Abstract
The ability to accurately diagnose malaria infections, particularly in settings where laboratory facilities are not well developed, is of key importance in the control of this disease. Rapid diagnostic tests (RDTs) offer great potential to address this need. Reports of significant variation in the field performance of RDTs based on the detection of Plasmodium falciparum histidine-rich protein 2 (HRP2) (PfHRP2) and of significant sequence polymorphism in PfHRP2 led us to evaluate the binding of four HRP2-specific monoclonal antibodies (MABs) to parasite proteins from geographically distinct P. falciparum isolates, define the epitopes recognized by these MABs, and relate the copy number of the epitopes to MAB reactivity. We observed a significant difference in the reactivity of the same MAB to different isolates and between different MABs tested with single isolates. When the target epitopes of three of the MABs were determined and mapped onto the peptide sequences of the field isolates, significant variability in the frequency of these epitopes was observed. These findings support the role of sequence variation as an explanation for variations in the performance of HRP2-based RDTs and point toward possible approaches to improve their diagnostic sensitivities.
The ability to reliably diagnose malaria infections is fundamental to both the management of individual patients as well as public health efforts to control the disease. Clinical diagnosis is often unreliable, while microscopic diagnosis, though sensitive and specific, is not universally available. Tests to identify parasite nucleic acids, principally by PCR, are widely available in research settings and are becoming increasingly used in diagnostic laboratories in developed countries. However, such tests are not currently suitable for use in most areas where malaria is endemic, as they are not amenable for point-of-care diagnosis and require sophisticated and expensive equipment and reagents, highly trained staff, and reliable power supplies. With the current impetus for the global distribution of increasingly expensive antimalarial drug combinations for multidrug-resistant parasites, the empirical use of antimalarial drugs following clinical diagnosis is a highly undesirable practice. Therefore, deployment of reliable rapid diagnostic tests (RDTs) remains a priority.
Since the development of the first RDT for malaria more than 10 years ago, over 25 products have been commercially marketed. Most are based on immunochromatographic antigen detection tests using monoclonal antibodies (MAB) raised against an abundant circulating protein of Plasmodium falciparum, termed histidine-rich protein 2 (HRP2). While these tests do not detect infection with other malaria species, RDTs targeting P. falciparum HRP2 (PfHRP2) have been reported to display high sensitivity and specificity for the diagnosis of P. falciparum infection. The sensitivities of these tests have been reported in some studies to be at least as good as that achieved by microscopic examination of thick blood films (∼100 parasites/μl) (3, 8, 20). However, in other studies, the sensitivities of these tests have been reported to be well below that required for operational use (6, 7, 9-11, 13, 17, 25, 27, 29). Variable test performance has been observed when panels of blood samples have been tested using different tests targeting PfHRP2 as well as when the same test has been evaluated in different locations (2-4, 6, 9, 13, 14, 18, 25, 27, 29). Although there have been reports of these RDTs failing to detect infections with high-level parasitemia (5, 9, 11, 27, 29), most of the variation has occurred with a relatively low level of parasitemia (100 to 500 parasites/μl) (3, 4, 10, 12, 17, 19, 22, 23, 26, 27), a level that nevertheless often results in symptomatic malaria in nonimmune individuals (7).
Possible device-related factors that may explain the variable performance of RDTs include poor manufacture, deterioration of the device, flawed techniques for carrying out the test, and misinterpretation of the test results. Possible parasite factors include the level of parasitemia, variability in the target epitopes of the parasite antigen, or quantity of parasite antigen produced by the parasite or present in the peripheral blood.
In previous work, we tested the unexplored hypothesis that polymorphisms in the PfHRP2 protein may explain some of the variability in RDT performance. We described significant genetic diversity in the PfHRP2 genes from a collection of 75 P. falciparum lines/isolates originating from 19 countries (1). Extensive diversity was observed in PfHRP2 sequences both within and between countries (1). We also demonstrated that the variation in the number and combination of repeats within PfHRP2 affected the sensitivity of two PfHRP2-based commercial RDTs (1). Therefore, there is a need to evaluate the effect of HRP2 sequence variation on the binding of MABs that are being used or that have the potential to be used in RDTs. In the present study, we sought to define the epitopes recognized by a panel of four MABs raised against PfHRP2 and to relate the number of PfHRP2 epitopes present in specific strains of P. falciparum to the recognition of parasite proteins.
MATERIALS AND METHODS
Parasite isolates.
The following eight P. falciparum parasite lines originating from four different countries, all with unique PfHRP2 gene sequences (1), were used in this study: AN143, TM91-C32B, FCQ41, INDO21, PH4, FCQ64, SJ15, and FCQ27-D10. (Table 1).
TABLE 1.
