Abstract
We prospectively studied the comparative epidemiology and risk factors for Clostridium difficile, Clostridium perfringens, and Staphylococcus aureus antibiotic-associated diarrhea (AAD). Four thousand six hundred fifty-nine inpatient fecal specimens (11 months) were tested for C. difficile cytotoxin, C. perfringens enterotoxin, and S. aureus by Vero cell assay, enzyme-linked immunosorbent assay, and growth on fresh blood agar, respectively. Two distinct age-, sex-, and location-matched control patient groups were used for multivariate logistic regression risk factor analyses: symptomatic patients who were AAD pathogen negative and asymptomatic patients with histories of recent antimicrobial therapy. All AAD pathogen isolates were DNA fingerprinted. In AAD cases, the prevalences of C. difficile cytotoxin, C. perfringens enterotoxin, and S. aureus were 12.7%, 3.3%, and 0.2%, respectively (15.8% overall). Age of >70 years was a common risk factor. Other risk factors for infective AAD and C. difficile AAD included length of hospital stay and use of feeding tubes (length of stay odds ratios [OR], 1.017 and 1.012; feeding tube OR, 1.864 and 2.808). Female gender and use of antacids were significantly associated with increased risk of C. perfringens AAD (OR, 2.08 and 2.789, respectively), but unlike what was found for C. difficile AAD, specific antibiotic classes were not associated with increased risk. A limited number of genotypes caused the majority of C. difficile and C. perfringens AAD cases. Similar to what was found for C. difficile AAD, there was epidemiological evidence of C. perfringens AAD case clustering and reinfection due to different strains. C. difficile AAD was approximately 4 and 60 times more common than C. perfringens AAD and S. aureus AAD, respectively. Risk factors for these AAD pathogens differed, highlighting the need to define specific control measures. There is evidence of nosocomial transmission in cases of C. perfringens AAD.
Antibiotic-associated diarrhea (AAD) is a significant cause of morbidity and mortality, particularly in the hospitalized elderly. The most common known infective cause of AAD is Clostridium difficile, but this organism accounts for only up to 33% of cases (42). Since the establishment of the association between C. difficile infection (CDI) and AAD in 1978 (3, 26), there has been considerable progress in understanding the etiology and epidemiology of CDI. However, C. difficile is not the sole known infective cause of AAD (4). C. perfringens was identified as a cause of AAD by Borriello et al., who identified C. perfringens enterotoxin in fecal samples from patients with nosocomial diarrhea (8). We previously found evidence of C. perfringens infection in up to 15% of AAD cases (38), a level comparable to that reported by Hancock (21). Various molecular fingerprinting techniques have been used to study C. perfringens (28, 33, 34, 40) but have not been employed in large-scale epidemiological studies of C. perfringens AAD. Staphylococcal enterocolitis associated with antibiotic administration was first described in the 1950s, but attention to this phenomenon then waned. Recently, however, a 2-year prospective hospital study in France identified 60 cases of Staphylococcus aureus AAD (19).
Multiple risk factors for AAD have been described, including increasing age (23, 30, 31), length of hospital stay (31), and administration of broad-spectrum cephalosporins, broad-spectrum penicillins, and clindamycin (16, 22, 30, 44). Risk-associated procedures include tube feeding (5, 31), enemas (30), endoscopy, and gastrointestinal surgery (32, 42). Administration of antineoplastics (20), proton pump inhibitors (10), and stool softeners (30) has also been implicated. However, in assessing the most common infective cause of AAD, previous studies have been biased towards or have concentrated solely on C. difficile and have generally not considered other infective causes. We therefore aimed to determine prospectively the relative prevalences of infective AAD due to C. difficile, C. perfringens, and S. aureus and to assess associated risk factors. Additionally, we performed molecular fingerprinting of bacterial isolates to elucidate the epidemiology of AAD.
MATERIALS AND METHODS
Specimen collection.
Four thousand six hundred fifty-nine consecutive inpatient fecal specimens submitted for routine C. difficile testing at Leeds Teaching Hospitals from June 2001 to April 2002 inclusively were included. Indications for testing were either clinical suspicion of AAD or diarrhea occurring ≥3 days postadmission (49). Only diarrheal fecal specimens (i.e., those that adopted the contours of the vessel) were included in the study. Samples from children of <2 years of age were excluded.
Microbiological analysis. (i) Toxin detection in feces.
All samples were tested for C. difficile cytotoxin and C. perfringens enterotoxin by Vero cell cytotoxicity assay as described previously (1). Also, a C. perfringens enterotoxin enzyme-linked immunosorbent assay (ELISA) kit (TechLab; distributed by BioConnections, Leeds, United Kingdom) was used according to the manufacturer's instructions.
(ii) C. difficile culture.
C. difficile cytotoxin-positive fecal specimens were inoculated onto cycloserine-cefoxitin egg yolk agar (CCEY; PHLS Media Services, Leeds, United Kingdom) with anaerobic incubation at 37°C for 48 h. Identification was based on colonial appearance, odor, and a Microscreen C. difficile latex slide agglutination test (Microgen Bioproducts Ltd., Surrey, United Kingdom). Isolates were stored on ProTect cryobeads (Technical Service Consultants, Heywood, United Kingdom) at −70°C.
(iii) C. perfringens culture.
Isolates were recovered from C. perfringens enterotoxin-positive fecal specimens as described previously (1). Identification was based on colonial appearance, Gram stain morphology, and reaction with C. perfringens type A antitoxin (TechLab). Isolates were stored on ProTect cryobeads at −70°C.
(iv) S. aureus culture and methicillin susceptibility testing.
