Abstract
A major class of plant disease resistance (R) genes encodes leucine-rich-repeat proteins that possess a nucleotide binding site and amino-terminal similarity to the cytoplasmic domains of the Drosophila Toll and human IL-1 receptors. In Arabidopsis thaliana, EDS1 is indispensable for the function of these R genes. The EDS1 gene was cloned by targeted transposon tagging and found to encode a protein that has similarity in its amino-terminal portion to the catalytic site of eukaryotic lipases. Thus, hydrolase activity, possibly on a lipid-based substrate, is anticipated to be central to EDS1 function. The predicted EDS1 carboxyl terminus has no significant sequence homologies, although analysis of eight defective eds1 alleles reveals it to be essential for EDS1 function. Two plant defense pathways have been defined previously that depend on salicylic acid, a phenolic compound, or jasmonic acid, a lipid-derived molecule. We examined the expression of EDS1 mRNA and marker mRNAs (PR1 and PDF1.2, respectively) for these two pathways in wild-type and eds1 mutant plants after different challenges. The results suggest that EDS1 functions upstream of salicylic acid-dependent PR1 mRNA accumulation and is not required for jasmonic acid-induced PDF1.2 mRNA expression.
Disease resistance in plants often is mediated by corresponding gene pairs in the plant (resistance or R gene) and pathogen (avirulence or avr gene) that condition specific recognition and activate plant defenses (1). The precise mechanisms controlling R-avr gene-specified resistance are poorly understood, although a requirement for salicylic acid (SA), a phenolic derivative, has been demonstrated in several plant-pathogen interactions (2, 3). Other studies suggest that the formation of reactive oxygen species, ion flux changes, and protein kinase activation are important early events in specific pathogen recognition (4–6).
R genes now have been cloned from several dicot and monocot species. The predominant class of predicted R gene products, specifying resistance to viral, bacterial, and fungal pathogens, possess sequences that constitute a nucleotide binding site (NB) and leucine-rich repeats (LRR) (5, 7). Therefore, recognition of different pathogen types may have common mechanistic features. The NB-LRR type R proteins have been further categorized based on different amino termini. One class, represented by the tobacco N, flax L6, and Arabidopsis thaliana RPP5 genes, has similarity to the cytoplasmic portions of the Drosophila Toll and mammalian interleukin 1 transmembrane receptors [referred to as the TIR (Toll, IL-1, resistance) domain], suggesting functional conservation with animal innate immunity pathways (1, 7, 8). A second class, comprising the Arabidopsis genes RPM1, RPS5, and RPS2, possesses a putative leucine zipper (the LZ domain), implicating a different signaling mechanism. Mutational analyses in Arabidopsis have led to the identification of other components that are required for R gene-specified resistance. Mutations in NDR1, a gene that encodes a small putatively membrane-associated protein (9), suppress resistance mediated by several R genes of the LZ-NB-LRR but not the TIR-NB-LRR class (10, 11). In contrast, mutations in EDS1 define an essential component of resistance specified by TIR-NB-LRR but not LZ-NB-LRR type R genes (10, 11). Thus, at least two different signaling pathways appear to be activated by particular R protein structural types. Here, we describe the cloning and characterization of EDS1 to investigate its role as a central component of a disease resistance pathway conditioned by TIR-NB-LRR type R genes.
METHODS
Plant Cultivation and Pathogenicity Tests.
Seeds of accession Columbia (Col-gl, containing the recessive mutation gl1) were obtained from J. Dangl (University of North Carolina, Chapel Hill). Landsberg-erecta (Ler) seed were from the Nottingham Arabidopsis Stock Centre (Nottingham, U.K.). The Ws-eds1–1 mutation has been described (10). The eds1 alleles (eds1–5, eds1–6, eds1–7, and eds1–8) were isolated from ethylmethane sulfonate (EMS)-mutagenized Ws-0 M2 seed obtained from Lehle Seeds (Tucson, AZ). Ler eds1 alleles eds1–2, eds1–3, and eds1–4 were isolated from fast neutron (FN)-bombarded Ler M2 seed (Lehle Seeds). eds1–4 was kindly provided by B. Staskawicz (University of California, Berkeley). Cultivation of seedlings for Peronospora parasitica and bacterial inoculations was as described (10). Ler Inhibitor/defective Suppressor (I/dSpm)-18 seed (12) was a kind gift from M. Aarts (CPRO, Wageningen, The Netherlands). Screens for susceptibility to P. parasitica isolate Noco2 were performed by spraying 9-day-old seedlings with conidiospore suspensions (4 × 104/ml) and incubating under appropriate conditions (10).
DNA Manipulations.