Isolates of Plasmodium falciparum cultured during the study and their origins
| Strain | Location |
|---|---|
| AN143 | Papua New Guinea |
| TM91-C32B | Thailand |
| FCQ41 | Papua New Guinea |
| INDO21 | Thailand |
| PH4 | Philippines |
| FCQ64 | Papua New Guinea |
| SJ15 | Solomon Islands |
| D10 | Papua New Guinea |
MABs.
Two MABs, 3A4 and 4A5, were kindly provided by David Sullivan, Department of Molecular Microbiology and Immunology, Johns Hopkins School of Public Health, Baltimore, MD, while MABs 2G12-1C12 and 1E1-A9 were kindly provided by Diane Taylor, Department of Biology, Georgetown University, Washington, D.C.
Preparation of parasite protein samples.
P. falciparum isolates were cultured in vitro using standard methods (28), synchronized repeatedly using a standard sorbitol protocol (15), and harvested at ring stages (100% rings in 1,000 erythrocytes). An aliquot of 1 × 107 parasite-infected erythrocytes was collected from the culture, and protein extracts were prepared by subjecting cells to three freeze-thaw cycles, followed by a 30-min incubation on ice in 50 μl phosphate-buffered saline (PBS) containing 1% Triton X-100 and a protease inhibitor cocktail (Proteinase Inhibitor Mini Cocktail tablets; Roche). Samples were centrifuged at 15,000 × g, and the supernatant was collected as a water-soluble protein extract and stored at −20°C in aliquots until needed.
Dot blot analysis.
A total parasite protein extract was prepared in PBS in doubling dilutions; 15 μl of each dilution was transferred onto a nitrocellulose membrane by vacuum aspiration in a dot blot apparatus. Membranes were blocked with 2% casein-Tris-buffered saline (TBS) and incubated with the MABs diluted 1:2,000 in TBS-2% casein. Following washing in TBS-Tween (TBS, 0.5% Tween 20), the alkaline phosphatase-conjugated goat anti-mouse immunoglobulin secondary antibody (Sigma) was added in a dilution of 1:5,000 in TBS-2% casein. Incubations with both primary and secondary antibodies were carried out for 1 h at room temperature. Detection of positive signals was undertaken by visualizing chemiluminescence with the CDP-STAR substrate (Amersham Bioscience) and captured on autoradiography film.
Western blot analysis.
Parasite proteins were separated by 8% sodium dodecyl sulfate-polyacrylamide gel electrophoresis under denaturing and reducing conditions using β-mercaptoethanol and transferred onto a polyvinylidene difluoride membrane using the Bio-Rad Mini-PROTEAN 3 cell system. Membranes were blocked as described above for dot blot experiments and cut into strips of 3- to 4-mm width. Strips were incubated with the MABs at a dilution of 1:4,000 in TBS-2% casein. This was followed by secondary antibody incubation and detection as outlined above for the dot blot analysis.
Epitope mapping.
The amino acid sequences encoded by the HRP2 genes were derived from the respective DNA sequences of the 46 different P. falciparum isolates reported in our previous publication (1). To cover all possible epitopes, a peptide library encompassing a total of 171 peptides, representing combinations of all 46 PfHRP2 sequences, each of 15 residues and overlapping sequentially by three amino acids, was synthesized. (Mimotopes Pty. Ltd., Clayton, Victoria, Australia). The peptides were synthesized with a biotin residue at the N terminus followed by an SGSG spacer. The peptides were coated onto streptavidin (5 μg/ml)-coated 96-well polycarbonate plates (Nunc Maxisorb). Plates were washed with TBS-Tween, blocked with PBS-1% casein, and incubated at room temperature for 1 h with 100 μl of each MAB (diluted 1:1,000 in PBS). Plates were then washed with PBS-0.1% Tween and incubated with a secondary antibody (horseradish peroxidase-conjugated anti-mouse immunoglobulin; Sigma) diluted 1:2,000 in PBS. Plates were developed using freshly prepared ABTS [2,2′-azinobis(3-ethylbenzthiazolinesulfonic acid)] substrate solution, and the optical density (OD) at 405 nm was measured after 35 min. The epitope recognized by each MAB was determined by arranging in hierarchical order the OD values obtained with each peptide such that peptides with the highest OD were determined to contain the optimal or complete epitope. Those peptides with a positive signal but a lower OD were judged to contain only partial or suboptimal epitopes.
Identification of the minimal epitope.
Once peptides that were reactive to each MAB were identified, they were subjected to further mapping by a glycine scan. Briefly, for each reactive peptide, a set of 10-mer peptides was synthesized, each of which had one amino acid replaced by a glycine residue. A total of 47 peptides were synthesized. All peptides carried biotin plus an SGSG spacer at the N terminus. The reactivity of each of these peptides with each MAB was tested as outlined above.
