Abstract
The frequencies of 21 competence genes were analyzed in 94 genotypes of Streptococcus mutans. These include those of a main regulatory system (comCDE), structural, and other regulatory orthologues identified in the genome of strain UA159. PCR and Southern blot analysis revealed that all genes are widespread within the species.
Streptococcus mutans are the major pathogens of dental caries, a biofilm-dependent infectious disease. These organisms are able to prevail in the complex microbial community of the oral biofilm in the presence of sucrose, under extremely low pHs responsible for tooth demineralization, and can physiologically adapt to the stressful conditions to which the cariogenic biofilm is exposed. Several of these processes involve two-component signal transduction systems (TCS). The most-studied TCS of S. mutans is the quorum-sensing system comCDE that regulates genetic competence (12) and also has been shown to play a role in biofilm formation and acid tolerance (10-12). Other regulatory (comX1, mecA, ciaH/R, and LuxS) genes appear to be involved in competence via a complex net of signals that may regulate structural genes involved in the early and late events of competence, including DNA binding, transport, processing, and recombination (16). These genes also may play a role in biofilm growth and structure (14, 20, 23, 27). The genetic diversity of the components of competence is unknown. In contrast to Streptococcus pneumoniae and many commensal streptococcal species of the oral cavity, e.g., Streptococcus gordonii, the frequency of S. mutans transformation is low, and the majority of isolates appear not to be transformable in vitro (17, 25). In the present study, we characterized the genetic organization of 11 chromosomal loci of regulatory and structural genes with known or putative roles in competence in a collection of S. mutans genotypes isolated from children during the initial phase of colonization.
A total of 94 S. mutans genotypes were analyzed. Fifty genotypes were isolated from 14 6- to 24-month-old children, 2 of whom presented with initial caries lesions (8). The other 44 genotypes were isolated from 35 12- to 30-month-old children in a separate study of the same population (13). Carious lesions were detected in 15 of these children. The genotypic identities were determined by arbitrarily primed PCR as described in previous studies (8, 13). Strains were grown from frozen stocks in Todd-Hewitt or brain heart infusion broth at 37°C in an atmosphere of 10% CO2 and 90% O2.
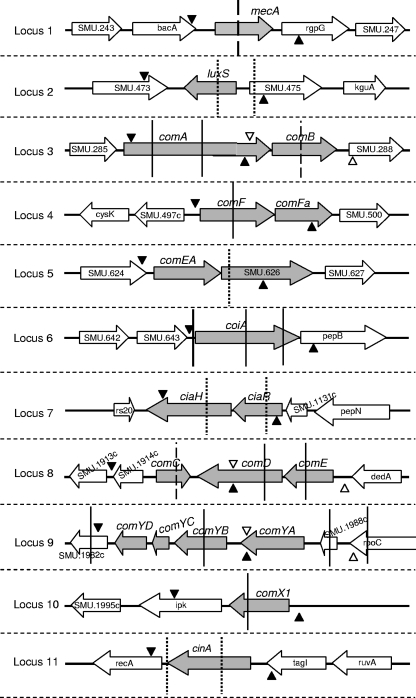
Genomic DNAs were purified from 1.5 ml of culture by using a MasterPure DNA purification kit (Epicenter Technologies, Madison, Wis.) as recommended by the manufacturer. A total of 21 genes from 11 loci within the genome of S. mutans strain UA159 (http://www.genome.ou.edu) were analyzed by PCR (Table 1). The gene organization within the UA159 chromosome and the position of the respective primer sets used for screening are shown in Fig. 1. Table 2 shows sequences of primers specific for each locus. PCRs were performed in volumes of 50 μl (200 μM deoxynucleoside triphosphates, 2.5 mM MgCl2, 0.3 μM concentrations of each upper and lower primer, and 1.25 U of Taq DNA polymerase [Invitrogen]). The thermal conditions varied slightly for each locus analyzed and included 35 cycles of denaturing at 95°C for 45s, annealing from 50 to 52°C for 1 min (Table 2), and extension at 72°C for 2 min. The genomic DNA of strain UA159 was used as a positive control in all PCR baths. Streptococcus sobrinus strain 15JP1 was used as a negative control. PCR products were resolved (8 V/cm in Tris-borate-EDTA) in 0.8% agarose gels and stained with ethidium bromide. To confirm gene absence in the PCR-negative genotypes, Southern blot assays were performed using the amplicons obtained from the control UA159 as probes. Restriction maps are shown in Fig. 1. After digestion of 3 μg of genomic DNA at the appropriated conditions, fragments were electrophoretically resolved (3 V/cm in 0.8% agarose gels) and transferred to Hybond+ membranes (Amersham Biosciences, Little Chalfont, Buckinghamshire, United Kingdom), as described elsewhere (22). Membranes were then probed and developed by using an enhanced chemiluminescence system (Amersham Biosciences), as recommended by the manufacturer.
