Abstract
Genetic and antigenic analyses of influenza B virus field strains isolated in Taiwan from 1998 to 2005 were performed. To investigate the molecular evolution of influenza B viruses, sequence analysis of the hemagglutinin (HA1 subunit) and neuraminidase genes was performed. All influenza B viruses isolated between 1998 and 2000 belonged to the B/Yamagata/16/88 lineage. The B/Victoria/2/87 lineage, which was cocirculating with the Yamagata lineage, was identified in Taiwan in March 2001. Concurrently, there was an increasing prevalence of this lineage in many parts of the world, including North America and Europe, during the 2001-2002 season. Since 2002, genetic reassortants of influenza B virus with the Victoria lineage of hemagglutinin and the Yamagata lineage of neuraminidase have been found at a rate of 46%. Therefore, in 2002, at least three sublineages of influenza B virus strains, the B/Shanghai/361/2002-like strain (Yamagata lineage), the B/Hong Kong/330/01-like strain (Victoria lineage), and the B/Hong Kong/1351/02-like strain (B reassortant lineage), were identified in Taiwan. The results showed that genetically distinct lineages can cocirculate in the population and that the reassortment among these strains plays a role in generating the genetic diversity of influenza B viruses. Interestingly, from January to April 2005, B reassortant viruses became dominant (73%) in Taiwan, which indicated that a mismatch had occurred between the influenza B vaccine strain recommended for the 2004-2005 season in the Northern hemisphere by the World Health Organization and the epidemic strain.
Infection with influenza viruses can cause severe morbidity and mortality in the elderly and in children. The relative prevalences of the viruses vary from year to year, as does their geographical distribution (6). The successful application of a vaccine for influenza control in a particular region depends on the circulating strains and seasonal patterns of influenza infection. Therefore, understanding the variation of influenza viruses can provide clues for viral prophylaxis.
While both influenza A and B viruses have high frequencies of point mutations on their hemagglutinin (HA) and neuraminidase (NA) genes, genetic reassortment is commonly seen in influenza A viruses and seldom in influenza B viruses (6, 14). Insertion and deletion of nucleotides in the HA gene is another strategy of evolution of the influenza B virus (7, 9). Since 1983, influenza B viruses have cocirculated with two evolutionary lineages that are distinct at the genetic and antigenic levels, the influenza B/Victoria/2/87 lineage and the B/Yamagata/16/88 lineage (12). Antibodies produced against these two distinct lineages of viruses have shown no cross-protection (5, 11). As a result, the World Health Organization recommended vaccine strains on the basis of locally circulating strains.
B/Victoria/2/87 lineage viruses first appeared in the 1980s, but B/Yamagata/16/88 lineage viruses became the dominant influenza B viruses in the 1990s (4). Between 1991 and 2000, B/Victoria/2/87 lineage viruses were identified only in eastern Asia. However, the reappearance of influenza B/Victoria/2/87 lineage viruses in North America and Europe occurred during the 2000-2001 and 2001-2002 seasons and then spread globally (13). Cocirculation of Victoria and Yamagata lineages of influenza B viruses were further reported in Italy, Europe, and Israel during the 2001-2002 season (1, 3). B reassortants with a Victoria lineage HA and a Yamagata lineage NA have been observed since February 2002 and have spread worldwide (3, 10).
Taiwan is located in close proximity to China, a country where new influenza strains frequently emerge. Not much is understood about the influenza B viruses that appear in Taiwan. The antigenic properties and phylogenetic relationships of HA and NA genes of influenza B virus isolates in Taiwan during 1998 to 2005 were examined in this study. Our results showed a greatly increasing reappearance of reassortant influenza B viruses in 2005.
MATERIALS AND METHODS
Viral isolation.
Throat and nasal swabs from patients with flu-like syndromes were collected by sentinel and National Cheng Kung University Hospital physicians and immediately placed in 2 ml of a homemade viral transport medium which contains 0.5% gelatin, Earle's minimum essential medium, 200 U/ml penicillin-streptomycin, 0.05 mg/ml gentamicin, and 1.25 μg/ml amphotericin B (Fungizone). Refrigerated samples were inoculated within 24 h into tubes containing Madin-Darby canine kidney (MDCK) cells. Influenza virus infection of cells was confirmed by the use of monoclonal antibodies for influenza A and B viruses (Chemicon, Inc.). Hemagglutinin inhibition (HI) assays for antigenic typing were carried out using the World Health Organization influenza reagent kit provided by the Centers for Disease Control and Prevention, Atlanta, GA.
Nucleotide sequencing analysis.