Statistical analysis.
Linear regression analysis was performed to test the relationship between epitope frequency and dot blot reactivity. Dot blot reactivity was assumed to be an ordinal variable based on the minimum dilution returning a positive test. The dilution categories ranged from 1 (1:32 dilution) to 7 (1:2,048 dilution). Potential differences in epitope frequencies in isolates from different geographic regions were tested using the Kruskal-Wallis test. When a significant result was obtained (P < 0.05), post hoc comparisons were conducted to determine which regions differed. Statistical analysis was performed using the SPSS software package (version 13.0; SPSS Inc.).
RESULTS
Dot blot analysis of recognition of parasite antigen by PfHRP2 MABs reveals significant variability.
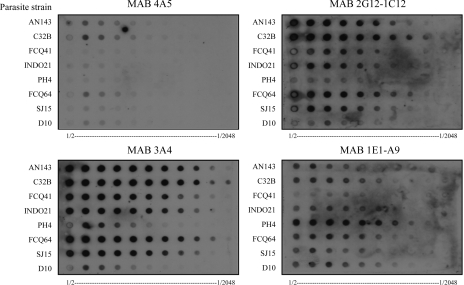
All four MABs, 3A4, 4A5, 2G12-1C12, and 1E1-A9, reacted with protein extracts of the eight parasite cultures bound to the nitrocellulose membrane (Fig. 1). No reactivity was seen in dot blot analysis with uninfected red blood cells (Fig. 2). A variation in the strength of reactivity to individual parasite lines was observed for each MAB tested (Fig. 1). MABs 3A4 and 2G12-1C12 reacted strongly with TM91-C32B; moderately with AN143, FCQ41, and FCQ64; and weakly with PH4, while MAB 1E1-A9 reacted weakly with all lines tested except PH4. MAB 4A5 reacted weakly with all parasite lines tested.
FIG. 1.
Dot blot analysis of protein extracts from eight P. falciparum isolates. Parasite protein extracts were prepared in doubling dilutions of 1/2 to 1/2,048 before application to each membrane as indicated at the bottom of each panel. Membranes were then incubated with the four monoclonal antibodies 4A5, 3A4, 2G12-1C12, and 1E1-A9.
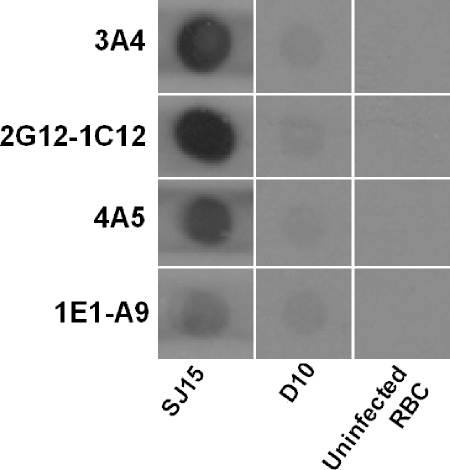
FIG. 2.
Control dot blot analysis of protein extracts from uninfected red blood cells (RBC) and two P. falciparum isolates. Protein extracts were prepared at a dilution of 1/8 before application onto each membrane, as indicated at the bottom of each row. Membranes were then incubated at a dilution of 1/2,000 with the four monoclonal antibodies 4A5, 3A4, 2G12-1C12, and 1E1-A9.
None of the MABs reacted strongly with FCQ27-D10, a strain that lacks the HRP2 gene, in accordance with previous reports (1, 15). However, as some low-level reactivity of each MAB to FCQ27-D10 was evident in the dot blot (Fig. 1 and 2), Western blot analysis was performed to investigate possible cross-reactivity to other P. falciparum proteins.
Western blot analysis of recognition of parasite antigens by PfHRP2 MABs indicates recognition of PfHRP2 and other HRPs under denatured and reduced conditions.
