Abstract
The transformed monocyte/macrophage cell line J774.2 undergoes apoptosis when treated for 48 h with competitive inhibitors of cyclooxygenase (COX) isoenzymes 1 and 2. Many of these nonsteroid antiinflammatory drugs (NSAIDs), but in particular diclofenac, induce during this time period a COX activity that coincides with a robust induction of COX-2 protein. Induction of this activity requires high, apoptosis-inducing concentrations of diclofenac (>100 μM). Prolonged treatment of J774.2 cells with lower doses of diclofenac inhibits COX activity, indicating that diclofenac is a time-dependent, pseudoirreversible inhibitor of COX-2. It is difficult to wash out the inhibition. However, the activity evoked by high concentrations of diclofenac has a profoundly distinct COX active site that allows diclofenac, its inducer, to be washed readily from its active site. The diclofenac-induced activity also has the unusual property of being more sensitive to inhibition by acetaminophen (IC50 = 0.1–1.0 mM) than COX-2 induced with bacterial lipopolysaccharide. Moreover, relative to COX-1 or COX-2, diclofenac-induced enzyme activity shows significantly reduced sensitivity to inhibition by diclofenac or other competitively acting nonsteroid antiinflammatory drugs (NSAIDs) and the enzyme activity is insensitive to aspirin. If the robust induction of COX-2 observed is responsible for diclofenac-induced COX enzyme activity, it is clear that COX-2 can, therefore, exist in two catalytically active states. A luciferase reporter–construct that contains part of the COX-2 structure and binds into the membrane showed that chronic diclofenac treatment of fibroblasts results in marked mobilization of the fusion protein. Such a mobilization could result in enzymatically distinct COX-2 populations in response to chronic diclofenac treatment.
The cloning of cyclooxygenase-2 (COX-2) in 1991 (1–3) explained many long-standing anomalies relating to the previous assumption of a single COX enzyme. However, not all of the problems have been solved by the characterization of two COX enzymes originating from different genes. One such problem is the effect of acetaminophen, which, unlike nonsteroid antiinflammatory drugs (NSAIDs), produces analgesia and antipyresis but has little effect on inflammation. The existence of additional COX enzymes was therefore postulated (4).
Acetaminophen is a weak inhibitor of COX-1 and COX-2, which displays some selectivity toward COX enzymes from different organs. Thus, it is a more potent inhibitor of COX from dog and rabbit brain than that of dog spleen (4). The activity of acetaminophen in vitro also depends on the addition of cofactors. In the presence of glutathione and hydroquinone, acetaminophen inhibited COX activity of bull seminal vesicle microsomes but, in the absence of these cofactors, stimulated that of bovine and ram seminal vesicle microsomes (5). After the recognition of COX-1 and COX-2, by using cultured whole cells, acetaminophen demonstrated a low inhibitory potency for COX enzymes. Only IC30 values could be obtained against COX-1 in bovine aortic endothelial cells and COX-2 in murine J774.2 macrophages stimulated with bacterial lipopolysaccharide (LPS), providing a COX-2/COX-1 ratio of 7.4 (6).
Micromolar (10–100 μM) concentrations of many NSAIDs cause apoptosis of chicken embryo fibroblasts in culture, at the same time inducing COX-2 RNA and protein (7). In the present study, we found that diclofenac and other NSAIDs in murine J774.2 macrophages in culture also induce COX-2. Furthermore, diclofenac concomitantly induces a COX whose activity is revealed when arachidonic acid is added, after the diclofenac inducer has been removed. This activity is more sensitive to inhibition with acetaminophen than COX-2 induced with LPS in the same cells. However, the NSAID-induced enzyme shows decreased sensitivity to inhibition by other NSAIDs compared with LPS-induced enzyme. Other differences in the properties of these two enzyme activities were also found.
MATERIALS AND METHODS
Cell Culture.
Murine monocyte/macrophages (J774.2; The European Collection of Animal Cell Culture, Salisbury, U.K.) were grown to confluence in 96-well culture plates with DMEM supplemented with 10% fetal calf serum and 4 mM l-glutamine.
Measurement of Prostaglandin E2 (PGE2) from Nonapoptotic Cells.