TABLE 1.
Competence genes identified in the genome of the strain UA159 that were analyzed in this study
| Gene | GenBank accession no.a | Assigned function |
|---|---|---|
| mecA | 24378754 | Negative regulator of genetic competence |
| luxS | 24378961 | Putative autoinducer-2 production protein LuxS |
| comA | 24378790 | ABC transporter, ATP-binding protein ComA |
| comB | 24378791 | ComB, accessory factor for ComA |
| comF | 24378982 | Late competence protein, required for DNA uptake |
| comFa | 24378983 | Late competence protein |
| comEA | 24378983 | DNA uptake protein and related DNA-binding proteins |
| comEC | 24379099 | DNA internalization-related competence protein ComEC/Rec2 |
| coiA | 24379117 | Putative competence protein, transcription factor |
| ciaH | 24379560 | Putative histidine kinase sensor CiaH |
| ciaR | 24379561 | Putative response regulator CiaR |
| comC | 24380265 | Competence stimulating peptide, precursor |
| comD | 24380266 | Putative histidine kinase of the competence regulon, ComD |
| comE | 24380267 | Putative response regulator of the competence regulon, ComE |
| comYD | 24380327 | Putative late competence protein ComYD, exogenous DNA-binding protein |
| comYC | 24380328 | Late competence protein, exogenous DNA-binding protein |
| comYB | 24380329 | Putative ABC transporter subunit ComYB; part of the DNA transport machinery |
| comYA | 24380330 | Putative ABC transporter, ATP-binding protein ComYA; late competence gene |
| SMU1988c | Putative DNA-binding protein | |
| comX1 | 24380340 | Transcriptional regulator of competence-specific gene |
| cinA | 24380420 | Putative competence and damage-inducible protein CinA |
Available online (http://www.ncbi.nlm.nih.gov/genomes/lproks.cgi).
FIG. 1.
Genomic organization of the 11 loci identified in the genome of the strain UA159 which contains 21 competence-related genes, as analyzed by PCR and Southern blotting. Shaded arrows indicate the open reading frame of the com genes and the direction of transcription. The locations of primers designed for PCR screening are indicated by bullets. Sets of primers designed for each amplicon are differentiated by color (black or white). Vertical lines indicate the restriction sites of the endonucleases selected for the Southern blot analysis within each amplicon: dotted line, HindIII; solid line, HaeIII; and dashed line, HhaI. Purified amplicons identified in PCRs with chromosomal DNA of the strain UA159 were applied as probes for Southern blot analysis.
TABLE 2.
