Abstract
The cell fate determinant Numb is a membrane-associated adaptor protein involved in both development and intracellular vesicular trafficking. It has a phosphotyrosine-binding (PTB) domain and COOH-terminal endocytic-binding motifs for α-adaptin and Eps15 homology domain-containing proteins. Four isoforms of Numb are expressed in vertebrates, two of which selectively associate with the cortical membrane. In this study, we have characterized a cortical pool of Numb that colocalizes with AP2 and Eps15 at substratum plasma membrane punctae and cortical membrane-associated vesicles. Green fluorescent protein (GFP)-tagged mutants of Numb were used to identify the structural determinants required for localization. In addition to the previously described association of the PTB domain with the plasma membrane, we show that the AP2-binding motifs facilitate the association of Numb with cortical membrane punctae and vesicles. We also show that agonist stimulation of G protein-coupled receptors (GPCRs) that are linked to phospholipase Cβ and protein kinase C (PKC) activation causes redistribution of Numb from the cortical membrane to the cytosol. This effect is correlated with Numb phosphorylation and an increase in its Triton X-100 solubility. Live-imaging analysis of mutants identified two regions within Numb that are independently responsive to GPCR-mediated lipid hydrolysis and PKC activation: the PTB domain and a region encompassing at least three putative PKC phosphorylation sites. Our data indicate that membrane localization of Numb is dynamically regulated by GPCR-activated phospholipid hydrolysis and PKC-dependent phosphorylation events.
INTRODUCTION
Numb is an adaptor protein that plays a well documented role in cell fate determination during Drosophila development (Uemura et al., 1989; Brewster and Bodmer, 1995; Spana and Doe, 1996; Ruiz Gomez and Bate, 1997). Of importance for this function is the ability of Numb to localize to the cell cortex, and, furthermore, to asymmetrically segregate to one pole during cell division such that it is only inherited by one daughter cell (Rhyu et al., 1994; Spana et al., 1995; Guo et al., 1996). The localization of dNumb is dependent on the evolutionarily conserved Par complex composed of Bazooka/Par-3, Par-6, and atypical protein kinase C (aPKC). Cell type-specific polarity cues direct the asymmetric localization of the Par complex, which in turn is responsible for phosphorylation of Lethal Giant Larvae (Lgl) and the subsequent localization of Numb at the opposite pole of the cell. However, the mechanism by which Numb is recruited to the cortex and then selectively localized to one pole of the cell is not known. Early work examining the localization of dNumb at the plasma membrane (PM), and subsequent asymmetric segregation, showed dependence on determinants within the N terminus of the protein (Knoblich et al., 1997; Jan and Jan, 1998). Deletion of the N-terminal 119 amino acids results in mainly cytosolic localization of dNumb. Association with other cortical membrane bound proteins, including PON and the transmembrane protein NIP, has also been proposed as a mechanism for localization of dNumb at the cortex (Lu et al., 1998; Qin et al., 2004). The asymmetric cortical localization of PON is dependent on the phosphorylation state of Lgl, although how this occurs is unclear (Betschinger et al., 2003; Langevin et al., 2005).
Studies examining the function of Numb-related genes identified in vertebrates suggest that these homologues function in a similar manner to Drosophila Numb with respect to cell fate selection (Verdi et al., 1996; Zhong et al., 1996; Wakamatsu et al., 1999). Indeed, ectopic expression of mNumb in Drosophila results in a similar phenotype to overexpression of dNumb. mNumb expression can rescue the dNumb null phenotype, and, like dNumb, mNumb is asymmetrically localized in progenitor cells of the peripheral and central nervous system (Verdi et al., 1996; Zhong et al., 1996). In vertebrates, mNumb is membrane localized and reported to be asymmetric in murine retinal cells and cortical progenitors (Knoblich et al., 1995; Zhong et al., 1996; Wakamatsu et al., 1999; Dooley et al., 2003). However, it is unknown how mammalian Numb is recruited to the PM and acquires asymmetric localization. Mammalian homologues of Par3/Par6/aPKC play an essential role in the establishment of polarity in epithelial cells (for review, see Ohno, 2001) and migrating cells (Etienne-Manneville and Hall, 2001). This, together with the observation that in Lgl knockout mice Numb fails to localize properly during the asymmetric cell divisions of neural progenitors (Klezovitch et al., 2004), supports the view that the signaling pathways involved in specifying Numb localization are conserved in Drosophila and mammals.
Numb is made up of a phosphotyrosine-binding (PTB) domain and a C-terminal region having conserved binding motifs for α-adaptin and Eps15 homology (EH) domain-containing proteins. Whereas only one form of Numb occurs in Drosophila, four isoforms of Numb are expressed in mammals. Notably, only two of the isoforms localize to the PM. Localization of the PM forms (isoforms p66 and p72) is facilitated by the presence of an alternatively spliced, highly basic insert within the PTB domain (PTBi) (Dho et al., 1999). Several studies have shown that Numb is localized to the PM and also to intracellular vesicles where it probably has a role in endocytic trafficking (Santolini et al., 2000; Cayouette and Raff, 2003; Nishimura et al., 2003; Smith et al., 2004). None of these studies, however, have identified the factor(s) that specify and/or regulate the subcellular localization of Numb.
In this study, we have characterized the cortical membrane pool of mammalian Numb by using fluorescently tagged mutants of Numb. We have delineated the structural determinants required for the localization of the cortical membrane forms of mammalian Numb, p66 and p72. Furthermore, we demonstrate that intracellular signals, elicited by G protein-coupled receptor (GPCR) activation of protein kinase C (PKC), cause a rapid redistribution of Numb proteins.
MATERIALS AND METHODS
DNA and Small Interfering RNA (siRNA) Constructs
All wild-type and deletion constructs were made by PCR amplification using mouse Numb cDNA template and ligated in frame into pEGFP-C1 or C2. PCR-based site-directed mutagenesis was used to mutate the NPF and DPF motifs as described previously (Smith et al., 2004). The expression vector pcDNA3.1HA-p115RGS, encoding the first 240 amino acids of human p115-RhoGEF spanning the Gα12/13-selective regulator of G protein signaling (RGS) domain, has been described previously (Hains et al., 2004). Expression vectors for the constitutively active mutants of human Gαq (Q209L) and Gα13 (Q226L) were obtained from the University of Missouri-Rolla cDNA Resource Center (Rola, MO) (www.cdna.org). The expression vector for green fluorescent protein (GFP)-RGS2 has been described previously (Roy et al., 2003) and was a kind gift from Dr. Peter Chidiac (University of Western Ontario, London, Ontario, Canada). The α-adaptin siRNA (target sequence GAGCAUGUGCACGCUGGC) was designed essentially as described previously (Hinrichsen et al., 2003; Motley et al., 2003) and was synthesized by Dharmacon RNA Technologies (Lafayette, CO), control siRNA was a nonfunctional, pooled, siRNA directed against α-adaptin.
Cell Culture, cDNA, and siRNA Transfections
All cells were grown in 10% fetal bovine serum in DMEM. SPR-HeLa cells, which stably overexpress the substance P receptor, were generated as described in Trejo and Coughlin (1999). For transfection, cells were grown on six-well dishes to ∼80% confluence. GFP cDNA constructs were transfected into the cells using 1.5 μl of Lipofectamine 2000 and 0.5 μg DNA per well in Opti-MEM. siRNA duplexes were introduced into the cells using 3 μl of Lipofectamine 2000 and 50–60 nM siRNA. Four hours after addition of nucleotide–Lipofectamine complex, the cells were trypsinized and reseeded in normal growth media onto glass coverslips for either immunocytochemistry or live imaging, which was carried out ∼24 h (cDNA) or 72 h (siRNA) later unless otherwise indicated.
Preparation of Triton X-100–soluble and –insoluble Cell Extracts
Cells grown to confluence on 10-cm2 culture plates were treated with either 0.5 μM 12-O-tetradecanoylphorbol-13-acetate (TPA) in 0.1% bovine serum albumin/phosphate-buffered saline (BSA/PBS) or 0.1% PBS alone for 10 min at 37°C. For each condition, 1 plate was lysed in 1 ml of CSK buffer (10 mM PIPES, pH 6.8, 50 mM NaCl, 300 mM sucrose, and 0.5% Triton X-100 plus protease inhibitors), extracted on ice for 30 min, and centrifuged at 14,000 × g for 15 min (this lysate is represented as the Triton-soluble fraction). The pellet was further extracted with NP-40/deoxycholate buffer [50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% NP-40, 0.25% (wt/vol) deoxycholate, and 1 mM EGTA plus protease inhibitors] and centrifuged (lysate is represented as the Triton-insoluble fraction). Numb was immunoprecipitated from each fraction by using the anti-Numb antibody anti-NbA, separated by SDS-PAGE, and visualized by Western blotting and enhanced chemiluminescence (ECL).
Alkaline Phosphatase Treatment of Immunoprecipitates
Cells grown to confluence on 10-cm2 culture plates were treated with either 0.5 μM TPA in 0.1% BSA/PBS or 0.1% BSA/PBS alone for 10 min at 37°C. Each plate was lysed in 1 ml of SAP lysis buffer (50 mM Tris, pH 8, 150 mM NaCl, 1.5 mM MgCl2, 1% Triton X-100, and 1 mM dithiothreitol [DTT] plus protease inhibitors), and the insoluble fraction removed by centrifugation at 14,000 × g. Numb was immunoprecipitated from each lysate with anti-NbA bound to protein A-Sepharose beads. The immunoprecipitates were washed with shrimp alkaline phosphatase (SAP) immunoprecipitation (IP) buffer (50 mM Tris, pH 8, 150 mM NaCl, 5 mM MgCl2, and 1 mM DTT plus added protease inhibitors), resuspended in 100 μl of SAP IP buffer with or without 10 units of SAP (Roche Diagnostics, Indianapolis, IN), and incubated for 30 min at 37°C. The immunoprecipitated Numb was then separated by SDS-PAGE and visualized by Western blotting and ECL.
