out with nuclear extracts from cells expressing SRF and 32P-labeled WKY, SHR, or ΔCArG oligonucleotides. A band shift was detected when nuclear extracts were incubated with WKY or SHR oligonucleotides, and a supershifted band was detected when anti-SRF antibodies were added to the reaction (Figure 6A). Moreover, the relative density of the signal from the SRF–DNA complex was more intense when incubated with SHR oligonucleotides than with WKY oligonucleotides. Titration competition assays with increasing amounts of nonradioactive competitor confirmed that the SRF–DNA complex is greater when incubated with SHR oligonucleotides than with WKY oligonucleotides. No signal was detected when the nuclear extracts were incubated with ΔCArG oligonucleotides.
Second, in vitro and in vivo ChIP assays were performed, using chromatin–DNA complexes isolated from VSMCs from SHR or WKY rats (Figure 6B) or aortas from SHR or WKY rats (Figure 6C). Even though the SRF enrichment is generally weaker in the in vivo assays, both ChIP assays clearly show that SRF enrichment is greater with intron 14 from SHR, in vitro and in vivo (Figure 6, B and C). Together, the EMSA and ChIP assays strongly support the idea that the 12-base pair insertion in the SHR intron is responsible for the increased SRF binding to the CArG box.
Inhibiting SRF Expression or Ras Signaling Blocked SRF-induced smMLCK Expression in VSMCs
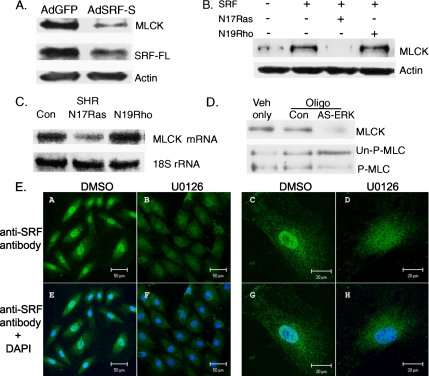
We next used an adenovirus (AdSRF-S) lacking exons 4 and 5 to express a truncated, dominant-negative form of SRF in VSMCs from WKY rats (Davis et al., 2002). Control VSMCs were infected with a virus expressing GFP (AdGFP) at the same multiplicity of infection. The cells were harvested 48 h after infection and analyzed by Western blotting. The results showed that the expression of smMLCK and full-length SRF decreased in VSMCs infected with the AdSRF-S virus compared with the cells infected with the control AdGFP virus (Figure 7A).
Figure 7.
Ras regulates smMLCK expression via SRF. (A) VSMCs from WKY rats were infected with adenoviruses expressing a short form of SRF (AdSRF-S), which acts as a dominant negative (Davis et al., 2002), or GFP (AdGFP) as a control. Western blot analyses were performed using antibodies to full-length SRF (SRF-FL), smMLCK, and actin (loading control). Dominant-negative SRF (AdSRF-S) down-regulates expression of smMLCK or full-length SRF (SRF-FL). (B) VSMCs were cotransfected with SRF and plasmids expressing dominant-negative Ras (N17Ras) or dominant-negative Rho (N19Rho). The cells were extracted and analyzed by Western blotting using antibodies to smMLCK and actin. SRF (lane 2) increased smMLCK expression compared with control VSMCs transfected with pcDNA 3.1 (lane 1). Dominant-negative Ras blocked this increase in expression, whereas dominant-negative Rho had no effect. (C) Northern blot analyses showed that N17Ras decreased smMLCK mRNA expression in VSMC from SHR, whereas N19Rho had no effect. (D) VSMCs from SHR were transiently transfected with vehicle (Veh only), scrambled oligonucleotides (Con), or antisense (AS) oligonucleotides to ERK. The antisense oligonucleotides decrease both smMLCK expression and MLC-P. Experiments in A–D were repeated three times and a representative blot is shown in each panel. (E) Inhibiting MEK decreases nuclear localization of SRF. A7r5 cells (A, B, E, and F) or VSMCs from SHR (C, D, G, and H) were serum starved overnight and treated with DMSO or U0126 for 1 d. Immunocytochemistry was performed using anti-SRF antibodies, and images were analyzed using a confocal microscope. SRF is shown in green (A–D), and nuclear staining is shown with 4,6-diamidino-2-phenylindole (DAPI) in blue. Superposition of green and blue channel is shown in E–H. Note that the nuclear localization of SRF is much less in cells treated with U0126 (B, D, F, and H) compared with the control cells (A, C, E, and G).
