Abstract
A method for the rapid and efficient identification of ligands to biological targets is reported. The combinatorial method does not require structural or mechanistic information and is accomplished in four straightforward steps. (i) A set of potential binding elements is prepared wherein each molecule incorporates a common chemical linkage group. (ii) The set of potential binding elements is screened to identify all binding elements that interact even weakly with the biological target. (iii) A combinatorial library of linked binding elements is prepared whereby the binding elements are connected by the common chemical linkage groups through a set of flexible linkers. (iv) The combinatorial library is screened to identify the tightest-binding ligands. The utility of the method was demonstrated by the identification of a potent and subtype-selective small molecule inhibitor of the non-receptor tyrosine kinase c-Src (IC50 = 64 nM). Because the method relies on connecting two distinct binding elements, the relative contributions of the two binding elements to the potency and selectivity of the inhibitor were readily determined. This information provides valuable insight into the molecular basis of inhibition.
Combinatorial methods for generating small molecule libraries coupled with high-throughput screening have become core technologies for the identification of small molecule ligands to receptors and enzymes (1–4). The identified ligands serve as powerful tools for pharmacological studies and are essential to drug development. Combinatorial approaches have been most successful when information has been used to design the library of molecules to be prepared and tested. In these efforts, libraries are designed by using knowledge of the mechanism or structure of the biological target (5–7), or by basing the library upon lead compound(s) that have previously been identified to bind to the biological target (8, 9). Unfortunately, for many biological targets structural or mechanistic information is not available or does not provide sufficient insight to enable productive library design. Additionally, for many targets, lead compounds have not yet been identified or novel motifs for binding are desired. Not surprisingly, under these circumstances the preparation and screening of libraries has been much less successful, because we can prepare and test only an infinitesimally small fraction of the more than 1060 small molecules that could theoretically be prepared (10).
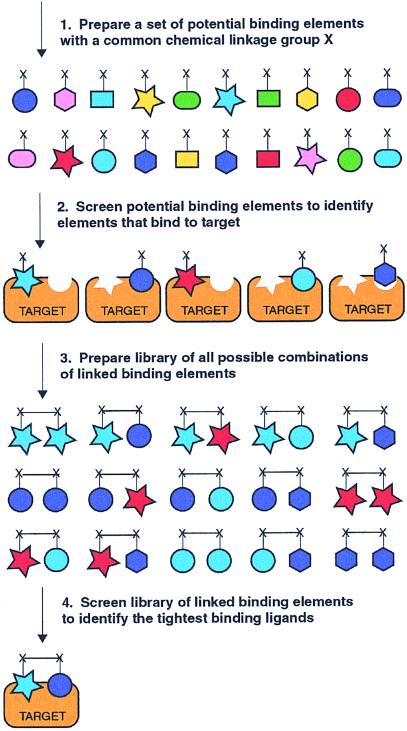
Herein, we report a powerful approach to rapidly identify small molecule ligands to biological targets. The method involves four sequential straightforward steps and does not rely on lead compounds, nor does it require knowledge of the mechanism or structure of the biological target (Fig. 1). (1) A set of potential binding elements is prepared wherein each molecule of the set must be soluble in aqueous solution at high concentrations and must incorporate a common chemical linkage group. (2) The set of potential binding elements is screened at high concentrations (≥1 mM) to identify all binding elements that interact even weakly with the biological target. (3) A combinatorial library of linked binding elements is prepared whereby the binding elements are connected by using the common chemical linkage groups through a set of flexible linkers. (4) The combinatorial library of linked binding elements is screened to identify the tightest-binding ligands.
Figure 1.
Combinatorial target-guided ligand assembly.
There are two important advantages of the reported method relative to traditional combinatorial small molecule library approaches. First, the number of compounds that must be prepared is greatly reduced because the binding elements used in the preparation of the combinatorial library are preselected by screening a much larger set of potential binding elements. Second, to increase the likelihood that both binding elements interact with the target, the binding elements are combined through a set of flexible linkers with different chain lengths instead of by using a single template as is often employed in traditional combinatorial methods.
Structure–activity relationships by nuclear magnetic resonance (SAR by NMR), developed by Fesik at Abbott Laboratories, is also based on the target-assisted selection of binding elements wherein compounds are screened by NMR for binding to an 15N-labeled protein (11, 12). Those compounds that are identified by NMR to bind in close proximity must then be chemically modified to link them together to provide high-affinity ligands. The method reported here, in contrast to SAR by NMR, does not require structural information. Instead, the common chemical linkage group present in each binding element enables a library of all combinations of binding elements and linkers to be rapidly and efficiently prepared and screened. This allows identification of both the most productive combinations of binding elements and the appropriate linkages necessary to achieve tight binding.
The utility of the described method is demonstrated by the identification of novel, potent, and subtype-selective inhibitors of the tyrosine kinase c-Src, which has wide-ranging biological functions and serves as the archetypal example of the large class of tyrosine kinases (13). Although a number of heterocyclic inhibitors have been developed that interact with the ATP-binding site of c-Src (14–17), achieving selectivity, particularly among Src family members, has proven to be a considerable challenge (18, 19). Notably, because this method relies on the appropriate linkage of distinct binding elements, the relative contributions of the individual binding elements to both the potency and selectivity of the identified inhibitor could readily be determined. This information provides significant insight into both the potency and the selectivity of the inhibitors.
Materials and Methods
Reagents and General Methods.
