Abstract
Polymorphisms along the hepatitis B virus (HBV) genome have an impact on disease outcome, sensitivity to antiviral treatment, escape from vaccination, and laboratory diagnosis. We have designed a diagnostic tool based on duplex amplification of the whole HBV genome and a high-density DNA chip designed to detect 245 mutations, 20 deletions, and 2 insertions at 151 positions and to determine the genotype of the virus in serum. Assay performances were evaluated with 170 samples, characterized by determination of viral load and sequencing of the Pol, S, and precore genes and the basal core promoter. One hundred fifty-three samples (90%) could be amplified and analyzed by the chip. Only two samples with more than 103 genome copies/ml could not be analyzed. Genotype had no impact on analytical sensitivity. Reproducibility studies showed no difference between repeats for codon and genotype determination. Genotype determination by sequencing and the chip were concordant in 148 of 151 samples. Twelve thousand one hundred sixty-one codons were analyzed by both techniques. Only 89.4% could be determined by sequencing, and among the remaining 11,335 codons, 92.8% were identical by sequencing and the chip. Failures to identify an amino acid by the chip were mainly due to reduced hybridization efficiency attributed to unexpected polymorphisms. Optimization of the chip-based reagent for the analysis of the HBV genome is ongoing. This first evaluation showed that DNA chip technology can provide important information in relation to the clinical management of chronic hepatitis B.
There are more than 350 million people living with chronic hepatitis B virus (HBV) infection, and approximately 500,000 to 1.2 million deaths worldwide per year are attributed to HBV infection (24). Although an effective vaccine exists, the number of new cases around the world has not dramatically decreased. The prevalence of chronically infected people may even be underestimated, as is the case in France, where an epidemiological study conducted in 2003 to 2004 showed that there are still 300,000 chronic HBV carriers, representing 0.68% of the population (20). Chronic HBV infection can evolve through different stages with variable severity, from asymptomatic infection to cirrhosis and hepatocellular carcinoma (HCC). Currently, alpha interferon, lamivudine, adefovir, and entecavir are approved for the treatment of HBV disease and are used by clinicians. However, alpha interferon has a limited effect on HBV replication, and resistance against the two nucleoside inhibitors is relatively frequent. Several other molecules are currently in clinical trials (emtricitabine, clevudine, telbivudine, L-deoxycytosine, and L-deoxyadenosine) and may complement the existing treatments or represent alternatives when resistance occurs (2).
The virologic monitoring of HBV infection and its treatment is critical for efficiently controlling the disease and improving patient management. Several diagnostic tools can be used to evaluate the immune and virological parameters of the patient: hepatitis B s antigen (HBsAg), antibodies to HBsAg, hepatitis B e antigen (HBeAg), antibodies to HBeAg, antibodies (immunoglobulin M [IgM] and IgG) to hepatitis B core antigen, and viral load and genotypic assays.
Currently, genotypic assays focus on the major mutations in the region of the Pol gene coding for the virus reverse transcriptase (RT) that confer resistance to antiviral treatment (4). However, many other polymorphisms have been shown to have effects on the level of viral replication and the pathogenesis of liver disease and on vaccine and interferon treatment failure. Mutations in the viral polymerase confer resistance to lamivudine, entecavir, and adefovir (reviewed in reference 30), and new mutations will probably also affect the efficiency of molecules currently in clinical trials. As is the case for human immunodeficiency virus (15), resistance testing will probably be recommended to monitor HBV-infected patients in the different settings where the presence of resistant variants is suspected. Both genotypic and phenotypic resistance tests currently exist (16). Mutations in the S region that encode the viral envelope proteins have been associated with vaccine, immune treatment, and diagnostic escape (5, 13). Mutations in the a determinant of the S antigen do not seem to increase over time and to seriously affect vaccination efficiency, as shown by a 15-year survey in Taiwan following nationwide vaccination (19). However, they have been detected in patients from other geographic origins, and these variant virus strains may escape current diagnostic tests (12, 17, 18). Polymorphisms in the precore-, core-, pre-S1-, and pre-S2-coding regions and in the basal core promoter (BCP) may affect virus replication and impact disease outcome. Mutations in the pre-core gene, especially the stop codon at position 28, which results in HBeAg seronegativity, are more often seen in patients with persistent viremia and active liver disease (8-12). Mutations in the BCP, especially at positions 1762 and 1764, down-regulate HBeAg synthesis. They have been associated with fulminant hepatitis (28), more-severe liver damage (23, 31), and HCC (3). The X gene has been associated with evolution of HBV infection toward HCC. Polymorphisms monitoring the X-coding region may be of interest (32, 36). There are eight HBV genotypes, from A to H, that have different geographical distributions. As for hepatitis C virus, different genotypes may have different disease outcomes and sensitivities to treatments. Patients infected with genotypes C and D have a faster progression to liver fibrosis and HCC than those infected by genotype A or B (11, 14, 21, 25, 33, 34, 35). The duration of the HBeAg-positive replicative phase varies for the different genotypes, being shorter for A and D, which dominate in Africa, the Mediterranean region, the Middle East, and India, than for B and C, which dominate in Southeast and East Asia. This may explain why horizontal transmission is favored in the former countries, while transmission is mostly vertical in the latter countries (27). Resistance to lamivudine seems to appear more frequently and rapidly in genotype A than in genotype D (37, 38).
The small size of the HBV genome (3.2 kilobases) makes genotyping and polymorphism analysis possible for all viral genes. This may provide clinically relevant information to adapt patient management and adjust treatment strategies. Currently, genotypic analysis is performed mainly by either sequencing or the reverse-hybridization assay. Sequencing of the complete HBV genome is expensive and time consuming and requires highly skilled personal. Reverse-hybridization assays are limited to a few viral genome nucleotide positions.
We have designed a prototype genotypic assay based on PCR and DNA microarrays (the HBV DNA chip, or “the Chip”) for the determination from patient serum of HBV genotype and the detection of polymorphisms at 151 positions in the Pol-, S-, precore-, core-, pre-S1-, pre-S2-, and X-coding regions and the BCP region. This assay has been evaluated for genotype determination and mutation detection in four genomic loci—BCP, precore, S, and Pol—using a panel of plasma samples collected through the HepBvar network from patients originating from four European countries and which were characterized by sequencing. This study provides detailed information on the performance of the HBV DNA chip as a tool for patient monitoring and for research. It also highlights technical aspects to be considered for the optimization of a chip-based reagent for the characterization of infectious agents.
MATERIALS AND METHODS
Patients and samples.
One hundred seventy samples were collected through the HepBvar network in France (81 samples), Poland (50 samples), Italy (31 samples), and the United Kingdom (8 samples). Ten patients were diagnosed as acutely infected and 160 were chronic HBsAg carriers at various stages of hepatitis B. Patients were either nontreated or treated with alpha interferon or lamivudine or both. Viral load was determined with either in-house assays or with the Amplicor HBV assay (Roche Diagnostics, Pleasanton, Calif.). Viral genomes from all 170 samples have been sequenced in the Pol- and S-coding regions. Among them, 81 also had a reference sequence in the precore-coding region and 50 in the BCP. The viral genotype in each patient was determined by phylogenetic analysis based on sequences in the Pol- and S-coding regions. An aliquot of each sample was sent to the bioMérieux laboratory for chip analysis without knowledge of the viral genome sequence data. HBV clones used to study mixture detection were generated as described by Brunelle et al. (7).