Fecal specimens were plated onto fresh blood agar and incubated aerobically for 24 h at 37°C. Predominant growth of S. aureus (≥90% of the total bacterial growth) was considered significant, as described previously (19). Methicillin susceptibility was determined using methicillin strips according to the manufacturer's instructions (Mast Diagnostics, Merseyside, United Kingdom). Isolates were stored on ProTect cryobeads at −70°C. Any nonfecal S. aureus isolates recovered from routine diagnostic sampling of patients with fecal samples positive for S. aureus were also stored.
(v) S. aureus toxin detection.
Production of staphylococcal enterotoxins A, B, C, and D and of staphylococcal toxic shock syndrome toxin was detected by reverse passive latex agglutination (Oxoid, Basingstoke, United Kingdom). Isolates were referred to the Prévost research group, France, for full toxin analysis (Panton-Valentine [PV] leucocidin, LukE-LukD, LukM-LukF′-PV-like, and exfoliative toxins A and B) (19).
Risk factor analysis. (i) Collection of patient data.
Approval for the collection of patient data was obtained from the Leeds (West) and Leeds (East) research ethics committees prior to study commencement. Data were collected for all patients with fecal samples positive for C. difficile toxin, C. perfringens toxin, or S. aureus (defined as infective AAD cases) and for controls (defined below); these data included demographics, admission details, lengths of hospital stay before diarrhea, and histories of nonantibiotic (antacids, antiperistaltics, cytotoxics, and gut stimulants) and antibiotic (aminoglycosides; narrow-, expanded-, and broad-spectrum cephalosporins; co-trimoxazole; fluoroquinolones; glycopeptides; macrolides; metronidazole; and broad- and narrow-spectrum penicillins) drug administration and procedures undertaken (endoscopy, enema, feeding tube insertion, and surgery) within the 28 days before the onset of symptoms.
(ii) Control patients.
Two sets of control group data were collected for the case control risk factor analyses. Control group 1 consisted of patients from whom diarrheal fecal samples were examined in the study but who were negative for C. difficile cytotoxin, C. perfringens enterotoxin, and S. aureus. Control group 2 consisted of hospitalized patients without diarrhea who had received antimicrobial therapy within the previous 28 days. All control patients were matched to cases by age (within stratified groups of 2 to 20, 21 to 40, 41 to 60, 61 to 70, 71 to 80, and >80 years of age), gender, date (within 2 months of the case becoming symptomatic), and ward location. For analysis of AAD risk factors, every third case was chosen for matching. Due to the predominance of CDI, it was possible to obtain sufficient data for analysis of pathogen-specific risk factors by extracting C. difficile-positive matched cases from the AAD data. However, all cases were matched with controls for analysis of C. perfringens AAD risk factors.
(iii) Statistical analysis.
Data were excluded from analyses if complete data sets were not obtained for each case and control. No diarrheal episode was included more than once. Subsequent diarrheal episodes were considered new episodes if the patient had been asymptomatic for at least 14 days. Statistical analyses were conducted using Stata (Stata Corporation, Texas) and SPSS (SPSS Inc., Chicago, Ill.) software. Age and gender data were analyzed by comparison of cases with the overall study population by use of multinomial logistic regression. Other risk factors were assessed using conditional backwards multivariate logistic regression with matched data.
DNA fingerprinting. (i) C. difficile.
C. difficile isolates were recovered from storage by inoculation of CCEY plus 5 mg/liter lysozyme agar without egg yolk supplementation (BioConnections, Leeds, United Kingdom) with anaerobic incubation at 37°C for 48 h (46). DNA was extracted from the colonies as described previously (13). For isolates that could not be recovered, 200 μl ProTect cryopreservative solution containing bacteria was centrifuged to pellet the bacterial cells. Cells were washed twice with phosphate-buffered saline, resuspended in 50 μl sterile distilled water, and DNA extracted as above. A random amplified polymorphic DNA molecular typing protocol was used as described previously (12). Two variants of the United Kingdom epidemic C. difficile strain (PCR ribotype 1) were used as internal controls; these differed in terms of clindamycin susceptibility and had distinct random amplified polymorphic DNA profiles (12).
(ii) C. perfringens.
C. perfringens isolates were recovered from storage by inoculation of fresh blood agar and anaerobic incubation for up to 48 h at 37°C. Amplified fragment length polymorphism was used as described previously with modifications (33). Briefly, brain heart infusion broth (Oxoid) was inoculated and incubated as described above. DNA was extracted using a Wizard genomic DNA purification kit (Promega, Southampton, United Kingdom) according to the manufacturer's instructions. Lysis was achieved using 10 mg/ml lysozyme, and DNA was rehydrated at 65°C for 1 h. DNA concentration and purity were analyzed spectrophotometrically. Extracted DNA was digested with HindIII with a reduced digestion time of 4 h. Adaptor oligonucleotides were ligated onto the digested DNA prior to PCR with primer HI-G (33). PCR products were resolved by 2% agarose gel electrophoresis in Tris-acetate-EDTA buffer. C. perfringens strain NCTC 8239 was included as an internal control.
(iii) S. aureus.
S. aureus isolates were recovered from storage by inoculation of fresh blood agar and aerobic incubation for 24 h at 37°C. Pulsed-field gel electrophoresis (PFGE) was used as described previously (2) with modifications. Bacterial cells were cultured overnight aerobically at 37°C in 2× yeast extract-tryptone plus 0.5% glycine broth (in-house). Agarose plugs containing bacterial DNA were incubated with SmaI restriction endonuclease for between 4 and 6 h at 30°C. In addition, all S. aureus isolates were subjected to phage typing as described previously (37).