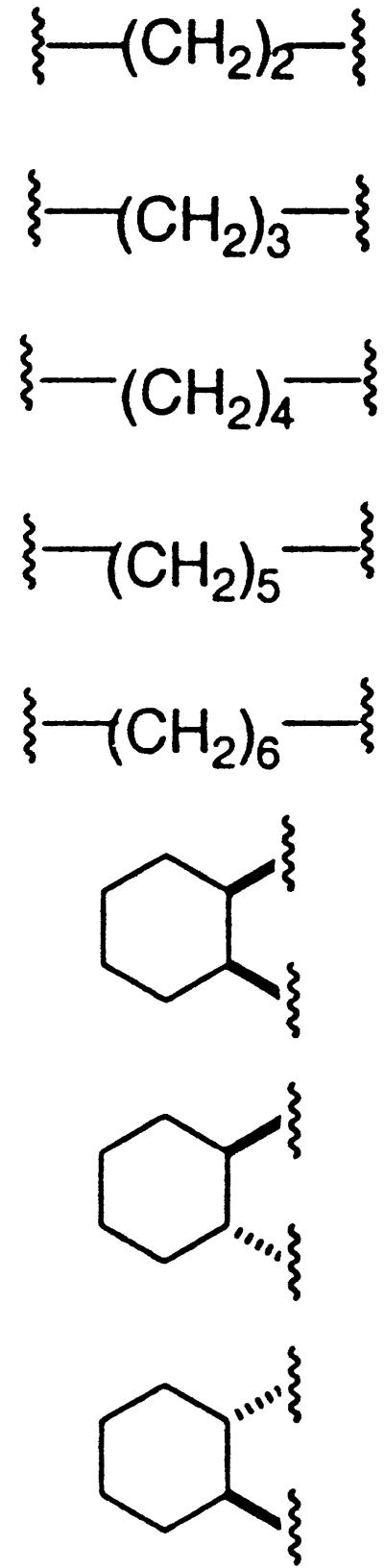
All starting materials and synthetic reagents were purchased from commercial suppliers unless otherwise noted. O,O′-Diaminoalkanediol linkers (ethyl, propyl, butyl, pentyl, hexyl) were prepared from the corresponding dibromoalkanes in two steps according to a modified procedure of Kim et al. (20). Constrained O,O′-diaminoalkanediol linkers [trans (1S,2S), trans (1R,2R), cis] were prepared from the optically pure alcohols according to a published procedure (21).
Potential Binding Element Synthesis.
A set of 305 aldehydes was selected for library synthesis on the basis of reaction compatibility, diversity, and cost. To each well of a 2-ml Beckman microtiter plate was added a DMSO stock solution of a unique aldehyde (190 μl, 0.15 M, 0.29 mmol), O-methylhydroxylamine (83 μl, 0.50 M, 0.42 mmol), and AcOH (23 μl, 0.50 M, 0.12 mmol). The plates were sealed and agitated overnight, after which they were stored at −20°C.
Linked Binding Element Synthesis (Mixture of 5 Linkers per Well).
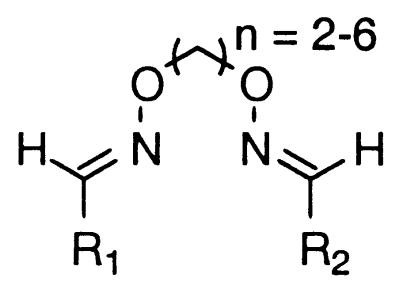
To each well of a 2-ml Beckman microtiter plate was added a DMSO stock solution of aldehydes A and B (45 μl, 0.15 M, 6.8 μmol each of A and B), an equimolar mixture of O,O′-diaminoalkanediol linkers (n = 2–6) (50 μl, 0.15 M, 1.4 μmol per linker), and AcOH (10 μl, 0.50 M, 5.0 μmol). The plates were sealed with Parafilm and agitated at room temperature overnight, after which they were stored at −20°C.
Oxime formation has previously been shown to be an efficient means of chemical ligation for biopolymer synthesis (22–27) and for combinatorial library preparation (28). To rigorously assess the synthesis procedure for preparing the library of linked binding elements, several control experiments were performed. According to the library synthesis procedure, the combination of two different aldehydes and five O,O′-diaminoalkanediol linkers should produce 15 different linked binding elements. The presence of all 15 linked binding elements was confirmed for five different aldehyde combinations employing aldehydes with different electronic properties, [27, 68, n = 2–6], [41, 68, n = 2–6], [68, 71, n = 2–6], [68, 265, n = 2–6], and [68, 281, n = 2–6] (see Figs. 4 and 5).† For each aldehyde combination flash chromatography was employed to separate the heterodimers (n = 2–6) from the two homodimers (n = 2–6). For each aldehyde combination the overall yield of the pure heterodimeric and the homodimeric binding elements exceeds 90%, confirming the high efficiency of the synthesis. In addition, for each aldehyde combination, a narrow range in the isolated yields of the heterodimers (n = 2–6) and homodimers (n = 2–6) was observed 42–52% and 22–29%, respectively. These yields correlate well with the expected 2:1:1 heterodimer:homodimer:homodimer statistical mixture. For every heterodimer mixture and homodimer mixture, a significant percentage of each of the five linker lengths (n = 2–6) was observed by electrospray mass spectrometry, clearly demonstrating that all 15 compounds were prepared for each aldehyde combination.
Figure 4.