Immunocytochemistry
Cells were fixed in 4% paraformaldehyde, 30 mM sucrose in PBS for 20 min at room temperature, permeablized with 0.1% Triton X-100, and blocked with 5% normal donkey serum (Jackson ImmunoResearch Laboratories, West Grove, PA). Primary antibodies were applied for 30 min at 37°C at the following dilutions: rabbit and guinea pig anti-NbC and rabbit anti-NbA, 1:100 (described Dho et al., 1999); anti-adaptin, 1:200 (Affinity Bioreagents, Golden, CO); anti-Eps15 (C20), 1:500 (Santa Cruz Biotechnology, Santa Cruz, CA); mouse anti-E-cadherin, 1:200 (BD Transduction Laboratories, Lexington, KY), rat anti-ZO-1, 1:200 (Zymed Laboratories, South San Francisco, CA); anti-Gαq (E-17), 1:200 (Santa Cruz Biotechnology); and anti-Gα13 (C terminus), 1:200 (catalog no. 371784; Chemicon International, Temecula, CA). Cells were then washed with 0.05% Triton X-100 in PBS and incubated with Alexa 488- (green; Invitrogen, Carlsbad, CA), Cy3- (red), or Cy5 (blue; Jackson ImmunoResearch Laboratories)-conjugated secondary antibodies. Confocal images were acquired using an LSM510 microscope (Carl Zeiss MicroImaging, Thornwood, NY) with a 100× oil immersion objective (numerical aperture 1.5). Images were collected at 8-bit depth, with a resolution of 1024 × 1024 pixels. LSM images were converted to TIFF files and figures were prepared in Adobe Illustrator (Adobe Systems, Mountain View, CA). In some figures, contrast enhancement was used to make the published images easier to view. When this was done, all images in the figure were treated equally.
Confocal Live-Cell Imaging
For live imaging, transfected SPR-HeLa cells on glass coverslips were mounted in 200 μl of RPMI 1640 medium containing HEPES/0.1% BSA and 1 mM added Ca2+. For time courses, images were acquired at 15-s intervals unless indicated otherwise. After at least 1 min to acquire baseline images, either TPA (final concentration 0.5 μM) or substance P (final concentration 100 nM) was added as a 20 μl 10× concentrated solution. Poststimulus image acquisition began within 5 s of its addition. Images were collected at 8-bits depth, 1024 × 1024 pixel resolution. Movies were converted to QuickTime format.
Quantification of GFP-Numb in Basal PM Punctae
The relative association of GFP-Numb wild-type and mutants in basal PM punctae was quantitated using ImageJ software (Rasband WS, Image J; National Institutes of Health, Bethesda, MD) (http://rsb.info.nih.gov/ij/, 1997–2006). Live-cell, 8-bit, TIFF images were manually thresholded to divide the image into puncta and background. Particle analysis was used to calculate the percentage of the basal PM area that is occupied by punctae.
RESULTS
Numb Is Localized at the Cell Cortex in Vesicles and PM Punctae
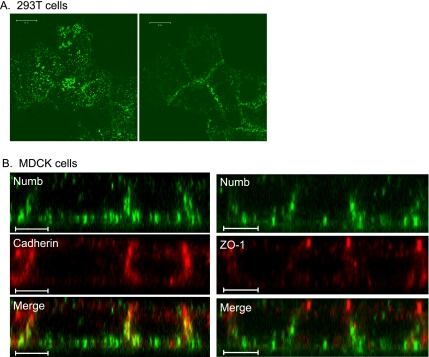
In previous work, we observed that in addition to the Numb that is localized to intracellular vesicles, some of which costains with EHD4 and/or ARF6, a significant proportion of Numb is discretely localized at the PM (Smith et al., 2004). Further examination, using anti-Numb serum, revealed strong cortical PM staining in many cell lines, including human embryonic kidney 293T (Figure 1A; our unpublished data), HeLa, Chinese hamster ovary (CHO), Cos1, A431, and CV1 lines. Particularly apparent was staining of basal membrane patches and vesicles closely apposed to the PM. Furthermore, in polarized Madin-Darby canine kidney (MDCK) cells, anti-NbC membrane staining was primarily restricted to the basal and basolateral cortical membrane (Figure 1B). Localization of Numb at both cytosolic vesicles and the cortical membrane is in keeping with a number of published observations (Santolini et al., 2000; Nishimura et al., 2003; Smith et al., 2004), including ours, which described the cortical membrane association of those Numb isoforms that include the alternatively spliced PTBi domain insert (Dho et al., 1999). In view of these observations, we were interested in characterizing this cortical pool of Numb and identifying the structural determinants of its subcellular localization.
Figure 1.
Numb is localized in basal PM punctae and on vesicles associated with the cell cortex. (A) 293T cells were grown on glass coverslips, fixed, permeablized, and stained with anti-NbC. The images shown were taken at the basal, substratum-associated membrane (left) and a midsection through the cell (right). (B) MDCK cells grown for 3 d on polycarbonate filters were fixed, permeablized, and colabeled with anti-NbC (green) and either anti E-cadherin (red; left) or anti ZO-1 (red; right). Bars, 10 μm.
Numb Colocalizes with Markers of Clathrin-coated Pits and Vesicles
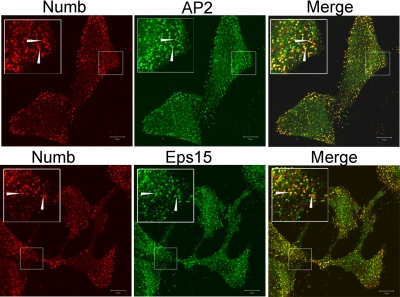
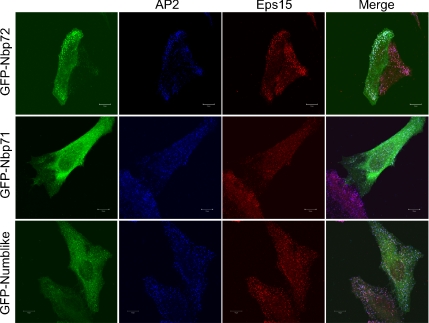
To characterize the substratum punctae, we examined the colocalization of Numb with markers of adhesive structures and clathrin-coated pits in HeLa cells. Pronounced colocalization was observed between Numb and both the α-adaptin subunit of AP2 and Eps15 (Figure 2). We did not observe colocalization with vinculin (denoting focal adhesions) nor caveolin (denoting caveoli) (our unpublished data). Our results are consistent with those of Santolini et al. (2000), although in their study the majority of Numb colocalization with AP2 and Eps15 was on intracellular vesicles, with only a small proportion colocalizing in clathrin-coated pits. This discrepancy may reflect differences in the antibodies used in these studies. Whereas we have used a Numb anti-serum directed at the C-terminal 15 amino acids of Numb (anti-NbC), the anti-serum used by Santolini et al. (2000) was directed at an internal sequence. Because anti-NbC could also recognize a similar sequence at the C terminus of the related Numb-like protein (Nbl), it was possible that the staining pattern observed could be due to Numblike protein reactivity. Therefore, we compared the localization of overexpressed GFP-tagged Numb and Numblike proteins in HeLa cells (Figure 3). GFP-Nbp72 was primarily PM associated with some cytosolic localization. Marked accumulation in basal PM puncta was observed, which costained with both anti-α-adaptin and anti-Eps15. GFP-Nbp66 was similarly membrane localized. In contrast, GFP-Nbl was largely localized within the cytosol and on cytosolic vesicles. Some small PM puncta were occasionally observed in some cells which, like Numb p72, colocalized with AP2 and Eps15. Indeed, the PM localization of Numblike was very similar to that of GFP-Numb p71. Thus, the anti-NbC staining observed at the PM represents primarily the forms of Numb containing the PTB domain having the alternatively spliced insert (PTBi).
Figure 2.
Numb colocalizes with α-adaptin and Eps15 in basal PM punctae. HeLa cells were fixed and colabeled with guinea pig anti-NbC (red) and either anti-adaptin or anti-Eps15 (green). Confocal microscopy was used to image the cell membrane attached to the glass coverslip. The boxed areas shown in each image are magnified to more clearly see punctae labeled with both anti-NbC and the indicated antibody (arrows indicate examples of colocalization). These images are representative of at least three separate experiments. Bars, 10 μm.
Figure 3.
GFP-tagged Numb isoforms and Numblike exhibit differential membrane localization. GFP-Nbp72 (PTBi isoform), GFP-Nbp71 (PTBo isoform), and GFP-Nbl were transiently expressed in HeLa cells. Approximately 24 h after transfection, transfected cells were fixed and labeled with anti-adaptin (Cy3-labeled anti-mouse secondary) and anti-Eps15 (Cy5-labeled anti-rabbit secondary). Cells were imaged at the basal membrane by using confocal microscopy. Bars, 10 μm.
AP2 and Adaptin-binding Motifs Are Required for the Localization of Numb at Substratum Punctae
The regions of Numb responsible for targeting Numb to the AP2/Eps15-positive membrane punctae and vesicles are not known. Putative localization motifs within the C-terminal region of Numb include two DPF motifs, which mediate AP2 binding (Santolini et al., 2000), and an NPF motif, which mediates binding to EH domain-containing proteins, including Eps15 and EHD4 (Salcini et al., 1997; Smith et al., 2004). Indeed, these motifs are conserved in all Numb isoforms and also Numblike (Smith et al., 2004).
