Abstract
A collection of 207 historically relevant Burkholderia pseudomallei isolates was analyzed by multilocus sequence typing (MLST). The strain collection contains environmental isolates obtained from a geographical distribution survey of B. pseudomallei isolates in Thailand (1964 to 1967), as well as stock cultures and colony variants from the U.S. Army Medical Research Unit (Malaysia), the Walter Reed Army Institute for Research, and the Pasteur Institute (Vietnam). The 207 isolates of the collection were resolved into 80 sequence types (STs); 56 of these were novel. eBURST diagrams predict that the historical-collection STs segregate into three complexes when analyzed separately. When added to the 760 isolates and 365 STs of the B. pseudomallei MLST database, the historical-collection STs cluster significantly within the main complex of the eBURST diagram in an ancestral pattern and alter the B. pseudomallei “population snapshot.” Differences in colony morphology among reference isolates were found not to affect the STs assigned, which were consistent with the original isolates. Australian ST84 is likely characteristic of B. pseudomallei isolates of Southeast Asia rather than Australia, since multiple environmental isolates from Thailand and Malaysia share this ST with the single Australian clinical isolate in the MLST database. Phylogenetic evidence is also provided suggesting that Australian isolates may not be distinct from those of Thailand, since ST60 is common to environmental isolates from both countries. MLST and eBURST are useful tools for the study of population biology and epidemiology, since they provide methods to elucidate new genetic relationships among bacterial isolates.
Burkholderia pseudomallei is a gram-negative organism endemic to Southeast Asia and Northern tropical Australia and is the causative agent of the disease melioidosis. The organism is an environmental saprophyte capable of long-term survival in soil or water (8). B. pseudomallei infection may be acquired due to inhalation, aspiration, or through direct contact of wounded or abraded skin with contaminated material (7). In areas to which it is endemic, individuals having frequent exposure to contaminated soil or stagnant waters are at the greatest risk of developing melioidosis; however, encountering B. pseudomallei in the environment can lead to one of three outcomes: no effect, asymptomatic seroconversion, or clinically apparent infection (2, 7, 30). The spectrum of disease ranges from asymptomatic infection to localized skin ulcers or abscesses, acute pulmonary infection, acute septicemic infection, or fulminant disease with abscesses throughout the body (3, 34). Many of the infections occur in individuals with preexisting compromising health conditions (3). Recrudescent melioidosis has been reported to occur as long as 62 years following initial exposure (22). Virulence factors of B. pseudomallei contributing to pathogenesis include capsular polysaccharide, type III secretion, and protease production (24, 25, 26, 32). B. pseudomallei is also able to survive in eukaryotic cell lines and professional phagocytic cells (18). A simple diagnostic technique to differentiate B. pseudomallei from the avirulent organism Burkholderia thailandensis is a lack of arabinose assimilation by B. pseudomallei (1, 20).
The epidemiology of melioidosis is complicated due to the environmental persistence of the organism and is subject to distinct differences in the organism's distribution in soil, disease presentation, and incidence rates among different areas of endemicity. Two important regions for comparison and contrast are Thailand and Australia. B. pseudomallei is commonly isolated from soil in animal paddocks or from water sources in Australia, which is in contrast to Southeast Asia, where in general B. pseudomallei is commonly isolated from cleared, cultivated, and irrigated agricultural sites (7). In both countries, melioidosis is primarily a rainy-season disease and is a well-recognized cause of community-acquired pneumonia and septicemia (2, 6, 30, 34). The incidence rates of melioidosis vary in the two regions. The annual incidence in the Northern Territories of Australia is 19.6 cases per 100,000 people, compared with 4.4 cases per 100,000 people in Thailand (5, 30). There are also unique disease presentations reported for each region. In the Northern Territories of Australia, genitourinary disease, prostatic disease, and encephalomyelitis are reported, whereas acute suppurative parotitis is more common in the pediatric melioidosis patients of Thailand (3, 4, 34). It is unknown why there are regional differences in melioidosis epidemiology. It was speculated that Australian isolates of B. pseudomallei are distinct from those of Thailand or Southeast Asia in general (4). The advent of multilocus sequence typing (MLST) provides a novel and beneficial scheme for the study of melioidosis epidemiology worldwide.
Maiden et al. developed MLST in 1998 as a relatively new typing scheme resulting in portable data easily shared and compared between laboratories worldwide via the Internet (19). The MLST scheme for B. pseudomallei was developed by Godoy et al. in 2003 in a similar manner using a strain collection of 147 isolates of B. pseudomallei, Burkholderia mallei, and B. thailandensis from wide geographical and temporal ranges (14). The typing method is based on sequence variation within seven housekeeping-gene fragments. Allele numbers are assigned to each of the seven housekeeping loci based on sequence differences and are then arranged into a string of seven integers to give the allele profile of an isolate. The B. pseudomallei MLST allele profile corresponds to the gene order ace-gltB-gmhD-lepA-lipA-narK-ndh (14). The sequence type (ST) of an isolate is defined specifically by the allele profile. The relationships among the STs may then be examined using various methods, such as eBURST (based upon related sequence types) (10, 15, 28). At the time of writing, the B. pseudomallei MLST database contained 760 isolates and 365 STs (http://bpseudomallei.mlst.net/). Approximately 63% of all B. pseudomallei isolates in the database are of clinical origin, and 18.6% are of environmental origin (including soil and water isolates). A larger proportion of environmental B. pseudomallei isolates in the MLST database would contribute to a greater understanding of melioidosis epidemiology.
In this study, we examined a significant historical B. pseudomallei strain collection of Southeast Asian isolates from predominantly environmental sources using MLST analysis. We then compared the MLST data with those of the B. pseudomallei MLST database using the eBURST method (10, 28) in order to determine the implications of the addition of the historical-strain data. The current study demonstrates the ancestral nature of the historical strains examined compared with the existing B. pseudomallei population data and highlights a potential epidemiological connection between environmental B. pseudomallei isolates of Thai and Australian origin. The addition of the historical-strain data to the B. pseudomallei MLST database also promotes a balance between the clinical and environmental isolates reported to date.
MATERIALS AND METHODS
Bacterial strains.
A total of 207 B. pseudomallei isolates were analyzed by MLST and are listed in Table 1. The isolates examined belong to a historical collection of strains collected in a geographic distribution survey of B. pseudomallei (then Pseudomonas pseudomallei) in Thailand by Finkelstein et al. during the period of 1964 to 1967 (11, 12). The historical collection is comprised of environmental B. pseudomallei isolates recovered from soil and water samples across Thailand by the hamster isolation technique (9, 11, 12). The collection also includes reference and stock cultures as well as colony variants from sources such as the U.S. Army Medical Research Unit (USAMRU) in Malaysia, the Walter Reed Army Institute for Research (WRAIR), and the Pasteur Institute in Vietnam (9, 12, 29). B. pseudomallei isolates obtained from soldiers serving in Vietnam were also included (12). The isolates were confirmed as B. pseudomallei based upon colony morphology on selective media, serology, and additional biochemical testing (12). Confirmed B. pseudomallei isolates were then lyophilized to preserve their characteristics. Upon receipt of the historical strain collection, each isolate was tested for growth on arabinose to exclude potential B. thailandensis species (1, 20). The strains were maintained as suspensions in 10% skim milk or 20% glycerol at −70°C.
TABLE 1.
