Abstract
Three-dimensional microfluidic systems were fabricated and used to pattern proteins and mammalian cells on a planar substrate. The three-dimensional topology of the microfluidic network in the stamp makes this technique a versatile one with which to pattern multiple types of proteins and cells in complex, discontinuous structures on a surface. The channel structure, formed by the stamp when it is in contact with the surface of the substrate, limits migration and growth of cells in the channels. With the channel structure in contact with the surface, the cells stop dividing once they form a confluent layer. Removal of the stamp permits the cells to spread and divide.
This paper describes a versatile technique for fabricating complex patterns of biological materials and cells on surfaces by using three-dimensional (3D) microfluidic systems (1, 2). The ability to design and create biologically relevant patterns on surfaces provides new capabilities for cell biology, the production of biosensors, and tissue engineering (3, 4). To pattern biomaterials and cells, we use the collection of techniques referred to as soft lithography. This collection uses a transparent elastomer—poly(dimethylsiloxane) (PDMS)—patterned in bas relief, as a stamp or mold with which to transfer patterns to the surface of a substrate (4, 5). Most soft lithographic techniques—for example, microcontact printing (μCP) (6, 7) and micromolding in capillaries (MIMIC) (8–12)—have been limited to procedures that pattern one substance at a time (although the resulting pattern can be complex, as exemplified in patterns generated by μCP; ref. 7), or to relatively simple, continuous patterns (which may contain multiple materials, as exemplified by structures created by MIMIC, ref. 10). These constraints are both topographical and practical. The surface of a stamp in μCP, or of a channel system in MIMIC, is effectively a two-dimensional structure. In μCP, this two-dimensionality of the stamp limits the types of patterns that can be transferred to a single “color” of ink, in the absence of a way of selectively “inking” different regions of the stamp with different materials. Patterning of multiple “colors” using reported methods requires multiple steps of registration. In MIMIC, the two-dimensional channel system limits patterning to relatively simple, continuous structures.
To overcome the topographical constraint imposed by the quasi-two-dimensionality of these existing methods, we have developed a technique we call 3D MIMIC for fabricating complex, discontinuous patterns that incorporate multiple biomaterials and cells on planar surfaces. This method is based on a procedure described by Anderson et al. (1) for fabricating 3D microchannel systems and is related to work described by Beebe et al. (2) and others for 3D microfabrication. Although the potential value of 3D microfluidic systems is intuitively obvious (especially for procedures that require patterning liquids or require patterning with liquids), the complexity in the fabrication of such systems has deterred their use. A topological analysis of the complexity in the system of channels required to fabricate arbitrary patterns has shown that two layers of interconnected channels in the stamp gives sufficient versatility for the generation of any arbitrary two-dimensional pattern (1). To summarize this analysis, we note that the projection of 3D microfluidic channels on a surface is analogous to the representation of knots in a plane. Any knot can be decomposed into a series of crossing of lines, in which one line goes either “over” or “under” the other. The ability to fabricate channel systems in which one channel can be made arbitrarily to go over or under a second therefore provides the capability for generating channel systems of arbitrary complexity. The advantage to using a 3D stamp (which requires two levels of fabrication and one step of registration) relative to multiple, registered steps of printing is that in the former, the registration step need be carried out only once, and the resulting stamp can be used many times to print patterns without registration, whereas in the latter, registration is required each time a substrate is printed. The fabrication of a 3D stamp requires only one more step of pattern fabrication, one step of alignment, and one step of sealing, relative to the fabrication of a conventional two-dimensional stamp. This process makes 3D MIMIC a powerful complement to and extension of the existing techniques of soft lithography.
Materials and Methods
Cell Cultures.
Bovine adrenal capillary endothelial cells (BCEs) were cultured as described (13). In brief, BCEs were grown in low glucose DMEM supplemented with 10% calf serum and 2 ng/ml basic fibroblast growth factor (bFGF), and kept in 10% CO2 atmosphere. Human bladder cancer cells (ECVs) from the ECV304 cell line were cultured in DMEM supplemented with 10% FBS and kept at 5% CO2. Cells from both cell types were labeled fluorescently before harvest at 37°C in the CO2 incubator. BCEs were incubated with 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine (DiI)-conjugated acetylated low-density lipoprotein at 4 μg/ml, which is actively taken up by BCEs and stored in endosomal granula (14). ECV304 cells were incubated with 5 μM 5-chloromethylfluorescein diacetate (CMFDA), which reacts with intracellular glutathione. Before patterning, cells were washed with PBS, dissociated from culture plates with trypsin/EDTA, washed with DMEM, and resuspended in the respective culture media at a density of ≈ 106 cells/ml. For culturing patterned cells (both BCEs and ECVs) after removal of the PDMS stamp, DMEM supplemented with 5% calf serum, 5% FBS, and 2 ng/ml bFGF was used, and the cells were kept at 10% CO2.