Oligonucleotides used for PCR screening of genes identified in the 11 loci of the S. mutans strain UA159 with a known or putative role in the phenotype of competence in S. mutans
| Locus | Amplicon (expected size in bp) | Primer sequence (forward and reverse) | Annealing temp (°C) | Competence gene(s) within the amplicon |
|---|---|---|---|---|
| 1 | MecA (1,083) | 5′-GCACGATTCTAACTGGTAGT-3′ | 52 | mecA |
| 5′-AGGAGTCAAAACAAGTGAAG-3′ | ||||
| 2 | LuxS (1,496) | 5′-GATCGTAGTCGAGTTCCTTA-3′ | 52 | luxS |
| 5′-GCTCATCAATATGCCTAGAT-3′ | ||||
| 3 | ComA (2,900) | 5′-GTCATAGCCGTTAACATTCT-3′ | 52 | comA |
| 5′-GAGAAATAAGACAGCAAAGC-3′ | ||||
| ComAB (1,425) | 5′-TTTTAGCTAAGCAAGGTTTC-3′ | 52 | comA, comB | |
| 5′-CTTTACGTCTGGACTGATTT-3′ | ||||
| 4 | ComFFa (1,776) | 5′-GTGATGGAGAATTTAGAGA-3′ | 50 | comF, comFa |
| 5′-GTTTTACACTACCATCTTCT-3′ | ||||
| 5 | ComEA (1,670) | 5′-CTGGAACCCAGAAAAGCATC-3′ | 50 | comEA, SMU.626 |
| 5′-AAAAGGGCAGCTGAATAGCA-3′ | ||||
| 6 | CoiA (1,367) | 5′-TGAAATCTACTTTAGTCCTT-3′ | 50 | coiA |
| 5′-AGCTTGTAATTCTTGATAGT-3′ | ||||
| 7 | CiaHR (1,385) | 5′-CGAGGTTGAATTTCTGTTAT-3′ | 52 | ciaH, ciaR |
| 5′-AAATTTTTGATCGTATCTGG-3′ | ||||
| 8 | ComCD (1,771) | 5′-TATCAGATGAGTTTGTCC-3′ | 52 | comC, comD |
| 5′-ATATCACCCAGTATAGTCAG-3′ | ||||
| ComDE (1,862) | 5′-AGAGATTCTATTTGCTGACT-3′ | 50 | comD, comE | |
| 5′-TATGTAGGAAGAGTTGAACA-3′ | ||||
| 9 | ComYDA (1,978) | 5′-TGACTCAACTGAGTAAGAAT-3′ | 52 | comYD, comYC |
| 5′-GTTTATGCTAGGATGTTAGA-3′ | comYB, comYA | |||
| ComYA1988 (1,399) | 5′-TCTAACATCCTAGCATAAAC-3′ | 50 | comYA, SMU1988c | |
| 5′-AGAGGACACAGTAGAAGAGT-3′ | ||||
| 10 | ComX (1,437) | 5′-GACAAAGTAGTCGCTAAAGG-3′ | 52 | comX |
| 5′-ACATACCCTGCTTTATCTTG-3′ | ||||
| 11 | CinA (1,789) | 5′-CAATATCAAGAGCCAGACTT-3′ | 52 | cinA |
| 5′-GTATACCTGCTCAAACGAAT-3′ |
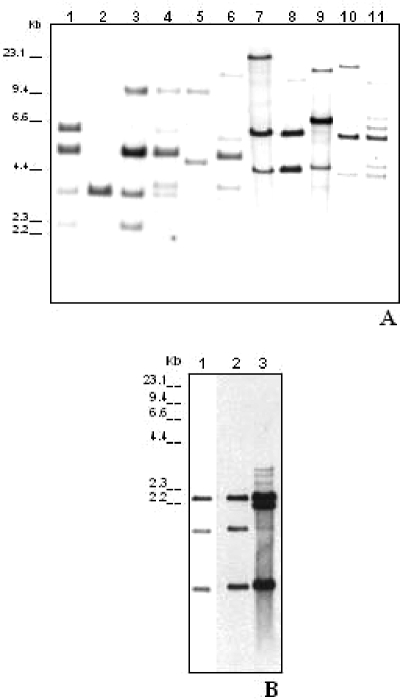
Several strains did not yield amplicons for one or more loci; however, Southern blot analysis revealed that all 11 gene loci were present in each of the tested strains (Table 3), although atypical restriction patterns were frequently observed (Table 3). Ten distinct classes of restriction (Fig. 2) were identified among the 19 strains showing a Southern blot pattern distinct from UA159 at the comCD locus (Table 3). Among the 26 genotypes that were PCR negative for comCD, 10 were also PCR negative for comDE, suggesting that polymorphism might be involved in at least comD or, perhaps, the whole comCDE locus. Southern blot analysis of three randomly selected clinical isolates that were PCR positive for comCD revealed a UA159-type pattern (data not shown). The lower number of atypical patterns in assays with comDE probe, compared to the comCD probe (Table 3), might be due to differences in the conservation of the restriction sites used. All of the strains yielded amplicons for ciaHR. It is possible that the primer set designed for these genes included highly conserved sequences because ciaHR primers have also generated amplicons for S. sobrinus strains, a species closely related to S. mutans that is also implicated in dental caries pathogenesis. BLAST analysis of ciaHR primer