Abstract
Currently, 51 human adenovirus (AdV) serotypes, which are divided into six species (A to F), are known. AdV infections are a major cause of morbidity and mortality in immunosuppressed individuals, particularly in allogeneic stem cell transplant (SCT) recipients. Any AdV species may cause life-threatening disease, but little information is available on the clinical relevance of individual serotypes. The use of serological testing for serotype identification is limited due to the impaired immune response during the posttransplant period. A new molecular approach to serotype identification is presented here that exploits variable regions within the hexon gene. All serotypes belonging to the species A, B, C, E, and F can be determined by fragment length analysis of a single PCR product. For species C, which is the most prevalent in many geographic regions, an alternative technique based on serotype-specific real-time quantitative PCR was established. Of 135 consecutive pediatric patients screened for AdV infections after allogeneic SCT, 40 tested positive. Detailed analysis revealed the presence of 10 different serotypes; serotypes 1 and 2 from species C (C01 and C02) showed the highest prevalence, accounting for 77% of the AdV-positive cases. Representatives of other species were observed less commonly: serotype A12 in 6.5%; serotype A31 in 4.5%; and B03, B16, C05, C06, D19, and F41 in 2%. The approach to rapid molecular serotype analysis presented here provides a basis for detailed studies on adenovirus epidemiology and on the transmission of nosocomial infections. Moreover, in view of the increasing importance of tailored therapy approaches, serotype identification may in the future have implications for the selection of the most appropriate antiviral treatment.
Adenoviruses (AdV) are pathogens causing serious infections in immunosuppressed patients, particularly in allogeneic stem cell transplant (SCT) recipients or in human immunodeficiency virus-positive individuals (2, 4, 22, 24, 28). Human AdV represent a large family, currently including 51 serotypes, which are divided into six species (A to F) (8, 26). We have recently demonstrated by a species-specific real-time quantitative PCR (RQ-PCR) approach covering the entire spectrum of human AdV that molecular detection of AdV in peripheral blood precedes the onset of life-threatening virus disease and provides a basis for early preemptive treatment (18). That study and a number of earlier reports revealed furthermore that any species can cause severe infections in immunosuppressed patients (12, 18). Rapid and reliable detection of all human AdV is therefore needed in order to permit early diagnosis of AdV infection and the timely onset of treatment. Individual AdV species are associated with certain clinical manifestations, and a species-specific response to antiviral therapy has been reported (21). However, little is known about the disease association of individual AdV serotypes and their sensitivity to antiviral therapy. Serotype-specific diagnosis may therefore be of interest in the context of clinical management of AdV-related disease. Detailed insight into the occurrence of specific serotypes and substrains could provide information important for the surveillance and control of AdV transmission within hospitals and individual wards. Moreover, serotype-specific analysis could be exploited for epidemiological studies to provide information on the incidence and distribution of infections by individual AdV serotypes, which might be of particular interest in the light of targeted immune therapy approaches.
Serological tests recognize serotype-specific epitopes in the hexon protein located on the surface of the virus particle (7). Currently available serological detection methods are rather time-consuming and do not permit the reliable detection of all AdV serotypes (7, 26). Moreover, the applicability of serological tests in immunosuppressed patients is limited. Other technical approaches to AdV serotype identification are therefore needed. The hexon protein comprises highly conserved portions and variable regions containing the serotype-specific domains (5). At the DNA level, these regions display significant variability in length among individual AdV serotypes. PCR assays facilitating amplification across variable regions of the hexon gene therefore provide a possible approach to molecular typing. Some of the existing type-specific PCR assays, however, only permit the detection of selected serotypes (11, 14, 20, 27, 29, 31); others are based on relatively complicated algorithms, including PCR amplification and subsequent digestion with different restriction enzymes (1). The lack of sequence information on all AdV serotypes and the existence of substrains within individual serotypes (6) have prevented the establishment of reliable and economic serotype-specific molecular assays.
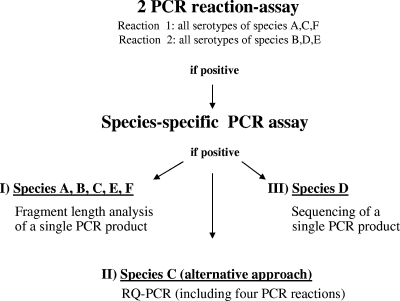
We have recently sequenced the hexon gene of all 34 hitherto-uncharacterized human adenovirus serotypes (9). The comprehensive sequence information has been exploited for the establishment of a recently published RQ-PCR assay that permits rapid detection of all human AdV serotypes but without identification of the species present (10). The AdV species (A to F) are determined by an assay including six RQ-PCRs, each of which specifically detects all serotypes belonging to a certain species (18) (see also Fig. 1). Based on the results of this assay, detailed serotype identification can be performed as described in the present study. (i) The identification of individual human AdV serotypes of the species A, B, C, E, and F is based on PCR amplification of a variable hexon gene region, and subsequent length analysis of the amplified PCR fragment by capillary electrophoresis coupled with fluorescence detection. (ii) For serotype detection within species C, which is most prevalent in many geographic regions, an alternative technical approach based on serotype-specific RQ-PCRs has been established. (iii) For serotype analysis within species D only, sequencing of a short fragment from the variable region V3, located on loop 2 of the hexon gene (9, 23), is necessary (Fig. 1). The approach to molecular typing described here focuses on PCR-fragment length analysis for the rapid identification of all serotypes belonging to the species A, B, C, E, and F. This approach covers the spectrum of AdV commonly observed in immunosuppressed patients after allogeneic stem cell or organ transplantation (15, 18, 28).