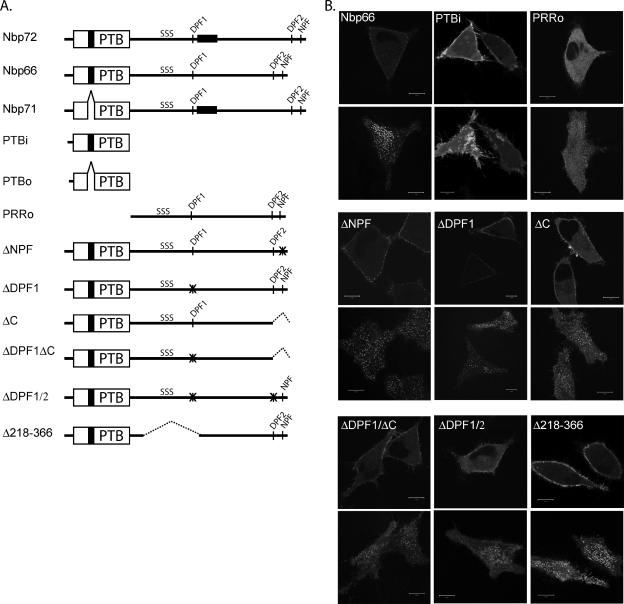
To identify the region(s) within Numb that are required for its association in AP2-positive punctae, we examined the localization of GFP-tagged Numb mutants in live HeLa cells stably expressing the substance P receptor (HeLa-SPR; Trejo and Coughlin, 1999). Each mutant was classified according to whether it was cytosolic, had generalized cortical membrane binding, or accumulated in basal membrane punctae or cortical membrane-associated vesicles. The extent of localization in the punctae was further quantitated by measuring the area occupied by GFP-positive punctae in relation to the total basal surface area of the cell attached to the glass (values are shown below in the text). The results are summarized in Table 1, and representative images are shown in Figure 4. We knew from previous work, and that shown in Figure 3, that the 11-amino acid insert found in the PTBi domain is needed to localize the full-length protein at the PM, likely via interactions with membrane phospholipids (Dho et al., 1999). However, whereas the full-length, wild-type Numb protein (GFP-Nbp66) localized to cortical punctae/vesicles (punctae represented 12.4% of the basal membrane area; 62 cells/10 experiments), the isolated N-terminal region of Numb, including the PTBi domain, was diffusely localized to the cortical membrane and did not accumulate in basal membrane punctae or cortical membrane-associated vesicles (Dho et al., 1999; Figure 4), suggesting that regions C-terminal to the PTB domain are required for localization to membrane puncta. Indeed, full-length Numb isoforms lacking the PTB insert (Nbp71; 4.2%, 10 cells) as well as the Numblike protein (5.2%, 6 cells), exhibited occasional punctate binding at the membrane. In addition, a Numb mutant possessing only the C-terminal half of Numb and lacking the PTB domain (GFP-PRRo) also showed some limited localization to cortical membrane-associated vesicles (3.6%, 20 cells). Together, these observations suggested that regions outside the PTB domain direct Numb localization at distinct cortical membrane sites.
Table 1.
Effect of Numb mutations on their cellular localization and response to TPA and substance P stimulation
| GFP-tagged mutant | Localization |
Response |
|||
|---|---|---|---|---|---|
| Cytosolic | Membrane | Puncta/vesicles | TPA | Substance P | |
| Nbp72 | + | + | ++++ | Yes | Yes |
| Nbp66 | + | + | ++++ | Yes | Yes |
| ΔNPF | + | + | +++ | Yes | Yes |
| ΔDPF1 | + | + | ++++ | Yes | Yes |
| ΔC | + | ++++ | ++ | Yes | Yes |
| ΔDPF1ΔC | + | ++++ | ++ | Yes | Yes |
| ΔDPF1/2 | + | ++++ | ++ | Yes | Yes |
| Nbp71 | ++++ | − | + | Yes | Yes |
| PRRo | ++++ | − | + | Yes | Yes |
| PTBo | ++++ | − | − | ||
| PTBi | + | ++++ | − | No | Transient |
| Δ218-366 | + | + | +++++ | No | Transient |
GFP-tagged Numb wild-type (p66, p71, and p72) and mutants were transiently expressed in SPR-HeLa cells. Confocal imaging of live cells was used to assess their localization and response to stimulation with 0.5 μM TPA and 100 nM substance P. Cellular localization was visually assessed under basal conditions, and the relative association of each protein in the cytosol or at the PM (either generalized binding or accumulation in punctae or vesicles) is indicated. Accumulation in basal PM punctae was further quantified as described in Materials and Methods (see text for values).
Figure 4.
Adaptin-binding motifs facilitate the localization of Numb at the PM. (A) Diagrammatic representation of GFP-tagged constructs used to identify regions within Numb involved in localization and stimulated mobilization. (B) GFP-tagged Numb wild type and mutants were transiently expressed in SPR-HeLa cells. Approximately 24 h after transfection, live cells were imaged by confocal microscopy to assess the distribution of the transfected protein. Two optical sections are shown for each cell, one section acquired at the basal membrane attached to the glass coverslip (bottom image of each pair) and a second section acquired in the mid-region of the cell (top image of each pair). Cells expressing low to mid levels of expression were chosen, because high expression was often associated with apparent mislocalization of Numb to all regions of the PM. Bars, 10 μm.
To further identify regions within Numb necessary for its association in AP2-positive punctae, we deleted the NPF and DPF motifs. Removal of the NPF motif by mutation to three alanine residues (GFP-ΔNPF) disrupts Numb’s interaction with EHD4 or Eps15 (Smith et al., 2004), but it had little effect on the localization of Numb (10.2%; 18 cells/2 experiments). Similarly, removal of the first DPF motif of Numb (DPF1 mutated to three alanine residues; GFP-ΔDPF1) did not change its membrane localization (13.1%; 11 cells/2 experiments). However, a Numb mutant in which the C-terminal 38 amino acids are deleted (ΔC), thereby removing the second DPF motif (DPF2), exhibited a decreased accumulation in substratum punctae (6.9%; 22 cells) and vesicles and more generalized cortical membrane binding. This effect was more dramatic in a Numb mutant combining the DPF1 mutation with the ΔC truncation (ΔDPF1ΔC; 4.8%, 26 cells), suggesting that, whereas the more C-terminal DPF motif (DPF2) of Numb is necessary for its accumulation in PM puncta and vesicles, the DPF1 motif may also play a facilitating role. Because truncation of the C terminus not only removes the second DPF motif but also possibly other potential protein–protein interaction sites, we also tested a Numb mutant in which only the two DPF motifs were mutated (GFP-ΔDPF1/2; each DPF motif was changed to 3 alanine residues). This mutant had a similar disruption in membrane localization (6.7%, 24 cells/2 experiments) to GFP-ΔDPF1ΔC, which indicates that other putative protein interaction motifs within the C-terminal 38 amino acids are not necessary for the recruitment of Numb into the AP2-positive punctae. Thus, although AP2-binding via the DPF motifs is likely the primary mechanism for localization in AP2/Eps15-positive punctae/vesicles, we cannot rule out the possibility that other protein–protein interactions are also involved, because the double mutant still exhibited low level recruitment into AP2/Eps15-positive membrane punctae. This residual localization may arise from Numb inclusion in a multi-adaptor complex, or alternatively, may result from the frank overexpression of the Numb mutants.
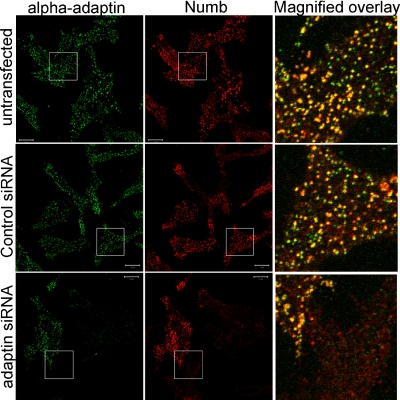
AP2 recruits a number of endocytic accessory proteins to clathrin-coated pits at the cell surface (e.g., Dab2, Epsin, Eps15, and AP180) via an interaction between the AP2 α-subunit appendage (α-ear) and DPF/W or FXDXF peptide sequences (Brett et al., 2002). To confirm the requirement for AP2 in the localization of Numb in PM punctae, we knocked down α-adaptin in SPR-HeLa cells by using siRNA interference (Hinrichsen et al., 2003; Motley et al., 2003). We confirmed efficient depletion of α-adaptin by immunocytochemistry using anti-α-adaptin (Figure 5). Costaining with anti-NbC indicated that when α-adaptin is knocked down, endogenous Numb no longer accumulates in PM punctae (Figure 5), indicating that Numb localization in membrane puncta requires association with α-adaptin and/or other adaptin-bound accessory proteins.
Figure 5.
PM α-adaptin is required for the localization of endogenous Numb in membrane punctae and vesicles. Endogenous Numb localization was assessed in SPR-HeLa cells treated with siRNA targeted against α-adaptin, control siRNA, or untreated as indicated. Cells were colabeled with anti-α-adaptin (red) and anti-NbC (green). The far right panels are the indicated boxed regions which were merged and magnified.
