Abstract
Like many eukaryotes, bacteria make widespread use of postreplicative DNA methylation for the epigenetic control of DNA-protein interactions. Unlike eukaryotes, however, bacteria use DNA adenine methylation (rather than DNA cytosine methylation) as an epigenetic signal. DNA adenine methylation plays roles in the virulence of diverse pathogens of humans and livestock animals, including pathogenic Escherichia coli, Salmonella, Vibrio, Yersinia, Haemophilus, and Brucella. In Alphaproteobacteria, methylation of adenine at GANTC sites by the CcrM methylase regulates the cell cycle and couples gene transcription to DNA replication. In Gammaproteobacteria, adenine methylation at GATC sites by the Dam methylase provides signals for DNA replication, chromosome segregation, mismatch repair, packaging of bacteriophage genomes, transposase activity, and regulation of gene expression. Transcriptional repression by Dam methylation appears to be more common than transcriptional activation. Certain promoters are active only during the hemimethylation interval that follows DNA replication; repression is restored when the newly synthesized DNA strand is methylated. In the E. coli genome, however, methylation of specific GATC sites can be blocked by cognate DNA binding proteins. Blockage of GATC methylation beyond cell division permits transmission of DNA methylation patterns to daughter cells and can give rise to distinct epigenetic states, each propagated by a positive feedback loop. Switching between alternative DNA methylation patterns can split clonal bacterial populations into epigenetic lineages in a manner reminiscent of eukaryotic cell differentiation. Inheritance of self-propagating DNA methylation patterns governs phase variation in the E. coli pap operon, the agn43 gene, and other loci encoding virulence-related cell surface functions.
INTRODUCTION
The word “epigenetics” is based on the Greek prefix “epi-,” denoting “on” or “in addition,” and “genetic,” meaning “pertaining to or produced from genes.” In the past, the term “epigenetics” has been used to describe the differentiation of genetically identical cells into distinct cell types to form tissues and organs during development of a multicellular organism. In current practice the word is used by biologists to describe heritable changes in gene expression that occur without changes in the DNA sequence. In the strict sense, epigenetic systems involve two or more heritable states, each maintained by a positive feedback loop. In a broader sense, however, any additional information superimposed to the DNA sequence (e.g., methylation of DNA) can be considered “epigenetic.” Here we review the current state of research in the field of bacterial epigenetics, with an emphasis on systems controlled by DNA methylation, which are the best known at the molecular level. We refer the reader to reviews covering other aspects of DNA methylation and related topics (16, 32, 51, 96, 143, 160, 172, 178, 202, 214, 264, 265, 285).
Epigenetic phenomena include prions, in which protein structure is heritably transmitted (223, 231, 235, 259); genomic imprinting, characterized by monoallelic repression of maternally or paternally inherited genes (52, 84, 128, 195, 213); histone modification, such as methylation of lysines by histone phase methyltransferases (MTases) that maintain active and silent chromatin states (132, 273); and DNA methylation patterns formed as a result of inhibition of methylation of specific DNA bases by protein binding (29, 41, 118, 262, 263). Each of these phenomena involve self-perpetuating states, be they protein or DNA related (116, 155, 230-232), and the particular state that the molecule is in affects gene expression.
Epigenetic regulation can enable unicellular organisms to respond rapidly to environmental stresses or signals. For example, the yeast prion PSI+ is generated by a conformational change of the Sup35p translation termination factor, which is then inherited by daughter cells. The PSI+ form of Sup35p allows readthrough of nonsense codons that can provide a survival advantage under adverse conditions such as growth in paraquat or caffeine (259). The PSI+ prion is a metastable element that is generated and lost spontaneously at low rates, and thus within a population of yeast, some yeast cells will carry the prion and others will not. This situation provides potential flexibility in the response of the yeast population to environmental changes, orchestrated through the ability of the PSI+ prion to act upon native Sup35p protein and convert it to prion protein (223).
Methylation of specific DNA sequences by DNA methyltransferases provides another mechanism by which epigenetic inheritance can be orchestrated. For example, in certain eukaryotes, including mammals, methylation of cytosine residues at 5′-CG-3′ (CpG) sequences facilitates binding of methyl-CpG binding proteins (134, 156, 187). In turn, methyl-CpG binding proteins affect the transcription state of a local DNA region through further interaction with chromatin-remodeling proteins (145). Methylation of CpG can affect gene expression, and the methylated state is usually correlated with transcriptional repression. The methylation pattern of a DNA region is defined as the collective presence or absence of methyl groups on specific target sites. DNA methylation patterns can vary between cells, tissues, and individuals. DNA methylation patterns are established via de novo methylation during the first stages of embryonic development (28, 81, 213). Such patterns are propagated by DNA methyltransferases known as maintenance methylases (Dnmt1), which are active on hemimethylated DNA substrates generated by DNA replication. Thus, if a DNA region contains methylated CpG sequences, they will be propagated in the methylated state. Nonmethylated CpG sequences, however, are not substrates for the maintenance DNA methylases. Thus, if a DNA region contains nonmethylated CpGs, they will tend to remain nonmethylated. A major area of research in eukaryotic epigenetic regulation is directed at understanding the mechanisms by which DNA methylation patterns are erased following cleavage of the fertilized egg and then established via de novo methylation (74, 81, 141, 180).
DNA methylation plays important roles in the biology of bacteria: phenomena such as timing of DNA replication, partitioning nascent chromosomes to daughter cells, repair of DNA, and timing of transposition and conjugal transfer of plasmids are sensitive to the methylation states of specific DNA regions (16, 160, 172, 178, 202, 285). All of these events use as a signal the hemimethylated state of newly replicated DNA, generated by semiconservative replication of a fully methylated DNA molecule. In the case of DNA replication, the protein SeqA binds preferentially to hemimethylated DNA target sites (GATC sequence) clustered in the origin of replication (oriC) and sequesters the origin from replication initiation. In addition, SeqA also transiently blocks synthesis of the DnaA protein, which is necessary for replication initiation, by binding to hemimethylated GATC sites in the dnaA promoter (36, 49, 100, 140, 146, 163, 179, 249). In DNA repair, the methyl-directed mismatch repair protein MutH recognizes hemimethylated DNA sites and cuts the nonmethylated daughter DNA strand, ensuring that the methylated parental strand will be used as the template for repair-associated DNA synthesis (8, 12, 25, 178, 227, 237). In transposition of Tn10, hemimethylated DNA plays two roles: enhancing binding of RNA polymerase to the transposase promoter and enhancing binding of transposase to its DNA target sites (144, 181, 219). DNA methylation appears to play similar roles in regulating Tn5 transposition (73, 161, 175, 217, 253, 292). None of these phenomena are heritable since the hemimethylated state of DNA is not heritable, occurring transiently in newly replicated DNA.
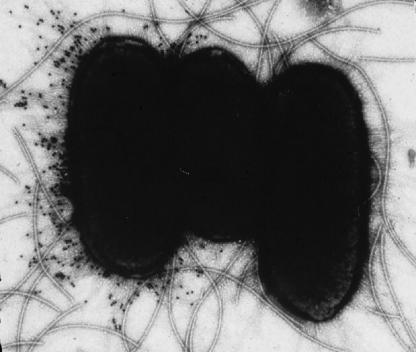
Phenomena involving inheritance of DNA methylation patterns are also known in bacteria, and the best-known examples involve phase variation. In phase variation, gene expression alternates between active (ON phase) and inactive (OFF phase) states. For example, uropathogenic Escherichia coli (UPEC) cells undergo pilus phase variation, which can be observed using immunoelectron microscopy with antipilus antibodies marked with colloidal gold (Fig. 1). Phase variation can occur through a variety of genetic mechanisms involving changes in nucleotide sequence (e.g., site-specific recombination and mutation) which result in heritably altered gene expression (1, 4, 26, 32, 33, 42, 53, 69, 75, 79, 86, 98, 113, 119, 122, 133, 164, 191, 229, 240, 244, 256, 265, 298). Bacteria also use epigenetic mechanisms to control phase variation. In all cases examined, these systems use DNA methylation patterns to pass information regarding the phenotypic expression state of the mother cell on to the daughter cells. A DNA methylation pattern is formed by binding of a regulatory protein(s) to a site that overlaps a methylation target, blocking methylation. This pattern can control gene expression if methylation, in turn, affects binding of the regulatory protein(s) to its DNA target site, which could occur by steric hindrance or alteration of DNA structure due to methylation (206, 207). Notably, most adhesin genes in E. coli are regulated by epigenetic mechanisms involving DNA methylation patterns (32, 115, 116, 262).
FIG. 1.
Pap phase variation in uropathogenic E. coli. Pap17 pilus phase variation of uropathogenic E. coli strain C1212 was visualized with anti-Pap17 antibodies labeled with 10-nm colloidal gold particles. The bacterium at the left is in the ON-phase state for Pap17 expression, whereas the two bacteria at the right are in the OFF phase. Note that these two bacteria express unmarked Pap21 pili, which are also under phase variation control but are not marked with the anti-Pap17 antiserum. In addition to the Pap pili (diameter of about 7 nm), flagella (diameter of about 20 nm) can also be seen.
Little is known concerning how widespread epigenetic control is in the bacterial world and the roles that epigenetic regulatory systems play in bacterial biology, including pathogenesis. Our main goal in writing this review is to introduce the reader to epigenetic regulatory control, focusing on the main features and unique aspects of the epigenetic control systems that have been studied. The list of examples discussed below can be grouped into several classes: (i) strict-sense epigenetic inheritance involving heritable transmission of DNA methylation states to daughter cells, as in the pap operon of uropathogenic E. coli; (ii) DNA methylation signals that generate distinct epigenetic states in DNA molecules coexisting in the same cell, as in IS10 transposition and in traJ regulation; and (iii) systems that are “epigenetic” in a broader sense, since DNA methylation provides a signal for temporal or spatial control of DNA-protein interactions but does not give rise to distinct lineages of cells or DNA molecules. Examples of the last class include the control of bacterial mismatch repair by DNA methylation and the coupling of promoters to distinct DNA methylation states during the cell cycle. We hope that this will be useful not only in understanding experiments carried out to date but also as a primer for future work in bacterial epigenetics.
FOUNDATIONS
Most epigenetic systems known in bacteria use DNA methylation as a signal that regulates a specific DNA-protein interaction. These systems are usually composed of a DNA methylase and a DNA binding protein(s) that bind to DNA sequences overlapping the target methylation site, blocking methylation of that site. Methylation of the target site, in turn, inhibits protein binding, resulting in two alternative methylation states of the target site, methylated and nonmethylated. The epigenetic regulatory methylases known in bacteria are designated “orphan” methylases since they lack a cognate restriction enzyme. We begin by discussing restriction-modification (R-M) systems, since they are likely the progenitors of the orphan methylases regulating epigenetic processes. Indeed, DNA methylation plays a regulatory role in some R-M systems, as described below.
Origins: R-M Systems
DNA methylation was originally discovered in the context of restriction-modification systems, in which a restriction endonuclease recognizes a specific target DNA sequence unless that sequence has been methylated by a cognate DNA methyltransferase (5, 27, 39, 153, 220, 260). Three main groups of R-M systems (types I, II, and III) have been described, based on whether the restriction and modification activities are within a single polypeptide (types I and III) or separate polypeptides (type II) and on whether the restriction enzymes cut at a site close to (types II and III) or far from (type I) the methylation target sequence (185, 221, 236, 238, 284). It has been postulated that R-M systems evolved as a form of cellular defense, targeting incoming viral and other foreign DNA sequences for degradation. Note that foreign DNAs would not be methylated at the appropriate target sites unless that sequence was derived from a bacterium with a cognate methylase of the same specificity (6, 77). In these systems, the restriction enzyme and cognate methylase are both expressed at levels that allow complete methylation of the genome, sufficient to block double-strand DNA cleavage by the restriction enzyme, a potentially fatal event. Incoming foreign DNA is efficiently destroyed, since the restriction enzyme has the upper hand over the methylase: for the DNA to survive, every restriction site it carries would have to be methylated before even a single site is cleaved by the cognate restriction enzyme, an unlikely event.
Work by Kobayashi and colleagues has suggested that R-M systems have attributes of selfish genes (148-150). Nakayama and Kobayashi showed that a plasmid containing the type II R-M EcoRV system could not be displaced from cells by an incompatible plasmid due to the death of cells that lost the EcoRV-containing plasmid, a form of postsegregational killing (186). In cells lacking the R-M gene complex, the levels of methylase and cognate restriction enzyme drop to a point where insufficient methylase is present to protect all chromosomal target sites; the restriction enzyme then cleaves one or more sites, killing the cell. This scenario is similar to that for addiction modules such as hok-sok, in which sok gene expresses an antisense RNA that inhibits translation of the hok toxin gene. When cells lose a plasmid containing hok-sok, they die; since hok mRNA is stable but sok RNA is unstable (half-life [t1/2]), <30 s), translation of hok ensues which leads to cell death (91, 92). Other addiction modules are made of two proteins, a toxin and an antitoxin (82, 90, 106).
Further analysis of the EcoRV system has shown that a regulatory gene designated “C,” sandwiched between the R and M genes, codes for a product that activates R gene expression (186). The C gene appears to be required for expression of the R gene, since postsegregational killing does not occur in C gene mutants. One function of the C gene is in establishment of an R-M system in a new host. In this case the M gene is immediately activated, allowing modification of host DNA sites. At the same time, C gene expression is also activated, building up the C protein level to a point that allows activation of R gene expression. This temporal delay in expression of the restriction enzyme is critical in allowing time for all chromosomal sites to be methylated and protected from digestion. In addition, C also functions as a suicide immunity gene, forcing expression of the R gene of an incoming closely related R-M complex with different restriction specificity, resulting in host cell death. This would be expected to prevent spread of a competing R-M complex of the same C gene immunity group (any R-M complex in which the resident C protein activates expression of an incoming R gene) within a bacterial population (250).
A second regulatory strategy used by R-M systems utilizes methylation of the cognate restriction site to control R-M transcription via a direct effect on RNA polymerase binding. For example, in the CfrBI system of Citrobacter freundii, methylation of a cytosine (underlined) within the 5′-CCATGG-3′ DNA restriction site decreases expression of the CfrBI methylase (CfrBIM) and concomitantly increases expression of the CfrBI restriction enzyme (CfrBIR) (18, 294). This appears to occur as a result of the location of the cfrBI site within the −35 RNA polymerase σ70 binding site of the cfrBIM gene. Since the cfrBIM promoter is stronger than that of cfrBIR, any bacterial cell receiving the CfrBI system will be methylated before restriction can occur. As the intracellular methylase level increases, the cfrBI site is methylated, decreasing expression of cfrBIM and enabling expression of cfrBIR. The latter may protect the cell from incoming foreign DNA lacking methylated sequences.
A third R-M regulatory mechanism utilizes the methylase itself as a feedback regulator. In a number of cases binding of the methylase to DNA occurs via an N-terminal extension containing a helix-turn-helix motif (142, 196, 197). For example, in the SsoII R-M system of Shigella sonnei, the SsoII methyltransferase (SsoIIM) represses its own synthesis and stimulates expression of the cognate restriction endonuclease (SsoIIR). Similar N-terminal extensions are present on a number of 5-methylcytosine methyltransferases, including those in the EcoRII, dcm, MspI, and LlaJI systems (142). The last system, present in Lactococcus lactis, encodes two methylases, M1.LlaJ1 and M2.LlaJ1, recognizing the complementary and asymmetric sequences 5′-GACGC-3′ and 5′-GCGTC-3′, respectively, with methylation of the internal cytosine in each case. Two LlaJI restriction sites are present 8 bp apart within the regulatory region of the llaJI operon, with one site overlapping the −35 RNA polymerase σ70 recognition site of the operon. Notably, methylation of both 5′-GCGTC-3′ sites by M2.LlaJ1 enhances binding of M1.LlaJ1, repressing transcription of the llaJI operon. The ability of the M1.LlaJ1 methylase to distinguish methylated and nonmethylated target sites provides a feedback mechanism by which expression of the llaJI operon is controlled by DNA methylation.
The analysis of regulation of the EcoRV, CfrBI and LlaJI R-M systems described above has provided insight into the evolution of epigenetic control systems that are predominantly controlled by “orphan” methyltransferases, including DNA cytosine methylase (Dcm) (202) in E. coli. It has been postulated that orphan methylases such as Dcm may have arisen by selection as vaccines against invasion of a restriction-modification complex (250). In the case of Dcm, which methylates the duplex sequence 5′-CCWGG-3′ (top strand shown; W = A or T) at the first cytosine, this methylation protects against cleavage by EcoRII. It was shown that postsegregational killing by the EcoRII R-M complex was diminished by the presence of dcm (250), which partially protected host chromosomal DNA from restriction attack. This function of Dcm as a possible molecular vaccine may be analogous to the function of cytosine methylation in certain eukaryotes, including mammals, where methylation has been postulated to inactivate transposons (293), although this hypothesis has been challenged (30). Dcm is not known to be involved in gene regulatory control. However, the other orphan methylase in E. coli, DNA adenine methylase (Dam), with homologues in other Alphaproteobacteria, does play an essential role in regulating epigenetic circuits. As well, Gammaproteobacteria have a cell cycle-regulated methylase (CcrM) which plays a major role in the control of chromosome replication and regulates expression of certain genes. In the next section we describe the biochemical properties of these DNA methylases and additional components of epigenetic switches before discussing specific epigenetic systems in detail.
Orphan DNA MTases
Dam.
Dam of E. coli is classified in the α group of DNA MTases based on the organization of 10 domains (167). The E. coli dam gene (accession no. J01600) is 834 bp and codes for a 32-kDa monomeric protein (114). Dam homologues are present in Salmonella spp., Haemophilus influenzae, and additional gram-negative bacteria (16, 204, 254). Dam binds to DNA nonspecifically as a monomer, moving by linear diffusion and specifically methylating 5′-GATC-3′ sequences. At GATC sites the adenine base is flipped out 180° into the active site of the enzyme, where it is stabilized by hydrophobic stacking with a tyrosine in the DPPY motif, which is conserved among adenine methyltransferases (123, 157). The methyl group donor, S-adenosyl-l-methionine (AdoMet), is required for stable binding of the flipped adenine in the active-site pocket of the enzyme and binds to Dam after the methylase binds DNA, transferring a methyl group to the exocyclic N6 nitrogen of adenine (261). AdoMet binds to two sites in the Dam protein: one is the catalytic center, and the other seems to be involved in an allosteric change that may increase specific binding of Dam to DNA (22). Dam appears to methylate only one of the adenosines of duplex GATC DNA sequence at a time (261). Notably, Dam shows high processivity for most DNAs; that is, after one methylation event, it slides on the same DNA molecule and carries out additional methylation events (turnovers). This high processivity effectively increases the rate of Dam methylation and may reflect the fact that there are few (<100) Dam molecules present in a single E. coli cell, yet there are about 19,000 GATC sites to methylate. Dam levels vary according to growth rate as a result of increased transcription from one of five dam gene promoters, designated P2 (158).
Based on the estimated numbers of Dam and GATC target sites per cell, each Dam molecule modifies between 20 and 100 GATC sites per minute (kcat) (261). This number is about 100-fold higher than the turnover number observed in vitro using an oligonucleotide substrate with one GATC site, indicating that there is likely some difference(s) in vivo that enables Dam to be more efficient at methylation (261). One possibility, suggested by Urig et al. (261), is that Dam is associated with the DNA polymerase III machine, scanning DNA for GATC sites as DNA replication proceeds and thus methylating DNA much more efficiently than it would in a random walk.
The processive nature of Dam contrasts sharply with DNA methylases associated with R-M systems, such as the EcoRV methylase (MEcoRV), which methylates its GATATC recognition sites distributively (95). In this case and for other R-M systems, incoming DNA needs to be restricted (cut) by the restriction enzyme before every site is methylated. The restriction enzyme has the advantage, since if just one restriction site in an incoming phage genome is left unmodified, the enzyme can cleave the DNA and block its replication. Note that restriction could be hampered if R-M DNA methylases were highly processive like Dam: processivity would increase the chances that all restriction sites in an incoming phage, for example, would be modified before restriction could occur.
Other gram-negative Gammaproteobacteria besides E. coli, including Salmonella spp., Serratia marcescens, Yersinia spp., Vibrio cholerae, Haemophilus influenzae, and Neisseria meningitidis, code for orphan MTases with significant sequence identity to EcoDam and which target adenosine of the GATC DNA sequence (162). Although Dam is not essential for growth of E. coli and Salmonella on laboratory media (14, 172, 254), the Dam homologues in Yersinia pseudotuberculosis, Yersinia enterocolitica, and Vibrio cholerae are essential gene products (135). However, a strain of Y. pseudotuberculosis in which dam mutations are viable has been described (252). It is not known what essential function(s) Dam plays in the pathogens in which it is essential, but it is provocative that both Yersinia and Vibrio contain two chromosomes, in contrast to the single chromosomes in E. coli and Salmonella spp., where Dam is not essential. A speculation is that Dam may be essential to coordinate DNA replication in bacteria with two or more chromosomes (78).
Dam homologues without a restriction enzyme counterpart are also present in bacteriophages, including Sulfolobus neozealandicus droplet-shaped virus (7), halophilic phage φCh1 (15), H. influenzae phage HP1 (204), phage P1 (61), phage T1 (9), and phage T4 (226). The last MTase, T4Dam, has been well characterized biochemically, primarily by Hattman and colleagues (123, 228). T4Dam, like EcoDam, is highly processive (169) and complements a dam mutant E. coli mutator phenotype (226). T4Dam and EcoDam may have a common evolutionary origin, sharing up to 64% sequence identity in four different regions (11 to 33 amino acids long) (105). After methylation with resulting formation of S-adenosyl-l-homocysteine, AdoMet binds to T4Dam without dissociating from the DNA duplex (299). Like EcoDam, T4Dam appears to flip out the adenosine of GATC sequence, facilitating its methylation (168).