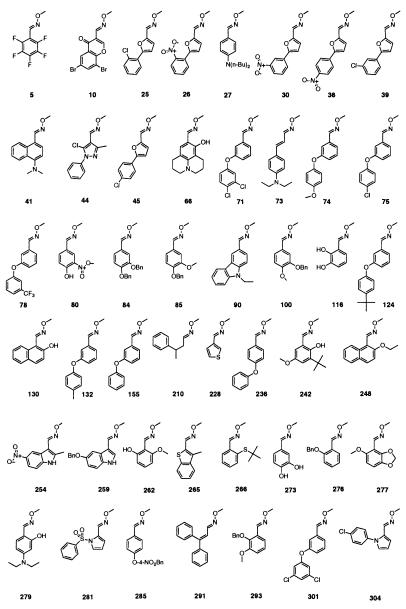
Structure of 47 binding elements that inhibit c-Src >70% at 500 μM. Bn, CH2C6H5.
Figure 5.
Comparison of individual binding elements to linked binding elements.
General Procedure for Scale-up Synthesis of Binding Elements.
To a 25-ml round-bottomed flask, aldehyde (1.0 mmol, 1.0 eq) was added, followed by the addition of 10 ml of anhydrous dimethylformamide. To this mixture was added O-methylhydroxylamine hydrochloride (3.0 mmol, 3.0 eq) followed by pyridine (3.0 mmol, 3.0 eq). The reaction solution was stirred under N2 at room temperature for 12 h. The solvent was evaporated under reduced pressure, and the solid residue was redissolved in EtOAc (50 ml), washed twice with 1 M HCl, and dried over Na2SO4. The organic extract was concentrated under reduced pressure, and the resultant solid was purified by silica gel column chromatography (EtOAc/hexanes).
O-Methyl oxime of 9-ethyl-3-carbazolecarboxaldehyde [90].
1H NMR (300 MHz, CDCl3) δ 1.44 (t, J = 7.2 Hz, 3H), 4.10 (s, 3H), 4.37 (q, J = 7.2 Hz, 2H), 7.26–7.32 (m, 1H), 7.36–7.43 (m, 2H), 7.48–7.53 (m, 1H), 7.74 (dd, J = 8.4, 1.6 Hz, 1H), 8.13 (d, J = 7.8 Hz, 1H), 8.27 (s, 1H), 8.29 (d, J = 1.6 Hz, 1H); 13C NMR (100.6 MHz, CDCl3) δ 13.9, 37.7, 61.9, 108.7, 108.8, 119.4, 119.8, 120.7, 122.9, 123.1, 123.2, 124.7, 126.2, 140.4, 140.9, 149.8. Analysis. Calcd for C16H16N2O: C, 76.16; H, 6.39; N, 11.1. Found: C, 76.49; H, 6.26; N, 11.01.
O-Methyl oxime of 3,4-dihydroxybenzaldehyde [273].
1H NMR (300 MHz, CDCl3) δ 3.94 (s, 3H), 5.62 (br s, 1H), 5.73 (br s, 1H), 6.85 (d, J = 8.2 Hz, 1H), 6.97 (dd, J = 8.2, 1.9 Hz, 1H), 7.19 (d, J = 1.9 Hz, 1H), 7.95 (s, 1H); 13C NMR (100.6 MHz, CDCl3) δ 61.7, 113.2, 115.5, 121.7, 124.3, 144.1, 146.3, 149.8. Analysis. Calcd for C9H9NO3: C, 57.48; H, 5.43; N, 8.38. Found: C, 57.60; H, 5.33; N, 8.46.
General Procedure for Scale-up Synthesis of Linked Binding Elements.
Aldehyde A (1.03 mmol) and aldehyde B (1.03 mmol) were added to a 10-ml flask, followed by 5 ml of dimethylformamide. To the reaction mixture was added O,O′-diaminoalkanediol linker (1.13 mmol) and AcOH (50 μl). The reaction mixture was stirred for 12 h at room temperature, after which the solvent was evaporated under reduced pressure. The heterodimer was separated from the homodimers by silica gel column chromatography.
[90, 273, n = 2].
1H NMR (300 MHz, CDCl3) δ 1.44 (t, J = 7.2 Hz, 3H), 4.37 (q, J = 7.2 Hz, 2H), 4.47–4.52 (m, 4H), 5.70 (br s, 1H), 5.83 (br s, 1H), 6.83 (d, J = 8.2 Hz, 1H), 6.91 (dd, J = 8.2, 1.9 Hz, 1H) 7.12 (d, J = 1.9 Hz, 1H), 7.23–7.28 (m, 1H), 7.36–7.41 (m, 2H), 7.46–7.52 (m, 1H), 7.72 (dd, J = 8.4, 1.6 Hz, 1H), 7.98 (s, 1H), 8.07 (d, J = 7.8 Hz, 1H), 8.25 (d, J = 1.6 Hz, 1H), 8.33 (s, 1H); 13C NMR (100.6 MHz, DMSO-d6) δ 14.2, 37.6, 72.2, 72.3, 109.9, 110.0, 113.4, 116.1, 119.8, 120.2, 120.4, 121.0, 122.5, 122.8, 123.2, 123.7, 124.9, 126.7, 140.4, 140.9, 146.1, 148.2, 149.6, 150.3. High-resolution mass spectrometry (fast-atom bombardment) [HRMS (FAB)] exact mass calcd for C24H23N3O4 (MH+) 418.1767, found 418.1777. Analysis. Calcd for C24H23N3O4: C, 69.05; H, 5.55; N, 10.07. Found: C, 69.11; H, 5.73; N, 9.93.