HBV DNA extraction.
Viral DNA was extracted from HBV-infected plasma using a QIAamp UltraSens virus kit from QIAGEN (QIAGEN, Hilden, Germany) according to the manufacturer's protocol.
Amplification of the HBV DNA by PCR.
After being extracted, the whole DNA genome was amplified by duplex PCR to generate amplicons 1,509- and 1,721-bp long. HBV DNA was amplified with primers A1, A2, and A3 for the 1.5-kb amplicon and primers A4 and A5 for the 1.7-kb amplicon (see primer sequences below). The PCR mixture consisted of 20 μl of extracted DNA, 0.2 μM (each) primer, 0.3 mM (each) deoxyribonucleotide triphosphate, and 2 U of Fast Start Taq DNA polymerase (Roche, Basel, Switzerland) in an amplification buffer containing 2 mM MgCl2. Amplification was performed with the following steps: denaturation for 4 min at 94°C; 40 cycles of 20 s at 93°C, 45 s at 52.5°C, and 2 min 30 s at 68°C; and an elongation step of 7 min at 68°C. Then 5 μl of PCR product was subjected to agarose gel electrophoresis, using a 1% agarose standard (Eurobio, Cortaboeuf, France) run in 0.5× Tris borate EDTA (Eurobio). The following primers were used for the 1.5-kb amplicon: TCCCCAACCTCCAATCAC (forward primer A1), TCCCAAATCTCCAGTCAC (forward primer A2, genotype B-specific), and AAAAAGTTGCATGGTGCTGGT (reverse primer A3). For the 1.72-kb amplicon, the following primers were used: CTTTTTCACCTCTGCCTAATCA (forward primer A4) and GGGACTGCGAATTTTGGC (reverse primer A5).
Labeling and cleavage.
Amplicons were biotinylated as previously described (6). Briefly, a 45-μl aliquot of each positive PCR product was biotin labeled by mixing it with 83 μl of RNase- and DNase-free water (Sigma) and 100 μl of 0.1 M meta-biotin-phenylmethyl-diazomethyl (m-bioPMdam; bioMérieux, Marcy L’Etoile, France) at 95°C for 25 min in a dry bath. Depurination-mediated cleavage was performed in 8.8 mM HCl (22 μl of 0.1 N HCl per assay) in a final volume of 250 μl at 95°C for 5 min. Fragmented, labeled DNA was purified with a QIAquick 8 PCR purification kit according to the manufacturer's protocol (QIAGEN, Hilden, Germany) and eluted in 120 μl of elution buffer.
DNA chip hybridization and reading.
A mixture of 100 μl of the purified biotin-labeled products was hybridized on a high-density DNA chip (designed by bioMérieux and manufactured by Affymetrix, Santa Clara, Calif.) in a final volume of 500 μl containing 400 μl of hybridization buffer (12× SSPE [1× SSPE is 0.18 M NaCl, 10mM NaH2PO4, and 1 mM EDTA, pH 7.7], 0.06% antifoam, 0.09% sodium azide, and 0.5 pmol/assay of control oligonucleotide), denatured at 95°C for 10 min and hybridized onto the array, using the automated Affymetrix GeneChip Fluidic Station 400, at 45°C for 1 h. After the hybridization step, the DNA chip was washed at 30°C in wash buffer type A containing 2× SSPE, 0.03% Triton X-100, 0.006% antifoam, and 0.027% sodium azide, then in wash buffer type B containing 6× SSPE, 0.03% Triton X-100, 0.006% antifoam, and 0.027% sodium azide at the same temperature. The DNA chip was stained for 15 min at 35°C in 510 μl of a solution containing 500 μl of staining buffer (100 mM Tris, 1 M NaCl, 0.05% Tween-20, 0.005% antifoam, and 0.09% sodium azide, [pH 7.2]), 5 μl of bovine serum albumin (0.5 mg/ml; Gibco-BRL, Gaithersburg, Md.), and 5 μl of streptavidin-R-phycoerythrin conjugate (3 μg/ml; Dako, Glostrup, Denmark). A final wash was then performed in wash buffer type B at 25°C. A Gene Array scanner (a confocal laser reader from Agilent, Palo Alto, Calif.) with a pixel resolution of 3 μm and a wavelength of 570 nm was used to detect the fluorescence signal emitted by the target bound to a given probe of the chip.
DNA chip design and results analysis.
Each base was identified using a set of four 20-mer oligonucleotide probes. All probes have the same sequence, matching the target sequence except at position 12, corresponding to the interrogated position, where they have one of the four possible nucleotides. The correct base was selected if the intensity of the fluorescence signal generated by the corresponding probe was at least 1.2 times higher than the intensity of any of the three other probes. To detect the presence of a mutant codon, at least 5 consecutive bases, including the codon bases, were resequenced. The probe sets for a given amino acid constitute a tile. Tiles of five bases have been used to identify wild-type codons: the three codon bases plus one upstream base and one downstream base. Tiles of seven bases are used to identify mutant codons: the mutated base, three upstream bases, and three downstream bases. Because natural polymorphisms and other mutations can affect the hybridization of probes, comprehensive sequence databases were built and different tiles containing polymorphisms significantly represented in these databases as well as described mutations were used to identify a wild-type or mutant codon. There are 83,048 probes on the surface of the chip. A percentage of the homology between the experimental sequence and the different reference sequences tiled on the array was assigned to each interrogated position (the base call). The ratio between the median intensity fluorescence signals of probes in a tile that correctly identify a base and the median background in the area of the chip where these probes are located (It/b) was also calculated. This parameter provides confidence in the robustness of tile selection. Probe array cell intensities, base calls, the It/b ratios, and reports were generated by GeneChip version 3.2 software developed by Affymetrix and Test Platform software developed by bioMerieux. The report sheet included the codon positions, amino acid substitutions, and corresponding nucleotide substitutions identified as different from those of the wild-type reference HBV sequence (genotype A; GenBank accession number X02763) in the region of interest.
RESULTS
The HBV DNA chip allows the identification of genotype and the detection of mutations in the pre-S1, pre-S2, S, precore, core, X, and Pol genes and the BCP region. A duplex amplification has been optimized to cover the whole HBV genome. A 1.5-kb amplicon covers half of pre-S2, Pol, S, and BCP and two-thirds of X. The other amplicon (1.7 kb) covers the remaining X and pre-S2, precore and core, and Pol regions coding for the terminal protein and small protein. Table 1 lists the positions and mutations for which probes have been included on the chip. One hundred sixty-one positions along the HBV genome have probes for resequencing; 245 mutations, 20 deletions, and 2 insertions were tiled.
TABLE 1.