General methods for DNA manipulation and DNA gel blotting were as described (13). Plant genomic DNA was extracted as in ref. 14. End probes were generated from P1 clones by using thermal assymetric interlaced–PCR (15). A Ler cDNA library was a kind gift from M. Coleman (University of East Anglia, Norwich, U.K.). A Ler genomic DNA library constructed in the binary cosmid vector, pCLD04541 (14) was provided by C. Lister and C. Dean (John Innes Centre, Norwich, U.K.). Cosmid clone DNA inserts were gel-purified and subcloned into pGEM3Zf(+) (Promega). I/dSpm-specific primers that annealed to the terminal repeat sequences (12) were used to amplify plant DNA flanking the element by inverse-PCR of gel-enriched HindIII-digested DNA.
EDS1 Mapping Analysis.
The FN-derived Ler eds1 allele, eds1–2 (11), was crossed with Col-gl to generate an F2 mapping population. F2 seedlings first were scored for resistance or susceptibility to P. parasitica isolate, Wand1 (16), that is recognized by two unlinked EDS1-dependent RPP loci (A.F. and J.E.P., unpublished data). Informative recombinants were further tested for resistance or susceptibility to P. parasitica isolate Noco2 that is recognized by a single EDS1-dependent RPP gene, RPP5 in Ler, and plants genotyped for RPP5, as described (14). The I18 marker comprises 186 bp of Ler genomic DNA that flanked a nonautonomous I/dSpm transposable element (12). Primers corresponding to I18 DNA sequence were used to identify positive clones from 96 pools of a P1 phage library containing Col-0 genomic DNA (17). The I18 marker and P1 end-probes hybridized with yeast artificial chromosome (YAC) clones (11D12, 3D2, and 7A9) from the CIC YAC library that were part of a YAC contig on the lower arm of chromosome 3 (information kindly provided by D. Bouchez, Institut National de la Recherche Agronomique, Versailles, France).
Nucleotide Sequence Determination and Computer Analyses.
Sequencing reactions were run on an Applied Biosystems 377 automatic sequencer. DNA sequences were assembled and analyzed by using the University of Wisconsin gcg computer packages. Computer-aided sequence similarity searches were made with the blast suite of programs at the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov). Sequence alignments were done by using pileup. Accession numbers at NCBI for the following sequences are as follows: Rhimi (P19515), Rhiniv (S39525), Penca (gi|298949), Humlan (gi|999873), Aspnig (gi|2760074), Isolog1 (gi|2344903), Isolog2 (gi|2245036), Isolog3 (gil|946364), Isolog4 (gi|l2832660), Ipomoea (gil|527001), Cael1 (gi|2291250), and Cael2 (gi|2736368). Motif searches were made by using prosite (http://expasy.hcuge.ch/sprot/prosite.html), tmpred (http://www.isrec.isb-sib.ch/software/software.html), and tmap (http://www.embl-heidelberg.de/tmap/tmap_sin.html). Predicted secondary structure for EDS1, Ipomoea lipase, and Isolog3 were obtained by using the predictprotein server (http://www.embl-heidelberg.de/predictprotein).
RNA Expression Analysis.
Pseudomonas syringae pv tomato strain DC3000 containing avrRps4 (18) in the broad host range vector pVSP61 (19) or DC3000 containing empty pVSP61 were cultured as described. For all treatments, 4-week-old plants grown in soil under an 8-hr photoperiod, were used. Plants were left untreated or whole leaves were infiltrated with suspensions of P. syringae at 107 colony-forming units/ml and incubated at 25°C and >70% humidity. For SA treatment, leaves were sprayed to imminent run-off with a 0.5 mM solution, containing 0.005% of the wetting agent Silwet L-77 (Union Carbide). Methyl jasmonate (JA; Bedoukian Research, Danbury, CT) was applied in the same way as a 1 mM solution. For wounding, leaves were pressed hard with tweezers. Total RNA was extracted from 6–8 leaves per treatment according to Reuber and Ausubel (20). RNA was separated on formaldehyde-agarose gels, transferred to nylon membranes, and probed with 32P-labeled EDS1 cDNA or PCR-amplified fragments of PR1 (21) and PDF1.2 (22).
RESULTS
Isolation of the EDS1 Gene.