CcrM.
The cell cycle-regulated DNA MTase family (CcrM) constitutes a second important group of orphan methyltransferases, classified in the β group of MTases and originally identified in Caulobacter crescentus (167, 242, 300). CcrM binds to and methylates adenosine in the sequence 5′-GANTC-3′, where “N” is any nucleotide (167, 300). Like EcoDam, CcrM is a functional monomer and acts processively (20), although evidence suggests that it is a dimer at physiologic concentration (234). However, unlike EcoDam, CcrM has a distinct preference for hemimethylated DNA as a substrate, based on the observation that the turnover rate for hemimethylated DNA containing a GANTC target site(s) was significantly higher than that for DNA containing nonmethylated sites (20). CcrM binds to and methylates adenosine in the sequence 5′-GANTC-3′, where “N” is any nucleotide. The GANTC sequence is also the target of HinfM methylase, which shares 49% identity with CcrM and whose cognate restriction enzyme HinfI from H. influenzae cuts at nonmethylated GANTC sites (300).
In Caulobacter, CcrM is an essential cell component and plays a crucial role in cell cycle regulation (20, 139, 170, 214-216, 242, 243, 300). CcrM homologues, which are likewise essential, have been found in Agrobacterium tumefaciens, the causative agent of crown gall disease in plants (137); in Rhizobium meliloti, the nitrogen-fixing symbiont of alfalfa and other legumes (286); and in the animal pathogen Brucella abortus (222). In B. abortus, aberrant CcrM expression impairs the pathogen's ability to proliferate in murine macrophages, raising the possibility that CcrM methylation might control the synthesis of virulence factors (222).
Regulation of Cellular Events by the Hemimethylated DNA State
Following passage of the DNA replication fork in E. coli, GATC sites methylated on the top and bottom strands in a mother cell (denoted as fully methylated) are converted into two hemimethylated DNA duplexes: one methylated on the top strand and nonmethylated on the bottom strand and one methylated on the bottom strand and nonmethylated on the top strand due to semiconservative replication (Fig. 2A). Most GATC sites are rapidly remethylated by Dam and exist in the hemimethylated state for only a fraction of the cell cycle (Fig. 2A). Exceptions are the DNA replication origin oriC, the dnaA promoter, and possibly additional GATC sites in the chromosome which bind SeqA (60). SeqA preferentially binds to clusters of two or more hemimethylated GATC sites spaced one to two helical turns apart (Fig. 2B). In the case of oriC, which contains a cluster of 13 GATC sites, sequestration delays remethylation and prevents binding of the DnaA protein, which controls the initiation of DNA replication. At other sites, binding of SeqA tetramers to hemimethylated GATC sites may organize nucleoid domains (100). Notably, the transcription profile of an E. coli SeqA− mutant was found to be similar to that of a Dam overproducer strain. Based on this observation, a model was developed in which Dam and SeqA compete for binding to hemimethylated DNA generated at the replication fork (159).
FIG. 2.
Generation of hemimethylated and nonmethylated GATC sites. (A) The vast majority of chromosomal GATC sites in E. coli are fully methylated until DNA replication generates two hemimethylated species, one methylated on the top strand and one methylated on the bottom strand. Within a short time after replication (less than 5 min), Dam methylates the nonmethylated GATC site, regenerating a fully methylated GATC site. (B) Two or more helically phased GATC sites (for example, in oriC) can be bound by SeqA when they are in the hemimethylated state. Binding of SeqA inhibits Dam methylation, maintaining the hemimethylated state for a portion of the cell cycle. Dissociation of SeqA allows Dam to methylate the hemimethylated DNAs, generating fully methylated DNA. (C) Certain GATC sites are present within or adjacent to regulatory protein binding sites. In some but not all cases, protein binding blocks DNA methylation over the entire cell cycle, stabilizing the hemimethylated state in the first generation and leading to a nonmethylated state in the second generation (only the second generation for the DNA methylated on the top strand is shown at the right).
The half-life of hemimethylated GATC sites not bound by SeqA has been estimated to be between 0.5 and 4 min, based on analysis of synchronized E. coli cells and monitoring the methylation status with restriction enzymes DpnI, which cuts fully methylated GATC sites; MboI, which cuts fully nonmethylated sites; and Sau3AI, which cuts GATC sites regardless of methylation state (50). In contrast, analysis of the origin of replication in the colicinogenic plasmid ColE1 indicated that remethylation of hemimethylated GATC sites occurs within a few seconds of passage of the replication fork (241). Notably, remethylation appeared to occur asynchronously, with methylation at GATC sites on the leading replication arm occurring more rapidly than GATC methylation on the lagging arm (about 2 seconds versus 4 seconds), suggesting that remethylation on the lagging arm occurs after ligation of Okazaki fragments. The reason for the discrepancy in estimation of the half-life of GATC sites is unclear but could reflect differences in chromosomal versus plasmid replication. For chromosomal replication the DNA polymerase III replication machinery is stationary, bound to the cytoplasmic membrane with DNA moving through it (154, 179). It is possible that Dam is present in a complex bound near the origin, methylating nascent DNA sequences as they arise.
The presence of hemimethylated GATC sites provides a signal that DNA replication has just occurred and plays a role in diverse cellular processes. For example, in methyl-directed mismatch repair the MutH protein binds to nonmethylated GATC sites and cleaves the nonmethylated DNA strand, ensuring that mutations in the daughter DNA strand are repaired using the parental strand as a template. In the absence of Dam, MutH can cleave the daughter strand, the parental strand, or both DNA strands. If the cell survives double-strand DNA breakage, 50% of the time the mutant daughter strand is used as a template to “repair” the parental strand, resulting in fixation of a mutation into the DNA (172, 285). Hemimethylated GATC sites are also used to control rates of transposition of insertion sequences IS3, IS10, IS50, and IS903 as well as transposons Tn5, Tn10, and Tn903 (73, 217, 219, 292). Elegant studies from Kleckner's laboratory showed that hemimethylated GATC sites control IS10 transposition in two different ways (181, 219). First, a GATC site present at bp −67 to −70 (here designated GATC-68) within the −10 module of the transposase promoter pIN controls transcription of the transposase gene. Full methylation of the GATC-68 inhibits RNA polymerase binding, reducing the level of tnp IS10 transcription. A second GATC site at bp 1320 to 1323 (GATC-1321) near the inner terminus of IS10 controls binding of transposase. Full methylation of GATC-1321 blocks transposition by inhibiting transposase binding. These two effects of DNA methylation on transposase expression and binding effectively limit IS10 transposition to a brief period immediately following DNA replication when GATC-68 and GATC-1321 are hemimethylated. Remarkably, the two hemimethylated IS10 DNAs have different transposition activities: IS10 methylated on the template strand is about 330 times more active than IS10 methylated on the nontemplate strand and 1,000 times more active than fully methylated IS10 (219). The majority of this difference is due to increased binding of transposase at the inner IS10 terminus; in addition, activation of the transposase promoter is more efficient in the IS10 hemimethylated species whose template strand is methylated. Since transposition of Tn10 does not involve the inner terminus, stimulation of Tn10 transposition following DNA replication is less efficient than for IS10 (219).
Like that of Tn10, transposition of IS50 and of Tn5 is stimulated by DNA replication (175). GATC sites are present within the inside end (IE) of IS50, similar to the case for IS10, and within the −10 region of the transposase regulatory region (73, 253, 292). In both IS50 and Tn5, Dam methylation represses tnp promoter activity and transposase binding to the IS50 IE (73, 253, 292). Increased transposition of IS50 and Tn5 in a Dam− host requires integration host factor (IHF), probably to compensate for a DNA conformational defect associated with the lack of Dam (165). In turn, binding of Fis (factor for inversion stimulation) to the IE inhibits IS50 transposition (276). Methylation of three GATC sites within the Fis recognition sequence inhibits Fis binding. Thus, immediately following DNA replication, Fis binds to the IE, inhibiting IS50 transposition, and counteracts the positive effects of the hemimethylated state on IS50 transposition. In contrast, Tn5 transposition is not inhibited by Fis, since it does not use IE (276).
DNA hemimethylation may regulate transcription of additional genes that contain GATC sites within their promoter regions. The list includes glnS, sulA, trpS, trpR, and tyrR of E. coli and cre of bacteriophage P1 (16, 172, 205, 246). Expression of these genes was increased in the absence of Dam, suggesting that GATC methylation may decrease binding of RNA polymerase. The possible physiologic significance of methylation of these sites is not known, but it could tie gene expression to the replication state of the cell, increasing transcription immediately after passage of the replication fork. In the case of the trpR gene, which encodes the repressor of the trp operon, an attractive speculation has been proposed by M. G. Marinus: because trpR is located between the origin of replication and the trp operon, a transient boost in trpR transcription might provide the increased concentration of repressor necessary to maintain repression when chromosome replication doubles trp operon dosage (171).
DNA Methylation Patterns
About 16 years ago, Blyn et al. discovered that one of two GATC sites within the regulatory region of the chromosomally encoded pyelonephritis-associated pilus (pap) operon of uropathogenic Escherichia coli (UPEC) was heritably nonmethylated, depending upon the pilus expression state of the cells (34). When DNA was isolated from cells expressing pyelonephritis-associated pili (Pap pili) (ON-phase cells), it was found that a GATC site proximal to the pap pilin promoter was methylated, whereas the promoter-distal GATC site was nonmethylated. This DNA methylation pattern characteristic of ON-phase cells differed from that of OFF-phase cells, which contained the converse pattern where the GATC site proximal to the pap pilin promoter was nonmethylated and the promoter-distal GATC site was methylated. The term “nonmethylated” is defined here as a state in which the GATC target of DNA adenine methylase is not methylated on either the top or bottom DNA strand, constituting a DNA methylation pattern analogous to those observed in mammalian cells (34). Since the term “unmethylated” might imply that an active demethylation has occurred, we prefer use of “nonmethylated” to describe DNA lacking a methyl group on both the top and bottom DNA strands. The phenomenon of demethylation, which occurs in eukaryotes to reset the DNA methylation pattern after zygote formation (88, 147), has not been reported to occur in prokaryotes. DNA methylation patterns are formed in bacteria by binding of a protein(s) at a DNA site(s) overlapping or near a GATC site(s), preventing methylation of that site(s) throughout the cell cycle (Fig. 2C). A direct role for DNA methylation patterns in the heritable control of gene expression in bacteria was first shown in the Pap system (41).
Further analysis of DNA methylation patterns in E. coli showed that multiple GATC sequences (ca. 36 sites) in the genome of E. coli K-12, which lack pap DNA sequences, were stably nonmethylated (218, 272). These sites were identified by digestion of chromosomal DNA with MboI, which cuts at nonmethylated GATC sites. Since nonmethylated GATC sites are rare, the DNA fragments generated by MboI digestion are too large to be resolved by conventional agarose gel electrophoresis. Pulsed-field gel electrophoresis was used to resolve these fragments; however, the DNA sequences flanking the nonmethylated GATC sites were not determined. Ringquist and Smith (218) also showed for the first time that a number of Dcm target sites [CC(A/T)GG; the second cytosine is methylated at the C-5 position] were stably nonmethylated.
Wang and Church analyzed Dam DNA methylation patterns to assess the binding of proteins to chromosomal DNA sites. Chromosomal DNA was digested with MboI and ClaI and cloned into pBluescript, which enabled the nonmethylated GATC sites to be sequenced (272). Since binding of proteins such as catabolite gene activator protein (CAP) is dependent upon environmental conditions via the secondary regulator cyclic AMP (cAMP), DNA methylation patterns within the regulatory regions of genes bound by cAMP-CAP and other regulatory factors were found to be environmentally controlled (218, 251). For example, a GATC sequence within the regulatory region of the car operon, controlling carbamoyl phosphate synthetase and involved in arginine and pyrimidine anabolism, was found to be protected from Dam methylation (272). This nonmethylated GATC site and others are listed in Table 1, with the chromosomal position (bp 29444 for the GATC near the carA gene) in E. coli MG1655 (a K-12 isolate) also shown. No protection of the car GATC site was detected in the absence of pyrimidines, consistent with the hypothesis that a pyrimidine repressor(s) binds to the car promoter region near or overlapping the GATC site, protecting it from methylation. Indeed, CarP and IHF were shown to bind in the regulatory region of carAB and protect GATC-207 (Table 1) from methylation (54).
TABLE 1.
Nonmethylated GATC sites in the E. coli K-12 chromosome
Location in the E. coli K-12 strain MG1655 chromosome. Analyses were carried out using Pattern Search and Fragment Viewer in ColiBase (http://colibase.bham.ac.uk/).
GATC sites protected from methylation are shown in boldface, with 9 base pairs of flanking sequence on each side.
The distance from the “G” in GATC to the “A” of the start codon for a particular gene is shown in base pairs. For regions where divergent transcription occurs, the distances to both genes/open reading frames is shown in base pairs.
Regulatory proteins known to protect methylation of a specific GATC site, based on in vitro methylation protection, are shown in boldface. Other potential methylation-blocking proteins are shown based on indirect data such as presence of a consensus regulatory protein binding site, observation of protein binding near the GATC sequence, or increased methylation of a GATC site observed in a regulatory mutant of E. coli.
Another nonmethylated GATC site identified was in the gut (also known as srl) operon, controlling uptake of the alcohol sugar glucitol (bp 2823768). A binding site for CAP was identified near the nonmethylated GATC site located at −44.5 (GATC-44.5) relative to the transcription start site (263), suggesting the possibility that binding of CAP to the gut promoter blocks methylation of the GATC −44.5 site (note that in Table 1 this GATC site is 86 bp upstream of the AUG start site for gutA and is thus labeled “−86”). Analysis of DNA methylation in E. coli containing a deletion of the crp gene, coding for CAP, showed that methylation protection of the GATC-44.5 was reduced from 95% in crp+ cells to 50% in Δcrp cells. These data supported the hypothesis that CAP contributes to methylation protection of GATC-44.5 in vivo. However, further analysis of the gut operon showed that although cAMP-CAP binds to sites overlapping GATC-44.5, CAP does not protect this site from Dam methylation (263). Instead, the GutR repressor, which also binds at GATC-44.5, blocks methylation of this site both in vitro and in vivo. GutR-dependent protection of methylation of GATC-44.5 in vivo was not observed in the presence of glucitol, an activator of gut transcription, indicating that under these conditions GutR was no longer bound at GATC-44.5, allowing methylation of this site by Dam. However, methylation of GATC-44.5 did not affect binding of GutR to the gut regulatory region. These results led to the conclusion that although methylation protection indicates the presence of a DNA binding site in vivo, the absence of methylation protection of a GATC site does not prove the absence of binding of a protein at that site (263).
Wang and Church also identified nonmethylated GATC sites within the mtl (mannitol, bp 3769597), cdd (deoxycytidine deaminase, bp 2229798), flh (flagellar synthesis, bp 1976481), psp (stress response, bp 1366007), and fep (iron transport, bp 621523) operons (272). Using a similar approach in which nonmethylated GATC sites in the E. coli chromosome were cloned by digestion with MboI and AvaI, Hale et al. identified four nonmethylated GATC sites in the regulatory regions of the ppiA (bp 3490085), yhiP (bp 3638351), rspA (bp 1653241), and b1776 (bp 1859455) genes (99). Protection of the ppiA GATC site was dependent upon growth phase and carbon source. Protection of a GATC site near yhiP required leucine-responsive regulatory protein (Lrp) and was leucine responsive, similar to the case for some operons controlled by this global regulator (44, 68, 188, 189). The other GATC sites were protected under all the environmental conditions examined (99). A more comprehensive approach to identification of nonmethylated GATC sites was undertaken by Tavoizoie and Church (251); this approach allowed 12 additional sites to be identified, all of which were located within 5′ noncoding regions of genes and open reading frames (Table 1).
Recent work by Blomfield's group on fim regulation controlling type 1 pili has identified two nonmethylated GATC sites at bp 4537512 and 4538525 in the E. coli chromosome near yjhA that are stably nonmethylated, separated from the fim locus by 1.4 kilobase pairs (80). These GATC sites are located near cis-active element regions 1 and 2, both of which play positive roles in transcription of the fimB recombinase gene, controlling type 1 pilus phase variation together with FimE (239). Binding of two regulatory proteins, the NanR sialic acid-responsive regulator and NagC, the N-acetylglucosamine-responsive regulatory protein, is required to activate fimB expression. Binding of NanR to region 1 blocks methylation of one adjacent GATC site, and binding of NagC to region 2 blocks methylation of the second GATC site. Only a fraction of the two GATC sites are nonmethylated after growth in glycerol minimal medium (239). Methylation protection of these GATC sites is not observed after addition of sialic acid (also known as N-acetyl-neuraminic acid). This likely occurs via inhibition of NanR binding, which is sensitive to sialic acid and inhibition by NagC via binding of N-acetylglucosamine-6-phosphate generated by sialic acid catabolism. Thus, binding of NanR and NagC controls methylation of two GATC sites adjacent to yjhA, likely by steric hindrance of Dam. However, mutation of the GATC site adjacent to region 1 did not affect fimB expression (239), indicating that methylation of this GATC site does not, in turn, modulate NagC binding. Moreover, in a dam mutant, expression of fimB is decreased, the opposite of what would be expected if GATC methylation inhibits NagC and NanR binding. These results indicate that the reported regulation of fim expression by Dam (199) does not occur via methylation of the GATC sites located near regions 1 and 2 adjacent to fim.
In summary, a small fraction of the approximately 20,000 GATC sites in the E. coli chromosome are totally or partially nonmethylated in any given growth state and environmental condition. The protection of GATC site methylation by Dam is dependent upon competition between Dam and specific DNA binding proteins. Dam appears to methylate most GATC sites in a highly processive manner, as discussed above. Recently, however, analysis of methylation of the regulatory GATC sites in the pap operon indicates that they are not methylated processively (32) . That is, Dam binds to pap DNA, methylates one GATC site, and then dissociates before methylating the second site. This effectively reduces the ability of Dam to compete with proteins that bind to DNA sequences containing one or more GATC sites. Bergerat et al. first proposed that DNA sequences surrounding GATC sites may dictate the avidity of Dam for its target sites (23). Mutation of the AT-rich flanking sequences of the pap GATC sites to CG sequences increased processivity, which appeared to be due to changes in the kinetics of methyl transfer and not in binding affinity (203). Analysis of known nonmethylated GATC sites tentatively suggests a trend toward having AT-rich flanking sequences, though this is not always the case (Table 1).
Since DNA methylation patterns are formed as a result of binding of proteins primarily at gene regulatory regions, they are altered by growth conditions that affect regulatory protein level(s) and/or DNA binding properties. As discussed above, identification of nonmethylated GATC sites has been used as a sort of natural in vivo footprint system to track binding of regulatory proteins under different environmental conditions (251, 272). In addition, it is clear that a subset of nonmethylated GATC sites (for example within the pap, sfa, daa, agn43, and other operons [see below]) play important roles in epigenetic regulation. In these systems, not only is a DNA methylation pattern established by protection of specific GATC sites by a regulatory protein(s), but methylation of the GATC site(s), in turn, modulates regulatory protein binding (263). This results in two heritable states: either the regulatory protein is bound to a specific DNA sequence containing a GATC site(s), protecting it from methylation, or the regulatory protein is not bound due to a reduction of binding affinity for target sequence(s) caused by GATC methylation. Clearly, only a subset of all nonmethylated GATC sites have these particular properties and are involved in epigenetic control systems. For example, as shown in Table 1, DNA methylation patterns have been shown to directly control expression of agn43 (111, 271) but do not control the gut (srl) operon (263) and do not appear to directly regulate fim (239). Further study will be necessary to determine if any of the other genes containing nonmethylated GATC sites in their regulatory regions are under methylation pattern control (Table 1).
DNA ADENINE METHYLATION-DEPENDENT REGULATORY SYSTEMS
In the sections below we describe the current state of knowledge regarding how DNA methylation controls bacterial gene expression. Our focus for each methylation-controlled operon is on aspects of regulation affected by methylation and not on complete descriptions of regulatory networks.
Pap Pili
Pyelonephritis-associated pili play an important role in attachment of UPEC to uroepithelial cells lining the upper urinary tract, facilitating colonization of the kidneys. Pap pilus expression switches on and off within individual cells in the bacterial population, a process known as phase variation. The biological role of Pap pilus phase variation is not known, but possibilities include (i) escape from immune detection; (ii) facilitation of a bind-release-bind series of events in which successive generations of bacteria ascend the urinary tract; and (iii) controlling growth of UPEC by modulating the effects of contact-dependent growth inhibition, a newly described bacterial phenomenon (3).
DNA adenine methylase controls Pap phase variation by methylation of two GATC sites, one proximal to the pap pilin promoter (GATCprox), located 53 bp from the papBA transcription start site, and the other located 102 bp upstream of GATCprox, designated GATCdist (Fig. 3A). Note that these two GATC sites are located within Lrp DNA binding site 2 and site 5, respectively. Methylation at these two pap GATC sites controls the binding of the global regulator Lrp (44, 189) and the coregulatory protein PapI (118, 138) to pap DNA sites 1, 2, and 3 proximal to the papBA pilin promoter and to sites 4, 5, and 6 distal to papBA. Lrp appears to bind cooperatively to sites 1, 2, and 3 or to sites 4, 5, and 6 (193). Binding to all six sites can be achieved in vitro by addition of sufficient Lrp but rarely occurs in vivo based on analysis of the methylation states of GATCprox and GATCdist (41). In ON-phase cells GATCdist is nonmethylated and GATCprox is methylated (41) (Fig. 3D). Protection of GATCdist from Dam methylation requires both Lrp and PapI based on the observation that GATCdist is fully methylated in either an lrp or a papI mutant (40, 41). In contrast, OFF-phase cells display the converse DNA methylation pattern in which GATCprox is nonmethylated and GATCdist is methylated (Fig. 3A). Protection of GATCprox requires Lrp but not PapI (41, 263). Based on these in vivo DNA methylation patterns together with in vitro studies of Lrp binding, it was concluded that in ON-phase cells PapI-Lrp binds to sites 4, 5, and 6, protecting GATCdist from Dam, and in OFF-phase cells Lrp binds to sites 1, 2, and 3, protecting GATCprox from Dam (41). These DNA methylation patterns result from competition between Dam and Lrp for binding at sites 1, 2, and 3 and at sites 4, 5, and 6, containing GATCprox and GATCdist, respectively, as discussed in detail below.