Positions and mutations for which probes have been included on the chip
| Gene | Position on codon | WT amino acida | Mutant amino acid(s) | Gene | Position on codon | WT amino acida | Mutant amino acid(s) | |
|---|---|---|---|---|---|---|---|---|
| Pol | tp17 | G | Del | s207 | V | I, M, L, E | ||
| tp18 | T | Del | s210 | S | T, R, N | |||
| sp8 | V | D, F, S, L | s213 | I | M, L | |||
| sp35 | A | D, V, S, T | s216 | L | -, S | |||
| rt70 | T | S | s220 | F | C, L | |||
| rt71 | N | T | s229 | L | V, M | |||
| rt86 | A | P | s306 | P | S | |||
| rt92 | P | L | Core | BCP 1752 | A | T, G, C | ||
| rt100 | H | R, N | BCP 1753 | T | A, C, G | |||
| rt102 | L | I, V | BCP 1754 | T | C, G | |||
| rt115 | L | V | BCP 1757 | G | A | |||
| rt128 | T | N, A | BCP 1762 | A | T | |||
| rt133 | H | Q | BCP 1764 | G | A | |||
| rt135 | S | H, Y, C | BCP 1766 | C | T, G | |||
| rt153 | W | Q, R | BCP 1768 | T | A | |||
| rt173 | V | L, G | BCP 1770 | G | T | |||
| rt177 | P | L | BCP 1802 | C | T | |||
| rt180 | L | M, L | BCP 1803 | G | T, C | |||
| rt181 | A | G | BCP 1809 | G | T | |||
| rt184 | T | N, S | BCP 1810 | C | T | |||
| rt190 | V | M, I | BCP 1811 | A | C, G | |||
| rt204 | M | V, I | BCP 1812 | C | T | |||
| rt207 | V | I, M, L, E | BCP 1748-1758 | Del | ||||
| rt212 | K | R | BCP 1750-1770 | Del | ||||
| rt213 | S | T, N | BCP 1753-1772 | Del | ||||
| rt217 | R | L | BCP 1755-1759 | Del | ||||
| rt229 | L | V, M | BCP 1755-1771 | Del | ||||
| rt331 | L | P | BCP 1755-1774 | Del | ||||
| rt332 | S | C | BCP 1766-1773 | Del | ||||
| rt336 | M | L, Q, I | BCP 1758-1765 | Del | ||||
| S | pre-S1 5 | S | Y, L, T | BCP 1759-1766 | Del | |||
| pre-S2 1 | M | V | BCP 1760-1765 | Del | ||||
| pre-S2 13 | Q | L, T, I | BCP 1757-1777 | Del | ||||
| s40 | N | S | BCP 1763-1770 | Del | ||||
| s85 | F | C | BCP 1765-1772 | Del | ||||
| s96 | V | G | BCP 1767 | Ins | ||||
| s98 | L | V | BCP 1767-1774 | Del | ||||
| s100 | Y | C, F | BCP 1768-1774 | Del | ||||
| s101 | Q | K, R | BCP 1770-1776 | Del | ||||
| s110 | I | T, L | pre-C 1 | M | -, L, P, K, T, R, I | |||
| s114 | T | S, Q | pre-C 2 | Q | -, L, R | |||
| s115 | T | A, Q | pre-C 9 | I | T, N | |||
| s118 | T | M, V | pre-C 11 | S | L | |||
| s120 | P | T, Q | pre-C 13 | T | T, S, L | |||
| s121 | C | R | pre-C 15 | P | S, L | |||
| s122 | K | R | pre-C 16 | T | I | |||
| s123 | T | I, N | pre-C 17 | V | F, L | |||
| s125 | T | M, L | pre-C 21 | K | - | |||
| s126 | T | I, S, A | pre-C 25 | G | G | |||
| s127 | P | S, T, L | pre-C 28 | W | R, - | |||
| s128 | A | V | pre-C 29 | G | S, D | |||
| s129 | Q | N, H, R, K, L | C2 | D | Ins | |||
| s130 | G | N, R, E, D, S | C5 | P | T, S, A | |||
| s131 | N | T | C27 | V | I, N | |||
| s134 | F | I, Y, L | C36 | A | Del | |||
| s138 | C | Y, W | C38 | Y | H, D, F | |||
| s140 | T | S, I | C40 | E | D, N, T | |||
| s141 | K | E | C60 | L | V, I | |||
| s142 | P | S | C 77 | E | D, N, Q, A | |||
| s143 | T | M, S, L | C 84 | L | S, A, Q | |||
| s144 | D | A, E | C 97 | I | L, F | |||
| s145 | G | R, K | C 130 | P | T, I, L, S, Q | |||
| s146 | N | S | X | x26 | R | C, S, H | ||
| s157 | A | D, V, G | x30 | L | V, F, I | |||
| s158 | F | L, S | x80 | E | R, K, A | |||
| s159 | A | G | x82 | T | A, S | |||
| s160 | K | R | x86 | H | S, P, R, N, D | |||
| s161 | Y | F | x87 | Q | R, H, W, M, L, G, - | |||
| s168 | V | A, I | x88 | I | N, H, S, G, F, V, A, M | |||
| s171 | S | Y | x94 | H | Y | |||
| s172 | W | - | x101 | P | S, F, A, T | |||
| s175 | L | S | x102 | A | G, V, S, T | |||
| s178 | P | Q | x103 | A | K, E, D | |||
| s189 | T | I | x116 | V | L | |||
| s192 | L | P | x123 | L | S, F | |||
| s196 | W | L | x125 | E | N, G, A | |||
| s198 | M | I | x127 | I | S, M, N, L, G, V, F, T | |||
| s199 | W | - | x130 | K | M | |||
| s204 | S | R, N, G, L | x131 | V | T, I, S | |||
| s205 | L | R | x144 | A | S, V, P |
The wild-type (WT) sequence is that of genotype A (GenBank accession number X02763).
Del, deletion; Ins, insertion; -, no amino acid.
Cutoff determination.
The base call and the It/b ratio were used to select tiles. Ideally, the base call should be 100%, but unexpected polymorphisms may be present in the test viral genome although they are not represented on the chip. In addition, hybridization efficiency may be low when using some of the probes of a given tile. Thus, tiles with less than a 100% base call were also considered. The following algorithm was tested: a tile was retained if at least five bases were correctly identified and the It/b was at least two. The amino acid assigned by the chip is the one which has the highest base call associated with the highest It/b ratio. If no tile meets these criteria or if a tile corresponding to an unexpected amino acid is detected as the first one, the result is considered indeterminate. Besides the tile with the highest base call and It/b, if a tile corresponding to another amino acid expected at that position meets the criteria, then a mixture between the two amino acids at that position is considered. This algorithm has been used to determine the cutoff value. Ten samples with viral loads ranging from 103 to 1010 copies of the HBV genome per ml were tested on the chip. Ten positions (rt173, rt180, rt204, rt207, s120, s129, s145, s171, preC28, and preC29) were used to establish the cutoff values. This panel contains all positions in S, RT, and the precore for which mutations have been described as playing a major role in resistance to treatment or in escape from vaccination or in impacting the course of HBV disease. The expected amino acid at these positions was determined by sequencing and considered as the reference. A total of 712 “positive” tiles (tiles corresponding to the expected amino acid) and 1,267 “negative” tiles (tiles corresponding to another amino acid) were analyzed. Using the algorithm, 236 tiles were retained. The mean It/b ratio was 41.3 (range, 2-211). Ninety-seven of one hundred codons were correctly identified. The three failures occurred in the samples with the lowest viral loads at positions rt204, s120, and s129. Only 16 among 1,267 “negative” tiles were retained, resulting in a specificity of 98.7%. None of these tiles were detected as the first tile. In six cases, these tiles corresponded to mixtures (s129Q/K, s145K or G/R, rt204I/V, and preC29G/D or G/S).