FIG. 1.
Algorithm of AdV typing. Previously described PCR assays were used to assess the presence of AdV infection and to determine the species (A to F) involved. AdV screening was performed by a two-reaction PCR assay permitting the detection of all serotypes of the AdV species A, C, and F and B, D, and E, respectively (10). In positive cases, the AdV species was identified by additional species-specific PCRs (18). Typing of AdV from the species A, B, C, E, and F was performed by PCR-based fragment length analysis, as described here. For species C, which is by far the most common in our and other geographic regions, an alternative approach to serotype identification based on RQ-PCR can be used, depending on the equipment available. Serotypes of species D can be identified by sequencing of a single ∼250-bp PCR product.
MATERIALS AND METHODS
Patients, virus strains, and isolation of DNA.
DNA was extracted from reference virus strains for all 51 AdV serotypes obtained from the American Type Culture Collection by using the QIAmp DNA blood minikit (QIAGEN, Hilden, Germany). Clinical specimens from 135 consecutive pediatric patients undergoing allogeneic SCT at our center (St. Anna Children's Hospital, Vienna, Austria) were screened for AdV infection after informed consent was obtained during the time period from January 2002 to October 2005. The screening was performed primarily in the absence of any signs of virus-related disease. The extraction protocol indicated above was used to isolate DNA for AdV analysis from specimens derived from peripheral blood. For investigation of AdV in stool samples, the QIAmp DNA Stool Minikit (QIAGEN) was used according to the manufacturer's recommendations.
PCR amplification and fragment analysis.
Primers for AdV species-specific PCR were placed within conserved hexon gene sequences flanking a variable region (nucleotide positions 411 to 689, based on the AdV C02 sequence) located on loop 1 of the hexon protein and displaying a highly variable length (9). A seminested PCR protocol was used to generate sufficient amounts of DNA for serotype analysis in samples with low AdV copy numbers (<104/ml in liquid specimens or per gram in solid specimens). The first-round PCRs were carried out in 25-μl reactions containing 1 U of QIAGEN HotStarTaq DNA polymerase (QIAGEN), 2.5 μl of 10× buffer, 2.5 mM MgCl2, 200 μM concentrations of each deoxynucleoside triphosphate (Invitrogen, Austria), 200 to 400 nM concentrations of the primers (Ingenetix, Austria; VBC Genomics Bioscience GmbH, Austria), and 6 μl of DNA template extracted as described above. The forward primers used were labeled with fluorescent dyes as indicated in Table 1. The amplification protocol included initial denaturation at 96°C for 10 min, followed by 30 cycles of denaturation at 95°C for 30 s, annealing at 50°C for 30 s, and extension at 72°C for 30 s, with a final elongation step at 60°C for 30 min. The set of forward primers indicated above was also used for the seminested PCR protocol in combination with different reverse primers (see Table 1). The seminested PCRs were performed under the conditions described above, except for the use of 1 μl of PCR product from the first reaction as a template and an annealing temperature of 55°C. For fragment length analysis of the amplicons, 1 μl of PCR product was diluted with 24 μl of HiDi-Formamide (Applied Biosystems, Foster City, CA) and 1 μl of internal lane standard 600 (ILS 600; Promega Corp., Madison, WI), and the DNA was denatured at 95°C for 3 min. Fragment analysis was performed by capillary electrophoresis with fluorescence-assisted detection using an ABI 3100 genetic analyzer (Applied Biosystems) with the injection parameters of 1 kV/5 to 10 s, 3 kV/15 s, and 6 kV/15 s. The sensitivity of the assay was determined by testing serial dilutions of DNA derived from AdV reference strains. Viral DNA was quantified by fluorometric analysis using the PicoGreen dsDNA quantitation kit (Molecular Probes, Inc., Eugene, OR) and an F-2500 fluorescence spectrophotometer (Hitachi, Japan). Sensitivity tests were performed by using AdV serotypes representative of each species, including A12, B07, C02, E04, and F40F. Serial dilutions of DNA corresponding to 102 to 107 virus particles per reaction were investigated by PCR and subsequent fragment length analysis using capillary electrophoresis and fluorescence detection. The reproducibility of the assay was documented by performing a minimum of five independent PCR amplification and fragment length analysis experiments for each serotype of the species A, B, C, E, and F.
TABLE 1.