Viral RNA was extracted from 140 μl of infected tissue culture fluid by using the QIAamp viral RNA mini kit. Five microliters of eluted RNA was used for amplification using a QIAGEN one-step reverse transcription (RT)-PCR reagent. Primers used for RT-PCR and sequencing (3) were as follows: B/HA98 (forward, 5′-ATAACATCGTCAAACTCACC-3′), B/HA836 (reverse, 5′-GCACCATGTAATCAACAACA-3′), B/NA1 (forward, 5′-GCTACCTTCAACTATACAAACG-3′), and B/NA2 (reverse, 5′-AACGAGGGTATGTCCACTCC-3′). Cycle conditions for RT-PCR for HA or NA gene amplification were as follows: 30 min at 50°C for reverse transcription, 1 cycle for 5 min at 95°C followed by 35 cycles of 30 s at 94°C, 30 s at 50°C (for HA) or 60°C (for NA), and 1 min at 72°C followed by 1 cycle for 10 min at 72°C, with subsequent holding at 4°C. Prior to sequencing, the PCR products were purified with a QIAquick PCR purification kit (QIAGEN). The purified DNA was sequenced using a BigDye Terminator cycle sequencing kit (Applied Biosystems, California) and a model 373A DNA sequencer (Perkin-Elmer). DNA sequence analysis was performed using software from the Wisconsin Genetics Computer Group (GCG). Phylogenetic comparisons were carried out using version 3.573c of the Phylogeny Inference Package (PHYLIP). Bootstrap values were obtained by performing 1,000 replicates.
Nucleotide sequence accession numbers.
Influenza virus sequences reported in this study have been deposited in the GenBank database under accession numbers DQ514388 to DQ514476.
RESULTS
Epidemiology of influenza B viruses in Taiwan.
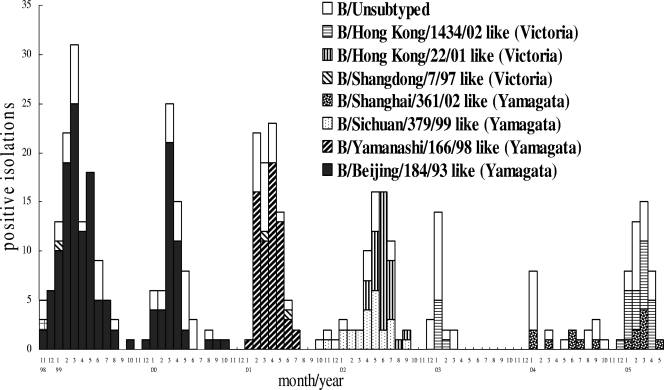
From January 1999 to May 2005, 404 influenza B viruses were isolated from 18,319 throat swab samples collected from patients with influenza-like symptoms. Influenza B virus epidemics occurred annually, with a peak in activity between March and May. Antigenic characterization of HA in circulating influenza B viruses was performed by using HI tests. The HI results showed that the majority of circulating influenza B viruses from 1998 to 2001 belonged to the Yamagata lineage. Most isolates from the 1998-1999 and 1999-2000 seasons were B/Beijing/184/93-like viruses, and most of the 2000-2001 season isolates were B/Yamanashi/166/98-like viruses (Fig. 1). In the 2001-2002 season, B/Hong Kong/22/01-like (Victoria lineage) and B/Sichuan/379/99-like (Yamagata lineage) viruses cocirculated. B/Hong Kong/1434/02-like (Victoria lineage) viruses were identified in the 2002-2003 season. The very mild influenza B activity (B/Shanghai/361/02-like strain, Yamagata lineage) seen during the 2003-2004 season was probably due to increased precautionary measures (such as isolating patients, wearing surgical masks in the hospital and public areas, and decreasing public gatherings) taken during the outbreak of severe acute respiratory syndrome. From December 2004 to April 2005, B/Hong Kong/1434/02-like viruses (Victoria lineage) reappeared, and some B/Shanghai/361/02-like strains (Yamagata lineage) also cocirculated between January and May 2005.
FIG. 1.
Monthly distribution of influenza B viruses based on HI tests in Taiwan from 1998 to 2005.
Phylogenetic analysis of influenza B viruses circulating in Taiwan.