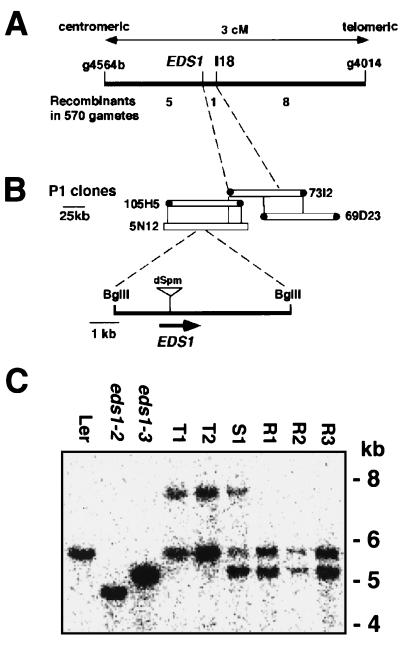
Previously, EDS1 was mapped between the markers m249 and BGL1 on the lower arm of chromosome 3 (10). In the present mapping analysis (see Methods), EDS1 was positioned <0.2 cM centromeric to the restriction fragment length polymorphism marker I18 (Fig. 1A). I18 was used to identify two overlapping clones, 73I2 and 69D3, from a P1 phage library containing Col-0 genomic DNA, and a P1 contig was extended in the direction of EDS1 (Fig. 1B). I18 was derived from plant DNA flanking a nonautonomous I/dSpm transposable element in Arabidopsis accession Ler (12). This accession contains the R gene, RPP5 conditioning resistance to isolate Noco2 of the oomycete pathogen, P. parasitica (14). We therefore selected 35 Ler lines that were homozygous for I/dSpm-18 and the stable Enhancer transposase source. These were selfed and their progeny were screened with Noco2 for insertional inactivation of EDS1. Five Noco2-susceptible plants (T1–T5) were rescued and shown to be defective in EDS1 function by crossing these with FN-derived eds1 mutant lines, eds1–2 or eds1–3 (ref. 11; data not shown). In two independently selfed progenies of these plants, reversion to resistance occurred at a frequency of ≈6%, indicating that the mutations were unstable and likely to be caused by a transposon insertion.
Figure 1.
High-resolution mapping and transposon tagging of EDS1. (A) Genetic map. Recombinant analysis placed EDS1 0.2 cM centromeric to I18, an restriction fragment length polymorphism (RFLP) marker derived from a I/dSpm transposon insertion in Ler. (B) P1 contig. I18 was used to identify two P1 phage clones, 73I2 and 69D23. An RFLP was detected between Ler and Col-0 DNA with the 73I2 centromeric end-probe, allowing orientation of P1 clones relative to EDS1. I/dSpm insertions into EDS1 were located in Ler DNA corresponding to a 5.7-kb internal BglII fragment of P1 clones 105H5 and 5N12. (C) A blot of BglII-digested genomic DNA was probed with a 32P-labeled inverse-PCR product derived from an I/dSpm insertion shared by eds1 lines T1–T5. The blot shows the wild-type Ler 5.7-kb band and deletions of ≈1 or ≈0.5 kb, respectively, in the FN-derived Ler mutants eds1–2 and eds1–3. Lines T1 and T2 possess an additional 7.9-kb band caused by insertion of a 2.2-kb I/dSpm element. In contrast to a Noco2-susceptible F1 plant (S1) derived from a cross between T1 and eds1–3, three independent Noco2-resistant (revertant) F1 plants (R1, R2, and R3) have lost the I/dSpm insertion.
Genomic DNA blot analysis showed that Noco2-susceptible plants T1–T5 had a I/dSpm-hybridizing band that was not present in Noco2-resistant siblings (data not shown). Plant DNA flanking this I/dSpm element was amplified by inverse-PCR and shown to hybridize to a 5.7-kb BglII internal fragment of P1 clones, 105H5 and 5N12 (Fig. 1B), and to a single 5.7-kb BglII DNA fragment in wild-type Ler (Fig. 1C). Probing DNA from the Ler mutant lines eds1–2 and eds1–3 with the inverse-PCR fragment revealed that both had extensive deletions in this fragment (Fig. 1C). I/dSpm-containing eds1 lines T1–T5 possessed a 7.9-kb hybridizing BglII band in addition to the wild-type 5.7-kb band, consistent with insertion of the 2.2-kb I/dSpm element and somatic excision events (Fig. 1C) (12). We examined DNA sequence footprints left by I/dSpm excision in Noco2-susceptible and Noco2-resistant (revertant) F1 progeny made from crosses between lines T1 or T2 and Ler eds1–3 (Fig. 1C). This analysis showed that I/dSpm excision had restored the EDS1 ORF in three independent revertant plants but had created sequence frame shifts in two independent Noco2-susceptible progeny (data not shown). Altogether, the data provided conclusive proof of EDS1 isolation.
The EDS Gene Structure.