Primer sequences for PCR-based AdV typing by fragment length analysis
| Primer (labeling)a | Oligonucleotide sequence (5′-3′) | Concn (μM) | Primer seminested PCR | Oligonucleotide sequence (5′-3′) | Concn (μM) |
|---|---|---|---|---|---|
| AdV A for (F) | CCAAAGGGCGCGCCYAATGC | 0.4 | |||
| AdV A rev 1 | CGAGCGTAGGAGCCATAGC | 0.2 | AdV A nested rev 1 | GCAGGGGTACATGGGTGTGG | 0.2 |
| AdV A rev 2 | CGGGCGTAAGACCCATAGC | 0.2 | AdV A nested rev 2 | GCACGGATACATGGGCGTAG | 0.2 |
| AdV B for 1 (F) | GCCCCAGCTTCAAACCCTAC | 0.2 | |||
| AdV B for 2 (F) | GTCCTAGCTTCAAGCCATAT | 0.2 | |||
| AdV B for 3 (F) | GTCCCAGTTTCAAACCCTAT | 0.2 | |||
| AdV B for 4 (F) | GCCCTAGCTTCAAGCCATAC | 0.2 | |||
| AdV B rev 1 | GTCTGGCAAATGAACCATAGC | 0.2 | AdV B nested rev 1 | CCATAGCAGGGTTTCATCTTTG | 0.2 |
| AdV B rev 2 | GTTTAGCAAAAGACCCATAGC | 0.2 | AdV B nested rev 2 | CCGTAGCAGGGTTTCATTTTAG | 0.2 |
| AdV B rev 3 | GCTTGGCGAAAGAACCATAGC | 0.2 | AdV B nested rev 3 | CCGTAGCATGGTTTCATCTTAG | 0.2 |
| AdV B rev 4 | GTTTGGCAAAGGACCCGTAGC | 0.2 | AdV B nested rev 4 | CCGTAGCATGGCTTCATGTTGG | 0.2 |
| AdV B rev 5 | GTCTCGCAAAAGACCCGTAGC | 0.2 | |||
| AdV C for (J) | ACTGCCTACAACGCTCTRGC | 0.4 | AdV C nested rev 1 | GTCTTTTTTAGTACTCTGCC | 0.2 |
| AdV C rev | CCRTAGCATGGTTTCATGGG | 0.4 | AdV C nested rev 2 | GTTTTTTTAAGCACTCTCCC | 0.2 |
| AdV C nested rev 3 | GTCTTTTTAAGAACCCTTCC | 0.2 | |||
| AdV C nested rev 4 | GTCTTTTTAAGGACTCTCCC | 0.2 | |||
| AdV E for (J) | CTGGCTCCCAAGGGAGCGCC | 0.4 | |||
| AdV E rev | AGTGTCCTTTAGAGCTCTGC | 0.4 | |||
| AdV F for (T) | TGCTGGATCGTGGTCCCAGC | 0.4 | AdV F nested rev 1 | CATGGCTGCATGGGCGTGG | 0.2 |
| AdV F rev | TGTTGGTTTAGCGTATGACC | 0.4 | AdV F nested rev 2 | CATGGTAACATTGGCGTGG | 0.2 |
Labeling: F, fluorescein; J, JO; T, TAMRA. for, forward; rev, reverse.
Detection of AdV species and selected serotypes by RQ-PCR.
Diagnostic tests for detection of all six AdV species were performed by RQ-PCR assays described previously (10, 18) (see Fig. 1). For type-specific RQ-PCR analysis of members of species C, primer and probe systems detecting the serotypes 1, 2, 5, and 6 in four separate reactions were designed by using Primer Express software (Applied Biosystems). All primers and probes bind within a variable region of the hexon gene and permit highly specific detection of the four serotypes (Table 2). The primer and probe sequences were carefully controlled for possible homology with other adenoviral and nonadenoviral sequences by using the BLAST software (National Center for Biotechnology Information). Moreover, each of the primer and probe sets was tested and proven not to cross-react with any AdV serotype from the same or other species. PCRs were set up in a total volume of 25 μl, including 12.5 μl of TaqMan universal master mix (Eurogentec, Seraing, Belgium), 1% formamide (Calbiochem, Darmstadt, Germany), 6 μl of DNA solution from patient samples, 400 nM concentrations of forward and reverse primers (VBC-GENOMICS Bioscience Research GmbH, Austria) and TaqMan probes at 200 nM. All TaqMan probes used were labeled with FAM (6-carboxyfluorescein) at the 5′ end and with 6-carboxyl-tetramethyl-rhodamine (TAMRA) as a quencher at the 3′ end (Eurogentec, Searing, Belgium). Amplifications were carried out by using a ABI Prism 7700 or 7900 (Applied Biosystems) for a total of 50 cycles. After an initial denaturation step for 10 min at 95°C, each cycle consisted of denaturation for 15 s at 95°C and annealing and primer extension for 60 s at 60°C. The reproducibility of identifying serotypes of species C by type-specific RQ-PCR was assessed by testing 10 replicate reactions for each of the serotypes. All patient specimens were analyzed in duplicate reactions. For the quantification of virus load, external standard curves were established for each serotype by using serial dilutions of fluorometrically quantified virus DNA preparations corresponding to defined virus particle equivalents, as described above.
TABLE 2.
Primer and probe sequences for real-time PCR detection of serotypes from AdV species C
| Primer or probe | Oligonucleotide sequence (5′-3′) | Nucleotide position (hexon gene)a | Concn (μM) |
|---|---|---|---|
| AdV1 forward | ATACCCAAACTG AAGGCAATCC | 592-614 | 0.4 |
| AdV1 probe (reverse) | ACTGAGATTCTCCAACCTGAGGTT | 620-643 | 0.2 |
| AdV1 reverse | ACTGAGATTCTCCAACCTGAGGTT | 645-668 | 0.4 |
| AdV2 forward | AACCTGTATACGCAGATCCTTCCTA | 607-632 | 0.4 |
| AdV2 probe (forward) | AGAACCTCAAATTGGCGAATCTCAGTGGA | 638-667 | 0.2 |
| AdV2 reverse | CCTGCCGCATTAGCATCAG | 675-694 | 0.4 |
| AdV5 forward | CACTCATATTTCTTACATGCCCACTATT | 890-918 | 0.4 |
| AdV5 probe (forward) | AGGAAGGTAACTCACGAGAACTAATGGGCCA | 920-951 | 0.2 |
| AdV5 reverse | GGCCTGTTGGGCATAGATTG | 953-973 | 0.4 |
| AdV6 forward | AAACCCTGCTATGGCTCATACG | 717-739 | 0.4 |
| AdV6 probe (forward) | CCACCAATTCCAACGGCGGACA | 747-769 | 0.2 |
| AdV6 reverse | TTTACCATTTTGTTCAACCATAACG | 772-797 | 0.4 |
All primer and probe sequences are located within the most variable region of the hexon gene, starting at nucleotide position 411.