J774.2 macrophages in culture were treated with LPS at 1 μg/ml for 12 h to induce COX-2. Culture medium was then changed, the cells were washed twice in serum-free medium, and acetaminophen (0.002–4 mM) was added for 30 min at 37°C. Arachidonic acid (30 μM) was then added, and the cells were incubated for a further 15 min at 37°C. The medium was removed, and a radioimmunoassay (RIA) (8) was used to measure the formation of PGE2. Antibodies to PGE2 were obtained from Sigma. [3H]PGE2 (specific activity 185 Ci/mM) was obtained from Amersham. In some cases, PGE2 synthesis was normalized for total cellular protein (9). Cells were solubilized in 0.62 N sodium hydroxide for 4 h, after which the solution was neutralized with an equal volume of 1.0 M Tris (pH 4.8). Protein concentration was measured by biuret assay (Bio-Rad) (9).
Induction of Apoptosis and COX-2 Activity.
NSAIDs (500 μM) were administered to J774.2 murine macrophages for 48 h (in tissue culture media containing 10% fetal calf serum) to induce apoptosis. Cell death was measured by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide assay (10) and by trypan blue exclusion. Trypan blue solution [0.2% trypan blue dye/0.06% potassium phosphate (dibasic)/0.8% NaCl] was added for 2 min to the cells adhering to the plate, after which it was gently aspirated and blue cells were counted. Induction of cell death through apoptosis (rather than through necrosis) was determined by visualization of nucleosomal ladders in which DNA was isolated and electrophoresed on agarose gels as described (7).
Induction of COX activity during the apoptotic process was determined by aspirating the medium containing the NSAID inducer of cell death, washing the cells twice in serum-free DMEM, and exposing the cells to 30 μM arachidonic acid for 15 min. PGE2 produced during this incubation was measured by RIA.
Testing of NSAID-Induced COX Activity Toward Inhibition by NSAIDs.
To test whether COX enzyme activity induced by NSAIDs was itself sensitive to NSAID inhibition, J774.2 cells were treated with 500 μM diclofenac to induce the enzyme activity. Cells were then washed twice with serum-free DMEM and exposed for 30 min to various NSAIDs at concentrations ranging from 1 μM to 2 mM. After this NSAID preincubation, arachidonic acid (30 μM) was added to the NSAID-containing medium for 15 min, and PGE2 generated by the apoptotic cells during this time was measured by RIA.
In parallel assays, J774.2 cells were treated with LPS (1 μg/ml) for the last 24 of the 48 h of NSAID incubation used to induce apoptosis and COX activity. In addition to this treatment, J774.2 cells treated with LPS (1 μg/ml) alone for 12 h were used here as a control for inhibition of COX-2.
Immunoblot (Western Blot) Analysis.
LPS- or NSAID-treated J774.2 macrophages cultured in 35-mm dishes were washed with PBS (pH 7.4) and incubated for 10 min with 2–3 ml of extraction buffer [50 mM Tris/10 mM EDTA/1% (vol/vol) Triton X-100/1 mM phenylmethylsulfonyl fluoride/50 μM pepstatin A/0.2 mM leupeptin] while being gently shaken. The cell extract was then boiled for 10 min with gel-loading buffer [50 mM Tris/10% (wt/vol) SDS/10% (vol/vol) glycerol/10% 2-mercaptoethanol/2 mg of bromophenol blue dye] in a ratio of 1:1 (vol/vol). Samples were loaded onto gradient gels (4–12% Tris glycine; Novex, British Biotechnology, Oxford) and separated by electrophoresis. After transfer to nitrocellulose, the blot was probed with a rabbit antibody raised to murine COX-2 (generously provided by Merck Frosst Labs, Pointe Claire, PQ, Canada). The blot was then incubated with a sheep anti-rabbit IgG linked to alkaline phosphatase conjugate, and the blot was developed with 5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium.
COX-2–Luciferase Reporter Construct (pCL-H5).
A reporter protein was constructed to track the subcellular localization of COX-2 in LA-24 cells, which is a subline of murine NIH 3T3 cells. This cell line was chosen because of the difficulty of obtaining stable transfection of J774.2 cells. The amino-terminal 127 aa of chicken COX-2 was appended to the amino terminus of luciferase.
The reporter was constructed with a luciferase cDNA, lacking the peroxisome-targeting signal-encoding sequence (pSP-luc+NF, Promega), which was inserted into the XbaI site of pRC/RSV (Invitrogen). This created pRSV-luc. pCL-H5 was then constructed by restricting pRSV-luc with HindIII and BstEII and inserting a HindIII-digested fragment encoding the first 381 nt of chicken COX-2. This region encodes the N-terminal, hydrophobic signal peptide for membrane insertion of COX-2, as well as the dimerization and membrane-binding domains. The construct lacks the COX-2 catalytic domain.