We then investigated the signaling pathway involved in the SRF-mediated smMLCK expression. Because the small GTPases Rho and Ras play important roles in regulating SRF-induced gene expression in VSMCs, we investigated the possibility that these GTPases are upstream regulators of SRF-induced smMLCK expression. First, VSMCs from WKY rats were cotransfected with SRF and plasmids expressing dominant-negative Ras (N17Ras) or dominant-negative Rho (N19Rho). Western blot analyses showed that SRF increased smMLCK protein expression (Figure 7B, lane 2), and cotransfection of dominant-negative Ras blocked the SRF-induced smMLCK expression (Figure 7B, lane 3). In contrast, Rho did not affect the SRF-induced increase in smMLCK expression (Figure 7B, lane 4).
To describe the relationship between Ras signaling and the regulation of smMLCK expression in more detail, VSMCs from SHR were transfected with plasmids expressing pcDNA, N17Ras, or N19Rho and Northern blot analyses were performed on total RNA prepared from the transfected cells. This analysis showed that dominant-negative Ras decreased smMLCK mRNA expression in VSMCs from SHR, whereas dominant-negative Rho had no effect (Figure 7C). To investigate downstream events of Ras signaling, VSMCs from SHR were treated with antisense oligonucleotides to ERK (Robinson et al., 1996). Antisense oligonucleotides to ERK also inhibited smMLCK expression and MLC-P in VSMCs from SHR (Figure 7D, lane 3), whereas scrambled oligonucleotides (control) had no effect (Figure 7D, lane 2). These data establish that smMLCK expression is regulated by Ras via ERK.
We then performed immunocytochemistry to determine whether Ras regulates smMLCK expression by changing the subcellular localization of SRF. We used U0126 to block the Ras–MEK–ERK cascade, because U0126 inhibits MEK (Favata et al., 1998), and ERK phosphorylation by MEK is an essential part of Ras signaling. Confocal microscopy (Figure 7E) performed on serum-starved A7r5 cells or VSMCs from SHR, with or without U0126 treatment, showed prominent SRF staining in the nucleus of control cells (DMSO). However, the signal was much less abundant in the nucleus of the cells treated with U0126. These data suggest that Ras signaling regulates SRF-dependent gene expression partly by regulating the nuclear localization of SRF.
Inhibiting MLCK or MEK Decreases Blood Pressure
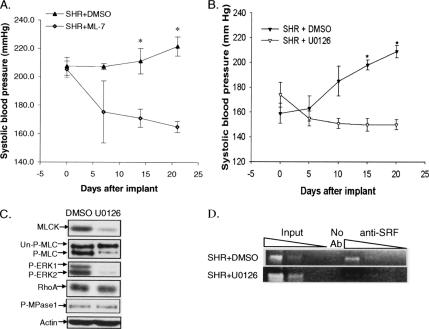
To determine the importance of the Ras–ERK–smMLCK pathway in vivo, we examined the effects of inhibiting MLCK on blood pressure in SHR. MLCK activity was inhibited with ML-7, a specific inhibitor of MLCK (Fazal et al., 2005). An osmotic pump was used to deliver ML-7 or vehicle (DMSO) continuously for 3 wk to SHR that were 16 wk old at the start of the experiment (Figure 8A). ML-7 resulted in a significant decrease in SBP (165.0 ± 3.9 mmHg) compared with SBP in SHR receiving DMSO (221.6 ± 6.8 mmHg).
Figure 8.
In vivo effects of inhibiting MLCK or MEK on blood pressure in SHR. (A) DMSO or ML-7 was continuously administered for 3 wk to adult SHR. SBP was measured every 5 d. Values are mean ± SE, n = 4, *SHR + ML-7 versus SHR + DMSO, p < 0.001. (B) Young SHR (8 wk old at the start of the experiment) were treated with U0126 for 3 wk. Values are mean ± SE, n = 6. *SHR + U0126 versus SHR + DMSO, p < 0.001. (C) Analysis of signaling molecules in the aortas from SHR in B demonstrated lower levels of smMLCK expression, MLC-P, and ERK-P compared with control. Actin was used as a loading control. (D) Cross-linked chromatin was isolated from aortas of DMSO or U0126-treated SHR and immunoprecipitated with antibodies to SRF. Input chromatin (1, 0.1, and 0.01%) and immunoprecipitated DNA (20, 2, and 0.2%) were amplified with primers specific for intron 14. Note that there is much less SRF associated with the intron in SHR treated with U0126 compared with control SHR. Experiments in C and D were repeated three times, and a representative blot is shown in each panel.