Sequencing of PCR products.
For serotype identification within species D, a fragment derived from a variable region of the hexon gene encompassing a length of about 250 bp was amplified by PCR using a single primer set (forward, 5′-GCGGTGGACAGCTATGATCC-3′; reverse, 5′-AACTCTTCCACAGGTTGGCCTG-3′), and the resulting PCR product was subjected to direct sequencing, as indicated below. The PCRs were set up in a total volume of 50 μl containing 6 μl of DNA solution, 2 U of QIAGEN HotStarTaq DNA Polymerase (QIAGEN, Hilden, Germany), the appropriate 10× buffer, 200 μM concentrations of each deoxynucleoside triphosphates (Invitrogen GmbH, Austria), and 400 nM concentrations of each primer. Amplification was performed with an initial denaturation step at 96°C for 10 min, followed by 35 cycles of denaturation at 95°C for 30 s, annealing at 50°C for 45 s, and extension at 72°C for 2 min. Direct sequencing of the purified PCR products was performed in both directions with sense and antisense primers at the Vienna Bio Center (VBC-GENOMICS Bioscience Research GmbH). The sequencing reactions were carried out with the same primers used for the PCR amplification.
Time requirement and costs of molecular serotyping.
The procedure from sample collection to serotype identification by fragment length analysis, including all of the steps indicated above, can be performed in one and a half working days. If serotype identification by fragment length analysis is replaced by type-specific RQ-PCR, which is feasible for the representatives of species C, the entire procedure can be completed within one working day.
The costs of consumables for the complete procedure are in the range of 200 euros, when serotype identification is performed by fluorescence-based fragment length analysis using the four-capillary sequencer ABI 3100 genetic analyzer. The costs are considerably lower for serotype identification within species C based on type-specific RQ-PCR using the ABI Prism 7900 (about 70 euros for the entire procedure). These calculations are based on parallel processing of four specimens and include the costs of all control experiments involved.
RESULTS
AdV typing by fragment length analysis of specific hexon gene-derived PCR products.
Species-specific PCRs permitting the detection of all serotypes belonging to the AdV species A, B, C, E, and F have been designed. For some PCRs, a mixture of primers was needed, owing to mismatches between serotypes belonging to the same species, in order to provide optimal specificity. The forward primers were labeled with fluorescent dyes (see Table 1) to permit analysis of the PCR products by fluorescence-based capillary electrophoresis. For clinical samples with a lower virus load, a seminested PCR approach was used (Table 1) to ensure adequate sensitivity of detection. Specific PCR products of each serotype within a species yielded amplicons of different sizes (Table 3), thus permitting reliable serotype identification by fragment length analysis. The PCR product sizes were determined by using a fluorescence-based capillary sequencer, as described above, and reference strains of all human AdV serotypes were used as an external standard. All fragment sizes of individual AdV serotypes range between 253 and 389 bp (Table 3). In a number of instances, the actual product size of PCR amplicons derived from individual AdV serotypes differed from that expected on the basis of sequence analysis. These observations are most likely attributable to the properties of the capillary electrophoresis apparatus used or to migration shifts during electrophoresis caused by the fluorescence dye component. Under standardized assay conditions, the migration patterns are nevertheless highly reproducible. The maximum deviation between multiple (≥5) independent repeats of electrophoresis runs were ±1 nucleotide, depending on the presence or absence of adenosine residue addition by the DNA polymerase (3). The analysis of serial dilutions of different AdV serotypes revealed a detection limit of capillary electrophoresis after single-round PCRs at the level of 103 virus particle equivalents, while the sensitivity of seminested PCR assays was reproducibly at 10 virus copies per reaction.
TABLE 3.
Fragment sizes of serotype-specific PCR products
| Serotype | Fragment size (bp)
|
|
|---|---|---|
| First-round PCR | Seminested PCR | |
| AdV A | ||
| AdV12 | 272 | 253 |
| AdV18 | 279 | 262 |
| AdV31 | 282 | 259 |
| AdV B | ||
| AdV03 | 354 | 348 |
| AdV07 | 346 | 329 |
| AdV11 | 378 | 369 |
| AdV14 | 382 | 372 |
| AdV16 | 337 | 330 |
| AdV21 | 379 | 375 |
| AdV34 | 388 | 253 |
| AdV35 | 389 | 383 |
| AdV50 | 360 | 351 |
| AdV C | ||
| AdV01 | 378 | 351 |
| AdV02 | 375 | 348 |
| AdV05 | 342 | 315 |
| AdV06 | 368 | 342 |
| AdV E | ||
| AdV04 | 259 | |
| AdV F | ||
| AdV40 | 328.4 | 300 |
| AdV41 | 338 (335)a | 309 (306)a |
In the current series, patients who tested AdV positive (n = 40) by the RQ-PCR assays described earlier (10, 18) were shown to have infections by the species A, B, C, D, or F. Positive samples were further analyzed by the single-round or, if necessary, by the seminested PCR assay described here, coupled with typing by fragment length analysis. The only exception was a patient who tested positive for AdV species D, in whom the serotype was identified by sequence analysis. Reference strains of all relevant serotypes (species A, serotypes 12, 18, and 31; species B, serotypes 3, 7, 11, 14, 16, 21, 34, 35, and 50; species C, serotypes 1, 2, 5, and 6; species E, serotype 4; species F, serotypes 40 and 41) were used as an external fragment size standard to facilitate accurate type identification in clinical specimens (Fig. 2). The inclusion of an appropriate external standard in each PCR assay eliminated the problem of shifted fragment migration and permitted reliable serotype assignment in all patient specimens. Detailed analysis of AdV-positive clinical samples showed the highest prevalence of serotype 2 (43%), followed by serotypes 1 (34%), 12 (6,5%), 31 (4,5%), 3 (2%), 16 (2%), 5 (2%), 6 (2%), and 41 (2%). In two cases, we observed contemporaneous infection with serotypes belonging to different species (patients 5 and 6; Table 4). The data of AdV typing by PCR fragment length analysis were controlled by sequencing of the hexon gene amplification products and subsequent comparison with known sequences of all AdV serotypes available from the National Center for Biotechnology Information) database and revealed identical results in all instances.