sequences against the unfinished genome of the strain S. sobrinus 6715 (http://www.tigr.org/tdb/mdb/mdbinprogress.html) did not reveal regions of homology that might account for amplification (data not shown). Except for ciaHR primers, all of the others appeared to be S. mutans specific, since they did not yield amplicons for the S. sobrinus strain 15JP1. In addition, during the course of the present study, a total of 11 strains previously defined as S. mutans species were negative in PCR for most other com genes analyzed (data not shown). Sequencing analysis of the 16S rRNA gene revealed that these 11 strains were S. sobrinus (data not shown) and were thus excluded from the analysis. The reason(s) for the significant differences in the capacities for genetic transformation between Streptococcus species and between strains within the same species are not understood. The ability to achieve competence in virulence is unclear. It is hypothesized that incorporation of foreign DNA may improve fitness to environmental stresses, providing competitive advantages (3, 7). There is evidence that some TCS that are involved in competence may regulate multiple virulence factors (10, 12, 24, 27, 28), and their components could be important targets for antibacterial therapy. Thus, it is important to establish the conservation of these systems within the S. mutans species.
TABLE 3.
Structural analysis of 21 competence-related genes organized in 11 loci identified in the chromosome of the strain UA159 in 94 clinical genotypes of S. mutans
| Amplicon | No. (%) of clinical genotypes
|
|||
|---|---|---|---|---|
| PCR screening in 94 genotypes
|
Southern blot analysis of PCR-negative genotypes
|
|||
| PCR positive | PCR negative | Polymorphic pattern | UA159 pattern | |
| MecA | 93 (98.9) | 1 (1.1) | 1 (1.1) | 0 |
| LuxS | 82 (87.2) | 12 (12.8) | 8 (8.5) | 4 (4.2) |
| ComA | 93 (98.9) | 1 (9.3) | 0 | 1 (1.1) |
| ComAB | 88 (93.6) | 6 (6.4) | 3 (3.2) | 3 (3.2) |
| ComFFa | 71 (75.5) | 23 (24.4) | 12 (12.8) | 11 (11.7) |
| ComEA | 89 (94.7) | 5 (5.3) | 5 (5.3) | 0 |
| CoiA | 92 (97.9) | 2 (2.1) | 0 | 2 (2.1) |
| CiaHR | 94 (100.0) | |||
| ComCD | 68 (72.3) | 26 (27.7) | 19 (20.2) | 7 (7.4) |
| ComDE | 73 (88.3) | 11 (11.7) | 2 (2.1) | 9 (9.6) |
| ComYDB | 90 (95.7) | 4 (4.3) | 1 (1.1) | 3 (3.2) |
| ComYA1988 | 80 (85.1) | 14 (14.9) | 0 | 14 (14.9) |
| ComX1 | 84 (89.4) | 10 (10.6) | 0 | 10 (10.6) |
| CinA | 90 (95.7) | 4 (4.3) | 1 (1.1) | 3 (3.2) |
FIG. 2.
Southern blot analysis of the comCDE locus in UA159 and clinical genotypes that were PCR-negative for comCD (A) and comDE (B) sequences. Lane 1 corresponds to the predicted Southern blot pattern obtained for the control strain UA159. (A) Lanes 2 to 11 correspond to strains representative of each one of the 10 distinct classes of Southern blot identified in 19 strains with atypical Southern blots probed with the comCD amplicon. (B) Lanes 2 and 3 are representative patterns of Southern blots probed with comDE amplicon in the comDE PCR-negative strains. Note that some PCR-negative strains, represented in lane 2, showed the same pattern as strain UA159.
