Abstract
The performance characteristics of the RealART and Molecular Beacons assays were compared with those of the Digene Hybrid Capture II assay (ultrasensitive). The results of the RealART and Digene Hybrid assays were related (r = 0.94; P < 0.001) and diverged by 2 orders of magnitude. The RealART assay can be used to effectively monitor serum hepatitis B virus DNA levels.
Hepatitis B virus (HBV) replication in patients with chronic hepatitis B (CHB) is typically monitored through measurement of serum HBV DNA levels. Commercial assays have included a solid-phase hybridization capture assay (Hybrid Capture II; Digene, Gaithersburg, MD) and a first-generation branched DNA signal amplification assay (Versant; Bayer Diagnostics, Emeryville, CA). However, these assays were limited by a lack of standardization and variability in their sensitivities, with a lower limit of detection of 105 to 106 copies/ml (5, 8, 11, 16). In addition to the lack of sensitivity, none of these commercial assays have detection ranges that can reliably detect the wide dynamic range of HBV DNA levels of more than 1010 copies/ml found in certain populations of HBV-infected patients.
With the recent advances in molecular biology techniques, the Versant HBV DNA 3.0 (Bayer Diagnostics, Emeryville, CA) and the COBAS AMPLICOR HBV MONITOR (Roche Diagnostics, Basel, Switzerland) assays have better sensitivities of 200 and 300 copies/ml, respectively, and have been used for the monitoring of HBV patients undergoing antiviral therapy and also of liver transplant recipients (3, 10, 12, 14, 15, 17, 18). Despite these improvements, the lower limits of detection of these two assays are still considered inadequate for measurement of the level of HBV replication during therapy, as they do not allow the detection of low-level virus replication in patients.
The use of a real-time fluorescence-based PCR system to detect nucleic acid from other viruses in plasma has recently been developed for the monitoring of HBV DNA levels (4, 6, 7, 13). Two such commercial assays are the RealART HBV LC PCR kit (Artus GmbH, Hamburg, Germany) and the Molecular Beacons assay (Molecular Diagnostics, Abbott Laboratories, Abbott Park, IL). In this study, we evaluated the performance characteristics of these two real-time PCR assays and compared them with those of the widely used Digene Hybrid Capture II assay (ultrasensitive) for the detection and quantification of HBV DNA in serial serum samples of patients undergoing antiviral therapy.
Serial serum samples from 14 hepatitis B virus e antigen-positive CHB patients who were enrolled in a randomized double-blind phase II study comparing emtricitabine plus adefovir dipivoxil combination therapy versus adefovir dipivoxil monotherapy were studied (9). Nine of these 14 patients were randomized to receive adefovir dipivoxil plus emtricitabine combination therapy, while 5 of the 14 patients were randomized to receive adefovir dipivoxil monotherapy. All patients were HBV DNA positive by the Digene Hybrid Capture II assay (ultrasensitive) at the baseline and were treated for 48 weeks. Serum samples were prospectively taken at the baseline (day 0) and at days 1, 3, 5, 7, 9, 11, and 14 and weeks 3, 8, 12, 16, 20, 24, 28, and 32 after the commencement of therapy; aliquoted; and stored at −70°C. In total, 224 serum samples were tested for HBV DNA. Both the RealART and the Molecular Beacons assays were complete kits with the exception of the consumables. All samples were tested in duplicate by all three assays. The mean value of the results was taken as the reference value.
Digene Hybrid Capture II assay (ultrasensitive).