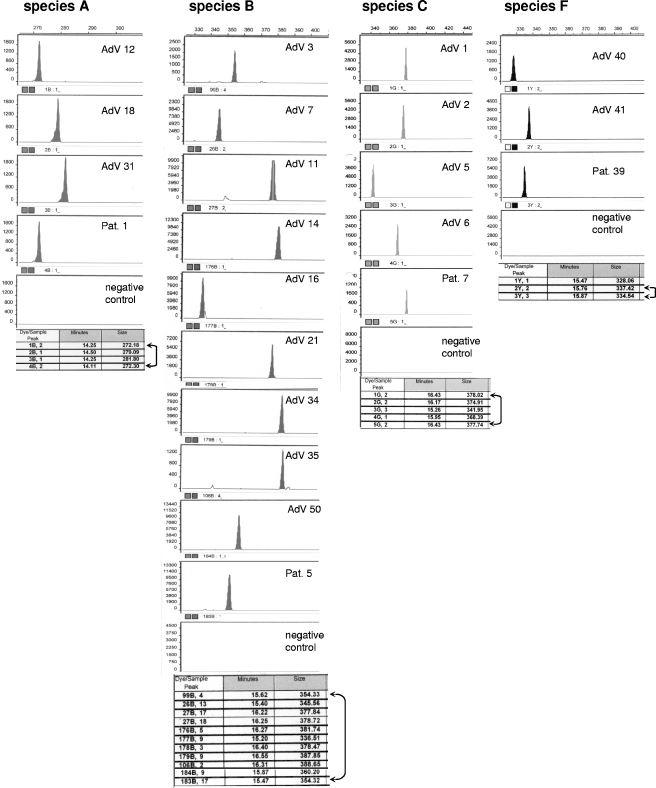
FIG. 2.
Examples of fragment analysis of AdV serotypes within species A, B, C, and F. The AdV serotypes present in patient specimens were determined by comparison of the respective fragment sizes detected with the external reference. Amplicons of ATCC strains representing all AdV serotypes belonging to species A, B, C, and F were used as an external reference for type identification in clinical samples. The examples shown include patient 1 (Pat. 1), who was positive for species A and revealed the presence of the serotype 12 (272 bp); patient 5, who had been diagnosed with an infection by AdV species B and who revealed the presence of serotype 3 (354 bp); patient 7, who was positive for AdV species C and showed the presence of serotype 1 (378 bp); and patient 39, who was positive for species F and who revealed the presence of serotype 41 (335 bp). The fragment size detected in this patient differed from the reference serotype by three nucleotides. This difference is attributable to the existence of two substrains of serotype 41 (see also Table 3).
TABLE 4.
Serotype analysis in AdV-positive specimens
| Patienta | AdV species | Specimen | Serotype |
|---|---|---|---|
| 1 | A | Stool | 12 |
| 2 | A | Stool | 12 |
| 3 | A | Stool | 12 |
| 4 | A | Stool | 31 |
| 5 | A | Stool | 31 |
| 5 | B | Stool | 3 |
| 6 | B | Stool | 16 |
| 5* | C | Stool | 1 |
| 6* | C | Stool | 1 |
| 7* | C | Stool | 2 |
| 8* | C | Stool | 2 |
| 9* | C | Peripheral blood | 1 |
| 10* | C | Stool | 1 |
| 11* | C | Stool | 2 |
| 12* | C | Stool | 1 |
| 13* | C | Stool | 1 |
| 14* | C | Stool | 1 |
| 15* | C | Stool | 2 |
| 16* | C | Stool | 2 |
| 17* | C | Stool | 2 |
| 18* | C | Stool | 1+2 |
| 19* | C | Stool | 2 |
| 20* | C | Stool | 2 |
| 21* | C | Stool | 1 |
| 22* | C | Stool | 1 |
| 23* | C | Stool | 2 |
| 24* | C | Stool | 2+5 |
| 25* | C | Stool | 2 |
| 26* | C | Stool | 2 |
| 27* | C | Stool | 2 |
| 28* | C | Stool | 1+2 |
| 29* | C | Stool | 2 |
| 30* | C | Stool | 2 |
| 32* | C | Stool | 2 |
| 33* | C | Stool | 1+2 |
| 34* | C | Stool | 1 |
| 35* | C | Stool | 1 |
| 36* | C | Stool | 6 |
| 37* | C | Stool | 1 |
| 38* | C | Stool | 2 |
| 39 | F | Stool | 41 |
| 40 | D | Stool | 19 |
*, Samples positive for any serotype of the AdV species C were tested using both fragment analysis and serotype-specific RQ-PCR assays.