We then investigated whether inhibiting Ras signaling could also decrease blood pressure in SHR. U0126, delivered for 3 wk by using an osmotic pump, significantly decreased blood pressure in SHR that were 16 wk old at the start of the experiment. The mean SBP was 181.4 ± 0.89 mmHg in SHR receiving U0126 compared with 221.6 ± 4.2 mmHg in SHR receiving vehicle at the end of the 3-wk treatment period (Supplemental Figure 2). U0126 resulted in a transient decrease in SBP in the normotensive WKY rats that returned to baseline within 2 wk of treatment (Supplemental Figure 2).
The experiments in Figures 8A and Supplemental Figure 2 show that ML-7 and U0126 can reverse hypertension in SHR. Therefore, we investigated whether U0126 could prevent the development of hypertension. SHR that were 8 wk old at the start of drug treatment, the age at which blood pressure starts to increase rapidly (Figure 1A), were treated with U0126 for 3 wk. SBP rose rapidly, from an average of 159.0 ± 8.3 mmHg at the start of the experiment to 208.9 ± 4.7 mmHg by the end of the experiment (Figure 8B), in SHR receiving vehicle. In contrast, U0126 lowered blood pressure to 150.4 ± 4.1 mmHg by the end of the treatment period. Furthermore, biochemical analyses showed that the levels of ERK-phosphorylation (ERK-P), smMLCK expression, and MLC-P were significantly decreased in aortas removed from U0126-treated SHR compared with control without significantly changing the levels of RhoA expression or MPase1-P (Figure 8C). To verify that U0126 decreases smMLCK expression via SRF-dependent gene transcription, ChIP assays were carried out on aortas isolated from DMSO- or U0126-treated SHR (Figure 8D). These assays showed that U0126 decreased SRF occupancy of the smMLCK promoter.
DISCUSSION
VSMCs are essential for maintaining homeostasis, and aberrations in VSMC contraction and proliferation are involved in many human diseases. Contraction of VSMCs directly affects blood pressure, and VSMC proliferation contributes in a significant way to hypertension and intimal hyperplasia associated with vascular injury. An important part of VSMC proliferation is the ability of VSMC to undergo phenotypic modulation in response to local stimuli. Differentiated VSMCs can switch from a contractile phenotype to a “proliferative” or “synthetic” phenotype that involves the down-regulation of smooth muscle-specific markers, increased proliferation and migration and the synthesis of collagen and matrix metalloproteinases (Owens, 1996). The SRF–CArG interaction is considered to be a critical convergence point for signals that determine the phenotypic characteristics of VSMCs. For example, after vascular injury, VSMCs proliferate rapidly and migrate to the intima, changing from a contractile to a proliferative phenotype. This is accompanied by a decrease in the expression of VSMC markers that are typical of the contractile phenotype, such as smooth muscle α-actin, MHC, and SM22α (Regan et al., 2000). Furthermore, PDGF-BB, which plays a central role in phenotypic modulation of VSMCs, down-regulates the expression of smooth muscle specific genes by a process that involves ERK activation and SRF binding to the CArG box (Wang et al., 2004).
Our demonstration of a SRF-dependent increase in smMLCK expression in hypertension is surprising in this context. Because smMLCK plays a crucial role in regulating smooth muscle contraction (de Lanerolle and Paul, 1991), one would predict that smMLCK expression would be down-regulated through ERK-dependent phosphorylation of ELK1 and subsequent competition with SRF and myocardin binding in a manner similar to that shown by Wang et al. (2004) for other smooth muscle marker genes. One possible explanation is that the regulation of VSMC gene expression is inherently different in hypertension than in vascular injury. For example, VSMC mass increases slowly and principally by enlargement or hypertrophy of preexisting VSMCs in hypertension (Owens and Schwartz, 1982), whereas rapid proliferation and migration of VSMCs from the media to the intima is a hallmark of vascular injury and atherosclerosis (Raines, 2004). Moreover, angiotensin II, not PDGF, seems to play the primary role in the process of VSMC hypertrophy in hypertension (Berk and Corson, 1997). Importantly, in contrast to the down-regulation of α-actin in vascular injury (Regan et al., 2000), angiotensin II increases the expression of α-actin (Yoshida et al., 2004). This suggests substantial specificity and subtlety in SRF-mediated gene regulation by various factors in individual pathological conditions. Further study is required to elucidate the precise mechanisms that are responsible for the specific effects.