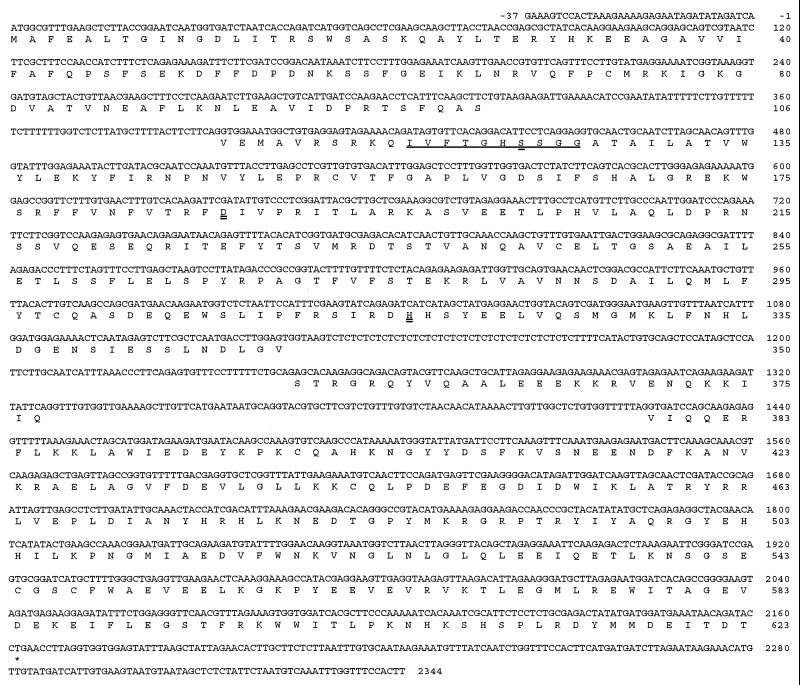
A 2.1-kb EDS1 cDNA clone was isolated from a Ler cDNA library, and this clone detected a ≈2-kb transcript on an RNA gel blot of Ler poly(A)+ RNA (data not shown). Sequence analysis of the cDNA and corresponding BglII genomic DNA fragment from a Ler cosmid library revealed that the EDS1 gene is comprised of four exons encoding a 623-aa protein with a predicted molecular mass of 71.6 kDa (Fig. 2). The presence of an in-frame stop (TAA) codon at position −27 relative to the ATG start codon indicated that the deduced ORF is correct. The I/dSpm insertion site was found to disrupt the first exon. No predicted signal peptide or obvious transmembrane regions were identified by using various motif-search programs (see Methods), suggesting that the protein is cytoplasmic. Inspection of the protein sequence revealed two possible bipartite nuclear localization signals (amino acid positions 366 and 440 in Fig. 2). These have spacers of 15 and 17 aa, respectively, between two blocks of basic amino acids (23). Scanning the prosite database also revealed two possible tyrosine kinase phosphorylation sites (amino acids 320 and 485 in Fig. 2). Genomic DNA sequence also was obtained for the wild-type EDS1 alleles in accessions Col-0 and Wassilewskija (Ws-0). All three alleles are highly conserved, exhibiting 98% identity at the amino acid level. Two Col-0 ESTs (T45498 and AA395521) were found in the Arabidopsis expressed sequence tag database (dbEST) that correspond to the Col-0 EDS1 cDNA.
Figure 2.
Nucleotide sequence of the EDS1 gene and derived amino acid sequence. The isolated EDS1 cDNA encodes a predicted ORF of 1,869 nt with untranslated 5′ and 3′ leader sequences of 37 and 180 nt, respectively. The L-family lipase consensus sequence around the predicted catalytic serine (S123) is underlined. The three predicted lipase catalytic residues, a serine (S123), an aspartate (D187), and a histidine (H317) are indicated by a double underline.
Assessment of EDS1 Homology to Eukaryotic Lipases.
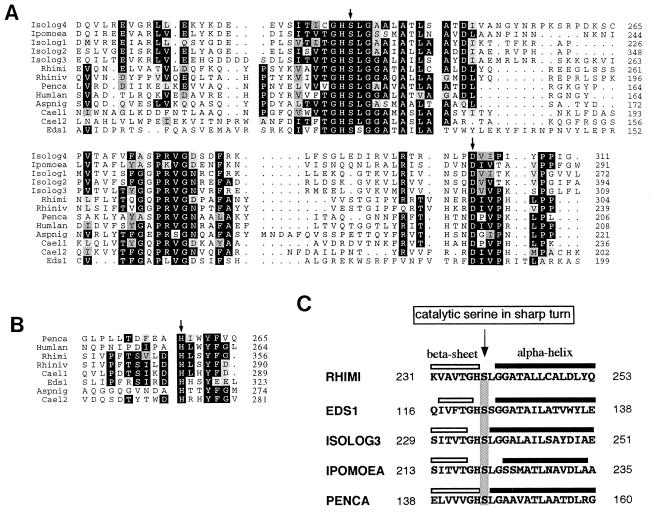
Database searches did not reveal sequences with extensive homology to EDS1, suggesting that EDS1 encodes a novel protein. However, discrete blocks of homology to eukaryotic lipases were identified within EDS1 exon 2 (Fig. 3), the highest scoring segment pairs being with the lipases of Rhizomucor miehei and Rhizopus niveus (24, 25). The regions of sequence similarity contain three amino acids: a serine, an aspartate, and a histidine, which form the lipase catalytic triad (Fig. 3). These fungal enzymes belong to the L-family of α/β hydrolases that comprises fungal triacylglycerol lipases (Rhimi and Rhiniv, Fig. 3A), a mono- and diacylglycerol lipase (Penca, Fig. 3A), mammalian hepatic and pancreatic lipases, and a number of phospholipase A1 (allergens from vespid and snake venom) and A2 enzymes (platelet-activating factor acetylhydrolase) (26–28). EDS1 conforms to the L-family amino acid signature: [LIV]-x-[LIVFY]-[LIVMST]-G-[HYWV]-S-x-G-[GSTAC] around the active site serine (Fig. 2) that flanks a more commonly occurring G-x-S-x-G motif in the α/β hydrolase fold. Several hypothetical proteins from Caenorhabditis elegans and Arabidopsis also possess discrete blocks of sequence conservation around the predicted lipase active site residues (Fig. 3A). Sequences are less conserved across the catalytic histidine (Fig. 3B). The Aspergillus niger protein faeA (Fig. 3A) contains the lipase consensus motifs but has been shown to be a ferulic acid esterase (29). Thus, although the sequence alignments suggest lipase function in EDS1, it is possible that EDS1 hydrolyzes a nonlipid ester bond.