In specimens positive for species D, the serotype was determined by sequencing.
Identification of AdV serotypes of species C by real-time PCR.
Earlier studies at our and other centers revealed a high prevalence of AdV species C (1, 15, 18). Within more than 100 consecutive patients investigated in our laboratory, 34 of 40 AdV-positive patients revealed the presence of species C. Due to the great predominance of this species in our geographic region, we have established a specific RQ-PCR detection system for each of the four serotypes of species C. Based on the serotype-specific sequences within variable regions of the hexon gene, primers and fluorescence-labeled TaqMan probes were designed to facilitate the specific detection of serotypes 1, 2, 5, and 6 in individual RQ-PCRs (Table 2). The primer and probe systems were tested for specificity using reference strains of all serotypes from species C. In a series of 10 replicate experiments, no cross-reactivity between the detection systems has been observed (data not shown). Standard curves established for each serotype of AdV species C permit virus quantification over a range of at least 7 logs (Fig. 3). For the assessment of virus copies per cell, a single-copy gene (β2 microglobulin) was tested in parallel by RQ-PCR, as described previously (30). In largely cell-free liquid specimens the virus number is indicated per milliliter of the sample, and in stool specimens the virus load is expressed as the number of particles per gram of material. The sensitivity of the assay permitted reliable detection of 102 virus particles per gram of the sample investigated. For reproducible quantification of virus load, however, the presence of ≥103 particles per gram were necessary.
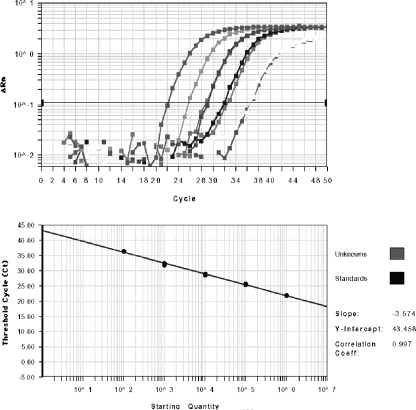
FIG. 3.
Amplification plot and standard curve for RQ-PCR detection of AdV serotype C01. The dynamic range of quantitative virus detection by RQ-PCR analysis is ≥7 logs, as demonstrated previously (10, 18). For the plots displayed, serial dilutions ranging from 102 to 106 virus particles per reaction were analyzed in duplicate by the RQ-PCR detection system specific for the serotype C01. The standard curves established for all other serotypes of species C, including C02, C05, and C06, were virtually identical to the curve displayed.
In patients positive for species C (n = 34), the results obtained by PCR fragment analysis were virtually identical to the serotype-specific RQ-PCR assays described herein. Only in two of the patients who displayed contemporaneous infection with two different AdV C serotypes (patients 18 and 24; Table 4), the subdominant serotype could only be identified by RQ-PCR, whereas the results of both assays were concordant for the dominant AdV serotype. This observation could be attributable to the >2-log difference between the copy numbers of the dominant and subdominant serotypes, which apparently precluded the detection of both viruses by fragment analysis due to competition.
Serotype identification within AdV species D by DNA sequencing.
Although species D, which comprises 32 serotypes, is the largest group within the AdV family, it has relatively rarely been observed in immunocompromised patients after allogeneic SCT (15). In the current cohort of patients studied, only one case positive for AdV species D has been observed. The hexon gene sequence displays very high similarity among serotypes of species D, thus precluding comprehensive serotype identification by fragment length analysis of PCR products derived from variable hexon gene regions. We have exploited a variable region of the hexon gene (V3) (9), permitting the identification of species D serotypes by sequence analysis of a short PCR product, as described previously (23). The only patient in this series who tested positive for species D was shown to carry AdV serotype D19 (see Table 4).
DISCUSSION
Molecular typing of human AdV in clinical specimens by PCR fragment analysis provides a rapid and economic approach to the identification of any serotype belonging to the AdV species A, B, C, E, and F. In view of the fact that AdV cause life-threatening infections in immunosuppressed patients, particularly in hematopoietic SCT recipients (13, 17, 24, 25), efficacious antiviral therapy is of paramount importance. Currently available antiviral agents with anti-AdV activity, including mainly cidofovir and ribavirin, have shown variable response rates in patients with invasive AdV infection (19). The recently introduced treatment with cytotoxic T cells is a very promising new approach to antiadenoviral therapy (16). However, the hexon gene, which carries the most important epitopes for immune recognition, displays significant differences between AdV serotypes, as revealed by a recent study performed in our laboratory (9). This implies that the efficacy of targeted immune therapy may, at least to some extent, be serotype dependent. Hence, rapid and reliable serotype identification could provide important information permitting the optimization of tailored antiadenovirus immune therapy.
Using the molecular typing approach described here, we have shown that the serotypes 1 and 2 from species C are by far the most frequently observed AdV types in our region. However, in view of the occurrence of various AdV species and serotypes in different areas, the availability of appropriate diagnostic methods is a prerequisite for epidemiological studies, which may in the future have an impact on the selection of the most effective antiviral treatment.