FIG. 3.
The Pap OFF- to ON-phase transition mechanism. The regulatory region of the pap operon is shown at the top, with six DNA binding sites for Lrp (gray rectangles) and GATCprox and GATCdist within Lrp binding sites 2 and 5, respectively. The divergent papI and papBA promoters are shown with arrows. Lrp (ovals), PapI (triangles), and PapB (diamonds) are shown. The methylation states of the top and bottom DNA strands of a GATC site are depicted by an open circle (nonmethylated) or closed circle (methylated). The OFF-to-ON switch is described in the text.
The Pap OFF- to ON-phase transition.
In Fig. 3A (lower section), pap regulatory DNA with the OFF-phase DNA methylation pattern is depicted: GATCdist is fully methylated, and GATCprox is fully nonmethylated as a result of binding of Lrp at pap sites 1, 2, and 3 overlapping GATCprox. Transcription from papBA is blocked by binding of Lrp at sites 1, 2, and 3 overlapping the promoter, likely as a result of steric hindrance of RNA polymerase binding (278). The OFF-phase state is stabilized by two main factors: mutual exclusion and DNA methylation. Binding of Lrp at sites 1, 2, and 3 reduces the affinity of Lrp for pap sites 4, 5, and 6 (overlapping GATCdist) by 10-fold via a phenomenon that has been denoted “mutual exclusion” (116). Mutual exclusion requires a supercoiled pap substrate by an unknown mechanism. One possibility is that Lrp could induce bending at sites 1, 2, and 3, propagating an alteration in twist to sites 4, 5, and 6. Methylation of GATCdist reduces the affinity of Lrp for sites 4, 5, and 6 by about 20-fold based on in vitro DNA binding measurements (118). In addition, there is an intrinsic twofold-higher affinity of Lrp for sites 1, 2, and 3 versus 4, 5, and 6. These factors contribute to stabilization of the OFF-phase Pap expression state (116).
The transition from the OFF to ON phase requires that GATCprox be methylated by Dam; either a dam mutant E. coli strain or a GCTCprox A-to-C transversion mutant that cannot be methylated by Dam but does not significantly alter the affinity of Lrp for sites 1, 2, and 3 is locked in the OFF phase (41). In contrast, methylation of GATCdist has an inhibitory effect on the OFF-to-ON switch: overexpression of Dam by just fourfold prevents the OFF-to-ON switch. Moreover, E. coli containing a GCTCdist mutation that blocks Dam methylation is locked in the ON phase, even under conditions of Dam overexpression (41). These data support the hypothesis that OFF-to-ON switching requires DNA replication to generate a hemimethylated GATCdist intermediate, which is bound by PapI-Lrp with a higher affinity than DNA with a fully methylated GATCdist (118). A low level of the coregulatory protein PapI, required for Pap pili expression (138, 193, 194), increases the affinity of Lrp for pap DNA hemimethylated at GATCdist but does not enhance binding of Lrp to pap DNA fully methylated at GATCdist (118). Notably, the hemimethylation state of pap matters: PapI increases Lrp's affinity for DNA methylated on the top strand at GATCdist about fourfold more than for DNA methylated on the bottom strand (118). These results raise the intriguing possibility that Pap phase switching may be biased: daughter cells receiving a DNA methylated on the top strand may have a higher probability of switching to the ON phase than cells receiving DNA methylated on the bottom strand.
PapI is a small (ca. 9-kDa) coregulatory protein expressed from the papI promoter divergent to the papBA pilin promoter (Fig. 3A, top). PapI increases the affinity of Lrp for pap site 5, and to a lesser extent site 2, but has no effect on binding of Lrp to any of the other four Lrp binding sites (118) (Fig. 3C). pap Lrp binding sites 5 and 2 share the sequence “ACGATC,” which differs from the other four pap Lrp binding sites and the ilvIH Lrp binding site 2 (65, 129, 138), which do not display PapI-dependent Lrp binding (118). All pap Lrp binding sites share the sequence “GNNNTTT” with the Lrp binding consensus determined by systematic evolution of ligands by exponential enrichment (64).
PapI does not appear to bind specifically to pap DNA by itself, based on gel shift analysis (138) and DNA cross-linking (118). DNA methylation interference indicated that methylation of bases in the sequence 5′-GNCGAT-3′ overlapping GATCdist in the top strand and 3′-TGCTAG-5′ in the bottom strand significantly reduced PapI-dependent Lrp binding compared with binding of Lrp alone. Methylation of the bottom-strand cytosine complementary to the guanine of “GATC” (meC9) blocked formation of the ternary PapI-Lrp-pap site 5 complex without affecting Lrp binding (118). These results support the hypothesis that enhancement of Lrp binding to site 5 occurs via formation of a PapI-dependent ternary complex with Lrp and pap DNA. Cross-linking with a photoactivatible 9-Å azidophenacyl cross-linker three bases from the presumptive PapI binding sequence “ACGATC” showed that PapI and Lrp were both cross-linked to pap DNA in the ternary complex with nonmethylated DNA, while only Lrp was cross-linked with DNA methylated at C9 (118). These results indicate that PapI is located near the pap ACGATC sequence in the PapI-Lrp-pap site 5 ternary complex and may directly contact this sequence.
The observation that PapI (100 nM) increases Lrp's affinity for pap site 2 (which contains the ACGATC PapI-specific sequence identical to site 5) (118) presents an apparent paradox, since this should block pap transcription due to its close proximity to the papBA pilin promoter (278). Further analysis showed that at low PapI levels significant enhancement of Lrp binding occurred at sites 4, 5, and 6 (CGATCdist) but not at sites 1, 2, and 3 (CGATCprox) (118). At 5 nM PapI, the affinity of Lrp was fourfold higher for pap sites 4, 5, and 6 (Kd = 0.25 nM) than for sites 1, 2, and 3 (Kd = 1.0 nM). Conversely, in the absence of PapI, the affinity of Lrp for sites 1, 2, and 3 (Kd = 1.2 nM) was about twofold higher than that for sites 4, 5, and 6 (Kd = 2.5 nM). Thus, binding of Lrp at sites 4, 5, and 6 should be favored at low PapI levels, resulting in activation of papBA transcription. This, in turn, would increase the PapI level via a PapB-mediated positive feedback loop whereby PapB binds upstream of the papI promoter and helps activate PapI expression (11, 85, 288) (Fig. 3B). High PapI levels could potentially shut off pap transcription by increasing the binding of PapI-Lrp complexes at promoter-proximal sites 1, 2, and 3. However, this is prevented by methylation of GATCprox by Dam, which specifically blocks PapI-dependent Lrp binding without affecting binding of Lrp alone (118).
To determine if the essential role of methylation of GATCprox in the OFF- to ON-phase transition is to specifically block PapI-dependent Lrp binding to sites 1, 2, and 3, the wild-type CGATCprox sequence was mutated to TGATCprox to specifically inhibit PapI-dependent Lrp binding. It was reasoned that under conditions in which PapI-dependent binding of Lrp to sites 1, 2, and 3 was blocked, switching from OFF to ON phase should occur in the absence of Dam. Analysis of the TGATCprox mutant showed that PapI-dependent Lrp binding to sites 1, 2, and 3 was inhibited but binding of Lrp was unaffected both in vitro and in vivo. Switch frequency analysis of E. coli containing the TGATCprox mutation showed that the OFF-to-ON rate (5.6 × 10−4/cell/generation) was about sevenfold higher than that of wild-type cells (8.2 × 10−5/cell/generation). Notably, in a dam null mutant background cells were locked in the ON-phase state, showing that methylation is not required for pap transcription under conditions in which PapI-dependent binding of Lrp to pap site 2 containing GATCprox is blocked. These results support the conclusion that methylation at GATCprox is required for the OFF- to ON-phase transition by specifically inhibiting PapI-dependent Lrp binding to sites 1, 2, and 3 (Fig. 3C, top).
Environmental mechanisms for switch control.
Binding of Lrp at sites 4, 5, and 6, together with binding of cAMP-CAP at −215.5 (relative to the papBA transcription start site) (277), enhances papBA transcription via contact between CAP activating region 1 and the αC-terminal domain of RNA polymerase (277). In this way, Pap pilus expression is environmentally controlled by carbon source via the cAMP level. The role of Lrp may be structural, bending pap DNA between the CAP binding site at −215.5 and the papBA promoter to facilitate contact between cAMP-CAP and the αC-terminal domain. This results in transcription initiation from papBA and expression of PapB, which has been reported to bind with highest affinity to a site between the papI promoter and the CAP binding site (85), stimulating papI transcription, which constitutes a positive feedback loop (Fig. 3D). The high PapI level ensures binding of PapI-Lrp to sites 4, 5, and 6, and methylation of GATCprox prevents binding of PapI-Lrp to sites 1, 2, and 3, which would shut off papBA transcription and turn the switch OFF (278). The fact that both PapI and PapB are required for switching from the OFF to ON phase raises a chicken-and-egg problem that has not been adequately addressed: which regulatory factor initiates the switch? We speculate that regulation is at the level of PapB expression and that a low level of papBA mRNA is made following DNA replication and Lrp/H-NS dissociation from sites 1, 2, and 3 (266). If this papBA mRNA is rapidly translated, it would induce papI transcription, initiating the OFF-to-ON switch cascade. There is indirect evidence to support the idea that there may be translational control involved in Pap pilus expression, since a rimJ mutation affects pap gene regulation (280-282). RimJ acetylates ribosomal protein S5 in the 30S subunit. Thus, it is possible that ultimately the initiation of the Pap OFF-to-ON switch may be dependent upon the translation of a basal level of papBA mRNA present immediately following DNA replication.
The global regulatory protein H-NS is not required for Pap phase variation (266), but it does modulate Pap gene expression and Pap switch rates. H-NS represses papBA transcription in response to low temperature (94), high osmolarity (283), and rich medium (283). This may occur by specific binding of H-NS to the pap regulatory region, as evidenced by blocking of methylation of both pap regulatory GATC sites in vitro and in vivo (279). Binding of H-NS near the papBA promoter could inhibit binding of RNA polymerase, repressing transcription. Notably, at 37°C H-NS appears to positively affect Pap phase variation, since the OFF-to-ON switch rate is reduced in an hns mutant (266, 283). This positive effect of H-NS on the OFF- to ON-phase transition could occur via competition with Lrp at sites 1, 2, and 3, which would help to move PapI-Lrp to sites 4, 5, and 6, analogous to the role of methylation of GATCprox (Fig. 3C).
Another environmental input into Pap phase variation is mediated by the CpxAR response regulatory system (117, 127). Under certain conditions that stress the cell envelope, including high pH, CpxA located in the inner membrane autophosphorylates and then transfers a phosphate group to CpxR to yield CpxR-phosphate (CpxR-P) (176, 211). CpxR-P binds to sites overlapping all six pap Lrp binding sites, competes with Lrp for binding to these sites, and shuts off papBA transcription and Pap pilus expression (115, 117). Notably, CpxR-P binding to pap sites 1 to 6 is not inhibited by DNA methylation, in contrast to Lrp, even though CpxR-P, like Lrp, binds at sites overlapping the pap GATCprox and GATCdist sites. The biological role of CpxAR regulation of Pap pilus expression is not fully clear. One possibility is that under conditions of envelope stress it makes sense to curtail pilus expression to prevent further damage to the membrane. Another provocative possibility is that under conditions of stress UPEC cells stop making Pap pili, making them susceptible to contact-dependent growth inhibition (3). The physiologic significance of this is unknown, but it might contribute to survival under harsh conditions by slowing bacterial metabolism and growth (3).
The Pap ON- to OFF-phase transition.
The Pap ON- to OFF-phase transition occurs at about a 100-fold-higher rate than the OFF- to ON-phase transition (35, 266). Notably, factors including H-NS, carbon source, and osmolarity do not affect the ON- to OFF-phase transition rate (35, 266, 283); therefore it appears that the ON- to OFF-phase transition is relatively constant under different environmental conditions. The ON- to OFF-phase transition has not been thoroughly examined, but based on knowledge of the OFF-to-ON switch mechanism (116-118) (see above), the following model is postulated. Starting with a cell in the ON-phase state (Fig. 4A), DNA replication is postulated to dissociate PapI-Lrp from sites 4, 5, and 6, enabling Dam to compete with Lrp for binding at GATCdist (Fig. 4C) Methylation of GATCdist is essential for the OFF-phase state (41). DNA replication also generates two hemimethylated GATCprox sites, one methylated on the top strand and one on the bottom strand (Fig. 4B). Whether a cell remains in the ON phase or transitions to the OFF state may be dictated by competition of Lrp for binding to pap promoter-proximal sites 1, 2, and 3 versus distal sites 4, 5, and 6 (Fig. 4B). Lrp has about a twofold-higher affinity for the proximal sites than for distal sites, and methylation of GATCprox does not affect Lrp binding to these proximal sites (118). In contrast, methylation of GATCdist inhibits binding of Lrp and PapI-Lrp to the distal sites (118, 194). These two factors should favor binding of Lrp to the proximal sites over the distal sites, which may account in part for the high ON-to-OFF rate observed. Following one additional round of DNA replication, the OFF-phase state is attained (Fig. 4D).
FIG. 4.
The Pap ON- to OFF-phase transition mechanism. See the legend to Fig. 3 for explanations of symbols. The ON-to-OFF switch mechanism is described in the text.
Clearly, the Pap epigenetic switch mechanism is complex, involving distinct DNA methylation and protein-DNA binding states. Therefore, it would be highly useful to have a mathematical model that could predict switch rates under a variety of conditions and identify the key regulatory step(s) determining switch outcome. Liao and coworkers have developed a model for Pap phase variation that takes into account many of the protein-protein and protein-DNA interactions of Lrp, PapI, and Dam described above (131, 297). To rigorously test a model, one would need to alter cellular levels of PapI, Lrp, and Dam and experimentally determine switch rates. In addition, a useful model should be able to predict switch outcomes when the affinities of PapI, Lrp, and Dam for pap DNA have been altered, for example. Although these types of analyses have not yet been carried out, preliminary data suggest that the Markov chain model for Pap may be useful in understanding Pap switch dynamics. However, the frequency of ON-state cells in the population was underestimated, for example (297). Reliable numbers for biochemical parameters of the Pap switch, such as association and dissociation binding constants for PapI-Lrp, Lrp, and Dam at sites 1, 2, and 3 and at sites 4, 5, and 6, and have not yet been obtained. This makes it difficult to determine if the Pap model does not accurately reflect experimental data due to incorrect biochemical parameters used in the model or because assumptions in the model are incorrect or incomplete. Recently, another Pap switch model was developed by Munsky and Khammash (183, 184). Further work as outlined above will be necessary to test these models and determine if they are useful in furthering our understanding of the Pap switch and other epigenetic switch systems (see below).
Pap-Related Systems
Analysis of pilus operons containing regulatory regions with homology to pap indicates that there are two groups: those that are positively regulated by PapI homologues, similar to the pap system, and those negatively regulated by PapI homologues.
PapI homologue acting as a positive regulator of pilus expression.
The regulatory regions of many pilus operons in E. coli, including Pap-related fimbriae (Prf), foo (F1651 pili), clp (CS41 pili), sfa (S pili), daa (F1845), fae (K88), and afa (afimbrial adhesin), share two GATC sites analogous to GATCprox and GATCdist and spaced 102 base pairs apart as in pap (151) (Fig. 5). Moreover, these GATC sites are present within additional conserved sequences, “CGATCdistTTTT” and “CGATCproxTT,” with the entire sequence called a “GATC box” (note the inverse orientations of the GATC boxes in the pilus regulatory sequences shown in Fig. 5). Since the GATC box sequence contains binding sites for Lrp and Dam, as well as a portion of the PapI response element “ACGATC,” this provides the means by which these various pilus operons are controlled by DNA methylation patterns.
FIG. 5.
DNA sequence alignment of the GATC box regions from pilus operons under DNA methylation pattern control. DNA base pairs conserved in all pap family regulatory regions are shaded black with light lettering. The distal and proximal regulatory GATC sites (GATCdist and GATCprox, respectively) are shown. Arrows show the inverted orientation of the two GATC box regions. The accession numbers for the sequences shown are as follows: pap, X14471; foo, AF109675; sfa, S59541; afa, X76688; daa, M98766; clp, L48184; fae, X77671; pef, L08613.
The sfa, daa, prf (pap-related fimbria), and afa-3 operons appear to be regulated by DNA methylation patterns, analogous to regulation of pap. Each of these pilus operons codes for a PapI and a PapB homologue, and cross-complementation between the PapB and PapI homologues between prf and sfa (182) and between pap and sfa and daa (267) was shown. The DaaF and SfaC proteins function similarly to PapI, positively regulating expression of daa and sfa, respectively, by facilitating binding of Lrp to promoter-distal binding sites overlapping GATCdist (267). Methylation of the pap-related GATC sites, in turn, controls binding of Lrp.
PapI homologue acting as a negative regulator of pilus expression.
Two methylation-controlled pilus operons in E. coli, clp (CS31A) and fae (K88), and one pilus operon in Salmonella enterica serovar Typhimurium, pef, share common regulatory features with pap but have distinct differences as well. The regulatory regions of clp, fae, and pef contain conserved GATC box sites and spacing identical to that in pap (Fig. 5). Also similar to pap, binding of Lrp to regulatory DNA is controlled by DNA methylation and a PapI homologue. However, all three methylation-controlled operons are carried on plasmids, and in each case PapI homologues negatively control phase variation and transcription.
K88 pili, expressed by enterotoxigenic E. coli infecting pigs, is not under phase variation control, in contrast to the case for all other Pap family members, (124). The fae regulatory region shares GATC box sequences with pap, spaced 102 bp apart, as well as a PapI homologue, FaeA, and a PapB homologue, FaeB (124). A third regulatory GATC site (GATC-III) is present 28 bp downstream (toward the faeB promoter) of GATCprox, and two IS1 sequences are present between faeB and faeA (Fig. 5). In contrast to the case for pap, FaeA and Lrp act to negatively control fae transcription. Data from Huisman et al. indicated that in the absence of FaeA, Lrp binds at sites overlapping GATCprox, protecting it from methylation by Dam (124, 125). However, in contrast to the case for pap, this Lrp binding has little effect on pilin transcription. In the presence of FaeA, the PapI homologue, additional binding of Lrp near GATC-III occurs, blocking methylation of both GATCprox and GATC-III and reducing fae transcription. This GATC-III site shares the “CGATCTTTTA” sequence of the pap and fae GATCdist sites, though in opposite orientation, possibly accounting for FaeA-mediated binding of Lrp to this region. However, FaeA-mediated binding of Lrp to GATCdist was not observed. In fact, mutation of the GATCdist site to GTTC sequence was lethal due to overproduction of K88 pili, indicating that methylation of GATCdist normally blocks binding of FaeA-Lrp. Whether FaeA-Lrp binds to GATCdist under normal physiologic conditions is not clear, but it is possible that binding to a hemimethylated GATCdist site might occur immediately following DNA replication, stimulating K88 expression under certain conditions. Another difference between regulation of fae and pap is in control of faeA and of papI transcription. In the case of pap, papI is regulated by PapB via a positive feedback mechanism (116), whereas in fae, an IS1 insertion apparently disrupts this positive feedback. Instead, FaeA may bind to its own promoter, acting as a positive autoregulator (125).
Regulation of the clp operon, coding for CS31A pili, which are expressed by enterotoxigenic E. coli, shares common regulatory features with pap but, like for fae and pef, has distinct differences as well. In E. coli isolate CS31A harboring clp, CS31A pili are under phase variation control, yet the plasmid-carried clp operon does not have a papI homologue associated with it (62, 173). It seems likely that a pap operon identified on the chromosome of E. coli CS31A supplies PapI in trans, but this has not been confirmed. Analysis of clp regulation in E. coli K-12 (no papI homologue present) showed that Lrp and the PapB homologue ClpB repressed clp transcription. However, even in the presence of Lrp and ClpB, a moderate level of clp pilin transcription was observed. In addition, in lrp+ clpB+ cells lacking Dam, transcription was almost maximally derepressed. Introduction of the PapI homologue AfaF resulted in phase variation of CS31A expression: instead of a normally distributed transcription of CS31A among the cell population, individual cells either transcribed (ON phase) or did not transcribe (OFF phase) the clp operon, with the methylation pattern of the former cells being GATCdist nonmethylated and GATCprox methylated and with the converse pattern for the latter cells. These results can be explained if Lrp and ClpB bind near the clp pilin promoter, moderately repressing transcription but still allowing some pilus expression to occur in the absence of the PapI homologue AfaF. The repressive effect of Dam on clp transcription could occur via methylation of GATCdist to block binding of Lrp to promoter-distal sites. Addition of AfaF should increase the affinity of Lrp for both GATCdist and GATCprox, similar to the case for pap. However, it may be that the affinity of AfaF-Lrp is marginally higher for GATCprox than GATCdist, the reverse of the case for pap, which could explain why only a small fraction of cells are in the ON phase in the presence of constitutively expressed AfaF. This could also explain why the transcription of AfaF+ OFF-phase cells appears to be lower than that of cells lacking AfaF (which do not show phase variation), since AfaF would increase Lrp's affinity for clp pilin promoter-proximal sites and more efficiently block transcription than Lrp alone.