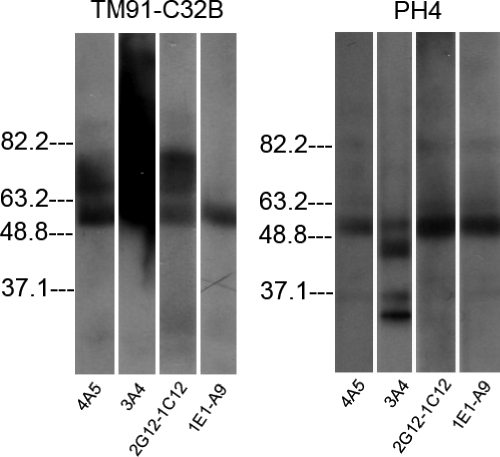
Western blot analysis of protein extracts from eight parasite lines indicated that all four MABs recognize parasite antigens in a denatured and reduced form. As was observed in the dot blot analysis, no reactivity was seen with uninfected red blood cells in the Western blot analysis (data not shown). A representative Western blot is shown in Fig. 3, and a summary of data for the recognition of bands of specific molecular weights is shown in Table 2. The previously reported apparent molecular masses upon polyacrylamide gel electrophoresis of three histidine-rich proteins, knob-associated HRP1 (KAHRP), HRP2, and HRP3, were 80 to 115 kDa, 60 to 115 kDa, and 40 to 55 kDa, respectively (24). As had previously been observed (24), multiple bands were observed by Western blot analysis using HRP-specific MABs. While the pattern of recognition of each of the four MABs was distinct, a band or bands were observed at ∼63 kDa for all four MABs except in D10. Bands were also consistently present at ∼37 kDa, 82 kDa, and 181 kDa (data not shown). We therefore assigned the identity of the bands at ∼63 kDa as HRP2, while the band at ∼37 kDa was identified as HRP3. The identities of the bands at 82 and 181 kDa are uncertain. The former may represent KAHRP or HRP2, while the identity of the band at 181 kDa is unknown. Of note, parasite line PH4 showed a quite distinct reactivity by Western blot compared with other strains, as represented by multiple bands at different sizes with all four antibodies (Fig. 3).
FIG. 3.
Western blot analysis of protein extracts of two P. falciparum isolates, TM91-C32B and PH4, using the four monoclonal antibodies 4A5, 3A4, 2G12-1C12, and 1E1-A9. Molecular masses (in kilodaltons) are shown at the left side of each panel.
TABLE 2.
Reactivities of MABs against parasite proteins determined by Western blot analysisa
| MAB | Molecular mass (kDa) (Ag) | Reactivity with parasite strain:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| AN143 | TM91-C32B | FCQ41 | INDO21 | PH4 | FCQ64 | SJ15 | D10 | ||
| 3A4 | 63 (HRP2) | + | + | + | + | + | + | + | |
| 37 (HRP3) | + | + | + | + | + | ||||
| 181 | + | + | |||||||
| 82 | |||||||||
| 4A5 | 63 (HRP2) | + | + | + | + | + | + | + | |
| 37 (HRP3) | + | + | |||||||
| 181 | + | + | |||||||
| 82 | + | + | |||||||
| 2G12-1C12 | 63 (HRP2) | + | + | + | + | + | + | + | |
| 37 (HRP3) | + | + | + | ||||||
| 181 | + | + | |||||||
| 82 | + | + | + | ||||||
| 1E1-A9 | 63 (HRP2) | + | + | + | + | + | + | + | |
| 37 (HRP3) | + | + | |||||||
| 181 | + | + | |||||||
| 82 | + | ||||||||
+ indicates that reactivity was present.
Epitope mapping.
In initial epitope mapping, three of the four MABs tested positive, each recognizing different epitopes. MAB 3A4 recognized DAHHAHHA as its major epitope, with possible substitutions of Y or V for the first and last amino acids. The optimal epitope for MAB 1E1-A9 was similar, recognizing YAHHAHHA as its major epitope and allowing possible substitutions of D or V for its first or last amino acid. MAB 2G12-1C12 recognized the major epitope DAHHAADAHH, with possible substitutions of H and V at the first and fifth amino acids (see the supplemental material). Attempts to define the epitope recognized by MAB 4A5 using the peptide library were unsuccessful.
To further define the composition and the minimum size of the epitopes recognized by the MABs, a glycine scan of the predicted epitopes was performed. The results indicated that the minimal epitope recognized by MAB 3A4 was AHHAHHA. Substitution of the first or last amino acid significantly decreased the OD reading (see the supplemental material). The minimal epitope recognized by MAB 1E1-A9 was AHHAHHV. Substitution of the first or last amino acid also significantly decreased the OD reading (see the supplemental material). The glycine scan of MAB 2G12-1C12 demonstrated no differences from background readings. It is suspected that the epitope may be longer than 10 amino acids.
Copy number of epitopes and correlation with dot blot reactivity in the eight test isolates.
The numbers of major epitopes of 3A4, 1E1, and 2G12 in the HRP2 and HRP3 proteins of the eight isolates tested by dot blot are listed in Table 3. For each MAB, the frequency of the epitopes in eight isolates was compared with the dot blot reactivity. For MAB 3A4, a moderate level of correlation between the copy number of epitopes in the sequence and the minimum dilution of the dot blot results was observed. A significant regression model (R2 = 0.522; P = 0.043) using the epitope frequency to predict dot blot reactivity was obtained: dilution category = 2.06 + 0.19 × number of AHHAHHA repeats in sequence. Hence, the number of times the epitope AHHAHHA appeared in the isolate sequence for the HRP2 and HRP3 proteins predicted the reactivity with MAB 3A4, with the maximum dilution increasing with increasing epitope frequency. For MAB1E1-A9, however, no significant association between the number of AHHAHHV epitopes in the sequence and the dot blot results using either the actual dilution or the dilution category was observed.