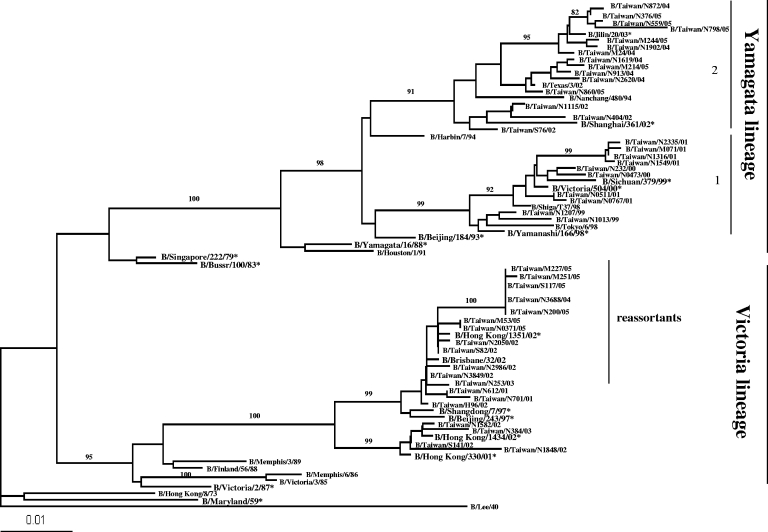
To determine the molecular evolution of the HA and NA genes, 44 and 45 strains, respectively, isolated in Taiwan from 1999 to 2005 were analyzed. Genetic analyses of the HA genes from 44 local isolates showed that 25 influenza B viruses belonged to the Yamagata lineage and 19 belonged to the Victoria lineage (Fig. 2). There were two clusters in the Yamagata lineage. Cluster 1 could be further divided into two clades, one of which included local isolates from 1999 which were similar to the B/Yamanashi/166/98 vaccine strain. The other clade included isolates from 2000 to 2001, which clustered with B/Sichuan/379/99 and B/Victoria/504/00 vaccine strains. Cluster 2 included isolates from 2002 and 2004 to 2005, which segregated with B/Shanghai/361/02 and B/Jilin/20/03 vaccine strains, respectively. B/Jilin/20/03 and B/Shanghai/361/02 vaccine strains (the vaccines recommended for the 2004-2005 season in the Northern hemisphere) were identified frequently during the 2003-2004 season globally. This B/Shanghai/361/02-like strain had appeared largely in Taiwan since 2002, indicating that this lineage occurred in Taiwan one to two seasons earlier than in other areas of the world. The B/Victoria/2/87 lineage, which cocirculated with the Yamagata lineage, was initially isolated and identified in Taiwan in March 2001. Nineteen local isolates of the Victoria lineage segregated into two clades. One clade contained four local isolates from 2002 and 2003 which clustered with the B/Hong Kong/1434/02 and B/Hong Kong/330/01 vaccine strains, and the second clade, which included two, five, one, one, and six isolates from 2001, 2002, 2003, 2004, and 2005, respectively, clustered with the B/Hong Kong/1351/02 and B/Brisbane/32/02 strains.
FIG. 2.
Phylogenetic analysis of HA genes of influenza B virus isolates obtained during the 1999-2005 influenza season. The dendrogram of the 44 influenza B virus outbreak strains, with 28 reference strains from GenBank, is based on 738 nucleotides (nucleotides 98 to 836) of the HA1 gene using the neighbor-joining method with the DNADIST distance measure program (PHYLIP, version 3.573c). The percentage of bootstrap frequency of each branch in the tree is indicated. B/Lee/40 was included as an outgroup.
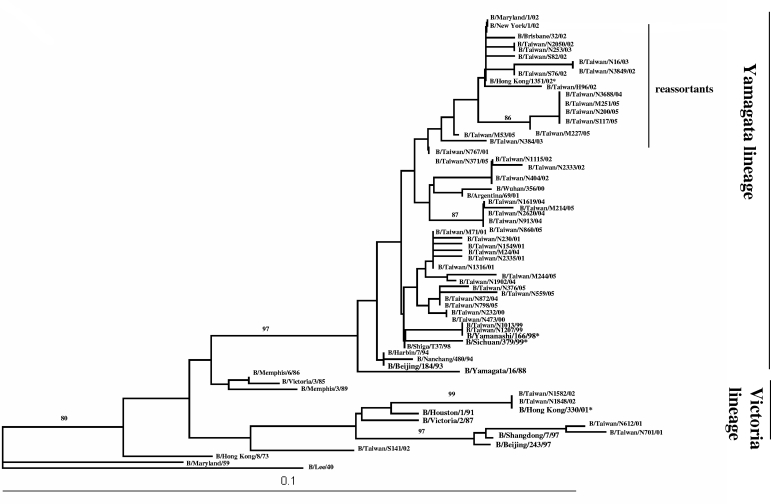
Sequence analysis of the NA genes from 45 Taiwan isolates showed that 40 influenza B viruses belonged to the Yamagata lineage and 5 belonged to the Victoria lineage (Fig. 3). One cluster among the Yamagata lineage included five, three, one, and five isolates from 2002, 2003, 2004, and 2005, respectively, which clustered with B/Hong Kong/1351/02 and B/Brisbane/32/02 and was recognized as a reassortant strain of viruses with an HA gene from the Victoria lineage but which had acquired an NA gene from a distinctly different lineage, the Yamagata lineage. The other cluster in the Yamagata lineage contained isolates from 1999 to 2005, which were similar to the B/Sichuan/379/99 vaccine strain. Four influenza B viruses of the Victoria lineage segregated into two clades. Two of the influenza B viruses from 2002 were similar to the B/Hong Kong/330/01 strain, and two viruses from 2001 were similar to the B/Shangdong/7/97 and B/Beijing/243/97 vaccine strains. The results showed that the B reassortant viruses were first identified in April 2002 and that there were three groups of influenza B viruses, including those belonging to the Yamagata lineage and the Victoria lineage and the B reassortant viruses which circulated in Taiwan in 2002.
FIG. 3.