Mechanistically, the presence of a single CArG box and a 12-base pair extension nearby in the SHR promoter may be important in SRF regulation of smMLCK expression. This notion is supported by the identification of CT dinucleotide repeats [(CT·GA)n] as a GAGA factor (GAF) binding site in Drosophila melanogaster (Wilkins and Lis, 1997). GAF is involved in chromatin remodeling and is thought to establish nucleosome-free DNase I-hypersensitive sites (Leibovitch et al., 2002). The open chromatin configuration created by GAF apparently allows other transcription factors to gain access to the regulatory regions of a gene. Similarly, it is possible that the 12-base pair insertion in the SHR promoter creates a more open chromatin configuration and allows greater access of SRF to the adjacent CArG box. Greater SRF binding to the CArG box of the SHR promoter than the WKY promoter (Figure 6) supports this idea. McDonald et al. (2006) have demonstrated the importance of epigenetic regulation of SRF binding by showing that SRF binding to CArG-containing regions of VSMC promoters plays a critical role in developmental control of VSMC gene expression. There is also support for the idea that changing the local environment of a promoter by adding CT dinucleotides changes the binding of transcription factors. For example, oligonucleotides sequences with 10-base pairs of GA (reverse CT) dinucleotide bind more nuclear proteins than six base pairs of GA repeats (Volpi et al., 2002). Thus, there are paradigms that support the idea that CT repeats provide structural motifs that promote the binding of transcription factors to promoter elements. However, a detailed description of how the CT repeats, generally, and the 12-base pair insertion in the smMLCK promoter, specifically, regulate SRF binding and gene expression requires further study.
It is of some note that the SRF-mediated smMLCK expression is regulated by Ras signaling. Because of its important roles in hypertension (Uehata et al., 1997) and gene regulation (Miralles et al., 2003), we expected that Rho would play an important role in regulating smMLCK expression. However, dominant-negative Rho did not decrease the SRF-stimulated increase in smMLCK protein (Figure 7B) or mRNA (Figure 7C) levels in VSMCs in vitro. Perhaps most telling, there were no changes in Rho A levels or the phosphorylation of MPase1 in blood vessels of SHR treated with U0126 (Figure 8C). The data imply that the Ras and Rho pathways contribute independently to hypertension. Our data also show that Ras regulates smMLCK expression and that this results in an increase in MLC-P and blood pressure. In contrast, Rho activation results in the phosphorylation and inactivation of MPase1, which, in turn, results in an increase in MLC-P (Uehata et al., 1997). Together, the data demonstrate the importance of the balance between smMLCK and MPase1 in determining the level of MLC-P and the central role of MLC-P in hypertension.
In summary, we have used SHR as a model to investigate the importance of SRF in regulating smMLCK expression. We have found that the smMLCK promoter is unique in that it contains a single CArG box (Figure 3B), whereas most other smooth muscle genes contain at least two CArG boxes (Miano, 2003). Our work has also revealed that an insertion in the promoter adjacent to the CArG box enhances SRF binding to the smMLCK promoter in SHR and promotes SRF-dependent reporter gene activity. We also demonstrated that smMLCK expression and MLC-P are increased in SHR and that blocking Ras signaling decreases blood pressure in SHR. These results provide new insights into gene regulation by the Ras/SRF pathways and lay the foundation for investigating the importance of smMLCK gene regulation and MLC-P in hypertension.