(iv) Molecular fingerprinting analysis.
Gels were imaged using GeneGenius (SYNGENE, Cambridge, United Kingdom) and analyzed using BioNumerics software (Applied Maths, Kortrijk, Belgium). Dendrograms were constructed by the unweighted paired group method with arithmetic mean clustering using Dice correlation coefficient. Isolates were assigned into groups using a percentage similarity cutoff based on the guidelines of Tenover et al. (43).
RESULTS
AAD pathogens.
At least one AAD pathogen was detected in 735 of 4,659 (15.8%) fecal samples; C. difficile cytotoxin, C. perfringens enterotoxin, and/or S. aureus were detected in 12.7% (n = 591), 3.3% (n = 155), and 0.2% (n = 10) of samples, respectively. Twenty-one samples (2.9% of positive specimens) had evidence of more than one AAD pathogen, typically C. difficile and C. perfringens (19/21). Of the 591 C. difficile cytotoxin-positive fecal samples, 566 were available for culturing. Of these, 310 (64%) yielded C. difficile. Of the 155 C. perfringens enterotoxin-positive fecal specimens, 130 were available for culturing, and C. perfringens was cultured from 113 (87%). Only 21 (13.5%) C. perfringens enterotoxin ELISA-positive specimens were positive by Vero cell cytotoxicity assay. All 10 S. aureus isolates were methicillin resistant. Five S. aureus isolates were enterotoxin C producers, one produced enterotoxins C and D, two produced enterotoxin A and toxic shock syndrome toxin 1, and two were nonenterotoxigenic. None of the S. aureus isolates produced detectable Panton-Valentine leucocidin, LukE-LukD, LukM-LukF′-PV-like, or exfoliative toxins A and B.
Analysis of risk factors.
Ninety-four percent of analyzed patients had documented evidence of diarrhea (≥3 episodes for at least 2 days). Eighty-five percent of the patients had received antibacterial therapy within the 28 days prior to the initiation of symptoms. Patients between 71 to 80 years of age and those of >80 years of age had a significantly increased risk of infective AAD (odds ratios [OR], 2.98 and 3.61, respectively; P ≤ 0.001). This age-related risk was consistent for both C. difficile (OR, 2.4 and 2.8, respectively; P values were ≤0.001 in both cases) and C. perfringens (OR, 10.7 and 13.7; P values were ≤0.05 and ≤0.01, respectively). Gender was not an independent risk factor for infective AAD (P = 0.607), but there was a significant age-sex interaction (P = 0.046). Compared with males, females of between 2 and 5 years of age had a reduced risk of infective AAD (OR = 0.27; P = 0.04), and females over 80 years of age had an increased risk (OR = 1.4; P = 0.03). Female gender was an independent risk factor for C. perfringens AAD (OR = 2.08; P ≤ 0.001).
Of 735 positive cases, 109 were removed from the data set either because they represented a repeat positive (without symptom resolution) (n = 93) or because complete data were not available (n = 16). Hence, complete risk factor data were analyzed for 626 patient AAD episodes (503 C. difficile, 132 C. perfringens, 8 S. aureus). Determination of risk factors associated with cases of infective AAD due to all causes, C. difficile-specific ADD, and C. perfringens-specific AAD in comparison with symptomatic controls involved the analysis of 155, 118, and 66 matched case control data sets, respectively; the corresponding numbers for asymptomatic controls were 142, 112, and 74, respectively. Due to the very low prevalence of S. aureus-associated AAD in this study, it was not possible to determine risk factors specifically associated with S. aureus AAD. Significant results from the conditional multiple logistic regression analyses are shown in Table 1. The major risk factors for infective AAD and CDI were administration of cytotoxic drugs and broad-spectrum cephalosporins; administration of penicillin was associated with reduced risk of AAD. Use of antacids was the sole risk factor identified for C. perfringens infection. Presence of a feeding tube was associated with increased risk of infective AAD and CDI and reduced risk of C. perfringens AAD.
TABLE 1.
Significant risk factors associated with infective AAD due to all causes and with C. difficile- and C. perfringens-specific AAD
| Risk factor | OR (95% confidence interval) for AAD due toa:
|
||
|---|---|---|---|
| All causes | C. difficile | C. perfringens | |
| Length of hospital stay | 1.017 (1.01-1.03)** | 1.012 (1.00-1.02)* | |
| 1.016 (1.01-1.03)** | 1.027 (1.01-1.04)** | ||
| Antacids | 2.789 (2.03-3.55)** | ||
| Cytotoxic drugs | 8.933 (7.28-10.59)** | 8.073 (6.36-9.79)* | |
| Feeding tubes | 1.864 (1.11-2.64)* | 2.808 (2.16-3.46)** | 0.130 (0.10-0.16)** |
| 2.511 (1.87-3.15)** | 5.630 (4.52-6.41)** | ||
| Penicillin V or G | 0.178 (0.04-0.32)* | 0.126 (0.07-0.18)* | |
| Broad-spectrum penicillins | 0.238 (0.14-0.34)** | ||
| 0.263 (0.16-0.31)** | |||
| Aminoglycosides | 0.197 (0.06-3.11)* | ||
| Narrow-spectrum cephalosporins | 2.277 (1.22-4.1)* | 2.760 (1.88-3.64)* | |
| Broad-spectrum cephalosporins | 3.594 (2.84-5.61)** | 3.810 (2.99-4.63)** | |
| Metronidazole | 0.271 (0.15-0.39)* | ||
| Macrolides | 3.565 (2.81-4.32)** | ||
| Sulfonamide-plus trimethoprim | 0.467 (0.40-0.53)* | ||
Data were analyzed by conditional multivariate logistic regression. Values in boldface are those for case control 1 analysis (cases versus symptomatic patients); values in lightface are those for case control 2 analysis (asymptomatic patients). *, P ≤ 0.05; **, P ≤ 0.01.