The clp operon and the closely related foo operon, coding for F1651 pili (24, 63, 101), have the distinction of being the only members of the Pap regulatory family controlled by the aliphatic amino acids leucine and alanine. Alanine and, to a lesser extent, leucine reduce the expression of CS31A pili (62, 173). This appears to occur as a result of diminished PapI homologue-dependent binding of Lrp to GATCdist and increased binding of Lrp to GATCprox, locking cells in the OFF-phase transcription state. Lrp has a binding site for aliphatic amino acids, which appears to modulate the multimeric state of Lrp between dimeric, octameric, and hexadecameric states (57, 59). If Lrp binding sites are phased such that they occur on the same DNA face, then an octameric Lrp could engage up to four sites, contributing to binding cooperativity. The reason that the transcription of certain operons, including clp, is modulated by alanine and leucine whereas that of other operons, such as pap, is not is unclear. However, recent results with the ilvIH operon, which is repressed by leucine, indicate that leucine inhibits long-range interactions between Lrp proteins bound to different sites in the ilvIH regulatory region (58).
The pef operon in Salmonella enterica serovar Typhimurium codes for plasmid-encoded fimbriae (Pef fimbriae) that appear to play a role in intestinal colonization (17, 126). Pef fimbriae are encoded on the pSLT virulence plasmid (87). Pef pili are expressed in vivo in bovine ligated ileal loops (126) but in the laboratory are expressed only in acidic (pH 5.1) rich broth in standing culture (190). Under these conditions, Pef pili are expressed under phase variation control. The pefI gene is located about 6 kb away from the pef regulatory region, and PefI acts negatively on Pef phase variation, blocking Pef pilus expression when expressed on a multicopy plasmid (190). This appeared to occur via increased affinity of Salmonella Lrp, which is almost identical to E. coli Lrp (one amino acid difference), for DNA sites overlapping GATCprox (previously denoted GATC II). Binding of Lrp at GATCdist appeared to correlate well with the ON-phase state, similar to the case for pap. Thus, a common theme for pef, clp, and fae is that in each case PapI homologues act negatively by increasing the binding of Lrp to pilin promoter-proximal sites, protecting GATCprox from methylation, and inhibiting transcription. The reason why PapI-Lrp binds with the highest affinity to sites around GATCdist in pap and closely related operons (see above) and to sites around GATCprox in pef, clp, and fae is not known. Analysis by Hernday et al. showed that the affinity of PapI-Lrp for Lrp binding site 5 containing GATCdist was significantly higher than its affinity for site 2 containing GATCprox (118). Analysis of site 2 and 5 regions in pap versus pef, clp, and fae does not provide any simple possible explanation for the mechanism by which PapI-Lrp affinity is reversed in these operons (Fig. 5). However, this regulatory distinction may explain the reason why the papI homologues in pef, clp, and fae have been disconnected from the positive feedback loop operating in other pap-related operons. If they were connected, one would expect that the consequence would be to turn off pilus expression entirely. Since pef and clp expression is under phase variation control, this shows that a positive feedback loop is not essential for phase variation. In fact, Pap phase variation occurs in papI-minus mutants containing PapI expressed constitutively on a plasmid, showing that disconnection of the feedback loop is not an essential feature of phase variation, although it likely contributes to signal-to-noise parameters. Although it is not clear why pef, clp, and fae display this regulatory difference from pap, it provides an additional means by which Pef, CS31A, and K88 pilus expression can be controlled by environmental and host factors via regulation of pefI, the resident pap operon(s), and faeA, respectively.
Phase-Variable Outer Membrane Protein Ag43
Besides the pap regulatory family of operons described above, the only other characterized phase variation system regulated by DNA methylation patterns is a gene originally designated by B. Diderichsen as flu for “fluffing,” based on the propensity of bacteria to “aggregate, fluff, and sediment” (71). Henderson et al. and Owen et al. (111, 200) later identified and characterized an autotransporter protein denoted antigen 43 (Ag43), which was shown to be identical to the flu product, and the gene was renamed agn43. The regulatory region of agn43 has a consensus binding site for the OxyR repressor (296) present on a number of genes regulated by oxidative stress, including mom in phage Mu (see below). In addition, three closely spaced GATC sites (GATC-I, GATC-II, and GATC-III) are present in the regulatory region within the OxyR binding sites (Fig. 6A). Transcription of agn43 begins at the “G” of the promoter-distal GATC-I site (269, 271) (Fig. 6A). Binding of OxyR to the agn43 regulatory region represses agn43 transcription in vivo based on the phase-locked ON phenotype of oxyR mutants (112). Based on these observations, it was proposed that the Ag43 phase switch is controlled by competition between OxyR and Dam for binding and methylation within the agn43 regulatory region. Methylation of any two of the three agn43 regulatory GATC sites was sufficient to inhibit binding of OxyR in vitro and allow phase variation to occur in vivo (269), although all three sites appear to be required for attaining normal phase variation rates (271). Binding of OxyR protected all GATC sites from methylation (60) and repressed agn43 transcription in vitro (271). This regulatory arrangement is similar in basic form to that of the pap regulatory family: in both systems binding of a global regulator to upstream regulatory sequences blocks methylation of GATC sites within the region and directly affects transcription. Methylation of these sites, in turn, inhibits regulatory protein binding. For Pap, the switch between the OFF and ON phases is facilitated by the coregulator PapI, which controls binding of Lrp between two GATC site regions by altering its affinity for pap DNA. For OxyR, which binds to one DNA region of about 60 bp encompassing all three GATC sites, it is not clear whether environmental inputs control phase switching as they do for pap. One possibility that has been considered is that the oxidative state of OxyR might be important in Ag43 regulation. This hypothesis is attractive since it would tie the oxidative stress response to biofilm formation, which is aided under certain conditions by Ag43 (66).
FIG. 6.
Model for phase variation of the outer membrane protein Ag43. The regulatory region of the agn43 operon is shown at the top (A). The three agn43 regulatory GATC sites, GATC-I (far left), GATC-II (middle), and GATC-III (right) are depicted as gray rectangles (A). The methylation states of the top and bottom DNA strands of a GATC site are depicted by an open circle (nonmethylated) or closed circle (methylated). “Rep” indicates a DNA replication event. The Ag43 switch model is discussed in the text.
OxyR exists in two redox states within cells, formed by disulfide bonding between cysteines 199 and 208. Disulfide bond reduction occurs enzymatically, primarily by glutaredoxin 1 (295). Data from Schembri and Klemm showed that expression of type 1 pili (fim) and P pili (pap) blocked Ag43 expression (225), which was proposed to occur via disulfide bridge formation in these pili, possibly driving OxyR toward the reduced state, repressing Ag43 expression. If this is correct, then transcription of other genes in the OxyR regulon, such as katG, should be affected, but this was not tested. Further analysis of the possible role of the redox state of OxyR in Ag43 regulation was done using OxyR(A233V) and OxyR(H198R) mutants, which are locked in the oxidative form and constitutively activate genes in the OxyR regulon (152). Neither mutant was found to repress Ag43 expression (112, 224), and it was concluded that only the reduced form of OxyR represses agn43 expression. However, Wallecha et al. showed that the affinity of OxyR(A233V) for nonmethylated agn43 regulatory DNA was at least fivefold lower than that of oxidized, wild-type OxyR and that the affinity of OxyR(H198R) was also lower than that of wild-type OxyR (270). Thus, the assumption that these mutants accurately reflect the role of oxidized wild-type OxyR does not appear to be valid. In vitro analysis showed that oxidized wild-type OxyR binds to agn43 DNA and represses agn43 transcription (270). Therefore, it appears that the redox state of OxyR does not control phase variation of Ag43 (270).
The mechanism(s) by which agn43 expression switches between the OFF and ON states is not known, though it likely requires DNA replication to generate a hemimethylated DNA intermediate (60). OxyR affinity for fully methylated agn43 regulatory DNA is too low to be measured by electrophoretic mobility shift, but the Kd of binding to nonmethylated agn43 is about 2 nM. Binding of OxyR to hemimethylated agn43 methylated on the top or bottom strand is similar, with at least a sixfold reduction in affinity compared to nonmethylated DNA (60). The intermediate affinity of OxyR for hemimethylated agn43 provides a switch transition mechanism: immediately following DNA replication OxyR presumably dissociates from agn43 DNA in OFF-phase cells, giving a window of opportunity for Dam to compete with OxyR due to the decreased affinity of OxyR for hemimethylated DNA (Fig. 6B). Full methylation of the agn43 GATC sites could occur in one step, preventing OxyR binding and repression, forming the ON-phase state. Similarly, hemimethylated DNA could facilitate the ON- to OFF-phase transition by providing an opportunity for OxyR to bind to hemimethylated agn43 GATC sites, blocking their methylation by Dam (Fig. 6C). After an additional round of replication, the OFF-phase DNA methylation pattern would be formed in half of the transitioning cells (Fig. 6D)
It is not clear if environmental or cellular factors directly regulate Ag43 switching, but it is possible that SeqA may play a role. SeqA binds to agn43 regulatory DNA containing hemimethylated GATC sites but does not bind to fully methylated or nonmethylated DNAs (60). The OFF- to ON-phase rate was reduced in a seqA mutant, but much of this effect could be accounted for by a reduction in the Dam/DNA ratio caused by increased asynchronous initiation of DNA replication that occurs in the absence of SeqA, which normally sequesters oriC and plays a critical role in timing of DNA replication (36). Under these conditions the balance is tipped toward repression, since OxyR more effectively competes with Dam.
VSP Repair
In enteric bacteria, very-short-patch (VSP) repair recognizes G-T mismatches and corrects them to G-C (25). VSP repair activity is partially redundant with Dam-directed mismatch repair, and the mechanisms that coordinate the use of either system are not fully understood (25). MutL and MutS are required for VSP repair, while MutH is not involved. Dam methylation is dispensable for VSP repair: mismatched duplexes containing GATC sites are repaired with similar efficiencies in methylated and nonmethylated DNA substrates. However, Dam− mutants of E. coli are defective in both Dam-directed mismatch repair and VSP repair (19), and their VSP repair defect appears to be caused by lack of Dam methylase. Synthesis of Vsr, the endonuclease that initiates VSP repair, is reduced in Dam− mutants, suggesting that Dam methylation regulates Vsr synthesis (19). The vsr gene is cotranscribed with dcm, the gene for Dcm methylase; however, synthesis of Dcm remains unaffected in a Dam− background (19). The absence of GATC sites in the dcm promoter (67) provides further evidence that Dam-mediated control of the Vsr level is not transcriptional. Because DNA modification cannot be expected to act directly at the posttranscriptional level, we are left with two alternative explanations: (i) the Dam methylase might have additional, hitherto unknown functions unrelated to DNA modification, or (ii) more likely, Dam methylation may regulate Vsr synthesis in an indirect fashion, by controlling transcription of one or more cell functions involved in posttranscriptional control. The case of vsr is unlikely to be unique, since evidence for posttranscriptional regulation by Dam methylation has been also found in the std fimbrial operon of Salmonella enterica (130). These examples raise the possibility that Dam methylation might regulate cell functions involved in RNA stability, mRNA translation, or protein turnover. However, the underlying molecular mechanisms remain to be identified.
Bacteriophage Infection
In the genomes of certain virulent phages of enteric bacteria, GATC sites are relatively scarce. Total E. coli DNA contains GATC sites at a frequency of one GATC site per 232 bp, which approaches the predicted random frequency of one GATC site per 256 bp (110). In contrast, bacteriophage T7 contains 6 GATC sites, while the predicted number is 141 (174). In the genomes of temperate phages such as E. coli lambda and Salmonella P22, the frequency of GATC sites is also lower than expected from their nucleotide composition, but the differences are not as spectacular as in the case of T7 (110, 174). Other phage genomes contain GATC sites at frequencies similar to that found in the host genome (31). It has been proposed that scarcity of GATC sites in the genomes of virulent phages may protect against DNA digestion by the host MutH endonuclease (70). Note that Dam-directed mismatch repair requires partial degradation of the daughter strand and resynthesis by host DNA polymerase I and DNA ligase, a laborious process that may not be feasible during the late stages of phage growth. On the other hand, T-even, P1, and other phages carry their own dam genes, which may ensure methylation of GATC sites during the lytic cycle (31). Aside from conferring protection from accidental MutHLS cleavage of concatemeric DNA, T4Dam may also protect T4 phage DNA from restriction by competing P1 phage (177).
Regulation of DNA packaging in bacteriophage P1.
Packaging of phage P1 DNA into capsids proceeds by a processive headful mechanism that uses concatemeric phage DNA molecules produced by rolling-circle replication during the late stages of phage infection (291). Packaging is initiated at the pac site, a 162-bp DNA sequence that contains seven GATC sites, a density 10-fold above random. The methylation state of these GATC sites affects packaging of P1 DNA into capsids, because the P1 packaging enzyme can cut pac only if most of its GATC sites are methylated in both DNA strands (245). The importance of Dam methylation in the regulation of P1 packaging is illustrated by the observation that growth of a P1 Dam− mutant on a Dam− E. coli strain causes a 20-fold reduction in phage progeny compared to infections carried out in the presence of either phage or host Dam methylase (245). Furthermore, the few phage produced in the absence of Dam methylation carry genomes which lack pac sequences at their ends (245).
Cutting phage genomes in a precise manner may optimize DNA packaging and facilitate circularization of phage DNA upon entry into the next recipient cell. However, the use of Dam methylation to label phage DNA ends is an enigmatic evolutionary acquisition. Because the DNA substrate for packaging is concatemeric DNA, methylation of all pac sites in a concatemer would permit multiple packaging initiations, disrupting the serial process of head filling. A model proposed by Yarmolinski and Sternberg in the late 1980s envisages that the P1 packaging enzyme (protein 9), which is the product of an early phage gene, might bind hemimethylated pac sites produced by theta replication and protect them from the host Dam methylase. P1 circular molecules with hemimethylated and nonmethylated pac sites would thus be produced (291). In the second stage of replication (rolling circle), P1 Dam methylase, the product of a late gene, would be allowed to methylate one and only one pac site per concatemer; the other pac sites would be protected (but not cut) by protein 9. This mechanism would permit headful packaging and avoid cutting of pac sites inside a concatemer (291). Note that every concatemer contains several P1 genomes, and cutting every pac site would prevent headful packaging and thus waste phage DNA.
Regulation of the cre gene in bacteriophage P1.
Cre is a site-specific recombinase involved in cyclization of P1 DNA upon injection into the host cytoplasm. The cre gene is driven by three promoters, and one of them (pCre1) contains two GATCs in its −35 module (246). Transcription from pCre1 is repressed by Dam methylation (246). The significance of this Dam dependence is unknown. Cre is expressed in cells lysogenized by P1 and may play a role in the partition of newly replicated prophages (291). Based on these observations, one may speculate that hemimethylation might cause transient derepression of the pCre1 promoter. The resulting boost in Cre synthesis might ensure proper partition of the daughter prophages.
Regulation of the mom operon in bacteriophage Mu.
The mom gene of bacteriophage Mu encodes a DNA modification enzyme that converts adenine to N6-carboxy-methyl-adenine (102, 248, 257). Mom-mediated modification of Mu DNA is postreplicative and protects Mu DNA from cleavage by a number of restriction endonucleases (103). Mom is not essential for phage growth but increases the host range of Mu within E. coli: if Mu infects a bacterial cell harboring restriction-modification systems different from those found in its last host, Mom-modified Mu DNA will be protected against nucleolytic attack (103). The mom gene is part of the mom operon, which includes a second gene, com, involved in translational regulation of the com-mom transcript (103). In turn, transcription of the mom operon requires a phage product, protein C, which binds the mom upstream activation sequence (UAS) −33 to −52 relative to the transcription start site (38). In the absence of protein C, RNA polymerase starts transcription at the opposite DNA strand, generating a transcript directed away from the mom gene (247). The DNA region upstream from the C binding site contains three GATC sites, spaced between −54 and −85 (103). This region serves as a binding site for a host-encoded protein, the redox-sensitive regulator OxyR, which acts as a repressor of mom transcription (103). However, OxyR can bind the mom UAS only if the GATCs therein are nonmethylated or hemimethylated (37, 104). The biological role of Dam methylation in the regulation of mom transcription is not fully understood. However, Mom− mutants have a subtle phenotype that may provide hints about the role of Dam in mom control: Mu DNA produced after infection is less modified by Mom than Mu DNA produced after prophage induction (258). A tentative explanation is that the mom promoter is fully methylated in a lysogen, thereby preventing OxyR-mediated repression (103). This may permit a level of synthesis of Mom product sufficient to modify phage DNA molecules produced upon induction. In an endogenous infection, however, the lag between phage DNA replication and Dam methylation will increase the chances that OxyR binds to a hemimethylated mom promoter, repressing transcription (103). Hence, phage DNA with a relatively low level of Mom modification will be introduced into capsids.
Conjugal Transfer in the Virulence Plasmid of Salmonella enterica
A decade ago, a screen for genes regulated by Dam methylation identified the transfer (tra) operon of the Salmonella virulence plasmid (pSLT) as a Dam-repressed locus (254). Derepression of tra in a Dam− background results in increased frequencies of conjugal transfer, a phenomenon also observed in other plasmids of the F-like family such as F and R100 (47, 255). In pSLT, Dam methylation does not act directly on the tra operon but acts on the regulatory genes traJ and finP (45, 255). Transcription of traJ, which encodes a transcriptional activator of tra, is repressed by Dam methylation (46). In turn, transcription of finP, which encodes a small RNA that antagonizes TraJ expression, is activated by Dam methylation (46, 255). This dual effect of Dam methylation accounts for the increase in tra operon expression observed in Dam− donors (48).
Regulation of traJ transcription.
Repression of traJ transcription by Dam methylation is a typical case of regulation of gene expression at the hemimethylated DNA state, reminiscent of Dam-mediated coupling of IS10 transposition to passage of the DNA replication fork (see above) (219). The traJ UAS contains two binding sites for Lrp, which is an activator of traJ transcription (45, 48). Both Lrp binding sites are necessary for transcriptional activation, and one of them (LRP-2) contains a GATC site whose methylation state affects Lrp binding. When the GATC is hemimethylated or nonmethylated, Lrp binds to LRP-2 with high affinity. If the GATC is methylated, however, the affinity of Lrp for LRP-2 is lowered. The binding pattern of Lrp at the traJ UAS is also different depending on the methylation state of LRP-2: DNase I footprinting reveals that Lrp protects the traJ UAS from −132 to −42 when the LRP-2 GATC site is nonmethylated and from −132 to −52 when the GATC site is methylated. Increased distance between the downstream end of the region bound by Lrp and the −35 module of the traJ promoter may explain the failure of Lrp to activate traJ transcription when the GATC within LRP-2 is methylated (48). Footprint analysis also shows that methylation of the LRP-2 GATC alters the distribution of DNase I-hypersensitive sites in the traJ UAS, providing further evidence that Lrp binding follows different patterns depending on the methylation state of the LRP-2 GATC (46). Lrp can also bind a hemimethylated traJ UAS (see below), suggesting that Dam methylation may serve as a sensor of plasmid replication: traJ transcription will be repressed in a nonreplicating plasmid, but repression will be lifted during the transient hemimethylation lapse that follows passage of the replication fork (46).
The affinity of Lrp for hemimethylated traJ UAS is influenced by the location of the methyl group within LRP-2. High-affinity Lrp binding occurs if the methylated GATC lies in the noncoding (template) strand of traJ. In contrast, Lrp binds to a hemimethylated DNA substrate containing a methyl group in the traJ coding strand with lower affinity. If these observations faithfully reproduce the scenario of a replicating plasmid, passage of the replication fork will permit Lrp binding to one daughter DNA molecule but not to the other, and traJ activation will occur in only one of the newly replicated plasmids. Electrophoretic migration of free, unbound traJ DNA is also different depending on the strand that contains N6-methyl-adenine (N6meA): a DNA fragment containing N6meA in the noncoding strand migrates like nonmethylated DNA, while a DNA fragment containing N6meA in the coding strand migrates like methylated DNA (46). A single methyl group is able to induce structural changes in a DNA fragment (72). Hence, subtle structural differences between the two hemimethylated traJ substrates may explain why Lrp is able to discriminate between “isomeric” DNA molecules.
If the above model is correct, Lrp-mediated activation of traJ transcription will be restricted to one hemimethylated daughter plasmid molecule (46). This epigenetic switch may be viewed as a mechanism to limit TraJ synthesis and hence to restrain activation of conjugal transfer. Higher TraJ levels might be superfluous, if not an energetic waste. Furthermore, because the pSLT strand transferred during conjugation is the noncoding strand, the active epigenetic state of traJ may be transmissible to the recipient cell: use of the incoming DNA strand as template will reproduce the methylation pattern that permits traJ activation, and the recipient cell will instantly become a donor if sufficient Lrp is available (Fig. 7). This infectious transmission of an epigenetic state may facilitate spread of the plasmid: as far as recipient cells are available, new donors will be formed by a positive feedback loop (46).
FIG. 7.
Epigenetic states of the traJ gene in the Salmonella virulence plasmid. In the donor cell, DNA hemimethylation permits traJ transcription only in the plasmid molecule that carries a methyl group in the noncoding DNA strand. As a consequence, plasmid replication generates two epigenetic states in the traJ gene and permits traJ transcription in only one daughter plasmid molecule. The methylation states of the top and bottom DNA strands of a GATC site are depicted by an open square (nonmethylated) or closed square (methylated). The possibility that the active epigenetic state of traJ can be transferred to the recipient cell is at this stage hypothetical.