TABLE 3.
Geographic distribution of repeats recognized by HRP-specific MABs
| Country | No. of samples | Mean no. of AHHAHHA repeats (min-max) | Mean no. of AHHAHHV repeats (min-max) |
|---|---|---|---|
| Africa | 12 | 14.0 (11-18) | 2.9 (1-6) |
| Indonesia/Malaysia | 3 | 13.0 (12-15) | 1.7 (1-3) |
| Philippines | 20 | 14.6 (10-17) | 2.1 (1-4) |
| Papua New Guinea | 9 | 15.0 (9-18) | 2.0 (1-4) |
| Cambodia/Vietnam | 5 | 16.4 (15-17) | 3.4 (2-6) |
| Solomon Islands | 7 | 16.0 (14-18) | 3.3 (2-6) |
| South America | 8 | 13.9 (13-15) | 3.6 (2-5) |
| Thailand | 7 | 16.0 (12-21) | 3.6 (1-5) |
Comparison of epitope frequency between countries.
When the frequencies of MAB 3A4 epitopes in the HRP2 and HRP3 sequences of 71 parasite isolates (1) from eight countries were classified according to geographic region, two distinct groups were identified. Group 1 (higher frequency of AHHAHHA) included Cambodia, Vietnam, Solomon Islands, Thailand, and Papua New Guinea. The second group (lower frequency of AHHAHHA) included Africa, South America, Indonesia, and Malaysia. Isolates from the Philippines had an intermediate value that differed significantly from those of Cambodia/Vietnam (highest frequency) and Indonesia/Malaysia (lowest frequency). However, the copy numbers in this group did not significantly differ from those of the isolates from the rest of the countries.
For MAB 1E1, multiple comparisons again indicated that the parasites segregated into two groups according to geographic origin. Group 1 (higher frequency of AHHAHHV) included South America, Thailand, Solomon Islands, Cambodia, and Vietnam. The second group (lower frequency of AHHAHHV) included the Philippines, Papua New Guinea, Indonesia, and Malaysia. For this epitope, Africa had an intermediate value that differed significantly from those from South America (highest frequency) and the Philippines, but it was not significantly different from those from the remaining countries.
DISCUSSION
We recently reported extensive diversity in PfHRP2 proteins among parasite isolates from around the world and demonstrated an association between this diversity and the sensitivity of two RDTs (1). This led us to explore possible approaches to improving the current RDTs by using MABs that are not affected by sequence diversity or by including several MABs that detect different B-cell epitopes of HRP. In this paper, we investigated the affinity of binding of four primary antibodies that could be used in HRP2-based RDTs.
Initial dot blot experiments were aimed at investigating the sensitivity of the four tested MABs against the eight different strains of P. falciparum HRP2 in a native and nondenatured and nonreduced form. The preparations used were thus parasitized erythrocyte protein preparations, preparations that mimic the physicochemical environment present in rapid diagnostic tests on the market. Therefore, the results could be an indication of the potential sensitivity of these antibodies when utilized in RDTs.
The results showed a variation between the four antibodies against each strain. The sensitivity of detection of some strains was significantly higher than those of other strains. For MABs 3A4 and 2G12, both antibodies were able to detect TM91-C32B at a very high intensity at a dilution of more than 1:1,024, while strain PH4 was detectable only to a dilution of 1:32. Opposite results were observed with antibody 1E1-A9. The antibody was able to detect strain PH4 with high sensitivity, while other strains were detected less strongly. This indicates that the low reactivity of MABs 3A4 and 2G12 with PH4 was not due to the degradation of proteins in some isolates and that the genetic variation of P. falciparum strains likely has a major effect on the binding affinity of the antibodies. The dot blot results demonstrate that if an RDT for malaria used one of these three antibodies, the RDT would likely be less readily able to detect infections with some strains at moderate or lower levels of parasitemia. Theoretically, a combination of 3A4 and 1E1-A9 or 2G12-1C12 and 1E1-A9 would facilitate the detection of all eight isolates at similar parasite levels.