The mean length of hospital stay prior to the onset of diarrheal symptoms was significantly longer for patients than for group 1 controls (29.2 ± 4.1 versus 19.5 ± 3.7 days; paired t test, t = 3.91; df = 154; P ≤ 0.001). A similar finding was obtained in a comparison with data from group 2 controls (28 ± 4.1 versus 20.4 ± 3.2 days; t = 2.99; df = 141; P ≤ 0.01).
Molecular fingerprinting.
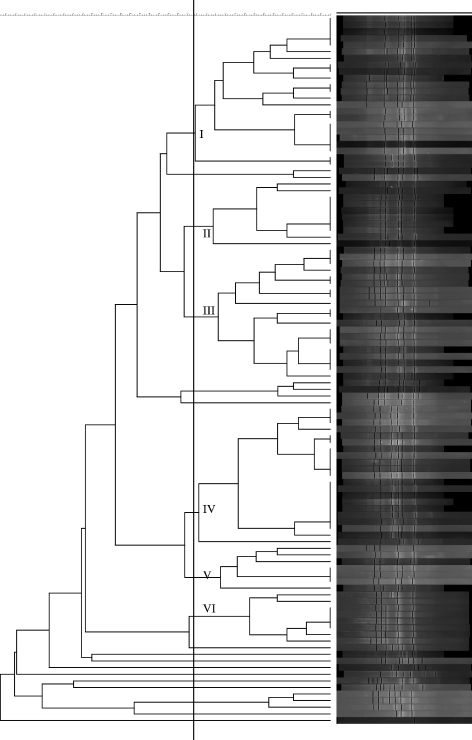
Seventy-five percent of C. difficile isolates had DNA fingerprints corresponding to the clindamycin-susceptible variant of the United Kingdom epidemic PCR ribotype 1 strain (11). The remaining isolates (n = 42) represented a wide variety of genotypes (n = 35). Nineteen distinct C. perfringens genotypes were identified; no one clone predominated, but six genotypes accounted for 83% of all isolates (Fig. 1). S. aureus isolates were distributed among three genotypes. A nonfecal S. aureus isolate was obtained from three AAD patients (one wound swab sample, one urine specimen, and one sputum specimen). Each of these isolates showed 100% DNA band homology with the corresponding fecal isolate. Phage typing results identified the two enterotoxin A-producing isolates as United Kingdom epidemic strains (16 EMRSA-16); all remaining isolates were EMRSA-15.
FIG. 1.
Dendrogram showing the phylogenetic relationship of C. perfringens isolates.
DISCUSSION
It is likely that a substantial proportion of AAD cases are not due to infective causes and instead represent either direct gut toxicity of antibiotics or other medical etiologies. Nevertheless, studies of the infective etiology of AAD have concentrated almost solely on C. difficile, and thus, in the great majority of cases, a pathogen has not been identified (42). We believe this is the first study to determine concurrently the relative prevalences of C. difficile, C. perfringens, and S. aureus AAD. C. difficile was the predominant infective cause of AAD and was approximately 4 and 60 times more common than C. perfringens and S. aureus AAD cases, respectively. For every 10 cases of C. difficile AAD, we identified 2.6 and 0.17 cases of C. perfringens and S. aureus AAD, respectively. The prevalence of infective AAD in the present study was lower than what we detected previously (15.8% versus 22%) (38). However, the earlier study included only those patients with clinical details suggestive of AAD. Conversely, here we also included fecal samples from those patients who developed diarrhea ≥3 days postadmission (49). Thus, more-frequent fecal testing may account for the lower proportion of pathogen-positive samples. Some AAD studies have defined a minimum frequency of diarrhea in the inclusion criteria. However, it is difficult to obtain accurate information regarding such factors because of poor documentation (44). We therefore opted to include all diarrheal fecal samples received by the laboratory in the microbiological evaluation. However, we emphasize that for cases where full data were obtained, almost all (94%) had recorded ≥3 episodes of diarrhea for at least 2 days. Notably, 85% of the cases had received antibacterial therapy within the 28 days before symptom onset. We are confident, therefore, that the great majority of cases represent truly symptomatic cases of AAD. We did not consider other putative infective causes of AAD, such as Candida and Klebsiella species, given the relatively weak evidence base for these organisms as pathogens (4).
Our data add to the evidence base that C. perfringens is a significant cause of nosocomial AAD (1, 21, 38). Laboratory identification of such cases can be justified in at least two ways. First, antibiotic treatment of C. perfringens AAD may aid symptom resolution (6). While there are no data to determine optimal therapy, it is plausible that either oral metronidazole or vancomycin could be effective. Further studies are clearly required here. Second, as discussed below, diagnosis of cases could be important to identify clusters and/or limit nosocomial transmission. Only 13.5% of C. perfringens enterotoxin ELISA-positive specimens were Vero cell assay positive. This reflects previous reports that the Vero cell cytotoxicity assay lacks sensitivity for the detection of C. perfringens enterotoxin (1, 9, 14). ELISA-based methods should therefore be employed to optimize the laboratory diagnosis of C. perfringens AAD.