Regulation of finP transcription.
Transcription of the pSLT finP gene occurs at reduced rates in Dam− mutants (46, 255). A combination of genetic evidence and gel retardation analysis has indicated that repression of finP transcription in a Dam− background is exerted by the nucleoid protein H-NS (46). However, the different expression levels of the finP gene in Dam+ and Dam− strains cannot be explained by a local effect of Dam methylation upon H-NS binding, because Dam-mediated repression is still observed in a mutant finP promoter lacking the GATC site that overlaps the −10 module (46). The involvement of upstream DNA sequences is likewise discarded by deletion analysis (46). Hence, H-NS-mediated repression of finP may reflect a condition or state that occurs in Dam− mutants but not in the wild type. Tentative explanations may be that a higher H-NS concentration exists in Salmonella Dam− mutants, as reported for E. coli (199), or that lack of N6meA favors a change in the pattern of H-NS association to the cell nucleoid. Because N6 methylation at individual GATC sites is known to influence local DNA structure (72), it seems conceivable that the methylation state of thousands of GATCs might influence nucleoid organization and potentially affect H-NS binding. Support for this hypothesis was obtained by microarray analysis of gene expression in E. coli overexpressing Dam (159).
Bacterial Virulence
In Salmonella, Haemophilus, and certain strains of Yersinia pseudotuberculosis, lack of Dam methylation causes attenuation of virulence in model animals (89, 93, 107, 201, 252, 274). In other pathogens, virulence attenuation is observed if Dam methylase is overproduced (56, 136). Albeit widespread, the involvement of Dam methylation in bacterial virulence is not universal; for instance, Dam− mutants of Shigella flexneri are not attenuated (121).
The involvement of Dam methylation in bacterial virulence may provide an example of a housekeeping function that has permitted adaptation to challenges associated with a pathogen lifestyle. One such challenge is the maintenance of genome integrity when the pathogen encounters DNA-damaging agents synthesized by the host (198, 268). In bacterial species that use Dam methylation as a strand discrimination signal for DNA mismatch repair, lack of Dam methylation leaves the cell at the mercy of the MutHLS system: if DNA lesions are produced, double-strand DNA breaks introduced by MutH can kill the cell (121).
Lack of mismatch repair is not the only virulence-related phenotype of Dam− mutants. Dam methylation regulates invasion of epithelial cells in Salmonella enterica (89) and Haemophilus influenzae (274), secretion of Yersinia outer membrane proteins (10, 136), and synthesis of Std fimbriae in Salmonella (13). It is intriguing to speculate that Dam methylation could provide a type of short-term memory for bacterial pathogens via formation of DNA methylation patterns that control expression of virulence genes. A potential advantage of such an epigenetic memory system is that information regarding environments that mother cells have encountered could be passed on to daughter cells, which might be useful in orchestrating appropriate temporal control of gene expression contributing to pathogenesis. Despite these examples and possibilities, the roles of Dam methylation in bacterial virulence are not fully understood, and their study might uncover hitherto unknown roles of N6meA in the bacterial cell.
Roles of Dam methylation in Salmonella virulence.
Dam− mutants of Salmonella enterica serovar Typhimurium are severely attenuated in the mouse model: the 50% lethal dose of a Dam− mutant is 10,000-fold higher than that of the wild type when administered by the oral route and 1,000-fold higher when administered intraperitoneally (89, 107). Attenuation by dam mutations is likewise observed in S. enterica serovar Enteritidis (93). Microscopic examination of murine ileal loops infected with Dam− salmonellae reveals a reduced ability of Dam− cells to interact with the intestinal epithelium. Furthermore, infection of epithelial cell lines indicates that Dam− strains have an invasion defect. This defect may be caused by reduced expression of genes in pathogenicity island 1 (SPI-1), including the main regulatory gene, hilA (13). The mechanisms by which Dam methylation activates gene expression in SPI-1 are not yet known. In silico examination of SPI-1 regulatory regions does not reveal the existence of any GATC clusters (13). However, this does not exclude the possibility that Dam methylation may activate SPI-1 expression at the transcriptional level, since the methylation state of a single GATC site can govern specific DNA-protein interactions (46, 118, 219). An additional defect of Salmonella Dam− mutants that may contribute to inefficient invasion of the intestinal epithelium is reduced motility, which may be caused by uncoordinated expression of flagellar genes (13).
Another relevant defect of S. enterica Dam− mutants is envelope instability, with release of outer membrane vesicles (210) and leakage of proteins (89). Vesicle release has been tentatively associated with impaired binding of Tol and PAL (210) proteins to peptidoglycan (210). Protein release may also be a side effect of envelope fragility. In addition, a fimbrial operon that is tightly repressed in the wild-type, stdABC, undergoes derepression in Dam− mutants (13). In a Dam− background, std mRNA increases over 100-fold (13), and the StdA protein becomes one of the most abundant proteins detected by two-dimensional gel electrophoresis in cell extracts (2). The presence of three GATC sites clustered in a 24-bp interval upstream from the stdABC promoter is reminiscent of genes in which Dam methylation regulates binding of a trans-acting regulator, for example, OxyR binding to agn43 (97, 111, 269) (see above), and raises the possibility that Dam methylation may control stdABC transcription (13). However, discrepancies between the std transcription rates and the levels of Std fimbrial proteins provide evidence for posttranscriptional control by Dam methylation (130), as previously described for the E. coli vsr gene (19). Production of Std fimbriae is tightly repressed in LB medium and becomes derepressed in ileal loops (126). Hence, the stdABC operon may provide an interesting example of the use of Dam methylation as a signal that is responsive to environmental cues. On the other hand, massive fimbrial expression on the cell surface, together with the envelope defects discussed above, may contribute to the avirulence of Dam− mutants by activating the host immune system. In fact, Dam− mutants of S. enterica have been shown to elicit animal immune responses with high efficiency (76, 108). The observation that Dam methylation often regulates cell surface functions (fimbriae, flagella, envelope structures, and secreted proteins) is intriguing and may suggest that certain gene families are more prone than others to fall under Dam control.
An additional defect of Salmonella Dam− mutants is sensitivity to bile (108, 210). Bile salts are detergents and DNA-damaging agents, and both activities appear to contribute to Dam− mutant killing during infection. Because of their envelope defects, Dam− mutants are more sensitive to the detergent activity of bile. In addition, in the absence of Dam methylation, exposure to bile salts triggers killing of Dam− cells by their own MutHLS system; every attempt to repair bile-induced DNA damage in the absence of DNA strand discrimination can result in a double-strand DNA break performed by the MutH endonuclease (208). A summary of the pleiotropic effects of a dam mutation on S. enterica serovar Typhimurium gene expression and physiology is shown in Fig. 8.
FIG. 8.
Multifactorial basis for attenuation in Dam− mutants of Salmonella enterica. The lack of strand discrimination for mismatch repair and altered gene expression patterns may explain some of the pleiotropic defects displayed by Salmonella Dam− mutants in the mouse model (13, 89, 208, 210).
Attenuation of bacterial virulence by Dam methylase overproduction.
Unlike for Salmonella, Shigella, and Haemophilus, analysis of Dam's role(s) in other pathogens has encountered the obstacle that Dam methylation is an essential function. An approach to overcome this hurdle was devised in M. Mahan's laboratory. Based on the previous finding that both lack of and overproduction of Dam methylase attenuated virulence in Salmonella enterica (107), the effects of Dam overproduction in Yersinia pseudotuberculosis and Vibrio cholerae, two species in which dam mutations are lethal, were tested (135). In both Yersinia and Vibrio, overproduction of Dam was tolerated and caused virulence attenuation (135). An independent study showed that overproduction of Dam methylase in Yersinia enterocolitica enhances invasion of epithelial cells yet results in decreased virulence (83).
Dam-overproducing strains of Yersinia pseudotuberculosis show increased secretion of Yersinia outer proteins (Yops), a group of virulence proteins that are injected in the host cytoplasm via a type III secretion apparatus (136). Yop secretion is tightly regulated by environmental signals such as temperature and calcium concentration (136). Upon Dam overproduction, synthesis of the YopE cytotoxin is insensitive to both temperature and calcium concentration, and YopE secretion becomes temperature independent (136). Synthesis of LcrV, a low-calcium-responsive virulence factor involved in Yop synthesis and translocation, is also altered in Dam-overproducing strains and may contribute to explaining the altered expression pattern of Yop proteins associated with Dam overproduction (10).
The success in attenuating virulence by overproduction of Dam methylase is intriguing and may indicate the existence of virulence genes regulated by stable undermethylation of critical GATC sites, in a fashion reminiscent of the pap operon or the agn43 gene. An alternative explanation is that Dam methylase overproduction might interfere with cellular processes which require SeqA binding to hemimethylated GATC sites, potentially disrupting organization of the nucleoid (159, 160). The latter view may be supported by the observation that SeqA− mutants of Salmonella enterica display virulence defects in the mouse model (209).
CcrM Methylation and Regulation of Cell Cycle in Alphaproteobacteria
Caulobacter is a dimorphic bacterium with two different cell types: the stalked cell and the swarmer cell (170). These cell types are formed by asymmetric cell division, and they differ in morphology and behavior. The swarmer cell is unable to divide and differentiates into a stalked cell which undergoes chromosome replication and cell division. Initiation of chromosome replication, which occurs only in the stalked cell, requires that the GANTC sites within the Caulobacter chromosomal origin (Cori) are methylated (170). Chromosome replication produces hemimethylated DNA, and the daughter chromosomes of the stalked cell remain hemimethylated until CcrM is produced during the late stage of chromosome replication (215). When CcrM is synthesized, methylation of the newly replicated chromosomes occurs. After cell division, the inheritance of a methylated Cori will permit the initiation of a new replication round in the daughter stalked cell (170, 215).
The fact that two independent bacterial lineages (Gammaproteobacteria and Alphaproteobacteria) use DNA adenine methylation as a signal for the initiation of chromosome replication is an interesting case of evolutionary convergence, which is strengthened by the evidence that the DNA methylases involved (Dam or CcrM) are also of independent origin.
Regulation of ccrM transcription.
Shortly after cell division, CcrM is degraded by a Lon-like protease in both daughter cells (170, 214). In the nondividing swarmer cell, initiation of chromosome replication is blocked by CtrA, a global regulator that binds the methylated Cori. In the stalked cell, CtrA is degraded and remains undetectable until chromosome replication has initiated (170). Because the ccrM gene is not transcribed until chromosome replication approaches the terminus, the origin (and most of the chromosome) will remain hemimethylated until the late stages of replication, when a burst in CcrM synthesis occurs (170). Transcription of the ccrM gene is activated by CtrA, which accumulates in the stalked cell as chromosome replication progresses. However, CtrA-mediated activation of ccrM transcription is inhibited by methylation of two GANTC sites located in the leader of the ctrA coding sequence (243). This inhibition may contribute to delay ccrM transcription until the replication fork reaches ccrM and may serve to prevent earlier activation by CtrA (215). If high levels of CcrM are present throughout the cell cycle, Caulobacter DNA is methylated all the time, the cell cycle is disrupted, and filaments made of polyploid cells are formed (287).
Regulation of ctrA transcription.
Synthesis of the cell cycle regulator CtrA is regulated by GANTC methylation in a fashion reminiscent of Dam-repressed genes such as tnp (IS10) and traJ (46, 219). One of the two ctrA promoters (P1) contains a GANTC site near its −35 module (216). Transcription starting at P1 is repressed when the GANTC is methylated. Passage of the replication fork renders the promoter hemimethylated and activates transcription (216). This mechanism may serve to boost ctrA gene transcription in response to replication progress. In turn, CtrA accumulation will activate transcription of the ccrM gene as soon as the replication fork renders the ccrM promoter hemimethylated. Note that the ability of the CtrA transcription factor to recognize hemimethylated ccrM DNA is a crucial factor to permit an orderly sequence of events during chromosome replication. The importance of hemimethylation in the Caulobacter cell cycle is supported by genetic experiments carried out in Shapiro's laboratory: if the ctrA gene is moved to an ectopic position near the replication terminus, ctrA transcription from the methylation-sensitive P1 promoter remains repressed for a longer lapse of the cell cycle, and CtrA accumulates more slowly (216). These elegant experiments provide further evidence that the hemimethylation wave associated with chromosome replication serves as a molecular clock for the Caulobacter cell cycle.
CONCLUDING REMARKS
DNA methyltransferases are widespread in bacteria, and most of them are part of restriction-modification systems. In addition, certain bacterial genomes contain solitary DNA methylases that are not involved in protecting DNA from a cognate restriction enzyme. Two of these enzymes, the Dam methylase of enteric bacteria and the CcrM methylase of Caulobacter crescentus, are paradigms of an evolutionary process in which DNA adenine methylation acts as a signaling mechanism that regulates DNA-protein interactions. In both Gamma- and Alphaproteobacteria, DNA adenine methylation regulates chromosome replication and couples transcription of certain genes to passage of the DNA replication fork. In some cases, regulatory protein binding inhibits DNA methylation, generating DNA methylation patterns that are hallmarks of alternative epigenetic states. DNA methylation patterns are modulated by environmental conditions via alterations in regulatory protein binding. Specific DNA methylation states can be propagated by positive feedback loops, and in certain cases they are clonally inherited by daughter cells. Protein binding prevents maintenance methylation, thereby generating sites that are stably hemimethylated or nonmethylated. Methylation-blocking factors include transcriptional regulators such as CAP, Lrp, OxyR, and other DNA binding proteins. Inheritance of DNA methylation patterns is a phenomenon reminiscent of eukaryotic imprinting of genes and may convey adaptive value: bacterial populations may use inherited DNA methylation patterns as a short-term memory of the metabolic conditions in which the former generation thrived and divided. DNA methylation also plays an essential role in diverse bacterial pathogens, raising the possibility of designing new antibacterial drugs that might inhibit DNA adenine methylation. A drug of this kind could be expected to inhibit the virulence of wild-type bacteria by transforming them into phenocopies of Dam− mutants.
Acknowledgments
We thank Bruce Braaten, Aaron Hernday, Stephanie Aoki, Brooke Trinh, and Marjan van der Woude for reading parts of the manuscript and/or helpful advice and Edward Robinson for work on Fig. 1.
Work in our laboratories is supported by grants BIO2004-3455-CO2-02 and GEN2003-20234-CO6-03 from the Spanish Ministry of Education and Science and the European Regional fund (to J.C.) and by National Institutes of Health grant AI23348 (to D.L.).
REFERENCES
- 1.Abraham, J. M., C. S. Freitag, J. R. Clements, and B. I. Eisenstein. 1985. An invertible element of DNA controls phase variation of type 1 fimbriae of Escherichia coli. Proc. Natl. Acad. Sci. USA 82:5724-5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alonso, A., M. G. Pucciarelli, N. Figueroa-Bossi, and F. García-del-Portillo. 2005. Increased excision of the Salmonella prophage ST64B caused by a deficiency in Dam methylase. J. Bacteriol. 187:7901-7911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aoki, S. K., R. Pamma, A. D. Hernday, J. E. Bickham, B. A. Braaten, and D. A. Low. 2005. Contact-dependent inhibition of growth in Escherichia coli. Science 309:1245-1248. [DOI] [PubMed] [Google Scholar]
- 4.Appelmelk, B. J., S. L. Martin, M. A. Monteiro, C. A. Clayton, A. A. McColm, P. Zheng, T. Verboom, J. J. Maaskant, D. H. van den Eijnden, C. H. Hokke, M. B. Perry, C. M. Vandenbroucke-Grauls, and J. G. Kusters. 1999. Phase variation in Helicobacter pylori lipopolysaccharide due to changes in the lengths of poly(C) tracts in α3-fucosyltransferase genes. Infect. Immun. 67:5361-5366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arber, W. 1974. DNA modification and restriction. Prog. Nucleic Acid Res. Mol. Biol. 14:1-37. [DOI] [PubMed] [Google Scholar]
- 6.Arber, W., and D. Dussoix. 1962. Host specificity of DNA produced by Escherichia coli. I. Host controlled modification of bacteriophage lambda. J. Mol. Biol. 5:18-36. [DOI] [PubMed] [Google Scholar]
- 7.Arnold, H. P., U. Ziese, and W. Zillig. 2000. SNDV, a novel virus of the extremely thermophilic and acidophilic archaeon Sulfolobus. Virology 272:409-416. [DOI] [PubMed] [Google Scholar]
- 8.Au, K. G., K. Welsh, and P. Modrich. 1992. Initiation of methyl-directed mismatch repair. J. Biol. Chem. 267:12142-12148. [PubMed] [Google Scholar]
- 9.Auer, B., and M. Schweiger. 1984. Evidence that Escherichia coli virus T1 induces a DNA methyltransferase. J. Virol. 49:588-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Badie, G., D. M. Heithoff, and M. J. Mahan. 2004. LcrV synthesis is altered by DNA adenine methylase overproduction in Yersinia pseudotuberculosis and is required to confer immunity in vaccinated hosts. Infect. Immun. 72:6707-6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baga, M., M. Goransson, S. Normark, and B. E. Uhlin. 1985. Transcriptional activation of a pap pilus virulence operon from uropathogenic Escherichia coli. EMBO J. 4:3887-3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bakker, A., and D. W. Smith. 1989. Methylation of GATC sites is required for precise timing between rounds of DNA replication in Escherichia coli. J. Bacteriol. 171:5738-5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Balbontin, R., G. Rowley, M. G. Pucciarelli, Y. Wormstone, S. Lucchini, F. Garcia-del-Portillo, J. C. D. Hinton, and J. Casadesus. Unpublished data. [DOI] [PMC free article] [PubMed]
- 14.Bale, A., M. d'Alarcao, and M. G. Marinus. 1979. Characterization of DNA adenine methylation mutants of Escherichia coli K12. Mutat. Res. 59:157-165. [DOI] [PubMed] [Google Scholar]
- 15.Baranyi, U., R. Klein, W. Lubitz, D. H. Kruger, and A. Witte. 2000. The archaeal halophilic virus-encoded Dam-like methyltransferase M.phiCh1-I methylates adenine residues and complements dam mutants in the low salt environment of Escherichia coli. Mol. Microbiol. 35:1168-1179. [DOI] [PubMed] [Google Scholar]
- 16.Barras, F., and M. G. Marinus. 1989. The great GATC: DNA methylation in E. coli. Trends Genet. 5:139-143. [DOI] [PubMed] [Google Scholar]
- 17.Baumler, A. J., R. M. Tsolis, F. A. Bowe, J. G. Kusters, S. Hoffmann, and F. Heffron. 1996. The pef fimbrial operon of Salmonella typhimurium mediates adhesion to murine small intestine and is necessary for fluid accumulation in the infant mouse. Infect. Immun. 64:61-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beletskaya, I. V., M. V. Zakharova, M. G. Shlyapnikov, L. M. Semenova, and A. S. Solonin. 2000. DNA methylation at the CfrBI site is involved in expression control in the CfrBI restriction-modification system. Nucleic Acids Res. 28:3817-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bell, D. C., and C. G. Cupples. 2001. Very-short-patch repair in Escherichia coli requires the Dam adenine methylase. J. Bacteriol. 183:3631-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berdis, A. J., I. Lee, J. K. Coward, C. Stephens, L. Wright, L. Shapiro, and S. J. Benkovic. 1998. A cell cycle-regulated adenine DNA methyltransferase from Caulobacter crescentus processively methylates GANTC sites on hemimethylated DNA. Proc. Natl. Acad. Sci. USA 95:2874-2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reference deleted.