Southeast Asian isolates of the historical B. pseudomallei strain collection analyzed by multilocus sequence typing
| Strain | Source | Country | Yr | ST | Allele profile
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ace | gltB | gmhD | lepA | lipA | narK | ndh | |||||
| K96243a | Human | Thailand | 1996 | 10 | 1 | 1 | 13 | 1 | 1 | 1 | 1 |
| Antigen 13 Smooth | 1965 | 46 | 3 | 1 | 2 | 1 | 1 | 3 | 3 | ||
| Antigen 25 Donutb | Malaysia | 1965 | 46 | 3 | 1 | 2 | 1 | 1 | 3 | 3 | |
| Antigen 25 HP Trans-mucb | Malaysia | 1965 | 46 | 3 | 1 | 2 | 1 | 1 | 3 | 3 | |
| Antigen 25 Medusab | Malaysia | 1965 | 46 | 3 | 1 | 2 | 1 | 1 | 3 | 3 | |
| Antigen 25 Smoothb | Malaysia | 1965 | 46 | 3 | 1 | 2 | 1 | 1 | 3 | 3 | |
| Antigen 1188HP-Rc | Human | Malaysia | 1965 | 99 | 1 | 1 | 4 | 1 | 1 | 4 | 1 |
| Antigen 1691d | Human | Malaysia | 1965 | 46 | 3 | 1 | 2 | 1 | 1 | 3 | 3 |
| Antigen 6066 | 35 | 1 | 6 | 14 | 2 | 8 | 8 | 4 | |||
| Chumphon 76 W-2 | Water | Thailand | 1965 | 377 | 3 | 30 | 11 | 3 | 1 | 4 | 3 |
| Hansene | Human | Vietnam | 1966 | 397 | 3 | 12 | 6 | 1 | 1 | 4 | 3 |
| J77e | Human | Vietnam | 1965 | 46 | 3 | 1 | 2 | 1 | 1 | 3 | 3 |
| Lavicte | Human | Vietnam | 1965 | 211 | 3 | 1 | 3 | 1 | 1 | 4 | 1 |
| Loei KK-S2 | Soil | Thailand | 1965 | 10 | 1 | 1 | 13 | 1 | 1 | 1 | 1 |
| Loei KK-S2(2) | Soil | Thailand | 1965 | 10 | 1 | 1 | 13 | 1 | 1 | 1 | 1 |
| Loei KK-W5(2) | Water | Thailand | 1965 | 365 | 3 | 2 | 4 | 1 | 1 | 3 | 1 |
| Nakhon Phanom 32-3 | Environment | Thailand | 1966 | 375 | 3 | 1 | 4 | 3 | 1 | 4 | 3 |
| Phangna 64 W | Water | Thailand | 1965 | 10 | 1 | 1 | 13 | 1 | 1 | 1 | 1 |
| Pasteur Institute 6068 | Vietnam | 1964 | 169 | 1 | 1 | 2 | 3 | 8 | 4 | 3 | |
| Pasteur Institute 6606 | Vietnam | 1964 | 35 | 1 | 6 | 14 | 2 | 8 | 8 | 4 | |
| Pasteur Institute 52237 | Vietnam | 1964 | 411 | 1 | 4 | 4 | 3 | 1 | 3 | 1 | |
| Pasteur Institute 63503 | Vietnam | 370 | 4 | 2 | 3 | 1 | 1 | 22 | 1 | ||
| Phattalung 49 W-1 | Water | Thailand | 1965 | 369 | 3 | 1 | 2 | 1 | 5 | 4 | 3 |
| Phattalung 49 W-2 | Water | Thailand | 1965 | 289 | 3 | 4 | 11 | 4 | 5 | 4 | 6 |
| Phattalung 51 W | Water | Thailand | 1965 | 369 | 3 | 1 | 2 | 1 | 5 | 4 | 3 |
| Phattalung 52 W-2 | Water | Thailand | 1965 | 164 | 3 | 1 | 2 | 3 | 5 | 4 | 3 |
| Phuket 3 W-1 | Water | Thailand | 1965 | 54 | 3 | 1 | 3 | 3 | 1 | 2 | 1 |
| Phuket 6 S-1 | Soil | Thailand | 1965 | 46 | 3 | 1 | 2 | 1 | 1 | 3 | 3 |
| Ramose | Human | Vietnam | 1966 | 211 | 3 | 1 | 3 | 1 | 1 | 4 | 1 |
| Ranong 8 | Environment | Thailand | 1965 | 409 | 3 | 1 | 36 | 1 | 1 | 4 | 1 |
| Ranong 70 W | Water | Thailand | 1965 | 38 | 1 | 12 | 2 | 1 | 1 | 1 | 1 |
| Ranong 73 W-1 | Water | Thailand | 1965 | 399 | 3 | 1 | 4 | 1 | 1 | 22 | 1 |
| Ranong 73 W-2 | Water | Thailand | 1965 | 227 | 1 | 1 | 2 | 1 | 5 | 4 | 1 |
| Smith 373 | 1966 | 99 | 1 | 1 | 4 | 1 | 1 | 4 | 1 | ||
| Smith 384 | 1966 | 211 | 3 | 1 | 3 | 1 | 1 | 4 | 1 | ||
| Smith 541 | 1967 | 410 | 1 | 1 | 11 | 1 | 1 | 4 | 1 | ||
| Smith 660 | 1966 | 211 | 3 | 1 | 3 | 1 | 1 | 4 | 1 | ||
| Smith 001963 | 1966 | 403 | 1 | 4 | 3 | 1 | 6 | 1 | 1 | ||
| Smith 002025 | 1966 | 84 | 3 | 1 | 11 | 4 | 5 | 4 | 6 | ||
| Smith 002026 | 1966 | 418 | 1 | 4 | 37 | 3 | 1 | 1 | 1 | ||
| Smith 002159 | 1966 | 419 | 3 | 4 | 37 | 3 | 1 | 1 | 1 | ||
| Smith 002559 | 1966 | 403 | 1 | 4 | 3 | 1 | 6 | 1 | 1 | ||
| Smith 22179 | 1967 | 387 | 1 | 12 | 6 | 1 | 1 | 1 | 1 | ||
| Smith 22294 | 1966 | 387 | 1 | 12 | 6 | 1 | 1 | 1 | 1 | ||
| Songkhla 11 W | Water | Thailand | 1965 | 168 | 3 | 1 | 2 | 1 | 5 | 4 | 1 |
| Songkhla 21 W-1 | Water | Thailand | 1965 | 289 | 3 | 4 | 11 | 4 | 5 | 4 | 6 |
| Songkhla 21 W-2 | Water | Thailand | 1965 | 228 | 1 | 2 | 3 | 1 | 1 | 4 | 1 |
| Songkhla 25 W-1 | Water | Thailand | 1965 | 382 | 1 | 1 | 3 | 4 | 5 | 1 | 1 |
| Songkhla 25 W-2 | Water | Thailand | 1965 | 382 | 1 | 1 | 3 | 4 | 5 | 1 | 1 |
| Songkhla 27 W-1 | Water | Thailand | 1965 | 369 | 3 | 1 | 2 | 1 | 5 | 4 | 3 |
| Songkhla 27 W-2 | Water | Thailand | 1965 | 369 | 3 | 1 | 2 | 1 | 5 | 4 | 3 |
| Songkhla 34 W-1 | Water | Thailand | 1965 | 414 | 3 | 1 | 2 | 1 | 8 | 4 | 3 |
| Songkhla 34 W-2 | Water | Thailand | 1965 | 84 | 3 | 1 | 11 | 4 | 5 | 4 | 6 |
| STW 1 | Water | Thailand | 1965 | 369 | 3 | 1 | 2 | 1 | 5 | 4 | 3 |
| STW 3 | Water | Thailand | 1965 | 366 | 3 | 1 | 2 | 3 | 8 | 4 | 3 |
| STW 4-2 | Water | Thailand | 1965 | 366 | 3 | 1 | 2 | 3 | 8 | 4 | 3 |
| STW 5-1 | Water | Thailand | 1965 | 289 | 3 | 4 | 11 | 4 | 5 | 4 | 6 |
| STW 7 | Water | Thailand | 1965 | 376 | 1 | 4 | 2 | 3 | 8 | 4 | 3 |
| STW 7-2 | Water | Thailand | 1965 | 376 | 1 | 4 | 2 | 3 | 8 | 4 | 3 |
| STW 10 | Water | Thailand | 1965 | 392 | 1 | 2 | 6 | 1 | 1 | 4 | 1 |
| STW 11-3 | Water | Thailand | 1965 | 288 | 3 | 2 | 3 | 2 | 1 | 3 | 1 |
| STW 22-1 | Water | Thailand | 1965 | 46 | 3 | 1 | 2 | 1 | 1 | 3 | 3 |
| STW 25 | Water | Thailand | 1965 | 46 | 3 | 1 | 2 | 1 | 1 | 3 | 3 |
| STW 26 | Water | Thailand | 1965 | 385 | 1 | 4 | 11 | 4 | 5 | 4 | 6 |
| STW 27-2 | Water | Thailand | 1965 | 70 | 3 | 4 | 11 | 3 | 5 | 4 | 6 |
| STW 28-2 | Water | Thailand | 1965 | 415 | 1 | 1 | 11 | 4 | 5 | 4 | 6 |
| STW 28-5 | Water | Thailand | 1965 | 376 | 1 | 4 | 2 | 3 | 8 | 4 | 3 |
| STW 32-1 | Water | Thailand | 1965 | 389 | 3 | 1 | 11 | 1 | 1 | 4 | 1 |
| STW 32-3 | Water | Thailand | 1965 | 407 | 3 | 1 | 3 | 3 | 1 | 2 | 18 |
| STW 33 | Water | Thailand | 1965 | 385 | 1 | 4 | 11 | 4 | 5 | 4 | 6 |
| STW 34 | Water | Thailand | 1965 | 407 | 3 | 1 | 3 | 3 | 1 | 2 | 18 |
| STW 35-1 | Water | Thailand | 1965 | 371 | 1 | 12 | 6 | 2 | 1 | 4 | 1 |
| STW 36-1 | Water | Thailand | 1965 | 366 | 3 | 1 | 2 | 3 | 8 | 4 | 3 |
| STW 38 | Water | Thailand | 1965 | 84 | 3 | 1 | 11 | 4 | 5 | 4 | 6 |
| STW 39 | Water | Thailand | 1965 | 371 | 1 | 12 | 6 | 2 | 1 | 4 | 1 |
| STW 42 | Water | Thailand | 1965 | 371 | 1 | 12 | 6 | 2 | 1 | 4 | 1 |
| STW 43-1 | Water | Thailand | 1965 | 371 | 1 | 12 | 6 | 2 | 1 | 4 | 1 |
| STW 44 | Water | Thailand | 1965 | 371 | 1 | 12 | 6 | 2 | 1 | 4 | 1 |
| STW 45-1 | Water | Thailand | 1965 | 289 | 3 | 4 | 11 | 4 | 5 | 4 | 6 |
| STW 55-2 | Water | Thailand | 1965 | 289 | 3 | 4 | 11 | 4 | 5 | 4 | 6 |
| STW 58-1 | Water | Thailand | 1965 | 289 | 3 | 4 | 11 | 4 | 5 | 4 | 6 |