In contrast to the results for C. perfringens ADD, the prevalence of S. aureus AAD was very low in our institution. As such, routine detection for S. aureus in AAD cases cannot be justified. All identified S. aureus strains were methicillin resistant, a result consistent with earlier studies (19, 29). By comparison, the prevalence of methicillin resistance in our nosocomial S. aureus isolates is 40% (comparison of percentages test value, 3.846; P < 0.001). The preponderance of methicillin-resistant S. aureus (MRSA) may be due to the relative endemicity of this phenotype and/or the greater potential for overgrowth by resistant strains under antimicrobial selective pressure. Notably, 8 of the 10 MRSA isolates phage typed as EMRSA-15 (one of the two main United Kingdom epidemic strains). The prevalence of EMRSA-15 among all our nosocomial MRSA isolates is approximately 73% (data not shown). Interestingly, in all three cases where MRSA was additionally isolated from a nonfecal source, each isolate pair was indistinguishable by PFGE, as seen previously (19). Thus, S. aureus fecal testing may be applicable for patients with AAD of unknown cause and/or when S. aureus, particularly MRSA, has been isolated from another site. Froberg et al. recently reported a case of simultaneous infections with C. difficile and S. aureus (MRSA) (17). The presence of discrete colonic pseudomembranes and confluent, loosely adherent ileal pseudomembranes suggested that separate pathological processes occur in these two types of AAD. The rarity of reports of S. aureus AAD and the relatively low incidence found in our study possibly may reflect the scarcity of pathogenic strains in susceptible hosts. Leucotoxin production was uncommon in our isolates, in contrast to the findings of the aforementioned French study (19).
Many AAD risk factors studies have been flawed due to limited size, poor choice of controls and thus potential for confounding, and choice of laboratory methodology (44). Also, most previous studies have focused solely on C. difficile as the etiological agent (32). Our study was designed to address these deficiencies. Confounding factors were reduced by matching cases and controls for age, gender, date, and location. Times and locations were matched to account for possible fluctuations in C. difficile endemicity, particularly the prevalence of environmental contamination and thus the risk of patient and healthcare worker exposure (13, 39). Some studies have used scoring systems, such as Horn's index, to assess the severity of underlying diseases (24). We chose not to use this approach due to the complexities involved in obtaining accurate data. However, matching of patient locations did provide some basic adjustment for this potential confounding variable. Controversy about optimal control populations for AAD risk factor studies was highlighted in a recent systematic review (44). Thomas et al. recommended that the control population should be selected from the source population and that the use of patients with diarrheal symptoms who prove negative for infection is not suitable (44). However, there are ethical and practical considerations in obtaining fecal specimens, particularly from elderly patients not suffering from diarrhea. In order to account for these dilemmas, we used two control groups. One control group comprised symptomatic patients without infective AAD from whom diarrheal fecal samples did not yield C. difficile cytotoxin, C. perfringens enterotoxin, or S. aureus. Case comparison with this group identified risk factors associated with these causes of infective AAD but not necessarily with nosocomial diarrhea per se (48). The second control group comprised asymptomatic hospitalized patients who had received antibiotics and who presumably would have been investigated for AAD had they become symptomatic. Comparison of these controls with cases identified risk factors associated with nosocomial diarrhea. Despite the use of two different control groups, the risk factors identified were broadly similar, as reported elsewhere (36). We believe that this discounts some of the concerns regarding choice of control population. Although we included patients with symptomatic recurrence in the risk factor analyses, in fact these amounted to a maximum of three cases in any one AAD group. It was unavoidable that a degree of data loss occurred prior to analysis, most commonly due to incomplete medical records.
It is recognized that the risk of AAD can be reduced by substituting the administration of high-risk antibiotics, such as broad-spectrum cephalosporins and clindamycin (16), with relatively low-risk drugs such as penicillin, trimethoprim, gentamicin (35), ciprofloxacin (27), and ureidopenicillins (41, 47). Identifying additional risk factors for AAD permits interventions to reduce disease burden. Risk factors identified for AAD are influenced greatly by risk factors specific to C. difficile AAD due to the predominance of this pathogen. Notably, each additional day of hospital stay equated to an ∼2%-increased risk of infection. Aminoglycosides were associated with a reduced risk of overall AAD, which is probably related to the innate resistance of anaerobic gut flora. Penicillin administration was also associated with reduced risk of AAD, which may reflect poor acid stability and therefore minimal effect on bowel flora. Many of the risk factors associated with AAD and CDI in this study, including cytotoxic drugs, feeding tubes, and broad-spectrum cephalosporins (5, 16, 20, 22, 30-32, 44), have been reported earlier. Other previously reported risk factors, such as broad-spectrum antibiotics, were not identified in this study (30, 44). Previous studies usually analyzed low numbers of risk factors, whereas our use of extensive multivariate analysis and two control groups likely increased the accuracy of results. Also, it is clear that CDI risk is not consistent across all broad-spectrum agents (16, 22, 27, 35). It is noteworthy that the risk factors associated with CDI differed from those associated with C. perfringens. The use of feeding tubes was associated with increased risk for infective AAD and CDI but with reduced risk for C. perfringens infection. It might be expected that the presence of a feeding tube would facilitate nosocomial transmission of enterotoxigenic C. perfringens. Possibly, the level of environmental contamination by C. perfringens here was too low to facilitate such cross-infection. Longitudinal studies similar to those performed for CDI are needed to determine the environmental burden of C. perfringens (12, 13, 39). It remains controversial whether antacids, particularly proton pump inhibitors, are a risk factor for AAD; conflicting results may relate to as-yet-unexplained confounding factors (10, 11). Intriguingly, we found that antacid therapy was a risk factor only for C. perfringens AAD. Other risk factors invariably associated with CDI, such as broad-spectrum cephalosporins (22), were found to have no association with C. perfringens AAD. Indeed, despite the large study size, administration of broad-spectrum cephalosporins was not significant by a simple logistic regression (P = 0.111). This is a strong indication that the risk factors associated with these two causes of AAD differ significantly. Recent data suggest that broad-spectrum cephalosporins may stimulate toxin production by C. difficile (15). Also, increasing evidence points to the importance of the antibiotic resistance of C. difficile (18). The roles of such factors in C. perfringens AAD remain to be elucidated.