The negative control clone of D10 displayed weak reactivity with all four antibodies in dot blot analyses, suggesting possible cross-reactivity with other parasite proteins. This cross-reactivity was confirmed by Western blot analysis, as all four MABs recognized both HRP2, a single band or multiple bands between the 63-kDa and 82-kDa markers, and additional bands around 37.1, 82, and 181 kDa. It is known that P. falciparum-infected erythrocytes synthesize at least three HRPs: KAHRP, which has an apparent molecular mass of 80 to 115 kDa by sodium dodecyl sulfate-polyacrylamide gel electrophoresis; HRP2, which has a molecular mass ranging from 60 to 105 kDa; and HRP3 (SHARP), with a molecular mass of 40 to 55 kDa (16, 24). Monoclonal antibodies generated against one HRP have been shown to cross-react with other HRPs (21, 24). Therefore, it is likely that HRP3 and HRP2 are the bands around 37 and 63 kDa, respectively, while the bands around 82 kDa are likely to represent KAHRP. The faint reactivity of the antibodies to D10 in dot blot analyses may be interpreted as HRP3 reactivity.
To understand the basis of the differences in reactivities to the eight isolates, we mapped linear epitopes using synthetic peptides. We demonstrated that MABs 3A4 and 1E1 recognized the closely related epitopes AHHAHHA and AHHAHHV, while 2G12-1C12 recognized distinct and longer epitopes. Attempts to the map the epitope of MAB 4A5 using peptides resulted in no significant reactivity above background readings. MAB 4A5 recognized parasite proteins under nonreduced (dot blot) and reduced (Western blot) conditions but failed to recognize any 15-mer peptides used in the epitope scan. Together, these observations suggest that the epitope for this MAB is linear or partially linear but that it requires more than 15 amino acids for significant binding.
It would be expected that an antigen with more epitopes would result in increased detection sensitivity compared to an antigen with fewer epitopes. Therefore, in principle, the reactivity of each of the three MABs to a given isolate should correlate with the number of epitopes present in HRP2. We observed a weak correlation between the reactivity of 3A4 on dot blot with the number of epitopes and no correlation for 1E1. A possible explanation for our observation of only a weak association is that the epitopes that we identified are the linear components of the epitope and that the optimal epitope may be conformational.
Interestingly, when we analyzed the number of 3A4 and 1E1-A9 epitopes in 71 isolates originating from 19 countries (1), we observed that the parasites segregated into two groups with different copy numbers of epitopes for each MAB. Of note, the geographic origin of the groups differed somewhat for the two MABs. Most notable is the discordance in South America, which had the highest frequency of AHHAHHV but had a lower number of AHHAHHA repeats. This result indicates that the combination of the two MABs may offer the potential to overcome the difference in sensitivities observed with HRP-based RDTs recognizing different epitopes.
The identities and specificities of the HRP2-specific MABs present in commercially available rapid diagnostic tests for malaria are not known to us. It is clear from this work that MABs specific for linear epitopes present in P. falciparum histidine-rich proteins recognize native parasite proteins. Furthermore, with a knowledge of the abundance of specific repeats and their geographic spread, it should be possible to optimize their sensitivities so that they work with acceptable sensitivity against a broad range of parasites from geographically distant regions. As noted above, if a universally abundant epitope cannot be readily identified, a combination of MABs may overcome the constraints imposed by the diversity of HRP2 repeats from different regions.
In summary, our findings provide further insight into the observed variability in sensitivities reported for the various HRP2-specific RDTs. The genetic diversity of PfHRP2 and its relationship with the ability of the MABs to bind present a serious challenge to the future testing and development of RDTs that target HRP2 for the diagnosis of P. falciparum infection.
Supplementary Material
Acknowledgments
We thank Diane Taylor, Department of Biology, Georgetown University, Washington, D.C., for providing MABs 2G12-1C12 and 1E1-A9 and David Sullivan, Department of Molecular Microbiology and Immunology, Johns Hopkins School of Public Health, Baltimore, MD, for MABs 3A4 and 4A5. We also thank the Australian Red Cross Blood Service (Brisbane) for providing human erythrocytes and serum used for in vitro cultivation of P. falciparum.
The project is partially funded by AusAID through the World Health Organization Roll Back Malaria Department and Regional Office for the Western Pacific.
The authors do not have a commercial or other association that might pose a conflict of interest. The opinions expressed herein are those of the authors and do not necessarily reflect those of the Defence Health Service or any extant policy of the Department of Defence, Australia, or the World Health Organization.