In the ultrasensitive format of the Digene Hybrid Capture II assay, 1 ml of a sample or a positive control supplied with the kit along with 50 μl of precipitation buffer was centrifuged at 33,000 × g at 4°C in a Hereaus Stratos Biofuge or a Jouan high-speed tabletop centrifuge for 110 min. This procedure yields a 30-fold increase in sensitivity and enhances the lower detection limit of the assay to approximately 4,700 HBV copies/ml. The assay relies on the hybridization of denatured HBV DNA to an HBV RNA transcript, which is subsequently captured by an immobilized anti-RNA-DNA antibody. Further amplification of the signal is achieved by the binding of more anti-RNA-DNA antibodies, each of which is conjugated to multiple alkaline phosphatase molecules. The alkaline phosphatase generates a chemiluminescent signal on the addition of a dioxetane substrate, and the signal generated is proportional to the input HBV DNA. The linear range of the assay is 4,700 to 5.6 × 109 copies/ml in the ultrasensitive format, whereas it is 1.42 × 105 to 1.7 × 109 copies/ml in the standard format.
RealART HBV LC PCR kit.
Serum samples of 200 μl were extracted by using the Roche Diagnostics (Mannheim, Germany) MagNA Pure system and total nucleic acid isolation kit. The extraction procedure uses the magnetic bead technology. Essentially, magnetized glass beads act as a binding medium for the DNA; beads were transferred by the device through various stringent washes to remove contaminating material before elution into 50 μl of the buffer supplied with the kit. RealART HBV LC PCR reagents constitute a ready-to use system for PCR amplification and the detection of HBV DNA on the LightCycler instrument (Roche Diagnostics, Basel, Switzerland). The RealART HBV LC PCR reagents master mixture contains reagents and enzymes for amplification of a 120-bp region of the HBV genome and for parallel detection of the specific amplification products. In addition, the RealART HBV LC PCR reagents contain a heterologous internal control for identification of possible PCR inhibition. For quantification, plasmids constructed and linearized in vitro were used as standards. The standards were included in the kit and were calibrated against an original stock solution of the World Health Organization international standard for HBV DNA. Five standard samples diluted 10-fold were used to generate the standard curve. The linear range of this assay was approximately 54 to 3.6 × 109 copies/ml.
Molecular Beacons assay.
Briefly, PCRs were carried out in a total volume of 100 μl with the following at the indicated concentrations: 1× PCR Gold buffer, 10 units of TaqGold DNA polymerase (Applied Biosystems, Foster City, CA), 0.4 mM of each of the four deoxynucleotide triphosphates, 3.5 mM MgCl2, 1.6 μM of HBV reverse primer, 0.2 μM 6-carboxyfluorescein-labeled HBV beacon probe, 0.2 μM VIC-labeled internal control beacon probe, and 1,000 copies of internal control target. The HBV primers and probes were selected from a highly conserved region of the HBV surface antigen gene. The HBV primers coamplify an unrelated internal control target DNA sequence. PCR conditions were as follows: TaqGold activation at 94°C for 10 min, followed by 45 cycles at 92°C for 30 s and 58°C for 1 min, when fluorescent readings were taken on an ABI PRISM 7000 sequence detection system (Applied Biosystems). The linear range of the assay was 300 to 109 copies/ml.
Statistical analysis.
All statistical analyses were performed by using the Statistical Program for Social Sciences (SPSS 12.0 for Windows; SPSS Inc., Chicago, IL). The Mann-Whitney U test was used for continuous variables that appeared to have a skewed distribution, and the chi-square test with Yates' correction factor or Fisher's exact test was used for categorical variables. A linear regression analysis (Pearson correlation) was used to define the relationship between the results of the different assays with the same serum samples. Statistical analysis was also performed as described by Bland and Altman (1). Continuous variables were expressed as median (range). Statistical significance was defined as a P value of <0.05.