PM Numb Is Mobilized by GPCR Agonists
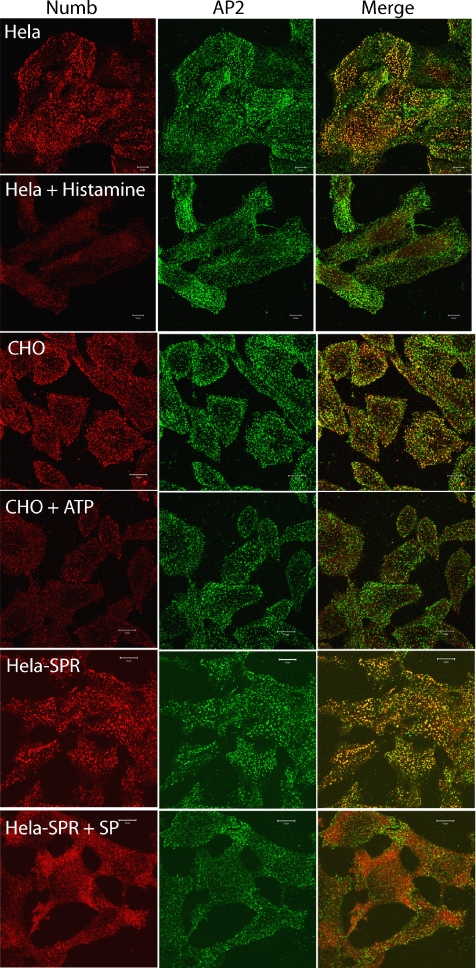
A number of endocytic adaptors are dynamically regulated in response to cell stimuli, either by direct phosphorylation or via their interaction with other proteins or membrane phospholipids. That Numb may be similarly regulated is suggested by the observations that the protein becomes asymmetrically localized in dividing neural progenitor cells in Drosophila and mammals (Rhyu et al., 1994; Knoblich et al., 1995; Spana et al., 1995; Guo et al., 1996; Zhong et al., 1996; Wakamatsu et al., 1999; Dooley et al., 2003) and that its intracellular localization is altered after stimulation with growth factors (Santolini et al., 2000). We did not observe a measurable change in anti-NbC staining at the cortex after epidermal growth factor (EGF) stimulation of HeLa, NIH-3T3, and A431 cells (our unpublished data). In contrast, we found that several GPCR ligands, including histamine (HeLa cells), ATP (CHO cells), SFLLRN peptide (PAR-1–expressing HeLa cells), and substance P (SPR-expressing HeLa cells), stimulated a marked decrease in anti-NbC staining at substratum punctae and cortical vesicles, which correlated with an increase in cytosolic staining (Figure 6; our unpublished data). The observed changes in Numb in response to GPCRs is unlikely to be a secondary consequence of stimulated loss of α-adaptin from the PM, because in general, changes in α-adaptin localization after stimulation were not observed (Figure 6). In some experiments, a slight decrease in AP2 was seen in cells stimulated with substance P, but this was not observed with other GPCRs tested.
Figure 6.
Endogenous Numb is rapidly lost from the PM after stimulation of G protein-coupled receptors. HeLa, CHO, and SPR-HeLa cells were grown on glass coverslips and stimulated for 5–10 min with the indicated GPCR agonist (200 μM histamine, 100 μM ATP, or 100 nM substance P). After fixation and permeablization, the cells were costained with anti-NbC (red) and anti-α adaptin (green). Bars, 10 μm.
GPCR signals are transduced via dissociation of receptor-associated Gα/Gβγ subunits and the subsequent modulation by activated Gα and/or freed Gβγ of downstream effectors such as ion channels, adenylyl cyclases, RGS domain-containing RhoGEFs, and phospholipases-Cβ, -Cε, and -D (for review, see McCudden et al., 2005) Numb mobilization was not observed after stimulation of β-adrenergic receptors with isoproterenol nor did the direct activation of adenylyl cyclase with forskolin affect Numb localization (our unpublished data). These observations suggest that Numb relocalization in response to GPCR activation is not dependent on cAMP-stimulated adenylyl cyclase activation. In addition, treatment of SPR-expressing HeLa cells for 20 h with 100 ng/ml pertussis toxin, which ADP-ribosylates Gi/o-subfamily heterotrimers and thus prevents functional coupling to their respective GPCRs, did not block the effect of subsequent stimulation with substance P (our unpublished data), suggesting that the GPCR signal to Numb is not transduced by Gi/o-coupled signaling pathways.
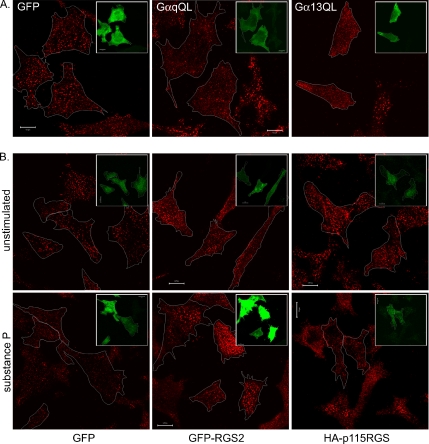
The substance P receptor is reported to be coupled to the activation of Gαq/11 (Kwatra et al., 1993; MacDonald et al., 1996) and Gα12/13 subunits (Barr et al., 1997). Similarly, other GPCRs shown in Figure 6 (ATP receptor, histamine receptor, or PAR1) have also been reported to signal through Gαq (Tilly et al., 1990; Raymond et al., 1991; Iredale and Hill, 1993; Strassheim and Williams, 2000). To address the involvement of Gαq and/or Gα12/13 in GPCR-stimulated Numb mobilization, SPR-expressing HeLa cells were transfected with constitutively active Gα subunits Gαq(Q209L) or Gα13(Q226L), fixed 4 h later, and costained with either anti-Gq or anti-G13 to identify overexpressing cells, and anti-NbC (Figure 7A). The amount of anti-NbC staining associated with basal membrane punctae was visually assessed by comparison of transfected cells with untransfected cells for each condition. The proportion of transfected cells showing a loss of Numb in membrane punctae was calculated as a percentage of total transfected cells counted. Expression of either GαqQL or G13αQL correlated with decreased anti-NbC staining at the PM in SPR cells: 69% (90 cells) and 87% (88 cells) of transfected cells, respectively, exhibited decreased staining, compared with 15% (107 cells) in GFP-transfected cells. Therefore, both Gαq and Gα13 are capable of activating the downstream pathways involved in Numb mobilization from the membrane. To confirm that these pathways mediate substance P-stimulated Numb mobilization, we used Gα subfamily-selective RGS proteins that bind to activated (GTP-loaded) Gα subunits and accelerate their inactivation via GTP hydrolysis (Hains et al., 2004). SPR cells were thus transfected with either GFP-RGS2, a potent GTPase-accelerating protein for activated Gαq (Heximer et al., 1997), or hemagglutinin (HA)-epitope-tagged p115RGS (the N-terminal RGS domain of p115 RhoGEF), which inactivates GTP-bound Gα12/13 subunits (Kozasa et al., 1998). Cells transfected with GFP alone were used as a control. After 24 h, cells were stimulated with substance P for 5 min, fixed, and stained with anti-NbC. Of the cells expressing GFP alone, 93% (total of 76 cells) of cells had decreased Numb localization at the basal membrane after substance P-stimulation. Of the cells expressing HA-p115RGS protein, 74% (48 cells) exhibited a substance P-stimulated loss of Numb. In contrast, expression of GFP-RGS2 inhibited Numb mobilization, because only 31% of cells (68 cells) exhibited decreased anti-NbC staining of the basal membrane after stimulation with substance P (Figure 7B). Thus, although activated Gαq and activated Gα13 are both capable of stimulating Numb movement, only the Gαq-mediated pathway is apparently involved downstream of the activated substance P receptor.
Figure 7.
GPCR-stimulated Numb mobilization is mediated by Gαq and Gα12/13. (A) SPR-HeLa cells transfected with either GFP, constitutively active forms of Gαq (GαqQL) or Gα13 (Gα13QL) were fixed and stained with anti-NbC (red). Transfected cells were identified using either anti-Gαq or anti-Gα13 and are shown in the insert of each image. Each transfected cell is outlined in the anti-NbC image. (B) SPR-HeLa cells transfected with GFP, GFP-RGS2, or HA-p115RGS were stimulated for 5 min with substance P, fixed, and stained with anti-NbC. Transfected cells are indicated as described in A and were identified by GFP fluorescence or labeling with anti-HA. Bars, 10 μm.
PKC Activation Stimulates PM Numb Mobilization
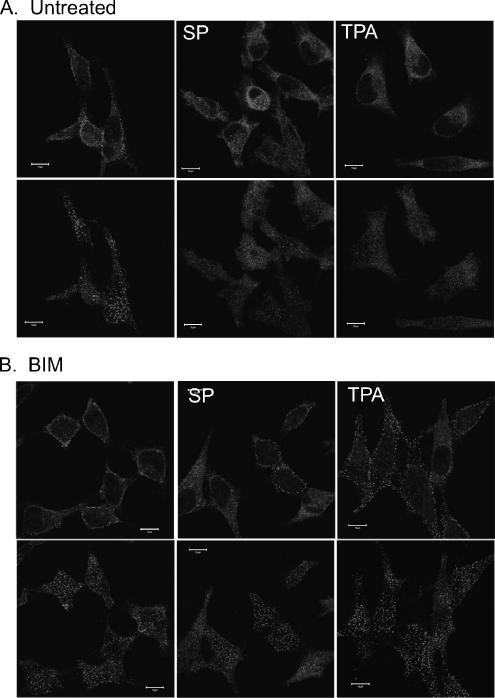
Activated Gαq and Gα12/13 are reported to stimulate PLCβ and PLCε activity, respectively (Taylor et al., 1991; Wu et al., 1992; Lopez et al., 2001; Harden and Sondek, 2006), leading to phosphatidylinositol bisphosphate (PIP2) hydrolysis, generation of diacylglycerol and 1,4,5 inositol trisphosphate second messengers, and the resultant downstream activation of PKC. Therefore, to assess the role of PKC in stimulating the mobilization of Numb, we treated HeLa cells with the phorbol ester TPA, an activator of PKC (Figure 8A). Stimulation of HeLa cells with 0.1 μM TPA caused a marked loss of Numb from the basal PM within 1 min of addition. However, TPA treatment did not decrease anti-α-adaptin staining at the PM. This effect was inhibited by pretreatment of cells for 30 min with the PKC inhibitor bisindolylmaleimide 1 (BIM; Figure 8B). In addition, pretreatment of cells with BIM also partially inhibited the loss of endogenous Numb from the PM after stimulation with substance P (Figure 8B) and histamine (our unpublished data). Therefore, the rapid mobilization of PM and vesicle-associated Numb seen upon GPCR activation seems to be mediated via PLC-dependent activation of PKC.
Figure 8.
Substance P- and TPA-stimulated loss of Numb from the PM is mediated by activation of PKC. HeLa cells grown on glass coverslips were pretreated for 1 h with (B) or without (A) the PKC inhibitor BIM (3 μM). Cells were then stimulated with either 100 nM substance P (SP) or 0.5 μM TPA for 5 min and fixed and stained with anti-NbC. The images shown were taken at the basal, substratum-associated membrane (bottom) and a midsection through the same cells (top). Bars, 10 μm.