[90, 273, n = 3].
1H NMR (300 MHz, CDCl3) δ 1.44 (t, J = 7.2 Hz, 3H), 2.18 (quint, J = 6.4 Hz, 2H), 4.29–4.41 (m, 6H), 5.41 (br s, 1H), 5.60 (br s, 1H), 6.85 (d, J = 8.2 Hz, 1H), 6.97 (dd, J = 8.2, 1.9 Hz, 1H), 7.20 (d, J = 1.9 Hz, 1H), 7.23–7.28 (m, 1H), 7.37–7.43 (m, 2H), 7.46–7.51 (m, 1H), 7.74 (dd, J = 8.4, 1.6 Hz, 1H), 7.98 (s, 1H), 8.10 (d, J = 7.8 Hz, 1H), 8.28 (d, J = 1.6 Hz, 1H), 8.31 (s, 1H); 13C NMR (100.6 MHz, CDCl3) δ 13.9, 29.2, 37.7, 70.9, 71.0, 108.8, 108.8, 112.9, 115.4, 119.5, 120.0, 120.8, 121.6, 122.9, 123.1, 123.2, 124.8, 125.4, 126.2, 140.5, 141.0, 143.8, 145.9, 148.6, 150.1. Analysis. Calcd for C25H25N3O4: C, 69.59; H, 5.84; N, 9.74. Found: C, 69.36; H, 5.96; N, 9.60.
[90, 273, n = 4].
1H NMR (300 MHz, CDCl3) δ 1.44 (t, J = 7.2 Hz, 3H), 1.86 (m, 4H), 4.21 (t, J = 6.2 Hz, 2H), 4.26 (t, J = 6.2 Hz, 2H), 4.37 (q, J = 7.2 Hz, 2H), 5.35 (br s, 1H), 5.55 (br s, 1H), 6.83 (d, J = 8.2 Hz, 1H), 6.96 (dd, J = 8.2, 1.9 Hz, 1H), 7.17 (d, J = 1.9 Hz, 1H), 7.23–7.28 (m, 1H), 7.36–7.43 (m, 2H), 7.46–7.51 (m, 1H), 7.74 (dd, J = 8.4, 1.6 Hz, 1H), 8.00 (s, 1H), 8.10 (d, J = 7.8 Hz, 1H), 8.27 (d, J = 1.6 Hz, 1H), 8.33 (s, 1H); 13C NMR (100.6 MHz, CDCl3) δ 13.9, 25.8, 25.9, 37.7, 73.8, 73.8, 108.8, 108.8, 112.9, 115.3, 119.4, 119.9, 120.7, 121.4, 122.9, 123.1, 123.2, 124.7, 125.4, 126.1, 140.4, 140.9, 143.9, 145.9, 148.2, 149.8. Analysis. Calcd for C26H27N3O4: C, 70.09; H, 6.11; N, 9.43. Found: C, 69.9; H, 6.03; N, 9.41.
[90, 273, n = 5].
1H NMR (300 MHz, CDCl3) δ 1.43 (t, J = 7.2 Hz, 3H), 1.52–1.62 (m, 2H), 1.73–1.88 (m, 4H), 4.16 (t, J = 6.6 Hz, 2H), 4.23 (t, J = 6.6 Hz, 2H), 4.36 (q, J = 7.2 Hz, 2H), 5.77 (br s, 1H), 5.86 (br s, 1H), 6.83 (d, J = 8.2 Hz, 1H), 6.94 (dd, J = 8.2, 1.8 Hz, 1H), 7.17 (d, J = 1.8 Hz, 1H), 7.22–7.29 (m, 1H), 7.36–7.43 (m, 2H), 7.46–7.52 (m, 1H), 7.73 (dd, J = 8.4, 1.5 Hz, 1H), 7.95 (s, 1H), 8.10 (d, J = 7.8 Hz, 1H), 8.27 (d, J = 1.5 Hz, 1H), 8.28 (s, 1H); 13C NMR (100.6 MHz, CDCl3) δ 13.9, 22.5, 29.0, 29.0, 37.7, 74.0, 74.0, 108.8, 108.8, 112.9, 115.3, 119.5, 119.9, 120.7, 121.4, 122.9, 123.0, 123.2, 124.7, 125.3, 126.2, 140.4, 140.9, 143.9, 145.9, 148.4, 150.0. Analysis. Calcd for C27H29N3O4: C, 70.57; H, 6.36; N, 9.14. Found: C, 70.36; H, 6.36; N, 8.98.
[90, 273, n = 6].
1H-NMR (300 MHz, CDCl3) δ 1.43 (t, J = 7.2 Hz, 3H), 1.43–1.52 (m, 4H), 1.70–1.80 (m, 4H), 4.14 (t, J = 6.6 Hz, 2H), 4.23 (t, J = 6.6 Hz, 2H), 4.34 (q, J = 7.2 Hz, 2H), 6.28 (br s, 2H), 6.84 (d, J = 8.2 Hz, 1H), 6.94 (dd, J = 8.2, 1.8 Hz, 1H), 7.19 (d, J = 1.8 Hz, 1H), 7.23–7.28 (m, 1H), 7.35–7.42 (m, 2H), 7.46–7.52 (m, 1H), 7.74 (dd, J = 8.4, 1.5 Hz, 1H), 7.95 (s, 1H), 8.09 (d, J = 7.8 Hz, 1H), 8.27 (d, J = 1.5 Hz, 1H), 8.29 (s, 1H); 13C NMR (100.6 MHz, CDCl3) δ 13.8, 25.7, 25.8, 29.1, 31.1, 37.7, 74.0, 74.0, 108.7, 108.7, 112.7, 115.2, 119.4, 119.8, 120.6, 121.3, 122.8, 123.0, 123.1, 124.6, 125.2, 126.1, 140.3, 140.8, 143.8, 145.7, 147.9, 149.7. Analysis. Calcd for C28H31N3O4: C, 71.01; H, 6.60; N, 8.87. Found: C, 70.92; H, 6.97; N, 8.87.