HBV DNA was detectable in all the pretreatment samples by the three assays. After the initiation of antiviral therapy, the serum HBV DNA levels were measurable in 215 of the 224 samples (96.0%) by the RealART assay, 167 of the 224 samples (74.6%) by the Digene Hybrid Capture II assay (ultrasensitive), and 188 of the 224 samples (83.9%) by the Molecular Beacons assay. For 47 of the 57 samples in which HBV DNA was undetectable (82.5%) by the Digene Hybrid Capture II assay (ultrasensitive), the DNA was detectable by the RealART assay. The median HBV DNA level in these 47 samples as determined by the RealART assay was 256 copies/ml (range, 85 to 3,514 copies/ml). On the other hand, HBV DNA was detectable in 25 of these 57 samples (43.9%) by the Molecular Beacons assay. The median HBV DNA levels in these 25 samples as determined by the Molecular Beacons assay was 947 copies/ml (345 to 4,210 copies/ml). The Digene Hybrid Capture II assay (ultrasensitive) did not detect HBV DNA in the samples in which HBV DNA was not detectable by either the RealART or the Molecular Beacon assays.
The RealART assay was able to detect HBV DNA in a higher number of samples than the Digene Hybrid Capture II assay (ultrasensitive) (P < 0.001) or the Molecular Beacons assay (P < 0.001). The Molecular Beacons assay was also able to detect a HBV DNA in a higher number of samples than the Digene Hybrid Capture II assay (ultrasensitive) (P = 0.01).
Two of the 224 samples (0.9%) had HBV DNA levels above the upper limit of detection by the Digene Hybrid Capture II assay (ultrasensitive), while none of the 224 samples (0%) had HBV DNA levels above the upper limit of detection by the RealART or Molecular Beacons assay.
Correlation between the results of the three assays.
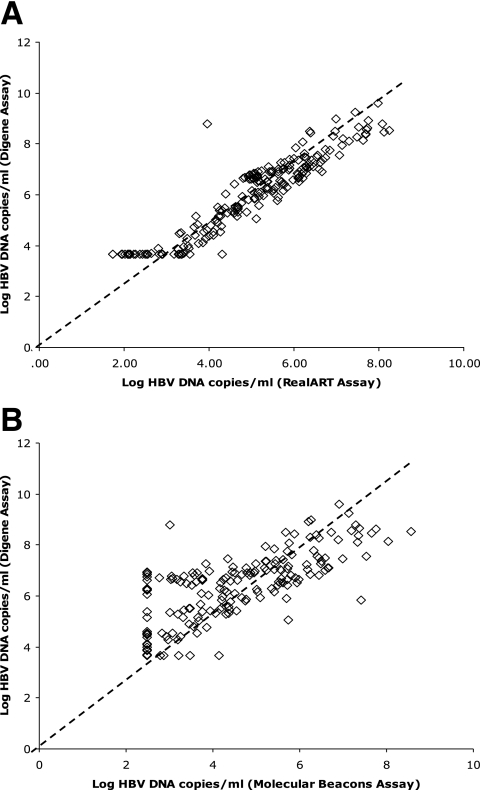
HBV DNA quantification by the RealART assay and the Digene Hybrid Capture II assay (ultrasensitive) in a given sample was significantly related (r = 0.94; P < 0.001) (Fig. 1A). The results of the Molecular Beacons assay were also related to those of the Digene Hybrid Capture II assay (ultrasensitive) (r = 0.79; P < 0.001) (Fig. 1B).
FIG. 1.
HBV DNA levels in all serum samples assessed by (A) the RealART assay and the Digene Hybrid Capture II assay (ultrasensitive) and (B) the Molecular Beacons assay and the Digene Hybrid Capture II assay (ultrasensitive) (the total number of datum points in panels A and B is 224).
Performance of the RealART assay versus that of the Digene Hybrid Capture II assay (ultrasensitive).