Supplementary Material
ACKNOWLEDGMENTS
We thank our colleague Wilma Hofmann for providing the purified TBP and for assistance with the ChIP assays. We also thank Dr. James Lessard (Children’s Hospital, Cincinnati, OH) for generously providing the C4 actin antibody, Dr. Tatyana A. Voyno-Yasenetskaya (Department of Pharmacology, University of Illinois at Chicago [UIC], Chicago, IL) for generously providing the dominant-negative Ras and Rho plasmids, Dr. Ken Byron (Loyola University Medical School, Chicago, IL) for providing A7r5 cells, Dr. Asgerally Fazleabas (Department of Physiology, UIC) for the use of a microscope, and M. L Chen (Research Resources Center, UIC) for assistance with the confocal microscopy. This work was supported, in part, by National Institutes of Health Grant HL-59618 (to P.d.L). Y.-J.H. and W.-Y.H. are supported by postdoctoral fellowships from the American Heart Association (0525686Z and 0425736Z, respectively).
Footnotes
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-04-0353) on July 5, 2006.
REFERENCES
- Berk B. R., Corson M. A. Angiotensin II signal transduction in vascular smooth muscle: role of tyrosine kinases. Circ. Res. 1997;80:607–616. doi: 10.1161/01.res.80.5.607. [DOI] [PubMed] [Google Scholar]
- Boorstein W. R., Craig E. A. Primer extension analysis of RNA. Methods Enzymol. 1989;180:347–369. doi: 10.1016/0076-6879(89)80111-9. [DOI] [PubMed] [Google Scholar]
- Buchwalter G., Gross C., Wasylyk B. Ets ternary complex transcription factors. Gene. 2004;324:1–14. doi: 10.1016/j.gene.2003.09.028. [DOI] [PubMed] [Google Scholar]
- Chen C. Y., Schwartz R. J. Recruitment of the tinman homolog Nkx2.5 by serum response factor activates cardiac alpha-actin gene transcription. Mol. Cell Biol. 1996;16:6372–6384. doi: 10.1128/mcb.16.11.6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis F. J., Gupta M., Pogwizd S. M., Bacha E., Jeevanandam V., Gupta M. P. Increased expression of alternatively spliced dominant-negative isoform of SRF in human failing hearts. Am. J. Physiol. 2002;282:H1521–H1533. doi: 10.1152/ajpheart.00844.2001. [DOI] [PubMed] [Google Scholar]
- de Lanerolle P., Adelstein R. S., Feramisco J. R., Burridge K. Characterization of antibodies to smooth muscle myosin kinase and their use in localizing myosin kinase in nonmuscle cells. Proc. Natl. Acad. Sci. USA. 1981;78:4738–4742. doi: 10.1073/pnas.78.8.4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lanerolle P., Paul R. J. Myosin phosphorylation/dephosphorylation and regulation of airway smooth muscle contractility. Am. J. Physiol. 1991;261:L1–L14. doi: 10.1152/ajplung.1991.261.2.L1. [DOI] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favata M. F., et al. Identification of a novel inhibitor of mitogen-activated protein kinase kinase. J. Biol. Chem. 1998;273:18623–18632. doi: 10.1074/jbc.273.29.18623. [DOI] [PubMed] [Google Scholar]
- Fazal F., et al. Inhibiting myosin light chain kinase induces apoptosis in vitro and in vivo. Mol. Cell. Biol. 2005;25:6259–6266. doi: 10.1128/MCB.25.14.6259-6266.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishkind D. J., Cao L. G., Wang Y. L. Microinjection of the catalytic fragment of myosin light chain kinase into dividing cells: effects on mitosis and cytokinesis. J. Cell Biol. 1991;114:967–975. doi: 10.1083/jcb.114.5.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher P. J., Herring B. P. The carboxyl terminus of the smooth muscle myosin light chain kinase is expressed as an independent protein, telokin. J. Biol. Chem. 1991;266:23945–23952. [PMC free article] [PubMed] [Google Scholar]
- Gallagher P. J., Herring B. P., Stull J. T. Myosin light chain kinases. J. Muscle Res. Cell Motil. 1997;18:1–16. doi: 10.1023/a:1018616814417. [DOI] [PubMed] [Google Scholar]
- Granok H., Leibovitch B. A., Shaffer C. D., Elgin S.C.R. Ga-ga over GAGA factor. Curr. Biol. 1995;5:238–241. doi: 10.1016/s0960-9822(95)00048-0. [DOI] [PubMed] [Google Scholar]