Sequence comparisons of comCDE genes between several species of the genus streptococcus have indicated that interspecies gene replacements may frequently occur between naturally competent streptococci within the Streptococcus mitis group (6). Differences in GC content within the comCDE locus in comparison with the whole streptococcal genome concur with this idea (7). A locus with homology to comCDE (named silCDE) was identified in the screening of virulence genes in the S. pyogenes genotype JS95 which was isolated from a subject with invasive infection. S. pyogenes is a species with a low rate of transformation and causes infections having a wide range of severity (7). Similar to the organization of comCDE genes in the genome of S. mutans UA159, the silC, silD, and silE genes are flanked by Blp bacteriocin genes and also by a transposable element IS1562. The silCDE locus was shown to confer competence in the virulent genotype (7), a trait that is not observed in the S. pyogenes strain M1 whose genome was sequenced and which does not have silCDE genes (7). M1 is defective in transformation, although it contains several com genes (4). Its inability to be transformed was attributed to the lack of comAB genes (4). In screening 214 strains of S. pneumoniae, comC and comA genes were shown to be widespread (21), and these genes were detected in all of the S. mutans strains (Table 3). The upper primer utilized for screening the comCD sequence targeted a small intergenic space between open reading frames SMU1914c and SMU1913c (Fig. 1) that encodes homologues of the immunity protein Blpl and a bacteriocin peptide, BlpJ, also identified in the S. pneumoniae strain TIGR4.
Because two distinct alleles of comC were detected among 42 strains of S. pneumoniae, the hypothesis that the combination of incompatible alleles of comC (encoding the CSP precursor) and comD (the respective receptor) (5) was raised to explain the deficiency in in vitro transformation observed among 50% of the S. pneumoniae isolates (19). However, 93% of 60 strains analyzed have shown only two distinct comC alleles matched with the respective comD alleles (26), arguing against the incompatible allele hypothesis. Thus, other factors may be involved in variations in the competence phenotype. It has been shown in S. pneumoniae that the effect of gene inactivation of components of TCS which are involved in virulence is dependent on the strain background (2). Since few S. mutans strains were identified as suitably transformable in vitro, most of the genetic studies of molecular mechanisms of virulence have been limited to a few strains, mainly GS5, NG8, and UA159 (and its variant LT11). The distribution and/or diversity of regulatory and structural genes implicated in competence might help to explain differences in competence. However, to our knowledge, there is no such information describing S. pneumoniae or other streptococcal species.
It has been estimated that transformation occurs in only 28% of clinical isolates of S. mutans in vitro (25), and it is not known whether this low frequency of transformation has a genetic basis or might be simply due to unsuitable lab conditions. The frequency of transformation is significantly variable among naturally competent strains (e.g., GS5, NG8, UA159, LT11, and UA140), and it has been suggested that variation in the genetic background (17) and/or in the expression of late competence genes may be associated with these variations (15). The alternate sigma factor comX1 that appears to regulate several genes involved in late events of competence was detected in all of the strains (Table 3). In contrast to S. pneumoniae, in which at least two alleles of comX1 were identified in the genome (9, 18), our analysis indicated that S. mutans genotypes contain only a single copy of comX1, as verified for the strain UA159 (1). All other structural and regulatory loci studied here have shown some degree of genetic diversity, although lower in frequency compared to the comCDE locus (Table 3). However, a more detailed analysis of these genes should be performed to allow comparisons in degree of conservation, since Southern blot assays were performed here only with PCR-negative strains, and a PCR-positive result does not imply an absence of polymorphisms. Apart from the modest diversity identified, the results indicated that all 11 loci that contain genes with a regulatory or structural role in competence were present in all of the S. mutans genotypes analyzed, suggesting that these genes each play a fundamental role in S. mutans biology. This knowledge may increase the interest in these genes as therapeutic targets. Furthermore, sequencing analysis of the polymorphic strains identified here, along with gene expression studies, might help to explain additional variation in the competence phenotype previously reported within the S. mutans species.