Phylogenetic analysis of NA genes of influenza B virus isolates obtained during the 1999-2005 influenza season. The dendrogram of the 45 influenza B virus outbreak strains, with 24 reference strains from GenBank, of the NA gene using the neighbor-joining method with the DNADIST distance measure program (PHYLIP, version 3.573c). The percentage of bootstrap frequency of each branch in the tree is indicated. B/Lee/40 was included as an outgroup.
Variations in HA and NA amino acid sequences of influenza B viruses.
Regions of the local influenza B virus isolates containing 248 and 66 amino acids, respectively, of HA and NA were compared with those of recent vaccine strains in order to understand the variations between the Yamagata and Victoria lineages of influenza B viruses (Table 1, Table 2, and Table 3). The isolates with HAs clustering within the Yamagata lineage can be segregated into two groups. Taiwan isolates from the 1999 to 2001 Yamagata lineage were similar to B/Sichuan/379/99, and isolates from 2002 to 2005 were similar to B/Shanghai/361/02 or B/Jilin/20/03. Ten signature amino acids in HA, R48, D56, T75, N116, D126, K149, N168, Y179, E183, and G184, of B/Shanghai/361/02, are all conserved in our 2002 to 2005 isolates. However, the HA of all Taiwan isolates from 1999 to 2005 had an asparagine (N) at position 197 instead of an aspartic acid (D), as had B/Sichuan/379/99 and B/Shanghai/361/02. Interestingly, there were two signature amino acids at positions H40Y and L131P of HA found between the B/Shanghai/361/02 strain and our 2004-2005 local isolates which were also found in B/Jilin/20/03 of the Yamagata lineage (Table 1). However, as found in B/Jilin/20/03, two common amino acid changes in HA, I180V and D233A, were seen in some local isolates from 2004 to 2005. The Victoria lineage strains were divided into two distinct groups, B/Hong Kong/330/01 and B/Hong Kong/1351/02 (B reassortant) (Table 2). Local isolates from 2002 to 2005 had an asparagine (N) at position 197 instead of a lysine (K) in HA, as seen in B/Hong Kong/1351/02. The reassortant B viruses from 2004 to 2005 had three notable amino acid changes from the strains of 2002 at K48E, K80R or K80E, and K129N in their HA genes. Notably, B reassortants from 2002 (strains B/Taiwan/S82/02, -H96/02, and -N3849/02) had all the same amino acids in their HA as the isolates from the 2001 Victoria lineage (B/Taiwan/N612/01 and -N701/01). In addition, one 2003 reassortant, B/Taiwan/N384/03, had the same amino acid in its HA as the Victoria isolate from 2002 (B/Taiwan/N1582/02). These results indicated that the HA genes of B reassortants were obtained from the Victoria lineage of the previous year.
TABLE 1.
Variations in HA amino acid sequences in recent influenza B viruses of the Yamagata lineage
| Amino acid at position:
|
||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Virus strain(s) | 29 | 40 | 48 | 56 | 75 | 87 | 88 | 116 | 126 | 131 | 149 | 164 | 168 | 175 | 179 | 180 | 183 | 184 | 197 | 220 | 230 | 233 |
| B/Yamanashi/166/98a | V | H | K | T | V | V | R | K | N | P | R | D | T | V | H | I | K | E | N | V | G | D |
| B/Sichuan/379/99a | A | I | I | K | K | D | I | |||||||||||||||
| B/Shanghai/361/02a | R | D | T | N | D | L | K | N | Y | E | G | D | ||||||||||
| B/Jilin/20/03a | Y | R | D | T | N | D | K | N | Y | V | E | G | A | |||||||||
| B/Taiwan/N1013/99 | T | D | N | |||||||||||||||||||
| B/Taiwan/N1207/99 | D | N | ||||||||||||||||||||
| B/Taiwan/N232/00 | A | I | K | I | ||||||||||||||||||
| B/Taiwan/N473/00 | A | T | K | I | ||||||||||||||||||
| B/Taiwan/N511/01, -N767/01 | A | I | K | I | I | |||||||||||||||||
| B/Taiwan/N1316/01, -M71/01, -N2335/01 | A | I | D | K | E | I | ||||||||||||||||
| B/Taiwan/N1549/01 | A | A | I | K | E | I | ||||||||||||||||
| B/Taiwan/S76/02 | R | D | T | N | D | L | K | N | Y | G | ||||||||||||
| B/Taiwan/N404/02, -N1115/02 | R | D | T | N | D | L | K | N | Y | E | G | |||||||||||
| B/Taiwan/M24/04 | Y | R | D | T | N | D | K | N | Y | E | G | A | ||||||||||
| B/Taiwan/N872/04, -N1902/04 | Y | R | D | T | N | D | K | N | Y | V | E | G | A | |||||||||
| B/Taiwan/N913/04, -N1619/04, -N2620/04 | Y | R | D | T | N | D | K | N | Y | E | G | |||||||||||
| B/Taiwan/N376/05, -N559/05, -N798/05, -M244/05 | Y | R | D | T | N | D | K | N | Y | V | E | G | A | |||||||||
| B/Taiwan/N860/05, -M214/05 | Y | R | D | T | N | D | K | N | Y | E | G | |||||||||||
Vaccine strain.