Figure 3.
Homology of EDS1 to eukaryotic lipases. (A) Alignment of EDS1 amino acids 95–205 containing the serine (S) and aspartic acid (D) residues that form part of a putative lipase catalytic triad, to lipases from R. miehei (Rhimi), R. niveus, (Rhiniv), P. camembertii (Penca) and Humicola lanuginosa (Humlan), an esterase from A. niger (Aspnig), and hypothetical lipases from A. thaliana (Isologs 1–4), Ipomoea nil (Ipomoea), and C. elegans (Cael1–2). Crystal structures for the Rhimi, Penca and Humlan lipases have been determined (26–28), and their active site S and D residues are indicated by an arrow above the sequence. Identical amino acids are shown in black boxes while conserved amino acid changes are shaded in gray. A pairwise alignment of the EDS1 and R. miehei lipase amino acid sequences shown here reveals overall identity of 29% and a similarity score of 46%. Numbers to the right refer to amino acid positions of the full-length proteins (see Methods). (B) Alignment of EDS1 amino acids 307–321 around the putative catalytic histidine (H) to fungal lipase/esterases (see A for details). The arrow marks the catalytic histidine determined from crystal structures for Rhimi, Penca, and Humlan (26–28). For the other putative plant lipases it was not possible to generate a consensus alignment around the known catalytic histidine of the above fungal lipases. (C) Conservation of secondary structure elements around the lipase catalytic serine. All sequences shown in A conform to this pattern. The arrow indicates the conserved conformational presentation of the catalytic serine in lipases. Amino acid positions are indicated on the right and left sides.
The crystal structures of several eukaryotic lipases, including R. miehei and Penicillium camembertii, show a strict conservation of the spatial presentation of the catalytic serine in a sharp turn between a β-sheet and an α-helix (26–28). In secondary structure predictions we found conservation of the β-sheet/active serine/α-helix pattern in EDS1 and the proteins aligned in Fig. 3A, as shown in Fig. 3C.
Characterization of eds1 Mutant Alleles.
Mutational screens of FN- or EMS-mutagenized seed of the accessions Ler or Ws-0 revealed eight independent eds1 alleles, the first of which, eds1–1, has been described (10). The eight mutant lines were inoculated with the P. parasitica isolate, Noco2, to assess the degree of disease susceptibility caused by the loss of RPP14-specified resistance in Ws-0 or RPP5-specified resistance in Ler (10, 11). These experiments showed that all eds1 alleles caused an equivalent, strong suppression of resistance to Noco2 (ref. 11; data not shown). The nature of the mutations in these lines is shown in Table 1. The formation of prematurely terminated proteins in EMS-derived alleles, eds1–6, eds1–7, and eds1–8, and the FN-generated allele, eds1–4, indicate that exons 1 and 2 alone are not sufficient for EDS1 function. It is notable that only one of the EMS-generated mutations, eds1–1, retains the predicted full-length ORF. Here, an amino acid exchange has occurred within the carboxyl-terminal portion of the predicted EDS1 protein. It is not known whether this mutation destroys an essential functional motif or causes a major alteration in the tertiary structure, possibly leading to protein instability.
Table 1.
Sequence changes in eds1 alleles
| Allele | Mutagen | Allele-specific DNA change | Change in EDS1 protein |
|---|---|---|---|
| Ws eds1–1 | EMS | G1688 ➛ A | E466 ➛ K |
| Ler eds1–2 | FN | Deletion 905–1844 | Truncated product S276 - stop |
| Ler eds1–3 | FN | Deletion ∼ 500bp of promoter and part exon 1 | No product |
| Ler eds1–4 | FN | Deletion 826–827 | Truncated product S259 - stop |
| Ws eds1–5 | EMS | G394 ➛ A | Alteration in 3′ splice acceptor site |
| Ws eds1–6 | EMS | C743 ➛ T | Q223 - stop |
| Ws eds1–7 | EMS | C950 ➛ T | Q292 - stop |
| WS eds1–8 | EMS | C1298 ➛ T | R368 - stop |
Numbering of nucleotides is according to the Ler DNA sequence in Fig. 2.