Footnotes
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Baker, J., J. McCarthy, M. Gatton, D. E. Kyle, V. Belizario, J. Luchavez, D. Bell, and Q. Cheng. 2005. Genetic diversity of Plasmodium falciparum histidine-rich protein 2 (PfHRP2) and its effect on the performance of PfHRP2-based rapid diagnostic tests. J. Infect. Dis. 192:870-877. [DOI] [PubMed] [Google Scholar]
- 2.Banchongaksorn, T., S. Prajakwong, W. Rooney, and P. Vickers. 1997. Operational trial of ParaSight-F (dipstick) in the diagnosis of falciparum malaria at the primary health care level. Southeast Asian J. Trop. Med. Public Health 28:243-246. [PubMed] [Google Scholar]
- 3.Beadle, C., G. W. Long, W. R. Weiss, P. D. McElroy, S. M. Maret, A. J. Oloo, and S. L. Hoffman. 1994. Diagnosis of malaria by detection of Plasmodium falciparum HRP-2 antigen with a rapid dipstick antigen-capture assay. Lancet 343:564-568. [DOI] [PubMed] [Google Scholar]
- 4.Bechem, N. N., R. F. Leke, F. Tietche, and D. W. Taylor. 1999. Evaluation of a rapid test for histidine rich protein 2 for diagnosis of Plasmodium falciparum infection in Cameroonian children. Trans. R. Soc. Trop. Med. Hyg. 93:46. [DOI] [PubMed] [Google Scholar]
- 5.Birku, Y., D. Welday, D. Ayele, and A. Shepherd. 1999. Rapid diagnosis of severe malaria based on the detection of Pf-Hrp-2 antigen. Ethiop. Med. J. 37:173-179. [PubMed] [Google Scholar]
- 6.Forney, J. R., A. J. Magill, C. Wongsrichanalai, J. Sirichaisinthop, C. T. Bautista, D. G. Heppner, R. S. Miller, C. F. Ockenhouse, A. Gubanov, R. Shafer, C. C. DeWitt, H. A. Quino-Ascurra, K. E. Kester, K. C. Kain, D. S. Walsh, W. R. Ballou, and R. A. Gasser, Jr. 2001. Malaria rapid diagnostic devices: performance characteristics of the ParaSight F device determined in a multisite field study. J. Clin. Microbiol. 39:2884-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forney, J. R., C. Wongsrichanalai, A. J. Magill, L. G. Craig, J. Sirichaisinthop, C. T. Bautista, R. S. Miller, C. F. Ockenhouse, K. E. Kester, N. E. Aronson, E. M. Andersen, H. A. Quino-Ascurra, C. Vidal, K. A. Moran, C. K. Murray, C. C. DeWitt, D. G. Heppner, K. C. Kain, W. R. Ballou, and R. A. Gasser, Jr. 2003. Devices for rapid diagnosis of malaria: evaluation of prototype assays that detect Plasmodium falciparum histidine-rich protein 2 and a Plasmodium vivax-specific antigen. J. Clin. Microbiol. 41:2358-2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia, M., S. Kirimoama, D. Marlborough, J. Leafasia, and K. H. Rieckmann. 1996. Immunochromatographic test for malaria diagnosis. Lancet 347:1549. [DOI] [PubMed] [Google Scholar]
- 9.Gaye, O., M. Diouf, E. F. Dansokho, G. McLaughlin, and S. Diallo. 1998. Diagnosis of Plasmodium falciparum malaria using ParaSight F, ICT malaria PF and malaria IgG CELISA assays. Parasite 5:189-192. [DOI] [PubMed] [Google Scholar]
- 10.Huong, N. M., T. M. Davis, S. Hewitt, N. V. Huong, T. T. Uyen, D. H. Nhan, and L. D. Cong. 2002. Comparison of three antigen detection methods for diagnosis and therapeutic monitoring of malaria: a field study from southern Vietnam. Trop. Med. Int. Health 7:304-308. [DOI] [PubMed] [Google Scholar]
- 11.Iqbal, J., P. R. Hira, A. Sher, and A. A. Al Enezi. 2001. Diagnosis of imported malaria by Plasmodium lactate dehydrogenase (pLDH) and histidine-rich protein 2 (PfHRP-2)-based immunocapture assays. Am. J. Trop. Med. Hyg. 64:20-23. [DOI] [PubMed] [Google Scholar]
- 12.Iqbal, J., N. Khalid, and P. R. Hira. 2002. Comparison of two commercial assays with expert microscopy for confirmation of symptomatically diagnosed malaria. J. Clin. Microbiol. 40:4675-4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jelinek, T., M. P. Grobusch, S. Schwenke, S. Steidl, F. Von-Sonnenburg, H. D. Nothdurft, E. Klein, and T. Loscher. 1999. Sensitivity and specificity of dipstick tests for rapid diagnosis of malaria in nonimmune travelers. J. Clin. Microbiol. 37:721-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jensen, M. D., M. Conley, and L. D. Helstowski. 1983. Culture of Plasmodium falciparum: the role of pH, glucose, and lactate. J. Parasitol. 69:1060-1067. [PubMed] [Google Scholar]