The proportion of cases on elderly medicine wards caused by the United Kingdom epidemic C. difficile strain (PCR ribotype 1) was 88%, which is consistent with previous findings (13). The lower overall prevalence of C. difficile PCR ribotype 1 (75%) reflects the inclusion of all hospital specialties in this study. The apparent predilection of the epidemic strain for elderly patients could reflect increased host susceptibility secondary to reduced anti-C. difficile toxin antibody response (25). Alternatively, the differing endemicity of C. difficile PCR ribotype 1 may be related to variation in antibiotic prescribing practice (12). Our data suggest that a limited number of C. perfringens genotypes cause the majority of AAD cases, although, unlike what is seen for C. difficile, there is no predominant genotype, at least in our institution. We did not examine the environment for C. perfringens, but it is feasible that spore contamination similar to that seen in areas of C. difficile endemicity occurs secondary to fecal soiling by incontinent patients. Strains with serotypes the same as those of fecal isolates were found in the hospital environment and on the hands of infected patients during an outbreak of C. perfringens diarrhea (7). A study from Japan found that of 60 C. perfringens isolates recovered from the feces of elderly hospitalized patients with sporadic diarrhea likely of nosocomial origin, 38 shared the same DNA PFGE pattern (45). During our study, we identified seven instances, involving 19 patients, where patients related spatially and temporally were infected with DNA fingerprint (amplified fragment length polymorphism)-indistinguishable C. perfringens strains. This is further evidence that C. perfringens may be acquired nosocomially. It is likely that measures to limit nosocomial dissemination of C. difficile, including case isolation and attention to environmental hygiene, will also be of benefit in the treatment of enterotoxigenic C. perfringens.
Acknowledgments
This study was supported by a research grant from the United Kingdom Hospital Infection Society.
We thank the staff of the Microbiology Departments at the Leeds Teaching Hospitals Trust and the Leeds Health Protection Agency. We also thank A. Scally, School of Health Studies, University of Bradford, United Kingdom, for statistical analysis. We are grateful to the research group of G. Prévost for the toxin typing of S. aureus isolates. N. J. Asha, D. Tompkins, and M. H. Wilcox have no conflicts of interest.
REFERENCES
- 1.Asha, N. J., and M. H. Wilcox. 2002. Laboratory diagnosis of Clostridium perfringens antibiotic-associated diarrhoea. J. Med. Microbiol. 51:891-894. [DOI] [PubMed] [Google Scholar]
- 2.Bannerman, T. L., G. A. Hancock, F. C. Tenover, and J. M. Miller. 1995. Pulsed-field gel electrophoresis as a replacement for bacteriophage typing of Staphylococcus aureus. J. Clin. Microbiol. 33:551-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartlett, J. G., T. W. Chang, M. Gurwith, S. L. Gorbach, and A. B. Onderdonk. 1978. Antibiotic-associated pseudomembranous colitis due to toxin-producing clostridia. N. Engl. J. Med. 298:531-534. [DOI] [PubMed] [Google Scholar]
- 4.Beaugerie, L., and J. C. Petit. 2004. Microbial-gut interactions in health and disease. Antibiotic-associated diarrhoea. Best Pract. Res. Clin. Gastroenterol. 18:337-352. [DOI] [PubMed] [Google Scholar]
- 5.Bliss, D. Z., S. Johnson, K. Savik, C. R. Clabots, K. Willard, and D. N. Gerding. 1998. Acquisition of Clostridium difficile-associated diarrhea in hospitalized patients receiving tube feeding. Ann. Intern. Med. 129:1012-1019. [DOI] [PubMed] [Google Scholar]
- 6.Boriello, S. P., and R. K. T. Williams. 1985. Treatment of Clostridium perfringens enterotoxin-associated diarrhoea with metronidazole. J. Infect. 10:65-67. [DOI] [PubMed] [Google Scholar]
- 7.Borriello, S. P., F. E. Barclay, A. R. Welch, et al. 1985. Epidemiology of diarrhoea caused by enterotoxigenic Clostridium perfringens. J. Med. Microbiol. 20:363-372. [DOI] [PubMed] [Google Scholar]
- 8.Borriello, S. P., H. E. Larson, A. R. Welch, F. Barclay, M. F. Stringer, and B. A. Bartholomew. 1984. Enterotoxigenic Clostridium perfringens: a possible cause of antibiotic-associated diarrhoea. Lancet i:305-307. [DOI] [PubMed] [Google Scholar]
- 9.Brett, M. M. 1998. Kits for the detection of some bacterial food poisoning toxins: problems, pitfalls and benefits. J. Appl. Microbiol. 84(Suppl.):110-118. [DOI] [PubMed] [Google Scholar]