Chemicals and Proteins.
BSA and PBS were obtained from Sigma. DMEM was purchased from JRH Biosciences (Lenexa, KS), basic fibroblast growth factor from Takeda (Osaka), and DiI-conjugated acetylated low-density lipoprotein from Biomedical Technologies (Stoughton, MA). FITC, fibrinogen, and CMFDA were obtained from Molecular Probes. PDMS prepolymer (Sylgard 184) was from Dow-Corning, Photoresist (SU 8–50) from MicroChem (Newton, MA), and silicon wafers from Silicon Sense (Nashua, NH).
Results and Discussion
Fabrication of 3D Microfluidic Stamps.
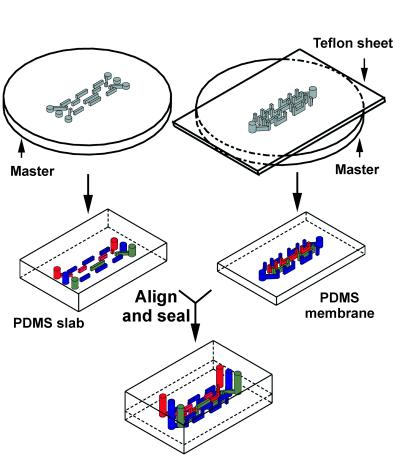
Our method for the fabrication of 3D microchannels has been described in detail elsewhere (1), and here we emphasize those features relevant to biological applications. The overall process requires five steps: (i) fabrication of silicon master for the top PDMS slab; this master has positive relief patterns of photoresist on Si and contains one plane of features, (ii) fabrication of a photoresist-patterned silicon master for the bottom of the PDMS membrane, using two-step photolithography (this master contains two planes of features), (iii) fabrication of the PDMS slab that forms the top layer of the structure, (iv) fabrication of PDMS membrane (bottom layer), and (v) alignment and sealing of the two PDMS structures fabricated in steps iii and iv. Once the masters for the top and bottom layers were made, fabrication of the 3D PDMS stamp requires repetition only of steps iii to v.
The top PDMS slab (left-hand side of Fig. 1) was fabricated by molding PDMS against the silicon master made in the first step. To fabricate the PDMS membrane, we sandwiched a drop of PDMS prepolymer between the master (made in step ii) and a Teflon sheet, and allowed it to cure under pressure at 70°C. Pressures in the range of 10 kPa to 50 kPa were required to ensure that the vias (the vertical channels) were not blocked by a thin underlayer of PDMS that seeped between the Teflon and the features during curing. Once the PDMS had cured, we peeled the Teflon sheet away, leaving the membrane remained attached to the wafer by van der Waals interactions. The useful difference between the strength of adhesion of the PDMS membrane to the Teflon and that to the silicon master rests on the very low interfacial free energy of Teflon. Because the membrane is never freestanding and is supported at all times during fabrication by the wafer, the features on the membrane were not distorted during handling.
Figure 1.
Scheme for the fabrication of 3D-microfluidic stamp. The master in the bottom layer required two steps of photolithography, whereas the master for the top layer was fabricated with a single step. The top layer and the bottom membrane were molded independently. To ensure the vias (vertical channels) in the membrane went through the entire thickness of the membrane, pressure (10–50 kPa) was applied to the Teflon sheet during curing of the PDMS. Once the two PDMS layers were cured, the two pieces were prealigned on an alignment stage, then oxidized for 40 sec in an oxygen plasma. Sealing was accomplished by bringing the oxidized surfaces of the two PDMS layers into contact.
The membrane has two levels of structures. One level provides one plane of channels; this plane is open for contact with the substrate that is to be patterned. The second level provides the vias that connect these channels in the membrane to those in the slab. There are, therefore, three planes of features in the final 3D-microfluidic stamps; this architecture is sufficient to represent any over and under crossover structures in the microchannels (1).