LA-24 cells containing this reporter protein in their membranes were treated for 24 h with 0.1% ethanol vehicle, 100 μM, or 500 μM diclofenac. Luciferase in the cytosol was measured and compared with total cellular luciferase as described below.
Luciferase Assay.
Luciferase assays were performed on extracts from either whole cells or the cytosolic fraction by applying the luciferase assay system (Promega) according to the manufacturer’s protocol. In these experiments, stable transfectants were grown to confluence. Cytosolic fractions were isolated by lysing cells scraped in PBS with 100 strokes in a tight-fitting Dounce homogenizer and then sonicated for 20 s. Nuclei and unbroken cells were then removed from the homogenate by centrifugation at 5,000 × g for 10 min. Supernatants were then centrifuged at 100,000 × g for 30 min, and luciferase activity was measured in the resulting cytosolic supernatant. Results were expressed as the percentage of luciferase activity present in the cytosolic fraction relative to the total luciferase activity present in the cell and have been normalized to protein concentrations (9).
Materials.
All compounds used were obtained from Sigma unless otherwise designated.
RESULTS
Morphological Effects of NSAIDs on J774.2 Cells.
Treatment of J774.2 cells with any one of 10 NSAIDs for 48 h showed that all but aspirin in doses ranging from 60 to 500 μM caused blebbing, production of apoptotic bodies, and nucleosomal ladders indicative of cell death. Doses of acetaminophen ranging from 0.5 to 2 mM were also effective inducers of cell death.
Induction of COX-2 Protein in J774.2 Cells.
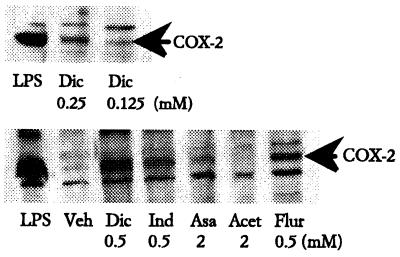
Examination of COX-1 and COX-2 protein levels showed that all NSAIDs, except aspirin and acetaminophen, at 100–500 μM induced COX-2 (Fig. 1). Aspirin induced some COX-2 at doses greater than 2 mM, but acetaminophen did not induce enzyme at any dose. Increasing concentrations of diclofenac showed dose-dependent induction of COX-2 in cells treated with doses as low as 125 μM (Fig. 1). This concentration is higher than those previously found to induce COX-2 in chicken embryo fibroblasts, in which induction was maximal at 25–50 μM (7). By contrast, COX-1 in both J774.2 cells and chicken embryo fibroblasts was only marginally induced by aspirin and diclofenac and was not affected by any of the other drugs (ref. 11; data not shown).
Figure 1.
Induction of COX-2 protein expression by NSAIDs. J774.2 cells were treated with LPS (1 μg/ml for 12 h), vehicle, or NSAIDs (for 48 h) at the doses indicated. Cellular proteins were electrophoresed and probed by Western blot with isoenzyme-specific anti-COX-2 sera. COX-2 was induced 10- to 30-fold by 0.5 mM indomethacin (Ind 0.5), 0.5 mM flurbiprofen (Flur 0.5), and 0.5 mM diclofenac (Dic 0.5) but was not detectably induced by 2 mM acetaminophen (Acet 2) and was only marginally induced by 2 mM aspirin (Asa 2). Induction of COX-2 by diclofenac was dose dependent (compare Upper and Lower).
Induction of COX Activity in J774.2 Cells.
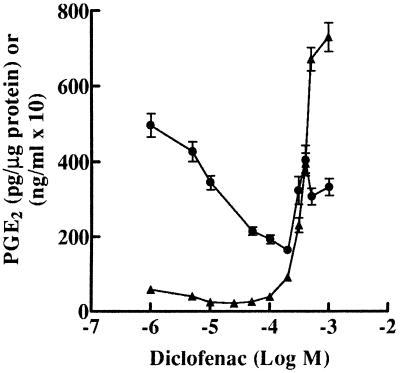
Cells were exposed to various doses of diclofenac for 48 h, after which the diclofenac-containing medium was removed and COX activity was measured by exposing the cells to 30 μM arachidonic acid. At low doses (5–50 μM), this lengthy diclofenac treatment inhibited COX activity in a dose-dependent and long-lasting fashion; however, at higher doses (100 μM–1 mM), diclofenac induced COX activity dose dependently (Fig. 2). This induction was strikingly obvious when COX activity was normalized to cellular protein concentrations (Fig. 2).
Figure 2.