| STW 61-2 | Water | Thailand | 1965 | 372 | 1 | 2 | 3 | 1 | 5 | 22 | 1 |
| STW 62 | Water | Thailand | 1965 | 84 | 3 | 1 | 11 | 4 | 5 | 4 | 6 |
| STW 64 | Water | Thailand | 1965 | 385 | 1 | 4 | 11 | 4 | 5 | 4 | 6 |
| STW 66 | Water | Thailand | 1965 | 384 | 1 | 12 | 3 | 3 | 1 | 4 | 1 |
| STW 67-1 | Water | Thailand | 1965 | 374 | 1 | 4 | 11 | 4 | 5 | 2 | 6 |
| STW 94-1 | Water | Thailand | 1965 | 164 | 3 | 1 | 2 | 3 | 5 | 4 | 3 |
| STW 95-1 | Water | Thailand | 1965 | 371 | 1 | 12 | 6 | 2 | 1 | 4 | 1 |
| STW 96-2 | Water | Thailand | 1965 | 371 | 1 | 12 | 6 | 2 | 1 | 4 | 1 |
| STW 97-1 | Water | Thailand | 1965 | 84 | 3 | 1 | 11 | 4 | 5 | 4 | 6 |
| STW 98-1 | Water | Thailand | 1965 | 51 | 3 | 1 | 2 | 3 | 1 | 4 | 3 |
| STW 99-2 | Water | Thailand | 1965 | 380 | 3 | 2 | 2 | 1 | 1 | 4 | 3 |
| STW 100-1 | Water | Thailand | 1965 | 15 | 1 | 2 | 2 | 2 | 1 | 3 | 1 |
| STW 101-1 | Water | Thailand | 1965 | 84 | 3 | 1 | 11 | 4 | 5 | 4 | 6 |
| STW 102-3 | Water | Thailand | 1965 | 84 | 3 | 1 | 11 | 4 | 5 | 4 | 6 |
| STW 104-1 | Water | Thailand | 1965 | 51 | 3 | 1 | 2 | 3 | 1 | 4 | 3 |
| STW 105-1 | Water | Thailand | 1965 | 371 | 1 | 12 | 6 | 2 | 1 | 4 | 1 |
| STW 106-1 | Water | Thailand | 1965 | 54 | 3 | 1 | 3 | 3 | 1 | 2 | 1 |
| STW 107-1 | Water | Thailand | 1965 | 372 | 1 | 2 | 3 | 1 | 5 | 22 | 1 |
| STW 110-1 | Water | Thailand | 1965 | 51 | 3 | 1 | 2 | 3 | 1 | 4 | 3 |
| STW 111-2 | Water | Thailand | 1965 | 164 | 3 | 1 | 2 | 3 | 5 | 4 | 3 |
| STW 114-1 | Water | Thailand | 1965 | 369 | 3 | 1 | 2 | 1 | 5 | 4 | 3 |
| STW 115-2 | Water | Thailand | 1965 | 51 | 3 | 1 | 2 | 3 | 1 | 4 | 3 |
| STW 116-2 | Water | Thailand | 1965 | 289 | 3 | 4 | 11 | 4 | 5 | 4 | 6 |
| STW 117-4 | Water | Thailand | 1965 | 371 | 1 | 12 | 6 | 2 | 1 | 4 | 1 |
| STW 120-1 | Water | Thailand | 1965 | 389 | 3 | 1 | 11 | 1 | 1 | 4 | 1 |
| STW 122 | Water | Thailand | 1965 | 168 | 3 | 1 | 2 | 1 | 5 | 4 | 1 |
| STW 152 | Water | Thailand | 1966 | 416 | 1 | 1 | 6 | 2 | 1 | 42 | 1 |
| STW 154 | Water | Thailand | 1965 | 371 | 1 | 12 | 6 | 2 | 1 | 4 | 1 |
| STW 157 | Water | Thailand | 1965 | 168 | 3 | 1 | 2 | 1 | 5 | 4 | 1 |
| STW 162-1 | Water | Thailand | 1965 | 289 | 3 | 4 | 11 | 4 | 5 | 4 | 6 |
| STW 168-3 | Water | Thailand | 1965 | 54 | 3 | 1 | 3 | 3 | 1 | 2 | 1 |
| STW 174 | Water | Thailand | 1965 | 402 | 3 | 1 | 2 | 3 | 1 | 2 | 1 |
| STW 175-1 | Water | Thailand | 1965 | 369 | 3 | 1 | 2 | 1 | 5 | 4 | 3 |
| STW 176 | Water | Thailand | 1965 | 401 | 3 | 3 | 2 | 1 | 1 | 4 | 1 |
| STW 181-1 | Water | Thailand | 1965 | 51 | 3 | 1 | 2 | 3 | 1 | 4 | 3 |
| STW 185 | Water | Thailand | 1965 | 46 | 3 | 1 | 2 | 1 | 1 | 3 | 3 |
| STW 185-1 | Water | Thailand | 1965 | 405 | 1 | 4 | 6 | 2 | 1 | 2 | 1 |
| STW 186-2 | Water | Thailand | 1965 | 366 | 3 | 1 | 2 | 3 | 8 | 4 | 3 |
| STW 187-3 | Water | Thailand | 1965 | 366 | 3 | 1 | 2 | 3 | 8 | 4 | 3 |
| STW 189-2 | Water | Thailand | 1965 | 396 | 3 | 2 | 2 | 3 | 5 | 3 | 1 |
| STW 197-1 | Water | Thailand | 1965 | 376 | 1 | 4 | 2 | 3 | 8 | 4 | 3 |
| STW 199-2 | Water | Thailand | 1965 | 376 | 1 | 4 | 2 | 3 | 8 | 4 | 3 |
| STW 200-1 | Water | Thailand | 1965 | 376 | 1 | 4 | 2 | 3 | 8 | 4 | 3 |
| STW 202-3 | Water | Thailand | 1965 | 366 | 3 | 1 | 2 | 3 | 8 | 4 | 3 |
| STW 204-2 | Water | Thailand | 1965 | 366 | 3 | 1 | 2 | 3 | 8 | 4 | 3 |
| STW 205-1 | Water | Thailand | 1965 | 51 | 3 | 1 | 2 | 3 | 1 | 4 | 3 |
| STW 208 | Water | Thailand | 1965 | 51 | 3 | 1 | 2 | 3 | 1 | 4 | 3 |
| STW 208-1 | Water | Thailand | 1965 | 51 | 3 | 1 | 2 | 3 | 1 | 4 | 3 |
| STW 214 | Water | Thailand | 1965 | 84 | 3 | 1 | 11 | 4 | 5 | 4 | 6 |
| STW 215-1 | Water | Thailand | 1965 | 408 | 3 | 1 | 2 | 3 | 8 | 4 | 24 |
| STW 216-2 | Water | Thailand | 1965 | 400 | 1 | 12 | 3 | 2 | 1 | 8 | 1 |
| STW 217-2 | Water | Thailand | 1965 | 383 | 1 | 4 | 6 | 2 | 3 | 1 | 1 |
| STW 219 | Water | Thailand | 1965 | 368 | 3 | 1 | 2 | 3 | 8 | 4 | 1 |
| STW 220-2 | Water | Thailand | 1965 | 289 | 3 | 4 | 11 | 4 | 5 | 4 | 6 |
| STW 221-1 | Water | Thailand | 1965 | 371 | 1 | 12 | 6 | 2 | 1 | 4 | 1 |
| STW 222-2 | Water | Thailand | 1965 | 378 | 18 | 12 | 3 | 2 | 1 | 2 | 1 |
| STW 224-1 | Water | Thailand | 1965 | 289 | 3 | 4 | 11 | 4 | 5 | 4 | 6 |
| STW 225-3 | Water | Thailand | 1965 | 84 | 3 | 1 | 11 | 4 | 5 | 4 | 6 |
| STW 230-1 | Water | Thailand | 1965 | 366 | 3 | 1 | 2 | 3 | 8 | 4 | 3 |
| STW 233-1 | Water | Thailand | 1965 | 371 | 1 | 12 | 6 | 2 | 1 | 4 | 1 |
| STW 235-1 | Water | Thailand | 1965 | 376 | 1 | 4 | 2 | 3 | 8 | 4 | 3 |
| STW 244-1 | Water | Thailand | 1965 | 378 | 18 | 12 | 3 | 2 | 1 | 2 | 1 |
| STW 305 | Water | Thailand | 1965 | 300 | 1 | 1 | 3 | 1 | 1 | 4 | 1 |
| STW 307-2 | Water | Thailand | 1965 | 395 | 1 | 3 | 36 | 1 | 1 | 4 | 3 |
| STW 312-1 | Water | Thailand | 1965 | 300 | 1 | 1 | 3 | 1 | 1 | 4 | 1 |
| STW 358-2 | Water | Thailand | 1965 | 312 | 1 | 4 | 2 | 1 | 1 | 3 | 1 |
| STW 359-1 | Water | Thailand | 1965 | 364 | 1 | 3 | 4 | 3 | 1 | 4 | 3 |
| STW 362 | Water | Thailand | 1965 | 364 | 1 | 3 | 4 | 3 | 1 | 4 | 3 |
| STW 364 | Water | Thailand | 1965 | 364 | 1 | 3 | 4 | 3 | 1 | 4 | 3 |
| STW 368-3 | Water | Thailand | 1965 | 364 | 1 | 3 | 4 | 3 | 1 | 4 | 3 |
| STW 402 | Water | Thailand | 1965 | 379 | 1 | 2 | 2 | 3 | 5 | 4 | 1 |
| STW 406 | Water | Thailand | 1965 | 164 | 3 | 1 | 2 | 3 | 5 | 4 | 3 |
| STW 414-1 | Water | Thailand | 1965 | 390 | 1 | 1 | 14 | 2 | 1 | 22 | 1 |
| STW 415 | Water | Thailand | 1965 | 290 | 3 | 4 | 11 | 3 | 5 | 4 | 1 |
| STW 420-2 | Water | Thailand | 1965 | 417 | 18 | 4 | 6 | 2 | 5 | 1 | 3 |
| STW 422 | Water | Thailand | 1965 | 391 | 1 | 2 | 10 | 2 | 1 | 8 | 1 |
| STW 424-1 | Water | Thailand | 1965 | 417 | 18 | 4 | 6 | 2 | 5 | 1 | 3 |
| STW 426-2 | Water | Thailand | 1965 | 409 | 3 | 1 | 36 | 1 | 1 | 4 | 1 |
| STW 429 | Water | Thailand | 1965 | 413 | 3 | 2 | 6 | 1 | 1 | 3 | 3 |
| STW 430 | Water | Thailand | 1965 | 51 | 3 | 1 | 2 | 3 | 1 | 4 | 3 |
| STW 447 | Water | Thailand | 1965 | 366 | 3 | 1 | 2 | 3 | 8 | 4 | 3 |
| STW 487-1 | Water | Thailand | 1965 | 3 | 1 | 1 | 2 | 2 | 5 | 3 | 1 |
| STW 539-1 | Water | Thailand | 1965 | 392 | 1 | 2 | 6 | 1 | 1 | 4 | 1 |
| STW 551 | Water | Thailand | 1965 | 385 | 1 | 4 | 11 | 4 | 5 | 4 | 6 |