In Vitro Kinase Inhibition Assay.
Tyrosine kinases c-Src, Fyn, Lyn, and Lck were purchased from Upstate Biotechnology. Kinase assays were performed with a protein tyrosine kinase ELISA kit purchased from Pierce. The assays were performed in neutravidin-coated 96-well microtiter plates containing the bound peptide p34cdc2(6–20) (biotin-KVEKIGEGTYGVVYK). Reactions were initiated by the addition of 10 μl of ATP and MnCl2 to produce a final 50-μl solution containing 5% DMSO, inhibitor, 125 μM ATP, 25 mM Tris (pH = 7.2), 30 mM MgCl2, 6 mM MnCl2, 500 μM EGTA, 1 mM DTT, and 0.6 unit of appropriate kinase. IC50 values for kinase inhibitors were determined in duplicate or triplicate at six inhibitor concentrations. IC50 values were determined from plots of Ai/Ao versus inhibitor concentration, where Ao is the absorbance in the absence of inhibitor and Ai is the absorbance with inhibitor. It should be noted that the presence of BSA in the assay buffer was found to significantly increase the IC50 values of the inhibitors.
Results
Selection of a Set of Potential Binding Elements.
Careful selection of the potential binding elements is essential for successful implementation of the outlined method. First, the common chemical linkage group present in each potential binding element must enable a uniform method for linking binding elements together. Second, the chemistry used to link the binding elements through the common chemical linkage group must be efficient and also chemoselective to allow the display of a wide variety of functionality. Third, the potential binding elements must be soluble in aqueous solution at high concentrations, because the potential binding elements may bind to their target only weakly and therefore must be screened at high micromolar to low millimolar concentrations. Finally, for expediency, the potential binding elements and linkers should be commercially available or readily accessible.
For this study, potential binding elements were selected with the O-methyl oxime as the chemical linkage group. A library of O-methyl oxime-based potential binding elements can easily be prepared by condensation of O-methylhydroxylamine with any of a large number of commercially available aldehydes (Fig. 2). Because excess reagents are not used and by-products are not produced, direct evaluation of compounds is possible without purification. Additionally, oxime formation is compatible with a wide range of functionality (22–28), including all of the functionalities present in proteins and oligonucleotides. This chemoselectivity enables compounds displaying a variety of functionality to be included in the set of O-methyl oxime potential binding elements. Finally, the O-alkyl oxime functionality is stable at physiological pH and is present in a number of approved drugs (29–31).
Figure 2.
General synthetic scheme for preparation of potential binding elements. rt, Room temperature.
Monomer Screening.
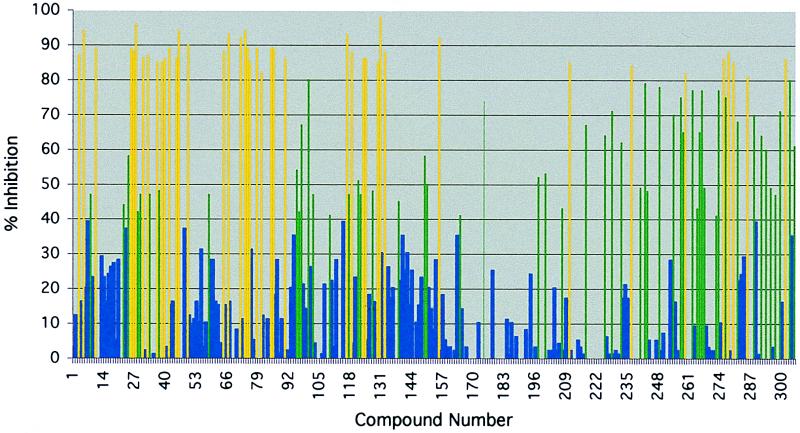
To screen the library of O-methyl oxime potential binding elements at high concentrations, a commercially available microtiter-based ELISA format based on the constitutively active form of c-Src was employed. A library of 305 discrete O-methyl oximes was first screened at 1 mM to ensure that even weak inhibitors of c-Src could be identified (Fig. 3). The 66 O-methyl oximes that showed greater than 60% inhibition of c-Src were rescreened at a concentration of 500 μM in duplicate. From the original 66 O-methyl oximes selected, 47 showed greater than 70% inhibition at a concentration of 500 μM (Fig. 4). The identified O-methyl oxime binding elements are diverse in structure, with specific structural motifs being observed. Of note are the related sets of phenyl ethers, indoles, phenols, phenyl-substituted furans, and anilines. Of the 47 binding elements, 37 were used to prepare the combinatorial library of linked binding elements. The remaining 10 binding elements, compounds 26, 30, 36, 75, 84, 100, 116, 124, 132, and 301, were not used because of their high structural similarity to binding elements that were included in library preparation.
Figure 3.