TABLE 2.
Variations in HA amino acid sequences in recent influenza B viruses of the Victoria lineage
| Virus strain(s) | Amino acid at position:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 48 | 75 | 80 | 116 | 121 | 127 | 129 | 164 | 184 | 197 | 217 | |
| B/Beijing/243/97a | K | N | K | H | I | A | K | D | G | T | A |
| B/Hong Kong/330/01a | R | N | E | S | |||||||
| B/Taiwan/N612/01, -N701/01 | T | N | |||||||||
| B/Taiwan/S141/02 | R | N | E | N | |||||||
| B/Taiwan/N1582/02 | R | N | E | E | N | ||||||
| B/Taiwan/N1848/02 | R | N | E | R | N | ||||||
| Reassortant isolates | |||||||||||
| B/Hong Kong/1351/02a | T | K | |||||||||
| B/Brisbane/32/02a | T | N | |||||||||
| B/Taiwan/N2050/02 | I | N | |||||||||
| B/Taiwan/N2986/02 | D | T | N | ||||||||
| B/Taiwan/S82/02, -H96/02, -N3849/02 | T | N | |||||||||
| B/Taiwan/N384/03 | R | N | E | E | N | ||||||
| B/Taiwan/N3688/04 | E | R | T | N | N | ||||||
| B/Taiwan/M53/05, -N371/05 | E | T | N | S | |||||||
| B/Taiwan/M251/05 | E | R | T | V | N | N | |||||
| B/Taiwan/S117/05 | E | R | T | N | N | ||||||
Vaccine strain.
TABLE 3.
Variations in NA amino acid sequences in recent influenza B viruses
| Virus strain(s) | Amino acid at position:
|
||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 27 | 36 | 40 | 41 | 42 | 44 | 45 | 46 | 49 | 50 | 51 | 53 | 54 | 55 | 60 | 62 | 65 | 69 | 70 | 71 | 73 | 74 | 75 | |
| Yamagata lineage | |||||||||||||||||||||||
| B/Sichuan/379/99a | S | I | F | S | P | E | I | T | T | M | P | D | C | A | A | A | R | K | G | V | L | L | L |
| B/Taiwan/N1013/99, -N1207/99 | N | V | P | ||||||||||||||||||||
| B/Taiwan/N232/00, -N473/00 | V | F | |||||||||||||||||||||
| B/Taiwan/N767/01 | V | ||||||||||||||||||||||
| B/Taiwan/N1316/01, -N2335/01 | T | V | |||||||||||||||||||||
| B/Taiwan/S76/02 | L | V | |||||||||||||||||||||
| B/Taiwan/N1115/02 | V | Q | V | ||||||||||||||||||||
| B/Taiwan/M24/04 | T | T | V | ||||||||||||||||||||
| B/Taiwan/N872/04 | T | V | F | ||||||||||||||||||||
| B/Taiwan/N1902/04 | T | I | V | ||||||||||||||||||||
| B/Taiwan/N1619/04, -N2620/04 | Q | I | V | ||||||||||||||||||||
| B/Taiwan/N559/05 | T | T | V | F | |||||||||||||||||||
| B/Taiwan/N376/05 | T | V | F | ||||||||||||||||||||
| B/Taiwan/M214/05, -N860/05 | Q | I | V | ||||||||||||||||||||
| B/Taiwan/M244/05 | T | I | V | F | |||||||||||||||||||
| B/Taiwan/N798/05 | T | V | F | ||||||||||||||||||||
| Reassortant isolates | |||||||||||||||||||||||
| B/Brisbane/32/02a | V | ||||||||||||||||||||||
| B/Taiwan/H96/02 | V | ||||||||||||||||||||||
| B/Taiwan/N2050/02, -S82/02 | V | T | |||||||||||||||||||||
| B/Taiwan/N3849/02 | L | V | |||||||||||||||||||||
| B/Taiwan/N253/03 | V | ||||||||||||||||||||||
| B/Taiwan/N384/03 | V | I | |||||||||||||||||||||
| B/Taiwan/N3688/04 | P | S | V | ||||||||||||||||||||
| B/Taiwan/M53/05, -N371/05 | V | ||||||||||||||||||||||
| B/Taiwan/N200/05, -M227/05, -M251/05 | P | S | V | ||||||||||||||||||||
| Victoria lineage | |||||||||||||||||||||||
| B/Beijing/243/97a | L | I | F | S | P | K | V | I | T | M | S | D | C | A | V | A | H | E | E | M | F | L | L |
| B/Taiwan/N612/01, -N701/01 | Y | T | |||||||||||||||||||||
| B/Taiwan/N1582/02 | I | T | S | F | K | ||||||||||||||||||
| B/Taiwan/S141/02 | S | V | E | I | T | P | K | ||||||||||||||||
Vaccine strain.