Analytical sensitivity.
One hundred seventy plasma samples, collected in four European countries through the HepBvar network, were analyzed. The viral load and genotype of all samples were known. The distribution of genotypes as determined by sequencing was representative of a European origin of patients, with a large majority of genotypes A (61 samples) and D (74 samples). No genotype G, only 1 genotype F (from Italy), and 2 genotypes H (from Poland) were included in the study.
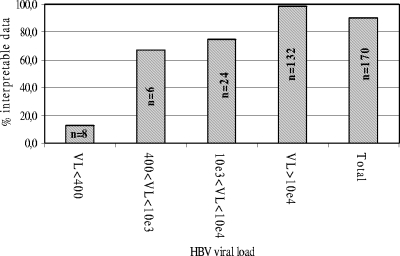
Among the 170 samples, 153 (90%) could be amplified by the duplex PCR protocol and subsequently analyzed using the chip; i.e., an amino acid could be identified in the majority of the positions and the genotype of the virus could be determined. Failure to amplify samples is likely to be due to low viral load (Fig. 1). All the samples which could not be analyzed had a viral load equal to or lower than 4.3 × 104 genome copies/ml and 9 of 17 (52.9%) had a viral load below 103 copies/ml. It is likely that difficulties in amplifying low-viral-load samples are due to the long lengths of the two amplicons (1.5 kb and 1.7 kb) that must be generated according to the PCR protocol. Amplification efficiency was not significantly associated with viral genotypes (data not shown).
FIG. 1.
Analytical sensitivity of the chip. Viral load (VL) is expressed as the number of HBV genome copies/ml for a serum sample. Data were considered interpretable when an amino acid could be identified in the majority of the positions and the genotype of the virus could be determined.
Reproducibility.
To assess reproducibility, an HBV-positive plasma sample was tested 11 times, using the complete process from DNA extraction to chip hybridization and reading. The sequence of the viral genome present in this sample was known, and its genotype was determined as D by phylogenetic analysis. Results obtained for the same 10 positions that were used for cutoff determination are shown in Table 2. In that sample, indeterminate results were obtained at positions rt204 and s129. Amino acids at other positions were correctly identified. For all positions, the same results were obtained in all repeats. The mean coefficient of variation (CV) of the It/b ratios was 18.6% (range, 14% to 23.7%). For all 90 positions that were analyzed, the mean coefficient of variation was 20.7% (range, 9.7% to 63%; data not shown). The highest CV was obtained at position preC25, where the correct amino acid was nevertheless identified in each repeat, with a mean It/b ratio of 42.7%.
TABLE 2.
Reproducibility of the chipa
| Codon | Expected amino acid | Amino acid determined by the chip in experimentb:
|
Mean It/b ratio | SD | CV (%) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | |||||
| rt173 | V | V | V | V | V | V | V | V | V | V | V | V | 45.81 | 7.43 | 16.2 |
| rt180 | L | L | L | L | L | L | L | L | L | L | L | L | 75.7 | 14.44 | 19.1 |
| rt204 | M | ind | ind | ind | ind | ind | ind | ind | ind | ind | ind | ind | 1.44 | 0.31 | 21.9 |
| rt207 | V | V | V | V | V | V | V | V | V | V | V | V | 51.87 | 7.48 | 14.4 |
| rt120 | P | P | P | P | P | P | P | P | P | P | P | P | 10.61 | 2.51 | 23.7 |
| rt129 | Q | ind | ind | ind | ind | ind | ind | ind | ind | ind | ind | ind | 1.05 | 0.17 | 16.2 |
| rt145 | G | G | G | G | G | G | G | G | G | G | G | G | 22.57 | 4.45 | 19.7 |
| rt171 | S | S | S | S | S | S | S | S | S | S | S | S | 75.70 | 14.44 | 19.1 |
| Precore 28 | W | W | W | W | W | W | W | W | W | W | W | W | 18.05 | 2.90 | 16.1 |
| Precore 29 | G | G | G | G | G | G | G | G | G | G | G | G | 16.50 | 3.26 | 19.8 |
Genotyping results showed similar reproducibility: The genotype determined by the chip in all 11 experiments was D, which was the expected genotype. For the genotyping experiments, the mean It/b ratio was 72.90% (standard deviation [SD], 1.13%) and the CV was 1.55%.
ind, indeterminate result.
In 11 repeats, the genotype determined by the chip was D, as expected from sequence analysis, with a very low CV for the base call (1.55%). The difference between the base call for genotype D and the second-highest genotype (genotype G; base call, 67%; standard deviation, 0.9%) insures robust genotyping.
Mixture detection.
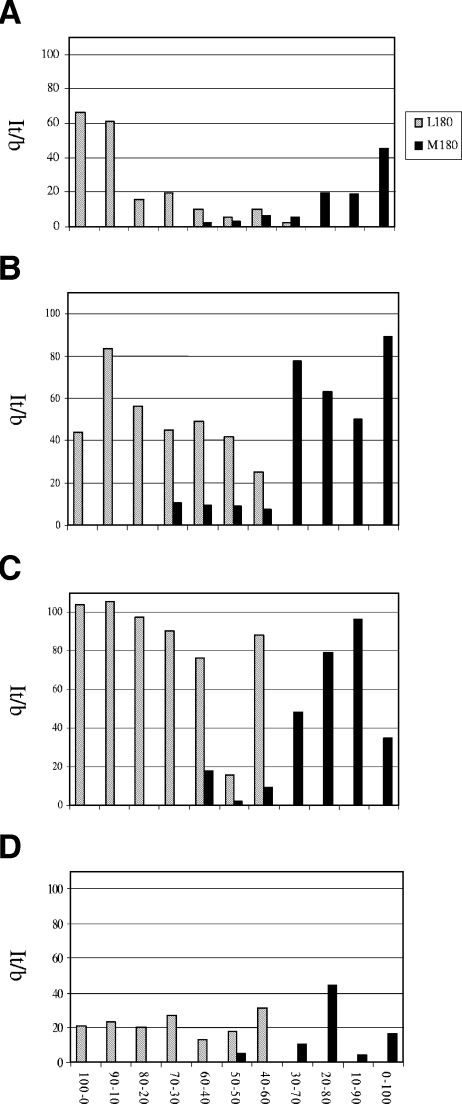
To demonstrate the capacity of the chip reagent to detect mixtures of variants, we prepared mixtures at different ratios of two cloned HBV genomes in HBV-negative plasma. One viral genome harbored the wild-type sequence, and the other has a mutation at position rt180 (L180M). The final virus concentration ranged from 104 to 107 copies of the HBV genome/ml. Figure 2 shows the results obtained at position rt180 for the It/b values of L180 and L180M. The values are those obtained for the first tile and for the second tile corresponding to the expected alternative amino acid. The minor population can generally be detected when it represents 30% to 40% of the mixture, except at the lowest virus concentration tested for which L180M could be detected when present as 50% of the whole viral population.