- 10.Cunningham, R., B. Dale, B. Undy, and N. Gaunt. 2003. Proton pump inhibitors as a risk factor for Clostridium difficile diarrhoea. J. Hosp. Infect. 54:243-245. [DOI] [PubMed] [Google Scholar]
- 11.Dial, S., J. A. C. Delaney, A. D. Barkun, and S. Suissa. 2005. Use of gastric acid-suppressive agents and the risk of community-acquired Clostridium difficile-associated disease. JAMA 294:2989-2995. [DOI] [PubMed] [Google Scholar]
- 12.Fawley, W. N., J. Freeman, and M. H. Wilcox. 2003. Evidence to support the existence of subgroups within the UK epidemic Clostridium difficile strain (PCR ribotype 1). J. Hosp. Infect. 54:74-77. [DOI] [PubMed] [Google Scholar]
- 13.Fawley, W. N., and M. H. Wilcox. 2001. Molecular epidemiology of endemic Clostridium difficile infection. Epidemiol. Infect. 126:343-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forward, L. J., D. S. Tompkins, and M. M. Brett. 2003. Detection of Clostridium difficile cytotoxin and Clostridium perfringens enterotoxin in cases of diarrhoea in the community. J. Med. Microbiol. 52:753-757. [DOI] [PubMed] [Google Scholar]
- 15.Freeman, J., F. J. O'Neill, and M. H. Wilcox. 2003. Effects of cefotaxime and desacetylcefotaxime upon Clostridium difficile proliferation and toxin production in a triple-stage chemostat model of the human gut. J. Antimicrob. Chemother. 52:96-102. [DOI] [PubMed] [Google Scholar]
- 16.Freeman, J., and M. H. Wilcox. 1999. Antibiotics and Clostridium difficile. Microbes Infect. 1:377-384. [DOI] [PubMed] [Google Scholar]
- 17.Froberg, M. K., E. Palavecino, R. Dykoski, D. N. Gerding, L. R. Peterson, and S. Johnson. 2004. Staphylococcus aureus and Clostridium difficile cause distinct pseudomembranous intestinal diseases. Clin. Infect. Dis. 39:747-750. [DOI] [PubMed] [Google Scholar]
- 18.Gerding, D. N. 2004. Clindamycin, cephalosporins, fluoroquinolones, and Clostridium difficile-associated diarrhea: this is an antimicrobial resistance problem. Clin. Infect. Dis. 38:646-648. [DOI] [PubMed] [Google Scholar]
- 19.Gravet, A., M. Rondeau, C. Harf-Monteil, F. Grunenberger, H. Monteil, J.-M. Scheftel, and G. Prévost. 1999. Predominant Staphylococcus aureus isolated from antibiotic-associated diarrhea is clinically relevant and produces enterotoxin A and bicomponent toxin LukE-LukD. J. Clin. Microbiol. 37:4012-4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Halim, H. A., G. M. Peterson, W. T. Friesen, and A. K. Ott. 1997. Case-controlled review of Clostridium difficile-associated diarrhoea in Southern Tasmania. J. Clin. Pharm. Ther. 22:391-397. [DOI] [PubMed] [Google Scholar]
- 21.Hancock, P. 1997. Antibiotic-associated diarrhoea: Clostridium difficile or C. perfringens? Rev. Med. Microbiol. 8(Suppl. 1):66-67. [Google Scholar]
- 22.Johnson, S., and D. N. Gerding. 1997. Clostridium difficile-associated diarrhea. Clin. Infect. Dis. 26:1027-1036. [DOI] [PubMed] [Google Scholar]
- 23.Kyne, L., C. Merry, B. O'Connell, A. Kelly, C. Keane, and D. O'Neill. 1999. Factors associated with prolonged symptoms and severe disease due to Clostridium difficile. Age Ageing 28:107-113. [DOI] [PubMed] [Google Scholar]
- 24.Kyne, L., S. Sougioultzis, L. V. McFarland, and C. P. Kelly. 2002. Underlying disease severity as a major risk factor for nosocomial Clostridium difficile diarrhea. Infect. Control Hosp. Epidemiol. 23:653-659. [DOI] [PubMed] [Google Scholar]
- 25.Kyne, L., M. Warny, A. Qamar, and C. P. Kelly. 2000. Asymptomatic carriage of Clostridium difficile and serum levels of IgG antibody against toxin A. N. Engl. J. Med. 342:390-397. [DOI] [PubMed] [Google Scholar]
- 26.Larson, H. E., A. B. Price, P. Honour, and S. P. Borriello. 1978. Clostridium difficile and the aetiology of pseudomembraneous colitis. Lancet i:1063-1066. [DOI] [PubMed] [Google Scholar]
- 27.Ludlam, H., N. Brown, O. Sule, C. Redpath, N. Coni, and G. Owen. 1999. An antibiotic policy associated with reduced risk of Clostridium difficile-associated diarrhoea. Age Ageing 28:578-580. [DOI] [PubMed] [Google Scholar]
- 28.Maslanka, S. E., J. G. Kerr, G. Williams, J. M. Barbaree, L. A. Carson, J. M. Miller, and B. Swaminathan. 1999. Molecular subtyping of Clostridium perfringens by pulsed-field gel electrophoresis to facilitate food-borne-disease outbreak investigations. J. Clin. Microbiol. 37:2209-2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McDonald, M., P. Ward, and K. Harvey. 1982. Antibiotic-associated diarrhoea and methicillin-resistant Staphylococcus aureus. Med. J. Aust. 1:462-464. [PubMed] [Google Scholar]
- 30.McFarland, L. V., C. M. Surawicz, and W. E. Stamm. 1990. Risk factors for Clostridium difficile carriage and C. difficile-associated diarrhea in a cohort of hospitalized patients. J. Infect. Dis. 162:678-684. [DOI] [PubMed] [Google Scholar]