Induction of COX activity by diclofenac in J774.2 cells. ●, J774.2 cells were treated with diclofenac at the concentrations indicated for 48 h. After treatment, cells were washed twice to remove drug and then exposed to 30 μM arachidonic acid for 15 min. PGE2 released was estimated by RIA and is expressed as ng of PGE2/ml of media times a factor of 10. ▴, Values have been recalculated as pg of PGE2/μg of protein to normalize for the large decrease in cell number and mass occurring at higher NSAID concentrations (200 μM or greater).
Effect of Acetaminophen on LPS and Diclofenac-Induced COX Activity.
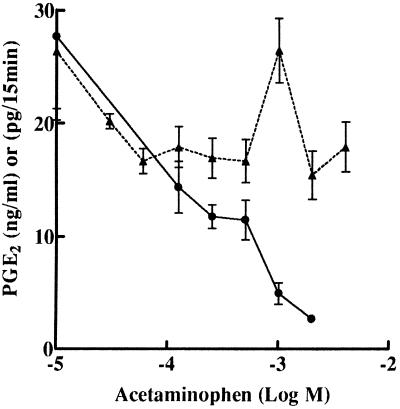
The increased COX activity in J774.2 cells induced by NSAIDs was more sensitive to acetaminophen. Diclofenac-induced activity in apoptotic cells was inhibited dose dependently by acetaminophen at concentrations greater than 0.06 mM (Fig. 3). However, under identical experimental conditions, COX activity in LPS-treated J774.2 cells was not appreciably inhibited by similar doses of acetaminophen (Fig. 3) as previously reported (6).
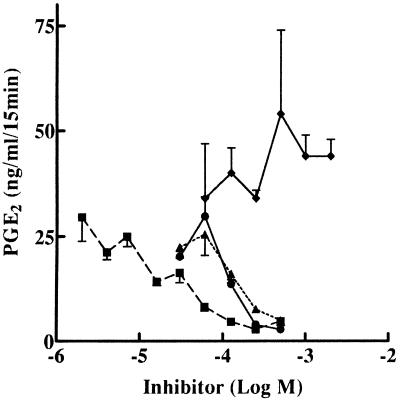
Figure 3.
Sensitivity of diclofenac-induced COX-2 but not LPS-induced COX-2 to inhibition by acetaminophen. ●, J774.2 cells were treated with 0.5 mM diclofenac for 48 h to induce COX-2 and apoptosis. After this treatment, diclofenac was removed by washing, and the cells were exposed to acetaminophen for 30 min at the doses shown. After acetaminophen treatment, cells were exposed to 30 μM arachidonic acid for 15 min and PGE2 released was measured as ng of PGE2/ml of media. ▴, J774.2 cells were treated with LPS (1 μg/ml for 12 h) to induce COX-2. Cells were treated with acetaminophen, and PGE2 release was measured as pg of PGE2/15 min.
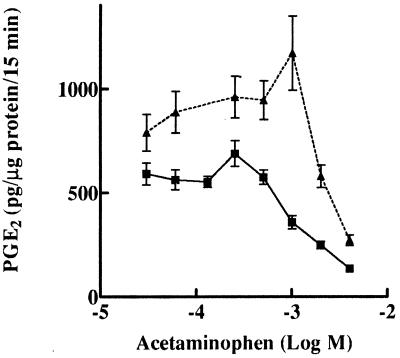
The high COX activity in J774.2 cells treated for 48 h with diclofenac was found to be additionally elevated by exposing the cells to LPS during the last 24 h of diclofenac treatment. This resulted in a 4-fold induction of COX-2, but not COX-1, protein above that obtained by diclofenac treatment alone (data not shown), suggesting that the increased COX activity observed was caused by elevated COX-2. In the presence of diclofenac, LPS-induced COX-2 activity in apoptotic cells was inhibited by acetaminophen to the same extent as the COX-2 activity induced by diclofenac alone (Fig. 4).
Figure 4.
Augmentation of diclofenac-induced COX-2 by LPS. J774.2 cells were treated for 48 h with 0.5 mM diclofenac (■) or diclofenac accompanied by LPS (1 μg/ml) for the last 24 h of the 48-h period (▴). Cells were treated for inhibition by acetaminophen as described in Fig. 3.
Effect of Other NSAIDs on Diclofenac-Induced COX-2 Activity.