- 15.Lambros, C., and J. P. Vanderberg. 1979. Synchronization of Plasmodium falciparum erythrocytic stages in culture. J. Parasitol. 65:418-420. [PubMed] [Google Scholar]
- 16.Lopez, R., M. Urquiza, H. Curtidor, C. J. Eduardo, H. Mora, A. Puentes, and M. E. Patarroyo. 2000. Plasmodium falciparum: red blood cell binding studies of peptides derived from histidine-rich KAHRP-I, HRP-II and HRP-III proteins. Acta Trop. 75:349-359. [DOI] [PubMed] [Google Scholar]
- 17.Mason, D. P., F. Kawamoto, K. Lin, A. Laoboonchai, and C. Wongsrichanalai. 2002. A comparison of two rapid field immunochromatographic tests to expert microscopy in the diagnosis of malaria. Acta Trop. 82:51-59. [DOI] [PubMed] [Google Scholar]
- 18.Mharakurwa, S., B. Manyame, and C. J. Shiff. 1997. Trial of the ParaSight-F test for malaria diagnosis in the primary health care system, Zimbabwe. Trop. Med. Int. Health 2:544-550. [DOI] [PubMed] [Google Scholar]
- 19.Mills, C. D., D. C. Burgess, H. J. Taylor, and K. C. Kain. 1999. Evaluation of a rapid and inexpensive dipstick assay for the diagnosis of Plasmodium falciparum malaria. Bull. W. H. O. 77:553-559. [PMC free article] [PubMed] [Google Scholar]
- 20.Palmer, C. J., J. F. Lindo, W. I. Klaskala, J. A. Quesada, R. Kaminsky, M. K. Baum, and A. L. Ager. 1998. Evaluation of the OptiMAL test for rapid diagnosis of Plasmodium vivax and Plasmodium falciparum malaria. J. Clin. Microbiol. 36:203-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Panton, L. J., P. McPhie, W. L. Maloy, T. E. Wellems, D. W. Taylor, and R. J. Howard. 1989. Purification and partial characterization of an unusual protein of Plasmodium falciparum: histidine-rich protein II. Mol. Biochem. Parasitol. 35:149-160. [DOI] [PubMed] [Google Scholar]
- 22.Playford, E. G., and J. Walker. 2002. Evaluation of the ICT malaria P.f/P.v and the OptiMal rapid diagnostic tests for malaria in febrile returned travellers. J. Clin. Microbiol. 40:4166-4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Proux, S., L. Hkirijareon, C. Ngamngonkiri, S. McConnell, and F. Nosten. 2001. Paracheck-Pf: a new, inexpensive and reliable rapid test for P. falciparum malaria. Trop. Med. Int. Health 6:99-101. [DOI] [PubMed] [Google Scholar]
- 24.Rock, E. P., K. Marsh, A. J. Saul, T. E. Wellems, D. W. Taylor, W. L. Maloy, and R. J. Howard. 1987. Comparative analysis of the Plasmodium falciparum histidine-rich proteins HRP-I, HRP-II and HRP-III in malaria parasites of diverse origin. Parasitology 95:209-227. [DOI] [PubMed] [Google Scholar]
- 25.Rubio, J. M., I. Buhigas, M. Subirats, M. Baquero, S. Puente, and A. Benito. 2001. Limited level of accuracy provided by available rapid diagnosis tests for malaria enhances the need for PCR-based reference laboratories. J. Clin. Microbiol. 39:2736-2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh, N., and N. Valecha. 2000. Evaluation of a rapid diagnostic test, ‘Determine malaria pf, ’ in epidemic-prone, forest villages of central India (Madhya Pradesh). Ann. Trop. Med. Parasitol. 94:421-427. [DOI] [PubMed] [Google Scholar]
- 27.Stow, N. W., J. K. Torrens, and J. Walker. 1999. An assessment of the accuracy of clinical diagnosis, local microscopy and a rapid immunochromatographic card test in comparison with expert microscopy in the diagnosis of malaria in rural Kenya. Trans. R. Soc. Trop. Med. Hyg. 93:519-520. [DOI] [PubMed] [Google Scholar]
- 28.Trager, W., and J. B. Jensen. 1976. Human malaria parasites in continuous culture. Science 193:673-675. [DOI] [PubMed] [Google Scholar]
- 29.Wongsrichanalai, C., N. Chuanak, S. Tulyayon, N. Thanoosingha, A. Laoboonchai, K. Thimasarn, T. G. Brewer, and D. G. Heppner. 1999. Comparison of a rapid field immunochromatographic test to expert microscopy for the detection of Plasmodium falciparum asexual parasitemia in Thailand. Acta Trop. 73:263-273. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.