Complete information on the AdV hexon gene sequence determined in our laboratory has paved the way for the establishment of a serotype-specific detection assay. A potential problem inherent in AdV typing by different methods (1, 20, 23, 27), including PCR fragment length analysis, is the occurrence of substrains displaying insertion and/or deletion (indel) mutations, which can lead to difficulties in serotype assignment. The available data indicate, however, that differences between substrains are usually limited to point mutations (6), which do not affect migration of PCR products during capillary electrophoresis and thus do not interfere with correct serotype identification. In rare instances, electrophoresis products differing in length from reference strains used as external controls have been observed. These products may represent known substrains of certain serotypes, which are deposited in public databases. It is conceivable, however, that sequencing of a PCR product may be required in some cases to identify a hitherto-unknown substrain of a serotype carrying an indel mutation. A recent report on the analysis of serotypes based on sequence analysis of variable DNA stretches of individual AdV serotypes (23) is a useful diagnostic approach, particularly for the identification of serotypes belonging to species D, which are highly homologous at the DNA level. For the more commonly occurring serotypes of other AdV species, particularly those of species C, the assays presented here offer an equally informative, but considerably more economic diagnostic approach. Hence, the assays described here provide a readily applicable diagnostic tool that permits the rapid identification of human AdV serotypes relevant in immunosuppressed patients after allogeneic stem cell or organ transplantation and will help to expand our current understanding of the role of AdV in human infections. The new insights made possible by the novel diagnostic options may ultimately contribute to optimized treatment and, hence, to a better outcome of AdV-associated disease in immunocompromised individuals.
Acknowledgments
This study was supported by the Jubiläumsfonds of the National Bank of Austria (grant 11168).
REFERENCES
- 1.Allard, A., B. Albinsson, and G. Wadell. 2001. Rapid typing of human adenoviruses by a general PCR combined with restriction endonuclease analysis. J. Clin. Microbiol. 39:498-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blanke, C., C. Clark, E. R. Broun, G. Tricot, I. Cunningham, K. Cornetta, A. Hedderman, and R. Hromas. 1995. Evolving pathogens in allogeneic bone marrow transplantation: increased fatal adenoviral infections. Am. J. Med. 99:326-328. [DOI] [PubMed] [Google Scholar]
- 3.Brownstein, M. J., J. D. Carpten, and J. R. Smith. 1996. Modulation of non-templated nucleotide addition by Taq DNA polymerase: primer modifications that facilitate genotyping. BioTechniques 20:1004-1010. [DOI] [PubMed] [Google Scholar]
- 4.Bruno, B., R. A. Zager, M. J. Boeckh, T. A. Gooley, D. H. Myerson, M. L. Huang, and R. C. Hackman. 2004. Adenovirus nephritis in hematopoietic stem-cell transplantation. Transplantation 77:1049-1057. [DOI] [PubMed] [Google Scholar]
- 5.Crawford-Miksza, L., and D. P. Schnurr. 1996. Analysis of 15 adenovirus hexon proteins reveals the location and structure of seven hypervariable regions containing serotype-specific residues. J. Virol. 70:1836-1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crawford-Miksza, L. K., R. N. Nang, and D. P. Schnurr. 1999. Strain variation in adenovirus serotypes 4 and 7a causing acute respiratory disease. J. Clin. Microbiol. 37:1107-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crawford-Miksza, L. K., and D. P. Schnurr. 1994. Quantitative colorimetric microneutralization assay for characterization of adenoviruses. J. Clin. Microbiol. 32:2331-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Jong, J. C., A. G. Wermenbol, M. W. Verweij-Uijterwaal, K. W. Slaterus, P. Wertheim-Van Dillen, G. J. Van Doornum, S. H. Khoo, and J. C. Hierholzer. 1999. Adenoviruses from human immunodeficiency virus-infected individuals, including two strains that represent new candidate serotypes Ad50 and Ad51 of species B1 and D, respectively. J. Clin. Microbiol. 37:3940-3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ebner, K., W. Pinsker, and T. Lion. 2005. Comparative sequence analysis of the hexon gene in the entire spectrum of human adenovirus serotypes: phylogenetic, taxonomic, and clinical implications. J. Virol. 79:12635-12642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ebner, K., M. Suda, F. Watzinger, and T. Lion. 2005. Molecular detection of the entire spectrum of human adenoviruses by a two-reaction RQ-PCR assay. J. Clin. Microbiol. 43:3049-3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erdman, D. D., W. Xu, S. I. Gerber, G. C. Gray, D. Schnurr, A. E. Kajon, and L. J. Anderson. 2002. Molecular epidemiology of adenovirus type 7 in the United States, 1966-2000. Emerg. Infect. Dis. 8:269-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hierholzer, J. C. 1992. Adenoviruses in the immunocompromised host. Clin. Microbiol. Rev. 5:262-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Howard, D. S., G. L. Phillips, I. I., D. E. Reece, R. K. Munn, J. Henslee-Downey, M. Pittard, M. Barker, and C. Pomeroy. 1999. Adenovirus infections in hematopoietic stem cell transplant recipients. Clin. Infect. Dis. 29:1494-1501. [DOI] [PubMed] [Google Scholar]