Analysis of EDS1 mRNA Expression.
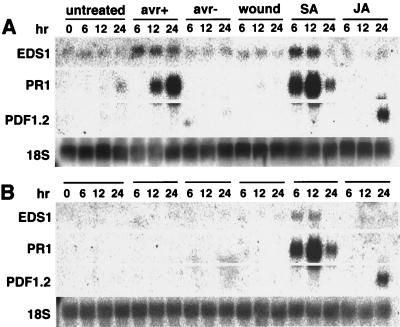
We examined the expression of EDS1 mRNA and two defense-related genes, PR1 and PDF1.2, in wild-type Ler and eds1–2 plants after various treatments (Fig. 4). PR1 mRNA is a marker for resistance responses that depend on SA, a phenolic signaling molecule that is required in several R-gene specified and systemic resistance responses (2, 21). In contrast, PDF1.2 encodes an antimicrobial defensin that is responsive to JA, a plant lipid-derived signal molecule with an essential role in the wound response (30, 31). JA also has been implicated in several plant-pathogen interactions (22, 32, 33). Plants were inoculated with a virulent P. syringae strain, DC3000, or avirulent DC3000 expressing avrRps4 that is recognized by an EDS1-dependent R gene, RPS4 (11). Inoculations of Ler plants with the avirulent bacterial pathogen or treatment with SA induced a 2- to 3-fold increase in EDS1 mRNA levels and a massive accumulation of PR1 mRNA (Fig. 4A). Inoculation with the virulent bacterial pathogen, wounding, or treatment with JA had no observable effect on EDS1 or PR1 mRNA levels. In eds1–2 plants, PR1 mRNA was undetectable after inoculation with the avirulent pathogen but fully inducible by SA (Fig. 4B). We conclude from these data that EDS1 operates upstream of PR1 mRNA accumulation. The observation that eds1–2 plants retain SA-induced activation of PR1 mRNA is consistent with placement of SA perception downstream of EDS1. This finding also is supported by the previous observation that SA application rescues resistance to P. parasitica in eds1 plants (10). However, SA appears to enhance EDS1 expression (Fig. 4), suggesting a possible role for SA and EDS1 in potentiating the defense response (34, 35).
Figure 4.
RNA gel blot of Arabidopsis Ler and eds1–2 plants after various treatments. Total RNA was extracted from wild-type Ler (A) and Ler eds1–2 (B) at indicated times: healthy leaves (untreated), leaves infiltrated with suspensions of avirulent P. syringae strain DC3000 expressing avrRps4 (avr+), or with virulent strain DC3000 containing no functional avr gene (avr−), wounded leaves (wound), and leaves sprayed with SA or JA. Blots were probed simultaneously with 32P-labeled EDS1, PR1, and PDF1.2 sequences and stripped before reprobing with an 18S ribosomal DNA fragment. A second, independent experiment gave similar results.
Increased expression of PDF1.2 mRNA was not observed except after application of JA in both wild-type and mutant eds1–2 plants (Fig. 4 A and B). Because applications of JA also failed to rescue disease resistance in eds1 plants (B.J.F. and J.E.P., unpublished data), we concluded that JA is not sufficient to restore the EDS1 pathway. However, these results do not discount the possibility that EDS1 could operate in a pathway leading to the elaboration of JA-related compounds or other lipid metabolites.
DISCUSSION
EDS1 encodes an essential component of disease resistance conferred by a subset of R genes that condition resistance to bacterial and oomycete pathogens (10, 11). The EDS1 protein therefore is likely to operate within a convergent pathway that is modulated through specific R-Avr protein recognition. Cloning EDS1 represents an important step toward unraveling the processes that are central to this resistance mechanism.
The discrete blocks of amino acid conservation between EDS1 and residues spanning the catalytic site of eukaryotic lipases suggest that EDS1 may function by hydrolyzing a lipid molecule. Our expression analysis shows that EDS1 functions upstream of SA-dependent PR1 mRNA accumulation in the plant response to an avirulent bacterial pathogen. The same resistance response did not lead to increased PDF1.2 mRNA expression, although PDF1.2 mRNA was induced by applications of the potent lipid-derived signaling molecule, JA. EDS1 may be involved in processing JA-related fatty acid intermediates (36, 37) or define an additional lipid-based signaling cascade. However, it is notable that a ferulic acid esterase from A. niger (29) possesses a similar pattern of conserved residues as EDS1 (Fig. 4), raising the possibility that EDS1 hydrolyzes a nonlipid substrate. Indeed, the serine-hydrolase fold has likely been recruited several times independently to derive distinct hydrolytic activities (28). The presence of EDS1 mRNA and protein (B.J.F. and J.E.P., unpublished data) in healthy tissues argues against tight control of expression. Therefore, we envisage that EDS1 may exist in the cell in an active conformation that can process a substrate elaborated specifically on R-Avr protein recognition. Alternatively, R-Avr protein recognition events could lead to posttranslational activation of EDS1 activity. Analysis of eds1 mutations (Table 1) reveals that the carboxyl-terminal 300 amino acids are essential for function and may regulate enzyme activity by exerting conformational constraints or associating with other proteins.