Percent inhibition of c-Src by 305 potential binding elements at 1 mM. Shown is the percent inhibition of enzyme activity at an inhibitor concentration of 1 mM as determined by the percent phosphorylation of the tyrosine kinase peptide substrate (biotin-KVEKIGEGTYGVVYK), [ATP] = 125 μM.
Preparation of a Combinatorial Library of Linked Binding Elements.
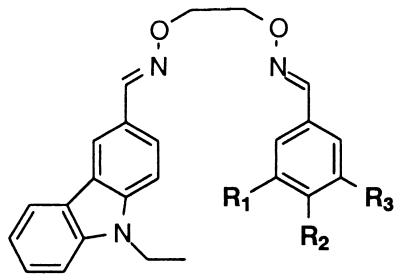
The linked binding elements were specifically designed such that the linker portion of the linked binding element is structurally related to the common chemical linkage group present in the potential binding elements (Fig. 5). This similarity in structure could prove to be important for those binding elements in which the chemical linkage group contributes to binding.
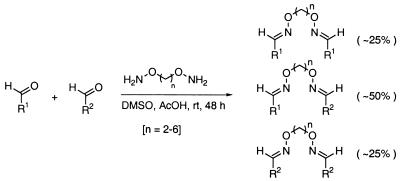
The linked binding elements are synthesized by condensing the aldehyde precursors of the binding elements with O,O′-diaminoalkanediol linkers. To prepare the library of linked binding elements, all possible precursor aldehyde combinations were arrayed in microtiter plates (666 wells). An equimolar mixture of five different O,O′-diaminoalkanediol linkers (n = 2–6) was then added to each aldehyde combination. An approximate 2:1:1 statistical mixture of heterodimeric binding elements to the two homodimeric binding elements would be expected, assuming that there is no cooperativity between the two reaction sites of the linkers (see Materials and Methods) (28) (Fig. 6). The pure homodimers were also prepared (37 wells) to serve as control compounds to differentiate between active homodimers and heterodimers in the screening step.
Figure 6.
General synthetic scheme for linked binding elements.
Screening the Library of Linked Binding Elements.
The library of linked binding elements was screened for inhibition of c-Src by using the microtiter-based ELISA format, but at a more stringent inhibitor concentration (5 μM) than was used to screen the potential binding elements (500 μM). The most active wells were rescreened at 1 μM in triplicate. Four of the wells showed greater than 50% inhibition at this concentration (entries 1–4, Table 1). Notably, all four of these wells contained linked binding elements that were derived from the catechol-containing binding element 273. In addition, linked binding elements derived from two different precursor aldehydes (entries 1–4, Table 1) showed greater activity than did linked binding elements derived from a single precursor aldehyde (entries 5–9, Table 1). Indeed, only the linked binding element [273, 273, n = 2–6] derived from the precursor aldehyde of catechol 273 showed appreciable activity, with 43% inhibition at 1 μM.
Table 1.
Percent inhibition of c-Src by linked binding elements at 1 μM
| Entry | Compound mixture* | % inhibition of c-Src |
|---|---|---|
 | ||
| 1 | [90, 273, n = 2–6] | 89 ± 5 |
| 2 | [73, 273, n = 2–6] | 87 ± 6 |
| 3 | [44, 273, n = 2–6] | 83 ± 8 |
| 4 | [39, 273, n = 2–6] | 63 ± 11 |
| 5 | [273, 273, n = 2–6] | 43 ± 20 |
| 6 | [90, 90, n = 2–6] | 13 ± 7 |
| 7 | [73, 73, n = 2–6] | 7 ± 6 |
| 8 | [44, 44, n = 2–6] | 9 ± 4 |
| 9 | [39, 39, n = 2–6] | 7 ± 11 |
Percent inhibition of enzyme activity at an inhibitor concentration of 1 μM as determined by the percent phosphorylation of the tyrosine kinase peptide substrate (biotin-KVEKIGEGTYGVVYK), [ATP] = 125 μM.
Compounds are denoted by listing the two linked binding elements followed by the linker length.
Characterization of the Most Potent Linked Binding Element.
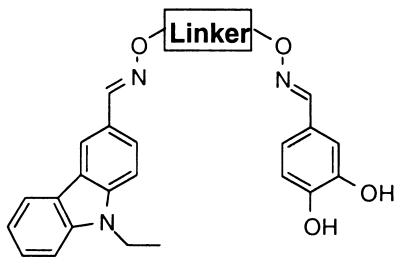
Each of the linked binding elements [90, 273, n = 2] to [90, 273, n = 6] from the most active well were prepared on large scale to obtain IC50 values on purified material. Dramatic linker length dependence was observed (entries 1–5, Table 2). The linked binding elements with the shortest linker length [90, 273, n = 2], IC50 = 64 nM, is almost two orders of magnitude more potent that the linked binding elements with the longest linker [90, 273, n = 6], IC50 = 5300 nM. A similar correlation of decreasing potency with increasing linker length is observed for linked binding elements [73, 273, n = 2–6] (data not shown). Several rigid analogs (entries 6–8, Table 2) of the most potent compound [90, 273, n = 2] were also prepared to evaluate the importance of linker structure and rigidity while maintaining a constant linker length. A significant drop in potency was observed for each of the rigid analogs. The strong dependence of inhibitor potency on linker length and structure demonstrates that the appropriate presentation of the two binding elements is critically important to achieve potency for this target.
Table 2.