- Hayashi K., Takahashi M., Kimura K., Nishida W., Saga H., Sobue K. Changes in the balance of phosphoinositide 3-kinase/protein kinase B (Akt) and the mitogen-activated protein kinases (ERK/p38MAPK) determine a phenotype of visceral and vascular smooth muscle cells. J. Cell Biol. 1999;145:727–740. doi: 10.1083/jcb.145.4.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann W. A., et al. Actin is part of pre-initiation complexes and is necessary for transcription by RNA polymerase II. Nat. Cell Biol. 2004;6:1094–1101. doi: 10.1038/ncb1182. [DOI] [PubMed] [Google Scholar]
- Holycross B. J., Blank R. S., Thompson M. M., Peach M. J., Owens G. K. PDGF-BB-induced suppression of smooth muscle cell differentiation. Circ. Res. 1992;71:1525–1532. doi: 10.1161/01.res.71.6.1525. [DOI] [PubMed] [Google Scholar]
- Klemke R. L., Cai S., Giannini A. L., Gallagher P. J., de Lanerolle P., Cheresh D. A. Regulation of cell motility by mitogen activated protein kinase. J. Cell Biol. 1997;137:481–492. doi: 10.1083/jcb.137.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibovitch B. A., Lu Q., Benjamin L. R., Liu Y., Gilmour D. S., Elgin S.C.R. GAGA factor and the TFIID complex collaborate in generating an open chromatin structure at the Drosophila melanogaster hsp26 promoter. Mol. Cell Biol. 2002;22:6148–6157. doi: 10.1128/MCB.22.17.6148-6157.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard J. L. Two monoclonal antibodies to actin: one muscle selective and one generally reactive. Cell Motil. Cytoskeleton. 1988;10:349–362. doi: 10.1002/cm.970100302. [DOI] [PubMed] [Google Scholar]
- Lu Q., Teare J. M., Granok H., Swede M. J., Xu J., Elgin S.C.R. The capacity to form H-DNA cannot substitute for GAGA factor binding to a (CT)n·(GA)n regulatory site. Nucleic Acids Res. 2003;31:2483–2494. doi: 10.1093/nar/gkg369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald O. G., Wamhoff B. R., Hoofnagle M. H., Owens G. K. Control of SRF binding to CArG box chromatin regulates smooth muscle gene expression in vivo. J. Clin. Investig. 2006;116:36–48. doi: 10.1172/JCI26505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manabe I., Owens G. K. CArG elements control smooth muscle subtype-specific expression of smooth muscle myosin in vivo. J. Clin. Investig. 2001;107:823–834. doi: 10.1172/JCI11385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miano J. M. Serum response factor: toggling between disparate programs of gene expression. J. Mol. Cell Cardiol. 2003;35:559–708. doi: 10.1016/s0022-2828(03)00110-x. [DOI] [PubMed] [Google Scholar]
- Miralles F., Posern G., Zaromytidou A., Treisman R. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell. 2003;113:329–342. doi: 10.1016/s0092-8674(03)00278-2. [DOI] [PubMed] [Google Scholar]
- Obara K., Takai A., Ruegg J. C., de Lanerolle P. Okadaic acid, a phosphatase inhibitor, produces a Ca2+ and calmodulin-independent contraction of smooth muscle. Pflugers Arch. 1989;414:134–138. doi: 10.1007/BF00580954. [DOI] [PubMed] [Google Scholar]
- Okamoto K., Aoki. K. Development of a strain of spontaneously hypertensive rats. Jpn. Circ. J. 1963;27:282–293. doi: 10.1253/jcj.27.282. [DOI] [PubMed] [Google Scholar]
- Owens G. K. Role of mechanical strain in regulation of differentiation of vascular smooth muscle cells. Circ. Res. 1996;79:1054–1055. doi: 10.1161/01.res.79.5.1054. [DOI] [PubMed] [Google Scholar]
- Owens G. K., Kimar M. S., Wamhoff B. R. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol. Rev. 2004;84:767–801. doi: 10.1152/physrev.00041.2003. [DOI] [PubMed] [Google Scholar]
- Owens G. K., Schwartz S. M. Alterations in vascular smooth muscle mass in the spontaneously hypertensive rat. Role of cellular hypertrophy, hyperploidy, and hyperplasia. Circ. Res. 1982;51:280–289. doi: 10.1161/01.res.51.3.280. [DOI] [PubMed] [Google Scholar]
- Peterson M. G., Tanese N., Pugh B. F., Tjian R. Functional domains and upstream activation properties of cloned human TATA binding protein. Science. 1990;248:1625–1630. doi: 10.1126/science.2363050. [DOI] [PubMed] [Google Scholar]