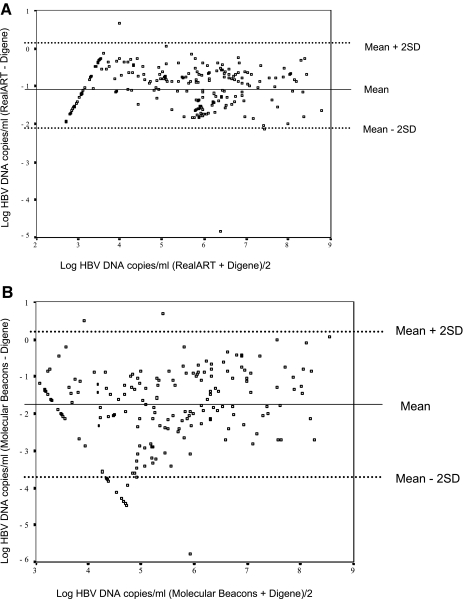
According to the statistical analysis, performed as described by Bland and Altman (1), there was good agreement between the RealART and the Digene assays (Fig. 2A). The mean difference was −1.05, as expected for good agreement, and the values for 95% of the data were between +0.12 and −2.19 after logarithmic transformation. Thus, the values differed by 2.31 orders of magnitude.
FIG. 2.
Difference against the mean for HBV DNA by (A) the RealART assay and the Digene Hybrid Capture II assay (ultrasensitive) (the total number of datum points in panel A is 218) and (B) the Molecular Beacons assay and the Digene Hybrid Capture II assay (ultrasensitive) (the total number of datum points in panel B is 209).
Performance of the Molecular Beacons assay versus that of the Digene Hybrid Capture II assay (ultrasensitive).
However, when the results of the Molecular Beacons assay were compared with those of the Digene assay, the mean difference was −1.73 and the limits of agreement (means ± 2× the standard deviations [SDs]) were +0.33 and −3.79, respectively (Fig. 2B). The values differed by +4.12 orders of magnitude.
In order to test the intra-assay variability, a high-titer serum sample was selected and tested five times by all three assays. The SDs of the means were 0.35 for the RealART assay, 0.39 for the Digene Hybrid Capture II assay (ultrasensitive), and 0.45 for the Molecular Beacons assay. This results in coefficients of variation of 5.33%, 6.03%, and 6.47%, respectively.
The results of the present study show a better correlation between the results of the RealART assay and those of the Digene Hybrid Capture II assay (ultrasensitive) for the quantification of serum HBV DNA levels over a wide range of levels of viremia. The RealART assay is more sensitive than the Digene Hybrid Capture II assay (ultrasensitive) or even the Molecular Beacons assay, particularly at the low-level viremia range.
As newer assays such as the COBAS AMPLICOR HBV MONITOR assay (Roche Diagnostics, Basel, Switzerland), the Versant HBV DNA 3.0 assay (Bayer Diagnostics), and the Roche HBV TaqMan assay (Roche Diagnostics, Basel, Switzerland), which are more sensitive than the Digene Hybrid Capture II assay (ultrasensitive) for the quantification of HBV DNA, have become available, more studies will be needed to compare the performance characteristics of the RealART assay with those of these more sensitive assays for the detection of low levels of HBV DNA in order to confirm the accuracy of the RealART assay for the detection of the very low levels of HBV DNA (7, 13, 15). When the 57 samples in which HBV DNA was undetectable by the Digene Hybrid Capture II assay (ultrasensitive) were retested with the COBAS AMPLICOR HBV MONITOR assay (Roche Diagnostics, Basel, Switzerland), the results of the COBAS AMPLICOR HBV MONITOR assay significantly correlated with those of the RealART assay (r = 0.52; P = 0.008) (data not shown).
The use of RealART assay in this study has demonstrated that those patients with HBV DNA levels below the detection limit of the Digene Hybrid Capture II assay (ultrasensitive) or the Molecular Beacons assay have low levels of viremia rather than complete clearance of the virus. The main advantage of the RealART assay is its reproducibility and high degrees of accuracy and precision, which are comparable to those of the Digene Hybrid Capture II assay (ultrasensitive).