Acknowledgments
This study was supported by the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP, proc. 02/07156-1; 03/01836-3) and the Public Health Service (NIDCR, grant DE-06153). S.B. was supported by Harvard Medical School (international fellowship). M.I.K. was supported by FAPESP (proc. 02/13473-0). R.O.M.-G. was supported by CAPES (ProDoc 029/03) and FAPESP (proc. 03/01836-3). D.J.S. was supported by NIH (DE-06153).
We thank R. A. Burne for help with the sequencing analysis of some of the isolates.
REFERENCES
- 1.Ajdic, D., W. M. McShan, R. E. McLaughlin, G. Savic, J. Chang, M. B. Carson, C. Primeaux, R. Tian, S. Kenton, H. Jia, S. Lin, Y. Qian, S. Li, H. Zhu, F. Najar, H. Lai, J. White, B. A. Roe, and J. J. Ferretti. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. USA 99:14434-14439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blue, C. E., and T. J. Mitchell. 2003. Contribution of a response regulator to the virulence of Streptococcus pneumoniae is strain dependent. Infect. Immun. 71:4405-4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dagkessamanskaia, A., M. Moscoso, V. Henard, S. Guiral, K. Overweg, M. Reuter, B. Martin, J. Wells, and J. P. Claverys. 2004. Interconnection of competence, stress and CiaR regulons in Streptococcus pneumoniae: competence triggers stationary phase autolysis of ciaR mutant cells. Mol. Microbiol. 51:1071-1086. [DOI] [PubMed] [Google Scholar]
- 4.Ferretti, J. J., W. M. McShan, D. Ajdic, D. J. Savic, G. Savic, K. Lyon, C. Primeaux, S. Sezate, A. N. Suvorov, S. Kenton, H. S. Lai, S. P. Lin, Y. Qian, H. G. Jia, F. Z. Najar, Q. Ren, H. Zhu, L. Song, J. White, X. Yuan, S. W. Clifton, B. A. Roe, and R. McLaughlin. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 98:4658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Havarstein, L. S., P. Gaustad, I. F. Nes, and D. A. Morrison. 1996. Identification of the streptococcal competence-pheromone receptor. Mol. Microbiol. 21:863-869. [DOI] [PubMed] [Google Scholar]
- 6.Havarstein, L. S., R. Hakenbeck, and P. Gaustad. 1997. Natural competence in the genus Streptococcus: evidence that streptococci can change pherotype by interspecies recombinational exchanges. J. Bacteriol. 179:6589-6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hidalgo-Grass, C., M. Ravins, M. Dan-Goor, J. Jaffe, A. E. Moses, and E. Hanski. 2002. A locus of group A streptococcus involved in invasive disease and DNA transfer. Mol. Microbiol. 46:87-99. [DOI] [PubMed] [Google Scholar]
- 8.Klein, M. I., F. M. Florio, A. C. Pereira, J. F. Hofling, and R. B. Goncalves. 2004. Longitudinal study of transmission, diversity, and stability of Streptococcus mutans and Streptococcus sobrinus genotypes in Brazilian nursery children. J. Clin. Microbiol. 42:4620-4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee, M. S., and D. A. Morrison. 1999. Identification of a new regulator in Streptococcus pneumoniae linking quorum sensing to competence for genetic transformation. J. Bacteriol. 181:5004-5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li, Y. H., M. N. Hanna, G. Svensater, R. P. Ellen, and D. G. Cvitkovitch. 2001. Cell density modulates acid adaptation in Streptococcus mutans: implications for survival in biofilms. J. Bacteriol. 183:6875-6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li, Y. H., P. C. Lau, J. H. Lee, R. P. Ellen, and D. G. Cvitkovitch. 2001. Natural genetic transformation of Streptococcus mutans growing in biofilms. J. Bacteriol. 183:897-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li, Y. H., N. Tang, M. B. Aspiras, P. C. Lau, J. H. Lee, R. P. Ellen, and D. G. Cvitkovitch. 2002. A quorum-sensing signaling system essential for genetic competence in Streptococcus mutans is involved in biofilm formation. J. Bacteriol. 