- Potier M., Chelot E., Perkasky Y., Gardner K., Rossier J., Turnell W. G. The human myosin light chain kinase (MLCK) from hippocampus: cloning, sequencing, expression, and localization to 3qcen-q21. Genomics. 1995;29:562–570. doi: 10.1006/geno.1995.9965. [DOI] [PubMed] [Google Scholar]
- Raines E. W. PDGF and cardiovascular disease. Cytokine Growth Factor Rev. 2004;15:237–254. doi: 10.1016/j.cytogfr.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Regan C. P., Adam P. J., Madsen C. S., Owens G. K. Molecular mechanisms of decreased smooth muscle differentiation marker expression after vascular injury. J. Clin. Investig. 2000;106:1139–1147. doi: 10.1172/JCI10522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson C. J., Scott P. H., Allan A. B., Jess T., Gould G. W., Plevin R. Treatment of vascular smooth muscle cells with antisense phosphorothioate oligodeoxynucleotides directed against p42 and p44 mitogen-activated protein kinases abolishes DNA synthesis in response to PDGF. Biochem. J. 1996;320:123–127. doi: 10.1042/bj3200123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross R. The smooth muscle cell. II. Growth of smooth muscle in culture and formation of elastic fibers. J. Cell Biol. 1971;50:172–186. doi: 10.1083/jcb.50.1.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touyz R. M. The role of angiotensin II in regulating vascular structural and functional changes in hypertension. Curr. Hypertens. Rep. 2003;5:155–164. doi: 10.1007/s11906-003-0073-2. [DOI] [PubMed] [Google Scholar]
- Treisman R. The serum response element. Trends Biochem. Sci. 1992;17:423–426. doi: 10.1016/0968-0004(92)90013-y. [DOI] [PubMed] [Google Scholar]
- Uehata M., et al. Calcium sensitization of smooth muscle mediated by a Rho-associated protein kinase in hypertension. Nature. 1997;389:990–994. doi: 10.1038/40187. [DOI] [PubMed] [Google Scholar]
- Volpi S., Rabadan-Diehl C., Cawley N., Aguilera G. Transcriptional regulation of the pituitary vasopressin V1b receptor involves a GAGA-binding protein. J. Biol. Chem. 2002;277:27829–27838. doi: 10.1074/jbc.M201508200. [DOI] [PubMed] [Google Scholar]
- Wang D., Olson E. N. Control of smooth muscle development by the myocardin family of transcriptional coactivators. Curr. Opin. Genet. Dev. 2004;14:558–566. doi: 10.1016/j.gde.2004.08.003. [DOI] [PubMed] [Google Scholar]
- Wang Z., Wang D. Z., Hockemeyer D., McAnally J., Nordheim A., Olson E. N. Myocardin and ternary complex factors compete for SRF to control smooth muscle gene expression. Nature. 2004;428:185–189. doi: 10.1038/nature02382. [DOI] [PubMed] [Google Scholar]
- Wilkins R. C., Lis J. T. Dynamics of potentiation and activation: GAGA factor and its role in heat shock gene regulation. Nucleic Acids Res. 1997;25:3963–3968. doi: 10.1093/nar/25.20.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A. K., Gorgas G., Claypool W. D., de Lanerolle P. An increase or a decrease in myosin II phosphorylation inhibits macrophage motility. J. Cell Biol. 1991;114:277–283. doi: 10.1083/jcb.114.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakita Y., Yamashiro S., Matsumura F. In vivo phosphorylation of regulatory light chain of myosin II during mitosis of cultured cells. J. Cell Biol. 1994;124:129–137. doi: 10.1083/jcb.124.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamori Y. Development of the Spontaneously Hypertensive Rat (SHR) and of various spontaneous rat models, and their implications. In: de Jong W., editor. Handbook of Hypertension, Vol. 4. Experimental and Genetic Models of Hypertension. Amsterdam, New York, Oxford: Elsevier; 1984. pp. 224–239. [Google Scholar]
- Yoshida T., Hoofnagle M. H., Owens G. K. Myocardin and Prx1 contribute to angiotensin II-induced expression of smooth muscle α-actin. Circ. Res. 2004;94:1075–1082. doi: 10.1161/01.RES.0000125622.46280.95. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.