Diclofenac-induced activity was inhibited by diclofenac itself and by other NSAIDs but at higher concentrations than those usually needed in LPS-treated cells (Fig. 5). COX-2 in J774.2 cells was inhibited by diclofenac, flurbiprofen, or tolfenamic acid with IC50 values of 1, 0.1, and 0.2 μM, respectively (6, 12). However, the COX activity in apoptotic, diclofenac-treated cells was inhibited with IC50 values for diclofenac, flurbiprofen, and tolfenamic acid of 10, 230, and 220 μM, respectively, or an increase of 1–5 orders of magnitude. Aspirin did not inhibit the diclofenac-induced enzyme activity (Fig. 5).
Figure 5.
Inhibition of diclofenac-induced COX-2 by NSAIDs. J774.2 cells were treated for 48 h with 0.5 mM diclofenac to induce COX-2. After diclofenac was removed by washing, cells were exposed for 30 min to diclofenac (■), flurbiprofen (▴), tolfenamic acid (●), and aspirin (⧫) at the doses shown. Inhibition of COX activity was measured in the presence of 30 μM arachidonic acid for 15 min as ng of PGE2/ml of media.
COX-2–Luciferase Reporter Construct.
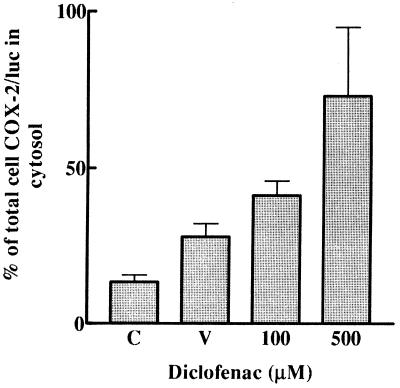
Subcellular localization of COX-2–luciferase activity was measured with the construct shown in Fig. 6. After treatment of LA 24 cells for 24 h with 100 or 500 μM diclofenac, a concentration-dependent increase of the COX-2–luciferase protein was found in the cytosol (Fig. 7); whereas, in untreated cells, only 5–15% of the COX-2–luciferase fusion protein was in the cytosol. In contrast, luciferase without COX-2 appended to its amino terminus did not localize to membrane fractions but was found almost entirely in the cytosol (data not shown). Treatment with the vehicle, 0.1% ethanol, for 24 h also caused some increase of COX-2–luciferase activity in the cytosol compared with total cellular luciferase (Fig. 7).
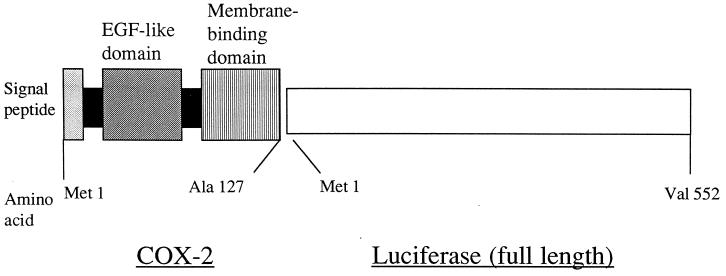
Figure 6.
Structure of a chicken COX-2–luciferase reporter construct. The first 381 nt of a full-length chicken COX-2 cDNA were fused in-frame to the 5′ end of a full-length firefly luciferase cDNA. As shown, the construct encoded the 17-aa, hydrophobic, N-terminal signal peptide (shaded box) of COX-2, as well as its epidermal growth factor (EGF)-like dimerization domain (diagonal stripes) and complete membrane-binding domain (vertical stripes).
Figure 7.
Subcellular localization of COX-2–luciferase activity. Localization of COX-2–luciferase was measured by the construct shown in Fig. 6. LA24 cells stably transfected with the COX-2–luciferase construct (control, C) were treated for 24 h with 0.1% ethanol vehicle (V) or 100 μM or 500 μM diclofenac. Luciferase in the cytosol was measured and compared with total cellular luciferase as described in Materials and Methods.
DISCUSSION
Many NSAIDs in high concentrations induce both apoptosis and COX-2 in chicken embryo fibroblasts and J774.2 cells in culture. However, induction of apoptosis and induction of COX-2 enzyme are not always linked. For example, in J774.2 cells, staurosporine or acetaminophen induce apoptosis but not COX-2. In low concentrations, the NSAIDs are time-dependent, pseudoirreversible inhibitors of COX-1 and COX-2. Such drugs frequently require 5 min or more of exposure to the enzyme to enter the narrow hydrophobic channel of the COX active site and to occlude it. When bound, these drugs often have such slow off-rates that their inhibition of COX activity is essentially irreversible. Diclofenac is such a drug (13–16).