184:2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mattos-Graner, R. O., Y. Li, P. W. Caufield, M. Duncan, and D. J. Smith. 2001. Genotypic diversity of mutans streptococci in Brazilian nursery children suggests horizontal transmission. J. Clin. Microbiol. 39:2313-2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Merritt, J., F. Qi, S. D. Goodman, M. H. Anderson, and W. Shi. 2003. Mutation of luxS affects biofilm formation in Streptococcus mutans. Infect. Immun. 71:1972-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Merritt, J., F. Qi, and W. Shi. 2005. A unique nine-gene comY operon in Streptococcus mutans. Microbiology 151:157-166. [DOI] [PubMed] [Google Scholar]
- 16.Morrison, D. A., and M. S. Lee. 2000. Regulation of competence for genetic transformation in Streptococcus pneumoniae: a link between quorum sensing and DNA processing genes. Res. Microbiol. 151:445-451. [DOI] [PubMed] [Google Scholar]
- 17.Murchison, H. H., J. F. Barrett, G. A. Cardineau, and R. Curtiss, III. 1986. Transformation of Streptococcus mutans with chromosomal and shuttle plasmid (pYA629) DNAs. Infect. Immun. 54:273-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peterson, S. N., C. K. Sung, R. Cline, B. V. Desai, E. C. Snesrud, P. Luo, J. Walling, H. Li, M. Mintz, G. Tsegaye, P. C. Burr, Y. Do, S. Ahn, J. Gilbert, R. D. Fleischmann, and D. A. Morrison. 2004. Identification of competence pheromone responsive genes in Streptococcus pneumoniae by use of DNA microarrays. Mol. Microbiol. 51:1051-1070. [DOI] [PubMed] [Google Scholar]
- 19.Pozzi, G., L. Masala, F. Iannelli, R. Manganelli, L. S. Havarstein, L. Piccoli, D. Simon, and D. A. Morrison. 1996. Competence for genetic transformation in encapsulated strains of Streptococcus pneumoniae: two allelic variants of the peptide pheromone. J. Bacteriol. 178:6087-6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qi, F., J. Merritt, R. Lux, and W. Shi. 2004. Inactivation of the ciaH Gene in Streptococcus mutans diminishes mutacin production and competence development, alters sucrose-dependent biofilm formation, and reduces stress tolerance. Infect. Immun. 72:4895-4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramirez, M., D. A. Morrison, and A. Tomasz. 1997. Ubiquitous distribution of the competence related genes comA and comC among isolates of Streptococcus pneumoniae. Microb. Drug Resist. 3:39-52. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook, J., E. F. Fritsch, and T. Maniatis. 2005. Molecular cloning: a laboratory guide. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 23.Wen, Z. T., and R. A. Burne. 2002. Functional genomics approach to identifying genes required for biofilm development by Streptococcus mutans. Appl. Environ. Microbiol. 68:1196-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wen, Z. T., P. Suntharaligham, D. G. Cvitkovitch, and R. A. Burne. 2005. Trigger factor in Streptococcus mutans is involved in stress tolerance, competence development, and biofilm formation. Infect. Immun. 73:219-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Westergren, G., and C. G. Emilson. 1983. Prevalence of transformable Streptococcus mutans in human dental plaque. Infect. Immun. 41:1386-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whatmore, A. M., V. A. Barcus, and C. G. Dowson. 1999. Genetic diversity of the streptococcal competence (com) gene locus. J. Bacteriol. 181:3144-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoshida, A., T. Ansai, T. Takehara, and H. K. Kuramitsu. 2005. LuxS-based signaling affects Streptococcus mutans biofilm formation. Appl. Environ. Microbiol. 71:2372-2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoshida, A., and H. K. Kuramitsu. 2002. Multiple Streptococcus mutans genes are involved in biofilm formation. Appl. Environ. Microbiol. 68:6283-6291. [DOI] [PMC free article] [PubMed] [Google Scholar]