Although the RealART assay shows a more extended dynamic range, it will still be unable to quantify HBV DNA in a large number of samples or in patients who have viral loads that exceed the upper limit of detection, as has been shown with an immunotolerant group of patients (2). These samples will need to be diluted and retested, leading to additional costs. The challenge is therefore to develop an assay that is capable of detecting HBV DNA over the entire dynamic range.
In conclusion, the RealART assay is a reliable and reproducible tool for the quantification of HBV DNA during antiviral therapy.
Acknowledgments
This project was supported with a grant from the Cheng Si-yuan (China International) Hepatitis Research Foundation (to the University of Hong Kong), a China National 973 research grant (G 1999 054105 to G. K. K. Lau), and Heung Chit Kau and family.
REFERENCES
- 1.Bland, J. M., and D. G. Altman. 1995. Comparing methods of measurement: why plotting difference against standard method is misleading. Lancet 346:1085-1087. [DOI] [PubMed] [Google Scholar]
- 2.Chu, C. M., P. Karyannis, M. J. F. Fowler, J. Monjardino, Y. F. Liaw, and H. C. Thomas. 1997. Natural history of chronic hepatitis B virus infection in Taiwan: studies of HBV DNA in serum. Hepatology 5:431-434. [DOI] [PubMed] [Google Scholar]
- 3.Dumortier, J., P. Chevallier, J. Y. Scoazec, F. Berger, and O. Boillot. 2003. Combined lamivudine and hepatitis B immunoglobulin for the prevention of hepatitis B recurrence after liver transplantation: long-term results. Am. J. Transplant. 3:999-1002. [DOI] [PubMed] [Google Scholar]
- 4.Hui, C. K., W. W. Cheung, W. Y. Au, A. K. W. Lie, H. Y. Zhang, Y. H. Yueng, B. C. Y. Wong, Y. K. Kwong, R. Liang, and G. K. K. Lau. 2005. Hepatitis B reactivation after withdrawal of preemptive lamivudine in patients with hematological malignancy upon completion of cytotoxic chemotherapy. Gut 154:1597-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hui, C. K., A. Lie, W. Y. Au, S. Y. Ma, Y. H. Leung, H. Y. Zhang, J. Sun, W. W. Cheung, C. S. Chim, Y. L. Kwong, R. Liang, and G. K. K. Lau. 2005. Effectiveness of prophylactic anti-HBV therapy in allogeneic hematopoietic stem cell transplantation with HBsAg positive donors. Am. J. Transplant. 5:1437-1445. [DOI] [PubMed] [Google Scholar]
- 6.Hui, C. K., J. Sun, W. Y. Au, A. K. W. Lie, Y. H. Yuen, H. Y. Zhang, N. P. Lee, J. L. Hou, R. Liang, and G. K. K. Lau. 2005. Occult hepatitis B virus infection in hematopoietic stem cell donors in a hepatitis B virus endemic area. J. Hepatol. 42:813-819. [DOI] [PubMed] [Google Scholar]
- 7.Hui, C. K., E. Lau, Wu, H., A. Monto, M. Kim, J. M. Luk, G. K. K. Lau, and T. L. Wright. 2006. Fibrosis progression in chronic hepatitis C patients with occult hepatitis B co-infection. J. Clin. Virol. 35:182-192. [DOI] [PubMed] [Google Scholar]
- 8.Krajden, M., J. Minor, L. Cork, and L. Comanor. 1998. Multi-measurement method comparison of three commercial hepatitis B virus DNA quantification assays. J. Viral Hep. 5:415-422. [DOI] [PubMed] [Google Scholar]