EDS1 is, as far as we know, the first plant L-family lipase representative to be cloned and assigned a function. Significantly, the EDS1 lipase motif highlights the existence of other lipase isologs in Arabidopsis and C. elegans with a similar catalytic signature (Fig. 3A), suggesting a broader relevance for this type of protein in multicellular organisms. Whatever its biochemical role, EDS1 is structurally different from other putative plant lipases that have been identified so far (38, 39). Further analysis of EDS1 expression and potential hydrolytic activity should clarify its role in plant disease resistance.
Acknowledgments
We thank Mark Aarts for provision of a I/dSpm18-containing line and Brian Staskawicz for eds1–4seed. We also thank R. Whittier and the Research Institute of Innovative Technology for the Earth (RITE) and the Mitsui Plant Biotechnology Research Institute for the Col-0 P1 library. We are grateful to Miguel Botella and Erik van der Biezen at SL for helpful discussions. This work was supported by the Gatsby Charitable Foundation, a British Biotechnology and Biological Sciences Research Council grant (B.J.F.), and the Swedish Council for Agriculture and Forestry Research (A.F.).
ABBREVIATIONS
- R
resistance
- avr
avirulence
- NB
nucleotide binding site
- LRR
leucine-rich repeats
- Ler
Landsberg-erecta
- EMS
ethylmethane sulfonate
- FN
fast neutron
- SA
salicylic acid
- JA
jasmonic acid
- I/dSpm
Inhibitor/defective Suppressor.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF128407).
References
- 1.Baker B, Zambryski P, Staskawicz B, Dinesh-Kumar S P. Science. 1997;276:726–733. doi: 10.1126/science.276.5313.726. [DOI] [PubMed] [Google Scholar]
- 2.Delaney T P, Uknes S, Vernooij B, Friedrich L, Weymann K, Negrotto D, Gaffney T, Gut-Rella M, Kessmann H, Ward E, Ryals J. Science. 1994;266:1247–1250. doi: 10.1126/science.266.5188.1247. [DOI] [PubMed] [Google Scholar]
- 3.Mauch-Mani B, Slusarenko A J. Plant Cell. 1996;8:203–212. doi: 10.1105/tpc.8.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou J, Loh Y-T, Bressan R A, Martin G B. Cell. 1995;83:925–935. doi: 10.1016/0092-8674(95)90208-2. [DOI] [PubMed] [Google Scholar]
- 5.Hammond-Kosack K E, Jones J D G. Plant Cell. 1996;8:1773–1791. doi: 10.1105/tpc.8.10.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lamb C, Dixon R A. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:251–275. doi: 10.1146/annurev.arplant.48.1.251. [DOI] [PubMed] [Google Scholar]
- 7.Jones D A, Jones J D G. Adv Bot Res. 1996;24:89–167. [Google Scholar]
- 8.Medzhitov R, Preston-Hurlburt P, Janeway C A., Jr Nature (London) 1997;388:394–397. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- 9.Century K S, Shapiro A D, Repetti P P, Dahlbeck D, Holub E, Staskawicz B J. Science. 1997;278:1963–1965. doi: 10.1126/science.278.5345.1963. [DOI] [PubMed] [Google Scholar]
- 10.Parker J E, Holub E B, Frost L N, Falk A, Gunn N D, Daniels M J. Plant Cell. 1996;8:2033–2046. doi: 10.1105/tpc.8.11.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aarts N, Metz M, Holub E, Staskawicz B J, Daniels M J, Parker J E. Proc Natl Acad Sci USA. 1998;95:10306–10311. doi: 10.1073/pnas.95.17.10306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aarts M G M, Corzaan P, Stiekema W J, Periera A. Mol Gen Genet. 1995;247:555–564. doi: 10.1007/BF00290346. [DOI] [PubMed] [Google Scholar]
- 13.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 14.Parker J E, Coleman M J, Szabò V, Frost L N, Schmidt R, van der Biezen E A, Moores T, Dean C, Daniels M J, Jones J D G. Plant Cell. 1997;9:879–894. doi: 10.1105/tpc.9.6.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y-G, Whittier R F. Genomics. 1995;25:674–681. doi: 10.1016/0888-7543(95)80010-j. [DOI] [PubMed] [Google Scholar]
- 16.Holub E B, Beynon J L. Adv Bot Res. 1997;24:228–273. [Google Scholar]