To align the two PDMS layers before sealing, we used a home-built micromanipulator stage: the two layers of PDMS were mounted on the alignment stage, facing each other, then brought into close proximity and aligned. After alignment, one membrane was backed away a few millimeters from the other by using the micromanipulator. The entire alignment stage was placed in a plasma cleaner (Anatech, model SP100 Plasma System, Springfield, VA) and oxidized for 40 sec in an oxygen plasma. The yield in sealing two pieces of PDMS after plasma oxidation (power: 60 W, duration: 40 sec, ≈ 0.2 Torr oxygen) was 100%. The millimeter gap between the two PDMS layers allowed for the efficient oxidation of the exposed PDMS surface. Sealing of the two layers was accomplished by removing the assembly from the plasma cleaner and immediately bringing the two oxidized PDMS surfaces into contact (15).
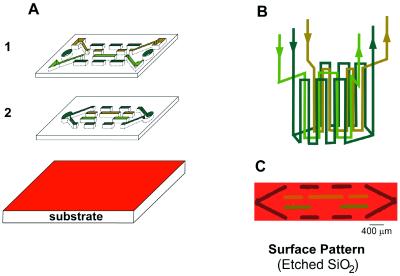
A Test System: Etching of a Si/SiO2 Wafer and Visualization Using Optical Interference Colors.
Before patterning biomaterials, we studied the etching of an oxidized Si wafer by buffered hydrofluoric acid (HF). We had two reasons for choosing to etch a Si/SiO2 wafer: (i) we could use optical interference colors to visualize easily the homogeneity in etching, and thus to infer qualitatively the flow profile near the surface, and (ii) we could use this selective etching of SiO2 surface to demonstrate the potential of using 3D MIMIC for altering the local topology of the substrate. Each thickness of the SiO2 layer on Si corresponds to a characteristic color, owing to interference between light reflected from the SiO2 surface and that from the SiO2/Si interface. This optical interference color is very sensitive to the thickness of the SiO2 layer: a difference in 30 nm (from 340 nm to 310 nm), for example, changes the color from light green to blue (16). This interference gave rise to the red-colored (red corresponds to ≈ 630 nm of SiO2 on Si) background in Fig. 2C. We used this optical interference to visualize the differences between the amount of SiO2 etched away in the three different sets of channels: ≈520 nm of SiO2 corresponds to green, ≈390 nm to yellow, and ≈70 nm to brown (16). The color from each set of channels remains constant in the pattern; this constancy indicates the etching was relatively homogeneous within each set of channels.
Figure 2.
(A and B) Schematic depicting the flow patterns for the 3D stamp of Fig. 1. To create the etched patterns in C, this stamp was first sealed conformally to a Si/SiO2 wafer with gentle pressure. Three different concentrations of buffered HF (10%, 5%, 3% of HF) were allowed to flow through the channels. Where the HF was in contact with the surface, it etched away the SiO2. The differences in the thickness of the SiO2 layer gave rise to the different interference colors in the etched pattern. These colors are caused by interference between light reflected from the air/SiO2 interface and that from the Si/SiO2 interface; they reveal the depth of etching.
To create the etched pattern shown in Fig. 2C, the microfluidic channel structure first was sealed conformally to the Si/SiO2 wafer with gentle pressure. Three aqueous solutions containing three different concentrations of HF (10%, 5%, and 3% HF, buffered at ≈ pH 5 with 6:1 ratio of NH4F/HF) were allowed to flow (<1 cm/sec in channels with a cross-sectional area of 500 μm2) in the three sets of channel structures (Fig. 2B). Where the HF solutions came into contact with the surface, they etched away the SiO2; the rate of etching SiO2 for 10% HF is ≈20 nm/min.
While filling the channels with aqueous solutions, we often observed bubble formation in the channels. The hydrophobic nature of the PDMS channel walls (θaH2O≈ 108o) required the application of vacuum at the outlet to assist solution flow; the reduced pressure caused air dissolved in the buffer to nucleate bubbles. We overcame this problem by absorbing onto the channel walls either charged polymers, poly(diallyldimethylammonium chloride) with an average molecular mass between ≈ 400 kDa and 500 kDa, or, for patterning cells, BSA. The walls of the channels also can be rendered hydrophilic by oxidation in a plasma cleaner for ≈ 1 min (15). This oxidation procedure, however, is not convenient for most applications because the oxidized PDMS surface slowly becomes hydrophobic again over several hours. Although degassing the solution also should overcome this bubble-formation problem, this procedure is not compatible with culture medium containing cells: both the degassing procedure and the lack of dissolved CO2 might compromise the viability of cells cultured in media containing bicarbonate buffers.