Numb is similar to a number of other endocytic adaptors and accessory proteins (e.g., Dab2 and Epsin) that have lipid-binding domains, AP2- and EH domain-binding motifs. To determine whether the effects of GPCRs and PKC are specific to Numb, or alternatively, are a general property of proteins associated with clathrin-coated pits, we examined the membrane distribution of Epsin-1, Dab2, and also Eps15, an AP2-associated, clathrin-coated pit-associated protein, after cell stimulation. We did not observe any marked changes in the membrane association of these proteins after stimulation of SPR-HeLa cells with either 100 nM substance P or 0.5 μM TPA, suggesting that the effect of these stimuli on Numb localization is specific (Supplemental Figure 1).
PKC Activation Stimulates the Phosphorylation of Numb and an Increase in Its Triton X-100 Solubility
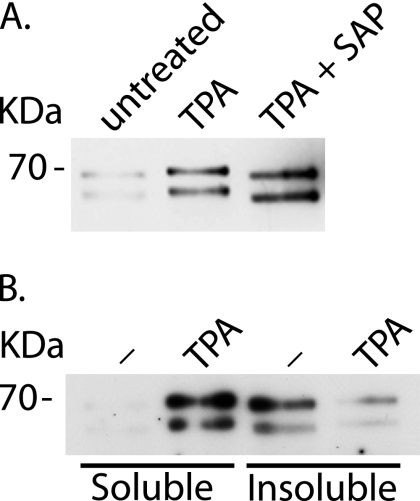
Coincident with the TPA-stimulated loss of Numb from the PM, there was also an upward mobility shift of all isoforms of Numb as resolved by SDS-PAGE (Figure 9A). This mobility shift could be collapsed by treatment of the immunoprecipitates with SAP, suggesting that TPA stimulates phosphorylation of Numb. It was also apparent that stimulation with TPA was associated with an increase in the amount of Numb in the soluble cell lysates (Figure 9A). We examined this further by carrying out a crude separation of the 0.5% Triton X-100–soluble and –insoluble fractions of SPR cells before and after stimulation with TPA and subjecting these lysates to immunoprecipitation with anti-NbC (Figure 9B). After stimulation with TPA for 10 min, there was a marked increase in Numb in the soluble fraction. Concomitant with this was a decrease in the amount of Numb immunoprecipitated from the insoluble fraction.
Figure 9.
PKC activation is associated with phosphorylation of endogenous Numb and an increase in Numbs Triton X-100 solubility. SPR-HeLa cells were stimulated with 0.5 μM TPA for 5 min followed by lysis and immunoprecipitation of Numb by using anti-NbA as described in Materials and Methods. Numb immunoprecipitates were further treated with or without SAP (A) or fractionated into Triton X-100–soluble and –insoluble pools (B). The soluble fraction represents half of the cellular input relative to the insoluble fraction.
Identification of Two Regions within Numb That Are Independently Responsive to GPCR-mediated Lipid Hydrolysis and PKC Activation
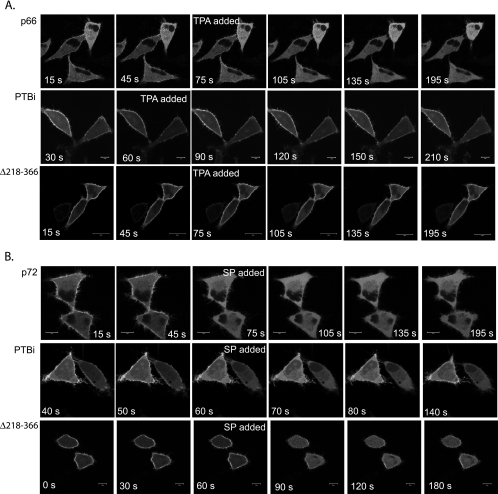
To determine which regions of Numb mediate the response to PKC activation, we carried out confocal imaging of live cells transiently expressing GFP-tagged Numb mutants and compared their response to TPA and substance P with wild-type GFP-Nbp66/p72 (Table 1; Figure 10). After addition of either substance P or TPA, we observed a rapid decrease in the GFP-Nbp66 and GFP-Nbp72 fluorescence associated with substratum punctae and cortical vesicles, indicating that the overexpressed GFP-tagged protein responded in the same way as the endogenous protein. Both GFP-Nbp66 and GFP-Nbp72 isoforms behaved similarly. We chose to follow the time course of changes in GFP-Nb bound to cortical membrane-associated vesicles in the midsection of the cell to allow for better visualization of changes in the intracellular pool of Numb. Optical slices were taken from the lower midsection of the cell at 15-s intervals. Within 15 s of addition of substance P (approximately 100 nM), a rapid loss of GFP-Numb from the PM and membrane-associated vesicles was observed (Figure 10B, top). Coincident with this was an increase in the intracellular fluorescence. In some cells, GFP-Numb fluorescence began to return to the PM within 3–5 min, although complete recovery was not observed in the time frame of these experiments (up to 10 min). The initial response of GFP-Numb to TPA was less rapid, occurring within 30–45 s, but there was a similar increase in cytosolic GFP-Numb (Figure 10A, top). However, in contrast to the response of GFP-Numb to stimulation with substance P, GFP-Numb did not return to the membrane over the 10 min of observation after stimulation with TPA. Preincubation with the protein kinase inhibitor BIM blocked the effects of TPA, confirming the involvement of PKC (our unpublished data). We then tested the response of Numb mutants (ΔC, ΔDPF1ΔC, and ΔDPF1/2) to substance P and TPA stimulation. As described above, these mutants lack the regions important for α-adaptin binding and the localization to punctae and vesicles (Figure 4). Despite their decreased ability to target to AP2-positive pits, these mutants were still mobilized in response to substance P and TPA (Table 1). These results suggested another region(s) was involved in mediating the responsiveness of Numb to these stimuli.
Figure 10.
TPA stimulates a sustained loss of GFP-Nbp66 from the PM that is prevented by deletion of a serine-rich region (GFP-Δ218-366). SPR-HeLa cells were transfected with wild-type GFP-Numb (p66 or p72), GFP-PTBi, or GFP-Δ218-366; trypsinized 4 h after transfection, and seeded onto glass coverslips. Twenty hours later, live cells were imaged by confocal microscopy. TPA (0.5 μM; A) or substance P (100 nM; B) was added where indicated, and images were acquired at 15-s intervals in the continued presence of the stimulus. Animation may be viewed in Supplemental VideoNbp66TPA.mov, VideoPTBiTPA.mov, Videodelta218–366TPA.mov, VideoNbp72SP.mov, VideoPTBiSP.mov, and Videodelta218-366SP.mov.
To delineate the stimulus-responsive regions, we tested a panel of Numb mutants, including the isolated N-terminal PTB domain, and examined the effect of TPA or GPCR stimulation on fusion protein localization. After addition of substance P, the membrane-bound GFP-PTBi protein was rapidly relocalized from the membrane into the cytosol (Figure 10B, middle). In contrast to the full-length protein, GFP-PTBi loss from the membrane was transient, beginning to reappear at the PM within 1–2 min after addition of substance P. TPA stimulation had no effect on GFP-PTBi localization (Figure 10A, middle). These results suggested that the PTBi domain, although not sensitive to activation of PKC, is responsive to other signals activated after stimulation with substance P, such as lipid hydrolysis. To elucidate the PKC-sensitive region in Numb, we examined the responsiveness of GFP-PRRo fusion, which contains the entire carboxy-terminal domain of Numb. When expressed in HeLa cells, this protein exhibits very limited association with the PM but is occasionally associated with vesicular structures close to the membrane (our unpublished data). However, after addition of TPA, the fraction of GFP-PRRo associated with the membrane was lost, indicating that the GFP-PRRo protein contains a PKC-responsive region. Because stimulation with either substance P or TPA is associated with apparent phosphorylation of Numb, coincident with its loss from the PM, we examined the Numb sequence for putative PKC phosphorylation sites. NetPhos 2.0 predicts 40 serine/threonine phosphorylation sites within the primary sequence of Numb p66 isoform. A cluster of 12 serine- and five threonine-putative phosphorylation sites is situated within the region bounded by the C terminus of the PTB domain and the first DPF motif, four of which are identified by Prosite as PKC phosphorylation sites. Included within this region is a previously identified Ca2+/calmodulin-kinase site (Tokumitsu et al., 2005). Therefore, we tested whether deletion of this serine-rich region would abrogate the effect of TPA on Numb membrane association. We examined the effect of TPA stimulation on the localization of a GFP-tagged Numb mutant lacking amino acids 218-366 (Δ218-366). In most unstimulated cells, this mutant exhibited greater association with basal punctae (15.1% of basal membrane area in 31 cells; Figure 5 and Table 1). GFP-Δ218-366 exhibited increased cortical membrane binding and decreased accumulation in the cytosol compared with wild-type Numb. In contrast with wild-type Numb, the effect of stimulation with TPA was severely abrogated, with only a small change membrane fluorescence observed (Figure 10A, bottom). Because the isolated PTB domain mediates a response to substance P stimulation via a PKC-independent mechanism, we tested whether GFP-Δ218-366 may still be responsive to substance P. We observed that GFP-Δ218-366 was still rapidly relocalized in response to substance P. However, in contrast to the wild-type GFP-Numb, GFP-Δ218-366 rapidly reassociated with the PM, with kinetics similar to that of the isolated GFP-PTBi.
DISCUSSION
This work defines the regions of mammalian Numb required for its localization to the cell cortex and demonstrates that extrinsic signals regulate this process. We show that both the PTB domain and the α-adaptin–binding motifs are required for localization of Numb to cortical membrane patches and vesicles. This cortical membrane pool of Numb is dynamically regulated by GPCR signaling through activated Gαq and Gα13 and by direct activation of PKC. Our results indicate that PKC-dependent phosphorylation events regulate the movement of Numb proteins between the cell cortex and the cytosol in mammalian cells.