- 17.Liu Y G, Mitsukawa N, Vasquez-Tello A, Whittier R F. Plant J. 1995;7:351–358. [Google Scholar]
- 18.Hinsch M, Staskawicz B. Mol Plant-Microbe Interact. 1996;9:55–61. doi: 10.1094/mpmi-9-0055. [DOI] [PubMed] [Google Scholar]
- 19.Innes R W, Bisgrove S R, Smith N M, Bent A F, Staskawicz B J, Liu Y-C. Plant J. 1993;4:813–820. doi: 10.1046/j.1365-313x.1993.04050813.x. [DOI] [PubMed] [Google Scholar]
- 20.Reuber T L, Ausubel F M. Plant Cell. 1996;8:241–249. doi: 10.1105/tpc.8.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uknes S, Mauch-Mani B, Moyer M, Potter S, Williams S, Dincher S, Chandler D, Slusarenko A, Ward E, Ryals J. Plant Cell. 1992;4:645–656. doi: 10.1105/tpc.4.6.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Penninckx I A M A, Eggermont K, Terras F R G, Thomma B P H J, De Samblanx G W, Buchala A, Métraux J-P, Manners J M, Broekhart W F. Plant Cell. 1996;8:2309–2323. doi: 10.1105/tpc.8.12.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dingwall C, Laskey R A. Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 24.Brady L, Brzozowski A M, Derewenda Z S, Dodson E, Dodson G, Tolley S, Turkenburg J P, Christiansen L, Huge-Jensen B, Norskov L, et al. Nature (London) 1990;343:767–770. doi: 10.1038/343767a0. [DOI] [PubMed] [Google Scholar]
- 25.Kugimiya W, Otani Y, Kohno M, Hashimoto Y. Biosci Biotechnol Biochem. 1992;56:716–719. doi: 10.1271/bbb.56.716. [DOI] [PubMed] [Google Scholar]
- 26.Derewenda Z. Adv Protein Chem. 1994;45:1–52. doi: 10.1016/s0065-3233(08)60637-3. [DOI] [PubMed] [Google Scholar]
- 27.Cousin X, Hotelier T, Giles K, Lievin P, Toutant J-P, Chatonnet A. Nucleic Acids Res. 1997;25:143–146. doi: 10.1093/nar/25.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schrag J D, Cyglar M. Methods Enzymol. 1997;284:107. [Google Scholar]
- 29.deVries R P, Michelsen B, Poulsen C H, Kroon P A, van den Heuvel R H, Faulds C B, Williamson G, van den Hombergh J P, Visser J. Appl Environ Microbiol. 1997;63:4638–4644. doi: 10.1128/aem.63.12.4638-4644.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Farmer E E, Ryan C A. Proc Natl Acad Sci USA. 1990;87:7713–7716. doi: 10.1073/pnas.87.19.7713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McConn M, Creelman R A, Bell E, Mullet J E, Browse J. Proc Natl Acad Sci USA. 1997;94:5473–5477. doi: 10.1073/pnas.94.10.5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bowling S A, Clarke J D, Liu Y, Klessig D F, Dong X. Plant Cell. 1997;9:1573–1584. doi: 10.1105/tpc.9.9.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vijayan P, Shockey J, Lévesque C A, Cook R J, Browse J. Proc Natl Acad Sci USA. 1998;95:7209–7214. doi: 10.1073/pnas.95.12.7209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shirasu K, Nakajima H, Krischnamachari Rajasekhar V, Dixon R A, Lamb C. Plant Cell. 1997;9:261–270. doi: 10.1105/tpc.9.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alvarez M E, Pennell R I, Meijer P J, Ishikawa A, Dixon R A, Lamb C. Cell. 1998;92:773–784. doi: 10.1016/s0092-8674(00)81405-1. [DOI] [PubMed] [Google Scholar]
- 36.Blechert S, Brodschelm W, Hölder S, Kammerer L, Kutchan T M, Mueller M J, Xia Z-Q, Zenk M H. Proc Natl Acad Sci USA. 1995;92:4099–4105. doi: 10.1073/pnas.92.10.4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weber H, Vick B A, Farmer E E. Proc Natl Acad Sci USA. 1997;94:10473–10478. doi: 10.1073/pnas.94.19.10473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brick D J, Brumlik M J, Buckley T, Cao J-X, Davies P C, Misra S, Tranbarger T J, Upton C. FEBS Lett. 1995;377:475–480. doi: 10.1016/0014-5793(95)01405-5. [DOI] [PubMed] [Google Scholar]
- 39.Baudouin E, Charpenteau M, Roby D, Marco Y, Ranjeva R, Ranty B. Eur J Biochem. 1997;248:700–706. doi: 10.1111/j.1432-1033.1997.t01-1-00700.x. [DOI] [PubMed] [Google Scholar]