Patterned Delivery of Proteins.
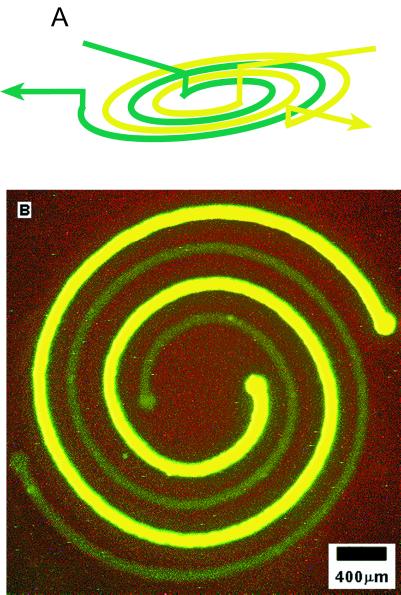
To demonstrate the capability of 3D MIMIC to generate complex patterns, we designed and fabricated a two-layer microfluidic system for delivering two proteins in a nested spiral arrangement. It would be difficult to create this pattern with μCP because it would be difficult to ink differentially the two spirals (which are separated by only 200 μm). The spiral that is more fluorescent in Fig. 3 is BSA, a protein that we and others use routinely to inhibit cell attachment to surfaces; the one that is less fluorescent is fibrinogen, a protein that promotes cell attachment. This nested spiral was patterned in four steps: (i) the stamp containing the channel system suggested by Fig. 3A was placed in contact with the surface of a polystyrene Petri dish; (ii) the two channels were filled with BSA and fibrinogen both labeled by reaction with FITC; (iii) the proteins were allowed to adsorb onto the surface of the polystyrene for 45 min; and (iv) the channels were flushed thoroughly with phosphate buffer, and the stamp was peeled away. The BSA spiral is more fluorescent because the concentration of BSA used (1 mg per ml of phosphate buffer) was higher than that of fibrinogen (0.1 mg per ml of phosphate buffer).
Figure 3.
Fluorescence photograph (B) showing nested spirals of two proteins on the surface of a polystyrene Petri dish patterned with the 3D stamp depicted in A. The bright green spiral is BSA; the light green one is fibrinogen. Both proteins were labeled with FITC. The BSA concentration used (1 mg/ml) was approximately 10 times higher than that of fibrinogen (0.1 mg/ml). To create this nested spiral pattern, the PDMS stamp (A) first was placed conformally to the polystyrene surface of the Petri dish. The two nested spiral channels then were filled with FITC-tagged BSA and fibrinogen in phosphate buffer (pH 7.4), respectively. The proteins were allowed to adsorb onto the polystyrene surface for ≈ 45 min. The channels were flushed thoroughly with phosphate buffer (pH 7.4); the stamp was peeled off; the picture was taken.
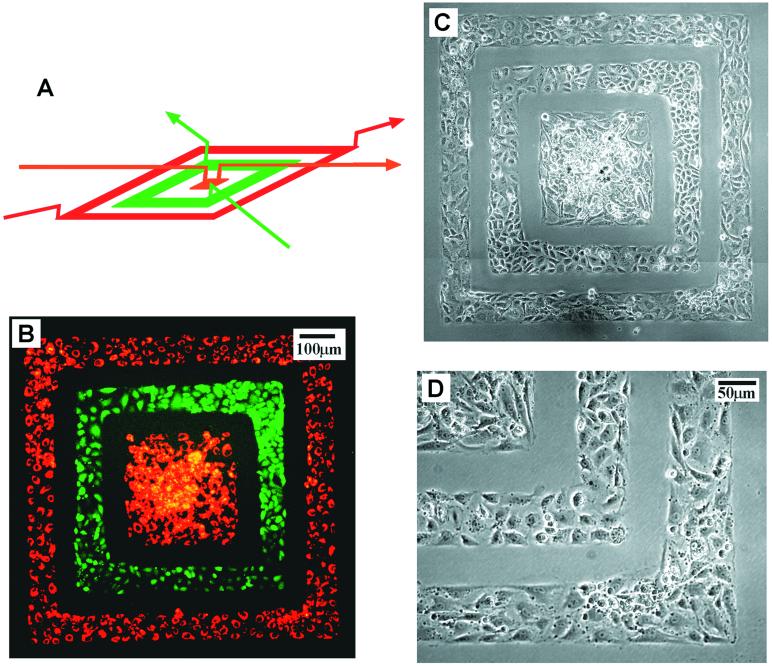
Patterning Cells.