- 22.Bergerat, A., W. Guschlbauer, and G. W. Fazakerley. 1991. Allosteric and catalytic binding of S-adenosylmethionine to Escherichia coli DNA adenine methyltransferase monitored by 3H NMR. Proc. Natl. Acad. Sci. USA 88:6394-6397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bergerat, A., A. Kriebardis, and W. Guschlbauer. 1989. Preferential site-specific hemimethylation of GATC sites in pBR322 DNA by Dam methyltransferase from Escherichia coli. J. Biol. Chem. 264:4064-4070. [PubMed] [Google Scholar]
- 24.Berthiaume, F., C. Crost, V. Labrie, C. Martin, E. B. Newman, and J. Harel. 2004. Influence of l-leucine and l-alanine on Lrp regulation of foo, coding for F1651, a Pap homologue. J. Bacteriol. 186:8537-8541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhagwat, A. S., and M. Lieb. 2002. Cooperation and competition in mismatch repair: very short-patch repair and methyl-directed mismatch repair in Escherichia coli. Mol. Microbiol. 44:1421-1428. [DOI] [PubMed] [Google Scholar]
- 26.Bhugra, B., L. L. Voelker, N. Zou, H. Yu, and K. Dybvig. 1995. Mechanism of antigenic variation in Mycoplasma pulmonis: interwoven, site-specific DNA inversions. Mol. Microbiol. 18:703-714. [DOI] [PubMed] [Google Scholar]
- 27.Bickle, T. A., and D. H. Kruger. 1993. Biology of DNA restriction. Microbiol. Rev. 57:434-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bird, A. 1999. DNA methylation de novo. Science 286:2287-2288. [DOI] [PubMed] [Google Scholar]
- 29.Bird, A. 2002. DNA methylation patterns and epigenetic memory. Genes Dev. 16:6-21. [DOI] [PubMed] [Google Scholar]
- 30.Bird, A. 1997. Does DNA methylation control transposition of selfish elements in the germline? Trends Genet. 13:469-472. [DOI] [PubMed] [Google Scholar]
- 31.Blaisdell, B. E., A. M. Campbell, and S. Karlin. 1996. Similarities and dissimilarities of phage genomes. Proc. Natl. Acad. Sci. USA 93:5854-5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blomfield, I. C. 2001. The regulation of pap and type 1 fimbriation in Escherichia coli. Adv. Microb. Physiol. 45:1-49. [DOI] [PubMed] [Google Scholar]
- 33.Blomfield, I. C., D. H. Kulasekara, and B. I. Eisenstein. 1997. Integration host factor stimulates both FimB- and FimE-mediated site-specific DNA inversion that controls phase variation of type 1 fimbriae expression in Escherichia coli. Mol. Microbiol. 23:705-717. [DOI] [PubMed] [Google Scholar]
- 34.Blyn, L. B., B. A. Braaten, and D. A. Low. 1990. Regulation of pap pilin phase variation by a mechanism involving differential dam methylation states. EMBO J. 9:4045-4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blyn, L. B., B. A. Braaten, C. A. White-Ziegler, D. H. Rolfson, and D. A. Low. 1989. Phase-variation of pyelonephritis-associated pili in Escherichia coli: evidence for transcriptional regulation. EMBO J. 8:613-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bogan, J. A., and C. E. Helmstetter. 1997. DNA sequestration and transcription in the oriC region of Escherichia coli. Mol. Microbiol. 26:889-896. [DOI] [PubMed] [Google Scholar]
- 37.Bolker, M., and R. Kahmann. 1989. The Escherichia coli regulatory protein OxyR discriminates between methylated and unmethylated states of the phage Mu mom promoter. EMBO J. 8:2403-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bölker, M., F. G. Wulczyn, and R. Kahmann. 1989. Role of bacteriophage Mu C protein in actvation of the mom gene promoter. J. Bacteriol. 171:2019-2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boyer, H. 1964. Genetic control of restriction and modification in Escherichia coli. J. Bacteriol. 88:1652-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Braaten, B. A., L. B. Blyn, B. S. Skinner, and D. A. Low. 1991. Evidence for a methylation-blocking factor (mbf) locus involved in Pap pilus expression and phase variation in Escherichia coli. J. Bacteriol. 173:1789-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Braaten, B. A., X. Nou, L. S. Kaltenbach, and D. A. Low. 1994. Methylation patterns in pap regulatory DNA control pyelonephritis-associated pili phase variation in E. coli. Cell 76:577-588. [DOI] [PubMed] [Google Scholar]
- 42.Cairns, J., and P. L. Foster. 1991. Adaptive reversion of a frameshift mutation in Escherichia coli. Genetics 128:695-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reference deleted.
- 44.Calvo, J. M., and R. G. Matthews. 1994. The leucine-responsive regulatory protein, a global regulator of metabolism in Escherichia coli. Microbiol. Rev. 58:466-490. (Erratum, 59:323, 1995.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Camacho, E. M., and J. Casadesus. 2002. Conjugal transfer of the virulence plasmid of Salmonella enterica is regulated by the leucine-responsive regulatory protein and DNA adenine methylation. Mol. Microbiol. 44:1589-1598. [DOI] [PubMed] [Google Scholar]
- 46.Camacho, E. M., and J. Casadesus. 2005. Regulation of traJ transcription in the Salmonella virulence plasmid by strand-specific DNA adenine hemimethylation. Mol. Microbiol. 57:1700-1718. [DOI] [PubMed] [Google Scholar]
- 47.Camacho, E. M., A. Serna, and J. Casadesus. 2005. Regulation of conjugal transfer by Lrp and Dam methylation in plasmid R100. Int. Microbiol. 8:279-285. [PubMed] [Google Scholar]
- 48.Camacho, E. M., A. Serna, C. Madrid, S. Marques, R. Fernandez, F. de la Cruz, A. Juarez, and J. Casadesus. 2005. Regulation of finP transcription by DNA adenine methylation in the virulence plasmid of Salmonella enterica. J. Bacteriol. 187:5691-5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Campbell, J. L., and N. Kleckner. 1990. E. coli oriC and the dnaA gene promoter are sequestered from dam methyltransferase following the passage of the chromosomal replication fork. Cell 62:967-979. [DOI] [PubMed] [Google Scholar]
- 50.Campbell, J. L., and N. Kleckner. 1988. The rate of Dam-mediated DNA adenine methylation in Escherichia coli. Gene 74:189-190. [DOI] [PubMed] [Google Scholar]
- 51.Casadesus, J., and R. D'Ari. 2002. Memory in bacteria and phage. Bioessays 24:512-518. [DOI] [PubMed] [Google Scholar]
- 52.Casadesus, J., and R. Maldonado. 1990. Genomic imprinting in microorganisms. Microbiologia 6:1-10. [PubMed] [Google Scholar]
- 53.Cerdeno-Tarraga, A. M., S. Patrick, L. C. Crossman, G. Blakely, V. Abratt, N. Lennard, I. Poxton, B. Duerden, B. Harris, M. A. Quail, A. Barron, L. Clark, C. Corton, J. Doggett, M. T. Holden, N. Larke, A. Line, A. Lord, H. Norbertczak, D. Ormond, C. Price, E. Rabbinowitsch, J. Woodward, B. Barrell, and J. Parkhill. 2005. Extensive DNA inversions in the B. fragilis genome control variable gene expression. Science 307:1463-1465. [DOI] [PubMed] [Google Scholar]
- 54.Charlier, D., D. Gigot, N. Huysveld, M. Roovers, A. Pierard, and N. Glansdorff. 1995. Pyrimidine regulation of the Escherichia coli and Salmonella typhimurium carAB operons: CarP and integration host factor (IHF) modulate the methylation status of a GATC site present in the control region. J. Mol. Biol. 250:383-391. [DOI] [PubMed] [Google Scholar]
- 55.Charlier, D., N. Huysveld, M. Roovers, and N. Glansdorff. 1994. On the role of the Escherichia coli integration host factor (IHF) in repression at a distance of the pyrimidine specific promoter P1 of the carAB operon. Biochimie 76:1041-1051. [DOI] [PubMed] [Google Scholar]
- 56.Chen, L., D. B. Paulsen, D. W. Scruggs, M. M. Banes, B. Y. Reeks, and M. L. Lawrence. 2003. Alteration of DNA adenine methylase (Dam) activity in Pasteurella multocida causes increased spontaneous mutation frequency and attenuation in mice. Microbiology 149:2283-2290. [DOI] [PubMed] [Google Scholar]
- 57.Chen, S., and J. M. Calvo. 2002. Leucine-induced dissociation of Escherichia coli Lrp hexadecamers to octamers. J. Mol. Biol. 318:1031-1042. [DOI] [PubMed] [Google Scholar]
- 58.Chen, S., M. Iannolo, and J. M. Calvo. 2005. Cooperative binding of the leucine-responsive regulatory protein (Lrp) to DNA. J. Mol. Biol. 345:251-264. [DOI] [PubMed] [Google Scholar]
- 59.Chen, S., M. H. Rosner, and J. M. Calvo. 2001. Leucine-regulated self-association of leucine-responsive regulatory protein (Lrp) from Escherichia coli. J. Mol. Biol. 312:625-635. [DOI] [PubMed] [Google Scholar]
- 60.Correnti, J., V. Munster, T. Chan, and M. Woude. 2002. Dam-dependent phase variation of Ag43 in Escherichia coli is altered in a seqA mutant. Mol. Microbiol. 44:521-532. [DOI] [PubMed] [Google Scholar]
- 61.Coulby, J. N., and N. L. Sternberg. 1988. Characterization of the phage P1 dam gene. Gene 74:191. [DOI] [PubMed] [Google Scholar]
- 62.Crost, C., A. Garrivier, J. Harel, and C. Martin. 2003. Leucine-responsive regulatory protein-mediated repression of clp (encoding CS31A) expression by l-leucine and l-alanine in Escherichia coli. J. Bacteriol. 185:1886-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Crost, C., J. Harel, F. Berthiaume, A. Garrivier, M. C. Tessier, H. Rakotoarivonina, and C. Martin. 2004. Influence of environmental cues on transcriptional regulation of foo and clp coding for F165(1) and CS31A adhesins in Escherichia coli. Res. Microbiol. 155:475-482. [DOI] [PubMed] [Google Scholar]
- 64.Cui, Y., M. A. Midkiff, Q. Wang, and J. M. Calvo. 1996. The leucine-responsive regulatory protein (Lrp) from Escherichia coli. Stoichiometry and minimal requirements for binding to DNA. J. Biol. Chem. 271:6611-6617. [DOI] [PubMed] [Google Scholar]
- 65.Cui, Y., Q. Wang, G. D. Stormo, and J. M. Calvo. 1995. A consensus sequence for binding of Lrp to DNA. J. Bacteriol. 177:4872-4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Danese, P. N., L. A. Pratt, S. L. Dove, and R. Kolter. 2000. The outer membrane protein, antigen 43, mediates cell-to-cell interactions within Escherichia coli biofilms. Mol. Microbiol. 37:424-432. [DOI] [PubMed] [Google Scholar]
- 67.Dar, M. E., and A. S. Bhagwat. 1993. Mechanism of expression of vsr repair gene vsr, an Escherichia coli gene that overlaps the DNA cytosine methylase gene, dcm. Mol. Microbiol. 9:823-833. [DOI] [PubMed] [Google Scholar]
- 68.D'Ari, R., R. T. Lin, and E. B. Newman. 1993. The leucine-responsive regulatory protein: more than a regulator? Trends Biochem. Sci. 18:260-263. [DOI] [PubMed] [Google Scholar]
- 69.Denison, A. M., B. Clapper, and K. Dybvig. 2005. Avoidance of the host immune system through phase variation in Mycoplasma pulmonis. Infect. Immun. 73:2033-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Deschavanne, P., and M. Radman. 1991. Counterselection of GATC sequences in enterobacteriophages by the components of the methyl-directed mismatch repair system. J. Mol. Evol. 33:125-132. [DOI] [PubMed] [Google Scholar]
- 71.Diderichsen, B. 1980. flu, a metastable gene controlling surface properties of Escherichia coli. J. Bacteriol. 141:858-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Diekmann, S. 1987. DNA methylation can enhance or induce DNA curvature. EMBO J. 6:4213-4217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dodson, K. W., and D. E. Berg. 1989. Factors affecting transposition activity of IS50 and Tn5 ends. Gene 76:207-213. [DOI] [PubMed] [Google Scholar]
- 74.Doerfler, W. 2006. De novo methylation, long-term promoter silencing, methylation patterns in the human genome, and consequences of foreign DNA insertion. Curr. Top. Microbiol. Immunol. 301:125-175. [DOI] [PubMed] [Google Scholar]
- 75.Dorman, C. J., and C. F. Higgins. 1987. Fimbrial phase variation in Escherichia coli: dependence on integration host factor and homologies with other site-specific recombinases. J. Bacteriol. 169:3840-3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dueger, E. L., J. K. House, D. M. Heithoff, and M. J. Mahan. 2003. Salmonella DNA adenine methylase mutants elicit early and late onset protective immune responses in calves. Vaccine 21:3249-3258. [DOI] [PubMed] [Google Scholar]
- 77.Dussoix, D., and W. Arber. 1962. Host specificity of DNA produced by Escherichia coli. II. Control over acceptance of DNA from infecting phage lambda. J. Mol. Biol. 5:37-49. [DOI] [PubMed] [Google Scholar]
- 78.Egan, E. S., S. Duigou, and M. K. Waldor. 2006. Autorepression of RctB, an initiator of Vibrio cholerae chromosome II replication. J. Bacteriol. 188:789-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Eisenstein, B. I. 1981. Phase variation of type 1 fimbriae in Escherichia coli is under transcriptional control. Science 214:337-339. [DOI] [PubMed] [Google Scholar]
- 80.El-Labany, S., B. K. Sohanpal, M. Lahooti, R. Akerman, and I. C. Blomfield. 2003. Distant cis-active sequences and sialic acid control the expression of fimB in Escherichia coli K-12. Mol. Microbiol. 49:1109-1118. [DOI] [PubMed] [Google Scholar]
- 81.El-Maarri, O., K. Buiting, E. G. Peery, P. M. Kroisel, B. Balaban, K. Wagner, B. Urman, J. Heyd, C. Lich, C. I. Brannan, J. Walter, and B. Horsthemke. 2001. Maternal methylation imprints on human chromosome 15 are established during or after fertilization. Nat. Genet. 27:341-344. [DOI] [PubMed] [Google Scholar]
- 82.Engelberg-Kulka, H., and G. Glaser. 1999. Addiction modules and programmed cell death and antideath in bacterial cultures. Annu. Rev. Microbiol. 53:43-70. [DOI] [PubMed] [Google Scholar]
- 83.Fälker, S., M. A. Schmidt, and G. Heusipp. DNA methylation in Yersinia enterocolitica: role of the DNA adenine methyltransferase in mismatch repair and regulation of virulence factors. Microbiology 151:2291-2299. [DOI] [PubMed]
- 84.Ferguson-Smith, A. C., and M. A. Surani. 2001. Imprinting and the epigenetic asymmetry between parental genomes. Science 293:1086-1089. [DOI] [PubMed] [Google Scholar]
- 85.Forsman, K., M. Goransson, and B. E. Uhlin. 1989. Autoregulation and multiple DNA interactions by a transcriptional regulatory protein in E. coli pili biogenesis. EMBO J. 8:1271-1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Foxall, P. A., B. S. Drasar, and C. J. Duggleby. 1990. Evidence for a DNA inversion system in Bordetella pertussis. FEMS Microbiol. Lett. 57:1-6. [DOI] [PubMed] [Google Scholar]
- 87.Friedrich, M. J., N. E. Kinsey, J. Vila, and R. J. Kadner. 1993. Nucleotide sequence of a 13.9 kb segment of the 90 kb virulence plasmid of Salmonella typhimurium: the presence of fimbrial biosynthetic genes. Mol. Microbiol. 8:543-558. [DOI] [PubMed] [Google Scholar]
- 88.Fulka, H., M. Mrazek, O. Tepla, and J. Fulka, Jr. 2004. DNA methylation pattern in human zygotes and developing embryos. Reproduction 128:703-708. [DOI] [PubMed] [Google Scholar]
- 89.Garcia-Del Portillo, F., M. G. Pucciarelli, and J. Casadesus. 1999. DNA adenine methylase mutants of Salmonella typhimurium show defects in protein secretion, cell invasion, and M cell cytotoxicity. Proc. Natl. Acad. Sci. USA 96:11578-11583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gazit, E., and R. T. Sauer. 1999. Stability and DNA binding of the phd protein of the phage P1 plasmid addiction system. J. Biol. Chem. 274:2652-2657. [DOI] [PubMed] [Google Scholar]
- 91.Gerdes, K., F. W. Bech, S. T. Jorgensen, A. Lobner-Olesen, P. B. Rasmussen, T. Atlung, L. Boe, O. Karlstrom, S. Molin, and K. von Meyenburg. 1986. Mechanism of postsegregational killing by the hok gene product of the parB system of plasmid R1 and its homology with the relF gene product of the E. coli relB operon. EMBO J. 5:2023-2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gerdes, K., J. S. Jacobsen, and T. Franch. 1997. Plasmid stabilization by post-segregational killing. Genet. Eng. 19:49-61. [DOI] [PubMed] [Google Scholar]
- 93.Giacomodonato, M. N., M. H. Sarnacki, R. L. Caccuri, D. O. Sordelli, and M. C. Cerquetti. 2004. Host response to a dam mutant of Salmonella enterica serovar Enteritidis with a temperature-sensitive phenotype. Infect. Immun. 72:5498-5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Goransson, M., B. Sonden, P. Nilsson, B. Dagberg, K. Forsman, K. Emanuelsson, and B. E. Uhlin. 1990. Transcriptional silencing and thermoregulation of gene expression in Escherichia coli. Nature 344:682-685. [DOI] [PubMed] [Google Scholar]
- 95.Gowher, H., and A. Jeltsch. 2000. Molecular enzymology of the EcoRV DNA-(adenine-N (6))-methyltransferase: kinetics of DNA binding and bending, kinetic mechanism and linear diffusion of the enzyme on DNA. J. Mol. Biol. 303:93-110. [DOI] [PubMed] [Google Scholar]
- 96.Guschlbauer, W. 1996. “Small is beautiful”: major modifications in DNA structure or dynamics by small substituents or ligands. Acta Biochim. Pol. 43:77-94. [PubMed] [Google Scholar]
- 97.Haagmans, W., and M. van der Woude. 2000. Phase variation of Ag43 in Escherichia coli: Dam-dependent methylation abrogates OxyR binding and OxyR-mediated repression of transcription. Mol. Microbiol. 35:877-887. [DOI] [PubMed] [Google Scholar]
- 98.Hagblom, P., E. Segal, E. Billyard, and M. So. 1985. Intragenic recombination leads to pilus antigenic variation in Neisseria gonorrhoeae. Nature 315:156-158. [DOI] [PubMed] [Google Scholar]
- 99.Hale, W. B., M. W. van der Woude, and D. A. Low. 1994. Analysis of nonmethylated GATC sites in the Escherichia coli chromosome and identification of sites that are differentially methylated in response to environmental stimuli. J. Bacteriol. 176:3438-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Han, J. S., S. Kang, H. Lee, H. K. Kim, and D. S. Hwang. 2003. Sequential binding of SeqA to paired hemi-methylated GATC sequences mediates formation of higher order complexes. J. Biol. Chem. 278:34983-34989. [DOI] [PubMed] [Google Scholar]
- 101.Harel, J., F. Daigle, C. Forget, M. C. Tessier, C. Crost, and C. Martin. 2000. Phase variation of F165(1) (Prs-like) fimbriae from Escherichia coli causing septicaemia in animals. Can. J. Microbiol. 46:1101-1107. [DOI] [PubMed] [Google Scholar]
- 102.Hattman, S. 1979. Unusual modification of bacteriophage Mu DNA. J. Virol. 32:468-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hattman, S. 1999. Unusual transcriptional and translational regulation of the bacteriophage Mu mom operon. Pharmacol. Ther. 84:367-388. [DOI] [PubMed] [Google Scholar]
- 104.Hattman, S., and W. Sun. 1997. Escherichia coli OxyR modulation of bacteriophage Mu mom expression in dam+ cells can be attributed to its ability to bind hemimethylated Pmom promoter DNA. Nucleic Acids Res. 25:4385-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hattman, S., J. Wilkinson, D. Swinton, S. Schlagman, P. M. Macdonald, and G. Mosig. 1985. Common evolutionary origin of the phage T4 dam and host Escherichia coli dam DNA-adenine methyltransferase genes. J. Bacteriol. 164:932-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hayes, F. 2003. Toxins-antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest. Science 301:1496-1499. [DOI] [PubMed] [Google Scholar]
- 107.Heithoff, D., R. L. Sinsheimer, D. A. Low, and M. J. Mahan. 1999. An essential role for DNA adenine methylation in bacterial virulence. Science 284:967-970. [DOI] [PubMed] [Google Scholar]
- 108.Heithoff, D. M., E. I. Enioutina, R. A. Daynes, R. L. Sinsheimer, D. A. Low, and M. J. Mahan. 2001. Salmonella DNA adenine methylase mutants confer cross-protective immunity. Infect. Immun. 69:6725-6730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Reference deleted.