| STW 561-1 | Water | Thailand | 1966 | 393 | 3 | 2 | 3 | 1 | 1 | 4 | 1 |
| STW 638-1 | Water | Thailand | 1966 | 398 | 1 | 2 | 3 | 1 | 1 | 22 | 1 |
| STW 640 | Water | Thailand | 1966 | 381 | 1 | 2 | 2 | 1 | 1 | 3 | 1 |
| STW 723 | Water | Thailand | 1966 | 388 | 1 | 1 | 4 | 1 | 1 | 22 | 1 |
| STW 729 | Water | Thailand | 1966 | 394 | 3 | 4 | 11 | 3 | 1 | 4 | 3 |
| STW 730-1 | Water | Thailand | 1966 | 404 | 1 | 1 | 4 | 3 | 1 | 4 | 3 |
| STW 753 | Water | Thailand | 1966 | 412 | 1 | 4 | 10 | 1 | 1 | 2 | 1 |
| STW 754 | Water | Thailand | 1966 | 386 | 3 | 1 | 3 | 1 | 1 | 2 | 1 |
| STW 760-2 | Water | Thailand | 1966 | 376 | 1 | 4 | 2 | 3 | 8 | 4 | 3 |
| STW 765 | Water | Thailand | 1966 | 70 | 3 | 4 | 11 | 3 | 5 | 4 | 6 |
| UB-8-Soil 1 | Soil | Thailand | 1965 | 167 | 1 | 1 | 4 | 1 | 1 | 3 | 1 |
| UB-8-Soil 2 | Soil | Thailand | 1965 | 60 | 3 | 1 | 12 | 1 | 1 | 3 | 1 |
| Ubol 6-1 | Environment | Thailand | 1965 | 373 | 3 | 1 | 11 | 2 | 5 | 4 | 1 |
| USAMRU Malaysia 1 | Malaysia | 1964 | 46 | 3 | 1 | 2 | 1 | 1 | 3 | 3 | |
| USAMRU Malaysia 3 | Malaysia | 1964 | 46 | 3 | 1 | 2 | 1 | 1 | 3 | 3 | |
| USAMRU Malaysia 4 | Malaysia | 1964 | 51 | 3 | 1 | 2 | 3 | 1 | 4 | 3 | |
| USAMRU Malaysia 5 | Malaysia | 1964 | 46 | 3 | 1 | 2 | 1 | 1 | 3 | 3 | |
| USAMRU Malaysia 7 | Malaysia | 1964 | 46 | 3 | 1 | 2 | 1 | 1 | 3 | 3 | |
| USAMRU Malaysia 8 | Malaysia | 1964 | 46 | 3 | 1 | 2 | 1 | 1 | 3 | 3 | |
| USAMRU Malaysia 10 | Malaysia | 1964 | 51 | 3 | 1 | 2 | 3 | 1 | 4 | 3 | |
| USAMRU Malaysia 11 | Malaysia | 1964 | 46 | 3 | 1 | 2 | 1 | 1 | 3 | 3 | |
| USAMRU Malaysia 12 | Malaysia | 1964 | 54 | 3 | 1 | 3 | 3 | 1 | 2 | 1 | |
| USAMRU Malaysia 13 | Malaysia | 1964 | 46 | 3 | 1 | 2 | 1 | 1 | 3 | 3 | |
| USAMRU Malaysia 14 | Malaysia | 1964 | 84 | 3 | 1 | 11 | 4 | 5 | 4 | 6 | |
| USAMRU Malaysia 15 | Malaysia | 1964 | 84 | 3 | 1 | 11 | 4 | 5 | 4 | 6 | |
| USAMRU Malaysia 16 | Malaysia | 1964 | 84 | 3 | 1 | 11 | 4 | 5 | 4 | 6 | |
| USAMRU Malaysia 17 | Malaysia | 1964 | 84 | 3 | 1 | 11 | 4 | 5 | 4 | 6 | |
| USAMRU Malaysia 18 | Malaysia | 1964 | 406 | 1 | 4 | 13 | 2 | 1 | 1 | 1 | |
| USAMRU Malaysia 19 | Malaysia | 1964 | 406 | 1 | 4 | 13 | 2 | 1 | 1 | 1 | |
| USAMRU Malaysia 22 | Malaysia | 1964 | 46 | 3 | 1 | 2 | 1 | 1 | 3 | 3 | |
| USAMRU Malaysia 23 | Malaysia | 1964 | 46 | 3 | 1 | 2 | 1 | 1 | 3 | 3 | |
| USAMRU Malaysia 24 | Malaysia | 1964 | 46 | 3 | 1 | 2 | 1 | 1 | 3 | 3 | |
| USAMRU Malaysia 25 | Malaysia | 1964 | 46 | 3 | 1 | 2 | 1 | 1 | 3 | 3 | |
| USAMRU Malaysia 26 | Malaysia | 1964 | 51 | 3 | 1 | 2 | 3 | 1 | 4 | 3 | |
| USAMRU Malaysia 27 | Malaysia | 1964 | 51 | 3 | 1 | 2 | 3 | 1 | 4 | 3 | |
| USAMRU Malaysia 28 | Malaysia | 1964 | 289 | 3 | 4 | 11 | 4 | 5 | 4 | 6 | |
| USAMRU Malaysia 29 | Malaysia | 1964 | 46 | 3 | 1 | 2 | 1 | 1 | 3 | 3 | |
| USAMRU Malaysia 30 | Malaysia | 1964 | 46 | 3 | 1 | 2 | 1 | 1 | 3 | 3 | |
| USAMRU Malaysia 31 | Malaysia | 1964 | 51 | 3 | 1 | 2 | 3 | 1 | 4 | 3 | |
| USAMRU Malaysia 32 | Malaysia | 1964 | 46 | 3 | 1 | 2 | 1 | 1 | 3 | 3 | |
| VW from infected rat | Rat | Malaysia? | 1967 | 367 | 3 | 1 | 11 | 1 | 1 | 3 | 1 |
| WRAIR 294 | Malaysia | 1964 | 51 | 3 | 1 | 2 | 3 | 1 | 4 | 3 | |
| WRAIR 1188 | Human | Malaysia | 1964 | 99 | 1 | 1 | 4 | 1 | 1 | 4 | 1 |
K96243 is the Sanger sequencing B. pseudomallei strain used as a positive control in this study (16).
Antigen 25 strains are colonial variants of USAMRU 25.
Antigen 1188HP-R is a rough colonial variant of WRAIR 1188.
Antigen 1691 is from a human isolate in Kuala Lumpur, 1921 (33).
Hansen, J77, Lavict and Ramos strains were isolated from soldiers in Vietnam.
The Thai culture collection, as well as antigen preparations and high-titer rabbit sera against representative strains, are also stored at the Centers for Disease Control and Prevention (Mindy Glass), Atlanta, Ga., and are available, on request, to qualified investigators.
Bacterial growth conditions and isolation of genomic DNA.
Live organisms were cultured, and genomic DNA was isolated in a category III biocontainment facility at the University of Calgary. Overnight bacterial cultures were inoculated from freezer stock and grown in Luria-Bertani (LB) broth (Invitrogen, Burlington, Ontario, Canada) at 37°C. Genomic DNA was isolated using the Wizard genomic DNA purification kit (Promega, Madison, Wis.) according to the manufacturer's instructions. Genomic DNAs were stored at −20°C.
Multilocus sequence typing.
MLST was carried out according to the methods of Godoy et al. (14) with modifications to promote consistency of product amplification and to improve sequencing quality. The primers used in the PCR amplification and sequencing of the seven housekeeping gene fragments are listed in Table 2. The following primer pairs were used in the PCR amplification of seven housekeeping-gene fragments from all B. pseudomallei strains analyzed: ace-up and ace-dn, gltB-up and gltB-dn, gmhD-up and gmhD-dn(outer), lepA-up and lepA-dn, lipA-up and lipA-dn, narK-up(outer) and narK-dn, and ndh-up and ndh-dn (Table 2).
TABLE 2.
Primers used in amplification and sequencing of seven housekeeping loci for multilocus sequence typing analysis of the historical B. pseudomallei strain collection
| Locus | Primer name | Primer sequence (5′→3′) | Annealing temp (°C) | Applicationa |
|---|---|---|---|---|
| ace | ace-up | GCT CGG CGC TTC TCA AAA CG | 61 | Amp & Seq |
| ace-dn | CAT GTC CGT GCC GAT GTA GC | |||
| gltB | gltB-up | GGC GGC AAG TCG AAC ACG G | 61 | Amp & Seq |
| gltB-dn | GCA GGC GGT TCA GCA CGA G | |||
| gmhD | gmhD-up | CTC GCG CAG GGC ACG CAG T | 63 | Amp & Seq |
| gmhD-dn | GTC AGG AAC GGC GCG TCG TA | Seq | ||
| gmhD-dn(outer) | GGC TGC CGA CCG TGA GAC C | Amp | ||
| lepA | lepA-up | CGC TTG ATC GGC ACT GAA TGG | 63 | Amp & Seq |
| lepA-dn | CGA ACC ACG AAT CGA TGA TGA G | |||
| lipA | lipA-up | CAT ACG GTG TGC GAG GAA GC | 61 | Amp & Seq |
| lipA-dn | CAG GAT CTC GTC GGT CGT CT | |||
| narK | narK-up(outer) | GCC GCG CAC GAC CAG CGC | 62 | Amp |
| narK-up | CGG ATT CGA TCA TGT CCA CTT C | Seq | ||
| narK-dn | CGG CAC CCA CAC GAA GCC C | Amp & Seq | ||
| ndh | ndh-up | GCA GTT CGT CGC GGA CTA TC | 61 | Amp & Seq |
| ndh-dn | GGC GCG GCA TGA AGC TCC A |
Amp, amplification; Seq, sequencing; Amp & Seq, both.