Correlation of linker structure with IC50 values for c-Src inhibition
Assays were performed with purified material. Enzyme inhibition was expressed as the IC50, the concentration of inhibitor required for half-maximal inhibition of phosphorylation of the biotinylated tyrosine kinase peptide substrate (biotin-KVEKIGEGTYGVVYK) at an ATP concentration of 125 μM. For each IC50 determination assays were performed in duplicate or triplicate at six inhibitor concentrations. For assay calibration, the IC50 of the known inhibitor PP1 was determined to be 0.32 μM (literature value = 0.17 μM) (32).
The inhibitory activity of the most potent linked binding element [90, 273, n = 2] toward several other closely related Src family members was next evaluated. Indeed, [90, 273, n = 2] showed greater than 75-fold selectivity over Lyn and Fyn and greater than 1000-fold selectivity over Lck (Table 3). The high selectivity that is observed for the novel inhibitor without any optimization is quite impressive, considering the general lack of selectivity that has been observed for inhibitors between Src family members.
Table 3.
IC50 values of individual binding elements of [90, 273, n = 2] for Src family kinases
| Compound | IC50, μM
|
|||
|---|---|---|---|---|
| c-Src | Fyn | Lyn | Lck | |
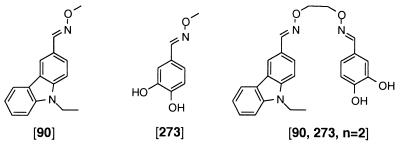 | ||||
| [273] | 41 ± 5 | >1000 | >1000 | >1000 |
| [90] | 40 ± 16 | 64 ± 50 | 400 ± 170 | >500 |
| [90, 273, n = 2] | 0.064 ± 0.038 | 5.0 ± 2.4 | 13 ± 3 | >250 |
See Table 2 for assay conditions.
Because of the process of linking selected binding elements together, the method provides a unique opportunity to rapidly assess the relative contributions of the constituent binding elements to the binding potency and specificity of [90, 273, n = 2]. As shown in Table 3, the individual binding elements 90 and 273 have comparable IC50 values of 40 μM and 41 μM, respectively. The considerable increase in inhibitory activity of the appropriately linked binding elements [90, 273, n = 2] over the individual binding elements 90 and 273 of 625- and 640-fold demonstrates the power of this method.
The relative contributions of the constituent binding elements to selectivity were also evaluated. Notably, the catechol binding element 273 showed minimal inhibition of Fyn, Lyn, and Lck and serves as the critical determinant for the >75-fold selectivity of [90, 273, n = 2]. The carbazole binding element 90 shows 1.5-, 10-, and >10-fold selectivity for c-Src over Fyn, Lyn, and Lck, respectively. Although the selectivity observed for 90 is more modest, the contributions of both binding elements 273 and 90 are apparent in the relative inhibitory activity of [90, 273, n = 2] to the four family members. The highest selectivity is observed for c-Src relative to Lck, which was inhibited poorly by both binding elements 90 and 273.
Because of the importance of the catechol binding element 273 to the specificity of [90, 273, n = 2], a series of analogs was prepared to define the key binding determinants provided by 273. As shown in Table 4, both hydroxyl groups are critical for inhibitory activity, although modulation of the pKa of the catechol moiety by substitution with the electronegative nitro group, [90, 313, n = 2], resulted in only a modest change in binding potency. It is notable that catechols have previously been determined to be important binding elements for a number of tyrosine kinase inhibitors derived from natural products (14–16).
Table 4.
IC50 values against c-Src for linked binding elements containing catechol analogs
See Table 2 for assay conditions.
Discussion
We have reported an efficient method to rapidly identify ligands to biological targets in four straightforward steps (Fig. 1). Furthermore, we have demonstrated the utility of the method by the identification of a potent inhibitor (IC50 = 64 nM) of the archetypal tyrosine kinase c-Src. Notably, the inhibitor shows greater than 75-fold selectivity for inhibition of c-Src relative to Fyn, Lyn, and Lck, which are family members with high sequence homology. Because the method relies on the appropriate linkage of distinct binding elements, the relative contributions of the individual binding elements to both potency and selectivity can readily be ascertained, thus providing new opportunities for both understanding and achieving affinity and selectivity.
While the O-methyl oxime was employed as the common chemical linkage group, clearly a wide range of alternative common chemical linkage groups and linkage chemistries could be considered. This straightforward method should also be applicable to diverse biological targets because the method does not rely on lead compounds or knowledge of the mechanism or structure of the biological target.
Acknowledgments
This work was supported by National Institutes of Health Grant GM50353.
Abbreviation
- HRMS (FAB)
high-resolution mass spectrometry (fast-atom bombardment)
Footnotes
Compound 68 corresponds to the aldehyde E-3,5-dimethoxy-4-hydroxycinnamaldehyde.