Within the 66-amino-acid region of NA, a position 42 substitution (P42T or P42Q) was noticed in nonreassortant strains of the Yamagata lineage from 2001 to 2005 (Table 3). Interestingly, the reassortant B viruses from 2004 to 2005 had two notable amino acid changes from the strains from 2002 to 2003 at S41P and P42S. The Victoria lineage strains from 2002 had three signature amino acid changes from the strains of B/Beijing/243/97, V45I, M50T, E69K (Table 3). It should be noted that B reassortants (B/Taiwan/H96/02, -N253/03, -M53/05, and -N371/05) were found to have the same amino acids in their NA genes as those found in the Yamagata lineage (B/Taiwan/N767/01). In addition, the B reassortant B/Taiwan/N3849/02 had the same amino acids in its NA gene as those in the previous Yamagata lineage strain (B/Taiwan/S76/02). The results indicated that the NA genes of B reassortants were obtained from the Yamagata strains of the previous season.
Comparison of HI test results and phylogenetic analysis of influenza virus.
The results of HI testing and phylogenetic analysis were compared (Table 4). The lineage of HI and phylogenetic analysis of the HA gene were very consistent. There were two peaks of reassortant B viruses circulating in Taiwan from April 2002 to April 2005; the first peak was in 2002, and the second was in 2005. Interestingly, these reassorted B viruses accounted for 46% (5/11) of influenza B viruses tested in 2002 and became dominant (73% [19/26]) in January to April 2005.
TABLE 4.
Comparison of HI test results and phylogenetic analyses of influenza B viruses
| Isolate(s) | HI result | Phylogenetic derivation ofe:
|
|
|---|---|---|---|
| HA | NA | ||
| B/Taiwan/N1013/99, -N1207/99 | B/Beijing/184/93-likea | Yam88 | Yam88 |
| B/Taiwan/N232/00, -N473/00 | B/Beijing/184/93-likea | Yam88 | Yam88 |
| B/Taiwan/M71/01, -N767/01, -N1316/01, -N1549/01, -N2335/01 | B/Yamanashi/166/98-likea | Yam88 | Yam88 |
| B/Taiwan/N612/01 | B/Shangdong/7/97-likeb | Vic87 | Vic87 |
| B/Taiwan/N701/01 | B/Beijing/243/97-likeb | Vic87 | Vic87 |
| B/Taiwan/N404/02, -S76/02, -N1115/02 | B/Sichuan/379/99-likea | Yam88 | Yam88 |
| B/Taiwan/N1582/02, -S141/02, -N1848/02 | B/Hong Kong/22/01-likeb | Vic87 | Vic87 |
| B/Taiwan/S82/02, -N2986/02,d -H096/02, -N2050/02 | B/Hong Kong/22/01-likeb | Vic87c | Yam88c |
| B/Taiwan/N3849/02 | Virus titer <4HA unit | Vic87c | Yam88c |
| B/Taiwan/N253, -N384/03 | B/Hong Kong/1434/02-likeb | Vic87c | Yam88c |
| B/Taiwan/M24/04 | B/Shichuan/379/99-likea | Yam88 | Yam88 |
| B/Taiwan/N913/04, -N1619/04, -N1902/04 | B/Shanghai/361/02-likea | Yam88 | Yam88 |
| B/Taiwan/N872/04, -N2620/04 | Virus titer <4HA unit | Yam88 | Yam88 |
| B/Taiwan/N3688/04 | B/Hong Kong/1434/02-likeb | Vic87c | Yam88c |
| B/Taiwan/N303/05,d -N376/05, -M214/05, -M244/05, -N860/05 | B/Shanghai/361/02-likea | Yam88 | Yam88 |
| B/Taiwan/N559/05, -N798/05 | Virus titer <4HA unit | Yam88 | Yam88 |
| B/Taiwan/M53/05, -N200/05, -N202/05,d -N253/05,d -N265/05,d -M75/05,d -N371/05, -N525/05,d -M178/05,d -M227/05, -M238/05,d -N239/05,d -N868/05, -S117/05, -M251/05, -M258/05,d -N309/05,d -N1153/05,d -N1212/05d | B/Hong Kong/1434/02-likeb | Vic87c | Yam88c |
Serotype similar to that of Yamagata lineage.
Serotype similar to that of Victoria lineage.
B reassortant virus of the B/Yamagata/16/88 (Yam88) or B/Victoria/2/87 (Vic87) lineage.
Derivations are indicated with the source lineage (Yamagata [Yam] or Vic [Vic]) and the year of isolation of the source.