FIG. 2.
Detection of L180 and L180M mixtures. Artificial mixtures of an HBV genome with a wild-type sequence and another one with a mutation at position rt180 (L180M). (A) Total HBV genome copy number for 107/ml of serum. (B) Total HBV genome copy number for 106/ml of serum. (C) Total HBV genome copy number for 105/ml of serum. (D) Total HBV genome copy for 104/ml of serum. It/b is the ratio between the median intensity fluorescence signals of probes in a tile that correctly identify a base and the median background in the area of the chip where these probes are located.
Genotyping.
Among the 153 plasma samples that could be analyzed, the chip identified a genotype in 151 cases. As no probes corresponding to genotype H had been included on the chip, the two samples that could not be genotyped could belong to H. The chip and sequencing were able to determine the same genotype in 148 of 151 samples (98%). All genotyping results for B, C, and F were concordant. One sample was identified as genotype D by sequencing and A by the chip, and one as genotype D by sequencing and C by the chip. A sample with a viral load of <200 copies/ml was determined as genotype E by sequencing and D by the chip. Apart from this sample, there was no evidence for a link between viral load and discrepancies between sequencing and the chip.
Detection of mutations.
All 153 samples which could be amplified and tested with the chip had a reference sequence in the Pol and S open reading frames (ORFs), 75 in the precore gene, and 46 in the BCP. Only 89.4% of the 12,681 codons analyzed in that panel had a sequence reference result. For the remaining 1,346 codons, sequencing gave an indeterminate result. A total of 11,335 codons were thus used for the comparison between the two techniques. Table 3 shows that the overall concordance between results found by sequencing and by the chip is 92.8%, with no significant variation according to the ORF. A large majority of the 815 discrepant results are due to the fact that no tile met the criteria of the selection algorithm described above for the chip because of weak hybridization.
TABLE 3.
Overall concordance between the chip and sequencing
| Gene | No. of codons identified by sequencing | No. of codons with concordant sequencing and chip results | % Concordance |
|---|---|---|---|
| RT | 3,179 | 2,990 | 94.1 |
| S | 7,264 | 6,686 | 92.0 |
| Precore | 892 | 844 | 94.6 |
| Total | 11,335 | 10,520 | 92.8 |
To demonstrate the capability of the chip to detect mutations and evaluate technical difficulties that might be encountered, we evaluated a sample of representative, clinically relevant codons and nucleotide positions located in several HBV ORFs. Twenty-six positions were analyzed by the chip in the Pol ORF, two in the terminal protein ORF, and two in the small protein ORF, corresponding respectively to 38 mutations, 2 deletions, and 8 mutations (Table 1). Table 4 shows the results obtained at positions rt173, rt180, rt204, and rt207, which are most often mutated when resistance to lamivudine develops. When that occurs, one of two amino acids, valine or isoleucine, replaces the wild-type methionine at position rt204. Ninety-three of 153 samples (60.8%) were found concordant following testing by chip analysis and by sequencing, including four samples that gave an indeterminate result in both cases. The chip correctly detected all 20 valine residues identified by sequencing but only 66 of 101 methionine and 3 of 18 isoleucine residues. Weak hybridization of the amplified material to the chip probably explains the undetermined results in the 50 samples.
TABLE 4.
Comparison of results for the chip and sequencing in determining the major nucleotide positions in RT, S, pre-core and BCP
| Gene and sequence | No. of specimens with indicated concordant resultsa
|
No. of specimens with indicated discrepancy in results (sequencing/chip)a
|
Correlation (%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Wild-type | Mutant | Mixture | Ind | Wild-type/mutant | Mutant/wild-type | Mixture/wild-type or mutant | Wild-type or mutant/mixture | Different mutant | Ind/mutant or wild-type | Wild-type or mutant/ind | ||
| RT | ||||||||||||
| V173L | 147 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 4 (1L and 3V) | 1 (1V) | 96.1 |
| L180M | 114 | 27 | 2 | 0 | 0 | 1 | 2 (1 L and 1 M) | 1 (1 M) | 0 | 5 (4L and 1 ML) | 1 (1M) | 93.5 |
| M204I or V | 66 | 23 (3 I and 20 V) | 0 | 4 | 1 (1 V) | 0 | 4 (1 IM/V and 3 VM/V) | 2 (2 I/VI) | 2 (2 I/V) | 1 (1 M) | 50 (35 M and 15 I) | 60.8 |
| V207L | 138 | 1 (1 L) | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 4 (4 V) | 9 (3 V and 6 I) | 91.5 |
| S | ||||||||||||
| P120S, Q or T | 138 | 1 (1 S) | 0 | 0 | 1 (1 T) | 1 | 0 | 0 | 0 | 4 (1 L and 3 V) | 1 (1 V) | 90.9 |
| Q129H | 128 | 1 | 2 | 0 | 0 | 1 | 2 (1 L and 1 M) | 1 | 0 | 5 (4 L and 1 ML) | 1 (1 M) | 85.6 |
| G145R K | 115 | 0 | 0 | 0 | 3 (3 K) | 1 (1 K) | 0 | 7 (1 RG and 6 KG) | 0 | 1 (1 M) | 26 (26 G) | 75.2 |
| Precore | ||||||||||||
| W28 | 40 | 29 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 2 (2 W) | 0 | 92.0 |
| G29D | 49 | 18 | 0 | 0 | 1 | 2 | 0 | 0 | 0 | 5 (3 G and 2 GD) | 0 | 89.3 |
| BCP | ||||||||||||
| 1762 | 24 | 3 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 17 | 58.7 |
| 1764 | 22 | 4 | 0 | 0 | 0 | 1 | 2 | 0 | 0 | 0 | 17 | 56.5 |
Ind, indeterminate result.
Fifty-one positions were analyzed in the S ORF, one in the pre-S1 ORF, and two in the pre-S2 ORF, corresponding to, respectively, 92, 3, and 4 mutations (Table 1). We selected three major positions where mutations have been shown to reduce vaccination efficiency: s120, s129, and s145 (Table 4). At position s145, concordance between sequencing and the chip was only 75.2% (115 of 153 samples). In 26 samples, the chip was not able to identify an amino acid because of weak hybridization.
Twenty-four positions in the BCP, 12 in the precore region, and 11 in the core gene can be analyzed by the chip, corresponding respectively to 42, 28, and 29 mutations (Table 1). In the precore ORF, mutations at two amino acid positions (28 and 29) are known to affect HBV pathogenesis, especially the stop codon mutation at position 28. Results obtained at these positions are shown in Table 4.
At positions 1762 and 1764 of the BCP, 24 specimens were identified to be the wild type (A1762 and G1764), as determined by sequencing. In two other samples, a mixture of wild-type and mutant bases (G and A) were detected by sequencing. Twenty specimens were determined to have the double mutation A1762T and G1764A. The chip correctly identified only three of these samples, due to weak hybridization.
Apart from codons rt204 and s145 and positions 1762 and 1764, for which hybridization problems due to unexpected polymorphism or to the simultaneous presence of two mutations were encountered, the number of indeterminate results was higher for sequencing (n = 29) than for the chip (n = 13). Twelve mixtures were detected by sequencing only and 11 by the chip only.