References
- 1.Thompson L A, Ellman J A. Chem Rev. 1996;96:555–600. doi: 10.1021/cr9402081. [DOI] [PubMed] [Google Scholar]
- 2.Balkenhohl F, von dem Bussche-Hünnefeld C, Lansky A, Zechel C. Angew Chem Int Ed Engl. 1996;35:2288–2337. [Google Scholar]
- 3.Dolle R E. Mol Diversity. 1998;3:199–233. doi: 10.1023/a:1009699413828. [DOI] [PubMed] [Google Scholar]
- 4.Dolle R E, Nelson K H. J Comb Chem. 1999;1:235–282. doi: 10.1021/cc9900192. [DOI] [PubMed] [Google Scholar]
- 5.Kick E K, Roe D C, Skillman A G, Liu G, Ewing T J, Sun Y, Kuntz I D, Ellman J A. Chem Biol. 1997;4:297–307. doi: 10.1016/s1074-5521(97)90073-9. [DOI] [PubMed] [Google Scholar]
- 6.Rockwell A, Melden M, Copeland R A, Hardman K, Decicci C P, DeGrado W F. J Am Chem Soc. 1996;118:10337–10338. [Google Scholar]
- 7.Gray N S, Wodicka L, Thunnissen A-M, Norman T C, Kwon S, Espinoza F H, Morgan D O, Barnes G, LeClerc S, Meijer L, et al. Science. 1998;281:533–538. doi: 10.1126/science.281.5376.533. [DOI] [PubMed] [Google Scholar]
- 8.Yang L, Berk S C, Rohrer S P, Mosley R T, Guo L, Underwood D J, Arison B H, Birzin E T, Hayes E C, Mitra S W, et al. Proc Natl Acad Sci USA. 1998;95:10836–10841. doi: 10.1073/pnas.95.18.10836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rohrer S P, Birzin E T, Mosley R T, Berk S, Hutchins S M, Shen D, Xiong Y, Hayes E C, Parmar R M, Foor F, et al. Science. 1998;282:737–740. doi: 10.1126/science.282.5389.737. [DOI] [PubMed] [Google Scholar]
- 10.Brown D. Mol Diversity. 1996;2:217–222. doi: 10.1007/BF01715637. [DOI] [PubMed] [Google Scholar]
- 11.Shuker S B, Hajduk P J, Meadows R P, Fesik S W. Science. 1996;274:1531–1534. doi: 10.1126/science.274.5292.1531. [DOI] [PubMed] [Google Scholar]
- 12.Fesik S W, Sheppard G, Nettesheim D G, Olejniczak E T, Shuker S B, Meadows R P, Steinman D H, Carrera G M, Marcotte P A, Severin J, et al. J Am Chem Soc. 1997;119:5818–5827. [Google Scholar]
- 13.Bolen J B, Rowley R B, Spana C, Tsygankov A Y. FASEB J. 1992;6:3403–3409. doi: 10.1096/fasebj.6.15.1281458. [DOI] [PubMed] [Google Scholar]
- 14.Levitzki A. Anticancer Drug Des. 1996;11:175–182. [PubMed] [Google Scholar]
- 15.Showalter H D, Kraker A J. Pharmacol Ther. 1997;76:55–71. doi: 10.1016/s0163-7258(97)00097-1. [DOI] [PubMed] [Google Scholar]
- 16.Lawrence D S, Niu J. Pharmacol Ther. 1998;77:81–114. doi: 10.1016/s0163-7258(97)00052-1. [DOI] [PubMed] [Google Scholar]
- 17.McMahon G, Sun L, Liang C, Tang C. Curr Opin Drug Disc Dev. 1998;1:131–146. [PubMed] [Google Scholar]
- 18.Bishop A C, Kung C-Y, Shah K, Witucki L, Shokat K M, Liu Y. J Am Chem Soc. 1999;121:627–631. [Google Scholar]
- 19.Bishop A C, Shokat K M. Pharmacol Ther. 1999;82:337–346. doi: 10.1016/s0163-7258(98)00060-6. [DOI] [PubMed] [Google Scholar]
- 20.Kim J N, Kim K M, Ryu E K. Synth Commun. 1992;22:1427–1432. [Google Scholar]
- 21.Choong I C, Ellman J A. J Org Chem. 1999;64:6528–6529. doi: 10.1021/jo990490h. [DOI] [PubMed] [Google Scholar]
- 22.Tuchscherer G. Tetrahedron Lett. 1993;34:8419–8422. [Google Scholar]
- 23.Rose K J. J Am Chem Soc. 1994;116:30–33. [Google Scholar]
- 24.Shao J, Tam J P. J Am Chem Soc. 1995;117:3893–3899. [Google Scholar]
- 25.Canne L E, Ferré-D'Amaré A R, Burley S K, Kent S B H. J Am Chem Soc. 1995;117:2998–3007. [Google Scholar]
- 26.Cornish V W, Hahn K M, Schultz P G. J Am Chem Soc. 1996;118:8150–8151. [Google Scholar]
- 27.Rodriguez E C, Marcaurelle L A, Bertozzi C R. J Org Chem. 1998;63:7314–7135. doi: 10.1021/jo981351n. [DOI] [PubMed] [Google Scholar]
- 28.Nazarpack-Kandlousy N, Zweigenbaum J, Henion J, Eliseev A V. J Comb Chem. 1999;1:199–206. [Google Scholar]
- 29.Galit J. Presse Med. 1999;26:16–21. [Google Scholar]
- 30.Cunha B A. Urology. 1993;41:249–282. doi: 10.1016/0090-4295(93)90568-u. [DOI] [PubMed] [Google Scholar]
- 31.Wilde M I, Plasker G L, Benfield P. Drugs. 1993;46:895–924. doi: 10.2165/00003495-199346050-00008. [DOI] [PubMed] [Google Scholar]
- 32.Hanke J H, Gardener J P, Dow R L, Changelian P S, Brisette W H, Weringer E J, Pollok B A, Connelly P A. J Biol Chem. 1996;271:695–701. doi: 10.1074/jbc.271.2.695. [DOI] [PubMed] [Google Scholar]