DISCUSSION
Circulating influenza B viruses can be divided into two antigenic and genetic groups represented by the B/Yamagata/16/88 and B/Victoria/2/87 reference strains. Analysis of influenza B virus isolates from Taiwan during the period 1998 to 2005 indicated that the circulating influenza B viruses prior to 2001 belonged to the Yamagata lineage. Yamagata and Victoria lineage viruses were cocirculating since March 2001. Interestingly, influenza B virus reassortants which had an HA gene from the Victoria lineage virus but had acquired an NA gene from a distinctly different lineage (the Yamagata lineage virus) have been identified in southern Taiwan since April 2002. Genetic analysis of the HA gene showed that the reassortant B viruses formed a separate branch from the Victoria lineage and were most closely related to the B/Hong Kong/1351/02 reference strain. These reassortant B viruses accounted for around 46% of influenza B viruses in 2002 and became dominant (73%) among influenza B virus strains isolated from January to April of 2005. The results indicated that a mismatch between the vaccine and epidemic strains had occurred in the Northern Hemisphere World Health Organization 2004 to 2005 influenza B virus vaccine recommendations, which was that B/Shanghai/361/2002-like (Yamagata lineage) should be the candidate strain. Among the Yamagata lineage strains, several isolates from 2002 were closely related to B/Shanghai/361/2002-like and B/Jilin/20/03-like viruses, which were recommended for the Northern hemisphere 2004-2005 season vaccine composition by the World Health Organization, demonstrating that these vaccine strains were circulating in Taiwan two seasons earlier. At least three lineages of influenza B virus strains, the B/Shanghai/361/2002-like strain (Yamagata lineage), the B/Hong Kong/330/01-like strain (Victoria lineage), and the B/Hong Kong/1351/02-like strain (B reassortant with Victoria-like HA and Yamagata-like NA genes), were cocirculating in 2002 in southern Taiwan.
From March to September of 2001 and during the 2001-2002 influenza season, Victoria lineage viruses were also detected in several countries, including Canada, the United States, Italy, The Netherlands, Norway, The Philippines, India, and Oman. This reemergence of the Victoria lineage led the World Health Organization to recommend B/Hong Kong/330/01 for the vaccine strain for the 2002-2003 influenza season. In addition, the Yamagata and Victoria lineages of influenza B viruses were found cocirculating in Europe and Israel in the 2001-2002 season (3). A number of the viruses in the B/Oman/16296/2001 group had also been found to be reassortants with a Victoria lineage HA and a Yamagata lineage NA gene (13). Similar to our results, phylogenetic analyses of HA and NA genes have found that the reassortant phenomenon occurred worldwide (2, 6, 13, 14). This group of reassortant viruses from the 2001-2002 season was most closely related to B/Hong Kong/1351/02 based on phylogenetic analysis. A previous report by Puzelli et al. (10) indicated that B reassortants in Italy were not the result of reassortment between those cocirculating strains, and no examples of viruses possessing mixtures of HA and NA from the Yamagata and Victoria lineages from the previous season were identified. In contrast, our results indicated that these B reassortants had resulted from the cocirculation of two lineages of preceding years. The HA sequence of three reassortant B viruses in 2002 (B/Taiwan/S82/02, -H96/02, and -N3849/02) were the same as those found for isolates from the previous year (2001) in Victoria lineage isolates (B/Taiwan/N612/01 and -701/01). In addition, the HA sequence of one B reassortant from 2003 (B/Taiwan/N384/03) seemed to have originated from a 2002 Victoria lineage virus (B/Taiwan/N1582/02). The NA genes of the 2002-2005 B reassortants were also closely related to those of isolates from the Yamagata lineage of the previous season, for example, B/Taiwan/767/01 and B/Taiwan/S76/02. The B reassortants discussed above appeared to be the result of reassortment between cocirculating strains of the previous season. Reassortant viruses also occurred previously in the early 1990s, except that their HA was related to a Yamagata lineage and their NA was related to a Victoria lineage. These reassortants circulated for several years in the 1990s (7). These two distinct genetic reassortant models demonstrate the strategies for the evolution of influenza B viruses in nature.
A notable variation in the amino acids defining the potential N-linked glycosylation site at amino acids 197 to 199 was found in Taiwan isolates of the Victoria lineage. Such an amino acid substitution is near the putative receptor-binding region and was suggested to play an important role in the determination of antigenicity (3, 13). In addition, all sequences from the Yamagata lineage contained the amino acid Asn at position 197, preserving the glycosylation site. In addition, predominant changes at positions 40 and 131 in the 2004-2005 season Taiwan isolates of the Yamagata lineage were also detected, which had not been reported previously. Furthermore, three novel signature amino acids, E48, R80, and N129, were found in the 2004-2005 local isolates of Victoria lineage.
Viral neuraminidase has been considered important in the effectiveness of vaccines; however, it is known that antibodies to viral neuraminidase play a lesser role in host immunity (8). The correspondence between the lineage of HI and the phylogenetic analysis of the HA gene was consistent in our data. This may represent the important role of the HA in the antigenic characteristics of HA over NA.
In conclusion, the phylogenetic analysis of both the HA and NA genes of local isolates revealed that the B reassortants had originated by reassortment between Victoria and Yamagata viruses cocirculating during the previous season. In addition, our findings suggest that Taiwanese influenza B virus strains were circulating one to two seasons ahead of the time at which the corresponding vaccine strains were recommended. Furthermore, the appearance of reassortant influenza B virus strains suggests that a virus typing scheme similar to that used for influenza A virus (e.g., H1N1, H3N2, etc.) may be useful and appropriate. The idea of a two-component typing scheme for influenza B virus may be a logical extension of this work. These results provide better understanding of the influenza B viruses in Taiwan and will contribute to the information necessary for continued worldwide surveillance of influenza viruses.