- 14.Kajon, A. E., and G. Wadell. 1996. Sequence analysis of the E3 region and fiber gene of human adenovirus genome type 7h. Virology 215:190-196. [DOI] [PubMed] [Google Scholar]
- 15.Kojaoghlanian, T., P. Flomenberg, and M. S. Horwitz. 2003. The impact of adenovirus infection on the immunocompromised host. Rev. Med. Virol. 13:155-171. [DOI] [PubMed] [Google Scholar]
- 16.Leen, A. M., U. Sili, E. F. Vanin, A. M. Jewell, W. Xie, D. Vignali, P. A. Piedra, M. K. Brenner, and C. M. Rooney. 2004. Conserved CTL epitopes on the adenovirus hexon protein expand subgroup cross-reactive and subgroup-specific CD8+ T cells. Blood 104:2432-2440. [DOI] [PubMed] [Google Scholar]
- 17.Leruez-Ville, M., V. Minard, F. Lacaille, A. Buzyn, E. Abachin, S. Blanche, F. Freymuth, and C. Rouzioux. 2004. Real-time blood plasma polymerase chain reaction for management of disseminated adenovirus infection. Clin. Infect. Dis. 38:45-52. [DOI] [PubMed] [Google Scholar]
- 18.Lion, T., R. Baumgartinger, F. Watzinger, S. Matthes-Martin, M. Suda, S. Preuner, B. Futterknecht, A. Lawitschka, C. Peters, U. Potschger, and H. Gadner. 2003. Molecular monitoring of adenovirus in peripheral blood after allogeneic bone marrow transplantation permits early diagnosis of disseminated disease. Blood 102:1114-1120. [DOI] [PubMed] [Google Scholar]
- 19.Ljungman, P. 2004. Treatment of adenovirus infections in the immunocompromised host. Eur. J. Clin. Microbiol. Infect. Dis. 23:583-588. [DOI] [PubMed] [Google Scholar]
- 20.Mitchell, S., H. J. O'Neill, G. M. Ong, S. Christie, P. Duprex, D. E. Wyatt, C. McCaughey, V. J. Armstrong, S. Feeney, L. Metwally, and P. V. Coyle. 2003. Clinical assessment of a generic DNA amplification assay for the identification of respiratory adenovirus infections. J. Clin. Virol. 26:331-338. [DOI] [PubMed] [Google Scholar]
- 21.Morfin, F., S. Dupuis-Girod, S. Mundweiler, D. Falcon, D. Carrington, P. Sedlacek, M. Bierings, P. Cetkovsky, A. C. Kroes, M. J. van Tol, and D. Thouvenot. 2005. In vitro susceptibility of adenovirus to antiviral drugs is species-dependent. Antivir. Ther. 10:225-229. [PubMed] [Google Scholar]
- 22.Runde, V., S. Ross, R. Trenschel, E. Lagemann, O. Basu, K. Renzing-Kohler, U. W. Schaefer, M. Roggendorf, and E. Holler. 2001. Adenoviral infection after allogeneic stem cell transplantation (SCT): report on 130 patients from a single SCT unit involved in a prospective multi center surveillance study. Bone Marrow Transplant. 28:51-57. [DOI] [PubMed] [Google Scholar]
- 23.Sarantis, H., G. Johnson, M. Brown, M. Petric, and R. Tellier. 2004. Comprehensive detection and serotyping of human adenoviruses by PCR and sequencing. J. Clin. Microbiol. 42:3963-3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schilham, M. W., E. C. Claas, W. van Zaane, B. Heemskerk, J. M. Vossen, A. C. Lankester, R. E. Toes, M. Echavarria, A. C. Kroes, and M. J. van Tol. 2002. High levels of adenovirus DNA in serum correlate with fatal outcome of adenovirus infection in children after allogeneic stem-cell transplantation. Clin. Infect. Dis. 35:526-532. [DOI] [PubMed] [Google Scholar]
- 25.Seidemann, K., A. Heim, E. D. Pfister, H. Koditz, A. Beilken, A. Sander, M. Melter, K. W. Sykora, M. Sasse, and A. Wessel. 2004. Monitoring of adenovirus infection in pediatric transplant recipients by quantitative PCR: report of six cases and review of the literature. Am. J. Transplant. 4:2102-2108. [DOI] [PubMed] [Google Scholar]
- 26.Shenk, T., and M. S. Horwitz. 2001. Adenoviridae: the viruses and their replication, p. 2265-2326. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 27.Takeuchi, S., N. Itoh, E. Uchio, K. Aoki, and S. Ohno. 1999. Serotyping of adenoviruses on conjunctival scrapings by PCR and sequence analysis. J. Clin. Microbiol. 37:1839-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teramura, T., M. Naya, T. Yoshihara, G. Kanoh, A. Morimoto, and S. Imashuku. 2004. Adenoviral infection in hematopoietic stem cell transplantation: early diagnosis with quantitative detection of the viral genome in serum and urine. Bone Marrow Transplant. 33:87-92. [DOI] [PubMed] [Google Scholar]
- 29.Vabret, A., S. Gouarin, M. Joannes, C. Barranger, J. Petitjean, S. Corbet, J. Brouard, F. Lafay, J. F. Duhamel, B. Guillois, and F. Freymuth. 2004. Development of a PCR- and hybridization-based assay (PCR adenovirus consensus) for the detection and the species identification of adenoviruses in respiratory specimens. J. Clin. Virol. 31:116-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watzinger, F., M. Suda, S. Preuner, R. Baumgartinger, K. Ebner, L. Baskova, H. G. Niesters, A. Lawitschka, and T. Lion. 2004. Real-time quantitative PCR assays for detection and monitoring of pathogenic human viruses in immunosuppressed pediatric patients. J. Clin. Microbiol. 42:5189-5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu, W., and D. D. Erdman. 2001. Type-specific identification of human adenoviruses 3, 7 and 21 by a multiplex PCR assay. J. Med. Virol. 64:537-542. [DOI] [PubMed] [Google Scholar]