Characteristics of NSAID-Induced COX Activity.
For the purposes of this discussion, we will assume that the mechanisms by which NSAIDs induce apoptosis and COX-2 are similar for all NSAIDs and will concentrate on diclofenac induction. There are several features of the diclofenac-induced COX-2 that distinguish it from the COX-2 induced in J774.2 cells by LPS.
1. Diclofenac washes out easily from its active site.
This occurs within a few minutes and suggests that this COX has a significantly different active site than either COX-1 or COX-2.
2. Acetaminophen is a more potent inhibitor.
Of particular interest was the inhibition of this enzyme activity by concentrations of acetaminophen too low to affect LPS-induced COX-2 in nonapoptotic J774.2 cells. Mitchell et al. (6) were able to measure only IC30 values for inhibition of LPS-induced COX-2 in J774.2 macrophages.
It is unlikely that the biochemical basis for the acetaminophen sensitivity of the enzyme is simply attributable to an increased permeability of acetaminophen into the cell. The majority of apoptotic cells in these cultures exclude trypan blue dye and, therefore, have intact periplasmic membranes. Moreover, homogenization of the apoptotic cells results in decreased rather than increased sensitivity to acetaminophen (data not shown). We postulate that acetaminophen has a selective inhibitory effect on the specific type of COX-2 activity found and induced in J774.2 cells undergoing apoptosis because of NSAID treatment. This inhibition by acetaminophen could occur either at the COX or peroxidase active site of the enzyme. If the enzyme activity identified in J774.2 cells is identified in vivo, it could represent a mechanism for the pharmacological mode of action of acetaminophen.
When J774.2 cells were treated initially with diclofenac alone for 24 h and the incubation was continued for a further 24 h in the presence of LPS and diclofenac, the total enzyme activity generated was higher than for treatment with diclofenac alone. This enzyme activity was totally sensitive to inhibition with acetaminophen.
3. Aspirin does not inhibit the diclofenac-induced variant enzyme.
This is a puzzling finding given that aspirin acetylates both COX-1 and COX-2. Normally, aspirin acetylates a serine in the COX active site (17). This finding of aspirin insensitivity further confirms that the composition of the active site of this variant enzyme has changed.
4. COX is less sensitive to inhibition by NSAIDs.
Flurbiprofen or tolfenamic acid, two of the most potent inhibitors of COX-1 and COX-2 in J774.2 cells, inhibited the diclofenac-induced enzyme only at high concentrations. Inhibition of the enzyme by diclofenac itself was also significantly reduced (approximately 10-fold).
Localization of NSAID-Induced COX-2 Variant.
The observed increase in acetaminophen-sensitive COX activity paralleled an increase in COX-2 protein, a phenomenon that was consistently observed in all of our studies. We have been unable to separate induction of the COX activity from induction of COX-2 protein, suggesting that the enzyme is a variant of COX-2. The physical changes responsible for such a variant are unknown but could include alterations in subcellular localization of COX-2, protein modification, protein–protein interactions, or redox state. We postulate that the COX-2 variant induced with NSAIDs has a different subcellular microenvironment from COX-2 induced with cytokines or LPS. Thus, when appended to luciferase, a COX-2–luciferase reporter protein is detected mainly in the cytosol in diclofenac-treated cells; whereas the reporter is localized to the nuclear or microsomal membranes in untreated cells. This translocation of the COX-2–luciferase protein into the cytosol does not involve the COX-2 catalytic site but reflects an action of diclofenac on the binding of the construct to nuclear or microsomal membranes. This translocation suggests that chronic treatment of J774.2 cells with diclofenac results in a mobilization of COX-2 in which the luciferase reporter has either lost affinity for its site in the endoplasmic reticulum or nascent COX–reporter was not translocated into the lumen of the endoplasmic reticulum. Recently, extraluminal COX-2 subpopulations have been identified within the nucleus, associated with nuclear matrix, and in the cytosol (18).
NSAID-Induced Apoptosis.
Diclofenac induction of COX activity occurred concomitantly with programmed cell death, although the induction of apoptosis per se was insufficient to induce the enzyme activity because induction of cell death by staurosporine (data not shown) did not induce any COX-2 activity in J774.2 cells. Apoptosis induced by NSAIDs was previously demonstrated in chicken embryo fibroblasts when treated with any one of 17 different COX inhibitors (7). This apoptotic response was accompanied by the generation of nucleosome-sized ladders of DNA fragments in treated cells and other apoptosis-associated biochemical events, such as induction of tissue transglutaminase and inhibition by bcl-2 (7, 11). COX-2 mRNA and protein were also induced by apoptotic NSAIDs, similar to these studies (7, 11).