- 9.Lau, G. K., H. Cooksley, R. M. Ribiero, S. Bowden, H. Mommeja-Marin, J. Sorbel, E. Mondou, F. Rousseau, S. Lewin, A. S. Perelson, S. Locarnini, and N. V. Naoumov. 2004. Randomized, double-blind study comparing adefovir dipivoxil (ADV) plus emtricitabine (FTC) combination therapy versus ADV alone in HBeAg (+) chronic hepatitis B: efficacy and mechanisms of treatment response. Hepatology 40(Suppl. 1):666A. [Google Scholar]
- 10.Lau, G. K., T. Piratvisuth, K. X. Luo, P. Marcellin, W. G. Cooksley, M. Fried, W. C. Chow, S. W. Paik, W. Y. Chang, T. Berg, R. Flisiak, P. McCloud, and N. Pluck. 2005. Peginterferon alfa-2a (40kD) (PEGASYS) monotherapy and in combination with lamivudine is more effective than lamivudine monotherapy in HBeAg positive chronic hepatitis B: results from a large, multinational study. N. Engl. J. Med. 352:2682-2695. [DOI] [PubMed] [Google Scholar]
- 11.Lindh, M., P. Horal, A. P. Dhillon, and G. Norkans. 2000. Hepatitis B virus DNA levels, precore mutations, genotypes and histological activity in chronic hepatitis B. J. Viral Hep. 7:258-267. [DOI] [PubMed] [Google Scholar]
- 12.Lo, C. M., C. K. Cheung, G. K. Lau, M. F. Yuen, C. L. Liu, S. C. Chan, S. T. Fan, and J. Wong. 2005. Significance of hepatitis B virus genotype in liver transplantation for chronic hepatitis B. Am. J. Transplant. 5:1893-1900. [DOI] [PubMed] [Google Scholar]
- 13.Loeb, K., K. Jerome, J. Goddard, M. Huang, A. Cent, and L. Corey. 2000. High-throughput quantitative analysis of hepatitis B serum using the TaqMan fluorogenic detection system. Hepatology 32:626-629. [DOI] [PubMed] [Google Scholar]
- 14.Marcellin, P., G. K. Lau, F. Bonino, P. Farci, S. Hadziyannis, R. Jin, Z. M. Lu, T. Piratvisuth, G. Germanidis, C. Yurdaydin, M. Diago, S. Gurel, M. Y. Lai, P. Button, N. Pluck and the Peginterferon Alfa-2a HBeAg-Negative Chronic Hepatitis B Study Group. 2004. Peginterferon alfa-2a alone, lamivudine alone, and the two in combination in patients with HBeAg-negative chronic hepatitis B. N. Engl. J. Med. 351:1206-1217. [DOI] [PubMed] [Google Scholar]
- 15.Marin, I. J., M. Poljak, K. Seme, J. Meglic-Volkar, M. Maticic, G. Lesnicar, and V. Brinoves. 2001. Comparative evaluation of semiautomated COBAS AMPLICOR hepatitis B virus (HBV) MONITOR test and manual microwell plate-based AMPLICOR HBV MONITOR test. J. Clin. Microbiol. 39:758-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pawlotsky, J. M., A. Bastie, C. Hezode, I. Lonjon, F. Carthuy, J. Remire, and D. Dhumeaux. 2000. Routine detection and quantification of hepatitis B virus DNA in clinical laboratories: performance of three commercial assays. J. Virol. Methods 85:11-21. [DOI] [PubMed] [Google Scholar]
- 17.Ronsin, C., A. Pillet, C. Bali, and G. A. Denoyel. 2006. Evaluation of the COBAS AmpliPrep-total nucleic acid isolation-COBAS TaqMan hepatitis B virus (HBV) quantitative test and comparison to the VERSANT HBV DNA 3.0 assay. J. Clin. Microbiol. 44:1390-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yao, J. D., M. G. Beld, L. L. Oon, C. H. Sherlock, J. Germer, S. Menting, S. Y. Se Thoe, L. Merrick, R. Ziermann, J. Surtihadi, and H. J. Hnatyszyn. 2004. Multicenter evaluation the VERSANT hepatitis B virus DNA 3.0 assay. J. Clin. Microbiol. 42:800-806. [DOI] [PMC free article] [PubMed] [Google Scholar]