- 110.Hénaut, A., T. Rouxel, A. Gleizes, I. Moszer, and A. Danchin. 1996. Uneven distribution of GATC motifs in the Escherichia coli chromosome, its plasmids and its phages. J. Mol. Biol. 257:574-585. [DOI] [PubMed] [Google Scholar]
- 111.Henderson, I. R., M. Meehan, and P. Owen. 1997. A novel regulatory mechanism for a novel phase-variable outer membrane protein of Escherichia coli. Adv. Exp. Med. Biol. 412:349-355. [DOI] [PubMed] [Google Scholar]
- 112.Henderson, I. R., and P. Owen. 1999. The major phase-variable outer membrane protein of Escherichia coli structurally resembles the immunoglobulin A1 protease class of exported protein and is regulated by a novel mechanism involving Dam and oxyR. J. Bacteriol. 181:2132-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Henderson, I. R., P. Owen, and J. P. Nataro. 1999. Molecular switches—the ON and OFF of bacterial phase variation. Mol. Microbiol. 33:919-932. [DOI] [PubMed] [Google Scholar]
- 114.Herman, G. E., and P. Modrich. 1982. Escherichia coli dam methylase. Physical and catalytic properties of the homogeneous enzyme. J. Biol. Chem. 257:2605-2612. [PubMed] [Google Scholar]
- 115.Hernday, A., B. Braaten, and D. Low. 2004. The intricate workings of a bacterial epigenetic switch. Adv. Exp. Med. Biol. 547:83-89. [DOI] [PubMed] [Google Scholar]
- 116.Hernday, A., M. Krabbe, B. Braaten, and D. Low. 2002. Self-perpetuating epigenetic pili switches in bacteria. Proc. Natl. Acad. Sci. USA 29:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hernday, A. D., B. A. Braaten, G. Broitman-Maduro, P. Engelberts, and D. A. Low. 2004. Regulation of the pap epigenetic switch by CpxAR: phosphorylated CpxR inhibits transition to the phase ON state by competition with Lrp. Mol. Cell 16:537-547. [DOI] [PubMed] [Google Scholar]
- 118.Hernday, A. D., B. A. Braaten, and D. A. Low. 2003. The mechanism by which DNA adenine methylase and PapI activate the pap epigenetic switch. Mol. Cell 12:947-957. [DOI] [PubMed] [Google Scholar]
- 119.High, N. J., M. P. Jennings, and E. R. Moxon. 1996. Tandem repeats of the tetramer 5′-CAAT-3′ present in lic2A are required for phase variation but not lipopolysaccharide biosynthesis in Haemophilus influenzae. Mol. Microbiol. 20:165-174. [DOI] [PubMed] [Google Scholar]
- 120.Holst, B., L. Sogaard-Andersen, H. Pedersen, and P. Valentin-Hansen. 1992. The cAMP-CRP/CytR nucleoprotein complex in Escherichia coli: two pairs of closely linked binding sites for the cAMP-CRP activator complex are involved in combinatorial regulation of the cdd promoter. EMBO J. 11:3635-3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Honma, Y., R. E. Fernandez, and A. T. Maurelli. 2004. A DNA adenine methylase mutant of Shigella flexneri shows no significant attenuation of virulence. Microbiology 150:1073-1078. [DOI] [PubMed] [Google Scholar]
- 122.Hood, D. W., M. E. Deadman, M. P. Jennings, M. Bisercic, R. D. Fleischmann, J. C. Venter, and E. R. Moxon. 1996. DNA repeats identify novel virulence genes in Haemophilus influenzae. Proc. Natl. Acad. Sci. USA 93:11121-11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Horton, J. R., K. Liebert, S. Hattman, A. Jeltsch, and X. Cheng. 2005. Transition from nonspecific to specific DNA interactions along the substrate-recognition pathway of dam methyltransferase. Cell 121:349-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Huisman, T. T., D. Bakker, P. Klaasen, and F. K. de Graaf. 1994. Leucine-responsive regulatory protein, IS1 insertions, and the negative regulator FaeA control the expression of the fae (K88) operon in Escherichia coli. Mol. Microbiol. 11:525-536. [DOI] [PubMed] [Google Scholar]
- 125.Huisman, T. T., and F. K. de Graaf. 1995. Negative control of fae (K88) expression by the ‘global’ regulator Lrp is modulated by the ‘local’ regulator FaeA and affected by DNA methylation. Mol. Microbiol. 16:943-953. [DOI] [PubMed] [Google Scholar]
- 126.Humphries, A. D., M. Raffatellu, S. Winter, E. H. Weening, R. A. Kingsley, R. Droleskey, S. Zhang, J. Figueiredo, S. Khare, J. Nunes, L. G. Adams, R. M. Tsolis, and A. J. Baumler. 2003. The use of flow cytometry to detect expression of subunits encoded by 11 Salmonella enterica serotype Typhimurium fimbrial operons. Mol. Microbiol. 48:1357-1376. [DOI] [PubMed] [Google Scholar]
- 127.Hung, D. L., T. L. Raivio, C. H. Jones, T. J. Silhavy, and S. J. Hultgren. 2001. Cpx signaling pathway monitors biogenesis and affects assembly and expression of P pili. EMBO J. 20:1508-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Jaenisch, R., and A. Bird. 2003. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 33(Suppl.):245-254. [DOI] [PubMed] [Google Scholar]
- 129.Jafri, S., S. Chen, and J. M. Calvo. 2002. ilvIH operon expression in Escherichia coli requires Lrp binding to two distinct regions of DNA. J. Bacteriol. 184:5293-5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Jakomin, M., and J. Casadesús. Unpublished data.
- 131.Jarboe, L. R., D. Beckwith, and J. C. Liao. 2004. Stochastic modeling of the phase-variable pap operon regulation in uropathogenic Escherichia coli. Biotechnol. Bioeng. 88:189-203. [DOI] [PubMed] [Google Scholar]
- 132.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 133.Jonsson, A. B., G. Nyberg, and S. Normark. 1991. Phase variation of gonococcal pili by frameshift mutation in pilC, a novel gene for pilus assembly. EMBO J. 10:477-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Jorgensen, H. F., and A. Bird. 2002. MeCP2 and other methyl-CpG binding proteins. Ment. Retard. Dev. Disabil. Res. Rev. 8:87-93. [DOI] [PubMed] [Google Scholar]
- 135.Julio, S. M., D. M. Heithoff, D. Provenzano, K. E. Klose, R. L. Sinsheimer, D. A. Low, and M. J. Mahan. 2001. DNA adenine methylase is essential for viability and plays a role in the pathogenesis of Yersinia pseudotuberculosis and Vibrio cholerae. Infect. Immun. 69:7610-7615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Julio, S. M., D. M. Heithoff, R. L. Sinsheimer, D. A. Low, and M. J. Mahan. 2002. DNA adenine methylase overproduction in Yersinia pseudotuberculosis alters YopE expression and secretion and host immune responses to infection. Infect. Immun. 70:1006-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kahng, L. S., and L. Shapiro. 2001. The CcrM DNA methyltransferase of Agrobacterium tumefaciens is essential, and its activity is cell cycle regulated. J. Bacteriol. 183:3065-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kaltenbach, L. S., B. A. Braaten, and D. A. Low. 1995. Specific binding of PapI to Lrp-pap DNA complexes. J. Bacteriol. 177:6449-6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kang, P. J., and L. Shapiro. 1994. Cell cycle arrest of a Caulobacter crescentus secA mutant. J. Bacteriol. 176:4958-4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kang, S., H. Lee, J. S. Han, and D. S. Hwang. 1999. Interaction of SeqA and Dam methylase on the hemimethylated origin of Escherichia coli chromosomal DNA replication. J. Biol. Chem. 274:11463-11468. [DOI] [PubMed] [Google Scholar]
- 141.Kapoor, A., F. Agius, and J. K. Zhu. 2005. Preventing transcriptional gene silencing by active DNA demethylation. FEBS Lett. 579:5889-5898. [DOI] [PubMed] [Google Scholar]
- 142.Karyagina, A., I. Shilov, V. Tashlitskii, M. Khodoun, S. Vasil'ev, P. C. Lau, and I. Nikolskaya. 1997. Specific binding of sso II DNA methyltransferase to its promoter region provides the regulation of sso II restriction-modification gene expression. Nucleic Acids Res. 25:2114-2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kessler, C., P. S. Neumaier, and W. Wolf. 1985. Recognition sequences of restriction endonucleases and methylases—a review. Gene 33:1-102. [DOI] [PubMed] [Google Scholar]
- 144.Kleckner, N., D. Morisato, D. Roberts, and J. Bender. 1984. Mechanism and regulation of Tn10 transposition. Cold Spring Harbor Symp. Quant. Biol. 49:235-244. [DOI] [PubMed] [Google Scholar]
- 145.Klose, R. J., and A. P. Bird. 2006. Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 31:89-97. [DOI] [PubMed] [Google Scholar]
- 146.Klungsoyr, H. K., and K. Skarstad. 2004. Positive supercoiling is generated in the presence of Escherichia coli SeqA protein. Mol. Microbiol. 54:123-131. [DOI] [PubMed] [Google Scholar]
- 147.Ko, Y. G., K. Nishino, N. Hattori, Y. Arai, S. Tanaka, and K. Shiota. 2005. Stage-by-stage change in DNA methylation status of Dnmt1 locus during mouse early development. J. Biol. Chem. 280:9627-9634. [DOI] [PubMed] [Google Scholar]
- 148.Kobayashi, I. 2001. Behavior of restriction-modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Res. 29:3742-3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kobayashi, I. 1998. Selfishness and death: raison d'etre of restriction, recombination and mitochondria. Trends Genet. 14:368-374. [DOI] [PubMed] [Google Scholar]
- 150.Kobayashi, I., A. Nobusato, N. Kobayashi-Takahashi, and I. Uchiyama. 1999. Shaping the genome—restriction-modification systems as mobile genetic elements. Curr. Opin. Genet. Dev. 9:649-656. [DOI] [PubMed] [Google Scholar]
- 151.Krabbe, M., N. Weyand, and D. Low. 2000. Environmental control of pilus gene expression, p. 305-321. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.
- 152.Kullik, I., M. B. Toledano, L. A. Tartaglia, and G. Storz. 1995. Mutational analysis of the redox-sensitive transcriptional regulator OxyR: regions important for oxidation and transcriptional activation. J. Bacteriol. 177:1275-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lederberg, S. 1965. Host-controlled restriction and modification of deoxyribonucleic acid in Escherichia coli. Virology 27:378-387. [DOI] [PubMed] [Google Scholar]
- 154.Lemon, K. P., and A. D. Grossman. 2000. Movement of replicating DNA through a stationary replisome. Mol. Cell 6:1321-1330. [DOI] [PubMed] [Google Scholar]
- 155.Levenson, J. M., and J. D. Sweatt. 2005. Epigenetic mechanisms in memory formation. Nat. Rev. Neurosci. 6:108-118. [DOI] [PubMed] [Google Scholar]
- 156.Lewis, J. D., R. R. Meehan, W. J. Henzel, I. Maurer-Fogy, P. Jeppesen, F. Klein, and A. Bird. 1992. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell 69:905-914. [DOI] [PubMed] [Google Scholar]
- 157.Liebert, K., A. Hermann, M. Schlickenrieder, and A. Jeltsch. 2004. Stopped-flow and mutational analysis of base flipping by the Escherichia coli Dam DNA-(adenine-N6)-methyltransferase. J. Mol. Biol. 341:443-454. [DOI] [PubMed] [Google Scholar]
- 158.Lobner-Olesen, A., E. Boye, and M. G. Marinus. 1992. Expression of the Escherichia coli dam gene. Mol. Microbiol. 6:1841-1851. [DOI] [PubMed] [Google Scholar]
- 159.Løbner-Olesen, A., M. G. Marinus, and F. G. Hanssen. 2003. Role of SeqA and Dam in Escherichia coli gene expression: a global/microarray analysis. Proc. Natl. Acad. Sci. USA 100:4672-4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Lobner-Olesen, A., O. Skovgaard, and M. G. Marinus. 2005. Dam methylation: coordinating cellular processes. Curr. Opin. Microbiol. 8:154-160. [DOI] [PubMed] [Google Scholar]
- 161.Lodge, J. K., K. Weston-Hafer, and D. E. Berg. 1991. Tn5 insertion specificity is not influenced by IS50 end sequences in target DNA. Mol. Gen. Genet. 228:312-315. [DOI] [PubMed] [Google Scholar]
- 162.Low, D. A., N. J. Weyand, and M. J. Mahan. 2001. Roles of DNA adenine methylation in regulating bacterial gene expression and virulence. Infect. Immun. 69:7197-7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Lu, M., J. L. Campbell, E. Boye, and N. Kleckner. 1994. SeqA: a negative modulator of replication initiation in E. coli. Cell 77:413-426. [DOI] [PubMed] [Google Scholar]
- 164.Lysnyansky, I., R. Rosengarten, and D. Yogev. 1996. Phenotypic switching of variable surface lipoproteins in Mycoplasma bovis involves high-frequency chromosomal rearrangements. J. Bacteriol. 178:5395-5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Makris, J. C., P. L. Nordmann, and W. S. Reznikoff. 1990. Integration host factor plays a role in IS50 and Tn5 transposition. J. Bacteriol. 172:1368-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Reference deleted.
- 167.Malone, T., R. M. Blumenthal, and X. Cheng. 1995. Structure-guided analysis reveals nine sequence motifs conserved among DNA amino-methyltransferases, and suggests a catalytic mechanism for these enzymes. J. Mol. Biol. 253:618-632. [DOI] [PubMed] [Google Scholar]
- 168.Malygin, E. G., A. A. Evdokimov, V. V. Zinoviev, L. G. Ovechkina, W. M. Lindstrom, N. O. Reich, S. L. Schlagman, and S. Hattman. 2001. A dual role for substrate S-adenosyl-l-methionine in the methylation reaction with bacteriophage T4 Dam DNA-[N6-adenine]-methyltransferase. Nucleic Acids Res. 29:2361-2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Malygin, E. G., B. Sclavi, V. V. Zinoviev, A. A. Evdokimov, S. Hattman, and M. Buckle. 2004. Bacteriophage T4 Dam DNA-(adenine-N(6))-methyltransferase. Comparison of pre-steady state and single turnover methylation of 40-mer duplexes containing two (un)modified target sites. J. Biol. Chem. 279:50012-50018. [DOI] [PubMed] [Google Scholar]
- 170.Marczynski, G. T., and L. Shapiro. 2002. Control of chromosome replication in Caulobacter crescentus. Annu. Rev. Microbiol. 56:625-656. [DOI] [PubMed] [Google Scholar]
- 171.Marinus, M. G. 1985. DNA methylation influences trpR promoter activity in Escherichia coli. Mol. Gen. Genet. 200:185-186. [DOI] [PubMed] [Google Scholar]
- 172.Marinus, M. G. 1996. Methylation of DNA, p. 782-791. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 173.Martin, C. 1996. The clp (CS31A) operon is negatively controlled by Lrp, ClpB, and l-alanine at the transcriptional level. Mol. Microbiol. 21:281-292. [DOI] [PubMed] [Google Scholar]
- 174.McClelland, M. 1985. Selection against dam methylation sites in the genomes of DNA of enterobacteriophages. J. Mol. Evol. 21:317-322. [DOI] [PubMed] [Google Scholar]
- 175.McCommas, S. A., and M. Syvanen. 1988. Temporal control of transposition in Tn5. J. Bacteriol. 170:889-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.McEwen, J., and P. Silverman. 1980. Genetic analysis of Escherichia coli K-12 chromosomal mutants defective in expression of F-plasmid functions: identification of genes cpxA and cpxB. J. Bacteriol. 144:60-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Miller, E. S., E. Kutter, G. Mosig, F. Arisaka, T. Kunisawa, and W. Ruger. 2003. Bacteriophage T4 genome. Microbiol. Mol. Biol Rev. 67:86-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Modrich, P. 1989. Methyl-directed DNA mismatch correction. J. Biol. Chem. 264:6597-6600. [PubMed] [Google Scholar]
- 179.Molina, F., and K. Skarstad. 2004. Replication fork and SeqA focus distributions in Escherichia coli suggest a replication hyperstructure dependent on nucleotide metabolism. Mol. Microbiol. 52:1597-1612. [DOI] [PubMed] [Google Scholar]
- 180.Morgan, H. D., F. Santos, K. Green, W. Dean, and W. Reik. 2005. Epigenetic reprogramming in mammals. Hum. Mol. Genet. 14(Spec. No. 1):R47-R58. [DOI] [PubMed] [Google Scholar]
- 181.Morisato, D., and N. Kleckner. 1987. Tn10 transposition and circle formation in vitro. Cell 51:101-111. [DOI] [PubMed] [Google Scholar]
- 182.Morschhauser, J., V. Vetter, L. Emody, and J. Hacker. 1994. Adhesin regulatory genes within large, unstable DNA regions of pathogenic Escherichia coli: cross-talk between different adhesin gene clusters. Mol. Microbiol. 11:555-566. [DOI] [PubMed] [Google Scholar]
- 183.Munsky, B. 2006. The finite state projection algorithm for the solution of the chemical master equation. University of California, Santa Barbara. [DOI] [PubMed]
- 184.Munsky, B., and M. Khammash. 2006. The finite state projection algorithm for the solution of the chemical master equation. J. Chem. Phys. 124:044104. [DOI] [PubMed] [Google Scholar]
- 185.Murray, N. E. 2000. Type I restriction systems: sophisticated molecular machines (a legacy of Bertani and Weigle). Microbiol. Mol. Biol. Rev. 64:412-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Nakayama, Y., and I. Kobayashi. 1998. Restriction-modification gene complexes as selfish gene entities: roles of a regulatory system in their establishment, maintenance, and apoptotic mutual exclusion. Proc. Natl. Acad. Sci. USA 95:6442-6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Nan, X., S. Cross, and A. Bird. 1998. Gene silencing by methyl-CpG-binding proteins. Novartis Found. Symp. 214:6-16. [DOI] [PubMed] [Google Scholar]
- 188.Newman, E. B., R. D'Ari, and R. T. Lin. 1992. The leucine-Lrp regulon in E. coli: a global response in search of a raison d'etre. Cell 68:617-619. [DOI] [PubMed] [Google Scholar]
- 189.Newman, E. B., and R. Lin. 1995. Leucine-responsive regulatory protein: a global regulator of gene expression in E. coli. Annu. Rev. Microbiol. 49:747-775. [DOI] [PubMed] [Google Scholar]
- 190.Nicholson, B., and D. Low. 2000. DNA methylation-dependent regulation of pef expression in Salmonella typhimurium. Mol. Microbiol. 35:728-742. [DOI] [PubMed] [Google Scholar]
- 191.Noormohammadi, A. H., P. F. Markham, A. Kanci, K. G. Whithear, and G. F. Browning. 2000. A novel mechanism for control of antigenic variation in the haemagglutinin gene family of mycoplasma synoviae. Mol. Microbiol. 35:911-923. [DOI] [PubMed] [Google Scholar]
- 192.Norregaard-Madsen, M., B. Mygind, R. Pedersen, P. Valentin-Hansen, and L. Sogaard-Andersen. 1994. The gene encoding the periplasmic cyclophilin homologue, PPIase A, in Escherichia coli, is expressed from four promoters, three of which are activated by the cAMP-CRP complex and negatively regulated by the CytR repressor. Mol. Microbiol. 14:989-997. [DOI] [PubMed] [Google Scholar]
- 193.Nou, X., B. Braaten, L. Kaltenbach, and D. A. Low. 1995. Differential binding of Lrp to two sets of pap DNA binding sites mediated by Pap I regulates Pap phase variation in Escherichia coli. EMBO J. 14:5785-5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Nou, X., B. Skinner, B. Braaten, L. Blyn, D. Hirsch, and D. Low. 1993. Regulation of pyelonephritis-associated pili phase-variation in Escherichia coli: binding of the PapI and the Lrp regulatory proteins is controlled by DNA methylation. Mol. Microbiol. 7:545-553. [DOI] [PubMed] [Google Scholar]
- 195.Obata, Y., and T. Kono. 2002. Maternal primary imprinting is established at a specific time for each gene throughout oocyte growth. J. Biol. Chem. 277:5285-5289. [DOI] [PubMed] [Google Scholar]
- 196.O'Driscoll, J., G. F. Fitzgerald, and D. van Sinderen. 2005. A dichotomous epigenetic mechanism governs expression of the LlaJI restriction/modification system. Mol. Microbiol. 57:1532-1544. [DOI] [PubMed] [Google Scholar]
- 197.O'Driscoll, J., F. Glynn, O. Cahalane, M. O'Connell-Motherway, G. F. Fitzgerald, and D. Van Sinderen. 2004. Lactococcal plasmid pNP40 encodes a novel, temperature-sensitive restriction-modification system. Appl. Environ. Microbiol. 70:5546-5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.O'Rourke, E. J., C. Chevalier, A. V. Pinto, J. M. Thiberge, L. Ielpi, A. Labigne, and J. P. Radicella. 2003. Pathogen DNA as target for host-generated oxidative stress: role for repair of DNA damage in Helicobacter pylori colonization. Proc. Natl. Acad. Sci. USA 100:2789-2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Oshima, T., C. Wada, Y. Kawagoe, T. Ara, M. Maeda, Y. Masuda, S. Hiraga, and H. Mori. 2002. Genome-wide analysis of deoxyadenosine methyltransferase-mediated control of gene expression in Escherichia coli. Mol. Microbiol. 45:673-695. [DOI] [PubMed] [Google Scholar]
- 200.Owen, P., M. Meehan, H. de Loughry-Doherty, and I. Henderson. 1996. Phase-variable outer membrane proteins in Escherichia coli. FEMS Immunol. Med. Microbiol. 16:63-76. [DOI] [PubMed] [Google Scholar]
- 201.Oza, J. P., J. B. Yeh, and N. O. Reich. 2005. DNA methylation modulates Salmonella enterica serovar Typhimurium virulence in Caenorhabditis elegans. FEMS Microbiol. Lett. 245:53-59. [DOI] [PubMed] [Google Scholar]
- 202.Palmer, B. R., and M. G. Marinus. 1994. The dam and dcm strains of Escherichia coli—a review. Gene 143:1-12. [DOI] [PubMed] [Google Scholar]
- 203.Peterson, S. N., and N. O. Reich. 2006. GATC flanking sequences regulate Dam activity: evidence for how Dam specificity may influence pap expression. J. Mol. Biol. 355:459-472. [DOI] [PubMed] [Google Scholar]
- 204.Piekarowicz, A., and J. Bujnicki. 1999. Cloning of the Dam methyltransferase gene from Haemophilus influenzae bacteriophage HP1. Acta Microbiol. Pol. 48:123-129. [PubMed] [Google Scholar]
- 205.Plumbridge, J., and D. Soll. 1987. The effect of dam methylation on the expression of glnS in E. coli. Biochimie 69:539-541. [DOI] [PubMed] [Google Scholar]
- 206.Polaczek, P., K. Kwan, and J. L. Campbell. 1998. GATC motifs may alter the conformation of DNA depending on sequence context and N6-adenine methylation status: possible implications for DNA-protein recognition. Mol. Gen. Genet. 258:488-493. [DOI] [PubMed] [Google Scholar]
- 207.Polaczek, P., K. Kwan, D. A. Liberies, and J. L. Campbell. 1997. Role of architectural elements in combinatorial regulation of initiation of DNA replication in Escherichia coli. Mol. Microbiol. 26:261-275. [DOI] [PubMed] [Google Scholar]
- 208.Prieto, A. I., F. Ramos-Morales, and J. Casadesús. 2004. Bile-induced DNA damage in Salmonella enterica. Genetics 168:1787-1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Prieto, A. I., I. Segura, F. Ramos-Morales, and J. Casadesús. Unpublished data.