The mechanism of NSAID-induced apoptosis in cultured cell lines is currently under debate as to whether COX enzymes are involved (19, 20). NSAIDs used in the present study inhibit COX-1- and COX-2-mediated synthesis of PGE2 in J774.2 cells at much lower doses than those required to produce apoptosis and to induce this COX-2-associated enzyme activity (6). This suggests that NSAID-mediated induction of COX-2 occurs by some other mechanism than simple interruption of a negative feedback loop through inhibition of PGE2 synthesis. In support of this concept, we have found PGE2 replacement to be ineffective at preventing NSAID induction of COX-2 in either chicken embryo fibroblasts or J774.2 cells (data not shown). However, we note that the activity of the diclofenac-induced enzyme described in these studies is inhibited by NSAIDs at concentrations that parallel their ability to induce apoptosis in cultured cells. Hence, this low-affinity COX target for NSAIDs may play a critical role in regulating apoptosis induced by NSAIDs. The low-affinity COX target may be present in many cells at significantly lower levels than the high-affinity forms of COX-1 or COX-2.
Induction of COX Activity by NSAIDs.
Until very recently, the only known shared pharmacological action of NSAIDs was inhibition of COX-1 and COX-2, for which the structural mechanism of inhibition is now defined at crystallographic resolution (21, 22). This common mode of action of NSAIDs is true when applied to low, physiologically relevant drug concentrations and constitutes the mechanism by which these drugs exert their physiological antiinflammatory effects. However, in tissue culture nonphysiological concentrations of NSAIDs can easily be administered to cells. Several laboratories in recent months have shown that many competitively acting NSAIDs, at concentrations greater than 10 μM, also bind and activate members of the peroxisome proliferator-activated receptor (PPAR) family of steroid orphan receptors (23–25). Aspirin does not activate these receptors (25). Significantly, PPAR agonists appear also to induce COX-2, just as NSAIDs do, during NSAID-induced apoptosis. For the PPAR agonist clofibrate, this induction of COX-2 occurs at drug doses that induce cell division, leading to the proposal that COX-2 induction through PPAR activation is critical to the proliferative and tumorigenic properties of PPAR agonists (24). It is also important to note that the enzymatic properties of COX-2 induced by PPAR receptors, although only partially characterized at present, are distinctly different from those of COX-2 induced by serum or other mitogens (24, 25). Specifically, the PPAR-induced enzyme does not result in increased cellular PGE2 synthesis, even though the enzyme is fully active when external arachidonic acid is added.
J774.2 cells were found to be unusual as to the degree to which this enzyme activity can be induced. For example, J774.1 cells derived from the same original tumor showed substantially reduced induction relative to J774.2 cells, and A549 cells were largely refractory to induction (data not shown). Hence, cell-specific factors or signaling pathways appear to be important to the induction of this enzyme activity.
It should be possible to design selective enzyme inhibitors for this diclofenac-induced activity whether or not this enzyme activity is a catalytic variant of the COX-2 enzyme or a third COX that is coregulated with COX-2 during NSAID-induced apoptosis. Such enzyme inhibitors may be particularly effective at inducing programmed cell death in neoplastic cells.
Acknowledgments
We thank Ivana Vojnovic for expert assistance with RIAs and Western blots and Elizabeth Wood for culturing the J774.2 cells. D.L.S. was supported by a National Institutes of Health Senior International Investigatorship (1FO6TW02180).