DISCUSSION
Although the size of the HBV genome is small, its organization is complex, with shifts in reading frames permitting the same genetic sequences to code for different proteins of the virus. Several key polymorphisms along this genome have been shown to strongly impact the pathogenesis of viral infection and the susceptibility of the virus to treatment.
As a consequence, HBV genome polymorphism analysis may be a major tool both for clinicians to monitor infection and treatment and for researchers to better understand viral genome variability and its consequences for disease outcome. The small size of the HBV genome is compatible with a strategy using whole-genome PCR amplification for the detection of sequence polymorphisms. We demonstrate in this study that a PCR protocol can be optimized and a genotyping tool designed to identify polymorphisms in 151 positions along the HBV genome, consisting of 245 mutations, 20 deletions, and 2 insertions, in a single test. Considering the high quantity of information that can be provided by a single test from one patient specimen, the short turnaround time, and limited hands-on time, there are strong advantages conferred by DNA chip technology. Obtaining the same information by sequencing would require the use of approximately 16 electrophoresis gels, substantially longer hands-on time at the bench, and analysis of electrophoregrams. Similarly, the number of nitrocellulose strips required to analyze 245 mutations would preclude the use of reverse-hybridization techniques for the extensive genotyping of viruses.
To efficiently amplify the whole 3.2-kb HBV genome, we designed two primer sets to generate two subgenomic fragments, one 1,509 bp and the other 1,721 bp in length. Using this protocol, we were able to amplify HBV DNA and subsequently obtain an interpretable chip result in 93.8% of samples with a detectable viral load. Ninety-five percent of samples with >1,000 copies/ml could be amplified, and only two samples with >10,000 copies/ml failed to be amplified. This analytical sensitivity is similar to the sensitivity generally admitted for sequencing and to that reported for INNO-LiPa (Innogenetics, Belgium) (1). In this study, only those samples that could be amplified using the PCR strategy and whose amplicons were sequenced were selected for chip testing. Nevertheless, 10.6% of the codons of viral genomes amplified from these samples could not be determined by sequencing. Given the extent of viral-load reduction achieved following efficient treatment, a PCR protocol with a sensitivity of 103 HBV genome copies/ml may be considered sufficient for practical purposes in the clinical context.
Based on a comprehensive HBV sequence database, amplification primers were selected from regions that showed high conservation among genotypes. An additional forward primer has also been included to accommodate genotype B-specific polymorphisms and more precisely amplify the 1,509-bp amplicon from this subtype. We did not detect any significant difference in amplification efficiency among genotypes A to F, although the non-European genotypes (B, C, E, F, G, and H) were poorly represented in our panel of samples.
Only 3 samples among 151 gave a genotype result that was discordant between sequencing and the chip. Among them was one that had a viral load of <200 copies of the HBV genome/ml. Thus, the performance of the chip reagent for HBV genotype determination can be considered similar to that by sequencing, which is so far the most widely used and is considered the gold standard approach to polymorphism analysis of viral genomes.
Overall, 11,335 codons have been determined by both sequencing and the chip. The correlation between the two techniques is 92.8%, with no significant difference for different regions of the genome. This result is slightly higher than the 88.9% concordance between INNO-LiPa and sequencing described by Lok et al. (26). The degree of concordance between the chip and sequencing would probably have been even higher upon retesting, which was not possible due to the distribution of discrepant codons among all samples. Although we have no data on the reproducibility of the sequencing protocols used in the different participating laboratories, our experiments have shown the chip approach to be highly reproducible.
As shown in this work, the chip is able to correctly identify wild-type and mutant codons. The photolithography technology developed by Affymetrix to produce high-density chips used in this study requires the manufacturing of dedicated masks, which are used for the synthesis of probes needed for the detection of specific target sequences. This manufacturing process is complex and does not allow frequent upgrades of the chip. Therefore, a prototype chip that could be compatible with PCR products generated from the optimized PCR protocol and that could identify sequence polymorphisms in positions of the amplicons that are difficult to resequence had to be synthesized. We were able to identify positions where difficulties were encountered, using this prototype chip. The genomic sequence of HBV, like that of human immunodeficiency virus, is highly polymorphic. The strategy chosen to consider natural polymorphism was to design probes with all the bases that are significantly represented in sequence databases, providing they are not distant from the central base of a wild-type codon or from a base leading to a mutation of interest by more than four nucleotides. However, at some positions, mismatches that are under-represented in the sequence database may affect hybridization, as exemplified by results obtained at position s145, for which weak hybridization efficiency was observed. This weakness is attributed to a polymorphism at the third base of codon rt144 (T to C), which is present in a significant proportion of viruses but was not retained for probe design, resulting in a systematic mismatch between the probes and target DNA. Adjunction of tiles containing probes with the T to C polymorphism at the third base of codon rt144 in a future version of the chip should correct this problem. Polymorphisms located more than four bases away from the resequenced base may also affect hybridization efficiency, as seen at position rt204, where weak hybridization is attributed to the presence of a natural polymorphism at the third base of codon rt202. Genotypes A and G contain a C (cytosine) at that position, whereas genotypes B, C, D, E, and F contain a T. This polymorphism is located more than five bases away from the central base of the wild-type codon ATG, specifying methionine, and more than six bases away from the third base of codon ATT, specifying isoleucine. Thus, probes that have been designed to resequence those two codons contain only a C as the third base of codon 202, introducing a mismatch with amplicons amplified from viruses with a T at that position. Although a polymorphism more than four bases away from the base of interest usually does not affect hybridization, in the case of position 204 of RT, it results in reduced hybridization efficiency and potentially prevents the chip from identifying an amino acid. All but one of the 20 discrepant samples did not belong to genotype A. In the next version of the chip, the region where polymorphisms are considered should be extended beyond four bases. We have also shown that linked mutations at close positions, like those occurring at positions 1762 and 1764 of the BCP, are difficult to detect if probes that hybridize to each of them are not simultaneously present on the chip.
Our study has shown that the chip reagent is able to identify samples with mixed viral populations. Using artificial mixtures, we have shown that for mixtures that carry >10,000 copies of the HBV genome/ml, the chip can detect L180M variants when they represent at least 30% to 40% of the whole viral population. This is better than the performances usually admitted for sequencing techniques (which can identify minority variants, if present, at about 50% of the total population). Other hybridization-based techniques (INNO-LiPa or low-density chips) can detect minority variants representing <10% of the viral population (22, 29). However, these reagents are able to analyze resistance mutations at positions rt180, rt204, and rt207 only, and the number of probes for each positions is limited, which makes them sensitive to hybridization problems due to polymorphisms.
The technique described in this study is intended to be used as a diagnostic as well as a research tool. Its protocol has been designed to be as simple as possible, and thus, amplification is performed on DNA directly extracted from serum samples. As a consequence, it detects individual polymorphisms only and does not provide any information on the linkage between multiple mutations that may be borne by the same HBV genome. It is nonetheless possible to obtain such linkage information using the chip if PCR cloning of constituents of the viral quasispecies present in the specimen is applied before submission for hybridization to the chip.