Previous studies have shown that the p66 and p72 isoforms of mammalian Numb are predominantly localized to the cortical membrane and intracellular vesicles. These two isoforms contain an alternatively spliced exon within the PTB domain (PTBi) possessing three lysine residues that, in addition to the five lysine residues located on either side, form a basic patch that could mediate electrostatic interactions with acidic membrane phospholipids. Indeed, we have previously shown that the PTBi domain preferentially binds to liposomes containing PI(4)P and PI(4,5)P2 (Dho et al., 1999). Lipid-binding regions are a common feature among other endocytic adaptors, including Dab1 and -2, AP180, epsin, and ARH (Mishra et al., 2002; Aguilar et al., 2003; Yun et al., 2003; Balla, 2005), and our analysis of a series of Numb localization mutants indicates that membrane localization of Numb p66 and p72 requires the PTBi domain.
Whereas the PTBi domain alone is sufficient for association with the PM, further compartmentalization of Numb into membrane patches and vesicles requires the adaptin-binding DPF motifs. This is mediated primarily by the DPF2 motif, because loss of the more amino-terminal DPF1 motif alone had no effect on membrane localization to AP2-positive puncta. Indeed, previous studies suggest that DPF1 does not bind to α-adaptin (Santolini et al., 2000). However, our studies indicate that DPF1-mediated interactions likely facilitate and/or stabilize Numb localization in cortical patches, because deletion of both DPF1 and DPF2 (either alone or in the context of a C-terminal truncation) has a more dramatic effect on the localization of Numb compared with loss of DPF2 alone. We have also shown that these motifs, which mediate the association of Numb with the AP2 complex, on their own are not sufficient for optimal localization of Numb to the cell cortex. Therefore, we propose that the PTBi domain promotes Numb association with membrane phospholipids and that additional protein–protein interactions through α-adaptin–binding motifs stabilize and promote Numb compartmentalization into membrane patches and vesicles that also contain AP2. Complete loss of Numb localization to membrane puncta occurred only when all sequences downstream of the PTBi domain were removed, suggesting that additional weak protein–protein interactions may further stabilize Numb localization. In support of this idea, several reports have suggested that Numb binds to Src homology (SH) 3 domain-containing proteins through a proline-rich region that contains putative binding sites for SH3 as well as WW domains (Verdi et al., 1996; Tang et al., 2005). A similar cooperative model of PM recruitment has recently been described, in which regulated protein–lipid and protein–protein interactions target and stabilize AP2 complexes in PM patches that mediate clathrin coat assembly (Honing et al., 2005).
The interaction between Numb and α-adaptin is conserved in Drosophila, and epistasis experiments have shown that α-adaptin acts downstream of dNumb in asymmetric cell divisions of the peripheral nervous system. Although Numb is not required for the cortical localization of α-adaptin in sensory organ precursor (SOP) cells undergoing asymmetric cell division, its polarized distribution is Numb dependent (Berdnik et al., 2002). In our analysis, Numb did not seem to play a role in localization of AP2 to the cortex, because expression of Numb localization mutants had no effect on the PM staining of α-adaptin. In addition, analysis of HeLa cells made Numb-deficient by RNA interference (RNAi), showed no change in α-adaptin localization (our unpublished data), as would be expected if Numb was important in the subcellular distribution of AP2. In contrast, in HeLa cells made AP2 deficient by α-adaptin RNAi, we observed a marked decrease in the association of Numb at the cortical membrane. Therefore, in nonpolarized mammalian cells, Numb is not required for localization of AP2 to the PM or cortical patches or vesicles. Our studies, however, cannot preclude the possibility that, in polarized or asymmetrically dividing mammalian cells, Numb may influence the distribution of AP2.
Many studies have shown that in Drosophila, Numb localization is dynamically regulated. In developing neuroblasts, dNumb is evenly distributed along the cell membrane until late prophase, when it forms a basal crescent overlying one of the centrosomes. dNumb remains asymmetric through mitosis when it is segregated into one of the daughter cells, after which it becomes homogeneously distributed again (Knoblich et al., 1995). Recent studies by Mayer et al. (2005) have shown that Drosophila Numb is rapidly exchanged between a cytoplasmic pool and the cell cortex, and during asymmetric cell division it is preferentially recruited to one side of the cell cortex. The formation of the Numb crescent is dependent on the activity of the PAR3/PAR6/aPKC polarity complex, which localizes to the opposite pole (Roegiers et al., 2001). In the neuroblast, this seems to be mediated in part through the aPKC-dependent phosphorylation of the cytoskeletal protein Lgl, which prevents localization of determinants such as Numb to the apical cell cortex (Betschinger et al., 2003). However, in SOP cells, Numb localization is independent of Lgl, suggesting a distinct mechanism of regulation by the PAR3/PAR6/aPKC polarity complex (Justice et al., 2003; Langevin et al., 2005).
Similarly, mammalian Numb proteins have been reported to localize asymmetrically in mitotic neuroepithelial cells in the chicken (Wakamatsu et al., 1999) and dividing mouse neural progenitor cells (Zhong et al., 1996). Polarized distribution of Numb not restricted to mitotic cells, because asymmetric localization of Numb is also observed in postmitotic retinal cells (Dooley et al., 2003), and as shown in our study, polarized MDCK cells. Although the factors that regulate asymmetric distribution of Numb in mammals have not been defined, Klezovitch et al. (2004) observed that in cortical progenitors, loss of the vertebrate homologue of Lgl results in a loss of asymmetric distribution of Numb. This finding suggests that the machinery responsible for directing asymmetric distribution of Numb may be conserved.
The cortical membrane pool of Numb is redistributed to the cytosol in response to activation of Gαq/Gα13-coupled GPCRs, and this cytosolic pool is subsequently recruited back to the cortical membrane. This effect is mediated by two distinct signaling events, which independently contribute to the loss of Numb association with the PM and have distinct effects on the rapidity with which it is subsequently recruited from the cytosol. First, the PTBi domain mediates a response with a rapid, transient time course, similar to that observed with the isolated PH domain of PLCδ, which is mobilized from the PM in response to hydrolysis of PI(4,5)P2 (Stauffer et al., 1998). Because the Numb PTBi domain binds PIP2, the PTB-dependent dissociation of Numb from the membrane is most likely a consequence of PLC-dependent PIP2 hydrolysis. Replenishment of PIP2 levels through the action of phosphatidylinositol phosphate kinases would allow redistribution of Numb back to the PM.
The region between amino acids 218 and 366 mediates the response of Numb to both GPCR stimulation and direct activation of PKC. This region encompasses at least 12 serine residues that are putative PKC phosphorylation sites, and it is required for the TPA-stimulated redistribution of Numb. Deletion of this serine-rich region not only blunts the response of Numb to TPA treatment but also enhances membrane association under basal conditions. This observation suggests that this region is not required for membrane binding but rather it plays a role in the relocalization of Numb in response to cellular signaling. In addition, the presence of this region slows the kinetics of redistribution of Numb from the cytosol to the membrane. Therefore, this region likely contains phosphorylation sites that could promote the binding of Numb to a cytosolic protein that facilitates its loss from the membrane or regulates membrane reassociation. In support of this model, we have used mass spectral analyses to identify nine sites of Numb phosphorylation, four of which lie within this region (Smith, Lau, Rahmani, Dho, Brothers, She, Berry, Bonneil, Thibault, Schweisguth, Le Borgne, and McGlade, unpublished data). In addition, a Ca2+ and calmodulin-dependent protein kinase 1 phosphorylation site was recently identified at amino acid 276, within the amino acid 218–366 region, and reported to mediate binding to 14-3-3 proteins in vitro (Tokumitsu et al., 2005), although the consequence of phosphorylation of this site on Numb localization was not evaluated. Together, these data suggest that, in mammalian cells, phosphorylation of Numb plays a role in regulating its distribution between a cytosolic pool and the membrane cortex. Phosphorylation may be a mechanism, analogous to that described for Lgl, by which Numb is asymmetrically restricted to specific regions of a cell, for example, the basalateral membrane of MDCK cells.
In addition to phosphorylation, a change in the solubility of Numb in Triton X-100 was also associated with TPA stimulation. A number of proteins bound to clathrin-coated pits, including Dab2, CALM, Epsin, and α-adaptin, are resistant to extraction with nonionic detergents such as Triton X-100 and saponin (Woodward and Roth, 1978; Tebar et al., 1999; Morris and Cooper, 2001). The observation that stimulation increases the solubility of Numb in detergent supports the view that Numb dissociates from a multimolecular complex associated with clathrin-coated pits.
Mammalian Numb functions as an endocytic adaptor protein and has been demonstrated to regulate the endocytosis and trafficking of surface molecules such as EGF receptor, Tac, L1, and Notch (Santolini et al., 2000; Nishimura et al., 2003; Smith et al., 2004; McGill and McGlade, unpublished data). Although we have shown that activation of GPCRs leads to changes in the subcellular distribution of Numb, we did not find any evidence that Numb was cotrafficked with these receptors, nor did we observe any effect of Numb expression or knockdown on the internalization or recycling of the substance P receptor (Dho, unpublished data). Therefore, we propose that the effects of GPCR activation on Numb localization are not secondary to its endocytic function but rather reflect a direct role for signaling events in controlling the distribution of Numb proteins. Heterotrimeric G proteins have emerged as important mediators of polarity and asymmetric cell divisions in Drosophila (Hampoelz and Knoblich, 2004; McCudden et al., 2005). In particular, overexpression of the activated form of the trimeric G protein Go (GoGTP) in Drosophila mitotic SOPs results in severe defects in Numb localization, including near-ubiquitous cortical Numb staining or loss from the PM (Katanaev and Tomlinson, 2006). Our results raise the possibility that activation of heterotrimeric G proteins is a conserved mechanism regulating the subcellular distribution of Numb. As such, the localization of Numb and, by extension its function, may be under the control of both extrinsic and intrinsic signaling cues, which regulate cell polarity.