Fifty-microliter-volume PCRs were carried out in a 96-well PCR plate format, allowing for two B. pseudomallei K96243 (Sanger sequencing strain) genomic DNA-positive controls and two sterile water negative controls per plate. PCRs contained the following: 1 μg genomic DNA template, 1× PCR buffer, 1.5 mM MgCl2, 0.2 mM mixed deoxynucleoside triphosphates, 0.8 μM mixed primers, 0.5× Q solution (QIAGEN, Mississauga, Ontario, Canada), and 2.5 U Taq DNA polymerase. All standard PCR reagents were acquired from Promega (Madison, Wis.). Initial denaturation at 95°C for 4 min was followed by 35 cycles of 95°C for 30 s, 61°C for 30 s (except for the gmhD and lepA primers, which require 63°C, and narK primers, which require 62°C), and 72°C for 1 min. Final extension was carried out at 72°C for 5 min, and the samples were maintained at 4°C. The amplified housekeeping-gene fragments were purified with 20% polyethylene glycol-8000-2.5 M NaCl precipitation according to the methods of Godoy et al. (14). The purified PCR products were analyzed via 1.5% agarose gel electrophoresis for sufficient product, correct size, and product purity following both PCR and purification steps. PCRs lacking sufficient product or displaying multiple products were discarded and repeated.
The purified housekeeping-DNA fragments were submitted in a 96-well PCR plate format for sequencing to the University of Calgary DNA Sequencing Core Service Facility. Each DNA fragment was sequenced on both strands using the primers indicated in Table 2. Seminested sequencing reactions were carried out for gmhD and narK gene fragments using the primers gmhD-dn and narK-up in place of gmhD-dn(outer) and narK-up(outer), respectively, to improve sequencing quality. All other housekeeping-gene products were sequenced using the same primers as for amplification (Table 2).
Data analysis.
For the sequence analysis of each DNA fragment, the forward and reverse sequences were aligned with a reference allele sequence obtained from the B. pseudomallei MLST website using the SeqManII module of Lasergene v. 6.0 software (DNAStar, Madison, Wis.). The resulting contig was trimmed and edited if required. Each sequence alignment was examined for sequencing quality. Sequences that were too short, of poor quality, or featured two nucleotide signals at the same position were discarded, and the samples were resequenced.
Allele numbers were assigned to each of the seven housekeeping loci by submission of the forward sequence of the alignment contig to the B. pseudomallei MLST website (http://bpseudomallei.mlst.net/). The allele numbers for all seven housekeeping gene fragments of each isolate were assembled into a string of seven integers corresponding to the gene order of ace-gltB-gmhD-lepA-lipA-narK-ndh, giving the allele profile for each isolate. The allele profiles were queried against the B. pseudomallei MLST website to obtain an ST number.
Novel allele sequences were confirmed with repeat PCR and sequencing reactions. Novel allele profiles were confirmed by repeated PCR and sequencing reactions for the locus that differed from an existing allele profile already cataloged in the B. pseudomallei MLST database. Novel allele sequences and novel allele profiles were then forwarded to the B. pseudomallei MLST website curator for allele number and ST assignment, respectively. The MLST analysis results for the 207 isolates of the historical B. pseudomallei collection were confirmed with repeat PCR and sequencing reactions for 10% of the remaining samples. The 5 novel allele sequences and 56 novel STs encountered during this study have been submitted to the B. pseudomallei MLST database, along with the strain information for all 207 isolates of the historical-strain collection.
In order to display the relatedness among the isolates of the historical B. pseudomallei collection, eBURST diagrams were generated. The eBURSTv3 Java application is available as a link from the B. pseudomallei MLST website. eBURST is based on a model of bacterial evolution whereby a single ancestral founding ST undergoes diversification to produce a subset of closely related STs (10, 28). Descendants of the founding ST diversify by accumulating point mutations or undergo recombination that eventually generates diversity among the MLST housekeeping-allele sequences, resulting in closely related but variant STs of the founding ST. Single locus variants (SLVs) of the group founding ST differ in their allele profiles by 1/7 housekeeping allele sequences. The SLVs diversify further into double locus variants where 2/7 housekeeping alleles differ from the original founding ST. A single spot on the eBURST diagram represents each individual ST, and the size of the spot is proportional to the number of isolates in the population that share that ST. SLVs are joined to the founding ST by a line, double locus variants are joined to the SLVs, and so on until a bacterial population is represented as a series of clonal complexes with the founding ST (defined as the ST with the most SLVs) located centrally with a series of variant STs radiating outward. Only the STs with similar allele profiles are grouped together with the default group definition of 6/7 shared alleles (10, 28).
The 207 isolates of the historical collection were analyzed separately by generating a “population snapshot” of all 80 STs encountered during the analysis by setting the group definition from 6/7 to 0/7 shared alleles. The 56 novel sequence types encountered were differentially highlighted by using the comparative function of eBURST. The existing STs encountered during the analysis (Reference) were compared to the novel STs encountered (Query). Since the novel STs do not appear in the Query data set, they are highlighted green. The comparative function of eBURST was then used to display the changes to the full-size (i.e., MLST database) B. pseudomallei “population snapshot” due to the addition of the historical-isolate MLST data. The database information excluding the novel STs encountered was loaded as the initial data set (Reference). The second data set (Query) included all STs pertaining to the historical-strain collection, including the 56 novel STs and the 24 existing STs that were already present in the database, which allowed differentiation of the specific historical-isolate STs encountered and all other STs of the B. pseudomallei MLST database. The novel STs do not appear in the Reference data set and are highlighted green. The existing STs do appear in the Reference data set and are highlighted magenta.
RESULTS
The historical collection of Southeast Asian B. pseudomallei isolates exhibits sequence type diversity.
The 207 isolates of the historical B. pseudomallei collection were analyzed by MLST (Table 1). Five new allele sequences were encountered: one (each) for ace (allele 18), gltB (allele 30), and ndh (allele 24) and two for gmhD (alleles 36 and 37). The strain collection was resolved into 80 different STs, 24 which were already present in the B. pseudomallei MLST database and 56 novel STs, which were designated ST364 through ST419 upon submission of the unique allele profiles to the database curator. Approximately half (102/207) of the isolates in the strain collection were assigned existing STs and are representative of B. pseudomallei MLST database isolates sourced from diverse geographical locations, including Thailand, Laos, Malaysia, Indonesia, Bangladesh, Singapore, China, Hong Kong, the Philippines, Ecuador, The United States, Fiji, and Australia (http://bpseudomallei.mlst.net/). The 56 novel STs are most similar to B. pseudomallei MLST database isolates sourced from Thailand, Malaysia, Vietnam, Cambodia, Bangladesh, Singapore, China, Hong Kong, the Philippines, the United States, Australia, and Burkina Faso in that they share five or six out of seven housekeeping alleles in common (i.e., single or double locus variants, respectively).
The historical-strain collection of B. pseudomallei isolates is biased towards the southern regions of Thailand. Isolates recovered from Phangna, Phattalung, Phuket, Ranong, Songkhla, and south Thailand water (STW) sources account for 69% (143/207) of the collection (Table 1). The greatest amount of ST diversity occurs among these isolates. Sixty-two percent (89/143) of the south Thailand isolates were assigned novel STs.
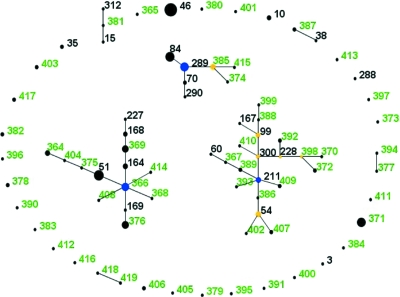
An eBURST diagram was generated to explore the relationships among the 207 isolates and 80 STs of the historical B. pseudomallei collection. A “population snapshot” was generated by setting the group definition in the “Analysis” panel from the default 6/7 alleles to 0/7 shared alleles and is shown in Fig. 1. The comparative function of eBURST was used to allow for the differential identification of the 56 novel STs encountered (shown in green text) from the 24 existing STs already present in the B. pseudomallei MLST database. The STs of the historical collection form three small, discontinuous complexes with ST366 as the predicted group founder for all 80 STs indicated in the figure. The majority of the STs appearing in the three complexes were isolated from the south of Thailand (i.e., STW isolates, Songkhla, Ranong, and Phuket), with Malaysian isolates among others interspersed. Multiple outlier STs surround the three complexes, since they are distinct from the predicted group founding ST and remain unlinked.
FIG. 1.
eBURST diagram displaying the relatedness of the 207 isolates and 80 sequence types of the historical B. pseudomallei strain collection. Predicted group founders are indicated in blue, and subgroup founders are indicated in yellow. The predicted group founder of all STs is ST366. The 56 novel STs encountered during the study are displayed in green text. The diagram was edited manually for clarity.
The STs of the historical collection of Southeast Asian B. pseudomallei isolates are ancestral compared to the B. pseudomallei population.
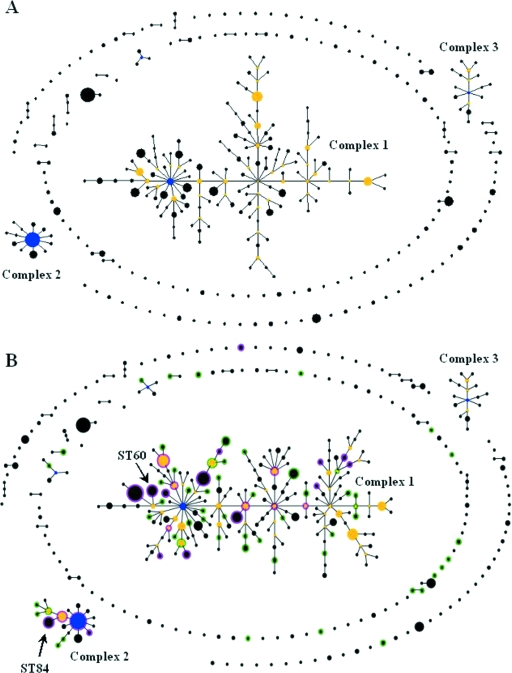
In order to determine the phylogenetic nature of the historical B. pseudomallei isolates, an eBURST diagram was generated to display the changes to the B. pseudomallei MLST database “population snapshot” upon the addition of the historical-isolate ST data. Figure 2A is the current “population snapshot” of all 760 isolates and 365 STs in the B. pseudomallei MLST database, excluding the historical-isolate data and novel sequence types submitted (ST364 to ST419). Figure 2B is the B. pseudomallei “population snapshot” upon the addition of the historical-strain collection MLST data. The comparative function of eBURST was used again to allow the differential identification of the 56 novel STs encountered (shown with green halos) and the 24 existing STs encountered (shown with magenta halos) from the remaining STs of the B. pseudomallei MLST database (Fig. 2B).