Acknowledgments
This study was supported by National Health Research Institutes grants and by grant CDC94-RM-012 from Department of Health, Taiwan Center for Disease Control.
REFERENCES
- 1.Ansaldi, F., P. D'Agaro, D. De Florentiis, S. Puzelli, Y. P. Lin, V. Gregory, M. Bennett, I. Donatelli, R. Gasparini, P. Crovari, A. Hay, and C. Campello. 2003. Molecular characterization of influenza B viruses circulating in northern Italy during the 2001-2002 epidemic season. J. Med. Virol. 70:463-469. [DOI] [PubMed] [Google Scholar]
- 2.Barr, I. G., N. Komadina, A. Hurt, R. Shaw, C. Durrant, P. Iannello, C. Tomasov, H. Sjogren, and A. W. Hampson. 2003. Reassortants in recent human influenza A and B isolates from South East Asia and Oceania. Virus Res. 98:35-44. [DOI] [PubMed] [Google Scholar]
- 3.Chi, X. S., T. V. Bolar, P. Zhao, R. Rappaport, and S.-M. Cheng. 2003. Cocirculation and evolution of two lineages of influenza B viruses in Europe and Israel in the 2001-2002 season. J. Clin. Microbiol. 41:5770-5773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hemphill, M. L., P. A. Rota, V. T. Ivanova, A. N. Slepushkin, and A. P. Kendal. 1993. Antigenic and genetic analyses of influenza type B viruses isolated in Russia, 1987-91. Epidemiol. Infect. 111:539-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levandowski, R. A., H. L. Regnery, E. Staton, B. G. Burgess, M. S. Williams, and J. R. Groothuis. 1991. Antibody responses to influenza B viruses in immunologically unprimed children. Pediatrics 88:1031-1036. [PubMed] [Google Scholar]
- 6.Lin, Y. P., V. Gregory, M. Bennett, and A. Hay. 2004. Recent changes among human influenza viruses. Virus Res. 103:47-52. [DOI] [PubMed] [Google Scholar]
- 7.McCullers, J. A., G. C. Wang, S. He, and R. G. Webster. 1999. Reassortment and insertion-deletion are strategies for the evolution of influenza B viruses in nature. J. Virol. 73:7343-7348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murphy, B. R., J. A. Kasel, and R. M. Chanock. 1972. Association of serum anti-neuraminidase antibody with resistance to influenza in man. N. Engl. J. Med. 286:1329-1332. [DOI] [PubMed] [Google Scholar]
- 9.Nerome, R., Y. Hiromoto, S. Sugita, N. Tanabe, M. Ishida, M. Matsumoto, S. E. Lindstrom, T. Takahashi, and K. Nerome. 1998. Evolutionary characteristics of influenza B virus since its first isolation in 1940: dynamic circulation of deletion and insertion mechanism. Arch. Virol. 143:1569-1583. [DOI] [PubMed] [Google Scholar]
- 10.Puzelli, S., F. Frezza, C. Fabiani, F. Ansaldi, L. Campitelli, Y. P. Lin, V. Gregory, M. Bennett, P. D'Agaro, C. Campello, P. Crovari, A. Hay, and I. Donatelli. 2004. Changes in the hemagglutinins and neuraminidases of human influenza B viruses isolated in Italy during the 2001-02, 2002-03, and 2003-04 seasons. J. Med. Virol. 74:629-640. [DOI] [PubMed] [Google Scholar]
- 11.Rota, P. A., M. L. Hemphill, T. Whistler, H. L. Regnery, and A. P. Kendal. 1992. Antigenic and genetic characterization of the haemagglutinins of recent cocirculating strains of influenza B virus. J. Gen. Virol. 73:2737-2742. [DOI] [PubMed] [Google Scholar]
- 12.Rota, P. A., T. R. Wallis, M. W. Harmon, J. S. Rota, A. P. Kendal, and K. Nerome. 1990. Cocirculation of two distinct evolutionary lineages of influenza type B virus since 1983. Virology 175:59-68. [DOI] [PubMed] [Google Scholar]
- 13.Shaw, M. W., X. Xu, Y. Li, S. Normand, R. T. Ueki, G. Y. Kunimoto, H. Hall, A. Klimov, N. J. Cox, and K. Subbarao. 2002. Reappearance and global spread of variants of influenza B/Victoria/2/87 lineage viruses in the 2000-2001 and 2001-2002 seasons. Virology 303:1-8. [DOI] [PubMed] [Google Scholar]
- 14.Xu, X., S. E. Lindstrom, M. W. Shaw, C. B. Smith, H. E. Hall, B. A. Mungall, K. Subbarao, N. J. Cox, and A. Klimov. 2004. Reassortment and evolution of current human influenza A and B viruses. Virus Res. 103:55-60. [DOI] [PubMed] [Google Scholar]