Supplementary Material
ACKNOWLEDGMENTS
We thank Mike Woodside and Paul Paroutis (Sickkids Imaging Facility) for help and advice and Donna Berry for help generating cDNA constructs. This work was supported by the National Cancer Institute of Canada with funds from the Canadian Cancer Society (to C.J.M.). D.P.S. was supported by National Institutes of Health Grant R01 GM-074268.
Footnotes

 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05.02-0097) on July 12, 2006.
REFERENCES
- Aguilar R. C., Watson H. A., Wendland B. The yeast Epsin Ent1 is recruited to membranes through multiple independent interactions. J. Biol. Chem. 2003;278:10737–10743. doi: 10.1074/jbc.M211622200. [DOI] [PubMed] [Google Scholar]
- Balla T. Inositol-lipid binding motifs: signal integrators through protein-lipid and protein-protein interactions. J. Cell Sci. 2005;118:2093–2104. doi: 10.1242/jcs.02387. [DOI] [PubMed] [Google Scholar]
- Barr A. J., Brass L. F., Manning D. R. Reconstitution of receptors and GTP-binding regulatory proteins (G proteins) in Sf9 cells. A direct evaluation of selectivity in receptor G protein coupling. J. Biol. Chem. 1997;272:2223–2229. doi: 10.1074/jbc.272.4.2223. [DOI] [PubMed] [Google Scholar]
- Berdnik D., Torok T., Gonzalez-Gaitan M., Knoblich J. A. The endocytic protein alpha-adaptin is required for numb-mediated asymmetric cell division in Drosophila. Dev. Cell. 2002;3:221–231. doi: 10.1016/s1534-5807(02)00215-0. [DOI] [PubMed] [Google Scholar]
- Betschinger J., Mechtler K., Knoblich J. A. The Par complex directs asymmetric cell division by phosphorylating the cytoskeletal protein Lgl. Nature. 2003;422:326–330. doi: 10.1038/nature01486. [DOI] [PubMed] [Google Scholar]
- Brett T. J., Traub L. M., Fremont D. H. Accessory protein recruitment motifs in clathrin-mediated endocytosis. Structure. 2002;10:797–809. doi: 10.1016/s0969-2126(02)00784-0. [DOI] [PubMed] [Google Scholar]
- Brewster R., Bodmer R. Origin and specification of type II sensory neurons in Drosophila. Development. 1995;121:2923–2936. doi: 10.1242/dev.121.9.2923. [DOI] [PubMed] [Google Scholar]
- Cayouette M., Raff M. The orientation of cell division influences cell-fate choice in the developing mammalian retina. Development. 2003;130:2329–2339. doi: 10.1242/dev.00446. [DOI] [PubMed] [Google Scholar]
- Dho S. E., French M. B., Woods S. A., McGlade C. J. Characterization of four mammalian numb protein isoforms. Identification of cytoplasmic and membrane-associated variants of the phosphotyrosine binding domain. J. Biol. Chem. 1999;274:33097–33104. doi: 10.1074/jbc.274.46.33097. [DOI] [PubMed] [Google Scholar]
- Dooley C. M., James J., Jane McGlade C., Ahmad I. Involvement of numb in vertebrate retinal development: evidence for multiple roles of numb in neural differentiation and maturation. J. Neurobiol. 2003;54:313–325. doi: 10.1002/neu.10176. [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S., Hall A. Integrin-mediated activation of Cdc42 controls cell polarity in migrating astrocytes through PKCzeta. Cell. 2001;106:489–498. doi: 10.1016/s0092-8674(01)00471-8. [DOI] [PubMed] [Google Scholar]
- Guo M., Jan L. Y., Jan Y. N. Control of daughter cell fates during asymmetric division: interaction of Numb and Notch. Neuron. 1996;17:27–41. doi: 10.1016/s0896-6273(00)80278-0. [DOI] [PubMed] [Google Scholar]
- Hains M. D., Siderovski D. P., Harden T. K. Application of RGS box proteins to evaluate G-protein selectivity in receptor-promoted signaling. Methods Enzymol. 2004;389:71–88. doi: 10.1016/S0076-6879(04)89005-0. [DOI] [PubMed] [Google Scholar]
- Hampoelz B., Knoblich J. A. Heterotrimeric G proteins: new tricks for an old dog. Cell. 2004;119:453–456. doi: 10.1016/j.cell.2004.10.025. [DOI] [PubMed] [Google Scholar]
- Harden T. K., Sondek J. Regulation of phospholipase C isozymes by ras superfamily GTPases. Annu. Rev. Pharmacol. Toxicol. 2006;46:355–379. doi: 10.1146/annurev.pharmtox.46.120604.141223. [DOI] [PubMed] [Google Scholar]
- Heximer S. P., Watson N., Linder M. E., Blumer K. J., Hepler J. R. RGS2/G0S8 is a selective inhibitor of Gqalpha function. Proc. Natl. Acad. Sci. USA. 1997;94:14389–14393. doi: 10.1073/pnas.94.26.14389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinrichsen L., Harborth J., Andrees L., Weber K., Ungewickell E. J. Effect of clathrin heavy chain- and alpha-adaptin-specific small inhibitory RNAs on endocytic accessory proteins and receptor trafficking in HeLa cells. J. Biol. Chem. 2003;278:45160–45170. doi: 10.1074/jbc.M307290200. [DOI] [PubMed] [Google Scholar]
- Honing S., Ricotta D., Krauss M., Spate K., Spolaore B., Motley A., Robinson M., Robinson C., Haucke V., Owen D. J. Phosphatidylinositol-(4,5)-bisphosphate regulates sorting signal recognition by the clathrin-associated adaptor complex AP2. Mol. Cell. 2005;18:519–531. doi: 10.1016/j.molcel.2005.04.019. [DOI] [PubMed] [Google Scholar]
- Iredale P. A., Hill S. J. Increases in intracellular calcium via activation of an endogenous P2-purinoceptor in cultured CHO-K1 cells. Br. J. Pharmacol. 1993;110:1305–1310. doi: 10.1111/j.1476-5381.1993.tb13960.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan Y. N., Jan L. Y. Asymmetric cell division. Nature. 1998;392:775–778. doi: 10.1038/33854. [DOI] [PubMed] [Google Scholar]
- Justice N., Roegiers F., Jan L. Y., Jan Y. N. Lethal giant larvae acts together with numb in notch inhibition and cell fate specification in the Drosophila adult sensory organ precursor lineage. Curr. Biol. 2003;13:778–783. doi: 10.1016/s0960-9822(03)00288-4. [DOI] [PubMed] [Google Scholar]
- Katanaev V. L., Tomlinson A. Dual roles for the trimeric G protein Go in asymmetric cell division in Drosophila. Proc. Natl. Acad. Sci. USA. 2006;103:6524–6529. doi: 10.1073/pnas.0601853103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klezovitch O., Fernandez T. E., Tapscott S. J., Vasioukhin V. Loss of cell polarity causes severe brain dysplasia in Lgl1 knockout mice. Genes Dev. 2004;18:559–571. doi: 10.1101/gad.1178004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblich J. A., Jan L. Y., Jan Y. N. Asymmetric segregation of Numb and Prospero during cell division. Nature. 1995;377:624–627. doi: 10.1038/377624a0. [DOI] [PubMed] [Google Scholar]
- Knoblich J. A., Jan L. Y., Jan Y. N. The N terminus of the Drosophila Numb protein directs membrane association and actin-dependent asymmetric localization. Proc. Natl. Acad. Sci. USA. 1997;94:13005–13010. doi: 10.1073/pnas.94.24.13005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozasa T., Jiang X., Hart M. J., Sternweis P. M., Singer W. D., Gilman A. G., Bollag G., Sternweis P. C. p115 RhoGEF, a GTPase activating protein for Galpha12 and Galpha13. Science. 1998;280:2109–2111. doi: 10.1126/science.280.5372.2109. [DOI] [PubMed] [Google Scholar]
- Kwatra M. M., Schwinn D. A., Schreurs J., Blank J. L., Kim C. M., Benovic J. L., Krause J. E., Caron M. G., Lefkowitz R. J. The substance P receptor, which couples to Gq/11, is a substrate of beta-adrenergic receptor kinase 1 and 2. J. Biol. Chem. 1993;268:9161–9164. [PubMed] [Google Scholar]
- Langevin J., Le Borgne R., Rosenfeld F., Gho M., Schweisguth F., Bellaiche Y. Lethal giant larvae controls the localization of notch-signaling regulators numb, neuralized, and Sanpodo in Drosophila sensory-organ precursor cells. Curr. Biol. 2005;15:955–962. doi: 10.1016/j.cub.2005.04.054. [DOI] [PubMed] [Google Scholar]
- Lopez I., Mak E. C., Ding J., Hamm H. E., Lomasney J. W. A novel bifunctional phospholipase c that is regulated by Galpha 12 and stimulates the Ras/mitogen-activated protein kinase pathway. J. Biol. Chem. 2001;276:2758–2765. doi: 10.1074/jbc.M008119200. [DOI] [PubMed] [Google Scholar]
- Lu B., Rothenberg M., Jan L. Y., Jan Y. N. Partner of Numb colocalizes with Numb during mitosis and directs Numb asymmetric localization in Drosophila neural and muscle progenitors. Cell. 1998;95:225–235. doi: 10.1016/s0092-8674(00)81753-5. [DOI] [PubMed] [Google Scholar]
- MacDonald D., Silberman S. C., Lowe J. A., 3rd, Drozda S. E., Leeman S. E., Boyd N. D. Photoaffinity labeling of the human substance P (neurokinin-1) receptor with [3H2]azido-CP-96,345, a photoreactive derivative of a nonpeptide antagonist. Mol. Pharmacol. 1996;49:808–813. [PubMed] [Google Scholar]
- Mayer B., Emery G., Berdnik D., Wirtz-Peitz F., Knoblich J. A. Quantitative analysis of protein dynamics during asymmetric cell division. Curr. Biol. 2005;15:1847–1854. doi: 10.1016/j.cub.2005.08.067. [DOI] [PubMed] [Google Scholar]