FIG. 2.
eBURST diagram displaying the “population snapshot” of the B. pseudomallei isolates of the MLST database before (A) and after (B) the addition of the MLST data of the historical B. pseudomallei collection. The predicted group founder of all isolates is ST48 in both panels. Panel A is an eBURST diagram demonstrating the relatedness of the 760 isolates and 365 STs of the B. pseudomallei MLST database. Panel B is a comparative eBURST diagram demonstrating the alteration to the original “population snapshot” upon the addition of the 207 isolates and 80 STs of the historical B. pseudomallei strain collection to the isolates of the B. pseudomallei MLST database. The 56 novel STs encountered during the study are indicated with green halos, and the 24 existing STs encountered during the study are indicated with magenta halos. The diagrams were edited manually, and the ST numbers were omitted for clarity. The clonal complex numbers are indicated. ST60 and ST84 are indicated with arrows.
The B. pseudomallei “population snapshot” contains a large complex at the center of the eBURST diagram (complex 1), as well as two smaller complexes in the bottom left (complex 2) and top right (complex 3) corners (Fig. 2A). Many outlying STs surround the main complex. The outlier STs are predominantly isolates of Australian origin as well as B. mallei and B. thailandensis spp. (http://bpseudomallei.mlst.net/). The predicted group founder for all STs in the diagram is ST48 (of Thai origin), shown in blue on the left-hand side of complex 1 (Fig. 2A). The addition of the historical B. pseudomallei strain collection MLST data to the “population snapshot” does not alter the predicted group founder (ST48) or the number of complexes present but alters the pattern of ST descent within complexes 1 and 2 but not 3. The majority (62/80) of the historical B. pseudomallei strain collection STs cluster within the main clonal complex in the center of the eBURST diagram, as well as within complex 2 (Fig. 2B). The novel STs, in particular, provide “branch points” or “linker” STs for descendant STs that are already cataloged in the MLST database and contribute to branch rearrangement within the main complex, suggesting an ancestral nature for the historical-strain collection STs.
Three isolates of the historical collection of B. pseudomallei isolates share ST10 with K96243.
B. pseudomallei K96243 is the Sanger sequencing strain and was previously assigned the MLST allele profile 1-1-13-1-1-1-1, which corresponds to ST10 (14, 16). There are currently two other clinical isolates in the B. pseudomallei MLST database with this ST. They are SID4350 and 2687. Three of the historical B. pseudomallei isolates sourced from environmental sampling in Thailand were found to share this ST. They are Loei KK-S2 (soil isolate), Loei KK-S2 (2) (soil isolate), and Phangna 64W (water isolate); all three were isolated in 1965. These are the first environmental B. pseudomallei isolates reported to be assigned ST10 according to the MLST database (http://bpseduomallei.mlst.net/). The Loei isolates were recovered in a geographical origin similar to that of the K96243 sequencing strain, which was isolated in 1996 from a female patient in Khon Kaen hospital in Northeast Thailand; however, the 31-year difference in the dates of isolation is considerable (16). The assignment of ST10 to the Phangna 64W strain indicates that ST10 may not be restricted to this region of Thailand alone and is also present in the southern regions of Thailand.
Variation in colony morphology does not affect the ST of B. pseudomallei isolates.
The reference strains included in the historical collection of B. pseudomallei isolates contain colonial variants. The four Antigen 25 strains in the collection (Antigen 25 Donut, HP Trans-muc, Smooth, and Medusa) are colonial variants of USAMRU Malaysia 25 (Table 1). All five isolates were assigned the allele profile 3-1-2-1-1-3-3, which corresponds to ST46. Additionally, Antigen 1188HP-R is a rough colonial variant of WRAIR 1188 (Table 1). Both isolates were assigned ST99 (allele profile 1-1-4-1-1-4-1). These results suggest that changes in colony morphology do not affect MLST typing of B. pseudomallei isolates.
The STs of the historical collection of Southeast Asian B. pseudomallei isolates indicate phylogenetic links between Thai and Australian STs.
A single isolate of the historical strain collection, UB-8-Soil 2, was assigned the allele profile 3-1-12-1-1-3-1, which corresponds to ST60. Compared with the isolates of the MLST database, it was noted that ST60 represents eight clinical and environmental isolates from Thailand, a clinical isolate from Fiji (isolated in 1992), and five environmental isolates from Australia (Table 3) (http://bpseudomallei.mlst.net/) (13, 14). This finding indicates that multiple environmental isolates from Thailand and Australia are very closely related as they share the same ST. ST60 also shares five out of seven alleles in common with ST48 (allele profile 3-1-2-1-1-4-1), the predicted group founder of Thai origin in the eBURST “population snapshot” of all B. pseudomallei isolates in the MLST database (Fig. 2).
TABLE 3.
B. pseudomallei isolates of the historical strain collection that share ST60 and ST84 in common with B. pseudomallei isolates of the MLST database
| Strain | ST | Source | Country | Yr | Reference |
|---|---|---|---|---|---|
| UB-8-Soil 2 | 60 | Soil | Thailand | 1965 | This study |
| 1244 | 60 | Human | Thailand | Unknown | http://bpseudomallei.mlst.net/ |
| 1248 | 60 | Human | Thailand | Unknown | http://bpseudomallei.mlst.net/ |
| 2708 | 60 | Human | Thailand | Unknown | http://bpseudomallei.mlst.net/ |
| 2820 | 60 | Human | Thailand | Unknown | http://bpseudomallei.mlst.net/ |
| E0013 | 60 | Environment | Thailand | Unknown | http://bpseudomallei.mlst.net/ |
| E0031 | 60 | Environment | Thailand | Unknown | http://bpseudomallei.mlst.net/ |
| E0378 | 60 | Environment | Thailand | Unknown | http://bpseudomallei.mlst.net/ |
| E0383 | 60 | Environment | Thailand | Unknown | http://bpseudomallei.mlst.net/ |
| 5892/339a | 60 | Human | Fiji | 1992 | http://bpseudomallei.mlst.net/ |
| D228a | 60 | Environment | Australia | Unknown | http://bpseudomallei.mlst.net/ |
| D260:53/30a | 60 | Environment | Australia | Unknown | http://bpseudomallei.mlst.net/ |
| D304:S3/40a | 60 | Environment | Australia | Unknown | http://bpseudomallei.mlst.net/ |
| 2002721631b | 60 | Environment | Australia | Unknown | http://bpseudomallei.mlst.net/ |
| 2002721632b | 60 | Environment | Australia | Unknown | http://bpseudomallei.mlst.net/ |
| Smith 002025 | 84 | Unknown | Unknown | 1966 | This study |
| Songkhla 34W-2 | 84 | Water | Thailand | 1965 | This study |
| STW 38 | 84 | Water | Thailand | 1965 | This study |
| STW 62 | 84 | Water | Thailand | 1965 | This study |
| STW 97-1 | 84 | Water | Thailand | 1965 | This study |
| STW 101-1 | 84 | Water | Thailand | 1965 | This study |
| STW 102-3 | 84 | Water | Thailand | 1965 | This study |
| STW 214 | 84 | Water | Thailand | 1965 | This study |
| STW 225-3 | 84 | Water | Thailand | 1965 | This study |
| USAMRU Malaysia 14 | 84 | Unknown | Malaysia | 1964 | This study |
| USAMRU Malaysia 15 | 84 | Unknown | Malaysia | 1964 | This study |
| USAMRU Malaysia 16 | 84 | Unknown | Malaysia | 1964 | This study |
| USAMRU Malaysia 17 | 84 | Unknown | Malaysia | 1964 | This study |
| 2002721162b | 84 | Human | Australia | 1970 | http://bpseudomallei.mlst.net/ |
Thirteen isolates of the historical-strain collection were assigned the allele profile 3-1-11-4-5-4-6, which corresponds to ST84 (Table 3). The 13 isolates were recovered from the southern region of Thailand and Malaysia between the years 1964 and 1966. ST84 represents a single clinical isolate from Australia in the MLST database, isolated in 1970 from a female patient with empyema whose travel history was unknown (13; J. Gee, personal communication) (Table 3). Within the eBURST “population snapshot,” of all B. pseudomallei isolates in the MLST database, ST84 clusters within complex 2 among other STs characteristic of isolates from Southeast Asia (Fig. 3). ST84 is a single locus variant of ST289 (allele profile 3-4-11-4-5-4-6), which is represented by a single clinical isolate from Thailand isolated in 1993. ST289 was also encountered in this study and was assigned to 11 isolates of the historical strain collection. These isolates were recovered in the southern region of Thailand (i.e., STW, Songkhla, and Phattalung) and Malaysia between 1964 and 1965 (Table 1). According to eBURST, ST84 is likely representative of B. pseudomallei isolates originating from Southeast Asia.
FIG. 3.
Clonal complex two of the altered eBURST “population snapshot” from panel B of Fig. 2. The group founder for the complex is ST70 and is indicated in blue. The novel STs encountered during the study are indicated with green text, and the existing STs encountered during the study are indicated with magenta text. The diagram was edited manually, and the origins of the STs were added.