ABBREVIATIONS
- COX
cyclooxygenase
- NSAID
nonsteroid antiinflammatory drug
- PG
prostaglandin
- LPS
lipopolysaccharide
- PPAR
peroxisome proliferator-activated receptor
References
- 1.Xie W, Chipman J G, Robertson D L, Erikson R L, Simmons D L. Proc Natl Acad Sci USA. 1991;88:2692–2696. doi: 10.1073/pnas.88.7.2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kujubu D A, Fletcher B S, Varnum B C, Lim R W, Herschman H R. J Biol Chem. 1991;266:12866–12872. [PubMed] [Google Scholar]
- 3.O’Banion M K, Sadowski J B, Winn J B, Young D A. J Biol Chem. 1991;266:23261–23267. [PubMed] [Google Scholar]
- 4.Flower R J, Vane J R. Nature (London) 1972;240:410–411. doi: 10.1038/240410a0. [DOI] [PubMed] [Google Scholar]
- 5.Robak J, Wieckowski A, Gryglewski R. Biochem Pharmacol. 1978;27:393–396. doi: 10.1016/0006-2952(78)90367-2. [DOI] [PubMed] [Google Scholar]
- 6.Mitchell J A, Akarasereenont P, Thiemermann C, Flower R J, Vane J R. Proc Natl Acad Sci USA. 1993;90:11693–11697. doi: 10.1073/pnas.90.24.11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu X, Xie W, Reed D, Bradshaw W S, Simmons D L. Proc Natl Acad Sci USA. 1995;92:7961–7965. doi: 10.1073/pnas.92.17.7961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salmon J A. Prostaglandins. 1978;15:383–397. doi: 10.1016/0090-6980(78)90122-3. [DOI] [PubMed] [Google Scholar]
- 9.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 10.Gross S, Jaffe E A, Levi R, Kilbourn R G. Biochem Biophys Res Commun. 1991;187:823–829. doi: 10.1016/0006-291x(91)90965-a. [DOI] [PubMed] [Google Scholar]
- 11.Lu X, Fairbairn D W, Bradshaw W S, O’Neill K L, Ewert D L, Simmons D L. Prostaglandins. 1997;54:549–568. doi: 10.1016/s0090-6980(97)00125-1. [DOI] [PubMed] [Google Scholar]
- 12.Akarasereenont, P., Mitchell, J. A., Thiemermann, C. & Vane, J. R. (1994) Br. J. Pharmacol112, Suppl., 183P. [DOI] [PMC free article] [PubMed]
- 13.Rome L H, Lands W E M. Proc Natl Acad Sci USA. 1975;72:4863–4865. doi: 10.1073/pnas.72.12.4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stanford N, Roth G J, Shen T Y, Majerus P W. Prostaglandins. 1977;13:669–675. doi: 10.1016/0090-6980(77)90237-4. [DOI] [PubMed] [Google Scholar]
- 15.Kulmacz R J, Lands W E. J Biol Chem. 1985;260:12572–12578. [PubMed] [Google Scholar]
- 16.Tobetto K, Yamamoto Y, Kataoka M, Ando T, Sugimoto K, Himeno M. Jpn J Pharmacol. 1997;75:371–379. doi: 10.1254/jjp.75.371. [DOI] [PubMed] [Google Scholar]
- 17.Roth G J, Majerus P W. J Clin Invest. 1975;56:624–632. doi: 10.1172/JCI108132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coffey R J, Hawkey C J, Damstrup L, Graves-Deal R, Daniel V C, Dempsey P J, Chinery R, Kirkland S C, DuBois R N, Jetton T L, et al. Proc Natl Acad Sci USA. 1997;94:657–662. doi: 10.1073/pnas.94.2.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piazza G A, Alberts D S, Hixson U, Paranka N S, Li H, Finn T, Bogert C, Guillen J M, Brendel K, Gross P H, et al. Cancer Res. 1966;57:2909–2915. [PubMed] [Google Scholar]
- 20.Herrmann C, Block C, Geisen C, Haas K, Weber C, Winde G, Moroy T, Muller O. Oncogene. 1998;17:1769–1776. doi: 10.1038/sj.onc.1202085. [DOI] [PubMed] [Google Scholar]
- 21.Loll P J, Picot D, Garavito R M. Nat Struct Biol. 1995;2:637–642. doi: 10.1038/nsb0895-637. [DOI] [PubMed] [Google Scholar]
- 22.Kurumbail R G, Stevens A M, Gierse J K, McDonald J J, Stegeman R A, Pak J Y, Gildehaus D, Miyashiro J M, Penning T D, Seibert K, et al. Nature (London) 1996;384:644–648. doi: 10.1038/384644a0. [DOI] [PubMed] [Google Scholar]
- 23.Green S. Mutat Res. 1995;333:101–109. doi: 10.1016/0027-5107(95)00136-0. [DOI] [PubMed] [Google Scholar]
- 24.Ledwith B J, Pauley C J, Wagner L K, Rokos C L, Alberts D W, Manam S. J Biol Chem. 1997;272:3707–3714. doi: 10.1074/jbc.272.6.3707. [DOI] [PubMed] [Google Scholar]
- 25.Lehmann J M, Lenhard J M, Oliver B B, Ringold G M, Kliewer S A. J Biol Chem. 1997;272:3406–3410. doi: 10.1074/jbc.272.6.3406. [DOI] [PubMed] [Google Scholar]