- 31.McFarland, L. V. 1995. Epidemiology of infectious and iatrogenic nosocomial diarrhea in a cohort of general medicine patients. Am. J. Infect. Control 23:295-305. [DOI] [PubMed] [Google Scholar]
- 32.McFarland, L. V. 1998. Epidemiology, risk factors and treatment for antibiotic-associated diarrhea. Dig. Dis. 16:292-307. [DOI] [PubMed] [Google Scholar]
- 33.McLauchlin, J., G. Ripabelli, M. M. Brett, and E. J. Threlfall. 2000. Amplified fragment length polymorphism (AFLP) analysis of Clostridium perfringens for epidemiological typing. Int J. Food Microbiol. 56:21-28. [DOI] [PubMed] [Google Scholar]
- 34.McLauchlin, J., J. E. Salmon, S. Ahmed, et al. 2002. Amplified fragment length polymorphism (AFLP) analysis of Clostridium novyi, C. perfringens and Bacillus cereus isolated from injecting drug users during 2000. J. Med. Microbiol. 51:990-1000. [DOI] [PubMed] [Google Scholar]
- 35.McNulty, C., M. Logan, I. P. Donald, D. Ennis, D. Taylor, R. N. Baldwin, M. Bannerjee, and K. A. Cartwright. 1997. Successful control of Clostridium difficile infection in an elderly care unit through use of a restrictive antibiotic policy. J. Antimicrob. Chemother. 40:707-711. [DOI] [PubMed] [Google Scholar]
- 36.Nelson, D. E., S. B. Auerbach, A. L. Baltch, et al. 1994. Epidemic Clostridium difficile-associated diarrhea: role of second- and third-generation cephalosporins. Infect. Control Hosp. Epidemiol. 15:88-94. [DOI] [PubMed] [Google Scholar]
- 37.Parker, M. T. 1972. Phage-typing of Staphylococcus aureus. p. 1-28. In J. R. Norris and D. W. Ribbons (ed.), Methods in microbiology, vol. 7B. Academic Press, London, United Kingdom. [Google Scholar]
- 38.Ransome, N. J, P. Fitzgerald, and M. H. Wilcox. 1998. Antibiotic-associated diarrhea and Clostridium perfringens enterotoxin, abstr. K-126. Program Abstr. 38th Intersci. Conf. Antimicrob. Agents Chemother., San Diego, Calif.
- 39.Samore, M. H., L. Venkataraman, P. C. DeGirolami, R. D. Arbeit, and A. W. Karchmer. 1996. Clinical and molecular epidemiology of sporadic and clustered cases of nosocomial Clostridium difficile diarrhea. Am. J. Med. 100:32-40. [DOI] [PubMed] [Google Scholar]
- 40.Schalch, B., J. Björkroth, H. Eisgruber, H. Korkeala, and A. Stolle. 1997. Ribotyping for strain characterization of Clostridium perfringens isolates from food poisoning cases and outbreaks. Appl. Environ Microbiol. 63:3992-3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Settle, C. D., M. H. Wilcox, W. N. Fawley, O. J. Corrado, and P. M. Hawkey. 1998. Prospective study of the risk of Clostridium difficile diarrhoea in elderly patients following treatment with cefotaxime or piperacillin-tazobactam. Aliment. Pharmacol. Ther. 12:1217-1223. [DOI] [PubMed] [Google Scholar]
- 42.Spencer, R. C. 1998. Clinical impact and associated costs of Clostridium difficile-associated disease. J. Antimicrob. Chemother. 41(Suppl. C):5-12. [DOI] [PubMed] [Google Scholar]
- 43.Tenover, F. C., R. D. Arbeit, and R. V. Goering. 1997. How to select and interpret molecular strain typing methods for epidemiological studies of bacterial infections: a review for healthcare epidemiologists. Infect. Control Hosp. Epidemiol. 18:426-439. [DOI] [PubMed] [Google Scholar]
- 44.Thomas, C., M. Stevenson, and T. V. Riley. 2003. Antibiotics and hospital-acquired Clostridium difficile-associated diarrhoea: a systematic review. J. Antimicrob. Chemother. 51:1339-1350. [DOI] [PubMed] [Google Scholar]
- 45.Wada, A., Y. Masuda, M. Fukayama, T. Hatakeyama, Y. Yanagawa, H. Watanabe, and T. Inamatsu. 1996. Nosocomial diarrhoea in the elderly due to enterotoxigenic Clostridium perfringens. Microbiol. Immunol. 40:767-771. [DOI] [PubMed] [Google Scholar]
- 46.Wilcox, M. H., W. N. Fawley, and P. Parnell. 2000. Value of lysozyme agar incorporation and alkaline thioglycollate exposure for the environmental recovery of Clostridium difficile. J. Hosp. Infect. 44:65-69. [DOI] [PubMed] [Google Scholar]
- 47.Wilcox, M. H., J. Freeman, W. Fawley, S. MacKinlay, A. Brown, K. Donaldson, and O. Corrado. 2004. Long-term surveillance of cefotaxime and piperacillin-tazobactam prescribing and incidence of Clostridium difficile diarrhoea. J. Antimicrob. Chemother. 54:168-172. [DOI] [PubMed] [Google Scholar]
- 48.Wilcox, M. H. 2001. Clarithromycin and risk of Clostridium difficile-associated diarrhoea. J. Antimicrob. Chemother. 47:357-368. [DOI] [PubMed] [Google Scholar]
- 49.Wood, M. 2001. When stool cultures from adult inpatients are appropriate. Lancet 357:901-902. [DOI] [PubMed] [Google Scholar]