- 210.Pucciarelli, M. G., A. I. Prieto, J. Casadesus, and F. Garcia-del Portillo. 2002. Envelope instability in DNA adenine methylase mutants of Salmonella enterica. Microbiology 148:1171-1182. [DOI] [PubMed] [Google Scholar]
- 211.Raivio, T. L., and T. J. Silhavy. 1999. The sigmaE and Cpx regulatory pathways: overlapping but distinct envelope stress responses. Curr. Opin. Microbiol. 2:159-165. [DOI] [PubMed] [Google Scholar]
- 212.Ramseier, T. M., and M. H. Saier, Jr. 1995. cAMP-cAMP receptor protein complex: five binding sites in the control region of the Escherichia coli mannitol operon. Microbiology 141:1901-1907. [DOI] [PubMed] [Google Scholar]
- 213.Reik, W., W. Dean, and J. Walter. 2001. Epigenetic reprogramming in mammalian development. Science 293:1089-1093. [DOI] [PubMed] [Google Scholar]
- 214.Reisenauer, A., L. S. Kahng, S. McCollum, and L. Shapiro. 1999. Bacterial DNA methylation: a cell cycle regulator? J. Bacteriol. 181:5135-5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Reisenauer, A., K. Quon, and L. Shapiro. 1999. The CtrA response regulator mediates temporal control of gene expression during the Caulobacter cell cycle. J. Bacteriol. 181:2430-2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Reisenauer, A., and L. Shapiro. 2002. DNA methylation affects the cell cycle transcription of the CtrA global regulator in Caulobacter. EMBO J. 21:4969-4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Reznikoff, W. S. 1993. The Tn5 transposon. Annu. Rev. Microbiol. 47:945-963. [DOI] [PubMed] [Google Scholar]
- 218.Ringquist, S., and C. L. Smith. 1992. The Escherichia coli chromosome contains specific, unmethylated dam and dcm sites. Proc. Natl. Acad. Sci. USA 89:4539-4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Roberts, D., B. C. Hoopes, W. R. McClure, and N. Kleckner. 1985. IS10 transposition is regulated by DNA adenine methylation. Cell 43:117-130. [DOI] [PubMed] [Google Scholar]
- 220.Roberts, R. J. 1978. Restriction and modification enzymes and their recognition sequences. Gene 4:183-194. [DOI] [PubMed] [Google Scholar]
- 221.Roberts, R. J., M. Belfort, T. Bestor, A. S. Bhagwat, T. A. Bickle, J. Bitinaite, R. M. Blumenthal, S. Degtyarev, D. T. Dryden, K. Dybvig, K. Firman, E. S. Gromova, R. I. Gumport, S. E. Halford, S. Hattman, J. Heitman, D. P. Hornby, A. Janulaitis, A. Jeltsch, J. Josephsen, A. Kiss, T. R. Klaenhammer, I. Kobayashi, H. Kong, D. H. Kruger, S. Lacks, M. G. Marinus, M. Miyahara, R. D. Morgan, N. E. Murray, V. Nagaraja, A. Piekarowicz, A. Pingoud, E. Raleigh, D. N. Rao, N. Reich, V. E. Repin, E. U. Selker, P. C. Shaw, D. C. Stein, B. L. Stoddard, W. Szybalski, T. A. Trautner, J. L. Van Etten, J. M. Vitor, G. G. Wilson, and S. Y. Xu. 2003. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 31:1805-1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Robertson, G. T., A. Reisenauer, R. Wright, R. B. Jensen, A. Jensen, L. Shapiro, and R. M. Roop II. 2000. The Brucella abortus CcrM DNA methyltransferase is essential for viability, and its overexpression attenuates intracellular replication in murine macrophages. J. Bacteriol. 182:3482-3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Satpute-Krishnan, P., and T. R. Serio. 2005. Prion protein remodelling confers an immediate phenotypic switch. Nature 437:262-265. [DOI] [PubMed] [Google Scholar]
- 224.Schembri, M. A., L. Hjerrild, M. Gjermansen, and P. Klemm. 2003. Differential expression of the Escherichia coli autoaggregation factor antigen 43. J. Bacteriol. 185:2236-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Schembri, M. A., and P. Klemm. 2001. Coordinate gene regulation by fimbriae-induced signal transduction. EMBO J. 20:3074-3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Schlagman, S. L., and S. Hattman. 1983. Molecular cloning of a functional dam+ gene coding for phage T4 DNA adenine methylase. Gene 22:139-156. [DOI] [PubMed] [Google Scholar]
- 227.Schlagman, S. L., S. Hattman, and M. G. Marinus. 1986. Direct role of the Escherichia coli Dam DNA methyltransferase in methylation-directed mismatch repair. J. Bacteriol. 165:896-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Schlagman, S. L., Z. Miner, Z. Feher, and S. Hattman. 1988. The DNA [adenine-N6]methyltransferase (Dam) of bacteriophage T4. Gene 73:517-530. [DOI] [PubMed] [Google Scholar]
- 229.Scott, T. N., and M. I. Simon. 1982. Genetic analysis of the mechanism of the Salmonella phase variation site specific recombination system. Mol. Gen. Genet. 188:313-321. [DOI] [PubMed] [Google Scholar]
- 230.Selker, E. U., M. Freitag, G. O. Kothe, B. S. Margolin, M. R. Rountree, C. D. Allis, and H. Tamaru. 2002. Induction and maintenance of nonsymmetrical DNA methylation in Neurospora. Proc. Natl. Acad. Sci. USA 99(Suppl. 4):16485-16490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Serio, T. R., A. G. Cashikar, A. S. Kowal, G. J. Sawicki, and S. L. Lindquist. 2001. Self-perpetuating changes in Sup35 protein conformation as a mechanism of heredity in yeast. Biochem. Soc. Symp. 68:35-43. [DOI] [PubMed] [Google Scholar]
- 232.Serio, T. R., and S. L. Lindquist. 2000. Protein-only inheritance in yeast: something to get [PSI+]-ched about. Trends Cell Biol. 10:98-105. [DOI] [PubMed] [Google Scholar]
- 233.Shea, C. M., and M. A. McIntosh. 1991. Nucleotide sequence and genetic organization of the ferric enterobactin transport system: homology to other periplasmic binding protein-dependent systems in Escherichia coli. Mol. Microbiol. 5:1415-1428. [DOI] [PubMed] [Google Scholar]
- 234.Shier, V. K., C. J. Hancey, and S. J. Benkovic. 2001. Identification of the active oligomeric state of an essential adenine DNA methyltransferase from Caulobacter crescentus. J. Biol. Chem. 276:14744-14751. [DOI] [PubMed] [Google Scholar]
- 235.Shorter, J., and S. Lindquist. 2005. Prions as adaptive conduits of memory and inheritance. Nat. Rev. Genet. 6:435-450. [DOI] [PubMed] [Google Scholar]
- 236.Sistla, S., and D. N. Rao. 2004. S-Adenosyl-l-methionine-dependent restriction enzymes. Crit. Rev. Biochem. Mol. Biol. 39:1-19. [DOI] [PubMed] [Google Scholar]
- 237.Smith, D. W., A. M. Garland, G. Herman, R. E. Enns, T. A. Baker, and J. W. Zyskind. 1985. Importance of state of methylation of oriC GATC sites in initiation of DNA replication in Escherichia coli. EMBO J. 4:1319-1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Smith, H. O., and D. Nathans. 1973. A suggested nomenclature for bacterial host modification and restriction systems and their enzymes. J. Mol. Biol. 81:419-423. [DOI] [PubMed] [Google Scholar]
- 239.Sohanpal, B. K., S. El-Labany, M. Lahooti, J. A. Plumbridge, and I. C. Blomfield. 2004. Integrated regulatory responses of fimB to N-acetylneuraminic (sialic) acid and GlcNAc in Escherichia coli K-12. Proc. Natl. Acad. Sci. USA 101:16322-16327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Srikhanta, Y. N., T. L. Maguire, K. J. Stacey, S. M. Grimmond, and M. P. Jennings. 2005. The phasevarion: a genetic system controlling coordinated, random switching of expression of multiple genes. Proc. Natl. Acad. Sci. USA 102:5547-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Stancheva, I., T. Koller, and J. M. Sogo. 1999. Asymmetry of Dam remethylation on the leading and lagging arms of plasmid replicative intermediates. EMBO J. 18:6542-6551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Stephens, C., A. Reisenauer, R. Wright, and L. Shapiro. 1996. A cell cycle-regulated bacterial DNA methyltransferase is essential for viability. Proc. Natl. Acad. Sci. USA 93:1210-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Stephens, C. M., G. Zweiger, and L. Shapiro. 1995. Coordinate cell cycle control of a Caulobacter DNA methyltransferase and the flagellar genetic hierarchy. J. Bacteriol. 177:1662-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Stern, A., and T. F. Meyer. 1987. Common mechanism controlling phase and antigenic variation in pathogenic neisseriae. Mol. Microbiol. 1:5-12. [DOI] [PubMed] [Google Scholar]
- 245.Sternberg, N., and J. Coulby. 1990. Cleavage of the bacteriophage P1 packaging site (pac) is regulated by adenine methylation. Proc. Natl. Acad. Sci. USA 87:8070-8074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Sternberg, N., B. Sauer, R. Hoess, and K. Abremski. 1986. Bacteriophage P1 cre gene and its regulatory region. Evidence for multiple promoters and for regulation by Dam methylation. J. Mol. Biol. 187:197-212. [DOI] [PubMed] [Google Scholar]
- 247.Sun, W., and S. Hattman. 1998. Bidirectional transcription in the mom promoter region of bacteriophage Mu. J. Mol. Biol. 284:885-892. [DOI] [PubMed] [Google Scholar]
- 248.Swinton, D., S. Hattman, P. F. Crain, C. S. Cheng, D. L. Smith, and J. A. McCloskey. 1983. Purification and characterization of the unusual nucleotide, a-N-(9-b-d-2′-deoxyribofuranosylpurin-6-yl)glycinamide, specified by the phage Mu modification function. Proc. Natl. Acad. Sci. USA 80:7400-7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Taghbalout, A., A. Landoulsi, R. Kern, M. Yamazoe, S. Hiraga, B. Holland, M. Kohiyama, and A. Malki. 2000. Competition between the replication initiator DnaA and the sequestration factor SeqA for binding to the hemimethylated chromosomal origin of E. coli in vitro. Genes Cells 5:873-884. [DOI] [PubMed] [Google Scholar]
- 250.Takahashi, N., Y. Naito, N. Handa, and I. Kobayashi. 2002. A DNA methyltransferase can protect the genome from postdisturbance attack by a restriction-modification gene complex. J. Bacteriol. 184:6100-6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Tavazoie, S., and G. M. Church. 1998. Quantitative whole-genome analysis of DNA-protein interactions by in vivo methylase protection in E. coli. Nat. Biotechnol. 16:566-571. [DOI] [PubMed] [Google Scholar]
- 252.Taylor, V. L., R. W. Titball, and P. C. F. Oyston. 2005. Oral immunization with a dam mutant of Yersinia pseudotuberculosis protects against plague. Microbiology 151:1919-1926. [DOI] [PubMed] [Google Scholar]
- 253.Tomcsanyi, T., and D. E. Berg. 1989. Transposition effect of adenine (Dam) methylation on activity of O end mutants of IS50. J. Mol. Biol. 209:191-193. [DOI] [PubMed] [Google Scholar]
- 254.Torreblanca, J., and J. Casadesus. 1996. DNA adenine methylase mutants of Salmonella typhimurium and a novel dam-regulated locus. Genetics 144:15-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Torreblanca, J., S. Marques, and J. Casadesus. 1999. Synthesis of FinP RNA by plasmids F and pSLT is regulated by DNA adenine methylation. Genetics 152:31-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Torres-Cruz, J., and M. W. van der Woude. 2003. Slipped-strand mispairing can function as a phase variation mechanism in Escherichia coli. J. Bacteriol. 185:6990-6994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Toussaint, A. 1976. The DNA modification function of temperate phage Mu-1. Virology 70:17-27. [DOI] [PubMed] [Google Scholar]
- 258.Toussaint, A. 1977. DNA modification of bacteriophage Mu-1 requires both host and phage modification functions. J. Virol. 23:825-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.True, H. L., I. Berlin, and S. L. Lindquist. 2004. Epigenetic regulation of translation reveals hidden genetic variation to produce complex traits. Nature 431:184-187. [DOI] [PubMed] [Google Scholar]
- 260.Uetake, H., S. Toyama, and S. Hagiwara. 1964. On the mechanism of host-induced modification. Multiplicity activation and thermolabile factor responsible for phage growth restriction. Virology 22:202-213. [DOI] [PubMed] [Google Scholar]
- 261.Urig, S., H. Gowher, A. Hermann, C. Beck, M. Fatemi, A. Humeny, and A. Jeltsch. 2002. The Escherichia coli dam DNA methyltransferase modifies DNA in a highly processive reaction. J. Mol. Biol. 319:1085-1096. [DOI] [PubMed] [Google Scholar]
- 262.van der Woude, M., B. Braaten, and D. Low. 1996. Epigenetic phase variation of the pap operon in Escherichia coli. Trends Microbiol. 4:5-9. [DOI] [PubMed] [Google Scholar]
- 263.van der Woude, M., W. B. Hale, and D. A. Low. 1998. Formation of DNA methylation patterns: nonmethylated GATC sequences in gut and pap operons. J. Bacteriol. 180:5913-5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.van der Woude, M. W. 2006. Re-examining the role and random nature of phase variation. FEMS Microbiol. Lett. 254:190-197. [DOI] [PubMed] [Google Scholar]
- 265.van der Woude, M. W., and A. J. Baumler. 2004. Phase and antigenic variation in bacteria. Clin. Microbiol. Rev. 17:581-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.van der Woude, M. W., L. S. Kaltenbach, and D. A. Low. 1995. Leucine-responsive regulatory protein plays dual roles as both an activator and a repressor of the Escherichia coli pap fimbrial operon. Mol. Microbiol. 17:303-312. [DOI] [PubMed] [Google Scholar]
- 267.van der Woude, M. W., and D. A. Low. 1994. Leucine-responsive regulatory protein and deoxyadenosine methylase control the phase variation and expression of the sfa and daa pili operons in Escherichia coli. Mol. Microbiol. 11:605-618. [DOI] [PubMed] [Google Scholar]
- 268.Vazquez-Torres, A., and F. C. Fang. 2001. Oxygen-dependent anti-Salmonella activity of macrophages. Trends Microbiol. 9:29-33. [DOI] [PubMed] [Google Scholar]
- 269.Waldron, D. E., P. Owen, and C. J. Dorman. 2002. Competitive interaction of the OxyR DNA-binding protein and the Dam methylase at the antigen 43 gene regulatory region in Escherichia coli. Mol. Microbiol. 44:509-520. [DOI] [PubMed] [Google Scholar]
- 270.Wallecha, A., J. Correnti, V. Munster, and M. van der Woude. 2003. Phase variation of Ag43 is independent of the oxidation state of OxyR. J. Bacteriol. 185:2203-2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Wallecha, A., V. Munster, J. Correnti, T. Chan, and M. van der Woude. 2002. Dam- and OxyR-dependent phase variation of Agn43: essential elements and evidence for a new role of DNA methylation. J. Bacteriol. 184:3338-3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Wang, M. X., and G. M. Church. 1992. A whole genome approach to in vivo DNA-protein interactions in E. coli. Nature 360:606-610. [DOI] [PubMed] [Google Scholar]
- 273.Wang, Y., W. Fischle, W. Cheung, S. Jacobs, S. Khorasanizadeh, and C. D. Allis. 2004. Beyond the double helix: writing and reading the histone code. Novartis Found. Symp. 259:3-21, 163-169. [PubMed] [Google Scholar]
- 274.Watson, M. E., J. Jarisch, and A. L. Smith. 2004. Inactivation of deoxyadenosine methyltransferase (dam) attenuates Haemophilus influenzae virulence. Mol. Microbiol. 55:651-654. [DOI] [PubMed] [Google Scholar]
- 275.Weiner, L., and P. Model. 1994. Role of an Escherichia coli stress-response operon in stationary-phase survival. Proc. Natl. Acad. Sci. USA 91:2191-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Weinreich, M. D., and W. S. Reznikoff. 1992. Fis plays a role in Tn5 and IS50 transposition. J. Bacteriol. 174:4530-4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Weyand, N. J., B. A. Braaten, M. van der Woude, J. Tucker, and D. A. Low. 2001. The essential role of the promoter proximal subunit of CAP in Pap phase variation: Lrp- and helical phase-dependent activation of papBA transcription by CAP from −215. Mol. Microbiol. 39:1504-1522. [DOI] [PubMed] [Google Scholar]
- 278.Weyand, N. J., and D. A. Low. 2000. Regulation of Pap phase variation. Lrp is sufficient for the establishment of the phase off pap DNA methylation pattern and repression of pap transcription in vitro. J. Biol. Chem. 275:3192-3200. [DOI] [PubMed] [Google Scholar]
- 279.White-Ziegler, C. A., M. L. Angus Hill, B. A. Braaten, M. W. van der Woude, and D. A. Low. 1998. Thermoregulation of Escherichia coli pap transcription: H-NS is a temperature-dependent DNA methylation blocking factor. Mol. Microbiol. 28:1121-1137. [DOI] [PubMed] [Google Scholar]
- 280.White-Ziegler, C. A., A. M. Black, S. H. Eliades, S. Young, and K. Porter. 2002. The N-acetyltransferase RimJ responds to environmental stimuli to repress pap fimbrial transcription in Escherichia coli. J. Bacteriol. 184:4334-4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.White-Ziegler, C. A., L. B. Blyn, B. A. Braaten, and D. A. Low. 1990. Identification of an Escherichia coli genetic locus involved in thermoregulation of the pap operon. J. Bacteriol. 172:1775-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.White-Ziegler, C. A., and D. A. Low. 1992. Thermoregulation of the pap operon: evidence for the involvement of RimJ, the N-terminal acetylase of ribosomal protein S5. J. Bacteriol. 174:7003-7012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.White-Ziegler, C. A., A. Villapakkam, K. Ronaszeki, and S. Young. 2000. H-NS controls pap and daa fimbrial transcription in Escherichia coli in response to multiple environmental cues. J. Bacteriol. 182:6391-6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Wilson, G. G. 1988. Type II restriction-modification systems. Trends Genet. 4:314-318. [DOI] [PubMed] [Google Scholar]
- 285.Wion, D., and J. Casadesus. 2006. N(6)-methyl-adenine: an epigenetic signal for DNA-protein interactions. Nat. Rev. Microbiol. 4:183-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Wright, R., C. Stephens, and L. Shapiro. 1997. The CcrM DNA methyltransferase is widespread in the alpha subdivision of proteobacteria, and its essential functions are conserved in Rhizobium meliloti and Caulobacter crescentus. J. Bacteriol. 179:5869-5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Wright, R., C. Stephens, G. Zweiger, L. Shapiro, and M. R. Alley. 1996. Caulobacter Lon protease has a critical role in cell-cycle control of DNA methylation. Genes Dev. 10:1532-1542. [DOI] [PubMed] [Google Scholar]
- 288.Xia, Y., K. Forsman-Semb, J. Jass, and B. E. Uhlin. 2000. Structural and functional studies of the fimbrial adhesin gene regulator PapB from uropathogenic Escherichia coli. Adv. Exp. Med. Biol. 485:123-126. [DOI] [PubMed] [Google Scholar]
- 289.Xu, J., and R. C. Johnson. 1995. Fis activates the RpoS-dependent stationary-phase expression of proP in Escherichia coli. J. Bacteriol. 177:5222-5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Yamada, M., S. Asaoka, M. H. Saier, Jr., and Y. Yamada. 1993. Characterization of the gcd gene from Escherichia coli K-12 W3110 and regulation of its expression. J. Bacteriol. 175:568-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Yarmolinski, M. B., and N. Sternberg. 1988. Bacteriophage P1, p. 782-791. In R. Calendar (ed.), The bacteriophages, vol. 1. Plenum Press, New York, N.Y. [Google Scholar]
- 292.Yin, J. C., M. P. Krebs, and W. S. Reznikoff. 1988. Effect of dam methylation on Tn5 transposition. J. Mol. Biol. 199:35-45. [DOI] [PubMed] [Google Scholar]
- 293.Yoder, J. A., C. P. Walsh, and T. H. Bestor. 1997. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 13:335-340. [DOI] [PubMed] [Google Scholar]
- 294.Zakharova, M., L. Minakhin, A. Solonin, and K. Severinov. 2004. Regulation of RNA polymerase promoter selectivity by covalent modification of DNA. J. Mol. Biol. 335:103-111. [DOI] [PubMed] [Google Scholar]
- 295.Zheng, M., F. Aslund, and G. Storz. 1998. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science 279:1718-1721. [DOI] [PubMed] [Google Scholar]
- 296.Zheng, M., X. Wang, B. Doan, K. A. Lewis, T. D. Schneider, and G. Storz. 2001. Computation-directed identification of OxyR DNA binding sites in Escherichia coli. J. Bacteriol. 183:4571-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Zhou, B., D. Beckwith, L. R. Jarboe, and J. C. Liao. 2005. Markov chain modeling of pyelonephritis-associated pili expression in uropathogenic Escherichia coli. Biophys. J. 88:2541-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Ziebuhr, W., V. Krimmer, S. Rachid, I. Lossner, F. Gotz, and J. Hacker. 1999. A novel mechanism of phase variation of virulence in Staphylococcus epidermidis: evidence for control of the polysaccharide intercellular adhesin synthesis by alternating insertion and excision of the insertion sequence element IS256. Mol. Microbiol. 32:345-356. [DOI] [PubMed] [Google Scholar]
- 299.Zinoviev, V. V., A. A. Evdokimov, E. G. Malygin, S. L. Schlagman, and S. Hattman. 2003. Bacteriophage T4 Dam DNA-(N6-adenine)-methyltransferase. Processivity and orientation to the methylation target. J. Biol. Chem. 278:7829-7833. [DOI] [PubMed] [Google Scholar]
- 300.Zweiger, G., G. Marczynski, and L. Shapiro. 1994. A Caulobacter DNA methyltransferase that functions only in the predivisional cell. J. Mol. Biol. 235:472-485. [DOI] [PubMed] [Google Scholar]