We fabricated a two-layer stamp for the deposition of two cell types in a concentric square pattern (Fig. 4). We designed this discontinuous, concentric pattern to demonstrate our ability to handle thin PDMS membranes without distortion, because the membrane was supported and attached to the wafer (the master with features) by van der Waals interactions at all times. It also would be difficult to use either μCP (6, 7) or MIMIC (8–12) to deliver multiple proteins or cell types in this discontinuous pattern. To deposit cells in this pattern, we: (i) autoclaved the PDMS stamp for 20 min, (ii) coated the channel walls with BSA, (iii) placed the stamp in contact with the substrate to be patterned, (iv) filled the channels with a suspension of cells (≈ 5 million cells per ml) and let the cells sediment and attach to the surface, and (v) let the cells grow and spread for 24–48 hr inside the PDMS microchannels. The cells we used were BCEs and a ECV line (ECV-304). Before being deposited, the BCEs were labeled with DiI-conjugated acetylated low-density lipoprotein, which was actively taken up by BCEs and stored in endosomal granula, and the ECVs with CMFDA, which reacted with intracellular glutathione.
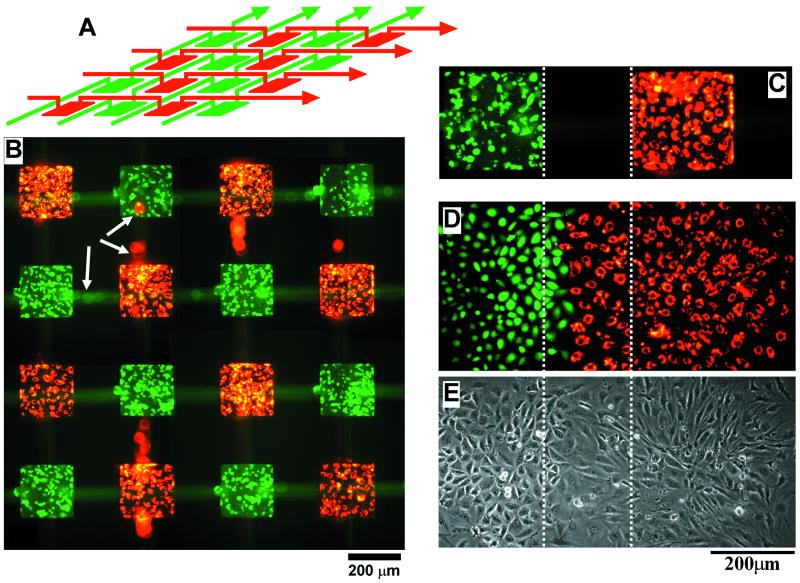
Figure 4.
Fluorescence (B) and phase-contrast (C and D) pictures of two cell types deposited on a tissue culture dish in a concentric square pattern by using the 3D stamp depicted in A. The cells that appear green are ECVs labeled with CMFDA; the cells that appear red are BCEs labeled with DiI-conjugated acetylated low density lipoprotein. These cells were labeled fluorescently before being deposited. Before patterning cells, the PDMS stamp first was autoclaved for 20 min, then the channel walls were coated with BSA (30 mg/ml in pH 7.4 phosphate buffer for 1 hr) for preventing cell attachment to the PDMS stamp. Suspensions of cells (≈ 5 million cells per ml) were introduced into the three sets of channels and were allowed to sediment and attach to the surface of the tissue culture dish. These cells were cultured with the stamp in place for ≈24 hr to grow and spread into a confluent layer. The pictures were taken immediately after the PDMS stamp was removed; these cells were immersed under media and were alive. An expanded view of the lower right corner of C is shown in D.