Finally, we had access only to samples characterized by sequencing in the S, Pol, precore, and core genes and the BCP. Although detection of polymorphisms in these coding regions is of major interest for the clinical management of patients, further studies are required to analyze the performance of the chip in probing for sequence polymorphisms in the X, pre-S1, pre-S2, and core genes.
To obtain maximal hybridization efficiency, it is very important to build and update high-quality sequence databases containing all naturally occurring polymorphisms. Similarly, the list of variants of interest for antiviral resistance or vaccine or diagnostic escape and for their impact on the course of the disease needs to be as complete as possible, including variants that have been found in vitro only. High-density chips produced by photolithography, like those used in this study, can accommodate the large number of probes required to cover all these polymorphisms. Improvement of hybridization at difficult positions, as well as the introduction of newly described polymorphisms, requires updates of chip design. The frequency with which chips are updated would depend on how well the updated chips performed and on the cost of each upgrade.
Although hybridization at difficult positions needs to be improved and newly described mutations have to be included in a new version of the assay, our data suggest that this prototype chip can already be used for clinical and research purposes. It can detect simultaneously in a single test a large number of clinically relevant mutations located along the whole genome of HBV and therefore constitutes a novel and valuable diagnostic tool for the management of chronic hepatitis B.
Acknowledgments
We thank B. Lacroix, Jérôme Maisetti, C. Drevet, and D. Robinot (bioMerieux, Lyon) for the design of the HBV probe array, the creation of the Test Platform software, and the maintenance of the Affymetrix system.
This work was supported by grants from the European Community (HepBvar, contract QLRT-2001-00977, and ViRgil, contract LSHM-CT-2004-503359).
REFERENCES
- 1.Aberle, S. W., J. Kletzmayr, B. Watschnger, B. Schmied, N. Vetter, and E. Puchhammer-Stöckl. 2001. Comparison of sequence analysis and the INNO-LiPA HBV DR line probe assay for detection of lamivudine-resistant hepatitis B virus strains in patients under various clinical conditions. J. Clin. Microbiol. 39:1972-1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asselah, T., M.-P. Ripault, C. Castelnau, N. Giuily, N. Boyer, and P. Marcellin. 2005. The current status of antiviral therapy of chronic hepatitis B. J. Clin. Virol. 34(Suppl. 1):S115-S124. [DOI] [PubMed] [Google Scholar]
- 3.Baptista, M., A. Kramvis, and M. C. Kew. 1999. High prevalence of 1762(T) 1764(A) mutations in the basic core promoter of hepatitis B virus isolated from black Africans with hepatocellular carcinoma compared with asymptomatic carriers. Hepatology 29:946-953. [DOI] [PubMed] [Google Scholar]
- 4.Bartholomeusz, A., B. G. Tehan, and D. K. Chalmers. 2004. Comparisons of the HBV and HIV polymerase, and antiviral resistance mutations. Antivir. Ther. 9:149-160. [PubMed] [Google Scholar]
- 5.Basuni, A. A., and W. F. Carman. 2004. HBV vaccine-escape variants. Methods Mol. Med. 95:115-124. [DOI] [PubMed] [Google Scholar]
- 6.Bernal-Mendez, E., C. Tora, I. Sothier, M. Kotera, A. Troesch, and A. Laayoun. 2003. Universal labeling chemistry for nucleic acid detection on DNA-arrays. Nucleosides Nucleotides Nucleic Acids 22:1647-1649. [DOI] [PubMed] [Google Scholar]
- 7.Brunelle, M.-N., A.-C. Jacquard, C. Pichoud, D. Durantel, S. Carrouée-Durantel, J.-P.Villeneuve, C. Trépo, and F. Zoulim. 2005. Susceptibility to antivirals of a human HBV strain with mutations conferring resistance to both lamivudine and adefovir. Hepatology 41:1391-1398. [DOI] [PubMed] [Google Scholar]
- 8.Brunetto, M. R., M. Stemler, F. Schodel, H. Will, A. Ottobrelli, M. Rizzetto, G. Verne, and F. Bonino. 1989. Identification of HBV variants which cannot produce precore derived HBeAg and may be responsible for severe hepatitis. Ital. J. Gastroenterol. 21:151-154. [Google Scholar]
- 9.Brunetto, M. R., E. Giarin, F. Oliveri, E. Chiaberge, M. Baldi, A. Alfarano, A. Serra, G. Saracco, G. Verme, H. Will, and F. Bonino. 1991. Wild-type and e antigen-minus hepatitis viruses and course of chronic hepatitis Proc. Natl. Acad. Sci. USA 88:4186-4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brunetto, M. R., M. Giarin, G. Saracco, F. Oliveri, P. L. Calvo, G. Capra, A. Randone, M. L. Abate, P. Manzini, M. Capalbo, P. Piantino, G. Verme, and F. Bonino. 1993. Hepatitis B virus unable to secrete e antigen and response to interferon in chronic hepatitis B. Gastroenterology 105:845-850. [DOI] [PubMed] [Google Scholar]
- 11.Buti, M., F. Rodriguez-Frias, R. Jardi, and R. Esteban. 2005. Hepatitis B virus genome variability and disease progression: the impact of pre-core mutants and HBV genotypes. J. Clin. Virol. 34(Suppl. 1):S79-S82. [DOI] [PubMed] [Google Scholar]
- 12.Carman, W. F., M. R. Jacyna, S. Hadziyannis, P. Karayiannis, M. J. McGarvey, A. Makris, and H. C. Thomas. 1989. Mutation preventing formation of hepatitis B e antigen in patients with chronic hepatitis B infection. Lancet ii:588-591. [DOI] [PubMed]
- 13.Carman, W. F., A. R. Zanetti, P. Karayiannis, J. Waters, G. Manzillo, E. Tanzi, A. J. Zuckerman, and H. C. Thomas. 1990. Vaccine-induced escape mutant of hepatitis B virus. Lancet 336:325-329. [DOI] [PubMed] [Google Scholar]
- 14.Chu, C. J., M. Hussain, and A. S. Lok. 2002. Hepatitis B virus genotype B is associated with earlier HbeAg seroconversion compared with hepatitis B virus genotype C. Gastroenterology 122:1756-1762. [DOI] [PubMed] [Google Scholar]
- 15. Department of Health and Human Services. 2005. Guidelines for the use of antiretroviral agents in HIV-1-infected adolescents and adults. Office of AIDS Research Advisory Council, Department of Health and Human Services, Washington, D.C. [Online.] http://aidsinfo.nih.gov/ContentFiles/AdultandAdolescentGL.pdf.