- McCudden C. R., Hains M. D., Kimple R. J., Siderovski D. P., Willard F. S. G-protein signaling: back to the future. Cell Mol. Life Sci. 2005;62:551–577. doi: 10.1007/s00018-004-4462-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra S. K., Watkins S. C., Traub L. M. The autosomal recessive hypercholesterolemia (ARH) protein interfaces directly with the clathrin-coat machinery. Proc. Natl. Acad. Sci. USA. 2002;99:16099–16104. doi: 10.1073/pnas.252630799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris S. M., Cooper J. A. Disabled-2 colocalizes with the LDLR in clathrin-coated pits and interacts with AP-2. Traffic. 2001;2:111–123. doi: 10.1034/j.1600-0854.2001.020206.x. [DOI] [PubMed] [Google Scholar]
- Motley A., Bright N. A., Seaman M. N., Robinson M. S. Clathrin-mediated endocytosis in AP-2-depleted cells. J. Cell Biol. 2003;162:909–918. doi: 10.1083/jcb.200305145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura T., Fukata Y., Kato K., Yamaguchi T., Matsuura Y., Kamiguchi H., Kaibuchi K. CRMP-2 regulates polarized Numb-mediated endocytosis for axon growth. Nat. Cell Biol. 2003;5:819–826. doi: 10.1038/ncb1039. [DOI] [PubMed] [Google Scholar]
- Ohno S. Intercellular junctions and cellular polarity: the PAR-aPKC complex, a conserved core cassette playing fundamental roles in cell polarity. Curr. Opin. Cell Biol. 2001;13:641–648. doi: 10.1016/s0955-0674(00)00264-7. [DOI] [PubMed] [Google Scholar]
- Qin H., Percival-Smith A., Li C., Jia C. Y., Gloor G., Li S. S. A novel transmembrane protein recruits numb to the plasma membrane during asymmetric cell division. J. Biol. Chem. 2004;279:11304–11312. doi: 10.1074/jbc.M311733200. [DOI] [PubMed] [Google Scholar]
- Raymond J. R., Albers F. J., Middleton J. P., Lefkowitz R. J., Caron M. G., Obeid L. M., Dennis V. W. 5-HT1A and histamine H1 receptors in HeLa cells stimulate phosphoinositide hydrolysis and phosphate uptake via distinct G protein pools. J. Biol. Chem. 1991;266:372–379. [PubMed] [Google Scholar]
- Rhyu M. S., Jan L. Y., Jan Y. N. Asymmetric distribution of numb protein during division of the sensory organ precursor cell confers distinct fates to daughter cells. Cell. 1994;76:477–491. doi: 10.1016/0092-8674(94)90112-0. [DOI] [PubMed] [Google Scholar]
- Roegiers F., Younger-Shepherd S., Jan L. Y., Jan Y. N. Bazooka is required for localization of determinants and controlling proliferation in the sensory organ precursor cell lineage in Drosophila. Proc. Natl. Acad. Sci. USA. 2001;98:14469–14474. doi: 10.1073/pnas.261555598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A. A., Lemberg K. E., Chidiac P. Recruitment of RGS2 and RGS4 to the plasma membrane by G proteins and receptors reflects functional interactions. Mol. Pharmacol. 2003;64:587–593. doi: 10.1124/mol.64.3.587. [DOI] [PubMed] [Google Scholar]
- Ruiz Gomez M., Bate M. Segregation of myogenic lineages in Drosophila requires numb. Development. 1997;124:4857–4866. doi: 10.1242/dev.124.23.4857. [DOI] [PubMed] [Google Scholar]
- Salcini A. E., Confalonieri S., Doria M., Santolini E., Tassi E., Minenkova O., Cesareni G., Pelicci P. G., Di Fiore P. P. Binding specificity and in vivo targets of the EH domain, a novel protein-protein interaction module. Genes Dev. 1997;11:2239–2249. doi: 10.1101/gad.11.17.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santolini E., Puri C., Salcini A. E., Gagliani M. C., Pelicci P. G., Tacchetti C., Di Fiore P. P. Numb is an endocytic protein. J. Cell Biol. 2000;151:1345–1352. doi: 10.1083/jcb.151.6.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. A., Dho S. E., Donaldson J., Tepass U., McGlade C. J. The cell fate determinant numb interacts with EHD/Rme-1 family proteins and has a role in endocytic recycling. Mol. Biol. Cell. 2004;15:3698–3708. doi: 10.1091/mbc.E04-01-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spana E. P., Doe C. Q. Numb antagonizes Notch signaling to specify sibling neuron cell fates. Neuron. 1996;17:21–26. doi: 10.1016/s0896-6273(00)80277-9. [DOI] [PubMed] [Google Scholar]
- Spana E. P., Kopczynski C., Goodman C. S., Doe C. Q. Asymmetric localization of numb autonomously determines sibling neuron identity in the Drosophila CNS. Development. 1995;121:3489–3494. doi: 10.1242/dev.121.11.3489. [DOI] [PubMed] [Google Scholar]
- Stauffer T. P., Ahn S., Meyer T. Receptor-induced transient reduction in plasma membrane PtdIns(4,5)P2 concentration monitored in living cells. Curr. Biol. 1998;8:343–346. doi: 10.1016/s0960-9822(98)70135-6. [DOI] [PubMed] [Google Scholar]
- Strassheim D., Williams C. L. P2Y2 purinergic and M3 muscarinic acetylcholine receptors activate different phospholipase C-beta isoforms that are uniquely susceptible to protein kinase C-dependent phosphorylation and inactivation. J. Biol. Chem. 2000;275:39767–39772. doi: 10.1074/jbc.M007775200. [DOI] [PubMed] [Google Scholar]
- Tang H., Rompani S. B., Atkins J. B., Zhou Y., Osterwalder T., Zhong W. Numb proteins specify asymmetric cell fates via an endocytosis- and proteasome-independent pathway. Mol. Cell. Biol. 2005;25:2899–2909. doi: 10.1128/MCB.25.8.2899-2909.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S. J., Chae H. Z., Rhee S. G., Exton J. H. Activation of the beta 1 isozyme of phospholipase C by alpha subunits of the Gq class of G proteins. Nature. 1991;350:516–518. doi: 10.1038/350516a0. [DOI] [PubMed] [Google Scholar]
- Tebar F., Bohlander S. K., Sorkin A. Clathrin assembly lymphoid myeloid leukemia (CALM) protein: localization in endocytic-coated pits, interactions with clathrin, and the impact of overexpression on clathrin-mediated traffic. Mol. Biol. Cell. 1999;10:2687–2702. doi: 10.1091/mbc.10.8.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilly B. C., Tertoolen L. G., Lambrechts A. C., Remorie R., de Laat S. W., Moolenaar W. H. Histamine-H1-receptor-mediated phosphoinositide hydrolysis, Ca2+ signalling and membrane-potential oscillations in human HeLa carcinoma cells. Biochem. J. 1990;266:235–243. doi: 10.1042/bj2660235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokumitsu H., Hatano N., Inuzuka H., Sueyoshi Y., Yokokura S., Ichimura T., Nozaki N., Kobayashi R. Phosphorylation of Numb family proteins. Possible involvement of Ca2+/calmodulin-dependent protein kinases. J. Biol. Chem. 2005;280:35108–35118. doi: 10.1074/jbc.M503912200. [DOI] [PubMed] [Google Scholar]
- Trejo J., Coughlin S. R. The cytoplasmic tails of protease-activated receptor-1 and substance P receptor specify sorting to lysosomes versus recycling. J. Biol. Chem. 1999;274:2216–2224. doi: 10.1074/jbc.274.4.2216. [DOI] [PubMed] [Google Scholar]
- Uemura T., Shepherd S., Ackerman L., Jan L. Y., Jan Y. N. numb, a gene required in determination of cell fate during sensory organ formation in Drosophila embryos. Cell. 1989;58:349–360. doi: 10.1016/0092-8674(89)90849-0. [DOI] [PubMed] [Google Scholar]
- Verdi J. M., Schmandt R., Bashirullah A., Jacob S., Salvino R., Craig C. G., Program A. E., Lipshitz H. D., McGlade C. J. Mammalian NUMB is an evolutionarily conserved signaling adapter protein that specifies cell fate. Curr. Biol. 1996;6:1134–1145. doi: 10.1016/s0960-9822(02)70680-5. [DOI] [PubMed] [Google Scholar]
- Wakamatsu Y., Maynard T. M., Jones S. U., Weston J. A. NUMB localizes in the basal cortex of mitotic avian neuroepithelial cells and modulates neuronal differentiation by binding to NOTCH-1. Neuron. 1999;23:71–81. doi: 10.1016/s0896-6273(00)80754-0. [DOI] [PubMed] [Google Scholar]
- Woodward M. P., Roth T. F. Coated vesicles: characterization, selective dissociation, and reassembly. Proc. Natl. Acad. Sci. USA. 1978;75:4394–4398. doi: 10.1073/pnas.75.9.4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D. Q., Lee C. H., Rhee S. G., Simon M. I. Activation of phospholipase C by the alpha subunits of the Gq and G11 proteins in transfected Cos-7 cells. J. Biol. Chem. 1992;267:1811–1817. [PubMed] [Google Scholar]
- Yun M., Keshvara L., Park C. G., Zhang Y. M., Dickerson J. B., Zheng J., Rock C. O., Curran T., Park H. W. Crystal structures of the Dab homology domains of mouse disabled 1 and 2. J. Biol. Chem. 2003;278:36572–36581. doi: 10.1074/jbc.M304384200. [DOI] [PubMed] [Google Scholar]
- Zhong W., Feder J. N., Jiang M. M., Jan L. Y., Jan Y. N. Asymmetric localization of a mammalian numb homolog during mouse cortical neurogenesis. Neuron. 1996;17:43–53. doi: 10.1016/s0896-6273(00)80279-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.