We found autoclaving was necessary to improve the viability of cells inside the PDMS stamp: without autoclaving, the cells often died after patterning and incubation; with autoclaving, the probability that the cells spread and grow into a confluent layer within the stamp was dramatically improved. Coating of the channels with 3% BSA in PBS prevented cell attachment to the PDMS wall during peel-off; BSA is a protein used commonly for rendering surfaces nonadhesive to cells. The cell types we used typically required 24–48 hr, depending on the initial cell density, to attach, spread, and grow into a confluent layer inside the channel patterns. For the concentric pattern (Fig. 4), we cultured the cells for ≈24 hr, with the PDMS stamp in place, to form a confluent layer of cells.
Fig. 5B shows two cell types deposited in a chessboard-like pattern. We designed this pattern as a simple demonstration of the potential to deposit multiple cell types in an array format appropriate for a biosensor or drug screening. In this type of array, the responding cells could be identified by their spatial locations. We believe that it would be possible, in principle, to pattern 200 or 300 different cell types in this array format. The fabrication of the 3D stamp for this large-area patterning, however, would be more complex than fabricating the stamp used for small-area patterning. PDMS is an elastomer, and different pieces of PDMS will shrink differently depending on the curing condition and the amount of cross-linking catalysts used. This difference in shrinkage between pieces of PDMS will make the alignment and sealing of PDMS membranes across a large area more complicated than across a small area. There are, however, approximately 250 basic cell types in human (17). For most applications, only a subset of these cell types would be required. Thirty-six cell types, for example, could be patterned in a 6-by-6 array; this type of array would require a fluidic stamp that would be within the capability of our current procedure to fabricate.
Figure 5.
Fluorescence picture (B) of CMFDA-labeled ECVs (green) and DiI-labeled BCEs (red) deposited in a chessboard-like pattern by using the stamp design in A. To show the overlaying weaving channel structures, the PDMS stamp was not removed before taking the picture in B. The white arrows indicate cells that were inside the channels in the top layer of the PDMS stamp; these cells appear blurred because they were above the focal plane of the microscope. The cells in B were cultured for 42 hr before the fluorescence picture was taken. (C) A picture of a confluent layer of cells before the PDMS stamp was removed; this image is slightly diffuse because the PDMS stamp might have caused optical aberrations. Pictures taken in fluorescence (D) and in phase contrast (E) show the spreading and growth of the two cell types after the removal of the PDMS stamp. The pictures were taken 20 hr after removal of the stamp. The three images in C–E are registered; the dotted lines show the relative orientation of the patterns. The BCEs spread more rapidly than the ECVs by a factor of 2–3.
The chessboard pattern (Fig. 5B) was produced with the same procedure used for patterning the concentric square pattern (Fig. 4). The two cell types used, BCEs and ECVs, were fluorescently labeled before being deposited. These cells were cultured for 42 hr before a confluent layer of cells were formed. The fluorescence picture (Fig. 5B) was taken with the PDMS stamp still in place to show the overlaying weaving channel structures. The blurred red and green spots (pointed to by white arrows in Fig. 5B) were cells in the channel structure (top layer) above the focal plane of the microscope.
Fig. 5 C–E demonstrates cell viability and spreading after removal of the PDMS stamp. We first let the patterned cells grow and spread into a confluent layer within the confines of the PDMS stamp (Fig. 5C). We then removed the PDMS stamp to let the two cell types grow and spread together (Fig. 5 D and E). Fig. 5 D and E shows pictures of the same region, one was taken in fluorescence (Fig. 5D) and the other in phase contrast (Fig. 5E). The fluorescence picture in Fig. 5C appears slightly blurred because the picture was taken with the PDMS stamp in place; this complexity in the optical field might have caused optical aberrations. We chose BCEs and ECVs for this spreading experiment to demonstrate the potential to study angiogenesis during tumor formation, because in vivo ECVs (tumor cells) attract and direct the growth of BCEs (capillary cells) to form new blood vessels to supply nutrients and oxygen for tumor growth. With a defined pattern of BCEs, ECVs, and other tumor cells, we might study the differential and competitive attraction of capillary endothelial cells to different tumor cell lines. This technique could lead to the development of a simple, standardized, and quantitative in vitro assay for comparing the angiogenic potential of tumor cells.