- 16.Durantel, D., M.-N. Brunelle, E. Gros, S. Carrouée-Durantel, C. Pichoud, S. Villet, C. Trépo, and F. Zoulim. 2005. Resistance of human hepatitis B virus to reverse transcriptase inhibitors: from genotypic to phenotypic testing. J. Clin. Virol. 34(Suppl. 1):S34-S43. [DOI] [PubMed] [Google Scholar]
- 17.Fortuin, M., V. Karthigesu, L. Allison, C. Howard, S. Hoare, M. Mendy, and H. C. Whittle. 1994. Breakthrough infections and identification of a novel variant in Gambian children immunized with hepatitis B vaccine. J. Infect. Dis. 169:1374-1376. [DOI] [PubMed] [Google Scholar]
- 18.Hsu, H. Y., M. H. Chang, Y. H. Ni, H. H. Lin, S. M. Wang, and D. S. Chen. 1997. Surface gene mutants of hepatitis B virus in infants who develop acute or chronic infections despite immunoprophylaxis. Hepatology 26:786-791. [DOI] [PubMed] [Google Scholar]
- 19.Hsu, H. Y., M. H. Chang, Y. H. Ni, and H. L. Chen. 2004. Survey of hepatitis B surface variant infection in children 15 years after a nationwide vaccination programme in Taiwan. Gut 53:1499-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.INVS. 2005. Estimation des taux de prévalence des anticorps anti-VHC et des marqueurs du virus de l'hépatite B chez les assurés sociaux du régime général de France métropolitaine 2003-2004. [Online.] http://www.invs.sante.fr/publications/2005/analyse_descriptive_140205/rapport_analyse_descriptive.pdf.
- 21.Ishikawa, K., T. Koyama, and T. Masuda. 2002. Prevalence of HBV genotypes in asymptomatic carrier residents and their clinical characteristics during long-term follow-up: the relevance to changes in the HBeAg/anti-HBe system. Hepatol. Res. 24:1. [DOI] [PubMed] [Google Scholar]
- 22.Jang, H., M. Cho, J. Heo, H. Kim, H. Jun, W. Shin, B. Cho, H. Park, and C. Kim. 2004. Oligonucleotide chip for detection of lamivudine-resistant hepatitis B virus. J. Clin. Microbiol. 42:4181-4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kao, J. H., P. J. Chen, M. Y. Lai, and D. S. Chen. 2000. Hepatitis B genotypes correlate with clinical outcomes in patients with chronic hepatitis B. Gastroenterology 118:554-559. [DOI] [PubMed] [Google Scholar]
- 24.Lavanchy, D. 2005. Worldwide epidemiology of HBV infection, disease burden and vaccine prevention. J. Clin. Virol. 34(Suppl. 1):S1-S3. [DOI] [PubMed] [Google Scholar]
- 25.Lee, C. M., C. H. Chen, S. N. Lu, H. D. Tung, W. J. Chou, J. H. Wang, T. M. Chen, C. H. Hung, C. C. Huang, and W. J. Chen. 2003. Prevalence and clinical implications of hepatitis B virus genotypes in southern Taiwan. Scand. J. Gastroenterol. 38:95-101. [DOI] [PubMed] [Google Scholar]
- 26.Lok, A. S. F., F. Zoulim, S. Locarnini, A. Mangia, G. Niro, H. Decraemer, G. Maertens, F. Hulstaert, K. De Vreese, and E. Sablon. 2002. Monitoring drug resistance in chronic hepatitis B virus (HBV)-infected patients during lamivudine therapy: evaluation of performance of INNO-LiPA HBV DR assay. J. Clin. Microbiol. 40:3729-3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Norder, H., A.-M. Couroucé, P. Coursaget, J.-M. Echevarria, S. D. Lee, I. K. Mushahwar, B. H. Robertson, S. Locarnini, and L. O. Magnius. 2004. Genetic diversity of hepatitis B virus strains derived worldwide: genotypes, subgenotypes, and HBsAg subtypes. Intervirology 47:289-309. [DOI] [PubMed] [Google Scholar]
- 28.Ogata, N., R. H. Miller, K. G. Ishak, and R. H. Purcell. 1993. The complete nucleotide sequence of a pre-core mutant of hepatitis B virus implicated in fulminant hepatitis and its biological characterization in chimpanzees. Virology 194:263-276. [DOI] [PubMed] [Google Scholar]
- 29.Pas, S. D., R. A. de Man, E. Fries, A. B. van Nunen, A. D. M. E. Osterhaus, and H. G. M. Niesters. 2002. The dynamics of mutations in the polymerase gene of hepatitis B virus during and after lamivudine treatment. J. Clin. Virol. 25:63-71. [DOI] [PubMed] [Google Scholar]
- 30.Pawlotsky, J.-M. 2005. The concept of hepatitis B virus mutant escape. J. Clin. Virol. 34(Suppl. 1):S125-S129. [DOI] [PubMed] [Google Scholar]
- 31.Sato, S., K. Suzuki, Y. Akahane, K. Akamatsu, K. Akiyama, K. Yunomura, F. Tsuda, T. Tanaka, H. Okamoto, Y. Miyakawa, and M. Mayumi. 1995. Hepatitis B virus strains with mutations in the core promoter in patients with fulminant hepatitis. Ann. Intern. Med. 122:241-248. [DOI] [PubMed] [Google Scholar]
- 32.Sirma, H., C. Giannini, K. Poussin, P. Paterlini, D. Kremsdorf, and C. Brechot. 1999. Hepatitis B virus, X mutants, present in hepatocellular carcinoma tissue abrogate both the antiproliferative and transactivation effects of HBx. Oncogene. 18:4848-4859. [DOI] [PubMed] [Google Scholar]
- 33.Sugauchi, F., A. Chutaputti, E. Orito, H. Kato, S. Suzuki, R. Ueda, and M. Mizokami. 2002. Hepatitis B virus genotypes and clinical manifestation among hepatitis B carriers in Thailand. J. Gastroenterol. Hepatol. 17:671-676. [DOI] [PubMed] [Google Scholar]
- 34.Sumi, H., O. Yokosuda, N. Seki, M. Arai, F. Imazeki, T. Kurihara, T. Kanda, K. Fukai, M. Kato, and H. Saiso. 2003. Influence of hepatitis B virus genotypes on the progression of chronic type B liver disease. Hepatology 37:19-26. [DOI] [PubMed] [Google Scholar]
- 35.Thakur, V., R. C. Guptan, S. N. Kazim, V. Malhotra, and S. K. Sarin. 2002. Profile, spectrum and significance of HBV genotypes in chronic liver disease patients in the Indian subcontinent. J. Gastroenterol. Hepatol. 17:165-170. [DOI] [PubMed] [Google Scholar]
- 36.Tu, H., C. Bonura, C. Giannini, H. Mouly, P. Soussan, M. Kew, P. Paterlini-Brechot, C. Brechot, and D. Kremsdorf. 2001. Biological impact of natural COOH-terminal deletions of hepatitis B virus X protein in hepatocellular carcinoma tissues. Cancer Res. 61:7803-7810. [PubMed] [Google Scholar]
- 37.Zollner, B., J. Petersen, M. Schroter, R. Laufs, V. Schoder, and H. H. Feucht. 2001. 20-fold increase in risk of lamivudine resistance in hepatitis B virus subtype adw. Lancet 57:934-935. [DOI] [PubMed] [Google Scholar]
- 38.Zollner, B., J. Petersen, E. Puchhammer-Stockl, J. Kletzmayr, M. Sterneck, L. Fischer, M. Schroter, R. Laufs, and H. H. Feucht. 2004. Viral features of lamivudine resistant hepatitis B genotypes A and D. Hepatology 9:42-50. [DOI] [PubMed] [Google Scholar]