DISCUSSION
MLST is an excellent tool for the study of bacterial populations and global epidemiology when paired with a phylogenetic analysis algorithm, such as eBURST, to display the relatedness of bacterial isolates as a phylogenetic network. MLST results in digital data that are easily shared between laboratories via the internet. Multiple methods for MLST data analysis are available. The unweighted-pair group method with arithmetic averages is the most commonly used but has many shortcomings. The unweighted-pair group method with arithmetic averages cannot detect zero-length branches and usually places the root of the tree based on an assumption that is usually false (15). Neighbor-joining dendrograms and minimum-evolution trees using concatenated allele sequences are also common in MLST analysis; however, eBURST is a more appropriate choice for phylogenetic analysis of MLST data, since it was designed specifically for that purpose (15). eBURST allows the researcher to view the bacterial population as a whole by generating a “population snapshot” and makes no attempt to display the relatedness between very different multilocus genotypes in a population (28). The eBURST method of phylogenetic analysis also provides predicted pathways of evolution and ancestry that are often absent from dendrograms (28).
Typing bacterial isolates by MLST has the potential to identify multiple different alleles, allele profiles, and therefore STs among strain collections, since variation between housekeeping-locus alleles is detected by sequencing internal PCR fragments directly. The population structure of B. pseudomallei is clonal at the rRNA level, although clinical isolates of B. pseudomallei exhibit genetic diversity and are thus excellent candidates for MLST (23). It was therefore expected that historical isolates of the B. pseudomallei strain collection would exhibit tremendous diversity according to MLST analysis. Significant ST diversity is evident among the 207 historical isolates. Five novel alleles were encountered during the study, along with 56 novel STs and 24 STs already cataloged in the MLST database. The majority of the isolates from the southern region of Thailand were assigned novel STs, indicating a great amount of genotypic diversity among environmental B. pseudomallei isolates from this region. The ST diversity encountered for southern Thai isolates in this study contrasts with previous research, where ribotyping analysis demonstrated a lack of genetic diversity among B. pseudomallei isolates from southern Thailand (23). eBURST analysis of the MLST data predicts that the isolates of the historical collection form three groups of related STs with many outlying, or unlinked, STs when analyzed separately as a population. This is a discontinuous pattern among isolates of a relatively clonal species as demonstrated by ribotyping (23). The epidemiological significance of the historical isolates is demonstrated upon the addition of the MLST data generated during this study to the current MLST data of the B. pseudomallei MLST database. Although a greater proportion of the STs assigned in this analysis were novel, the majority of the historical-isolate STs clustered within the main complex as well as a smaller complex of the eBURST “population snapshot” and contributed to a rearrangement of branches within the main complex. The historical-isolate MLST data also support the eBURST assignment of ST48 of Thai origin as the group founder for all STs in the diagram. Many of the historical-isolate STs serve as branch points or linker STs in the phylogenetic eBURST network. The placement of the historical isolate STs within the eBURST diagram suggests an ancestral nature of the B. pseudomallei isolates obtained in an environmental survey of Thailand 40 years ago compared with the more recently obtained isolates of the MLST database.
The historical collection of B. pseudomallei isolates is heavily biased towards the southern region of Thailand, and therefore, isolates from this region are overrepresented in the collection. In the original geographic distribution study of B. pseudomallei isolates in Thailand carried out by Finkelstein, the southern region was sampled more extensively than the central, northern, and northeastern regions and resulted in the highest isolation rates of B. pseudomallei from water samples using the hamster isolation technique (11, 12). An early Malaysian study also reported the highest isolation rate of B. pseudomallei from water samples (29). Isolates were also recovered from sampling in Ubon, Nakhon Phanom, and Loei in the survey, but the high isolation rate for B. pseudomallei from the south of Thailand is in disagreement with clinical melioidosis data (11, 12). Other studies involving environmental sampling for B. pseudomallei isolation have reported contradictory isolation rates for B. pseudomallei from soil, which have been found to be the highest in the northeast region, where clinical cases of melioidosis are prevalent, as well as in the south, where there are fewer clinical cases reported (21, 27, 30, 31). The annual incidence of melioidosis in the northeast region of Thailand is 137.9 cases per 100,000 hospital inpatients, whereas the annual incidence in the southern region is 14.4 cases per 100,000 hospital inpatients (31). The number of CFU per ml of soil/water suspensions was found to be highest in the northeast region of Thailand and is probably related to the risk of disease in that region (27, 31). B. pseudomallei is reportedly unevenly distributed in soil, and the differences in isolation rates may be explained by regional variations in temperature, rainfall, sunlight, soil composition, and relative humidity (7, 27, 31). Although the collection itself is regionally biased towards the south, the addition of the historical-isolate data to the B. pseudomallei MLST database will provide balance, representing the geographical distribution and population biology of B. pseudomallei isolates in Thailand, and will shift the ratio of clinical isolates to environmental isolates, promoting increased support for environmental strains of this soil saprophyte and human pathogen in the MLST database.
MLST analysis of the historical collection of B. pseudomallei isolates revealed an epidemiological connection between the first environmental isolates reported to be assigned the same ST as Sanger sequencing strain K96243 (14, 16). Two of the three isolates were recovered from soil samples in Loei, which is in close geographical proximity to Khon Kaen, where strain K96243 was isolated from a female diabetic patient in 1996 (16). The 31-year difference in the dates of isolation is considerable and provides support for persistence of closely related B. pseudomallei isolates in the environment. ST10 is not restricted to the northeast region of Thailand. The third historical isolate assigned ST10 was isolated from a water sample obtained in Phangna in the southern region of Thailand. The occurrence of ST10 in the northeast and southern regions of Thailand suggests there is a lack of regional specificity for B. pseudomallei STs.
The colonial variants of the historical collection of B. pseudomallei isolates were found to retain the same allele profile and thus ST as their parent isolates. The Antigen 25 colonial variants Donut, HP Trans-muc, Medusa, and Smooth of USAMRU 25 and the Antigen 1188 HP-R rough colonial variant of WRAIR 1188 are genetically similar to their parent isolates by MLST definition despite phenotypic variation. MLST allows an organism to be typed in a manner representative of the genome and does not account for genomic rearrangement or discrete changes outside of the chosen housekeeping loci. Since MLST analysis cannot detect a correlation between ST and disease presentation, it would not be expected that a change in phenotype would affect the allele profile of an isolate (4).
The epidemiology of melioidosis in Australia is unique compared with that of other areas of endemicity. It has been speculated that Australian B. pseudomallei isolates are unique and distinct from those of Southeast Asia based on the geographical separation of Australia as well as epidemiological differences noted for each area (4). There have been longstanding reports in the literature regarding the different aspects of epidemiology specific to Australia and Thailand, including differences in B. pseudomallei distribution in soil, disease presentation, and annual incidence rates; however, there are aspects of epidemiology that are shared between the two regions, such as disease correlation with rainfall and B. pseudomallei infection as an important cause of community-acquired pneumonia and septicemia (2, 5, 6, 17, 27, 30, 31, 34). Interestingly, two Australian STs were encountered during the study, ST60 and ST84. ST84 was encountered among 13 strains of the historical collection and was assigned to isolates from the southern region of Thailand and Malaysia. According to the MLST database, ST84 is represented by a single clinical isolate from Australia obtained in 1970 from an Australian woman with empyema and an unknown travel history (13; J. Gee, personal communication). eBURST analysis places ST84 within complex 2, which includes STs representative of Southeast Asia. ST84 is also a single locus variant of ST289, an ST that is representative of B. pseudomallei isolates from Thailand and Malaysia. It is most likely that the patient traveled to Thailand or Malaysia and acquired B. pseudomallei abroad, suggesting that ST84 is representative of isolates from Southeast Asia rather than Australia.
A single isolate of the historical collection, UB-8-Soil 2, was isolated in 1965 from the northeast region of Thailand and was assigned ST60 in this study. According to the B. pseudomallei MLST database, ST60 is shared by four clinical isolates and four environmental isolates from Thailand, a single clinical isolate from Fiji, and five environmental isolates from Australia (http://bpseudomallei.mlst.net/) (13). Further investigation with eBURST revealed that ST60 shares five out of seven alleles in common with ST48, the predicted group founder of Thai origin in the “population snapshot” of all B. pseudomallei isolates in the MLST database, and clusters within the main clonal complex among other Thai and Southeast Asian STs. This finding is profoundly significant to melioidosis epidemiology and research, since it suggests an environmental association between Thai and Australian B. pseudomallei isolates specifically. Australian B. pseudomallei isolates were recently reported as being distinct from other isolates sourced elsewhere in the world based on MLST analysis (4). This is no longer the case. MLST analysis of the historical strain collection has highlighted an important epidemiological link between environmental isolates of Thai and Australian origin.
The development of MLST by Maiden et al. in 1998 provided a significant contribution of an effective typing scheme with great resolving power to the study of population biology and epidemiological study of pathogens. MLST databases for many different human pathogens are currently available for international access (http://www.mlst.net/ and http://pubmlst.org/). The B. pseudomallei MLST database is no exception. Although relatively young, the B. pseudomallei MLST database continues to grow at a rapid pace, facilitating the study of melioidosis epidemiology. MLST has allowed the epidemiological investigation of historical B. pseudomallei isolates collected in a nationwide geographical survey in Thailand 40 years prior to this study. The collection was found to be genetically diverse, and the historical isolates were ancestral in nature compared to the B. pseudomallei population even though isolates from the southern region of Thailand were overrepresented. The first environmental isolates possessing ST10, the same ST as K96243, were reported. Finally, an environmental epidemiological link was discovered between B. pseudomallei isolates originating from Thailand and Australia, which has important implications for future studies in melioidosis epidemiology.
Acknowledgments
This work was funded by the Canadian Institutes of Health Research grant MOP-36343 to D.E.W., a Canada Research Chair in Microbiology.