We expect 3D MIMIC to complement existing soft lithographic techniques for the patterning of cells and other biomaterials. Although fabrication of the PDMS stamp is more complex for 3D MIMIC than fabrication of simpler structures for other soft lithographic methods, the capability to deposit multiple cell types or proteins in complex, discontinuous patterns makes 3D MIMIC a powerful technique in situations where multiple cell types are required. Tissues of higher organism exhibit a distinct micro-architecture defined by the topological relationship between different cell types. The ability to pattern deposit different cell types, in close proximity and in well-defined structures, will pave the way for studying the functional significance of tissue architecture at the resolution of individual cells, and the molecular interactions between cell types that underlie processes such as embryonic morphogenesis, formation of the blood–brain barrier, and tumor angiogenesis.
Acknowledgments
C.J.W. thanks the Materials Research Science and Engineering Centers for a summer undergraduate internship. This work is supported by Defense Advanced Research Planning Agency/Air Force Research Laboratory/Space and Naval Warfare Systems Command and National Science Foundation Grant ECS-9729405 and National Science Foundation Grant DMR 9809363 MRSEC.
Abbreviations
- 3D
three-dimensional
- μCP
microcontact printing
- MIMIC
micromolding in capillaries
- BCE
bovine capillary endothelial
- DiI
1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine
- PDMS
poly(dimethylsiloxane)
- CMFDA
5-chloromethylfluorescein diacetate
- HF
hydrofluoric acid
- ECV
human bladder cancer cell
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.040562297.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.040562297
References
- 1.Anderson, J. R., Chiu, D. T., Jackman, R. J., Cherniavskaya, O., McDonald, J. C., Wu, H., Whitesides, S. H. & Whitesides, G. M. (2000) Anal. Chem, in press. [DOI] [PubMed]
- 2.Jo, B. H., VanLerberghe, L. M., Motsegood, K. M. & Beebe, D. J. (2000) J. Microelectromech. Syst., in press.
- 3.Kovacs G T A. Micromachined Transducers Sourcebook. Boston: WCB/McGraw-Hill; 1998. [Google Scholar]
- 4.Kane R S, Takayama S, Ostuni E, Ingber D E, Whitesides G M. Biomaterials. 1999;20:2363–2376. doi: 10.1016/s0142-9612(99)00165-9. [DOI] [PubMed] [Google Scholar]
- 5.Xia Y, Whitesides G M. Angew Chem Int Ed Engl. 1998;37:551–575. doi: 10.1002/(SICI)1521-3773(19980316)37:5<550::AID-ANIE550>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 6.Chen C S, Mrksich M, Huang S, Whitesides G M, Ingber D E. Science. 1997;276:1425–1428. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- 7.Bernard A, Delamarche E, Schmid H, Michel B, Bosshard H R, Biebuyck H. Langmuir. 1998;14:2225–2229. [Google Scholar]
- 8.Jeon N L, Choi I S, Xu B, Whitesides G M. Adv Mat. 1999;11:946–949. [Google Scholar]
- 9.Delamarche E, Bernard A, Schmid H, Michel B, Biebuyck H. Science. 1997;276:779–781. doi: 10.1126/science.276.5313.779. [DOI] [PubMed] [Google Scholar]
- 10.Delamarche E, Bernard A, Schmid H, Bietsch A, Michel B, Biebuyck H. J Am Chem Soc. 1998;120:500–508. [Google Scholar]
- 11.Folch A, Ayon A, Hurtado O, Schmidt M A, Toner M. J Biomech Eng. 1999;121:28–34. doi: 10.1115/1.2798038. [DOI] [PubMed] [Google Scholar]
- 12.Folch A, Toner M. Biotech Prog. 1998;14:388–392. doi: 10.1021/bp980037b. [DOI] [PubMed] [Google Scholar]
- 13.Folkman J, Haudenschild C C, Zetter B R. Proc Natl Acad Sci USA. 1982;76:5217–5221. doi: 10.1073/pnas.76.10.5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Voyta J C, Via D P, Butterfield C E, Zetter B R. J Cell Biol. 1984;99:2034–2040. doi: 10.1083/jcb.99.6.2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duffy D C, McDonald J C, Schueller O J A, Whitesides G M. Anal Chem. 1998;70:4974–4984. doi: 10.1021/ac980656z. [DOI] [PubMed] [Google Scholar]
- 16.Anner G E. Planar Processing Primer. New York: Van Nostrand Reinhold; 1990. [Google Scholar]
- 17.Alberts A, Bray D, Lewis J, Raff M, Roberts K, Watson J D. Molecular Biology of the Cell. New York: Garland; 1983. [Google Scholar]