We thank Sharon Peacock and Mongkol Vesaratchavest of the Wellcome Trust, Faculty of Tropical Medicine, Mahidol University in Bangkok, Thailand, for providing MLST protocols and training in MLST techniques. We also thank Daniel Godoy of the Department of Infectious Disease Epidemiology, Faculty of Medicine, Imperial College in London, England, for his assistance in assigning allele numbers to the novel allele sequences as well as assigning sequence types to the novel allele profiles discovered in this study.
REFERENCES
- 1.Brett, P. J., D. DeShazer, and D. E. Woods. 1998. Burkholderia thailandensis sp. nov., a Burkholderia pseudomallei-like species. Int. J. Syst. Bacteriol. 48:317-320. [DOI] [PubMed] [Google Scholar]
- 2.Chaowagul, W., N. J. White, D. A. Dance, Y. Wattanagoon, P. Naigowit, T. M. Davis, S. Looareesuwan, and N. Pitakwatchara. 1989. Melioidosis: a major cause of community-acquired septicemia in northeastern Thailand. J. Infect. Dis. 159:890-899. [DOI] [PubMed] [Google Scholar]
- 3.Cheng, A. C., and B. J. Currie. 2005. Melioidosis: epidemiology, pathophysiology and management. Clin. Microbiol. Rev. 18:383-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng, A. C., D. Godoy, M. Mayo, D. Gal, B. G. Spratt, and B. J. Currie. 2004. Isolates of Burkholderia pseudomallei from northern Australia are distinct by multilocus sequence typing, but strain types do not correlate with clinical presentation. J. Clin. Microbiol. 42:5477-5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Currie, B. J., S. P. Jacups, A. C. Cheng, D. A. Fisher, N. M. Anstey, S. E. Huffam, and V. L. Krause. 2004. Melioidosis epidemiology and risk factors from a prospective whole-population study in northern Australia. Trop. Med. Int. Health 9:1167-1174. [DOI] [PubMed] [Google Scholar]
- 6.Currie, B. J., and S. P. Jacups. 2003. Intensity of rainfall and severity of melioidosis, Australia. Emerg. Infect. Dis. 9:1538-1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dance, D. A. B. 2000. Ecology of Burkholderia pseudomallei and the interactions between environmental Burkholderia spp. and human-animal hosts. Acta Trop. 74:159-168. [DOI] [PubMed] [Google Scholar]
- 8.Dance, D. A. B. 1991. Melioidosis: the tip of the iceberg? Clin. Microbiol. Rev. 4:52-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellison, D. W., H. J. Baker, and M. Mariappan. 1969. Melioidosis in Malaysia. I. A method for isolation of Pseudomonas pseudomallei from soil and surface water. Am. J. Trop. Med. Hyg. 18:694-697. [PubMed] [Google Scholar]
- 10.Feil, E. J., B. C. Li, D. M. Aanensen, W. P. Hanage, and B. G. Spratt. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finkelstein, R. A. 1966. Investigation of melioidosis in Thailand. SEATO Lab Annu. Rep. 1966:228-234. [Google Scholar]
- 12.Finkelstein, R. A., P. Atthasampunna, and M. Chulasamaya. 2000. Pseudomonas (Burkholderia) pseudomallei in Thailand, 1964-1967: geographic distribution of the organism, attempts to identify cases of active infection, and presence of antibody in representative sera. Am. J. Trop. Med. Hyg. 62:232-239. [DOI] [PubMed] [Google Scholar]
- 13.Gee, J. E., C. T. Sacchi, M. B. Glass, B. K. De, R. S. Weyant, P. N. Levett, A. M. Whitney, A. R. Hoffmaster, and T. Popovic. 2003. Use of 16S rRNA gene sequencing for rapid identification and differentiation of Burkholderia pseudomallei and B. mallei. J. Clin. Microbiol. 41:4647-4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Godoy, D., G. Randle, A. J. Simpson, D. M. Aanensen, T. L. Pitt, R. Kinoshita, and B. G. Spratt. 2003. Multilocus sequence typing and evolutionary relationships among the causative agents of melioidosis and glanders, Burkholderia pseudomallei, and Burkholderia mallei. J. Clin. Microbiol. 41:2068-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hall, B. G., and M. Barlow. 2006. Phylogenetic analysis as a tool in molecular epidemiology of infectious diseases. Ann. Epidemiol. 16:157-169. [DOI] [PubMed] [Google Scholar]
- 16.Holden, M. T. G., R. W. Titball, S. J. Peacock, A. M. Cerdeño-Tárraga, T. Atkins, L. C. Crossman, T. Pitt, C. Churcher, K. Mungall, S. D. Bentley, M. Sebaihia, N. R. Thamson, N. Bason, I. R. Beacham, K. Brooks, K. A. Brown, N. F. Brown, G. L. Challis, I. Cherevach, T. Chillingworth, A. Cronin, B. Crossett, P. Davies, D. DeShazer, T. Feltwell, A. Fraser, Z. Hance, H. Hauser, S. Holroyd, K. Jagels, K. E. Keith, M. Maddison, S. Moule, C. Price, M. A. Quail, E. Rabbinowitsch, K. Rutherford, M. Sanders, M. Simmonds, S. Songsivilai, K. Stevens, S. Tumapa, M. Vesaratchavest, S. Whitehead, C. Yeats, B. G. Battell, P. C. F. Oyston, and J. Parkhill. 2004. Genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei. Proc. Natl. Acad. Sci. USA 101:14240-14245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inglis, T. J. J., N. F. Foster, D. Gal, K. Powell, M. Mayo, R. Norton, and B. J. Currie. 2004. Preliminary report on the northern Australia melioidosis environmental surveillance project. Epidemiol. Infect. 132:813-820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones, A. L., T. J. Beveridge, and D. E. Woods. 1996. Intracellular survival of Burkholderia pseudomallei. Infect. Immun. 64:782-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Cougant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moore, R. A., S. Reckseidler-Zenteno, H. Kim, W. Nierman, Y. Yu, A. Tuanyok, J. Warawa, D. DeShazer, and D. E. Woods. 2004. Contribution of gene loss to the pathogenic evolution of Burkholderia pseudomallei and Burkholderia mallei. Infect. Immun. 72:4172-4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nachiangmai, N., P. Patamasucon, B. Tipayamonthein, A. Kongpon, and S. Nakaviroj. 1985. Pseudomonas pseudomallei in southern Thailand. Southeast Asian J. Trop. Med. Public Health 16:83-87. [PubMed] [Google Scholar]
- 22.Ngauy, V., Y. Lemeshev, L. Sadkowski, and G. Crawford. 2005. Cutaneous melioidosis in a man who was taken as a prisoner of war by the Japanese during World War II. J. Clin. Microbiol. 43:970-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pitt, T. L., S. Trakulsomboon, and D. A. B. Dance. 2000. Molecular phylogeny of Burkholderia pseudomallei. Acta Trop. 74:181-185. [DOI] [PubMed] [Google Scholar]
- 24.Reckseidler, S. L., D. DeShazer, P. A. Sokol, and D. E. Woods. 2001. Detection of bacterial virulence genes by subtractive hybridization: identification of capsular polysaccharide of Burkholderia pseudomallei as a major virulence determinant. Infect. Immun. 69:34-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reckseidler-Zenteno, S. L., R. DeVinney, and D. E. Woods. 2005. The capsular polysaccharide of Burkholderia pseudomallei contributes to survival in serum by reducing complement factor C3b deposition. Infect. Immun. 73:1106-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sexton, M. M., A. L. Jones, W. Chaowagul, and D. E. Woods. 1994. Purification and characterization of a protease from Pseudomonas pseudomallei. Can. J. Microbiol. 40:903-910. [DOI] [PubMed] [Google Scholar]
- 27.Smith, M. D., V. Wuthiekanun, A. L. Walsh, and N. J. White. 1995. Quantitative recovery of Burkholderia pseudomallei from soil in Thailand. Trans. R. Soc. Trop. Med. Hyg. 89:488-490. [DOI] [PubMed] [Google Scholar]
- 28.Spratt, B. G., W. P. Hanage, B. Li, D. M. Aanensen, and E. J. Feil. 2004. Displaying the relatedness among isolates of bacterial species—the eBURST approach. FEMS Microbiol. Lett. 241:129-134. [DOI] [PubMed] [Google Scholar]
- 29.Strauss, J. M., M. G. Groves, M. Mariappan, and D. W. Ellison. 1969. Melioidosis in Malaysia. II. Distribution of Pseudomonas pseudomallei in soil and surface water. Am. J. Trop. Med. Hyg. 18:698-702. [PubMed] [Google Scholar]
- 30.Suputtamongkol, Y., A. J. Hall, D. A. Dance, W. Chaowagul, A. Rajchanuvong, M. D. Smith, and N. J. White. 1994. The epidemiology of melioidosis in Ubon Ratchatani, northeast Thailand. Int. J. Epidemiol. 23:1082-1090. [DOI] [PubMed] [Google Scholar]
- 31.Vuddhakul, V., P. Tharavichitkul, N. Na-Ngam, S. Jitsurong, B. Kunthawa, P. Noimay, P. Noimay, A. Binla, and V. Thamlikitkul. 1999. Epidemiology of Burkholderia pseudomallei in Thailand. Am. J. Trop. Med. Hyg. 60:458-461. [DOI] [PubMed] [Google Scholar]
- 32.Warawa, J., and D. E. Woods. 2005. Type III secretion system cluster 3 is required for maximal virulence of Burkholderia pseudomallei in a hamster infection model. FEMS Microbiol. Lett. 242:101-108. [DOI] [PubMed] [Google Scholar]
- 33.Wetmore, P. W., and W. S. Gochenour, Jr. 1956. Comparative studies of the genus Malleomyces and selected Pseudomonas species. I. Morphological and cultural characteristics. J. Bacteriol. 72:79-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.White, N. J. 2003. Melioidosis. Lancet 361:1715-1722. [DOI] [PubMed] [Google Scholar]